- Department of Respiratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Background: Current understanding of the link between microbiota imbalance and immune function in non-small cell lung cancer (NSCLC) has not been fully elucidated. This study aims to explore the link between dysbiotic lung microbiota and immunity in NSCLC, which may provide valuable information for disease progression monitoring and prognosis prediction.
Methods: Lung microbial communities from both the tumor-affected (n = 43) and contralateral healthy sides (n = 38) of lung cancer patients were analyzed by 16S rRNA sequencing. The association between microbial abundance and tumor stages, metastasis or not, nodule size, PD-L1 expression, as well as Ki-67 levels was conducted. Mann-Whitney tests were used to evaluate differences in the systemic immune-inflammation index (SII), T cell subsets (CD3+, CD4+, CD8+), as well as the CD4+/CD8+ ratio between different microbial expression patterns of Prevotella and Veillonella.
Results: Significant β-diversity differences were observed between the tumor-bearing and contralateral normal lungs in individuals diagnosed with lung carcinoma. A notable increase in Prevotella (P = 0.044) and Veillonella (P = 0.02) was detected within NSCLC-affected lungs, whereas Pseudomonas (P = 0.008) as well as Staphylococcus (P = 0.033) were significantly reduced. Increased levels of Veillonella were detected in NSCLC patients at stage IIIB-IV and were positively correlated with Ki-67 expression. Furthermore, patients with higher abundance of Prevotella and Veillonella exhibited a significantly elevated systemic immune-inflammation index (SII) compared to the lower-abundance group (P = 0.0329), while their CD8+ T cell levels were significantly decreased in the higher abundance group (P = 0.0027).
Conclusion: Microbial composition differed significantly between the tumor-affected and healthy sides in lung cancer patients. Veillonella was more abundant NSCLC patients at stage IIIB-IV, while increased Prevotella and Veillonella abundance correlated positively with SII but negatively with CD8+ T cell levels. These findings provide valuable insights into tumor-associated microbiota for monitoring disease advancement, treatment stratification and prognostic assessment.
1 Introduction
As the deadliest malignancy worldwide, lung cancer has a poor prognosis, with a 5-year survival probability of just 27% (1–3). Given its high mortality, lung cancer warrants the refinement of monitoring and therapeutic strategies. Beyond genetic and environmental factors, the lung microbiome has gained attention for its influence on lung cancer progression and immune modulation. It has been shown that dysbiotic commensal microbial communities may exist in the respiratory tract of lung cancer (4–7). For example, several studies have reported that lung cancer cases are associated with an increased presence of Capnocytophaga, Selenomonas, and Veillonella in both saliva and sputum (8–10). The dysbiotic microbial landscape associated with lung cancer remains inadequately explored.
Numerous studies established immune cell dysregulation is closely linked to the prognosis of lung cancer (11, 12). Peripheral blood immune cells have been utilized for biomarkers in disease monitoring and prognosis evaluation, offering a feasible, prompt, and non-invasive method (13, 14). The Systemic Immune-Inflammation Index (SII) is recognized as a potential parameter in disease prognosis, determined by the formula: SII = platelet count × neutrophil count/lymphocyte count (15). SII can reflect systemic inflammation and immune status, which is associated with worse overall survival (OS) and progression-free survival (PFS) of lung cancer patients (16, 17). CD8+ T cells serve as the primary cytotoxic effectors in antitumor immunity, and their depletion or functional exhaustion is closely linked to tumor progression and immune evasion (18–20).
Using 16S rRNA sequencing, this study investigated lung microbiota dysbiosis in lung cancer by comparing bronchoalveolar lavage fluid (BALF) from tumor-affected and contralateral healthy lungs. Additionally, we analyzed the association between dysbiotic lung microbiota and key immune and disease progression markers, including SII, CD8+ T cell levels, Ki-67, and tumor stage. This research sought to investigate the connection of lung microbiota imbalance with immune dysregulation in lung cancer, providing insights for cancer progression monitoring, treatment stratification and prognosis prediction.
2 Materials and methods
2.1 Study participant recruitment
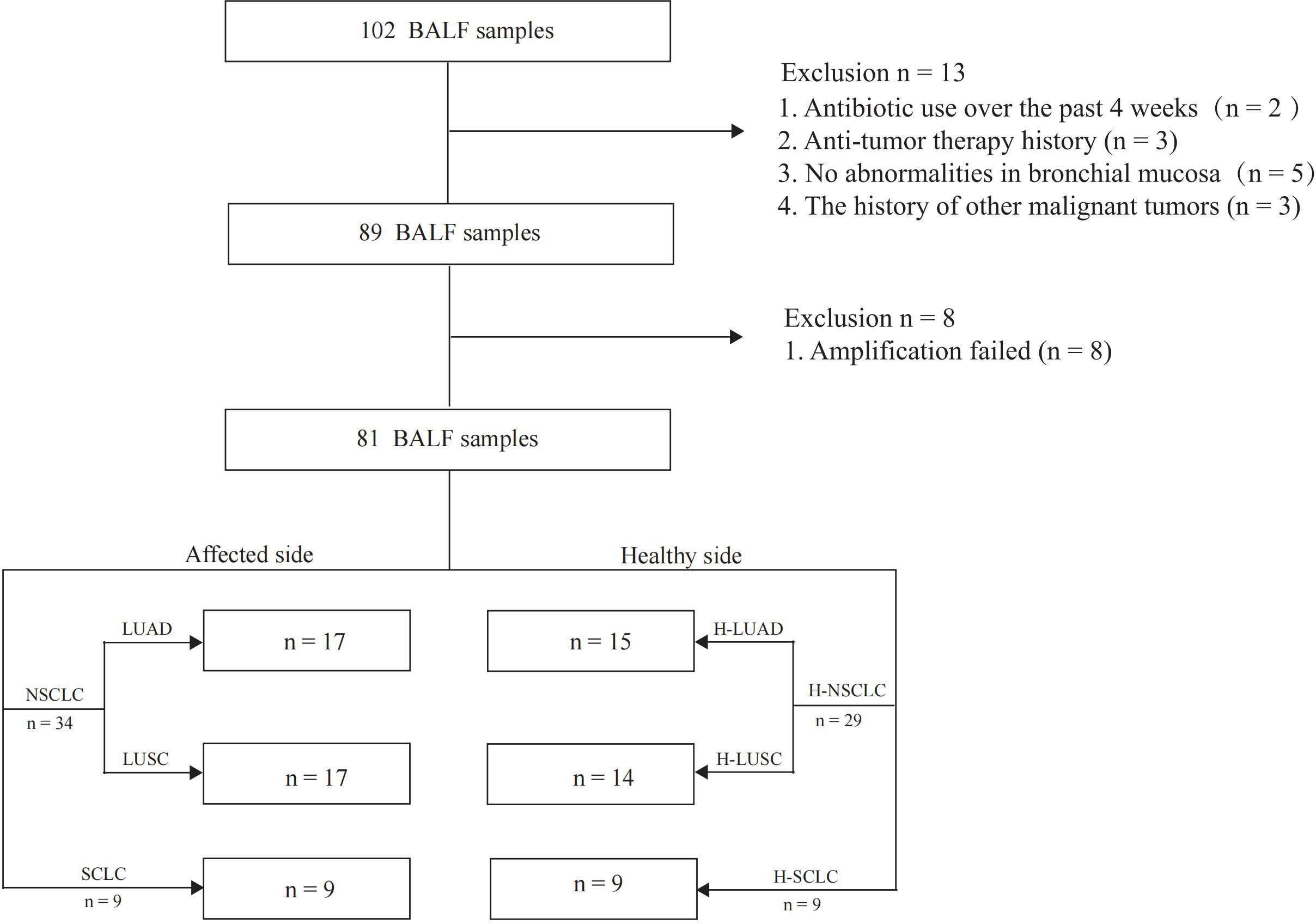
Between November 2021 and June 2022, we collected 102 BALF samples at Zhengzhou University’s First Affiliated Hospital. After quality control, bronchoalveolar lavage fluid samples of both the tumor-affected (n = 43) and contralateral healthy sides (n = 38) from lung cancer patients were selected for 16S rRNA sequencing analysis. Eight samples were excluded due to 16S rRNA amplification failure. All enrolled participants were newly diagnosed with lung cancer, had no history of cancer treatment, and had not used antibiotics in the last four weeks. Additionally, individuals with a prior diagnosis of cancer were removed from this study (21). We collected clinical data, such as pathological diagnosis and tumor stage. Additionally, we assessed nodule features (size and location) and laboratory parameters, such as blood routine tests, inflammatory markers, tumor markers, and absolute counts of T-cell subsets. Immunohistochemical markers and gene mutation profiles were analyzed in paraffin-embedded lung tissue specimens. The research received ethical approval from the Ethics Committee of Scientific Research and Clinical Trials of the First Affiliated Hospital of Zhengzhou University (No. 2024-KY-0348-003), and all enrolled patients signed an informed consent form.
2.2 Sample collection
BALF samples were collected by an experienced clinician following standard fiberoptic bronchoscopy protocols (22, 23), with strict precautions to minimize oral contamination. Samples were obtained from both the tumor-affected and corresponding healthy lung lobes, with 10–15 mL collected from each side per patient (24). The tumor-affected side was designated as the lung cancer group, while the contralateral healthy side was defined as the control group.
2.3 Microbial 16S rRNA analysis
DNA was isolated from BALF and subsequently subjected to 16S rRNA sequencing. For 16S rRNA sequencing, the hypervariable V3-V4 regions were subjected to PCR using primers 341F and 806R (CCTAYGGGRBGCASCAG, GGACTACNNGGGTATCTAAT). Following PCR product purification, the library quality was evaluated prior to sequencing (25).
Paired-end reads were processed into raw tags with FLASH (Version 1.2.11) (26) to generate raw tags. Quality filtering was performed under specific conditions using the fastp (v0.23.1) quality control procedure to acquire high-quality filtered tags (27). Using the UCHIME algorithm, clean tags were aligned to the Silva database to filter out chimeric sequences, yielding the final effective tags.
2.4 Sequencing analysis
Taxonomic annotation was performed using QIIME2 with the Silva 138.1 reference database. Community richness, diversity, and evenness were measured by QIIME2. β-diversity was quantified by calculating both weighted and unweighted UniFrac distances. Significant differences in species composition across taxonomic levels were identified utilizing MetaStat and t-tests in R (v4.0.3). LEfSe analysis was employed to identify biomarkers, with an LDA score threshold set to 4 in the LEfSe software.
2.5 Data analysis and statistical methods
Statistical tests were performed using SPSS (IBM SPSS 26.0, SPSS Inc.), applying the Mann-Whitney U test or t-test for two-group comparisons of continuous variables, while categorical variables were assessed using the χ² test. Results were expressed as mean ± SD. Abundance, α-diversity, and β-diversity analyses between groups were assessed via the Mann-Whitney test. Heat maps and relationship heat maps were generated using R Version 4.3.2 software. Correlation analysis was conducted using Spearman correlation methods. A threshold of P-value less than 0.05 was defined as statistically significant.
3 Results
3.1 Patient features
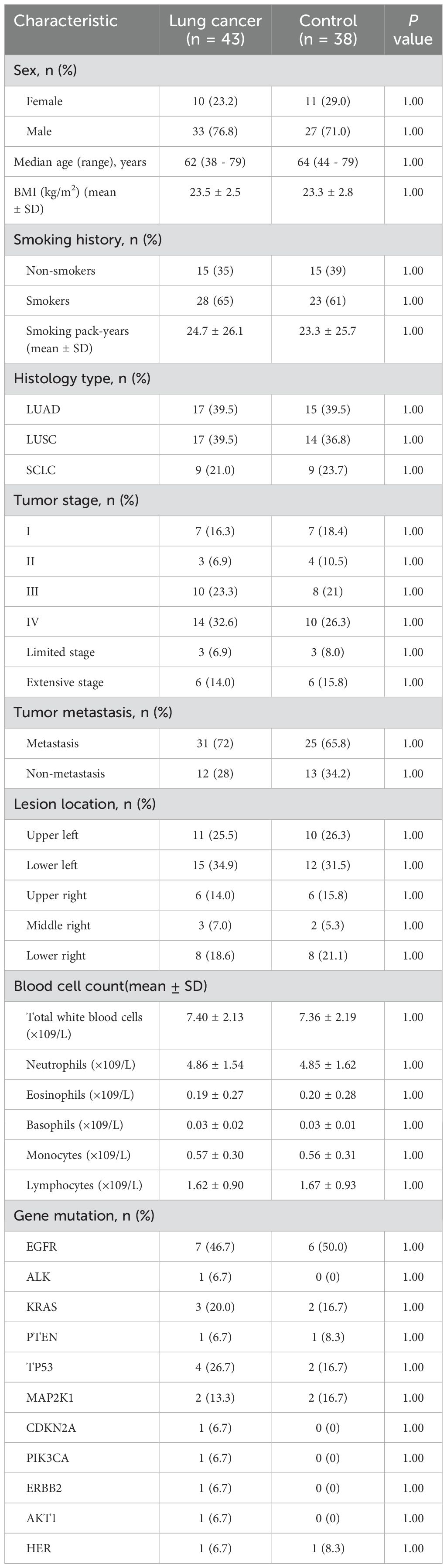
81 BALF samples were included in the final analysis, with the study flowchart depicted in Figure 1. The median age of the lung cancer group was 62 (38–79) years, while it was 64 (44–79) years in the control group. The enrolled patients included 17 with LUAD, 17 with LUSC, and 9 with SCLC. Among the enrolled patients, 7 were at stage I, 3 at stage II, 10 at stage III, and 14 at stage IV. Three patients were diagnosed with limited-stage and six with extensive-stage SCLC. Distant metastases were detected in 31 patients, whereas 12 had non-metastatic disease. Genomic mutation data were retrieved from 23 patients (Table 1).
3.2 Microbial composition of BALF in lung cancer
A total of 23,994 ASVs were identified, comprising 16,445 ASVs from the affected lung segment and 13,146 ASVs from the contralateral healthy lung segment (Figure 2A). Microbial communities in the lower airway differ between the tumor-affected and contralateral healthy sides in lung cancer patients. The top 10 genera, Prevotella, Streptococcus, Ralstonia, Alloprevotella, Veillonella, Pseudomonas, Neisseria, and Fusobacterium, were prevalent on both affected and contralateral healthy sides. Prevotella, Veillonella, Corynebacterium, and Muribaculaceae were notably abundant in cancer-affected individuals, while Pseudomonas, Porphyromonas, and Actinobacillus were predominant in the contralateral healthy group (Figures 2B, C, Supplementary Figure S1).
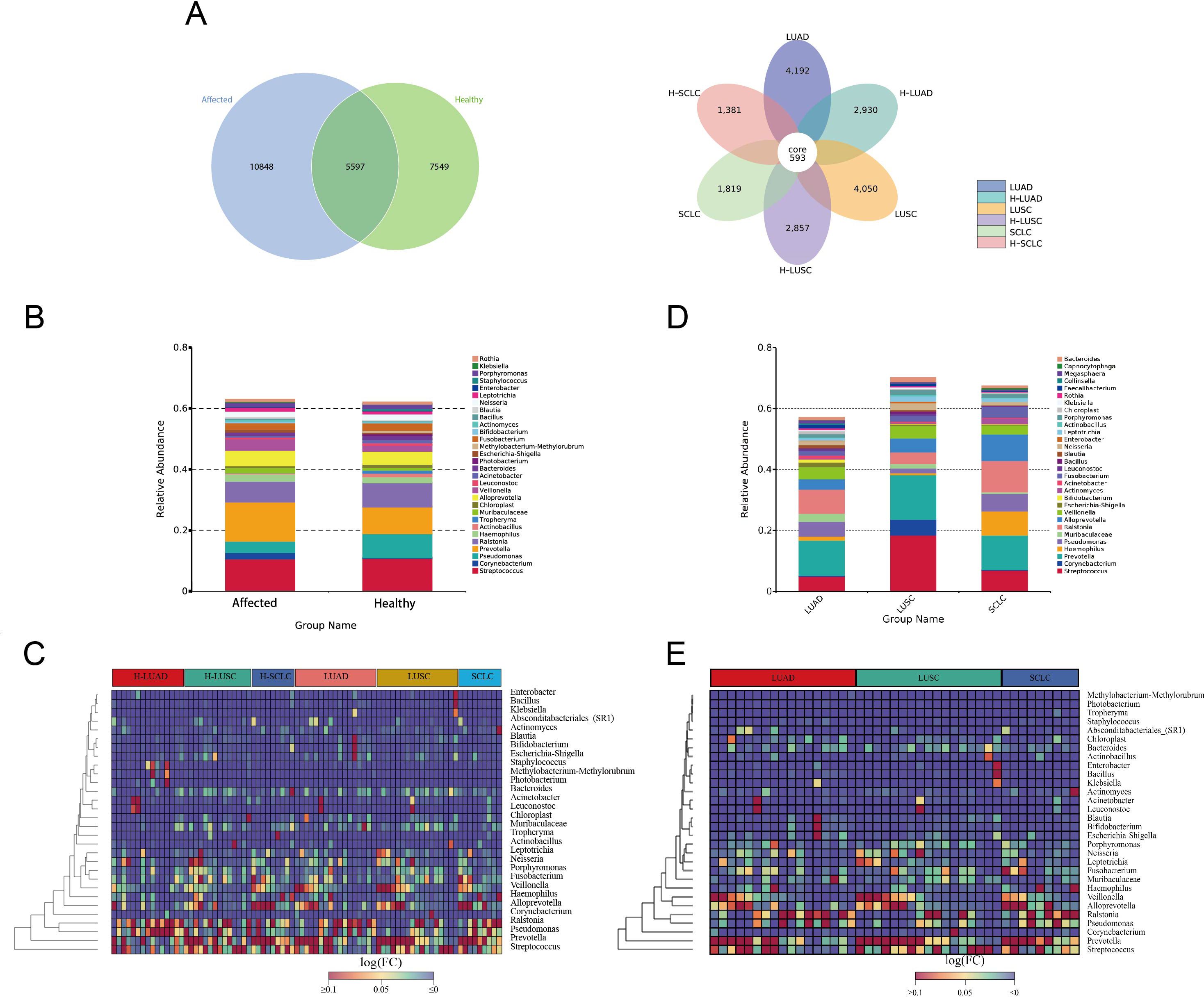
Figure 2. Microbial composition of BALF in lung cancer. (A) Venn diagrams illustrate the shared and unique ASVs (Amplicon Sequence Variants) between the tumor-affected and healthy sides across different pathological subtypes of lung cancer. (B, C) Genus-Level Taxonomic Profile of the BALF Microbiome, comparing the affected and healthy sides of lung cancer. (D, E) Taxonomic profiles of BALF microbiomes across different pathological subtypes of lung cancer. The affected side includes NSCLC, LUAD, LUSC, and SCLC. The corresponding healthy sides are denoted as H-NSCLC, H-LUAD, H-LUSC, and H-SCLC, respectively.
The microbial composition differs significantly across various pathological subtypes of lung cancer. In individuals diagnosed with LUAD and LUSC, the ten taxa with the highest prevalence are Prevotella, Ralstonia, Streptococcus, Pseudomonas, Veillonella, Alloprevotella, Muribaculaceae, Fusobacterium, Escherichia-Shigella, and Neisseria. In contrast, the top 10 genera in small cell lung cancer (SCLC) are Prevotella, Ralstonia, Alloprevotella, Haemophilus, Streptococcus, Pseudomonas, Fusobacterium, Veillonella, Actinomyces, and Leptotrichia (Figures 2D, E).
3.3 Biodiversity analysis demonstrates divergent microbial community compositions between the tumor-affected and contralateral healthy sides
The tumor-affected and contralateral healthy sides exhibited no significant differences in α-diversity, as evaluated by Chao1, Observed OTUs, Shannon, and Simpson indices (Figure 3A). However, distinct β-diversity patterns were observed between tumor-affected and contralateral healthy sides in NSCLC, supported by weighted UniFrac and Bray-Curtis indices (Figure 3B). These observations highlight the microbial composition and community structure of the tumor-affected lung that differ significantly from those of the contralateral healthy lung segment.
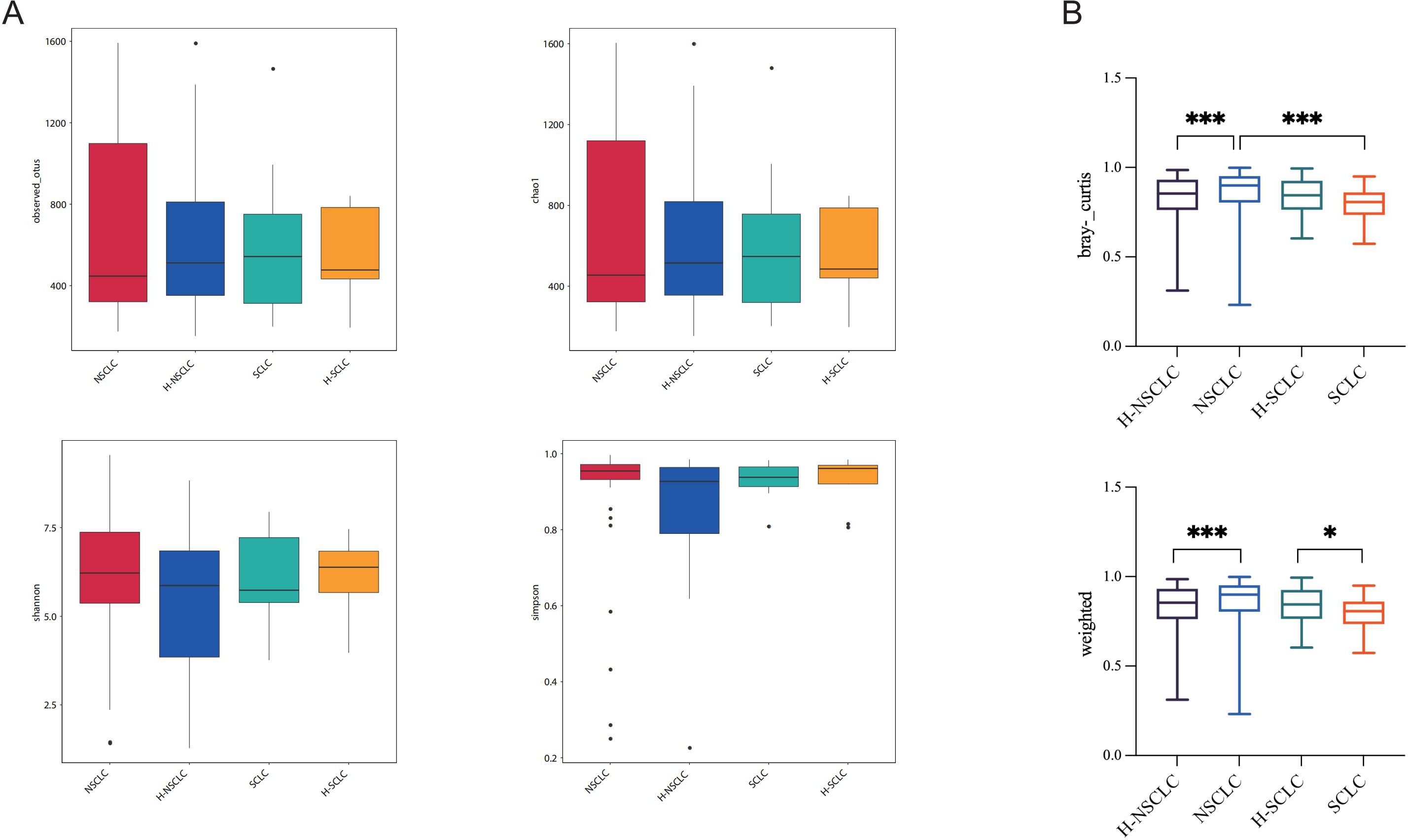
Figure 3. Biodiversity analysis demonstrates divergent microbial community compositions between the tumor-affected and contralateral healthy sides. (A) Observed OTUs, Chao1, Shannon, and Simpson indices were used to analyze microbial α-diversity. (B) β-diversity comparison of BALF microbiota between the tumor-affected and healthy sides in NSCLC and SCLC, as measured by weighted UniFrac distances and Bray-Curtis dissimilarity indices. *P-value < 0.05; ***P-value < 0.001.
3.4 Differentially abundant microbes in lung cancer dysbiosis
We investigated microbial differences between the tumor-affected and contralateral healthy sides in NSCLC to characterize lung cancer-associated dysbiosis. Compared to the contralateral healthy side, the NSCLC-affected side displayed an enrichment of Prevotella (P = 0.044) and Veillonella (P = 0.02), whereas Pseudomonas (P = 0.008) as well as Staphylococcus (P = 0.033) displayed a notable decrease (Figure 4A).
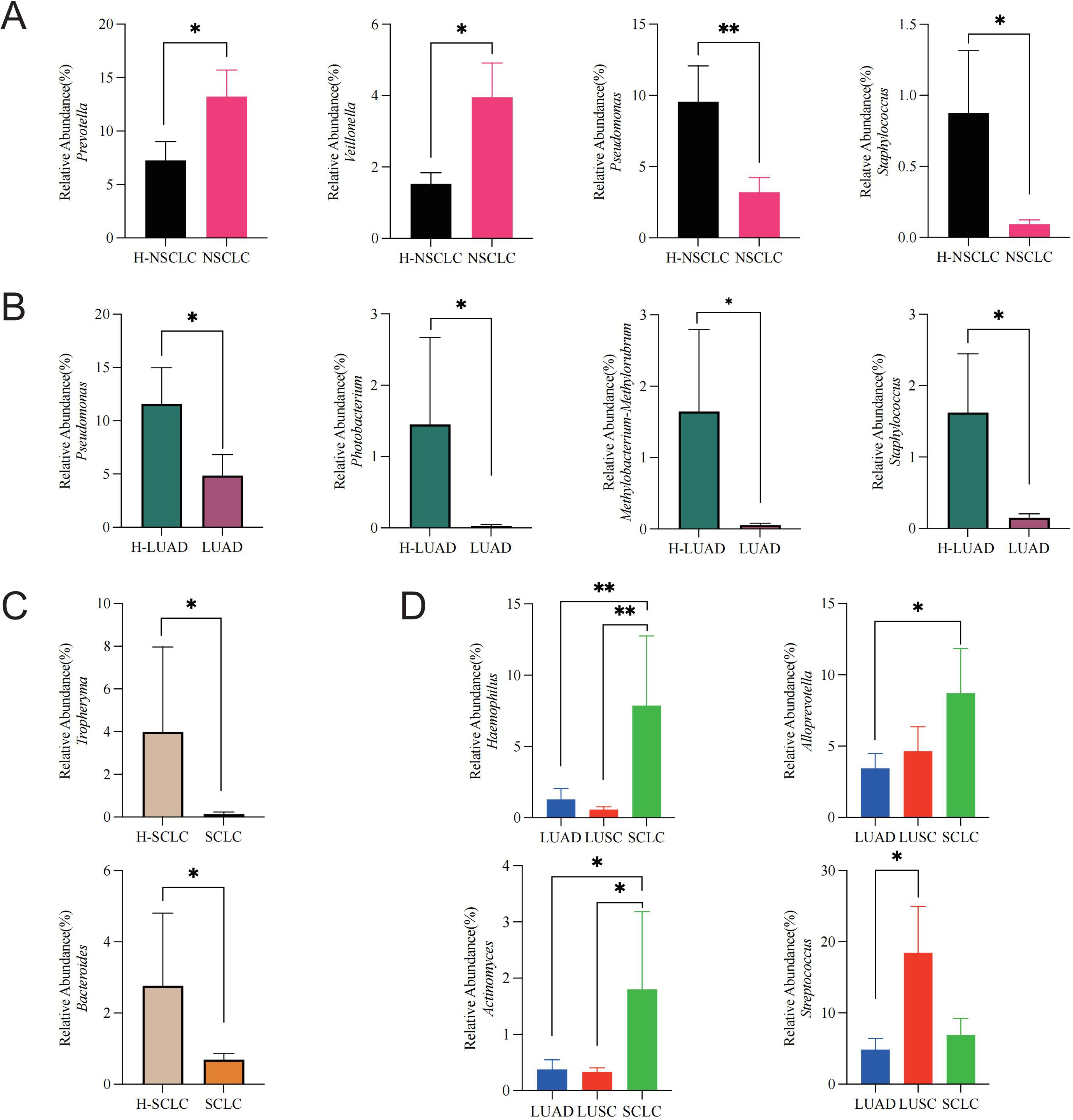
Figure 4. Lung cancer microbiome dysbiosis profiles. (A) Comparison between the tumor-affected and healthy sides in NSCLC, (B) LUAD, (C) SCLC, and (D) across different pathological subtypes of lung cancer. *P-value < 0.05; **P-value < 0.01.
To assess subtype-specific dysbiosis, we further analyzed microbial differences between the tumor-affected and contralateral healthy sides across distinct pathological subtypes. In LUAD, Pseudomonas (P = 0.042), Photobacterium (P = 0.049), Methylobacterium-Methylorubrum (P = 0.02), and Staphylococcus (P = 0.003) were markedly more abundant on the healthy side (Figure 4B). Similarly, in SCLC, Tropheryma (P = 0.035) and Bacteroides (P = 0.048) exhibited higher abundances on the healthy side (Figure 4C).
Furthermore, to explore inter-subtype differences in dysbiosis, we compared microbial profiles among tumor-affected lung segments of different pathological subtypes. Haemophilus was significantly enriched in SCLC compared to LUAD (P = 0.008) and LUSC (P = 0.003). Alloprevotella abundance was higher in SCLC compared to LUAD (P = 0.041). Actinomyces was markedly enriched in SCLC relative to LUAD (P = 0.017) and LUSC (P = 0.014). Additionally, Streptococcus was significantly enriched in LUSC relative to LUAD (P = 0.022) (Figure 4D).
3.5 Association between Veillonella abundance and tumor stage, proliferation, and PD-L1
To explore the clinical relevance of microbial alterations, we analyzed their associations with key tumor-related indices and Veillonella abundance. The relative abundance of Veillonella was markedly elevated in NSCLC patients (stage IIIB-IV) relative to patients (stage I-IIIA) (P = 0.0266). Notably, the Veillonella-low group had a higher fraction of early-stage patients, whereas the Veillonella-high group was predominantly composed of late-stage patients (Figure 5A). Furthermore, when the relative abundance of Veillonella was categorized into high and low groups, the high group demonstrated a significantly greater Ki-67 expression level compared to the low group (Figure 5C). Additionally, a trend of higher PD-L1 expression was detected in Veillonella-rich subjects (Figure 5D), and Veillonella abundance appeared greater in NSCLC patients with distant metastases compared to the M0 group. However, neither difference reached statistical significance (Figure 5B). Lung nodule size and Veillonella abundance were not notably related (Figure 5E).
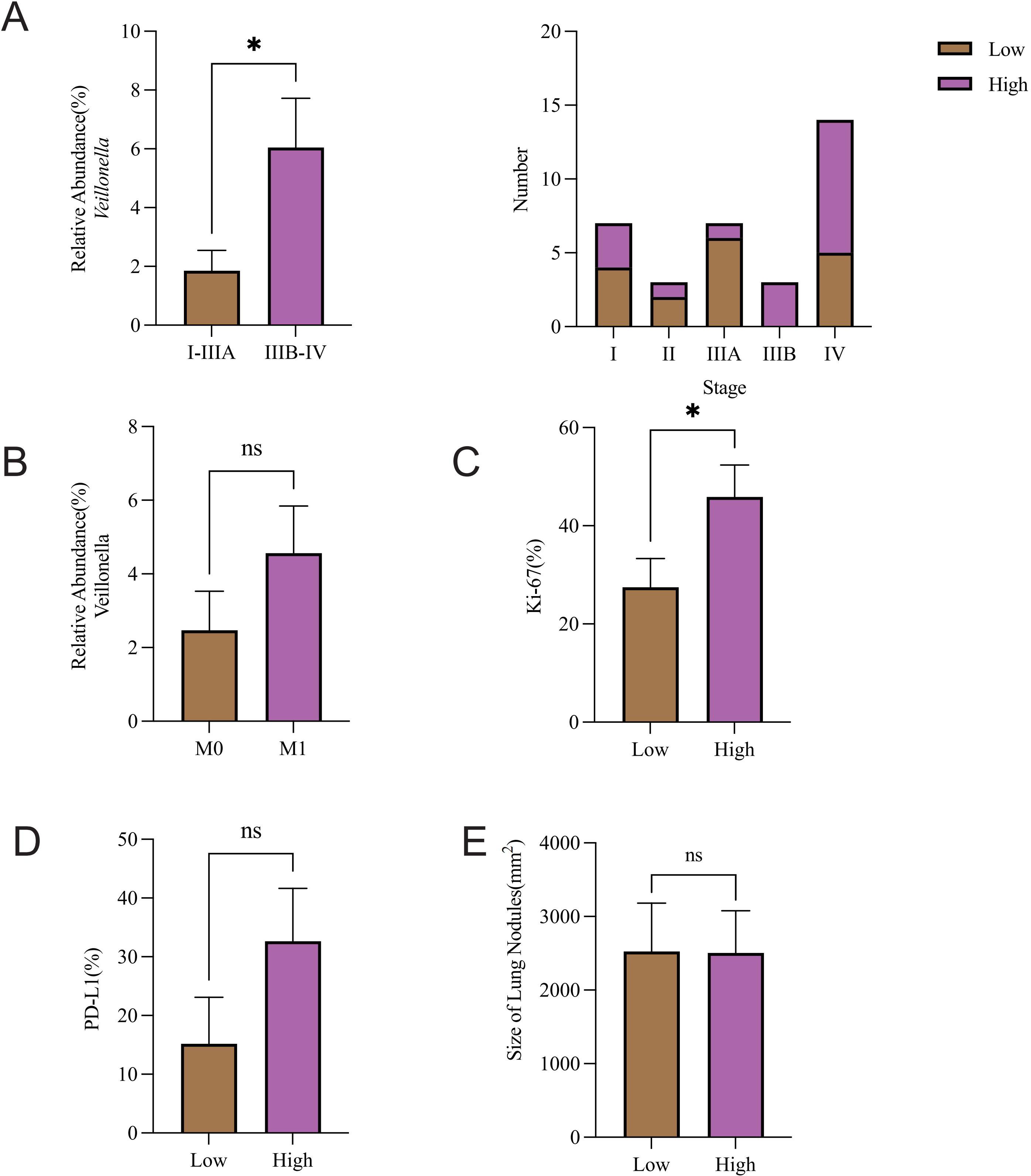
Figure 5. Association between Veillonella abundance and tumor stage, proliferation, and PD-L1. Association of Veillonella relative abundance with (A) pathological stage and (B) metastasis. Comparative analysis of (C) Ki-67 expression, (D) PD-L1 expression, and (E) Lung nodule size across Veillonella expression groups was evaluated. *P-value < 0.05, ns P-value > 0.05.
3.6 Altered commensal microbiota is linked to immune system disturbances and systemic inflammation in NSCLC
To investigate the relationship between microbial abundance and immune-inflammatory responses, we analyzed the immune profiles of patients stratified by Prevotella and Veillonella abundance. Spearman correlation analysis of the top 30 microbial taxa revealed that Prevotella exhibited a substantial positive association with Veillonella (Figure 6A). According to the median abundance of Prevotella and Veillonella, patients were grouped into high and low abundance categories; SII was markedly elevated in the high-abundance group relative to the low-abundance group (Figure 6B). Furthermore, CD8+ T cell levels were significantly lower in the high-abundance Prevotella and Veillonella group compared to the low-abundance group (Figure 6E). Analysis revealed no significant differences between the two groups in terms of CD3+ T cells, CD4+ T cells, or the CD4+/CD8+ T cell ratio (Figure 6F).
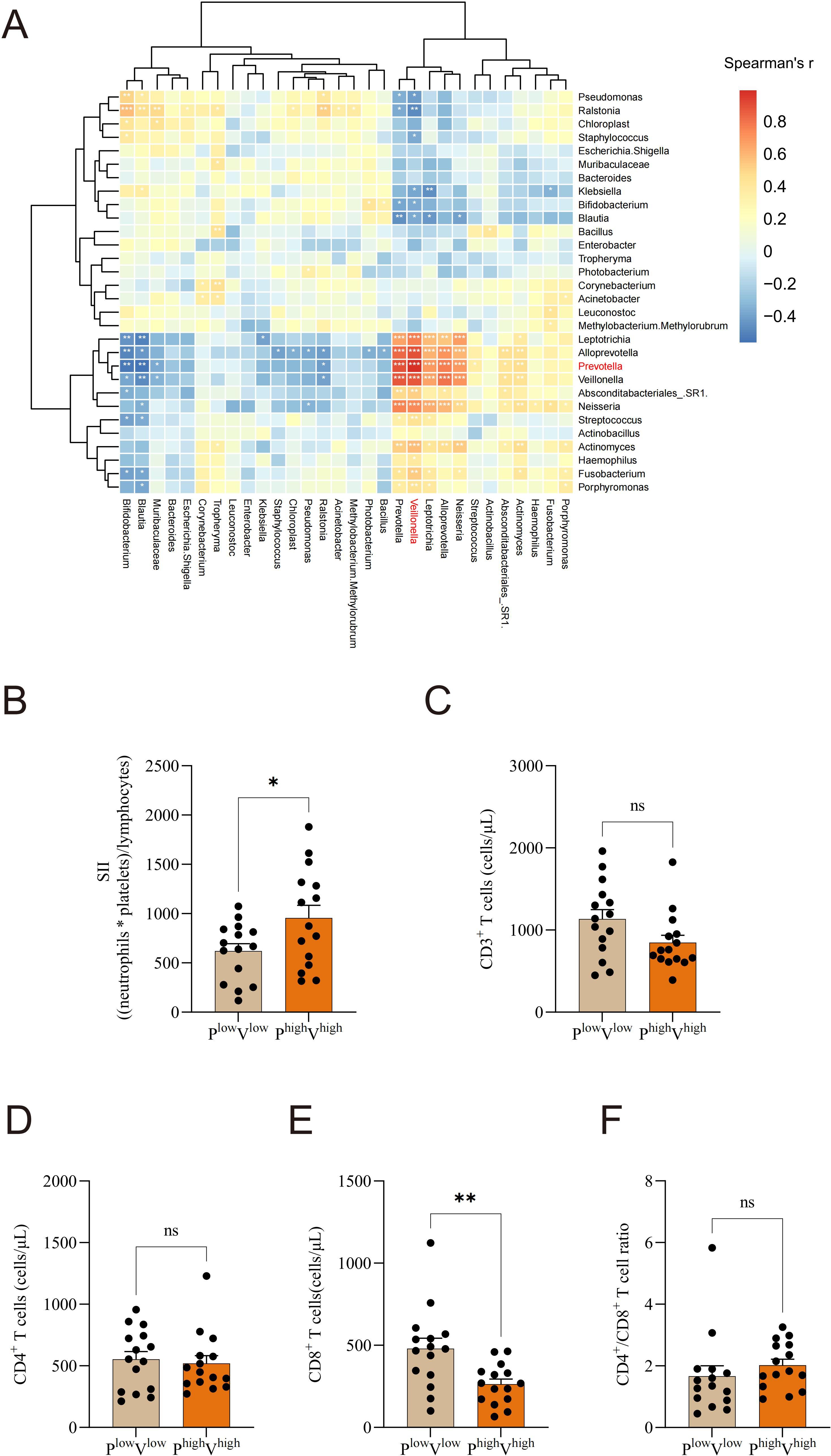
Figure 6. Altered commensal microbiota is linked to immune system disturbances and systemic inflammation in NSCLC. (A) Spearman correlation analysis of the top 30 microbial taxa, (B) Based on Prevotella and Veillonella median levels, individuals were classified into high- and low-abundance groups. The systemic immune-inflammation index (SII) was compared between the high-abundance Prevotella and Veillonella group and the low-abundance group. (C) CD3+ T cells, (D) CD4+ T cells, (E) CD8+ T cells, and (F) CD4+/CD8+ T cell ratio levels were compared between the high-abundance Prevotella and Veillonella group and the low-abundance group. *P-value < 0.05; **P-value < 0.01, ***P-value < 0.001, ns P-value > 0.05.
4 Discussion
Lung microbial dysbiosis has been implicated in lung cancer (28), yet its relationship with tumor progression and immunity remains incompletely understood. Investigating this interplay is essential for elucidating the potential role of microbial alterations in cancer development. In this study, we collected BALF from both the tumor-affected and contralateral healthy lungs of newly diagnosed lung cancer patients and performed 16S rRNA sequencing to profile microbial communities. We further analyzed microbial diversity, identified dysbiotic microbial taxa, and explored their associations with tumor stage, metastasis, and systemic immune status. Our findings contribute to a more profound understanding of lung cancer-associated microbial dysbiosis and its potential relevance to disease progression and immune modulation.
In this study, instead of using healthy individuals as controls, we selected the contralateral healthy lung of the same patient as an internal control, thereby minimizing inter-individual heterogeneity (29–31). Consistent with the findings from other studies (29, 32), our results confirm that no notable variable in α-diversity was found between the tumor-affected and contralateral healthy sides from lung cancer patients. However, distinct microbial β-diversity patterns between the tumor-affected and contralateral healthy sides underscore the importance of analyzing both sites to understand disease-associated microbial alterations.
Previous studies have demonstrated that bacterial composition and abundance differ among different pathological types (33–35). In our study, we observed that the top 10 most abundant bacterial genera were largely similar between LUAD and LUSC, whereas SCLC exhibited distinct microbial characteristics. The results emphasize the clinical significance of microbiome-based stratification in lung cancer subtypes.
Accumulating evidence highlights microbial dysbiosis is closely linked to cancer onset, progression, and patient outcomes (36–38). In our study, we observed an enrichment of Prevotella and Veillonella from the tumor-affected BALF of lung cancer patients. Similarly, Wen Zeng et al. reported a marked rise in Prevotella and Veillonella from lung cancer patients, further supporting our findings (39). Previous studies have demonstrated that Prevotella facilitates tumorigenesis in diverse malignancies, including lung cancer through upregulation of pro-inflammatory cytokines (e.g., IL-1β) and oral squamous cell carcinoma via suppression of tumor suppressor gene expression and tumor microenvironment remodeling (21, 40). Additionally, it accelerates breast cancer progression through the depletion of the host’s intrinsic indole-3-pyruvic acid (41). Notably, experimental evidence also supports these mechanisms, as Prevotella copri-colonized mice develop marked inflammation and immune dysregulation (42), reinforcing the link between Prevotella, immunity and cancer progression. While Veillonella has been recognized as a promising marker for disease assessment and classification (8, 43). Furthermore, Veillonella has been associated with activation of tumor-promoting pathways, including PI3K (21, 44–47) and the Nod2/CCN4/NF-κB axis, which may contribute to inflammatory responses and proliferation in non-small cell lung adenocarcinoma. V. parvula mediates activation of the Nod2/CCN4/NF-κB signaling pathway to promote non-small cell lung adenocarcinoma progression (48). Additionally, V. parvula has been linked to induce B cells in the tumor microenvironment, potentially promoting colorectal tumor development (49). While our study cannot fully resolve the causal direction between microbial dysbiosis and immune dysregulation, experimental models (e.g., Prevotella-colonized mice developing inflammation) and their tumor-promoting mechanisms (e.g., NF-κB-mediated cytokine upregulation) suggest that dysbiosis may contribute to immune dysregulation in lung cancer. Further longitudinal studies are warranted to validate this hypothesis.
In contrast, we noted a substantial decline in Pseudomonas and Staphylococcus abundance in the tumor-affected lung compared to the contralateral healthy side. Interestingly, recent studies have highlighted the potential anticancer properties of Pseudomonas aeruginosa, particularly through azurin, a protein known to inhibit tumor growth (50). Genetically modified P. aeruginosa strains have been shown to induce cancer cell death, inhibit proliferative signaling pathways, and activate anti-tumor immune responses (51–58). This study raises the intriguing possibility that the depletion of Pseudomonas in lung cancer patients may alter the tumor microenvironment in a way that facilitates cancer progression. However, more research is essential to illuminate the underlying biological pathways. While the reduced Staphylococcus abundance challenges its previously reported pro-tumorigenic role (59, 60), it highlights the complexity of host-microbiome interactions in lung cancer progression.
Microbiota significantly contribute to tumor progression. Our study report that Veillonella abundance is significantly higher in lung cancer patients at stage IIIB-IV (typically not surgery-eligible), compared with those at stage I-IIIA (usually surgery-eligible). This discovery offers a new potential biological marker for helping assess surgical eligibility in NSCLC patients, enabling more precise guidance for clinical treatment strategies. Similarly, previous research has demonstrated significant differences in fecal microbiota between early- and late-stage melanoma patients (61). In our study, NSCLC patients with distant metastases (M1) exhibited higher Veillonella abundance than the M0 group, though the difference wasn’t statistically significant, probably owing to the limited sample size. This warrants verification through large-cohort studies. Furthermore, our study identified that patients with higher Veillonella abundance exhibited a significant increase in Ki-67 expression, a well-established marker of cellular proliferation (62, 63). Veillonella is known to be involved in lactate metabolism (64), and excessive lactate accumulation has been shown to trigger the activation of the HIF-1α, leading to enhanced tumor proliferation (65, 66). These findings suggest that Veillonella may potentially contribute to a tumor-promoting microenvironment by modulating lactate metabolism and hypoxia-related signaling pathways. From a translational medicine viewpoint, detecting specific microbial taxa in BALF, like Prevotella and Veillonella, could allow for helping develop treatment plans. Moreover, these microbial biomarkers have prognostic potential. For example, the correlation between Veillonella abundance and tumor stage/Ki-67 expression indicates it could monitor disease progression and predict prognoses.
Systemic inflammation is a hallmark of cancer progression (67), and an elevated systemic immune-inflammation index (SII) is associated with poor prognosis in NSCLC (68–72). In our study, Prevotella and Veillonella abundance was positively correlated with SII, suggesting a link to a pro-inflammatory state. CD8+ T cells are key to anti-tumor immunity (73). The observed negative correlation between Prevotella and Veillonella abundance and CD8+ T cell levels suggests potential immune suppression. Further research is needed to determine whether Prevotella and Veillonella actively promote tumor progression or serve as biomarkers of disease severity. The link between Prevotella, Veillonella, systemic inflammation, and immune suppression suggests that these microbial features might predict immune therapy responses, helping clinicians guide patient stratification and develop personalized treatment regimens.
This study has several limitations. First, while 16S rRNA sequencing provided taxonomic profiling of BALF microbiota, its resolution is insufficient to identify microbial species or strains, which may obscure functionally distinct subgroups. Future studies should employ shotgun metagenomic sequencing (mNGS) or culture-based strain isolation to resolve microbial genomic heterogeneity and validate their immunomodulatory roles. Second, the small cohort of SCLC patients limits the statistical power to detect clinically meaningful associations. Multi-center studies with expanded sample sizes are warranted to confirm the generalizability of our findings. Third, although significant correlations were observed between dysbiosis (e.g., Prevotella/Veillonella enrichment) and immune dysregulation, these associations do not establish causality. Mechanistic validation using germ-free murine models colonized with patient-derived microbiota or in vitro co-culture systems is critical to delineate microbial-immune crosstalk in the future. Lastly, while microbiota-targeted interventions (e.g., probiotics, phage therapy, or precision antibiotics) represent promising adjuvant strategies for NSCLC, their clinical efficacy and safety require rigorous evaluation.
In conclusion, our study demonstrates that tumor-affected lower airways experience a significant disruption in microbial homeostasis, relative to the contralateral healthy lower airway in lung cancer patients. By utilizing the contralateral lung as an internal control, we provide a refined analysis of microbial alterations, revealing bacterial taxa that may be associated with tumor progression through inflammation and immune modulation. Specifically, Prevotella and Veillonella were enriched in tumor-affected lungs and correlated with systemic inflammation and immune suppression. These results provide a framework for upcoming studies on microbiome-based biomarkers for disease monitoring and prognostic assessment in lung cancer.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1134686.
Ethics statement
The studies involving humans were approved by the Research and Clinical Trial Ethics Committee of the First Affiliated Hospital of Zhengzhou University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
JC: Data curation, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. HW: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 81972182) and the Health Commission of Henan Province (No. 201100310600).
Acknowledgments
We appreciate the support provided by the Translational Medical Center at The First Affiliated Hospital of Zhengzhou University in accessing analytic instruments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1589843/full#supplementary-material
References
1. Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. (2025) 75:10–45. doi: 10.3322/caac.21871
2. Wu Y, He S, Cao M, Teng Y, Li Q, Tan N, et al. Comparative analysis of cancer statistics in China and the United States in 2024. Chin Med J (Engl). (2024) 137:3093–100. doi: 10.1097/CM9.0000000000003442
3. Houston KA, Mitchell KA, King J, White A, Ryan BM. Histologic lung cancer incidence rates and trends vary by race/ethnicity and residential county. J Thorac Oncol. (2018) 13:497–509. doi: 10.1016/j.jtho.2017.12.010
4. Meng Y, Mao Y, Tang Z, Qiu X, Bajinka O, Tan Y, et al. Crosstalk between the lung microbiome and lung cancer. Microb Pathog. (2023) 178:106062. doi: 10.1016/j.micpath.2023.106062
5. Madapoosi SS, Cruickshank-Quinn C, Opron K, Erb-Downward JR, Begley LA, Li G, et al. Lung microbiota and metabolites collectively associate with clinical outcomes in milder stage chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2022) 206:427–39. doi: 10.1164/rccm.202110-2241OC
6. Khan FH, Bhat BA, Sheikh BA, Tariq L, Padmanabhan R, Verma JP, et al. Microbiome dysbiosis and epigenetic modulations in lung cancer: From pathogenesis to therapy. Semin Cancer Biol. (2022) 86:732–42. doi: 10.1016/j.semcancer.2021.07.005
7. Saeedi P, Salimian J, Ahmadi A, Imani Fooladi AA. The transient but not resident (TBNR) microbiome: a Yin Yang model for lung immune system. Inhal Toxicol. (2015) 27:451–61. doi: 10.3109/08958378.2015.1070220
8. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. (2015) 5:3111–22.
9. Jiang X, Liu S, Zhang Y, Ji Y, Sohail A, Cao C, et al. Free-flow isoelectric focusing for comprehensive separation and analysis of human salivary microbiome for lung cancer. Anal Chem. (2020) 92:12017–25. doi: 10.1021/acs.analchem.0c02627
10. Zhang W, Luo J, Dong X, Zhao S, Hao Y, Peng C, et al. Salivary microbial dysbiosis is associated with systemic inflammatory markers and predicted oral metabolites in non-small cell lung cancer patients. J Cancer. (2019) 10:1651–62. doi: 10.7150/jca.28077
11. Teng JJ, Zhang J, Zhang TY, Zhang S, Li BS. Prognostic value of peripheral blood lymphocyte-to-monocyte ratio in patients with solid tumors: a meta-analysis. Onco Targets Ther. (2016) 9:37–47. doi: 10.2147/OTT.S94458
12. Liu YY, Yang QF, Yang JS, Cao RB, Liang JY, Liu YT, et al. Characteristics and prognostic significance of profiling the peripheral blood T-cell receptor repertoire in patients with advanced lung cancer. Int J Cancer. (2019) 145:1423–31. doi: 10.1002/ijc.32145
13. Cui X, Jia Z, Chen D, Xu C, Yang P. The prognostic value of the C-reactive protein to albumin ratio in cancer: An updated meta-analysis. Med (Baltimore). (2020) 99:e19165. doi: 10.1097/MD.0000000000019165
14. Qiang G, Liang C, Xiao F, Yu Q, Wen H, Song Z, et al. Prognostic significance of platelet-to-lymphocyte ratio in non-small-cell lung cancer: a meta-analysis. Onco Targets Ther. (2016) 9:869–76. doi: 10.2147/OTT.S96804
15. Guo D, Zhang J, Jing W, Liu J, Zhu H, Fu L, et al. Prognostic value of systemic immune-inflammation index in patients with advanced non-small-cell lung cancer. Future Oncol. (2018) 14:2643–50. doi: 10.2217/fon-2018-0285
16. Huang W, Luo J, Wen J, Jiang M. The relationship between systemic immune inflammatory index and prognosis of patients with non-small cell lung cancer: A meta-analysis and systematic review. Front Surg. (2022) 9:898304. doi: 10.3389/fsurg.2022.898304
17. Liu J, Li S, Zhang S, Liu Y, Ma L, Zhu J, et al. Systemic immune-inflammation index, neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio can predict clinical outcomes in patients with metastatic non-small-cell lung cancer treated with nivolumab. J Clin Lab Anal. (2019) 33:e22964. doi: 10.1002/jcla.22964
18. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. (2009) 114:1537–44. doi: 10.1182/blood-2008-12-195792
19. Prado-Garcia H, Romero-Garcia S, Aguilar-Cazares D, Meneses-Flores M, Lopez-Gonzalez JS. Tumor-induced CD8+ T-cell dysfunction in lung cancer patients. Clin Dev Immunol. (2012) 2012:741741. doi: 10.1155/2012/741741
20. Thommen DS, Schreiner J, Muller P, Herzig P, Roller A, Belousov A, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res. (2015) 3:1344–55. doi: 10.1158/2326-6066.CIR-15-0097
21. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. (2018) 198:1188–98. doi: 10.1164/rccm.201710-2118OC
22. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184:957–63. doi: 10.1164/rccm.201104-0655OC
23. Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. (2012) 186:536–45. doi: 10.1164/rccm.201204-0693OC
24. Masuhiro K, Tamiya M, Fujimoto K, Koyama S, Naito Y, Osa A, et al. Bronchoalveolar lavage fluid reveals factors contributing to the efficacy of PD-1 blockade in lung cancer. JCI Insight. (2022) 7:e157915. doi: 10.1172/jci.insight.157915
25. Baranova E, Druzhinin V, Matskova L, Demenkov P, Volobaev V, Minina V, et al. Sputum microbiome composition in patients with squamous cell lung carcinoma. Life (Basel). (2022) 12:1365. doi: 10.3390/life12091365
26. Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
27. Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. (2013) 10:57–9. doi: 10.1038/nmeth.2276
28. Thapa R, Magar AT, Shrestha J, Panth N, Idrees S, Sadaf T, et al. Influence of gut and lung dysbiosis on lung cancer progression and their modulation as promising therapeutic targets: a comprehensive review. MedComm (2020). (2024) 5:e70018. doi: 10.1002/mco2.70018
29. Zhuo M, An T, Zhang C, Wang Z. Characterization of microbiota in cancerous lung and the contralateral non-cancerous lung within lung cancer patients. Front Oncol. (2020) 10:1584. doi: 10.3389/fonc.2020.01584
30. Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer. (2018) 142:769–78. doi: 10.1002/ijc.31098
31. Wu Z, Tang J, Zhuang R, Meng D, Zhang L, Gu C, et al. The microbiome of lower respiratory tract and tumor tissue in lung cancer manifested as radiological ground-glass opacity. Front Bioeng Biotechnol. (2022) 10:892613. doi: 10.3389/fbioe.2022.892613
32. Cheng C, Wang Z, Wang J, Ding C, Sun C, Liu P, et al. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl Lung Cancer Res. (2020) 9:693–704. doi: 10.21037/tlcr-19-590
33. Sun Y, Liu Y, Li J, Tan Y, An T, Zhuo M, et al. Characterization of lung and oral microbiomes in lung cancer patients using culturomics and 16S rRNA gene sequencing. Microbiol Spectr. (2023) 11:e0031423. doi: 10.1128/spectrum.00314-23
34. Huang D, Su X, Yuan M, Zhang S, He J, Deng Q, et al. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res. (2019) 9:2047–63.
35. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep. (2019) 9:12838. doi: 10.1038/s41598-019-49195-w
36. Wang KK, He KY, Yang JY, Liu MJ, Guo JR, Liang JY, et al. Lactobacillus suppresses tumorigenesis of oropharyngeal cancer via enhancing anti-tumor immune response. Front Cell Dev Biol. (2022) 10:842153. doi: 10.3389/fcell.2022.842153
37. Zhu Z, Huang J, Li X, Xing J, Chen Q, Liu R, et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes. (2020) 12:1788891. doi: 10.1080/19490976.2020.1788891
38. Yu MR, Kim HJ, Park HR. Fusobacterium nucleatum accelerates the progression of colitis-associated colorectal cancer by promoting EMT. Cancers (Basel). (2020) 12:2728. doi: 10.3390/cancers12102728
39. Zeng W, Zhao C, Yu M, Chen H, Pan Y, Wang Y, et al. Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered. (2022) 13:6665–77. doi: 10.1080/21655979.2022.2045843
40. Zhou Y, Qin Y, Ma J, Li Z, Heng W, Zhang L, et al. Heat-killed Prevotella intermedia promotes the progression of oral squamous cell carcinoma by inhibiting the expression of tumor suppressors and affecting the tumor microenvironment. Exp Hematol Oncol. (2024) 13:33. doi: 10.1186/s40164-024-00500-y
41. Su J, Lin X, Li D, Yang C, Lv S, Chen X, et al. Prevotella copri exhausts intrinsic indole-3-pyruvic acid in the host to promote breast cancer progression: inactivation of AMPK via UHRF1-mediated negative regulation. Gut Microbes. (2024) 16:2347757. doi: 10.1080/19490976.2024.2347757
42. Qian X, Zhang HY, Li QL, Ma GJ, Chen Z, Ji XM, et al. Integrated microbiome, metabolome, and proteome analysis identifies a novel interplay among commensal bacteria, metabolites and candidate targets in non-small cell lung cancer. Clin Transl Med. (2022) 12:e947. doi: 10.1002/ctm2.947
43. Rybojad P, Los R, Sawicki M, Tabarkiewicz J, Malm A. Anaerobic bacteria colonizing the lower airways in lung cancer patients. Folia Histochem Cytobiol. (2011) 49:263–6. doi: 10.5603/fhc.2011.0036
44. Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. (2010) 2:26ra5. doi: 10.1126/scitranslmed.3000251
45. Kadara H, Shen L, Fujimoto J, Saintigny P, Chow CW, Lang W, et al. Characterizing the molecular spatial and temporal field of injury in early-stage smoker non-small cell lung cancer patients after definitive surgery by expression profiling. Cancer Prev Res (Phila). (2013) 6:8–17. doi: 10.1158/1940-6207.CAPR-12-0290
46. Kadara H, Fujimoto J, Yoo SY, Maki Y, Gower AC, Kabbout M, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst. (2014) 106:dju004. doi: 10.1093/jnci/dju004
47. Tsay JC, Li Z, Yie TA, Wu F, Segal L, Greenberg AK, et al. Molecular characterization of the peripheral airway field of cancerization in lung adenocarcinoma. PloS One. (2015) 10:e0118132. doi: 10.1371/journal.pone.0118132
48. Zeng W, Wang Y, Wang Z, Yu M, Liu K, Zhao C, et al. Veillonella parvula promotes the proliferation of lung adenocarcinoma through the nucleotide oligomerization domain 2/cellular communication network factor 4/nuclear factor kappa B pathway. Discov Oncol. (2023) 14:129. doi: 10.1007/s12672-023-00748-6
49. Qian Y, Kang Z, Zhao L, Chen H, Zhou C, Gao Q, et al. Berberine might block colorectal carcinogenesis by inhibiting the regulation of B-cell function by Veillonella parvula. Chin Med J (Engl). (2023) 136:2722–31. doi: 10.1097/CM9.0000000000002752
50. Choi JK, Naffouje SA, Goto M, Wang J, Christov K, Rademacher DJ, et al. Cross-talk between cancer and Pseudomonas aeruginosa mediates tumor suppression. Commun Biol. (2023) 6:16. doi: 10.1038/s42003-022-04395-5
51. Xiu D, Cheng M, Zhang W, Ma X, Liu L. Pseudomonas aeruginosa-mannose-sensitive hemagglutinin inhibits chemical-induced skin cancer through suppressing hedgehog signaling. Exp Biol Med (Maywood). (2020) 245:213–20. doi: 10.1177/1535370219897240
52. Qi JL, He JR, Jin SM, Yang X, Bai HM, Liu CB, et al. P. aeruginosa mediated necroptosis in mouse tumor cells induces long-lasting systemic antitumor immunity. Front Oncol. (2020) 10:610651. doi: 10.3389/fonc.2020.610651
53. Zhao X, Wu X, Yu W, Cai X, Liu Q, Fu X, et al. PA-MSHA inhibits proliferation and induces apoptosis in human non-small cell lung cancer cell lines with different genotypes. Mol Med Rep. (2016) 14:5369–76. doi: 10.3892/mmr.2016.5869
54. Wei Y, Liu D, Jin X, Gao P, Wang Q, Zhang J, et al. PA-MSHA inhibits the growth of doxorubicin-resistant MCF-7/ADR human breast cancer cells by downregulating Nrf2/p62. Cancer Med. (2016) 5:3520–31. doi: 10.1002/cam4.938
55. Wang C, Hu Z, Zhu Z, Zhang X, Wei Z, Zhang Y, et al. The MSHA strain of Pseudomonas aeruginosa (PA-MSHA) inhibits gastric carcinoma progression by inducing M1 macrophage polarization. Tumour Biol. (2016) 37:6913–21. doi: 10.1007/s13277-015-4451-6
56. Chang L, Xiao W, Yang Y, Li H, Xia D, Yu G, et al. Pseudomonas aeruginosa-mannose-sensitive hemagglutinin inhibits epidermal growth factor receptor signaling pathway activation and induces apoptosis in bladder cancer cells in vitro and in vivo. Urol Oncol. (2014) 32:36 e11–8. doi: 10.1016/j.urolonc.2013.02.013
57. Liu ZB, Hou YF, Zhu J, Hu DL, Jin W, Ou ZL, et al. Inhibition of EGFR pathway signaling and the metastatic potential of breast cancer cells by PA-MSHA mediated by type 1 fimbriae via a mannose-dependent manner. Oncogene. (2010) 29:2996–3009. doi: 10.1038/onc.2010.70
58. Cao Z, Shi L, Li Y, Wang J, Wang D, Wang G, et al. Pseudomonas aeruginosa: mannose sensitive hemagglutinin inhibits the growth of human hepatocarcinoma cells via mannose-mediated apoptosis. Dig Dis Sci. (2009) 54:2118–27. doi: 10.1007/s10620-008-0603-5
59. Qi JL, He JR, Liu CB, Jin SM, Gao RY, Yang X, et al. Pulmonary Staphylococcus aureus infection regulates breast cancer cell metastasis via neutrophil extracellular traps (NETs) formation. MedComm (2020). (2020) 1:188–201. doi: 10.1002/mco2.22
60. Hattar K, Reinert CP, Sibelius U, Gokyildirim MY, Subtil FSB, Wilhelm J, et al. Lipoteichoic acids from Staphylococcus aureus stimulate proliferation of human non-small-cell lung cancer cells in vitro. Cancer Immunol Immunother. (2017) 66:799–809. doi: 10.1007/s00262-017-1980-4
61. Witt RG, Cass SH, Tran T, Damania A, Nelson EE, Sirmans E, et al. Gut microbiome in patients with early-stage and late-stage melanoma. JAMA Dermatol. (2023) 159:1076–84. doi: 10.1001/jamadermatol.2023.2955
62. Peng Y, Chen FF, Ge J, Zhu JY, Shi XE, Li X, et al. miR-429 inhibits differentiation and promotes proliferation in porcine preadipocytes. Int J Mol Sci. (2016) 17:2047. doi: 10.3390/ijms17122047
63. Wang J, Wu M, Zheng D, Zhang H, Lv Y, Zhang L, et al. Garcinol inhibits esophageal cancer metastasis by suppressing the p300 and TGF-beta1 signaling pathways. Acta Pharmacol Sin. (2020) 41:82–92. doi: 10.1038/s41401-019-0271-3
64. Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. (2019) 25:1104–9. doi: 10.1038/s41591-019-0485-4
65. Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. (2015) 21:65–80. doi: 10.1016/j.cmet.2014.12.005
66. Certo M, Tsai CH, Pucino V, Ho PC, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol. (2021) 21:151–61. doi: 10.1038/s41577-020-0406-2
67. Muhammed A, Fulgenzi CAM, Dharmapuri S, Pinter M, Balcar L, Scheiner B, et al. The systemic inflammatory response identifies patients with adverse clinical outcome from immunotherapy in hepatocellular carcinoma. Cancers (Basel). (2021) 14:186. doi: 10.3390/cancers14010186
68. Li W, Ma G, Deng Y, Chen W, Liu Z, Chen F, et al. Systemic immune-inflammation index is a prognostic factor for breast cancer patients after curative resection. Front Oncol. (2021) 11:570208. doi: 10.3389/fonc.2021.570208
69. Zhang Y, Chen Y, Guo C, Li S, Huang C. Systemic immune-inflammation index as a predictor of survival in non-small cell lung cancer patients undergoing immune checkpoint inhibition: A systematic review and meta-analysis. Crit Rev Oncol Hematol. (2025) 210:104669. doi: 10.1016/j.critrevonc.2025.104669
70. Huai Q, Luo C, Song P, Bie F, Bai G, Li Y, et al. Peripheral blood inflammatory biomarkers dynamics reflect treatment response and predict prognosis in non-small cell lung cancer patients with neoadjuvant immunotherapy. Cancer Sci. (2023) 114:4484–98. doi: 10.1111/cas.15964
71. Fu F, Deng C, Wen Z, Gao Z, Zhao Y, Han H, et al. Systemic immune-inflammation index is a stage-dependent prognostic factor in patients with operable non-small cell lung cancer. Transl Lung Cancer Res. (2021) 10:3144–54. doi: 10.21037/tlcr-21-267
72. Tong YS, Tan J, Zhou XL, Song YQ, Song YJ. Systemic immune-inflammation index predicting chemoradiation resistance and poor outcome in patients with stage III non-small cell lung cancer. J Transl Med. (2017) 15:221. doi: 10.1186/s12967-017-1326-1
Keywords: non-small cell lung cancer, lung microbiota, Prevotella, Veillonella, systemic immune-inflammation index, CD8 + T cell
Citation: Cheng J and Wang H (2025) Lung microbiome alterations correlate with immune imbalance in non-small cell lung cancer. Front. Immunol. 16:1589843. doi: 10.3389/fimmu.2025.1589843
Received: 08 March 2025; Accepted: 24 April 2025;
Published: 14 May 2025.
Edited by:
Beiwen Zheng, Zhejiang University, ChinaReviewed by:
Chong Chen, Xuzhou Medical University, ChinaJie Yu, Jiangxi Provincial People’s Hospital, China
Copyright © 2025 Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaqi Wang, d2FuZ2h1YXFpMjAyMkAxNjMuY29t
 Jiuling Cheng
Jiuling Cheng Huaqi Wang
Huaqi Wang