- 1Institute for Organic Chemistry and Biochemistry, Technical University of Darmstadt, Darmstadt, Germany
- 2Goethe University, Frankfurt am Main, Germany
- 3Frankfurt Cancer Institute (FCI), Goethe University, Frankfurt am Main, Germany
- 4German Cancer Consortium (DKTK), Partner Site Frankfurt/Mainz and German Cancer Research Center (DKFZ), Heidelberg, Germany
- 5Mildred Scheel Career Center (MSNZ), Hospital of the Goethe University Frankfurt, Frankfurt am Main, Germany
- 6Centre for Synthetic Biology, Technical University of Darmstadt, Darmstadt, Hesse, Germany
Therapeutic monoclonal antibodies (mAbs) constitute cornerstone therapeutics in oncology, yet their clinical utility is often limited by on-target, off-tumor toxicity due to shared antigen expression in both tumor and healthy tissues. To counteract this issue, various approaches, including pH-dependent, as well as affinity-based and steric hindrance-based masked antibodies, have been developed. Several steric hindrance-based masking strategies have been proposed utilizing non-human proteins, potentially leading to an immunogenic response. To address this challenge, we engineered a modular protein-based masking platform leveraging the high-affinity interaction between human calmodulin (CaM) and a calmodulin-binding peptide (CBP). This strategy enables conditional activation of antibodies via tumor microenvironment (TME)-associated proteases (e.g., MMP-9), minimizing systemic off-tumor binding. The CaM-CBP peptide clamp, composed exclusively of human-derived protein domains, was fused to the amino termini of heavy and light chains of trastuzumab and cetuximab. On-cell binding assays demonstrated up to a 410-fold reduction in EC50 for masked constructs across multiple antigen-antibody systems. Functional validation using a reporter-cell-based antibody-dependent cellular cytotoxicity (ADCC) assay confirmed that masking abrogated effector cell activation, leading to up to 78-fold reduction of EC50 and no ADCC activation at concentrations corresponding to the onset of maximal ADCC activation by unmodified antibodies. Demasking via MMP-9-mediated linker hydrolysis restored antigen binding and ADCC potency. Structural optimization revealed that linker length and clamp positioning critically influenced masking efficiency. This human-derived, modular masking platform mitigates immunogenicity risks while enabling tumor-selective antibody activation. Its adaptability across antibody scaffolds underscores broad applicability for improving the therapeutic index of antibodies.
1 Introduction
Cancer therapy has undergone a profound transformation in recent decades, evolving from surgery and radiotherapy to the advent of chemotherapy and, more recently, to highly targeted biological agents such as monoclonal antibodies (mAbs) and personalized immunotherapies including chimeric antigen receptor (CAR) T cell therapies (1–3). mAbs, in particular, have revolutionized oncology by offering the potential for selective tumor targeting while minimizing collateral damage to healthy tissues. However, this specificity is not absolute; tumor-associated antigens (TAAs) recognized by therapeutic mAbs are often also expressed on normal cells, leading to on-target, off-tumor toxicity that can be dose-limiting and compromise therapeutic efficacy (4–6). To address this challenge, innovative strategies have been developed to mask antibody binding domains and enable conditional activation within the TME (7, 8). These approaches typically involve the attachment of a masking domain via a flexible linker to the N-terminus of the antibody heavy or light chain, thereby sterically blocking antigen engagement in healthy tissues. Upon exposure to TME-specific stimuli, such as elevated protease activity or altered pH, the mask is removed or rendered non-functional, restoring antibody binding and effector functions (7, 9–11). Current masking strategies can be broadly categorized into two mechanistic classes: steric hindrance-based and affinity-based masks. Affinity-based masks employ domains such as anti-idiotypic single-chain variable fragments (scFvs), nanobodies®, or affinity peptides, which bind specifically to the antibody paratope and require custom engineering for each target (9, 12–15). In contrast, steric hindrance-based masks function independently of specific antibody-antigen interactions, offering modularity and facilitating the transfer of masking domains between different antibody scaffolds. This modularity streamlines the development of masked antibody therapeutics and expands their applicability across diverse oncology targets.
We recently masked the therapeutic antibody 6G11 (BI-1206) by isolating an anti-idiotypic scFv from a chicken immunization library and fusing the molecule to the N-terminus of the VL. This modification effectively blocked the interaction between 6G11 and Fcγ receptor IIB. However, the affinity of the masking scFv required rational engineering to ensure that antigen binding could be fully restored following protease-mediated demasking, specifically via MMP-9 cleavage (15). A prominent example of an affinity peptide-based approach is the cetuximab probody PB1 by CytomX. By fusing a blocking peptide identified from a bacterial peptide display library to the amino terminus of VL of cetuximab, binding to EGFR was diminished and could be restored upon incubation with proteases (10). This affinity peptide-based masking approach was then used to generate a conditional PD-1/PD-L1 probody and a masked anti-CD166 ADC (16). In Phase I/II trials, the masked ADC demonstrated translational and clinical activity in a variety of different tumor types (17). While effective, such affinity-based masks are always antibody-specific and require custom optimization for each target.
In contrast, steric hindrance-based masking approaches offer broader applicability. For instance, Seagen engineered a generic masking platform by fusing heterodimeric coiled-coil domains to the amino termini of the heavy and light chains of therapeutic antibodies, separated by MMP-2/-9-cleavable linkers. Masking HER2 and CD-19 targeting antibodies with a WinZip-A1B1-derived coiled-coil from a library approach resulted in a 753-fold reduction in on-cell EC50 for HER2-positive SK-BR-3 cells and led to diminished internalization and complement-dependent cellular cytotoxicity (CDC) in CD19-positive Raji cells (18). This masking approach was then transferred to an anti-CD47 antibody, generating the masked antibody SGN-CD47M. Based on promising results in tolerability studies in cynomolgus monkeys and anti-tumor activity in xenograft mouse models, this therapeutic has recently advanced into phase I dose-escalation clinical trials (19). Beyond protease activation, the unique biochemical features of the TME, such as acidic pH, can also be leveraged for conditional antibody activation. For example, engineering a pH-sensitive variant of the Mycoplasma genitalium-derived protein M enabled the reversible masking of trastuzumab. The engineered protein M binds the Fv light chain domain with high affinity at physiological pH, blocking antigen binding, but dissociates at lower pH, partially restoring binding to HER2-positive cells in a time-dependent manner (11).
Steric hindrance-based masking strategies for therapeutic antibodies offer distinct advantages over affinity-based masking, including broad modularity, accelerated development timelines, and simplified manufacturing processes. However, current implementations of such approaches often employ masking domains of non-human origin, which could elicit undesirable immunogenic responses in patients. To address this limitation, we sought to develop a generic masking platform using exclusively human-derived proteins. Our strategy exploits the high-affinity interaction between calmodulin (CaM) and a calmodulin-binding peptide (CBP) derived from the carboxy-terminal domain of skeletal muscle myosin light chain kinase (20). Calmodulin, a highly conserved 17 kDa calcium sensor, adopts a dumbbell-like structure with two globular domains connected by a central α-helix, each domain containing two EF-hand calcium-binding motifs (21). Upon binding to the α-helical CBP, CaM undergoes a dramatic conformational change, wrapping around the peptide to form a compact globular complex (Figure 1A) (22). By genetically fusing CaM and CBP to the amino-terminal domains of antibody heavy and light chains, we engineered a modular peptide clamp capable of sterically occluding antibody paratopes. The resulting peptide clamp is linked to the antibody via a protease-cleavable sequence sensitive to matrix metalloproteinases (MMPs), particularly MMP-2 and MMP-9, which are frequently overexpressed in the tumor microenvironment. This design enables conditional, tumor-selective activation of the antibody, minimizing systemic toxicity and on-target, off-tumor effects (Figure 1B).
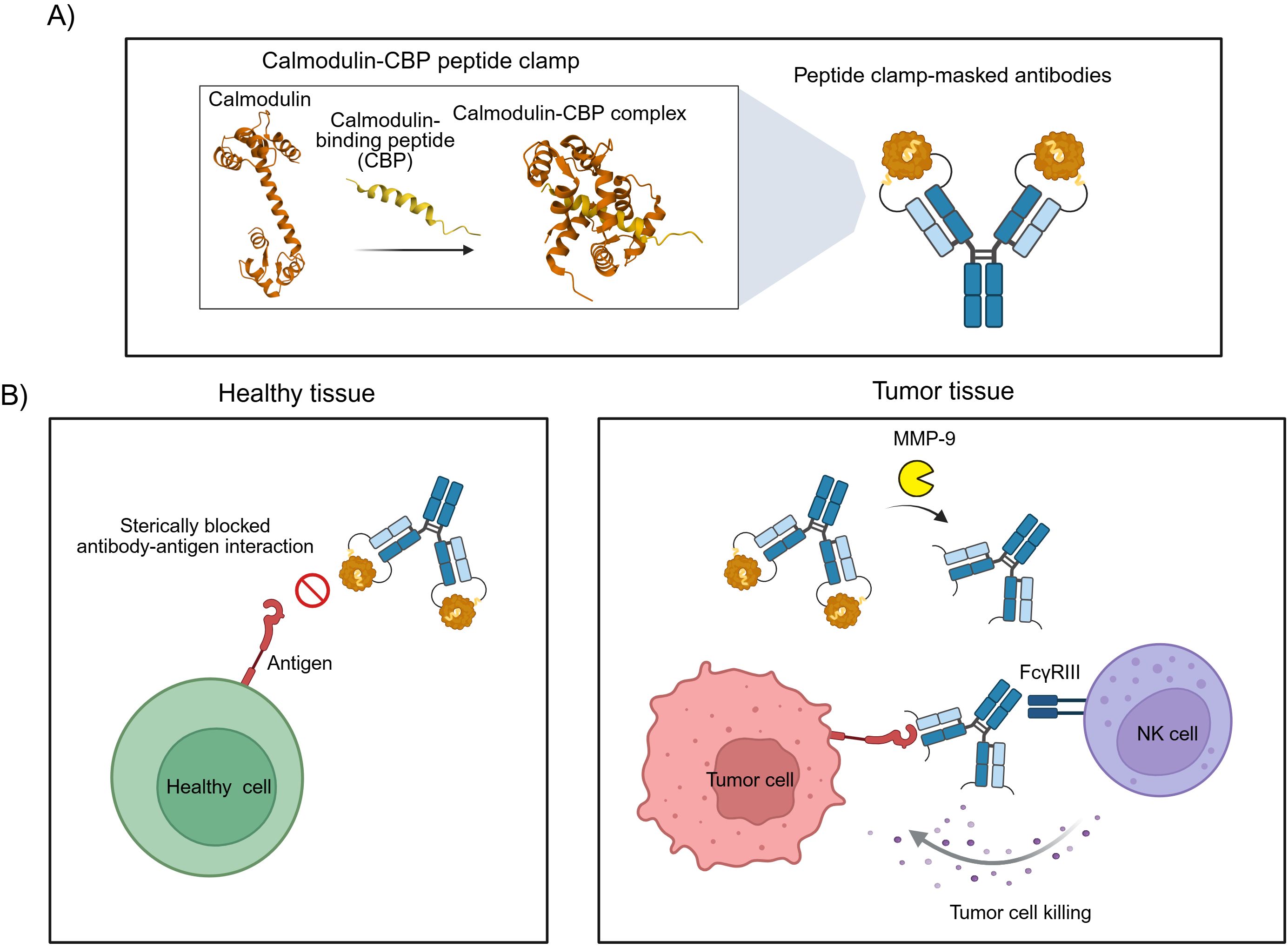
Figure 1. Design and mode of action of antibodies masked with a peptide clamp. (A) Scheme for the masking of therapeutic antibodies with a peptide clamp to generate conditionally active antibodies. By fusing a peptide clamp consisting of calmodulin and calmodulin-binding peptide (CBP) to the N-terminal domain of the heavy and light chain of an antibody via an MMP-9 cleavable linker, masked antibodies are generated. Calmodulin binds to calmodulin-binding peptide, forming a globular CaM-CBP complex. (B) Conceptual illustration of the mode of action of peptide clamp-masked antibodies. In healthy tissue, the binding of the antibody to its antigen is prevented due to steric hindrance in the antibody-antigen interaction introduced by the peptide clamp. Thereby, on-target, but off-tumor side effects are reduced (left panel). Entering the tumor microenvironment, matrix metalloproteinases cleave off the peptide clamp. Removal of the mask restores antibody binding and can enable FcγRIII-mediated killing of tumor cells by natural killer (NK) cells (right panel). Structures used in Figure A: Calmodulin (PDB: 1CLL) and CaM-CBP complex (PDB: 2LV6). Created with Biorender.com.
To validate our approach, we selected as a model for CaM-dependent functional blocking two clinically relevant monoclonal antibodies with known on-target, off-tumor toxicity: cetuximab (anti-EGFR) and trastuzumab (anti-HER2). Trastuzumab showed a 26% response rate in HER2-positive breast cancer patients (23). But in early trials, congestive heart failures were reported. Especially when patients were treated concurrently with trastuzumab and anthracyclines, cardiotoxic effects were observed in 27% of patients (24, 25). As a second example, we chose the monoclonal antibody cetuximab. In Phase II trials of cetuximab treatment in patients with epidermal growth factor receptor (EGFR) positive refractory colorectal cancer, more than 86% of patients developed acne-like skin rash, including 18% developing grade 3 symptoms (26). In healthy tissue, EFGR is commonly expressed in the basal layer of the epidermis and regulates growth, differentiation, and wound healing. By inhibiting EGFR functions, additional side effects arise as changes in the scalp and hair growth, xerosis and pruritus, as well as nail and eye disorders (27–29). In both breast cancer and colorectal cancer, overexpression of MMPs is reported, and increased MMP expression is associated with poor survival rates and tumor progression (30–32). CX-2051, an anti-EpCAM antibody-drug conjugate developed by CytomX Therapeutics using their PROBODY® platform, is designed for protease-dependent activation within the tumor microenvironment. Recent interim results from Phase 1 dose escalation studies have demonstrated promising efficacy and a favorable safety profile (33).
In this study, we demonstrate the efficacy of our CaM-CBP peptide clamp in masking trastuzumab and cetuximab. We investigated the impact of linker length and peptide clamp orientation on masking efficiency, as assessed by on-cell binding and antibody-dependent cell-mediated cytotoxicity (ADCC) reporter assays. Masked antibodies exhibited significantly reduced binding and effector function, which was restored upon in vitro incubation with MMP-9. Additionally, we evaluated the thermal stability and aggregation propensity of masked constructs using fluorescence-based thermal shift assays and analytical size exclusion chromatography, respectively.
2 Materials and methods
2.1 Cell lines
SK-BR-3 and A431 cells were cultivated at 37°C and 5% CO2 and humid atmosphere in Dulbecco’s Eagle Medium (DMEM, Thermo Fisher Scientific) supplemented with 10% FBS (Merck Millipore, Burlington, MA, USA) and 1% Penicillin–Streptomycin (Sigma Aldrich, St. Louis, MI, USA) in T75 cell culture flasks. Sub-culturing was performed every 3–4 days. Expi293F™ cells (Gibco, Thermo Fisher Scientific: A14527) were cultivated at 37°C and 5% CO2 and humid atmosphere in Expi293™ Expression Medium (Thermo Fisher Scientific). Sub-culturing was performed every 3–4 days.
2.2 Cloning, protein expression and purification of masked antibodies
DNA coding for masked antibodies was cloned into pTT5 plasmids via Golden Gate assembly. Therefore, 100 ng of pTT5 plasmid was incubated with a 7-molar excess of insert, BsaI (NEB: R3733), and T4 DNA ligase (NEB: M0202). After the correct plasmid sequence was verified via sequencing, it was used for the transfection of Expi293F™ cells (Gibco, Thermo Fisher Scientific: A14527) following the manufacturer’s protocol, followed by purification via Protein A (1 mL HiTrap Protein A HP, Cytiva: 29048576) on an ÄKTA start chromatography system (Cytiva) as described before (34). Fractions were dialyzed against TBS buffer (150 mM NaCl, 50 mM TRIS pH 7.5).
2.3 Thermal shift assay
The thermal stability of antibodies was determined using a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories GmbH). Briefly, 18 μL of protein solution (0.1-1 μg/μL) was mixed with 2 μL SYPRO® Orange Protein Gel Stain (Sigma-Aldrich) diluted 1:10 with dH2O. Samples were measured in a Hard-Shell 96-well PCR plate. A temperature gradient of 0.5°C per minute was applied. Changes in fluorescence were measured in the FRET channel. The calculation of the thermal stability was performed using Bio-Rad CFX Manager Software version 3.0. Melting curves were visualized in GraphPad Prism 9.
2.4 On-cell binding assay
The on-cell EC50 of masked antibodies was determined by antibody titration using SK-BR-3 cells for masked trastuzumab and A431 cells for masked cetuximab. Therefore, cells were detached using Trypsin-EDTA (Gibco™) and washed once using PBS-B (0.1% BSA). Subsequently, 60.000 to 100.000 cells per well were transferred to a 96 well plate, washed once with PBS-B (0.1% BSA) and then incubated with antibodies in varying concentrations (333 nM to 0.05 nM, 1:3 serial dilution for trastuzumab constructs; 200 nM to 0.03 nM, 1:3 diluton for cetuximab constructs). The cells were incubated for 45 minutes on ice, afterwards washed twice using PBS-B, and then incubated with a 1:75 dilution of goat anti-human Fc PE secondary antibody (Thermo Scientific) for 15 minutes on ice. Afterwards, cells were washed three times before analysis by flow cytometry using a CytoFLEX S (Beckman Coulter). Binding curves were compiled from the geometric mean of the fluorescence in the PE channel and plotted in GraphPad Prism 9 using a sigmoidal 4-parameter logistic regression model to determine EC50 values. Fold reduction values were calculated by dividing the EC50 of masked antibodies by the EC50 of the unmodified parental antibody.
2.5 Antibody-dependent cell-mediated cytotoxicity assay
ADCC activity was assessed using the ADCC Reporter Bioassay Kit (G7010, Promega) according to the manufacturer’s guidelines. Briefly, 7,500 SK-BR-3 cells (for trastuzumab constructs) or A431 cells (for cetuximab constructs) were seeded per well in white, flat-bottom 96-well assay plates (Corning, Cat. #3917) and allowed to adhere by incubating for 20–24 h at 37°C with 5% CO2. Serial dilutions of antibody constructs were prepared (33 nM to 5 pM for trastuzumab; 10 nM to 1.5 pM for cetuximab; 1:3 serial dilution) and added to target cells. Effector cells were then introduced at effector-to-target cell ratios of 15:1 (trastuzumab) or 10:1 (cetuximab), followed by incubation for 6 h at 37°C and 5% CO2. Following incubation and addition of luciferin substrate solution, luminescence intensity was measured using a CLARIOstar Plus plate reader. Background luminescence, determined from a control well lacking antibody, was subtracted from each sample. The resulting luminescence values were plotted as a function of antibody concentration and analyzed by non-linear regression using a sigmoidal 4-parameter logistic model in GraphPad Prism 9 to determine the EC50. To quantify the impact of antibody masking, fold reduction values were calculated by dividing the EC50 of the masked antibody constructs by the EC50 of the corresponding unmodified parental antibody.
2.6 Size exclusion chromatography
The aggregation content of antibody samples was assessed using analytical SEC. Briefly, 13–20 μg of antibody was loaded onto a TSKgel SuperSW3000 column (Tosoh Bioscience) equilibrated in PBS (pH 7.4) using an Agilent 1260 Infinity HPLC system (Agilent Technologies). Chromatographic separation was performed at a constant flow rate of 0.35 mL/min, and protein elution was monitored by absorbance at 220 nm. SEC elution profiles were visualized in GraphPad Prism 9.
2.7 Demasking of antibodies by MMP-9 hydrolysis
To achieve demasking of antibodies, 0.1 μg of human MMP-9 (Acro Biosystems) was added per 0.1 mg of antibody, and the mixture was incubated at 37°C for 48 to 72 hours. Pro-MMP-9 was activated by incubation with 1 mM 4-aminophenylmercuric acetate (AMPA) overnight at 37°C. Following incubation of antibodies with MMP-9, the extent of proteolytic cleavage was assessed by SDS-PAGE under reducing conditions, allowing visualization of the released antibody fragments and confirming successful removal of the masking domain. The Color Prestained Protein Standard, Broad Range (10–250 kDa) (NEB) was used as a molecular weight marker.
2.8 Analysis of protein structures
For structural analysis, protein models were retrieved from the RCSB Protein Data Bank (PDB). All structures were visualized and analyzed using the built-in PDB viewer available through the RCSB PDB web interface. Distance measurements between selected residues were performed directly within the viewer’s measurement tool. Measurements were taken for the distance of the N-terminal domains of the heavy and light chain in the trastuzumab Fab (PDB: 6BGT), the distance of the N-terminal domains of the heavy and light chain in the cetuximab Fab (PDB: 1YY8), and the distance of the C-terminal domains of human CaM and CBP in the CaM-CBP complex (2LV6).
2.9 Isolation of PBMCs
Buffy coats from healthy donors were obtained from the German Red Cross Blood Donation Service Baden-Württemberg-Hessen, Frankfurt am Main, Germany. Density gradient centrifugation was used to isolate peripheral blood mononuclear cells, as previously described (35). All donors provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki. Distribution of immune cell populations was validated via flow cytometry. The following antibodies were utilized for staining: CD3-BUV395 (Clone: SK7), CD14-BV711 (Clone: M5E2), CD45-BV510 (Clone: HI30), and CD56-BV421 (Clone: NCAM16.2) (antibodies were obtained from BD Biosciences, San Jose, California, USA), CD19-BB515 (Clone: HIB19) and CD16-PE (Clone: 3G8) (both antibodies were obtained from BioLegend, San Diego, California, USA). Analysis of samples was carried out with the BD FACSCelesta instrument (36). For cryopreservation of PBMCs 5 x 106 cells were collected by centrifugation (500 x g, 4 min) and resuspended in 500 µL medium A (RPMI 1640 GlutaMAX™ + 40% (v/v) FCS). Subsequently, 500 µL of medium B (RPMI 1640 GlutaMAX™ + 20% (v/v) DMSO) were added and the vial was gently inverted three times. The vials were transferred into a pre-cooled Mr. Frosty™ Freezing Container at 4°C, which was stored at -80°C for 24h. For long term storage, the cells were transferred into the vapor phase of liquid nitrogen.
2.10 PBMC-based killing assay
The PBMC-based killing assay was performed as described in Yamashita et al. (37). Briefly, A431 target cells were labeled with 5 μM carboxyfluorescein succinimidyl ester (CFSE, eBioscience) for 5 minutes, then seeded in growth medium into 96-well F-bottom culture plates and incubated for 24 hours at 37°C, 5% CO2. The following day, human PBMCs thawed from cryostock were added to the target cells at effector:target ratio of 7.5:1. Antibody constructs (cetuximab and variants) were added in a concentration range of 200 nM to 0.03 nM in RPMI 1640 supplemented with 10% heat-inactivated FBS and 1% PenStrep. Co-cultures were incubated for 24 hours at 37°C, 5% CO2. After incubation, cells were harvested and washed once, followed by staining with Fixable Viability Dye (FVD, eBioscience) for 20 minutes on ice in the dark. Cells were washed again and resuspended for flow cytometry analysis using a CytoFLEX S (Beckman Coulter). Target cells were gated based on CFSE positivity in the FITC channel, and dead target cells were identified via FVD staining in the APC-A750 channel.
3 Results
3.1 Rational design of a human-derived peptide clamp for antibody masking
To sterically block antigen binding, we engineered a peptide clamp fused to the amino terminus of the heavy and light chain of an antibody. By utilizing the combination of human CaM and CBP, we aimed to minimize immunogenicity issues (Figure 1A). The peptide clamp was connected to the antibody via MMP-2/-9 cleavable linkers (Supplementary Table S1). By introducing a peptide clamp with a MMP-2/-9 cleavable linker that imposes steric hindrance on antigen binding, the inhibition is subsequently reversed through proteolytic cleavage of the linker, thereby enabling selective restoration of antigen recognition predominantly within the tumor microenvironment (TME). This controlled demasking is anticipated to minimize off-target binding to healthy cells while enhancing specific interaction with tumor cells (Figure 1B). We hypothesized that the length of the linker connecting the antibody and the components of the peptide clamp could influence masking efficiency. Structural analysis of trastuzumab and cetuximab indicated that the distance between the amino termini of the VH and VL domains is approximately 34 Å (Supplementary Figure S1). In parallel, the distance between the carboxy termini of CaM and CBP in the CaM-CBP complex was calculated to be 41.12 Å (Supplementary Figure S2A), supporting the feasibility of positioning the clamp in close proximity to the paratope. To systematically investigate the impact of linker properties on masking efficiency, we designed two distinct linker variants: a 32 amino acid (aa) linker incorporating an MMP-2/-9 cleavage site flanked by two G4S repeat units (referred to as “long”), and a 12 aa linker with a single GS repeat flanking the cleavage site (referred to as “short” linker) (9, 38). We postulated that a shorter, less flexible linker would position the peptide clamp closer to the antibody, thereby increasing steric hindrance and enhancing masking efficacy. Furthermore, we considered that the orientation of the peptide clamp, specifically, the fusion of CaM or CBP to either the heavy or light chain, could influence masking efficiency due to the asymmetric molecular dimensions of the CaM-CBP complex. Projection of a line connecting the carboxy termini of CaM and CBP revealed that the complex is more voluminous near the carboxy terminus of CaM (Supplementary Figure S2B). Consequently, distinct masking effects were anticipated depending on the arrangement of the clamp components. To test these hypotheses, we generated a series of constructs for both trastuzumab and cetuximab. For each antibody, four variants were produced, differing in the orientation of the peptide clamp and the linker length: TrHC(CaM)LC(CBP)_long, TrHC(CaM)LC(CBP)_short, TrHC(CBP)LC(CaM)_long, and TrHC(CBP)LC(CaM)_short. The nomenclature reflects the antibody (Tr for trastuzumab), the fusion orientation with CaM fused to the heavy chain and CBP fused to the light chain (HC(CaM)LC(CBP)) or vice versa (HC(CBP)LC(CaM)), and the linker length (long or short). To assess the steric hinderance generated by only fusing CaM to the antibody, we also generated constructs in which only CaM was fused to either the heavy or light chain using the same linker variants: TrHC(CaM)_long, TrHC(CaM)_short, TrLC(CaM)_long, and TrLC(CaM)_short. To demonstrate the broad applicability of our approach, we transferred the same peptide clamp variants to cetuximab, resulting in the following constructs: CetHC(CaM)LC(CBP)_long, CetHC(CaM)LC(CBP)_short, CetHC(CBP)LC(CaM)_long, and CetHC(CBP)LC(CaM)_short. Additionally, we generated the corresponding CaM-only variants: CetHC(CaM)_long, CetHC(CaM)_short, CetLC(CaM)_long, and CetLC(CaM)_short. This comprehensive panel of constructs enabled a detailed evaluation of the contribution of linker properties, and clamp orientation to the efficiency of antibody masking.
3.2 Masking trastuzumab using the CaM-CBP-based peptide clamp
Peptide clamp-masked antibodies were produced in HEK293 cells, followed by Protein A purification. Analysis by analytical SEC was applied to examine aggregation behavior. Masked antibodies display low aggregation propensities with 3.9 to 7.6% aggregates, while unmodified trastuzumab displays an aggregation content of 1.3% (Supplementary Figures S3A–D). Next, we tested the influence of the peptide clamp on the thermal stability of the construct in a fluorescence-based thermal shift assay. For unmodified trastuzumab, three melting events can be observed at 64°C, 68°C, and 80°C. Peptide clamp-masked trastuzumab variants mostly display two melting events at Tm 61 to 65°C and 77 to 78°C (Supplementary Figures S4A–D). To simulate demasking in the TME, masked antibodies were incubated with MMP-9 in vitro. Peptide clamp removal was verified using SDS PAGE (Supplementary Figures S5A–D). All four antibody constructs exhibit shifts in the molecular weight of the heavy and light chains to molecular weights of the unmodified trastuzumab upon linker hydrolysis. Further, after cleavage, CaM was detected at a molecular weight of approximately 15 kDa or 21 kDa. After confirming linker cleavage, we investigated the blocking efficiency in an on-cell binding assay on SK-BR-3 cells (Figure 2). TrHC(CaM)LC(CPB)_long exhibited a 35-fold increase in EC50 compared to unmodified trastuzumab, with presence of CaM and CBP enhancing masking efficiency by approximately 3-fold comparing TrHC(CaM)LC(CBP)_long to TrHC(CaM)_long (Figure 2A). Upon MMP-9-mediated cleavage of the linker, antigen binding was restored to a level within 2-fold of the EC50 measured for unmodified trastuzumab. Altering the peptide clamp orientation to TrHC(CBP)LC(CaM)_long further increased masking efficiency, resulting in a 65-fold elevation in EC50 relative to trastuzumab. Again, presence of CaM and CBP increased masking efficiency, comparing the peptide clamp masked construct to the construct partially masked with only CaM fused to the light chain, and binding was restored after MMP-9-mediated linker cleavage. Decreasing linker length and flexibility (TrHC(CaM)LC(CBP)_short) further improved masking efficiency, with an over 257-fold increase in EC50 compared to trastuzumab and an 55-fold enhancement in masking efficiency relative to the corresponding non-clamp variant (Figure 2B). Whereas in the TrHC(CBP)LC(CaM)_short orientation, masking efficiency was decreased around 4-fold, and presence of CaM and CBP led to a decrease in masking efficiency. Upon linker cleavage, binding was restored to within 1.1- to 1.3-fold EC50 of that observed for unmodified trastuzumab. To investigate the additional steric hindrance introduced by an alpha-helical peptide not bound by calmodulin, we generated a construct in which CaM was fused to the heavy chain of trastuzumab via a short linker, and a 13-amino acid alpha-helical peptide was fused to the light chain of trastuzumab via a long linker, resulting in the construct TrHC(CaM)LC(helix)_short (Supplementary Figure S6A). Compared to TrHC(CaM)LC(CBP)_short, this construct exhibited more than a 2-fold reduction in masking efficiency (Supplementary Figure S6B, Supplementary Table 2). To assess whether reduced on-cell binding also translated to reduced effector function of peptide clamp-masked antibodies, we performed a reporter-cell-based ADCC assay using HER2-positive SK-BR-3 cells (Figure 3). Masked antibodies show significantly reduced ADCC. Trends in relative masking efficiency regarding linker length and peptide clamp orientation observed in on-cell binding were reflected in the ADCC assay. The construct TrHC(CaM)LC(CBP)_long showed a 25-fold increased EC50 (Figure 3A). While again TrHC(CaM)LC(CBP)_short displayed the greatest reduction in ADCC with a 43-fold increased EC50 (Figure 3B). Whereas trastuzumab induced maximal ADCC activation at 1 nM, TrHC(CaM)LC(CBP)_short required approximately 10 nM for detectable activation. Demasked antibodies, as well as antibodies only masked with CaM, and peptide clamp-masked antibodies exhibiting low masking efficiency, demonstrated enhanced maximum ADCC activation. Notably, the demasked TrHC(CBP)LC(CaM)_short variant exhibited a 2-fold higher Emax than unmodified trastuzumab at maximal activation. Importantly, this enhancement in maximal ADCC activation was not accompanied by a corresponding decrease in EC50 values.
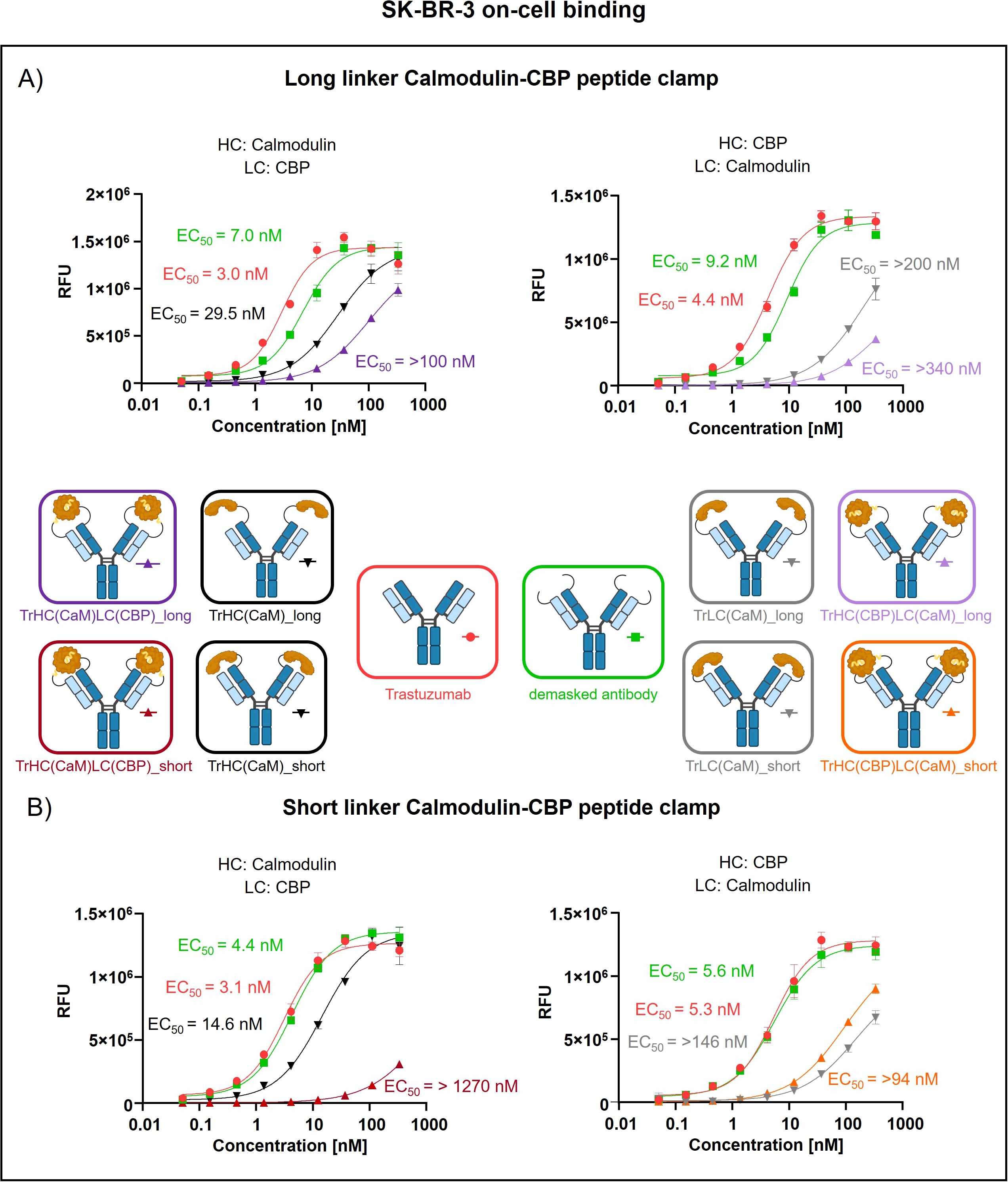
Figure 2. Evaluation of on-cell binding of trastuzumab masked with a calmodulin-CBP peptide clamp in different orientations. Analysis of concentration-dependent on-cell binding on HER2-positive SK-BR-3 cells using flow cytometry. Antibody concentration (nM) is plotted on the x-axis, and fluorescence intensity, measured in relative fluorescence units (RFU), is displayed on the y-axis. (A) Trastuzumab was masked using the CaM-CBP peptide clamp connected to the antibody via a long MMP-9 cleavable linker. The peptide clamp is engineered in two orientations: CaM fused to the N-terminus of the heavy chain and CBP to the N-terminus of the light chain (left), or the reverse configuration (right). (B) The linker used to connect trastuzumab and the peptide clamp was changed to a short MMP-9 cleavable linker. Moreover, the two orientations with CaM either fused to the N-terminus of the heavy chain and CBP to the N-terminus of the light chain (left), or the reverse configuration (right) was tested. Antibody binding was visualized using a PE-conjugated anti-human IgG detection antibody. Binding curves were fitted using a sigmoidal four-parameter logistic regression model. Error bars indicate the standard deviation of experimental triplicates. Experiments were repeated several times, showing similar results. Created with Biorender.com.
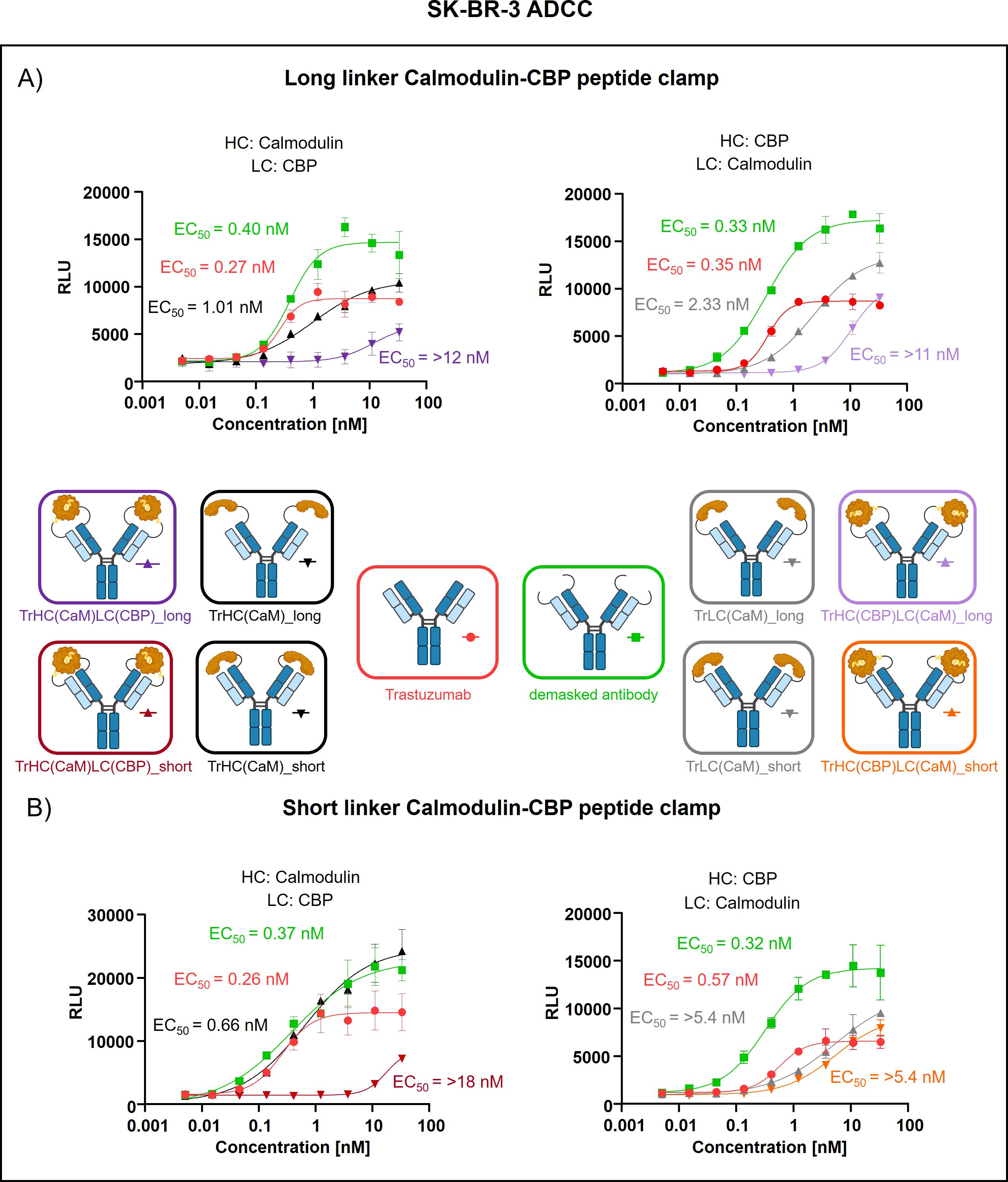
Figure 3. Evaluation of ADCC of trastuzumab masked with a calmodulin-CBP peptide clamp in different orientations. ADCC activity was assessed by a cell-based reporter assay utilizing HER2-overexpressing SK-BR-3 cells as target cells. Antibody concentration (nM) is plotted on the x-axis, and luminescence intensity, measured in relative luminescence units (RFU), is displayed on the y-axis. (A) By attaching the CaM-CBP peptide clamp to the N-terminal domains of trastuzumab via a long linker cleavable by MMP-9, the antibody was masked. The orientation of the peptide clamp was modified. Either CaM was fused to the N-terminus of the heavy chain and CBP to the N-terminus of the light chain (left), or vice versa (right). (B) By applying a short MMP-9 cleavable linker, the influence of the linker length on masking efficiency was tested. Again, the influence of the peptide clamp orientation on masking efficiency was tested by either fusing CaM to the N-terminus of the HC and CBP to the N-terminus of the LC (left), or fusion of the peptide clamp in the inverse orientation (right). Curves were fitted using a sigmoidal four-parameter logistic regression model. Error bars indicate the standard deviation of experimental duplicates. Experiments were repeated several times, showing similar results. Created with Biorender.com.
3.3 Transferring the CaM-CBP-based peptide clamp to cetuximab
To assess the generalizability of our peptide clamp masking strategy, we extended its application to cetuximab, a clinically relevant anti-EGFR monoclonal antibody. Analytical SEC revealed that masked cetuximab constructs exhibited low aggregation propensities, with aggregate content ranging from 3.9% to 6.8%, whereas unmodified cetuximab did not display detectable aggregation (Supplementary Figures S3E–H). Thermal stability analysis by fluorescence-based thermal shift assay revealed that unmodified cetuximab undergoes two distinct melting transitions at approximately 60°C and 72°C (Supplementary Figure S4). Masked cetuximab variants with CaM fused to the heavy chain exhibited melting transitions at 61-62°C and 67-68°C. In contrast, fusion of CaM to the light chain via a long linker resulted in transitions at 66°C and 72°C. Substitution of the long linker with a short linker in the light chain fusion yielded a single melting transition at 67°C. After confirming protein stability, proteolytic linker cleavage was achieved by in vitro incubation with MMP-9, as confirmed by SDS-PAGE analysis (Supplementary Figures S5E–G). Complete removal of the peptide clamp was observed, with demasked heavy and light chains migrating at positions consistent with unmodified cetuximab, while CaM was detected as a distinct band at approximately 21 kDa. On-cell binding assays using EGFR-positive A431 cells revealed that fusion of the peptide clamp via a long linker resulted in a modest 6-fold increase in EC50 independent on peptide clamp orientation (Figure 4A). Fusion of CaM alone to either the heavy or light chain resulted in a marginal 2-fold shift in EC50, regardless of orientation. Importantly, removal of the masking moiety restored binding of the antibody, with EC50 values only 1- to 1.4-fold higher than those of unmodified cetuximab. Substitution with the short linker and fusion of CaM to the light chain and CBP to the heavy chain (CetHC(CaM)LC(CBP)_short), yielded a 5-fold increase in EC50 relative to cetuximab, with binding largely restored following linker hydrolysis (1.2-fold EC50 compared to cetuximab; Figure 4B). Fusion of CaM solely to the heavy chain using the short linker resulted in a 1.5-fold increase in EC50. Strikingly, inverting the orientation of the peptide clamp with the short linker substantially enhanced masking efficiency. The construct CetHC(CBP)LC(CaM)_short demonstrated an EC50 increase of more than 140-fold compared to unmodified cetuximab. In contrast, fusion of CaM alone to the heavy chain with the short linker led to a 4-fold increase in EC50. Upon cleavage of the linker in CetHC(CBP)LC(CaM)_short, antigen binding was mostly restored, with EC50 remaining 7.2-fold higher than that of cetuximab. Functional validation of the observed effect was performed via an ADCC reporter assay utilizing EGFR-positive A431 target cells (Figure 5). For the majority of constructs evaluated, only a modest reduction in ADCC activation was detected, consistent with the trends observed in the on-cell binding assay. The construct CetHC(CaM)LC(CBP)_long only shows 5-fold higher EC50 compared to cetuximab (Figure 5A). Notably, the construct with the same orientation of the peptide clamp but connected with the short MMP-9 cleavable linker, CetHC(CBP)LC(CaM)_short, exhibited a pronounced impairment in ADCC activation, as evidenced by a greater than 78-fold increase in EC50 (Figure 5B). Cetuximab achieved maximal ADCC activation at approximately 0.4 nM, whereas CetHC(CBP)LC(CaM)_short required concentrations of around 1 nM to initiate detectable ADCC activation. Following linker hydrolysis, ADCC activation was restored, with only a 3-fold increase in EC50 relative to cetuximab. Fusion of CaM to the light chain via the short linker resulted in a 6-fold shift in EC50 and an accompanied decrease in maximal ADCC activation compared to cetuximab. Analyzing Emax values, an increase relative to unmodified cetuximab was observed for peptide clamp-masked antibodies with low masking efficiency (except CetHC(CaM)LC(CBP)_short), in partially masked antibodies containing only CaM fused to the antibody (except CetLC(CaM)_short), and in demasked antibodies. Notably, Emax was increased by up to 2.8-fold in demasked CetHC(CBP)LC(CaM)_long compared to unmodified cetuximab. To further assess the functional activity of CetHC(CBP)LC(CaM)_short, we evaluated its capacity to mediate ADCC in a PBMC-based killing assay of HER2-overexpressign A431 cells (Supplementary Figure S7). Consistent with the results obtained in the reporter cell-based ADCC assay, only minimal activity, was observed for the masked construct at concentrations around 10 nM, where maximal killing begins. Upon demasking, however, the antibody restored cytotoxic activity to levels comparable to unmodified cetuximab.
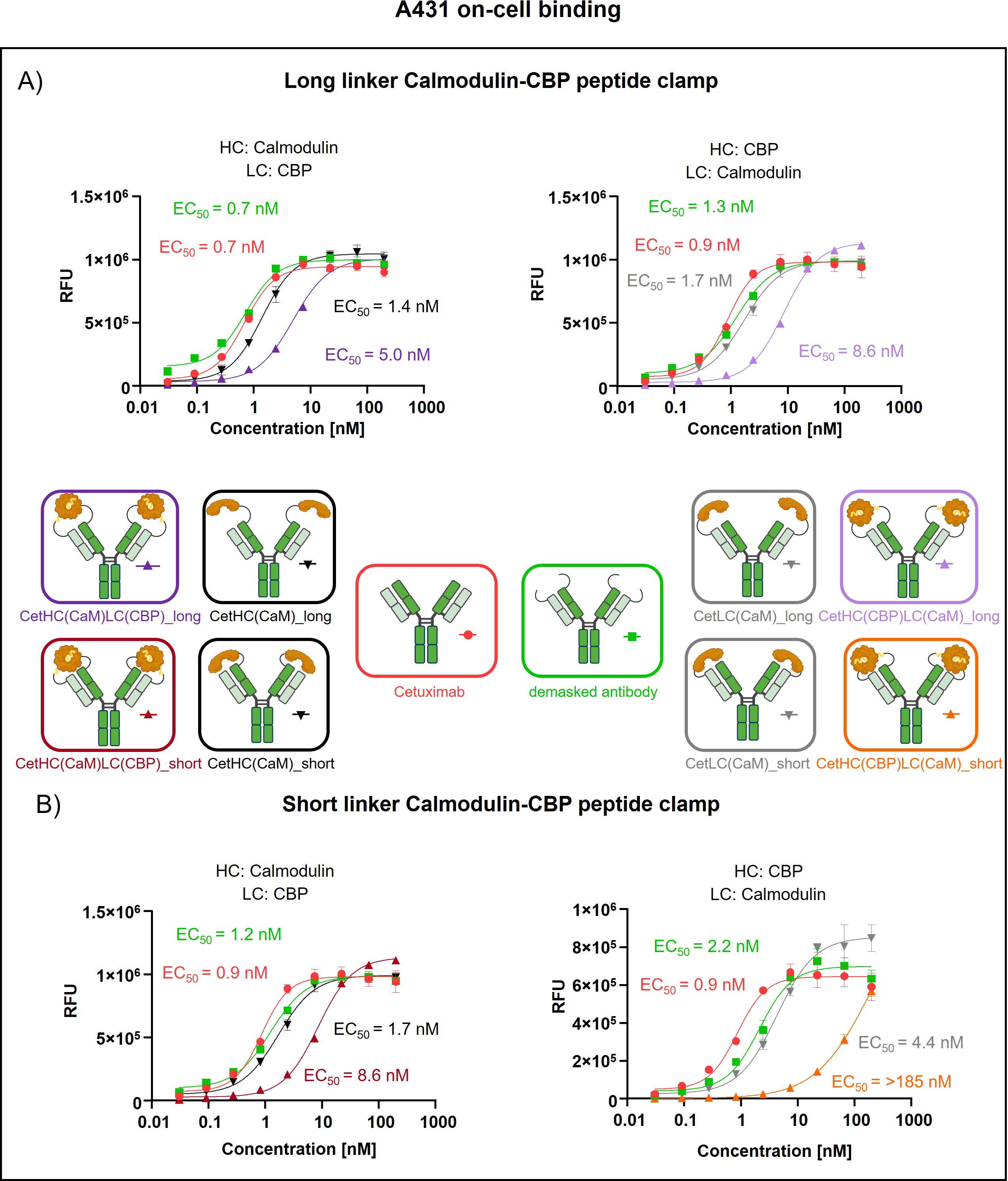
Figure 4. Evaluation of on-cell binding of cetuximab masked with a calmodulin-CBP peptide clamp in different orientations. Analysis of concentration-dependent on-cell binding on EGFR-positive A431 cells using flow cytometry. Antibody concentration (nM) is plotted on the x-axis, and fluorescence intensity, measured in relative fluorescence units (RFU), is displayed on the y-axis. (A) Cetuximab is masked using the CaM-CBP peptide clamp connected to the antibody via a long MMP-9 cleavable linker. The peptide clamp configuration is altered by fusing CaM to the N-terminus of the heavy chain and CBP to the N-terminus of the light chain (left) or employing the reciprocal arrangement (right). (B) The influence of the linker length connecting cetuximab and the peptide clamp is investigated by including a short MMP-9 cleavable linker. In addition, the influence of the peptide clamp orientation is tested by either fusing CaM to N-terminus of the heavy chain and CBP to N-terminus of the light chain (right) or the reverse configuration (left). Antibody binding is visualized using a PE-conjugated anti-human IgG detection antibody. Binding curves were fitted using a sigmoidal four-parameter logistic regression model. Error bars indicate the standard deviation of experimental triplicates. Experiments were repeated several times, showing similar results. Created with Biorender.com.
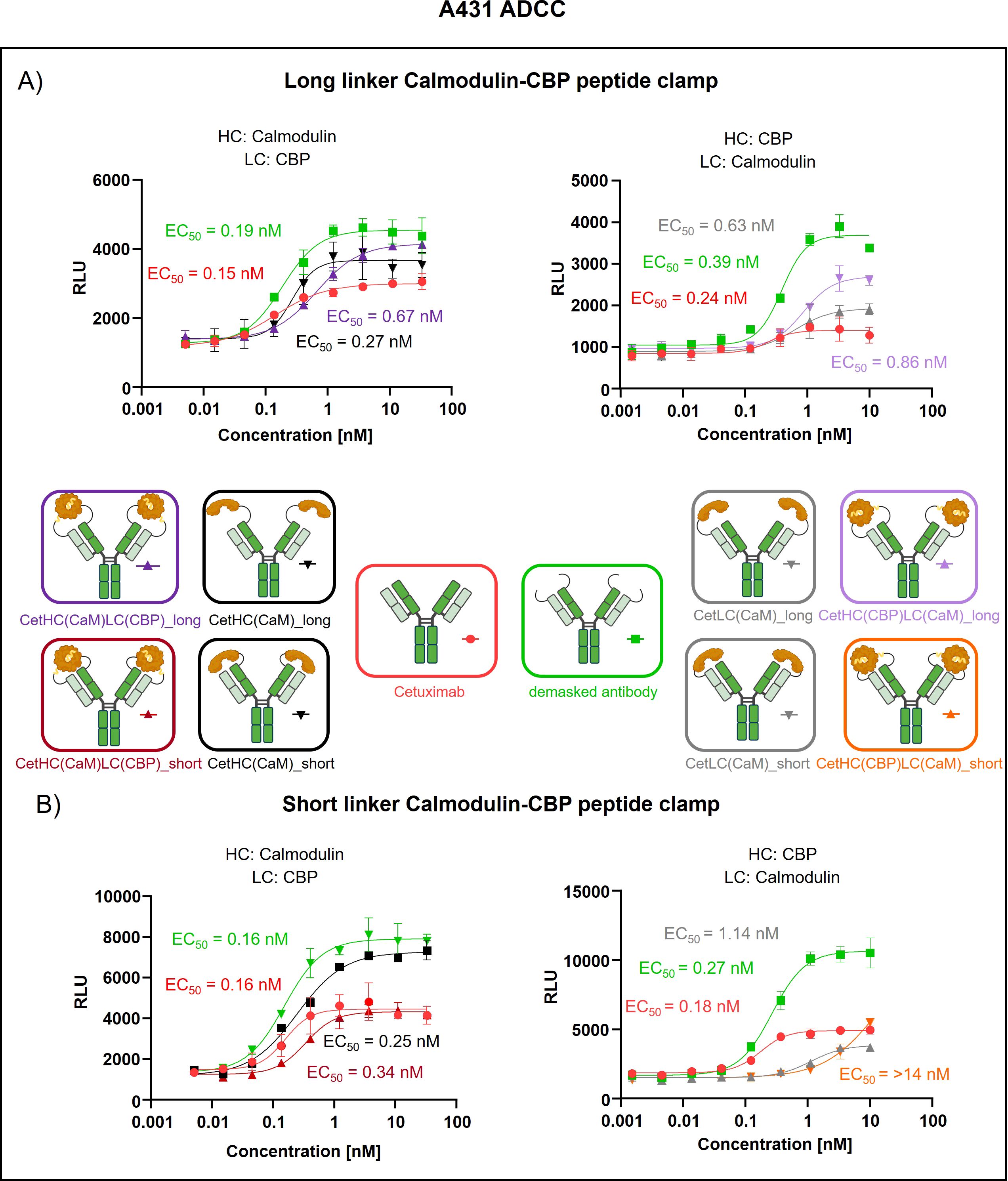
Figure 5. Evaluation of ADCC of cetuximab masked with a calmodulin-CBP peptide clamp in different orientations. ADCC was evaluated using a cell-based reporter assay with EGFR-positive A431 cells as the target cell line. Antibody concentration (nM) is plotted on the x-axis, and luminescence intensity, measured in relative luminescence units (RFU), is displayed on the y-axis. (A) Masking of cetuximab was achieved by connecting the CaM-CBP peptide clamp to the antibody via a long MMP-9 cleavable linker. The influence of the configuration of the peptide clamp on masking was assessed by either fusing CaM to the N-terminus of the heavy chain and CBP to the N-terminus of the light chain (left) or employing the reciprocal arrangement (right). (B) To assess the influence of the linker length connecting the antibody and the peptide clamp, a short MMP-9 cleavable linker is applied. Furthermore, CaM is either fused to N-terminus of the HC and CBP is fused N-terminus of the LC (left) or inversely (right). Curves were fitted using a sigmoidal four-parameter logistic regression model. Error bars indicate the standard deviation of experimental duplicates. Experiments were repeated several times, showing similar results. Created with Biorender.com.
4 Discussion
In order to reduce on-target, but off-tumor side effects in cancer treatment using monoclonal antibodies, several masking strategies that rely on activation of the masked antibody in the tumor microenvironment have been proposed. For example, Adagene`s SAFEbody technology used to mask the anti-CTLA 4 antibody ADG126, showed promising results in Phase 1b/2 trials (39). Additionally, the steric-hinderance-based coiled-coil masking unit approach by Pfizer was used to generate the masked anti-CD47 antibody SGN-CD47M. Based on promising results in tolerability studies in cynomolgus monkeys and anti-tumor activity in xenograft mouse models, this therapeutic has recently advanced into phase I dose-escalation clinical trials (19).
While generic, steric hindrance-based masking strategies have demonstrated robust masking efficiencies, the use of non-human masking moieties raises concerns regarding potential immunogenicity (11, 18). In this study, we introduce a novel, potentially generic, and fully human peptide clamp masking strategy based on the CaM-CBP interaction. This system enables conditional antibody activation via tumor-associated protease cleavage (MMP-9), thereby restoring antigen binding selectively in a conditional manner. We demonstrate that the peptide clamp can be modularly transferred between two different therapeutic antibodies. Utilizing rational design, we investigated the influence of linker length on masking efficiency. When applied to trastuzumab, the masking strategy resulted in a 257-fold reduction in on-cell binding EC50 on HER2-positive SK-BR-3 cells (Supplementary Table S2). Similarly, transferring the mask to cetuximab yielded a 140-fold decrease in on-cell binding EC50 on EGFR-positive A431 cells. Our data further indicate that masking efficiency is influenced by both the orientation and the linker length of the peptide clamp. Functionally, the reduction in on-cell binding correlated with diminished effector function. In a reporter cell-based antibody-dependent cellular cytotoxicity (ADCC) assay, masked trastuzumab and cetuximab exhibited 43-fold and 78-fold reductions in ADCC activation, respectively. Notably, at concentrations corresponding to the onset of maximal ADCC activation by unmodified antibodies, the masked variants did not elicit detectable ADCC activity. In vitro removal of the masking moiety via MMP-9-mediated hydrolysis resulted in the restoration of both on-cell binding and ADCC effector function to levels comparable to those observed with the unmodified antibody.
When investigating the influence of the linker length connecting the peptide clamp and the orientation of the peptide clamp, we found fusion of CaM and CBP to the antibodies only mildly increased aggregate formation by up to 6.8%. The content of aggregates was around 2-fold higher when CaM was fused to the heavy chain of the antibody (Supplementary Figure S3). Aggregate formation could be due to the homodimerization of masked antibodies when CaM on one antibody binds the CBP on another antibody in proximity. In general, masked antibodies displayed decreased retention time compared to the unmodified antibody. This is most likely caused by the increase in hydrodynamic radius due to the peptide clamp. All SEC chromatograms exhibited a peak at around 12 minutes, including the unmodified antibodies, suggesting an artifact possibly attributable to buffer components such as TRIS, despite their expected elution at a later timepoint. Following MMP-9 cleavage of the linker, the amount of aggregates decreases, and the retention time is similar to the unmodified antibody (Supplementary Figure S8). Next, melting temperatures were determined. For unmodified trastuzumab, three melting events were observed at 64°C, 68°C, and 80°C (Supplementary Figure S4). In a structural investigation of IgG1 antibodies, it was reported that melting of the Fc fragment results in two transitions with melting temperatures around 66°C and 82°C corresponding to melting of the CH2 and CH3 domain, while melting of the Fab fragment results in one transition with a Tm around 70°C (31). Fusing the peptide clamp to trastuzumab and cetuximab slightly decreased the thermal stability of the antibodies by a maximum of 7°C. The strongest decrease in stability was seen in the second melting event for masked trastuzumab variants, probably corresponding to the stability of the Fab region. While unmodified showed a Tm of 68°C, masked variants showed 67°C to 61°C, with some displaying overlapping melting of the CH2 domain and Fab. The decrease in thermal stability could be induced by disrupted domain packing or weakening stabilizing interactions at the heavy and light chain interface introduced by fusion of the peptide clamp to the antibody. Upon MMP-9 cleavage, Tm values did not change, indicating a structural difference in the Fab region. The decrease in thermal stability should not hinder developability, as in developability case studies, an IgG antibody is considered to display good thermodynamic stability with an onset of thermal transition at 56°C (40). Following demasking, SDS-PAGE analysis of the antibodies revealed expected shifts in the apparent molecular masses of both the heavy and light chains, consistent with those observed for the unmodified antibody (Supplementary Figure S5). CaM was detected at a molecular weight of approximately 15 kDa or 21 kDa, resulting from different folding states that could arise from deviations in incubation time at 98°C before SDS PAGE, as shown previously (31).
Not only does the orientation and linker length of the peptide clamp slightly influence aggregation behavior and thermal stability, but it also affects masking efficiency. Modifying TrHC(CaM)LC(CBP)_long, decreasing the linker length, strongly enhanced masking efficiency (Figures 2A, B, 3A, B). In contrast, decreasing the linker length with the orientation of the peptide clamp switched (TrHC(CBP)LC(CaM)_long), decreased masking efficiency (Figures 2A, B, 3A, B). Investigating the masking of cetuximab, only CetHC(CBP)LC(CaM)_short showed efficient masking (Figures 4B, 5B). In both trastuzumab and cetuximab variants, constructs featuring the shortest linker exhibited the greatest extent of antigen masking. Notably, for trastuzumab, the variant with CaM fused to the heavy chain and CBP fused to the light chain exhibited the greatest extent of masking, while for cetuximab, the variant with CaM fused to the light chain and CBP fused to the heavy chain exhibited the greatest extent of masking. Differences in masking efficiency could result from the positioning of the peptide clamp relative to the complementarity-determining regions (CDRs) of the antibody. As the CaM-CBP complex is asymmetric in molecular mass near the carboxy termini of CaM and CBP, changing the orientation of the peptide clamp influences the steric hindrance induced in the antibody-antigen interaction (Supplementary Figure S2B). The distance and positioning of the N-termini to the main interaction points in the antibody-antigen interaction could further influence the masking efficiency. In both the trastuzumab-HER2-complex and the cetuximab-EGFR-complex, the distance from the N-termini of the light chains to the CDR3 of the heavy and light chains is shorter than the distance from the N-termini of the HC to the CDR3s (Supplementary Figure S9). For both antibodies, the CDR3 mainly contributes to antigen-binding (41, 42). In performing reporter-cell-based ADCC assays, we showed, as expected, that reduced on-cell binding also leads to reduced ADCC activation. The reduction in EC50 upon masking observed in on-cell binding studies was stronger than in the ADCC assay. This could result from differences in assay sensitivity. While in on-cell binding, every bound antibody leads to a signal, the ADCC assay relies on pathway activation. Already, a small number of bound antibodies can trigger effector cell pathway activation, thereby increasing sensitivity. Analyzing demasked antibodies, we saw an increase in Emax compared to unmodified antibodies. This phenomenon could have different reasons. It could arise from increased koff of demasked antibodies resulting from linker residues still left after cleavage. By faster koff, antibodies could increase the local abundance of the effector cells near the cancer cell due to a shorter time of interaction. Another explanation could be increased calcium concentration by CaM. The reporter cell-based ADCC assay quantifies activation of the nuclear factor of activated T-cells (NFAT) pathway by employing a gene reporter system in which the NFAT response element drives luciferase expression. Activation of the NFAT pathway relies on an intracellular increase in Ca2+ concentration, leading to activation of Calcineurin, a phosphatase that dephosphorylates NFAT proteins (43). This increase in intracellular Ca2+ is achieved by depletion of calcium stores in the endoplasmic reticulum (ER), followed by calcium influx from the extracellular space through store-operated calcium channels. Internalization of antibodies containing CaM via receptor-mediated endocytosis could increase the Ca2+ concentration in the ER. Following lysosomal degradation of these complexes, liberated Ca2+ might further amplify the intracellular Ca2+ signal, followed by potentially enhanced NFAT pathway activation. When testing the CetHC(CBP)LC(CaM)_short in a PBMC-based killing assay, the enhanced Emax was not observed, pointing toward a bias in the reporter cell based assay (Supplementary Figure S8).
We showed that the peptide clamp is transferable between antibodies by efficiently masking trastuzumab (Figures 2, 3) and cetuximab (Figures 4, 5). Therefore, this newly developed masking strategy could potentially be transferable to mask any antibody. To test this, the peptide clamp should be applied to a broader range of therapeutic antibodies. As we showed, finding the optimal orientation and linker length of the mask is critical; therefore, building a tool to predict the ideal orientation and linker length on each antibody could be cost-saving and time-saving. However, such a tool remains to be established. When incorporating the CaM-CBP peptide clamp into antibody constructs, we recommend using the short linker to connect the peptide clamp to the antibody. Additionally, it is advisable to empirically test both orientations of the clamp attachment to identify the variant that provides the most effective masking of antibody-antigen interaction. Previously, using molecular dynamics simulations, Chen et al. showed that the relative positioning of a steric-hinderance-based mask to the antibody CDRs strongly influences masking efficiency (44). Demasking of antibodies restored target binding. But the kinetics of the demasking reaction and the influence of the linker length and MMP-9 cleavage sequence are still to be determined. We propose that native mass spectrometry represents a robust analytical platform for investigating these processes. Specifically, by in-line activation of inactive pro-MMP-9 through the addition of 4-aminophenylmercuric acetate (APMA), followed by real-time monitoring of immunocomplex formation between the unmasked antibody and its antigen at defined time intervals, it should be possible to interrogate the kinetics of linker hydrolysis (45, 46). By now, only in vitro experiments have been performed, but to further characterize linker stability and, most importantly, demasking of peptide clamp-masked antibodies in the TME, in vivo studies should be considered. Recently, in animal studies, it was shown that steric-hinderance-based masking enhances the tolerability of antibodies with on-target, but off-tumor side effects. Furthermore, after 72 hours post-dose, 50% of the masked antibody is found demasked in the tumor (19). The demasking of the antibody in the TME remains one major limitation of masked antibodies. Whether our approach can achieve efficient demasking within the TME remains to be determined and should be addressed in future studies, for example using tumor graft models. Additionally, as we concluded for TrHC(CaM)LC(CBP)_short and CetHC(CBP)LC(CaM)_short, formation of the peptide clamp can be necessary for efficient masking, and only fusing CaM to the heavy chain of trastuzumab using the short linker did not result in efficient masking. Furthermore additionally fusing an alpha-helical peptide to the N-terminus of the light chain using the long MMP-9 cleavable linker TrHC(CaM)LC(helix)_short) lead to 2-fold lower EC50 compared to TrHC(CaM)LC(CBP)_short (Supplementary Figure S7). Whether the observed change in masking efficiency is due to peptide-clamp formation or additional steric hindrance remains unclear. At present, we lack direct evidence for peptide-clamp formation. Future studies should therefore aim to directly demonstrate such interactions, for example by employing native mass spectrometry or high-resolution structural approaches such as cryo-EM. Converting the peptide clamp into a logic gate approach by utilizing different stimuli for linker cleavage could be conceivable. This could be especially useful for bispecific antibodies, where each binding site is restored under certain conditions. Another topic to be investigated is whether the CaM-CBP peptide clamp can be applied to antibody-drug conjugates. In conclusion, we have developed a novel and promising masking strategy based on a human CaM-CBP peptide clamp. This approach offers the advantage of potentially being generic and, due to its human-derived components, is unlikely to elicit immunogenicity. However, further studies, particularly in vivo studies, are required to validate this strategy and to explore its full range of potential applications. Additionally, the CaM–CBP peptide clamp has so far only been evaluated on two therapeutic antibodies, where it showed efficient blocking. To establish whether this strategy is generically applicable, further studies should explore its masking capacity across a wider range of antibodies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethics Committee of Goethe University, Frankfurt am Main, Germany (Approval number 329/10). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired were acquired as gifts from another research group. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
AB: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft. JZ: Investigation, Methodology, Writing – review & editing. JH: Conceptualization, Methodology, Writing – review & editing. EU: Conceptualization, Methodology, Writing – review & editing. HK: Conceptualization, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported in part by grants from Deutsche Forschungsgemeinschaft (DFG) (KO1390/14-1).
Acknowledgments
We thank Janine Becker for support in antibody expression. We acknowledge support by the Deutsche Forschungsgemeinschaft (DFG) and the Open Access Publishing Fund of the Technical University of Darmstadt. Open access funding is enabled and organized by Projekt DEAL. Figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Perplexity AI (Version 2025, using models: OpenAI GPT-4 Omni; Claude 3 Sonnet, Opus, and Haiku; Sonar Large 32k; Grok-2; DeepSeek R1) was used for assistance in manuscript language editing.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1640427/full#supplementary-material
References
1. DeVita VT and Chu E. A history of cancer chemotherapy. Cancer Res. (2008) 68:8643–53. doi: 10.1158/0008-5472.CAN-07-6611
2. Nadler LM, Stashenko P, Hardy R, Kaplan WD, Button LN, Kufe DW, et al. Serotherapy of a patient with a monoclonal antibody directed against a human lymphoma-associated antigen. Cancer Res. (1980) 40:3147–54.
3. Eshhar Z, Waks T, Gross G, and Schindler DG. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci U S A. (1993) 90:720–4. doi: 10.1073/pnas.90.2.720
4. Hansel TT, Kropshofer H, Singer T, Mitchell JA, and George AJT. The safety and side effects of monoclonal antibodies. Nat Rev Drug Discov. (2010) 9:325–38. doi: 10.1038/nrd3003
5. Segaert S and van Cutsem E. Clinical signs, pathophysiology and management of skin toxicity during therapy with epidermal growth factor receptor inhibitors. Ann Oncol. (2005) 16:1425–33. doi: 10.1093/annonc/mdi279
6. Kuramochi Y, Guo X, and Sawyer DB. Neuregulin activates erbB2-dependent src/FAK signaling and cytoskeletal remodeling in isolated adult rat cardiac myocytes. J Mol Cell Cardiol. (2006) 41:228–35. doi: 10.1016/j.yjmcc.2006.04.007
7. Lucchi R, Bentanachs J, and Oller-Salvia B. The masking game: design of activatable antibodies and mimetics for selective therapeutics and cell control. ACS Cent Sci. (2021) 7:724–38. doi: 10.1021/acscentsci.0c01448
8. Lin W-W, Lu Y-C, Chuang C-H, and Cheng T-L. Ab locks for improving the selectivity and safety of antibody drugs. J BioMed Sci. (2020) 27:76. doi: 10.1186/s12929-020-00652-z
9. Geiger M, Stubenrauch K-G, Sam J, Richter WF, Jordan G, Eckmann J, et al. Protease-activation using anti-idiotypic masks enables tumor specificity of a folate receptor 1-T cell bispecific antibody. Nat Commun. (2020) 11:3196. doi: 10.1038/s41467-020-16838-w
10. Desnoyers LR, Vasiljeva O, Richardson JH, Yang A, Menendez EEM, Liang TW, et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci Transl Med. (2013) 5:207ra144. doi: 10.1126/scitranslmed.3006682
11. Biewenga L, Vermathen R, Rosier BJHM, and Merkx M. A generic antibody-blocking protein that enables pH-switchable activation of antibody activity. ACS Chem Biol. (2024) 19:48–57. doi: 10.1021/acschembio.3c00449
12. Coelho M, Gauthier P, Pugnière M, Roquet F, Pèlegrin A, and Navarro-Teulon I. Isolation and characterisation of a human anti-idiotypic scFv used as a surrogate tumour antigen to elicit an anti-HER-2/neu humoral response in mice. Br J Cancer. (2004) 90:2032–41. doi: 10.1038/sj.bjc.6601825
13. Alvarez-Rueda N, Ladjemi MZ, Béhar G, Corgnac S, Pugnière M, Roquet F, et al. A llama single domain anti-idiotypic antibody mimicking HER2 as a vaccine: Immunogenicity and efficacy. Vaccine. (2009) 27:4826–33. doi: 10.1016/j.vaccine.2009.05.067
14. Elter A, Yanakieva D, Fiebig D, Hallstein K, Becker S, Betz U, et al. Protease-activation of Fc-masked therapeutic antibodies to alleviate off-tumor cytotoxicity. Front Immunol. (2021) 12:715719. doi: 10.3389/fimmu.2021.715719
15. Habermann J, Happel D, Bloch A, Shin C, and Kolmar H. A competition-based strategy for the isolation of an anti-idiotypic blocking module and fine-tuning for conditional activation of a therapeutic antibody. Biotechnol J. (2024) 19:e202400432. doi: 10.1002/biot.202400432
16. Assi HH, Wong C, Tipton KA, Mei L, Wong K, Razo J, et al. Conditional PD-1/PD-L1 probody therapeutics induce comparable antitumor immunity but reduced systemic toxicity compared with traditional anti-PD-1/PD-L1 agents. Cancer Immunol Res. (2021) 9:1451–64. doi: 10.1158/2326-6066.CIR-21-0031
17. Boni V, Fidler MJ, Arkenau H-T, Spira A, Meric-Bernstam F, Uboha N, et al. Praluzatamab ravtansine, a CD166-targeting antibody-drug conjugate, in patients with advanced solid tumors: an open-label phase I/II trial. Clin Cancer Res. (2022) 28:2020–9. doi: 10.1158/1078-0432.CCR-21-3656
18. Trang VH, Zhang X, Yumul RC, Zeng W, Stone IJ, Wo SW, et al. A coiled-coil masking domain for selective activation of therapeutic antibodies. Nat Biotechnol. (2019) 37:761–5. doi: 10.1038/s41587-019-0135-x
19. Levengood MR, Carosino CM, Zhang X, Lucas S, Ortiz DJ, Westendorf L, et al. Preclinical development of SGN-CD47M: protease-activated antibody technology enables selective tumor targeting of the innate immune checkpoint receptor CD47. Mol Cancer Ther. (2025) 24:471–84. doi: 10.1158/1535-7163.MCT-24-0371
20. Linse S, Helmersson A, and Forsén S. Calcium binding to calmodulin and its globular domains. J Biol Chem. (1991) 266:8050–4. doi: 10.1016/S0021-9258(18)92938-8
21. Chattopadhyaya R, Meador WE, Means AR, and Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. (1992) 228:1177–92. doi: 10.1016/0022-2836(92)90324-d
22. Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, and Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. (1992) 256:632–8. doi: 10.1126/science.1585175
23. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. (2002) 20:719–26. doi: 10.1200/JCO.2002.20.3.719
24. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. (2001) 344:783–92. doi: 10.1056/NEJM200103153441101
25. Qiu Y, Jiang P, and Huang Y. Anthracycline-induced cardiotoxicity: mechanisms, monitoring, and prevention. Front Cardiovasc Med. (2023) 10:1242596. doi: 10.3389/fcvm.2023.1242596
26. Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, and Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. (2004) 22:1201–8. doi: 10.1200/JCO.2004.10.182
27. Segaert S, Tabernero J, Chosidow O, Dirschka T, Elsner J, Mancini L, et al. The management of skin reactions in cancer patients receiving epidermal growth factor receptor targeted therapies. J Dtsch Dermatol Ges. (2005) 3:599–606. doi: 10.1111/j.1610-0387.2005.05058.x
28. Roé E, García Muret MP, Marcuello E, Capdevila J, Pallarés C, and Alomar A. Description and management of cutaneous side effects during cetuximab or erlotinib treatments: a prospective study of 30 patients. J Am Acad Dermatol. (2006) 55:429–37. doi: 10.1016/j.jaad.2006.04.062
29. Lacouture ME and Melosky BL. Cutaneous reactions to anticancer agents targeting the epidermal growth factor receptor: a dermatology-oncology perspective. Skin Ther Lett. (2007) 12:1–5.
30. Scorilas A, Karameris A, Arnogiannaki N, Ardavanis A, Bassilopoulos P, Trangas T, et al. Overexpression of matrix-metalloproteinase-9 in human breast cancer: a potential favourable indicator in node-negative patients. Br J Cancer. (2001) 84:1488–96. doi: 10.1054/bjoc.2001.1810
31. González LO, Pidal I, Junquera S, Corte MD, Vázquez J, Rodríguez JC, et al. Overexpression of matrix metalloproteinases and their inhibitors in mononuclear inflammatory cells in breast cancer correlates with metastasis-relapse. Br J Cancer. (2007) 97:957–63. doi: 10.1038/sj.bjc.6603963
32. Yu J, He Z, He X, Luo Z, Lian L, Wu B, et al. Comprehensive analysis of the expression and prognosis for MMPs in human colorectal cancer. Front Oncol. (2021) 11:771099. doi: 10.3389/fonc.2021.771099
33. CytomX Therapeutics. CytomX announces positive interim data from phase 1 dose escalation study of epCAM antibody drug conjugate (CX-2051) candidate in patients with advanced colorectal cancer (CRC)(2025). Available online at: https://ir.cytomx.com/news-releases/news-release-details/cytomx-announces-positive-interim-data-phase-1-dose-escalation (Accessed May 30, 2025).
34. Bogen JP, Hinz SC, Grzeschik J, Ebenig A, Krah S, Zielonka S, et al. Dual function pH responsive bispecific antibodies for tumor targeting and antigen depletion in plasma. Front Immunol. (2019) 10:1892. doi: 10.3389/fimmu.2019.01892
35. Graafen L, Heinze A, Albinger N, Salzmann-Manrique E, Ganß F, Hünecke S, et al. Immune profiling and functional analysis of NK and T cells in ataxia telangiectasia. Front Immunol. (2024) 15:1377955. doi: 10.3389/fimmu.2024.1377955
36. Reindl LM, Jalili L, Bexte T, Harenkamp S, Thul S, Hehlgans S, et al. Precision targeting of rhabdomyosarcoma by combining primary CAR NK cells and radiotherapy. J Immunother Cancer. (2025) 13(7):e011330. doi: 10.1136/jitc-2024-011330
37. Yamashita M, Kitano S, Aikawa H, Kuchiba A, Hayashi M, Yamamoto N, et al. A novel method for evaluating antibody-dependent cell-mediated cytotoxicity by flowcytometry using cryopreserved human peripheral blood mononuclear cells. Sci Rep. (2016) 6:19772. doi: 10.1038/srep19772
38. Turk BE, Huang LL, Piro ET, and Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. (2001) 19:661–7. doi: 10.1038/90273
39. Adagene Inc. Update of phase 1b/2 study of muzastotug (ADG126, an anti-CTLA-4 SAFEbody®) in combination with pembrolizumab in advanced/metastatic MSS CRC(2025). Available online at: https://netshare.adagene.com/pdf/2025-ASCO-GI-ADG126-P001-Combo-Poster-Jan13.pdf (Accessed May 22, 2025).
40. Yang X, Xu W, Dukleska S, Benchaar S, Mengisen S, Antochshuk V, et al. Developability studies before initiation of process development: improving manufacturability of monoclonal antibodies. MAbs. (2013) 5:787–94. doi: 10.4161/mabs.25269
41. Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. (2003) 421:756–60. doi: 10.1038/nature01392
42. Li S, Schmitz KR, Jeffrey PD, Wiltzius JJW, Kussie P, and Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. (2005) 7:301–11. doi: 10.1016/j.ccr.2005.03.003
43. Hogan PG, Chen L, Nardone J, and Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. (2003) 17:2205–32. doi: 10.1101/gad.1102703
44. Chen I-J, Chuang C-H, Hsieh Y-C, Lu Y-C, Lin W-W, Huang C-C, et al. Selective antibody activation through protease-activated pro-antibodies that mask binding sites with inhibitory domains. Sci Rep. (2017) 7:11587. doi: 10.1038/s41598-017-11886-7
45. Ogata Y, Itoh Y, and Nagase H. Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)-tissue inhibitor of metalloproteinases-1 complex by 4-aminophenylmercuric acetate and proteinases. J Biol Chem. (1995) 270:18506–11. doi: 10.1074/jbc.270.31.18506
Keywords: antibody engineering, antibody masking, conditional antibody activation, off-target toxicity, MMP-9
Citation: Bloch A, Zimmermann JF, Habermann J, Ullrich E and Kolmar H (2025) A masking clamp for conditional activation of therapeutic antibodies. Front. Immunol. 16:1640427. doi: 10.3389/fimmu.2025.1640427
Received: 03 June 2025; Accepted: 21 October 2025;
Published: 30 October 2025.
Edited by:
Chun-Wai Mai, IMU University, MalaysiaReviewed by:
Dafne Müller, University of Stuttgart, GermanyAndré Luiz Lourenço, Fluminense Federal University, Brazil
Andreas Stengl, Ludwig Maximilian University of Munich, Germany
Copyright © 2025 Bloch, Zimmermann, Habermann, Ullrich and Kolmar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harald Kolmar, SGFyYWxkLktvbG1hckBUVS1EYXJtc3RhZHQuZGU=
 Adrian Bloch1
Adrian Bloch1 Jan Habermann
Jan Habermann Evelyn Ullrich
Evelyn Ullrich Harald Kolmar
Harald Kolmar