- 1Department of Epidemiology and Health Statistics, School of Public Health, Southeast University, Nanjing, China
- 2Yinzhou District Center for Disease Control and Prevention, Ningbo, China
- 3School of Public Health, Xinjiang Medical University, Urumqi, China
- 4Department of Infectious Disease, the Second Hospital of Nanjing, Nanjing University of Chinese Medicine, Nanjing, China
Objective: This systematic review and meta-analysis aims to characterize the distribution of HIV-1 viral tropism at diagnosis among people living with HIV (PLWH) and examine its association with baseline CD4+ T lymphocyte counts, thereby providing an evidence base for optimizing clinical interventions.
Method: Observational studies reporting viral tropism prevalence and/or baseline CD4+ T cell counts stratified by tropism were retrieved from PubMed, Web of Science, Embase, and Cochrane Library. A random-effects model was employed for pooled prevalence estimation and mean difference calculation. Heterogeneity was quantified using I² statistics, with subgroup analyses and sensitivity tests to identify heterogeneity sources.
Results: 27 articles (N = 9372) were included in this study to analyze the distribution of viral tropism, and the prevalence of Non-R5 tropism was 15.68%. Subgroup analysis showed that the prevalence of Non-R5 IDU (27.86%) was significantly higher than that of sexual transmission (15.29%) and other routes (4.62%). The prevalence of Non-R5 tropism in the CRF01_AE subtype group (30.02%) was significantly higher than that of the B subtype (15.33%) and other subtypes (3.44%) (P ≤ 0.05). A comparison of CD4+ T cell counts (17 articles) showed a difference of −97.77 cells/μL for the Non-R5 tropic group relative to the R5 group.
Conclusion: Our study find that PLWH with Non-R5 virus had more severe immune damage at diagnosis compared to PLWH with R5 virus. This can update the baseline status of patients in clinical practice. since this is a cross-sectional study, future cohort studies should be conducted to verify the relationship between tropism and changes in immunological indicators.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/, identifier CRD420251088996.
1 Introduction
The human immunodeficiency virus (HIV) induces progressive immune destruction by infecting CD4+ T lymphocytes. HIV’s pathogenicity is closely associated with the interaction between viral envelope protein gp120 and host chemokine receptors (1). According to viral preference for chemokine receptor utilization, HIV-1 tropism is classified into three categories: 1) R5 tropic viruses that utilize C-C motif chemokine receptor type 5 (CCR5) for cellular entry (2), 2) X4 tropic viruses that utilize C-X-C motif chemokine receptor 4 (CXCR4) (3), and 3) R5/X4 tropic viruses capable of utilizing both CCR5 and CXCR4 receptors (4). HIV-1 transmitted/founder (T/F) viruses are almost exclusively R5 tropic and can efficiently infect CD4+ T cells with physiological CCR5 expression (5). In chronic progression, approximately 50~70% of people living with HIV (PLWH) experience viral tropism switching that results in X4 tropic viruses emergence. This process is typically associated with accelerated disease progression, CD4+ T cell depletion, and poorer clinical outcomes after antiretroviral treatment (ART) (6).
HIV tropism switching is recognized as a critical event in the natural history of infection, although the exact timing and driving mechanisms remain poorly characterized. Notably, some patients demonstrate completed tropism switching upon initial diagnosis. Current consensus indicates this phenotypic transition is associated with amino acid mutations in the V3 loop region (7). Viral chemokine receptor usage demonstrates subtype-dependent patterns: approximately 50% of subtype B infections develop X4 tropic variants (8), whereas CRF01_AE displays a higher prevalence of R5/X4 viruses (9). In contrast, Ndung’u et al. demonstrated that subtype C-infected PLWH rarely harbor R5/X4 tropic viruses at any disease stage (10). The population-level prevalence of R5/X4 tropic viruses has not been fully determined (11). Geographical variations in tropism conversion rates, which may reflect regional subtype distributions (e.g., subtype C predominance in Africa versus CRF01_AE prevalence in Southeast Asia), await systematic evaluation. Furthermore, epidemiological data concerning R5/X4 tropic virus prevalence at HIV diagnosis still require comprehensive synthesis.
Compared to R5 viruses, X4 tropic variants exhibit higher pathogenicity (12), and are associated with accelerated CD4+ T cell depletion in PLWH (11, 13). PLWH infected with X4 viruses show reduced sensitivity to ART (14). The impact of tropism on virological responses to ART remains controversial (15). CCR5 inhibitors serve as salvage therapy for R5 tropic infections (16), but become contraindicated upon emergence of R5/X4 tropic variants (11). Baseline CD4+ T cell count represents another critical determinant of ART outcomes, with higher baseline levels correlating with maximal potential for complete immune reconstitution (17). Elucidating both viral tropism and CD4+ T cell counts at diagnosis thus provides key evidence for formulating personalized antiretroviral therapy regimens and determining optimal timing for immune recovery interventions.
Given the lack of studies summarizing viral tropism characteristics at HIV diagnosis in PLWH, this study conducted a meta-analysis through systematic retrieval and integration of multicenter medical data. We updated the prevalence of R5 and Non-R5 tropism to 2025 and analyzed differences in baseline CD4+ T lymphocyte counts among PLWH infected with different tropism variants. By exploring the association between virological characteristics and immunological profiles at diagnosis, this work provides evidence-based guidance for optimizing the timing of ART and developing personalized immune reconstitution strategies.
Given the current lack of systematic studies on the prevalence of HIV viral tropism, this study aims to fill this gap. This study conducted a meta-analysis through systematic retrieval and integration of multicenter medical data. Our study extends the prevalence estimates of R5 and Non-R5 tropism through 2025, while examining baseline CD4+ T lymphocyte count disparities across tropism variants. Through elucidating virological-immunological correlations at diagnosis, this investigation offers evidence to refine ART initiation timelines.
2 Methods
2.1 Search strategy
The review protocol has been registered on PROSPERO (CRD420251088996). Data sources encompassed Web of Science, PubMed, Cochrane Library, Embase, and Scopus. Search terms included: HIV, human immunodeficiency virus, HIV-1, tropism, coreceptor usage, R5 tropic, X4 tropic, CCR5 receptor, CXCR4 receptor, CD4 Lymphocyte Count, CD4 count, CD4+ T cell, CD4 cell, and immune status. Prior to formal retrieval, all search terms were systematically searched within the MeSH thesaurus, supplemented with common expressions. The search timeframe spanned from the inception year of each database to November 4, 2025.
2.2 Inclusion/exclusion criteria
Inclusion criteria: 1) Cross-sectional studies published in English prior to November 4, 2025; 2) Participants were newly diagnosed, treatment-naïve HIV-1-infected individuals; 3) Studies reporting the number of participants stratified by viral tropism; 4) Sample size ≥ 20; 5) Additional criteria for CD4+ T cell analysis; 6) Reported CD4+ T cell count stratified according to viral tropism.
Exclusion criteria: 1) Non-original studies e.g., reviews, brief reports); 2) Participants aged < 15 years; 3) Pregnant or peripartum women; 4) Unclear methodology for viral tropism detection; 5) Viral tropism or CD4+ T cell count not assessed at diagnosis; 6) In vitro, animal, or mechanistic studies; 7) Insufficient data extraction or full-text inaccessibility. Only the earliest published studies were included for studies using the same cohort or data.
2.3 Data extraction and quality assessment
Literature management was conducted using Zotero 7 software. Data extraction and entry were independently conducted by two researchers using Excel software. Discrepancies were resolved through discussion until consensus was reached or by consulting a third author. The following information was extracted: first author, publication year, title, sample source, study period, sample collection, study population, number of R5 tropic and Non-R5 tropic infected individuals, and CD4+ T cell counts stratified by viral tropism. If studies included participants with distinct subtypes or genders and provided precise data, these subgroups were extracted as separate data points. Quality assessment criteria were adapted from the AHRQ guidelines for cross-sectional studies, incorporating the STROME-ID statement and study-specific characteristics, resulting in 11 assessment items for two predefined topics.
Quality Assessment Criteria (11 items): 1) Clearly defined study design (e.g., cohort study, cross-sectional study, or designs referenced to relevant protocols); 2) Specific inclusion and exclusion criteria; 3) Explicit statement of the study’s timeframe and sample sources; 4) Specification of sample types used in the study typically including plasma); 5) Clarification of the viral tropism detection methods employed; 6) Description of the threshold for genotypic prediction of tropism; 7) Documentation of the amplification success rate in the env region; 8) Description of any quality assurance tests performed; 9) Explanation of reasons for excluding study data or subjects; 10) Specification of measures to control potential laboratory contamination or confounding factors; 11) Explanation of strategies to address missing data in the analysis.
2.4 Subgroup settings
Gender: Male and Female; Area: China, European countries, and Other areas; Transmission route (Route 1): Sex, Inject Drugs Users (IDU), and Other. Sex transmission route (Route 2): MSM and Hetero; HIV-1 gene subtype: CRF01_AE; B Subtype and Non-CRF01_AE/Non-B’s other subtype; Recent infection: Unknown and Yes. Tropism detection: G2P and SVM; FPR threshold: 2% and 10%.
2.5 Meta-analysis and data statistics
Viral tropism was categorized into R5 tropic, X4 tropic, and R5/X4 tropic. Considering that in many studies, R5/X4 viruses exhibit characteristics of X4 viruses, the presence of R5/X4 viruses in an infected individual, like X4 viruses, may lead to the failure of CCR5 inhibitor therapy. X4 and R5/X4 viruses are combined as Non-R5 viruses. Accordingly, we also merged these two groups in this manner and established R5 virus and Non-R5 virus groups. In addition, only one included study reported R5/X4 virus data separately, and combining X4 and R5/X4 viruses also facilitated the analysis. Data management and cleaning were performed using Excel software, while statistical analyses were conducted with the meta package in R version 4.5.0. This study evaluated the pooled prevalence of Non-R5 viruses at HIV-1 diagnosis and the pooled effect size of CD4+ T cell counts between different tropism groups. If the distribution of rates does not follow a normal distribution, the arcsine square root transformation method is applied to make the data conform to a normal distribution. Heterogeneity was assessed using Cochran’s Q-test and I2 statistics. τ2 is more robust for small samples or extreme heterogeneity, avoiding negative variance estimates, and this statistic is used to jointly assess heterogeneity. Significant heterogeneity was defined as P ≤ 0.10 or I2 ≥ 25%. Random-effects models were employed when P ≤ 0.10 or I2 ≥ 50%; otherwise, fixed-effects models were applied. Publication bias was examined via Egger’s regression test (P > 0.05 indicating no substantial bias), with the trim-and-fill method used for adjustment if required. Sensitivity analysis was performed by sequentially excluding individual studies to assess result stability. Subgroup analyses were conducted to explore potential sources of heterogeneity.
3 Result
3.1 Study selection
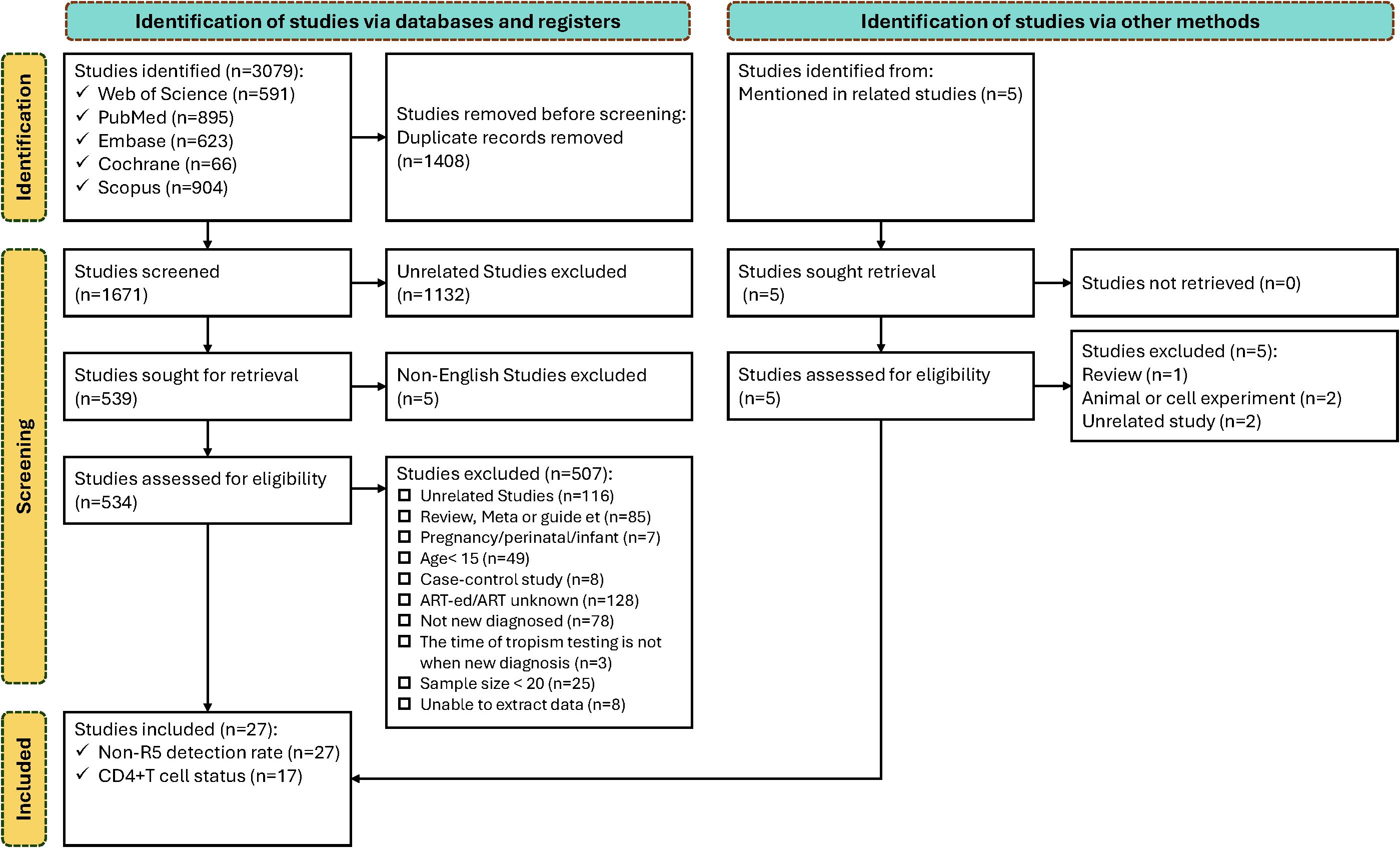
A total of 3,079 references were retrieved from literature databases. After removing duplicates, 1,671 articles underwent title/abstract screening, with 1,137excluded (1,132 irrelevant to the topic; 5 non-English publications). Full-text review was conducted for 534 articles. A total of 507 articles were excluded based on predefined criteria: 116 irrelevant studies; 85 reviews/meta-analyses/guidelines/case reports; 7 involving pregnancy/perinatal women/vertically infected infants; 49 articles with participants aged <15 years; 8 case-control studies with uncalculable prevalence; 128 articles including antiretroviral therapy-experienced/undefined treatment status populations; 78 articles with non-newly diagnosed/undefined diagnosis status; 3 articles without baseline tropism testing; 25 articles with sample size <20; and 8 articles with unavailable data.
27 articles were ultimately included in the analysis, among which 17 reported CD4+ T cell counts at diagnosis stratified by HIV tropism (R5 tropic vs. Non-R5 tropic). The study selection process is presented in Figure 1.
3.2 General characteristic
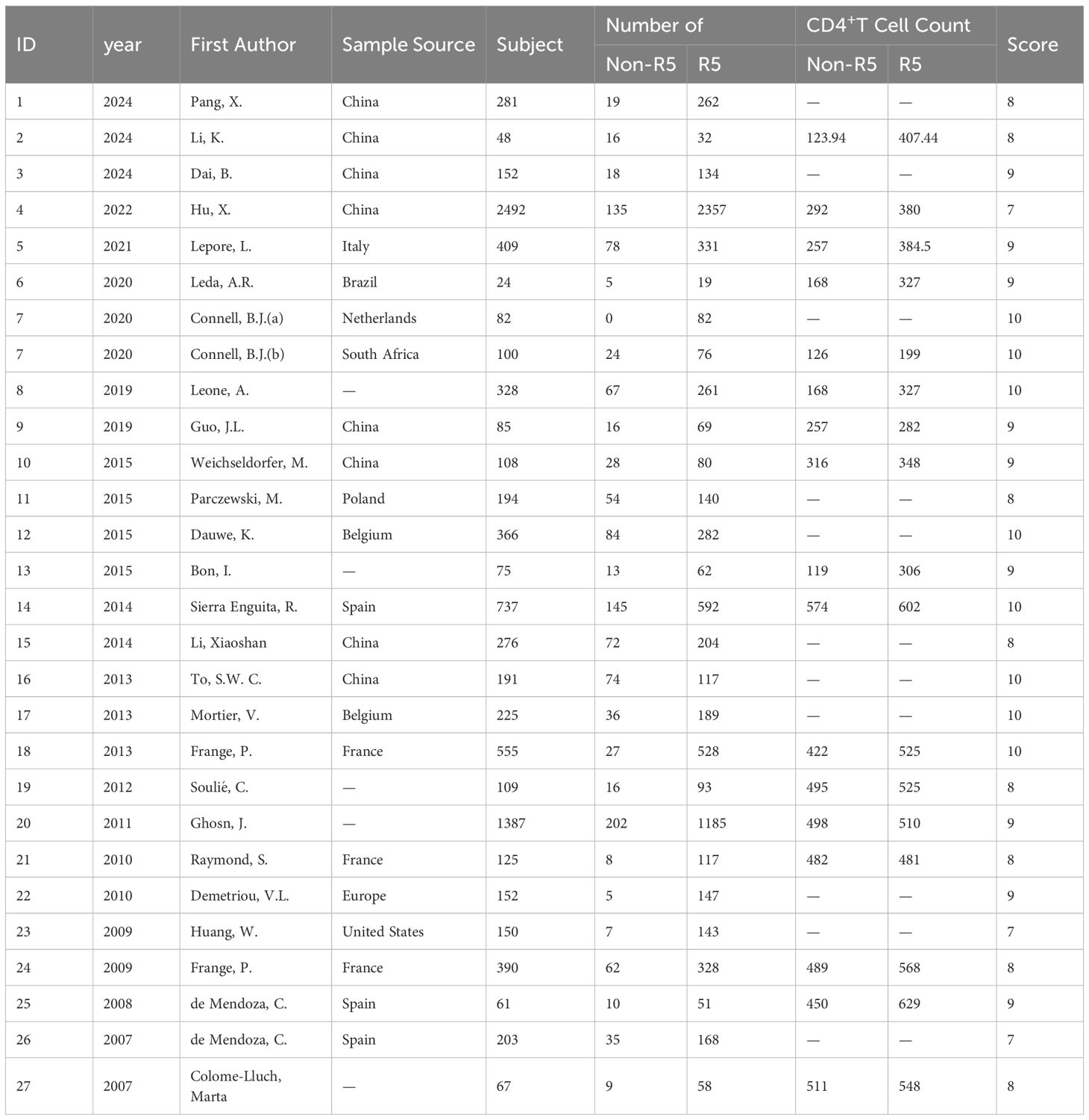
This study included 27 articles (27 data points) for meta-analysis of prevalence across different viral tropisms. The meta-analysis of CD4+ T cell count differences among individuals with distinct tropisms comprised 17 data points. Table 1 presents characteristics of the 27 included studies, encompassing 9,372 HIV-1-infected individuals primarily distributed in China, France, Poland, and Spain. Three data points exclusively involved MSM populations, while the remaining data points were derived from general population studies. Quality assessment scores for each included study are provided in the Supplementary Material.
3.3 Tropism prevalence
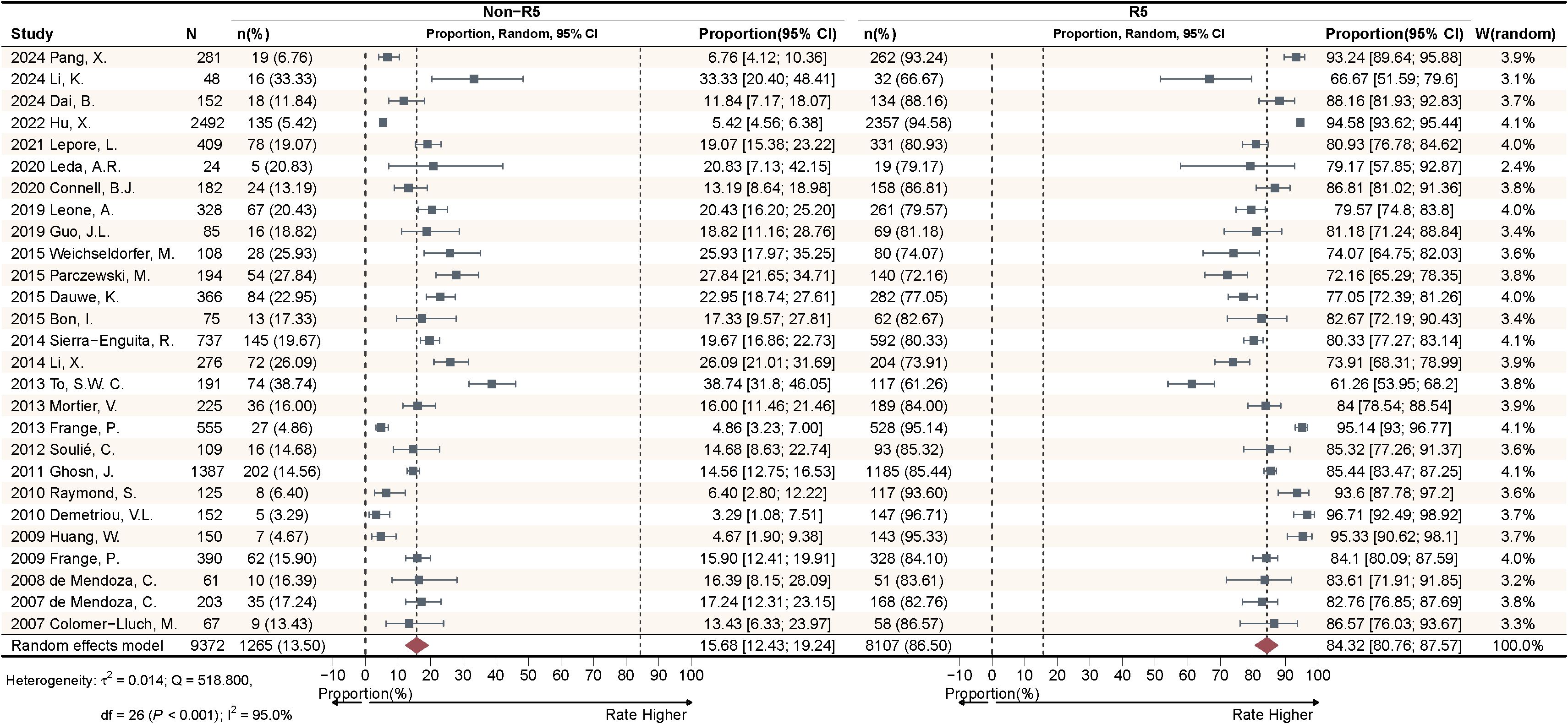
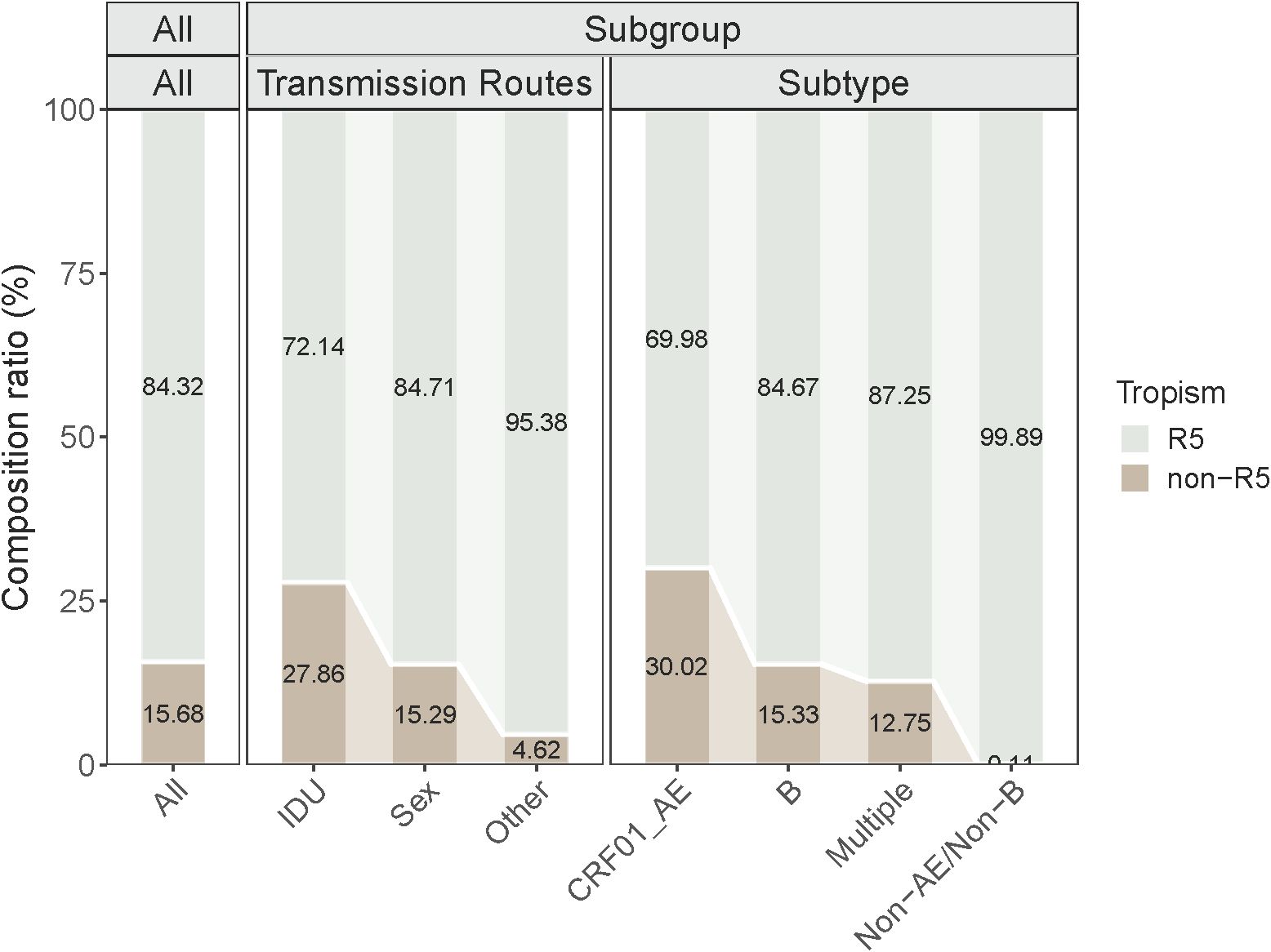
This study included a total of 27 articles to calculate the pooled effect size of Non-R5 and R5 tropism virus prevalence among HIV-1 PLWH. The meta-analysis (Figure 2) demonstrated that the random-effects model yielded a pooled prevalence of 15.68% [95% confidence interval (CI): 12.43-19.24%] for Non-R5 tropism and 84.32% (95% CI: 80.76-87.57%) for R5 tropism. Substantial heterogeneity was observed with I²=95.0%, accompanied by significant between-study variation (QB = 518.800, P < 0.001).
3.4 Subgroup analysis
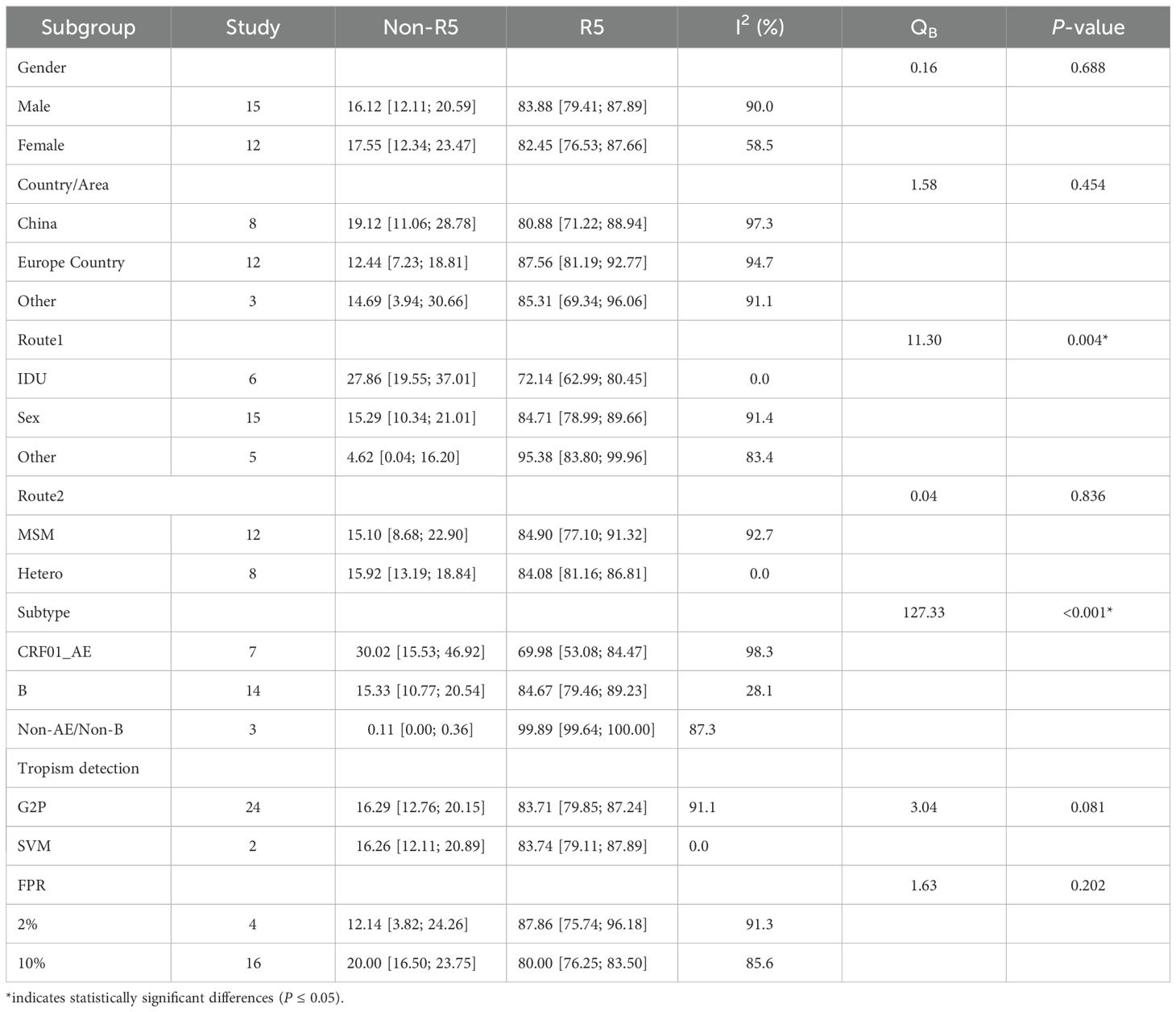
Area subgroup (23 data points): 1 article contributed 2 data points (Europe Country and Other), while 5 articles did not report sample sources. Transmission Route 1 subgroup: 6 articles included 2 data points (IDU and Sex), 2 articles reported 2 data points (Sex and Other), and 2 articles provided 3 data points (IDU, Sex, and Other). 12 articles lacked extractable subgroup information. Transmission Route2 subgroup (Sex transmission, 20 data points): 8 articles reported 2 data points (MSM and Hetero), and 15 articles lacked relevant data. HIV gene subtype subgroup (24 data points), 3 articles reported 3 data points (CRF01_AE, B, and other subtypes), 1 article provided 2 data points (CRF01_AE and B), 10 articles had no relevant data.
No statistically significant differences in Non-R5 tropism prevalence were observed among newly diagnosed PLWH across gender, geographic region, or Route2 subgroups (P > 0.05). However, significant differences were identified in Non-R5 tropism prevalence among PLWH stratified by Transmission Route1 and HIV gene subtypes (P ≤ 0.05) (Table 2).
Figure 3 illustrates the prevalence of Non-R5 tropic and R5 tropic viruses across subgroups of PLWH at diagnosis. The Non-R5 tropic virus prevalence was significantly higher among IDU (27.86%) compared to those infected through sexual transmission (15.29%) and other routes (4.62%). In the CRF01_AE subtype group, the proportion of Non-R5 tropic viruses (30.02%) exceeded that observed in subtype B (15.33%) and other subtypes (3.44%). These inter-subgroup differences in tropism prevalence showed statistically significant variations (P ≤ 0.05).
3.5 Differences in immune characteristics
This study included 17 articles comparing the pooled effect sizes of CD4+ T cell counts at diagnosis among PLWH with different viral tropisms. The pooled effect sizes of CD4+ T cells in PLWH with R5 tropic and Non-R5 tropic HIV were 431.82 and 329.02 cells/μL, respectively (Figure 4). The meta-analysis demonstrated that patients harboring Non-R5 tropic viruses had significantly lower CD4+ T cell counts at diagnosis compared to those with R5-tropic viruses, with a mean difference of −97.77 cells/μL (95% CI: -140.88 to -54.67) (Figure 5).
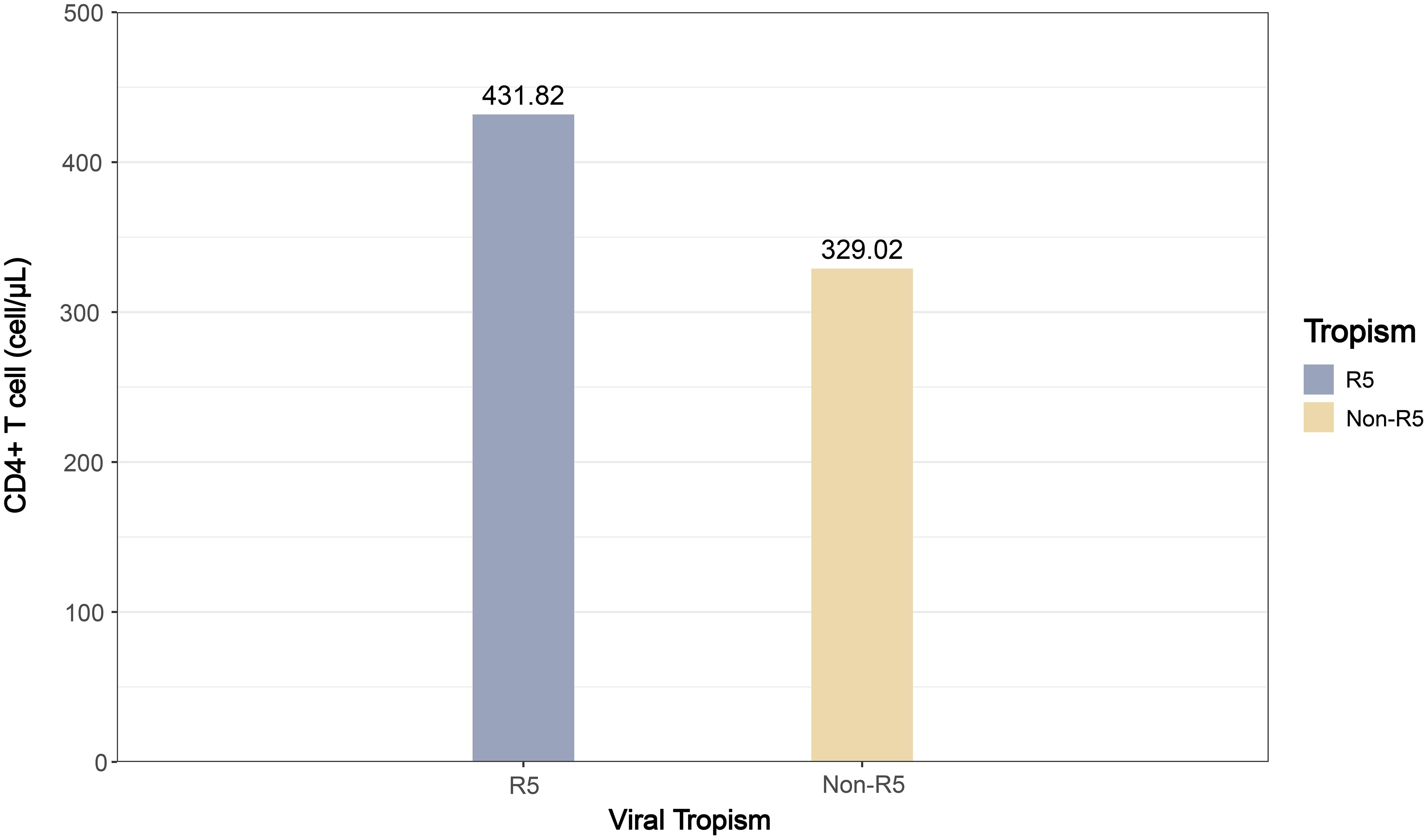
Figure 4. The bar plot of pooled effect sizes for CD4+ T cell counts by viral tropism (R5 vs. Non-R5) among PLWH.
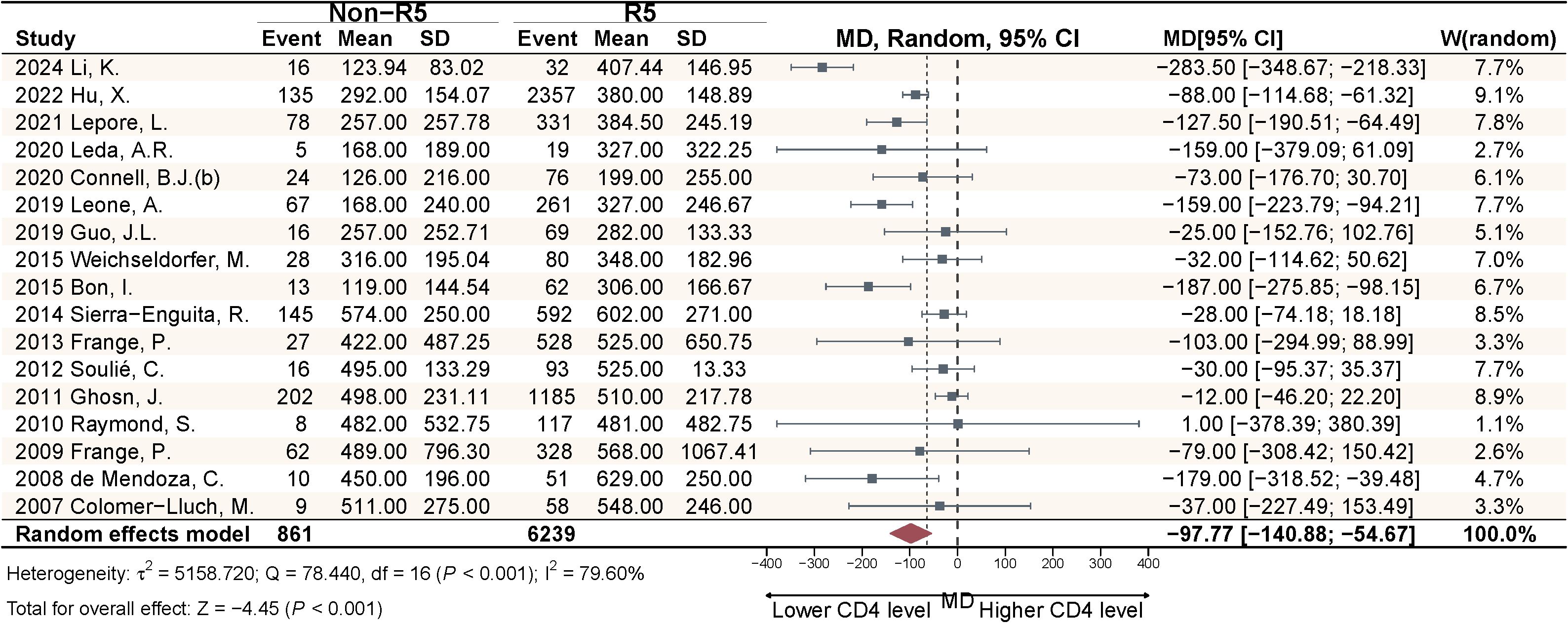
Figure 5. Meta-analysis of CD4+ cell count differences at diagnosis between viral tropism groups in PLWH.
3.6 Publication bias and sensitivity analysis
This study constructed funnel plots and performed asymmetry tests to evaluate potential bias, with results presented in the Supplementary Figures S1, S2. Egger’s test indicated no significant publication bias for either component (P1 = 0.026; P2 = 0.431). To assess the robustness of our findings, we performed a leave-one-out sensitivity analysis. The results demonstrated that the pooled effect size remained stable without significant changes, indicating that our results are robust. For detailed results, please refer to Supplementary Figure S3.
4 Discussion
This meta-analysis and systematic review provides the most comprehensive contemporary evidence on HIV-1 tropism distribution among antiretroviral-naïve PLWH at diagnosis. Compared to previous comprehensive analyses, our study has updated through 2025 and incorporated published multicenter datasets with expanded geographic coverage. While focusing on inter-subtype differences in tropism prevalence, we have also conducted preliminary investigations into the association between viral tropism and pre-treatment immune status. The results demonstrate a Non-R5 viruses prevalence of 15.68% (95% CI: 12.43%-19.24%). Subgroup analysis revealed two distribution patterns: with a higher Non-R5 virus prevalence associated with IDU; marked inter-subtype variation in Non-R5 virus prevalence (CRF01_AE > B > Other subtypes). In addition, the significant difference in CD4+ T lymphocyte counts at diagnosis between Non-R5 virus carriers and R5 virus carriers (−97.77 cells/μL; 95% CI: −140.88 - −54.67) provides population-level validation.
This study revealed a 15.68% prevalence of Non-R5 viruses among newly diagnosed PLWH, suggesting these individuals may have passed the early/acute infection phase and represent advanced-stage diagnoses. Globally, late diagnosis rates among newly diagnosed patients ranges from 15.00% to 43.00% (18), with high-income countries reporting up to 50% late presentation (19). Specifically, European countries report rates of 22.20%~74.30%, Japan 71.12%, while recent data from Chinese indicate 57.60%~70.20% (20). These findings underscore the imperative to optimize diagnostic timelines, emphasizing enhanced screening coverage and innovative nucleic acid testing technologies for early detection. Timely diagnosis facilitates prompt ART initiation, a strategy endorsed by international consensus guidelines for its clinical benefits (19). Notably, expanded PrEP implementation has increased detection during seroconversion phase. Transient CD4+ T lymphocyte counts declines during seroconversion (21) may inflate late-presentation estimates. Comprehensive diagnostic evaluation integrating viral tropism assessment is therefore recommended to improve staging accuracy.
The subgroup analysis revealed a significant association between the IDU transmission route and higher prevalence of Non-R5 viruses (P = 0.004). This phenomenon may originate from the selective disadvantage of Non-R5 viruses in sexual transmission routes (22). Our study revealed that the prevalence of Non-R5 viral strains in CRF01_AE (30.02%) and subtype B (15.33%) was significantly higher than in other subtypes (3.44%). Li et al.’s (23) previously reported a 45.5% X4 viruses prevalence among treatment-naïve populations with CRF01_AE, while Zhang et al. (24) demonstrated an even higher X4 viruses prevalence of 61.30% in CRF01_AE. In contrast, X4 variants were virtually absent in CRF07_BC subtypes (0.00%). Notably, CRF01_AE has become the predominant subtype in Asia (84.00%) (25). Clinically, the 30.02% Non-R5 prevalence in CRF01_AE populations challenges CCR5 antagonists utility across East/Southeast Asia, while subtype B subtype’s 15.33% rate warrants cautious implementation in Western countries. prevalent regions. These geographically stratified findings underscore the necessity of tropism testing prior to coreceptor antagonist regimens, particularly in high-prevalence subtypes.
This study estimated the pooled effect size of baseline immune status across viral tropism groups. At diagnosis, PLWH with Non-R5 viral infections exhibited lower CD4+ T cell count (329.02 cells/μL) compared to those with R5 viruses (431.82 cells/μL), remained above 200 cells/μL. Conventional perspectives posit that Non-R5 tropic variants predominantly emerge during advanced disease stages (CD4<200 cells/μL) (26, 27). However, our findings revealed that tropism switching had already occurred when the disease had not yet progressed to the late stage. In conjunction with previous reports of higher Non-R5 prevalence in CRF01_AE and, these data suggest accelerated switching kinetics within this subtype, warranting further investigation into underlying drivers. The clinical implications of viral tropism characteristics are particularly relevant for treatment-experienced patients with potential multidrug resistance receiving CCR5 antagonists as salvage therapy, given their association with baseline immune profiles and therapeutic outcomes. Given that Non-R5 tropic variants predominantly emerge during advanced disease stages and considering the substantial proportion of late-presenters, this finding underscores the critical importance of early HIV detection for interrupting transmission chains and improving clinical outcomes. Based on these findings, it is recommended that clinicians implement more intensive immune monitoring for patients with Non-R5 tropic infections due to their accelerated progression to AIDS.
This study has several limitations. We were unable to incorporate data from ongoing investigations or unpublished sources, although publication bias assessment and sensitivity analyses were conducted. The insufficient data available to analyze tropism characteristics across additional subtypes and to establish further subgroup analyses examining the association between tropism and CD4+ T cell counts necessitate updates upon publication of future studies. Despite the non-significant findings in country/region subgroup analysis, the high representation of Chinese studies may introduce a regional selection bias, necessitating caution when generalizing conclusions to global context. Although subgroup analyses were conducted to attempt to address heterogeneity, there may remain unmeasured confounding variables or insufficiently reported data that precluded comprehensive analysis. The cross-sectional design of this study precludes establishing temporal associations between tropism and immunological parameters. Prospective longitudinal studies are warranted to elucidate these dynamics. These limitations underscore the necessity to update the analyses as more robust evidence emerges.
5 Conclusion
This study analyzed the distribution of viral tropism and its association with baseline immune status in newly diagnosed PLWH. providing critical evidence for optimizing clinical testing strategies. Non-R5 viruses were found to be prevalent among newly infected individuals, exhibiting subtype-specific distribution patterns. This study findings highlight the necessity of early detection of Non-R5 infections and provide critical evidence for guiding individualized clinical management. Future studies should validate these patterns across broader geographical regions and explore tropism evolution mechanisms associated with long-term immune reconstitution.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YL: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review & editing. DY: Formal analysis, Methodology, Writing – review & editing. YY: Data curation, Methodology, Writing – review & editing. QH: Data curation, Methodology, Writing – review & editing. MZ: Data curation, Writing – review & editing. HM: Data curation, Writing – review & editing. PW: Conceptualization, Investigation, Writing – review & editing. YG: Investigation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX24_0484), the SEU Innovation Capability Enhancement Plan for Doctoral Students (CXJH_SEU 24042), and the Zhejiang Provincial Natural Science Foundation of China (LQ23H260002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1701028/full#supplementary-material
References
1. Zhang C, Lan Y, Li L, He R, Meng Y, Li J, et al. HIV-1 tropism in low-level viral load HIV-1 infections during HAART in Guangdong, China. Front Microbiol. (2023) 14:1159763. doi: 10.3389/fmicb.2023.1159763
2. Tuttle DL, Harrison JK, Anders C, Sleasman JW, and Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. (1998) 72:4962–9. doi: 10.1128/JVI.72.6.4962-4969.1998
3. Yi Y, Isaacs SN, Williams DA, Frank I, Schols D, De Clercq E, et al. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J Virol. (1999) 73:7117–25. doi: 10.1128/JVI.73.9.7117-7125.1999
4. Kotokwe K, Moyo S, Zahralban-Steele M, Holme MP, Melamu P, Koofhethile CK, et al. Prediction of coreceptor tropism in HIV-1 subtype C in Botswana. Viruses. (2023) 15:403. doi: 10.3390/v15020403
5. Marichannegowda MH, Setua S, Bose M, Sanders-Buell E, King D, Zemil M, et al. Transmission of highly virulent CXCR4 tropic HIV-1 through the mucosal route in an individual with a wild-type CCR5 genotype. eBioMedicine. (2024) 109:105410. doi: 10.1016/j.ebiom.2024.105410
6. Cho Y-K, Kim J-E, and Lee J. Korean Red Ginseng slows coreceptor switch in HIV-1 infected patients. J Ginseng Res. (2023) 47:117–22. doi: 10.1016/j.jgr.2022.06.003
7. Rosen O, Sharon M, Quadt-Akabayov SR, and Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion. Proc Natl Acad Sci USA. (2006) 103:13950–5. doi: 10.1073/pnas.0606312103
8. Crous S, Shrestha RK, and Travers SA. Appraising the performance of genotyping tools in the prediction of coreceptor tropism in HIV-1 subtype C viruses. BMC Infect Dis. (2012) 12:203. doi: 10.1186/1471-2334-12-203
9. Ge Z, Feng Y, Li K, Lv B, Zaongo SD, Sun J, et al. CRF01_AE and CRF01_AE cluster 4 are associated with poor immune recovery in Chinese patients under combination antiretroviral therapy. Clin Infect Dis. (2021) 72:1799–809. doi: 10.1093/cid/ciaa380
10. Ndung’u T, Sepako E, McLane MF, Chand F, Bedi K, Gaseitsiwe S, et al. HIV-1 subtype C in vitro growth and coreceptor utilization. Virology. (2006) 347:247–60. doi: 10.1016/j.virol.2005.11.047
11. Connell BJ, Hermans LE, Wensing AMJ, Schellens I, Schipper PJ, van Ham PM, et al. Immune activation correlates with and predicts CXCR4 co-receptor tropism switch in HIV-1 infection. Sci Rep. (2020) 10:15866. doi: 10.1038/s41598-020-71699-z
12. Pérez-Álvarez L, Delgado E, Vega Y, Montero V, Cuevas T, Fernández-García A, et al. Predominance of CXCR4 tropism in HIV-1 CRF14_BG strains from newly diagnosed infections. J Antimicrob Chemother. (2014) 69:246–53. doi: 10.1093/jac/dkt305
13. Li K, Chen H, Li J, Feng Y, Liang S, Rashid A, et al. Distinct genetic clusters in HIV-1 CRF01_AE-infected patients induced variable degrees of CD4+ T-cell loss. Mbio. (2024) 15:e0334923. doi: 10.1128/mbio.03349-23
14. Kaleebu P, Nankya IL, Yirrell DL, Shafer LA, Kyosiimire-Lugemwa J, Lule DB, et al. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D. J Acquir Immune Defic Syndr. (2007) 45:28–33. doi: 10.1097/QAI.0b013e3180385aa0
15. Saracino A, Monno L, Cibelli DC, Punzi G, Brindicci G, Ladisa N, et al. Co-receptor switch during HAART is independent of virological success. J Med Virol. (2009) 81:2036–44. doi: 10.1002/jmv.21598
16. Judicate GP, Barabona G, Kamori D, Mahiti M, Tan TS, Ozono S, et al. Phenotypic and genotypic co-receptor tropism testing in HIV-1 epidemic region of Tanzania where multiple non-B subtypes co-circulate. Front Microbiol. (2021) 12:703041. doi: 10.3389/fmicb.2021.703041
17. Khandu L, Dhakal GP, and Lhazeen K. Baseline CD4 count and the time interval between the initial HIV infection and diagnosis among PLHIV in Bhutan. Immun Inflammation Dis. (2021) 9:883–90. doi: 10.1002/iid3.444
18. Simon B, Grabmeier-Pfistershammer K, Rieger A, Sarcletti M, Schmied B, and Puchhammer-Stöckl E. HIV coreceptor tropism in antiretroviral treatment-naive patients newly diagnosed at a late stage of HIV infection. AIDS. (2010) 24:2051–8. doi: 10.1097/QAD.0b013e32833c93e6
19. Clemente T, Papaioannu-Borjesson R, Castagna A, and Spagnuolo V. Addressing late HIV presentation: the current knowledge and unmet needs. Expert Rev Anti-Infect Ther. (2025) 23:1–17. doi: 10.1080/14787210.2025.2530632
20. Yang Z, Chen Y, Tang H, Qin Q, Fei L, Wang S, et al. Trend analysis of late presentation of patients with HIV/AlDS withdifferent characteristics in China between 2013 and 2023. Chin J AIDS STD. (2025) 31:17–23. doi: 10.13419/j.cnki.aids.2025.01.02
21. Lodi S, Phillips A, Touloumi G, Geskus R, Meyer L, Thiébaut R, et al. Time from human immunodeficiency virus seroconversion to reaching CD4+ Cell count thresholds <200, <350, and <500 cells/mm3: assessment of need following changes in treatment guidelines. Clin Infect Dis. (2011) 53:817–25. doi: 10.1093/cid/cir494
22. Moore JP, Kitchen SG, Pugach P, and Zack JA. The CCR5 and CXCR4 coreceptors—Central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. In: AIDS Research and Human Retroviruses. (2004) 20:111–126. doi: 10.1089/088922204322749567
23. Li K, Chen H, Li J, Feng Y, Lan G, Liang S, et al. Immune reconstruction effectiveness of combination antiretroviral therapy for HIV-1 CRF01_AE cluster 1 and 2 infected individuals. Emerg Microbes Infect. (2022) 11:158–67. doi: 10.1080/22221751.2021.2017755
24. Zhang C, Xu S, Wei J, and Guo H. Predicted co-receptor tropism and sequence characteristics of China HIV-1 V3 loops: implications for the future usage of CCR5 antagonists and AIDS vaccine development. Int J Infect Dis. (2009) 13:e212–6. doi: 10.1016/j.ijid.2008.12.010
25. Hashempour A, Akbarinia S, Khodadad N, Safari F, and Mehrabi Z. Prediction of the Co-receptor usage of the main worldwide HIV-1 subtypes, CRF, and CRF35-AD in Iranian patients via the five genotypic tools. Biochem Biophysics Rep. (2025) 41:101939. doi: 10.1016/j.bbrep.2025.101939
26. Burger H, Hoover D, Zhang Y, de Lara C, Worth A, Hegedus A, et al. Accelerated in vivo proliferation of memory phenotype CD4+ T-cells in human HIV-1 infection irrespective of viral chemokine co-receptor tropism. PloS Pathog. (2008) 9:e1003310. doi: 10.1371/journal.ppat.1003310
Keywords: HIV-1, viral tropism, immune, CD4+T cell, epidemiology
Citation: Liu Y, Yuan D, Yin Y, He Q, Zhao M, Ma H, Wei P and Ge Y (2025) High prevalence of baseline Non-R5 viral tropism in PLWH is associated with immune damage: a systematic review and meta-analysis. Front. Immunol. 16:1701028. doi: 10.3389/fimmu.2025.1701028
Received: 08 September 2025; Accepted: 19 November 2025; Revised: 11 November 2025;
Published: 02 December 2025.
Edited by:
Marcelo A. Soares, National Cancer Institute (INCA), BrazilReviewed by:
M. Victoria Delpino, CONICET Instituto de Investigaciones Biomédicas en Retrovirus y SIDA (INBIRS), ArgentinaAisuluu Kubatova, FHI 360, United States
Copyright © 2025 Liu, Yuan, Yin, He, Zhao, Ma, Wei and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You Ge, Z2V5b3VfNTIxQDEyNi5jb20=; Pingmin Wei, bXB3XzE5NjNAMTI2LmNvbQ==
 Yangyang Liu
Yangyang Liu Defu Yuan
Defu Yuan Yueqi Yin
Yueqi Yin Qian He3
Qian He3 Meng Zhao
Meng Zhao Hongfei Ma
Hongfei Ma You Ge
You Ge