- 1Institute for Food Safety and Hygiene, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland
- 2Institute of Food, Nutrition and Health, Department of Health Science and Technology, ETH Zürich, Zurich, Switzerland
Staphylococcus aureus is a leading cause for clinical infections and food intoxications, causing over 100,000 yearly cases of bacteremia in the United States and 434 food-borne outbreaks in the European Union. Approximately 30% of the population permanently carry S. aureus asymptomatically in their nasal cavity. The risk of infection and transmission to food items or the environment is higher in individuals that are nasally colonized. In addition, S. aureus can acquire various antimicrobial resistances leading to therapeutic failure, additional medical costs, and fatalities. Methicillin-resistant S. aureus (MRSA) cause a considerable burden of disease in humans and animals. MRSA carriage has been associated with animal and in particular livestock contact. Extensive current data on the virulence gene profiles, as well as data on antimicrobial resistance determinants is crucial in developing effective strategies to mitigate the burden of disease. To this end, we screened the anterior nares of 160 test subjects (87 pupils and 73 members of farmer families) in Switzerland for S. aureus carriage. A total of 73 S. aureus isolates were obtained. Factors such as exposure to farm or companion animals and personal medical history were recorded using a questionnaire. Using a DNA microarray, isolates were assigned to clonal complexes (CCs), and virulence and resistance gene profiles were determined. The collected strains were assigned to 20 CCs, among others CC1, CC7, CC8, CC15, CC30, CC45, CC97, and CC398. Two MRSA strains and one multiresistant isolate carrying genes blaZ/I/R, InuA, aadD, tetK, and fosB were isolated from farmers with intensive exposure to animals. Strains carrying pvl, causing severe skin lesions and necrotizing pneumonia, as well as tetracycline, erythromycin, and kanamycin resistance genes were found in individuals that had taken antibiotics during the last year. A variety of superantigenic toxin genes was detected, including among others, the toxic shock syndrome toxin (tst1), and various enterotoxins (sea, sec, sel, and the egc cluster). Contact to chickens was identified as a significant factor contributing to S. aureus colonization.
Introduction
Staphylococcus aureus is a human pathogen, resulting in infections and foodborne intoxications. It caused over 100,000 cases of bacteremia in the United States in 2017 and 434 food-borne outbreaks in the European Union in 2015 (Tong et al., 2015; EFSA, 2016; Kourtis et al., 2019). Furthermore, it can colonize human anterior nares as a commensal bacterium. Around 20–30% of the population permanently carry S. aureus asymptomatically in their nose (Sakr et al., 2018), another 20–60% can be intermittent carriers (Kluytmans et al., 1997; Wertheim et al., 2005). Initial colonization may take place during the first days of life via transmission from mother to child (Maayan-Metzger et al., 2017). After birth the hands have been identified as the main source of transmission for the pathogen to colonize the nose (Wertheim et al., 2005). The nasal habitat may act as a reservoir for pathogen spread and can pose a threat to the carrier itself (Nouwen et al., 2006). An elevated risk of infection associated with persistent nasal carriage of S. aureus in surgical or dialysis patients emphasizes the importance of characterizing nasal colonization and associated risk factors (Kluytmans et al., 1997; Nouwen et al., 2006). In addition, antimicrobial resistance among S. aureus is rapidly emerging globally and rendering treatment of chronic and acute S. aureus infections increasingly difficult. The alarming prospect of a post-antibiotic era highlights the importance of identifying new therapeutic agents (Johler, 2017; Vestergaard et al., 2019). Specific virulence factors and/or master virulence regulators have been suggested as promising targets (Maura et al., 2016; Ansari et al., 2019). In particular, methicillin-resistant S. aureus (MRSA) have been studied in recent years with regard to their transmission dynamics (Christopher et al., 2011; Davis et al., 2012). Forms of transmission include hospital-acquired, community-acquired, and livestock-acquired with the MRSA in question being designated as HA-MRSA, CA-MRSA, and LA-MRSA, respectively (Deurenberg and Stobberingh, 2008; Knox et al., 2015; Smith, 2015; Kwok et al., 2018). Intensive contact to animals has been shown to increase the risk of LA-MRSA carriage among farmers, veterinarians, and animal owners (Graveland et al., 2011; Davis et al., 2012; Fluit, 2012). Several studies have investigated S. aureus colonization of veterinarians (Huber et al., 2011; Rosenkranz Wettstein et al., 2014), on-farm and food-chain epidemiology (Kraemer et al., 2017; Leuenberger et al., 2019; Morach et al., 2019), or colonization in the general population in Switzerland (Mégevand et al., 2010; Olearo et al., 2016). Here, we aim to provide insight into colonization patterns in asymptomatic carriers with varying animal exposure. We screened 160 individuals for nasal S. aureus and collected data on animal contact and medical history using a questionnaire. The obtained isolates were characterized by DNA microarray, which allows for assignment of clonal complexes (CCs), and determines the presence of resistance and virulence genes.
Materials and Methods
Questionnaire
A questionnaire was used to collect metadata. We assessed exposure of participants to farm and companion animals, medical history including antibiotic prescriptions and hospital or doctor visits. Answers concerning exposure to animals ranged from never to daily (0, 1, 2, 3, 4, 5). Medical history was recorded to assess, whether the individual had been to a doctor/hospital and/or received antibiotics in the last year (0/1). Raw data can be examined in Supplementary Table S2.
Sampling, Bacterial Isolation, Species Identification, and DNA Extraction
Each volunteer (87 pupils and 73 members of farmer families) provided a written declaration of consent and took his/her own nasal swab after prior demonstration of the sampling procedure. All nasal swabs were streaked on EASY Staph® plates (BIOKAR Diagnostics, Beauvais, France) and incubated at 37°C for 48 h. Presumptive S. aureus colonies were confirmed using matrix assisted laser desorption/ionization—time of flight mass spectrometry (MALDI-TOF MS, Bruker, Fällanden, Switzerland). After cell lysis with Lysostaphin (Sigma–Aldrich, Buchs, Schweiz), DNA was extracted with the Blood & Tissue Kit (Qiagen, Hombrechtikon, Switzerland).
DNA Microarray Analysis
DNA microarray was performed using Staphytype genotyping kit 2.0 (Alere, Wädenswil, Switzerland) following the manufacturer’s instructions. An ArrayMate reader (Alere) was used for signal acquisition. In addition, the similarity of the virulence and resistance gene profiles was visualized using SplitsTree41 (Huson and Bryant, 2006) as previously described (Wattinger et al., 2012). Readouts can be found in Supplementary Table S1.
Statistical Analysis
Binomial and multinomial logistic regression analyses were performed to ascertain the effects of age, gender, profession, medical history, and exposure to farm and companion animals on the likelihood that participants are colonized with S. aureus, with certain CCs or harbor MRSA strains or enterotoxin gene carrying isolates (SPSS, v26.0.0.0). P-values < 0.05 were considered significant (Supplementary Table S4).
Results
S. aureus Prevalence and Clonal Complexes in Sampled Populations
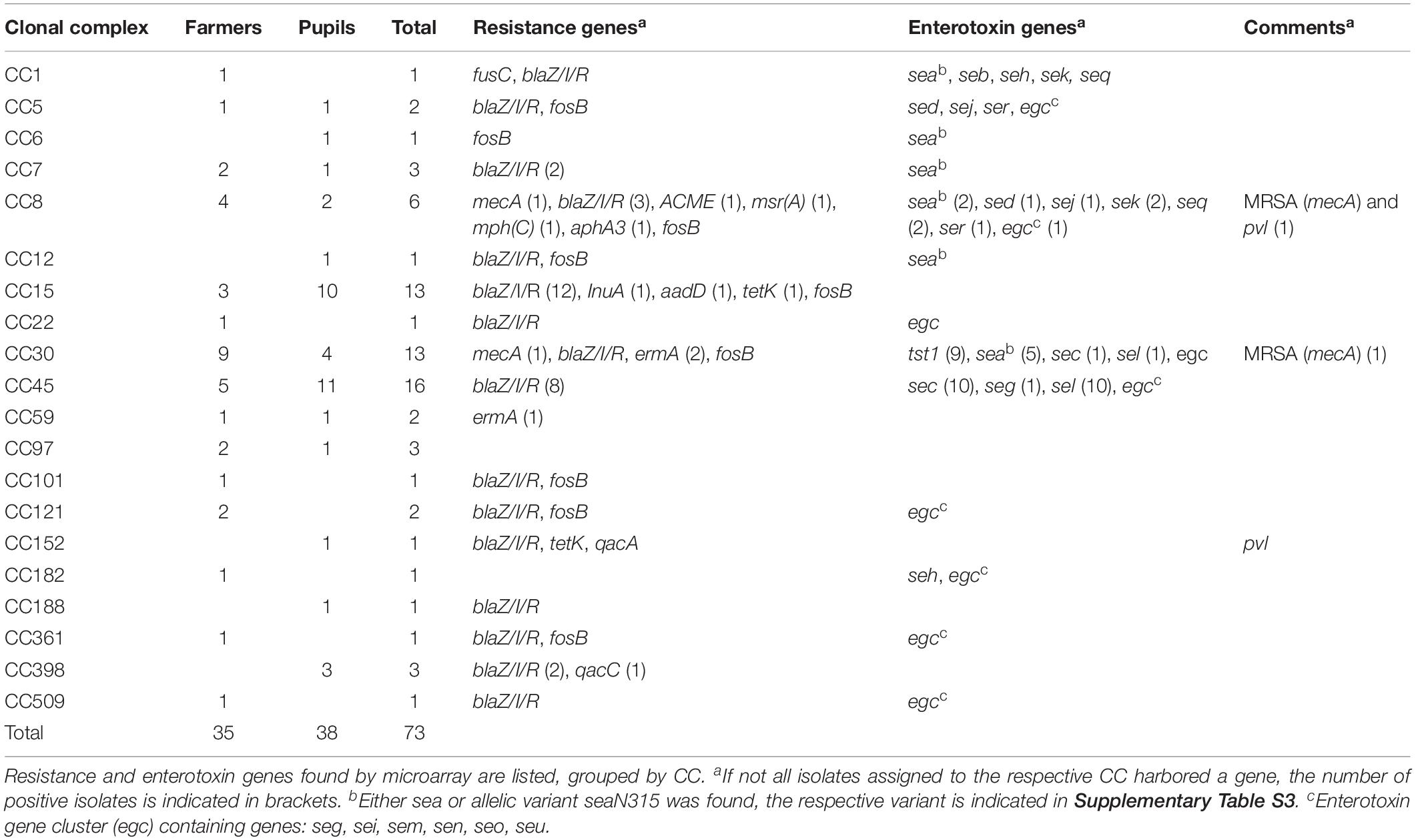
Of the 160 nasal swabs from asymptomatic carriers 46% (n = 73, confidence interval = 7.72) were positive for S. aureus. In the sampled 87 pupils and 73 family members of farmers colonization rates slightly differed with 44% in pupils and 48% in family members of farmers. The most common CCs for pupils were CC45 (n = 11) and CC15 (n = 10), while for family members of farmers, it were mainly CC30 (n = 9) and CC45 (n = 5) (Table 1). Some CC were exclusively found in one of the two groups, e.g., CC1, CC22, CC101, CC121, CC182, CC361, and CC509 in family members of farmers, while CC6, CC12, CC152, CC188, and CC398 were only found in pupils (Figure 1). All other strains belonged to CC5, CC7, CC8, CC59, and CC97.
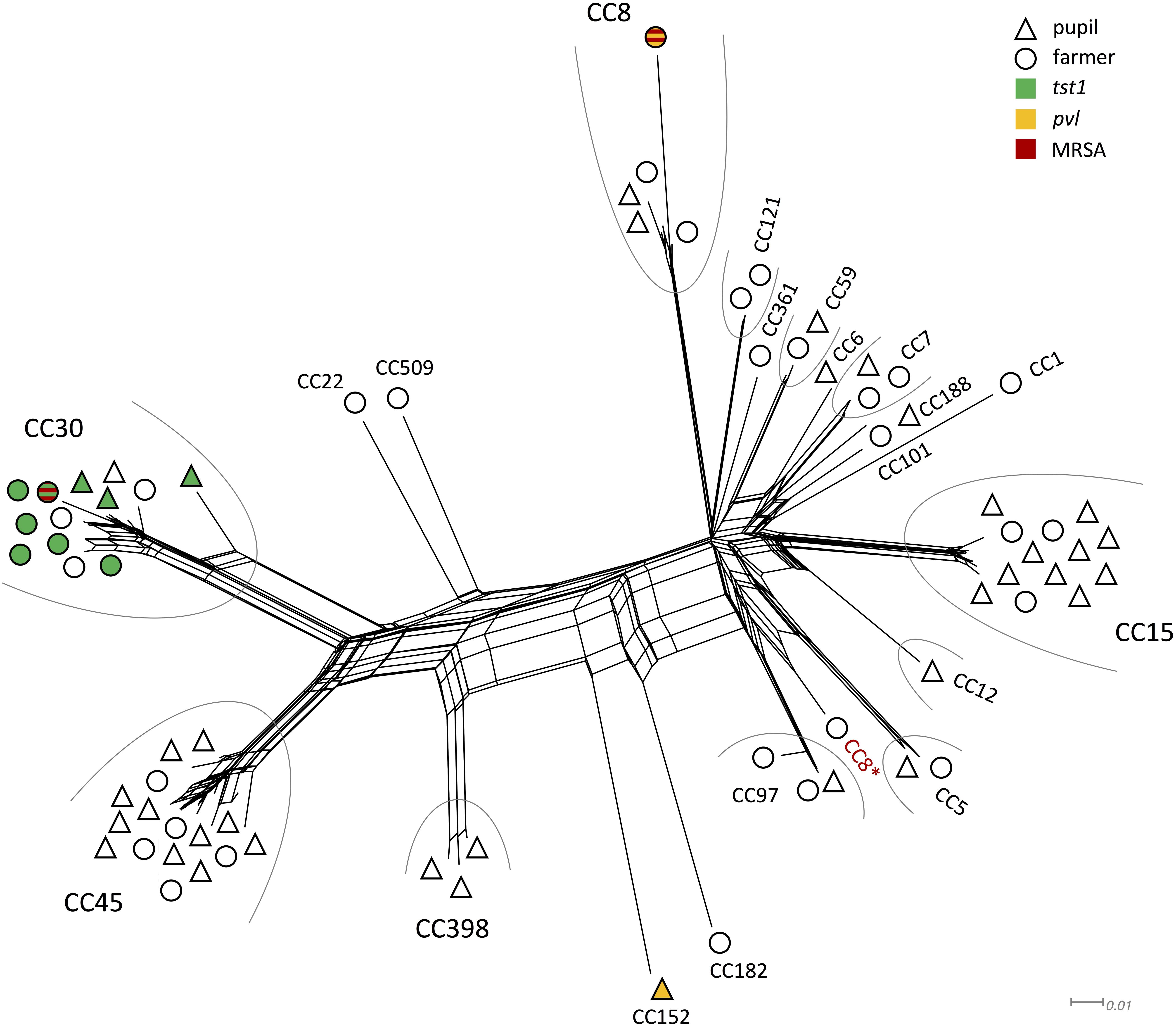
Figure 1. Splitstree showing the similarity of genomic profiles determined by microarray. Clonal complexes are grouped by an arc, if there were at least two isolates of the same CC. Symbol shape denominates whether the isolate originated from a farmer (O) or pupil (Δ). The fill color indicates presence of tst1 (green), pvl (yellow), or MRSA (red). ∗CC8 strain that grouped differently than other CC8 strains.
Exposure to Animals and Medical History of Participants
Overall animal exposure varied greatly between the two populations. Family members of farmers had an average exposure value of 2.4, corresponding to monthly-to-weekly exposure. Pupils on the other hand only reached a value of 0.8, corresponding to never-to-yearly exposure. Almost all family members of farmers were in contact with cattle on a daily basis (∅ = 4.8). Binomial logistic regression analysis revealed that exposure to chickens correlated significantly with whether an individual was colonized with S. aureus or not (p = 0.019). No association between exposure to certain animals and the occurrence of specific CC could be found. Medical history including visits to doctors or hospitals, as well as antibiotic prescription was not significantly associated with neither S. aureus carriage, nor occurrence of resistance in this study.
Similarity of Genomic Profiles
All isolates grouped according to their respective CC, with the exception of one CC8 strain that clustered closely with CC5 and CC97 strains (Figure 1). The two MRSA strains grouped very far from each other. The same applies for the two pvl positive strains. Genomic profiles of CC1, CC6, CC7, CC59, CC101, CC121, CC188, and CC361 all clustered closely.
Resistance Genes
A broad variety of resistance genes was found among isolates. The most common resistance genes were blaZ/I/R (n = 52) conferring β-lactam resistance and fosB (n = 40) coding for a metallothiol transferase, followed by ermA (n = 3) an erythromycin resistance gene, and tetK (n = 2) coding for tetracycline resistance. Two strains were identified as MRSA (CC8, CC30), both harboring the mecA cassette. Both were isolated from family members of farmers (Figure 1).
Other resistance genes included the arginine catabolic mobile element (ACME) that provides multiple immune modulating functions including resistance to polyamines (Joshi et al., 2012). A fusidic acid resistance protein fusC, and the genes msr(A) and mph(C) both coding for macrolide efflux pumps (Ross et al., 1995; Matsuoka et al., 2003) were observed. Genes aadD and aphA3 conferring resistance to aminoglycosides such as kanamycin and gentamycin were found. Additionally, InuA a gene responsible for lincosamide modification, and qacA and caqC encoding antiseptic resistance were discovered (Supplementary Table S3).
Enterotoxin Genes and Other Virulence Factors
Of the 73 isolates 48 isolates (66%) carried at least one enterotoxin gene (Table 1). Two isolates (CC8, CC152) carried the virulence factor panton-valentine leukocidin (pvl) gene that is responsible for severe skin lesions and necrotizing pneumonia (Lina et al., 1999; Kaneko and Kamio, 2004). One of the obtained pvl isolates was identified as an MRSA strain (Figure 1). All strains harboring the tst1 gene (n = 9) were assigned to CC30 (Figure 1). The most common enterotoxin genes were the genes grouped in the enterotoxin gene cluster egc: seg, sei, sem, sen, seo, seu (n = 38), sea (n = 13), sec (n = 11), and sel (n = 11). Other enterotoxin genes included seb (n = 1), sed (n = 3), seh (n = 2), sej (n = 3), sek (n = 3), and seq (n = 3).
Discussion
The observed colonization frequency of 46% (44% in pupils and 48% in family members of farmers) is in accordance with current estimates for nasal carriage rates (Sakr et al., 2018). When we compared the genomic profiles of all isolates, there were no overlaps, meaning that all isolates were unique in their genomic profile and therefore not clonal. All big clusters of CCs with more than three isolates were found in both family members of farmers and pupils. The large clusters of CC8, CC15, CC30, and CC45 comprised 66% of isolates and all represent community or hospital associated complexes (Pérez-Montarelo et al., 2017). One CC8 strain differed from all other detected CC8 strains in view of its enterotoxin gene profile. While other CC8 strains harbored diverse enterotoxins, the outlier strain only carried egc. Strains carrying egc were shown to be more common in asymptomatic carriers than in infections (Van Belkum et al., 2006). Similar distributions have been reported (Van Belkum et al., 2006; Mohamed et al., 2019). Surprisingly, strains of CC398 were exclusively found in pupils with very limited farm animal contact (once a year or less), although this CC has regularly been linked to livestock (Verkade and Kluytmans, 2014). This may be a first indicator for a possible shift from livestock-acquired to community-acquired transmission. Most other complexes were associated with isolates from both populations. This is consistent with findings of previous studies for various CCs including CC5, CC7, CC59, and CC97 (Nulens et al., 2008; Coombs et al., 2010; Budd et al., 2015; Wang et al., 2017; Challagundla et al., 2018).
Chickens were identified as a significant factor for whether an individual was colonized with S. aureus or not. No significant factors that correlated with colonization of certain CC, resistance genes, or toxin profiles could be determined. This was likely due to limitations in sample size and overall low number of strains exhibiting resistance/toxin genes. It is, however, noteworthy that one of the two MRSA strains was isolated from an individual that had taken antibiotics during the last year. The same was the case for both strains harboring pvl, both strains harboring qacA/C, and one of the strains carrying ermA. Increased occurrence of these genes in HA-strains has been reported (Lina et al., 1999; Schmitz et al., 2000; Mayer et al., 2001). An unusual accumulation of CC152 pvl strains has been reported in African refugees in Switzerland, possibly representing an emerging genotype (Jaton et al., 2016). One of two MRSA strains (CC8) that was isolated from a family member on a farm with intensive contact to cattle, pigs, and chicken possessed additional genes coding for pvl, ACME, blaZ/I/R, msr(A), mph(C), aphA3, and fosB. This represents a combination very similar to USA300 MRSA, a CA-MRSA strain often encountered in the US and considered a “superbug” (Monecke et al., 2011; Smith, 2015; Planet, 2017). Similar strains were found in Swiss hospital patients accounting for approximately 10% of the encountered MRSA (Seidl et al., 2014). The second MRSA strain featured genetic elements that are typical for CA- or HA-acquired MRSA of CC30, including the presence of blaZ/I/R, ermA, fosB, egc, and tst1 (Monecke et al., 2011). Another strain belonging to CC15 was classified as multiresistant according to international standard definitions for acquired resistance (Magiorakos et al., 2012) by carrying blaZ/I/R, InuA, aadD, tetK, and fosB. All three multiresistant strains were isolated from individuals regularly exposed to animals. While the DNA microarray data presented in this study provide new insights into the genetic structure of the tested isolates, it should not be used to extrapolate phenotypic traits.
This study provides an overview of S. aureus isolates found in asymptomatic carriers with varying exposure to animals. A diverse set of strains was characterized in terms of population structure and presence of virulence and resistance determinants. Our findings suggest that exposure to chicken may increase the risk of S. aureus nasal colonization and that intensive animal exposure may represent a risk factor for acquisition of multiresistant strains.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: https://www.ebi.ac.uk/arrayexpress/. Accession number: E-MTAB-8960.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SC and RS contributed to conception and design of the study. DE analyzed the data and wrote the first draft of the manuscript. SS, SC, and NC were responsible for data acquisition. DE, SJ, and RS wrote sections of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge all participants for filling out the questionnaire and providing nasal samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01289/full#supplementary-material
TABLE S1 | Microarray profile raw data. The outcome of every hybridization reaction is shown as either positive, negative, or ambiguous as determined by the ArrayMate reader (Alere).
TABLE S2 | Questionnaire raw data. Answers to the questionnaire are provided as raw data.
TABLE S3 | Strains. List of all isolates with corresponding CCs, resistance genes, and toxin profile.
TABLE S4 | Statistics. Binomial regression analysis output from SPSS.
Footnotes
References
Ansari, S., Jha, R. K., Mishra, S. K., Tiwari, B. R., and Asaad, A. M. (2019). Recent advances in Staphylococcus aureus infection: focus on vaccine development. Infect. Drug Resist. 12, 1243–1255. doi: 10.2147/IDR.S175014
Budd, K. E., McCoy, F., Monecke, S., Cormican, P., Mitchell, J., and Keane, O. M. (2015). Extensive genomic diversity among bovine-adapted Staphylococcus aureus: evidence for a genomic rearrangement within CC97. PLoS One 10:e0134592. doi: 10.1371/journal.pone.0134592
Challagundla, L., Reyes, J., Rafiqullah, I., Sordelli, D. O., Echaniz-Aviles, G., Velazquez-Meza, M. E., et al. (2018). Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the Western Hemisphere. Front. Microbiol. 9:1901. doi: 10.3389/fmicb.2018.01901
Christopher, S., Verghis, R. M., Antonisamy, B., Sowmyanarayanan, T. V., Brahmadathan, K. N., Kang, G., et al. (2011). Transmission dynamics of methicillin-resistant Staphylococcus aureus in a medical intensive care unit in India. PLoS One 6:e0020604. doi: 10.1371/journal.pone.0020604
Coombs, G. W., Monecke, S., Ehricht, R., Slickers, P., Pearson, J. C., Tan, H. L., et al. (2010). Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob. Agents Chemother. 54, 1914–1921. doi: 10.1128/AAC.01287-09
Davis, M. F., Iverson, S. A., Baron, P., Vasse, A., Silbergeld, E. K., Lautenbach, E., et al. (2012). Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 12, 703–716. doi: 10.1016/S1473-3099(12)70156-1
Deurenberg, R. H., and Stobberingh, E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. doi: 10.1016/j.meegid.2008.07.007
EFSA (2016). The Euroean Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 13:3991.
Fluit, A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. doi: 10.1111/j.1469-0691.2012.03846.x
Graveland, H., Wagenaar, J. A., Bergs, K., Heesterbeek, H., and Heederik, D. (2011). Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS One 6:e0016830. doi: 10.1371/journal.pone.0016830
Huber, H., Giezendanner, N., Stephan, R., and Zweifel, C. (2011). Genotypes, antibiotic resistance profiles and microarray-based characterization of methicillin-resistant Staphylococcus aureus strains isolated from livestock and veterinarians in Switzerland. Zoonoses Public Health 58, 343–349. doi: 10.1111/j.1863-2378.2010.01353.x
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Jaton, L., Pillonel, T., Jaton, K., Dory, E., Prod’hom, G., Blanc, D. S., et al. (2016). Common skin infection due to panton–valentine leucocidin-producing Staphylococcus aureus strains in asylum seekers from Eritrea: a genome-based investigation of a suspected outbreak. Clin. Microbiol. Infect. 22, 739.e5–739.e5. doi: 10.1016/j.cmi.2016.05.026
Johler, S. (2017). A glimpse into the future–new therapeutic targets could transform the way we treat staphylococcal infections. Virulence 8, 1508–1510. doi: 10.1080/21505594.2017.1393138
Joshi, G., Spontak, J., Klapper, D., and Richardson, A. (2012). ACME encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyami. Mol. Microbiol. 82, 9–20. doi: 10.1111/j.1365-2958.2011.07809
Kaneko, J., and Kamio, Y. (2004). Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci. Biotechnol. Biochem. 68, 981–1003. doi: 10.1271/bbb.68.981
Kluytmans, J., Van Belkum, A., and Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520. doi: 10.1128/cmr.10.3.505
Knox, J., Uhlemann, A.-C., and Lowy, F. D. (2015). Staphylococcus aureus infections: transmission within households and the community. Trends Microbiol. 23, 437–444. doi: 10.1016/j.tim.2015.03.007
Kourtis, A. P., Hatfield, K., Baggs, J., Mu, Y., See, I., Epson, E., et al. (2019). Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections - United States. Morb. Mortal. Wkly. Rep. 68, 214–219. doi: 10.15585/mmwr.mm6809e1
Kraemer, J. G., Pires, J., Kueffer, M., Semaani, E., Endimiani, A., Hilty, M., et al. (2017). Prevalence of extended-spectrum (β-lactamase-producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus in pig farms in Switzerland. Sci. Total Environ. 603–604, 401–405. doi: 10.1016/j.scitotenv.2017.06.110
Kwok, K. O., Read, J. M., Tang, A., Chen, H., Riley, S., and Kam, K. M. (2018). A systematic review of transmission dynamic studies of methicillin-resistant Staphylococcus aureus in non-hospital residential facilities. BMC Infect. Dis. 18:188. doi: 10.1186/s12879-018-3060-6
Leuenberger, A., Sartori, C., Boss, R., Resch, G., Oechslin, F., Steiner, A., et al. (2019). Genotypes of Staphylococcus aureus: on-farm epidemiology and the consequences for prevention of intramammary infections. J. Dairy Sci. 102, 3295–3309. doi: 10.3168/jds.2018-15181
Lina, G., Piemont, Y., Godail-Gamot, F., Bes, M., Peter, M.-O., Gauduchon, V., et al. (1999). Involvement of panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. doi: 10.1086/313461
Maayan-Metzger, A., Strauss, T., Rubin, C., Jaber, H., Dulitzky, M., Reiss-Mandel, A., et al. (2017). Clinical evaluation of early acquisition of Staphylococcus aureus carriage by newborns. Int. J. Infect. Dis. 64, 9–14. doi: 10.1016/j.ijid.2017.08.013
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Matsuoka, M., Inoue, M., Endo, Y., and Nakajima, Y. (2003). Characteristic expression of three genes, msr(A), mph(C) and erm(Y), that confer resistance to macrolide antibiotics on Staphylococcus aureus. FEMS Microbiol. Lett. 220, 287–293. doi: 10.1016/S0378-1097(03)00134-4
Maura, D., Ballok, A. E., and Rahme, L. G. (2016). Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46. doi: 10.1016/j.mib.2016.06.001
Mayer, S., Boos, M., Beyer, A., Fluid, A. C., and Schmitz, F.-J. (2001). Distribution of the antiseptic resistance genes. J. Antimicrob. Chemother. 47, 896–897.
Mégevand, C., Gervaix, A., Heininger, U., Berger, C., Aebi, C., Vaudaux, B., et al. (2010). Molecular epidemiology of the nasal colonization by methicillin-susceptible Staphylococcus aureus in Swiss children. Clin. Microbiol. Infect. 16, 1414–1420. doi: 10.1111/j.1469-0691.2009.03090.x
Mohamed, N., Timofeyeva, Y., Jamrozy, D., Rojas, E., Hao, L., Silmon de Monerri, N. C., et al. (2019). Molecular epidemiology and expression of capsular polysaccharides in Staphylococcus aureus clinical isolates in the United States. PLoS One 14:e0208356. doi: 10.1371/journal.pone.0208356
Monecke, S., Coombs, G., Shore, A. C., Coleman, D. C., Akpaka, P., Borg, M., et al. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936
Morach, M., Käppeli, N., Hochreutener, M., Johler, S., Julmi, J., Stephan, R., et al. (2019). Microarray based genetic profiling of Staphylococcus aureus isolated from abattoir byproducts of pork origin. PLoS One 14:e0222036. doi: 10.1371/journal.pone.0222036
Nouwen, J., Schouten, J., Schneebergen, P., Snijders, S., Maaskant, J., Koolen, M., et al. (2006). Staphylococcus aureus carriage patterns and the risk of infections associated with continuous peritoneal dialysis. J. Clin. Microbiol. 44, 2233–2236. doi: 10.1128/JCM.02083-05
Nulens, E., Stobberingh, E. E., Van Desse, H., Sebastian, S., Van Tiel, F. H., Beisser, P. S., et al. (2008). Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch university hospital between 1999 and 2006. J. Clin. Microbiol. 46, 2438–2441. doi: 10.1128/JCM.00808-08
Olearo, F., Albrich, W. C., Vernaz, N., Harbarth, S., and Kronenberg, A. (2016). Staphylococcus aureus and methicillin resistance in Switzerland: regional differences and trends from 2004 to 2014. Swiss Med. Wkly. 146:w14339. doi: 10.4414/smw.2016.14339
Pérez-Montarelo, D., Viedma, E., Murcia, M., Muñoz-Gallego, I., Larrosa, N., Brañas, P., et al. (2017). Pathogenic characteristics of Staphylococcus aureus endovascular infection isolates from different clonal complexes. Front. Microbiol. 8:917. doi: 10.3389/fmicb.2017.00917
Planet, P. J. (2017). Life after USA300: the rise and fall of a superbug. J. Infect. Dis. 215, S71–S77. doi: 10.1093/infdis/jiw444
Rosenkranz Wettstein, K., Rothenanger, E., Brodard, I., Collaud, A., Overesch, G., Bigler, B., et al. (2014). Nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA) among Swiss veterinary health care providers: detection of livestock- and healthcare-associated clones. Schweiz. Arch. Tierheilkd. 156, 317–325. doi: 10.1024/0036-7281
Ross, J. I., Eady, E. A., Cove, J. H., and Baumberg, S. (1995). Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene 153, 93–98. doi: 10.1016/0378-1119(94)00833-E
Sakr, A., Brégeon, F., Mège, J. L., Rolain, J. M., and Blin, O. (2018). Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 9:2419. doi: 10.3389/fmicb.2018.02419
Schmitz, F. J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Koöhrer, K., et al. (2000). Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45, 891–894. doi: 10.1093/jac/45.6.891
Seidl, K., Leimer, N., Palheiros Marques, M., Furrer, A., Senn, G., Holzmann-Bürgel, A., et al. (2014). USA300 methicillin-resistant Staphylococcus aureus in Zurich, Switzerland between 2001 and 2013. Int. J. Med. Microbiol. 304, 1118–1122. doi: 10.1016/j.ijmm.2014.08.005
Smith, T. C. (2015). Livestock-associated Staphylococcus aureus: the United States experience. PLoS Pathog. 11:e1004564. doi: 10.1371/journal.ppat.1004564
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Van Belkum, A., Melles, D. C., Snijders, S. V., Van Leeuwen, W. B., Wertheim, H. F. L., Nouwen, J. L., et al. (2006). Clonal distribution and differential occurrence of the enterotoxin gene cluster, egc, in carriage- versus bacteremia-associated isolates of Staphylococcus aureus. J. Clin. Microbiol. 44, 1555–1557. doi: 10.1128/JCM.44.4.1555-1557.2006
Verkade, E., and Kluytmans, J. (2014). Livestock-associated Staphylococcus aureus CC398: animal reservoirs and human infections. Infect. Genet. Evol. 21, 523–530. doi: 10.1016/j.meegid.2013.02.013
Vestergaard, M., Frees, D., and Ingmer, H. (2019). Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 7, 1–23. doi: 10.1128/microbiolspec.gpp3-0057-2018
Wang, X. L., Li, L., Li, S. M., Huang, J. Y., Fan, Y. P., Yao, Z. J., et al. (2017). Phenotypic and molecular characteristics of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in slaughterhouse pig-related workers and control workers in Guangdong Province, China. Epidemiol. Infect. 145, 1843–1851. doi: 10.1017/S0950268817000085
Wattinger, L., Stephan, R., Layer, F., and Johler, S. (2012). Comparison of Staphylococcus aureus isolates associated with food intoxication with isolates from human nasal carriers and human infections. Eur. J. Clin. Microbiol. Infect. Dis. 31, 455–464. doi: 10.1007/s10096-011-1330-y
Keywords: nasal colonization, MRSA, pvl, farmers, livestock, community, hospital, companion animals
Citation: Etter D, Corti S, Spirig S, Cernela N, Stephan R and Johler S (2020) Staphylococcus aureus Population Structure and Genomic Profiles in Asymptomatic Carriers in Switzerland. Front. Microbiol. 11:1289. doi: 10.3389/fmicb.2020.01289
Received: 06 April 2020; Accepted: 20 May 2020;
Published: 24 June 2020.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Frieder Schaumburg, University of Münster, GermanyPeter Kinnevey, Dublin Dental University Hospital, Ireland
Copyright © 2020 Etter, Corti, Spirig, Cernela, Stephan and Johler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danai Etter, ZGFuYWkuZXR0ZXJAaGVzdC5ldGh6LmNo
 Danai Etter
Danai Etter Sabrina Corti1
Sabrina Corti1 Roger Stephan
Roger Stephan Sophia Johler
Sophia Johler