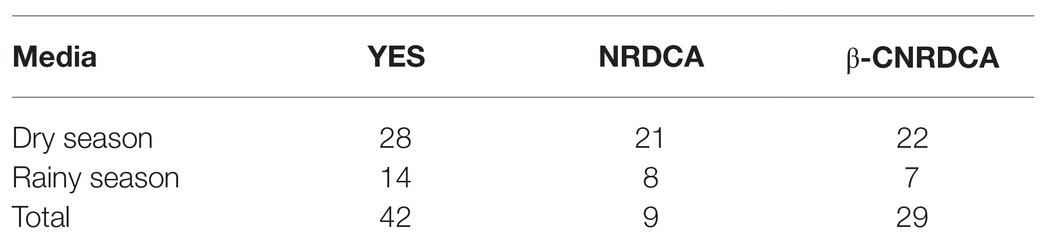- 1Department of Animal Health, Northwest University, Mmabatho, South Africa
- 2Department of Applied Biology and Biochemistry, National University of Science and Technology, Bulawayo, Zimbabwe
- 3Food Security and Food Safety Niche Area, Northwest University, Mmabatho, South Africa
- 4Department of Biological Sciences, Trinity University, Lagos, Nigeria
The presence of molds, especially certain species of Aspergillus, in food commodities may contribute to aflatoxin contamination. The aim of this study was to determine the biodiversity of Aspergillus species in dairy feeds from farms in select locations in Zimbabwe and assess their aflatoxin production potential using a polyphasic approach. A total of 96 feed samples were collected, which consisted of dairy feed concentrate, mixed ration, brewers’ spent grain, and grass from 13 farms during the dry season (August–October, 2016) and the following rainy season (January–March, 2017). A total of 199 presumptive isolates representing four sections from genus Aspergillus (Nigri, Fumigati, Flavi, and Circumdati) were recovered from the feeds. Section Flavi, which includes several aflatoxin producers, constituted 23% (n = 46) of the isolates. Species from this section were A. flavus, A. nomius, A. oryzae, A. parasiticus, and A. parvisclerotigenus, and 39 (84.4%) of these showed evidence of aflatoxin production in plate assays. Of the 46 section Flavi isolates examined, some lacked one or more of the five targeted aflatoxin cluster genes (aflD, aflR, aflS, aflM, and aflP). The presence of the five genes was as follows: aflD (76.9%), aflR (48.7%), aflS (74.4%), aflM (64.1%), and aflP (79.5%). This study highlights the species diversity of aflatoxigenic fungi that have the potential to contaminate different types of feed for dairy cows. Our findings underscore the importance of preventing contamination of feedstuffs by these fungi so that aflatoxins do not end up in the diets of consumers.
Introduction
Worldwide mycotoxin contamination of agricultural commodities by members of the Aspergillus genus, especially A. flavus and A. parasiticus, has been reported (Dutta and Das, 2001; Ghiasian and Maghsood, 2011; Ibrahim et al., 2016). These two species, together with A. nomius, are well known producers of the mycotoxin, aflatoxin, a secondary metabolite (Rodrigues et al., 2011; Monson et al., 2015). The ability of aflatoxin production by other species from this section such as A. pseudotamarii, A. bombycis, A. toxicarius, A. parvisclerotigenus, A. minisclerotigenes, and A. arachidicola have been documented (Ito et al., 2001; Pildain et al., 2008; Matumba et al., 2015). Aflatoxin contamination of food and feeds has gained global attention due to their negative effects on the health of both humans and animals (Fink-Grernmels, 1999; Zain, 2011; Arapcheska et al., 2015; Kumar et al., 2017). The more serious mycotoxins produced by Aspergillus species include aflatoxin (AF) B1, B2, G1, and G2 (Kumar et al., 2017). AFB1 is the most potent of them all and has been implicated as a causative agent of human hepatocarcinoma (Liu and Wu, 2010; Baranyi et al., 2013; Patel et al., 2015). Biosynthesis of aflatoxins involves participation of several enzymes and regulatory proteins whose genes are found in a single cluster of 70–75 kb in size. The order of these genes in Aspergillus is highly conserved (Woloshuk and Prieto, 1998; Yu et al., 2004; Adhikari et al., 2016). Not all section Flavi species produce aflatoxins (Atanda et al., 2006, 2011; Degola et al., 2007; Yazdani et al., 2010). The inability to produce aflatoxins by some species in this section is mainly due to mutations in at least one of the genes in the aflatoxin biosynthetic gene cluster (Degola et al., 2007; Adhikari et al., 2016).
Accurate identification of members of the Aspergillus genus using a single method is difficult due to their morphological similarities (Moslem et al., 2010; Zulkifli and Zakaria, 2017). Also, use of macro- and micro-morphological characters alone for identification is tedious and time consuming (Henry et al., 2000). To accelerate the species identification process, molecular markers such as the internal transcribed spacer (ITS) region have become the primary locus for fungal identification (Schoch et al., 2012; Raja et al., 2017). Extrolite profiling (e.g., types of aflatoxin produced) has also been used to aid in the identification and classification of Aspergilli though less frequently (Frisvad et al., 2007). Authors have proposed several media for cultural identification of aflatoxin producers (Bothast and Fennell, 1974; Lin and Dianese, 1976; Davis et al., 1987; Atanda et al., 2006, 2011). These media have chemical additives that enhance aflatoxin production for easy detection under UV radiation at 365 nm (Fente et al., 2001; Midorikawa et al., 2008). However, there are also limitations with using these types of plate assays (Suzuki and Iwahashi, 2016). Researchers report circumventing culture-based limitations by amplifying genes coding for key enzymes in the aflatoxin biosynthetic pathway (aflD, aflR, aflS, aflM, aflP) for identification of aflatoxigenic species (OBrian et al., 2007; Abdel-Hadi et al., 2011a; Mejía-Teniente et al., 2011; Baranyi et al., 2013; Zhi et al., 2013; Verheecke et al., 2015). Thus, for precise identification of aflatoxigenic Aspergillus isolates, a polyphasic approach is encouraged (Adetunji et al., 2019).
Animal feeds are formulated using agricultural commodities such as cereal grains and oil seed cakes. Cereal grain stovers and grass as silage can also be used as animal feed (Pleadin, 2015; Morrison et al., 2017). Moreover, the high cost of feed has led to the addition of stale bread, kitchen, and bakery wastes to extend its use, while the scarcity of protein sources for animal feeds has led to amendments with items such as brewers’ spent grain (BSG; Li et al., 2014). These waste products are usually tainted with fungi which may be a contributing factor in mycotoxin contamination in cattle feed. Upon ingestion by the cow, AFB1 is biotransformed into its hydroxylated metabolite aflatoxin M1 (AFM1), which is secreted in the milk (Britzi et al., 2013; Giovati et al., 2015) and becomes a route through which aflatoxins can be introduced into humans. Therefore, it is important to assess the quality of the feed fed to dairy cows in order to curb the transfer of the aflatoxins into milk. In this study, the diversity of Aspergillus species in dairy feeds, as well as their potential contamination with aflatoxins, was assessed using cultural-based and molecular methods.
Materials and Methods
Sample Collection
A total of 96 feed samples consisting of dairy feed concentrate (CN, n = 32), mixed ration (MR, n = 35), brewers’ spent grain (BSG, n = 7), and grass (GR, n = 22) were collected from 13 farms during the dry season (August–October, 2016) and the following rainy season (January–March, 2017). Samples were collected in sterile polythene zip-lock bags, which were transported in cooler boxes to the laboratory where they were ground to a fine powder using an IKA® M20 universal batch mill (Germany) and stored in the freezer at −20°C until time for analysis.
Isolation and Presumptive Identification of Aspergillus Fungi
Fungal isolation from feed samples followed the method of Aliyu et al. (2012) with some modifications as follows: feed samples weighing 1 g were suspended in 9 ml of sterile distilled water and mixed through vortexing. The sample was allowed to settle and 1 ml of the supernatant transferred into a test tube containing 9 ml of distilled water. The 10-fold serial dilution was carried out up to 10−6. This was followed by spread plating 0.1 ml of each dilution onto potato dextrose agar (PDA) supplemented with chloramphenicol (100 μg/ml). The plates were incubated at 25 ± 2°C for 3–7 days. Pure cultures were obtained through the single spore method (Almoammar et al., 2013). Since closely related Aspergillus species within each section have similar colony colors, we used this macro-morphological phenotype to identify our presumptive Aspergillus isolates to section level (Almoammar et al., 2013; Mostafa and Amer, 2013).
Screening for Aflatoxin-Producing Isolates
Each presumptive Aspergillus isolate was screened for the production of aflatoxin using multiple plating methods. Yeast Extract Sucrose (YES) agar (2% yeast extract, 15% sucrose, and 1.5% agar) shows the presence of aflatoxin through an induced color change plum red when exposed to ammonium hydroxide vapors (Yazdani et al., 2010). Aflatoxin-producing Aspergilli grown on Neutral Red Desiccated Coconut Agar (NRDCA; 20% desiccated coconut, 2% agar, and 0.03% neutral red stain; Atanda et al., 2011) and Neutral Red Desiccated Coconut Agar with β-cyclodextrin (β-CNRDCA; 20% desiccated coconut, 2% agar, 0.03% neutral red stain, and 0.03% β-cyclodextrin; Adetunji et al., 2019) will display a yellow/orange ring around the growing colony that will fluoresce on the reverse side of the plate when exposed to long-wave UV light (365 nm).
Molecular Identification of Isolates
For molecular investigations, we first inoculated each presumptive isolate on PDA and incubated them at 28°C for 72 h. DNA extraction utilized a Zymo Research Quick-DNA™ Fungal/Bacterial Miniprep Kit (The Epigenetics Company™, United States), and purity was assessed using a Nano-200 Microspectrometer (Allsheng Instruments Co., Ltd). DNA was stored at −20°C until time for use (Egbuta et al., 2015).
To refine our presumptive isolate identifications to species level, we amplified the internal transcribed spacer (ITS) region of each isolate’s DNA (ITS1-5.8S-ITS2) following the methods of Egbuta et al. (2015) with some modifications. Briefly, universal primers, ITS1 (5'-CTTGGT CAT TTA GAG GAA GTA A-3') and ITS4 (5'-TCC TCC GCT TAT TGA TATGC-3') synthesized by Inqaba Biotec (Pretoria, South Africa) were used. The PCR reaction consisted of 12.5 μl of 2× PCR master mix, 0.5 μl of each 25 μM primer, and 5 μl of DNA and constituted to a final volume of 25 μl with nuclease-free water and was carried out in a T100™ thermal cycler (Bio-Rad, Singapore) under the following conditions as follows: pre-dwelling at 95°C for 3 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 45 s, extension at 72°C for 1 min 30 s, post-dwelling at 72°C for 10 min, and holding at 4°C until samples were retrieved (Egbuta et al., 2015).
PCR amplification of key regulatory and structural genes from the aflatoxin pathway (aflD, aflR, aflS, aflM, aflP) involved primer pairs and cycling parameters previously described in other studies (Table 1). All PCR products were electrophoresed on a 1% agarose gel and visualized using a Bio-Rad Molecular Image® Gel Doc™ XR+ with Image Lab™ software (Bio-Rad, CA, United States). The amplicons were sent to Inqaba Biotec (South Africa) for sequencing. Analysis and cleaning of chromatograms were done using Finch TV software version 1.4.0 (Frickmann et al., 2015). Our Aspergillus sequences were queried against previously accessioned sequences in GenBank (National Centre for Biotechnology and Information) using the Basic Local Alignment Search Tool (BLAST; Magnani et al., 2005; Castrillo et al., 2012; Zarrin and Erfaninejad, 2016). Species identification was based on the best score (≥99% similarity; Gajjar et al., 2013; Frickmann et al., 2015; Rossi-Tamisier et al., 2015; Beye et al., 2017).
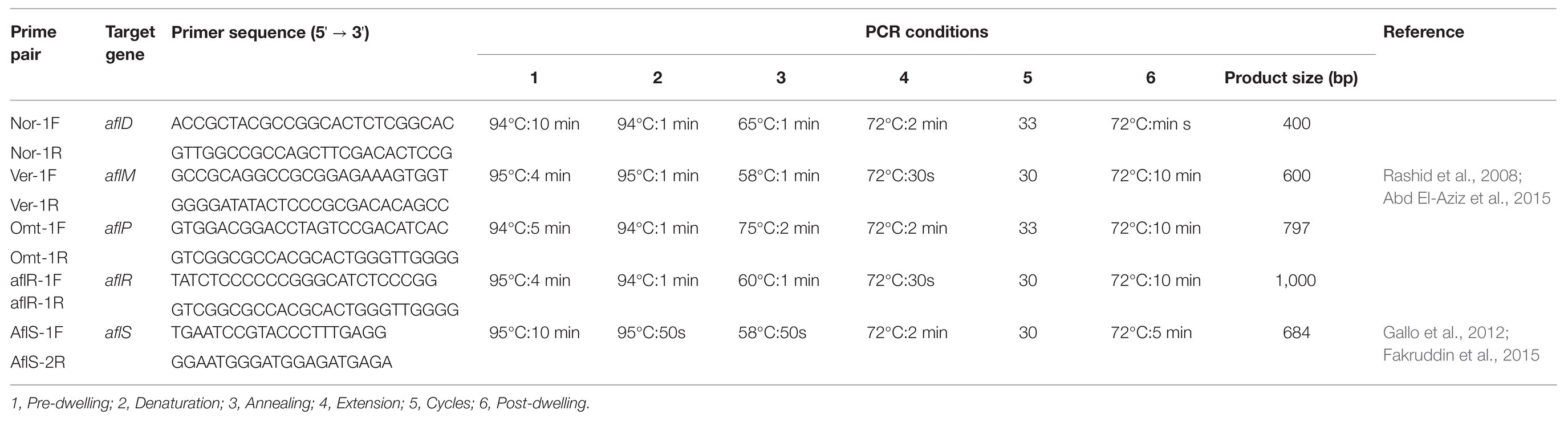
Table 1. Primer sequences and PCR conditions used for the detection of aflatoxigenic Aspergillus isolated from dairy feeds.
Results
Identification and Characterization of Isolates
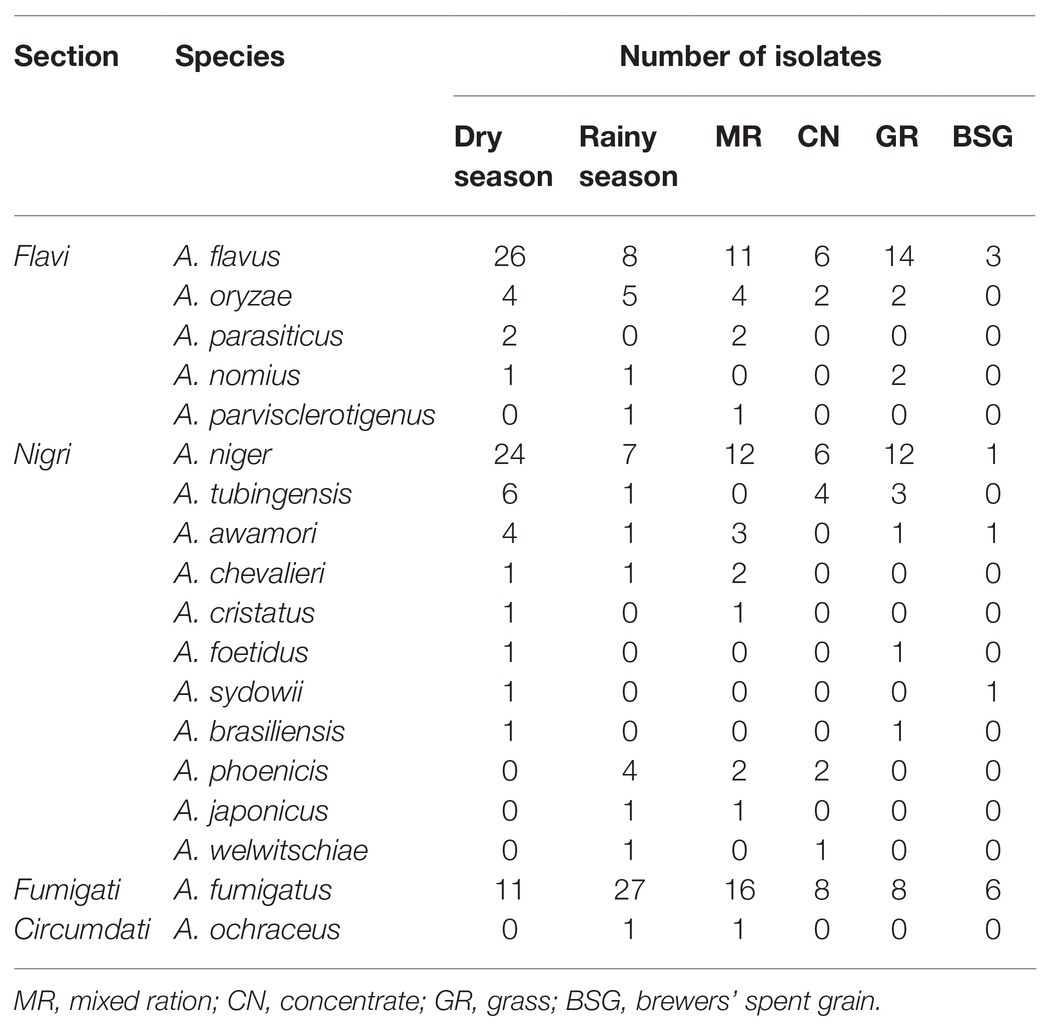
A sample population of 123 dry-season fungal isolates and 78 rainy-season fungal isolates were presumptively identified as Aspergillus and segregated by colony color into two main subgenera, namely, Circumdati and Fumigati, and four sections: Fumigati (blue colonies), Flavi (green colonies), Circumdati (yellow colonies), and Nigri (black colonies). The relative abundances of isolates from each section during the dry season and rainy season, as well as in the different feeds, are shown in Figure 1.
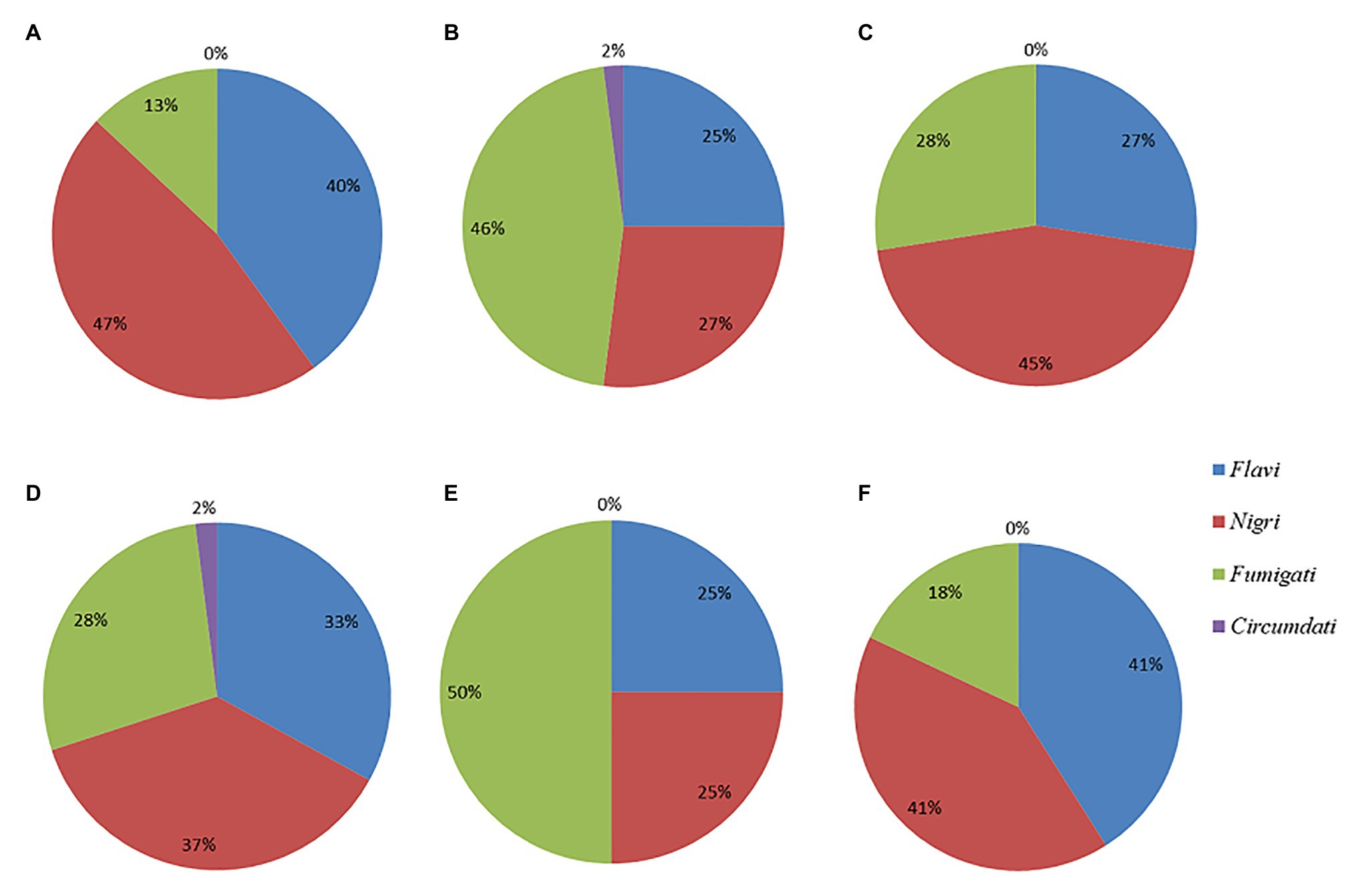
Figure 1. Pie charts showing the percent distributions of the four Aspergillus sections represented by the (A) dry season isolates (B) rainy season isolates (C) CN isolates (D) MR isolates (E) BSG isolates, and (F) GR isolates.
The ITS PCR amplicons were of the expected size for the region (600 bp). A total of 107 dry season and 78 rainy season sequences were of good enough quality to BLAST query. The remaining amplicons (n = 14 isolates) yielded poor sequences; therefore, these were not accessioned or subjected to further analysis. BLAST query of the ITS sequences of 185 presumptive Aspergillus isolates revealed that 83 of the isolates from the dry season had 99–100% similarity to the following species: A. niger, A. awamori, A. tubingensis, A. flavus, A. nomius, A. oryzae, A. parasiticus, A. fumigatus, A. foetidius, A. chavalieri, A. sydowii, A. brasiliensis, and A. cristatum. The remaining 24 belonged to other genera such as Byssochlamys, Cladosporium, Penicillium, Talaromyces, Eurotium, and Didymella. From the rainy season, 59 showed the same percentage similarity to A. niger, A. awamori, A. tubingensis, A. flavus, A. nomius, A. oryzae, A. fumigatus, A. ochraceus, A. welwitschiae, A. phoenicis, A. parvisclerotigenus, and A. japonicus. The other 17 isolates belonged to Byssochlamys, Cladosporium, Penicillium, Talaromyces, Alternaria, Paecilomyces, Rhizopus, and Sarocladium. The isolates were accessioned to GenBank and given numbers MG659595 to MG659694 for dry season isolates and MH270529 to MH270615 for rainy season isolates (Supplementary Table S1).
Out of the 142 isolates identified as members of the Aspergillus genus, 48 belonged to section Flavi. The distribution of the species in the various feeds is shown in Table 2. A. flavus (section Flavi) had the highest number of isolates in the dry season and were mainly from grass, whereas A. fumigatus (section Fumigati) was more predominant in the rainy season being isolated most frequently from the mixed ration samples. Section Nigri was dominated by A. niger, and most of these isolates were from the mixed ration and grass feeds. Only one isolate of A. ochraceus represented section Circumdati and it was isolated from a rainy season mixed ration feed sample (Table 2).
Findings Relating to Aflatoxin Production
Assessment of aflatoxin-producing ability was carried out on all the isolates belonging to the Aspergillus genus. Results for plate assays are shown in Table 3. Out of the 142 isolates, 42 tested positive for aflatoxin on YES agar, 27 from the dry season, and 15 from the rainy season (Supplementary Table S2). From section Flavi, A. flavus dominated for both seasons. Other members of the same section included A. oryzae, A. parasiticus, and A. nomius. Some isolates outside section Flavi, such as A. tubingensis ND2 and ND9, A. ochraceus NR2, A. fumigatus NR37, and A. japonicus NR67 also tested positive on YES. For NRDCA and β-CNRDCA, some isolates only showed the yellow ring with no fluorescence under UV light, others had no yellow ring but showed some fluorescence, and others showed both a yellow ring and the fluorescence in the reverse (Supplementary Table S2). Only isolates showing both yellow ring and fluorescence were considered as positive for aflatoxin production. Distribution of the aflatoxin biosynthesis cluster genes across the 142 Aspergillus isolates analyzed is given in Table 4. The percentage occurrences of the aflatoxin biosynthetic genes were as follows: (51%) aflD (14%) aflR (29%) aflS (14%) aflM, and (32%) aflP. Most of the isolates harbored at least one of the genes, and eight possessed all the five genes analyzed (Supplementary Table S3). Confirmation of aflatoxin-producing isolates using combined methods identified 30 positive isolates all belonging to section Flavi, namely, A. flavus, A. oryzae, A. parasiticus, A. nomius, and A. parvisclerotigenus (Table 5). The majority of the isolates were from the dry season and isolated from mixed ration and grass (Figure 2) and the least from brewers’ spent grain (Figure 3).
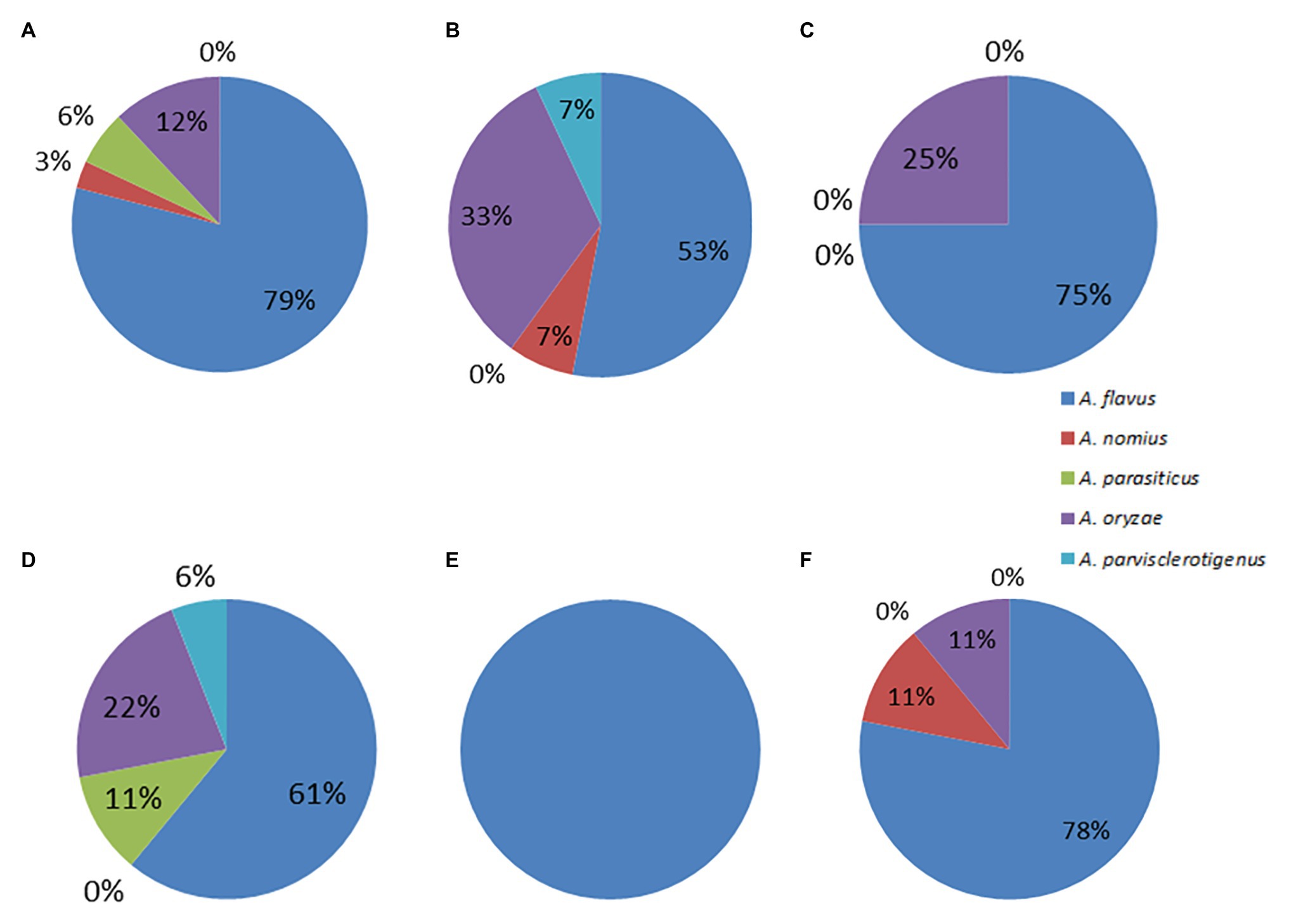
Figure 2. Pie charts showing the percent distributions of species within section Flavi represented by the (A) dry season isolates (B) rainy season isolates (C) CN isolates (D) MR isolates (E) BSG isolates, and (F) GR isolates.
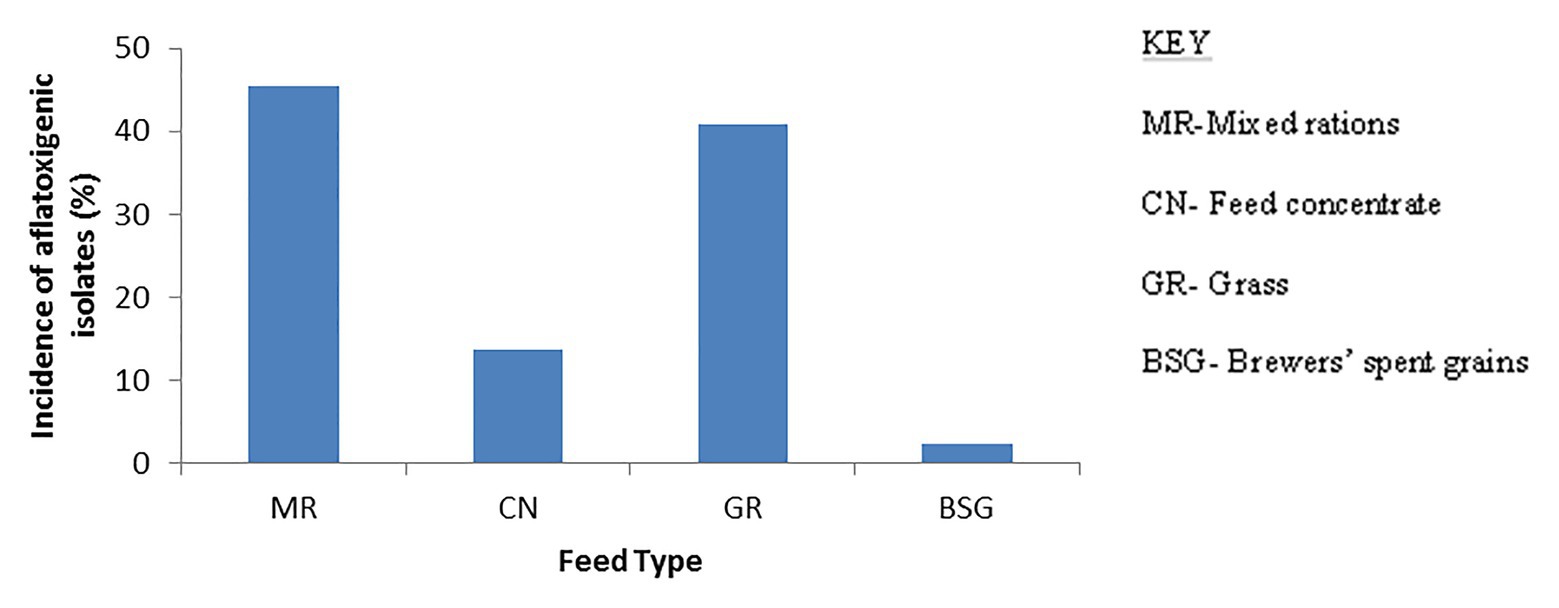
Figure 3. Bar charts showing occurrence of aflatoxigenic Aspergillus strains in the different types of feeds. The mixed ration harbored most of the aflatoxigenic strains and BSG the least.
Discussion
It has been reported that ingredients used in animal feed formulations can be contaminated by molds that are capable of producing aflatoxins (Gelven, 2010). This has led to extensive research in order to understand the diversity of Aspergillus species in feeds and feedstuffs (Variane et al., 2018). Findings from previous studies have indicated the presence of a wide range of Aspergillus species in feeds and feedstuffs. However, in Zimbabwe no work has reported on Aspergillus diversity and levels of aflatoxin contamination in animal feed.
Our findings showed that most of the isolates from our feed samples belonged to subgenera Fumigati (section Fumigati) and Circumdati (sections Nigri, Circumdati, and Flavi) based on observed morphological characteristics. The characteristic feature used in the identification of the isolates was the colony color that classified the isolates as follows: section Fumigati (blue), Nigri (black), Circumdati (yellow), and Flavi (green). However, use of only macro-morphological features has limitations since it does not guarantee accurate identification to species level. In order to identify the species with greater confidence, colony color should be augmented by examination of micro-morphological features of the isolates such as conidiophore, vesicle, metulae, phialides, and conidia, which was not done in our case. Alteratively, molecular analyses were performed on the isolates in order to resolve the limitations of the cultural methods. ITS sequence analysis using the BLAST search tool identified the black isolates as A. niger, A. brasiliensis, A. tubigensis, A. foetidus, A. phoenicis, A. welwitschiae, A. awamori, A. cristatus, A. sydowii, A. chevaliieri, and A. japonicus. The green isolates were identified as A. flavus, A. oryzae, A. nomius, A. parasiticus, and A. parvisclerotigenus. This shows the limitation of using morphological features for characterization; besides the process being time consuming and laborious (Henry et al., 2000) it can lead to misidentification. Therefore, there is a need for a polyphasic approach for the proper identification of the isolates (Adetunji et al., 2019). This work is limited in the sense that only the ITS region of the Aspergillus isolates was amplified. The use of ITS alone for the identification of some fungal genera has received criticism, especially in the discrimination of closely related species for genera such as Aspergillus, Cladosporium, Fusarium, Penicillium, and Trichoderma (Raja et al., 2017). The inability of the ITS region to differentiate between identical species such as A. flavus and A. oryzae has also been highlighted by Adetunji et al. (2019, 2020). Since amplification of ITS alone is not sufficient, further analysis of the isolates using other gene markers such as calmodulin (caM) and β-tubulin (benA) is therefore recommended in order to improve species identifications.
Some Aspergillus species outside section Flavi, such as those from sections Nidulantes and Ochraceorosei, have been reported to have the ability to produce aflatoxins (Cary et al., 2005; Varga et al., 2009); therefore, all 142 isolates were screened for aflatoxin producing potential (Supplementary Table S3). Sudini et al. (2015) indicated chances of having false-positive and false-negative results when using cultural methods as some non-aflatoxigenic species can produce yellow pigmentation and also fluoresce under UV light. This has been previously highlighted by Stark (2009) and Atanda et al. (2011). In this study, one A. fumigatus isolate NR12 exhibited some fluorescence and presence of yellow ring on β-CNRDCA, whereas for YES and molecular analysis the results were negative for the same isolate for aflatoxin production. Identification of these isolates based on the use of plate assays classified them as aflatoxin producers (Supplementary Table S2) while they are non-aflatoxigenic strains. Similarly, A. ochraceus isolate NR2 tested positive in both plate assays but lacked all aflatoxin cluster genes examined. This species is known for ochratoxin A production, which is able to fluoresce under UV light (Mohamed et al., 2013; Bueno et al., 2017); hence, using cultural methods would have classified it inappropriately as an aflatoxin producer. Similar observations were observed for isolates A. oryzae NR57 and A. fumigatus NR37, which indicated aflatoxin-positive results for both plate assays yet lacked one or more of the genes examined during molecular analysis. On the other hand, classification of isolates A. flavus NR3 and A. oryzae NR35 using only the coconut-based media suggested both as non-aflatoxigenic strains, yet the YES plate assay suggested they were aflatoxin positive, and both contained all the genes examined. Thus, it is important to classify organisms using combined methods. Another isolate, A. parvislerotigenus NR40, gave negative results for plate assays but had all major aflatoxin cluster genes analyzed in this study. It could have been that the isolate produced the toxins in low enough quantities to escape detection by culture assays. Therefore, in addition to the plate assays, the presence of aflatoxin could have been confirmed by chromatographic methods such as thin layer chromatography, high performance liquid chromatography (HPLC) or liquid chromatography with tandem mass spectrometry (LC-MS/MS).
Use of enhancing agents like β-cyclodextrin to improve fluorescence of aflatoxins has been reported (Abbas et al., 2004; Stark, 2009; Degola et al., 2012). In this study, only one isolate, A. japonicus NR67, had enhanced fluorescence on β-cyclodextrin-supplemented NRDCA. This may suggest that the use of plain media or supplemented media does not affect the results. Although this isolate gave positive results on YES and exhibited some fluorescence under UV light on β-cyclodextrin-supplemented media, it was classified as a non-aflatoxigenic strain as none of the aflatoxin cluster genes were amplified. There are no reports of aflatoxin production by species from sections Nigri and Fumigati, which is also in agreement with our observation from this study. The fluorescence under UV light exhibited by the isolates suggests possibility of production of other metabolites that can fluoresce. Investigations into the identities of these extrolites need to be carried out in the future.
Characterization of the isolates using molecular methods showed the presence of the aflD gene in 52% of the isolates. According to previous studies (Cleveland et al., 2009; Abdel-Hadi et al., 2011a,b; Yu, 2012), aflD codes for an enzyme that converts norsolorinic acid, the first stable intermediate in the aflatoxin biosynthetic pathway, to averantin. The other two structural genes, aflM and aflP, had a frequency of 14 and 25%, respectively. The aflatoxin biosynthetic pathway is said to be regulated by the aflR gene which encodes for transcriptional activators for the structural genes, however the discovery of yet another gene, aflJ, showed that aflR does not solely regulate the biosynthesis of aflatoxins since its activities were affected by aflS (Cleveland et al., 2009). Therefore, in this study, we used aflD, aflR, and aflS as the key markers for discriminating aflatoxigenic species from non-aflatoxigenic strains. In this study, detection of aflR was low with only 17% of the isolates showing the presence of the gene. This is in agreement with Abdel-Hadi et al. (2011b) who highlighted failure of aflR in discriminating aflatoxin producers from non-aflatoxin producing isolates. Similarly, Adetunji et al. (2019) observed the presence of the aflR gene in some A. oryzae isolates from cashew nuts even though these isolates tested negative for aflatoxin production. A much higher percentage of the isolates (23%) showed the presence of aflS. This suggests that aflS may be the main regulatory gene in the isolates found in Zimbabwe. The inability of amplification by some of the genes in the isolates could be due to the fact that they could not be expressed under the lab conditions and there is a need to provide conditions that can lead to the expression of the genes.
Isolates from section Flavi had an occurrence frequency of 34% across the feeds (Supplementary Table S4). This is a cause of concern as this section is well known for its aflatoxin producing species, A. flavus (Rodrigues et al., 2007). From this study, 88% of the isolates from section Flavi tested positive for aflatoxin production. A. flavus constituted 71% of the isolates in this section, and 56% of these isolates showed the potential to produce aflatoxin using both the cultural and molecular methods of validation. Aspergillus oryzae has not been associated with aflatoxin production and has been used in the production of fermented foods (Chang et al., 2006); however, six out of the nine isolates identified as A. oryzae exhibited aflatoxin production potential in our plate assays. Reconfirmation of the identities of these six isolates should involve BLAST query of other conserved genomic loci such as calmodulin (caM) and β-tubulin (benA). Aspergillus oryzae is the domesticated form of A. flavus, and the duo are morphologically identical (Payne et al., 2006); hence, there is a possibility that the accessioned sequences in GenBank were misidentified by their submitters. However, the presence of aflatoxin regulatory gene, aflR has been reported in A. oryzae (Kusumoto et al., 1998; Lee et al., 2006; Adetunji et al., 2019), but they were found to be non-aflatoxin producers. The presence of aflR cannot be readily associated with aflatoxin production, since any of 25 genes in the aflatoxin pathway could be present and non-functional.
This study reports for the first time the biodiversity of aflatoxigenic Aspergillus species in feeds used in dairy farming in Zimbabwe, and the findings showed the presence of four sections of Aspergilli with the predominating section being Nigri. Nigri species are non-aflatoxigenic; however, some are capable of producing ochratoxin A, another regulated mycotoxin. This wide diversity could be attributed to use of contaminated ingredients. Omeiza et al. (2018) highlighted strain diversity as a risk associated with unregulated trade routes used by farmers in Africa when acquiring ingredients for animal feeds. Aflatoxin-producing fungi have a preference for oil seed crops such as maize, cotton, and peanut. The most contaminated feed type was mixed ration, which is usually made by mixing mainly waste products of oil extraction from seeds and other ingredients. Cottonseed cake was the main waste product used by most farmers for the mixed ration. Therefore, farmers are encouraged to avoid contaminated ingredients for formulating mixed rations. The fact that feed producers could be using potentially contaminated ingredients is a serious issue. The local government should regulate these cottonseed meal producers and force them to assess the presence of aflatoxin in their product before they can sell it to growers.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MG659595 to MG659694 https://www.ncbi.nlm.nih.gov/, MH270529 to MH270615.
Author Contributions
NN did the isolation and characterization of the isolates from the samples. MA assisted with the assaying for aflatoxin production by the isolates, reviewing and proof reading of the article. LN assisted with the molecular characterization. MM helped with the supervision of the whole research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funds from the National Research Foundation (NRF) Grant number 105882 and the Research Board, NUST (Grant number RB No. 43/16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to appreciate the farmers and the personnel from the different farms where the feed samples were collected. “The content of this manuscript has been published [in Chapter 3] as part of the thesis of Nleya (2019).”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.599605/full#supplementary-material
References
Abbas, H. K., Zablotowicz, R., Weaver, M., Horn, B., Xie, W., and Shier, W. (2004). Comparison of cultural and analytical methods for determination of aflatoxin production by Mississippi Delta Aspergillus isolates. Can. J. Microbiol. 50, 193–199. doi: 10.1139/W04-006
Abd El-Aziz, A. R., Mahmoud, M. A., Al-Othman, M. R., and Al-Gahtani, M. F. (2015). Use of selected essential oils to control aflatoxin contaminated stored cashew and detection of aflatoxin biosynthesis gene. Sci. World J. 2015:958192. doi: 10.1155/2015/958192
Abdel-Hadi, A. M., Caley, D. P., Carter, D. R., and Magan, N. (2011a). Control of aflatoxin production of Aspergillus flavus and Aspergillus parasiticus using RNA silencing technology by targeting aflD (nor-1) gene. Toxins 3, 647–659. doi: 10.3390/toxins3060647
Abdel-Hadi, A., Carter, D., and Magan, N. (2011b). Discrimination between aflatoxigenic and non-aflatoxigenic Aspergillus section Flavi strains from Egyptian peanuts using molecular and analytical techniques. World Mycotoxin J. 4, 69–77. doi: 10.3920/WMJ2010.1223
Adetunji, M. C., Ezeokoli, O. T., Ngoma, L., and Mwanza, M. (2020). Phylogenetic diversity and prevalence of mycoflora in ready-to-eat supermarket and roadside-vended peanuts. Mycologia 1–11. doi: 10.1080/00275514.2020.1804235
Adetunji, M. C., Ngoma, L., Atanda, O. O., and Mwanza, M. (2019). A polyphasic method for the identification of aflatoxigenic aspergilla from cashew nuts. World J. Microbiol. Biotechnol. 35:15. doi: 10.1007/s11274-018-2575-8
Adhikari, B. N., Bandyopadhyay, R., and Cotty, P. J. (2016). Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America. AMB Express 6:62. doi: 10.1186/s13568-016-0228-6
Aliyu, R., Abubakar, M., Adamu, A., and Egwu, E. (2012). Mycological quality of commercially prepared and self-compounded poultry feeds in Sokoto Metropolis, Sokoto, Nigeria. Afr. J. Microbiol. Res. 6, 7314–7318. doi: 10.5897/AJMR12.761
Almoammar, H., Bahkali, A. H., and Abd-Elsalam, K. A. (2013). A polyphasic method for the identification of aflatoxigenic Aspergillus species isolated from camel feeds. Aust. J. Crop. Sci. 7:1707.
Arapcheska, M., Jovanovska, V., Jankuloski, Z., Musliu, Z., and Uzunov, R. (2015). Impact of aflatoxins on animal and human health. Int. J. Innov. Sci. Eng. Technol. 2, 156–161.
Atanda, O., Akpan, I., and Enikuomehin, O. (2006). Palm kernel agar: an alternative culture medium for rapid detection of aflatoxins in agricultural commodities. Afr. J. Biotechnol. 5, 1029–1033. doi: 10.5897/AJB05.460
Atanda, O., Ogunrinu, M., and Olorunfemi, F. (2011). A neutral red desiccated coconut agar for rapid detection of aflatoxigenic fungi and visual determination of aflatoxins. World Mycotoxin J. 4, 147–155. doi: 10.3920/WMJ2010.1241
Baranyi, N., Kocsubé, S., Vágvölgyi, C., and Varga, J. (2013). Current trends in aflatoxin research. Acta Biologica Szegediensis 57, 95–107.
Beye, M., Fahsi, N., Raoult, D., and Fournier, P. -E. (2017). Careful use of 16S rRNA gene sequence similarity values for the identification of Mycobacterium species. New Microbes New Infect. 22, 24–29. doi: 10.1016/j.nmni.2017.12.009
Bothast, R., and Fennell, D. (1974). A medium for rapid identification and enumeration of Aspergillus flavus and related organisms. Mycologia 66, 365–369.
Britzi, M., Friedman, S., Miron, J., Solomon, R., Cuneah, O., Shimshoni, J. A., et al. (2013). Carry-over of aflatoxin B1 to aflatoxin M1 in high yielding Israeli cows in mid-and late-lactation. Toxins 5, 173–183. doi: 10.3390/toxins5010173
Bueno, D., Munoz, R., and Marty, J. (2017). “Low cost and optical detection from fluorescence of Ochratoxin A” in 2017 Global Medical Engineering Physics Exchanges/Pan American Health Care Exchanges. GMEPE/PAHCE: IEEE, 1–4.
Cary, J., Klich, M., and Beltz, S. (2005). Characterization of aflatoxin-producing fungi outside of Aspergillus section Flavi. Mycologia 97, 425–432. doi: 10.3852/mycologia.97.2.425
Castrillo, M., Fonseca, M., Bich, G., Jerke, G., Horianski, M., and Zapata, P. (2012). Taxonomy and phylogenetic analysis of Aspergillus section Nigri isolated from yerba mate in Misiones (Argentina). J. Basic Appl. Genet. 23, 19–27.
Chang, P. -K., Ehrlich, K. C., and Hua, S. -S. T. (2006). Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. Int. J. Food Microbiol. 108, 172–177. doi: 10.1016/j.ijfoodmicro.2005.11.008
Cleveland, T. E., Bhatnagar, D., and Yu, J. (2009). “Elimination and control of aflatoxin contamination in agricultural crops through Aspergillus flavus genomics” in Mycotoxin prevention and control in agriculture. eds. M. Appell, D. F. Kendra, and M. W. Trucksess (Washington, DC: ACS Division of Agriculture and Food Chemistry), 93–106.
Davis, N., Iyer, S., and Diener, U. (1987). Improved method of screening for aflatoxin with a coconut agar medium. Appl. Environ. Microbiol. 53, 1593–1595. doi: 10.1128/AEM.53.7.1593-1595.1987
Degola, F., Berni, E., Dall’Asta, C., Spotti, E., Marchelli, R., Ferrero, I., et al. (2007). A multiplex RT-PCR approach to detect aflatoxigenic strains of Aspergillus flavus. J. Appl. Microbiol. 103, 409–417. doi: 10.1111/j.1365-2672.2006.03256.x
Degola, F., Dall’Asta, C., and Restivo, F. (2012). Development of a simple and high-throughput method for detecting aflatoxins production in culture media. Lett. Appl. Microbiol. 55, 82–89. doi: 10.1111/j.1472-765X.2012.03264.x
Dutta, T., and Das, P. (2001). Isolation of aflatoxigenic strains of Aspergillus and detection of aflatoxin B1 from feeds in India. Mycopathologia 151, 29–33. doi: 10.1023/a:1010960402254
Egbuta, M. A., Mwanza, M., Njobeh, P. B., Phoku, J. Z., Chilaka, C. A., and Dutton, M. F. (2015). Isolation of filamentous fungi species contaminating some Nigerian food commodities. J. Food Res. 4, 38–50. doi: 10.5539/jfr.v4n1p38
Fakruddin, M., Chowdhury, A., Hossain, M. N., and Ahmed, M. M. (2015). Characterization of aflatoxin producing Aspergillus flavus from food and feed samples. Springerplus 4:159. doi: 10.1186/s40064-015-0947-1
Fente, C., Ordaz, J. J., Vazquez, B., Franco, C., and Cepeda, A. (2001). New additive for culture media for rapid identification of aflatoxin-producing Aspergillus strains. Appl. Environ. Microbiol. 67, 4858–4862. doi: 10.1128/AEM.67.10.4858-4862.2001
Fink-Grernmels, J. (1999). Mycotoxins: their implications for human and animal health. Vet. Q. 21, 115–120. doi: 10.1080/01652176.1999.9695005
Frickmann, H., Dekker, D., Schwarz, N. G., Hahn, A., Boahen, K., Sarpong, N., et al. (2015). 16S rRNA gene sequence-based identification of bacteria in automatically incubated blood culture materials from tropical sub-Saharan Africa. PLoS One 10:e0135923. doi: 10.1371/journal.pone.0135923
Frisvad, J. C., Larsen, T. O., de Vries, R., Meijer, M., Houbraken, J., Cabañes, F., et al. (2007). Secondary metabolite profiling, growth profiles and other tools for species recognition and important Aspergillus mycotoxins. Stud. Mycol. 59, 31–37. doi: 10.3114/sim.2007.59.04
Gajjar, D. U., Pal, A. K., Ghodadra, B. K., and Vasavada, A. R. (2013). Microscopic evaluation, molecular identification, antifungal susceptibility, and clinical outcomes in Fusarium, Aspergillus and, Dematiaceous keratitis. Biomed. Res. Int. 2013:605308. doi: 10.1155/2013/605308
Gallo, A., Stea, G., Battilani, P., Logrieco, A. F., and Perrone, G. (2012). Molecular characterization of an Aspergillus flavus population isolated from maize during the first outbreak of aflatoxin contamination in Italy. Phytopathol. Mediterr. 51, 198–206. doi: 10.14601/Phytopathol_Mediterr-9461
Gelven, R. E. (2010). Approaches for alleviating toxic effects of aflatoxin in lactating dairy cows and weanling pigs. University of Missouri-Columbia.
Ghiasian, S., and Maghsood, A. (2011). Occurrence of aflatoxigenic fungi in cow feeds during the summer and winter season in Hamadan, Iran. Afr. J. Microbiol. Res. 5, 516–521. doi: 10.5897/AJMR10.600
Giovati, L., Magliani, W., Ciociola, T., Santinoli, C., Conti, S., and Polonelli, L. (2015). AFM1 in milk: physical, biological, and prophylactic methods to mitigate contamination. Toxins 7, 4330–4349. doi: 10.3390/toxins7104330
Henry, T., Iwen, P. C., and Hinrichs, S. H. (2000). Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 38, 1510–1515. doi: 10.1128/JCM.38.4.1510-1515.2000
Ibrahim, F., Jalal, H., Khan, A., Asghar, M., Iqbal, J., Ahmed, A., et al. (2016). Prevalence of aflatoxigenic Aspergillus in food and feed samples from Karachi, Pakistan. JIMB 4, 1–8. doi: 10.14737/journal.jimb/2016/4.1.1.8
Ito, Y., Peterson, S. W., Wicklow, D. T., and Goto, T. (2001). Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 105, 233–239. doi: 10.1017/S0953756200003385
Kumar, P., Mahato, D. K., Kamle, M., Mohanta, T. K., and Kang, S. G. (2017). Aflatoxins: a global concern for food safety, human health and their management. Front. Microbiol. 7:2170. doi: 10.3389/fmicb.2016.02170
Kusumoto, K. -I., Yabe, K., Nogata, Y., and Ohta, H. (1998). Aspergillus oryzae with and without a homolog of aflatoxin biosynthetic gene ver-1. Appl. Microbiol. Biotechnol. 50, 98–104. doi: 10.1007/s002530051262
Lee, C. -Z., Liou, G. -Y., and Yuan, G. -F. (2006). Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology 152, 161–170. doi: 10.1099/mic.0.27618-0
Li, X., Zhao, L., Fan, Y., Jia, Y., Sun, L., Ma, S., et al. (2014). Occurrence of mycotoxins in feed ingredients and complete feeds obtained from the Beijing region of China. J. Anim. Sci. Biotechnol. 5:37. doi: 10.1186/2049-1891-5-37
Lin, M., and Dianese, J. (1976). A coconut-agar medium for rapid detection of aflatoxin production by Aspergillus spp. Phytopathology 66, 1466–1469.
Liu, Y., and Wu, F. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ. Health Perspect. 118, 818–824. doi: 10.1289/ehp.0901388
Magnani, M., Fernandes, T., Prete, C. E. C., Homechim, M., Ono, E. Y. S., Vilas-Boas, L. A., et al. (2005). Molecular identification of Aspergillus spp. isolated from coffee beans. Sci. Agric. 62, 45–49. doi: 10.1590/S0103_90162005000100009
Matumba, L., Sulyok, M., Njoroge, S. M., Ediage, E. N., Van Poucke, C., De Saeger, S., et al. (2015). Uncommon occurrence ratios of aflatoxin B1, B2, G1, and G2 in maize and groundnuts from Malawi. Mycotoxin Res. 31, 57–62. doi: 10.1007/s12550-014-0209-z
Mejía-Teniente, L., Chapa-Oliver, A. M., Vazquez-Cruz, M. A., Torres-Pacheco, I., and Guevara-González, R. G. (2011). “Aflatoxins biochemistry and molecular biology-biotechnological approaches for control in crops” in Aflatoxins-detection, measurement and control. ed. I. Torres-Pacheco (InTech), 317–354.
Midorikawa, G., Pinheiro, M., Vidigal, B., Arruda, M., Costa, F., Pappas, G. Jr., et al. (2008). Characterization of Aspergillus flavus strains from Brazilian Brazil nuts and cashew by RAPD and ribosomal DNA analysis. Lett. Appl. Microbiol. 47, 12–18. doi: 10.1111/j.1472-765X.2008.02377.x
Mohamed, S., Flint, S., Palmer, J., Fletcher, G., and Pitt, J. (2013). An extension of the coconut cream agar method to screen Penicillium citrinum isolates for citrinin production. Lett. Appl. Microbiol. 57, 214–219. doi: 10.1111/lam.12102
Monson, M. S., Coulombe, R. A., and Reed, K. M. (2015). Aflatoxicosis: lessons from toxicity and responses to aflatoxin B1 in poultry. Agriculture 5, 742–777. doi: 10.3390/agriculture5030742
Morrison, D. M., Ledoux, D. R., Chester, L. F., and Samuels, C. A. (2017). A limited survey of aflatoxins in poultry feed and feed ingredients in Guyana. Vet. Sci. 4:60. doi: 10.3390/vetsci4040060
Moslem, M., Mashraqi, A., Abd-Elsalam, K., Bahkali, A., and Elnagaer, M. (2010). Molecular detection of ochratoxigenic Aspergillus species isolated from coffee beans in Saudi Arabia. Genet. Mol. Res. 9, 2292–2299. doi: 10.4238/vol9-4gmr943
Mostafa, A. A., and Amer, S. M. (2013). Molecular characterization of toxigenic Aspergillus flavus strains isolates from animal feed stuff in Egypt. Life Sci. J. 10, 20–30.
Nleya, N. (2019). Study of toxigenic Aspergillus in feed and impact of bovine breed on aflatoxins carryover in milk and urine in dairy cows: a case of Bulawayo, Zimbabwe. dissertation/master’s thesis. South Africa: North-West University.
OBrian, G., Georgianna, D., Wilkinson, J., Yu, J., Abbas, H., Bhatnagar, D., et al. (2007). The effect of elevated temperature on gene transcription and aflatoxin biosynthesis. Mycologia 99, 232–239. doi: 10.3852/mycologia.99.2.232
Omeiza, G., Kabir, J., Kwaga, J., Kwanashie, C., Mwanza, M., and Ngoma, L. (2018). A risk assessment study of the occurrence and distribution of aflatoxigenic Aspergillus flavus and aflatoxin B1 in dairy cattle feeds in a central northern state, Nigeria. Toxicol. Rep. 5, 846–856. doi: 10.1016/j.toxrep.2018.08.011
Patel, S. V., Bosamia, T. C., Bhalani, H. N., Singh, P., and Kumar, A. (2015). Aflatoxins: causes & effects. J. Agric. Biol. Sci. 13, 140–141.
Payne, G., Nierman, W., Wortman, J., Pritchard, B., Brown, D., Dean, R., et al. (2006). Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44(Suppl. 1), S9–S11. doi: 10.1080/13693780600835716
Pildain, M. B., Frisvad, J. C., Vaamonde, G., Cabral, D., Varga, J., and Samson, R. A. (2008). Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 58, 725–735. doi: 10.1099/ijs.0.65123-0
Pleadin, J. (2015). Mycotoxins in grains and feed-contamination and toxic effect in animals. BAH 31, 441–456. doi: 10.2298/BAH1504441P
Raja, H. A., Miller, A. N., Pearce, C. J., and Oberlies, N. H. (2017). Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Prod. 80, 756–770. doi: 10.1021/acs.jnatprod.6b01085
Rashid, M., Khalil, S., Ayub, N., Ahmed, W., and Khan, A. G. (2008). Categorization of Aspergillus flavus and Aspergillus parasiticus isolates of stored wheat grains in to aflatoxinogenics and non-aflatoxinogenics. Pak. J. Bot. 40, 2177–2192.
Rodrigues, P., Santos, C., Venâncio, A., and Lima, N. (2011). Species identification of Aspergillus section Flavi isolates from Portuguese almonds using phenotypic, including MALDI-TOF ICMS, and molecular approaches. J. Appl. Microbiol. 111, 877–892. doi: 10.1111/j.1365-2672.2011.05116.x
Rodrigues, P., Soares, C., Kozakiewicz, Z., Paterson, R., Lima, N., and Venâncio, A. (2007). “Identification and characterization of Aspergillus flavus and aflatoxins” in Communicating current research and educational topics and trends in applied microbiology. 527–534.
Rossi-Tamisier, M., Benamar, S., Raoult, D., and Fournier, P. -E. (2015). Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 65, 1929–1934. doi: 10.1099/ijs.0.000161
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Stark, A. -A. (2009). “Molecular mechanism of detection of aflatoxins and other mycotoxins” in Mycotoxins in food, feed and bioweapons. eds. M. Rai and A. Varma (Springer), 21–37.
Sudini, H., Srilakshmi, P., Vijay Krishna Kumar, K., Njoroge, S. M., Osiru, M., Seetha, A., et al. (2015). Detection of aflatoxigenic Aspergillus strains by cultural and molecular methods: a critical review. Afr. J. Microbiol. Res. 9, 484–491. doi: 10.5897/AJMR2014.7309
Suzuki, T., and Iwahashi, Y. (2016). Addition of carbon to the culture medium improves the detection efficiency of aflatoxin synthetic fungi. Toxins 8:338. doi: 10.3390/toxins8110338
Varga, J., Frisvad, J., and Samson, R. (2009). A reappraisal of fungi producing aflatoxins. World Mycotoxin J. 2, 263–277. doi: 10.3920/WMJ2008.1094
Variane, A. C. F., dos Santos, F. C., de Castro, F. F., Barbosa-Tessmann, I. P., dos Santos, G. T., and dos Santos Pozza, M. S. (2018). The occurrence of aflatoxigenic Aspergillus spp. in dairy cattle feed in southern Brazil. Braz. J. Microbiol. 49, 919–928. doi: 10.1016/j.bjm.2018.05.005
Verheecke, C., Liboz, T., Anson, P., Diaz, R., and Mathieu, F. (2015). Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces. Microbiology 161, 967–972. doi: 10.1099/mic.0.000070
Woloshuk, C., and Prieto, R. (1998). Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160, 169–176. doi: 10.1111/j.1574-6968.1998.tb12907.x
Yazdani, D., Zainal, A. M., Tan, Y., and Kamaruzaman, S. (2010). Evaluation of the detection techniques of toxigenic Aspergillus isolates. Afr. J. Biotechnol. 9, 7654–7659. doi: 10.5897/AJB10.1128
Yu, J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 4, 1024–1057. doi: 10.3390/toxins4111024
Yu, J., Chang, P. -K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. doi: 10.1128/aem.70.3.1253-1262.2004
Zain, M. E. (2011). Impact of mycotoxins on humans and animals. J. Saudi Chem. Soc. 15, 129–144. doi: 10.1016/j.jscs.2010.06.006
Zarrin, M., and Erfaninejad, M. (2016). Molecular variation analysis of Aspergillus flavus using polymerase chain reaction-restriction fragment length polymorphism of the internal transcribed spacer rDNA region. Exp. Ther. Med. 12, 1628–1632. doi: 10.3892/etm.2016.3479
Zhi, Q., Xie, Y., and He, Z. (2013). Genome mining for aflatoxin biosynthesis. Fungal Genet. Biol. 3:2. doi: 10.4172/2165-8056.1000108
Keywords: Aspergillus, biodiversity, feeds, aflatoxins, morphological, molecular
Citation: Nleya N, Ngoma L, Adetunji MC and Mwanza M (2021) Biodiversity of Aflatoxigenic Aspergillus Species in Dairy Feeds in Bulawayo, Zimbabwe. Front. Microbiol. 11:599605. doi: 10.3389/fmicb.2020.599605
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Geromy G. Moore, Southern Regional Research Center (USDA-ARS), United StatesAhmed Abdel-Hadi, Majmaah University, Saudi Arabia
Copyright © 2021 Nleya, Ngoma, Adetunji and Mwanza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy Nleya, bmRhbmd3YUBnbWFpbC5jb20=, bmFuY3kubmxleWFAbnVzdC5hYy56dw==
 Nancy Nleya
Nancy Nleya Lubanza Ngoma
Lubanza Ngoma Modupeade C. Adetunji
Modupeade C. Adetunji Mulunda Mwanza
Mulunda Mwanza