- 1MS Degree Program in Applied Microbiology, Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 2Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 3Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 4Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Agaricus is a saprophytic mushroom genus widely distributed throughout the world. In this study, a survey of the Agaricus species carried out around Chiang Mai University in northern Thailand from 2018 to 2019 yielded 12 collections. Morphological characteristics and phylogenic analyses based on the internal transcribed spacers (ITS) and a fragment of the large subunit (LSU) of the nuclear ribosomal DNA (rDNA), and a fragment of the translation elongation factor 1-alpha (tef1) genes were investigated. The results revealed that these collections belong to six species including Agaricus erectosquamosus, Agaricus pallidobrunneus, Agaricus subrufescens, and three new species. Agaricus thailandensis sp. nov. was found to belong to Agaricus sect. Minores, which is placed in Agaricus subg. Minores. Aagricus pseudoerectosquamosus sp. nov. was placed in Agaricus sect. Brunneopicti within Agaricus subg. Pseudochitonia. Furthermore, Agaricus lannaensis remains an incertae sedis in Agaricus subg. Pseudochitonia. Additionally, this study was proposed that A. pallidobrunneus was discovered in Thailand for the first time. Full descriptions, color photographs, illustrations, and phylogenetic trees are provided.
Introduction
The genus Agaricus L. with Agaricus campestris L. as the type species was first proposed by Donk (1962). This genus belongs to the family Agaricaceae of the order Agaricales. It is distributed worldwide and is commonly found in grasslands and forests (Kerrigan et al., 2005, Kerrigan, 2016; Parra, 2008, 2013; Zhao et al., 2012a, 2016; Chen et al., 2015; He et al.,2018a,b). Agaricus is a saprophytic fungal genus and is characterized by its white to pink free lamellae when young, which becomes brown at maturity, the presence of an annulus on the stipe, and brown to dark brown spore prints (Kerrigan et al., 2005; Zhao et al., 2016; He et al., 2017, 2018a). In the global market, Agaricus bisporus (J.E. Lange) Imbach (white bottom mushroom), and Agaricus subrufescens Peck (almond mushroom) are edible mushrooms that are commercially cultivated. Medicinal properties have been reported for both species (He et al., 2017, 2018a; Zhang et al., 2017). Many other Agaricus species such as Agaricus agrinferus (Kerrigan and Callac), Agaricus augustus Fr., Agaricus bitorquis (Quél.) Sacc., A. campestris, Agaricus flocculosipes (Zhao, Desjardin, Guinb, and Hyde), Agaricus sinodeliciosus (Zhao, Wang, and Zhao), and Agaricus taeniatus (Sai, Li, Shao, Li, and Wen) have been reported as edible (Kerrigan et al., 2008; Zhao et al., 2012b; Li et al., 2014; Thongklang et al., 2014b; Wang et al., 2015; Karunarathna et al., 2016; Zhang et al., 2017). In contrast, Agaricus species in Agaricus sect. Xanthodermatei and Agaricus sect. Hondenses have been described as poisonous as they contain toxic phenolic compounds that can cause gastrointestinal symptoms (Kerrigan et al., 2005; Petrova et al., 2007; Parra, 2013; Asef et al., 2016).
There are approximately 6,000 records of Agaricus in the Species Fungorum (accessed in April 2021). However, these records have been found to include synonyms, some misidentifications, and a number of species that have not yet been well-documented. To date, a total of over 500 species of Agaricus are currently recognized. These include many new species from America, Asia, Australia, and Europe (Wang et al., 2015; Kerrigan, 2016; He et al., 2017; Callac and Chen, 2018) and few new species from Africa (Hama et al., 2010; Zhao et al., 2012b; Ling et al., 2021) and Oceania (Geml et al., 2007; Lebel and Syme, 2012; Lebel, 2013). Among the approximately 200 new species described since 2000, more than half have been reported to be from Asia, and a quarter have been reported in America (particularly North America) (Zhao et al., 2011, 2016; Thongklang et al., 2014a; Liu et al., 2015; Karunarathna et al., 2016; Kerrigan, 2016; Chen et al., 2017; Callac and Chen, 2018; Hyde et al., 2018). Morphological characteristics, odor, and the chemical reactions of Schäffer’s reagent and KOH have mainly been used in the traditional identification of the Agaricus species (Parra, 2008; Karunarathna et al., 2014; Chen et al., 2015, 2017; Zhou et al., 2016). However, high variations of phenotypes, varying environmental factors, differing geographic conditions, and the separate developmental stages of basidiomata may influence the morphological identification process. This could make it difficult to distinguish this particular species from other closely related Agaricus species (Heinemann, 1978; Kerrigan, 1986; Singer, 1986; Callac et al.,1998a,b; Challen et al., 2003; Parra, 2008). Therefore, the application of molecular tools that are based on DNA analyses has proven to be essential in identifying the Agaricus species (Challen et al., 2003; Kerrigan et al., 2005, 2008; Zhao et al., 2011; Chen et al., 2015; Thongklang et al., 2016).
Zhao et al. (2016) classified Agaricus into five subgenera and 20 sections based on morphological characteristics, multigene molecular phylogeny (ITS, LSU, and tef1), and the divergence time. The revised classification of Agaricus by Chen et al. (2017), and Parra et al. (2018) resulted in six subgenera (Agaricus subg. Agaricus, Agaricus subg. Flavoagaricus, Agaricus subg. Minores, Agaricus subg. Minoriosis, Agaricus subg. Pseudochitonia and Agaricus subg. Spissicaules) with 23 sections. Subsequently, a new section of Agaricus subg. Pseudochitonia was introduced by He et al. (2018a). Therefore, Agaricus now contains six subgenera and 24 sections.
Over the last decade, the study of Agaricus has expanded rapidly, especially in tropical regions. Notably, Thailand is proving to be a hot spot for the discovery of novel species. This is evidenced by the discovery of many new species of macrofungi, 45 of which are new Agaricus species that have been discovered since 2011 (Zhao et al., 2011, 2012a, 2016; Karunarathna et al., 2014; Thongklang et al.,2014a,b; Ariyawansa et al., 2015; Chen et al., 2015, 2017; Liu et al., 2015; Li et al., 2016; Hyde et al., 2017, 2018; He et al., 2018a). This study outlines how we found twelve Agaricus specimens during the course of our investigation of macrofungi in northern Thailand. Among these, we describe three new species and one new record. This investigation introduces the taxa based on studies of morphology and multigene analyses of combined ITS, LSU, and tef1 sequences. Additionally, this study included a mini-review of Agaricus species that are found in Thailand.
Materials and Methods
Sample Collection
Agaricus were surveyed at Chiang Mai University, Chiang Mai Province, Thailand during the rainy seasons of the years 2018 and 2019. Photographs were immediately taken in the field. Basidiomata were collected and wrapped in aluminum foil and kept in plastic boxes while being transferred to the laboratory within 24 h of collection. Specimens were dried in a hot air oven at 45°C until they were completely dried. They were then kept in a plastic zip-locked bag and deposited in the Biology Department’s Herbarium (CMUB) along with the Herbarium of Sustainable Development of Biological Resources (SDBR-CMU), Faculty of Science, Chiang Mai University, Thailand. Facesoffungi and MycoBank numbers have also been provided (Robert et al., 2013; Jayasiri et al., 2015).
Morphological Observation
Macroscopic descriptions were made based on fresh specimens. Color, name, and codes were given according to the methods employed by Kornerup and Wanscher (1978). Chemical reactions were determined following the methods described by Chen et al. (2015) and He et al. (2017) including Melzer’s reagent, 10% potassium hydroxide (KOH) in water, and Schäffer’s reaction. Microscopic characteristics, including basidiospores, basidia, cystidia, and pileipellis, were observed from dried specimens that had been rehydrated in 95% ethanol followed by distilled water, 5% aqueous KOH, or Melzer’s reagent. A minimum of 50 basidiospores, 20 basidia, and cystidia were measured using a compound light microscope (Olympus CX31, Japan). Basidiospores are presented in the following format: (a)b–c–d(e), for which “c” represents the average, “b” and “d” represent the average + and - standard deviation (SD), respectively, and “a” and “e” represent the minimum and maximum values, respectively. For spore statistics, Q represents the ratio of length divided by the width of the basidiospore, and Qm represented the average Q of all specimens ± standard deviation.
DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted from the fresh tissue of each specimen using the FAVOGEN Genomic DNA Extraction Mini Kit (Taiwan) by following the manufacturer’s instructions. The ITS region of the rDNA was amplified using the ITS4/ITS5 primers (White et al., 1990) by polymerase chain reaction (PCR). The LSU of the rDNA gene was amplified with LR5/LROR primers (Vilgalys and Hester, 1990) and the tef1 gene was amplified with primers EF1-983F/EF1-1567R (Rehner and Buckley, 2005). The PCR programs of ITS, LSU, and tef1 genes were established by following the methods employed by He et al. (2017). PCR products were checked by electrophoresis on 1% agarose gels stained with ethidium bromide and observed under UV light. PCR products were purified using NucleoSpin Gel and a PCR Clean-up Kit (Macherey-Nagel, Germany). PCR products were then sent to a commercial sequencing provider (1ST BASE Company, Kembangan, Malaysia). The obtained sequences were ultimately subjected to BLASTn search in GenBank.1
Sequence Alignment and Phylogenetic Analyses
Newly generated sequences were assembled using the Sequencher program. Details of the sequences used for phylogenetic analysis obtained from this study and previous other studies are provided in Supplementary Table 1. Two sequence datasets were prepared for phylogenetic analyses. The first dataset was comprised of sequences of two subgenera, namely Agaricus subg. Flavoagaricus and Agaricus subg. Minores. The second dataset contains sequences of six sections in Agaricus subg. Pseudochitonia namely, Agaricus sect. Bohusia, Agaricus sect. Brunneopicti, Agaricus sect. Floculenti, Agaricus sect. Nigrobrunnescentes, Agaricus sect. Rubricosi, and Agaricus sect. Sanguinolenti. The datasets were then aligned using MAFFT version 7 (Katoh and Standley, 2013). The first and second aligned datasets were deposited in TreeBASE under the numbers 27426 and 28087, respectively. Maximum Likelihood (ML) phylogenetic tree inference was performed for each dataset using RAxML-HPC2 version 8.2.10 (Stamatakis, 2006) on the CIPRES web portal (Miller et al., 2009). The phylogenetic tree was inferred from a four-partitions (ITS, LSU, tef1 exons, and tef1 introns) combined dataset using the GTRCAT model with 25 categories. A. campestris LAPAG370 was used as an outgroup for both datasets. Statistical support of the clades was obtained with 1,000 rapid bootstrap replicates. For Bayesian Inference (BI), the best-fit model of substitution amongst those implementable in MrBayes was estimated separately for each region using jModelTest 2 (Darriba et al., 2012) on the CIPRES portal based on the Bayesian Information Criterion (BIC). The selected models that are similar in both data sets, were HKY + I + G for ITS, SYM + G for tef1 exons, K80 + G for introns of tef1. While for LSU, were K80 + I + G in the first dataset, and K80 + I in the second dataset. Partitioned BI was performed with MrBayes 3.2.6 software for Windows (Ronquist et al., 2012). Two runs of five chains were conducted for eleven million (first dataset) and three hundred thousand generations (second dataset), which were sampled every 200 generations. At the end of the runs, the average deviations of split frequencies were 0.007058 (first dataset) and 0.009527 (second dataset). The potential scale reduction factor values of all parameters were close to 1. The burn-in phase (25%) was estimated by checking the stationarity in the plot generated by the sump command.
Results
Phylogenetic Analyses Results
A total of 36 sequences from 12 specimens were newly obtained from this study and deposited in GenBank (Supplementary Table 1). The combined ITS, LSU, tef1 exons, and tef1 introns sequence datasets consisted of 81 and 54 taxa in the first and second datasets, respectively. The first and second aligned datasets were comprised of 2257 (ITS: 1–778, LSU: 779–1674, tef1 exons: 1675–2140 and tef1 introns: 2141–2257) and 2204 (ITS: 1–784, LSU: 785–1689, tef1 exons: 1690–2093 and tef1 introns: 2094–2204) characters including gaps, respectively. ML and BI trees of both datasets revealed similar topologies without any supported conflict (BS ≥ 70% and PP ≥ 0.90). Phylogenetic results were based on the first dataset (Figure 1) which contained Agaricus subg. Flavoagaricus and Agaricus subg. Minores. Selected Agaricus species formed two main clades namely Agaricus subg. Flavoagaricus (BS = 100%, PP = 1) and Agaricus subg. Minores (BS = 87%, PP = 0.90). Agaricus thailandensis (vouchers SDBR-CMU-CJ118 and SDBR-CMU-CJ225) formed a monophyletic clade (BS = 100%, PP = 1) in Agaricus sect. Minores of Agaricus subg. Minores and closely related to Agaricus sp. voucher CA935 with the high support values (BS = 100%, PP = 1). Furthermore, they formed a sister clade to Agaricus flammicolor (Chen et al., 2017) and A. badioniveus (Chen et al., 2017) (BS = 71%, PP = 0.92). Another obtained specimen voucher SDBR-NK0079 was placed within the cluster of the known species clade of A. subrufescens (BS = 100%, PP = 1) in Agaricus sect. Arvenses of Agaricus subg. Flavoagaricus.
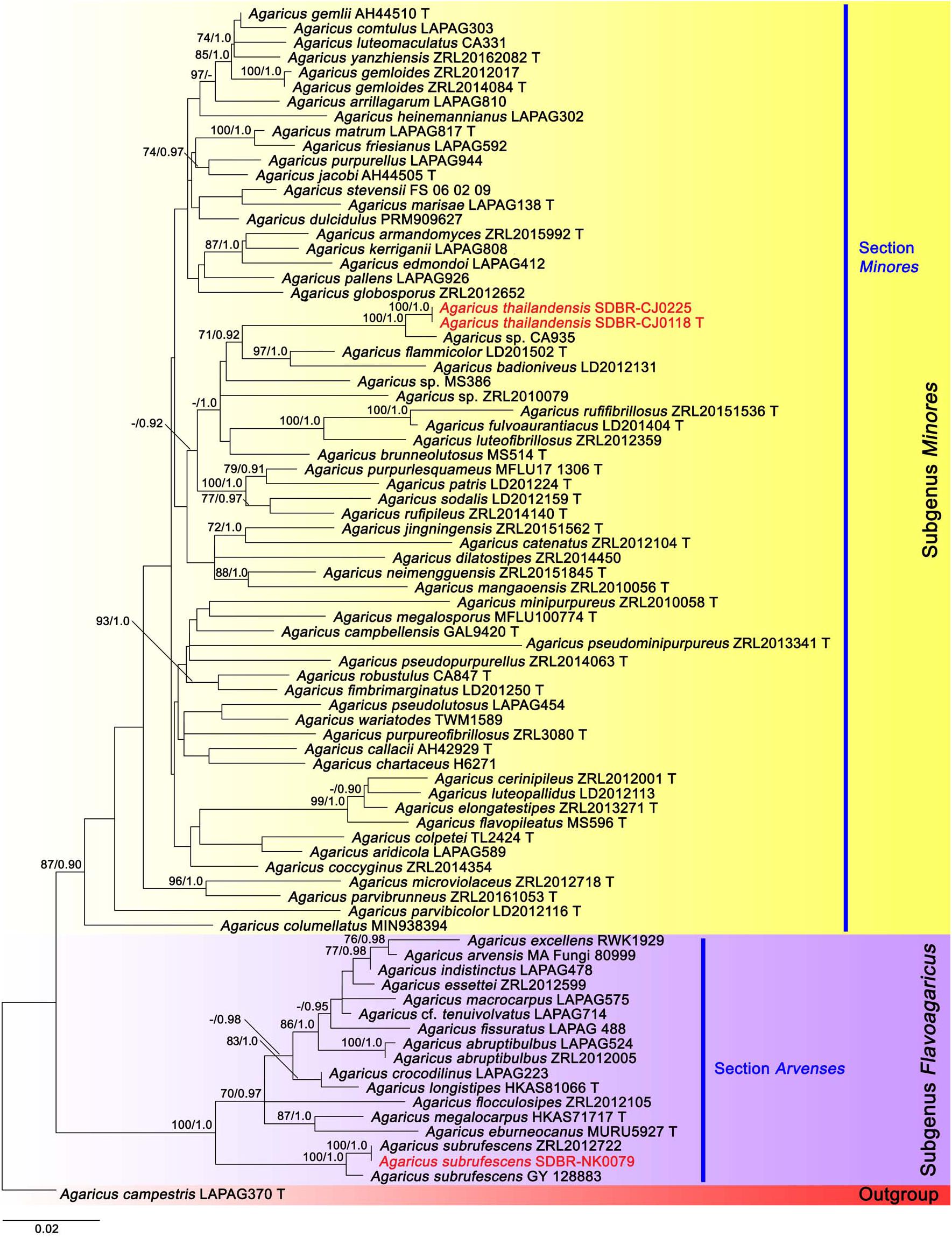
Figure 1. Phylogenetic tree of the genus Agaricus generated from maximum likelihood based on multigene sequences (ITS, LSU, tef1 exons, and tef1 introns) of Agaricus subg. Flavoagaricus and Agaricus subg. Minores. Agaricus campestris was used as an outgroup. The vouchers from this study are in red and “T” means type specimen. Bootstrap support values (BS ≥ 70%) and posterior probabilities (PP ≥ 0.90) are presented above the supported branches.
The second tree was based on sequences of six sections of Agaricus subg. Pseudochitonia (Figure 2). Six main clades were assigned including, Agaricus sect. Bohusia (BS = 92%, PP = 0.99), Agaricus sect. Brunneopicti (BS = 92%, PP = 1), Agaricus sect. Floculenti (BS = 62%, PP = 0.93), Agaricus sect. Nigrobrunnescentes (BS = 57%, PP = 0.93), Agaricus sect. Rubricosi (BS = 100%, PP = 1) and Agaricus sect. Sanguinolenti (BS = 100%, PP = 1). Two new species, namely Agaricus lannaensis and Aagricus pseudoerectosquamosus formed two different species clades within Agaricus subg. Pseudochitonia. Agaricus lannaensis (vouchers SDBR-CJ192, SDBR-NK0584, and SDBR-NK0564) formed a highly supported clade with Agaricus sp. voucher CA820 (BS = 100%, PP = 1) and they formed a sister clade to unnamed species Agaricus voucher LD2012162 (BS = 92%, PP = 1). This clade was closely related to the clade of Agaricus sect. Flocculenti. Another new species, A. pseudoerectosquamosus (vouchers SDBR-CJ108 and SDBR-NK0064) formed a clade (BS = 100%, PP = 0.99) that was closely related to Agaricus sp. voucher NTT117 with the high support values (BS = 100%, PP = 1), in Agaricus sect. Brunneopicti (BS = 92%, PP = 1). Additionally, the other four Agaricus collections obtained in this study were clustered within two known species clades, in which Agaricus vouchers SDBR-NK0080, SDBR-CJ0032, and SDBR-CJ0131 were clustered with Aagricus erectosquamosus Linda J. Chen, K.D. Hyde & R.L. Zhao. Agaricus voucher SCBR-NK0368 formed a clade with Agaricus pallidobrunneus R.L. Zhao. with high value supports (BS = 100%, PP = 1).
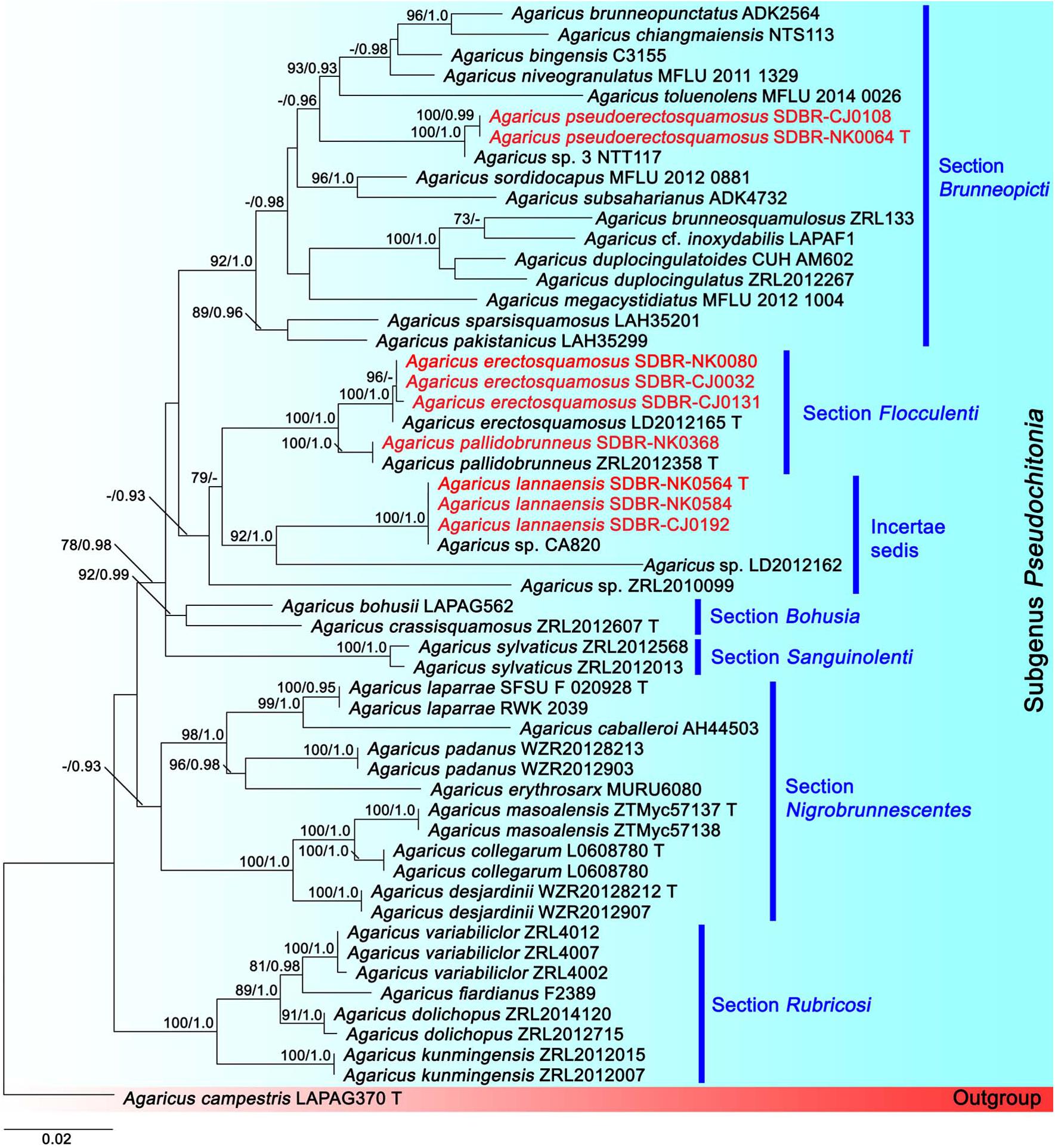
Figure 2. Phylogenetic tree of the genus Agaricus generated from maximum likelihood based on multigene sequences (ITS, LSU, tef1 exons, and tef1 introns) of six sections in Agaricus subg. Pseudochitonia namely, Agaricus sect. Bohusia, Agaricus sect. Brunneopicti, Agaricus sect. Floculenti, Agaricus sect. Nigrobrunnescentes, Agaricus sect. Rubricosi and Agaricus sect. Sanguinolenti. Agaricus campestris was used as an outgroup. The vouchers from this study are in red and “T” means type specimen. Bootstrap support values (BS ≥ 70%) and posterior probabilities (PP ≥ 0.90) are presented above the supported branches.
Taxonomic Description of New Species
Agaricus lannaensis N. Suwannarach, J. Kumla & S. Lumyong sp. nov. Figure 3
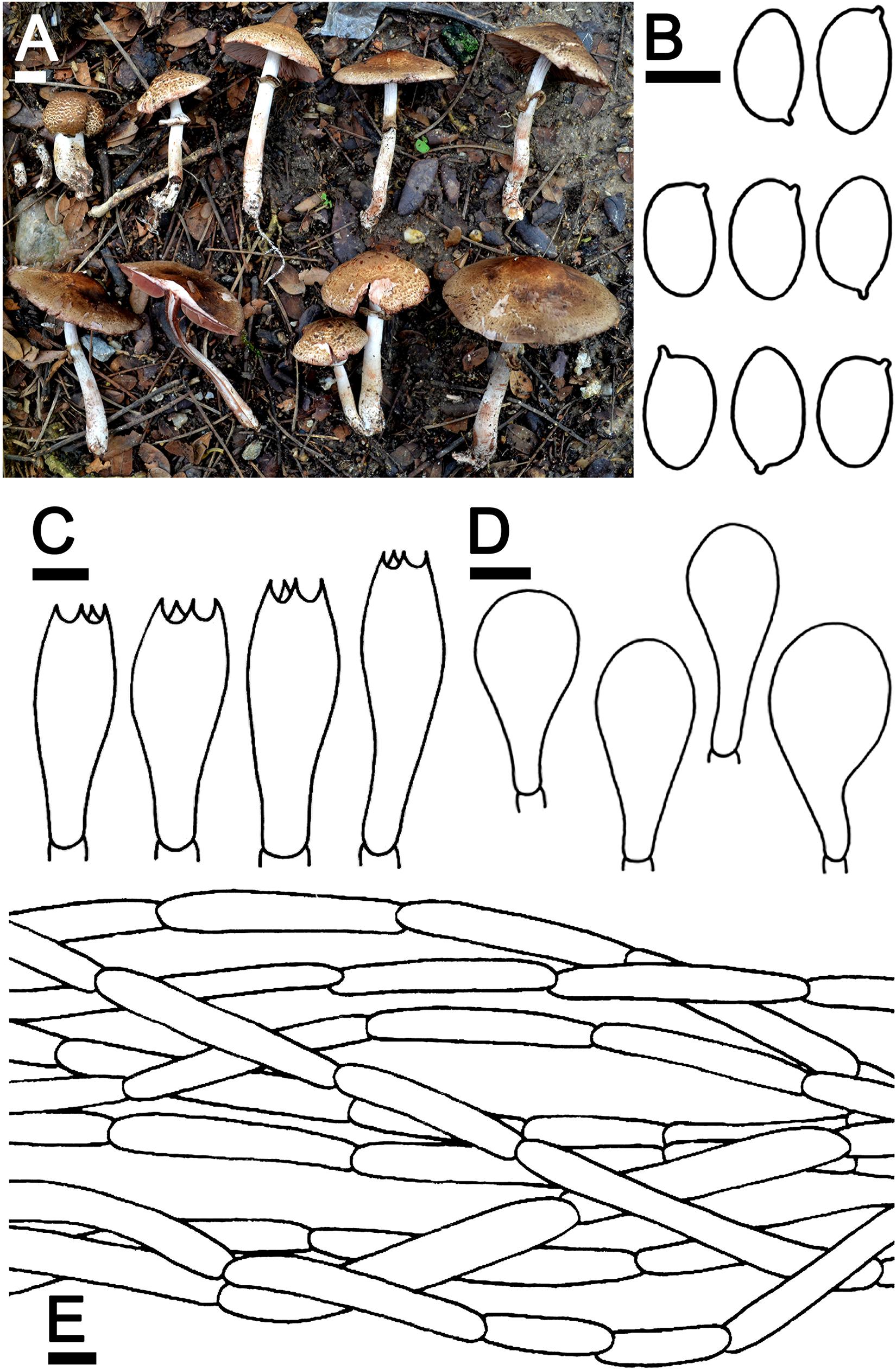
Figure 3. Agaricus lannaensis SDBR-NK0564 (holotype). (A) Basidiomata on the field, (B) Basidospores, (C) Basidia, (D) Cheilocystidia, (E) Pileipellis. Scale bars (A) = 1 cm and (B–E) = 5 μm.
MycoBank: MB 838052
Facesoffungi number: FoF 09468
Etymology: “lannaensis” referring to the Kingdom of Lanna, the historic name of northern Thailand, where the new species was found.
Holotype: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°47′33.6″N 98°57′30.2″E, elevation 331 m, June 2, 2019 (N. Suwannarach and J. Kumla) SDBR-NK0564, CMUB 39945.
Description: Basidiomata medium-sized, stipitate-pileate with lamellate hymenophore. Pileus (2.4) 3.2–7.4 cm in diameter, at first spherical, campanulate, then subumbonate with straight margins, and whole cap covered by fibrils and broken into small squamules. The surface background is pale orange (6A3) at the center and turns gradually orange white (5A2) at the margins, covered by innate scales. The pileus is at first covered, then gradually sprayed, denser at pileus, partially at margins when aged, appressed innately scaled, brown (7E8) at the top of the pileus, and gradually brown (7E7) at margins, then grayish red (8C5) when cut or touched. Pileus context soft white (1A1) then reddish brown (8E7) when cut, pluteoid. Lamellae emarginate, crowded, thin, regular, 3–4 different lengths of lamellae, at first pastel red (8A4), becoming dark brown (8F8). Stipe (4.3)4.6–8.8 × 0.6–1.2 cm, cylindrical, smooth above annulus until under lamellae, covered by small squamules which fibrilloses white (1A1) under annulus until the base, then grayish red (8C5) when cut or touched, scales reddish brown (8D5) at the base. Stipe context soft, fistulose, orange-white (6A2), then reddish brown (8E7) when cut. Annulus are membranous, thin, pendant, simple, stick above the middle of the stipe, at first white (1A1), dark brown (7F4) at edges, then light brown (7D5) when aged. Spores print dark brown (8F5) and the odor is phenol-like. Macrochemical reactions; KOH reaction yellow and Schäffer’s reactions negative.
Basidiospores (5.5)6.3–6.8–7.3(8.0) × (4.0)4.1–4.3–4.4(4.5) μm (n = 50), Q = 1.22–2.00, Qm = 1.62 ± 0.19, broadly ellipsoid to elongate, smooth, thin-walled, brown in water and KOH, inamyloid. Basidia 19–26 × 5.5–8.5 μm, clavate, 4-spored, hyaline, sterigmata up to 2.5 μm long. Cheilocystidia 13–47 × 6.5–24 μm, clavate to broadly clavate, often with a short peduncle, hyaline. Pleurocystidia absent. Pileipellis composed of 3.8–7.5 μm wide cutis hyaline hyphae, smooth, cylindrical, occasionally branched. Annulus composed of 1–3 μm wide hyaline hyphae, cylindrical with rounded apex, branched. Stipitipellis composed of cutis hyphae wide up to 2–5 μm, cylindrical, occasionally branched, hyaline. Clamp connections absent.
Ecology and distribution: Fruiting solitary or gregarious on sandy loam soil during the rainy season (mid-May to October). Known only from Thailand.
Additional specimens examined: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°47′33.6″N 98°57′30.2″E, elevation 331 m, June 2, 2019, J. Kumla, SDBR-NK0584; 18°47′45″N 98°57′2″E, elevation 340 m, October 9, 2019, C. Jaichaliaw, SDBR-CJ0192.
Note: In the field, A. lannaensis is morphologically similar to Agaricus brunneopileatus Callac & R.L. Zhao. However, A. brunneopileatus differs from A. lannaensis by the fact that it had a negative reaction in KOH and Schäffer’s reactions, smaller basidiospores (4.9–5.8 × 2.5–3.6 μm) and shorter basidia (12.6–18 × 6.2–8.8 μm) (Zhao et al., 2016). The phylogeny also supports the determination that they are different species, for which A. brunneopileatus was assigned to Agaricus sect. Subrutilescentes in Agaricus subg. Spissicaules (Zhao et al., 2016). Phylogenetically, A. lannaensis formed a clade with the unnamed species Agaricus sp. voucher CA820 collected from Thailand, and they formed a sister clade to Agarius sect. Flocculenti comprised with A. erectosquamosus and A. pallidobrunneus (Figure 2). Agaricus erectosquamosus were clearly distinguished from A. lannaensis by the orange KOH reaction and the presence of erect squamules on the pileus that appeared dense at the disc and brown against a dirty white background (Zhao et al., 2016). A. pallidobrunneus has brownish orange to dark brown pileus, but A. lannaensis has a pale orange pileus (Zhao et al., 2016).
Agaricus pseudoerectosquamosus J. Kumla, N. Suwannarach, and S. Lumyong sp. nov. (Figure 4)
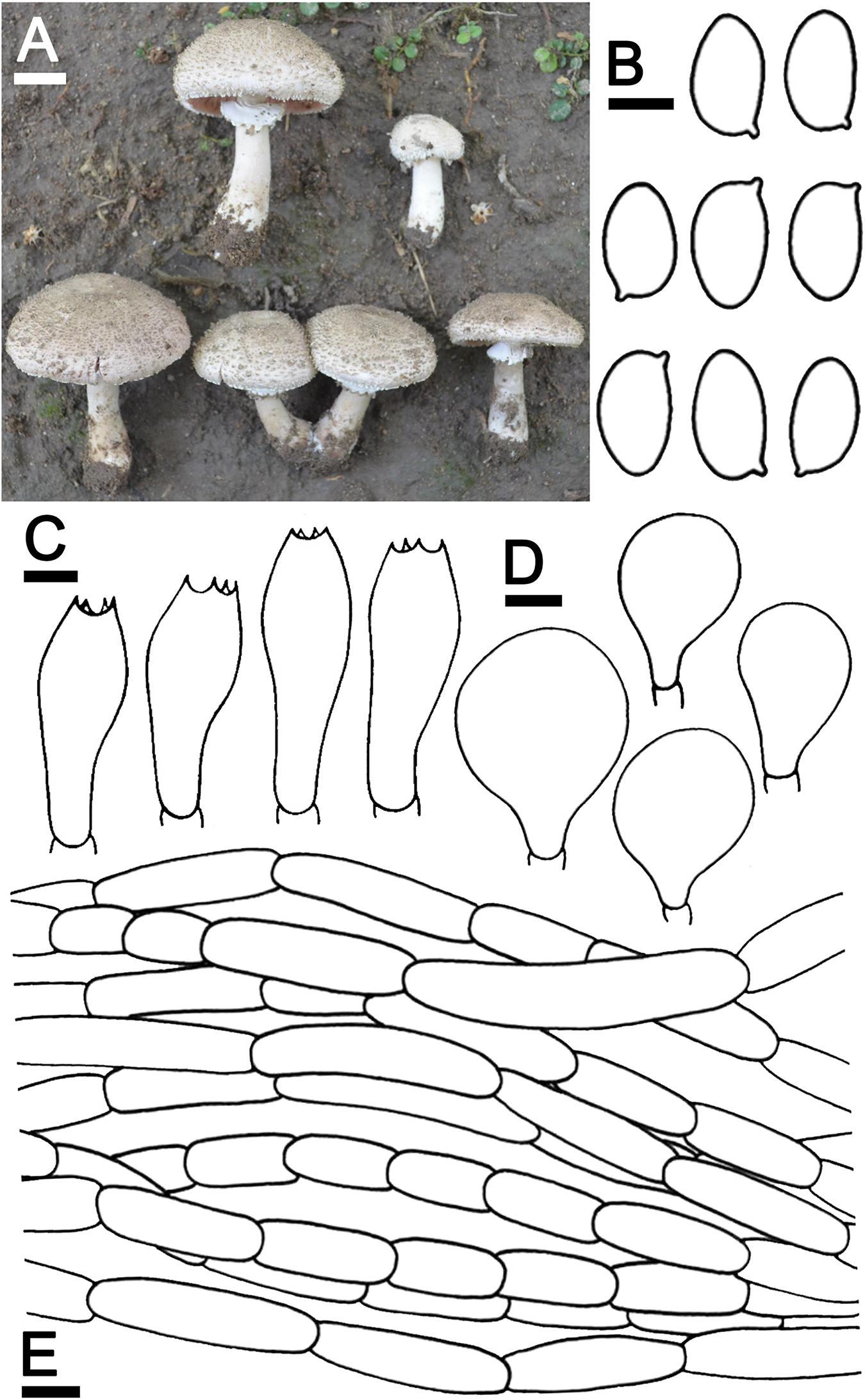
Figure 4. Agaricus pseudoerectosquamosus SDBR-NK0064 (holotype). (A) Basidiomata on the field, (B) Basidiospores, (C) Basidia, (D) Cheilocystidia, (E) Pileipellis. Scale bars (A) = 2 cm and (B–E) = 5 μm.
MycoBank: MB 838053
Facesoffungi number: FoF 09469
Etymology: “pseudo” = false, referring to the morphological characteristics that are easily mistaken for A. erectosquamosus
Holotype: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°48′6.4″N 98°57′23″E, elevation 330 m, September 20, 2018, J. Kumla, SDBR-NK0064, CMUB 39944
Description: Basidiomata medium-sized, stipitate-pileate with lamellate hymenophore. Pileus (3)5.2–7.5 cm in diameter, at first umbonate, broadly conical, then hemispherical with inflexed margin, rimos; surface background white (1A1), innately squamulose, denser at the center, poor at the margins with depressed at the center, uplifted, brownish orange (5C4) at the center, pale to white (1A1) at the edges. Pileus context soft, white (1A1). Lamellae free, crowded, even, 4–5 different lengths of lamellullae, pale red (8A3). Stipe 6.7–10 × 1.2–2.1 cm, subclavate, inserted or caespitose, surface skin covered pale yellow near lamellae to dark brown (4A3 to 6F8) at the base by the fibrillose, denser, white (1A1). Annulus thick, double, stick above the middle of stipe, white (1A1), and often cogwheel-like. Stipe context fistulose, white (1A1). Spores print dark brown (8F5). Odor phenol-like. Macrochemical reactions; KOH reaction yellow and Schäffer’s reactions negative.
Basidiospores (8.0)8.2–8.5–8.8(9.0) × (4.5)4.7–5.0–5.4(5.5) μm (n = 50), Q = 1.45–2.00, Qm = 1.69 ± 0.14, ellipsoid to elongate, smooth, thin-walled, brown in water and KOH, inamyloid. Basidia 19.5–26 × 8–9.5 μm, clavate, 4-spored, hyaline, sterigmata up to 2 μm long. Cheilocystidia 19.5–40 × 11.5–22.5 μm, clavate to broadly clavate, often with a long peduncle, hyaline. Pileipellis composed of fibrilloses 5–20 × 4–5.6 μm hyaline hyphae, smooth, short cylindrical, occasionally branched, and cutis hyphae 2–5 μm, wide, occasionally branched, hyaline. Annulus composed of 2–4.5 μm wide hyaline hyphae, cylindrical with a rounded apex and branched. Stipitipellis composed of cutis hyphae up to 3–7 μm wide, cylindrical, occasionally branched, hyaline. Clamp connections absent.
Ecology and distribution: Fruiting solitary or gregarious on sandy loam soil during the rainy season (mid-May to October). Known only from Thailand.
Additional specimens examined: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°48′17″N 98°57′13″E, elevation 340 m, September 22, 2019, C. Jaichaliaw, SDBR-CJ0108.
Note: Morphological characteristics and phylogenetic analysis assigned A. pseudoerectosquamosus to Agaricus sect. Brunneopicti in Agaricus subg. Pseudochitonia. Morphologically, A. pseudoerectosquamosus is similar to A. erectosquamosus (Zhao et al., 2016). However, the orange associated with the KOH reaction, the dark brown squamules on the surface of the pileus, the shorter basidiospores (6.6–7.6 × 4.1–5.0 μm) and the narrower basidia (15–22 × 6.5–7.5 μm) in A. erectosquamosus clearly distinguished it from A. pseudoerectosquamosus. Moreover, the phylogeny placed A. erectosquamosus within Agaricus sect. Flocculenti of Agaicus subg. Pseudochitonia, (Zhao et al., 2016). Phylogenetically, A. pseudoerectosquamosus is a monophyletic clade that formed a sister clade to the unnamed species Agaricus sp. 3 voucher NTT117 collected from Thailand, and they formed a sister clade to A. bingensis (Heinem., Bull. Jard. Bot. État Bruxelles), A. brunneopunctatus Linda (J. Chen, Callac, & Parra), A. chiangmaiensis (Karunarathna, Guinb. & Hyde), A. niveogranulatus (Linda J. Chen, R.L. Zhao, Callac & Hyde), and A. toluenolens (Callac, Linda J. Chen & K.D. Hyde) (Figure 2). Notably, these have all been found in Thailand except for A. bingensis and A. brunneopunctatus, which has been identified in Africa (Benin, Congo, Togo, and Uganda) (Pegler, 1977; Chen et al., 2015). However, A. bingensis has a larger pileus (8–25 cm in diameter), a longer stipe (11–20 × 1.2–4 cm), and the narrower basidia (16–21 × 5–6 μm) than A. pseudoerectosquamosus (Pegler, 1977; Chen et al., 2015). While, A. brunneopunctatus has mostly shorter basidiospores (7.3–8.45 μm) than A. pseudoerectosquamosus (Chen et al., 2015). Agaricus chiangmaiensis differs from A. pseudoerectosquamosus by its larger pileus (10–17 cm in diameter), narrower basidiospores (7–8.5 × 3–4 μm), and smaller cheilocystidia (15–18 × 8–10 μm) (Karunarathna et al., 2014). The pileus (10–16 cm in diameter) and basidia (16–24 × 6.8–8.5 μm) of A. niveogranulatus were larger and mostly narrower than A. pseudoerectosquamosus, respectively (Chen et al., 2015). Additionally, A. toluenolens mostly has a narrower stipe (3.5 × 0.6–1 cm) and basidia (16–21 × 7–8.5 μm) than A. pseudoerectosquamosus (Chen et al., 2015).
Agaricus thailandensis Jaichliaw & S. Lumyong sp. nov. Figure 5
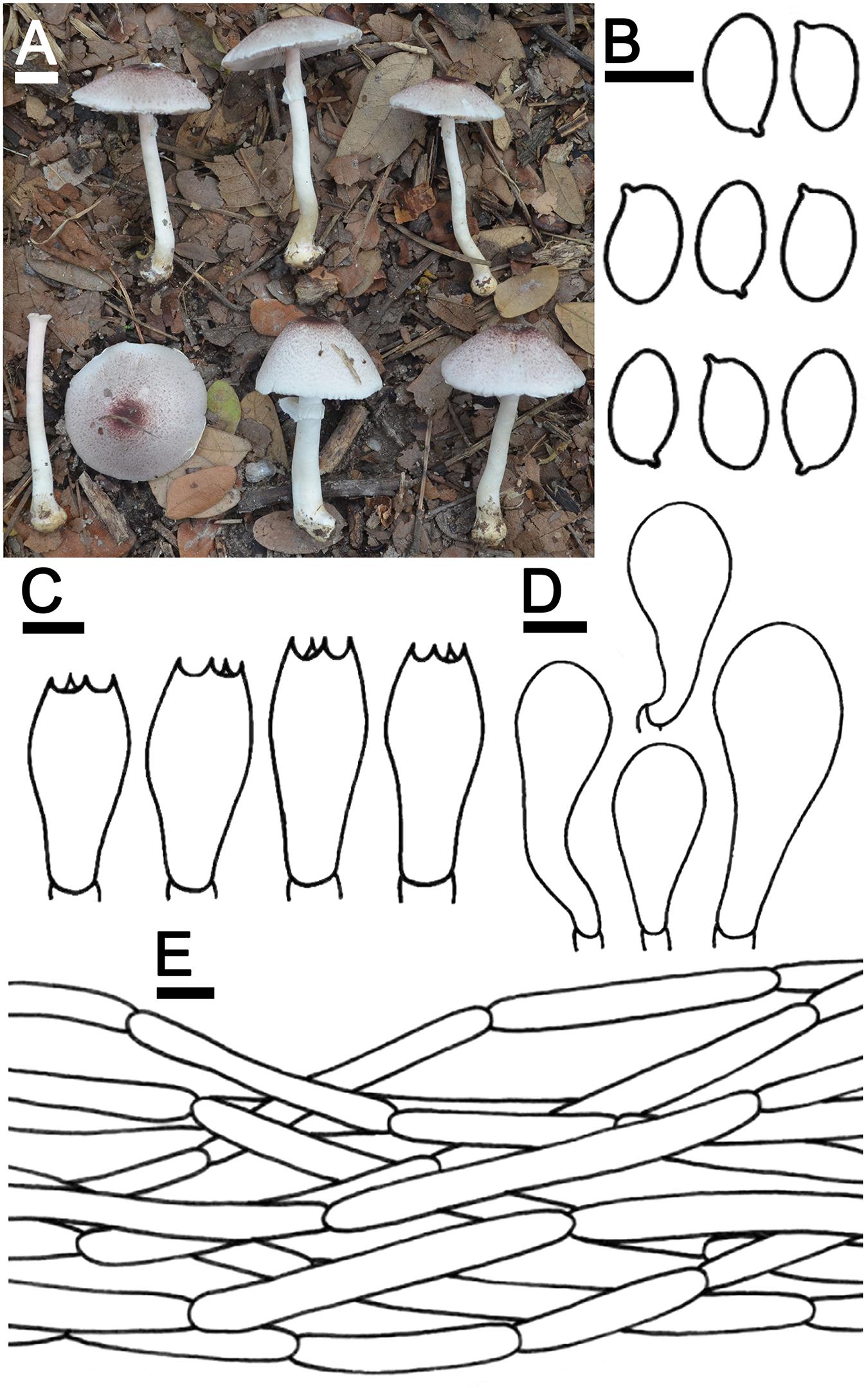
Figure 5. Agaricus thailandensis SDBR-CJ0118A (holotype). (A) Basidiomata on the field, (B) Basidiospores, (C) Basidia, (D) Cheilocystidia, (E) Pileipellis. Scale bars (A) = 1 cm, (B–D) = 5 μm, and (E) = 10 μm.
MycoBank: MB 838054
Facesoffungi number: FoF 09470
Etymology: “thailandensis” referring to Thailand, where the new species was found.
Holotype: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°48′31″N 98°57′15″E, elevation 340 m, September 16, 2019, C. Jaichaliaw. SDBR-CJ0118, CMUB 39946
Description: Basidiomata medium-sized, stipitate-pileate with lamellate hymenophore. Pileus 2.6–5.8 cm in diameter, at first obtusely conical, then convex or plano-convex with straight margins; surface background white (1A1) to pinkish-white (13A2) at the center, covered by dark ruby (12F8) at the center, pale to grayish magenta (13D3) at the margins of the fibrillose scales, appressed, denser at the center of pileus and poor at the margin. Pileus context soft, white (1A1). Lamellae free, crowded, even, regular, thin, 2–3 different lengths of lamellae, concolorous, at first white (1A1), then reddish gray (12B2). Stipe 5.2–5.9 × 0.3–0.7 cm, cylindrical with subbulbous, inserted; surface smooth, background white (1A1) to dark yellow (4C8) at the base, covered by small fibrilloses, minutely, white (1A1). Annulus thin, simple, stick out above the middle of the stipe and at the margins of the pileus, pendant, white (1A1). Stipe context soft, fistulose, white (1A1). Spores print dark brown (7F8). The odor smells like almonds. Macrochemical reactions; KOH reaction yellow and Schäffer’s reactions orange.
Basidiospores (6.0)6.1–6.3–6.4(6.5) × (3.5)3.7–4.0–4.3(4.5) μm (n = 50), Q = 1.44–1.86, Qm = 1.58 ± 0.15, ellipsoid to elongate, smooth, thin-walled, brown in water and KOH, inamyloid. Basidia 13–20 × 6–8 μm, clavate, 4-spored, hyaline, sterigmata up to 4 μm long. Cheilocystidia 12.5–50 × 7–15.5 μm, clavate to broadly clavate, often with a long peduncle, hyaline. Pleurocystidia absent. Pileipellis cylindrical hyphae 1.5–8.5 μm wide, elongate, constricted, smooth, occasionally branched, hyaline. Annulus composed of 2.5–21.5 μm wide, cylindrical with rounded apex, branched hyaline hyphae. Stipitipellis composed of 2.5–22.5 μm wide, cylindrical with rounded apex, constricted, hyaline. Clamp connections absent.
Ecology and distribution: Fruiting solitary or gregarious on sandy loam soil during the rainy season (mid-May to October). Known only from Thailand.
Additional specimens examined: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°47′45″N 98°57′2″E, elevation 340 m, September 27, 2019 (C. Jaichaliaw) SDBR-CJ0225.
Note: Agaricus thailandensis was placed within Agaricus sect. Minores of Agaricus subg. Minores based on the morphological and molecular data. The grayish magenta to dark ruby fibrils on the pileus surface and the reaction of KOH Schäffer’s reactions of A. thailandensis are morphologically similar to those of A. purpureofibrillosus (Linda J. Chen, R.L. Zhao & K.D. Hyde) in Agaricus sect. Minores of Agaricus subg. Minores (Fr.) (Chen et al., 2017). In contrast, A. purpureofibrillosus has shorter basidiospores (4.5–5.3 × 2.7–3 μm) than A. thailandensis and their clear separation was supported by the phylogeny (Chen et al., 2017). Phylogenetically, A. thailandensis formed a sister clade to the unnamed species Agaricus sp. voucher CA935 collected from Thailand, and they formed a sister clade to A. flammicolor and A. badioniveus (Figure 1). Both species have been collected from China and Thailand (Chen et al., 2017). Morphologically, A. flammicolor differed from A. thailandensis by the presence of orange bright fibrils on the pileus, smaller basidiospores (4.4–6.2 × 2.5–3.2 μm) and narrower basidia (12–16 × 5–6 μm) (Chen et al., 2017). Agaricus badioniveus is different from A. thailandensis by the yellowish-brown fibrils on the pileus and the mostly smaller basidiospores (5–6.2 × 3.1–3.8 μm) (Chen et al., 2017).
Taxonomic Description of New Record
Agaricus pallidobrunneus R.L. Zhao., Fungal Divers.: 33, 2016 Figure 6
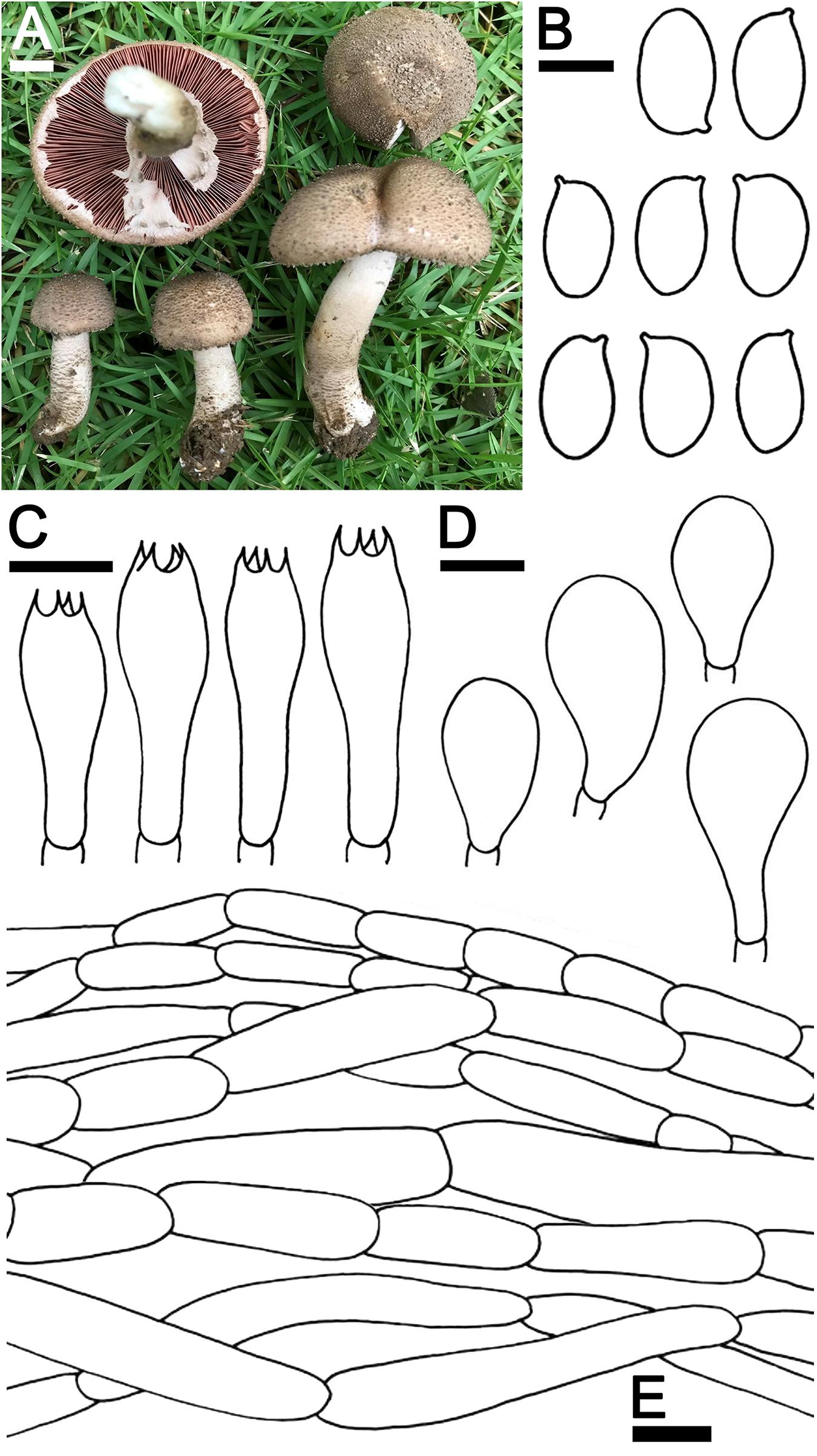
Figure 6. Agaricus pallidobrunneus SDBR-NK0368. (A) Basidiomata on the field, (B) Basidiospores, (C) Basidia, (D) Cheilocystidia, (E) Pileipellis. Scale bars (A) = 1 cm, (B) = 5 μm, and (C–E) = 10 μm.
Description: Basidiomata medium-sized, stipitate-pileate with lamellate hymenophore. Pileus (2)3.6–8 cm in diameter, paraboloid with inflexed margins, becoming applanate to plano-concave, depressed at the center with reflexed margins; surface background white (1A1), partially or entirely covered by brownish orange (5C5) to dark brown to (7F7) scales, the scales at first appressed then uplifted and innately scaled with age. Pileus context soft, white (1A1) grayish red (10D5) when cut. Lamellae free, crowded, thin, regular, 4–5 different lengths of lamellae, 3–4 mm wide half-way to margins, at first grayish rose (12B6) then grayish brown (7F3) when aged. Stipe 9–10 × 0.7–1.3 cm, tapering upwards, covered with small brown scales at the base, moist, white (1A1) near the cap, gradually light brown (5D4) at the base. Annulus thin, double, sticking up above the middle of the stipe and at margins, pendant, white (8A1) becoming brownish orange (5C5) when aged. Stipe context soft, fistulose, white (1A1), grayish red (10D5) when cut. Spores print dark brown (7F5). Odor is pleasant. Macrochemical reactions; KOH reaction yellow and Schäffer’s reactions negative.
Basidiospores (7)7.3–7.6–7.9(8.5) × (4.0)4.3–4.5–4.7(5.0) μm (n = 50), Q = 1.65–1.70, Qm = 1.67 ± 0.08, ellipsoid to elongate, smooth, thin-walled, brown in water and KOH, inamyloid. Basidia 15–30 × 5–9 μm, clavate, 4-spored, hyaline, sterigmata up to 2.5 μm long. Cheilocystidia 18–29 × 10–17 μm, clavate to broadly clavate, hyaline. Pleurocystidia absent. Pileipellis composed of hyaline hyphae, smooth, short cylindrical, with occasionally branched and cutis hyphae 5–12.5 μm wide, occasionally branched, hyaline. Annulus composed of 5–9.5 μm wide hyaline hyphae, cylindrical with rounded apex, branched. Stipitipellis composed of cutis hyphae wide up to 2.5–7 μm, cylindrical, occasionally branched, hyaline. Clamp connections absent.
Ecology and distribution: Fruiting solitary or gregarious on sandy loam soil during the rainy season (mid-May to October). Known from China and Thailand.
Material examined: THAILAND, Chiang Mai Province, Mueang Chiang Mai District, Chiang Mai University, 18°48′5″N 98°57′24″E, elevation 330 m, October 31, 2018, J. Kumla, SDBR-NK0368.
Note: Morphological characteristics and phylogenetic analysis of Thai specimens were used to assign A. pallidobrunneus to Agaricus sect. Brunneopicti in Agaricus subg. Pseudochitonia, according to Zhao et al. (2016). Agaricus pallidobrunneus is morphologically similar to A. erectosquamosus. However, A. erectosquamosus differs from A. pallidobrunneus by its erect, dark brown squamules on the pileus and stipe surface. The phylogeny also supports the determination that they are different species (Zhao et al., 2016).
Discussion
Agaricus is widely distributed in both temperate and tropical areas throughout the world (Kerrigan et al., 2005, Kerrigan, 2016; Chen et al., 2015; Zhao et al., 2016; He et al., 2017, 2018a,b; Callac and Chen, 2018; Ling et al., 2021). Morphological characteristics have been traditionally used in the identification of specimens of the Agaricus species. However, identification can be difficult as some species have similar features. Thus, identification can be limited by the morphological characteristics as well as the different environmental conditions that affect those morphological characteristics (Heinemann, 1978; Kerrigan, 1986; Singer, 1986; Callac et al.,1998a,b; Parra, 2008). Over the last two decades, molecular phylogeny has been an essential tool in the identification of Agaricus (Challen et al., 2003; Kerrigan et al., 2005, 2008; Zhao et al., 2011; Parra, 2013; Chen et al., 2015). The current classification of the genus Agaricus consists of six subgenera and twenty-four sections based on the combined data of morphological characteristics, the multigene phylogenetic analysis, and an estimation of divergence times (Zhao et al., 2016; Chen et al., 2017; He et al., 2018a; Parra et al., 2018).
In Thailand, the fourteen Agaricus species that were recorded by Thai taxonomists were identified only by their morphological characteristics. Of these, most have previously been found in temperate areas. However, there has been a lack of available molecular data on this species (Chandrasrikul et al., 2011) and there may have been incidences of mis-classification because tropical microflora are poorly understood. Thus, we are not sure these fourteen species have been accurately identified, expect A. subrufescens (Wisitrassameewong et al., 2012). Agaricus species richness is high in Thailand but many species have not yet been described based on a single collection and some of them have not been confirmed by phylogenetic data. Zhao et al. (2011) classified Agaricus collected from temperate and tropical regions, while ten Agaricus species collected from Thailand have been described based on their morphological and molecular characteristics.
During the period of 2012 to 2014, seven new species and three new records of Agaricus were found in Thailand (Chen et al., 2012; Wisitrassameewong et al., 2012; Zhao et al.,2012a,b; Karunarathna et al., 2014; Thongklang et al.,2014a,b). Thirty-seven new species of Agaricus were described from 2015 to 2017 (Ariyawansa et al., 2015; Chen et al., 2015, 2017; Liu et al., 2015; Thongklang et al., 2016; Zhao et al., 2016; Zhou et al., 2016; Hyde et al., 2017). In 2018, a new species of Thai Agaricus was reported by He et al. (2018a). Prior to this study, Agaricus in Thailand were classified in five subgenera (Agaricus subg. Agaricus, Agaricus subg. Flavoagaricus, Agaricus subg. Minores, Agaricus subg. Pseudochitonia and Agaricus subg. Spissicaules), thirteen sections and fifty-eight species based on morphological and molecular data.
In this study, six species of Agaricus including three new species (A. lannaensis, A. pseudoerectosquamosus, and A. thailandensis), one new record (A. pallidobrunneus), and two previously known species (A. erectosquamosus and A. subrufescens) collected from Chiang Mai Province, Thailand were identified based on their morphological characteristics and a multigene phylogenetic analysis. Agaricus lannaensis and A. pseudoerectosquamosus belong to Agaricus subg. Pseudochitonia in an incertae sedis clade and Agaricus sect. Brunneopicti, respectively. Agaricus thailandensis has been placed in Agaricus sect. Minores of Agaricus subg. Minores. Based on the phylogenetic analyses, A. lannaensis (including Agaricus sp. CA820) formed a sister clade to unnamed species Agaricus voucher LD2012162 (Figure 2); however, they cannot be assigned to Agaricus sect. Flocculenti according to the classification system of Agaricus (Zhao et al., 2016). Agaricus sp. CA820 collected from Thailand should be recognized in A. lannaensis, however, its morphological characteristics should be further confirmed. Agaricus pseudoerectosquamosus and A. thailandensis are closely related to the unnamed species Agaricus sp. voucher NTT117 and CA935, respectively which have been collected in Thailand (Figures 1, 2). However, there is a lack of available information on the morphological characteristics of these unnamed species. Thus, their species definition will be required in future studies. Notably, A. subrufescens and A. erectosquamosus have been previously reported to be from Thailand by Wisitrassameewong et al. (2012) and Zhao et al. (2016), respectively. Agaricus pallidobrunneus has been reported from China (Zhao et al., 2016), however, it has now been found for the first time in Thailand. Thus, to our knowledge, the Agaricus species recorded in Thailand has been raised to 62 species, 13 sections in five subgenera by the morphological and molecular evidence. Nevertheless, 13 Agaricus species listed by Chandrasrikul et al. (2011) require further confirmation by molecular data.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CJ, JK, and NS: conceptualization and resources. CJ, JK, SV, and NS: methodology, formal analysis, and writing-review and editing. SV and CJ: software. NS and SL: validation. CJ, JK, and SV: investigation, data curation, and writing-original draft. NS and SL: supervision. All authors read, revised, and approved the final manuscript.
Funding
This work was supported by Chiang Mai University, Thailand.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledged the Department of Biology and Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University for providing the laboratories and molecular work. We are grateful to Russell Kirk Hollis for assisting with English language editing of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.650513/full#supplementary-material
Supplementary Table 1 | Names, voucher numbers, countries and the corresponding GenBank accession numbers of the taxa used in the phylogenetic analyses. The sequences obtained in this study are in bold and superscript “T” refers to a type.
Footnotes
References
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. T., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252 taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s13225-015-0346-5
Asef, R., Hosseini, S. M. N., Callac, P., Goltapeh, E. M., Safaie, N., and Mahdizadeh, V. (2016). Agaricus section Xanthodermatei in Iran. Phytotaxa 247, 181–196. doi: 10.11646/phytotaxa.247.3.2
Bashir, H., Hussain, S., Khalid, A. N., Niazi, A. R. K., Parra, L. A., Callac, P., et al. (2018). First report of Agaricus sect. Brunneopicti from Pakistan with descriptions of two new species. Phytotaxa 357, 167–178. doi: 10.11646/phytotaxa.357.3.1
Bates, S. T., Chapman, R. M., Islam, M. B., Schwabe, A., Wardenaar, E. C., and Evenson, V. S. (2016). Phylogenetic placement of the secotioid fungus Araneosa columellata within Agaricus. Mycotaxon 131, 103–110. doi: 10.5248/131.103
Callac, P., and Chen, J. (2018). “Tropical species of Agaricus,” in Updates on Tropical Mushrooms. Basic and Applied Research, eds J. E. Sánchez, G. Mata, and D. J. Royse, (San Cristobal de Las Casas: Chiapas), 25–38.
Callac, P., Hocquart, S., Imbernon, M., Desmerger, C., and Olivier, J. M. (1998a). Bsn–t alleles from French field strains of Agaricus bisporus. Appl. Environ. Microbiol. 64, 2105–2110. doi: 10.1128/AEM.64.6.2105-2110.1998
Callac, P., Moquet, F., Imbernon, M., Guedes-Lafargue, M. R., Mamoun, M., Olivier, J. M., et al. (1998b). Evidence for PPC1, a determinant of the Pilei–Pellis Color of Agaricus bisporus Fruitbodies. Fungal Genet. Biol. 23, 181–188. doi: 10.1006/fgbi.1998.1035
Challen, M. P., Kerrigan, R. W., and Callac, P. (2003). A phylogenetic reconstruction and emendation of Agaricus section Duploannulatae. Mycologia 95, 61–73. doi: 10.1080/15572536.2004.11833132
Chandrasrikul, A., Suwanarit, P., Sangwanit, U., Lumyong, S., Payapanon, A., Sanoamuang, N., et al. (2011). Checklist of Mushrooms (Basidiomycetes) in Thailand. Bangkok: Office of Natural Resources and Environmental Policy and Planning.
Chen, J., Callac, P., Parra, L. A., Karunarathna, S. C., He, M. Q., Moinard, M., et al. (2017). Study in Agaricus subgenus Minores and allied clades reveals a new American subgenus and contrasting phylogenetic patterns in Europe and Greater Mekong Subregion. Persoonia 38, 170–196. doi: 10.3767/003158517X695521
Chen, J., Zhao, R., Parra, L. A., Guelly, A. K., Kesel, D. A., Rapior, S., et al. (2015). Agaricus section Brunneopicti: a phylogenetic reconstruction with descriptions of four new taxa. Phytotaxa 192, 145–168. doi: 10.11646/phytotaxa.192.3.2
Chen, J., Zhao, R. L., Karunarathna, S. C., Callac, P., Raspé, O., Bahkali, A. H., et al. (2012). Agaricus megalosporus: a new species in section Minores. Cryptogam. Mycol. 33, 145–155. doi: 10.7872/crym.v33.iss2.2012.145
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dui, K. D. H., and Zhao, R. (2015). Edible species of Agaricus (Agaricaceae) from Xinjiang Province (Western China). Phytotaxa 202, 185–197. doi: 10.11646/phytotaxa.202.3.2
Foulongne-Oriol, M., Spataro, C., Moinard, M., Cabannes, D., Callac, P., and Savoie, J. M. (2012). Development of polymorphic microsatellite markers issued from pyrosequencing technology for the medicinal mushroom Agaricus subrufescens. FEMS Microbiol. Lett. 334, 119–126. doi: 10.1111/j.1574-6968.2012.02627.x
Geml, J., Geiser, D. M., and Royse, D. J. (2004). Molecular evolution of Agaricus species based on ITS and LSU rDNA sequences. Mycol. Prog. 3, 157–176. doi: 10.1007/s11557-006-0086-8
Geml, J., Laursen, G. A., Nusbaum, H. C., and Taylor, D. L. (2007). Two new species of Agaricus from the Subantarctic. Mycotaxon 100, 193–208.
Gui, Y., Zhu, G. S., Callac, P., Hyde, K. D., Parra, L. A., Chen, J., et al. (2015). Agaricus section Arvenses: three new species in highland subtropical Southwest China. Fungal Biol. 119, 79–94. doi: 10.1016/j.funbio.2014.10.005
Hama, O., Maes, E., Guissou, M. L., Ibrahim, D., Baragé, M., Parra, L. A., et al. (2010). Agaricus subsaharianus, une nouvelle espèce comestible et consommée au Niger, au Burkina Faso et en Tanzanie. Cryptogam. Mycol. 31, 221–234.
He, M. Q., Chen, J., Zhou, J. L., Ratchadawan, C., Hyde, K. D., and Zhao, R. L. (2017). Tropic origins, a dispersal model for saprotrophic mushrooms in Agaricus section Minores with descriptions of sixteen new species. Sci. Rep. 7, 1–31. doi: 10.1038/s41598-017-05203-5
He, M. Q., Chuankid, B., Hyde, K. D., Cheewangkoon, R., and Zhao, R. L. (2018a). A new section and species of Agaricus subgenus Pseudochitonia from Thailand. MycoKeys 40, 53–67. doi: 10.3897/mycokeys.40.26918
He, M. Q., Hyde, K. D., Wei, S. L., Xi, Y. L., Cheewangkoon, R., and Zhao, R. L. (2018b). Three new species of Agaricus section Minores from China. Mycosphere 9, 189–201. doi: 10.5943/mycosphere/9/2/3
He, M. Q., and Zhao, R. L. (2015). A new species of Agaricus section Minores from China. Mycology 6, 182–186. doi: 10.1080/21501203.2015.1121931
Heinemann, P. (1978). Essai d’une clé de determination des genres Agaricus et Micropsalliota. Sydowia 30, 6–37.
Hyde, K. D., Norphanphoun, C., Abreu, V. P., Bazzicalupo, A., Chethana, K. T., Clericuzio, M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. doi: 10.1007/s13225-017-0391-3
Hyde, K. D., Norphanphoun, C., Chen, J., Dissanayake, A. J., Doilom, M., Hongsanan, S., et al. (2018). Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 93, 215–239. doi: 10.1007/s13225-018-0415-7
Ibrahim, D. M., Barage, M., Marafa, D. I., Sanchez, L. A. P., Raspe, O., Kesel, A. D., et al. (2010). Agaricus subsaharianus, une nouvelle espèce comestible et consommée au Niger, au Burkina Faso et en Tanzanie. Cryptogam. Mycol. 31, 221–234.
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L. D., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Karunarathna, S. C., Chen, J., Mortimer, P. E., Xu, J. C., Zhao, R. L., Callac, P., et al. (2016). Mycosphere essay 8: a review of genus Agaricus in tropical and humid subtropical regions of Asia. Mycosphere 7, 417–439. doi: 10.5943/mycosphere/7/4/3
Karunarathna, S. C., Guinberteau, J., Chen, J., Vellinga, E. C., Zhao, R. L., Chukeatirote, E., et al. (2014). Two new species in Agaricus tropical clade I. Chiang Mai J. Sci. 41, 771–780.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kerrigan, R. W., Callac, P., Guinberteau, J., Challen, M. P., and Parra, L. A. (2005). Agaricus section Xanthodermatei: a phylogenetic reconstruction with commentary on taxa. Mycologia 97, 1292–1315. doi: 10.1080/15572536.2006.11832737
Kerrigan, R. W., Callac, P., and Parra, L. A. (2008). New and rare taxa in Agaricus section Bivelares (Duploannulati). Mycologia 100, 876–892. doi: 10.3852/08-019
Lebel, T. (2013). Two new species of sequestrate Agaricus (section Minores) from Australia. Mycol. Prog. 12, 699–707. doi: 10.1007/s11557-012-0879-x
Lebel, T., and Syme, A. (2012). Sequestrate species of Agaricus and Macrolepiota from Australia: new species and combinations and their position in a calibrated phylogeny. Mycologia 104, 496–520. doi: 10.3852/11-092
Li, G. J., Hyde, K. D., Zhao, R. L., Hongsanan, S., Abdel-Aziz, F. A., Abdel-Wahab, M. A., et al. (2016). Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78, 1–237. doi: 10.1007/s13225-016-0366-9
Li, S. F., Xi, Y. L., Qi, C. X., Liang, Q. Q., Wei, S. L., Li, G. J., et al. (2014). Agaricus taeniatus sp. nov., a new member of Agaricus sect. Bivelares from northwest China. Mycotaxon 129, 187–196. doi: 10.5248/129.187
Ling, Z. L., Zhou, J. L., Parra, L. A., Kesel, A. D., Callac, P., Zhao, R. L., et al. (2021). Four new species of Agaricus subgenus Spissicaules from China. Mycologia 113, 476–491. doi: 10.1080/00275514.2020.1852808
Liu, J. K., Hyde, K. D., Jones, E. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Miller, M. A., Holder, M. T., Vos, R., Midford, P. E., Liebowitz, T., Chan, L., et al. (2009). The CIPRES Portals. CIPRES. Available online at: http://www.phylo.org/portal2/home (accessed October 10, 2020).
Parra, L. A. (2008). Agaricus L. Allopsalliota Nauta & Bas. I. Fungi Europaei, Vol. 1. Italy: Edizione Candusso.
Parra, L. A. (2013). Fungi Europaei: Agaricus: Allopsalliota, Nauta and Bas, Vol. 2. Italy: Edizione Candusso.
Parra, L. A., Angelini, C., Ortiz-Santana, B., Mata, G., Billette, C., Rojo, C., et al. (2018). The genus Agaricus in the Caribbean. nine new taxa mostly based on collections from the Dominican Republic. Phytotaxa 345, 219–271. doi: 10.11646/phytotaxa.345.3.2
Parra, L. A., Muñoz, G., and Callac, P. (2014). Agaricus caballeroi sp. nov., a new species of the section Nigrobrunnescentes collected in Spain. Micol. Veget. Medit. 29, 21–38.
Parra, L. A., Wisman, J., Guinberteau, J., Wilhelm, M., Weholt, Ø, and Musumeci, E. (2015). Agaricus collegarum and Agaricus masoalensis, two new taxa of the section Nigrobrunnescentes collected in Europe. Micol. Vegetazione Mediterr. 30, 3–26.
Pegler, D. N. (1977). A Preliminary Agaric Flora of East Africa. London: Her Majesty’s Stationery Office.
Petrova, A., Alipieva, K., Kostadinova, E., Antonova, D., Lacheva, M., Gjosheva, M., et al. (2007). GC–MS studies of the chemical composition of two inedible mushrooms of the genus Agaricus. Chem. Cent. J. 1:33. doi: 10.1186/1752-153X-1-33
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1–α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.1080/15572536.2006.11832842
Robert, V., Vu, D., Amor, A. B., Wiele, N. V. D., Brouwer, C., Jabas, B., et al. (2013). MycoBank gearing up for new horizons. IMA Fungus 4, 371–379. doi: 10.5598/imafungus.2013.04.02.16
Ronquist, F., Teslenko, M., Mark, V. D. P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schoch, C. L., Robbertse, B., Robert, V., Vu, D., Cardinali, G., Zhang, N., et al. (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. DataBase. 2014, 1–21. doi: 10.1093/database/bau061
Singer, R. (1986). The Agaricales in Modern Taxonomy, 4th Edn. Koenigstein: Koeltz Scientific Books.
Stamatakis, A. (2006). RAxML–VI–HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Tarafder, E. N. T. A. J., Dutta, A. K., Sarkar, J., and Acharya, K. (2018). A new species of Agaricus sect. Brunneopicti from Eastern India. Phytotaxa 374, 139–146. doi: 10.11646/phytotaxa.374.2.5
Thongklang, N., Chen, J., Bandara, A. R., Hyde, K. D., Raspé, O., Parra, L. A., et al. (2016). Studies on Agaricus subtilipes, a new cultivatable species from Thailand, incidentally, reveal the presence of Agaricus subrufescens in Africa. Mycoscience 57, 239–250. doi: 10.1016/j.myc.2016.02.003
Thongklang, N., Nawaz, R., Khalid, A. N., Chen, J., Hyde, K. D., Zhao, R. L., et al. (2014a). Morphological and molecular characterization of three Agaricus species from tropical Asia (Pakistan, Thailand) reveals a new group in section Xanthodermatei. Mycologia 106, 1220–1232. doi: 10.3852/14-076
Thongklang, N., Sysouphanthong, P., Callac, P., and Hyde, K. D. (2014b). First cultivation of Agaricus flocculosipes and a novel Thai strain of A. subrufescens. Mycosphere 5, 814–820. doi: 10.5943/mycosphere/5/6/11
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. Res. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, Z. R., Parra, L., Callac, P., Zhou, J. L., Fu, W. J., Dui, S. H., et al. (2015). Edible species of Agaricus (Agaricaceae) from Xinjiang Province (Western China). Phytotaxa 202, 185–197. doi: 10.11646/phytotaxa.202.3.2
White, T. J., Burns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. Whitish, (San Diego, CA: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wisitrassameewong, K., Karunarathna, S. C., Thongklang, N., Zhao, R. L., Callac, P., Chukeatirote, E., et al. (2012). Agaricus subrufescens: new to Thailand. Chiang Mai J. Sci. 39, 281–291.
Zhang, M. Z., Li, G. J., Dai, R. C., Xi, Y. L., Wei, S. L., and Zhao, R. L. (2017). The edible wide mushrooms of Agaricus section Bivelares from Western China. Mycosphere 8, 1640–1652. doi: 10.5943/mycosphere/8/10/4
Zhao, R. L., Desjardin, D. E., Callac, P., Parra, L. A., Guinberteau, J., Soytong, K., et al. (2012a). Two species of Agaricus sect. Xanthodermatei from Thailand. Mycotaxon 122, 187–195. doi: 10.5248/122.187
Zhao, R. L., Hyde, K. D., Karunarathna, S. C., Desjardin, D. E., Raspé, O., Soytong, K., et al. (2012b). Agaricus flocculosipes sp. nov., a new potentially cultivatable species from the palaeotropics. Mycoscience 53, 300–311. doi: 10.1007/s10267-011-0169-5
Zhao, R. L., Karunarathna, S., Raspé, O., Parra, L. A., Guinberteau, J., Moinard, M., et al. (2011). Major clades in tropical Agaricus. Fungal Divers. 51, 279–296. doi: 10.1007/s13225-011-0136-7
Zhao, R. L., Zhou, J. L., Chen, J., Margaritescu, S., Sánchez-Ramírez, S., Hyde, K. D., et al. (2016). Towards standardizing taxonomic ranks using divergence times–a case study for reconstruction of the Agaricus taxonomic system. Fungal Divers. 78, 239–292. doi: 10.1007/s13225-016-0357-x
Keywords: Agaricales, new taxa, phylogeny, saprophytic mushroom, taxonomy
Citation: Jaichaliaw C, Kumla J, Vadthanarat S, Suwannarach N and Lumyong S (2021) Multigene Phylogeny and Morphology Reveal Three Novel Species and a Novel Record of Agaricus From Northern Thailand. Front. Microbiol. 12:650513. doi: 10.3389/fmicb.2021.650513
Received: 07 January 2021; Accepted: 19 April 2021;
Published: 21 June 2021.
Edited by:
Peter Edward Mortimer, Kunming Institute of Botany, Chinese Academy of Sciences, ChinaReviewed by:
Philippe Callac, INRAE, MycSA, FranceSamantha Chandranath Karunarathna, Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, China
Copyright © 2021 Jaichaliaw, Kumla, Vadthanarat, Suwannarach and Lumyong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nakarin Suwannarach, c3V3YW5fNDYxQGhvdG1haWwuY29t; Saisamorn Lumyong, c2Nib2kwMDlAZ21haWwuYWMudGg=
 Chanyawat Jaichaliaw1,2
Chanyawat Jaichaliaw1,2 Jaturong Kumla
Jaturong Kumla Santhiti Vadthanarat
Santhiti Vadthanarat Nakarin Suwannarach
Nakarin Suwannarach Saisamorn Lumyong
Saisamorn Lumyong