- Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Pseudomonas cannabina pv. alisalensis (Pcal) causes bacterial leaf spot and blight of Brassicaceae and Poaceae. We previously identified several potential Pcal virulence factors with transposon mutagenesis. Among these a trpA mutant disrupted the tryptophan synthase alpha chain, and had an effect on disease symptom development and bacterial multiplication. To assess the importance of TrpA in Pcal virulence, we characterized the trpA mutant based on inoculation test and Pcal gene expression profiles. The trpA mutant showed reduced virulence when dip- and syringe-inoculated on cabbage and oat. Moreover, epiphytic bacterial populations of the trpA mutant were also reduced compared to the wild-type (WT). These results suggest that TrpA contributes to bacterial multiplication on the leaf surface and in the apoplast, and disease development. Additionally, several Brassicaceae (including Japanese radish, broccoli, and Chinese cabbage) also exhibited reduced symptom development when inoculated with the trpA mutant. Moreover, trpA disruption led to downregulation of bacterial virulence genes, including type three effectors (T3Es) and the phytotoxin coronatine (COR), and to upregulation of tryptophan biosynthesis genes. These results indicate that a trade-off between virulence factor production and Pcal multiplication with tryptophan might be regulated in the infection processes.
Introduction
The foliar plant bacterial pathogen Pseudomonas syringae causes economically important diseases in a wide range of plants (Agrios, 2005). P. syringae colonizes leaf surfaces (epiphytic) of host plants, enters natural opening sites, including stomata, and then multiplies in the leaf interior (apoplast) (Xin and He, 2013). During infection processes, P. syringae suppresses plant basal defense by using virulence factors, such as specialized protein secretion systems, toxins, plant hormones, bacterial surface attachment factors, flagella, and siderophores (Xin and He, 2013). P. syringae pv. tomato (Pst) DC3000 also infects Arabidopsis (Whalen et al., 1991), and is used as a model pathogen to study plant-bacterial interactions. Pst DC3000 is a highly aggressive pathogen once inside host tissue, and uses many type three effectors (T3Es) and the phytotoxin coronatine (COR; Buell et al., 2003; Feil et al., 2005). The function of these two Pst DC3000 virulence factors have been well characterized at the molecular level. However, Boch et al. (2002) demonstrated that a wide range of plant-induced loci in Pst DC3000 included not only virulence associated genes such as hrp genes and COR biosynthesis genes, but also genes involved in stress tolerance, polysaccharide synthesis, nutrient uptake, amino acid assimilation, and carbon metabolism. Although virulence associated genes related to type three secretion system (T3SS) and COR have been investigated well in the P. syringae virulence, functions of other genes including amino acid metabolism remain largely unclear.
Pseudomonas cannabina pv. alisalensis (Pcal) causes bacterial leaf spot and blight of Brassicaceae and Poaceae (Takikawa and Takahashi, 2014). Pcal was formally classified as P. syringae pv. maculicola (Psm). Although Pcal and Psm have similar characteristics, these two pathogens are defined by some bacteriological characteristics, genetic traits, and their ability to infect monocot plants such as oat (Avena sativa) and timothy (Phleum pretense) (Cintas et al., 2002; Bull et al., 2010; Takikawa and Takahashi, 2014). Our recent study identified several potential Pcal virulence factors, including T3SS, membrane transporters, transcriptional factors, and amino acid metabolism (Sakata et al., 2019). Among these, the mutants which were disrupted in amino acid metabolism showed no pathogenicity, similar to a T3SS mutant (Sakata et al., 2019). A trpA (encoding tryptophan synthase alpha chain) mutant exhibited reduced disease symptom development and bacterial multiplication (Sakata et al., 2019). Helmann et al. (2019) conducted a genome wide screening to identify P. syringae pv. syringae (Pss) B728a virulence factors, and identified that trpA contributes to bacterial fitness on both the leaf surface and in the apoplast. Tryptophan is the least abundant amino acid in leaf exudates from many plant species (Morgan and Tukey, 1964; Rico and Preston, 2008). Although amino acid metabolism is essential for bacterial growth, the function of amino acid metabolism in plant bacterial virulence has not been investigated.
Here, we showed that TrpA contributes to Pcal virulence in successful infection processes. Moreover, trpA mutant expression profiles analysis showed downregulation of virulence related genes, including T3Es and COR, and upregulation of tryptophan biosynthesis related genes compared to WT. These results suggest that trade-off between virulence factor production and tryptophan biosynthesis might be present in bacterial infection processes.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are described in Supplementary Table 1. P. cannabina pv. alisalensis strain KB211 (Pcal KB211) was used as the pathogenic strain to inoculate cabbage, oat, Japanese radish, broccoli, and Chinese cabbage. Pcal wild-type (WT) was grown on King’s B (KB; King et al., 1954) medium at 28°C. NB35, NF2, NF34, NI13, NH11, NM37, and NN31 were grown on KB containing kanamycin (10 μg/ml) (Km). trpA mutant complemented with pDSK-trpA was grown on KB containing Km (10 μg/ml) and gentamicin (Gen) (25 μg/ml) (Supplementary Table 1). Before Pcal inoculation, bacteria were suspended in sterile distilled H2O, and the bacterial cell densities at 600 nm (OD600) were measured using a Biowave CO8000 Cell Density Meter (Funakoshi, Tokyo, Japan).
Bacterial in vitro Growth Measurements
Wild-type, the trpA mutant, and the trpA mutant complemented with pDSK-trpA were grown at 28°C on Luria-Bertani (LB; Sambrook et al., 1989) medium. The bacterial suspensions were standardized to an OD600 of 0.01 with LB, and bacterial growth was measured at OD600 for 24 h. WT and the trpA mutant were also grown at 28°C in mannitol-glutamate (MG; Keane et al., 1970) medium. The bacterial suspensions were standardized to an OD600 of 0.1. L-tryptophan (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) was used to co-incubate with MG medium at 1, 10, and 100 μM, respectively. Bacterial growth was measured at OD600 for 24 h.
Plant Materials
Plants used for Pcal virulence assays include cabbage (Brassica oleracea var. capitate) cv. Kinkei 201, oat (Avena strigosa) cv. Hayoat, Japanese radish (Raphanus sativus var. longipinnatus) cv. Natsutsukasa, Chinese cabbage (Brassica rapa var. pekinensis) cv. Akimeki, and broccoli (Brassica oleracea var. italica) cv. Midoribue. All plants were grown from seed at 23–25°C with a light intensity of 200 μE/m2/s and a 16 h light/8 h dark photoperiod. Plants were used for dip- and spray-inoculation assays around two weeks after germination, and for syringe-inoculation assays around 3 weeks after germination. For flood-inoculation, cabbage seeds were germinated and grown on 1/2 strength Murashige and Skoog (MS) medium (0.3% phytagel) with Gamborg vitamins (Sigma-Aldrich, St. Louis, MO, United States). Cabbage seedlings were incubated in a growth chamber at 24°C with a light intensity of 200 μE/m2/s and a 12 h light/12 h dark photoperiod, and used for the inoculation assays 2 weeks after germination.
Bacterial Inoculation
To assay for disease on cabbage, oat, Japanese radish, broccoli, and Chinese cabbage plants, dip-inoculations were conducted by soaking seedlings in bacterial suspensions (5 × 107 CFU/ml) containing 0.025% Silwet L-77 (OSI Specialities, Danbury, CT, United States). The seedlings were then incubated in growth chambers at 85–95% RH for the first 24 h, then at 80–85% RH for the rest of the experimental period. Disease symptoms were photographed at 5 days post-inoculation (dpi). To assess bacterial growth in all plants, the internal bacterial population was measured after dip-inoculation. Inoculated seedlings were collected, and two inoculated leaves were measured chronologically. The leaves were surface-sterilized with 10% H2O2 for 3 min. After washing three times with sterile distilled water, the leaves were homogenized in sterile distilled water, and diluted samples were plated onto solid KB agar medium. Two or three days after dilution sample plating, the bacterial colony forming units (CFUs) were counted and normalized as CFU per gram, using the total leaf weight. The bacterial populations at 0 dpi were estimated using leaves harvested 1-hour post-inoculation (hpi) without surface-sterilization. The bacterial populations were evaluated in at least three independent experiments.
For syringe-inoculation, cabbage and oat leaves were syringe-inoculated with bacterial suspensions (5 × 105 or 5 × 107 CFU/ml) with a 1-ml blunt syringe. L-tryptophan was co-inoculated with the trpA mutant at 100 μM, 1 mM, 10 mM, and 50 mM, respectively. The plants were then incubated at 70–80% RH for the rest of the experimental period. Leaves were removed and photographed at 5 dpi. To assess bacterial growth in cabbage, the internal bacterial population was measured after syringe-inoculation. Leaf disks were harvested using a 3.5 mm-diameter cork-borer from syringe-infiltrated leaf zones. To assess bacterial growth in oat, leaf pieces were cut from syringe-infiltrated leaf zones and the area (cm2) measured. The bacterial CFUs were counted and normalized as CFU per cm2 of leaf tissue. The bacterial populations were evaluated in at least three independent experiments.
Epiphytic Bacterial Growth Assay
Flood-inoculation was conducted as described previously (Ishiga et al., 2011). Briefly, 50 ml of bacterial suspension (1 × 108 CFU/ml) made in sterile distilled H2O containing 0.025% Silwet L-77 (OSI Specialities, Danbury, CT, United States) was dispensed onto a plate containing 2-week-old cabbage seedlings grown on 1/2 strength MS medium for uniform inoculation, then the plates were incubated for 2–3 min at room temperature. After the bacterial suspension was decanted from the plates, they were sealed with 3M Microspore 2.5 cm surgical tape (3M, St. Paul, MS, United States) and incubated at 24°C with a light intensity of 200 μE/m2/s and a 12 h light/12 h dark photoperiod. To assess epiphytic bacterial population on cabbage, the leaves were washed with washing buffer (50 mM PBS buffer) in an ultrasonic bath for 7 min, and then dilutions were plated to KB agar medium at 1 and 2 dpi. The bacterial CFUs were counted and normalized as CFU per gram, using the total leaf weight. The bacterial populations were evaluated in at least three independent experiments.
Monitoring Bacterial Gene Expression in Culture
For expression profiles in culture conditions, Pcal was grown in KB medium for 24 h, then adjusted to an OD600 of 0.1 with fresh KB medium and grown for 3 h. The bacterial cells in the suspension were harvested by centrifugation at 12,000 rpm for 2 min, and cell pellets were resuspended with HS medium optimized for COR production (HSC; Palmer and Bender, 1993) and grown for 30 min. The bacterial cells in the suspension were harvested by centrifugation at 12,000 rpm for 2 min, and cell pellets were used for subsequent purification. Total RNA was extracted using Reliaprep (Promega, Madison, WI, United States) according to the manufacturer’s protocol. Two micrograms of total RNA were treated with gDNA Remover (TOYOBO, Osaka, Japan) to eliminate genomic DNA, and the DNase-treated RNA was reverse transcribed using the ReverTra Ace qPCR RT Master Mix (TOYOBO). The cDNA (1:10) was then used for RT-qPCR using the primers shown in Supplementary Table 2 with THUNDERBIRD SYBR qPCR Mix (TOYOBO) on a Thermal Cycler Dice Real Time System (Takara Bio). Pcal KB211 outer membrane porin F (oprF) and recombinase A (recA) were used to normalize gene expression. Gene expression of the trpA mutant was calculated as a relative value of WT expression. The reagent blank (no-template) controls were used to detect contamination. The expression profiles were evaluated in four independent samples.
Monitoring Bacterial Gene Expression in planta
To analyze Pcal gene expression profiles during infection, we syringe-inoculated cabbage plants with Pcal at 5 × 107 CFU/ml, and at 3 and 6 hrs the total RNAs including plant and bacterial RNAs were extracted from infected leaves and purified. Total RNA extraction and real-time quantitative RT-PCR (RT-qPCR) were done as described previously (Ishiga and Ichinose, 2016). Two micrograms of total RNA were treated with gDNA Remover (TOYOBO) to eliminate genomic DNA, and the DNase-treated RNA was reverse transcribed using the ReverTra Ace qPCR RT Master Mix (TOYOBO). The cDNA (1:10) was then used for RT-qPCR using the primers shown in Supplementary Table 2 with THUNDERBIRD SYBR qPCR Mix (TOYOBO) on a Thermal Cycler Dice Real Time System (Takara Bio). Pcal KB211 outer membrane porin F (oprF) and recombinase A (recA) were used to normalize gene expression. Gene expression of the trpA mutant was calculated as a relative value of WT 3 h expression. The reagent blank (no-template) controls were used to detect contamination. The expression profiles were evaluated in at least six independent samples.
Results
TrpA Is Involved in Pcal Pathogenicity
To investigate functions of amino acid metabolism in Pcal virulence, we conducted inoculation assay using six amino acid metabolism mutants (Sakata et al., 2019). These six mutants (NF2, NF34, NH11, NI13, NM37, and NN31; see Supplementary Table 1) and a T3SS mutant (NB35) were syringe-inoculated into cabbage to investigate which mutants were most involved in Pcal virulence. NH11, NM37, and NN31 showed necrosis similar to WT. NB35, NF34, and NI13 only showed chlorosis. Only the NF2 mutant, which is mutated in the trpA (encoding tryptophan synthase alpha chain), showed no symptoms (Figure 1A). Moreover, trpA mutant bacterial populations were severely reduced among all mutants, including the T3SS mutant (Figure 1B).
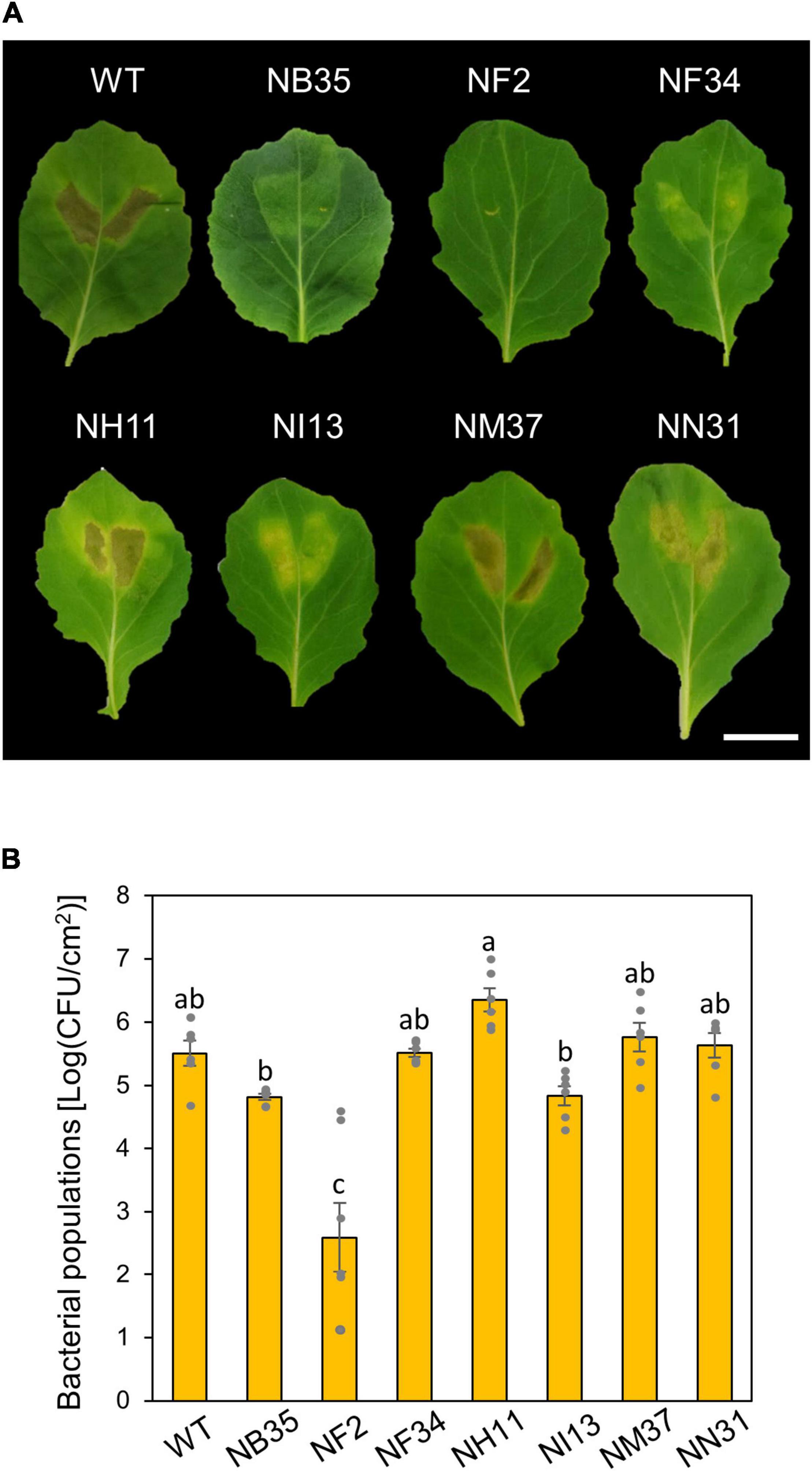
Figure 1. Disease phenotypes and bacterial populations of Pseudomonas cannabina pv. alisalnesis KB211 WT and amino acid mutants in cabbage after syringe-inoculation. Disease symptoms (A) and bacterial populations (B) on cabbage leaves syringe-inoculated with WT, a type three secretion system (T3SS) mutant (NB35) and six amino acid metabolism mutants (NF2, NF34, NH11, NI13, NM37, and NN31) described in Supplementary Table 1. Cabbage plants were syringe-inoculated with 5 × 105 CFU/ml of inoculum. The bacterial concentrations in the plant leaves were evaluated at 5 dpi. The leaves were photographed at 5 dpi. Scale bar shows 2 cm. Vertical bars indicate the standard error for at least six independent experiments. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (p < 0.05).
TrpA Contributes to Pcal Multiplication in the Apoplast to Cause Disease
trpA is apparently dispensable for Pcal growth in rich LB medium, since no growth difference was observed between the WT and the trpA mutant (Supplementary Figure 1). Cabbage inoculated with WT showed severe chlorosis, but the trpA mutant induced no symptoms (Figure 2A). Although bacterial populations of the WT and the trpA mutant complemented with pDSK-trpA reached around 1 × 108 CFU/ml when dip-inoculated onto cabbage, trpA mutant populations were 104 times less (Figure 2B). Moreover, the trpA mutant multiplication defect was observed within 1 dpi (Figure 2B). To further investigate whether the trpA mutant is impaired in apoplastic growth and pathogenicity, syringe-infiltration was used to bypass stomatal defense. When infiltrated directly into the cabbage apoplast, trpA mutant disease development and bacterial populations were decreased compared to the WT (Figures 2C,D).
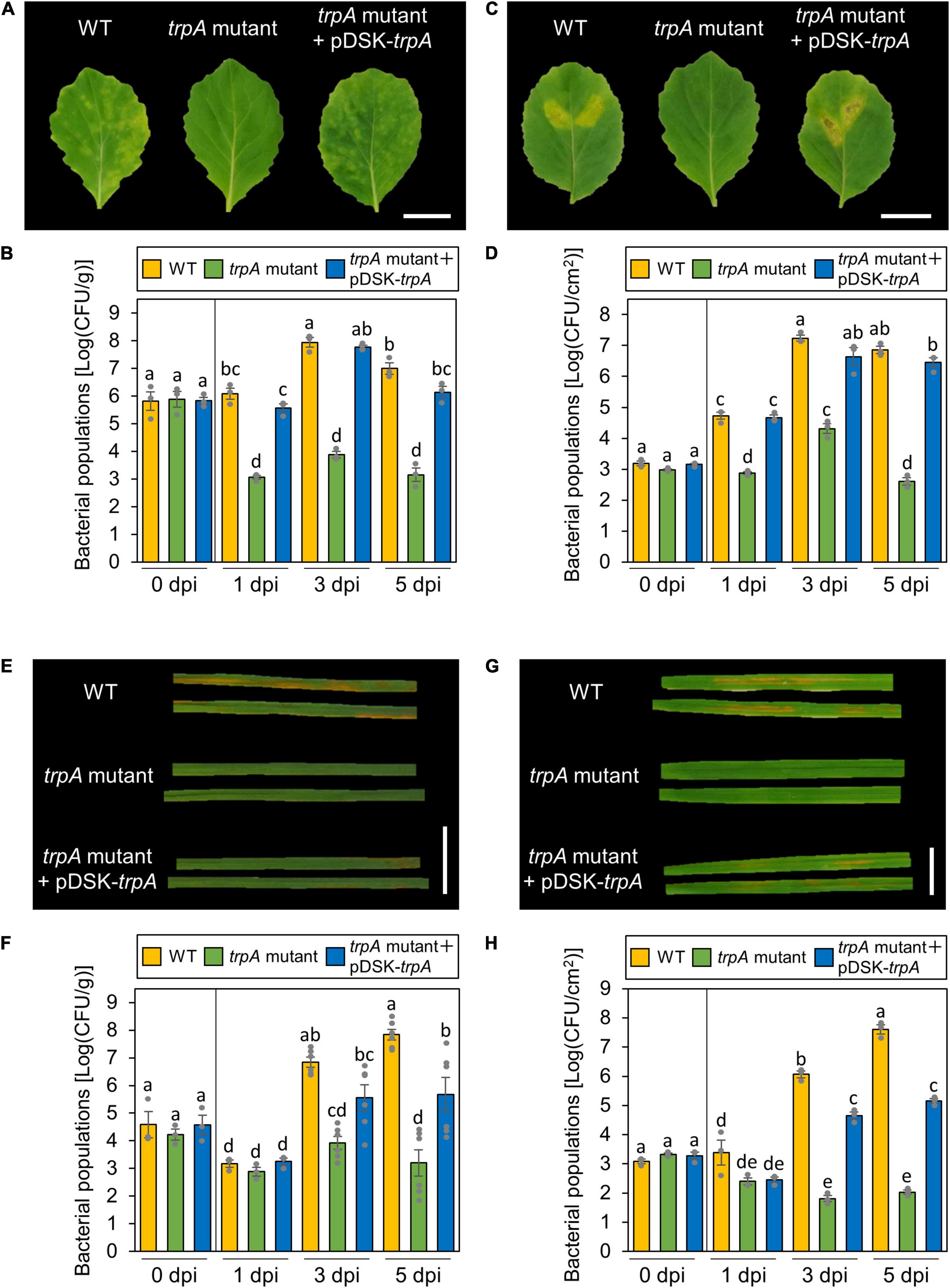
Figure 2. Disease phenotypes and bacterial populations of Pseudomonas cannabina pv. alisalnesis KB211 WT and the trpA mutant in cabbage and oat after dip- and syringe-inoculation. Disease symptoms (A) and bacterial populations (B) in cabbage dip-inoculated with WT, the trpA mutant, and the trpA mutant complemented with pDSK-trpA. Disease symptoms (C) and bacterial populations (D) in cabbage syringe-inoculated with WT, the trpA mutant, and the trpA mutant complemented with pDSK-trpA. Disease symptoms (E) and bacterial populations (F) in oat dip-inoculated with WT, the trpA mutant, and the trpA mutant complemented with pDSK-trpA. Disease symptoms (G) and bacterial populations (H) in oat syringe-inoculated with WT, the trpA mutant, and the trpA mutant complemented with pDSK-trpA. Cabbage and oat were dip-inoculated with 5 × 107 CFU/ml of inoculum containing 0.025% SilwetL-77 and syringe-inoculated with 5 × 105 CFU/ml of inoculum, respectively. Bacterial concentrations in the plant leaves were evaluated at 0, 1, 3, and 5 dpi. The leaves were photographed 5 dpi. Vertical bars indicate the standard error for at least three independent experiments. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (p < 0.05). Scale bar shows 2 cm.
Importantly, Pcal is pathogenic on monocot plants, such as oat, as well as cruciferous species (Ishiyama et al., 2013). Oat plants inoculated with the trpA mutant also showed reduced symptoms as well as bacterial populations for both dip- and syringe-inoculation methods (Figures 2E–H).
TrpA Contributes to Multiplication on the Leaf Surface and Disease
Globally, the microbiota on plant leaf surfaces is estimated to around 1026 bacterial cells (Lindow and Brandl, 2003). The plant surface is generally considered to be suboptimal for microbes (Lindow and Brandl, 2003). Because the trpA mutant is auxotrophic, and does not grow on minimal medium (MG medium) (Sakata et al., 2019), the trpA mutant is likely to be affected by other plant microbiomes, which compete for limited nutrients. Thus, to rule out the possibility that the trpA mutant showed reduced virulence because of a microbiome effect, we conducted flood-inoculation onto cabbage, which can assay the plant-bacterial interaction with a sterility test (Ishiga et al., 2011), and investigated epiphytic bacterial populations. The trpA mutant showed reduced population via flood-inoculation (Figure 3).
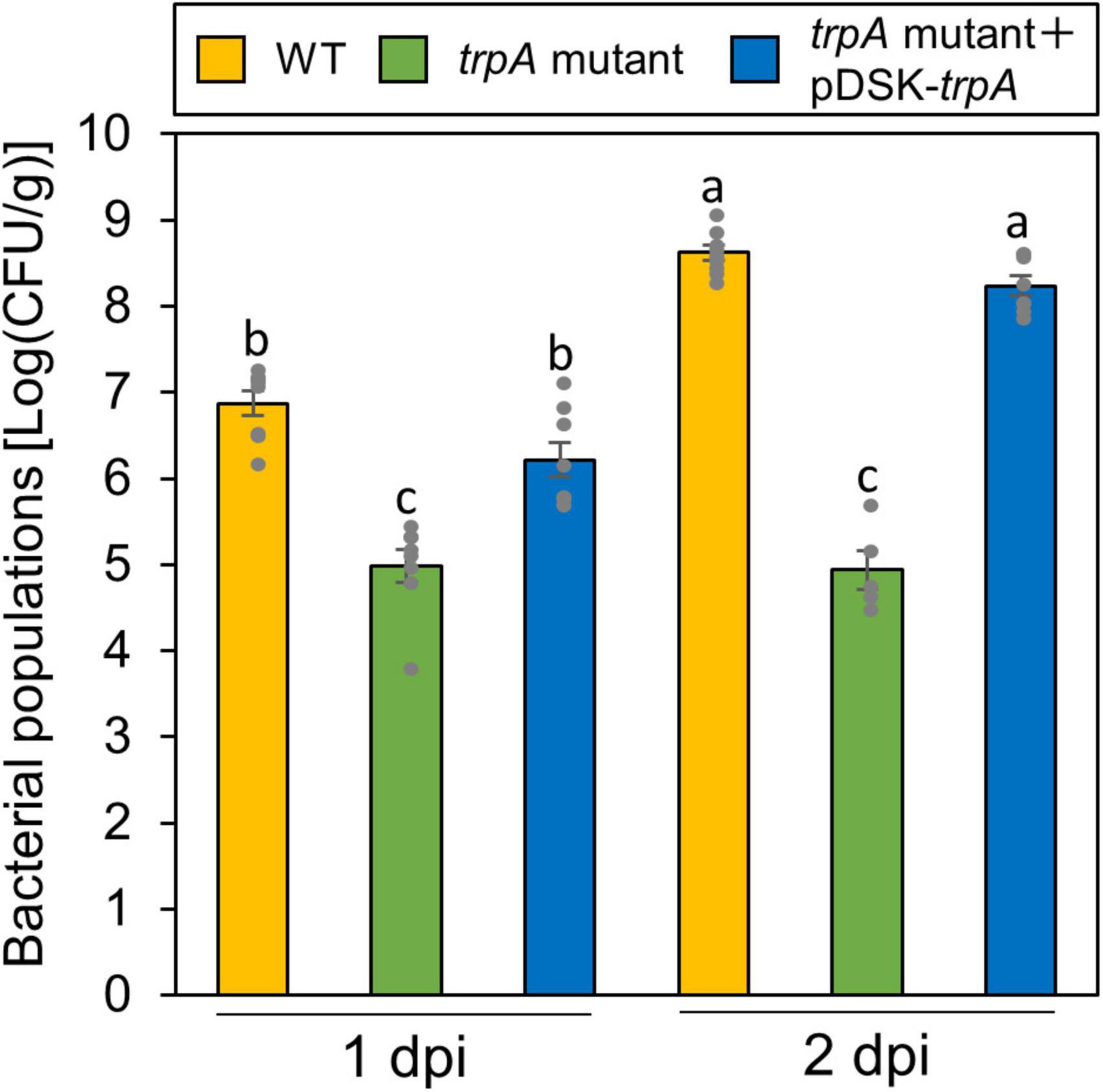
Figure 3. Epiphytic bacterial populations of Pseudomonas cannabina pv. alisalnesis KB211 WT and the trpA mutant on cabbage after flood-inoculation. Cabbage seedlings were flood-inoculated with 1 × 108 CFU/ml of WT, the trpA mutant, and the trpA mutant complemented with pDSK-trpA containing 0.025% SilwetL-77. Epiphytic bacterial populations were determined by washing the leaves with washing buffer (50 mM PBS buffer) in an ultrasonic bath for 7 min, and then by plating dilutions to KB agar medium. Bacterial concentrations on the leaf surface were evaluated at 1 and 2 dpi. Vertical bars indicate the standard error for at least five biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (p < 0.05).
Tryptophan Is an Essential Growth Resource
To investigate whether exogenous tryptophan application restores this virulence impairment, we first measured bacterial growth in MG medium with tryptophan. The trpA mutant was growth deficient in MG medium (Figure 4A). Co-incubation with only 10 μM tryptophan restored trpA mutant bacterial growth to the WT level in vitro (Figure 4A).
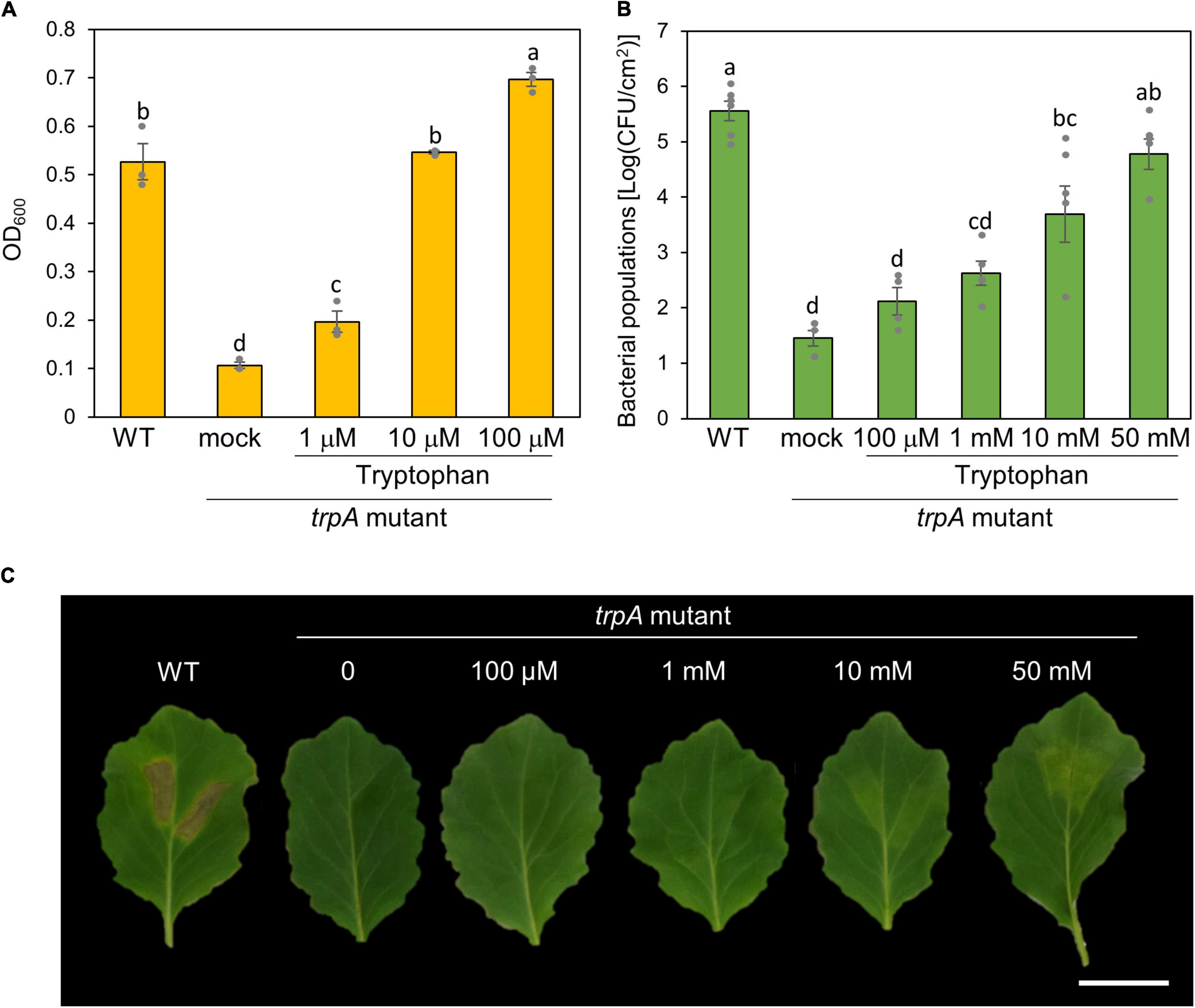
Figure 4. Effect of exogenous tryptophan application on trpA mutant growth. (A) Growth of Pseudomonas cannabina pv. alisalnesis KB211 WT and the trpA mutant in MG medium. WT and the trpA mutant were adjusted to an OD600 of 0.1 in MG medium. Additionally, the trpA mutant was co-incubated with tryptophan; 1, 10, and 100 μM, respectively. Then, cultures were incubated with shaking at 28°C. Bacterial populations were quantified at 24 h. (B) Bacterial populations in cabbage syringe-inoculated with WT and the trpA mutant. Cabbage plants were syringe inoculated with 5 × 105 CFU/ml of WT and the trpA mutant. The trpA mutant was co-inoculated with tryptophan; 100 μM, 1 mM, 10 mM, and 50 mM, respectively. Bacterial concentrations in the plant leaves were evaluated at 5 dpi. (C) Disease symptoms on cabbage co-inoculated with tryptophan. Cabbage plants were syringe-inoculated with 5 × 105 CFU/ml of WT and the trpA mutant. The trpA mutant was co-inoculated with 100 μM, 1, 10, and 50 mM tryptophan, respectively. The leaves were photographed at 5 dpi. Scale bar shows 2 cm.
We further tested whether exogenous tryptophan application restores trpA mutant virulence on cabbage. Co-inoculation of the trpA mutant with tryptophan restored virulence in a dose dependent manner (Figures 4B,C). Although only 10 μM tryptophan restored trpA mutant bacterial growth to the WT level in vitro, at least 50 mM tryptophan was required for the trpA mutant to multiply to a similar level as WT in cabbage by 5 dpi (Figure 4B). Although bacterial multiplications were recovered, 50 mM exogenous application could not fully recover disease symptoms (Figure 4C and Supplementary Figure 2A). To further investigate symptom development and bacterial populations, we conducted inoculation with or without tryptophan (50 mM) in a time-dependent manner. Exogenous tryptophan application partially recovered trpA mutant multiplication at 1 and 3 dpi (Supplementary Figure 2B).
trpA Mutant Virulence Genes Show Reduced Expression During Infection
Ten μM exogenous tryptophan application recovered bacterial growth in vitro, while at least 50 mM of tryptophan was required for growth recovery in planta (Figures 4A,B). Therefore, we assumed that tryptophan is not only a nutrient but also involved in regulating bacterial virulence gene expression. We firstly examined Pcal gene expression profiles in culture medium. As we expected, T3Es genes (including avrPto, hopM1, and avrE1) and COR biosynthesis related genes (including cmaA, cfl, and corR) showed reduced expression in the trpA mutant compared to WT (Supplementary Figures 3A–F). trpB showed significantly greater expression in the trpA mutant (Supplementary Figure 3G).
Then, we next investigated these gene expression profiles during infection. The trpA mutant showed significant bacterial population reduction compared to the WT within 1 dpi. To determine the time point at which the WT and trpA mutant had the same bacterial populations, we syringe-inoculated these into cabbage at 5 × 107 CFU/ml and examined earlier time points including 3, 6, and 9 hpi. trpA mutant bacterial populations were decreased compared to WT at 6 hpi, and this defect was greater at 9 hpi (Supplementary Figure 4). Therefore, we conducted syringe-inoculation into cabbage with these strains at 5 × 107 CFU/ml and extracted the total RNAs at 3 and 6 hpi. T3Es genes (Figures 5A–C) and COR biosynthesis related genes (Figures 5D–F) exhibited reduced expression in the trpA mutant compared to WT. We also investigated tryptophan biosynthesis gene expression profiles. trpB, trpE, trpG, and trpI showed greater expression in the trpA mutant than in the WT (Figures 5G–J).
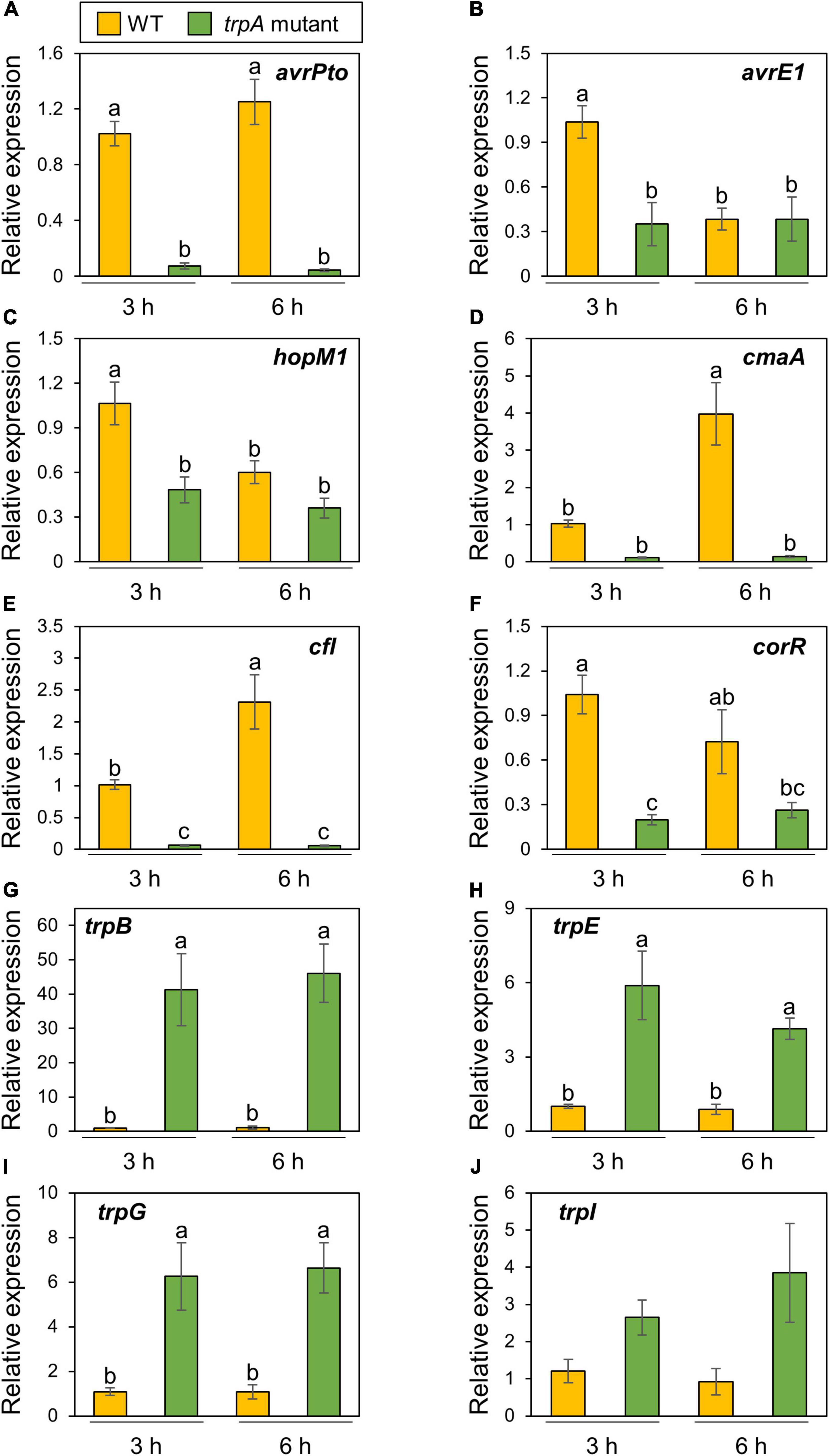
Figure 5. Expression profiles of bacterial virulence genes and tryptophan biosynthesis genes during Pseudomonas cannabina pv. alisalnesis KB211 WT and trpA mutant infection. Cabbage plants were syringe-inoculated with 5 × 107 CFU/ml of WT and the trpA mutant and total RNAs were collected 3 and 6 h after inoculation. Expression profiles of type three effector (T3Es) genes [including avrPto (A), avrE1 (B), and hopM1 (C)] and COR biosynthesis related genes [including cmaA (D), cfl (E), and corR (F)] were investigated. Additionally, expression profiles of tryptophan biosynthesis related genes [including trpB (G), trpE (H), trpG (I), and trpI (J)], were also investigated. Total RNA was extracted for use in real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) with gene-specific primer sets (Supplementary Table 2). Expression was normalized using oprE and recA. Vertical bars indicate the standard error for at least six biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (p < 0.05).
TrpA Contributes to Disease on Multiple Host Plants
Since the trpA mutant showed no pathogenicity on both cabbage and oat, we hypothesized that TrpA was required for pathogenesis on multiple host plants. We therefore inoculated the WT and trpA mutant onto several host plants, including Japanese radish, broccoli, and Chinese cabbage, and measured bacterial populations. The trpA mutant showed reduced symptom development and bacterial multiplication in all host plants (Figure 6 and Supplementary Figure 5).
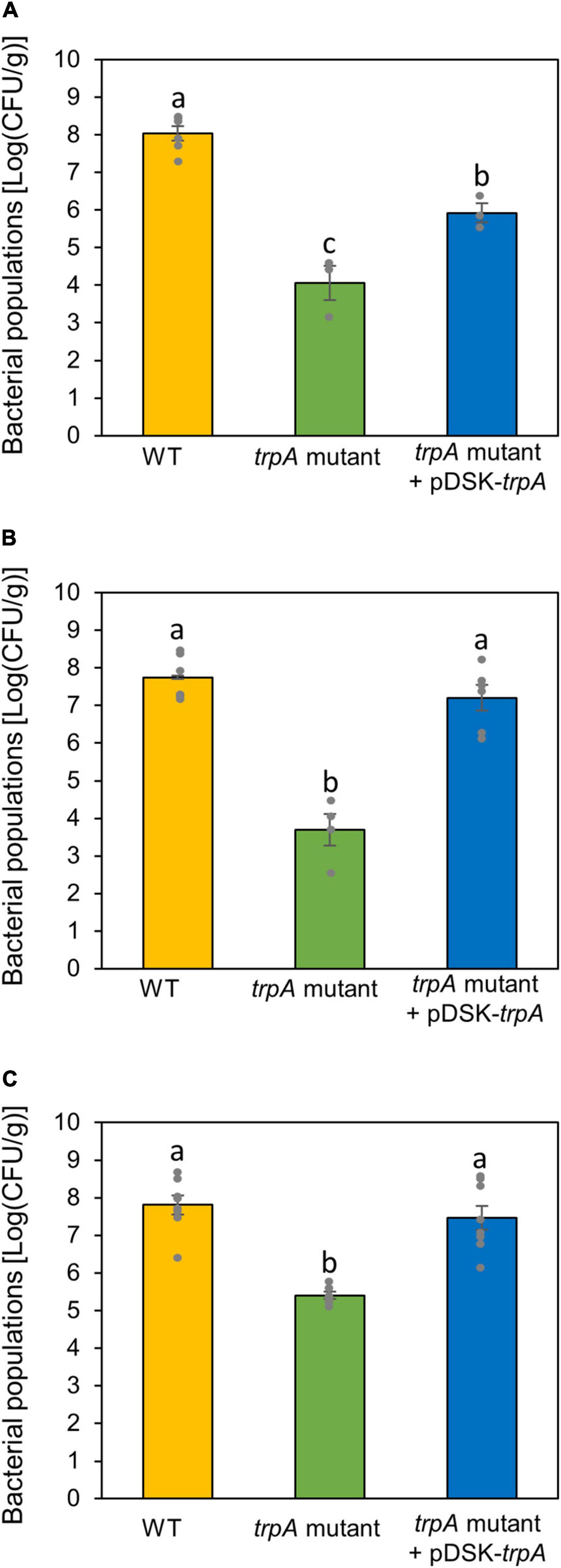
Figure 6. Bacterial populations of Pseudomonas cannabina pv. alisalnesis KB211 WT and the trpA mutant in Japanese radish (A), broccoli (B), and Chinese cabbage (C). All plants were spray inoculated with 5 × 107 CFU/ml of inoculum containing 0.025% Silwet L-77. Bacterial concentrations in the plant leaves were evaluated at 5 dpi. Vertical bars indicate the standard error for at least three biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (p < 0.05).
Discussion
Plant pathogens deploy multiple virulence factors for successful infection. This study was performed to understand the role of TrpA in Pcal virulence. Overall, this study showed that TrpA contributes to Pcal pathogenesis and is important for successful infection in multiple host plants. Importantly, trpA disruption leads to downregulation of bacterial virulence genes, including T3Es and COR, and to upregulation of tryptophan biosynthesis related genes, demonstrating the importance of tryptophan biosynthesis in bacterial pathogenesis.
Among six amino acid metabolism mutants we identified (Sakata et al., 2019), the trpA mutant showed no pathogenesis (Figure 1). These results indicate that tryptophan biosynthesis is critical for Pcal virulence. Schreiber et al. (2012) conducted a high-throughput forward genetic screen and demonstrated the nutritional requirements of Psm ES4326 (Pcal ES4326) in Arabidopsis. Amino acid auxotrophs showed dramatically reduced bacterial populations following spray inoculation (Schreiber et al., 2012), suggesting that free amino acids are limited on leaf surfaces and are important for growth. Together, these results underscore the importance of amino acid metabolism in plant bacterial pathogen virulence.
Our data showed that TrpA contributes to multiplication on the leaf surface and in the apoplast, causing disease (Figures 2, 3). Helmann et al. (2019) conducted RB-TnSeq to define the fitness contributions of Pss B728a genes. Consistent with our results, they demonstrated that genes within the tryptophan biosynthetic pathway had the greatest effect on fitness both on the leaf surface and in the apoplast (Helmann et al., 2019). Generally, the plant surface is considered suboptimal for microbes, which provides limited nutrient resources to bacterial colonists (Lindow and Brandl, 2003). Morgan and Tukey (1964) demonstrated that tryptophan was not detected in several plant species among 20 amino acids detected in plant leachates, suggesting a pressing need for its synthesis by bacterial colonists. Moreover, the ability of Pcal to synthesize tryptophan strongly influences bacterial proliferation in multiple host plants (Figure 6). Together, tryptophan biosynthesis is an essential process for successful Pcal infection on multiple host plants.
Bacterial multiplication of the trpA mutant was restored to WT levels with only 10 μM tryptophan in vitro (Figure 4A). Exogenous tryptophan application rescued the trpA mutant bacterial multiplication defect in planta, but at least 50 mM tryptophan was required (Figure 4B). Therefore, we assumed that the reduced virulence of the trpA mutant is involved in plant-bacterial interactions, as well as lack of nutrition. Transcripts of T3Es and COR related genes were reduced during trpA mutant infection compared to WT (Figure 5). Conversely, tryptophan biosynthesis related genes showed greater expression during trpA mutant infection than WT (Figure 5). The tryptophan biosynthesis pathway and genes have been proposed to be highly conserved in proteobacteria (Bae et al., 1989; Essar et al., 1990; Gussin, 2004). In P. syringae, the trpBA operon is regulated by TrpI, a LysR-type transcriptional activator whose gene is transcribed divergently (Auerbach et al., 1993). When tryptophan concentration is low, indoleglycerol phosphate (InGP) accumulates and TrpI assumes its active conformation, where it is able to bind at two operator sites in the trpI-trpBA intergenic region (Chang and Crawford, 1990; Merino et al., 2008). Our results demonstrated that these genes showed greater expression in the trpA mutant than in the WT (Figure 5). Taken together, our results suggest that trpA mutation directly affects tryptophan biosynthesis genes, and indirectly affects virulence factor related genes. One possible explanation how mutation in trpA affects T3Es and COR related genes is trade-off between nutrition acquisition and virulence. Several studies have demonstrated that nutrient assimilation during host infection is critical for pathogenesis (Brown et al., 2008; Eisenreich et al., 2010; Barbier et al., 2011; Schoen et al., 2014). In the plant pathogen Ralstonia solanacearum, exopolysaccharide (EPS) (which are critical for disease symptom production) biosynthesis and secretion represent a significant energetic cost for the pathogen, resulting in reduced bacterial growth (Peyraud et al., 2016). Furthermore, the animal pathogen Salmonella also balances the trade-off between fast growth and T3SS production (Ackermann et al., 2008; Diard et al., 2013). Taken together, the trade-off between growth and virulence should be considered in plant and bacterial pathogen interactions. Although further investigation will be necessary to understand how mutation in trpA affect the bacterial virulence factors, trade-off between virulence factor production and bacterial proliferation with nutrients such as tryptophan might be present in Pcal infection processes.
In conclusion, these data strongly suggest that TrpA contributes to bacterial multiplication on the leaf surface and in the apoplast, and contributes to disease in multiple host plants. Furthermore, trpA mutation leads to downregulation of virulence genes related to T3Es and COR. Since most amino acids are apparently present at relatively low apoplastic concentrations (O’Leary et al., 2016), it is expected that lack of amino acid metabolites function causes reduced virulence in addition to growth defects. Our findings suggest that amino acid metabolites can be targeted for developing new disease control strategies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/s/753ec4995e4086b047da.
Author Contributions
NS and YI designed the experiments and wrote the manuscript. All authors performed the experiments.
Funding
This work was supported, in part, by Japan Society for the Promotion of Science (JSPS), Grant Number: 19K06045 (YI), by Sustainable Food Security Research Project in the form of an operational grant from the National University Corporation (YI), and by Japan Science and technology Agency (JST), Exploratory Research for Advanced Technology (ERATO) NOMURA Microbial Community Control Project, Grant Number: JPMJER1502 (YI).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Christina Baker for editing the manuscript. Pcal was kindly given from the Nagano vegetable and ornamental crops experiment station, Nagano, Japan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.659734/full#supplementary-material
References
Ackermann, M., Stecher, B., Freed, N. E., Songhet, P., Hardt, W.-D., and Doebeli, M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990. doi: 10.1038/nature07067
Auerbach, S., Gao, J., and Gussin, G. N. (1993). Nucleotide sequences of the trpI, trpB, and trpA genes of Pseudomonas syringae: positive control unique to fluorescent pseudomonads. Gene 123, 25–32. doi: 10.1016/0378-1119(93)90534-a
Bae, Y. M., Holmgren, E., and Crawford, I. P. (1989). Rhizobium meliloti anthranilate synthase gene: cloning, sequence, and expression in Escherichia coli. J. Bacteriol. 171, 3471–3478. doi: 10.1128/jb.171.6.3471-3478.1989
Barbier, T., Nicolas, C., and Letesson, J. J. (2011). Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 585, 2929–2934. doi: 10.1016/j.febslet.2011.08.011
Boch, J., Joardar, V., Gao, L., Robertson, T. L., Lim, M., and Kunkel, B. N. (2002). Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44, 73–88. doi: 10.1046/j.1365-2958.2002.02877.x
Brown, S. A., Palmer, K. L., and Whiteley, M. (2008). Revisiting the host as a growth medium. Nat. Rev. Microbiol. 6, 657–666. doi: 10.1038/nrmicro1955
Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 100, 10181–10186. doi: 10.1073/pnas.1731982100
Bull, C. T., Manceau, C., Lydon, J., Kong, H., Vinatzer, B. A., and Fischer-Le Saux, M. (2010). Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Šutič & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Syst. Appl. Microbiol. 33, 105–115. doi: 10.1016/j.syapm.2010.02.001
Chang, M., and Crawford, I. P. (1990). The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 18, 979–988. doi: 10.1093/nar/18.4.979
Cintas, N. A., Koike, S. T., and Bull, C. T. (2002). A new pathovar, Pseudomonas syringae pv. alisalensis pv. nov., proposed for the causal agent of bacterial blight of broccoli and broccoli raab. Plant Dis. 86, 992–998. doi: 10.1094/PDIS.2002.86.9.992
Diard, M., Garcia, V., Maier, L., Remus-Emsermann, M. N. P., Regoes, R. R., Ackermann, M., et al. (2013). Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494, 353–356. doi: 10.1038/nature11913
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. doi: 10.1038/nrmicro2351
Essar, D. W., Eberly, L., Han, C. Y., and Crawford, I. P. (1990). DNA sequences and characterization of four early genes of the tryptophan pathway in Pseudomonas aeruginosa. J. Bacteriol. 172, 853–866. doi: 10.1128/jb.172.2.853-866.1990
Feil, H., Feil, W. S., Chain, P., Larimer, F., DiBartolo, G., Copeland, A., et al. (2005). Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 102, 11064–11069. doi: 10.1073/pnas.0504930102
Gussin, G. N. (2004). Activation of transcription initiation and regulation of tryptophan biosynthesis in fluorescent Pseudomonads. Pseudomonas 3, 293–322. doi: 10.1007/978-1-4419-9088-4_11
Helmann, T. C., Deutschbauer, A. M., and Lindow, S. E. (2019). Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. U.S.A. 116, 18900–18910. doi: 10.1073/pnas.1908858116
Ishiga, Y., and Ichinose, Y. (2016). Pseudomonas syringae pv. tomato OxyR is required for virulence in tomato and Arabidopsis. Mol. Plant Microbe Interact. 29, 119–131. doi: 10.1094/MPMI-09-15-0204-R
Ishiga, Y., Ishiga, T., Uppalapati, S. R., and Mysore, K. S. (2011). Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7:32. doi: 10.1186/1746-4811-7-32
Ishiyama, Y., Yamagishi, N., Ogiso, H., Fujinaga, M., and Takikawa, Y. (2013). Bacterial brown spot on Avena storigosa schereb. caused by Pseudomonas syringae pv. alisalensis. J. Gen. Plant Pathol. 79, 155–157. doi: 10.1007/s10327-013-0427-9
Keane, P. J., Kerr, A., and New, P. B. (1970). Crown gall of stone fruit ii. identification and nomenclature of agrobacterium isolates. Aust. J. Bio. Sci. 23, 585–596. doi: 10.1071/bi9700585
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307.
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/aem.69.4.1875-1883.2003
Merino, E., Jensen, R. A., and Yanofsky, C. (2008). Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 11, 78–86. doi: 10.1016/j.mib.2008.02.005
Morgan, J. V., and Tukey, H. B. (1964). Characterization of leachate from plant foliage. Plant Physiol. 39, 590–593. doi: 10.1104/pp.39.4.590
O’Leary, B. M., Neale, H. C., and Geilfus, C. M. (2016). Early changes in apoplast composition associated with defense and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae pv. phaseolicola. Plant Cell Environ. 39, 2172–2184. doi: 10.1111/pce.12770
Palmer, D. A., and Bender, C. L. (1993). Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl. Environ. Microbiol. 59, 1619–1626. doi: 10.1128/AEM.59.5.1619-1626.1993
Peyraud, R., Cottret, L., Marmiesse, L., Gouzy, J., and Genin, S. (2016). A resource allocation trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog. 12:e1005939. doi: 10.1371/journal.ppat.1005939
Rico, A., and Preston, G. M. (2008). Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant. Microbe. Interact. 21, 269–282. doi: 10.1094/MPMI-21-2-0269
Sakata, N., Ishiga, T., Masuo, S., Hashimoto, Y., and Ishiga, Y. (2021). Coronatine contributes to Pseudomonas cannabina pv. alisalensis virulence by overcoming both stomatal and apoplastic defenses in dicot and monocot plants. Mol. Plant Microbe Interact. doi: 10.1094/MPMI-09-20-0261-R [Epub ahead of print].
Sakata, N., Ishiga, T., Saito, H., Nguyen, V. T., and Ishiga, Y. (2019). Transposon mutagenesis reveals Pseudomonas cannabina pv. alisalensis optimizes its virulence factors for pathogenicity on different hosts. PeerJ. 7:e7698. doi: 10.7717/peerj.7698
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold spring harbor, NY: Cold spring harbor laboratory press.
Sawada, T., Eguchi, M., Asaki, S., Kashiwagi, R., Shimomura, K., Taguchi, F., et al. (2018). MexEF-OprN multidrug efflux pump transporter negatively controls N-acyl-homoserine lactone accumulation in Pseudomonas syringae pv. tabaci 6605. Mol. Genet. Genomics 293, 907–917. doi: 10.1007/s00438-018-1430-9
Schoen, C., Kischkies, L., Elias, J., and Ampattu, B. J. (2014). Metabolism and virulence in Neisseria meningitidis. Front. Cell. Infect. Microbiol. 4:114. doi: 10.3389/fcimb.2014.00114
Schreiber, K. J., Ye, D., Fich, E., Jian, A., Lo, T., and Desveaux, D. (2012). A high-throughput forward genetic screen identifies genes required for virulence of Pseudomonas syringae pv. maculicola ES4326 on Arabidopsis. PLoS One 7:e41461. doi: 10.1371/journal.pone.0041461
Takikawa, Y., and Takahashi, F. (2014). Bacterial leaf spot and blight of crucifer plants (Brassicaceae) caused by Pseudomonas syringae pv. maculicola and P. cannabina pv. alisalensis. J. Gen. Plant Pathol. 80, 466–474. doi: 10.1007/s10327-014-0540-4
Whalen, M. C., Innes, R. W., Bent, A. F., and Staskawicz, B. J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. doi: 10.1105/tpc.3.1.49
Keywords: Pseudomonas cannabina pv. alisalensis, tryptophan, type three secretion system, coronatine, cabbage, oat
Citation: Sakata N, Ishiga T and Ishiga Y (2021) Pseudmonas cannabina pv. alisalensis TrpA Is Required for Virulence in Multiple Host Plants. Front. Microbiol. 12:659734. doi: 10.3389/fmicb.2021.659734
Received: 28 January 2021; Accepted: 22 March 2021;
Published: 20 April 2021.
Edited by:
Carmen R. Beuzón, University of Malaga, SpainReviewed by:
Jose Sebastian Rufian, Chinese Academy of Sciences (CAS), ChinaAmanda May Vivian Brown, Texas Tech University, United States
Copyright © 2021 Sakata, Ishiga and Ishiga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuhiro Ishiga, aXNoaWdhLnlhc3VoaXJvLmttQHUudHN1a3ViYS5hYy5qcA==
 Nanami Sakata
Nanami Sakata Takako Ishiga
Takako Ishiga