- National Botulism Laboratory, Enteric Diseases Laboratory Branch, Division of Foodborne, Waterborne and Environmental Diseases, National Center for Emerging Zoonotic and Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
Three cases of infant botulism were reported in a small Colorado town between 1981 and 1984. The first two cases occurred in 1981, 6 months apart, and the third case occurred in 1984. Clostridium botulinum type A was isolated from stool of all three case patients and from environmental samples of the patient’s homes. An epidemiological investigation and follow-up study were conducted from 1981 to 1986 and concluded the cases were likely related. In this study, we sought to determine whether the C. botulinum type A clinical isolates were related to each other and to isolates obtained from environmental samples. We performed whole genome sequencing (WGS) for 17 isolates associated with this potential cluster of infant botulism. Fifteen isolates were confirmed to be C. botulinum type A(B) and contained botulinum toxin gene subtypes A1 and B5 by WGS; these strains formed a monophyletic cluster in a phylogeny and were considered closely related to each other (0–18 high-quality single-nucleotide polymorphisms), but distinct from other C. botulinum type A(B) in Colorado and elsewhere in the United States. Results of our study suggest that the three infant botulism cases could have represented a cluster due to a C. botulinum type A(B) strain present in the environment.
Introduction
Clostridium botulinum are spore-forming Gram-positive bacteria that produce a potent neurotoxin responsible for the severe paralytic disease botulism. In the United States, there are on average 153 cases of botulism each year, and the majority (110 cases per year) are attributed to infant botulism.1 C. botulinum strains produce seven well-characterized botulinum toxin types (A-G), although human illness in the United States is primarily caused by toxin types A and B, followed by E and F. Some C. botulinum strains, denominated bivalent, can produce more than one toxin type (Sobel, 2005; Raphael et al., 2014). C. botulinum is a diverse species and has historically been characterized into four metabolic groups designated Groups I, II, III, and IV, based on biochemical and microbiological traits (Raphael et al., 2014; Smith et al., 2015; Williamson et al., 2016). Group I consists of proteolytic strains that produce botulinum toxin types A, B, and F (Smith et al., 2015; Williamson et al., 2016). Group I strains are most often associated with human clinical cases and are the most common cause of botulism cases in the United States. Group I also includes bivalent strains (Ab, Af, Ba, and Bf), and strains designated type A(B), which carry types A and B botulinum neurotoxin (bont) genes, but only produce type A toxin (Franciosa et al., 1994; Hill et al., 2007). These C. botulinum A(B) strains have been discovered to be common among the US botulism cases (Raphael et al., 2014; Halpin et al., 2017).
The disease botulism can be classified into four natural-occurring forms: foodborne botulism (through ingestion of foods contaminated with botulinum toxin), wound botulism (through spores that germinate leading to growth in wounds, and toxin production in situ), infant botulism (through intestinal colonization in infants less than 1 year old), and adult intestinal colonization (through intestinal colonization of patients older than 1 year; Sobel, 2005). Infant botulism is the most common type of botulism in the United States, and it occurs in persons under 1 year old, through ingestion of C. botulinum spores that germinate in the intestines, leading to growth and production of botulinum toxin (Sobel, 2005). Spores of C. botulinum are commonly found in soil, and therefore, it has been proposed as the most probable source of spores in infant botulism cases (Koepke et al., 2008).
Here, we present a follow-up study, to characterize C. botulinum strains isolated from clinical and environmental samples associated with the investigation conducted by Istre et al. (1986), these cases were of interest since transmission of botulism is not considered communicable (does transmit from person to person) and the incidence of infant botulism is extremely low. The first two cases occurred in 1981, 6 months apart, while the third case occurred in September of 1984. All three infants lived in the same neighborhood, within 800 m of each other. The three cases were confirmed by detection of botulinum neurotoxin type A in patient stools using the mouse bioassay (Istre et al., 1986). C. botulinum type A was isolated from all three of the patient’s stools. C. botulinum type A was also present in cultures of environmental samples taken in 1982 and 1985, following the second and third cases. Specifically, C. botulinum type A was found in soil and vacuum dust samples taken from the homes of the second and third cases and also from the crib of the second case patient (Istre et al., 1986). C. botulinum type A was also found in soil of the first case patient’s home after the fact, but no house dust samples were obtained as the family moved prior to the onset of the second case (Istre et al., 1986). This investigation prompted a study in 1986, conducted by CDC, to test additional environmental samples from homes of infants in the community who were considered healthy control infants, as they were presumed not to be experiencing symptoms of botulism. Stool samples from these healthy infants were also collected. Ten stool samples, 22 soil samples, and 13 vacuum dust samples were collected: 91% (20/22) of soil samples from the homes of control infants yielded C. botulinum type A, and 77% (10/13) of vacuum dust samples from the homes of control infants also yielded C. botulinum type A. C. botulinum type A was not identified in the stool of any control infants; however, C. botulinum type B was found in the stools of two control infants (unpublished data). In this study, we employed a whole genome sequencing (WGS) approach to further characterize the clinical and environmental isolates associated with this investigation and to determine (1) whether the three cases represented a cluster of infant botulism cases and (2) whether the C. botulinum type A clinical isolates were related to the isolates found in environmental samples.
Materials and Methods
Strains
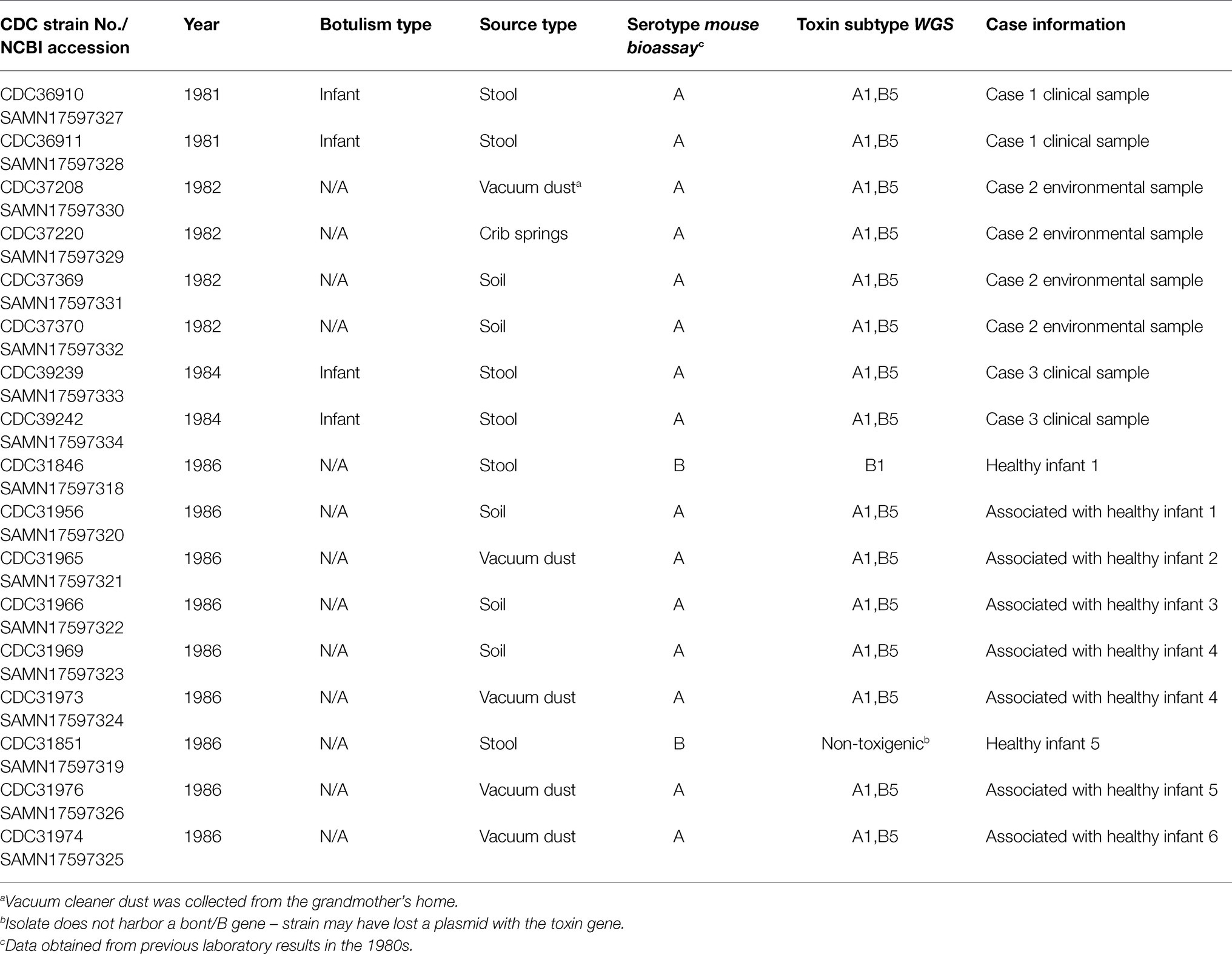
Seventeen C. botulinum isolates were selected from the CDC strain collection for further characterization (refer to Table 1): four type A isolates from patient stool samples associated with cases one and three (unfortunately, the isolate from case 2 was not available in CDC’s strain collection), two type B isolates from stools of two healthy infants, and 11 type A isolates from environmental samples (from cases two and three and the homes of six healthy infants). Eight of the isolates were from the original investigation by Istre et al., 1986, and nine isolates were from the unpublished study in 1986. Additional isolates for geographical perspective were also included in the study, based on availability of WGS data (refer to Supplementary Table 1). Strains were grown at 35°C under anaerobic conditions in Trypticase Peptone Yeast Extract (Remel, Lenexa, KS). All strains were coded prior to the study to comply with a human subjects research protocol. DNA was extracted using a modified MasterPure DNA extraction protocol (Epicenter, Madison, WI; Halpin et al., 2019).
Sequencing
Isolates were sequenced on either the Ion Torrent S5 or Illumina MiniSeq. DNA libraries were prepared for WGS using the Nextera DNA Flex kit (Illumina, San Diego, CA) and sequenced on the MiniSeq (Illumina, San Diego, CA) with 2 × 150 bp chemistry. DNA libraries were also prepared using the Ion Torrent Chef (Thermo Fisher Scientific, Waltham, MA) and sequenced on the Ion Torrent S5 (Thermo Fisher Scientific, Waltham, MA) using previously established methods (Halpin et al., 2019).
Genome Assembly and Quality Filtering
Genome sequences were downloaded from NCBI or assembled from raw reads using SPAdes. SPAdes version 3.14.0 and the option --careful were used for Illumina reads, while SPAdes version 3.13.0 and the options -sc, --iontorrent, and --careful were used for Ion Torrent reads (Bankevich et al., 2012). Short contigs (<500 bp) were removed using CG-Pipeline script run_assembly_filterContigs.pl (Kislyuk et al., 2010). Sequence accession numbers are associated with NCBI Bioproject PRJNA428620; these can be found in Table 1.
Toxin Gene Location and Type Determination
bont gene subtypes were determined with CLC Genomics Workbench v10 Map to Reference feature (CLC bio, Aarhus, Denmark). Location of bont genes was determined using a command-line nucleotide blast with Blast+ version 2.9.0 (Camacho et al., 2009), and their location on either the chromosome or a plasmid was predicted using nucleotide blast of the C. botulinum database on NCBI. Seven-gene Multi-Locus Sequence Typing (MLST) types were determined by querying the C. botulinum PubMLST database3 (Jacobson et al., 2008).
WGS Analysis
A single-nucleotide polymorphism (SNP) analysis was performed using only the highest quality SNPs. An appropriate reference genome for a high-quality SNP analysis was identified by first querying our in-house genome sequence repository. These in-house sequences were then compared to additional genome sequences on NCBI, using Mashtree (Katz et al., 2019). An appropriate reference was selected based on two criteria: closeness to the study strains in the Mashtree and completeness of the reference assembly. Plasmid sequences were identified in assemblies using PLSDB (Galata et al., 2019) and removed from the reference sequence to avoid erroneous SNP counts due to plasticity of some strains. Read pairs were preprocessed with Lyve-SET version 1.1.4f scripts prior to performing SNP analyses; read pairs were first interleaved with shuffleSplitReads.pl. and then cleaned with run_assembly_trimClean.pl. with the options --min_avg_quality 24 and --nosingletons (Katz et al., 2017). Lyve-SET version 1.1.4f was used for the SNP analysis with the following settings and the cleaned reads: --allowedFlanking 5 --min_alt_frac 0.75 --min_coverage 10 --mask-phages --mask-cliffs --mapper smalt --numcpus 10 --numnodes 50 (Katz et al., 2017). Lyve-SET quality filters for SNP coverage, percent consensus, and flanking distance ensure that only the highest quality SNPs are used in the analysis and that low-quality regions susceptible to recombination and horizontal exchange are not included (Katz et al., 2017). External reference genome sequences were also placed in the Lyve-SET asm folder for WGS read simulation as needed to root the tree. Phylogenetic trees were visualized using Mega7 (Kumar et al., 2016), and SNP distance matrices were visualized using a text editor or Microsoft Excel (Microsoft, Albuquerque, NM).
Results
Toxin Gene Location and Type Determination
All 15 C. botulinum type A clinical and environmental isolates were confirmed to be A(B), bont gene subtypes A1 and B5 by WGS, and MLST type ST6. The bont genes were predicted to be on the chromosome for all C. botulinum type A(B) isolates associated with this investigation. Interestingly, the type A and type B toxin genes were located on a single 311,258 bp contig in one of the strains (CDC31973). The presence of both type A and type B toxin genes on the chromosome has been seldomly reported in C. botulinum type A(B) strains (Franciosa et al., 2009; Gonzalez-Escalona and Sharma, 2020). One C. botulinum type B isolate (CDC31846) from healthy infant stool was confirmed to harbor bont/B1 gene and was predicted to be on a plasmid. CDC31851, originally classified as C. botulinum type B by mouse bioassay in 1986, did not carry a bont gene and was determined as non-toxigenic in this study. The absence of the toxin gene in this isolate CDC31851 could have been due to loss of a plasmid. Both CDC31846 and CDC31851 were confirmed to be MLST type ST96.
WGS Analysis
The C. botulinum type A(B) study strains in Table 1 clustered together in the Mashtree and also with other unrelated C. botulinum type A(B) from the United States (Figure 1; Supplementary Table 1). However, they did not cluster with C. botulinum type A(B) isolated from other infant and foodborne botulism cases in Colorado (CDC61017, CDC64216, CDC64218, and CDC64223; Figure 1; Supplementary Table 1).
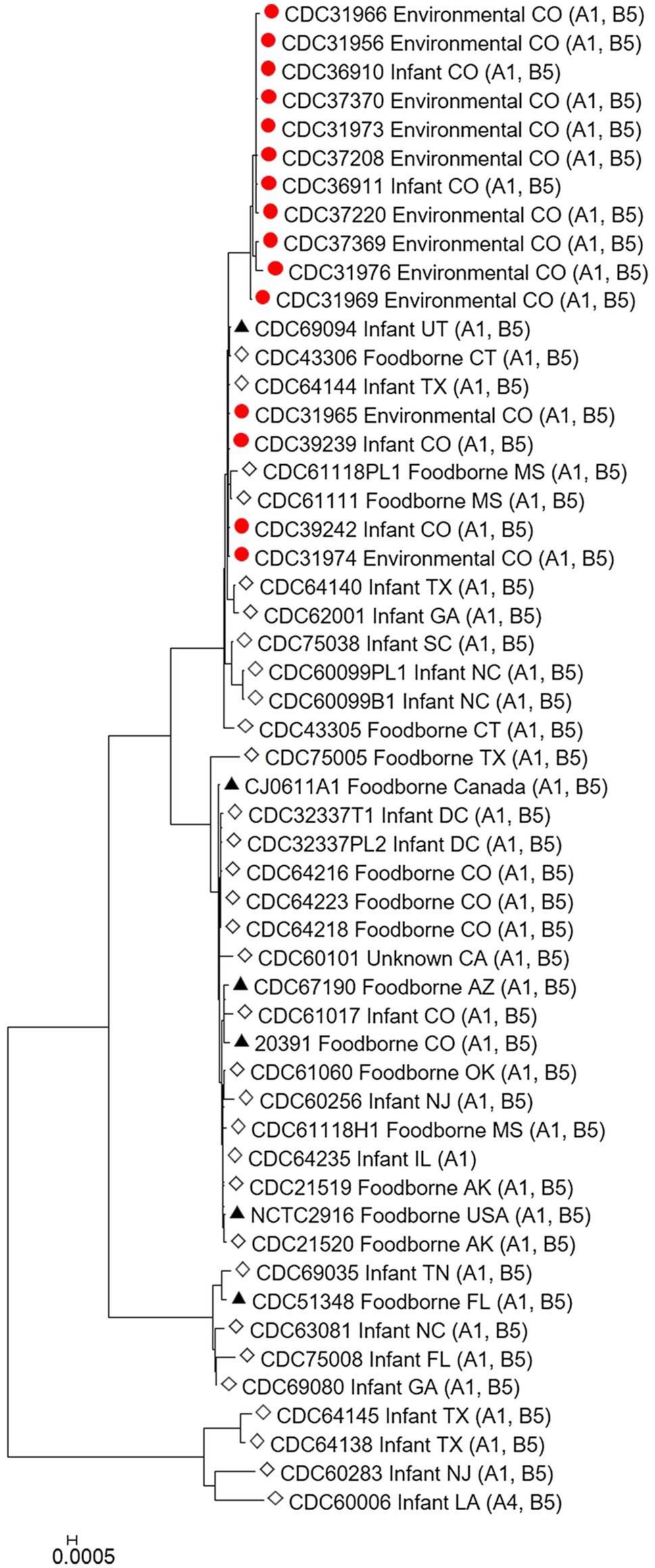
Figure 1. Neighbor-joining tree of Clostridium botulinum type A(B) isolates. Tree was drawn using Mashtree and includes study strains harboring both bont/A1 and bont/B5 genes as described in Table 1 (red circles), reference sequences from NCBI (black triangles), and other unpublished sequences from the CDC reference collection (white diamonds), as described in Supplementary Table 1. Scale bar represents the Mash distance.
The C. botulinum type A(B) study strains formed a monophyletic cluster in a Lyve-SET phylogeny and are considered closely related to each other (2–18 high-quality SNPs), but distinct from other closely related C. botulinum type A(B) from the United States (CDC43305, CDC43306, CDC69094, and CDC64144; Figures 2A,B). Interestingly, the clinical isolates (CDC36910, CDC36911, CDC39239, and CDC39242) had fewer SNP differences between isolates (5–7 high-quality SNPs), highlighting that the additional diversity (up to 18 SNP differences) in the C. botulinum type A(B) cluster is due to the environmental isolates, which were taken at different time points (see Table 1; Figures 2A,B). The two isolates recovered from healthy infant stools (C. botulinum type B CDC31846 and the non-toxigenic isolate CDC31851) were 28 SNPs different from each other and were considered unrelated (data not shown).
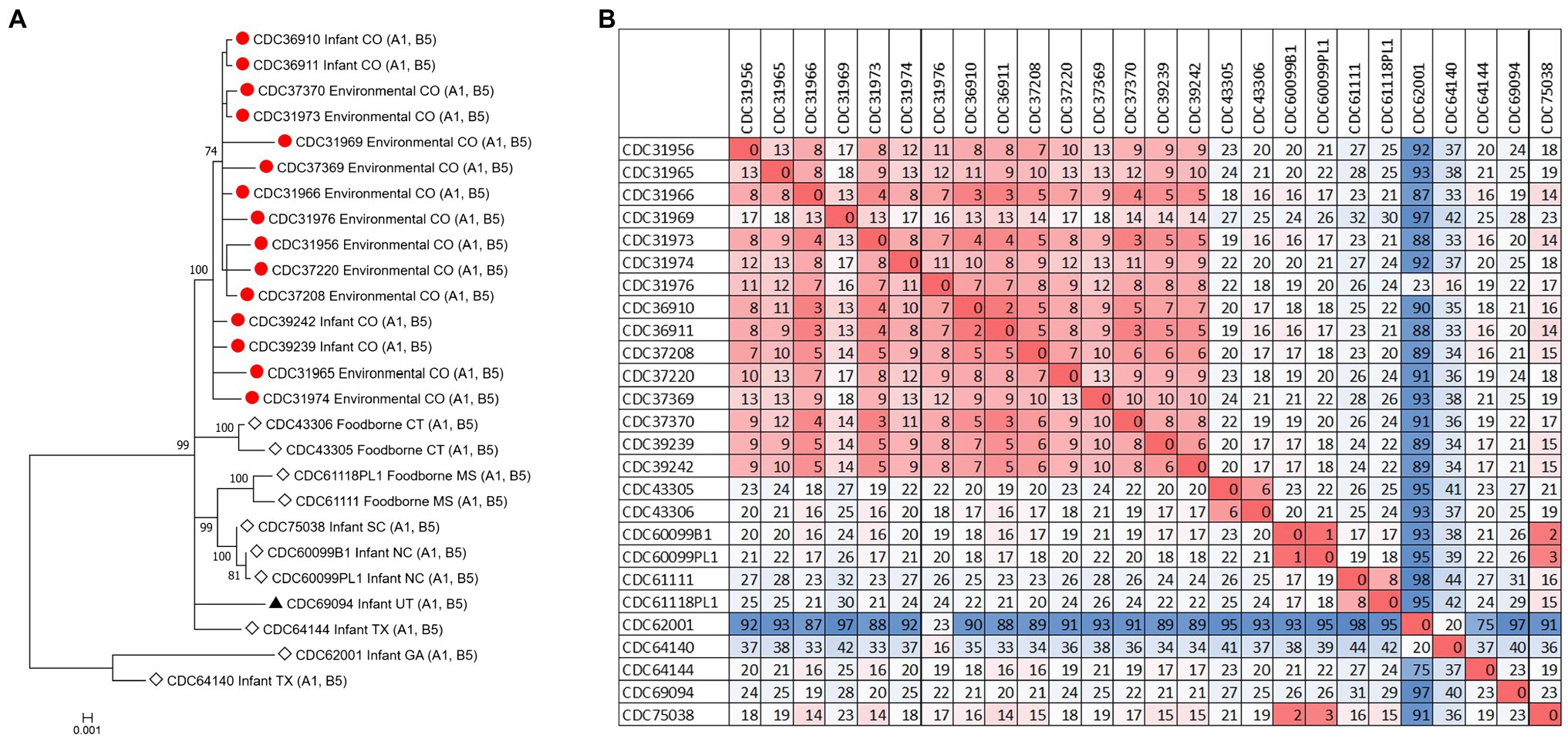
Figure 2. (A) Maximum-likelihood whole genome high-quality SNP tree of C. botulinum type A(B) isolates harboring both bont/A1 and bont/B5 genes. Tree was drawn using Lyve-SET and includes study strains as described in Table 1 (red circles), reference sequences from NCBI (black triangles), and other unpublished sequences from C. botulinum type A(B) strains from the CDC reference collection (white diamonds). Scale bar represents the number of changes per 100 sites. Bootstrap values greater than 70 are shown. (B) Distance matrix of SNP counts from Lyve-SET. The matrix was drawn in Excel using conditional formatting.
Discussion
In this study, we sought to characterize C. botulinum type A isolated from clinical specimens from infant botulism cases reported in Colorado between 1981 and 1984 (Istre et al., 1986), and other C. botulinum type A isolated from environmental samples. Unfortunately, the clinical isolate associated with the second case was not available in CDC’s strain collection and could not be included as part of this study; while this is a limitation of the study, we did include environmental isolates associated with the second case as part of this study. In a subsequent study done by CDC in 1986, stool samples were collected from 10 healthy control infants as well as soil and dust samples from their homes. The control stools were negative for C. botulinum type A, but two infants had C. botulinum type B in their stool (unpublished data). It is unclear how the two presumed healthy infants had C. botulinum type B in their stool, and to our knowledge, there are no reports of healthy infants harboring C. botulinum (Dowell et al., 1977). Additionally, C. botulinum type A was found in the environmental samples collected from homes of the healthy control infants. Our findings confirm this and add more to the investigation – the study strains also had a silent bont/B5 gene (Franciosa et al., 1994).
The source of C. botulinum spores in infant botulism cases is rarely determined, but exposure is thought to be through the environment, by ingestion of microscopic dust particles, or by consumption of honey (Dabritz et al., 2014). Furthermore, our results suggest that soil or dust may have been the probable source of spores in these three infant botulism cases, since the C. botulinum type A(B) strains isolated from environmental samples were closely related to the C. botulinum type A(B) strains isolated from stool samples. Some diversity (up to 18 SNPs) was observed among the environmental isolates, while very little diversity was observed in the clinical isolates (5 to 7 SNPs). This finding is interesting because the number of SNP differences is similar to what we would expect to see between unrelated clusters of C. botulinum A(B) in point source outbreaks (e.g., foodborne outbreaks). Raphael et al. reported SNP ranges of 3–8 between epidemiologically linked C. botulinum A(B) isolates of clinical and food pairs in six foodborne outbreaks using a reference-free SNP analysis (Raphael et al., 2014). Halpin et al. reported a range of 2–6 SNPs among C. botulinum clinical isolates and an epidemiologically linked contaminated powder infant formula isolate harboring the bont/B7 gene using a reference approach in Lyve-SET (Halpin et al., 2019). The differences observed among the environmental isolates in this study may be a result of the diversity of C. botulinum spores in the environment where they are collected.
Our study demonstrates the utility of WGS analysis and its enhanced resolution for resolving clusters of C. botulinum, in particular of C. botulinum type A(B) strains, which have been historically challenging to differentiate (Hill et al., 2007; Raphael et al., 2014; Smith et al., 2015; Halpin et al., 2017). By using Lyve-SET high-quality SNP analysis, we were able to differentiate the study isolates from other closely related C. botulinum type A(B) isolates from Colorado and the United States that were not a part of this cluster, but otherwise appear related in a simple distance-based Mashtree; for example, CDC69094, CDC43306, CDC64144, CDC61118PL1, and CDC61111 which cluster in the Mashtree with study strains were unrelated to the study strains in the Lyve-SET phylogeny (see Figures 1, 2A). Katz et al. previously showed the usefulness of Lyve-SET for investigating closely related strains within outbreaks of Listeria monocytogenes, Escherichia coli, Salmonella enterica, and Campylobacter jejuni (Katz et al., 2017). At the time of the original investigation, the mouse bioassay was used for confirming the infant botulism cases, and for identifying the botulinum toxin serotype. The mouse bioassay does not, and it is not intended to provide resolution to resolve clusters of closely related strains nor identify an unexpressed botulinum toxin gene. Additional subtyping methods, such as pulsed-field gel electrophoresis, seven-gene MLST, multi-loci variable number of tandem repeat analysis, and amplified fragment length polymorphism analysis, have been utilized to subtype C. botulinum isolates in the last 25 years, but none of these subtyping tools can provide the level of resolution needed for resolving clusters of C. botulinum type A(B) strains (Raphael et al., 2014; Smith et al., 2015; Halpin et al., 2017). For instance, our study showed that all of the type A(B) isolates shared seven-gene MLST type ST6 with other type A(B) isolates that were not part of the cluster of infant botulism cases (data not shown). Our study builds on previous work to show the utility of reference-based whole genome SNP typing to successfully resolve clusters of C. botulinum (Gonzalez-Escalona et al., 2014; Raphael et al., 2014; Williamson et al., 2016; Gonzalez-Escalona and Sharma, 2020). In conclusion, results of this study suggest that these C. botulinum type A(B) isolates obtained from infant botulism cases are genetically related to each other and to the environmental isolates obtained from soil and dust samples.
Data Availability Statement
The datasets generated for this study can be found online at the NCBI Sequence Read Archive (SRA): https://www.ncbi.nlm.nih.gov/sra/PRJNA428620.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
LG, JH, and CL wrote this manuscript and conceived the project study design. LG and JH selected the datasets. LG analyzed the sequence data and generated figures and tables. JH prepared DNA and sequenced the strains in this study. All authors contributed to the article and approved the submitted version.
Funding
Centers for Disease Control and Prevention and Center for Preparedness and Response provided support for this project. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Charles L. Hatheway and Loretta M. McCroskey for their previous work on this outbreak and contributions to the investigation. We would also like to thank our reviewers for thoughtful critique of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmicb.2021.688240/full#supplementary-material
Footnotes
References
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10:421. doi: 10.1186/1471-2105-10-421
Dabritz, H. A., Hill, K. K., Barash, J. R., Ticknor, L. O., Helma, C. H., Dover, N., et al. (2014). Molecular epidemiology of infant botulism in California and elsewhere, 1976–2010. J. Infect. Dis. 210, 1711–1722. doi: 10.1093/infdis/jiu331
Dowell, V. R. Jr., McCroskey, L. M., Hatheway, C. L., Lombard, G. L., Hughes, J. M., and Merson, M. H. (1977). Coproexamination for botulinal toxin and Clostridium botulinum. A new procedure for laboratory diagnosis of botulism. JAMA 238, 1829–1832. doi: 10.1001/jama.1977.03280180033021
Franciosa, G., Ferreira, J. L., and Hatheway, C. L. (1994). Detection of type A, B, and E botulism neurotoxin genes in Clostridium botulinum and other clostridium species by PCR: evidence of unexpressed type B toxin genes in type A toxigenic organisms. J. Clin. Microbiol. 32, 1911–1917. doi: 10.1128/JCM.32.8.1911-1917.1994
Franciosa, G., Maugliani, A., Scalfaro, C., and Aureli, P. (2009). Evidence that plasmid-borne botulinum neurotoxin type B genes are widespread among Clostridium botulinum serotype B strains. PLoS One 4:e4829. doi: 10.1371/journal.pone.0004829
Galata, V., Fehlmann, T., Backes, C., and Keller, A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202. doi: 10.1093/nar/gky1050
Gonzalez-Escalona, N., and Sharma, S. K. (2020). Closing Clostridium botulinum group I genomes using a combination of short- and long-reads. Front. Microbiol. 11:239. doi: 10.3389/fmicb.2020.00239
Gonzalez-Escalona, N., Timme, R., Raphael, B. H., Zink, D., and Sharma, S. K. (2014). Whole-genome single-nucleotide-polymorphism analysis for discrimination of Clostridium botulinum group I strains. Appl. Environ. Microbiol. 80, 2125–2132. doi: 10.1128/AEM.03934-13
Halpin, J. L., Dykes, J. K., Katz, L., Centurioni, D. A., Perry, M. J., Egan, C. T., et al. (2019). Molecular characterization of Clostridium botulinum harboring the bont/B7 gene. Foodborne Pathog. Dis. 16, 428–433. doi: 10.1089/fpd.2018.2600
Halpin, J. L., Joseph, L., Dykes, J. K., McCroskey, L., Smith, E., Toney, D., et al. (2017). Pulsotype diversity of Clostridium botulinum strains containing serotypes A and/or B genes. Foodborne Pathog. Dis. 14, 494–501. doi: 10.1089/fpd.2017.2280
Hill, K. K., Smith, T. J., Helma, C. H., Ticknor, L. O., Foley, B. T., Svensson, R. T., et al. (2007). Genetic diversity among botulinum neurotoxin-producing clostridial strains. J. Bacteriol. 189, 818–832. doi: 10.1128/JB.01180-06
Istre, G. R., Compton, R., Novotny, T., Young, J. E., Hatheway, C. L., and Hopkins, R. S. (1986). Infant botulism. Three cases in a small town. Am. J. Dis. Child. 140, 1013–1014. doi: 10.1001/archpedi.1986.02140240059027
Jacobson, M. J., Lin, G., Whittam, T. S., and Johnson, E. A. (2008). Phylogenetic analysis of Clostridium botulinum type A by multi-locus sequence typing. Microbiology 154, 2408–2415. doi: 10.1099/mic.0.2008/016915-0
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., Zhang, S., den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4:1762. doi: 10.21105/joss.01762
Katz, L. S., Griswold, T., Williams-Newkirk, A. J., Wagner, D., Petkau, A., Sieffert, C., et al. (2017). A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front. Microbiol. 8:375. doi: 10.3389/fmicb.2017.00375
Kislyuk, A. O., Katz, L. S., Agrawal, S., Hagen, M. S., Conley, A. B., Jayaraman, P., et al. (2010). A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics 26, 1819–1826. doi: 10.1093/bioinformatics/btq284
Koepke, R., Sobel, J., and Arnon, S. S. (2008). Global occurrence of infant botulism, 1976–2006. Pediatrics 122, e73–e82. doi: 10.1542/peds.2007-1827
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Raphael, B. H., Shirey, T. B., Luquez, C., and Maslanka, S. E. (2014). Distinguishing highly-related outbreak-associated Clostridium botulinum type A(B) strains. BMC Microbiol. 14:192. doi: 10.1186/1471-2180-14-192
Smith, T. J., Hill, K. K., and Raphael, B. H. (2015). Historical and current perspectives on Clostridium botulinum diversity. Res. Microbiol. 166, 290–302. doi: 10.1016/j.resmic.2014.09.007
Keywords: Clostridium botulinum, botulism, infant botulism cluster, single-nucleotide polymorphism, high-quality SNP typing
Citation: Gladney L, Halpin JL and Lúquez C (2021) Genomic Characterization of Strains From a Cluster of Infant Botulism Type A in a Small Town in Colorado, United States. Front. Microbiol. 12:688240. doi: 10.3389/fmicb.2021.688240
Edited by:
Fabrizio Anniballi, Istituto Superiore di Sanità, ItalyReviewed by:
Michael William Peck, Quadram Institute, United KingdomConcetta Scalfaro, National Institute of Health (ISS), Italy
Copyright © 2021 Gladney, Halpin and Lúquez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lori Gladney, bGdsYWRuZXlAY2RjLmdvdg==
 Lori Gladney
Lori Gladney Jessica L. Halpin
Jessica L. Halpin Carolina Lúquez
Carolina Lúquez