- 1Centre for Bioinnovation, University of the Sunshine Coast, Sippy Downs, QLD, Australia
- 2School of Science, Technology and Engineering, University of the Sunshine Coast, Sippy Downs, QLD, Australia
- 3School of Chemistry and Molecular Biosciences, The University of Queensland, St Lucia, QLD, Australia
- 4Australian Infectious Diseases Research Centre, The University of Queensland, St Lucia, QLD, Australia
- 5BioCifer Pty Ltd., Auchenflower, QLD, Australia
Introduction: Zika virus (ZIKV) is a re-emerging flavivirus that poses a significant public health threat. ZIKV exhibits a wide array of non-vector borne human transmission routes, such as sexual transmission, transplacental transmission and blood transfusion. Detection and surveillance of ZIKV is considered paramount in prevention of major outbreaks. With the majority of cases reported in low-resource locations, simple, low-cost detection methods are considered highly desirable.
Materials and Methods: Here we have developed a sensitive and specific ZIKV diagnostic using reverse transcription recombinase-aided amplification (RT-RAA) coupled with lateral flow detection (LFD) targeting a highly conserved region of the ZIKV NS1 gene.
Results: We show our rapid, isothermal-ZIKV-diagnostic (Iso-ZIKV-Dx) can detect 500 copies of synthetic ZIKV RNA/μL in under 30 min at a constant 39°C. Using simulated urine samples, we observed that Iso-ZIKV-Dx also detects as low as 34.28 RNA copies/reaction of ZIKV (MR766 strain). Specificity testing confirmed that our test does not detect any co-circulating flaviviruses (dengue, West Nile, Japanese encephalitis, Murray Valley encephalitis and yellow fever viruses) or chikungunya virus. Sample processing results show complete inactivation of ZIKV (MR766 strain) in 5 min at room temperature using our novel viral RNA sample preparation reagent. Furthermore, lateral flow strips testing demonstrates positive diagnoses in as little as 5 min in running buffer.
Discussion: Contrary to conventional RT-qPCR, our Iso-ZIKV-Dx does not require expensive machinery, specialised laboratory settings or extensively trained personnel. Pre-clinical evaluation demonstrates that our test exhibits robust, in-field capabilities without compromising sensitivity or specificity. When compared to the gold-standard RT-qPCR, our Iso-ZIKV-Dx test offers an array of applications that extend beyond diagnostics alone, including potential for surveillance and monitoring of ZIKV vector competency.
Highlights
• Development of a low-resource rapid Zika virus diagnostic test.
• Rapid one-step sample processing protocol inactivating Zika virus in 5 min.
• Test format utilising isothermal amplification coupled with lateral flow detection.
• Achieved detection of highly virulent ZIKV MR766 in under 30 min.
• Rapid Zika virus test 4 times faster than RT-qPCR.
Introduction
Zika virus (ZIKV), a member of the Flaviviridae family, was first isolated in 1947 from the serum of a rhesus monkey in Uganda (Dick et al., 1952). In 2007, an outbreak on Yap Island (Hayes, 2009) resulting in the first instance of ZIKV transmission outside of Africa and Asia was reported. ZIKV outbreaks have subsequently been reported in multiple locations including French Polynesia (Roth et al., 2014), Papua New Guinea (Chang et al., 2016), New Caledonia (Dupont-Rouzeyrol et al., 2015), and Brazil (Wen et al., 2017). Spanning throughout nearly all seven continents, ZIKV remains a priority disease by World Health Organisation (WHO) (TwistDx™, 2023b). Although predominantly transmitted via mosquitos (Aedes albopictus and aegypti) (Azar and Weaver, 2019), blood transfusion and sexual transmission have also been reported (Gregory et al., 2017). Historically ZIKV transmission was primarily observed in remote, developing countries (Paixão et al., 2016). However, in 2019, the first locally acquired cases of ZIKV transmission were reported in southern Europe (Brady and Hay, 2019), initiating the prevalence and spread of ZIKV in developed nations. Most ZIKV infections manifest in mild, flu like symptoms (Mumtaz et al., 2016). In severe cases, medical complications such as Guillain Barre (Hendel-Paterson et al., 2016) and severe thrombocytopenia (Sharp et al., 2016) have been reported. ZIKV infection mechanisms also allow for cross placental infections, resulting in microcephaly in developmental infants (Wen et al., 2017).
With no approved vaccine or therapeutics (Da Silva et al., 2018), rapid and accurate detection of ZIKV is a crucial component in predicting and monitoring potential outbreaks (Heukelbach et al., 2016). The majority of ZIKV-positive individuals, including pregnant women are also asymptomatic (Paixao et al., 2018). Crucially delayed diagnosis among pregnant women is particularly concerning due to the inherent risk of congenital abnormalities. Current ZIKV detection and diagnostic strategies utilise both nucleic acid amplification tests (NAATs) (Gourinat et al., 2015) or antibody-based detection-based techniques (Kadkhoda et al., 2017). Due to antigenic cross-reactivity between Zika antigens and other flaviviruses (Stettler et al., 2016), serology based testing such as Enzyme Linked Immunosorbent Assay (ELISA) are less favoured. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) therefore, still remains the ‘gold standard’ for detection and diagnostics among arboviruses (Dong et al., 2012). While accurate and sensitive, a draw-back for RT-qPCR is the need for specialised equipment and trained personnel that restricts these tests to centralised laboratories.
A diagnostic platform that enables in-field or point-of-care (POC) detection without the need for highly trained personnel or specialised equipment offers beneficial attributes to both clinical diagnostics and surveillance of not just ZIKV, but arboviruses alike (Ahmed et al., 2022). Isothermal NAAT’s address all these issues and have been described as potential alternatives to RT-qPCR for viruses such as Zika (Cheikh Tidiane Diagne et al., 2020). As of recent years, innovation and proliferation of various isothermal amplification tests have taken significant strides within the field of rapid diagnostics (Xue et al., 2020). Recombinase aided amplification (RAA) (Jiangsu Qitian Gene Biotechnology Co., Ltd, 2023) is a promising isothermal technique that utilises similar molecular mechanisms as Recombinase Polymerase Amplification (RPA) (TwistDx™, 2023a). For RNA viruses such as ZIKV, a reverse transcriptase enzyme is added to the RAA reaction. Comparable to RT-qPCR, reverse transcriptase-recombinase aided amplification (RT-RAA) has been shown to be both rapid (Xue et al., 2020) and clinically sensitive (Wang et al., 2020). However, NAAT test uptake for low-resource detection of disease is hampered by the lack of field-friendly sample preparation techniques, instead requiring purification of RNA using magnetic beads or column-based laboratory technologies (Pollak et al., 2022).
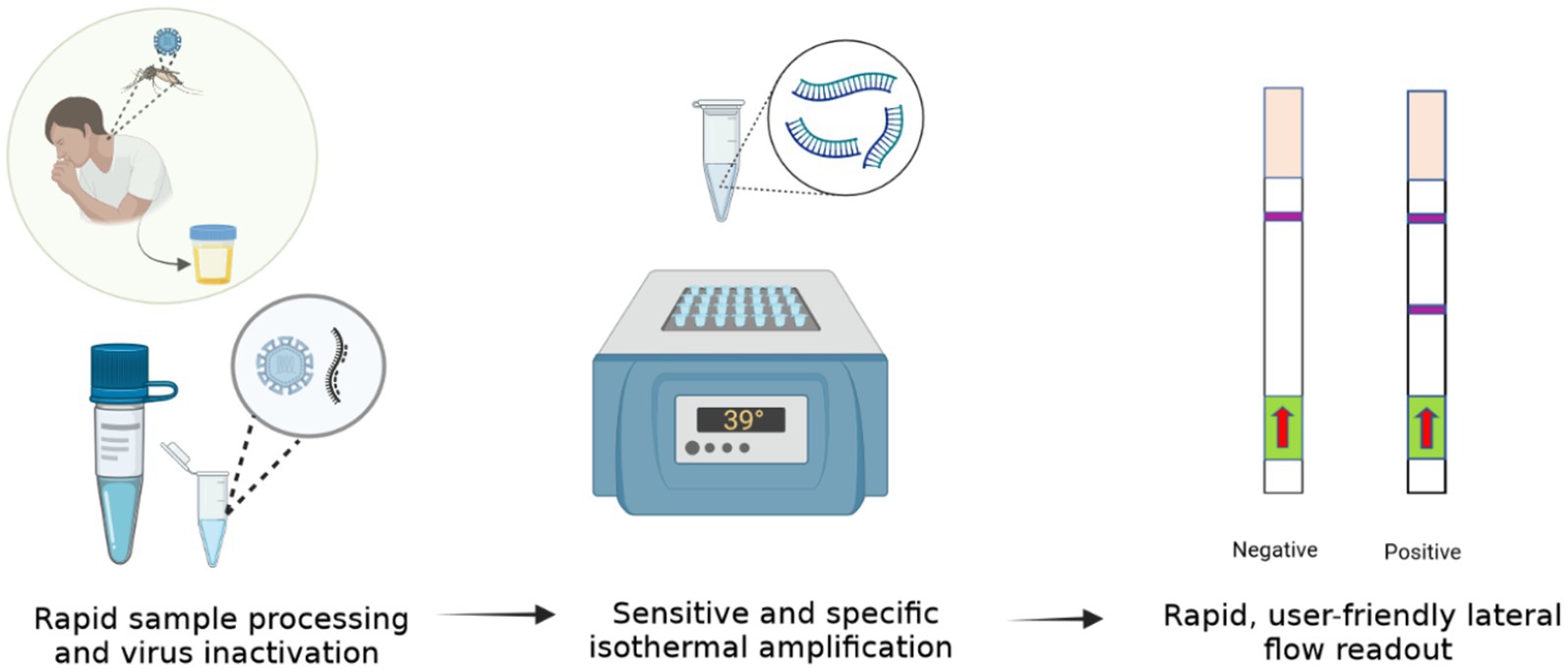
Here we describe a rapid, isothermal ZIKV diagnostic (Iso-ZIKV-Dx). Our test format combines a unique low-resource sample preparation reagent with RT-RAA amplification, and lateral flow detection (LFD) to enable ZIKV detection in urine in under 30 min without the need for expensive nor advanced instrumentation. As such, our Iso-ZIKV-Dx exhibits promising diagnostic capabilities suitable for low-resource settings.
Materials and methods
Plasmids and RNA template preparation
Plasmids (pBIC-A) containing regions of the ZIKV Envelope (E) gene fragment (OM964568.1, 901–2,412 nt) and non-structural 1 (NS1) gene fragment (MW015936, 249–3,545 nt) of ZIKV were obtained from Bioneer Pacific Pty Ltd., Victoria, AUS. The pBIC-A- NS1 and E gene vector was transformed into E. coli. A single colony of E. coli containing the pBIC-A- NS1 plasmid was streaked onto LB broth agar supplemented with 100 μg/mL of ampicillin and incubated at 37°C. The selected colony was grown in liquid LB broth supplemented with 100 μg/mL of ampicillin for plasmid isolation using the ‘NucleoBond Xtra Midi’ kit (Machery-Nagel, GER). MEGAscript (Ambion, Austin USA) in vitro transcription kit was used to yield RNA transcript from pBIC-A- NS1 plasmids. XhoI was the chosen restriction enzyme to digest the linearize plasmid in preparation for in vitro transcription. Transcribed RNA from pBIC-A- NS1 plasmid was quantified using Quibit RNA Hs Kit. Using software, a calculation of an approximate numerical quantity of RNA copies/μL from extracted RNA was established (Pollak et al., 2023c). RNA transcripts were then stored at −80°C as working stocks and used as reference RNA standards for qRT-PCR.
RAA primer and probe design
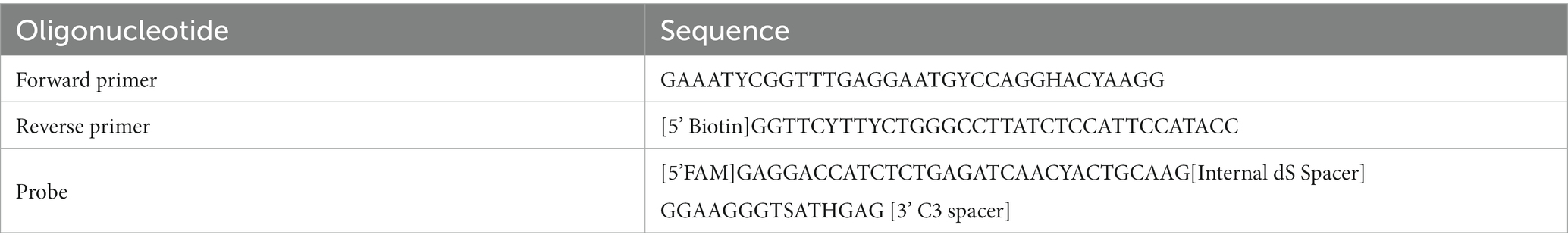
A total of 967 ZIKV NS1 genes obtained from NCBI’s databank representing both the African and Asian were aligned using Geneious Prime (version 2023.0.4) (Kearse et al., 2012). The first phylogenetic tree was created to isolate only unique ZIKV NS1 sequences. A total of 105 unique sequences were isolated and then imported to Geneious Prime for re-alignment using MAFFT (Nakamura et al., 2018). To assess the range of ZIKV sequences targeted by the primers and probes, a maximum likelihood (ML) phylogenetic tree (Figure 1) was constructed using IQTREE2 (Minh et al., 2020), utilising 10,000 bootstrap replicates and the TEST function. A consensus sequence of all unique ZIKV NS1 gene sequences was created using BioEdit 7.2 (Informer Technologies, INC, GER). Primers and probes were then designed using the most conserved region of the NS1 gene within the consensus sequence. Primer and probe design were further evaluated (primer dimers, secondary structures and GC content) using an oligoevaluator (Sigma-Aldrich Co. LLC, 2014). ALL primers and probes were bioinformatically evaluated to ensure no non-specific annealing to other flavivirus NS1 sequences occurred. Probes and primers (Table 1) were synthesised by (Bioneer Pacific Pty Ltd., Victoria, AUS) and purified via PAGE and HPLC, respectively.
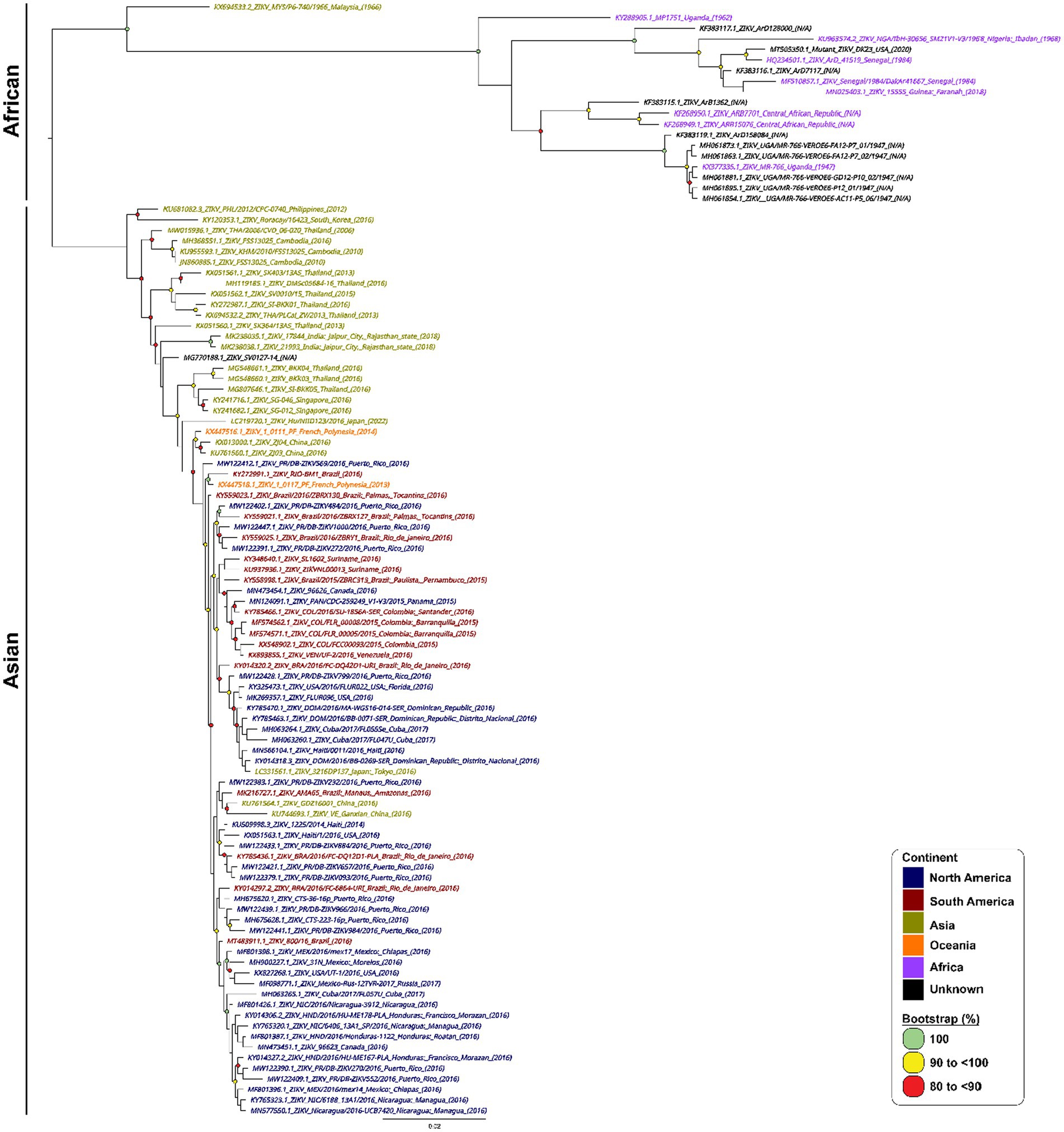
Figure 1. Phylogenetic tree of 105 unique ZIKV NS1 gene sequences. Phylogenetic relationships among 105 ZIKV NS1 sequences. ZIKV NS1 sequences are named by their corresponding GenBank accession number/strain/country of origin, with text being coloured according to their continent of isolation. The midpoint-rooted maximum likelihood phylogenetic trees were constructed using IQTREE2, which automatically incorporates the most appropriate nucleotide substitution model (TN + F + G4). The scale bar indicates the number of nucleotide substitutions per site. Bootstrap values above 80% are displayed next to the node of each clade and are coloured according to the figure legend.
Iso-ZIKV-Dx test
Rapid sample processing
ZIKV MR766 culture was mixed into urine and RPMI media samples at a ratio of 1:1 with TNA-Cifer Reagent E (BioCifer, Auchenflower, AUS) at room temperature for 5 min. Urine and RPMI media samples were diluted 1:5 in nuclease free H20.
RT-RAA amplification
Each RT-RAA test was performed using the RAA kit (Jiangsu Qitian Gene Biotechnology Co. Ltd., Wuxi City China) with final reaction conditions of 0.83x RAA rehydration buffer, 1/5 RAA pellet, forward primer (350 nM), reverse primer (350 nM), probe (200 nM), Endonuclease IV (2U; New England Biolabs, Victoria, AUS), Moloney Murine Leukemia virus reverse transcriptase enzyme (M-MLV, 60U; Biocifer Pty Ltd., Auchenflower, AUS), magnesium acetate (MgOAc, 23.33 mM) and 2 μL template in a final reaction volume of 12 μL. MgOAc was added to the cap of each 0.2 mL PCR tube. Once template had been added, tubes were centrifuged and incubated at 39°C for 20 min using a heating block.
Lateral flow strip detection
HybriDetect lateral flow strips (LFS) (Milenia Biotec, Giessen, GER) were treated with 8 μL of 0.4% casein blocking buffer for pre-activation (Rames and Macdonald, 2019). To each strip, 2 μL of amplicon was pipetted on to the sample pad. The LFS strips were placed into 2 mL Eppendorf tubes containing 100 μL of LFS Running Buffer (Li et al., 2019) for 5 min. LFS were scanned using an Epson Perfection V39 Flatbed Scanner (Epson, New South Wales, AUS). The scanned images were converted to greyscale using Irfan View 64 and then imported to ImageJ for analysis. Band intensity analysis and statistical quantification were conducted as previously described (Pollak et al., 2023b).
Sensitivity and specificity testing
Analytical sensitivity testing for Iso-ZIKV-Dx tests were performed using a 10-fold serial dilution of purified, synthetic RNA transcripts coding for ZIKV E and NS1 gene fragments. Analytical specificity testing utilised purified RNA of various virus strains (Table 2). Synthetic ZIKV RNA transcript (1 × 106 copies/μL) was used as a positive control for validation of specificity.
Simulated-infected urine
ZIKV culture 3.56 × 108 TCID50/ml was spiked into Pickering Laboratories #1700–0018 Artificial urine medium for growing urological pathogens (Walker Scientific Pty Ltd., Joondalup DC, AUS).
Viruses and cell culture
All viral strains used in this study are listed in Table 2. Excluding ZIKV, all viruses were obtained from Hobson-Peters lab (University of Queensland, St Lucia, QLD) and were cultured at high titres as previously described (Hobson-Peters et al., 2013).
Cell culture
Aedes albopictus larvae cells C6/36 (ATCC-CRL-1660™) were obtained from the ATCC. Cell lines were cultured in Gibco (USA) 1640 RPMI supplemented with 5% Foetal Bovine Serum (FBS), 2 mM L-glutamine, Gibco (USA) 1x Antibiotic/Antimycotic at 28°C in a 5% C02 incubator until approximately 80% confluent (Pollak et al., 2023b).
ZIKV infection and titre determination
Virus culture
ZIKV MR766 culture (accession MK105975) was used to infect C6/36 cells at a multiplicity of Infection (MOI) of 0.01 for 5 and 7 days, respectively (Pollak et al., 2023a). Virus culture media was harvested after centrifugation at 4°C for 10 min at 130 × g and then stored at −80°C.
Titre determination
ZIKV titre determination was evaluated by standard TCID50 assays and fixed-cell ELISAs using C6/36 cells in a 96 well plate as previously described (Pollak et al., 2023b). Titres were calculated using the Reed and Muench method (Reed and Muench, 1938).
ZIKV inactivation testing
ZIKV inactivation testing was performed using ZIKV culture (3.56 × 108 TCID50/ml) mixed with TNA-Cifer Reagent E (TCE; BioCifer, Auchenflower, AUS) at different ratios (1:1 and 2:1, ZIKV culture to TCE) and incubated from 0 min up to 10 min. Titre was determined as previously described (Pollak et al., 2023b).
RNA purification
RNA from viral stocks were purified using TRIzol™ (Invitrogen by Thermo Fisher Scientific Pty Ltd., Victoria, AUS) or column-based kit (NucleoSpin RNA Virus Isolation Mini kit, Machery-Nagel, Duren, GER) following protocols outlined by the manufacturer. Viral RNA was eluted into 150 μL of nuclease free H2O and stored at −80°C.
ZIKV RT-qPCR
For a ZIKV RT-qPCR comparative, TaqMan™ Fast Virus 1-Step Master Mix (Thermo Fisher Pty Ltd., Victoria, AUS) was used in conjunction with primers and probes previously described (de Moraes et al., 2018). Parameters for RT-qPCR were implemented as per manufacturer’s instructions.
Results
In designing our primers and probes, we performed stringent bioinformatic analysis to ensure our primers and probes target as many strains and isolates of ZIKV from all existing lineages as possible. We mapped out all known ZIKV sequences from NCBI covering both Asian and African lineages of ZIKV and constructed a phylogenetic tree using the most unique 105 sequences (Figure 1).
Analytical sensitivity
In developing a rapid, low-resource reliant diagnostic for ZIKV, we strategically designed RT-RAA primers and probes to target a highly conserved region of the NS1 gene. To test analytical sensitivity, serial dilutions of RNA transcripts were assessed. Our RT-RAA tests exhibited a limit of detection (LOD) of 500 RNA copies/μL (Figure 2).
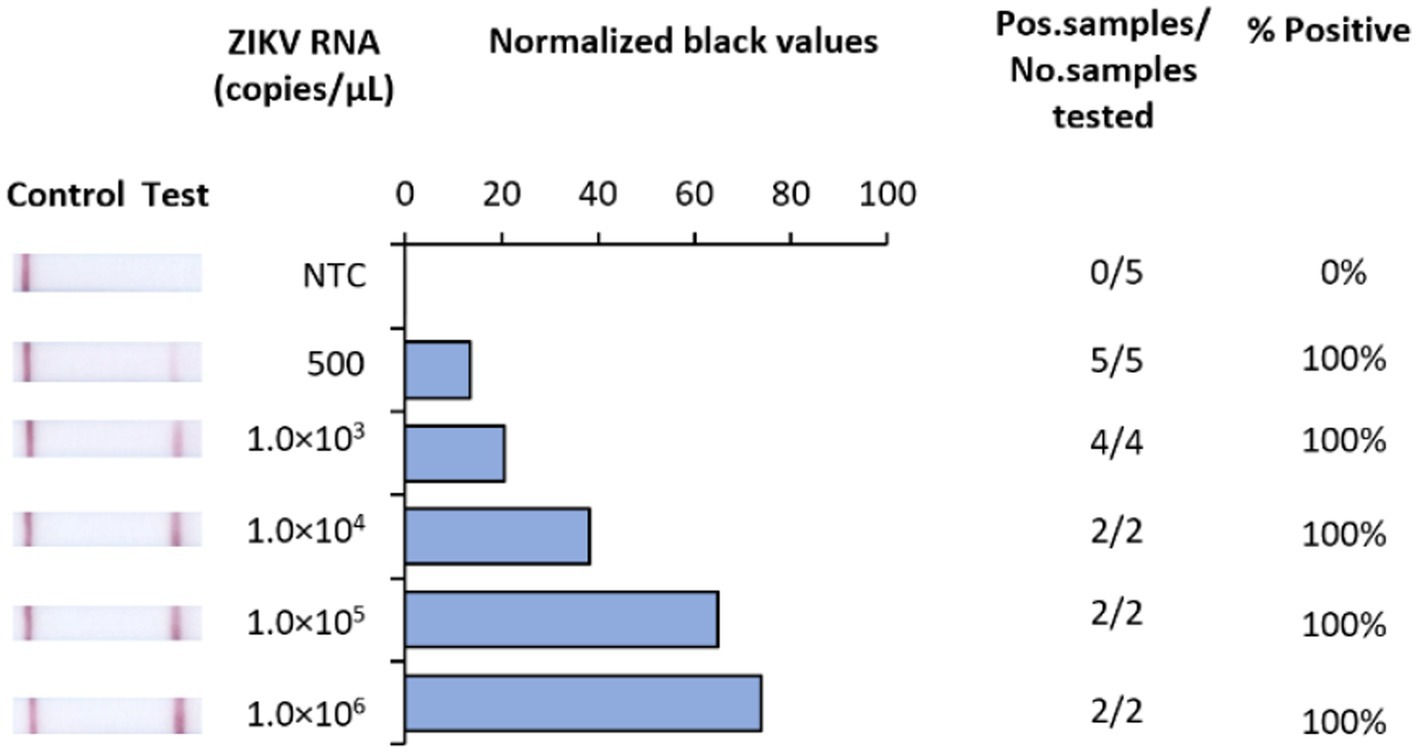
Figure 2. Analytical sensitivity of Iso-ZIKV-Dx using synthetic RNA transcripts. Sensitivity tests used 10-fold serially diluted synthetic ZIKV RNA. Scanned images of lateral flow strips with two lines (control and test) indicating positive detection of ZIKV and one being negative; no template control (NTC) consisted of nuclease free H2O in place of RNA (left). Normalised pixel density (black shifted) contrasted with the white space on lateral flow strips (middle). Number of positive samples detected over number of samples tested along with calculated percentage accuracy of positive results for each dilution series (right).
Analytical specificity
The analytical specificity of Iso-ZIKV-Dx was assessed against RNA from eight common flaviviruses that co-circulate with ZIKV (Table 2). Alphavirus chikungunya virus (CHIKV) (Table 2) was also included due to its common co-circulation with ZIKV. Our results showed that none of the common co-circulating viruses were detected using Iso-ZIKV-Dx (Figure 3). In addition, bioinformatic analysis of different ZIKA isolates indicated that the designed primers and probes are homologous to all ZIKV strains (African and Asian lineages) (Supplementary Figure 1).
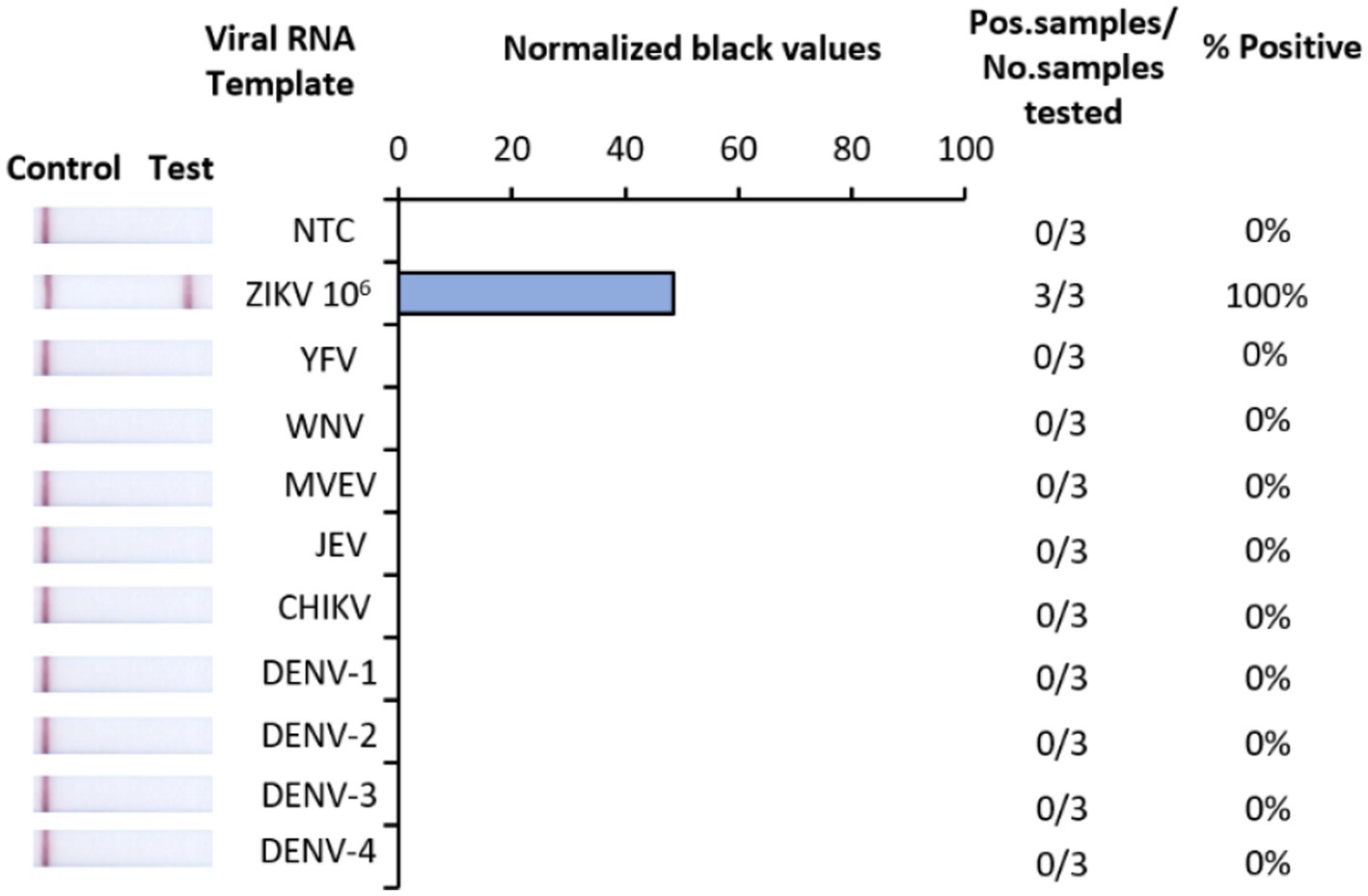
Figure 3. Analytical specificity of Iso-ZIKV-Dx using co-circulating flaviviruses and chikungunya virus. Specificity tests used synthetic RNA from ZIKV (PTC) and TRIzol extracted RNA from yellow fever virus (YFV), West NileKunjin virus (WNVKUN), Murray Valley Encephalitis virus (MVEV), Japanese Encephalitis virus (JEV), chikungunya virus (CHIKV), dengue 1, 2, 3 and 4 virus (DENV-1, 2, 3, and 4). Scanned images of lateral flow strips with two lines (control and test) indicating positive detection of ZIKV and one being negative; no template control (NTC) consisted of nuclease free H2O in place of RNA (left). Normalised pixel density (black shifted) contrasted with the white space on lateral flow strips (middle). Number of positive samples detected over number of samples tested along with calculated percentage accuracy of positive results (right).
Sample preparation inactivates ZIKV
A rapid diagnostic suitable for low-resource implementation requires a simple sample preparation procedure. Here we assessed the capacity of TNA-Cifer Reagent E (TCE) (BioCifer, Auchenflower, QLD, AUS) as a sample preparation reagent for rapid inactivation and sample preparation of ZIKV. Our results showed that ZIKV MR766 (3.56 × 108 TCID50/mL) was completely inactivated after 5 min when using a 1:1 ratio of sample to TCE, and 10 min when using a 2:1 ratio (Figure 4).
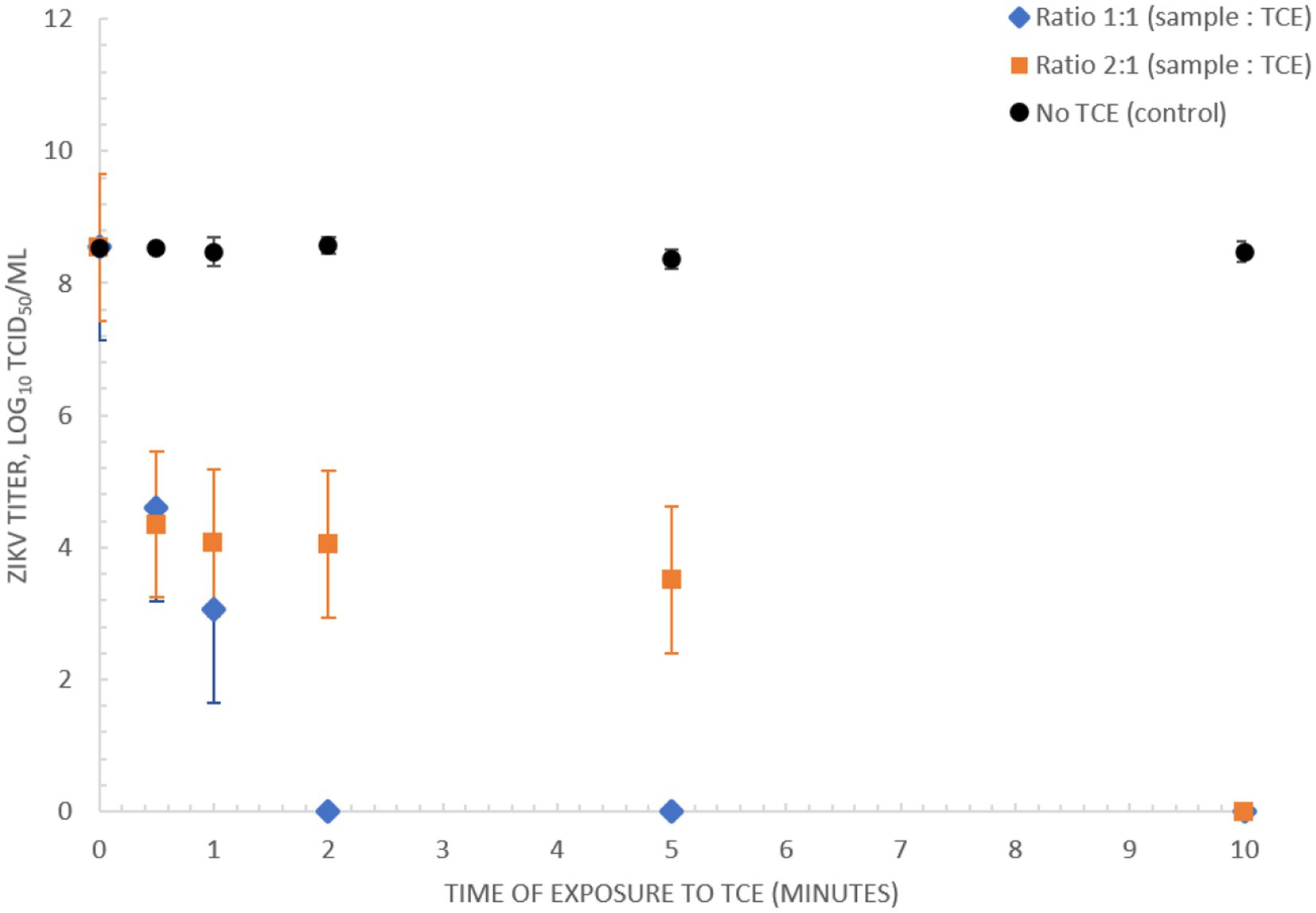
Figure 4. Inactivation of ZIKV (MR766) culture using TNA-Cifer Reagent E. Inactivation of 3.56 × 108 TCID50/mL ZIKV (MR766) culture using TNA-Cifer Reagent E (TCE) at 1:1 and 2:1 ratio (sample to TCE) incubated for 0, 0.5, 1, 2, 5, and 10 min at room temperature.
Detection of ZIKV in synthetic urine and RPMI medium
A key principle of our rapid Iso-ZIKV-Dx is operation in a manner that is not dependant on intricate resources or involve complex methodology. As such, the use of urine as a clinical matrix (Gourinat et al., 2015; Bonaldo et al., 2016; Lamb et al., 2016) eliminates the requirement for excessive sample collection procedures such as phlebotomy and/or serum and plasma extraction. Here we trialled synthetic urine and RPMI media spiked with MR766 ZIKV culture as the simulated clinical matrix for our rapid Iso-ZIKV-Dx. Using the same spiked samples, we performed our Iso-ZIKV-Dx test concurrently with virus isolation and TaqMan RT-qPCR for a comprehensive comparison. The rapid Iso-ZIKV-Dx test exhibited a LOD of 3.56 × 107 TCID50/mL of ZIKV in both synthetic urine (Figure 5) and RPMI media (Supplementary Figure 2). In RPMI culture media, this was quantified by TaqMan RT-qPCR to be equivalent to a Ct value of 35.91 and 449 copies/reaction. In urine, our LOD was quantified by TaqMan RT-qPCR to be equivalent to a Ct value of 37.71 and 34.28 copies/reaction.
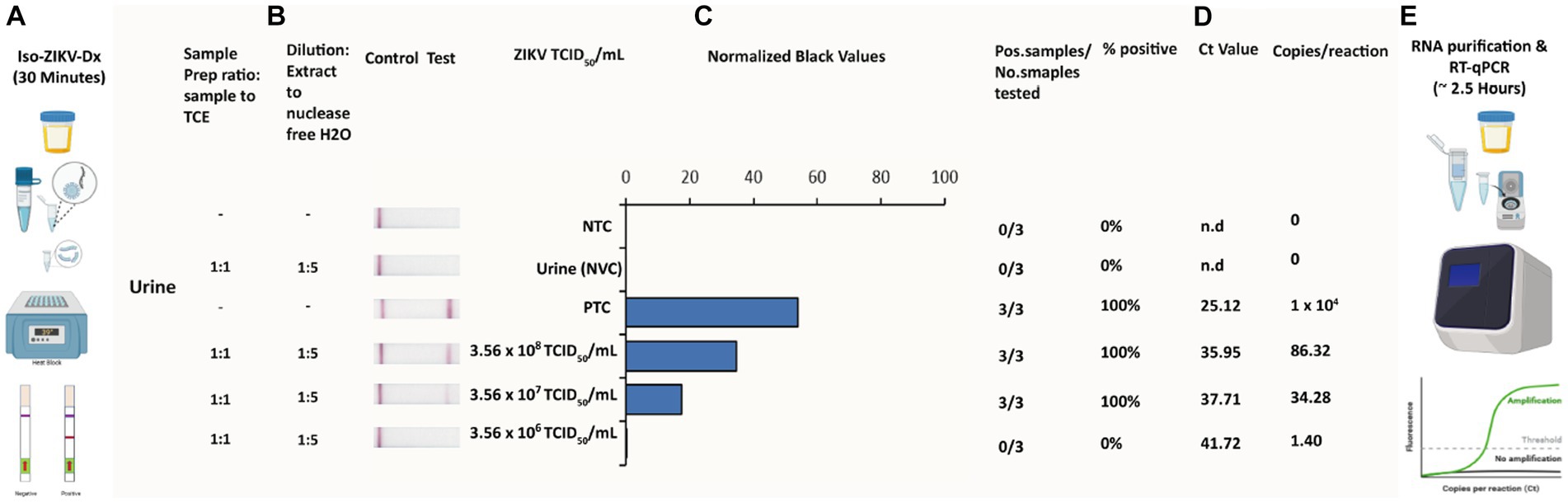
Figure 5. Rapid Iso-ZIKV-Dx of simulatedurine samples spiked with ZIKV (MR766). (A) Workflow, equipment needed and time frame of Iso-ZIKV-Dx. (B) Sample processing conditions including sample to reagent ratio and processed sample dilution ratio. (C) Sample description and quantities (NTC, non-template control; PTC, positive template control, synthetic ZIKV RNA transcripts 106 copies/μl); NVC, no virus control, urine. Scanned lateral flow strips showing test and control bands observable by naked eye. Normalised pixel densities (black values) from the displayed lateral flow strips. (D) Comparative Ct values and copies/reaction quantified via TaqMan RT-qPCR. (E) Workflow, equipment and time frame involved in conventional ZIKV RT-qPCR diagnostic.
Discussion
Due to the lack of approved vaccines or specific therapeutics for ZIKV, rapid detection remains crucial in predicting and controlling potential outbreaks. ZIKV outbreaks often occur in rural and remote areas, making it necessary for ZIKV diagnostics to be suitable for use in resource-limited settings, whilst still meeting the requirement for sensitive and accurate detection of ZIKV infection. Isothermal NAAT techniques are a potential solution, especially when combined with safe and simple sample preparation methods, as they could be easily deployed in low-resource settings with minimal training requirements for healthcare workers. In this study, we evaluated a novel rapid ZIKV diagnostic (Iso-ZIKV-Dx) which combines a low-resource sample preparation, RT-RAA test, and LFD. We used a phylogenetically divergent and highly virulent strain (MR766) of ZIKV (Shao et al., 2017). In this study we demonstrated the analytical sensitivity of our RT-RAA test to be 500 copies/μL when using synthetic ZIKV RNA, and confirmed the test did not detect other co-circulating viruses. It should be noted that our synthetic ZIKV RNA generated from RNA transcripts of the NS1 gene whilst the co-circulating viruses consisted of total viral RNA. The rapid Iso-ZIKV-Dx detected 3.56 × 107 TCID50/mL, equivalent to 34.28 copies/reaction RT-qPCR in synthetic urine spiked with ZIKV MR766 and offers improved safety for low-resource testing as the virus is inactivated in the very first step of the procedure. The novel sample preparation reagent, TNA-Cifer Reagent E, has previously been shown to inactivate other pathogens, such as DENV (Calvert et al., 2017), Nipah virus (Rossini et al., 2017) and Hendra virus (Jiangsu Qitian Gene Biotechnology Co., Ltd, 2023), ensuring operator safety in the event of other potential pathogens within the clinical sample. Our data emphasises the benefits of our uncomplicated sample preparation protocol, which proved effective in detecting ZIKV in urine samples. It is noteworthy that ZIKV RNA is known to have limited stability at room temperature, making it challenging to detect within a specific time frame (Tan et al., 2017). Of note, a comparative analysis on current RT-qPCR tests for ZIKV reported discrepancies in detection sensitivity and specificity amongst Asian versus African lineages (de Moraes et al., 2018) with particular emphasis on the difficulty observed among African lineages specifically. Therefore, the primers and probes used in this study were strategically designed to target highly conserved regions of the NS1 gene among both ZIKV lineages as illustrated via our bioinformatic alignment analysis of the most unique NS1 gene sequences.
RT-qPCR is still considered the most reliable technique for diagnosing ZIKV in clinical pathology, and is widely recognised as the gold standard method for detection. However, due to the RNA stability issues at room temperature reported in urine (Tan et al., 2017), methods such as RT-qPCR that require sample storage favour clinical matrices such as plasma, serum or whole blood. For comparison, in serum and plasma, ZIKV RT-qPCRs typically demonstrate detection limits between 1 and 64 copies/reaction (n = 31) (Pessôa et al., 2016). A comparative study of RT-qPCR versus RT-LAMP using urine to detect ZIKV, however, demonstrated a detection limit of 6 copies/reaction (n = 8) for RT-qPCR (Calvert et al., 2017). Our rapid Iso-ZIKV-Dx test showed similar levels of sensitivity, detecting as low as 34 copies/reaction in ZIKV-spiked synthetic urine. The Range of reported ZIKV concentration in urine was observed to be between 0.7 and 2206 copies/mL (Gourinat et al., 2015). The vastness of this range is thought to be due to the variability in ZIKV RNA stability, further supporting the need for a rapid sample processing method. Non-invasive clinical sample collections such as urine (Lamb et al., 2016) offer significant advantages for isothermal, POC compatible methods such as RT-RAA, RT-RPA, and RT-LAMP, due to the prolonged period of ZIKV RNA detectable throughout the infection period (Rossini et al., 2017). As such, our rapid Iso-ZIKV-Dx test could be valuable in overcoming the obstacle of testing urine samples by potentially detecting ZIKV directly at the point of sample collection. One study using RT-LAMP reported a LOD of 6.6 copies/reaction in urine (n = 63) (Castro et al., 2018), whilst another study achieved 6 copies/reaction in urine (n = 178) (Calvert et al., 2017). Although both RT-qPCR and RT-LAMP offer slightly improved sensitivity compared to the Iso-ZIKV-Dx test, their application as POC diagnostics in low-resource settings is limited. RT-qPCR requires highly skilled personnel, expensive machinery and laboratory-based sample preparation. While RT-LAMP is known for its simplicity, sensitivity and speed which resulted in wide scale application in low resource settings, it still needs heating machines capable of attaining temperatures of 55–65°C. In contrast, RT-RAA, similarly to RT-RPA can be performed at near-ambient temperature (37–40°C) and moreover has a higher tolerance to PCR inhibitors (Li et al., 2020). Both have previously required laboratory-based purification of RNA, whereas our Iso-ZIKV-Dx test does not require any sophisticated laboratory equipment, providing potential the entire procedure, from sample to result, be performed in low-resource near-patient settings. The ideal rapid POC test should provide a range of detection and diagnostic capabilities, as defined by the REASSURED criteria – Real-time connectivity, Ease of specimen collection, Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, and Deliverable to end-users (Land et al., 2019). Our data suggests that the Iso-ZIKV-Dx (sample preparation, RT-RAA, LFD) has potential to meet the REASSURED criteria, providing real-time connectivity through direct analogue reading of the result which could be performed at or near the patient in a low-resource clinic. Insofar as our assay has met pre-clinical and REASSURED criteria, our next phase of evaluation will involve clinical and in-field evaluation of the assay to evaluate clinical efficacy and performance among pathology of ZIKV infections. As our study only involved live MR766 strain of ZIKV, our hope would be to evaluate our ZIKV test with multiple strains within these clinical trials.
Due to manual steps in our assay, it should be acknowledged that screening of large-scale clinical samples would not occur as rapidly as standard RT-PCR assays. However, unlike RT-PCR, our rapid ZIKV test has been designed to be field deployable, requiring minimal resources and minimally trained staff. As a result, the time from sample preparation to diagnostic result is considerably faster than when compared to transportation and processing within central laboratory. Moreover, this particular attribute of our ZIKV test renders it a potential early warning screening tool in the face of a sudden outbreaks. A second potential challenge for our ZIKV test, and indeed many in-field nucleic acid amplification assays (Tang et al., 2016), is the risk of post amplification cross-contamination. A potential solution to this issue is the substitution of dTTP with dUTP in the amplification reactions. This approach has been shown to reduce cross-contamination in LAMP assays (Kil et al., 2015). However, to the authors knowledge, the feasibility of implementing such measure in recombinase-aided amplification assays has yet to be assessed. Therefore, a more feasible alternative is to utilise ‘U-Star’ disposable cartridges (TwistDx™, 2023b), which would reduce cross contamination occurrences by limiting external exposure of lateral flow strips to the environment.
Rapid detection of arboviruses such as ZIKV play a crucial role in limiting outbreaks that have already emerged or are currently on going. Additionally, surveillance and monitoring can serve to identify early signs of the pathogen and thereby prevent outbreaks through vector control. The study of vector competency and transovarial transmission of arboviruses such as ZIKV thus remain a particular area of focus. Due to factors such as climate change and increased urbanisation (Li et al., 2017) vector competency continues to broaden, further implicating the emergence of novel and existing viruses. A tool with in-field capabilities that provides robust and rapid results could be highly beneficial in epidemiology for monitoring ZIKV. To that end, further studies are warranted to determine if our Iso-ZIKV-Dx is compatible for detection of virus in mosquitoes, as was similarly performed for the TNA-Cifer Reagent E combined with RPA-LFD for detection of dengue virus in Aedes aegpytii mosquitoes. This would allow our Iso-ZIKV-Dx test to be extended beyond diagnostics alone, in potentially offering strategies toward aiding in surveillance and monitoring of viral vectors (Pollak et al., 2023b) and possibly reservoir hosts for arboviruses such as ZIKV.
Conclusion
In conclusion, we developed a rapid, isothermal test for ZIKV that only requires incubation at 39°C and produces a real-time result in 30 min. Pre-clinical evaluation suggests that our Iso-ZIKV-Dx could offer promising innovations for diagnostics in ZIKV endemic areas. We demonstrate that our ZIKV test was able to detect as low as 500 copies/μL of synthetic ZIKV RNA and also, does not detect any other co-circulating arbovirus. We successfully established a simple sample preparation procedure which demonstrated that we were able to completely inactivate one of the highest virulent strains of ZIKV (MR766, African lineage) in 5 min at room temperature, whilst at the same time, extracting sufficient quantities of viral RNA for detection. By combining our simple sample preparation procedure with RT-RAA and Lateral Flow Strip detection, we successfully developed a robust rapid isothermal ZIKV diagnostic. Our data illustrated that our ZIKV test operates five times faster than ‘gold standard’ RT-qPCR and exhibited a detection limit of 34 RNA copies/reaction in spiked synthetic urine. Furthermore, due to the simplistic nature of our ZIKV test, we surmise that the applications of our Iso-ZIKV-Dx extend beyond detection alone, and could potentially provide innovations toward surveillance and monitoring for ZIKV and arboviruses alike.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RB: conceptualization, data curation, formal analysis, investigation, methodology, resources, supervision, validation, visualization, writing – original draft, and writing – review and editing. NP, JM, and DM: conceptualization, formal analysis, funding acquisition, methodology, project administration, resources, supervision, and writing – review and editing. JH-P: resources and writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Research Training Program (RTP) Commonwealth Research Scholarship (https://www.education.gov.au/research-block-grants/research-training-program), in accordance with the Higher Education Support act 2003 with the University of the Sunshine Coast (QLD, Australia). This work was also supported, in part by the Bill & Melinda Gates foundation (OPP1140133).
Acknowledgments
We wish to thank BioCifer Pty Ltd. for generously providing TNA-Cifer Reagent E and Reverse Transcriptase used to develop the ZIKV diagnostic test. We also wish to thank Vasilli Kasimov (Centre for Bioinnovation, University of the Sunshine Coast) for assisting in the bioinformatic analysis and formulation of the phylogenetic tree for our primer and probe design.
Conflict of interest
RB reports supplies were provided by BioCifer Pty Ltd. JM reports financial support was provided by Bill and Melinda Gates Foundation. JM reports a relationship with BioCifer Pty Ltd. that includes: board membership and equity or stocks.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1214148/full#supplementary-material
References
Ahmed, M., Nath, N. S., Hugo, L. E., Devine, G. J., Macdonald, J., and Pollak, N. M. (2022). Rapid detection of kdr mutation F1534C in Aedes aegypti using recombinase polymerase amplification and lateral flow dipsticks. Pestic. Biochem. Physiol. 187:105209. doi: 10.1016/j.pestbp.2022.105209
Azar, S. R., and Weaver, S. C. (2019). Vector competence: what has Zika virus taught us? Viruses 11:867. doi: 10.3390/v11090867
Bonaldo, M. C., Ribeiro, I. P., Lima, N. S., dos Santos, A. A. C., Menezes, L. S. R., da Cruz, S. O. D., et al. (2016). Isolation of infective Zika virus from urine and saliva of patients in Brazil. PLoS Negl. Trop. Dis. 10:e0004816. doi: 10.1371/journal.pntd.0004816
Brady, O. J., and Hay, S. I. (2019). The first local cases of Zika virus in Europe. Lancet 394, 1991–1992. doi: 10.1016/S0140-6736(19)32790-4
Calvert, A. E., Biggerstaff, B. J., Tanner, N. A., Lauterbach, M., and Lanciotti, R. S. (2017). Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP). PLoS One 12:e0185340. doi: 10.1371/journal.pone.0185340
Castro, T., Sabalza, M., Barber, C., Abrams, W., da Costa, A. C., de Pádua Milagres, F. A., et al. (2018). Rapid diagnosis of Zika virus through saliva and urine by loop-mediated isothermal amplification (LAMP). J. Oral Microbiol. 10:1510712. doi: 10.1080/20002297.2018.1510712
Chang, C., Ortiz, K., Ansari, A., and Gershwin, M. E. (2016). The Zika outbreak of the 21st century. J. Autoimmun. 68, 1–13. doi: 10.1016/j.jaut.2016.02.006
Cheikh Tidiane Diagne, M. F., Lopez-Jimena, B., El Wahed, A. A., Loucoubar, C., Fall, C., Mencatelli, G., et al. (2020). Comparative analysis of Zika virus detection by RT-qPCR, RT-LAMP, and RT-RPA, in methods in molecular biology. Methods Mol. Biol. 2142, 165–179. doi: 10.1007/978-1-0716-0581-3_14
Da Silva, S., Martins, D. O. S., and Jardim, A. C. G. (2018). A review of the ongoing research on Zika virus treatment. Viruses 10:255. doi: 10.3390/v10050255
de Moraes, F. M., Espósito, D. L. A., Klein, T. M., and da Fonseca, B. A. L. (2018). Searching for the best real-time RT-PCRs to detect Zika virus infections: the importance of comparing several protocols. Braz. J. Med. Biol. Res. 51:e7221. doi: 10.1590/1414-431x20187221
Dick, G. W. A., Kitchen, S. F., and Haddow, A. J. (1952). Zika virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 46, 509–520. doi: 10.1016/0035-9203(52)90042-4
Dong, D., Fu, S. H., Wang, L. H., Lv, Z., Li, T. Y., and Liang, G. D. (2012). Simultaneous detection of three arboviruses using a triplex RT-PCR enzyme hybridization assay. Virol. Sin. 27, 179–186. doi: 10.1007/s12250-012-3246-9
Dupont-Rouzeyrol, M., O’Connor, O., Calvez, E., Daurès, M., John, M., Grangeon, J. P., et al. (2015). Co-infection with Zika and dengue viruses in 2 patients, New Caledonia, 2014. Emerg. Infect. Dis. 21, 381–382. doi: 10.3201/eid2102.141553
Gourinat, A. C., O’Connor, O., Calvez, E., Goarant, C., and Dupont-Rouzeyrol, M. (2015). Detection of Zika virus in urine. Emerg. Infect. Dis. 21, 84–86. doi: 10.3201/eid2101.140894
Gregory, C. J., Oduyebo, T., Brault, A. C., Brooks, J. T., Chung, K. W., Hills, S., et al. (2017). Modes of transmission of Zika virus. J. Infect. Dis. 216, S875–S883. doi: 10.1093/infdis/jix396
Hayes, E. B. J. E. I. D. (2009). Zika virus outside. Africa 15, 1347–1350. doi: 10.3201/eid1509.090442
Hendel-Paterson, B., Anderson, K., Fabrizius, R. G., Walker, P. F., Maalim, S., and Kaiser, R. M. (2016). Guillain-Barré syndrome associated with Zika virus infection in a traveler returning from Guyana. Am J Trop Med Hyg 95, 1161–1165. doi: 10.4269/ajtmh.16-0397
Heukelbach, J., Alencar, C. H., Kelvin, A. A., de Oliveira, W. K., and Pamplona de Góes Cavalcanti, L. (2016). Zika virus outbreak in Brazil. J. Infect. Dev. Ctries. 10, 116–120. doi: 10.3855/jidc.8217
Hobson-Peters, J., Yam, A. W. Y., Lu, J. W. F., Setoh, Y. X., May, F. J., Kurucz, N., et al. (2013). A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 8:e56534. doi: 10.1371/journal.pone.0056534
Jiangsu Qitian Gene Biotechnology Co., Ltd. (2023). Recombinase aided amplification. 2012–2023. Available at: http://en.qt-bio.com/.
Kadkhoda, K., Gretchen, A., and Racano, A. (2017). Evaluation of a commercially available Zika virus IgM ELISA: specificity in focus. Diagn. Microbiol. Infect. Dis. 88, 233–235. doi: 10.1016/j.diagmicrobio.2017.04.002
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kil, E. J., Kim, S., Lee, Y. J., Kang, E. H., Lee, M., Cho, S. H., et al. (2015). Advanced loop-mediated isothermal amplification method for sensitive and specific detection of tomato chlorosis virus using a uracil DNA glycosylase to control carry-over contamination. J. Virol. Methods 213, 68–74. doi: 10.1016/j.jviromet.2014.10.020
Lamb, L. E., Bartolone, S. N., Kutluay, S. B., Robledo, D., Porras, A., Plata, M., et al. (2016). Advantage of urine based molecular diagnosis of Zika virus. Int. Urol. Nephrol. 48, 1961–1966. doi: 10.1007/s11255-016-1406-9
Land, K. J., Boeras, D. I., Chen, X. S., Ramsay, A. R., and Peeling, R. W. (2019). REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 4, 46–54. doi: 10.1038/s41564-018-0295-3
Li, C.-X., Guo, X. X., Deng, Y. Q., Xing, D., Sun, A. J., Liu, Q. M., et al. (2017). Vector competence and transovarial transmission of two Aedes aegypti strains to Zika virus. Emerg. Microbes Infect. 6, 1–7. doi: 10.1038/emi.2017.8
Li, J., Macdonald, J., and von Stetten, F. (2020). Correction: review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 145, 1950–1960. doi: 10.1039/C9AN90127B
Li, J., Pollak, N. M., and Macdonald, J. J. A. O. (2019). Multiplex detection of nucleic acids using recombinase polymerase amplification and a molecular colorimetric 7-segment display 4, 11388–11396. doi: 10.1021/acsomega.9b01097,
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mumtaz, N., van Kampen, J. J. A., Reusken, C. B. E. M., Boucher, C. A. B., and Koopmans, M. P. G. (2016). Zika virus: where is the treatment? Curr. Treat. Options Infect. Dis. 8, 208–211. doi: 10.1007/s40506-016-0083-7
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Paixão, E. S., Barreto, F., da Glória Teixeira, M., da Conceição N. Costa, M., and Rodrigues, L. C. (2016). History, epidemiology, and clinical manifestations of Zika: a systematic review. Am. J. Public Health 106, 606–612. doi: 10.2105/AJPH.2016.303112
Paixao, E. S., Leong, W. Y., Rodrigues, L. C., and Wilder-Smith, A. (2018). Asymptomatic prenatal Zika virus infection and congenital Zika syndrome. Open Forum Infect. Dis. 5:ofy073. doi: 10.1093/ofid/ofy073
Pessôa, R., Patriota, J. V., Lourdes de Souza, M., Felix, A. C., Mamede, N., and Sanabani, S. S. (2016). Investigation into an outbreak of dengue-like illness in Pernambuco, Brazil, revealed a Cocirculation of Zika, chikungunya, and dengue virus type 1. Medicine 95, –e3201. doi: 10.1097/MD.0000000000003201
Pollak, N. M., Fais, O., Kristoffersen, J., Phuthaworn, C., Knibb, W., and Macdonald, J. (2022). Rapid sample preparation and low-resource molecular detection of hepatopancreatic parvoviruses (HPV) by recombinase polymerase amplification lateral flow detection assay in shrimps (Fenneropenaeus merguiensis). PLoS One 17:e0276164. doi: 10.1371/journal.pone.0276164
Pollak, N. M., Marsh, G. A., Olsson, M., McMillan, D., and Macdonald, J. (2023a). Rapid, sensitive, and specific, low-resource molecular detection of Hendra virus. One Health 16:100504. doi: 10.1016/j.onehlt.2023.100504
Pollak, N. M., Olsson, M., Ahmed, M., Tan, J., Lim, G., Setoh, Y. X., et al. (2023b). Rapid diagnostic tests for the detection of the four dengue virus serotypes in clinically relevant matrices. Microbiol. Spectr. 11:e0279622. doi: 10.1128/spectrum.02796-22
Pollak, N. M., Olsson, M., Marsh, G. A., Macdonald, J., McMillan, D., et al. (2023c). Evaluation of three rapid low-resource molecular tests for Nipah virus. Front. Microbiol. 13:1101914. doi: 10.3389/fmicb.2022.1101914
Rames, E. K., and Macdonald, J. (2019). Rapid assessment of viral water quality using a novel recombinase polymerase amplification test for human adenovirus. Appl. Microbiol. Biotechnol. 103, 8115–8125. doi: 10.1007/s00253-019-10077-w
Reed, L. J., and Muench, H. J. A. J. O. E. (1938). A simple method of estimating fifty per cent endpoints 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408,
Rossini, G., Gaibani, P., Vocale, C., Cagarelli, R., and Landini, M. P. (2017). Comparison of Zika virus (ZIKV) RNA detection in plasma, whole blood and urine – case series of travel-associated ZIKV infection imported to Italy, 2016. J. Infect. 75, 242–245. doi: 10.1016/j.jinf.2017.05.021
Roth, A., Mercier, A., Lepers, C., Hoy, D., Duituturaga, S., Benyon, E., et al. (2014). Concurrent outbreaks of dengue, chikungunya and Zika virus infections – an unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012–2014. Eur. Secur. 19:20929. doi: 10.2807/1560-7917.ES2014.19.41.20929
Shao, Q., Herrlinger, S., Zhu, Y. N., Yang, M., Goodfellow, F., Stice, S. L., et al. (2017). The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Development 144, 4114–4124. doi: 10.1242/dev.156752
Sharp, T. M., Muñoz-Jordán, J., Perez-Padilla, J., Bello-Pagán, M. I., Rivera, A., Pastula, D. M., et al. (2016). Zika virus infection associated with severe thrombocytopenia. Clin. Infect. Dis. 63, 1198–1201. doi: 10.1093/cid/ciw476
Sigma-Aldrich Co. LLC (2014). OligoEvaluator. Available at: http://www.oligoevaluator.com/LoginServlet
Stettler, K., Beltramello, M., Espinosa, D. A., Graham, V., Cassotta, A., Bianchi, S., et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection 353, 823–826. doi: 10.1126/science.aaf8505,
Tan, S. K., Sahoo, M. K., Milligan, S. B., Taylor, N., and Pinsky, B. A. (2017). Stability of Zika virus in urine: specimen processing considerations and implications for the detection of RNA targets in urine. J. Virol. Methods 248, 66–70. doi: 10.1016/j.jviromet.2017.04.018
Tang, Y., Chen, H., and Diao, Y. (2016). Advanced uracil DNA glycosylase-supplemented real-time reverse transcription loop-mediated isothermal amplification (UDG-rRT-LAMP) method for universal and specific detection of Tembusu virus. Sci. Rep. 6:27605. doi: 10.1038/srep27605
TwistDx™. (2023a). Recombinase polymerase amplification, or RPA, is the breakthrough, isothermal replacement to PCR. Available at: https://www.twistdx.co.uk/rpa/.
TwistDx™. (2023b). U-star disposable nucleic acid lateral flow detection units products. Available at: https://www.twistdx.co.uk/product/u-star-disposable-nucleic-acid-lateral-flow-detection-units-pack-of-20/
Wang, Y., Cui, Y., Yu, Z., Li, Y., Bai, C., Sun, P., et al. (2020). Development of a recombinase-aided amplification assay for detection of orf virus. J. Virol. Methods 280:113861. doi: 10.1016/j.jviromet.2020.113861
Wen, Z., Song, H., and Ming, G. L. (2017). How does Zika virus cause microcephaly? Genes Dev. 31, 849–861. doi: 10.1101/gad.298216.117
Keywords: Zika virus, NS1 gene, recombinase aided amplification, lateral flow detection, point-of-care, rapid test
Citation: Balea R, Pollak NM, Hobson-Peters J, Macdonald J and McMillan DJ (2023) Development and pre-clinical evaluation of a Zika virus diagnostic for low resource settings. Front. Microbiol. 14:1214148. doi: 10.3389/fmicb.2023.1214148
Edited by:
Ke Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Diogo Antonio Tschoeke, Federal University of Rio de Janeiro, BrazilKim-Kee Tan, University of Malaya, Malaysia
Fabiana Feitosa-Suntheimer, Boston University, United States
Copyright © 2023 Balea, Pollak, Hobson-Peters, Macdonald and McMillan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanne Macdonald, am1hY2RvbjFAdXNjLmVkdS5hdQ==; Nina M. Pollak, bnBvbGxha0B1c2MuZWR1LmF1; David J. McMillan, ZG1jbWlsbDFAdXNjLmVkdS5hdQ==
 Rickyle Balea
Rickyle Balea Nina M. Pollak
Nina M. Pollak Jody Hobson-Peters
Jody Hobson-Peters Joanne Macdonald
Joanne Macdonald David J. McMillan1,2*
David J. McMillan1,2*