- 1Department of Biochemistry, Faculty of Science, University of Tabuk, Tabuk, Saudi Arabia
- 2Department of Biology, Faculty of Science, University of Tabuk, Tabuk, Saudi Arabia
- 3Microbial Activity Unit, Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center, Giza, Egypt
- 4Department of Computer Science, Faculty of Science and Engineering, University of Manchester, Manchester, United Kingdom
The increasing demand for novel bioactive compounds is driving research into natural sources, particularly through the valorization of agricultural residues. Endophytic fungi have emerged as a promising source of bioactive metabolites with diverse therapeutic potential. We employed a novel approach utilizing the isolated Trichoderma asperellum WNZ-21 [identified morphologically and molecularly (ITS: OR857252, tef1 gene: PP069312)] in semi-solid-state fermentation to bioconvert common bean biomass residue (RCBB) into a rich source of metabolites with significant therapeutic potential. First, T. asperellum efficiently degraded 67.19% RCBB, exhibiting robust enzymatic activity (cellulase: 18.067 units per gram (U/g), xylanase: 15.967 U/g, protease: 5.550 U/g). The fermented RCBB filtrate exhibited a rich chemical profile, including (mg/g RCBB) amino acids (13.51), phenolics (55.22), flavonoids (11.22), tannins (18.90), and several secondary metabolites. FT-IR analysis indicated the presence of a complex mixture of amino acids, polysaccharides, proteins, and other metabolites. GC-MS analysis identified 17 compounds. The fermented biomass possesses multi-biological activities, including anticancer against hepatocellular carcinoma (IC50 = 35.62 μg/mL) and mammary gland breast cancer (IC50 = 59.20 μg/mL) cell lines, with reduced toxicity toward normal human lung fibroblast cells (IC50 = 76.33 μg/mL). It also exhibits anti-hemolytic activity in erythrocyte protection assays, reducing oxidative stress-induced hemolysis to 4.40%. Additionally, the filtrate demonstrates anti-inflammatory activity in a carrageenan-induced paw edema model in rats, providing 36.50% protection after 5 h of treatment. Our findings establish RCBB valorization as a promising route to produce multi-bioactive fungal metabolites. Future research should focus on isolating active compounds, optimizing production, and conducting in vivo studies to fully realize therapeutic potential.
1 Introduction
The ever-increasing generation of plant biomass residues demands innovative solutions, not only for sustainable resource management but also for the discovery of novel drugs with diverse therapeutic properties (El-Naggar et al., 2014). In the face of growing antibiotic resistance and chronic illnesses, researchers are actively exploring natural sources for new drugs. Among these, endophytic fungi have arisen as an auspicious resource due to their bioactive metabolites with significant therapeutic potential (De Silva et al., 2019; Sridharan et al., 2021). These fungi, which reside within plant tissues, possess a unique enzymatic arsenal capable of degrading complex organic matter, making them ideal candidates for the valorization of agricultural residues.
Millions of tons of plant protein residues are generated annually, and their disposal poses significant environmental and economic challenges. Traditional methods of biomass degradation, such as acid hydrolysis, are often inefficient and can lead to the destruction of valuable compounds, including amino acids (Moussa et al., 2023). In contrast, microbial degradation, particularly through semi-solid-state fermentation (SSSF), offers a sustainable and efficient alternative for converting these residues into bioactive metabolites. This less commonly employed technique falls between solid and liquid-state fermentation, and provides several advantages, including reduced energy requirements, increased productivity, and higher concentrations of target molecules (El-Metwally et al., 2023; Elsayed et al., 2021).
Phaseolus vulgaris L. (common bean), a key global crop, which accounts for 85% of the world’s bean production, generates substantial biomass residues rich in protein content (Nasar et al., 2023). The residue of common bean biomass (RCBB) is recalcitrant and poses challenges for biotechnological applications. However, they represent an untapped resource to produce bioactive compounds through microbial fermentation (Al-Askar et al., 2022; Moussa et al., 2023). While RCBB can be composted for traditional uses, its potential as a basis of cost-effective multi-bioactive metabolites for medical applications remains largely unexplored.
This study addresses this gap by exploring the potential of endophytic fungi to valorize RCBB through SSSF. Endophytic fungi, particularly those belonging to Trichoderma spp., are known to produce a diverse array of enzymes and secondary metabolites with significant bioactivities (Al-Askar et al., 2022; De Silva et al., 2019). By leveraging the enzymatic capabilities of these fungi, we aim to transform RCBB into valuable bioactive compounds with potential therapeutic applications.
Recent advances in microbial biomass valorization have highlighted the potential of endophytic fungi to produce bioactive metabolites from agricultural residues. For instance, studies have demonstrated the ability of Trichoderma species to degrade lignocellulosic biomass and synthesize enzymes such as cellulase, xylanase, and protease, which are essential for breaking down complex plant materials (Al-Askar et al., 2022; Moussa et al., 2023). Additionally, the use of untargeted metabolomics and bioassay-guided fractionation has enabled the identification of novel bioactive compounds from fungal filtrates, further underscoring the potential of these microorganisms in drug discovery (Elattar et al., 2024; Salem et al., 2022; Wang et al., 2023).
Despite advancements in bioconversion, a significant gap exists in the direct utilization of proteinaceous residues such as RCBB by endophytic fungi for the synthesis of bioactive metabolites. To address this gap and provide a sustainable waste valorization strategy, this study aimed to identify and select an endophytic fungus with potent enzymatic activity for the efficient bioconversion of RCBB via a less common technique (semi-solid-state fermentation). Following the selection of Trichoderma asperellum WNZ-21 based on its superior hydrolytic capabilities, the next novel step was to produce and comprehensively profile the multi-biological activity of the resulting fungal filtrate, including the identification of various bioactive compounds, and to evaluate its potential for anticancer, antihemolytic, and anti-inflammatory applications. This novel integrated approach bridges two interconnected research areas: (1) Trichoderma’s capacity to degrade plant waste through, and (2) the multi-bioactive potential of the resulting metabolite-rich fermentation filtrate.
2 Materials and methods
2.1 Isolation of the endophytic fungi
The endophytic fungi were isolated from common bean plants. Plant samples were collected from different locations in the Tabuk region, Saudi Arabia, from an area of 50 × 50 m around each sampling site in a random zigzag pattern. The surveyed area lies between latitudes of 28°23′59″ N, and longitudes of 36°34′17″ E, and elevation above sea level is 768 m. The collected plants were kept in polyethylene bags at 4°C and transferred to the laboratory for the isolation process using the method reported by Kuo et al. (2021) with some modifications.
Leaves and stems were washed with tap water to remove surface particles. The epiphytic microbes were removed through surface sterilization by immersing in ethyl alcohol (70%/10 s), then soaking in sodium hypochlorite (4%/30 s), before washing twice in sterile distilled water. Plants were cut into pieces 0.5–1.0 cm long. The segments were placed on 2% malt extract agar (BD Biosciences, Franklin Lakes, NJ, United States) containing 0.5 g/L penicillin G and streptomycin sulfate (Sigma-Aldrich, St. Louis, MO, United States), at 5 mg/L each. The inoculated plates were incubated at 25 ± 2°C under an alternating 12 h light/darkness cycle of cool white, fluorescent light. The developing fungal colonies were observed daily for 1 month using both stereo and compound microscopes to aid in the identification of the recovered fungi. Fungi with different mycelial morphotypes were isolated using single-spore and/or hyphal-tip techniques and subcultured on malt extract agar. The purified isolates were preserved on potato dextrose agar plates (BD Difco, Becton, Dickinson and Company, United States).
2.2 Fungal identification
2.2.1 Morphological identification
All fungi were morphologically identified according to their fungal morphology, cultural features, and microscopic characteristics. Four fungal genera were specified. The general morphological characteristics of Trichoderma spp. were performed with the aid of Asis et al. (2021) and Siddiquee and Siddiquee (2017). Aspergillus species were identified according to Diba et al. (2007), Domsch et al. (1980), Samson and Pitt (2000). General characteristics of Penicillium spp. were identified compared with (de Hoog et al., 2005; Samson et al., 2014; St-Germain and Summerbell, 2003; Sutton et al., 1998). Morphological characteristics of the genus Mucor spp. were identified according to de Hoog et al. (2005) and Sutton et al. (1998).
2.2.2. Scanning electron microscopy
Fungal mycelium was fixed with 2% (w/v) osmium tetroxide vapor for 20 h, dehydrated through a graded ethanol series (30 –100%), and critical point dried using liquid CO2. Samples were mounted on copper stubs, sputter-coated with gold-palladium (15 nm thickness), and imaged using a JEOL JSM-7600F scanning electron microscope at 10 kV (Bozzola and Russell, 1999; Goldstein et al., 2017).
2.2.3 Molecular identification (ITS and tef1 gene sequencing)
The isolation and purification of genomic DNA from the selected fungus were performed using Norgen’s Plant/Fungi DNA Isolation Kit (Norgen Biotek Corp., Thorold, ON, Canada). The study focused on amplifying two specific regions: the internal transcribed spacer (ITS) region and the translation elongation factor 1-alpha (tef1) gene. The primer pairs used were ITS-1 F (5′-TCCGTAGGTGAACCTGCGG-3′)/ITS-4 R (5′-TCCTCCGCTTATTGATATGC-3′) for amplifying the ITS region (∼ 600 bp) and tef1-F (5′-CATCGAGAAGTTCGAGAAGG-3′)/tef1-R (5′-GCCATCCTTGGGAGATACCAGC-3′) for amplifying the tef1 gene (∼730 bp) (Cai and Druzhinina, 2021; White et al., 1990).
The PCR was conducted in a 50 μL mixture, comprising 25 μL of Master Mix (Sigma), 3 μL of each primer (10 pmol/μL), 3 μL of template DNA (10 ng/μL), and 16 μL of distilled water. Thermal cycling conditions consisted of an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 40 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min, with a final step at 72°C for 7 min. The PCR products underwent electrophoresis on a 1.5% agarose gel stained with ethidium bromide (0.5 μg/mL) in 1X TBE buffer at 95 V for 45 min, followed by visualization under UV light. A 100-nucleotide DNA ladder (Thermo Scientific, Germany) served as the molecular size standard. Subsequently, the PCR products were purified using an EZ-10 spin column PCR purification kit (Bio Basic Inc., Ontario, Canada). The PCR segments were sequenced bidirectionally using the same PCR amplification primers, and sequenced. The sequence was deposited in the GenBank of the National Center for Biotechnology Information after being aligned and compared to the database using the BLASTn algorithm tool.1 The evolutionary analyses of the nucleotide sequences were performed using MEGA11. For each sequence pair, the ambiguous positions were eliminated and inferred using the Neighbor-Joining technique (Saitou and Nei, 1987). Jukes-Cantor distances (Jukes and Cantor, 1969) were employed, and the optimal tree was presented with branch lengths reflecting evolutionary units and bootstrap support values (1000 replicates) (Felsenstein, 1985).
2.3 Fungal fermentation of RCBB
The previously collected common bean plants were used to prepare RCBB before being used as a fermentation substrate during SSSF. The RCBB was dried (70°C) overnight and ground into pieces, measuring 1–2 mm. The chemical composition of RCBB was carried out according to AOAC (2019).
The procedure of SSSF was carried out in 250 mL Erlenmeyer flasks. The RCBB-based medium was supported with 10 mL salt solution (composed of NaH2PO4; 12.8, KH2PO4; 3, NaCl; 0.5, MgSO4.7H2O; 0.5, NH4Cl; 1.0, and CaCl2.2H2O; 0.01 g/L, pH 6) (Tunga et al., 1998) per 1 g RCBB. The media were autoclaved (15 min/121°C) before being inoculated with 1 ml of 10∧6 spore suspension (from the 7-day-old culture) and incubated in a CGI-400P growth chamber (Taisite Lab.) under cool white, fluorescent light with a 12-hour light/darkness cycle (28 ± 2°C, 65% humidity, and illumination set to 4,000 lux.). A medium without inoculation was used as a control. After 10 days of incubation, 10 mL of Tween 80 (0.01%) was added, and the flasks were shaken (30 min/150 rpm), then filtered and centrifuged (3,354 g/20 min) to separate the fungal filtrate. The residual RCBB was weighted to calculate the remaining biomass (%).
2.4 Biochemical analysis of the fermented fungal filtrate
2.4.1 Protease assay and total free amino acids content
The proteolytic activity and TFAAs were determined, utilizing casein as a substrate and Folin-Ciocalteu reagent (Fluka, Biochemical Inc., Romania). Tyrosine was used as a standard (Cupp-Enyard, 2008). One unit (U) of protease is defined as the quantity of enzyme that liberates 1 μmol of tyrosine/gram/min (1 μmol/g/min) under the given conditions.
2.4.2 Cellulase assay
Cellulase activity was determined using microcrystalline cellulose as the substrate. Briefly, the reaction mixture contained 1.5 mL of 1% (w/v) microcrystalline cellulose in 0.05 M sodium citrate buffer (pH 4.8) and 0.5 mL of fungal filtrate. The mixture was incubated at 50°C for 1 h (Saber et al., 2015). Next, the released reducing sugars were quantified using the 3,5-dinitrosalicylic acid (DNS) method (Miller, 1959). Glucose was used as the standard for calibration. One cellulase U was defined as the enzyme amount required to release one μmol of glucose/min/g RCBB under specified assay conditions.
2.4.3 Xylanase assay
Xylanase activity was measured using 1% (w/v) oat spelt xylan (Sigma-Aldrich). Briefly, the reaction mixture comprised 0.5 mL of fungal filtrate and 1.5 mL of substrate solution in 0.1 M phosphate buffer (pH 6.0), then incubated at 50°C for 15 min (Abdelwahed et al., 2011). The released reducing sugars were quantified via the DNS method (Miller, 1959), with xylose (Sigma-Aldrich) as the standard. One xylanase U was defined as the enzyme quantity required to release 1 μmol of xylose/min/g RCBB under the assay conditions.
2.4.4 High-performance liquid chromatography
The amino acids (AAs) profile of the fungal filtrate was detected with an Agilent 1260 series HPLC system equipped with an Eclipse Plus C18 column (4.6 × 250 mm i.d., 5 μm). A mobile phase composed of sodium phosphate dibasic/sodium borate buffer (pH 8.2), acetonitrile, methanol, and water (45:45:10 v/v/v) was employed at a flow rate of 1.5 mL/min. The filtrate (1.25 mL) was acid hydrolyzed by mixing with 1.25 mL of water and 2.5 mL of 6 M HCl. The resulting mixture was incubated at 100°C for 24 h and subsequently filtered. One milliliter of the processed filtrate was injected into the HPLC system for analysis (Jajić et al., 2013). Identification of AAs was achieved by comparing retention times of the calibration curve generated from a standard under identical conditions.
2.4.5 Gas chromatography-mass spectrometry analysis
The lyophilized fungal filtrate was resuspended in methoxyamine hydrochloride (20 mg/mL pyridine) for oximation (90 min). Bis(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane was then added for derivatization (70 °C, 30 min) to TMS derivatives suitable for GC-MS. An Agilent Technologies 7890B GC coupled to a 5977A MS was used for analysis. The HP-5MS column (30 m × 0.25 mm × 0.25 μm) with the carrier gas (hydrogen, 1.0 mL/min) and a 2 μL injection volume separated the metabolites. The oven program started at 50°C and ramped to 300°C at 10°C/min. The detector and injector temperatures were kept at 250°C. Electron ionization (70 eV) generated mass spectra (m/z 30–700) after a 9-min solvent delay. The temperatures of the mass and transfer line were 230 and 150°C, respectively. Metabolite identification utilized the Wiley and NIST Mass Spectral Libraries.
2.4.6 Phytochemical content in the fungal filtrate
The photochemical content in the fungal filtrate, resulting from the degradation of plant biomass, was determined. The total of phenolics was measured using the Folin-Ciocalteu reagent, using gallic acid as a standard (Sánchez-Rangel et al., 2013). The total flavonoids were measured, applying catechin as a standard (Zhishen et al., 1999). The tannin content was measured using the vanillin-hydrochloride method (Aberoumand, 2009; Burlingame, 2000). Tannic acid was utilized as a standard to determine the total tannin content.
2.4.7 Fourier transform infrared spectroscopy
The fungal filtrate (5 mg) was mixed with 100 mg potassium bromide, and the mixture was analyzed using a Bruker Vertex 70 RAM II FT-IR spectrometer. Spectra were recorded in the 4,000–500–1 range at 4 cm–1, with 32 scans averaged per sample. Peaks were analyzed for functional groups to characterize the chemical composition.
2.5 Biological activity of the fermented fungal filtrate
2.5.1 Cytotoxicity and anticancer tests
The MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay is largely used for measuring cell viability and cytotoxicity of a compound. Normal (human lung fibroblast; WI38) and tumor (hepatocellular carcinoma; HePG2, and mammary gland breast cancer; MCF7) cell lines were obtained from ATCC through VACSERA, Cairo, Egypt. Doxorubicin served as the standard chemotherapeutic anticancer (control). Cells were cultured in RPMI-1640 medium (Sigma Co., Louis, MO, United States) supplemented with fetal bovine serum (10%) (Thermo Fisher Scientific, GIBCO, United Kingdom), streptomycin (100 μg/mL), and penicillin (100 units/mL) under 5% CO2 conditions at 37°C. After seeding individually in a 96-well plate at a density of 104 cells/well, the cells were incubated (48 h/37°C with 5% CO2), then exposed to various concentrations (1.56–100 μg/mL) of the lyophilized fungal filtrate for an additional 24 h. Next, 20 μL (5 mg/mL) of MTT was mixed and incubated for 4 h. Hundred microliter of dimethyl sulfoxide was added to the wells to dissolve purple formazan (Denizot and Lang, 1986). The absorbance (A at λ = 570 nm) of the developed color intensity was measured utilizing a plate reader (EXL 800, Cranston, RI, United States). Relative cell viability was calculated (Equation 1), and the IC50 against the cell line was determined.
2.5.2 Animal studies
Male Wistar rats (6 weeks old, 164–176 g), gifted from the Animal House of the National Research Center, Cairo, Egypt, were acclimatized for 1 week under standard laboratory conditions. Animals were provided with standard rodent chow and tap water ad libitum and kept on a 12 h light/12 h dark cycle at 24 ± 1°C and humidity of 50 ± 10%.
2.5.2.1 Anti-hemolytic assay
The erythrocyte hemolysis assay was used to assess the antihemolytic activity of the fungal filtrate to protect red blood cells (RBCs) from lysis induced by oxidative stress. The blood samples were collected into heparinized tubes from rats (n = 6 per group) through cardiac puncture (Yang et al., 2005). Erythrocytes were isolated from the plasma by centrifugation. The buffy coat was then washed thoroughly 3 times with sterile saline (0.89% w/v NaCl, pyrogen-free) to eliminate residual plasma. The washed erythrocytes were centrifuged at 834 g for 10 min to obtain a standardized erythrocyte suspension. The fungal filtrate (5 mg/mL) was added to a 10% erythrocyte suspension in phosphate-buffered saline (pH 7.4) and incubated for 45 min. at 37°C. A control sample containing L-ascorbic acid was incubated with the erythrocyte suspension without the fungal filtrate. Distilled water served as the maximal hemolytic control. Following incubation, erythrocytes were separated by centrifugation. Hemoglobin release, indicative of lysed RBCs, was quantified at 540 nm. The percent hemolysis was calculated by subtracting the value obtained from the saline control-treated group. The hemolysis percentage was calculated using Equation (2):
2.5.2.2 Anti-inflammatory assay
The anti-inflammatory activity of the fungal filtrate was appraised in vivo by applying carrageenan-induced paw edema in a rat model (Morris, 2003). Briefly, carrageenan, a polysaccharide derived from red seaweed (1% in sterile saline, Sigma Co., United States), was prepared by heating to 90°C without boiling and cooling to room temperature. Rats (18 individuals) were randomly assigned to three groups (n = 6/group) using a blinded randomization procedure to minimize bias. The right hind paw of each rat received a sub-plantar injection of carrageenan solution (0.1 mL). The treatment group received an intraperitoneal injection of the fungal filtrate (5 mg/mL), while the positive control group received indomethacin (a nonsteroidal anti-inflammatory drug, NSAID) at 5 mg/kg. Paw thickness was measured using a vernier caliper at baseline and 0.5, 1, 2-, 3-, 4-, and 5 h post-injection. Paw edema was calculated for each rat by subtracting the baseline paw thickness from the thickness measured at each time point. The anti-inflammatory protection (reduction in paw edema (swelling) induced by carrageenan) was calculated (Equation 3):
No euthanasia was used. All protocols complied with the guidelines and regulations of the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no.85-23, revised 1996) (Morris, 2003). The study followed the guidelines of Animal Research: Reporting of In Vivo Experiments (ARRIVE).2
2.6 Data analysis
CoStat (version 6.45, CoHort Software, Birmingham, UK) software was utilized. One-way ANOVA of a completely randomized design was applied to analyze the data mean (± standard deviation, SD). Tukey’s HSD (Honestly Significant Difference) post-hoc test (α ≤ 0.05) was used for pairwise comparisons of means, applying minimum significant difference.
3 Results
3.1 The recovered fungal isolates
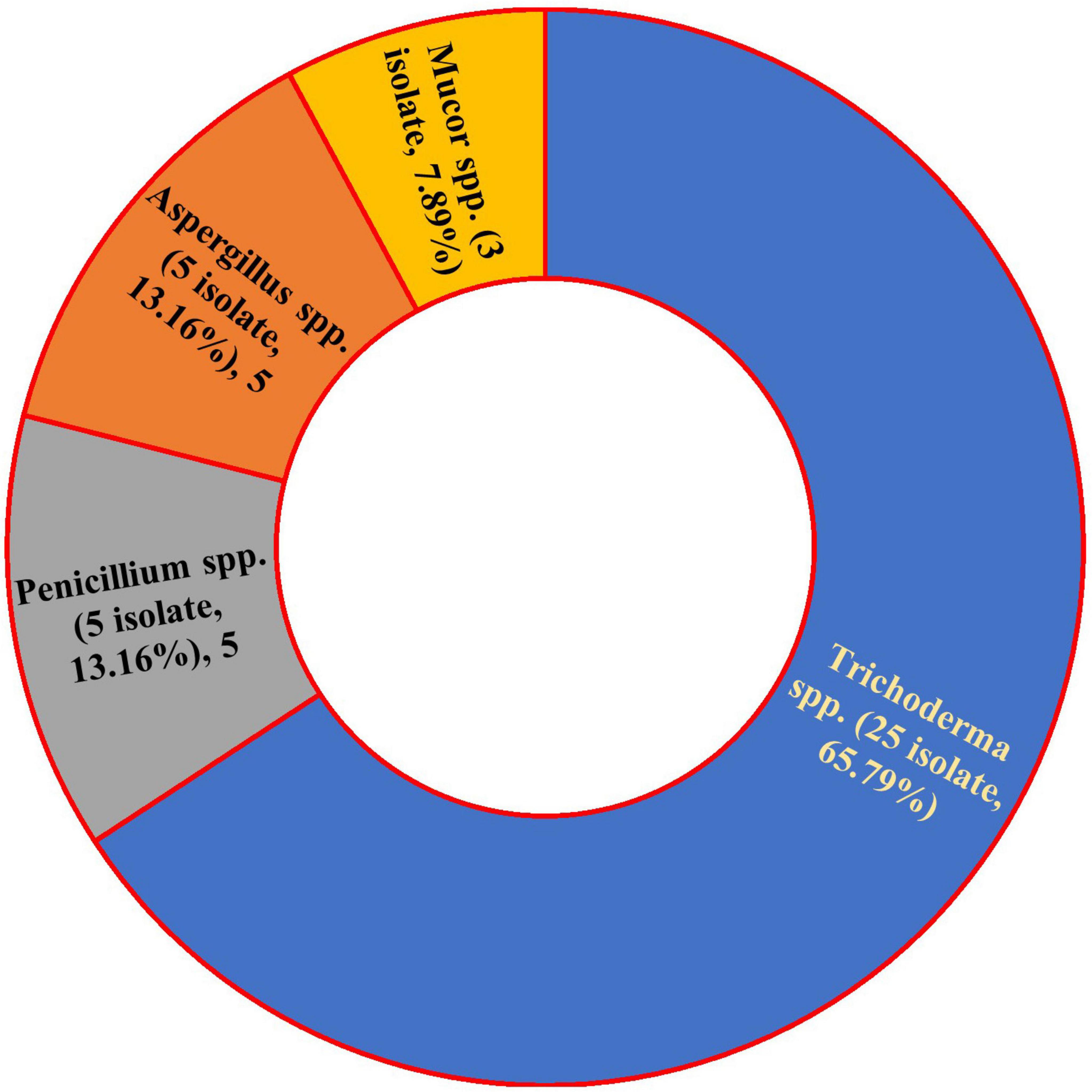
This study isolated 38 endophytic fungi from common bean plants (Figure 1). The fungal isolates were initially identified to belong to four genera: Trichoderma spp. (25 species, 65.8%), Aspergillus spp. (5 species, 13.2%), Penicillium spp. (5 species, 13.2%), and Mucor (3 species, 7.9%). Trichoderma spp. exhibited the highest prevalence among the isolated endophytes. The macroscopic and microscopic features of the four isolated fungal species were determined (Supplementary File).
3.2 Screening the hydrolytic activity
First, the chemical composition of RCBB consists of 90.0% dry matter, with a 6.42% ash content. The organic matter content is 85.58%, which includes 15.80% crude protein and 37.50% crude fiber. The ether extract is 1.520%. The organic carbon percentage is calculated to be 38.51%, and the total nitrogen (N) content is 2.53%. This results in a C/N ratio of 15.22, indicating the balance of carbon and nitrogen in RCBB.
The ability of the endophytic fungi to degrade RCBB using SSSF (Table 1 and Supplementary Figure 1) was tested. Among the fungi studied, Trichoderma sp. WNZ-21 was selected as a notably robust profile of hydrolytic enzymes, including cellulase (18.067 U), xylanase (15.967 U), and protease (5.550 U), and yielded the highest levels of TFAAs (12041.97 μg/g RCBB), achieving the greatest biomass degradation (32.81 ± 0.65% remaining biomass) compared to others.
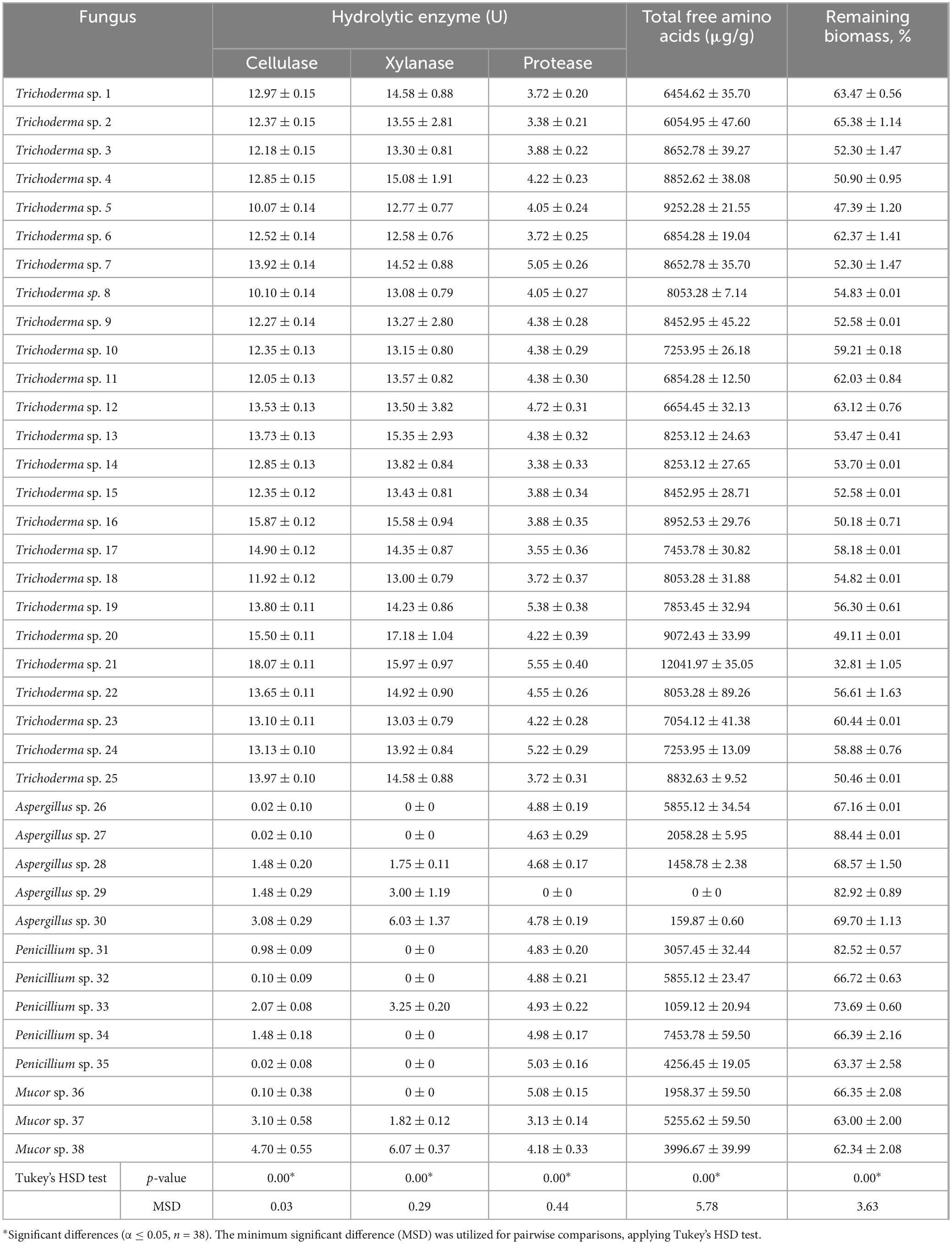
Table 1. Hydrolytic potentiality, total free amino acids production, and the remaining biomass as a result of the fungal activity on semi-solid-state fermentation medium containing the residue of common bean biomass as substrate.
3.3 Identification of Trichoderma sp. WNZ-21
3.3.1 Morphological and microscopic features
The macroscopic characteristics of the front and reverse views of Trichoderma sp. WNZ-21 (Figures 2A,B) shows the development of dense conidia, with a green center and white conidia towards the periphery. The reverse of the colony is creamy and often folded or convoluted. No pigment diffusion is observed on the PDA plate. There is neither aerial mycelium nor sweet coconut odor.
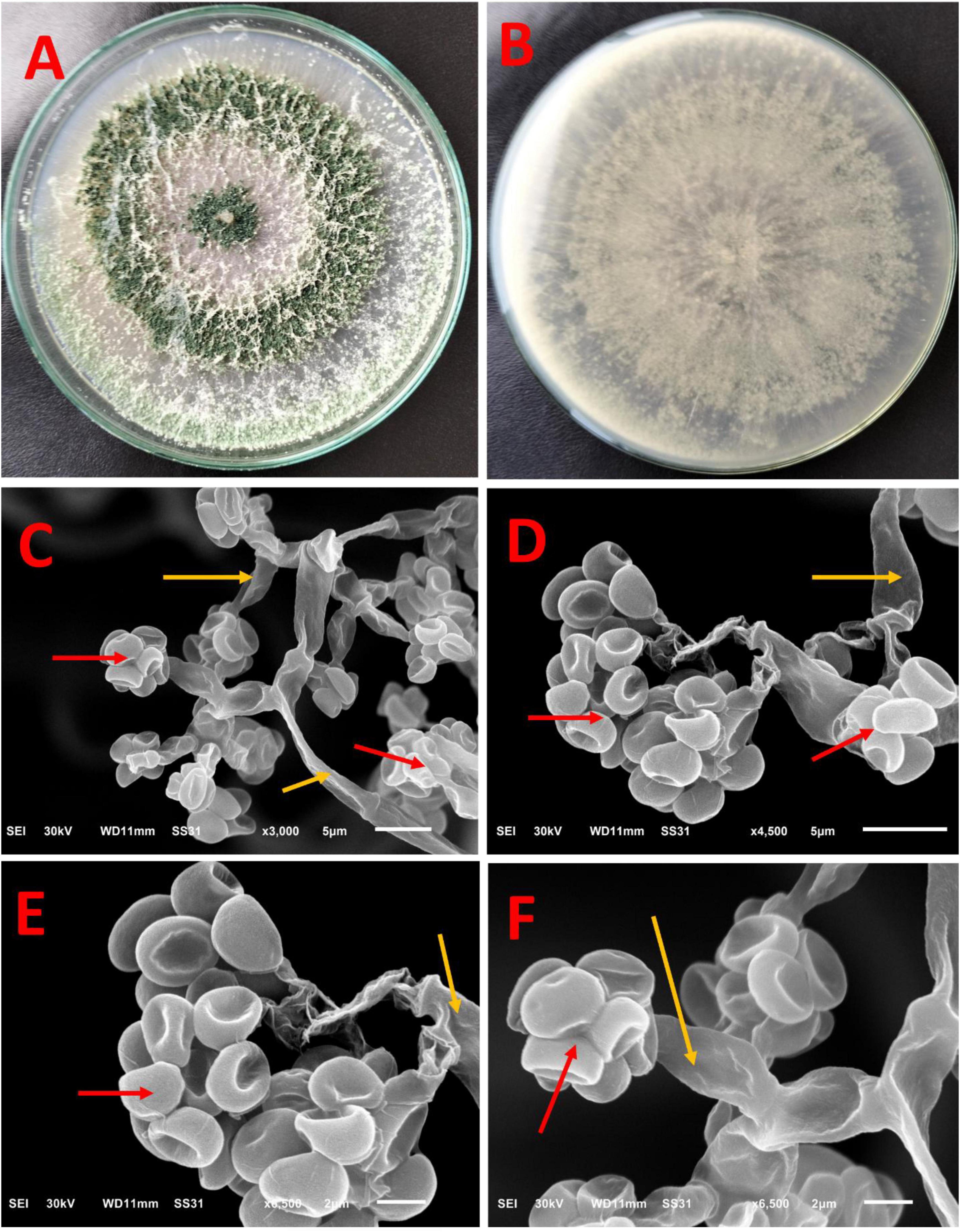
Figure 2. Macroscopic and ultrastructure of Trichoderma sp. WNZ-21 after 5 days of growth at 28 ± 2°C. Macroscopic view: (A) front view and (B) reverse view. Scanning electron microscopy under different magnifications (C–F) shows conidia (red arrows) and phialides (yellow arrows).
The SEM (Figures 2C–F) shows phialides, produced at the tips of these branches and arranged in whorls of 2–4, are straight, ampulliform, and slightly enlarged in the middle, measuring an average of 9–12 micrometers long and 1–3 micrometers wide. This isolate does not contain any intercalary phialides. Additionally, dark green conidia were observed. These conidia, globose to sub-globose or ovoidal in shape, measure between 3 and 3.5 micrometers long and 2–2.5 micrometers wide. The red arrows refer to conidia, while the yellow arrows refer to phialides.
3.3.2 Molecular identification
The BLAST analysis of the ITS region against the NCBI nucleotide database (Supplementary Table 1) revealed a high degree of similarity among the analyzed sequences with a consistently significant E-value of zero across all comparisons. Notably, eight out of the twelve most similar sequences corresponded to T. asperellum, while the remaining four belonged to the same genus (Trichoderma sp.) but lacked species-level identification. This data strongly supports the fungus as T. asperellum based on the ITS region. Further analysis identified T. asperellum strain MF632083.1 as the closest match, exhibiting a significantly higher sequence identity (97.88%) compared to other strains (94.42 to 94.62%). Importantly, all strains, including WNZ-21, displayed 100% query coverage, further solidifying the identification. The phylogenetic tree (Figure 3) depicts the relationship between T. asperellum WNZ and other related strains based on genetic markers. It is positioned closely to T. asperellum strain MF632083. This visual representation corroborates with BLAST analysis (Supplementary Table 1), highlighting a closer genetic affinity between these two strains compared to others. However, the phylogenetic analysis, together with ITS and BLAST, is consistent with the previous morphological characteristics, confirming the identification as T. asperellum.
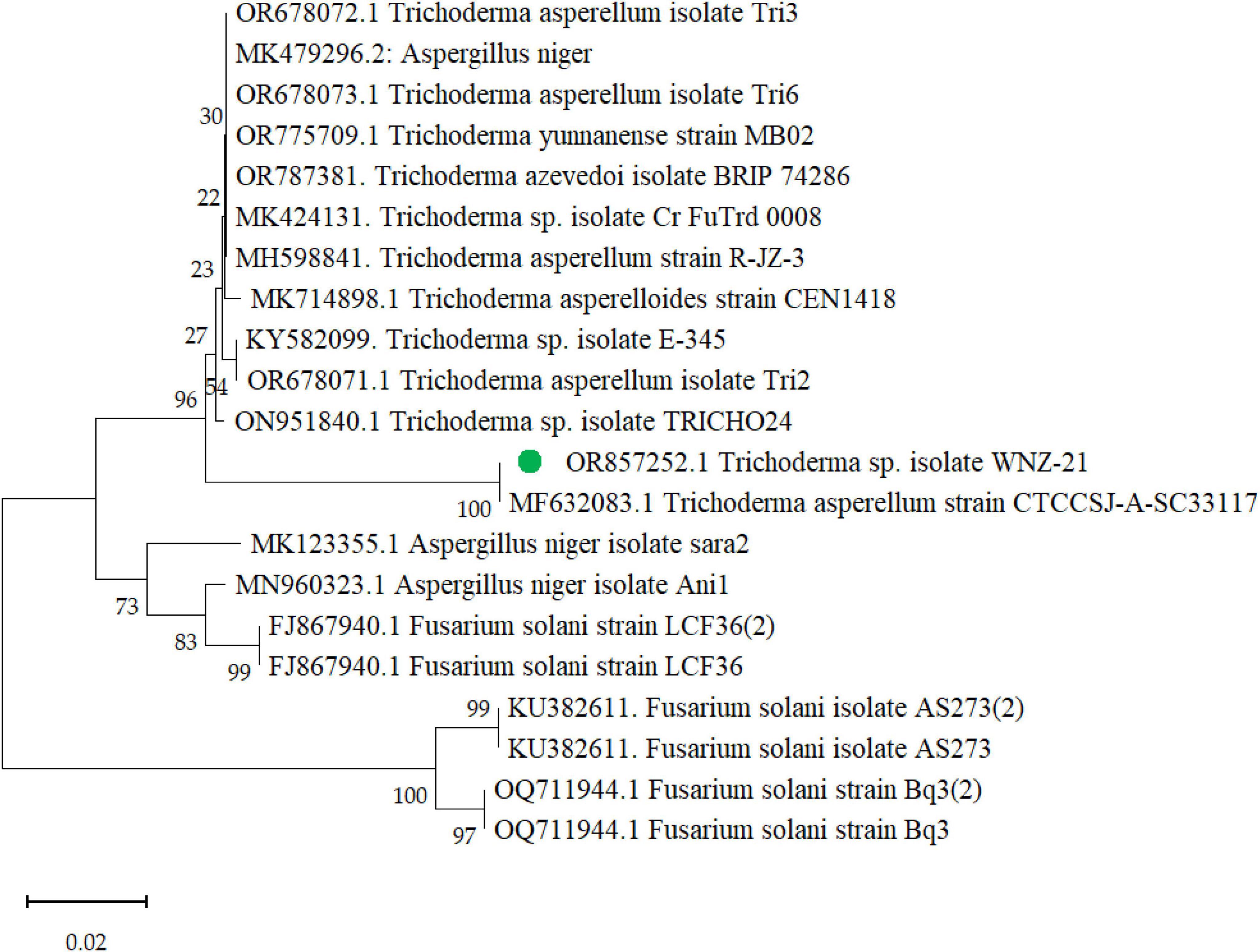
Figure 3. Phylogenetic tree of Trichoderma asperellum WNZ-21 (designated in green dot) and related strains based on ITS region sequences with accession number OR857252.
Furthermore, the phylogenetic exploration based on the tef1 gene confirmed the identification of our isolate (PP069312) as T. asperellum. BLAST analysis (Supplementary Table 2) showed high sequence similarity (99–100%) to other T. asperellum sequences in GenBank. Phylogenetic analysis (Figure 4) placed our isolate within a monophyletic T. asperellum clade with strong bootstrap support, indicating a close evolutionary relationship with other members of this species. Our isolate formed a distinct subclade within the T. asperellum group, suggesting potential genetic divergence and potentially unique characteristics warranting further investigation.
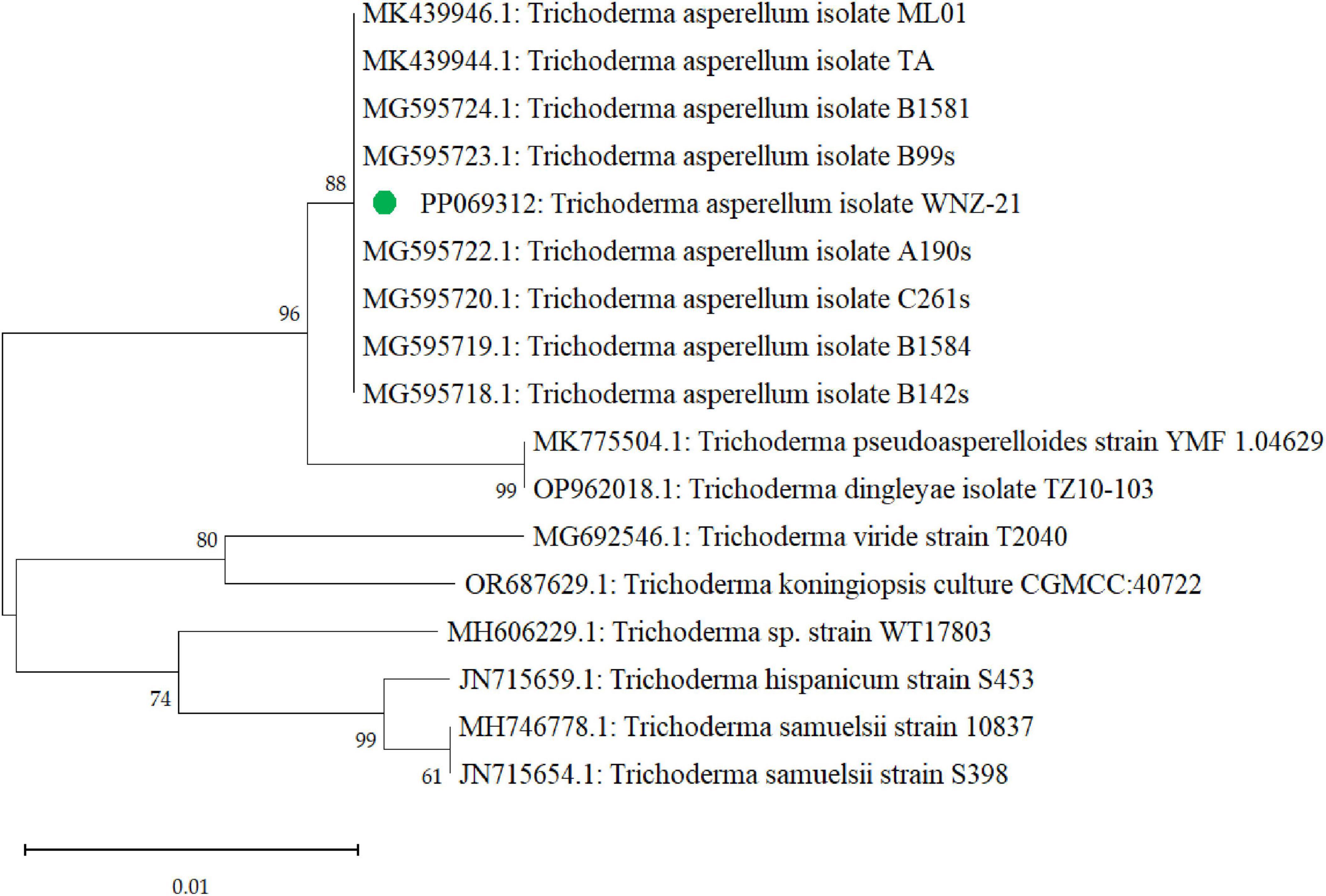
Figure 4. A phylogenetic tree that was generated through sequence analysis of the tef1 gene illustrates the positioning of the Trichoderma asperellum isolate identified via the gene sequence (PP069312, green dot) among the related sequences.
3.4 Analysis of T. asperellum filtrate
3.4.1 Amino acid profile
The degradation of RCBB by T. asperellum released 16 AAs (Figure 5), with a total amount being 13509.21 μg/g RCBB. Common AAs include glutamic acid, aspartic acid, serine, glycine, threonine, arginine, histidine, alanine, tyrosine, and methionine. Other notable AAs included cystine, valine, isoleucine, phenylalanine, leucine, lysine, and proline. Lysine was the most abundant individual compound, accounting for almost 30.76% of the total area. Eight of these AAs are essential for human health, while eight are non-essential. Four of the non-essential AAs are conditionally essential. The highest concentrations of AAs were found in lysine (8324.55 μg/g RCBB), glutamic acid (874.14 μg/g RCBB), proline (842.95 μg/g RCBB), tyrosine (396.68 μg/g RCBB), serine (392.89 μg/g RCBB), and threonine (386.65 μg/g RCBB).
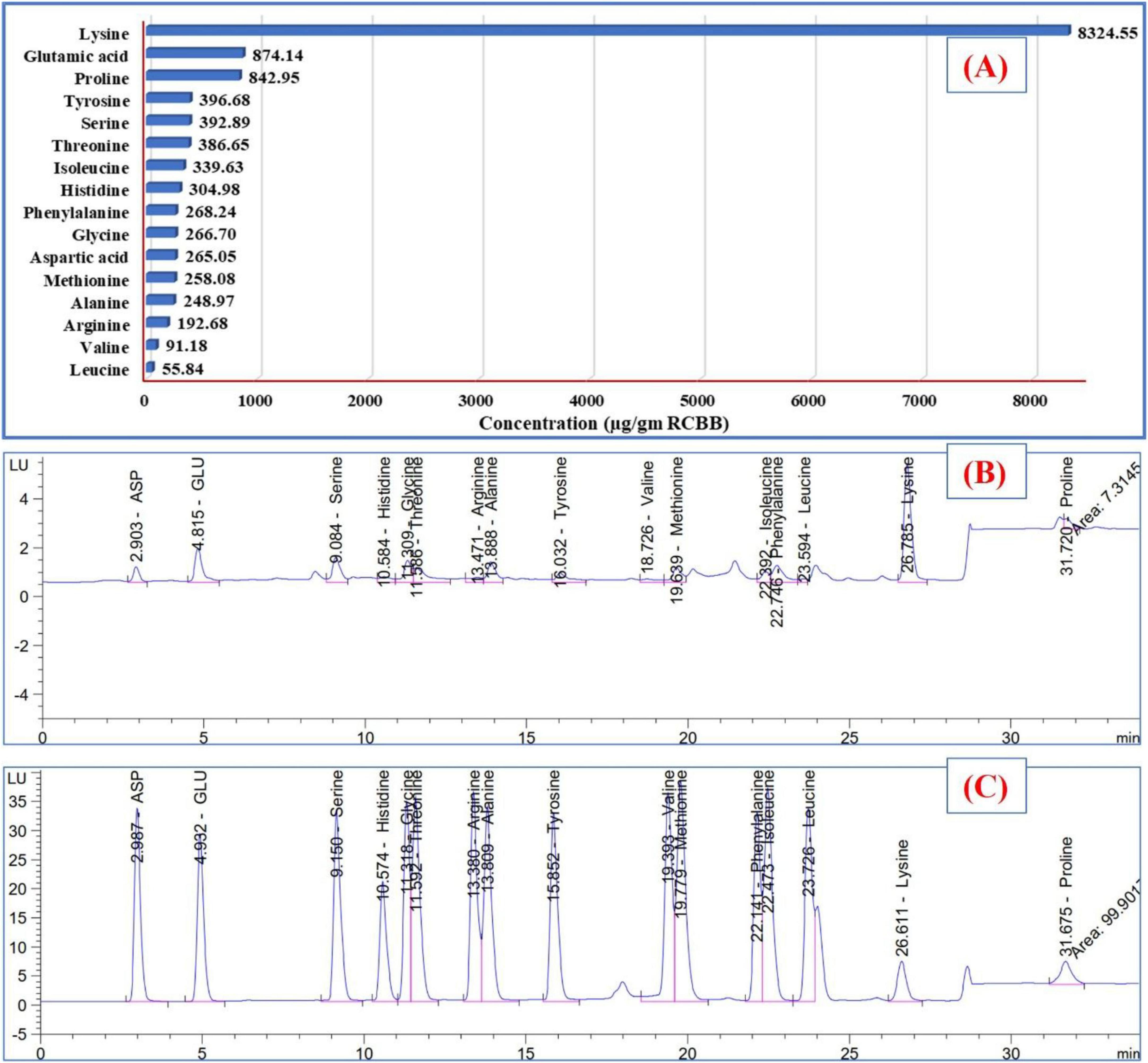
Figure 5. The amounts (A) of amino acids detected in the filtrate of the fungus grown on the residue of common bean biomass, and (B) HPLC chart, and amino acids standard chart (C).
3.4.2 GC-MS analysis of the fungal filtrate
GC-MS confirmed RCBB degradation by T. asperellum WNZ-21, identifying 17 metabolites (Table 2, Supplementary Table 3, and Supplementary Figure 2). Key bioactive entities included D-Pinitol (47.04%) and erythro-pentonic acid derivatives (18.65%). The remaining components include a diverse array of trace fatty acids and ester derivatives. These findings highlight the fungus’ capacity to transform plant biomass into structurally diverse metabolites with therapeutic potential. However, Future studies will prioritize isolation and HR-MS/NMR characterization.
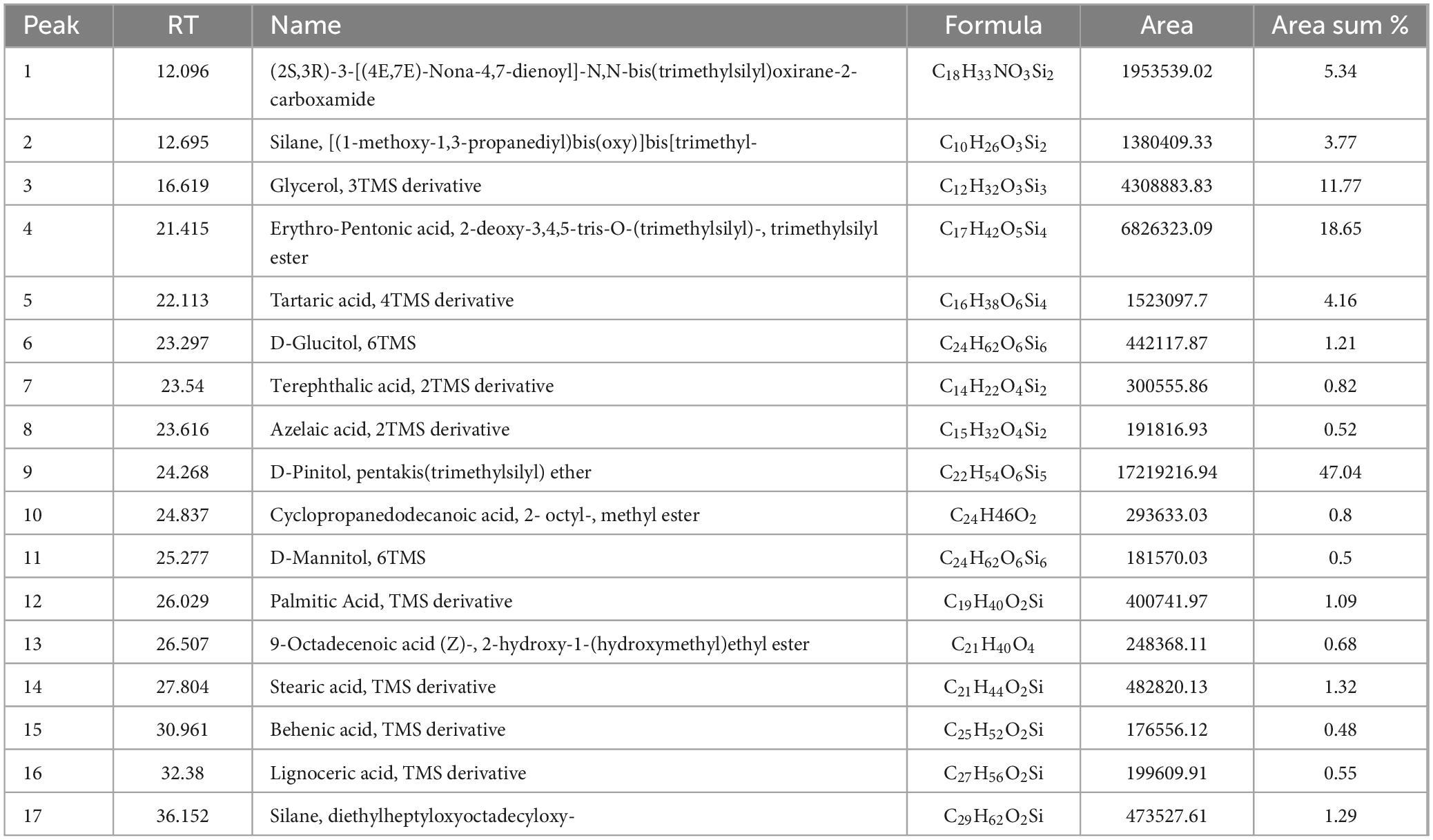
Table 2. GC-MS analysis of Trichoderma asperellum filtrate grown on the residue of common bean biomass.
3.4.3 Phytochemical and FT-IR spectral analysis
Finally, the fungal filtrate exhibited significant phytochemical content (Supplementary Figure 3), including phenolics (55.22 mg/g), flavonoids (11.22 mg/g), and tannins (18.90 mg/g). FT-IR analysis (Supplementary Figure 4 and Supplementary Table 4) corroborated these findings, revealing NH/C-N stretches (3362–1231 cm−1) indicative of amino acids and proteins, C = O bands (1,713–1,645 cm–1) from carboxylic acids and peptides, and polysaccharide-associated C-O stretches (1,066 cm–1). These results align with GC-MS-detected metabolites (e.g., D-Pinitol, fatty acids) and underscore the filtrate’s chemical complexity.
3.5 Biological activity of the fungal filtrate
3.5.1 Cytotoxic study on cell lines
The MTT assay was applied, and doxorubicin was utilized as a standard drug (Supplementary Figure 5). As a general rule, there was a negative correlation (r) between cell line viability and the concentration of the fungal filtrate. The values of r were −0.9380, −0.8967, and −0.8567 (p ≤ 0.05) for WI38, HePG2, and MCF7, respectively.
The average of relative viability reached 47.5 ± 1.6, 29.8 ± 1.2, and 23.4 ± 0.4 at 100 μg/mL of fungal filtrate for WI38, HePG2, and MCF7 cell lines, respectively. Lower concentrations resulted in higher cell viability. No cytotoxicity was recorded at 6.25 (WI38), 3.125 (HePG2), and 1.56 (MCF7) μg/mL.
Furthermore, the cytotoxic potency of the fungal filtrate against the three cell lines was calculated (Figure 6), where the IC50 values were 76.33 ± 3.9, 35.62 ± 2.1, and 59.20 ± 1.7 μg/mL for WI38 (normal cell line), HePG2, and MCF7 (tumor cell lines), respectively. Notably, the fungal filtrate was weak against the normal cell line, whereas it exhibited selective cytotoxicity against tumor cell lines.
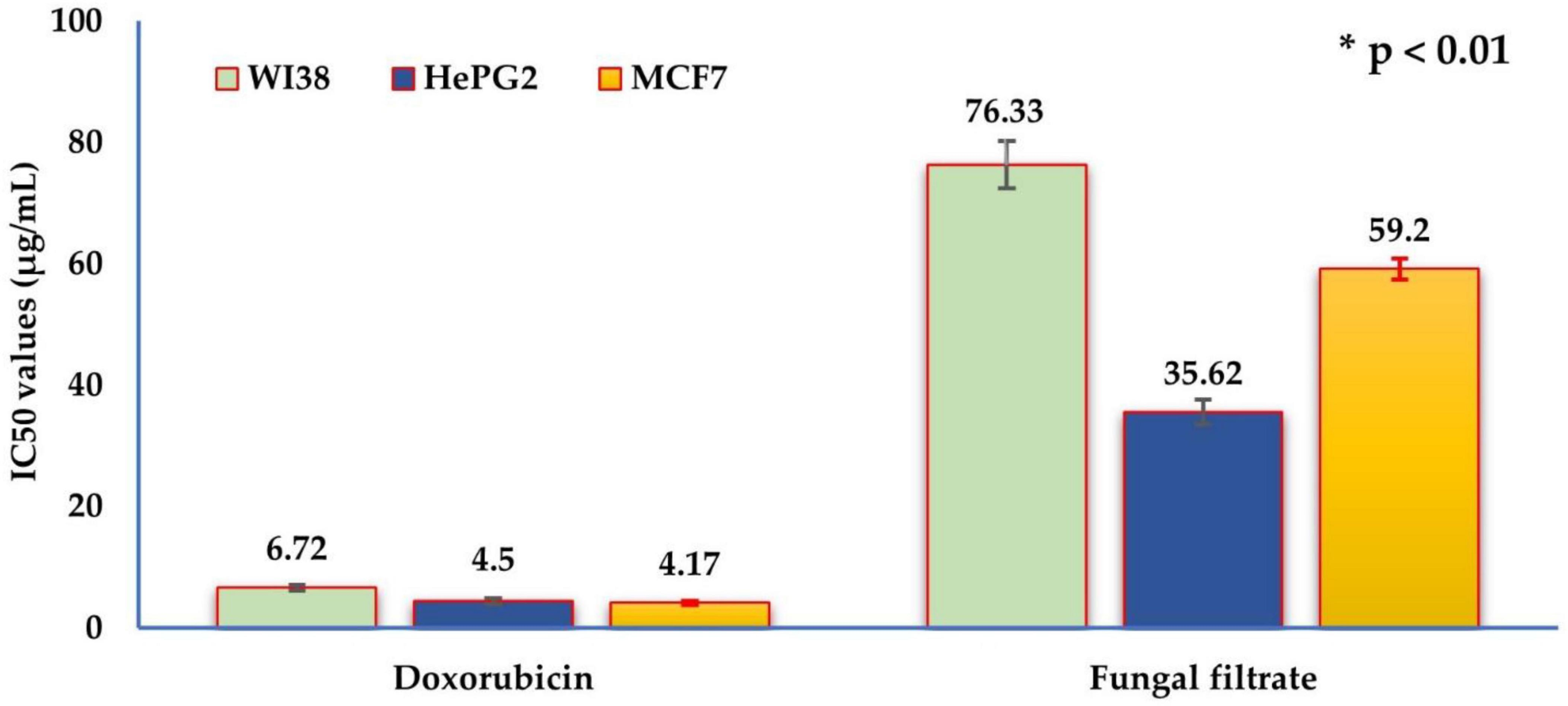
Figure 6. The IC50 values of the in vitro cytotoxic activity of Trichoderma asperellum filtrate and standard doxorubicin drug against the three human cell lines: human lung fibroblast (WI38), hepatocellular carcinoma (HePG2), and mammary gland breast cancer (MCF7). The half maximal inhibitory concentration (μg/mL) categories are very strong (1–10), strong (11–20), moderate (21–50), weak (51–100), and non-cytotoxic (>100).
3.5.2 Anti-hemolytic activity
The results of the erythrocyte hemolysis activity of blood samples (Table 3) indicated a relatively low hemolysis percentage of fungal filtrate (4.40 ± 0.69%), indicating reasonably anti-hemolytic activity. However, L-ascorbic acid showed lower hemolysis as an antioxidant and can protect cells, including red blood cells, from oxidative damage.
3.5.3 Anti-inflammatory activity on rats
The carrageenan-induced paw edema assay on rats (Figure 7) demonstrated a time-dependent anti-inflammatory response elicited by both indomethacin and the tested fungal filtrate. At zero time, a baseline level of protection is evident in both groups, with the fungal filtrate showing marginally higher protection (indomethacin: 0.71%, fungal filtrate: 1.18%). As the experiment progresses, both substances exhibit an increase in protection percentages. Notably, indomethacin demonstrates a consistent and pronounced rise in protection, reaching 58.41 % after 5 h. T. asperellum WNZ-21 filtrate also exhibits anti-inflammatory action, although at a lower level of protection compared to indomethacin. At the 5-h point, the fungal filtrate demonstrates a protection percentage of 36.50%.
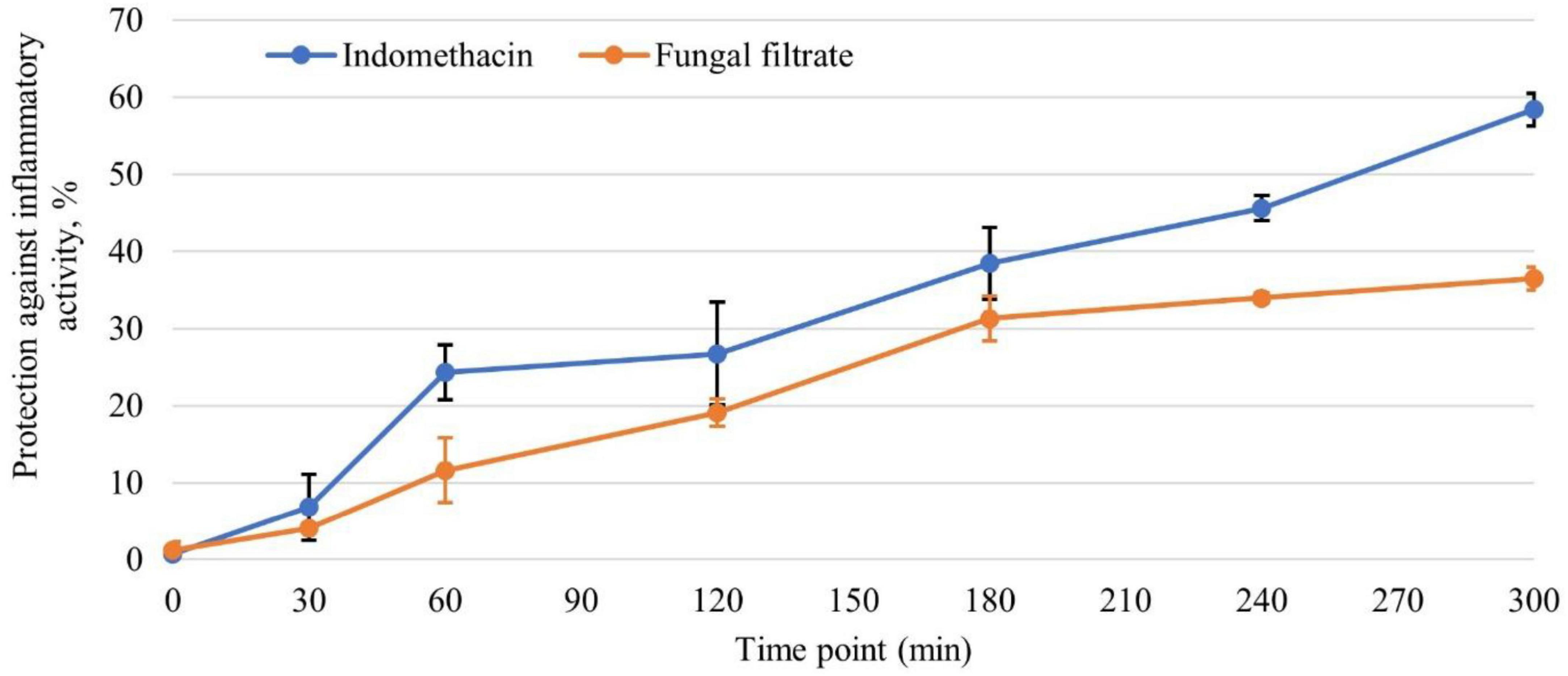
Figure 7. The efficacy of Trichoderma asperellum filtrate in protecting against inflammatory activity at different time points, in comparison with the standard indomethacin drug (n = 6).
4 Discussion
Endophytes, nonharmful microbes residing within plant tissues throughout part or all of their life cycle, represent a significant and often underexplored source of diverse enzymes and secondary metabolites with unique features and therapeutic potential (Al-Askar et al., 2022; Alrdahe et al., 2024; Moussa et al., 2023). Recognizing this vast potential and the pressing need for sustainable strategies for agricultural residue valorization, the central aim of this study was to explore a novel approach: the bioconversion of RCBB into valuable bioactive compounds. We specifically investigated the potential of using SSSF mediated by a selected potent endophytic fungus to achieve this goal and identify multi-bioactive metabolites.
Initially, the chemical composition of RCBB used in the current study aligned with previous studies (de Castro et al., 2016; de castro et al., 2021; Ferro et al., 2020; Santos et al., 2018) with slight differences. These differences may be attributed to the variety, growing season, and the ratio of plant parts.
Our initial investigation into the endophytic fungal community of common bean revealed the predominance of Trichoderma spp., consistent with their known metabolic versatility and prevalence in plant hosts, further supporting our focus on this genus. In comparison, 26 fungal species were recovered from the peanut plants, and some of the isolates produced active metabolites, including enzymes and citric acid (Al-Askar et al., 2022).
All isolates grew well and produced protease and released AAs, which are the key enzymes/factors in the biomass breakdown (El-Hersh et al., 2016; El-Hersh et al., 2014; Negm et al., 2021). The ability of the endophytic fungi to degrade RCBB using SSSF was confirmed. The SSSF technique is rarely used, especially with endophytes. It was chosen here due to its simplicity, low energy needs, and ability to mimic the natural growth environment of the fungi (AbdAl-Aziz et al., 2012; Negm et al., 2021).
Compared to the other isolates, which showed moderate degradation, the exceptional biomass degradation efficiency of isolate WNZ-21 underscores its role in biomass breakdown and the synthesis of secondary metabolites.
Trichoderma sp. WNZ-21 had an efficient enzyme profile and degraded RCBB, thus selected for further studies. RCBB is a complex substrate rich in hemicellulose, cellulose, and protein, its degradation requires several hydrolytic enzymes (cellulase, xylanase, and protease). While enzyme activity varied among isolates, most produced the three enzymes. The significant variation observed in the hydrolytic profile among different fungi may be attributed to differences in strain types and genetic variations (Al-Askar et al., 2022). Cellulose is broken down by cellulases into individual glucose monomers (Al-Askar et al., 2022). Likewise, xylanase facilitates the breakdown of hemicellulose (xylan), releasing xylose units (Abdelwahed et al., 2011; Bailey et al., 1992). Proteases break down the protein component of plant tissues into free AAs and peptides (Al-Askar et al., 2021) using endopeptidases and exopeptidases. Exopeptidases catalyze the terminal peptide bonds, while endopeptidases catalyze the non-terminal peptide bonds (El-Hersh et al., 2014). The synergistic action of these hydrolytic enzymes resulted in the breakdown of plant tissues into free AAs (Al-Askar et al., 2021; Al-Askar et al., 2022; Moussa et al., 2021).
Trichoderma sp. WNZ-21 was identified as T. asperellum based on morphological, cultural, and SEM examination. While these methods provide valuable insights, definitive fungal identification typically relies on a combination of morphological analysis and molecular techniques. The ITS region is a well-established fungal barcode due to its high sequence variability and conserved flanking regions, enabling accurate and reliable species-level identification (AbdAl-Aziz et al., 2012; Raja et al., 2017; Saber et al., 2010; Tretter et al., 2012).
Molecular identification via ITS (GenBank: OR857252) and tef1 gene (GenBank: PP069312) sequencing confirmed the isolate as T. asperellum WNZ-21. Phylogenetic analysis resolved species-level ambiguities, aligning with prior taxonomic frameworks (De Bruyn et al., 2014; Hermosa et al., 2004). ITS is a valuable molecular marker for identifying Trichoderma species, highlighting the resolution and accuracy at the species level, while tef1 is often employed in multiplex PCR assays for the precise differentiation of Trichoderma species (Hermosa et al., 2004; Letunic and Bork, 2021).
The fungal filtrate was analyzed for bioactive metabolites. The presence of these compounds indicated the successful degradation of plant biomass during fermentation.
The phytochemical content of the filtrate displayed levels of phenolics comparable to or exceeding values reported for T. harzianum filtrates (Lakhdari et al., 2023) and surpasses those of T. harzianum and Glomus versiforme (Omomowo et al., 2020), suggesting the antioxidant potential of phenolic compounds (Lakhdari et al., 2023). Flavonoid content aligns with the level documented for T. harzianum filtrates (Lakhdari et al., 2023). Flavonoids have health-promoting properties with antioxidant capacity (Elattar et al., 2024; Lakhdari et al., 2023). Tannin of the present isolate is considerably higher than those reported for T. harzianum filtrates and the Calotropis procera filtrate, where tannins can have diverse effects depending on their type and interaction with other components (Nofal et al., 2023). The fungal filtrate under study demonstrates a promising phytochemical profile, suggesting potential for further exploration.
The release of AAs and biomass reduction (%) provides further evidence of the degradation during fungal fermentation. However, the type and the amounts of AAs vary depending on the substrate, microbe, and fermentation settings. For instance, 14 AAs were released during the fermentation of chicken feathers by Bacillus paramycoides, where proline and aspartic acid had the highest concentrations of the released AAs (Moussa et al., 2021). Thrane et al. (2001) identified aspartic acid, glutamic acid, threonine, and alanine. However, the variation of the AAs profile suggests potential strain-specific variation in metabolic pathways. Furthermore, several AAs are reported to have antifungal activity, suggesting potential biocontrol capabilities (Moo-Koh et al., 2022).
While native RCBB contains 15.80% crude protein and 37.50% crude fiber, its recalcitrant lignocellulosic matrix prevents the spontaneous release of bioactives, as confirmed by the non-inoculated control. Fungal degradation via robust hydrolytic unlocks structural components, generating monomers that fuel secondary metabolism. While this study does not present time-course enzyme activity data, the 10-day SSSF protocol aligns with established fungal bioprocess timelines, where peak hydrolytic activity typically occurs within 3–5 days (Alrdahe et al., 2024; El-Naggar et al., 2014; Saber et al., 2015). The biomass degradation by T. asperellum WNZ-21 (67.19% RCBB), for instance, positively correlates with its enzymatic activity and TFAA yield. For instance, AAs and phenolic liberation are direct consequences of protease- and ligninolytic-driven degradation, respectively, which concluded that fungal enzyme activity and substrate degradation are interdependent processes (De Silva et al., 2019; Moussa et al., 2023). Future studies will incorporate kinetic enzyme assays and substrate degradation profiles to optimize bioprocessing parameters, building on the current findings.
The genus Trichoderma exhibits a diverse metabolite profile under standard growth conditions (25°C, pH 6.0, glucose-based media), characterized predominantly by peptaibols, polyketides, terpenes, and volatile organic compounds (Moussa et al., 2023; Reino et al., 2008). Key metabolites include 6-pentyl-α-pyrone, harzianic acid, harzianolide, and various trichothecenes that contribute to their biocontrol capabilities (Vinale et al., 2014). Peptaibols such as alamethicin and trichokonin demonstrate significant antimicrobial activity through membrane permeabilization (Zhishen et al., 1999). The secondary metabolome also features gliotoxin, gliovirin, and koninginins, which vary in concentration depending on species and strain (Zeilinger et al., 2016). Environmental factors substantially influence metabolite production patterns, with carbon source availability and nitrogen limitations particularly affecting the biosynthesis of antifungal compounds (Mukherjee et al., 2013). Recent metabolomic analyses have revealed over 300 unique compounds in T. harzianum alone, highlighting the chemical complexity underlying Trichoderma’s ecological success as both a saprophyte and mycoparasite (Contreras-Cornejo et al., 2016).
However, our GC-MS metabolite profile of T. asperellum WNZ-21 during RCBB fermentation diverges significantly from this baseline. This difference might be attributable to the variation in the growth media and the fungal origin. Endophytes surpass other fungi by generating a unique range of secondary metabolites (peptaibols, gliotoxin, gliovirin, polyketides, pyrones, and terpenes) with varied bioactivities on minimal nutrients, offering diverse applications, e.g., pharmaceuticals, and plant growth regulation (De Silva et al., 2019; Sridharan et al., 2021; Vinale et al., 2008; Vizcaino et al., 2005). The GC-MS analysis indicated that only four components were reported in previous studies. Palmitic acid TMS derivative is present in brown seaweeds and the associated endophytic fungi (Teixeira et al., 2019). D-Pinitol, pentakis (trimethylsilyl), the major component of the fermented filtrate, is present in carob syrups (Christou et al., 2019). D-Mannitol, 6TMS was reported in olive leaf extract (Islamčević Razboršek et al., 2015). Silane, diethylheptyloxyoctadecyloxy- was reported in the leaves extract of Murraya koenigii (Prabaharan et al., 2023). The last three compounds identified in this filtrate are, to our knowledge, unreported in other microbial filtrates, while the remaining 13 are neither discovered in microbes nor plants. This may be due to the unique features of the endophytic fungus and suggesting a wide range of potential bioactivities. While structural confirmation via NMR/HR-MS is warranted, their bioactivities (anticancer, anti-hemolytic) provide functional evidence for prioritizing follow-up research. This aligns with the study’s objective to establish a bioprocess framework for valorizing agricultural waste, with structural studies reserved for later phases.
FT-IR signatures of the T. harzianum filtrate revealed the presence of various aliphatic chains and amino functional groups characteristic of AAs. Recent literature indicated the presence of AAs and other nitrogenous compounds by Trichoderma species, aligning with our observations of NH, CH, C = O, and C-N stretches in the FT-IR spectrum, suggesting the production of metabolites and secondary metabolites, which could contribute to the diverse peaks beyond those indicative of amino acids in the FT-IR spectrum (Abdel-Kareem and Zohri, 2018; Alrdahe et al., 2024; Elattar et al., 2024; Konappa et al., 2021; Tomah et al., 2020). Furthermore, FT-IR signatures (e.g., N-H bends, C = C stretches) suggest potential contributions from alkaloids, terpenoids, and peptides. FT-IR analysis of the T. harzianum culture filtrate revealed the presence of polysaccharides (Saravanakumar et al., 2021). Another study observed notable changes in the total protein of Trichoderma spp.-treated tomato seed radicles, accompanied by a parallel reduction in pectin and/or xyloglucan levels (Vukelić et al., 2024).
This study utilized the SSSF system integrating T. asperellum WNZ-21 with RCBB and mineral salts. This system was specifically designed to mimic natural fungal degradation processes and prioritize the synergistic interplay between fungal enzymes, such as cellulase, xylanase, and protease, and substrate breakdown, rather than isolating individual contributors. Crucially, non-inoculated controls confirmed that mineral salts and unfermented RCBB exhibited no hydrolytic activity or bioactivity, thereby validating fungal metabolism as the direct driver for the observed effects and the accumulation of diverse metabolites, including amino acids and a broad spectrum of secondary metabolites. Furthermore, the compounds identified via GC-MS were unambiguously determined to be of fungal origin, distinguishing them from any pre-existing plant-derived phytochemicals like phenolics or flavonoids. Beyond elucidating the fungal degradation pathways within this SSSF framework, this research comprehensively examines the biological properties of the resultant filtrate, whose bioactivity suggests potential therapeutic and health benefits derivable from these transformed plant residues. Consequently, the focus on the holistic fungal-plant biomass interaction and the characterization of its multi-bioactive products not only underscores the novelty and broader implications of valorizing agricultural waste but also highlights the complex and multifaceted nature of natural bioactivity for sustainable biotechnology and diverse applications.
The filtrate exhibited selective cytotoxicity against tumor cell lines (HePG2 and MCF7) than normal cells (WI38). AAs play a complex role in cancer, impacting energy production, redox balance, epigenetic regulation, and immune responses (Lieu et al., 2020). While AA restriction shows promise in some contexts (Guillén-Mancina et al., 2023), AAs also support healthy vasculature (Durante, 2020) and can contribute to treatment resistance (Yoo and Han, 2022). Notably, specific AAs like lysine (Abd El-Aal et al., 2023), glutamic acid (Dutta et al., 2013), and proline (Yin et al., 2022) exhibit anti-cancer potential, highlighting the need for further research into manipulating AA metabolism for effective cancer treatment. Furthermore, some GC-MS-detected components, such as stearic acid, a saturated fatty acid, have been reported to hinder the growth of human breast cancer cells in vitro (Khan et al., 2013), whereas palmitic acid has anticancer activity against prostate and breast cancer (Zafaryab et al., 2019; Zhu et al., 2021).
In the realm of anticancer activities, both AAs and secondary metabolites play crucial roles through diverse mechanisms. Lysine not only exhibits antibacterial effects but also contributes to anticancer effects by modulating AAs metabolism within cancer cells, effectively starving tumors of essential nutrients (Yoo and Han, 2022). Glutamic acid also demonstrates anticancer potential by interfering with glutamine metabolism, a pathway vital for cancer cell proliferation (Dutta et al., 2013). Building upon these mechanisms, phenolic compounds exhibit anticancer effects through multiple pathways, including reactive oxygen species (ROS) generation, DNA damage induction, and apoptosis activation in tumor cells (Lakhdari et al., 2023; Lieu et al., 2020; Yin et al., 2022). Flavonoids contribute to anticancer activity by inducing cell cycle arrest at the G2/M phase and inhibiting angiogenesis via downregulation of vascular endothelial growth factor signaling (Lakhdari et al., 2023; Saravanakumar et al., 2021). Tannins demonstrate anticancer potential through suppression of the NF-κB signaling pathway, which reduces tumor-associated inflammation and metastatic potential (Ekambaram et al., 2022). Similarly, tyrosine derivatives, particularly tyrosine kinase inhibitors, exert anticancer effects by blocking critical oncogenic pathways (Yin et al., 2022; Yoo and Han, 2022). Finally, methionine inhibits cancer cell proliferation by altering redox homeostasis through glutathione depletion and oxidative stress amplification (Durante, 2020; Wu et al., 2003).
Antioxidant properties are another significant feature, particularly of some AAs. Proline acts as a direct antioxidant by scavenging free radicals, thereby protecting cells from oxidative damage and indirectly supporting anticancer effects by reducing oxidative stress-driven mutations (Yin et al., 2022). Tyrosine, with its phenolic hydroxyl group, also neutralizes ROS and contributes to antioxidant defense (Cui et al., 2019). Methionine, with its sulfur-containing structure, regenerates glutathione, a key cellular antioxidant, further solidifying its role in combating oxidative stress (Wu et al., 2003). The provided information on secondary metabolites emphasizes their antioxidant properties, such as ROS, suggesting an indirect modulation of the cellular redox balance (Cui et al., 2019; Wu et al., 2003; Yin et al., 2022).
Various other molecules also exhibit significant anticancer activities. Fatty acid derivatives and related compounds, cyclopropanedodecanoic acid, 2- octyl-, methyl ester, have shown the potential to induce apoptosis in cancer cells (Zafaryab et al., 2019). Both palmitic acid, TMS derivative, and Stearic acid, TMS derivative, demonstrate anticancer effects, with palmitic acid inhibiting cancer cell proliferation and stearic acid derivatives inhibiting tumor growth (Zafaryab et al., 2019). These diverse compounds, working through various mechanisms, highlight the complexity of anticancer strategies derived from natural sources.
The GC-MS-detected compounds also showed antioxidant properties, contributing to cellular defense against oxidative stress. (2S,3R)-3-[(4E,7E)-Nona-4,7-dienoyl]-N,N-bis(trimethylsilyl)oxirane-2-carboxamide, D-Pinitol, pentakis(trimethylsilyl) ether, and D-Mannitol, 6TMS all exhibit radical scavenging activity (Copini et al., 2020; Ramos-Escudero et al., 2021). Glycerol, TMS derivative, acts as a hydrogen donor to neutralize ROS (Ramos-Escudero et al., 2021), while tartaric acid, TMS derivative, scavenges ROS and enhances cellular defense mechanisms (Saffari and Saffari, 2020). Erythro-Pentonic acid, 2-deoxy-3,4,5-tris-O-(trimethylsilyl)-, trimethylsilyl ester chelates metal ions to reduce oxidative stress, and D-Glucitol, TMS stabilizes free radicals (Ramos-Escudero et al., 2021). This diverse array of compounds underscores the multifaceted nature of antioxidant defenses.
However, while the studied fungal filtrate exhibits promisingly lower toxicity against normal cells, its cytotoxic mechanism against WI38, HePG2, and MCF7 cells remains unclear. Further, in vivo studies are warranted to investigate potential allergenic and immunomodulatory effects.
Anti-hemolytic activity, another biological assay, evaluates a material’s ability to protect RBCs from lysis. Hemolysis, if uncontrolled, can lead to complications such as anemia, jaundice, gallstones, and, in severe cases, organ failure (Fattizzo and Barcellini, 2022; Weisel and Litvinov, 2019). Mechanistically, anti-hemolytic activity may arise from free radical scavenging, oxidative stress reduction, RBC membrane stabilization, and inhibition of hemolytic agents (Karim et al., 2020; Yang et al., 2005). The observed anti-hemolytic activity of the T. asperellum WNZ-21 filtrate (4.40 ± 0.69% hemolysis) is attributed, at least in part, to glutamic acid derivatives and lysine, both of which exhibit protective effects (Neubauer et al., 2020). Specifically, glutamic acid derivatives (Elsayed et al., 2023) prevent RBC breakdown, while lysine (Wan et al., 2022) inhibits RBC lysis by stabilizing cell membranes. These findings align with the current filtrate’s rich amino acid profile, underscoring its potential to mitigate oxidative damage and hemolytic pathology.
Anti-inflammatory activities represent another important facet of bioactivity. Several of the mentioned compounds demonstrate anti-inflammatory effects through various mechanisms. (2S,3R)-3-[(4E,7E)-Nona-4,7-dienoyl]-N,N-bis(trimethylsilyl)oxirane-2-carboxamide, Azelaic acid, TMS derivative, and D-Pinitol, pentakis(trimethylsilyl) ether may modulate cytokine pathways or inhibit pro-inflammatory mediators (Manivannan et al., 2017). 9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl)ethyl ester suppresses pro-inflammatory cytokines (Manivannan et al., 2017), while Stearic acid, TMS derivative reduces inflammation via lipid mediator regulation (Pan et al., 2010). These diverse compounds offer various avenues for mitigating inflammatory responses.
The anti-inflammatory activity observed in the T. asperellum WNZ-21 filtrate may be attributed to its biomolecules, including lysine and glutamic acid, as well as palmitic acid. AAs are known to effectively neutralize pro-inflammatory bacterial components like lipopolysaccharides and lipoteichoic acid. This neutralization likely contributes to the observed anti-inflammatory effect by downregulating the expression of key mediators of inflammation, such as cytokines, enzymes, and transcription factors (Brunetti et al., 2020; Das and Gavel, 2020; Wan et al., 2022; Yang et al., 2020; Younis and Saleh, 2021). Some fatty acids, such as stearic acid, have demonstrated anti-inflammatory action that could potentially mitigate the liver damage caused by inflammation associated with cholestasis (Pan et al., 2010). 9-Octadecenoic acid (Z)- derivatives exhibited anti-inflammatory properties in lipopolysaccharide-activated macrophages (Manivannan et al., 2017). This anti-inflammatory activity might be linked to modulation of the immune cell function and scavenging of reactive oxygen species (Yoshitomi and Nagasaki, 2014). While T. asperellum WNZ-21 filtrate exhibits anti-inflammatory activity, albeit less potent than indomethacin, its discernible protective effect warrants further investigation into underlying mechanisms and optimized dosage regimens for the potential development of novel anti-inflammatory treatments.
The fungal filtrate was found to be rich in enzymes. The therapeutic potential of proteases as anti-inflammatory agents is well-established, with specific variants being utilized as adjunctive treatments or supplements in clinical practice. While proteases have demonstrated anticancer properties in experimental models through mechanisms such as extracellular matrix modulation (Chobotova et al., 2010), they have not yet achieved widespread clinical application as primary anticancer therapeutics compared to specialized enzymes targeting metabolic pathways like L-asparaginase (Naser et al., 2020; Othman et al., 2022). Cellulases and xylanases, which hydrolyze structural cellulose and xylan specific to plant cell walls, have limited direct therapeutic relevance as anticancer, anti-inflammatory, or anti-hemolytic drugs due to the absence of their specific substrates within animal tissues.
Given the promising anti-hemolytic and anti-inflammatory activities demonstrated by the fungal filtrate, particularly considering that these properties have not, to our knowledge, been previously reported for fermented plant biomass residues, further investigation is warranted to elucidate the underlying mechanisms and fully assess the therapeutic potential.
Future research must prioritize in vivo toxicity, safety, and clinical trial assessments to confirm therapeutic potential. Cytotoxicity assessments, although showing selectivity, require expansion to a broader range of cell lines to fully understand mechanisms and refine applications. Furthermore, in vivo studies should address potential allergenic and immunomodulatory effects. The SSSF method, chosen for its advantages, presents scalability challenges concerning heat and mass transfer in industrial-scale vessels, necessitating optimization for process efficiency. Chemical characterization, while identifying 17 compounds, demands further structural validation and multi-biological activity analysis of these entities. Future research should therefore focus on: (1) purification and structural identification of active, (2) mechanistic elucidation of bioactivities; (3) SSSF process optimization and scale-up; and (4) investigation of host and environmental factors influencing metabolite production to enhance broader applicability.
5 Conclusion
This study addressed the critical need for bioactive compounds with multi-biological activities by exploring the potential of endophytic fungi to valorize proteinaceous agricultural waste, specifically the protein-rich RCBB. We investigated the ability of T. asperellum WNZ-21 to bioconvert RCBB through SSSF, achieving 67.19% biomass degradation, and profiled the multi-biological activity of the resulting filtrate.
Our findings demonstrate that T. asperellum WNZ-21, identified morphologically and molecularly (GenBank ITS: OR857252; tef1 gene: PP069312), efficiently degraded RCBB via SSSF, yielding a metabolite-rich filtrate. This filtrate contained a significant amount of amino acids (total: 13,509.21 μg/g RCBB), notably lysine (8,324.55 μg/g RCBB), alongside high levels of phenolics (55.22 mg/g RCBB), flavonoids (11.22 mg/g RCBB), and tannins (18.90 mg/g RCBB). Importantly, GC-MS analysis revealed 17 compounds, highlighting the potential for discovering new chemical structures.
Biologically, the filtrate exhibited promising selective anticancer activity against HePG2 (IC50: 35.62 ± 2.1 μg/mL) and MCF7 (IC50: 59.20 ± 1.7 μg/mL) cells, showing reduced toxicity toward normal WI38 cells (IC50: 76.33 ± 3.9 μg/mL). It also demonstrated significant anti-hemolytic effects, resulting in only 4.40 ± 0.69% hemolysis, and notable anti-inflammatory effects (36.50% protection in a carrageenan-induced paw edema model). These observed activities are consistent with the presence of identified amino acids and secondary metabolites known for membrane stabilization and oxidative stress modulation.
This work successfully demonstrates the biotechnological potential of using endophytic fungi to convert agricultural waste into multi-bioactive compounds, advancing sustainable biotechnology and drug discovery efforts. This study provides a strong foundation, and future research will be crucial to isolate the reported compounds and confirm their structure, in-depth elucidate their specific mechanisms of action, optimize the fermentation process for scalability, and conduct comprehensive safety and efficacy studies for potential therapeutic applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal studies were approved by Institutional Review Board Statement. The animal study protocols were approved by the Ethics Committee of the Agricultural Research Center, Institutional Animal Care and Use Committee (ARC-IACUC), Giza, Egypt, under protocol approval code; ARC-SWERI-3-24. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
YA: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Writing – original draft, Writing – review & editing. SA: Conceptualization, Data curation, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. ZM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Writing – original draft, Writing – review & editing. DD: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. HA: Data curation, Formal Analysis, Software, Visualization, Writing – original draft, Writing – review & editing. WS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia, for funding this research work through the project number (0202-1443-S).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1609361/full#supplementary-material
Footnotes
1. ^National Center for Biotechnology Information. BLAST® (Basic Local Alignment Search Tool). Retrieved Jan 3, 2025 from http://www.ncbi.nlm.nih.gov/BLAST
2. ^The ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. Retrieved Jan 3, 2025 from https://arriveguidelines.org
References
Abd El-Aal, A. A. A., Jayakumar, F. A., Lahiri, C., Tan, K. O., and Reginald, K. (2023). Novel cationic cryptides in Penaeus vannamei demonstrate antimicrobial and anti-cancer activities. Sci. Rep. 13:14673. doi: 10.1038/s41598-023-41581-9
AbdAl-Aziz, S. A., El-Metwally, M. M., and Ismail Ali Saber, W. (2012). Molecular identification of a novel inulinolytic fungus isolated from and grown on tubers of Helianthus tuberosus and statistical screening of medium components. World J. Microbiol. Biotechnol. 28, 3245–3254. doi: 10.1007/s11274-012-1134-y
Abdel-Kareem, M. M., and Zohri, A. A. (2018). Extracellular mycosynthesis of gold nanoparticles using Trichoderma hamatum: Optimization, characterization and antimicrobial activity. Lett. Appl. Microbiol. 67, 465–475. doi: 10.1111/lam.13055
Abdelwahed, N., and El-Naggar, N. E.-A., and Saber, W. (2011). Factors and correlations controlling cellulase-free xylanase production by Streptomyces halstedii NRRL B-1238 in submerged culture. Aus. J. Basic Appl. Sci. 5, 45–53.
Aberoumand, A. (2009). Nutritional evaluation of edible Portulaca oleracia as plant food. Food Anal. Methods 2, 204–207. doi: 10.1007/s12161-008-9049-9
Al-Askar, A. A., Rashad, E. M., Ghoneem, K. M., Mostafa, A. A., Al-Otibi, F. O., and Saber, W. I. A. (2021). Discovering Penicillium polonicum with high-lytic capacity on Helianthus tuberosus tubers: Oil-based preservation for mold management. Plants (Basel) 10:413. doi: 10.3390/plants10091905
Al-Askar, A. A., Rashad, E. M., Moussa, Z., Ghoneem, K. M., Mostafa, A. A., Al-Otibi, F. O., et al. (2022). A novel endophytic Trichoderma longibrachiatum WKA55 with biologically active metabolites for promoting germination and reducing mycotoxinogenic fungi of peanut. Front. Microbiol. 13:772417. doi: 10.3389/fmicb.2022.772417
Alrdahe, S. S., Moussa, Z., Alanazi, Y. F., Alrdahi, H., Saber, W. I., and Darwish, D. B. E. (2024). Optimization of biodegradation of common bean biomass for fermentation using Trichoderma asperellum WNZ-21 and artificial neural networks. Fermentation 10:354. doi: 10.3390/fermentation10070354
AOAC (2019). Official methods of analysis of the association of official analytical chemists: Official methods of analysis of AOAC international, 21th Edn. Rockville, MD: AOAC.
Asis, A., Shahriar, S. A., Naher, L., Saallah, S., Fatihah, H. N. N., Kumar, V., et al. (2021). Identification patterns of Trichoderma strains using morphological characteristics, phylogenetic analyses and lignocellulolytic activities. Mol. Biol. Rep. 48, 3285–3301. doi: 10.1007/s11033-021-06321-0
Bailey, M. J., Biely, P., and Poutanen, K. (1992). Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23, 257–270. doi: 10.1016/0168-1656(92)90074-J
Bozzola, J. J., and Russell, L. D. (1999). Electron microscopy: Principles and techniques for biologists. Burlington, MA: Jones & Bartlett Learning.
Brunetti, J., Carnicelli, V., Ponzi, A., Di Giulio, A., Lizzi, A. R., Cristiano, L., et al. (2020). Antibacterial and anti-inflammatory activity of an antimicrobial peptide synthesized with D amino acids. Antibiotics (Basel) 9:840. doi: 10.3390/antibiotics9120840
Cai, F., and Druzhinina, I. S. (2021). In honor of john bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 107, 1–69. doi: 10.1007/s13225-020-00464-4
Chobotova, K., Vernallis, A. B., and Majid, F. A. A. (2010). Bromelain’s activity and potential as an anti-cancer agent: Current evidence and perspectives. Cancer Lett. 290, 148–156. doi: 10.1016/j.canlet.2009.08.001
Christou, C., Poulli, E., Yiannopoulos, S., and Agapiou, A. (2019). GC-MS analysis of D-pinitol in carob: Syrup and fruit (flesh and seed). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1116, 60–64. doi: 10.1016/j.jchromb.2019.04.008
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Del-Val, E., and Larsen, J. (2016). Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: Interactions with plants. FEMS Microbiol. Ecol. 92:fiw036. doi: 10.1093/femsec/fiw036
Copini, S., Micheletti, A. C., Lima, D. P. D., Gomes, R. S., Meza, A., and Beatriz, A. (2020). Synthesis and antioxidant and antimicrobial properties of β-hydroxy sulfides, sulfoxides, and sulfones derived from cardanol and glycerol derivatives. J. Braz. Chem. Soc. 31, 2569–2582. doi: 10.21577/0103-5053.20200136
Cui, H., Hayat, K., and Zhang, X. (2019). Antioxidant activity in vitro of N-(1-deoxy-α-d-xylulos-1-yl)-Phenylalanine: Comparison among maillard reaction intermediate, end-products and xylose-phenylalanine. J. Food Sci. 84, 1060–1067. doi: 10.1111/1750-3841.14579
Cupp-Enyard, C. (2008). Sigma’s non-specific protease activity assay-casein as a substrate. J. Vis. Exp. 17:899. doi: 10.3791/899
Das, A. K., and Gavel, P. K. (2020). Low molecular weight self-assembling peptide-based materials for cell culture, antimicrobial, anti-inflammatory, wound healing, anticancer, drug delivery, bioimaging and 3D bioprinting applications. Soft. Matter 16, 10065–10095. doi: 10.1039/d0sm01136c
De Bruyn, A., Martin, D. P., and Lefeuvre, P. (2014). Phylogenetic reconstruction methods: An overview. Methods Mol. Biol. 1115, 257–277. doi: 10.1007/978-1-62703-767-9_13
de Castro, W. J. R., de Moura Zanine, A., de Souza, A. L., de Jesus Ferreira, D., Geron, L. J. V., Leão, A. G., et al. (2016). Inclusion of different levels of common-bean residue in sheep diets on nutrient intake and digestibility. Sem. Ciências Agrárias 37, 369–380. doi: 10.5433/1679-0359.2016v37n1p369
de castro, W. J. R., Zanine, A. D. M., Ferreira, D. D. J., de Souza, A. L., Geron, L. J. V., Santos, E. M., et al. (2021). Impact of increased bean residue levels on the feed efficiency and ingestive behaviour of sheep. Biol. Rhythm Res. 52, 41–49. doi: 10.1080/09291016.2019.1583503
de Hoog, G. S., Guarro, J., Gené, J., and Figueras, M. (2005). Atlas of clinical fungi. Netherlands: CBS Publishers.
De Silva, N. I., Brooks, S., Lumyong, S., and Hyde, K. D. (2019). Use of endophytes as biocontrol agents. Fungal Biol. Rev. 33, 133–148. doi: 10.1016/j.fbr.2018.10.001
Denizot, F., and Lang, R. (1986). Rapid colorimetric assay for cell growth and survival. modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89, 271–277. doi: 10.1016/0022-1759(86)90368-6
Diba, K., Kordbacheh, P., Mirhendi, S., Rezaie, S., and Mahmoudi, M. (2007). Identification of Aspergillus species using morphological characteristics. Pak. J. Med. Sci. 23:867.
Domsch, K. H., Gams, W., and Anderson, T.-H. (1980). Compendium of soil fungi. London: Academic Press Ltd.
Durante, W. (2020). Amino acids in circulatory function and health. Adv. Exp. Med. Biol. 1265, 39–56. doi: 10.1007/978-3-030-45328-2_3
Dutta, S., Ray, S., and Nagarajan, K. (2013). Glutamic acid as anticancer agent: An overview. Saudi Pharmaceut. J. 21, 337–343. doi: 10.1016/j.jsps.2012.12.007
Ekambaram, S. P., Aruldhas, J., Srinivasan, A., and Erusappan, T. (2022). Modulation of NF-κB and MAPK signalling pathways by hydrolysable tannin fraction from Terminalia chebula fruits contributes to its anti-inflammatory action in RAW 264.7 cells. J. Pharmacy Pharmacol. 74, 718–729. doi: 10.1093/jpp/rgab178
Elattar, K. M., Ghoniem, A. A., Al-Askar, A. A., Bhgat El-Gazzar, U., El-Hersh, M. S., Elsherbiny, E. A., et al. (2024). Melanin synthesized by the endophytic Aureobasidium pullulans AKW: A multifaceted biomolecule with antioxidant, wound healing, and selective anti-cancer activity. Curr. Top. Med. Chem. 24, 2141–2160. doi: 10.2174/0115680266300091240730111333
El-Hersh, M. S., Saber, W., El-Fadaly, H. A., and Mahmoud, M. K. (2016). Lysine and glutamic acids as the end products of multi-response of optimized fermented medium by Mucor mucedo KP736529. Pak. J. Biol. Sci. PJBS 19, 279–288. doi: 10.3923/pjbs.2016.279.288
El-Hersh, M. S., Saber, W. I., and El-Fadaly, H. A. (2014). Amino acids associated with optimized alkaline protease production by Bacillus subtilis ATCC 11774 using statistical approach. Biotechnology 13, 252–262. doi: 10.3923/biotech.2014.252.262
El-Metwally, M. M., Abdel-Fattah, G. M., Al-Otibi, F. O., Khatieb, D., Helmy, Y. A., Mohammed, Y. M. M., et al. (2023). Application of artificial neural networks for enhancing Aspergillus flavipes lipase synthesis for green biodiesel production. Heliyon 9:e20063. doi: 10.1016/j.heliyon.2023.e20063
El-Naggar, N. E.-A., Abdelwahed, N. A., Saber, W. I., and Mohamed, A. A. (2014). Bioprocessing of some agro-industrial residues for endoglucanase production by the new subsp.; Streptomyces albogriseolus subsp. cellulolyticus strain NEAE-J. Braz. J. Microbiol. 45, 743–751. doi: 10.1590/S1517-83822014005000049
Elsayed, E. M., Farghali, A. A., Zanaty, M. I., Abdel-Fattah, M., Alkhalifah, D. H. M., Hozzein, W. N., et al. (2023). In vitro cytotoxicity and hemolysis effect of poly-gamma-glutamic acid nano-polymer biosynthesized using some isolates of Bacillus spp. Microbiol. Res. 14, 1720–1735. doi: 10.3390/microbiolres14040118
Elsayed, M. S., Eldadamony, N. M., Alrdahe, S. S. T., and Saber, W. I. A. (2021). Definitive screening design and artificial neural network for modeling a rapid biodegradation of date palm fronds by a new Trichoderma sp. PWN6 into citric acid. Molecules 26:5048. doi: 10.3390/molecules26165048
Fattizzo, B., and Barcellini, W. (2022). Autoimmune hemolytic anemia: Causes and consequences. Exp. Rev. Clin. Immunol. 18, 731–745. doi: 10.1080/1744666X.2022.2089115
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Ferro, M. M., De Moura Zanine, A., De Souza, A. L., De Jesus Ferreira, D., Santos, E. M., Ribeiro Alves, G., et al. (2020). Residue from common bean in substitution of cottonseed cake in diets for sheep. Biol. Rhythm Res. 51, 471–480. doi: 10.1080/09291016.2018.1535566
Goldstein, J. I., Newbury, D. E., Michael, J. R., Ritchie, N. W., Scott, J. H. J., and Joy, D. C. (2017). Scanning electron microscopy and X-ray microanalysis. Berlin: Springer.
Guillén-Mancina, E., Jiménez-Alonso, J. J., Calderón-Montaño, J. M., Jiménez-González, V., Díaz-Ortega, P., Burgos-Morón, E., et al. (2023). Artificial diets with selective restriction of amino acids and very low levels of lipids induce anticancer activity in mice with metastatic triple-negative breast cancer. Cancers 15:1540. doi: 10.3390/cancers15051540
Hermosa, M. R., Keck, E., Chamorro, I., Rubio, B., Sanz, L., Vizcaíno, J. A., et al. (2004). Genetic diversity shown in Trichoderma biocontrol isolates. Mycol. Res. 108, 897–906. doi: 10.1017/S0953756204000358
Islamčević Razboršek, M., Pavlovič, I., Knez, Ž, and Škerget, M. (2015). GC-MS determination of glucose and mannitol after oximation and trimethylsilylation in olive leaves extracts. Technol. Acta 8, 9–18.
Jajić, I., Krstović, S., Glamočić, D., Jakšić, S., and Abramović, B. (2013). Validation of an HPLC method for the determination of amino acids in feed. J. Serb. Chem. Soc. 78, 839–850. doi: 10.2298/JSC120712144J
Jukes, T. H., and Cantor, C. R. (1969). Evolution of protein molecules. Mamm. Protein Metab. 3, 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Karim, M. A., Islam, M. A., Islam, M. M., Rahman, M. S., Sultana, S., Biswas, S., et al. (2020). Evaluation of antioxidant, anti-hemolytic, cytotoxic effects and anti-bacterial activity of selected mangrove plants (Bruguiera gymnorrhiza and Heritiera littoralis) in Bangladesh. Clin. Phytosci. 6:8. doi: 10.1186/s40816-020-0152-9
Khan, A. A., Alanazi, A. M., Jabeen, M., Chauhan, A., and Abdelhameed, A. S. (2013). Design, synthesis and in vitro anticancer evaluation of a stearic acid-based ester conjugate. Anticancer Res. 33, 2517–2524.
Konappa, N., Udayashankar, A. C., Dhamodaran, N., Krishnamurthy, S., Jagannath, S., Uzma, F., et al. (2021). Ameliorated antibacterial and antioxidant properties by Trichoderma harzianum mediated green synthesis of silver nanoparticles. Biomolecules 11:535. doi: 10.3390/biom11040535
Kuo, J., Chang, C.-F., and Chi, W.-C. (2021). Isolation of endophytic fungi with antimicrobial activity from medicinal plant Zanthoxylum simulans Hance. Folia Microbiol. 66, 385–397. doi: 10.1007/s12223-021-00854-4
Lakhdari, W., Benyahia, I., Bouhenna, M. M., Bendif, H., Khelafi, H., Bachir, H., et al. (2023). Exploration and evaluation of secondary metabolites from Trichoderma harzianum: GC-MS analysis, phytochemical profiling, antifungal and antioxidant activity assessment. Molecules 28:5025. doi: 10.3390/molecules28135025
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lieu, E. L., Nguyen, T., Rhyne, S., and Kim, J. (2020). Amino acids in cancer. Exp. Mol. Med. 52, 15–30. doi: 10.1038/s12276-020-0375-3
Manivannan, P., Muralitharan, G., and Balaji, N. P. (2017). Prediction aided in vitro analysis of octa-decanoic acid from Cyanobacterium lyngbya sp. as a proapoptotic factor in eliciting anti-inflammatory properties. Bioinformation 13, 301–306. doi: 10.6026/97320630013301
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Moo-Koh, F. A., Cristobal-Alejo, J., Andres, M. F., Martin, J., Reyes, F., Tun-Suarez, J. M., et al. (2022). In vitro assessment of organic and residual fractions of nematicidal culture filtrates from thirteen tropical Trichoderma strains and metabolic profiles of most-active. J. Fungi (Basel) 8:82. doi: 10.3390/jof8010082
Morris, C. J. (2003). Carrageenan-induced paw edema in the rat and mouse. Inflamm. Protocols 225, 115–121. doi: 10.1385/1-59259-374-7:115
Moussa, Z., Alanazi, Y. F., Khateb, A. M., Eldadamony, N. M., Ismail, M. M., Saber, W. I. A., et al. (2023). Domiciliation of Trichoderma asperellum suppresses Globiosporangium ultimum and promotes pea growth, ultrastructure, and metabolic features. Microorganisms 11:198. doi: 10.3390/microorganisms11010198
Moussa, Z., Darwish, D. B., Alrdahe, S. S., and Saber, W. I. A. (2021). Innovative artificial-intelligence- based approach for the biodegradation of feather keratin by Bacillus paramycoides, and cytotoxicity of the resulting amino acids. Front. Microbiol. 12:731262. doi: 10.3389/fmicb.2021.731262
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Ann. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Nasar, S., Shaheen, H., Murtaza, G., Tinghong, T., Arfan, M., and Idrees, M. (2023). Socioeconomic evaluation of common bean (Phaseolus vulgaris L.) cultivation in providing sustainable livelihood to the mountain populations of Kashmir Himalayas. Plants 12:213. doi: 10.3390/plants12010213
Naser, S., Saber, W., El-Metwally, M., Moustafa, M., and El-Kott, A. (2020). Fungal assembly of L-asparaginase using solid-state fermentation: A review. Biocell 44, 147–155. doi: 10.32604/biocell.2020.09522
Negm, S., El-Metwally, M., Eldin Saber, W., Abo-Neima, S., Moustafa, M., and El-Kott, A. (2021). Nanoparticles induce the biosynthesis and activity of the new possible therapeutic proteinase source, Talaromyces purpureogenus KJ584844. Biocell 45, 119–127. doi: 10.32604/biocell.2021.012011
Neubauer, D., Jaskiewicz, M., Bauer, M., Golacki, K., and Kamysz, W. (2020). Ultrashort cationic lipopeptides-effect of N-Terminal amino acid and fatty acid type on antimicrobial activity and hemolysis. Molecules 25:257. doi: 10.3390/molecules25020257
Nofal, A. M., Hamouda, R. A., Rizk, A., El-Rahman, M. A., Takla, A. K., Galal, H., et al. (2023). Polyphenols-Rich extract of Calotropis procera alone and in combination with trichoderma culture filtrate for biocontrol of cantaloupe wilt and root rot fungi. Molecules 29:139. doi: 10.3390/molecules29010139
Omomowo, I., Fadiji, A., and Omomowo, O. (2020). Antifungal evaluation and phytochemical profile of Trichoderma harzianum and Glomus versiforme secondary metabolites on cowpea pathogens. Asian Jr. Microbiol. Biotech. Env. Sci. 22, 265–272.
Othman, S. I., Mekawey, A. A., El-Metwally, M. M., Gabr, S. A., Alwaele, M. A., Al Fassam, H., et al. (2022). Rhizopus oryzae AM16; a new hyperactive L-asparaginase producer: Semi solid-state production and anticancer activity of the partially purified protein. Biomed. Rep. 16:15. doi: 10.3892/br.2022.1498
Pan, P. H., Lin, S. Y., Ou, Y. C., Chen, W. Y., Chuang, Y. H., Yen, Y. J., et al. (2010). Stearic acid attenuates cholestasis-induced liver injury. Biochem. Biophys. Res. Commun. 391, 1537–1542. doi: 10.1016/j.bbrc.2009.12.119
Prabaharan, J., Prabakaran, M., Prabhakaran, M., Abinaya, K., Krishnan, N., Karen, D. S., et al. (2023). Comparison on extracted metabolites from different regions grown Murraya koenigii and validation by antibacterial, antioxidant, and molecular docking studies. Biomass Conv. Bioref. 1–29. doi: 10.1007/s13399-023-04105-z
Raja, H. A., Miller, A. N., Pearce, C. J., and Oberlies, N. H. (2017). Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 80, 756–770. doi: 10.1021/acs.jnatprod.6b01085
Ramos-Escudero, F., Casimiro-Gonzales, S., Fernández-Prior, Á, Chávez, K. C., Gómez-Mendoza, J., de la Fuente-Carmelino, L., et al. (2021). Colour, fatty acids, bioactive compounds, and total antioxidant capacity in commercial cocoa beans (Theobroma cacao L.). LWT 147:111629. doi: 10.1016/j.lwt.2021.111629
Reino, J. L., Guerrero, R. F., Hernandez-Galan, R., and Collado, I. G. (2008). Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 7, 89–123. doi: 10.1007/s11101-006-9032-2
Saber, W., El-Naggar, N. E., and AbdAl-Aziz, S. (2010). Bioconversion of lignocellulosic wastes into organic acids by cellulolytic rock phosphate-solubilizing fungal isolates grown under solid-state fermentation conditions. Res. J. Microbiol. 5, 1–20. doi: 10.3923/jm.2010.1.20
Saber, W. I., El-Naggar, N. E., El-Hersh, M. S., and El-Khateeb, A. Y. (2015). An innovative synergism between Aspergillus oryzae and Azotobacter chroococcum for bioconversion of cellulosic biomass into organic acids under restricted nutritional conditions using multi-response surface optimization. Biotechnology 14, 47–57. doi: 10.3923/biotech.2015.47.57
Saffari, V. R., and Saffari, M. (2020). Effects of EDTA, citric acid, and tartaric acid application on growth, phytoremediation potential, and antioxidant response of Calendula officinalis L. in a cadmium-spiked calcareous soil. Int. J. Phytoremed. 22, 1204–1214. doi: 10.1080/15226514.2020.1754758
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salem, M., El-Metwally, M., Saber, W., Negm, S., El-Kott, A., Mazroua, Y., et al. (2022). Secondary antiviral metabolites from fungi with special reference to coronaviruses. Biocell 46, 1–10. doi: 10.32604/biocell.2022.019301
Samson, R. A., and Pitt, J. I. (2000). Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Boca Raton, FL: CRC Press.
Samson, R. A., Visagie, C. M., Houbraken, J., Hong, S. B., Hubka, V., Klaassen, C. H., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78, 141–173. doi: 10.1016/j.simyco.2014.07.004
Sánchez-Rangel, J. C., Benavides, J., Heredia, J. B., Cisneros-Zevallos, L., and Jacobo-Velázquez, D. A. (2013). The Folin–Ciocalteu assay revisited: Improvement of its specificity for total phenolic content determination. Anal. Methods 5, 5990–5999. doi: 10.1039/C3AY41125G
Santos, K. C. D., Magalhães, A. L. R., Conceição, M. G. D., Carvalho, F. F. R. D., and Ferreira, G. D. G. (2018). Common bean residue as additive in sugarcane silage. Rev. Ciência Agronômica 49, 159–166. doi: 10.5935/1806-6690.20180018
Saravanakumar, K., Park, S., Sathiyaseelan, A., Mariadoss, A. V. A., Park, S., Kim, S.-J., et al. (2021). Isolation of polysaccharides from Trichoderma harzianum with antioxidant, anticancer, and enzyme inhibition properties. Antioxidants 10:1372. doi: 10.3390/antiox10091372
Shi, M., Chen, L., Wang, X. W., Zhang, T., Zhao, P. B., Song, X. Y., et al. (2012). Antimicrobial peptaibols from Trichoderma pseudokoningii induce programmed cell death in plant fungal pathogens. Microbiology 158, 166–175.
Siddiquee, S., and Siddiquee, S. (2017). “Morphology-based characterization of Trichoderma species,” in Practical handbook of the biology and molecular diversity of Trichoderma species from tropical regions. Fungal Biology series, eds. S. Siddiquee, V. K. Gupta, and M. G. Tuohy (Springer), 41–73.
Sridharan, A. P., Sugitha, T., Karthikeyan, G., Nakkeeran, S., and Sivakumar, U. (2021). Metabolites of Trichoderma longibrachiatum EF5 inhibits soil borne pathogen, Macrophomina phaseolina by triggering amino sugar metabolism. Microb. Pathog 150:104714. doi: 10.1016/j.micpath.2020.104714
St-Germain, G., and Summerbell, R. (2003). Identifying filamentous fungi: A clinical laboratory handbook. Rev. Instituto Med. Trop. São Paulo 45, 152–152. doi: 10.1590/S0036-46652003000300017
Sutton, D. A., Fothergill, A. W., and Rinaldi, M. G. (1998). Guide to clinically significant fungi. Philadelphia, PA: Williams & Wilkins.
Teixeira, T. R., Santos, G. S., Turatti, I. C. C., Paziani, M. H., von Zeska Kress, M. R., et al. (2019). Characterization of the lipid profile of Antarctic brown seaweeds and their endophytic fungi by gas chromatography–mass spectrometry (GC–MS). Polar Biol. 42, 1431–1444. doi: 10.1007/s00300-019-02529-w
Thrane, U., Poulsen, S. B., Nirenberg, H. I., and Lieckfeldt, E. (2001). Identification of Trichoderma strains by image analysis of HPLC chromatograms. FEMS Microbiol. Lett. 203, 249–255. doi: 10.1111/j.1574-6968.2001.tb10849.x
Tomah, A. A., Alamer, I. S. A., Li, B., and Zhang, J.-Z. (2020). Mycosynthesis of silver nanoparticles using screened Trichoderma isolates and their antifungal activity against Sclerotinia sclerotiorum. Nanomaterials 10:1955. doi: 10.3390/nano10101955
Tretter, E., Wang, Y., Johnson, E. M., and White, M. M. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U. S. A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Tunga, R., Banerjee, R., and Bhattacharyya, B. (1998). Optimizing some factors affecting protease production under solid state fermentation. Bioprocess Eng. 19, 187–190. doi: 10.1007/s004490050504
Vinale, F., Sivasithamparam, K., Ghisalberti, E., Marra, R., Barbetti, M., Li, H., et al. (2008). A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86. doi: 10.1016/j.pmpp.2008.05.005
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Woo, S. L., Nigro, M., Marra, R., et al. (2014). Trichoderma secondary metabolites active on plants and fungal pathogens. Open Mycol. J. 8, 127–139. doi: 10.2174/1874437001408010127
Vizcaino, J. A., Sanz, L., Basilio, A., Vicente, F., Gutierrez, S., Hermosa, M. R., et al. (2005). Screening of antimicrobial activities in Trichoderma isolates representing three trichoderma sections. Mycol. Res. 109(Pt 12), 1397–1406. doi: 10.1017/s0953756205003898
Vukelić, I., Radić, D., Pećinar, I., Lević, S., Djikanović, D., Radotić, K., et al. (2024). Spectroscopic investigation of tomato seed germination stimulated by Trichoderma spp. Biology 13:340. doi: 10.3390/biology13050340
Wan, Y., Wang, X., Bai, T., Zheng, X., Yang, L., Li, Q., et al. (2022). Lysine inhibits hemolytic activity of Staphylococcus aureus and its application in food model contaminated with Staphylococcus aureus. Toxins (Basel) 14:867. doi: 10.3390/toxins14120867
Wang, X., Zhang, C., and Bao, N. (2023). Molecular mechanism of palmitic acid and its derivatives in tumor progression. Front. Oncol. 13:1224125. doi: 10.3389/fonc.2023.1224125
Weisel, J. W., and Litvinov, R. I. (2019). Red blood cells: The forgotten player in hemostasis and thrombosis. J. Thromb. Haemost. 17, 271–282. doi: 10.1111/jth.14360
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press).
Wu, H.-C., Shiau, C.-Y., Chen, H.-M., and Chiou, T.-K. (2003). Antioxidant activities of carnosine, anserine, some free amino acids and their combination. J. Food Drug Anal. 11:13. doi: 10.38212/2224-6614.2720
Yang, P., Wang, W., Chi, S., Mai, K., Song, F., and Wang, L. (2020). Effects of dietary lysine on regulating GH-IGF system, intermediate metabolism and immune response in largemouth bass (Micropterus salmoides). Aquaculture Rep. 17:100323. doi: 10.1016/j.aqrep.2020.100323
Yang, Z. G., Sun, H. X., and Fang, W. H. (2005). Haemolytic activities and adjuvant effect of Astragalus membranaceus saponins (AMS) on the immune responses to ovalbumin in mice. Vaccine 23, 5196–5203. doi: 10.1016/j.vaccine.2005.06.016
Yin, Y., Sheng, L., Zhang, J., Zhang, L., Liu, J., Wen, X., et al. (2022). Synthesis and in vitro/in vivo anticancer evaluation of pentacyclic triterpenoid derivatives linked with l-phenylalanine or l-proline. Bioorg. Chem. 126:105865. doi: 10.1016/j.bioorg.2022.105865
Yoo, H. C., and Han, J. M. (2022). Amino acid metabolism in cancer drug resistance. Cells 11:140. doi: 10.3390/cells11010140
Yoshitomi, T., and Nagasaki, Y. (2014). Reactive oxygen species-scavenging nanomedicines for the treatment of oxidative stress injuries. Adv. Healthc. Mater. 3, 1149–1161. doi: 10.1002/adhm.201300576
Younis, A., and Saleh, H. (2021). Phytochemical screening and assessment of antioxidant and antimicrobial potentialities of two egyptian medicinal plants. Egypt. J. Pure Appl. Sci. 59, 49–57. doi: 10.21608/ejaps.2021.97655.1008
Zafaryab, M., Fakhri, K. U., Khan, M. A., Hajela, K., and Rizvi, M. M. A. (2019). In vitro assessment of cytotoxic and apoptotic potential of palmitic acid for breast cancer treatment. Int. J. Life Sci. Res. 7, 166–174. doi: 10.13140/RG.2.2.33419.13606
Zeilinger, S., Gruber, S., Bansal, R., and Mukherjee, P. K. (2016). Secondary metabolism in Trichoderma–chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Zhishen, J., Mengcheng, T., and Jianming, W. (1999). The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 64, 555–559. doi: 10.1016/S0308-8146(98)00102-2
Keywords: common bean, semi-solid-state fermentation, Trichoderma sp., amino acids, multibiological activity
Citation: Alanazi YF, Alrdahe SS, Moussa Z, Darwish DBE, Alrdahi H and Saber WIA (2025) Endophytic Trichoderma asperellum WNZ-21 produces novel bioactives with anticancer, anti-inflammatory, and hemoprotective properties from fermented bean biomass. Front. Microbiol. 16:1609361. doi: 10.3389/fmicb.2025.1609361
Received: 10 April 2025; Accepted: 16 May 2025;
Published: 10 June 2025.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Yaqian Li, Shanghai Jiao Tong University, ChinaGuo Qingfeng, Huanghe University of Science and Technology, China
Ping-Hsiu Huang, Jiangsu Food and Pharmaceutical Science College, China
Copyright © 2025 Alanazi, Alrdahe, Moussa, Darwish, Alrdahi and Saber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salma Saleh Alrdahe, c2FscmRhaGVAdXQuZWR1LnNh; WesamEldin I. A.Saber, d2VzYW1lbGRpbi5zYWJlckBhcmMuc2NpLmVn; d2lhc2FiZXJAZ21haWwuY29t
 Yasmene F. Alanazi1
Yasmene F. Alanazi1 Salma Saleh Alrdahe
Salma Saleh Alrdahe Zeiad Moussa
Zeiad Moussa Doaa Bahaa Eldin Darwish
Doaa Bahaa Eldin Darwish WesamEldin I. A. Saber
WesamEldin I. A. Saber