Abstract
Introduction:
Lactic acid bacteria (LAB) have attracted significant interest as natural antimicrobials for food safety and preservation. Some LAB strains effectively inhibit food-borne molds and yeasts responsible for spoilage and mycotoxin contamination. However, the antifungal effect of Lactococcus garvieae remains largely unexplored.
Methods:
This study investigates the properties of L. garvieae ZB15 isolated from Zhenba bacon, emphasizing its genetic safety and antimicrobial potential. The genomic information and antifungal properties of Lactococcus garvieae strain ZB15 were comprehensively analyzed using whole-genome sequencing and in vitro experiments.
Results:
Genomic analysis confirmed the absence of virulence and antibiotic resistance genes, along with minimal prophage presence, underscoring its safety for application. The strain exhibited strong self-aggregation, co-aggregation, and hydrophobic properties, suggesting its potential for intestinal adhesion. The cell-free supernatant (CFS) of L. garvieae ZB15 demonstrated notable antifungal activity, achieving over 50% inhibition against four fungal strains at 60 mg/mL. The antifungal compounds in the CFS retained high activity after thermal treatment at 100°C for 1 hour, highlighting their thermal stability. Additionally, enzymatic treatments with trypsin, pepsin, and proteinase K, along with neutralization of organic acids, significantly reduced the antifungal activity, indicating the involvement of proteinaceous compounds and organic acids. Catalase treatment partially affected antifungal activity, suggesting hydrogen peroxide as a contributing factor.
Discussion:
Lactococcus garvieae ZB15 possessed probiotic characteristics, including strong adhesion properties and pathogen exclusion capabilities. The strain’s genetic safety, lack of antibiotic resistance genes, and ability to produce heat-stable antifungal compounds highlighted its potential as a natural food preservative.
1 Introduction
Lactic acid bacteria (LAB) represent a diverse group of Gram-positive, non-spore-forming, facultative anaerobes that predominantly produce lactic acid through carbohydrate fermentation (Navarré et al., 2024). Ubiquitously distributed, LAB can be isolated from a wide variety of sources, including fermented foods such as pickles (Seddik et al., 2017), dairy products (Taye et al., 2021; Yu et al., 2011), vegetables (Bringel et al., 2005; Kim et al., 2008), meat products (Li et al., 2024), silage (Ellis et al., 2016), and wine (Ambe et al., 2024). Moreover, LAB form a critical component of the human and animal gastrointestinal and vaginal microbiota (Liu et al., 2021; Zhang et al., 2024; Zhong et al., 2022). The metabolic products of LAB have extensive applications in industries ranging from animal feed and aquaculture to food processing and storage, where their preservative effects and contributions to product stability and flavor are highly valued. To date, LAB comprises over 18 genera and more than 200 species, with prominent members including Lactobacillus, Enterococcus, Lactococcus, and Leuconostoc (Goldstein et al., 2015).
Lactococcus garvieae is a facultative anaerobic, Gram-positive bacterium found in diverse environments (Jantrakajorn et al., 2024). Initially identified as a fish pathogen, certain strains exhibit probiotic properties. For example, L. garvieae B301 reduces diarrhea and mortality in poultry while enhancing weight gain, suggesting its potential as a feed additive (Gradoville et al., 2018). L. garvieae DCC43, isolated from wild duck intestines, produces a circular bacteriocin with antibacterial properties (Kawada et al., 2016). Another strain from Mongolian sheep intestines improves gut barrier integrity and mitigates gut dysbiosis in mice infected with Clostridium perfringens (Wang et al., 2024). In poultry farming, L. garvieae supplementation enhances immune function, promotes growth, increases secretory IgA (SIgA) levels, and supports gut microbial balance by reducing Escherichia coli populations while increasing LAB abundance (Zhang et al., 2016).
Lactic acid bacteria exhibit antifungal activity through competitive exclusion, organic acid production, and secretion of antifungal peptides and metabolites such as hydrogen peroxide and bacteriocins (Rabaoui et al., 2023). Some LAB strains effectively inhibit food-borne molds and yeast responsible for spoilage and mycotoxin contamination. The introduction of Lactobacillus kefiri M4 into apple juice suppresses fungal growth via nutrient competition with fungal species (Sadiq et al., 2019). Fatty acids and acetate produced by Lactiplantibacillus, Furfurilactobacillus milii, and Lentilactobacillus parabuchneri inhibit the growth of Candida sake, Saccharomyces bayanus, and Torulaspora delbrueckii in milk by suppressing fungal hyphal development (Nasrollahzadeh et al., 2022). However, the antifungal effect of L. garvieae remain largely unexplored.
Zhenba bacon, a traditional fermented meat product from Shaanxi Province, China, is known for its distinct color and flavor, achieved through salting and smoking processes. These processes foster complex microbial communities typical of Chinese cured meats (Rubio et al., 2014). Within the microbiota of Zhenba bacon, we identified the L. garvieae ZB15 strain, which exhibits a high growth rate, robust acid production, and antifungal potential. However, its genomic properties and antifungal activity remain underexplored.
This study aims to characterize the genome of L. garvieae ZB15 and assess its antifungal properties through in vitro experiments. The findings will enhance understanding of its probiotic properties, contributing to the development of sustainable, safe, and effective food preservation strategies.
2 Materials and methods
2.1 Bacterial strain and growth conditions
Lactococcus garvieae ZB15 was isolated from Zhenba bacon during the curing process with characteristics of high growth rate and robust acid production (data not shown). The strain was preserved as a glycerol stock at −80°C, streaked onto MRS agar plates and incubated overnight at 37°C. A single colony was picked and incubated overnight at 37°C in 2 mL sterile MRS broth until the culture reached an OD600 of 2.0, after which it was stored at 4°C for subsequent use.
2.2 Genomic DNA extraction, whole genome sequencing, and assembly
Genomic DNA was extracted from L. garvieae ZB15 using the HiPure Bacterial DNA Kit (Magen, Guangzhou, China) following the manufacturer’s protocol (Khan et al., 2016). The extracted DNA was quantified using a NanoDrop spectrophotometer and subjected to quality control by gel electrophoresis. DNA libraries were prepared and sequenced using an Illumina/ONT platform at Genedenovo Biotechnology Co., Ltd (Guangzhou, China). Raw sequencing reads were assembled de novo using Falcon (v0.3.0) and further refined with Flye (v2.8.1-b1676). Gene annotation was conducted using multiple databases, including NCBI-Nr, UniProt/Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and Clusters of Orthologous Groups (COG). Non-coding RNA elements were identified using rRNAmmer (v1.2), tRNAscan-SE (v1.3.1), and cmscan (v1.1.2). Gene islands and prophages were predicted using Island Path-DIMOB (v1.0.0) and PHAST (v2.0), respectively.
2.3 Carbohydrate fermentation
Carbohydrate metabolism of L. garvieae ZB15 was assessed using the API 50 CHL test kit (bioMérieux, Marcy l’Etoile, France) as previously described (Yetiman et al., 2024). The bacterial suspension was prepared in API 50 CHL medium and adjusted to the turbidity standard recommended by the manufacturer. The inoculated test strip was incubated at 37°C under anaerobic conditions, and metabolic activity was determined by color change due to pH shift. No treated medium was taken as the negative control.
2.4 Antibiotic susceptibility testing
The Kirby–Bauer disk diffusion method was used to assess the antibiotic susceptibility of L. garvieae ZB15 as previously described (Khan et al., 2019; Sirichoat et al., 2020). Briefly, a standardized bacterial suspension was spread onto Mueller-Hinton agar, and 15 antibiotic-impregnated disks (Hangzhou Microbial Reagent Company, Hangzhou, China) were placed on the surface. After incubation at 37°C for 24 h, inhibition zone diameters were measured and interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (Hindler and Stelling, 2007).
2.5 Auto-aggregation, co-aggregation and hydrophobicity
2.5.1 Auto-aggregation assay
Auto-aggregation ability was assessed as described by Vasiee et al. (2019) and El-Deeb et al. (2020). L. garvieae ZB15 was cultured in MRS broth until reaching an OD600 of 2.0. The culture was centrifuged at 4,000 × g for 10 min. After discarding the supernatant, the cell pellets were washed three times with sterile phosphate-buffered saline (PBS; pH 7.2) and resuspended in PBS to an initial OD600 of 1.0 (A0). The bacterial suspension was incubated statically at 37°C for 5 h. The upper phase was gently transferred into a 96-well plate (100 ul per well) for the measurement of OD600 (A1). Auto-aggregation percentage was calculated as:
2.5.2 Co-aggregation assay
Co-aggregation (CA) assay was assessed as previously described (Mohanty et al., 2019). Briefly, L. garvieae ZB15 culture was prepared in MRS broth, and Escherichia coli ATCC 25922 (E. coli ATCC 25922), Escherichia coli K88 (E. coli K88), Staphylococcus aureus CMCC 26002 (S. aureus CMCC 26002), and Listeria monocytogenes CICC 21635 (L. monocytogenes CICC 21635) were cultured in LB medium. Upon reaching an OD600 of 2.0, bacterial cells were harvested by centrifuge (4,000 g, 10 min). The cell pellets were washed three times and resuspended in PBS. Pathogen suspensions were adjusted to an OD600 of 0.4 (A2), while the L. garvieae ZB15 suspension was adjusted to an OD600 of 0.6 (A3). Equal volumes of the pathogen and L. garvieae ZB15 suspensions were mixed at a 1:1 (v/v) ratio in 1 mL sterile tubes and incubated statically at 37°C for 5 h. The upper phase was gently transferred into a 96-well plate (100 μL per well) for the measurement of OD600 (A4), and CA percentage was calculated as:
2.5.3 Hydrophobicity ability detection
Hydrophobicity was measured based on bacterial affinity to chloroform, following Steinberg et al. (2022) with modifications. The bacterial cells were then harvested by centrifugation, washed with sterile PBS, and resuspended in PBS to an OD600 of 0.4 (A5). Subsequently, 3 mL of the cell suspension was mixed with 1 mL of chloroform, vortexed vigorously for 30 s, and allowed to stand undisturbed to allow phase separation. After separation, 1 mL of the upper aqueous phase was carefully removed and transferred into a 96-well plate (100 μL per well), and the optical density of the remaining aqueous phase was measured at 600 nm (A6). Hydrophobicity was calculated as:
2.6 Characterization of fungal inhibition
2.6.1 Fungal growth inhibition assay
Referring to the method Ali and Reddy described (Ali and Reddy, 2000), the antifungal activity of the cell-free supernatant (CFS) from L. garvieae ZB15 was evaluated against Penicillium grisofulvum Dierckx (P. grisofulvum Dierckx), Talaromyces verruculosus (T. verruculosus), Fusarium verticillioides (F. verticillioides), and Aspergillus ochraceus (A. ochraceus) (BeNa Culture Collection, Xinyang, Henan, China). Fungal spores were harvested and adjusted to an OD600 of 1.0 with PBS. Ten microliter fungal spores were inoculated into a well of 96-well plates containing 130 uL PDA medium with 60 ul serial dilutions of CFS (20–100 mg/mL). Plates were incubated at 28°C for 48 h, and OD600 was measured (AE). In the control group, an equivalent volume (60 μL) of PDA medium was supplemented to take the place of CFS for measurement of OD600 (Ac). For fungal inhibition rate (%) determination and calculated as:
2.6.2 Thermal stability of CFS
Cell-free supernatant (60 mg/mL) was subjected to heat treatment at 60°C, 80°C, and 100°C for 1 h. Subsequently, heat-treated CFS were used for fungal growth inhibition assay, and untreated CFS served as the control.
2.6.3 pH neutralization assay
To evaluate the role of organic acids, the pH of CFS (initial pH 4.5) was adjusted to pH 5.8 using 5 mol/L NaOH. Then, fungal growth inhibition assay was performed, and untreated CFS (pH 4.5) served as the control.
2.6.4 Protease sensitivity assay
To assess proteinaceous antifungal compounds, CFS was treated with protease (Trypases, Pepsin and Protease K, 2 mg/mL) for 2 h, followed by enzyme inactivation at 100°C for 5 min. Then, fungal growth inhibition assay was performed, and untreated CFS taken as the control.
2.6.5 Hydrogen peroxide sensitivity assay
To determine the role of hydrogen peroxide, CFS was treated with 10 mg/mL catalase for 2 h, followed by enzyme inactivation at 100°C for 5 min. Then, fungal growth inhibition assay was performed, and untreated CFS served as the control.
2.6.6 Scanning electron microscopy (SEM) of fungal morphology
Fungal morphology following CFS treatment was examined using SEM (Hitachi Reguius 8100, Hangzhou Feyman Biotechnology Co., Hangzhou, China). Samples treated with 60 mg/mL CFS were prepared for imaging, and untreated CFS served as the control.
2.6.7 Data analysis
All experiments were conducted independently in triplicate. Data were presented as mean values ± standard deviation (SD). Statistical analyses, including significance testing and Pearson correlation coefficients, were performed using SPSS 23 (IBM, Armonk, NY, USA). A p-value of less than 0.05 was considered statistically significant.
3 Result
3.1 Whole-genome sequencing and functional genomic characterization
The genomic features of L. garvieae ZB15 are presented in Table 1. Its genome consists of a single circular chromosome with a total length of 1,996,386 base pairs and a GC content of 38.94%. In addition, it contains genetic elements such as CRISPR arrays, prophages, repeat sequences, and gene islands. These genetic factors may contribute to the genomic plasticity of the strain and play a crucial role in its probiotic and antifungal potential.
TABLE 1
| Features | Values | Features | Values |
| Genome size | 1,996,386 bp | Prophage | 2 |
| GC Content | 38.94% | CRISPR array region | 6 |
| Number of protein coding sequences | 1,902 | Repeated sequence | 72 |
| Number of tRNAs | 62 | Tandem repeat | 40 |
| Number of rRNAs | 16 | Gene islands | 6 |
| Number of non-coding RNA | 78 |
General genomic information of the strain L. garvieae ZB15.
The complete circular genome map of L. garvieae ZB15 was illustrated in Figure 1a. The outermost ring represents the genome size, with each scale mark denoting 0.2 Mb. The second and third rings displayed protein-coding sequences (CDS) on the positive and negative strands, respectively, with color-coded classifications based on COG. The fourth ring highlighted the locations of rRNA and tRNA genes, while the fifth ring showed GC content variations. Regions with GC content exceeding the genome-wide average were marked in red, with peak heights corresponding to the degree of deviation. The innermost ring represented the GC skew, calculated as (G − C) / (G + C), where a positive value suggested a bias toward CDS transcription on the positive strand, whereas a negative value indicated preference for the negative strand.
FIGURE 1
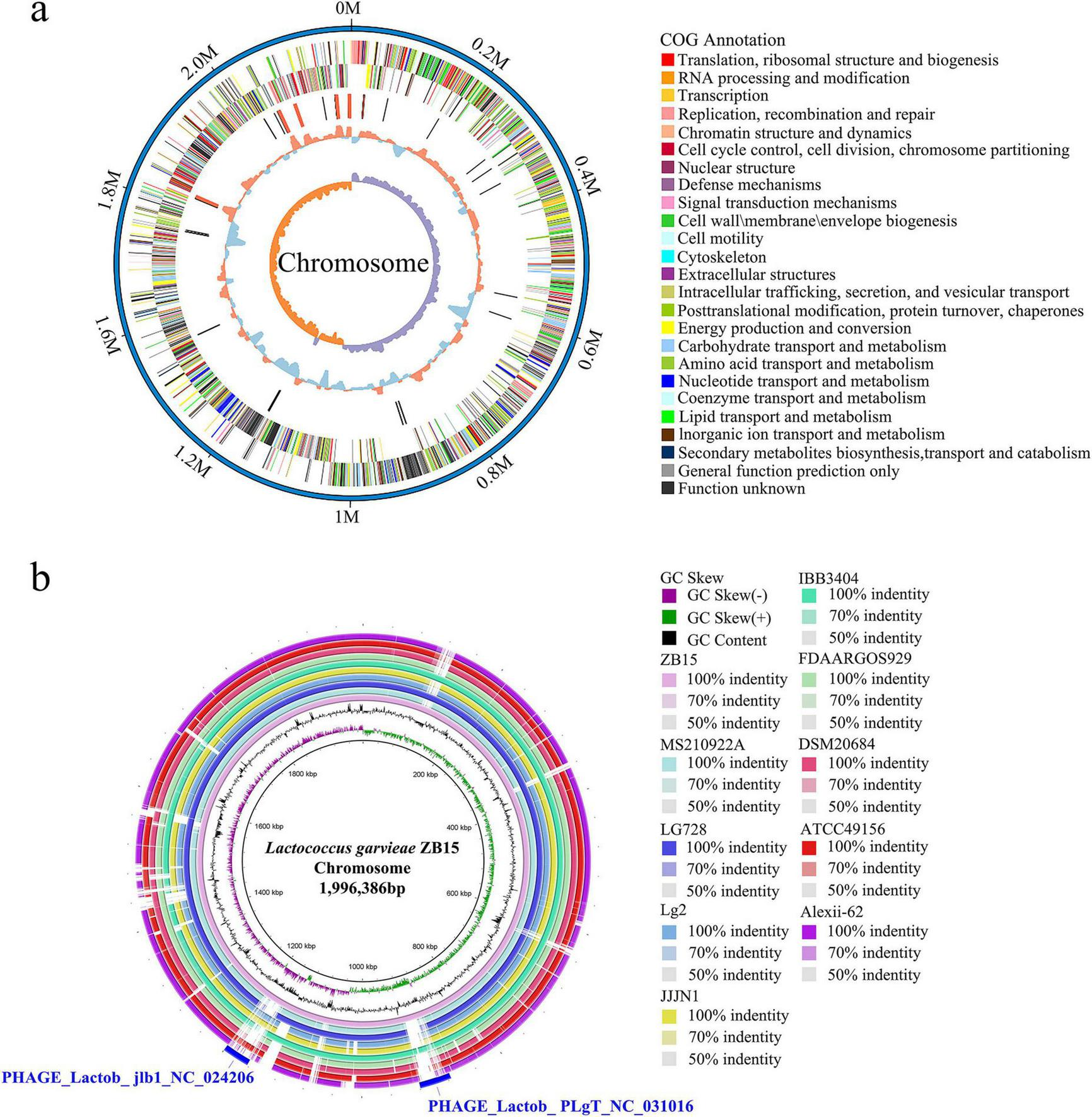
Circular genome map of L. garvieae ZB15. (a) The complete circular genome map of L. garvieae ZB15. (b)L. garvieae ZB15 genome BLAST comparison ring map.
Comparative genomic analysis of L. garvieae ZB15 with other L. garvieae strains was presented in Figure 1b. The BLAST ring comparison included L. garvieae strains Alexi-62, ATCC49156, DSM20684, FDAARGOS929, IBB3404, JJJN1, Lg2, 931, LG728, and MS210922A, with the outermost ring representing L. garvieae ZB15. This analysis revealed two distinct prophage regions: one classified as an intact phage and the other as a putative prophage. The intact prophage region exhibited a high degree of similarity to PHAGE_Lactob_ jlb1_NC024206 (46.7 kb), while the putative prophage region shared homology with PHAGE_Lactob_PLgT_NC031016 (43.4 kb) (Figure 1b). The intact prophage region encoded 58 proteins, including 34 hypothetical proteins, 17 phage-associated proteins, 3 portal proteins, 2 head proteins, 1 structural protein, and 1 domain-containing protein (Supplementary Table 1). In contrast, the putative prophage region encoded 66 proteins, comprising 51 hypothetical proteins, 5 phage-associated proteins, 3 portal proteins, 2 capsid decoration proteins, 1 ABC transporter, 1 DNA helicase, 1 tail-measuring protein, 1 minor tail protein, and 1 putative DnaC protein (Supplementary Table 2). The presence of these phage-associated genes suggested potential roles in horizontal gene transfer, genomic stability, and adaptation to environmental challenges.
To assess the genomic relatedness of L. garvieae ZB15 to other L. garvieae strains, an average nucleotide identity (ANI) analysis was conducted (Figure 2). The OrthoANI values indicated a high level of genomic similarity between L. garvieae ZB15 and FDAARGOS929 (98.49%), MS210922A (98.5%), DSM20684 (98.5%), LG728 (99.92%), JJJN1 (98.23%), Lg2 (98.23%), IBB3403 (98.18%), Alexi-62 (96.98%), and ATCC49156 (98.25%). Given that an ANI value above 96% was generally indicative of species-level classification, these results confirmed that L. garvieae ZB15 belonged to the L. garvieae species and shared high genetic similarity with these reference strains.
FIGURE 2
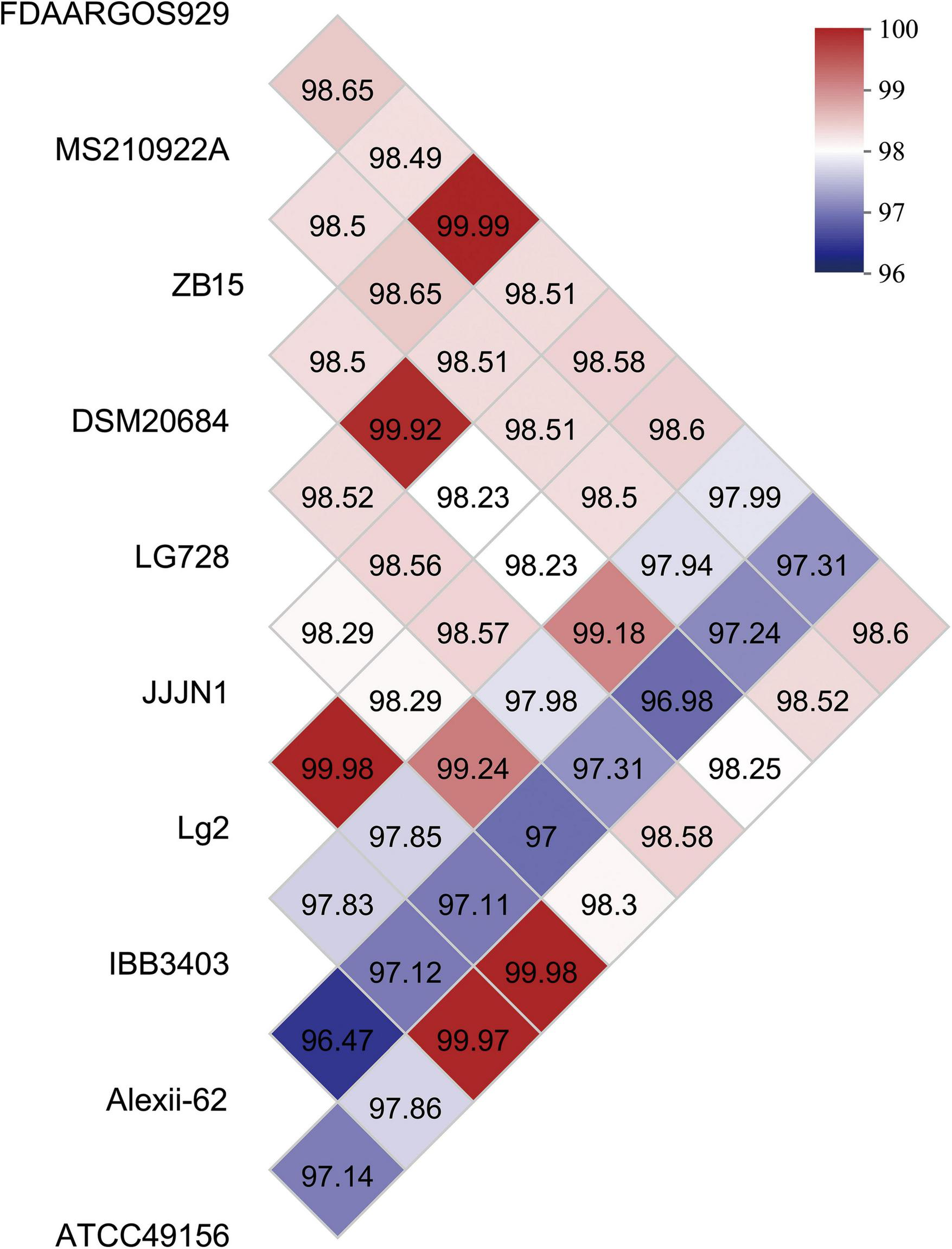
Average nucleotide identity (ANI) values of L. garvieae ZB15 and other compared well-known L. garvieae species.
Functional annotation of L. garvieae ZB15 revealed 1,551 gene functions based on GO classification (Supplementary Figure 1). These functions were primarily distributed across three major categories: biological processes, cellular components, and molecular functions. The most enriched GO terms include cellular processes, metabolic processes, localization, response to stimuli, and biological regulation. Furthermore, molecular functions related to catalytic activity, binding, transporter activity, ATP-dependent activity, and transcriptional regulation were significantly represented.
Pathway enrichment analysis using the KEGG database identified 23 significantly enriched pathways (Figure 3a). The most prominent pathways included carbohydrate metabolism, amino acid metabolism, and the metabolism of cofactors and vitamins. Additionally, genes involved in translation and membrane transport were significantly enriched, underscoring the strain’s potential metabolic versatility and environmental adaptability.
FIGURE 3
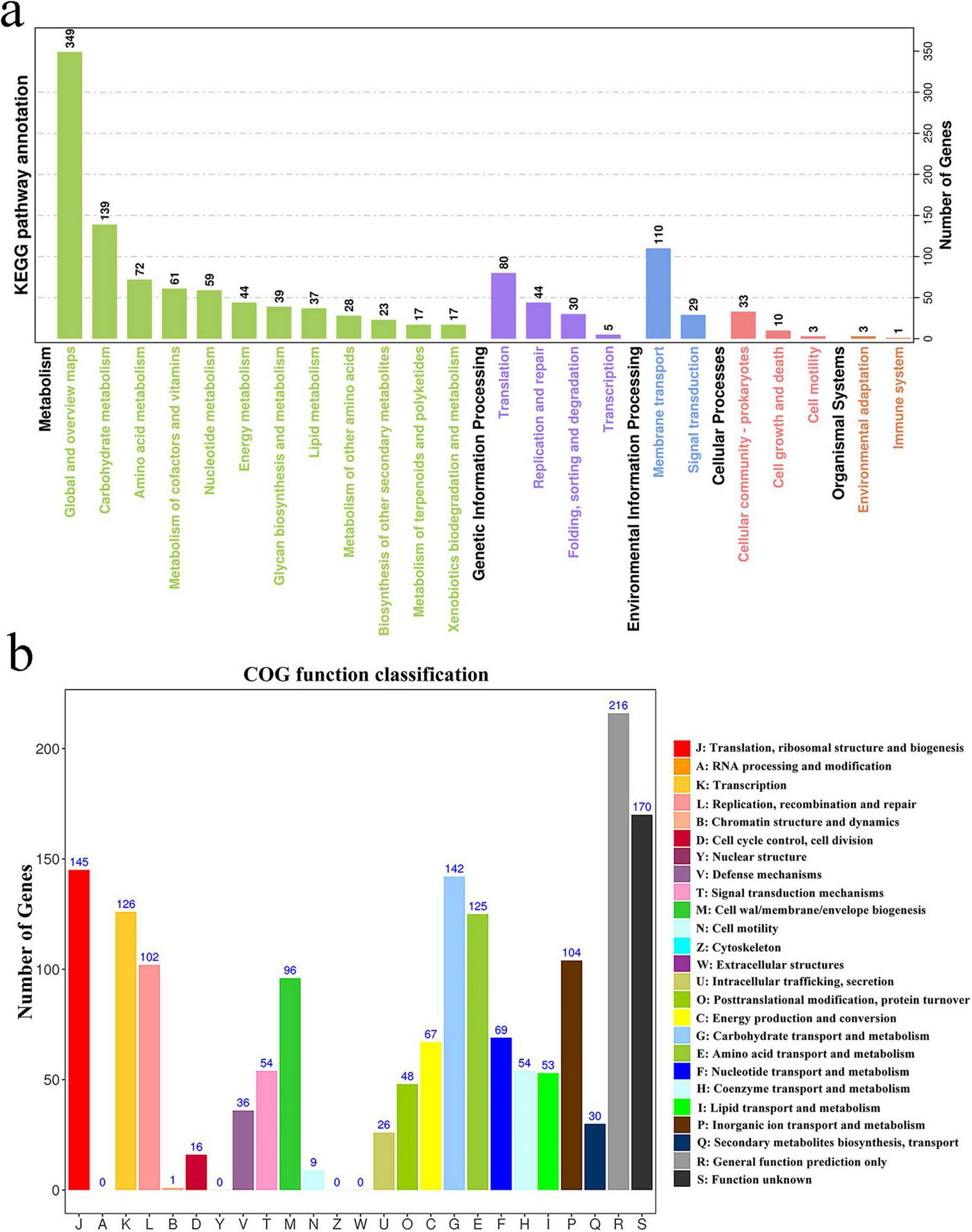
Whole-genome analysis of selected strains. (a) Enrichment of KEGG pathways for the L. garvieae ZB15 genome; (b) Functional classification of the L. garvieae ZB15 genome according to the Clusters of Orthologous Groups (COG) of proteins.
Cluster of Orthologous Groups classification further elucidated the functional profile of L. garvieae ZB15 (Figure 3b). The most highly represented functional categories include: translation, ribosomal structure, and biogenesis (J), RNA processing and modification (A), DNA replication, recombination, and repair (L).
These findings collectively provided insights into the genomic and functional characteristics of L. garvieae ZB15, suggesting its potential as a probiotic strain. The presence of phage-derived elements, diverse metabolic capabilities, and genetic similarity to other L. garvieae strains further highlighted its ecological adaptability and possible biotechnological applications.
3.2 Probiotic properties
Generally, the adhesion capacity (AC) of LAB encompasses cell surface hydrophobicity (CSH), auto-aggregation and CA capabilities. Auto-aggregation and CSH are crucial factors influencing the colonization ability of LAB in the intestinal environment. L. garvieae ZB15 exhibited an auto-aggregation percentage of 33.6% and a CSH of 79% (Table 2), suggesting a strong ability to adhere to intestinal epithelial cells, which is an important trait for effective probiotic function.
TABLE 2
| L. garvieae ZB15 | Percentage/% |
| Auto-aggregation | 33.6% ± 0.33% |
| Cell surface hydrophobicity | 79% ± 2.14% |
The Auto-aggregation capacity and cell surface hydrophobicity of L. garvieae ZB15.
In addition, the CA ability of L. garvieae ZB15 was evaluated, as it plays a key role in inhibiting pathogenic bacteria by preventing their adhesion and biofilm formation. The strain exhibited a CA rate of 62.96% with Escherichia coli ATCC25922, 14.44% with Escherichia coli K88, 35.37% with Staphylococcus aureus CMCC26002, and 32.43% with Listeria monocytogenes CICC21635 (Table 3). These results indicate that L. garvieae ZB15 has a substantial ability to co-aggregate with multiple pathogens, reinforcing its potential as a probiotic strain with antibacterial properties.
TABLE 3
| Pathogenic bacterium | Co-aggregation capacity |
| Escherichia coli ATCC25922 | 62.96% ± 1.33% |
| Escherichia coli K88 | 14.44% ± 0.88% |
| Staphylococcus aureus CMCC26002 | 35.37% ± 0.98% |
| Listeria monocytogenes CICC21635 | 32.43% ± 1.27% |
The Co-aggregation capacity of L. garvieae ZB15.
3.3 Antibiotic resistance
The antibiotic susceptibility profile of L. garvieae ZB15 was determined using the disc diffusion method (Supplementary Table 3). The strain displayed resistance to kanamycin (30 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), enrofloxacin (10 μg), vancomycin (30 μg), and sulfafurazole (300 μg). However, it was susceptible to penicillin G (10 U), ampicillin (10 μg), and amoxicillin (20 μg). Intermediate susceptibility was observed for cefotaxime sodium (30 μg), gentamicin (10 μg), erythromycin (15 μg), tetracycline (30 μg), and minocycline (30 μg).
To elucidate the genetic basis of these resistance phenotypes, whole-genome sequencing data were analyzed using the Comprehensive Antibiotic Resistance Database (CARD). No known antibiotic resistance genes were identified, suggesting that the observed resistance was likely mediated by intrinsic mechanisms rather than horizontally acquired resistance determinants. These findings indicated that L. garvieae ZB15 did not harbor mobile genetic elements associated with antibiotic resistance, supporting its potential safety as a probiotic candidate.
3.4 Carbohydrate fermentation patterns
Genomic analysis revealed that L. garvieae ZB15 encoded a total of 294 carbohydrate-active enzymes (CAZymes), classified into 117 glycosyltransferases, 123 glycoside hydrolases, 39 carbohydrate-binding modules, 12 carbohydrate esterases, 2 auxiliary activity enzymes, and 1 polysaccharide lyase (Supplementary Figure 2a). This extensive repertoire of CAZymes suggested a broad capacity for carbohydrate metabolism and structural modification.
Carbohydrate utilization was experimentally evaluated using the API 50 CHL assay, demonstrating that L. garvieae ZB15 could metabolize 17 out of 49 tested carbohydrates (Table 4 and Supplementary Figure 2b). The strain efficiently utilized D-ribose, D-galactose, D-glucose, D-fructose, D-mannose, D-mannitol, N-acetylglucosamine, amygdalin, arbutin, esculin ferric citrate, salicin, D-cellobiose, D-maltose, D-trehalose, amidon, gentiobiose, D-tagatose, and gluconate. These results highlighted the strain’s metabolic versatility and its potential adaptability to diverse nutritional environments.
TABLE 4
| Number | Sugars | Utilization | Number | Sugars | Utilization |
| 0 | Control | − | 25 | Esculin ferric citrate | + |
| 1 | Glycerol | − | 26 | Salicin | + |
| 2 | Erythritol | − | 27 | D-Cellobiose | + |
| 3 | D-Arabinose | − | 28 | D-Maltose | + |
| 4 | L-Arabinose | − | 29 | D-Lactose | − |
| 5 | D-Ribose | + | 30 | D-Melibiose | − |
| 6 | D-Xylose | − | 31 | D-Sucrose | − |
| 7 | L-Xylose | − | 32 | D-Trehalose | + |
| 8 | Adonitol | − | 33 | Inulin | − |
| 9 | Methyl-β-D-xylopyranoside | − | 34 | D-Melezitose | − |
| 10 | D-Galactose | + | 35 | D-Raffinose | − |
| 11 | D-Glucose | + | 36 | Amidon | + |
| 12 | D-Fructose | + | 37 | Glycogen | − |
| 13 | D-Mannose | + | 38 | Xylitol | − |
| 14 | L-Sorbose | − | 39 | Gentiobiose | + |
| 15 | L-Rhamnose | − | 40 | D-Turanose | − |
| 16 | Dulcitol | − | 41 | D-Lyxose | − |
| 17 | Inositol | − | 42 | D-Tagatose | + |
| 18 | D-Mannitol | + | 43 | D-Fucose | − |
| 19 | D-Sorbitol | − | 44 | L-Fucose | − |
| 20 | Methyl-α-D-mannopyranoside | − | 45 | D-Arabitol | − |
| 21 | Methyl-α-D-glucopyranoside | − | 46 | L-Arabitol | − |
| 22 | N-Acetylglucosamine | + | 47 | Gluconate | + |
| 23 | Amygdalin | + | 48 | 2-Keto-gluconate | − |
| 24 | Arbutin | + | 49 | 5-Keto-gluconate | − |
Statistics on carbohydrate metabolism ability of L. garvieae ZB15.
“+” means that the carbon sources can be fermented by L. garvieae ZB15, while “−” means that the carbon sources cannot be used.
3.5 Evaluation of antifungal activity
3.5.1 Effect of cell-free supernatant (CFS) concentration on fungal growth
The antifungal activity of L. garvieae ZB15 CFS exhibited a concentration-dependent effect (Figure 4 and Supplementary Figure 3). At 20 mg/mL, the inhibition rate ranged from 16.8 to 24.3%, increasing to 36.1%–40% at 40 mg/mL. A concentration of 60 mg/mL resulted in inhibition rates between 50.1 and 66.2%, while 80 mg/mL further enhanced inhibition to 60.9%–75.41%. At the highest concentration of 100 mg/mL, fungal inhibition exceeded 85%, indicating potent antifungal activity. Based on these findings, subsequent experiments were conducted using CFS at 60 mg/mL, which consistently demonstrated approximately 50% inhibition. Furthermore, Pearson correlation analysis revealed a significant positive correlation between CFS concentration and antifungal activity.
FIGURE 4
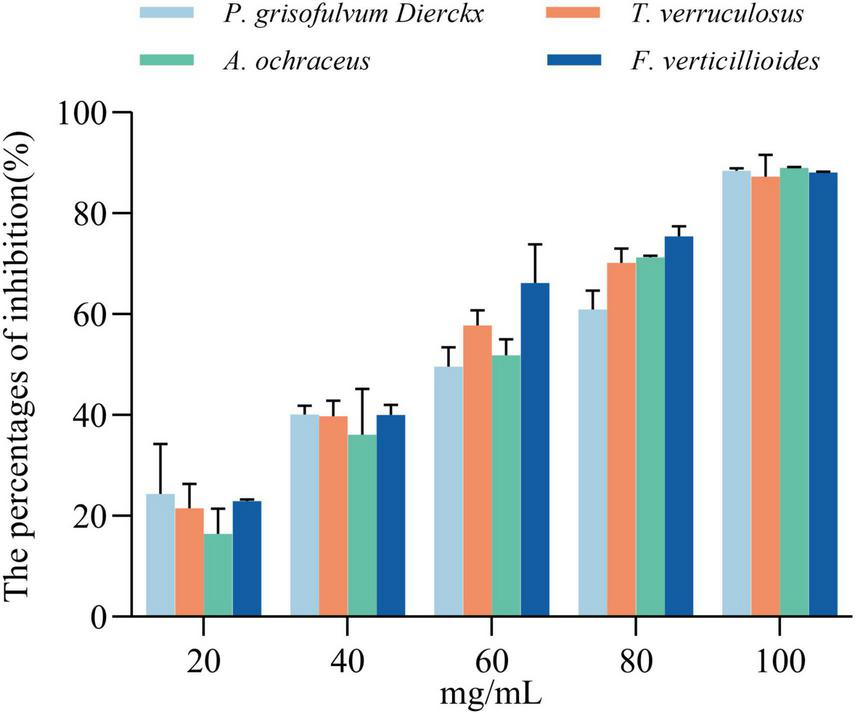
Inhibition rates of cell-free supernatant (CFS) at different concentrations against four fungal strains at 48 h. Statistical significance was determined by compared to the untreated controls, and “ns” indicates no significant difference.
Further characterization of CFS stability showed that antifungal activity remained largely unaffected after heating at 100°C for 1 h, with only minor reductions in inhibition rates (Figure 5a). These results suggested that the antifungal compounds are heat-stable and potentially resistant to thermal degradation.
FIGURE 5
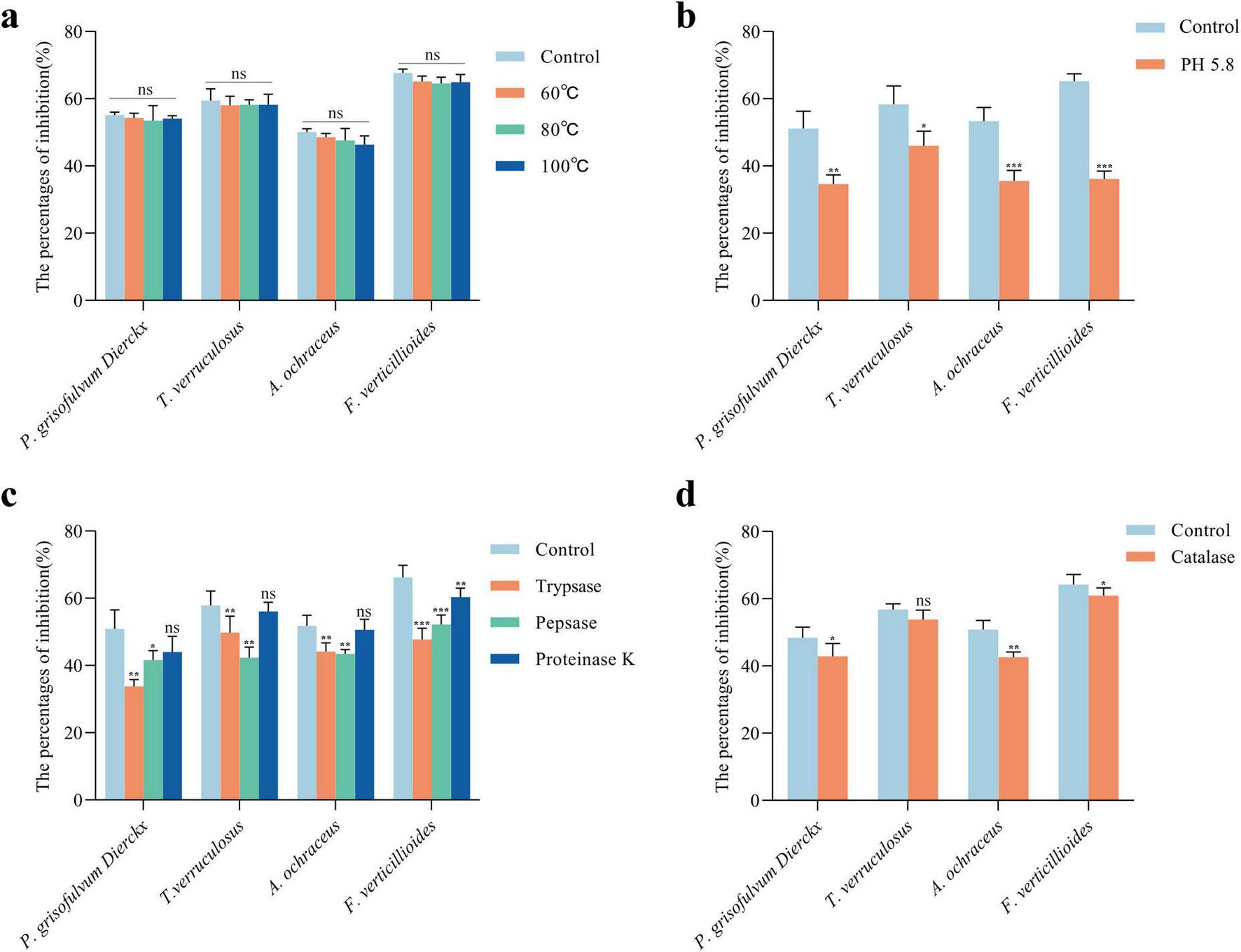
Characterization of antifungal active substances in L. garvieae ZB15 CFS. (a) Thermal stability of the CFS. (b) Antifungal activity of the CFS after organic acid neutralization. (c) Antifungal activity of the CFS following protease treatment. (d) Antifungal activity of the CFS after hydrogen peroxide treatment. Statistical significance was determined by compared to untreated controls. P < 0.05 is denoted by *, P < 0.01 by **, P < 0.001 by ***, and “ns” indicates no significant difference.
pH adjustment experiments indicated that neutralization of organic acids significantly reduced antifungal activity (Figure 5b). Increasing the CFS pH from 4.5 to 5.8 led to significant reductions in inhibition rates against P. grisofulvum Dierckx (33%, P < 0.01), T. verruculosus (20%, P < 0.05), A. ochraceus (33.2%, P < 0.001), and F. verticillioides (44.6%, P < 0.001). These findings indicated that organic acids played a key role in antifungal activity.
Protease treatments further elucidated the nature of antifungal compounds in the CFS (Figure 5c). Trypsin and pepsin treatments significantly reduced inhibition rates (P < 0.01), suggesting the involvement of proteinaceous components, whereas proteinase K treatment did not showed significant changes, indicating that partial proteins contributed to the antifungal effect.
Hydrogen peroxide (H2O2) degradation by catalase treatment resulted in a moderate reduction in antifungal activity (Figure 5d). The inhibition of P. grisofulvum, A. ochraceus, and F. verticillioides decreased after catalase treatment, with a notable 16.22% reduction in A. ochraceus. However, T. verruculosum remained unaffected, suggesting strain-specific susceptibility to H2O2.
In addition, Pearson correlation analysis revealed a significant negative correlation between treated CFS and antifungal activity. These results indicated that the antifungal activity of L. garvieae ZB15 CFS was mediated by a combination of organic acids, proteinaceous compounds, and hydrogen peroxide, each contributing differentially to the inhibition of fungal growth.
3.5.2 Fungal morphological changes induced by CFS
The effects of L. garvieae ZB15 CFS (60 mg/mL) on fungal morphology were examined using scanning electron microscopy (SEM) (Figure 6). Untreated control samples exhibited normal fungal hyphal structures, characterized by smooth, elongated, and intact morphology. In contrast, CFS-treated samples showed extensive morphological disruptions, including hyphal twisting, fragmentation, and surface roughening. These alterations were evident in all tested fungal species, with varying degrees of structural damage.
FIGURE 6
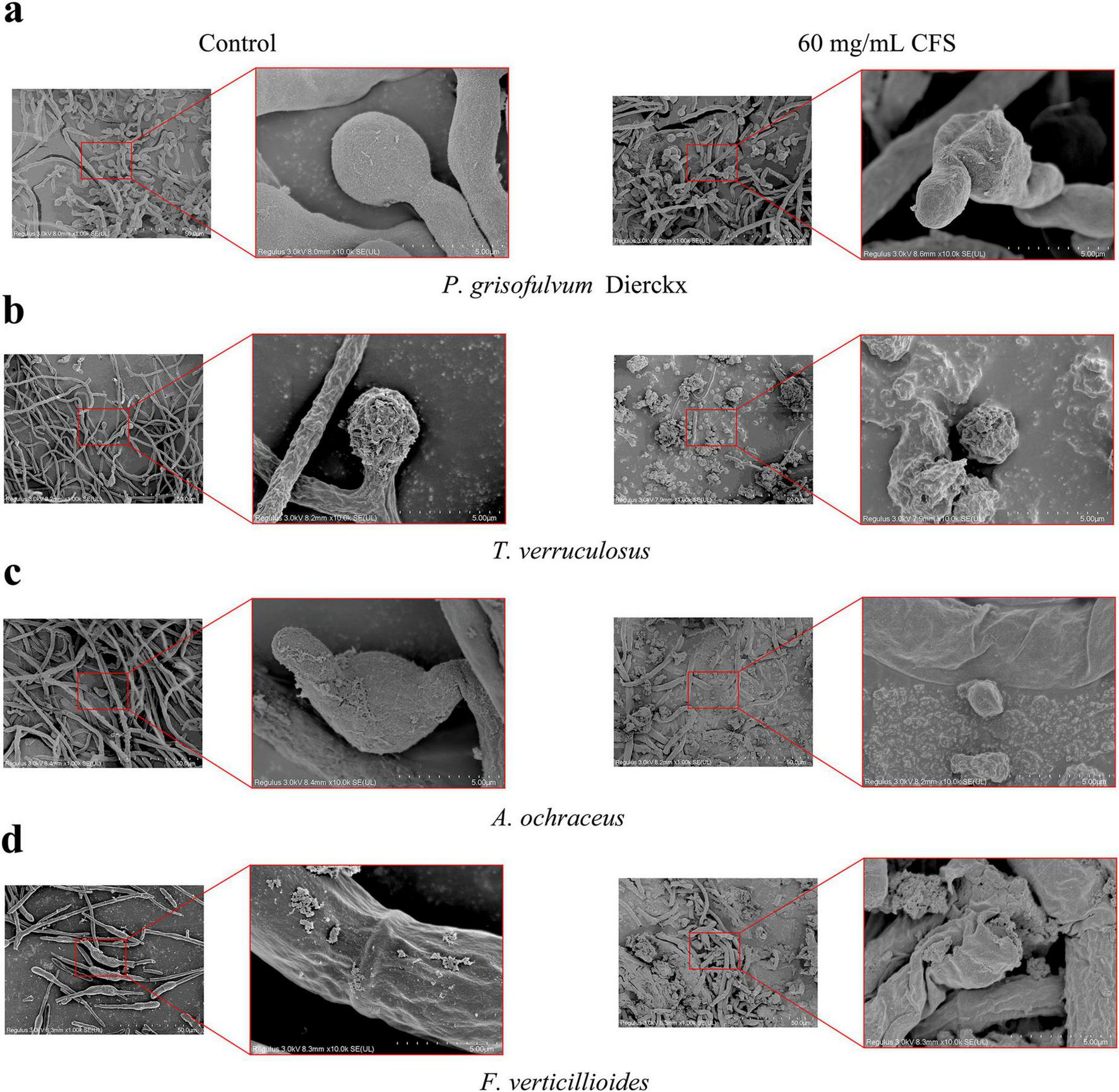
Fungal morphological changes induced by L. garvieae CFS. (a) The morphological influence of L. garvieae CFS on P. grisofulvum Dierckx. (b) The morphological influence of L. garvieae CFS on T. verruculosus. (c) The morphological influence of L. garvieae CFS on A. ochraceus. (d) The morphological influence of L. garvieae CFS on F. verticillioides.
Hyphal integrity loss was particularly pronounced in P. grisofulvum and F. verticillioides, where severe deformation and cell wall collapse were observed. A. ochraceus and T. verruculosum displayed milder but significant morphological changes, suggesting varying degrees of susceptibility among fungal species. These findings provided strong evidences that L. garvieae ZB15 CFS exerted antifungal effects through direct structural disruption of fungal cells, further supporting its potential as a biological control agent.
4 Discussion
Lactic acid bacteria have long been recognized as safe microorganisms with a history of use in fermented foods, extending their role beyond flavor enhancement to health promotion and food preservation(Wang et al., 2021). Growing research interest has focused on their ability to inhibit pathogenic microorganisms, with particular attention to the antifungal properties of LAB CFSs as potential natural preservatives.
Advancements in whole-genome sequencing have enabled comprehensive safety assessments of LAB strains. L. garvieae ZB15 exhibited no known virulence or antibiotic resistance genes, reinforcing its suitability as a probiotic. This finding aligns with studies on other LAB strains. For instance, Lactobacillus paracasei DTA93 was found to be free of virulence factors, prophages, and antibiotic resistance genes (Tarrah et al., 2020). Similarly, Limosilactobacillus fermentum AGA52 contained two intact prophages but lacked antibiotic resistance genes (Yetiman et al., 2024). The genome of L. garvieae ZB15 contained two prophages (one intact, one incomplete) but no evidence of horizontal gene transfer from different bacterial species, further confirming its genetic stability and safety for potential applications.
The ability of LAB to adhere to intestinal epithelial cells is crucial for colonization and competitive exclusion of pathogens. This property is influenced by auto-aggregation (AC), CA, and CSH (Pino et al., 2022). The 2002 Guidelines for Evaluation of Probiotics in Food, issued by the United Nations, state that the adhesive capacity of probiotics should encompass auto-aggregation, CA, and hydrophobicity. A high auto-aggregation capacity in LAB enables strains to attain a high density within the intestinal tract, which enhances their adhesion to the gut mucosa (Espeche et al., 2012). In this study, L. garvieae ZB15 exhibited an AC rate of 33.6%, comparable to previously reported values for LAB strains such as Lactococcus lactis and Enterococcus species, which ranged from 27.86 to 38.84% (Nasrollahzadeh et al., 2022).
Co-aggregation is another key probiotic trait that facilitates the inhibition of pathogen adhesion. This process facilitates the CA of pathogens with probiotic cells. Subsequently, the aggregated complexes are eliminated from the body through the gastrointestinal tract, thereby mitigating the hazards associated with pathogenic contamination (Espeche et al., 2012). Lactococcus garvieae ZB15 demonstrated CA with E. coli ATCC25922 (62.96%), E. coli K88 (14.44%), S. aureus CMCC26002 (35.37%), and L. monocytogenes CICC21635 (32.43%). These values fall within the reported range of LAB-pathogen CA rates (9.03%–96.78%) (Reuben et al., 2020), supporting the potential of L. garvieae ZB15 to competitively exclude pathogenic bacteria.
Cell-surface hydrophobicity represents a key physicochemical property of microorganisms. High cell-surface hydrophobicity facilitates non-specific adhesion to biotic or abiotic surfaces. Consequently, LAB exhibiting strong hydrophobicity demonstrate enhanced colonization potential within the intestinal tract (Vadillo-Rodríguez et al., 2005). CSH, a critical determinant of bacterial adhesion, was considered high when exceeding 50% (Steinberg et al., 2022). Compared to Latilactobacillus curvatus (13.65%) and Weissella cibaria (17.32%) (Zhang et al., 2022), L. garvieae ZB15 exhibited remarkably high hydrophobicity (79%), further underscoring its strong adhesive potential and probiotic viability.
Previous research has demonstrated that LAB-derived CFS inhibits both fungal growth and mycotoxin production. Organic acids (e.g., lactic acid, acetic acid) within CFS diffuse across the cell membrane in their hydrophobic, undissociated forms. Subsequent intracellular dissociation releases H+ ions, acidifying the cytosol and disrupting the proton gradient, thereby inhibiting fungal growth. Compounds such as propionic acid further reduce fungal proliferation by inhibiting amino acid synthesis or upregulating the phiA protein (Moradi et al., 2020). Beyond organic acids, antifungal peptides represent another key class of LAB metabolites. Studies indicate that antifungal peptides induce fungal cell death by binding to chitin, causing cell wall rupture, or damaging the plasma membrane, leading to leakage of intracellular constituents (Chen et al., 2023). In our study, the CFS of L. garvieae ZB15 exhibited concentration-dependent antifungal activity. A dose of 60 mg/mL inhibited over 50% fungal growth, while 100 mg/mL achieved suppression exceeding 80%. These findings align with results observed for Lactiplantibacillus MYS44, which significantly inhibited Fusarium parasiticum growth and aflatoxin production (Ström et al., 2002). Consistently, Gerez et al. (2010) assessed the antifungal activity of various LAB cultivated in wheat flour hydrolysate (WFH), noting inhibition of mycelial growth in Aspergillus niger CH 101 and Fusarium graminearum CH 103 by CFS from L. plantarum CRL 778 and L. brevis CRL 796.
Thermal stability is an essential characteristic of effective antifungal agents for food preservation. The antifungal activity of L. garvieae ZB15 CFS remained high even after heating at 100°C for 1 h, with inhibition rates ranging from 46 to 67%. This aligns with reports that Lactococcus gasseri and Limosilactobacillus fermentum CFS retained antimicrobial activity after exposure to 100°C (Scillato et al., 2021), highlighting the potential of L. garvieae ZB15 CFS for heat-processed food applications.
The antifungal activity of LAB CFS was attributed to multiple biologically active ingredients, including organic acids, proteins, and hydrogen peroxide (De Vuyst and Leroy, 2007). In this study, protease treatments (trypsin, pepsin, and proteinase K) significantly reduced the antifungal activity of L. garvieae ZB15 CFS, indicating that proteinaceous components play a key role. However, antifungal activity was not completely abolished, suggesting the presence of additional active compounds.
Acid neutralization significantly reduced inhibition rates against all tested fungal strains, confirming that organic acids mainly contributed to the antifungal effect. This was consistent with findings on plantarum QS494 and Lactiplantibacillus pentosus QS530, where acid removal diminished antifungal activity (de Lira et al., 2023). Research indicates that most LAB produce hydrogen peroxide during fermentation. However, these bacteria inherently lack the ability to produce catalase. Consequently, hydrogen peroxide accumulates in the fermented product. This compound inhibits fungal growth by oxidizing both the cytoplasmic membrane and proteins within fungal cells (Sadiq et al., 2019). Furthermore, catalase treatment reduced inhibition, indicating a role for hydrogen peroxide, similar to previous observations.
Scanning electron microscopy (SEM) revealed that L. garvieae ZB15 CFS caused substantial structural alterations in fungal hyphae, including fragmentation, surface roughening, and deformation. These findings were consistent with previous reports showing that LAB CFS induced severe morphological damage in fungal pathogens. Lactiplantibacillus MYS6, for example, was shown to cause hyphal twisting and breakage in Fusarium proliferatum (Deepthi et al., 2016). Similarly, Lactiplantibacillus YML007 disrupted fungal cell walls, leading to reduced hyphal integrity (Ahmad Rather et al., 2013). However, the precise biochemical mechanisms underlying these morphological changes remained unclear and warrant further investigations.
5 Conclusion
This study demonstrated that L. garvieae ZB15 possessed probiotic characteristics, including strong adhesion properties and pathogen exclusion capabilities. Its CFS exhibited significant antifungal activity, mediated by organic acids, proteins, and hydrogen peroxide (Figure 7). The strain’s genetic safety, lack of antibiotic resistance genes, and ability to produce heat-stable antifungal compounds highlighted its potential as a natural food preservative. Future studies would focus on characterizing the specific biologically active ingredients responsible for antifungal activity and elucidating their molecular mechanisms.
FIGURE 7
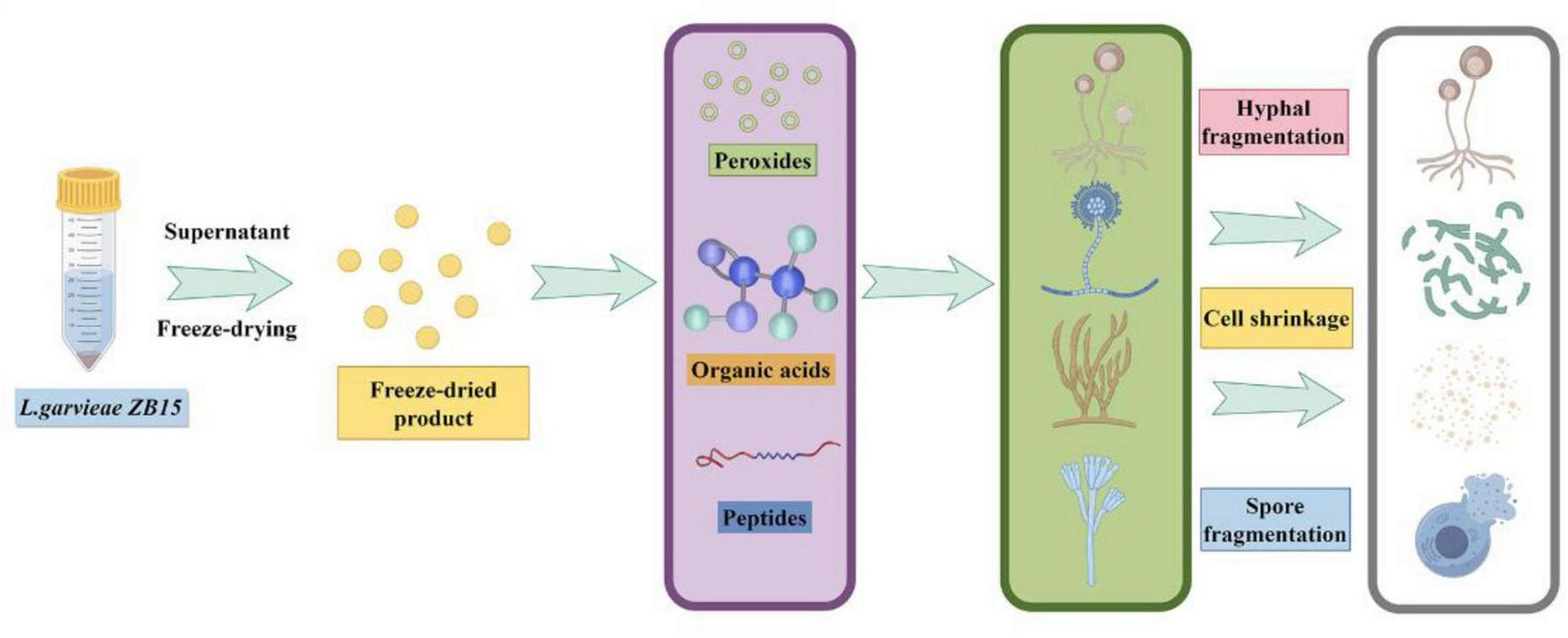
The antifungal effect of a novel L. garvieae derived from bacon.
Statements
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
GL: Conceptualization, Formal Analysis, Investigation, Methodology, Writing – original draft. XH: Data curation, Investigation, Validation, Writing – review and editing. JL: Formal Analysis, Supervision, Writing – original draft. YJ: Investigation, Resources, Writing – original draft. LZ: Investigation, Methodology, Writing – review and editing. SW: Investigation, Resources, Writing – review and editing. WZ: Conceptualization, Resources, Supervision, Writing – review and editing. TZ: Formal Analysis, Investigation, Writing – review and editing. HL: Formal Analysis, Investigation, Writing – review and editing. LW: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Key Scientific Research Project of Education Department of Shaanxi Provincial Government (24JR046, 24JR045) and college student innovation and entrepreneurship project (S202410720053).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1610971/full#supplementary-material
References
1
Ahmad RatherI.SeoB.Rejish KumarV.ChoiU.ChoiK.LimJ.et al (2013). Isolation and characterization of a proteinaceous antifungal compound from Lactobacillus plantarum YML007 and its application as a food preservative.Lett. Appl. Microbiol.5769–76. 10.1111/lam.12077
2
AliG.ReddyA. (2000). Inhibition of fungal and bacterial plant pathogens by synthetic peptides: In vitro growth inhibition, interaction between peptides and inhibition of disease progression.Mol. Plant Microbe Interact.13847–859. 10.1094/MPMI.2000.13.8.847
3
AmbeN.BobgaT.Toukam TatsinkouL.Sotoing TaiweG.FossiB. (2024). Ameliorative effects of probiotic Limosilactobacillus fermentum and Enterococcus lactis isolated from cameroonian traditionally processed milk and palm wine against chronic constriction injury induced neuropathic pain in mice.J. Ethnopharmacol.334:118560. 10.1016/j.jep.2024.118560
4
BringelF.CastioniA.OlukoyaD.FelisG.TorrianiS.DellaglioF. (2005). Lactobacillus plantarum subsp. argentoratensis subsp. nov., isolated from vegetable matrices.Int. J. Syst. Evol. Microbiol.551629–1634. 10.1099/ijs.0.63333-0
5
ChenH.YanX.DuG.GuoQ.ShiY.ChangJ.et al (2023). Recent developments in antifungal lactic acid bacteria: Application, screening methods, separation, purification of antifungal compounds and antifungal mechanisms.Crit. Rev. Food Sci. Nutr.632544–2558. 10.1080/10408398.2021.1977610
6
de LiraF.TanakaF.RiosE.CarrilhoS.de AbreuS.FerreiraG.et al (2023). Identification of lactic acid bacteria with anti-listeria activity. Characterization and application of a bacteriocinogenic strain in the control of Listeria monocytogenes in cheese.J. Dairy Res.90318–323. 10.1017/S0022029923000584
7
De VuystL.LeroyF. (2007). Bacteriocins from lactic acid bacteria: Production, purification, and food applications.J. Mol. Microbiol. Biotechnol.13194–199. 10.1159/000104752
8
DeepthiB.Poornachandra RaoK.ChennapaG.NaikM.ChandrashekaraK.SreenivasaM. (2016). Antifungal attributes of Lactobacillus plantarum MYS6 against Fumonisin producing Fusarium proliferatum associated with poultry feeds.PLoS One11:e0155122. 10.1371/journal.pone.0155122
9
El-DeebW.FayezM.ElsohabyI.GhoneimI.Al-MarriT.KandeelM.et al (2020). Isolation and characterization of vaginal Lactobacillus spp. in dromedary camels (Camelus dromedarius): In vitro evaluation of probiotic potential of selected isolates.PeerJ8:e8500. 10.7717/peerj.8500
10
EllisJ.HindrichsenI.KlopG.KinleyR.MiloraN.BanninkA.et al (2016). Effects of lactic acid bacteria silage inoculation on methane emission and productivity of Holstein Friesian dairy cattle.J. Dairy Sci.997159–7174. 10.3168/jds.2015-10754
11
EspecheM.PellegrinoM.FrolaI.LarriestraA.BogniC.Nader-MacíasM. (2012). Lactic acid bacteria from raw milk as potentially beneficial strains to prevent bovine mastitis.Anaerobe18103–109. 10.1016/j.anaerobe.2012.01.002
12
GerezC.CarbajoM.RollánG.Torres LealG.Font de ValdezG. (2010). Inhibition of citrus fungal pathogens by using lactic acid bacteria.J. Food Sci.75M354–M359. 10.1111/j.1750-3841.2010.01671.x
13
GoldsteinE.TyrrellK.CitronD. (2015). Lactobacillus species: Taxonomic complexity and controversial susceptibilities.Clin. Infect. Dis.60S98–S107. 10.1093/cid/civ072
14
GradovilleM.CrumpB.HäseC.WhiteA. (2018). Environmental controls of oyster-pathogenic vibrio spp. in oregon estuaries and a shellfish hatchery.Appl. Environ. Microbiol.84e2156–e2117. 10.1128/AEM.02156-17
15
HindlerJ.StellingJ. (2007). Analysis and presentation of cumulative antibiograms: A new consensus guideline from the clinical and laboratory standards institute.Clin. Infect. Dis.44867–873. 10.1086/511864
16
JantrakajornS.SuyapohW.WongtavatchaiJ. (2024). Characterization of Lactococcus garvieae and Streptococcus agalactiae in cultured red tilapia Oreochromis sp. in Thailand.J. Aquat. Anim. Health36192–202. 10.1002/aah.10217
17
KawadaY.YokoyamaS.YanaseE.NiwaT.SuzukiT. (2016). The production of S-equol from daidzein is associated with a cluster of three genes in Eggerthella sp. YY7918.Biosci. Microbiota Food Health35113–121. 10.12938/bmfh.2015-023
18
KhanM.HabibiN.ShaheedF.MustafaA. (2016). Draft genome sequences of five clinical strains of brucella melitensis isolated from patients residing in kuwait.Genome Announc.4e1144–e1116. 10.1128/genomeA.01144-16
19
KhanZ.SiddiquiM.ParkS. (2019). Current and emerging methods of antibiotic susceptibility testing.Diagnostics (Basel)9:10. 10.3390/diagnostics9020049
20
KimN.MoonP.KimS.ChoiI.AnH.MyungN.et al (2008). Lipid profile lowering effect of Soypro fermented with lactic acid bacteria isolated from Kimchi in high-fat diet-induced obese rats.Biofactors3349–60. 10.1002/biof.5520330105
21
LiZ.ZhouH.LiuW.WuH.LiC.LinF.et al (2024). Beneficial effects of duck-derived lactic acid bacteria on growth performance and meat quality through modulation of gut histomorphology and intestinal microflora in Muscovy ducks.Poult. Sci.103:104195. 10.1016/j.psj.2024.104195
22
LiuZ.XuC.TianR.WangW.MaJ.GuL.et al (2021). Screening beneficial bacteriostatic lactic acid bacteria in the intestine and studies of bacteriostatic substances.J. Zhejiang Univ. Sci. B22533–547. 10.1631/jzus.B2000602
23
MohantyD.PandaS.KumarS.RayP. (2019). In vitro evaluation of adherence and anti-infective property of probiotic Lactobacillus plantarum DM 69 against Salmonella enterica.Microb. Pathog.126212–217. 10.1016/j.micpath.2018.11.014
24
MoradiM.KoushehS.AlmasiH.AlizadehA.GuimarãesJ.YılmazN.et al (2020). Postbiotics produced by lactic acid bacteria: The next frontier in food safety.Compr. Rev. Food Sci. Food Saf.193390–3415. 10.1111/1541-4337.12613
25
NasrollahzadehA.MokhtariS.KhomeiriM.SarisP. (2022). Antifungal preservation of food by lactic acid bacteria.Foods11:395. 10.3390/foods11030395
26
NavarréA.NazarethT.LuzC.MecaG.EscriváL. (2024). Characterization of lactic acid bacteria isolated from human breast milk and their bioactive metabolites with potential application as a probiotic food supplement.Food Funct.158087–8103. 10.1039/d4fo02171a
27
PinoA.VaccalluzzoA.CaggiaC.BalzarettiS.VanellaL.SorrentiV.et al (2022). Lacticaseibacillus rhamnosus CA15 (DSM 33960) as a candidate probiotic strain for human health.Nutrients14:4902. 10.3390/nu14224902
28
RabaouiG.Sánchez-JuanesF.TebiniM.NaghmouchiK.BellidoJ.Ben-MahrezK.et al (2023). Potential probiotic lactic acid bacteria with anti-Penicillium expansum activity from different species of tunisian edible snails.Probiotics Antimicrob. Proteins1582–106. 10.1007/s12602-021-09882-5
29
ReubenR.RoyP.SarkarS.Rubayet Ul AlamA.JahidI. (2020). Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties.J. Dairy Sci.1031223–1237. 10.3168/jds.2019-17092
30
RubioR.JofréA.MartínB.AymerichT.GarrigaM. (2014). Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages.Food Microbiol.38303–311. 10.1016/j.fm.2013.07.015
31
SadiqF.YanB.TianF.ZhaoJ.ZhangH.ChenW. (2019). Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: A comprehensive review.Compr. Rev. Food Sci. Food Saf.181403–1436. 10.1111/1541-4337.12481
32
ScillatoM.SpitaleA.MongelliG.PriviteraG.ManganoK.CianciA.et al (2021). Antimicrobial properties of Lactobacillus cell-free supernatants against multidrug-resistant urogenital pathogens.Microbiologyopen10:e1173. 10.1002/mbo3.1173
33
SeddikH.BendaliF.GancelF.FlissI.SpanoG.DriderD. (2017). Lactobacillus plantarum and its probiotic and food potentialities.Probiotics Antimicrob. Proteins9111–122. 10.1007/s12602-017-9264-z
34
SirichoatA.FlórezA.VázquezL.BuppasiriP.PanyaM.LulitanondV.et al (2020). Antibiotic susceptibility profiles of lactic acid bacteria from the human vagina and genetic basis of acquired resistances.Int. J. Mol. Sci.21:2594. 10.3390/ijms21072594
35
SteinbergR.SilvaL.de SouzaM.ReisR.BicalhoA.NunesJ.et al (2022). Prospecting of potentially probiotic lactic acid bacteria from bovine mammary ecosystem: Imminent partners from bacteriotherapy against bovine mastitis.Int. Microbiol.25189–206. 10.1007/s10123-021-00209-6
36
StrömK.SjögrenJ.BrobergA.SchnürerJ. (2002). Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and 3-phenyllactic acid.Appl. Environ. Microbiol.684322–4327. 10.1128/AEM.68.9.4322-4327.2002
37
TarrahA.da Silva DuarteV.PakrooS.CorichV.GiacominiA. (2020). Genomic and phenotypic assessments of safety and probiotic properties of Streptococcus macedonicus strains of dairy origin.Food Res. Int.130:108931. 10.1016/j.foodres.2019.108931
38
TayeY.DeguT.FessehaH.MathewosM. (2021). Isolation and identification of lactic acid bacteria from cow milk and milk products.ScientificWorldJournal2021:4697445. 10.1155/2021/4697445
39
Vadillo-RodríguezV.BusscherH.van der MeiH.de VriesJ.NordeW. (2005). Role of lactobacillus cell surface hydrophobicity as probed by AFM in adhesion to surfaces at low and high ionic strength.Colloids Surf. B Biointerfaces4133–41. 10.1016/j.colsurfb.2004.10.028
40
VasieeA.MortazaviS.SankianM.YazdiF.MahmoudiM.ShahidiF. (2019). Antagonistic activity of recombinant Lactococcus lactis NZ1330 on the adhesion properties of Escherichia coli causing urinary tract infection.Microb. Pathog.133:103547. 10.1016/j.micpath.2019.103547
41
WangX.MengF.ZhangM.LiF.LeiY.MaZ.et al (2024). Gut Lactococcus garvieae promotes protective immunity to foodborne Clostridium perfringens infection.Microbiol. Spectr.12:e0402523. 10.1128/spectrum.04025-23
42
WangY.WuJ.LvM.ShaoZ.HungweM.WangJ.et al (2021). Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry.Front. Bioeng. Biotechnol.9:612285. 10.3389/fbioe.2021.612285
43
YetimanA.HorzumM.BaharD.AkbulutM. (2024). Assessment of genomic and metabolic characteristics of cholesterol-reducing and GABA producer Limosilactobacillus fermentum AGA52 isolated from lactic acid fermented shalgam based on in silico and in vitro approaches.Probiotics Antimicrob. Proteins16334–351. 10.1007/s12602-022-10038-2
44
YuJ.WangW.MengheB.JiriM.WangH.LiuW.et al (2011). Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia.J. Dairy Sci.943229–3241. 10.3168/jds.2010-3727
45
ZhangA.YangY.LiY.ZhengY.WangH.CuiH.et al (2024). Effects of wheat-based fermented liquid feed on growth performance, nutrient digestibility, gut microbiota, intestinal morphology, and barrier function in grower-finisher pigs.J. Anim. Sci.102:skae229. 10.1093/jas/skae229
46
ZhangQ.WangM.MaX.LiZ.JiangC.PanY.et al (2022). In vitro investigation on lactic acid bacteria isolatedfrom Yak faeces for potential probiotics.Front. Cell. Infect. Microbiol.12:984537. 10.3389/fcimb.2022.984537
47
ZhangT.XieJ.ZhangM.FuN.ZhangY. (2016). Effect of a potential probiotics Lactococcus garvieae B301 on the growth performance, immune parameters and caecum microflora of broiler chickens.J. Anim. Physiol. Anim. Nutr.100413–421. 10.1111/jpn.12388
48
ZhongY.FuD.DengZ.TangW.MaoJ.ZhuT.et al (2022). Lactic acid bacteria mixture isolated from wild pig alleviated the gut inflammation of mice challenged by Escherichia coli.Front. Immunol.13:822754. 10.3389/fimmu.2022.822754
Summary
Keywords
Lactococcus garvieae, genome, probiotic, antifungal effect, lactic
Citation
Li G, He X, Li J, Jian Y, Zhu L, Wang S, Zeng W, Zhang T, Lu H and Wang L (2025) Assessment of genomic and antifungal properties of Lactococcus garvieae ZB15 isolated from Zhenba bacon. Front. Microbiol. 16:1610971. doi: 10.3389/fmicb.2025.1610971
Received
13 April 2025
Accepted
04 June 2025
Published
26 June 2025
Volume
16 - 2025
Edited by
Joaquin Bautista-Gallego, University of Extremadura, Spain
Reviewed by
Giuseppe Perri, University of Bari Aldo Moro, Italy
Iftikhar Younis Mallhi, Minhaj University Lahore, Pakistan
Updates
Copyright
© 2025 Li, He, Li, Jian, Zhu, Wang, Zeng, Zhang, Lu and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongzhao Lu, zl780823@snut.edu.cnLing Wang, wangling619@163.com
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.