- 1Department of Critical Care Medicine, The Affiliated Hospital of Guizhou Medical University, Guiyang, China
- 2Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education and School/Hospital of Stomatology, Guizhou Medical University, Guiyang, China
- 3Clinical Medical College, Guizhou Medical University, Guiyang, China
- 4School/Hospital of Stomatology, Guizhou Medical University, Guiyang, Guizhou, China
- 5Key Laboratory of Microbiology and Parasitology of the Education Department of Guizhou, Guizhou Medical University, Guiyang, China
- 6Collaborative Innovation Center for Prevention and Control of Endemic and Ethnic Regional Diseases Co-Constructed by the Province and Ministry, Guiyang, China
- 7CAS Key Laboratory of Biofuels, Shandong Provincial Key Laboratory of Synthetic Biology, Shandong Engineering Laboratory of Single Cell Oil, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 8Shandong Energy Institute, Qingdao, China
- 9Qingdao New Energy Shandong Laboratory, Qingdao, China
Background: Clostridioides difficile infection (CDI) represents a significant global public health concern. The Phosphatidylinositol Glycan Class L (pig-L) gene in C. difficile encodes an enzyme critical for the biosynthesis of Glycosylphosphatidylinositol (GPI) anchor, which play a vital role in bacterial surface protein localization and function.
Methods: To investigate the role of pig-L in C. difficile pathogenesis, we utilized CRISPR-Cas9 gene editing to generate a pig-L knockout strain and a complementation strain in the wild-type (WT) background. Phenotypic characterization of these strains was performed through a suite of assays, including virulence assays, biofilm formation assays, oxidative stress sensitivity testing, and antimicrobial susceptibility testing. Proteomics analysis was conducted to identify differentially expressed proteins in the knockout strain.
Results: Deletion of the pig-L gene resulted in a significant reduction in C. difficile virulence, decreased biofilm formation, and increased susceptibility to oxidative stress. Proteomic analysis revealed significant alterations in protein expression, with 170 proteins exhibiting upregulation and 101 proteins demonstrating downregulation in the knockout strain. Complementation of the pig-L gene partially restored the phenotypes observed in the deletion strain.
Conclusion: These findings demonstrate that the pig-L gene functions as a crucial regulator of C. difficile virulence, biofilm formation, and peroxide resistance. Targeting the pig-L gene or its downstream effectors represents a promising avenue for the development of novel therapeutic strategies to effectively control C. difficile infection.
1 Introduction
Clostridioides difficile (C. difficile) is an intestinal pathogen known for its production of toxins and is the etiological agent of C. difficile infection (CDI) (Schäffler and Breitrück, 2018). It possesses flagella for motility and reproduces through sporulation, primarily spreading via the fecal-oral route (Zeng et al., 2022). This bacterium is a primary causative agent of antibiotic-associated diarrhea, accounting for about 50–75% of such cases. Moreover, it plays a significant role in antibiotic-associated colitis and pseudomembranous colitis, responsible for 50–75% and 95–100% of these conditions, respectively (Nakao et al., 2020).
The primary clinical symptoms of CDI include diarrhea, abdominal pain, and fever. Severe cases may lead to pseudomembranous colitis and potentially progress to toxic megacolon, bowel perforation, septic shock, or even death (Abbas and Zackular, 2020). CDI is associated with prolonged hospital stays, high recurrence and mortality rates, posing a severe risk to hospitalized patients. The emergence of hypervirulent strains such as ribotype 027/BI/NAP1 has intensified these challenges. According to data from the U.S. Centers for Disease Control and Prevention, there are approximately 453,000 CDI cases annually in the United States, resulting in nearly 29,000 deaths and healthcare expenditures reaching $4.8 billion [Centers for Disease Control and Prevention (U.S.), 2019]. The European Union faces an estimated annual economic burden of up to €300 million due to CDI (Wiegand et al., 2012). In China, research on C. difficile has significantly increased over the past decade. Meta-analyses reveal that up to 14% of patients with diarrhea are found to carry toxigenic C. difficile (Wen et al., 2023). Additionally, cross-regional studies indicate that 10% of patients with diarrhea are affected by CDI (Jin et al., 2017). These findings underscore that CDI is a critical pathogen that significantly impacts human health and imposes substantial burdens on healthcare systems.
The Glycosylphosphatidylinositol (GPI) anchor is a complex glycolipid compound that covalently anchors proteins to the outer surface of the cell membrane, playing roles in various biological and pathological processes (Guo and Kundu, 2024). The common core structure of GPI includes phosphatidylinositol, GlcN, three mannose molecules, and an ethanolamine phosphate. The biosynthesis of GPI occurs on the membrane of the endoplasmic reticulum (ER) (Watanabe et al., 1999). In eukaryotic cells, proteins on the cell surface are connected to the cell membrane via GPI anchors. In yeast, the biosynthesis of the GPI anchor involves 11 steps (Müller, 2011; Komath et al., 2018). The gene phosphatidylinositol glycan class L (pig-L) encodes the N-deacetylase enzyme Gpi12p, which catalyzes the second step in GPI anchor biosynthesis: the deacetylation of N-acetylglucosaminyl-phosphatidylinositol (GlcNAc-PI) to form glucosaminyl (GlcN)-PI (Watanabe et al., 1999). Mutations in the pig-L gene in eukaryotes can lead to defects in the GPI anchor biosynthetic pathway, affecting the precise localization and function of cell surface proteins, thereby altering cellular functions. In CHO-K1 cells, pig-L encodes a membrane protein with 252 amino acids, predominantly located on the cytoplasmic side (Nakamura et al., 1997). Abnormalities in pig-L gene function are associated with various genetic disorders, such as CHIME syndrome, Fryns syndrome, and syndromes related to intellectual disability and epilepsy. These diseases present symptoms including delayed psychomotor development, seizures, and possibly elevated alkaline phosphatase levels (Ceroni et al., 2018).
In prokaryotes, studies on homologous genes of pig-L are relatively limited. In Pseudomonas aeruginosa, the dnpA gene from the pig-L family deacetylase is associated with fluoroquinolone tolerance (Liebens et al., 2014). The receptor for Clostridium septicum alpha toxin, a GPI-anchored protein, is also involved (Gordon et al., 1999). In Clostridioides difficile, a gene sequence homologous to pig-L is present (CD23150), but its function in this organism and its impact on virulence remain unclear. This study plans to use CRISPR-Cas9 gene editing technology to obtain Δpig-L mutant strains and their complemented strain (::pig-L), comparing the phenotypes of wild-type and mutant strains to elucidate the function of the pig-L gene in Clostridioides difficile.
2 Materials and methods
2.1 Strains and culture conditions
The Clostridioides difficile CD630 strain was acquired from the American Type Culture Collection (ATCC BAA-1382). The ∆pig-L mutant and the complemented ::pig-L strains of C. difficile were constructed in this study. For culturing, C. difficile strains were grown in Brain Heart Infusion (BHI) agar medium supplemented with yeast extract in a COY anaerobic workstation (Model: COY-7200220). Escherichia coli CA434 and NEBExpress (New England BioLabs, Beijing) were cultured using Luria-Bertani (LB) medium. Vero cells were maintained in DMEM complete culture medium under 5% CO2 conditions in a CO2 incubator. To prepare the BHI agar medium, brain heart infusion powder (38.5 g/L), L-cysteine (1 g/L), and yeast extract (5 g/L) were dissolved in 1 liter of double-distilled water, with agarose at 1.5% added as needed to obtain a solid medium (Zhou et al., 2022). For LB medium preparation, tryptone (10 g/L), sodium chloride (NaCl, 10 g/L), and yeast extract (5 g/L) were dissolved in 1 liter of double-distilled water. To prepare the solid medium, agarose at 1.5% was added to this solution. Reagents such as L-cysteine, yeast extract, tryptone, sodium chloride, tetracycline, amoxicillin, ampicillin, vancomycin, erythromycin, metronidazole, clindamycin, and norfloxacin were sourced from Solarbio Biotechnology Co., Ltd. (Beijing). Restriction enzymes Btg ZI (R0703S), Not I-HF (R3189S), and Bam HI-HF (R3136V) were obtained from New England BioLabs (Beijing). DNA recovery kits, plasmid extraction kits, and ClonExpress Multis One Step Cloning reagents were purchased from Vazyme Biotech Co., Ltd. (Nanjing). Gene sequencing services were provided by Sangon Biotechnology Co., Ltd. (Shanghai).
2.2 Sequence homology analysis
The protein multiple sequence alignment process was conducted as follows: Initially, the target gene (e.g., Clostridium difficile AM180355.1, CD630_23150) was searched in NCBI1 to obtain its amino acid sequence. Subsequently, Protein BLAST2 was used with the refseq_select database to identify homologous sequences, filtering for sequences originating from typical strains and downloading all FASTA files. The integrated sequences were submitted to CLUSTALW3 (Thompson et al., 2003) for multiple sequence alignment, generating an *.aln file. This was followed by using Jalview 2.11.4.0 software (Waterhouse et al., 2009) to adjust the layout and output SVG vector-editable sequence comparison results. Further analysis utilized TMHMM4 for predicting transmembrane domains of the target protein. Finally, Affinity Designer 2.4.0 was employed to adjust image quality, producing a visualization PDF map that includes transmembrane structure domains (TM), conserved residues (marked by asterisks/black shadow), and similarity (indicated by asterisks/colons/grey shadows).
2.3 Plasmid construction
The targeting plasmid for the pig-L gene contains a segment driven by the small RNA promoter (PsRNA) that recognizes pig-L crRNA, flanked by homologous arms upstream and downstream of the pig-L gene (Hong et al., 2018). The construction steps are as follows: (1) Using pJZ23 as the backbone vector, the PsRNA promoter+crRNA fragment amplified from pSH12 was ligated to Btg ZI linearized pJZ23 to obtain plasmid pHTY4. (2) Employing the C. difficile CD630 genome as a template, PCR amplification of the upstream and downstream homologous arms of the pig-L gene was performed, followed by their ligation into Not I linearized pHTY4 backbone, resulting in plasmid pHTY5 (Zhang et al., 2020). The pHTY5 plasmid, facilitating pig-L gene knockout, was purified using a plasmid extraction kit. To construct the pig-L complementation strain, primers HW2125/HW2156 were used to amplify the pig-L promoter+gene fragment. This amplified segment was then recombined with BamH I linearized pMTL82151 vector to generate the pig-L gene complementation plasmid pHTY6 (Yang et al., 2025).
2.4 Bacterial growth rate determination
Bacterial strains WT, Δpig-L, and :: pig-L were individually streaked for single colonies on solid agar plates. Single colonies were then inoculated into Brain Heart Infusion Supplemented (BHIS) liquid medium and anaerobically cultured at 37 °C until reaching the logarithmic growth phase (OD600 ≈ 0.6). Cultures were subsequently transferred to fresh BHIS medium with a 1% inoculum, with three biological replicates per strain. Growth was monitored by measuring OD600 at 3-h intervals using a spectrophotometer (Varioskan LUX, Thermo Fisher Scientific, United States). Growth curves were generated by plotting OD600 values against incubation time (h), and growth rate characteristics were analyzed.
2.5 Antibiotic susceptibility testing
Antibiotic susceptibility of strains WT, Δpig-L, and :: pig-L to metronidazole, vancomycin, amoxicillin, clindamycin, cefoxitin, norfloxacin, ampicillin, and erythromycin was determined using the plate dilution method according to the guidelines of Clinical and Laboratory Standards Institute (CLSI) (Lewis et al., 2025). Antibiotics were solubilized in a proper solution and sterile filtered (e.g., tetracycline, norfloxacin, and cefoxitin were solubilized in hydrochloric acid, acetic acid, and dimethyl sulfoxide, respectively. Other antibiotics were dissolved in sterile water) BHIS medium, melted and cooled to 45 ~ 50 °C, was mixed with the stock antibiotic solutions to achieve final concentrations of 128 μg/mL, 64 μg/mL, 32 μg/mL, 16 μg/mL, 8 μg/mL, 4 μg/mL, 2 μg/mL, 1 μg/mL, 0.5 μg/mL, and 0.25 μg/mL. A negative control, consisting of BHIS medium without antibiotic addition, was also included. Colonies of each mutant strain were inoculated into 5 mL of BHIS liquid medium and incubated at 37 °C under anaerobic conditions until the optical density at 600 nm (OD600) reached 0.5. Following this, 2 μL of the bacterial culture was spotted onto the surface of the agar plates, with three replicates per strain. The plates were incubated for 18–24 h, and the resulting growth was assessed (Zhao et al., 2020).
2.6 Vero cell cytotoxicity assay
Strains WT, Δpig-L, and :: pig-L were cultured in Brain Heart Infusion Supplemented (BHIS) medium at 37 °C under anaerobic conditions until OD600 reached 1.0. African green monkey kidney cells (Vero cells) served as the experimental model, grown in Dulbecco’s Modified Eagle Medium (89% DMEM) supplemented with 10% fetal bovine serum and 1% antibiotics (10 mg/mL streptomycin and 10,000 U/mL penicillin). Cells were cultured to approximately 60% confluence. Vero cells were then washed three times with phosphate-buffered saline (PBS), followed by a 24-h incubation in DMEM at 37 °C. For each strain, 1 mL of culture was centrifuged at 8,000 rpm for 6 min. The supernatants were filter-sterilized using a 0.22 μm membrane and serially diluted in BHIS medium from 5 × 101 to 5 × 108-fold. Both diluted and undiluted supernatants (200 μL each) were added to Vero cells, which were then incubated overnight at 37 °C. Cell morphology was examined using an optical microscope to identify the highest dilution causing cytopathic effects in Vero cells (Yang et al., 2025).
2.7 Biofilm formation assay
Overnight cultures were diluted to a 1:200 concentration in fresh BHIS medium. Following thorough mixing, 2 mL of the diluted culture was dispensed into each well of a 24-well plate, with four replicates per sample and a blank control consisting of BHIS medium alone. The plate was then incubated at 37 °C under anaerobic conditions for 48 h after sealing. Upon incubation completion, the surface growth was carefully removed from each well using a micropipette, avoiding contact with the well walls. Subsequently, 1 mL of 0.2% crystal violet solution was added to each well for fixation and staining, followed by a 1-h incubation at 37 °C. After removal of the stain, the wells were washed three times with PBS until the wash solution was nearly colorless. The plate was then air-dried at room temperature and photographed. Finally, 1 mL of anhydrous ethanol was added to each well and incubated at room temperature for 15 min to fully dissolve the bound crystal violet.
3 Results
3.1 Multiple sequence alignment
The pig-L gene (CD23150) is 801 base pairs (bp) in length and encodes a 267 amino acid (aa) protein. It is flanked by the genes CD23140 and CD23160, which encode a membrane protein and a response regulator transcription factor, respectively (Figure 1A). To investigate the potential structural and functional features of the PIG-L (WP_004454616.1) protein from Clostridioides difficile, its amino acid sequence was aligned with homologous proteins from related bacterial species using Clustal Omega (Sievers et al., 2011), Romboutsia weinsteinii (PIG-LRW_WP_094368733.1), Faecalimonas dakanense (PIG-LFD_WP_052356827.1), Metaclosteridium mangenotii (PIG-LMM_WP_209455428.1), and Terrisporobacter petrolearius (PIG-LTP_WP_228109113.1). The multiple sequence alignment (Figure 1B) revealed a high degree of amino acid conservation among these homologous proteins, particularly in several distinct regions indicated by shaded residues and asterisks. This high conservation suggests that these regions are functionally and structurally important across these species. A predicted transmembrane (TM) region was identified near the N-terminus of the protein (amino acids ~22–35, highlighted with a grey background in Figure 1B), indicating its likely insertion into a cellular membrane. Despite overall conservation, some variations in amino acid sequences were observed between the different bacterial species, which may account for species-specific functional adaptations. The consistent presence of the TM domain across these clostridial and related species implies a conserved membrane-associated role for this protein family.
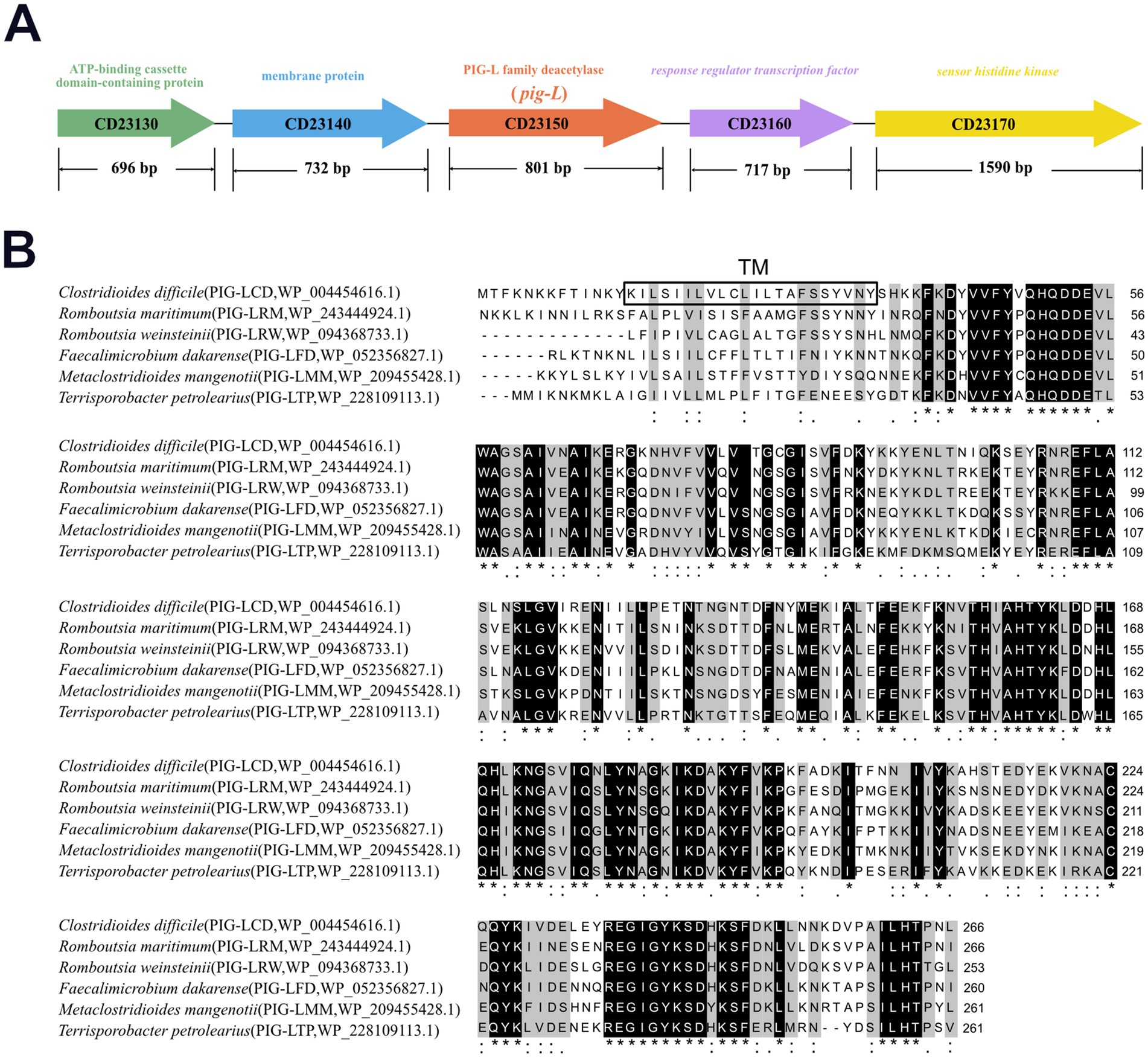
Figure 1. Multiple sequence alignment of Clostridioides difficile PIG-L with PIG-L family proteins from other bacteria. (A) Genomic context of the pig-L gene. (B) The PIG-L protein from Clostridioides difficile was aligned with homologous sequences from Romboutsia maritima, Romboutsia weinsteinii, Faecalimicrobium dakarense, Metaclostridioides mangenoti, and Terrisporobacter petrolearius using Clustal Omega. The boxed region indicates the predicted transmembrane (TM) domain from Clostridioides difficile PIG-L. Black-shaded areas and asterisks denote identical amino acids, while colons, single dots, and gray-shaded areas represent similar amino acids. An asterisk (*) signifies fully conserved sites, a colon (:) indicates strongly similar residues, and a single dot (.) denotes weakly similar residues.
3.2 Plasmids construction
The [PsRNA promoter + crRNA] sequence was amplified from pSH12, yielding a 381 bp target fragment. This fragment was then ligated into the pJZ23 vector (Zhang et al., 2020) at the Btg ZI single restriction site. Gel electrophoresis (821 bp) confirmed successful recombination and ligation, showing successful construction of pHTY4. Subsequently, using the wild-type (WT) genome as a template, PCR amplification yielded a 645 bp upstream homologous arm and a 722 bp downstream homologous arm. This homologous arm fragment was ligated into the Not I-HF linearized pHTY4 to generate pig-L targeting plasmid pHTY5 (Figure 2A). Next, BamH I linearized the pMTL82151 vector, and the pig-L gene, along with its native promoter, were combined by using the TEDA method (Xia et al., 2019), yielding the pig-L complementation vector, pHTY6 (Figure 2B).
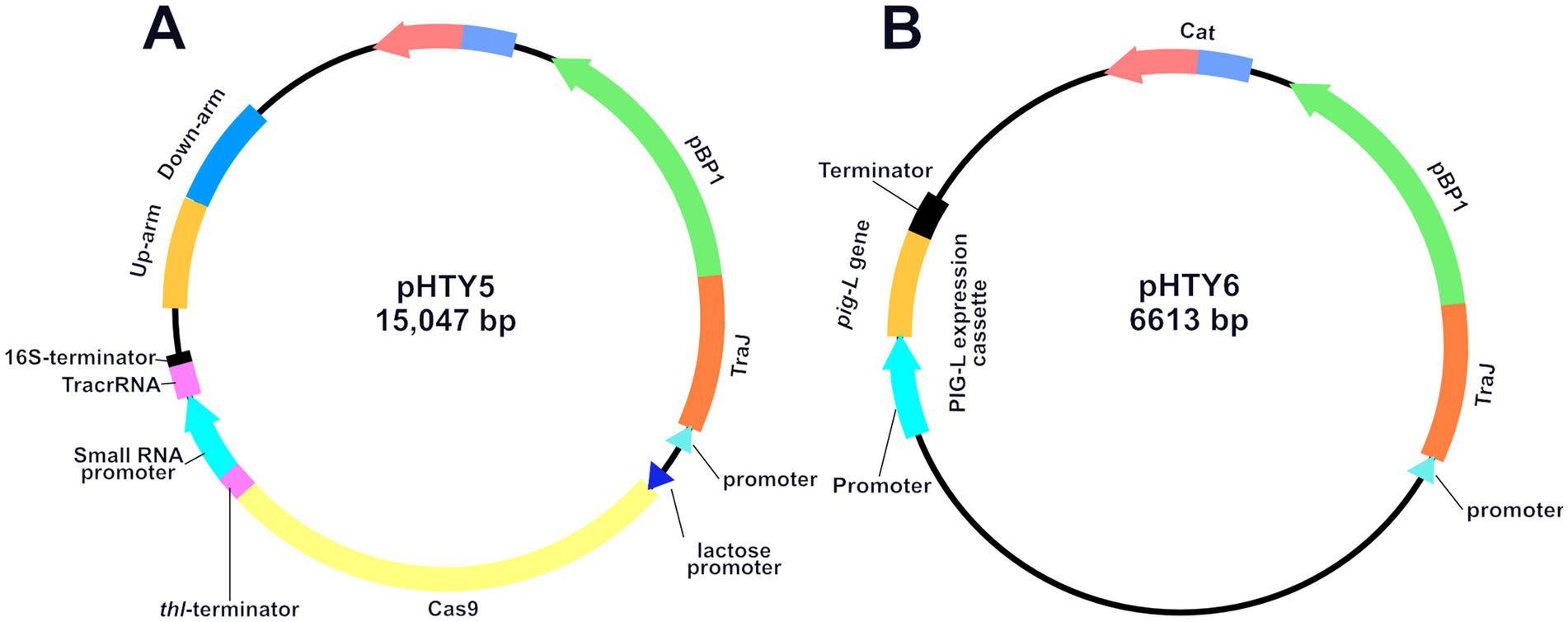
Figure 2. Plasmid Maps of pHTY5 (A) and pHTY6 (B). Schematic diagrams of the Clostridioides difficile pig-L gene knockout plasmid pHTY5 and complementation plasmid pHTY6.
3.3 Screening for pig-L gene mutant and plasmid curing
Wild-type (WT) C. difficile was utilized as the mother strain. Transformants harboring the recombinant pHTY5 plasmid were selected on Lac-BHIS medium supplemented with thiamphenicol (Tm) (Figures 3A–D). PCR identification of 16 randomly selected single colonies was performed using primers HW2066/HW2067 (Supplementary Table S1). All tested strains exhibited a reduction in PCR product size consistent with the designed knockout sequence (801 bp). Further gene sequencing confirmed the successful deletion of the pig-L gene sequence from the Clostridioides difficile chromosome, resulting in a gene knockout efficiency of 100% (Figure 3E). Following selection of the mutant strain in Lac-BHI-Tm medium, the pHTY5 plasmid was cured from the deletion mutant by continuous subculturing in BHIS medium, yielding a stable ∆pig-L mutant strain (Figure 3F).
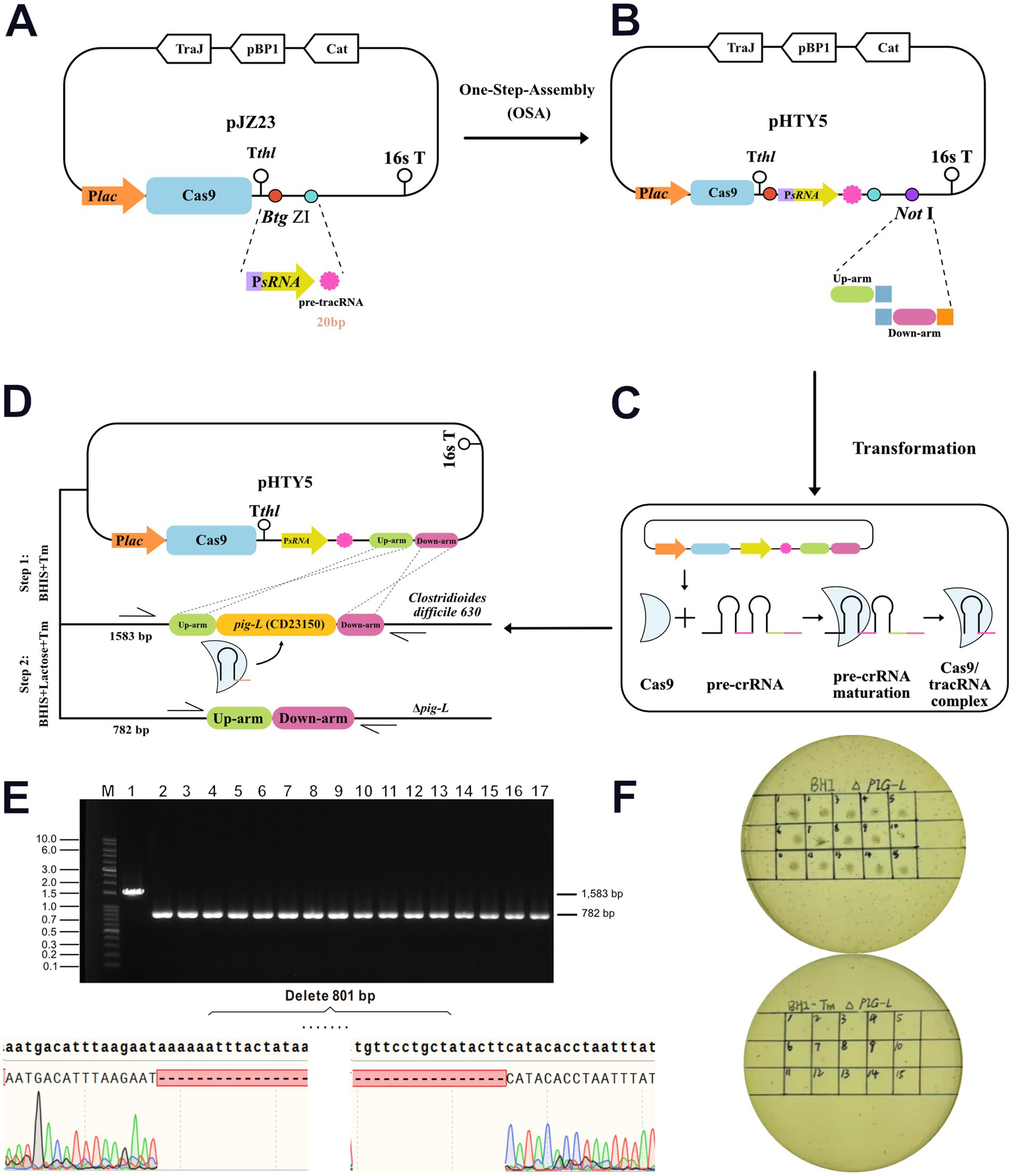
Figure 3. Generation and validation of the ∆pig-L mutant strain. (A) The pre-tracRNA expression cassette was inserted into the pJZ23 plasmid backbone. (B) The pHTY5 targeting plasmid was constructed by incorporating the upstream and downstream homology arms of the pig-L gene into the vector from the previous step. (C) Expression and assembly of the pHTY5 plasmid into the targeting effector complex. (D) A two-step selection process to isolate the Δpig-L mutant in C. difficile. Step 1, C. difficile transformants harboring a pHTY5 targeting plasmid were cultured in BHIS medium supplemented with thiamphenicol (Tm) (BHIS+Tm). The presence of Tm maintained the pHTY5 plasmid within the transformants. Step 2: The transformants were plated onto solid BHIS+Lactose+Tm agar. Lactose induced Cas9 protein expression, leading to the formation of a mature Cas9/tracrRNA ribonucleoprotein complex that specifically targeted the pig-L gene. Homologous recombination between the cleaved genomic DNA and the pHTY5 homology arms resulted in the generation of Δpig-L knockout mutants. Transformants that failed to undergo homologous recombination succumbed to a genomic DNA double break. (E) Verification of the ∆pig-L mutant was conducted via colony PCR. All examined colonies (n = 16, upper panel) exhibited the ∆pig-L gene deletion, evidenced by the amplicon size difference (1814 bp for wild-type, 875 bp for ∆pig-L). Subsequent gene sequencing confirmed the deletion of the pig-L gene within the ∆pig-L mutant (lower panel). (F) Curing of the targeting plasmid from the ∆pig-L mutant was assessed. Thiamphenicol resistance was tested for the first generation (upper panel) and again after 10 consecutive passages (lower panel).
3.4 Analysis of pig-L gene expression and growth kinetics
To confirm the deletion of the pig-L gene at the transcriptional level and to assess its impact on the growth and autolysis of C. difficile, we first quantified pig-L transcript levels in the WT, ∆pig-L, and :: pig-L strains. The results demonstrated that pig-L expression was undetectable in the ∆pig-L mutant, whereas the complemented strain (::pig-L) exhibited high levels of expression; both were significantly different compared to the WT strain (Figure 4A). We then characterized the growth curves for the three strains. No significant differences in growth rate were observed among the WT, ∆pig-L, and :: pig-L strains during the initial growth phases (0–15 h). However, in the decline phase (15–33 h), the rate of autolysis in the ∆pig-L strain was markedly reduced compared to the WT strain (p < 0.0001). This attenuated autolysis phenotype was also observed in the :: pig-L strain. Specifically, at the 21-h time point, the OD600 values for WT, ∆pig-L, and :: pig-L were 0.6, 1.2, and 1.4, respectively. These values correspond to a cellular autolysis of 65% in the WT, but only 29% in ∆pig-L and 18% in :: pig-L (Figure 4B). To elucidate the attenuated autolysis observed in the Δpig-L mutant, we quantified the expression levels of the known autolysin genes acd (Figure 4C), cwp19 (Figure 4D), and cwp66 (Figure 4E) across different mutant strains. Our results demonstrate a significant reduction in the expression of these genes in the Δpig-L mutant (Dhalluin et al., 2005; Wydau-Dematteis et al., 2018; Zhou et al., 2022).
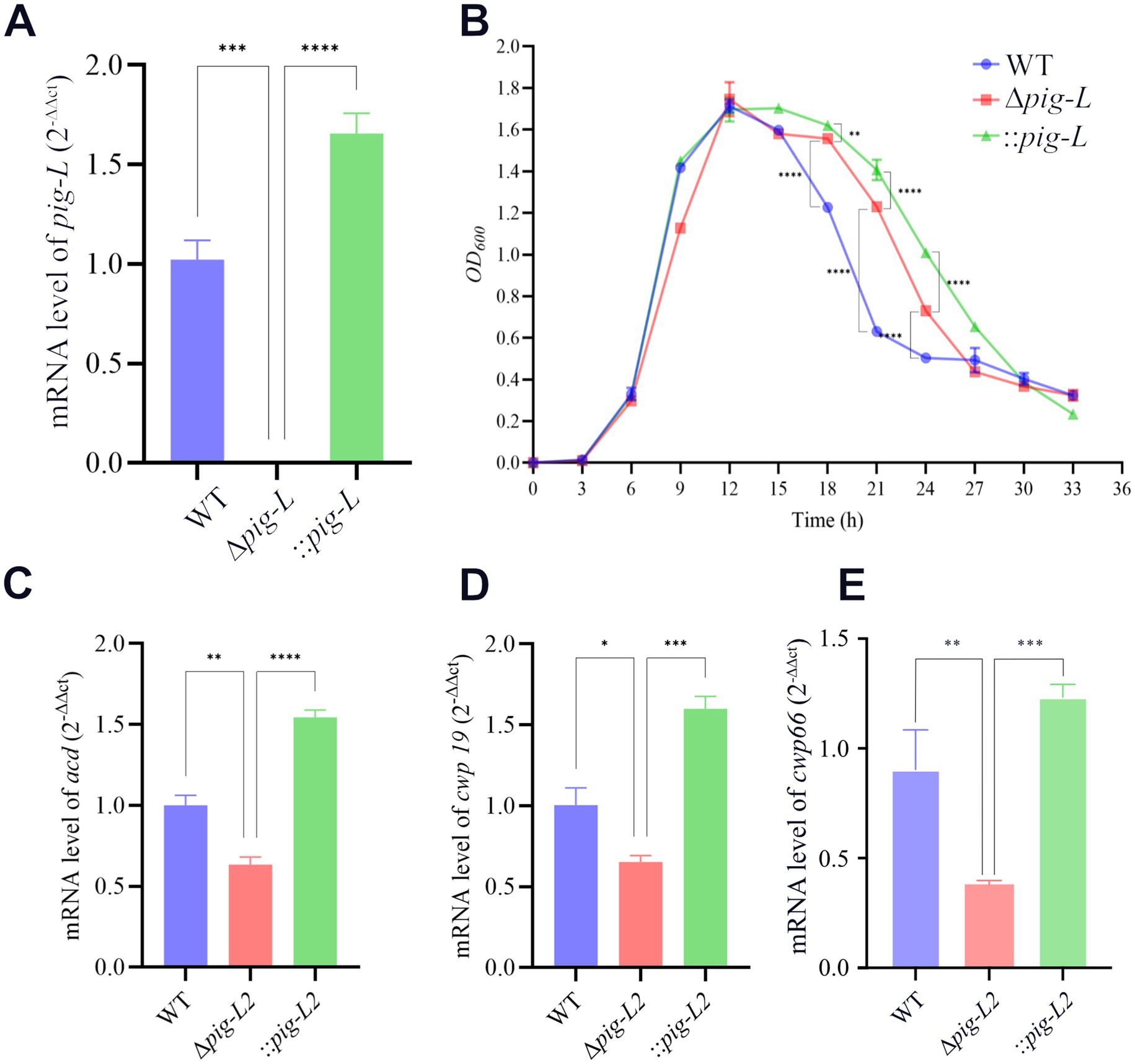
Figure 4. Expression of the pig-L gene and its effect on bacterial growth in WT, ∆pig-L, and :: pig-L strains. (A) The expression level of the pig-L gene in WT, ∆pig-L, and :: pig-L strains. (B) Determination of the growth curves. The horizontal coordinate represents time in hours, and the vertical coordinate represents the optical density at OD600 nm. (C–E) The expression levels of the acd, cwp19, and cwp66 genes are in WT, Δpig-L, and :: pig-L strains. Statistical significance is denoted as follows: *: p <0.05: **: p < 0.01; ***: p < 0.001; ****: p < 0.0001.
3.5 Toxin gene expression and cytotoxicity assay
To investigate the impact of the pig-L gene on the cytotoxicity of C. difficile, we quantified the expression levels of tcdA and tcdB toxin genes in the WT, ∆pig-L, and:: pig-L strains using RT-qPCR. Results demonstrated a significant decrease in tcdA (Figure 5A) and tcdB (Figure 5B) expression in the ∆pig-L mutant compared to the WT strain. Conversely, the :: pig-L complemented strain exhibited significantly increased tcdA and tcdB expression levels relative to the WT strain. Subsequent cytotoxicity assays revealed the highest supernatant dilutions that induced morphological changes (corneoid to round) in Vero cells were 0.5 × 10-7 (Figure 5C-WT-Right), 0.5 × 10-5 (Figure 5C-∆pig-L-Right), and 0.5 × 10-6 (Figure 5C-::pig-L-Right) for WT, Δpig-L, and :: pig-L, respectively. Consequently, the Δpig-L strain exhibited a 100-fold decrease in cytotoxicity compared to the WT, while the :: pig-L strain displayed cell cytotoxicity intermediate between WT and Δpig-L, with enhanced cytotoxicity compared to Δpig-L. These findings suggest that deletion of the pig-L gene substantially reduces the cytotoxicity of C. difficile.
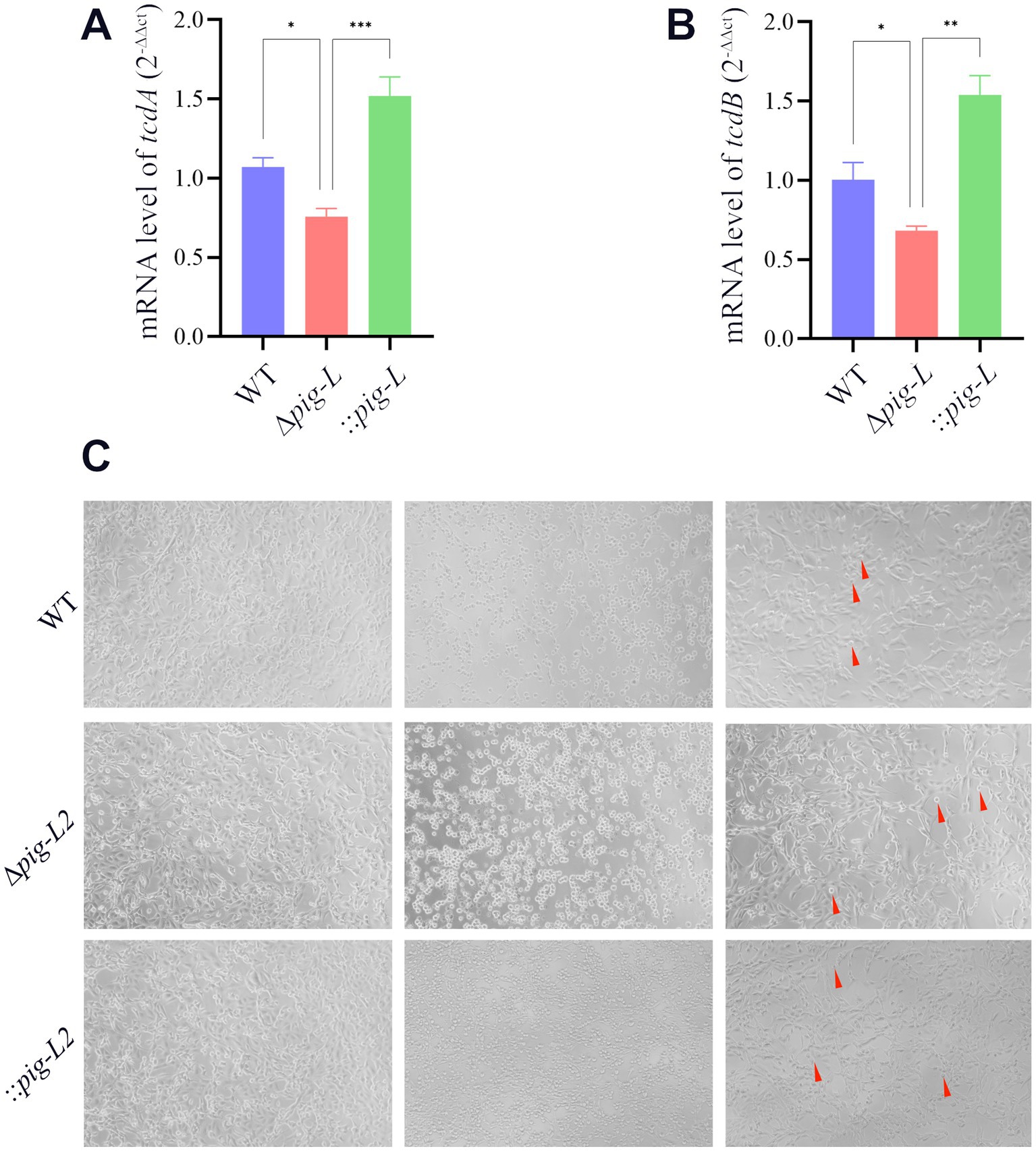
Figure 5. Characterization of WT, ∆pig-L, and :: pig-L strains. (A,B) Quantitative RT-qPCR analysis of tcdA and tcdB gene expression in the WT, ∆pig-L, and :: pig-L strains. (C) Cytotoxicity assays of the WT, ∆pig-L, and :: pig-L strains. Vero cells were exposed to PBS (left), undiluted culture supernatants (middle), or diluted supernatants (right) from each strain: WT at 10−7 dilution, ∆pig-L at 10−5 dilution, and :: pig-L at 10−6 dilution. Red arrows indicate rounded cells. Statistical significance is denoted as follows: *: p < 0.05; **: p < 0.01; ***: p < 0.001.
3.6 Biofilm formation
The pig-L gene encodes an N-deacetylase, a key enzyme in the synthesis of Glycosylphosphatidylinositol (GPI) anchors. GPI anchors are complex glycolipids that covalently attach proteins to the outer cell membrane. Cell surface proteins play a critical role in biofilm formation in pathogenic microorganisms (Brauer et al., 2021). Therefore, we investigated the effect of the pig-L gene on biofilm formation in C. difficile. Results showed that the ∆pig-L mutant exhibited reduced biofilm formation compared to the WT strain (Figure 6A). Overexpression of the pig-L gene resulted in significantly enhanced biofilm formation in C. difficile compared to the WT strain (Figure 6B).
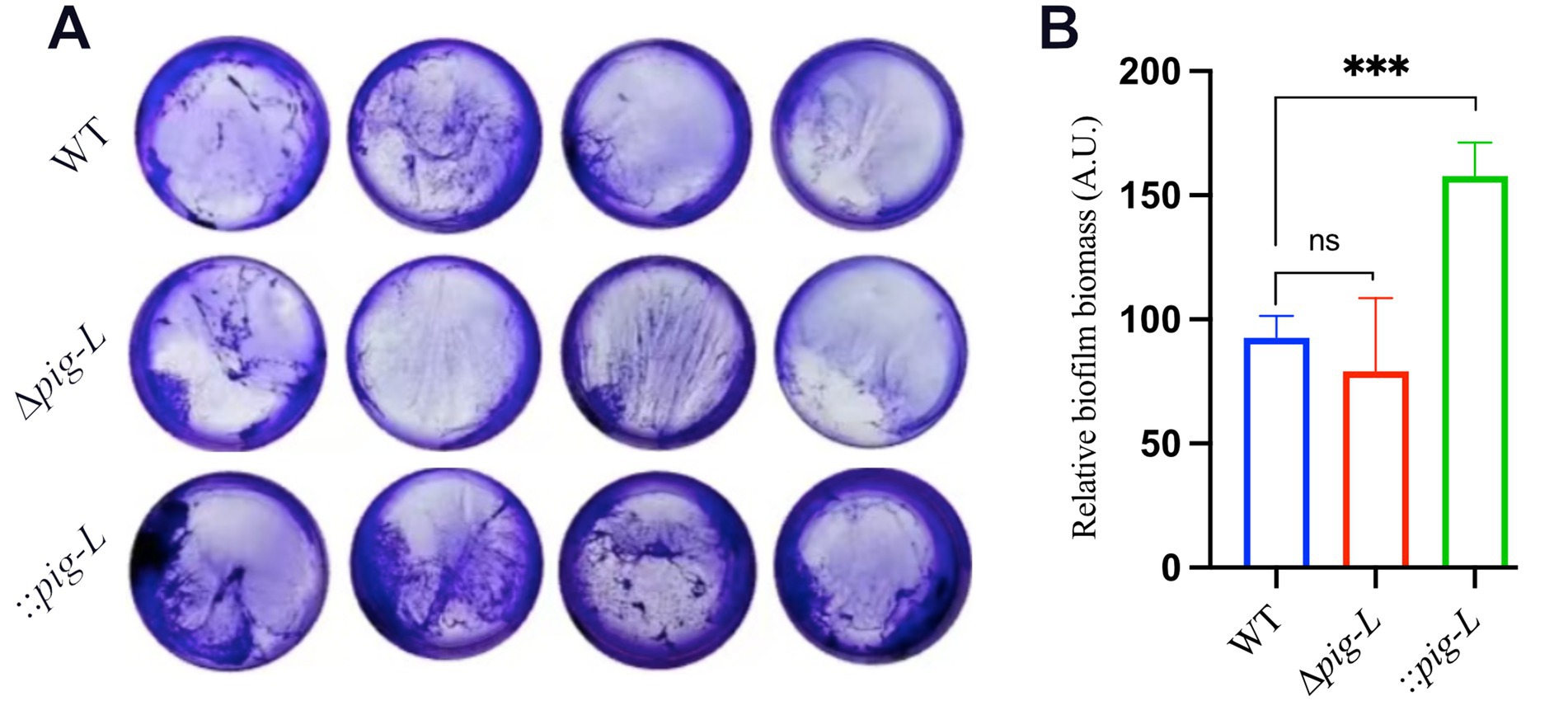
Figure 6. Biofilm formation assays of WT, ∆pig-L, and:: pig-L strains. (A) Biofilm formation of the WT, ∆pig-L, and :: pig-L strains. (B) Quantitative analysis of biofilm formation by the WT, ∆pig-L, and :: pig-L strains. Statistical significance is denoted as follows: ***: p < 0.001; ns: p > 0.05.
3.7 Peroxide tolerance assay
The role of the pig-L gene in conferring oxidative stress tolerance to pathogens remains unclear (Watanabe et al., 1999; Pascual and Planas, 2021). To deepen our understanding of the pig-L gene’s impact on C. difficile’s resistance to reactive oxygen species (ROS), we assessed its influence on peroxide tolerance. Results indicated that compared to WT strains, the ∆pig-L mutant showed significantly reduced hydrogen peroxide tolerance; the :: pig-L strain exhibited markedly higher peroxide tolerance than WT, particularly at a concentration of 25 nM. Additionally, at higher peroxide concentrations (50 to 55 nM), the :: pig-L complemented strain also demonstrated increased hydrogen peroxide tolerance (Figure 7).
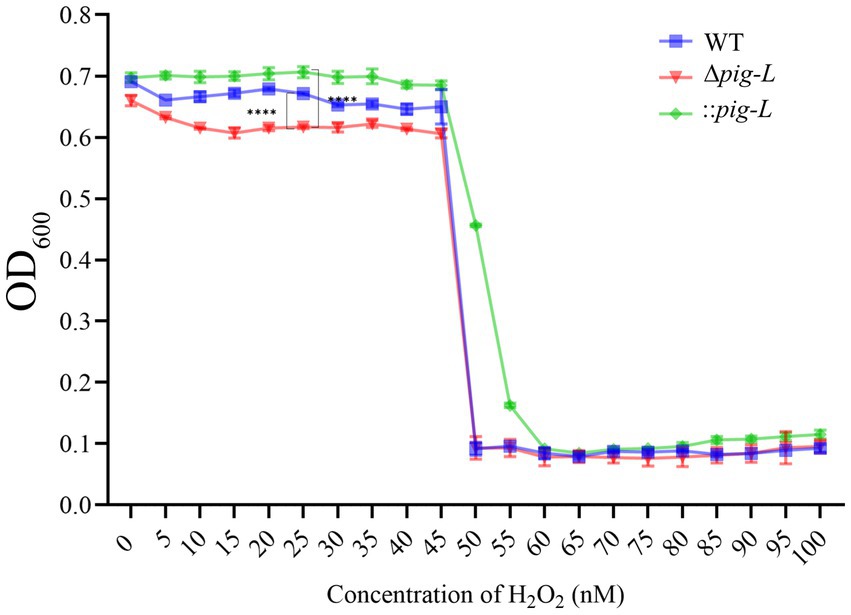
Figure 7. Peroxide tolerance of WT, ∆pig-L, and :: pig-L Strains. Changes in optical density at 600 nm (OD600) for WT, ∆pig-L, and :: pig-L strains under various hydrogen peroxide concentrations. Statistical significance is denoted as follows: ****: p < 0.0001.
3.8 Antibiotic susceptibility analysis
The susceptibility of WT, ∆pig-L, and :: pig-L strains to various commonly used antibiotics is depicted in Figures 8A–I. Compared with the WT strain, the ∆pig-L and:: pig-L mutant exhibited no statistically significant alterations in antibiotic resistance across the range of assays performed (Figures 8A–I; Supplementary Table S3).
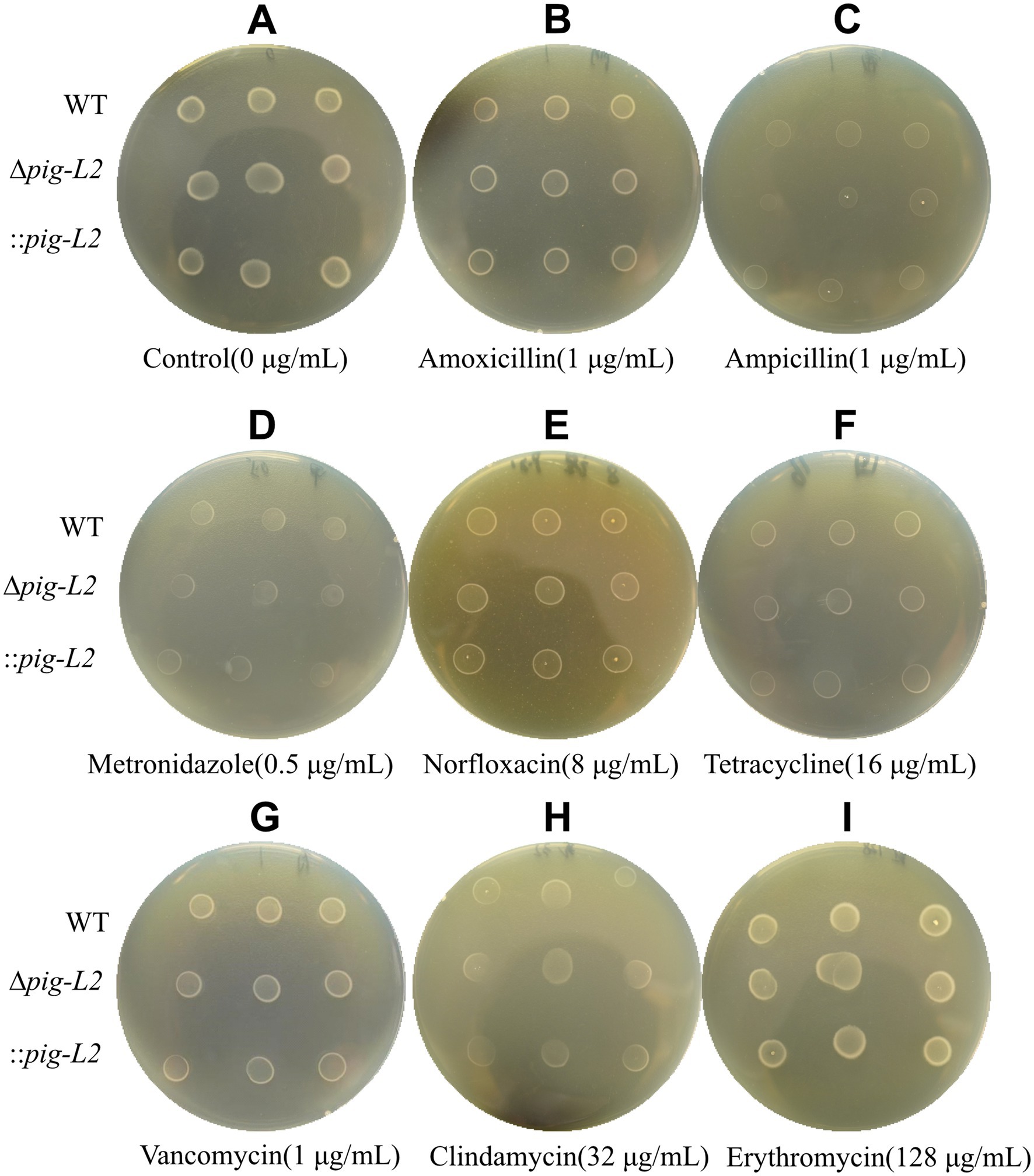
Figure 8. Antibiotic susceptibility test for WT, ∆pig-L, and :: pig-L Strains. The antibiotics tested include Control (A), Amoxicillin (B), Ampicillin (C), Metronidazole (D), Norfloxacin (E), Tetracycline (F), Vancomycin (G), Clindamycin (H), and Erythromycin (I).
3.9 Protein expression differences
Proteomic analysis was used to compare gene expression between the ∆pig-L mutant strain and the wild type (WT). The volcano plot shows that the ∆pig-L group had 170 proteins significantly upregulated, 101 proteins significantly downregulated, and 2,357 proteins with unchanged expression (adjusted p-value <0.05, |log₂FC| > 1) (Figure 9). The top 10 notably upregulated proteins include Spore coat-associated protein CotJA, Endoglucanase (Cellulase), ABC-type transport system/bacitracin/multidrug-family permease, Cation:proton antiporter, Putative membrane protein, PTS system beta-glucoside-specific EIIBCA component, FeoA domain, Ferrous iron transport protein B, ABC transporter ribose-specific ATP-binding protein, and D-alanine—D-alanine ligase. The top 10 significantly downregulated proteins include Cholera toxin secretion protein EpsF, DUF3795 domain-containing protein, Phosphatidylglycerol lysyltransferase, Uncharacterized protein, ABC transporter ATP-binding protein, DegV family protein, Spo0E-like sporulation regulatory protein, Ethanolamine transport protein, ABC-type transport system permease, and Uncharacterized protein OS=Clostridioides difficile (Supplementary Table S2).
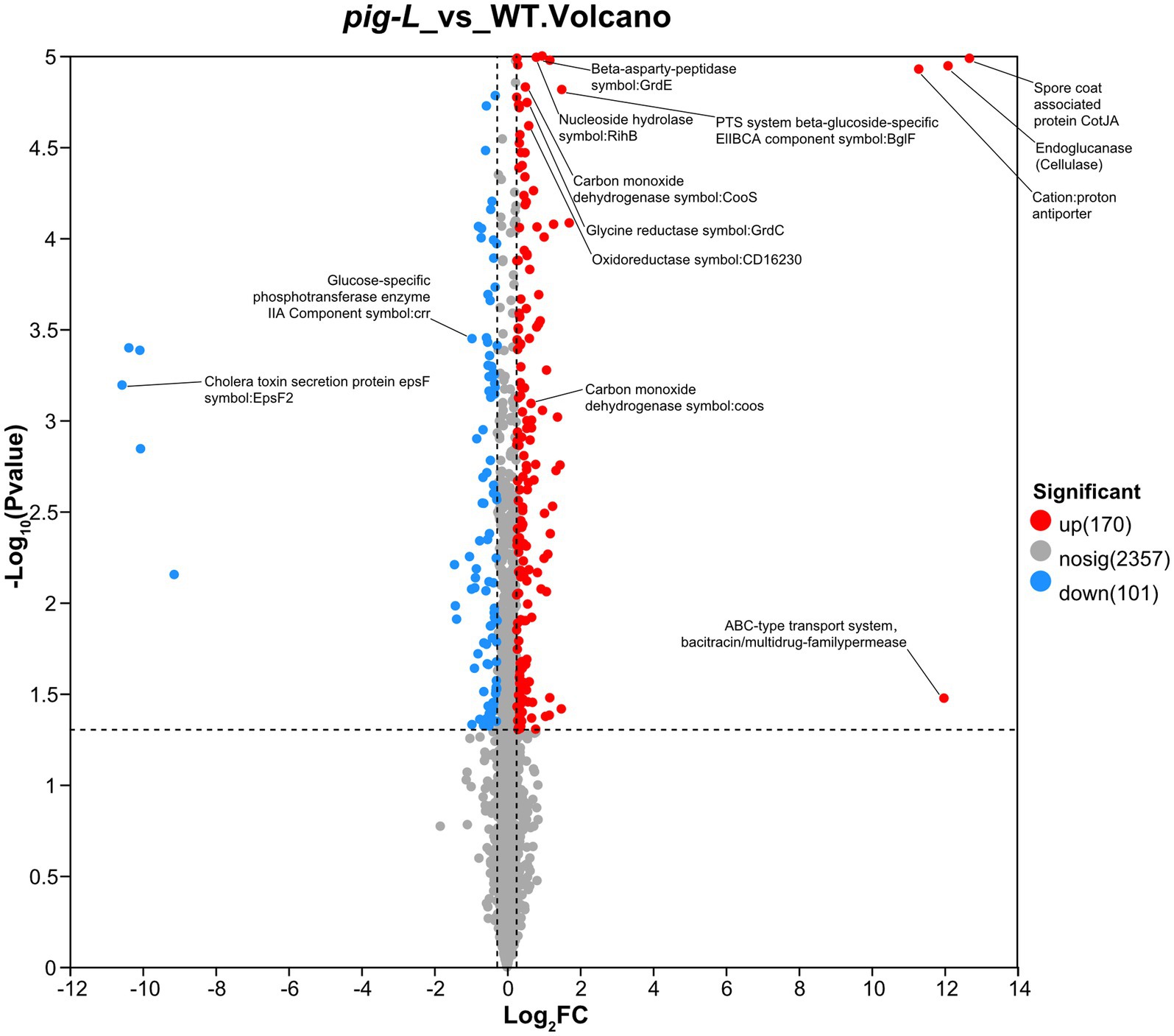
Figure 9. Volcano plot of differentially expressed genes between ∆pig-L and WT. Each point represents a gene. The horizontal axis displays the log₂ fold change values (∆pig-L/WT), while the vertical axis shows the -log₁₀ of adjusted p-values. Red points indicate 170 significantly upregulated genes; blue points represent 101 significantly downregulated genes; gray points denote 2,357 genes with no significant difference in expression between ∆pig-L and WT strains.
4 Discussion
GPI anchors are complex glycolipid structures that covalently tether proteins to the extracellular surface of cellular membranes. The pig-L gene encodes an enzyme involved in catalyzing the second step of GPI anchor biosynthesis, playing a role in various biological and pathological processes associated with GPI anchors. This study first analyzed the homology of the CD23150 (pig-L) gene with GPI anchors using sequence alignment methods. Subsequently, CRISPR-Cas9 technology was employed to construct ∆pig-L and its complementation strain, ::pig-L. Gene and expression level assays confirmed successful construction of the knockout and complementation strains. Growth rate analysis revealed no significant differences in lag, exponential, or stationary phases between ∆pig-L, :: pig-L mutants, and WT strains; however, during the decline phase, mutant strains exhibited significantly reduced autolysis rates compared to WT. The ∆pig-L strain showed a marked decrease in toxin gene expression levels and cytotoxicity, with no significant change in biofilm formation ability. This strain also exhibited significantly reduced resistance to reactive oxygen species (ROS). Proteomic analysis revealed that, compared to the WT strain, 170 proteins were significantly upregulated, and 101 proteins were downregulated in ∆pig-L mutants. These strains exhibited noticeable metabolic remodeling in fatty acid utilization, endospore resistance, polysaccharide degradation, and anaerobic fermentation. Downregulation of ion homeostasis and nucleotide metabolism suggested altered stress response strategies.
Toxin gene expression assays and cytotoxicity experiments indicated that the ∆pig-L mutant strain had significantly reduced toxin expression and cytotoxicity levels. This phenotype is consistent with observations from CD27900 (another PIG-L family member) knockout strains, which showed decreased autolysis rates, virulence, and tolerance to acid and antibiotics (Yang et al., 2023). Moreover, we observed a 1511.49-fold decrease in the expression of Cholera toxin secretion protein E (EpsF), an essential inner membrane platform protein in Vibrio cholerae’s T2SS involved in cholera toxin secretion (Abendroth et al., 2009; Shannon et al., 2025). This drastic reduction may correlate with decreased extracellular toxin levels in C. difficile.
Biofilm formation experiments showed that the ∆pig-L mutant had a slightly lower biofilm formation capacity than WT, without significant differences. Interestingly, the :: pig-L complementation strain significantly enhanced its biofilm formation ability compared to WT, suggesting pig-L positively regulates C. difficile biofilm formation. Proteomic analysis revealed a downregulation of about 2-fold in sugar PTS system EIIA component (crr, K02777) protein. Crr, as part of the Sugar PTS system, typically upregulates Klebsiella pneumoniae biofilm formation through the cAMP-CRP signaling pathway by increasing the synthesis of Type 3 fimbriae. The ∆crr mutant shows reduced Type 3 fimbriae expression and significantly decreased biofilm formation due to weakened cAMP-CRP signaling.
The ∆pig-L strain also demonstrated significantly reduced tolerance to oxidative stress compared to WT. Proteomic analysis showed a decrease of 1.57- and 1.30-fold in cooS (carbon monoxide dehydrogenase catalytic subunit) and gcvH (GCSH oxidoreductase complex), respectively, both possessing oxidoreductase activity (Qiuying et al., 2022; Lingna et al., 2021). These proteins collectively form a “carbon-electron-antioxidant” defense system against ROS damage (Lu and Imlay, 2021). The decreased expression of these proteins may explain the reduced H2O2 tolerance in ∆pig-L mutants.
Despite successfully constructing ∆pig-L and :: pig-L strains via CRISPR-Cas9, this study has limitations. Firstly, while it is speculated that pig-L affects biofilm formation possibly through crr gene regulation, direct experimental validation, such as crr knockout or overexpression assays, was lacking. Secondly, while key proteins like cooS, acsA, and gcvH were downregulated in oxidative stress tolerance studies, their precise roles and impacts require further investigation. Lastly, experimental results mainly focused on in vitro conditions, lacking in vivo validation (e.g., animal models), which limits comprehensive understanding and application.
The findings of this study offer new insights into the biological characteristics and pathogenic mechanisms of C. difficile, with potential applications. Firstly, elucidating pig-L’s role in toxin production and biofilm formation provides a basis for developing novel therapeutic strategies against C. difficile infections, such as designing small-molecule inhibitors targeting pig-L or its downstream regulatory pathways to reduce biofilm formation and toxin production. Secondly, research on pig-L’s involvement in oxidative stress tolerance presents potential targets to boost bacterial resistance to oxidative stress, enhancing their application value in biotechnology. Finally, this study deepens our understanding of pig-L’s gene function, laying a foundation for further control of CDI infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, PXD067439.
Author contributions
YC: Writing – review & editing, Writing – original draft, Formal analysis, Data curation. HH: Writing – original draft, Investigation. TH: Investigation, Writing – original draft. XL: Investigation, Writing – original draft. FC: Writing – original draft, Investigation. PS: Writing – original draft, Investigation. WM: Writing – original draft, Investigation. YL: Investigation, Writing – original draft. SL: Investigation, Writing – original draft. GC: Writing – original draft, Writing – review & editing, Supervision. XQ: Writing – original draft, Resources, Writing – review & editing, Supervision. Y-JL: Writing – review & editing, Funding acquisition. WH: Funding acquisition, Visualization, Resources, Validation, Formal analysis, Project administration, Writing – original draft, Supervision, Investigation, Data curation, Writing – review & editing, Methodology, Conceptualization, Software.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (nos. 32170134, 32560029); the Excellent Young Talents Plan of Guizhou Medical University (2022)101; Taishan Scholars Program tsqn202306289. Innovation and Entrepreneurship Training Programs for University Students (2024106600498, 26252030741); Project for Scientific Research-Driven Talent Cultivation and Scientific Research Feedback to Teaching of the Affiliated Hospital of Guizhou Medical University (gyfykj-2025-y27).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1691769/full#supplementary-material
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/
2. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Abbas, A., and Zackular, J. P. (2020). Microbe–microbe interactions during Clostridioides difficile infection. Curr. Opin. Microbiol. 53, 19–25. doi: 10.1016/j.mib.2020.01.016
Abendroth, J., Mitchell, D. D., Korotkov, K. V., Johnson, T. L., Kreger, A., Sandkvist, M., et al. (2009). The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J. Struct. Biol. 166, 303–315. doi: 10.1016/j.jsb.2009.03.009
Brauer, M., Lassek, C., Hinze, C., Hoyer, J., Becher, D., Jahn, D., et al. (2021). What’s a biofilm?—how the choice of the biofilm model impacts the protein inventory of Clostridioides difficile. Front. Microbiol. 12:682111. doi: 10.3389/fmicb.2021.682111
Centers for Disease Control and Prevention (U.S.). 2019 Antibiotic resistance threats in the United States, (2019).
Ceroni, J. R., Yamamoto, G. L., Honjo, R. S., Kim, C. A., Passos-Bueno, M. R., and Bertola, D. R. (2018). Large deletion in PIGL: a common mutational mechanism in CHIME syndrome? Genet. Mol. Biol. 41, 85–91. doi: 10.1590/1678-4685-gmb-2017-0172
Dhalluin, A., Bourgeois, I., Pestel-Caron, M., Camiade, E., Raux, G., Courtin, P., et al. (2005). Acd, a peptidoglycan hydrolase of Clostridium difficile with N-acetylglucosaminidase activity. Microbiology 151, 2343–2351. doi: 10.1099/mic.0.27878-0
Gordon, V. M., Nelson, K. L., Buckley, J. T., Stevens, V. L., Tweten, R. K., Elwood, P. C., et al. (1999). Clostridium septicum alpha toxin uses glycosylphosphatidylinositol-anchored protein receptors. J. Biol. Chem. 274, 27274–27280. doi: 10.1074/jbc.274.38.27274
Guo, Z., and Kundu, S. (2024). Recent research progress in glycosylphosphatidylinositol-anchored protein biosynthesis, chemical/chemoenzymatic synthesis, and interaction with the cell membrane. Curr. Opin. Chem. Biol. 78:102421. doi: 10.1016/j.cbpa.2023.102421
Hong, W., Zhang, J., Cui, G., Wang, L., and Wang, Y. (2018). Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection. ACS Synth. Biol. 7, 1588–1600. doi: 10.1021/acssynbio.8b00087
Jin, D., Luo, Y., Huang, C., Cai, J., Ye, J., Zheng, Y., et al. (2017). Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J. Clin. Microbiol. 55, 801–810. doi: 10.1128/JCM.01898-16
Komath, S. S., Singh, S. L., Pratyusha, V. A., and Sah, S. K. (2018). Generating anchors only to lose them: the unusual story of glycosylphosphatidylinositol anchor biosynthesis and remodeling in yeast and fungi. IUBMB Life 70, 355–383. doi: 10.1002/iub.1734
Lewis, J. S., Kirn, T. J., Weinstein, M. P., Limbago, B., Bobenchik, A. M., Mathers, A. J., et al. (n.d.). CLSI publishes M100—Performance standards for antimicrobial susceptibility testing, 35th Edition, CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. (Accessed March, 2025).
Liebens, V., Defraine, V., Van Der Leyden, A., De Groote, V. N., Fierro, C., Beullens, S., et al. (2014). A putative de- N -acetylase of the PIG-L superfamily affects fluoroquinolone tolerance in Pseudomonas aeruginosa. Patho. Dis. 71, 39–54. doi: 10.1111/2049-632X.12174
Lingna, Z., Li, D., and Hongmei, L. (2021). Survival strategies of bacteria in response to excessive reactive oxygen species: a review. Microbiology China 48, 1249–1259. doi: 10.13344/j.microbiol.china.200636
Lu, Z., and Imlay, J. A. (2021). When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat. Rev. Microbiol. 19, 774–785. doi: 10.1038/s41579-021-00583-y
Müller, G. (2011). Novel applications for glycosylphosphatidylinositol-anchored proteins in pharmaceutical and industrial biotechnology. Mol. Membr. Biol. 28, 187–205. doi: 10.3109/09687688.2011.562557
Nakamura, N., Inoue, N., Watanabe, R., Takahashi, M., Takeda, J., Stevens, V. L., et al. (1997). Expression cloning of PIG-L, a CandidateN-Acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272, 15834–15840. doi: 10.1074/jbc.272.25.15834
Nakao, S., Hasegawa, S., Shimada, K., Mukai, R., Tanaka, M., Matsumoto, K., et al. (2020). Evaluation of anti-infective-related Clostridium difficile -associated colitis using the Japanese adverse drug event report database. Int. J. Med. Sci. 17, 921–930. doi: 10.7150/ijms.43789
Pascual, S., and Planas, A. (2021). Carbohydrate de-N-acetylases acting on structural polysaccharides and glycoconjugates. Curr. Opin. Chem. Biol. 61, 9–18. doi: 10.1016/j.cbpa.2020.09.003
Qiuying, L., Zhang Jingyang, X., Jinxiu, S. T., Xuepeng, L., and Jianrong, L. (2022). Genome-wide identification and analysis of two-component signal transduction Systems in Shewanella putrefaciens from aquatic products. J. Chin. Inst. Food Sci. Technol. 22, 290–297. doi: 10.16429/j.1009-7848.2022.06.030
Schäffler, H., and Breitrück, A. (2018). Clostridium difficile – from colonization to infection. Front. Microbiol. 9:646. doi: 10.3389/fmicb.2018.00646
Shannon, A., Johnson, T., Roberts, C. S., Chaton, C. T., Korotkov, K. V., and Sandkvist, M. (2025). The PDZ domain of EpsC is required for extracellular secretion of VesB by the type II secretion system in Vibrio cholerae. J. Bacteriol. 207:e0014425. doi: 10.1128/jb.00144-25
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. CP in Bioinformatics 2.3:1–22. doi: 10.1002/0471250953.bi0203s00
Watanabe, R., Ohishi, K., Maeda, Y., Nakamura, N., and Kinoshita, T. (1999). Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 339, 185–192. doi: 10.1042/bj3390185
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wen, B.-J., Dong, N., Ouyang, Z.-R., Qin, P., Yang, J., Wang, W.-G., et al. (2023). Prevalence and molecular characterization of Clostridioides difficile infection in China over the past 5 years: a systematic review and meta-analysis. Int. J. Infect. Dis. 130, 86–93. doi: 10.1016/j.ijid.2023.03.009
Wiegand, P. N., Nathwani, D., Wilcox, M. H., Stephens, J., Shelbaya, A., and Haider, S. (2012). Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J. Hosp. Infect. 81, 1–14. doi: 10.1016/j.jhin.2012.02.004
Wydau-Dematteis, S., El Meouche, I., Courtin, P., Hamiot, A., Lai-Kuen, R., Saubaméa, B., et al. (2018). Cwp19 is a novel lytic Transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. MBio 9. doi: 10.1128/mBio.00648-18
Xia, Y., Li, K., Li, J., Wang, T., Gu, L., and Xun, L. (2019). T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 47:e15. doi: 10.1093/nar/gky1169
Yang, J., Bao, J., Shao, R., Zhang, T., Jian, L., Yumei, C., et al. (2023). CD630_27900 gene deletion significantly reduces autolysis rate and virulence of Clostridioides difficile and the tolerance to acids and antibiotics. Acta Microbiol Sin., 63:417–432. doi: 10.13343/j.cnki.wsxb.20220731
Yang, Y., Huang, T., Yang, J., Shao, R., Shu, L., Ling, P., et al. (2025). The sigma factor σ54 (rpoN) functions as a global regulator of antibiotic resistance, motility, metabolism, and virulence in Clostridioides difficile. Front. Microbiol. 16:1569627. doi: 10.3389/fmicb.2025.1569627
Zeng, J., Wang, H., Dong, M., and Tian, G.-B. (2022). Clostridioides difficile spore: coat assembly and formation. Emerg. Microb. Infect. 11, 2340–2349. doi: 10.1080/22221751.2022.2119168
Zhang, J., Hong, W., Guo, L., Wang, Y., and Wang, Y. (2020). Enhancing plasmid transformation efficiency and enabling CRISPR-Cas9/Cpf1-based genome editing in Clostridium tyrobutyricum. Biotechnol. Bioeng. 117, 2911–2917. doi: 10.1002/bit.27435
Zhao, K., Huang, T., Lin, J., Yan, C., Du, L., Song, T., et al. (2020). Genetic and functional diversity of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Front. Microbiol. 11:598478. doi: 10.3389/fmicb.2020.598478
Keywords: Clostridioides difficile, phosphatidylinositol glycosyltransferase L class gene (pig-L gene), gene deletion, virulence, biofilm, oxidative stress tolerance
Citation: Cheng Y, Hu H, Huang T, Luo X, Chen F, Shi P, Ma W, Lu Y, Lan S, Cui G, Qi X, Liu Y-J and Hong W (2025) Pig-L mediates virulence, biofilm formation, and oxidative stress tolerance in Clostridioides difficile. Front. Microbiol. 16:1691769. doi: 10.3389/fmicb.2025.1691769
Edited by:
Maria Westerholm, Swedish University of Agricultural Sciences, SwedenReviewed by:
Shaohua Wang, Ohio University, United StatesYangchun Cao, Northwest A&F University, China
Copyright © 2025 Cheng, Hu, Huang, Luo, Chen, Shi, Ma, Lu, Lan, Cui, Qi, Liu and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hong, aG9uZ3dlaV8yMDE1QGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Yumei Cheng1†
Yumei Cheng1† Guzhen Cui
Guzhen Cui Xiaolan Qi
Xiaolan Qi Wei Hong
Wei Hong