- 1CSIR-Central Salt and Marine Chemicals Research Institute (CSIR-CSMCRI), Council of Scientific and Industrial Research (CSIR), Bhavnagar, Gujarat, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
Polyethylene (PE), a type of plastic, is the primary contributor to persistent and prolonged environmental contamination. Plastic biodegradation is considered a promising approach to addressing current environmental issues. In this study, 300 marine isolates were evaluated for their ability to biodegrade PE. Based on total cellular fatty acid profiling and 16S rRNA gene sequence homology, RS120 was identified as Metabacillus niabensis (M. niabensis) among positively tested marine strains. Furthermore, early bacterial attachment to the PE surface was observed during the biotreatment. Then, after, chemical structural changes before and after the biodegradation were shown by the disappearance of the larger hydrocarbon tetra-tetracontane (C44H20) and the emergence of the smaller hydrocarbon heptadecane (C17H36), tetracosane, benzene, 1,1′-(cyclobutanediayl) (C16H16), and 2-tert-butyl-4-6-bis (3-5-di-tert-butyl). After that, Fourier Transform Infrared Spectroscopy (FTIR) spectra designated two new, shouldered peaks at 2590 cm−1 and 2,610 cm−1 along with a longer, sharper, and pointed peak at 2361 cm−1 in weathered PE, which substantiates significant alteration in chemical structure. Moreover, significant variation in thermal stability and crystallinity of plastic was also investigated. According to the enzymatic studies, bacterial peroxidase and dehydrogenase activities during treatment were higher at 30 days of treatment. In the meantime, the weight loss of the film was regularly monitored, and within 30 days of co-incubation with RS120, about 3.3% of its initial weight had been lost. These findings showed that the strain partially degrades the PE film when used as the sole carbon source. The results revealed that M. niabensis is reported here for the first time as an efficient PE degrader.
1 Introduction
In many applications, plastic materials have been preferred over metal, wood, and leather products in recent decades. Many facets extensively utilize it due to its durability and stability (Andrady and Neal, 2009; Ganesh et al., 2020). A study by Ritchie and Roser (2018) predicts that by 2034, annual plastic production will exceed 320 million tons. The amount of plastic produced worldwide has increased from 2 million tons in 1950 to 450 million tons in 2019, and it is projected to reach 507 to 537 million tons by 2025 (Bahl et al., 2020). India has also become a hotspot for plastic pollution, largely due to its expanding economy, which produces approximately 9.4 million tons of plastic annually (Joshi et al., 2025). Ironically, they constituted a thoughtful ecological issue due to their xenobiotic and resistant characteristics. Plastic garbage can now be disposed of in three primary ways: recycling, incineration, or landfill disposal (Ignatyev et al., 2014). In addition to releasing hazardous chemicals like hydrogen chloride and hydrogen cyanide, which are harmful to both animals and the environment, burning plastic also releases poisonous gases such as furans and dioxins (Shahnawaz et al., 2016). Between 60 and 80% of all marine garbage in the marine ecosystem is composed of plastic (Derraik, 2002). The water is disposed of, along with almost one million tons of plastic, per year. Conversely, landfilling plastic waste has a detrimental effect on the fertility of the surrounding area and the quality of life for the local population (North and Halden, 2013). Furthermore, a lot of birds are killed by strangling or ingesting plastic debris from rivers and city drainage systems that get into the ocean, even though plastic products have been found in human tissue such as brain, fat, nasal secretions, urine, and serum, indicating widespread human exposure to them (Harray et al., 2024).
Polymer degradation is caused by a combination of chemical, biological, and physical processes. According to Ojha et al. (2017), microbial-mediated degradation is an environmentally benign method that involves reducing the average molecular weight by microorganisms, backbone chain breaking, changes in surface character, and a loss of mechanical strength. Consequently, energy is generated from the products of microbial breakdown of plastics (Peixoto et al., 2017). In biodegradation, enzymes play a crucial role and belong to two enzyme classes: intracellular and extracellular depolymerases (Tournier et al., 2023). Large complex polymers are first broken down into smaller molecules, such as monomers, dimers, and oligomers, and then they are mineralized. These are also too small to pass through the exterior, semi-permeable gate and can be metabolized after being taken up and biodegraded within the microbial cell. After that, the fatty acid undergoes oxidation within the cell. The process is called depolymerization. When CO₂, H₂O, or CH₄ are the end products, the degradation is referred to as mineralization (Lv et al., 2024).
Enzymes and genes that break down resistant pollutants, such as plastic waste, are found in marine microorganisms. The biodegradation of plastics facilitated by microorganisms involves microbial attachment, in which a hydrophobic surface becomes more hydrophilic, leading to early physical and chemical breakdown through the introduction of hydrophilic groups. The microbe begins to develop by consuming the degraded PE as a source of carbon. The ocean contains a vast array of microorganisms that may contribute to the decomposition of synthetic plastic materials. Among the microorganisms implicated in the biodegradation of polyethylene (PE) are fungi, bacteria, and actinomycetes. The marine-sourced Pseudomonas, Moraxella, Micrococcus, Streptococcus, and Staphylococcus were identified as plastic degraders, with degradation rates ranging from 2.19 to 20.54% (Chandra and Singh, 2020). Furthermore, numerous studies on marine bacteria have demonstrated that they can break down plastic; some of these bacteria include Bacillus cereus and Bacillus sphericus (Sudhakar et al., 2008). Furthermore, Bravibacillus borstelensis and Rhodococcus ruber have also been shown in studies to be able to colonize and subsequently break down PE (Sivan et al., 2006; Hadad et al., 2005). However, degradation can be achieved by enhancing the biodegradability of plastics by combining them with cellulose or starch and pro-oxidants to facilitate their easy breakdown and, secondly, by enhancing microorganisms that efficiently break down polymers.
The genus Metabacillus is a reclassified group of bacilli. To better reflect the variety within the original Bacillus group, Patel and Gupta (2020) proposed the Metabacillus as one among six new genera. There were numerous Bacillus species, including previously reported species, that were moved to the new genus. Metabacillus fastidiosus was the first isolate of the Metabacillus genus. The original name of this species was Bacillus fastidiosus (Dooren and de Jong, 1929). Following the 2021 reclassification of the genus Metabacillus, the Bacillus niabensis 4T19T was also renamed as M. niabensis, which was originally isolated from cotton-waste compost used in mushroom growth (Kwon et al., 2007). A variety of habitats, including soil, hypersaline lakes, and marine coastal areas, were used to isolate the Metabacillus species. The cellular proteins that distinguish M. niabensis from its other species include the flagellar M-ring protein, the 50S ribosomal protein, the ATP phosphoribosyltransferase regulatory subunit, and others (Patel and Gupta, 2020). Some species can withstand moderate levels of salt (0–5%), and they can be grown at temperatures between 4 and 45 °C, with 25–37 °C being the ideal/optimum range for growth. In summary, M. niabensis is an aerobic, motile, Gram-negative bacterium with a genomic size of 4,987,608 bp and 4,969 recognized genes (Kangale et al., 2021). Bacteria from marine sources typically exhibit shorter-chain fatty acids to counteract the freezing effect of cold on long-chain fatty acids, enabling them to survive in harsh climates. Short-chain fatty acids preserve the physiological fluidity of the outer membrane (Sardar et al., 2015).
Since only a tiny fraction of the PE will be absorbed into humus, bacterial cellular biomass, and other ordinary products, the complete bio-deterioration and biodegradation of plastics is not yet feasible (Ghatge et al., 2020). Changes in physical properties and chemical structure of the shredded polymers were carried out using scanning electron microscopy (SEM), gas chromatography mass spectroscopy (GC–MS), Fourier transform infrared (FTIR) spectroscopy, X-ray diffraction (XRD), differential scanning colorimetry (DSC), and weight loss recorded in order to uncover the possible degradation mechanism of the bacteria that break down PE. Additionally, the activity of enzymes in the biological degradation of PE polymers after bacterial treatment was hypothesized.
The goal of the current investigation was to identify a new, potent strain of marine isolate capable of degrading PE. The further aim is also to assess and characterize the degree of PE degradation using the discovered marine isolate. Finally, these studies tested the ability of culturable marine bacteria to degrade PE. After identifying the putative marine bacterial isolate, their 30 day PE-degrading capacity was evaluated. M. niabensis is the first species of the genus Metabacillus to be investigated and shows the ability to degrade PE.
2 Methods
2.1 Polyethylene
The polymer used as the carbon source in this study was a medium-density polyethylene (MDPE) plastic film, 10 μm (0.010 mm) thick, purchased commercially from Good Fellow Cambridge Ltd. in Huntingdon, England, to serve as the foundational material. The MDPE is a moderately branched polymer with a molecular weight comparable to that of LDPE and HDPE plastics. MDPE’s density ranges from 0.926 to 0.940 g/cm3. Following a 1 mm × 1 mm incision, the polymer films were weighed, disinfected with 70% ethanol, and dried.
2.2 Marine bacteria
The bacterial strain RS120 used in the PE plastic biodegradation study was among the marine bacteria (300 marine bacteria) used for screening for biodegradation. These bacterial strains were received from the laboratory culture collection repository at the Applied Phycology and Biotechnology Division, CSIR-CSMCRI, Bhavnagar, which were earlier collected from salt water from Sikka, Gujarat, on the Arabian Sea coast (latitude N 22° 25′98.0″ and longitude E 69° 49′3.85″) in India. Following a 24–48-h incubation at 30 °C, these isolates were revived on Zobell Marine Agar medium (Himedia). After isolating and transferring pure bacterial colonies into Zobell Marine Broth, they were allowed to grow and then purified. These cultures were then stored at −80 °C in 10% glycerol.
2.3 Preliminary screening of PE degrading marine bacteria
Before the biodegradation experiment, tiny 1 cm x 1 cm pieces of PE film were cut, cleaned, weighed, and sterilized. Before use in any experiment, these tiny strips were sterilized by immersing them in 70% alcohol, then exposed to UV light and dried in a sterile condition. Then after this, sterilized PE films (weight 100 mg) were added to 20 mL of carbon-deficient Bushnell Hass (BH) minimum broth media supplemented with 3% NaCl (Eriksson et al., 2000). In additional culture tubes, 20 μL of 2, 3, 5-triphenyl tetrazolium chloride (TTC) indicator and 1% fresh bacterial inoculum of the RS120 strain were added to assess the survival of bacteria in BH media devoid of carbon content. The color of the media altered over the incubation period, according to Tan et al. (2021), suggesting that bacteria were using the carbon from PE for growth. Similarly, one treatment served as the control, with no bacterial inoculation, and three duplicates of each treatment were used to produce the blanks. Sets of every tube were incubated for 30 days at 180 rpm and 30 °C in an aseptic environment. Every 7 days, the plastic films were removed in triplicate from the culture vessels and subjected to a range of analytical measurements to illustrate bacterial attachment and PE film degradation.
2.4 Identification of selected performant strain
2.4.1 Total cellular fatty acids profiling
Whole-cell fatty acid contents of strain RS120 were transformed into methyl ester and analyzed by gas chromatography (GC). For the Fatty Acid Methyl Ester (FAME) study, the sample was prepared in five steps, viz., harvesting, saponification, methylation, extraction, and base washing, following the MIDI FAME protocol. The prepared methyl ester extract was analyzed using a GC 6850 series (Agilent Technology, Hewlett-Packard, Wilmington, DE, USA), which detected straight-chain saturated fatty acids extending in length from nine to twenty carbons, as well as five hydroxyl acids, along with a calibration standard (Supleco). The GC 6850 series was equipped with a 30 m × 0.25 mm × 0.25 mm fused silica capillary column, utilizing very high-purity hydrogen as the carrier gas, and a flame-ionization detector (FID). The initial oven temperature was 170 °C, increased by 5 °C min−1 to 260 °C, then by 30 °C min−1 and held constant at 300 °C for 2 min. Helium gas was applied as a carrier at a flow rate of 1.0 mL min−1 (Sasser, 2006; Veum et al., 2021). Retention durations for fatty acids were compared to a bacterial fatty-acid reference mix (bacterial acid methyl ester mix CP, 4–7,080; Supleco, Inc., Bellafonte, PA). Agilent Technology uses the RTBS6 library database for matching the FA.
2.4.2 Molecular identification and phylogenetic analysis
Marine strain RS120 was selected for its high potential to degrade PE compared to other screened microorganisms that grow on PE and underwent genotypic identification by 16S rRNA gene sequencing. The total genomic deoxyribonucleic acid of strain RS120 was extracted using the conventional phenol-chloroform technique (Weisburg et al., 1991). For amplification of the 16S rRNA gene, the universal primers 27F (50-AGAGTTTGATCCTGGCTCAG-30) and 1492R (50-CTACGGCTACCTTGTTACGA-30) were utilized (Galkiewicz and Kellogg, 2008). Each primer (10 pmol) was added to a 50 μL reaction mixture together with dNTPs (400 μM), Taq polymerase (2.5 U), MgCl₂ (1.5 mM), DNA template (40–50 ng), and 10X PCR buffer (5 μL). Amplification of the 16S rRNA gene was conducted using the following PCR program: initial denaturation at 94 °C for 8 min; 30 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 2 min; and a closing extension at 72 °C for 10 min. Afterwards, the amplified product was purified using the PCR Purification Kit (Qiagen), and sequencing was facilitated by M/S Microgen Inc., Seoul, South Korea. Neighbor-joining (NJ), maximum-likelihood (ML), and maximum-parsimony (MP) approaches were employed to construct phylogenetic trees based on unambiguously matched sequences (Sardar, 2025) using MEGA version 11 software (Tamura et al., 2021). For each of the three techniques, 1000 bootstrap trials were carried out to assess the robustness of tree topologies.
2.5 Microbial colonization on PE film
SEM images visualized the initial adherence of strain RS120 to the PE surface during incubation. PE films were prepared for SEM imaging using the method illustrated by Joshi et al. (2022). Films were removed from broth, and attached particles and precipitates were achieved using phosphate buffer. Then, the films were rinsed lightly, treated with 2% glutaraldehyde, and dehydrated three times: once in 50% ethanol for 30 min, once in 70% ethanol overnight, and once in 100% ethanol for 30 min. After the samples were fixed, vacuum-dried, and coated with gold, a field emission scanning electron microscope (JSM-7100 F, Jeol Ltd., USA) was used to analyze them.
2.6 Characterization of PE-degradation
2.6.1 GC–MS analysis
The culture filtrate (untreated and M. niabensis-treated) was removed after 30 days of treatment using diethyl ether solvent, following the extraction procedure. The cell-free supernatant was dissolved in diethyl ether for the extraction of the residual material with a 1:1 (v/v) ratio. With periodic shaking, this mixture was maintained at 100 °C in the water bath for six hours. During the incubation period, the mixture was removed from the water bath after 3 h, cooled to room temperature, and then returned to the water bath for an additional 3 h. The next step involved vortexing the content for 2 min and then holding it static for 10 min to allow the two phases to disperse. Using a glass Pasteur pipette, the organic phase was extracted from the bottom layer and analyzed by GC–MS (Shimadzu QP2020, Japan) to determine the breakdown products of PE. An AOC-20i autoinjector was used to inject 1 μL of the sample. At a pace of 6 °C per minute, the oven’s temperature rose from 50 °C to 280 °C. At a threshold of 1,000, the ion source was kept at 200 °C. A scan period of 0.30 s was established, spanning 50 to 500 m/z (Liu et al., 2022).
2.6.2 FTIR analysis of PE films
Alterations in the PE structure were investigated using Fourier Transform Infrared (FTIR) analysis (PerkinElmer, Spectrum EX) during the ongoing bacterial incubation. After a month of incubation with bacterial treatment, both treated and untreated PE film samples were processed using the method described by Joshi et al. (2022). Dried and clean PE films were used for analysis by spectroscopy from the 4,000–400 cm−1 frequency band, which was employed with a 1 cm−1 resolution (Albertsson and Karlsson, 1990).
2.6.3 XRD and DSC analysis
The X-ray diffractometer (Philips X’pert MPD, The Netherlands) was used to examine the crystalline structure of both treated and untreated PE. The θ/θ range at ambient temperature was 2° to 60°. With a 240 mm goniometer radius and a receiving slit size of 0.1 mm, the irradiation length and specimen length were both 10 mm. The focus and divergence slits were separated by 100 mm. Utilizing an h/h geometry with Cu Kα (k = 1.5406 Å) radiation, the system is equipped with an Ultima IV (Tokyo, Japan) goniometer. The operational parameters were I = 30 mA and U = 40 kV. Cleansed and dried PE thin film was placed on quartz substrata, and continuous measurements were made at d-spacings according to the diffracted X-ray at each θ value, with a step time of 1 s (Wu et al., 2023). Furthermore, the percentage crystallinity of both sets of PE film was calculated by using OriginPro 8.5 and the following formula to process the peaks at 2θ in the XRD data.
Thermal analysis of PE films was performed in nitrogen conditions (N₂, 20.0 mL/min/N₂, 70.0 mL/min) using a differential scanning calorimetry (DSC) machine (NETZSCH DSC 204F1 Phoenix 240–12-0239-L, Germany). Around 5.0 mg of plastic was weighed into an aluminum crucible pan covered with a pierced lid and heated from 24 °C to 250 °C (10.0 K/min). PE samples were cooled to 30 °C and then heated to 180 °C, 220 °C, or 300 °C, respectively (heating/cooling rates were 10 °C/min). Three observations, including glass transition temperature (Tg), melting point (Tm), and cold crystallization temperature (Tc), were obtained from the DSC thermograms (Beltrán-Sanahuja et al., 2021).
2.6.4 Determination of weight loss
The biodegradation of PE was assessed using a weight-loss assay. Every 7 days, the PE films treated with and without strain RS120 were removed from the culture tubes and weighed on a weighing balance with a readability of 0.1 mg (Shimadzu Analytical Pvt. Ltd., India). After three Milli-Q water washes, the plastic films were soaked in a 2% (w/v) SDS solution for 2 h. After the films were dried for the whole night at 50 °C in an oven, the molecular mass loss was computed as follows: Weight of treated PE minus weight of untreated PE (blank) equals total mass reduction (Sardar, 2025).
2.6.5 Peroxidase and dehydrogenase activity
Since microbial enzymes play a role in plastic biodegradation, peroxidase assays were conducted using cell-free supernatants. At 7, 14, 21, and 30 days of treatment, culture supernatant was collected by centrifuging broth aliquots containing films at 5000 rpm for 5 min at room temperature. To test peroxidase activity, a mixture of 1 mL of crude enzyme, 4.0 mM hydrogen peroxide, 0.1 mM potassium phosphate buffer (pH 7.0), and 1.0 mM levodopa was used as the substrate (Zhao et al., 2004).
To conduct a dehydrogenase assay, a mixture consisting of crude cell lysate (150 μL), 1% triphenyl tetrazolium chloride (50 μL), and phosphate buffer (300 μL) was prepared and incubated for 15 min at 37 °C, then 500 μL of icy methanol was added to stop the reaction. The color shift was measured and noted in a spectrophotometer at 470 nm for peroxidase and 480 nm for dehydrogenase. The quantity of enzyme required to elevate 1.0 absorbance units per minute is equivalent to one unit of enzyme activity (Mayer and Arnold, 2002).
2.7 Statistical analysis
The experiment included five replications. Statistical analysis was performed using SigmaPlot 12. All values were presented as the mean ± SD. Differences between groups that were statistically significant (p < 0.05) were found using Duncan’s test and one-way analysis of variance (ANOVA).
3 Results
3.1 Screening of PE-degrading bacteria
Biological degradation of the PE film was anticipated by the marine bacterial strain RS120, as indicated by the TTC reduction assay. The colorless TTC turns into the red insoluble triphenyl formazan (TPF) when bacteria use the released carbon from the PE sheet as their sole energy source for growth in the BH Media (Supplementary Figure S1). The electron transport chain drives this activity. As indicated by the tube’s screening results, the capable strain RS 120 broth containing (tube A) turned red (Supplementary Figure S1) under the PE-supplemented condition, indicating that it was the only carbon source. While tube C does not exhibit redness when PE is absent. As shown in the tube (B), the absence of redness indicates that no microorganisms were present during the experiment, which verifies the lack of contamination. Therefore, during the screening process, the tested positive strain grew only when using free carbon elements released by PE films in BH broth, which turned the broth red, and was chosen as a bacterium that degrades plastic.
3.2 Identification of PE-degrading bacteria
3.2.1 Total cellular fatty acids profiling
Whole cell fatty acid profiling of the PE-degrading strain RS120 exhibited the majorly branched fatty acids C15:0 ante-iso (51.55%), C15:0 iso (13.77%), and C17:0 ante-iso (8.74%), accompanied by some straight-chain C16:0 (4.69%) and C14:0 (1.20%), which are mentioned in Table 1. Their corresponding GC–MS peaks are also reflected in Supplementary Figure S2. In the hostile oceanic environment, the abundance of branched fatty acids and shorter chains in marine bacteria helps mitigate the environmental stress caused by temperature, pH, and salt concentrations.
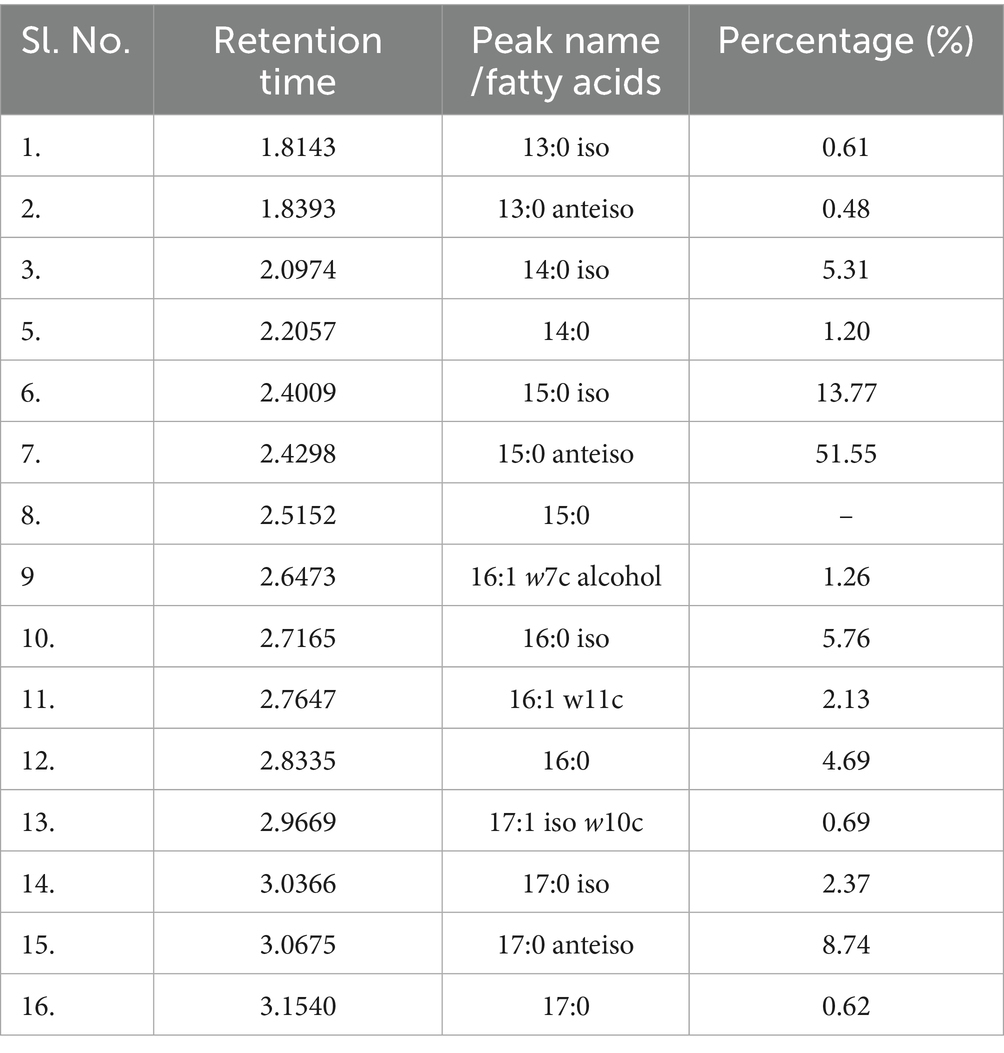
Table 1. GC–MS: total cellular fatty acid profile of M. niabensis strain RS120 using method: RTSBA6.
3.2.2 16S rRNA gene and phylogenetic analysis
A performant strain was identified and confirmed following completion of the BLAST study of the 16S rRNA gene. The organism was identified as a gram-positive M. niabensis strain 120 with a 99% homology match. Furthermore, the partial gene sequence of 16S rRNA (1,370 nucleotides) was submitted to the National Center for Biotechnology Information (NCBI) database, and the GenBank accession number is PP858188. According to a phylogenetic study with the NJ, ML, and MP algorithms, strain RS120 and M. niabensis (AY998119) formed a phylogenetic lineage within the genus Bacillus, as shown in Figure 1. The M. niabensis strain 120 showed taxonomically very close with the moderately halophilic bacterium M. halosaccharovorans, as depicted in phylogenetic analysis Figure 1, which was isolated from a hypersaline lake. The GenBank databases provided the reference sequences used for strain comparison. According to FAME and 16S rDNA sequence analysis, the isolate is native to the marine environment, consistent with other investigations (Romano et al., 2020).
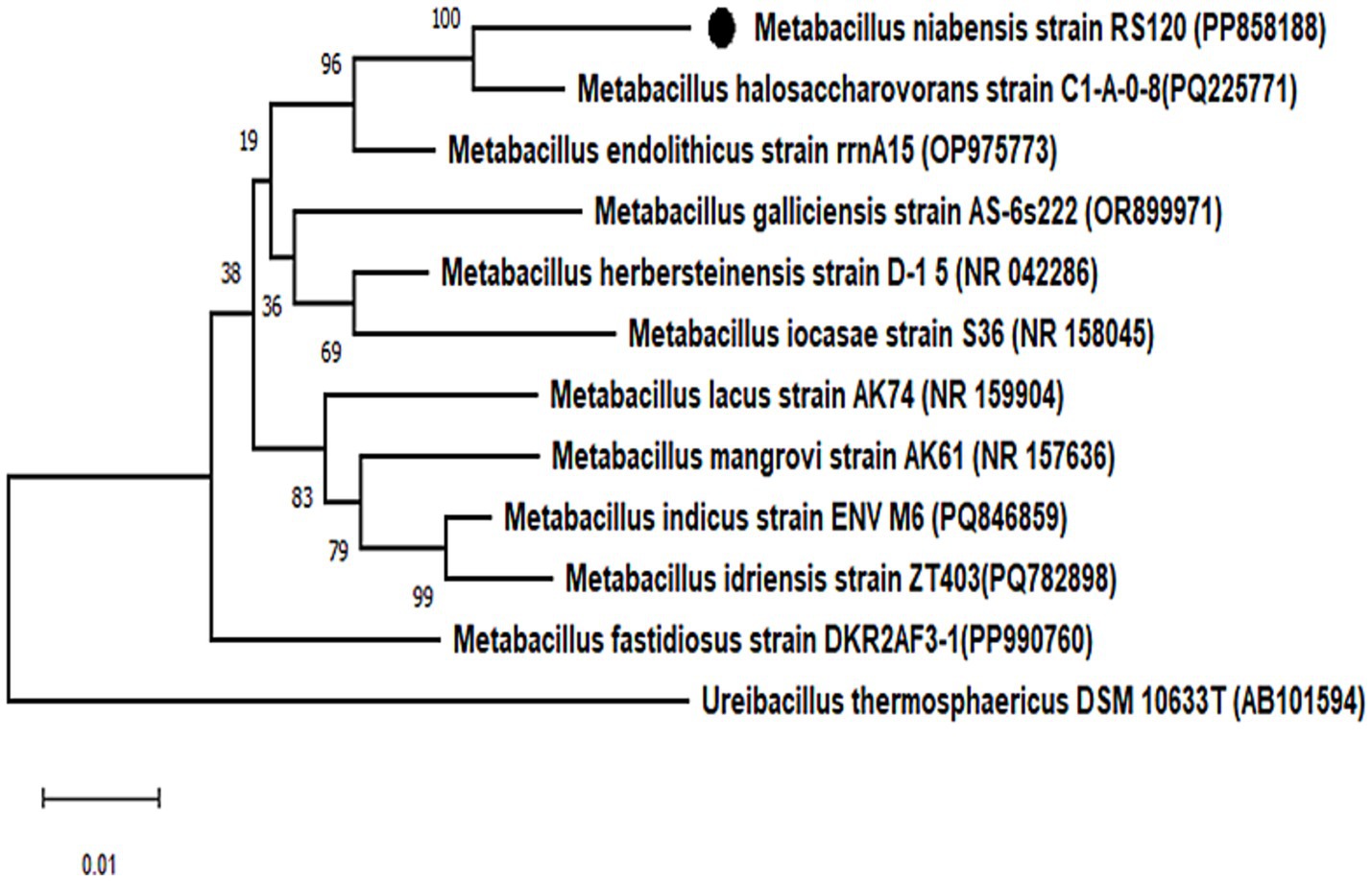
Figure 1. Phylogenetic analysis of bacterial strains. Phylogenetic tree based on 16S rRNA gene sequence showing the relationship between the isolated bacterial strain Metabacillus niabensis RS120 and other relatives within the genus. Bootstrap values greater than 60% are shown on branch nodes, which are based on 1,000 replicates. Ureibacillus thermosphaericus DSM 10633T, accession no. AB101594 served as the outgroup. Evolutionary analyses were conducted in MEGA 11.
3.2.3 Microbial adherence using SEM
SEM scans recorded during biotreatment of PE film exhibited microbial colonization by strain RS120 on the surface, as shown in Figure 2. Microbial aggregates and dissemination were observed on the PE surface during the co-incubation. Conversely, the PE surface treated without strain showed no signs of aggregation and was used as a control for comparison (Figure 2A). As a result, the attachment of the bacterial strain M. niabensis RS120 was the cause of these highly apparent observed changes in visibility. The bacterium B. niabensis, also known as M. niabensis, produces surfactants that make plastic more accessible to microbial strains for colonization (Alemán-Vega et al., 2020). This can lead to a higher breakdown efficiency.
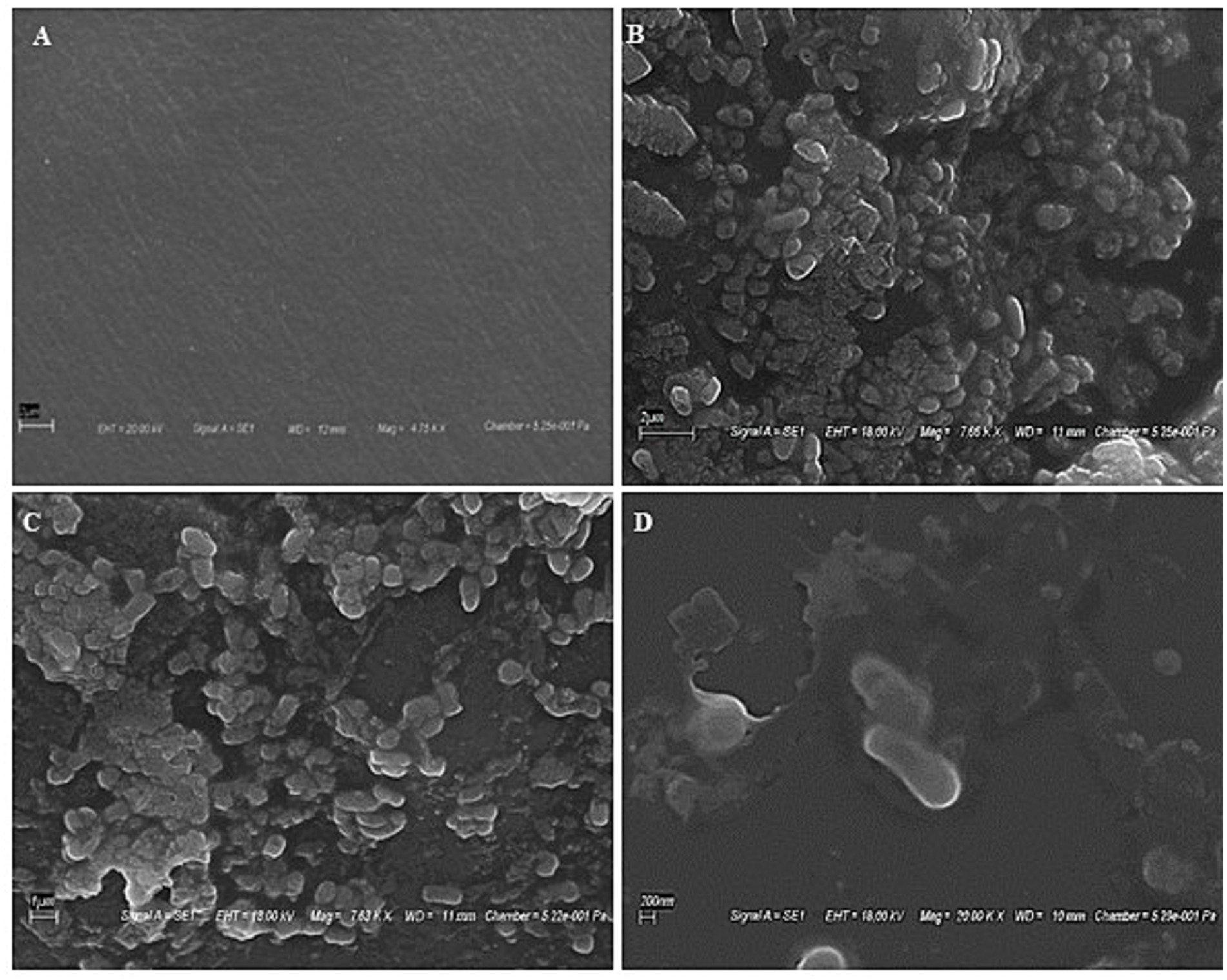
Figure 2. SEM images showing bacterial colonization over plastic films (A) Untreated film, (B) film at 2 μm, (C) film at 1 μm, and (D) film at 200 nm showing the presence of bacterial strain RS120 over the surface.
3.3 Characterization of PE degradation
3.3.1 GC–MS analysis
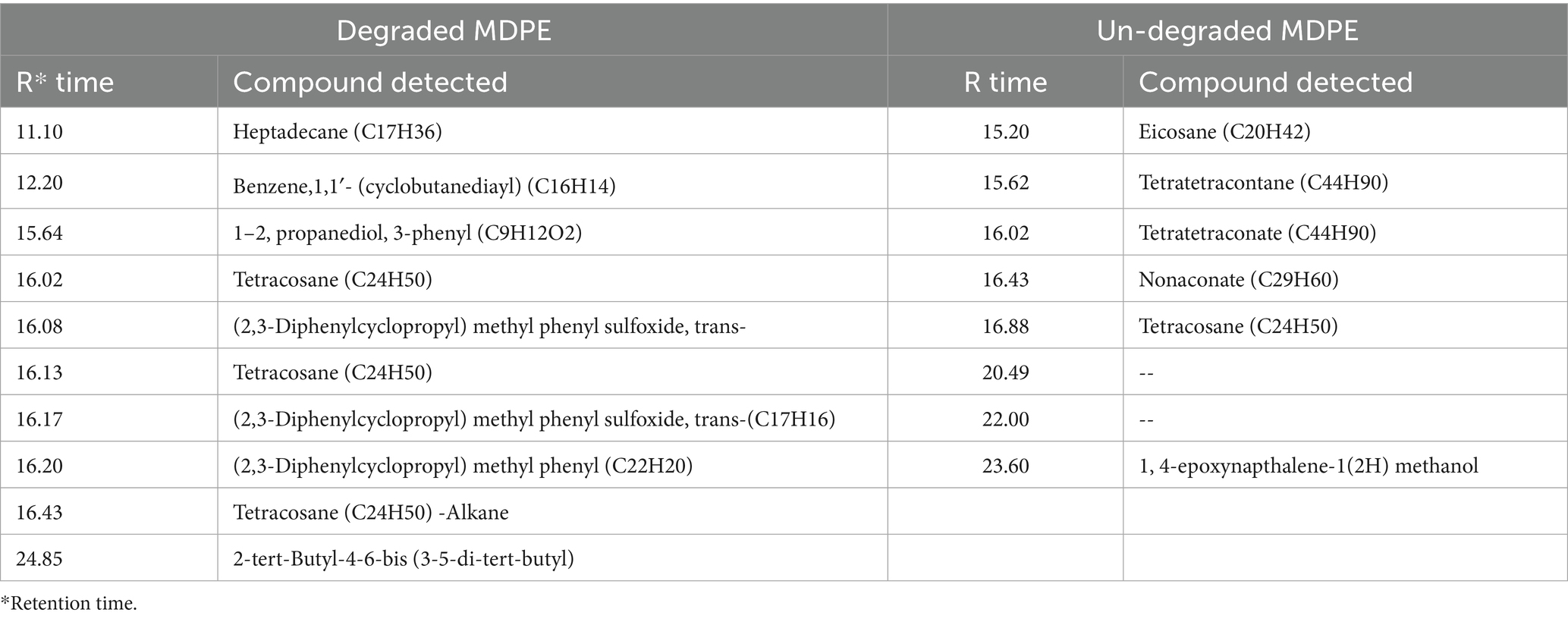
The components of the shredded PE differed significantly from those of the untreated control, as determined by GC–MS analysis. Larger polymeric compounds nonaconate (C29H60) and tetra-tetracontane (C44H90) at retention times 15.62 and 16.02 were found in the untreated (Figure 3A). However, as a result of M. niabensis breaking down PE, the extract contained short chains of (1–2, propanediol, 3-phenyl (C9H12O2), and benzene, 1, 1′-(cyclobutanediayl) (C16H16) alkanes, carboxylic acids, and other compounds (listed in Table 2). Furthermore, several byproducts, including short-chain hydrocarbon compounds ranging from C9 to C17, were also found in the degraded PE extract; these may result from biodegradation.
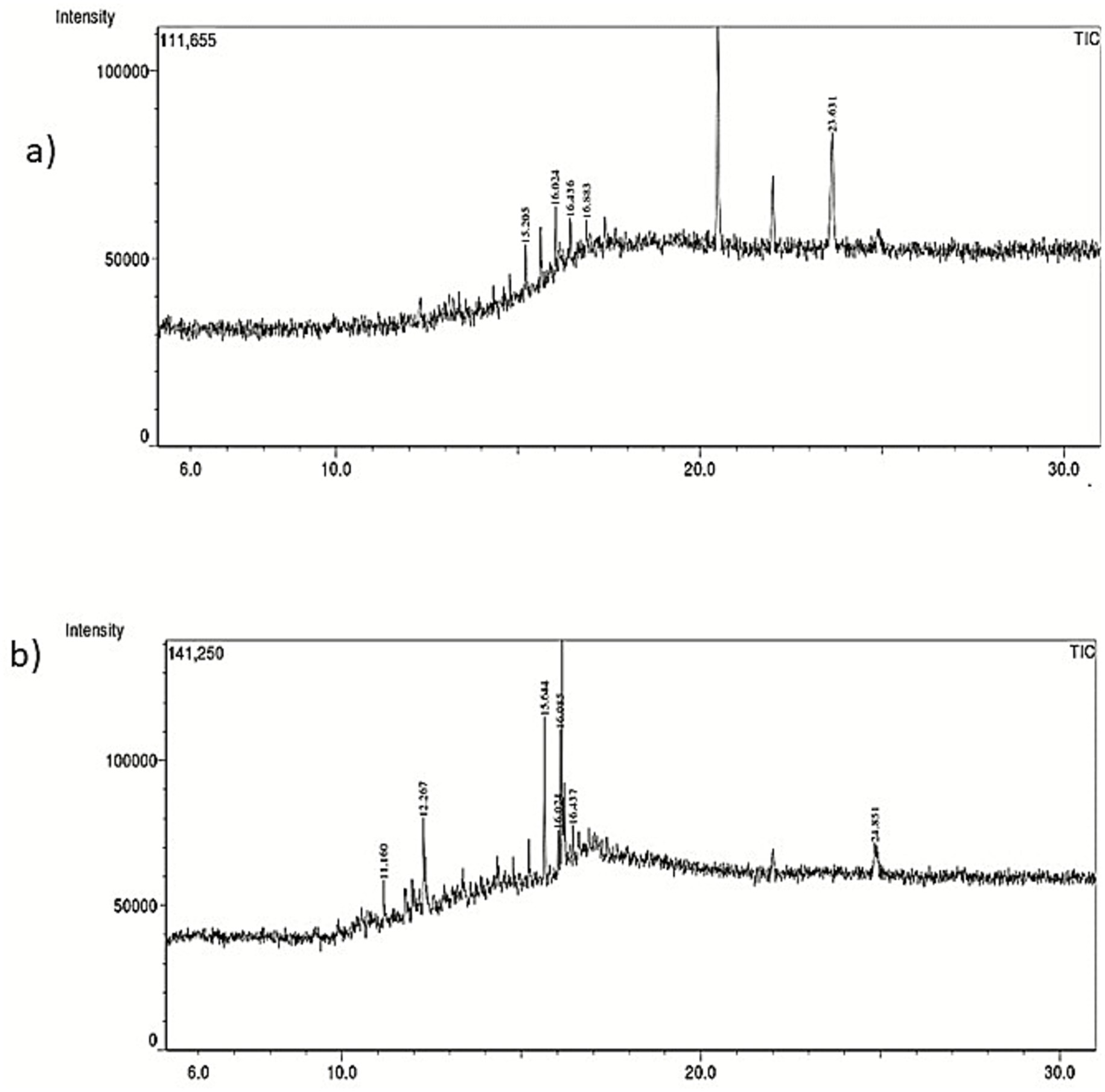
Figure 3. GC–MS chromatogram indicating chemical structure changes in (a) untreated and (b) treated with M. niabensis RS120.
3.3.2 FTIR study
FT-IR analysis revealed alterations to the side chain and changes in functional groups induced by microbial enzymatic activity. According to Shah et al. (2007), the peaks at 2921 cm−1 and 2,851 cm−1 that are discovered to be common in both the degraded and undegraded film lie within the absorbance range of 2,800 to 3,000 cm−1, which is comparable to C–H stretching, the presence of alkanes, and the peak that is brought on by vibrations in the C=C bond stretching. Instead of a single peak at 2610 cm−1, the weakened PE film displayed two shouldered peaks at 2590 cm−1 and 2,610 cm−1 (Figure 4). In degraded PE, a peak at 2361 cm−1 showed longer and had a sharp, pointed appearance rather than being tiny in undegraded PE. The biotreatment of M. niabensis was thought to be the cause of these alterations in functional group and chemical shifts.
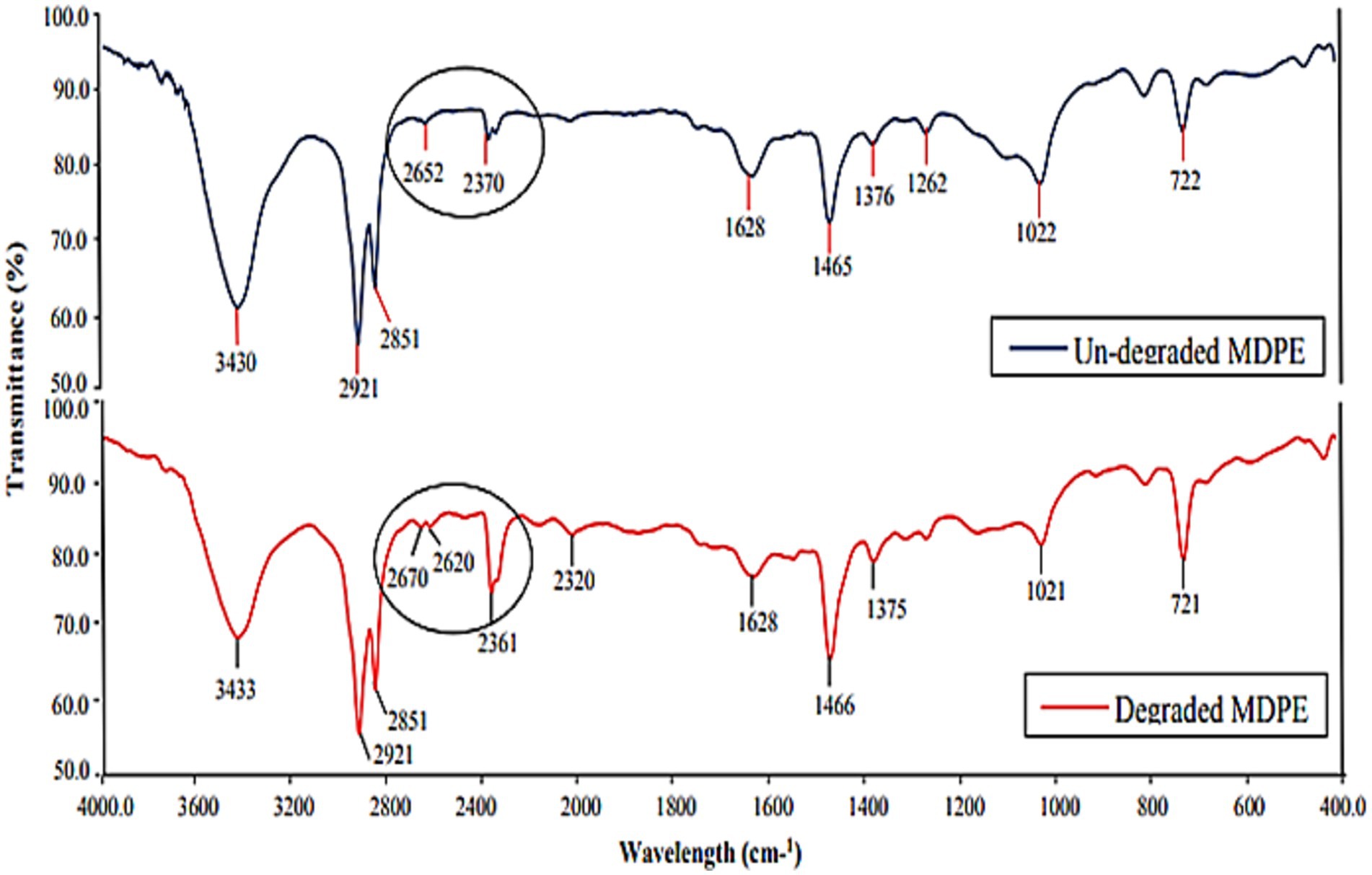
Figure 4. Infrared tests of the untreated and the treated group of PE after 30 days of treatment with strain M. niabensis RS120.
3.3.3 XRD and DSC analysis
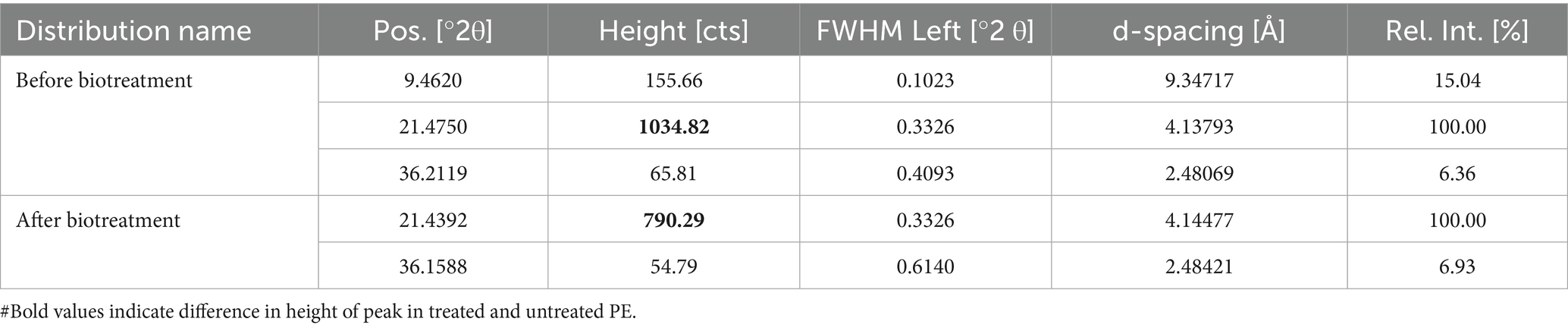
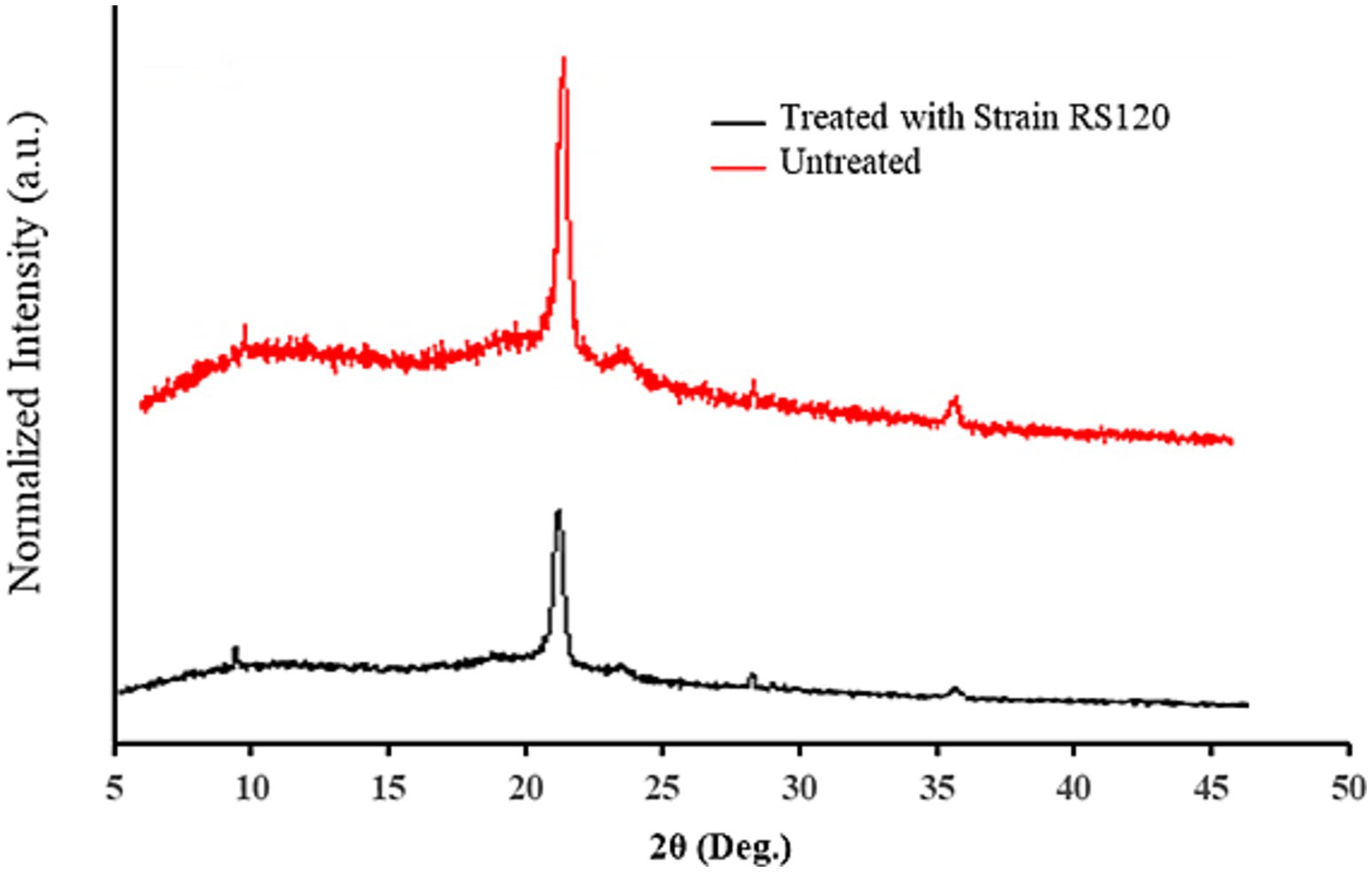
XRD spectra showed a sharp, distinguished, large peak at 21.47 along with two minor peaks at 9.46 and 36.21 of the angular position 2θ in the undegraded PE film. Conversely, small shifts in the peak locations at 21.43 and 36.15 were observed in the degraded PE film, while the peak at 9.46 completely disappeared. The results specified that the intensity of the peaks illustrated in Table 3 and indicated as vertical coordinates in Figure 5 in degraded PE was significantly diminished after 30 days of incubation with M. niabensis in liquid mineral medium. The significant decrease in the relative height counts (Table 3) of the peaks indicated a decrease in the percent crystallinity (6.23%). Thus, it may be inferred that there may have been a change in the sample’s microstructure, including preferred orientation, crystallite size, and atomic arrangement in the films. These observed differences indicate a significant change in biodegradation caused by the M. niabensis.
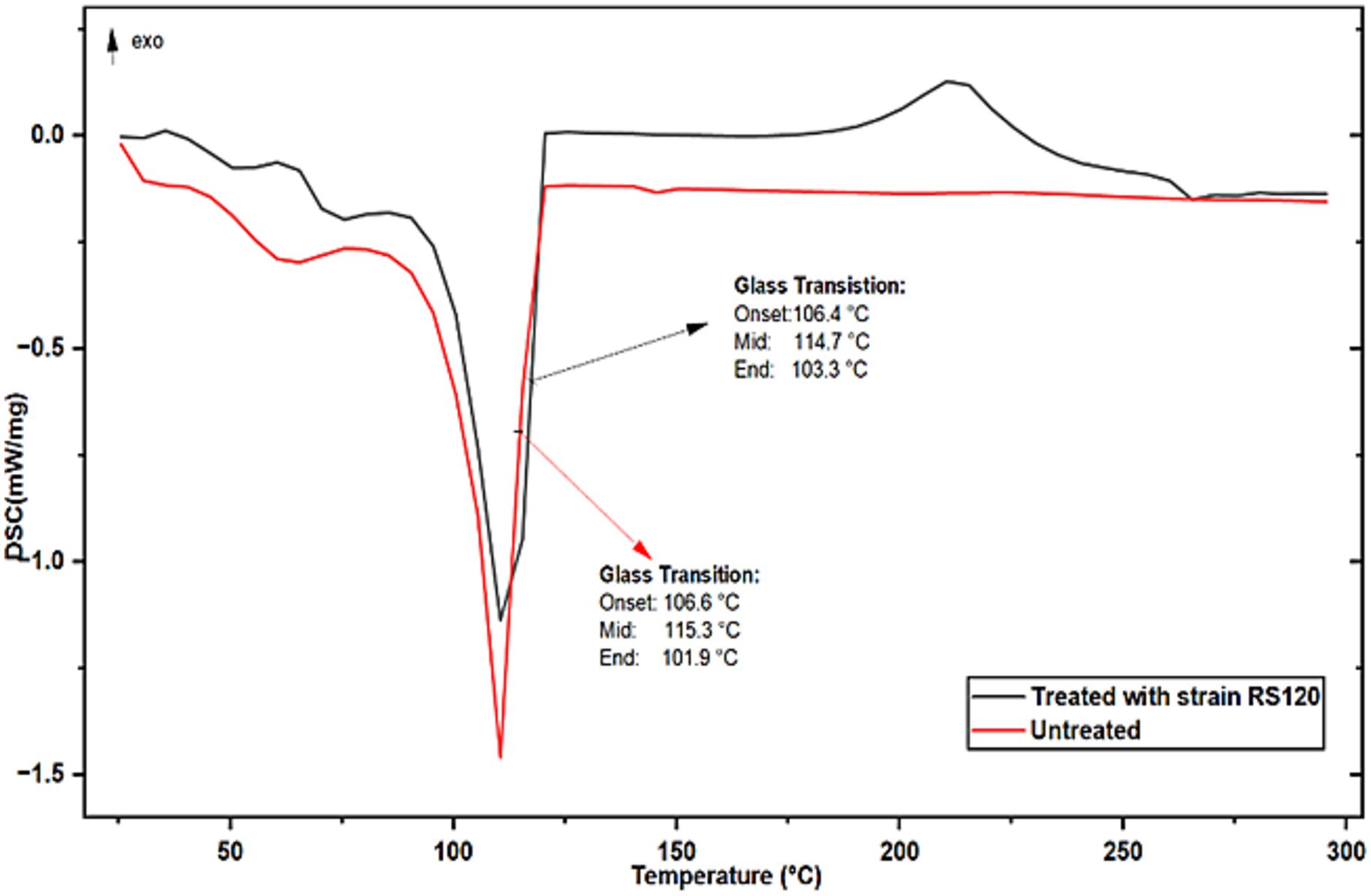
The biologically treated and untreated PE strips were analyzed by DSC over a wide temperature range (200 °C to 300 °C) to determine their melting points. Results are shown in Figure 6, which represent the DSC curves of the thermally homogeneous material, as indicated by a single sharp exothermic melting peak at 114.70 °C. This value is somewhat lower than that of the control (Tm = 115.30 °C) PE. As reported in the literature, the cyanobacterial Nostoc carneum degrades low-density polyethylene (LDPE) and produces similar results (Sarmah and Rout, 2019). Both the melting temperature and the glass transition depend on the mobility of the polymer chains, which in turn depends on the chemical structure. Therefore, these properties can be used to identify a polymer with a specific chemical structure.
3.4 Determination of weight loss percent
Weight loss was recorded at 7 day intervals for a maximum of thirty days of biotreatment. The significant reduction in the total quantity of PE films demonstrates how effectively the marine bacteria biodegrade the incredibly strong polymer structure when the plastic serves as the sole carbon and energy source. Following the 7th, 14th, 21th, and 30th days of treatment, the mean dry weight loss ± SD% was 0.78 ± 0.01, 1.58 ± 0.02, 2.45 ± 0.03, and 3.30 ± 0.03, correspondingly. The possibility that the bacteria took advantage of the carbon released from PE films during the incubation period is a reasonable explanation (Sardar, 2025). The observed weight decrease is thought to result from the oxidative fragmentation of the polymeric chain’s carbon backbone, generating volatile or gaseous species.
3.5 Enzyme activity
3.5.1 Peroxidase and dehydrogenase
Peroxidase activity increased during the 7 and 21 day biotreatment periods, as shown in Figure 7a. However, comparatively sluggish increases were also observed at 14 and 30 days. Microbial oxidative enzymes, such as peroxidases, can catalyze the oxidative cleavage of carbon–carbon bonds in various organic molecules, thereby enhancing the biodegradation process. Consequently, the altered shape and form of PE with weight loss demonstrate the effectiveness of peroxidase in breakdown using M. niabensis. For the effective biodegradation of waste PE, using a strain that generates peroxidase is typically a feasible biotechnique. Moreover, it was verified that the dehydrogenase enzyme activity was high during the biotreatment until the first 14 days of incubation; at that point, it declined and then gradually increased (Figure 7b). According to Elumalai et al. (2021) and Abbasian et al. (2015), the enzyme dehydrogenase breaks down large hydrocarbon molecules into smaller ones.
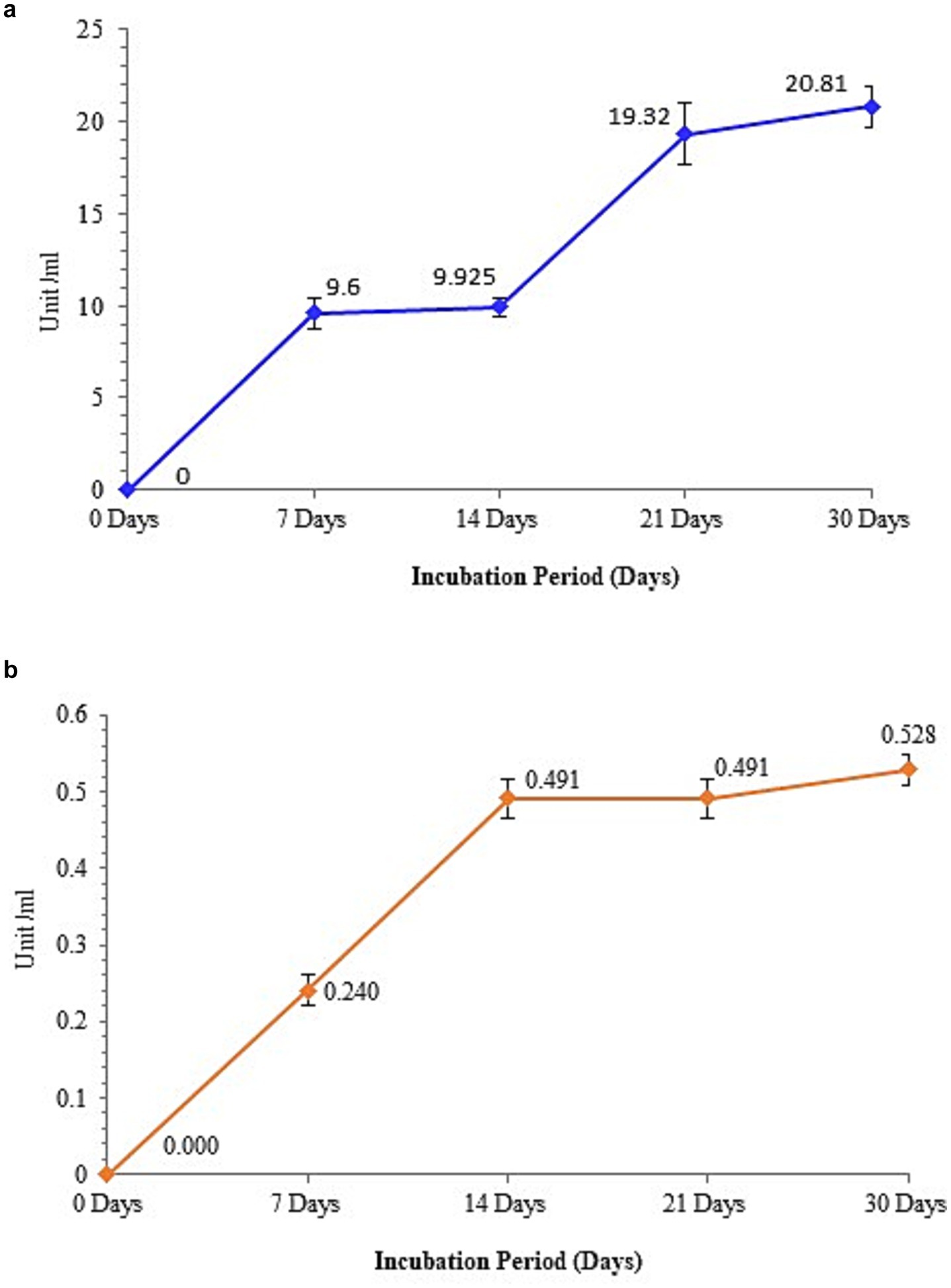
Figure 7. Enzyme assay showing (a) peroxidase activity and (b) dehydrogenase activity by strain RS120.
4 Discussion
The rapid production of plastic waste in the environment has led to several environmental risks. The sea has an enormous reservoir of bioactive compounds that can be used to clean up contaminants from human activity. The conversion of TTC into TPF by marine bacteria, which is essential for their survival and growth during biodegradability tests of PE in the absence of carbon, confirmed this potential. Besides this, some marine bacteria screened for the biodegradation of HDPE include Bacillus spp. and Pseudomonas sp. (Devi et al., 2019), as well as Pseudomonas sp. and Arthrobacter sp. (Balasubramanian et al., 2010).
Total cellular fatty acid profiling of the PE-degrading strain RS120 discloses that the majorly branched fatty acids recorded are in agreement with the previously informed strain M. niabensis 4T19T of the same genus. The common major fatty acid components were C15:0 ante-iso, C16:0 iso, C17:0 ante-iso, C16:0, and C14:0 iso, as reported by Kwon et al. (2007).
The phylogenetic analysis also revealed proximity to Metabacillus halosaccharovorans C-1-0-8 and a distance from the outgroup, Ureibacillus thermosphaericus DSM 10633 T, among a dozen retrieved 16S gene sequence data sets for the genus Metabacillus. Although many marine-sourced microorganisms involved in plastic biodegradation have been reported earlier, M. niabensis is added to this list.
For a view of microbial attachment on the surface of PE, marine bacteria Pseudomonas sp. and Lysinibacillus sp. (Oliveira et al., 2021) and Micrococcus flavus (Sardar, 2025), strain RS120, also showed a similar strategy. Microbial cell attachment to the film surface may be driven by the secretion of substances with maximum early activity, such as sugars, proteins, enzymes, and organic acids. For the initiation of biodegradation, biofilm formation is observed as a key factor. Plastic’s hydrophobicity, roughness, topography, and electrostatic interactions all contribute to its adsorption, desorption, and deterioration.
GC–MS analysis reveals the emergence of low molecular n-alkane compounds like heptadecane (C17H36) and tetracosane (C24H50) with benzene and phenol-containing intermediate products in the weathered PE shown in Figure 3. A recent study by Liu et al. (2022) described the degradation of LDPE films in 3 months by Bacillus velezensis and also showed the intermediate product tetracosane (C24H50) among C24–C29 n-alkanes, which corroborates my findings. The presence of these by-products after PE biodegradation and their detection provide important evidence that PE undergoes depolymerization. Moreover, the creation of alkane compounds such as eicosane, tetratriacontane, nonaconate, and tetracosane in degraded PE (Tiago et al., 2023) explicated the direction of the breakage of the polymer chain, originating carbonyl radicals that can react with an alkoxy radical (R–O) on the PE chain (Norrish I-type reaction). According to a previous report by Awasthi et al. (2017), Rhizopus oryzae’s breakdown of LDPE releases alkanes and carboxylic acids. Furthermore, another study reported that GC–MS analysis of PE showed the production of fatty acids due to enzymatic activity. The degradation of plastic is also attributed to extracellular enzymes produced by microorganisms, such as laccase, lignin peroxidase, and esterase. According to a prior study, there are three stages in the biodegradation of PE. Initially, PE is degraded into oligomers, dimers, or monomers through biological oxidation or enzyme-mediated processes. Due to further oxidation reactions, the polymer chains are broken down, forming fatty acids. Finally, fatty acids are metabolized by microorganisms into the small molecules CO₂ and H₂O (Gao et al., 2024).
FTIR reveals structural destabilization and changes in the chemical structure of plastic treated with M. niabensis, precisely depicting the biodegradation of PE. This study demonstrates that shifts and changes in peak transmittance, as well as a decrease in side-chain peak and functional group transmittance, were achieved due to the enzymatic degradation of PE. A similar result was also depicted by Bacillus weihenstephanensis. The shift from wave number 1634 to 1,629 cm−1 in the treated film shown in Figure 4 corresponds to the carbonyl group, which is related to oxidation of the PE sample (Rajandas et al., 2012). At the same time, these peaks suggest that the metabolic activity of these strains destroys the chemical inertness of PE by inducing unstable and reactive bonds with hydroxyl groups (Matsunaga and Whitney, 2000; Ng et al., 2018). Furthermore, as outlined by Sen and Raut (2015), some extracellular enzymes can introduce oxygen atoms into the chemical structure of PE, suggesting that strain RS120 may also exhibit a similar hydrolase activity. Therefore, GC–MS and FTIR analysis revealed the emergence of a novel molecule and a modified peak in degraded PE, which may be attributed to the activity of enzymes released under bio-stress conditions.
XRD can be used to evaluate the degree of crystallinity of the polymers. PE’s orthorhombic crystal structure is characterized by reflections at 2θ peaks at 21.43° and another peak created at 2θ = 36.15° (Morancho et al., 2006). Consequently, biodegradation processes that shrink the crystallites and ultimately expand the amorphous region decrease crystallinity. Balasubramanian et al. (2010) reported a comparable decrease in crystallinity following incubation with bacterial strains. According to the reported data, bacterial enzymes may penetrate and disrupt crystalline regions, leading to a loss of chain order due to the disordering of crystalline domains, which results in an overall decrease in crystallinity. A further drop in crystallinity occurs due to the attack on smaller crystals. A similar observation was also noted by Wu et al. (2023) in the biodegradation of PE by Bacillus paramycoides.
The observed melting peak of shredded PE (Tm = 114 °C) and the shift in unshredded PE (Tm = 115 °C) may be due to the breakdown of branches and changes in PE density. According to Menczel and Prime (2008), the characteristic phase transition temperature can be influenced by the magnitude of polymer branching, as well as by additives, contaminants, and particle size. According to published research, high-density PE exhibits somewhat larger melting peak temperatures due to its greater degree of branching (Camacho and Karlsson, 2001; Sorolla-Rosario et al., 2022). Since the complex, mixed branching structure of LDPE molecules influences their crystallization during the cooling phase of the DSC experiment, resulting in a wide range of lamellar thicknesses, these results make sense. The density of the polymer also affects its correlation with the melting temperature of PE; higher-density PE exhibits a higher melting temperature, and vice versa.
The degradation efficiency can be calculated by measuring the weight loss of PE films (Yang et al., 2020). Here, the weight loss rate of the PE film gradually increased with increasing co-culture time. A different study by Sudhakar et al. (2008) found that after a prolonged incubation, B. sphericus caused a dry weight loss of approximately 3.5%, while B. cereus caused a dry weight loss of around 2% in treated PE films. The observed weight loss is close to my findings. Moreover, Exiguobacterium and B. subtilis colonize and degrade PE by 4.7 and 12.1%, respectively (Muthukumar et al., 2014). The deteriorating surface area resulting from direct contact of bacterial strains with the polymer and subsequent consumption was linked to the weight loss percentage of DPE. Low molecular weight chains in PE are consumed, and chain scission arises at the end or branch of the polymer chain rather than the center of the molecule. Such disintegration of PE releases free carbon from HDPE sheets throughout the incubation in the liquid broth, resulting in a weight reduction. However, the long hydrophobic polymer chains cannot be transported directly into microorganisms via outer membranes.
A hydrolytic breakdown of the polymer is triggered by the release of extracellular enzymes that attach to it (Wilkes and Aristilde, 2017; Sheel and Pant, 2018). As previously noted, the periplasmic dehydrogenase from Gram-negative bacteria plays a crucial role in the biodegradation of polyvinyl alcohol (PVA) (Kawai and Hu, 2009). Periplasmic PVA dehydrogenase, an active enzyme that hydrolyzes PVA, was shown to be present in Sphingopyxis sp. 113P3. To obtain energy, microorganisms speed up chemical reactions that release electrons from polymers. Among bacterial oxidative enzymes, peroxidase exhibited the capacity to catalyze the oxidative cleavage of carbon–carbon bonds, hence increasing the rate of biodegradation (Mohammadi et al., 2022). Hydroquinone peroxidase, the first microbial enzyme identified as being involved in the breakdown of polystyrene, was isolated from Azotobacter beijerinckii HM121 (Oyewole et al., 2022). Discovering bacteria and enzymes that can break down plastic polymers opens the door to bioremediation, an environmentally beneficial, natural way to reduce pollution. In addition to minimizing damage to marine life and restoring marine biodiversity, it also helps to prevent the accumulation of microplastics in marine ecosystems. It converts plastics into biomass, carbon dioxide, and water.
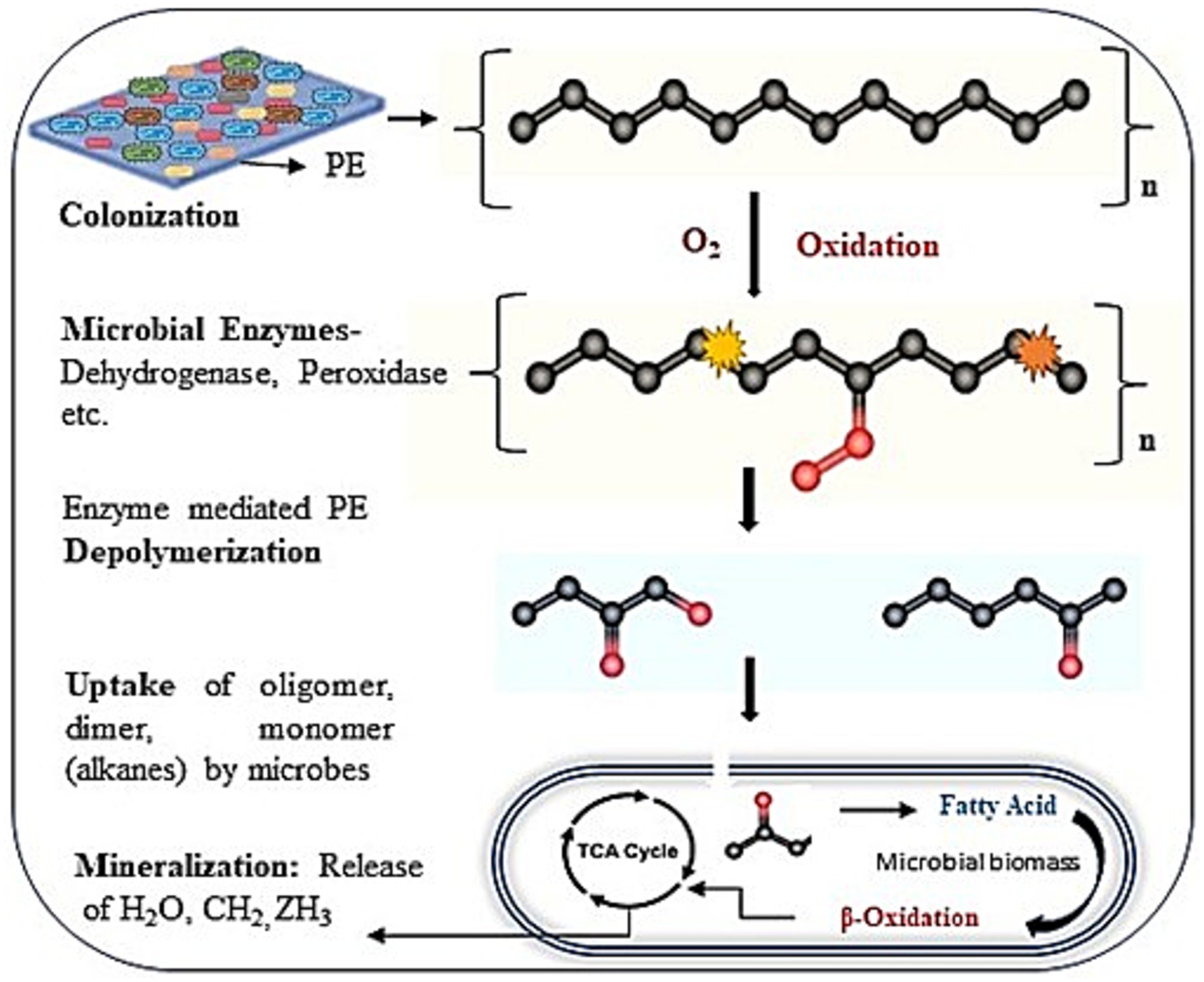
Here, Figure 8 illustrates a proposed mechanism for the biodegradation of PE through microbial attack. The four steps of PE biodegradation, including colonization, depolymerization, assimilation, and mineralization, are described (Restrepo-Flórez et al., 2014; Ali et al., 2021). Microbial aggregates, composed of cells and extracellular polymers, can adhere to plastic surfaces or to one another during the colonization phase, forming a complex biofilm that is visualized in Figure 2 (Flemming et al., 2016). The polymer surface’s chemical inertness is decreased, and its hydrophobicity is compromised as a result of microorganisms interacting with different extracellular enzymes (peroxidase, hydrolases) generated by microbial strains (Gaur et al., 2022). After that, several enzymes released by the microbial strain attack the plastic’s molecular chain, breaking the hydrolysable chemical bond or the terminus of the chain (Othman et al., 2021; Show et al., 2021). As a result, monomers and dimers would develop, lowering the plastic’s molecular weight (600 Da) (Santacruz-Juarez et al., 2021; Santo et al., 2013; Spina et al., 2021). Oxygen-containing functional groups, mentioning hydroxyl and carbonyl groups, were formed concurrently by further oxidation under the influence of oxygen. The generated oligomers and broken molecular chains are discharged into the environment. The microbial enzyme system (such as oxidoreductase, esterase, and lipase) can more readily identify and target the degradation intermediates because some of the oligomers with lower molecular weights can penetrate the strain’s cell membrane and enter the microbe (Shah et al., 2008). Considering the chemical similarity between PE and alkanes, it is believed that the metabolic pathways of alkanes and polymer breakdown are quite comparable when the sizes of polymer molecules are reduced to an appropriate range for enzyme attack (Jeon and Kim, 2015). According to Van Beilen et al. (2003), terminal oxidase later catalyzes the breakdown of PE intermediates to produce alcohols, which are then further oxidized by alcohol and aldehyde dehydrogenase, and the resultant fatty acids enter the oxidation cycle. Second, subterminal oxidation monooxygenase catalyzes the breakdown of PE intermediates to form secondary alcohols, which are then oxidized to ketones by alcohol dehydrogenase. This is followed by β-oxidation, which turns oxidized carboxylic molecules with an even number of carbon atoms into either propionyl CoA or acetyl CoA (if the number of carbons is odd). Propionyl-CoA carboxylase is responsible for carboxylating propionyl-CoA into succinyl-CoA (Gravouil et al., 2017). The tricarboxylic acid (TCA) cycle is entered by acetyl-CoA and succinyl-CoA. The reducing power (NADH and CoQ10H2) produced by this cycle is utilized in the respiratory chain to generate ATP, which is required for the production of new microbial biomass through replication mechanisms. Certain short-chain, water-soluble intermediates created by depolymerization are detected by receptors during absorption and subsequently transferred to microorganisms across the membrane, where they participate in a range of metabolic processes and stimulate cell division. Ultimately, a few metabolites and unassimilated products were fully absorbed, utilized, and transformed into energy, carbon sources, carbon dioxide, and water molecules during the mineralization process.
5 Conclusion
Microbes from marine sources can bioremediate untreated PE, as this study shows. Prolonged incubation caused by the newly found M. niabensis of 22 known species of the genus, which adhere to the surface as a biofilm, further altered the PE structure and shifted peak absorbance. This resulted in the creation of a novel intermediate by-product, which was examined using various analytical tools. The biotreatment method is an environmentally beneficial approach to removing plastic from the environment and converting it into biomass in a laboratory setting. These findings demonstrated the potential of marine bacteria in waste management, and future studies will focus on tackling plastic waste, promoting a circular economy, and optimizing microbial populations for various plastics and practical applications.
Data availability statement
The data presented in this study are publicly available. The data can be found at: https://www.ncbi.nlm.nih.gov/genbank, accession PP858188.
Author contributions
RS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The entire facility for conducting research was provided by the CSIR-CSMCRI Marine Biotechnology and Ecology division. Prof. B. Jha, the chief scientist and discipline chair (retd.), is greatly appreciated for giving the lab space. The author is thankful for funding from the Council of Scientific and Industrial Research (CSIR), New Delhi, under a project entitled CSC0120-Waste to Wealth. RS expresses gratitude for the financial support provided by the CSIR, New Delhi, India, through the project Junior Research Fellowship. The institute’s Analytical Science Discipline is appropriately acknowledged for providing instrumentation facilities.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1693690/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Screening of PE-degrading bacteria using Bushnell and Haas broth (minimal media) showed (a) MDPE film with the RS120 strain, (b) MDPE film only, and (c) Strain RS120 without MDPE.
SUPPLEMENTARY FIGURE S2 | GC-MS Chromatogram of total cellular fatty acids of M. niabensis strain RS120.
References
Abbasian, F., Lockington, R., Mallavarapu, M., and Naidu, R. (2015). A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 176, 670–699. doi: 10.1007/s12010-015-1603-5
Albertsson, A. C., and Karlsson, S. (1990). The influence of biotic and abiotic environments on the degradation of polyethylene. Prog. Polym. Sci. 15, 177–192.
Alemán-Vega, M., Sánchez-Lozano, I., Hernández-Guerrero, C. J., Hellio, C., and Quintana, E. T. (2020). Exploring antifouling activity of biosurfactants producing marine bacteria isolated from gulf of California. Int. J. Mol. Sci. 21:6068. doi: 10.3390/ijms21176068
Ali, S. S., Elsamahy, T., Al-Tohamy, R., Zhu, D. C., Mahmoud, Y. A., Koutra, E., et al. (2021). Plastic wastes biodegradation: mechanisms, challenges and future prospects. Sci. Total Environ. 780:146590. doi: 10.1016/j.scitotenv.2021.146590
Andrady, A. L., and Neal, M. A. (2009). Applications and societal benefits of plastics. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 364, 1977–1984. doi: 10.1098/rstb.2008.0304
Awasthi, S., Srivastava, P., and Singh, P. (2017). Biodegradation of thermally treated high-density polyethylene (HDPE) by Klebsiella pneumoniae CH001. 3 Biotech 7:332. doi: 10.1007/s13205-017-0969-2
Bahl, S., Dolma, J., Singh, J. J., and Sehgal, S. (2020). Biodegradation of plastics: a state-of-the-art review. Mater Today Proc 39, 31–34. doi: 10.1016/j.matpr.2020.04.722
Balasubramanian, V., Natarajan, K., and Hemambika, B. R. (2010). High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of gulf of Mannar, India. Lett. Appl. Microbiol. 51, 205–211. doi: 10.1111/j.1472-765X.2010.02883.x
Beltrán-Sanahuja, A., Benito-Kaesbach, A., Sánchez-García, N., and Sanz-Lázaro, C. (2021). Degradation of conventional and biobased plastics in soil under contrasting environmental conditions. Sci. Total Environ. 787:147678. doi: 10.1016/j.scitotenv.2021.147678
Camacho, W., and Karlsson, S. (2001). NIR, DSC, and FTIR as quantitative methods for compositional analysis of blends of polymers obtained from recycled mixed plastic waste. Polym. Eng. Sci. 41, 1626–1635. doi: 10.1002/pen.10839
Chandra, P., and Singh, D. P. (2020). Microplastic degradation by bacteria in aquatic ecosystem. In. Microorganisms for sustainable environment and health (pp. 431–467). Elsevier.
Derraik, J. G. B. (2002). The pollution of the marine environment by plastic debris: a review. Mar. Pollut. Bull. 44, 842–852. doi: 10.1016/S0025-326X(02)00220-5
Devi, R. S., Ramya, R., and Kannan, K. (2019). Investigation of biodegradation potentials of high-density polyethylene degrading marine bacteria isolated from the coastal regions of Tamil Nadu, India. Mar. Pollut. Bull. 138, 549–560. doi: 10.1016/j.marpolbul.2018.12.052
Dooren, D., and de Jong, L. E. (1929). Über Bacillus fastidiosus. Zentbl. Bakteriol. Parasitenkd 79, 344–357.
Elumalai, P., Parthipan, P., Huang, M., Muthukumar, B., Cheng, L., Govarthanan, M., et al. (2021). Enhanced biodegradation of hydrophobic organic pollutants by the bacterial consortium: impact of enzymes and biosurfactants. Environ. Pollut. 289:117956. doi: 10.1016/j.envpol.2021.117956
Eriksson, M., Dalhammar, G., and Borg-Karlson, A. K. (2000). Biological degradation of selected hydrocarbons in an old PAH/creosote contaminated soil from a gas work site. Appl. Microbiol. Biotechnol. 53, 619–626.
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Galkiewicz, J. P., and Kellogg, C. A. (2008). Cross-kingdom amplification using bacteria-specific primers: complications for studies of coral microbial ecology. Appl. Environ. Microbiol. 74, 7828–7831. doi: 10.1128/AEM.01303-08
Ganesh, K. A., Anjana, K., Hinduja, M., Sujitha, K., and Dharani, G. (2020). Review on plastic wastes in marine environment—biodegradation and biotechnological solutions. Mar. Pollut. Bull. 150:110733. doi: 10.1016/j.marpolbul.2019.110733
Gao, W., Xu, M., Zhao, W., Yang, X., Xin, F., Dong, W., et al. (2024). Microbial degradation of (micro) plastics: mechanisms, enhancements, and future directions. Fermentation 10:441. doi: 10.3390/fermentation10090441
Gaur, V. K., Gupta, S., Sharma, P., Gupta, P., Varjani, S., Srivastava, J. K., et al. (2022). Metabolic cascade for remediation of plastic waste: a case study on microplastic degradation. Curr. Pollut. Rep. 8, 30–50. doi: 10.1007/s40726-021-00210-7
Ghatge, S., Yang, Y., Ahn, J. H., and Hur, H. G. (2020). Biodegradation of polyethylene: a brief review. Appl. Biol. Chem. 63:27. doi: 10.1186/s13765-020-00511-3
Gravouil, K., Ferru-Clément, R., Colas, S., Helye, R., Kadri, L., Bourdeau, L., et al. (2017). Transcriptomics and lipidomics of the environmental strain Rhodococcus ruber point out consumption pathways and potential metabolic bottlenecks for polyethylene degradation. Environ. Sci. Technol. 51, 5172–5181. doi: 10.1021/acs.est.6b06095
Hadad, D., Geresh, S., and Sivan, A. (2005). Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 98, 1093–1100. doi: 10.1111/j.1365-2672.2004.02593.x
Harray, A., Herrmann, S., Papendorf, H., Miller, C., Vermeersch, A., Smith, T., et al. (2024). Plastics in human diets: development and evaluation of the 24-h dietary recall—plastic exposure and the dietary plastics score. Front. Nutr. 11:1443792. doi: 10.3389/fnut.2024.1443792
Ignatyev, I. A., Thielemans, W., and Beke, B. V. (2014). Recycling of polymers: a review. ChemSusChem 7, 1579–1593. doi: 10.1002/cssc.201300898
Jeon, H. J., and Kim, M. N. (2015). Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 103, 141–146. doi: 10.1016/j.ibiod.2015.04.024
Joshi, G., Goswami, P., Jha, D. K., Vinithkumar, N., Dharani, G., Vinithkumar, N. V., et al. (2025). Microplastics under siege: biofilm-forming marine bacteria from the microplastisphere and their role in plastic degradation. Sci. Total Environ. 980:179539. doi: 10.1016/j.scitotenv.2025.179539
Joshi, G., Goswami, P., Verma, P., Prakash, G., Simon, P., Vinithkumar, N. V., et al. (2022). Unraveling the plastic degradation potentials of the plastisphere-associated marine bacterial consortium as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 425:128005. doi: 10.1016/j.jhazmat.2021.128005
Kangale, L. J., Levasseur, A., Raoult, D., Ghigo, E., and Fournier, P. E. (2021). Draft genome of Metabacillus niabensis strain 4T19T isolated from cotton-waste composts for mushroom cultivation. New Microb. New Infect. 42:100894. doi: 10.1016/j.nmni.2021.100894
Kawai, F., and Hu, X. (2009). Biochemistry of microbial polyvinyl alcohol degradation. Appl. Microbiol. Biotechnol. 84, 227–237.
Kwon, S. W., Lee, S. Y., Kim, B. Y., Weon, H. Y., Kim, J. B., Go, S. J., et al. (2007). Bacillus niabensis sp. nov., isolated from cotton-waste composts for mushroom cultivation. Int. J. Syst. Evol. Microbiol. 57, 1909–1913. doi: 10.1099/ijs.0.64874-0
Liu, X., Zhang, Y., Sun, Q., Liu, Z., Zhao, Y., Fan, A., et al. (2022). Rapid colonization and biodegradation of untreated commercial polyethylene wrap by a new strain of Bacillus velezensis C5. J. Environ. Manag. 301:113848. doi: 10.1016/j.jenvman.2021.113848
Lv, S., Li, Y., Zhao, S., and Shao, Z. (2024). Biodegradation of typical plastics: from microbial diversity to metabolic mechanisms. Int. J. Mol. Sci. 25:593. doi: 10.3390/ijms25010593
Matsunaga, M., and Whitney, P. J. (2000). Surface changes brought about by corona discharge treatment of polyethylene film and the effect on subsequent microbial colonization. Polym. Degrad. Stab. 70, 325–332. doi: 10.1016/S0141-3910(00)00125-2
Mayer, K. M., and Arnold, F. H. (2002). A colorimetric assay to quantify dehydrogenase activity in crude cell lysates. J. Biomol. Screen. 7, 213–215. doi: 10.1177/108705710200700223
Menczel, J. D., and Prime, R. B. (2008). Thermal analysis of polymers: Fundamentals and applications. Hoboken, NJ: John Wiley & Sons.
Mohammadi, S., Moussavi, G., and Rezaei, M. (2022). Enhanced peroxidase-mediated biodegradation of polyethylene using the bacterial consortia under H2O2-biostimulation. Polymer 240:124508. doi: 10.1016/j.polymer.2021.124508
Morancho, J. M., Ramis, X., Fernández, X., Cadenato, A., Salla, J. M., Vallés, A., et al. (2006). Calorimetric and thermogravimetric studies of UV-irradiated polypropylene/starch-based materials aged in soil. Polym. Degrad. Stab. 91, 44–51. doi: 10.1016/j.polymdegradstab.2005.04.049
Muthukumar, T., Aravinthana, A., and Dineshram, R. (2014). Biodegradation of starch blended high-density polyethylene using marine bacteria associated with biofilm formation and its isolation characterization. J. Microb. Biochem. Technol. 6, 116–122. doi: 10.4172/1948-5948.1000127
Ng, C., Le, T. H., Goh, S. G., Liang, L., Kim, Y., Rose, J. B., et al. (2018). A comparison of microbial water quality and diversity for ballast and tropical harbor waters. PLoS One 13:e0201388. doi: 10.1371/journal.pone.0201388
North, E. J., and Halden, R. U. (2013). Plastics and environmental health: the road ahead. Rev. Environ. Health 28, 1–8. doi: 10.1515/reveh-2012-0030
Ojha, N., Pradhan, N., Singh, S., Barla, A., Khatua, P., Rai, V., et al. (2017). Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Sci. Rep. 7:39515. doi: 10.1038/srep39515
Oliveira, M. M., Proenca, A. M., and Moreira-Silva, E. (2021). Biofilms of Pseudomonas and Lysinibacillus marine strains on high-density polyethylene. Microb. Ecol. 81, 833–846. doi: 10.1007/s00248-020-01618-3
Othman, A. R., Hasan, H. A., Muhamad, M. H., Ismail, N. I., and Abdullah, S. R. S. (2021). Microbial degradation of microplastics by enzymatic processes: a review. Environ. Chem. Lett. 19, 3057–3073. doi: 10.1007/s10311-021-01197-9
Oyewole, O. A., Saidu, M. M., Idris, A. D., Yakubu, J. G., and Bello, A. B. (2022). “Communities of microbial enzymes and biodegradation of persistent environmental pollutants” in Ecological interplays in microbial enzymology. eds. N. R. Maddela, A. S. Abiodun and R. Prasad. (Singapore: Springer Nature Singapore), 247–277.
Patel, S., and Gupta, R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. Nov., Cytobacillus gen. Nov., Mesobacillus gen. Nov., Neobacillus gen. Nov., Metabacillus gen. Nov. and Alkalihalobacillus gen. Nov. Int. J. Syst. Evol. Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Peixoto, J., Silva, L. P., and Kruger, R. H. (2017). Brazilian Cerrado soil reveals an untapped microbial potential for unpretreated polyethylene biodegradation. J. Hazard. Mater. 324, 634–644. doi: 10.1016/j.jhazmat.2016.11.039
Rajandas, H., Parimannan, S., and Sathasivam, K. (2012). A novel FTIR–ATR spectroscopy-based technique for the estimation of low-density polyethylene biodegradation. Polym. Test. 31, 1094–1099. doi: 10.1016/j.polymertesting.2012.07.016
Restrepo-Flórez, J. M., Bassi, A., and Thompson, M. R. (2014). Microbial degradation and deterioration of polyethylene – a review. Int. Biodeterior. Biodegrad. 88, 83–90. doi: 10.1016/j.ibiod.2013.12.014
Ritchie, H., and Roser, M. 2018. Plastic pollution. Our world in data. Oxford: Oxford Martin School. Available online at: https://ourworldindata.org/plastic-pollution (Accessed October 5, 2023).
Romano, R., Raddadi, N., and Fava, F. (2020). Mediterranean Sea bacteria as a potential source of long-chain polyunsaturated fatty acids. FEMS Microbiol. Lett. 367:fnaa132.
Santacruz-Juarez, E., Buendia-Corona, R. E., Ramirez, R. E., and Sanchez, C. (2021). Fungal enzymes for the degradation of polyethylene: molecular docking simulation and biodegradation pathway proposal. J. Hazard. Mater. 411:125118. doi: 10.1016/j.jhazmat.2021.125118
Santo, M., Weitsman, R., and Sivan, A. (2013). The role of the copper-binding enzyme laccase in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 84, 204–210. doi: 10.1016/j.ibiod.2012.03.001
Sardar, R. K. (2025). Identification and biodegradation characterization of high-density polyethylene using marine bacteria isolated from the coastal region of the Arabian Sea, at Gujarat, India. World J. Microbiol. Biotechnol. 41:74. doi: 10.1007/s11274-025-03786-0
Sardar, R. K., Kavita, K., and Jha, B. (2015). Lipopolysaccharide of Marinobacter litoralis inhibits swarming motility and biofilm formation in Pseudomonas aeruginosa PA01. Carbohydr. Polym. 123, 468–475. doi: 10.1016/j.carbpol.2014.12.071
Sarmah, P., and Rout, J. (2019). Cyanobacterial degradation of low-density polyethylene (LDPE) by Nostoc carneum isolated from submerged polyethylene surface in domestic sewage water. Energy Ecol. Environ. 4, 240–252. doi: 10.1007/s40974-019-00133-6
Sasser, M. (2006). Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). Newark, NY: Microbial ID.
Sen, S. K., and Raut, S. (2015). Microbial degradation of low-density polyethylene (LDPE): a review. J. Environ. Chem. Eng. 3, 462–473. doi: 10.1016/j.jece.2014.12.006
Shah, A. A., Hasan, F., and Hameed, A. (2007). Isolation and characterization of poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading bacteria and purification of PHBV depolymerase from newly isolated Bacillus sp. Int. Biodeterior. Biodegrad. 60, 109–115. doi: 10.1016/j.ibiod.2006.12.002
Shah, A. A., Hasan, F., Hameed, A., and Ahmed, S. (2008). Biological degradation of plastics: a comprehensive review. Biotechnol. Adv. 26, 246–265. doi: 10.1016/j.biotechadv.2007.12.005
Shahnawaz, M., Sangale, M. K., and Ade, A. B. (2016). Rhizosphere of Avicennia marina (Forsk.) Vierh. As a landmark for polythene degrading bacteria. Environ. Sci. Pollut. Res. 23, 14621–14635. doi: 10.1007/s11356-016-6616-0
Sheel, A., and Pant, D. (2018). “Chemical depolymerization of polyurethane foams via glycolysis and hydrolysis” in Recycling of polyurethane foams eds. S. Thomas, A. V. Rane, V. K. Abitha, and M. G. Thomas. (Norwich, NY: William Andrew Publishing), 67–75.
Show, P. L., Thangalazhy-Gopakumar, S., and Foo, D. C. (2021). Sustainable technologies for waste reduction and pollutants removals. Clean Techn. Environ. Policy 23, 1–2. doi: 10.1007/s10098-020-02001-y
Sivan, A., Szanto, M., and Pavlov, V. (2006). Biofilm development of the polyethylene-degrading bacterium Rhodococcus ruber. Appl. Microbiol. Biotechnol. 72, 346–352. doi: 10.1007/s00253-005-0259-4
Sorolla-Rosario, D., Llorca-Porcel, J., Pérez-Martínez, M., Lozano-Castelló, D., and Bueno-López, A. (2022). Study of microplastics with semicrystalline and amorphous structure identification by TGA and DSC. J. Environ. Chem. Eng. 10:106886. doi: 10.1016/j.jece.2021.106886
Spina, F., Tummino, M. L., Poli, A., Prigione, V., Ilieva, V., Cocconcelli, P., et al. (2021). Low density polyethylene degradation by filamentous fungi. Environ. Pollut. 274:116548. doi: 10.1016/j.envpol.2021.116548
Sudhakar, M., Doble, M., and Murthy, P. S. (2008). Marine microbe-mediated biodegradation of low- and high-density polyethylene. Int. Biodeterior. Biodegrad. 61, 203–213. doi: 10.1016/j.ibiod.2007.07.011
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, K. M., Fauzi, N. A. M., Kassim, A. S. M., Razak, A. H. A., and Kamarudin, K. R. (2021). Isolation and identification of polystyrene degrading bacteria from Zophobas morio’s gut. Walailak J. Sci. Technol. 18, 9118–9111. doi: 10.48048/wjst.2021.9118
Tiago, G. A., Mariquito, A., Martins-Dias, S., and Marques, A. C. (2023). The problem of polyethylene waste – recent attempts for its mitigation. Sci. Total Environ. 892:164629. doi: 10.1016/j.scitotenv.2023.164629
Tournier, V., Duquesne, S., Guillamot, F., Cramail, H., Taton, D., Marty, A., et al. (2023). Enzymes’ power for plastics degradation. Chem. Rev. 123, 5612–5701. doi: 10.1021/acs.chemrev.2c00644
Van Beilen, J. B., Li, Z., Duetz, W. A., Smits, T. H., and Witholt, B. (2003). Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci. Technol. 58, 427–440. doi: 10.2516/ogst:2003027
Veum, K. S., Acosta-Martinez, V., Lehman, R. M., Li, C., Cano, A., and Nunes, M. R. (2021). “PLFA and EL-FAME indicators of microbial community composition” in Soil health series: Volume 2 laboratory methods for soil health analysis Eds. D. L. Karlen, D. E. Stott and M. M. Mikha. ACSESS (American Society of Agronomy) and Wiley-VCH GmbH: USA. 251–288.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol 173, 697–703.
Wilkes, R. A., and Aristilde, L. (2017). Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: capabilities and challenges. J. Appl. Microbiol. 123, 582–593. doi: 10.1111/jam.13472
Wu, H., Liu, Q., Sun, W., Lu, Y., Qi, Y., and Zhang, H. (2023). Biodegradability of polyethylene mulch film by Bacillus paramycoides. Chemosphere 311:136978. doi: 10.1016/j.chemosphere.2022.136978
Yang, Y., Wang, J., and Xia, M. (2020). Biodegradation and mineralization of polystyrene by plastic-eating superworms Zophobas atratus. Sci. Total Environ. 708:135233. doi: 10.1016/j.scitotenv.2019.135233
Keywords: Metabacillus niabensis, SEM, FTIR, GC–MS, dehydrogenase, peroxidase, polyethylene
Citation: Sardar RK (2025) Discovery and biodegradation characterization of polyethylene by Metabacillus niabensis. Front. Microbiol. 16:1693690. doi: 10.3389/fmicb.2025.1693690
Edited by:
Jinwei Zhang, University of Exeter, United KingdomReviewed by:
Murat Ozdal, Atatürk University, TürkiyeGajendra Joshi, National Institute of Ocean Technology, India
Copyright © 2025 Sardar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raj Kumar Sardar, cmFhai5zYXJkYXIxMTVAZ21haWwuY29t
 Raj Kumar Sardar
Raj Kumar Sardar