- 1Key Laboratory of Agricultural Microbiomics and Precision Application (MARA), Key Laboratory of Agricultural Microbiome (MARA), Guangdong Microbial Culture Collection Center (GDMCC), Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, State Key Laboratory of Applied Microbiology Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
- 2Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (South China), Ministry of Agriculture and Rural Affairs, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Guangdong Engineering Research Center for Litchi, College of Horticulture, South China Agricultural University, Guangzhou, China
Denitrifying bacteria with flocculation capacity were dual-function microorganisms that can simultaneously remove nitrogen (N) and reduce suspended particles in wastewater, providing a sustainable bioremediation strategy. In this study, a novel denitrifying bacterium capable of producing bioflocculants, Thauera sp. JM12B12, was isolated and investigated. The results confirmed that this strain could completely remove NO3−-N and NO2−-N under microaerobic conditions with a low C/N ratio of 5, using lactate as the optimal carbon source. Notably, no other harmful inorganic N species were produced during denitrification, and total N removal efficiency consistently exceeded 93.0%. Optimal denitrification conditions include a pH range of 7–9, salinity of 0–1.5%, temperature of 25–40 °C, and static incubation. Remarkably, this strain synthesized extracellular bioflocculants during NO3−-N removal, achieving 91.4% flocculation efficiency with cell-free supernatant. Genome analyses revealed a complete denitrification pathway (possessing napA, two nirS, norB, nosZ) and 80 bioflocculant-related genes (polysaccharide production and protein secretion), highlighting its dual capacity for N and suspended particle removal. PCR also confirmed key denitrification genes. Therefore, JM12B12 could be a multifunctional microbial agent for N removal and flocculation, offering a sustainable solution for low C/N wastewater treatment, particularly valuable in recirculating aquaculture systems.
1 Introduction
Recirculating aquaculture system (RAS) represented a promising direction for future aquaculture development, offering its energy conservation, water preservation, land efficiency, and no seasonal constraints (Wang F. et al., 2025). With the long-term operation of RASs, the concentrations of nitrate (NO3−-N) in aquaculture water could increase to 100 mg/L or more, and the high concentration of NO3−-N threatened aquatic animals and aquatic ecosystems (Li et al., 2023; Ma et al., 2024). Suspended particles (mainly from residual feed and animal feces) were also a major pollutant in aquaculture water, which directly heightened water turbidity and influenced the survival of aquatic animals (Zhang K. et al., 2025). For instance, an overabundance of suspended particles in aquatic systems could induce mortality events in juvenile chum salmon, primarily attributed to respiratory failure caused by gill clogging from particles (Kishi et al., 2025). Therefore, removal of NO3−-N and suspended particles was of great significance for enhancing the quality of aquatic products.
Biological nitrogen (N) removal has emerged as a cost-effective and environmentally friendly approach to treating N pollution, and traditional N removal relies on nitrifying bacteria and denitrifying bacteria, which require strictly aerobic and anoxic conditions, respectively (Gu et al., 2022). However, to ensure the health and growth of reared animals, the aquatic water usually needs to be maintained in aerobic conditions, which is detrimental to the traditional denitrifying bacteria (Li et al., 2023). Fortunately, the aerobic denitrifying bacteria, which defied traditional metabolic paradigms by reducing NO3−-N or nitrite (NO2−-N) to N2 under aerobic conditions, have been uncovered. These functional bacteria, such as Paracoccus, Pseudomonas, Acinetobacter, and Alcaligenes, have been isolated from a variety of natural environments (Chen Z. et al., 2024; Lan et al., 2023; Li et al., 2025; Wang H. et al., 2025). Moreover, aquatic water has a kind of relatively low carbon-to-N (C/N) ratio, resulting in many aerobic denitrifying bacteria that hardly adapt to the aquatic water environment (Pan et al., 2024). Because of this, the aerobic denitrifying bacteria that achieve efficient denitrification at a low C/N ratio exhibit great application value.
Bioflocculants could reduce the water turbidity by binding suspended particles, and they offered advantages of biodegradability and non-toxicity compared to other flocculants (Show et al., 2024). Bioflocculants were the macromolecular metabolites with flocculation activity produced by microorganisms, primarily consisting of extracellular polymeric substances (EPS), including polysaccharides, proteins, and nucleic acids (Chen W. et al., 2024). Some bioflocculant-producing bacteria have been reported, such as Bacillus, Azoarcus, and Pseudomonas (Alias et al., 2024; Chen L. et al., 2024; Christiaens et al., 2023). Even though several potential bioflocculant-producing strains have already been investigated, studies on denitrifiers possessing this function are still limited at present. Further exploration of bioflocculant-producing denitrifying bacteria was necessary to improve the aquatic water quality.
Thauera was the dominant genus in various types of wastewater treatment systems, such as the sequencing batch biofilm reactor, the sequencing batch reactor, and the moving bed biofilm reactor (Yu et al., 2024; Yuan et al., 2023; Zhang et al., 2024). In recent years, several Thauera strains have been isolated. For instance, Thauera sp. SND5 was capable of removing phosphorus and N through simultaneous nitrification–denitrification and phosphate accumulation (Wang and He, 2020); Thauera sp. RT1901 could achieve denitrification and phosphorus accumulation in both microaerobic and anaerobic environments (Ren T. et al., 2024); Thauera sp. AutoDN2 could remove nitrate and oxidize sulfide to elemental sulfur under autotrophic conditions (Zhang Y. et al., 2025). Strains of Thauera with denitrification capability were commonly reported, however, those simultaneously exhibiting high efficiency denitrification and flocculation functions remain scarce.
In this study, we presented a novel bacterium, Thauera sp. JM12B12, isolated from aquaculture water, exhibited the remarkable ability to perform aerobic denitrification, even at a low C/N ratio, and concurrently produced bioflocculants during the process of NO3−-N removal. The denitrification performance of this strain under various conditions and its flocculation activity during the process of denitrification were investigated. Additionally, JM12B12’s aerobic denitrification pathway and flocculation characteristics were conjectured by analyzing the related functional genes according to the genome annotations. Our findings demonstrated that strain JM12B12 could be a promising candidate for simultaneously removing N and suspended particles from aquaculture water with a low C/N ratio.
2 Materials and methods
2.1 Medium
For preliminary isolation of the aerobic denitrifying bacteria, a denitrification screening medium with low N (DM-L) was employed. The composition of DM-L medium included the following components per liter: sodium succinate 0.25 g, sodium citrate dihydrate 0.25 g, Na2HPO4 1.6 g, KH2PO4 1.0 g, NaCl 0.5 g, NaNO2 0.07 g, KNO3 0.1 g, (NH4)2SO4 0.066 g, MgSO4·7H2O 0.2 g, 0.2% (v/v) trace element solution (TES), and 0.1% (v/v) mixed carbon source solution (CSS). The pH was adjusted to pH 7.2. Solid plates were prepared by supplementing DM-L with 15.0 g/L agar. Before use, the medium (excluding MgSO4·7H2O, TES, and CSS) was sterilized by autoclaving at 121 °C (0.11 MPa) for 30 min. Filter-sterilized MgSO4·7H2O, TES, and CSS were aseptically supplemented to the autoclaved medium. The formulations of TES and CSS followed protocols established in our previous work (Zhang et al., 2022).
2.2 Isolation and identification of aerobic denitrifying bacteria
Samples collected from Penaeus vannamei aquaculture water in Jiangmen City, Guangdong Province, China, were diluted (10−1 to 10−4), and 0.1 mL from 10−2 to 10−4 dilutions was spread on DM-L agar, incubated aerobically at 30 °C for 2–7 days. Colonies with distinct morphology were purified and stored at −80 °C in sterile physiological saline supplemented with 25.0% (v/v) glycerol. For denitrifying bacteria isolation, selected strains were grown statically at 30 °C in DM-L medium with 5.0 mg/L NO2−-N, and residual NO2−-N was measured using the Griess reaction.
Strain JM12B12 was cultured on LB agar at 30 °C for 48 h, with colonies observed macroscopically and cells via electron microscopy (H7650, Hitachi). Bacterial genomic DNA was extracted using the HiPure Bacterial DNA Kit (Magen Biotech., China). The 16S rRNA genes were PCR-amplified with primers 27F and 1492R, sequenced by GENEWIZ (Suzhou, China), and aligned against the EzBioCloud database (Chalita et al., 2024). Strain JM12B12 showed the highest NO2−-N removal efficiency and was further analyzed for morphology, phylogeny, and genome. The 16S rRNA sequences were aligned using MAFFT v7.526 under the L-INS-i iterative refinement (Rozewicki et al., 2019). Maximum-likelihood (ML) phylogenetic tree was reconstructed with IQ-TREE v2.1.2 with integrated ModelFinder for evolutionary model selection, selecting the best model via Bayesian Information Criterion and assessing node support with 1,000 bootstrap replicates (Hoang et al., 2018; Minh et al., 2020). The phylogenetic tree was visualized using MEGA 11 (Tamura et al., 2021).
2.3 Nitrogen removal characteristics
Our results showed that JM12B12 exhibited more effective capability in the removal of NO2−-N compared to other isolates. Therefore, the effects of various conditions on the N removal efficiency of JM12B12 were investigated. Each variable was adjusted independently, and the optimal conditions were applied in subsequent experiments.
A single colony of JM12B12 was grown in LB medium for 12 h, washed, and resuspended to OD600 of 1.0. The suspension was inoculated into media to study N removal, testing sodium acetate, citrate, succinate, lactate, glucose, and sucrose as carbon sources at C/N ratios of 1 to 20. The basal medium (BM) without carbon and N sources was formulated as (per liter): Na2HPO4 1.6 g, KH2PO4 1.0 g, NaCl 0.5 g, MgCl2·6H2O 0.1 g, D-biotin 5.0 mg, cobalamin 5.0 mg. Furthermore, the effects of various culture conditions on the denitrification of JM12B12 were investigated, including initial pH (5–11), concentrations of NaCl (0–3.0%), temperature (20–45 °C), and shaking speeds (0–200 rpm). The BM was supplemented with NO3−-N and sodium lactate as sole N and carbon sources, and the C/N ratio was adjusted to 10. After 48 h of incubation, samples were analyzed for OD600, and the relevant culture supernatants were used for the measurement of N concentrations (NO2−-N, NO3−-N, and NH4+-N). Uninoculated media were used as controls.
To evaluate JM12B12’s denitrification process and N balance, sodium lactate-supplemented BM with either NO2−-N (BM1) or NO3−-N (BM2) was used. During incubation, samples were taken to measure OD600, and the relevant supernatants were used for detecting concentrations of NO2−-N, NO3−-N, and NH4+-N. Additionally, total N (TN-N) and intracellular N (CN-N) concentrations were measured at 0 h and 48 h. Uninoculated media were used as controls.
2.4 Flocculating activity
Strain JM12B12 was cultured in the BM with NO3−-N and sodium lactate as the sole N and carbon source, respectively. After 48 h of incubation, the bacterial suspension (BS) was centrifuged at 12,000 g for 10 min to obtain cell-free supernatant (CFS) and cells (CE). The flocculation capabilities of BS, CFS, and CE were determined by the kaolin suspension method (Chen L. et al., 2024). Briefly, the experimental flocculation system comprised 2.0 mL sample (cells were resuspended with the physiological saline solution), 3.0 mL 1.0% CaCl2 (w/v), and 95.0 mL 4.0 g/L kaolin suspension, with pH maintained at 7.5 via HCl and/or NaOH adjustment. The mixture was stirred at 200 rpm for 2 min, then stirred at 50 rpm for 5 min, and then allowed to settle for 5 min. The 3 mL of the supernatant was aspirated from a consistent depth of 1 cm below the air-liquid interface. The optical density measurement was conducted immediately post-collection using a calibrated spectrophotometer at 550 nm wavelength. The flocculating efficiency (FloE) was calculated according to the equation: FloE (%) = (A-B)/A × 100, where A represented the absorbance values of samples (BS, CFS, and CE) at 550 nm, and B represented the reference absorbance values (sterile culture medium for BS and CFS; physiological saline for CE) at 550 nm.
2.5 Complete genome sequencing, annotations, and analysis
The complete genome of strain JM12B12 was sequenced at Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (China) using a hybrid approach combining Nanopore PromethION (Oxford Nanopore, Oxford, UK) and Illumina HiSeq 2,500 (Illumina, Inc., San Diego, CA, USA). For Illumina sequencing, genomic DNA was fragmented to 400–500 bp using Covaris M220, and libraries were prepared with the NEXTFLEX Rapid DNA-Seq kit. For Nanopore sequencing, DNA fragments were repaired, purified, and ligated with sequencing adapters from the SQK-LSK kit before library preparation and sequencing. Raw Illumina reads were filtered using fastp v0.23.0, while Nanopore reads were processed (basecalling, demultiplexing, trimming) with a minimum Q score of 7. Hybrid assembly was performed using Unicycler v0.4.8, and Pilon v1.22 was used for error correction. The complete genome was reconstructed by integrating data from both platforms.
GeneMarkS v4.3, tRNA-scan-SE v2.0.12, and barrnap v0.9 were used to predict coding sequences (CDS), tRNA, and rRNA, respectively. The predicted CDS were annotated using COG (202006) and KEGG (202209) databases via sequence alignment tools. Core genome circular maps, COG, and KEGG analyses were conducted on Majorbio’s Cloud platform (Han et al., 2024). The genome was also annotated with RAST v2.0 under the Classic RAST scheme. Digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) values were calculated using Genome-to-Genome Distance Calculator 3.0 (formula 2) and FastANI (Jain et al., 2018; Meier-Kolthoff et al., 2013). The phylogenomic tree was constructed using Up-to-date Bacterial Core Gene sets (UBCGs) (Riesco and Trujillo, 2024).
2.6 Detection of the key genes for denitrification
Genomic DNA of strain JM12B12 was extracted using the method described in Section 2.2. Primer sets napA_5F/ napA_3R, nirS1_5F/ nirS1_3R, nirS2_5F/ nirS2_3R, norB_5F/ norB_3R, and nosZ_5F/ nosZ_3R were utilized to amplify the denitrification genes from JM12B12 (Supplementary Table S2). PCR amplification was performed using 2 × Phanta Max Master Mix (Vazyme Biotech, Nanjing, China) in a total reaction volume of 20 μL, comprising the following components: 10 μL of 2 × Phanta Max Master Mix, 0.4 μL of primers (10 μM each), 1 μL of DNA template (50 ng/μL), and 8.2 μL of ddH2O. PCR reaction program was as follows: 95 °C for 5 min; 30 cycles consisting of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 2 min 30 s; followed by 72 °C for 10 min. The PCR products were verified by comparing with Trans2K® Plus II DNA Marker (TransGen Biotech, Beijing, China) electrophoretically using a 1.5% agarose gel.
2.7 Analytical methods
NO2−-N, NO3−-N, and NH4+-N concentrations were quantified using N-(1-naphthyl) ethylenediamine dihydrochloride, ultraviolet spectrophotometry, and Nessler’s reagent photometry at the wavelength of 420 nm, respectively (Baird et al., 2017). TN-N and CN-N concentrations were quantified using alkaline potassium persulfate digestion followed by spectrophotometric detection at 220 nm and 275 nm, respectively (Baird et al., 2017). Standard curves for the determination of N concentrations were shown in Supplementary Figure S1. OD600 was determined at a wavelength of 600 nm. N removal efficiency was calculated by the following formula: N removal efficiency (%) = ([Nx]i - [T3N]f)/[Nx]i × 100, where [Nx]i is the initial NO3−-N, NO2−-N, and NH4+-N concentration, respectively; [T3N]f is the sum of NO3−-N, NO2−-N, and NH4+-N concentrations. All experiments were carried out in quadruplicate, and results were expressed as the mean of four replicates ± standard deviation (mean ± SD).
3 Results and discussion
3.1 Isolation and identification of JM12B12
There were 10 isolates obtained, and strain JM12B12 exhibited excellent performance in removing NO2−-N (Supplementary Table S1). The colonies of strain JM12B12 appeared off-white, opaque, with a smooth surface, regularly circular, and neat-edged, measuring approximately 1 mm in diameter (Figure 1a). Cells of this strain were observed to be rod-shaped (0.7–0.9 × 1.9–2.5 μm) with a single polar flagellum (Figure 1b). The 16S rRNA gene sequence of JM12B12 obtained through PCR amplification (1,400 bp) showed complete identity with the corresponding sequences extracted from its genomic DNA (1,537 bp). The complete 16S rRNA gene sequence was deposited in the NCBI GenBank database under accession number PP716610. The sequence comparison showed that JM12B12 shared the highest similarity with T. chlorobenzoica 3CB-1 (99.2%), followed by strains T. selenatis ATCC 55363 (99.2%), T. aminoaromatica S2 (99.1%), T. phenylacetica B4P (99.0%), and T. mechernichensis TL1 (98.9%). Phylogenetic reconstruction based on the 16S rRNA sequences revealed that JM12B12 formed a distinct clade within the genus Thauera cluster, supported by 73% bootstrap values (Figure 1c). Therefore, JM12B12 was taxonomically assigned to the genus Thauera.

Figure 1. Comprehensive morphological, phylogenetic, and comparative genomic characterization of Thauera sp. JM12B12. (a) Colonies. (b) Cell morphology. (c) ML tree from 16S rRNA sequences of JM12B12 and related type strains. (d,e) ANI and dDDH similarities between JM12B12 and strains (orange-circled) with high 16S rRNA sequence similarity.
3.2 Nitrogen removal characteristics of JM12B12
3.2.1 Effect of carbon source and C/N on nitrogen removal efficiency
To evaluate N removal efficiencies, strain JM12B12 was tested with different carbon sources at a fixed C/N ratio of 10. As shown in Figure 2a, sodium lactate supported the highest cell growth across all N sources (NO3−-N, NO2−-N, and NH4+-N). The maximum NO3−-N removal efficiency (98.9%) was achieved using acetate or lactate as the carbon source, consistent with the performance of Thauera sp. SND5 under identical carbon conditions (Wang and He, 2020). Complete NO2−-N removal occurred with succinate or lactate, but the NO2−-N removal efficiency was only 24.3% when acetate served as the carbon source. The results indicated that JM12B12 exhibited distinct optimal carbon source preferences in denitrification processes aimed at removing NO2−-N as opposed to NO3−-N. This metabolic divergence was likely attributed to differential enzymatic activities and energy-yielding efficiencies associated with the two N sources during the dissimilatory nitrate reduction pathway (Lu et al., 2024). Unfortunately, JM12B12 exhibited relatively weak NH4+-N removal capacity, achieving a maximum removal efficiency of only 30.7% when lactate served as the carbon source. Notably, no inorganic N accumulation was observed when any single N source was removed. Based on these results, sodium lactate was selected as the optimal carbon source for subsequent experiments.
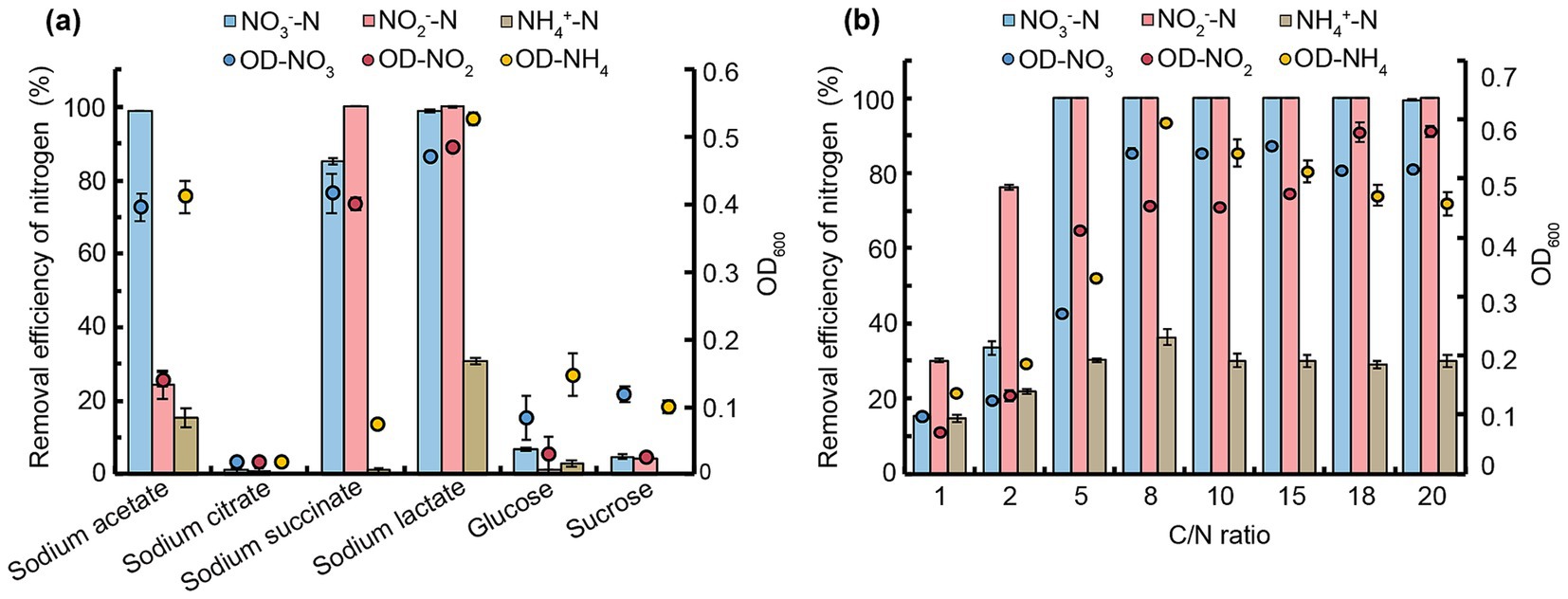
Figure 2. Impact of carbon sources and C/N ratios on NO3−-N, NO2−-N, and NH4+-N removal by strain JM12B12. (a) Carbon sources. (b) C/N ratios. Data shown as mean ± SD (n = 4). OD-NO3, OD-NO2, and OD-NH4 represent OD600 values with NO3−-N, NO2−-N, and NH4+-N as sole nitrogen sources, respectively.
The C/N ratio significantly influenced bacterial growth, denitrification efficiency, and environmental adaptability (Gu et al., 2022). To determine the optimal ratio for JM12B12, we tested C/N ratios from 1 to 20 (Figure 2b). For NO3−-N removal, the efficiency improved from 15.3 to 100% when the ratio increased from 1 to 5. Complete NO3−-N removal occurred at ratios ≥8, with 99.5% efficiency maintained at a ratio of 20. No NO2−-N or NO3−-N accumulated during this process. For NO2−-N removal, the efficiency improved from 30.0 to 100% when the ratio increased from 1 to 5. And complete NO2−-N removal was consistently observed across C/N ratios of 8 to 20. No NO3−-N or NH₄⁺-N accumulation during this process. In general, the majority of aerobic denitrifiers exhibited high denitrification efficiency at C/N ratios of 5 or above, such as P. balearica strain RAD-17 (optimal ratio of 7.5), P. denitrificans XW11 (optimal ratio of 10), and Pseudomonas sp. Y15 (optimal ratio of 15) (Chen J. et al., 2024; Wu Y. et al., 2024; Zhang et al., 2023b). The required C/N ratio of JM12B12 was significantly lower than that of these strains, indicating that JM12B12 possessed efficient denitrification capability under low C/N ratio conditions. For NH4+-N removal, the efficiency rose slightly from 14.7 to 30.2% as the C/N ratio increased from 1 to 5, remaining stable (ranging from 29.0 to 36.3%) at ratios of 5 to 20. These results suggested that JM12B12 exhibited inefficient NH4+-N removal. It was suggested that this strain removed NH4+-N primarily through assimilation rather than via the heterotrophic nitrification-aerobic denitrification pathway, a finding consistent with the behavior observed in denitrifiers such as Klebsiella sp. TSH15, P. denitrificans R-1, and P. mosselii 9–1 (Cai et al., 2023; Ren J. et al., 2024; Sun et al., 2024).
3.2.2 Effect of various environmental factors on denitrification
Initial pH critically influenced bacterial denitrification by regulating enzyme activity (Yue et al., 2023). As shown in Figure 3a, JM12B12 achieved near-complete NO3−-N removal (100, 100, 99.3%) at pH 7 to 9, but efficiency plummeted to 5.5 and 5.2% at pH 6 and 10, respectively, with negligible removal observed at pH 5 and 11. It was suggested that the slightly acidic and strongly alkaline environment negatively affected denitrification enzymic activities and caused the N metabolic imbalance of bacteria (Lan et al., 2023; Pan et al., 2023). JM12B12 exhibited optimal NO3−-N removal efficiency at pH 7–9 (neutral to slightly alkaline), consistent with P. mendocina A4 and Pelomonas puraquae WJ1 (Shu et al., 2024; Wu T. et al., 2024).
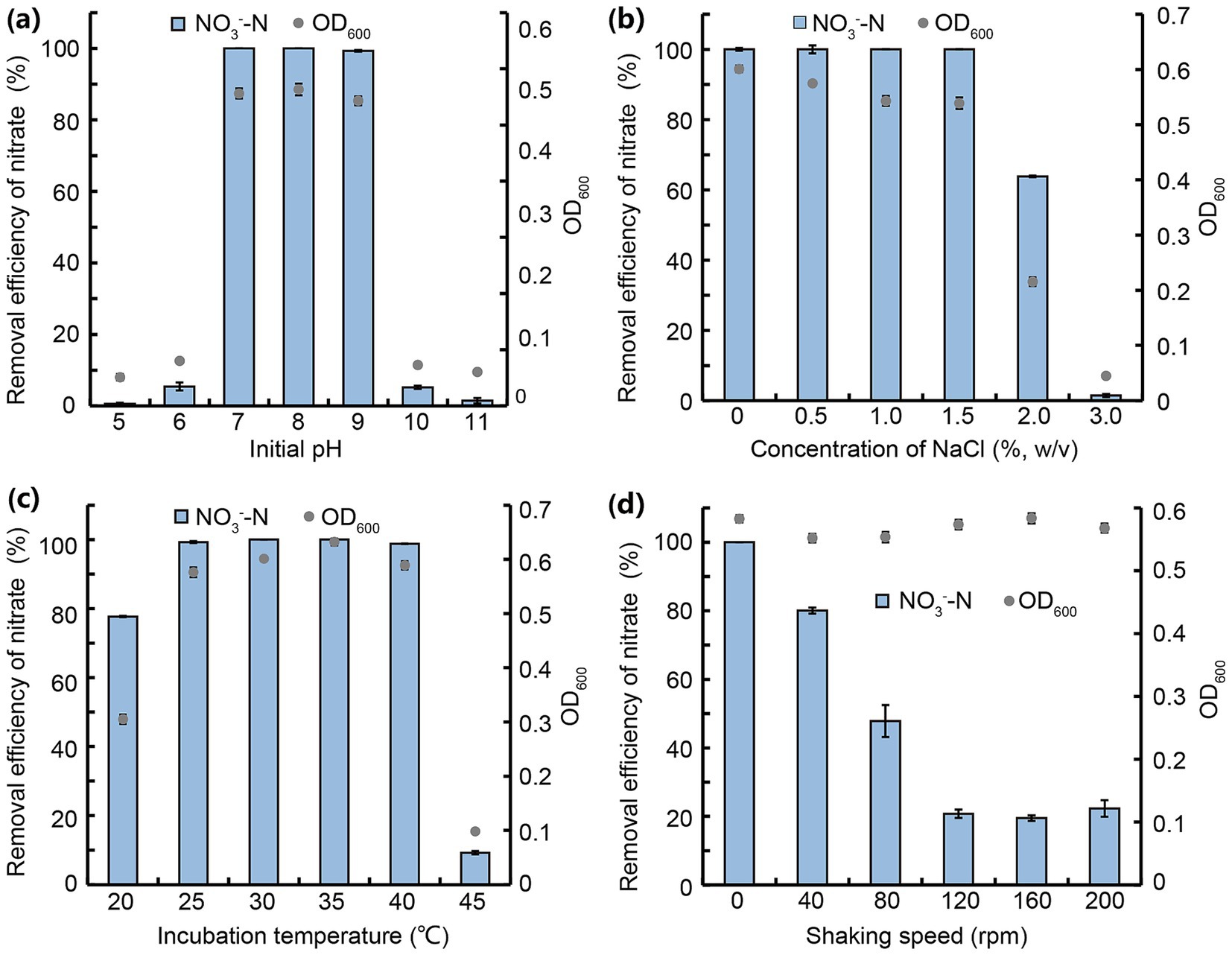
Figure 3. NO3−-N removal by strain JM12B12 in denitrification media across varied conditions. (a) Initial pH. (b) Concentration of NaCl. (c) Incubation temperature. (d) Shaking speed. Data shown as mean ± SD.
Salinity significantly influenced cellular osmotic pressure and microbial enzymatic activity (Hu et al., 2023). As shown in Figure 3b, JM12B12 exhibited complete NO3−-N removal (0–1.5% NaCl) without detectable NO2−-N or NH4+-N accumulation, a performance contrasting with other reported denitrifiers that exhibit metabolic inhibition under similar high-salinity (1.3%) conditions (Li et al., 2022). However, the NO3−-N removal efficiency was reduced to 63.8% when NaCl concentration was elevated to 2.0% (also without detectable NO2−-N or NH4+-N accumulation), and it was completely inhibited at 3.0%. For JM12B12, high salt might inhibit the activity of nitrate reductase rather than that of nitrite reductase. Based on the OD600 values, JM12B12 was unable to grow at 3.0% NaCl. Therefore, we hypothesized that high salt might lead to the death of JM12B12 by altering cellular osmotic pressure, ultimately resulting in its poor denitrification performance. These results indicated that JM12B12 might be tolerant of a certain salinity, but the high salinity (above 2.0%) had a significant and negative impact on the NO3−-N reduction capability of this strain. Based on farmers’ experiences and researchers’ findings, pond water for breeding shrimp should maintain proper salinity. For instance, Kumar et al. (2023) reported that shrimp cultured at a salinity of 15 g/L exhibited better growth, survival, and feed efficiency. Therefore, JM12B12 possessed promising potential for removing excessive N from the culture water of shrimp.
Temperature, as an important influencing factor on denitrification, delayed the regulation of denitrification key genes and inhibited enzymatic activity (Yang et al., 2020). As shown in Figure 3c, JM12B12 completely removed NO3−-N at 30 °C and 35 °C with no NO2−-N or NH4+-N accumulation. It also achieved 99.3 and 98.8% removal efficiencies at 25 °C and 40 °C, respectively. But the NO3−-N removal efficiencies dropped significantly to 77.7% at 20 °C and 9.2% at 45 °C. This phenomenon might be attributed to the suppression of denitrifying bacterial enzyme activities, cell proliferation, and metabolic processes under conditions of excessively low or high temperature (Liao et al., 2021). Our results indicated that JM12B12 adapted effectively to a certain temperature range (25–40 °C), consistent with the performance of most denitrifying bacteria (Wang et al., 2024; Yang et al., 2023).
To investigate the effect of DO concentration on the denitrification of JM12B12, the NO3−-N removal performance of this strain under varying shaking speed conditions was evaluated. As shown in Figure 3d, JM12B12 completely removed NO3−-N with no NO2−-N or NH4+-N accumulation at 0 rpm. At 40 and 80 rpm, removal efficiencies dropped to 80.0 and 47.8%, with trace NO2−-N accumulation. At 120–200 rpm, shaking speed had no further impact on NO3−-N removal, but efficiency fell to 20.9%, accompanied by slight NO2−-N accumulation (~6.6 mg/L). Our results demonstrated that the DO concentration influenced the denitrification process of JM12B12. However, this influence was less pronounced compared to that on traditional denitrifying bacteria, which are strictly anaerobic (Lou et al., 2023). Therefore, JM12B12 was a microaerobic denitrifying bacterium that exhibited high denitrification efficiency under low concentrations of DO, which was inconsistent with most aerobic denitrifiers, such as P. versutus JUST-3, Stutzerimonas stutzeri os3, and Bacillus sp. L2 (Li et al., 2024; Wang F. et al., 2025; Yan et al., 2025). Strain JM12B12 could potentially be utilized for N removal from the aquaculture wastewater in biofilters (without aeration devices) within RASs.
3.2.3 Denitrification process and nitrogen balance
Growth and denitrification of JM12B12 were investigated JM12B12, showing a strong correlation between its growth rates and N substrate removal rates. As shown in Figure 4a, the OD600 values increased from 0.1 to 0.7, and the NO2−-N was completely removed after 20 h of incubation. Correspondingly, the average NO2−-N removal rate of this strain was determined to be 4.2 mg/L/h, exceeding that of Halomonas sp. DN3 (0.95 mg/L/h), Peribacillus sp. EM-C3 (2.2 mg/L/h), and Glutamicibacter arilaitensis EM-H8 (3.9 mg/L/h) (Chen et al., 2023; Liang et al., 2023; Xie et al., 2023), but lower than that of A. johnsonii EN-J1 (6.8 mg/L/h) (Zhang et al., 2023a). As shown in Table 1, when NO2−-N served as the sole N source, the TN-N concentration decreased from 84.0 mg/L to 5.2 mg/L, achieving a TN-N removal efficiency of 93.8% (ΔTN-N = 78.8 mg/L). Concurrently, the CN-N concentration increased from 2.9 mg/L to 12.8 mg/L (ΔCN-N = 9.9 mg/L), implying that 11.8% of the initial TN-N (9.9 mg/L of 84.0 mg/L) was assimilated by JM12B12 for biomass synthesis. 82.0% of the initial TN-N (68.8 mg/L) might be converted to gaseous N products (N2, N2O, NO), accounting for the observed loss from the culture medium. This suggested that the N removal process was dominated by the denitrification pathway, with a minor fraction diverted to microbial growth.
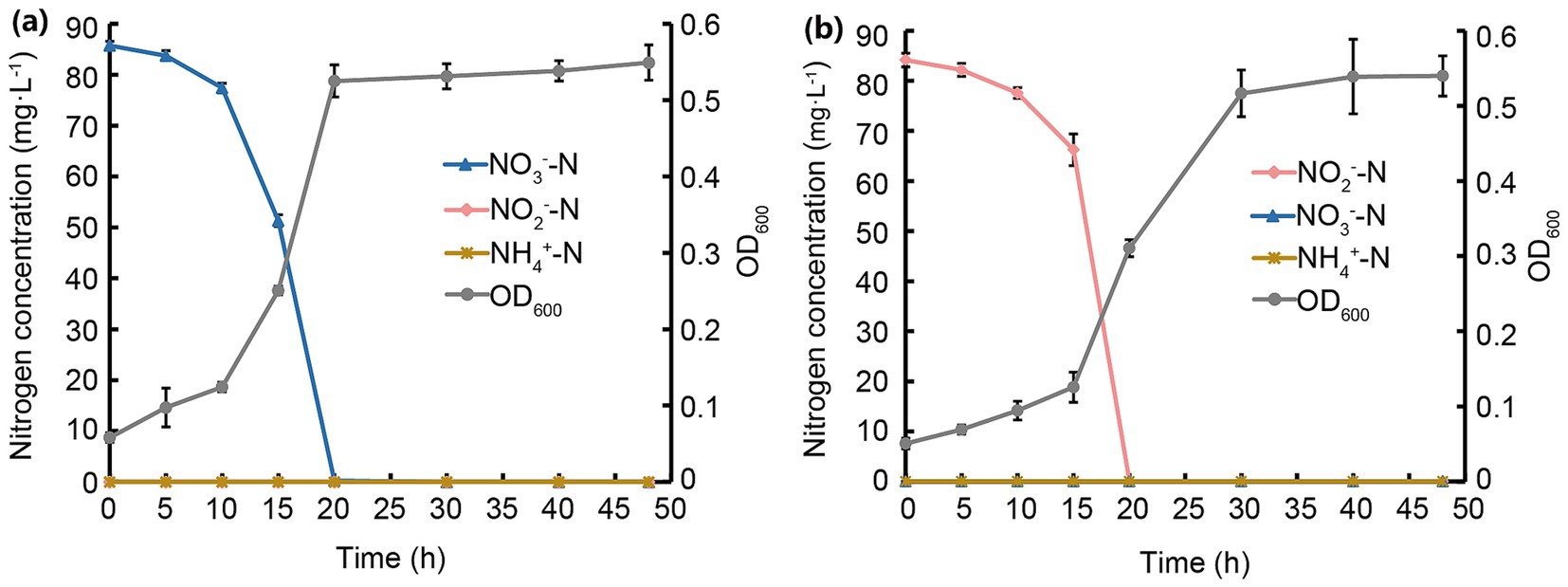
Figure 4. Processes of NO3−-N and NO2−-N removal and growth of strain JM12B12. (a) NO3−-N. (b) NO2−-N. Data shown as mean ± SD.
As shown in Figure 4b, JM12B12 entered the stationary growth phase after 20 h of incubation (OD600 = 0.9), coinciding temporally with the complete removal of NO3−-N. The average NO3−-N removal rate was 4.3 mg/L/h during the first 20 h, which was similar to that of G. arilaitensis EM-H8, and significantly higher than that of Halomonas sp. DN3 (1.9 mg/L/h), Pseudomonas sp. G16 (2.9 mg/L/h), and Stutzerimonas sp. X87 (3.8 mg/L/h) (Gao et al., 2023; Huang et al., 2025; Xie et al., 2023). Notably, no significant NO2−-N or NH4+-N accumulation was observed throughout the N removal process, a distinguishing feature of JM12B12 that underscores its performance of no secondary pollution. This phenomenon differed from that observed in some aerobic denitrifiers, which typically accumulated NO2−-N or NH4+-N during NO3−-N removal process (Huang et al., 2023; Mao et al., 2025). As shown in Table 1, when NO3−-N served as the sole N source, the TN-N concentration decreased from 84.1 mg/L to 5.8 mg/L, achieving a TN-N removal efficiency of 93.1% (ΔTN-N = 78.3 mg/L). The CN-N concentration increased from 3.0 mg/L to 13.3 mg/L (ΔCN-N = 10.3 mg/L), implying that 12.2% of the initial TN-N was assimilated by JM12B12 for biomass synthesis. 80.9% of the initial TN-N (68.4 mg/L) might be converted to gaseous N products, accounting for the observed loss from the culture medium.
Overall, Thauera sp. JM12B12, a micro-aerobic denitrifying bacterium, demonstrated higher removal efficiencies for NO2−-N, NO3−-N, and TN-N during denitrification compared to other denitrifying bacteria, such as P. puraquae WJ1, A. johnsonii EN-J1, and Pseudomonas sp. B-1 (Lan et al., 2023; Wu T. et al., 2024; Zhang et al., 2023a). Excess NO3−-N was prevalent in the intensive RASs, necessitating cost-effective and eco-friendly wastewater treatment for sustainable aquaculture. Therefore, JM12B12 exhibited significant potential as a denitrifying bacterium for the treatment of aquaculture effluents, especially within RASs.
3.3 Characterization of genes in the denitrification pathway of JM12B12
Denitrifying bacteria require an array of reductases to accomplish the denitrification pathway. According to RAST annotation of genome, 36 genes were associated with denitrification and nitrite/nitrate assimilation pathways (Figure 5a). Dissimilatory nitrate reduction to nitrite was catalyzed by two types of enzymes: the membrane-bound nitrate reductase (Nar) and the periplasmic nitrate reductase (Nap), and Nar and Nap reductases were often associated with anaerobic and aerobic denitrification, respectively (Hu et al., 2023). A napFDAGHBC gene cluster (gene_3698/3697/3696/3695/3694/3693/3692) encoding NapA and relevant enzymes was identified in JM12B12, which was consistent with Shewanella oneidensis MR-1 (Liu et al., 2021). Dissimilatory nitrite reduction to nitric oxide was catalyzed by two structurally distinct enzymes: a cytochrome cd1-dependent nitrite reductase (encoded by nirS) and a copper-containing nitrite reductase (encoded by nirK). Interestingly, two nirS gene (gene_1573 and gene_1693) sequences with a similarity of 66.2% were found in the genome of JM12B12, which was inconsistent with Bradyrhizobium diazoefficiens (Pacheco et al., 2022). Previous studies have demonstrated that nirS-type denitrifying bacteria possessed robust metabolic systems for energy conservation, facilitating their survival under environmental stresses (Ming et al., 2024). The genome of JM12B12 harbored a functionally enriched cluster of energy metabolism-associated genes, demonstrating evolutionary adaptations critical for environmental persistence. The key functional genes for reducing NO to nitrous oxide (N2O) (norB, gene_1714) and reducing N2O to N2 (nosZ, gene_0523) were also identified.
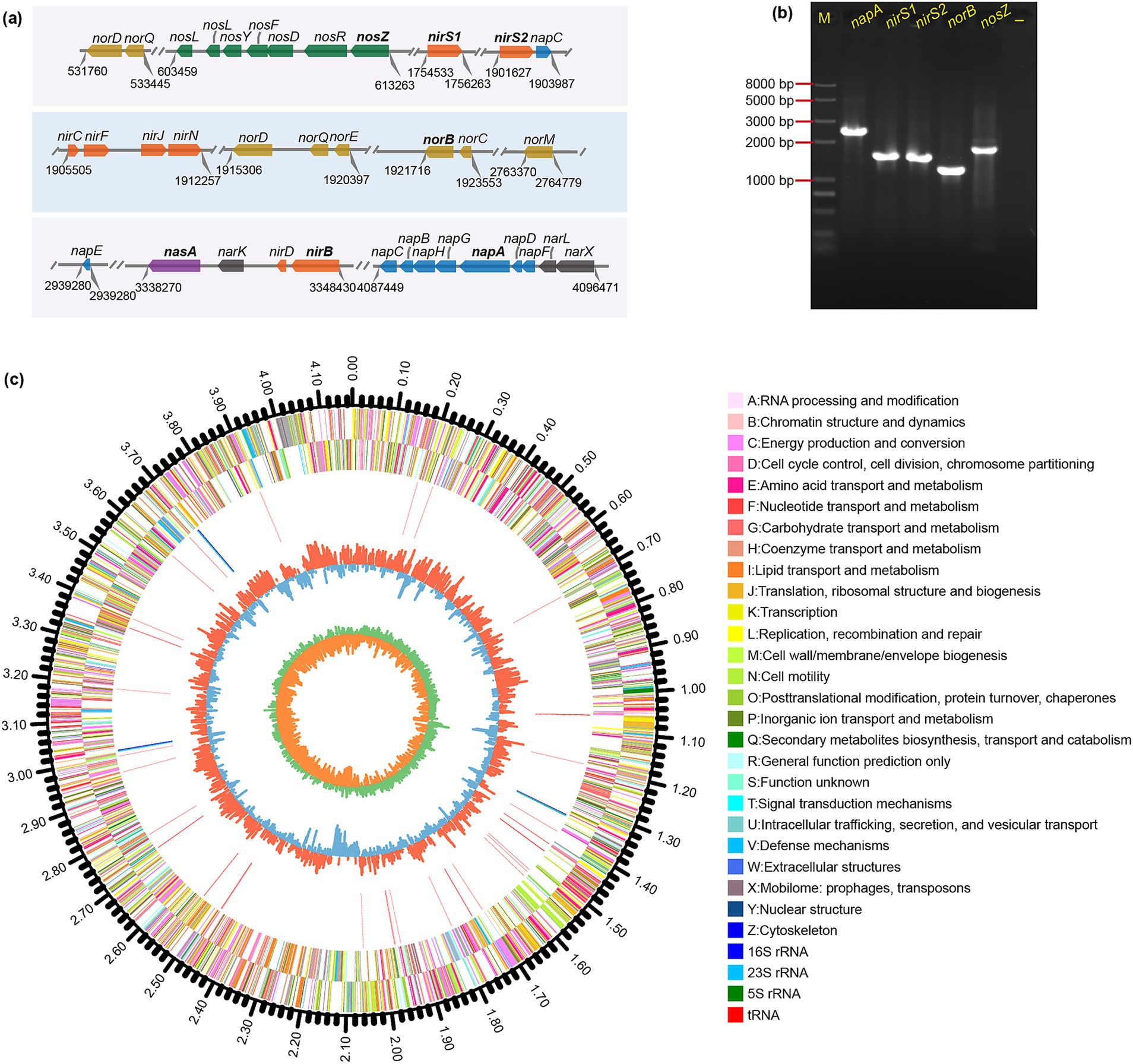
Figure 5. Genomic circular map and functional genes associated with aerobic denitrification in strain JM12B12. (a) Denitrification gene layout, with parallel lines marking sequence breaks and numbers indicating loci. (b) PCR results for aerobic denitrification key genes. -, negative control. (c) Circos plot of the closed circular genome, featuring concentric layers: genome size scale, CDS on both strands (color-coded by COG categories), rRNA/tRNA positions, GC content, and GC skew.
Furthermore, the key genes involved in denitrification were amplified via PCR from the genomic DNA of JM12B12. As shown in Figure 5b, the napA, nirS1, nirS2, norB, and nosZ genes were determined via agarose gel electrophoresis. The results were consistent with the predicted sizes of the relevant functional genes within the JM12B12 genome assembly. Overall, the strain JM12B12 harbored all the denitrification genes, which indicated its potential for complete N removal through the denitrification pathway: NO3−-N → NO2−-N → NO→N2O → N2.
3.4 Flocculation characterization of JM12B12
Bioflocculants produced by different bacteria were various in their distribution. Some were secreted into the extracellular environment, and others were tightly adhered to the cellular surface. To investigate the flocculation characterization of JM12B12, the BS, CFS, and CE were obtained after it grew in the denitrification medium. As shown in Figure 6, the flocculation efficiencies of BS, CFS, and CE were 83.7, 91.4, and 21.8%, respectively. The results indicated that the flocculants produced by JM12B12 were found mainly in the cell-free supernatant, consistent with that of strains Stenotrophomonas pavanii GXUN74707, Pseudomonas sp. XD-3, and Providencia huaxiensis OR794369.1 (Chen L. et al., 2024; Qin et al., 2024; Selepe and Maliehe, 2024). Bioflocculants produced by bacteria were found to be EPS, which were mainly composed of polysaccharides, protein polymers, glycoproteins, etc. (Selepe and Maliehe, 2024). Therefore, JM12B12 was a novel denitrifying bacterium capable of producing bioflocculants, suggesting that it possesses important features in practical applications of bioremediation and wastewater treatment.
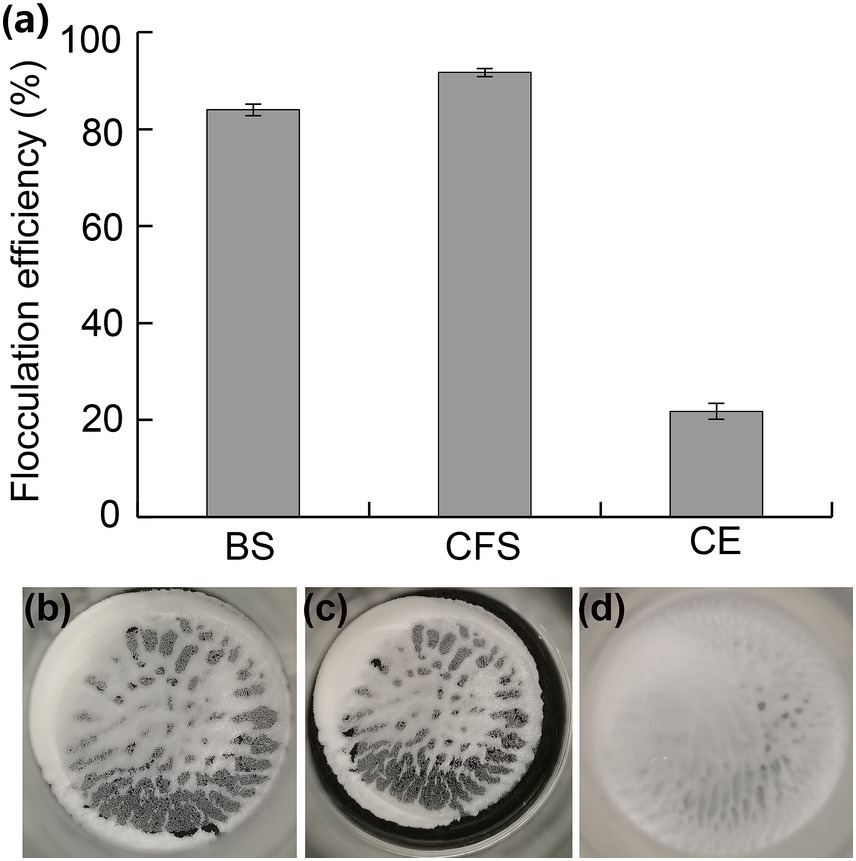
Figure 6. Flocculating activity of bacterial suspension (BS), cell-free supernatant (CFS), and cells (CE) of strain JM12B12. (a) Flocculation efficiencies. (b–d) Images of kaolin flocs with BS, CFS, and CE, respectively. Data shown as mean ± SD.
3.5 Profiling of potential flocculation genes based on the genome of JM12B12
Our results indicated that JM12B12 was capable of producing extracellular bioflocculants. In general, bacterial bioflocculants were composed of EPS, which included polysaccharides, proteins, extracellular nucleic acids, and lipids, and extracellular polysaccharides (exopolysaccharides) constituted the dominant component of EPS (Flemming et al., 2025; Vandana and Das, 2023). Therefore, the genes associated with exopolysaccharide biosynthesis and protein secretion systems were identified by analyzing the genome annotations of JM12B12. Exopolysaccharides production was a multistage process where polysaccharides were intracellularly synthesized and exported outside (Stephens et al., 2023). According to analyses of annotations, there were 31 genes related to polysaccharide biosynthetic, polysaccharide export, and biopolymer transport processes, such as genes alg, rfb, udg, wbp, eps, exb, and tol, etc. (Supplementary Table S3). These genes might be essential for exopolysaccharide production of JM12B12. Extracellular proteins, one of the key amphiphilic macromolecules within EPS, played a crucial role in governing flocculating ability (He et al., 2024). Secretion systems were protein export machines that enable bacteria to exploit their environment through the release of protein effectors (Lauber et al., 2024). There were 49 genes identified as components of the bacterial protein secretion machineries, encompassing the Sec (general secretory pathway) and Tat (twin-arginine translocation) systems, along with Type I, II, III, IV, and VI secretion systems (Supplementary Table S4). Overall, the genes involved in exopolysaccharides production and protein secretion systems might be the potential functional genes associated with the flocculating activity of JM12B12. However, the mechanism of bioflocculant synthesis in this bacterium remains to be fully clarified in the future.
3.6 Comparative genomic analysis and metabolic reconstruction of JM12B12
The circus map of genome characteristics of JM12B12 was comprehensively displayed in Figure 5c. The genomic analysis of JM12B12 revealed a single circular chromosome spanning 4,171,389 bp, characterized by a GC content of 67.9%. Notably, no plasmid was identified in this bacterium. The absence of plasmid reduced the probability of genes (such as genes related to antimicrobial resistance) transfer within strain JM12B12, potentially resulting in enhanced safety compared to the plasmid-harboring bacteria. Genome data has been deposited in the NCBI GenBank database under the accession number CP154859. Compared with related strains in the genus Thauera, JM12B12 had ANI values of 84.2–87.8% and dDDH values of 26.3–31.4% (Figures 1d,e), which were all below the classical standard species delineation threshold values of 95.0 and 70%, respectively (Goris et al., 2007; Richter and Rosselló-Móra, 2009). The phylogenomic tree constructed using UBCGs indicated that JM12B12 fell in a large clade with members of the genus Thauera, which was consistent with the phylogenetic relationships inferred from 16S rRNA gene sequence analysis (Supplementary Figure S2). Moreover, JM12B12 together with T. aminoaromatica S2 and T. phenylacetica B4P formed a clade supported by the bootstrap value of 100%. Phylogenomic analyses incorporating dDDH values, ANI values, and phylogenetic reconstruction conclusively confirmed that strain JM12B12 was a novel species in the genus Thauera (Riesco and Trujillo, 2024).
Aggregate annotation results indicated that a total of 3,771 genes, including 3,707 protein-coding sequences (CDS), 55 tRNA genes, and 9 rRNA genes, were identified. The metabolic pathways were analyzed based on genome annotations. The RAST annotation showed that there were 363, 301, 300, and 277 genes associated with ‘Amino acids and derivatives’, ‘Cofactors, vitamins, prosthetic groups, pigments’, and ‘Carbohydrates’, respectively (Supplementary Figure S3). The KEGG annotation showed that the top four pathways with the most enriched number of genes were ‘Global and overview maps’, ‘Energy metabolism’, and ‘Amino acid metabolism’, respectively (Supplementary Figure S4). The COG annotation revealed that 287, 282, 282, and 259 genes were categorized under the functional classes ‘Energy production and conversion’, ‘Amino acid transport and metabolism’, and ‘Signal transduction mechanisms’, respectively (Supplementary Figure S5). The reconstruction of this bacterium’s carbon metabolic pathways based on genome annotation revealed that the tricarboxylic acid cycle (TCA), glyoxylate cycle (GAC), glycolysis pathway (Embden-Meyerhof-Parnas, EMP), and gluconeogenesis pathway (GNG) were all complete. These carbon metabolic pathways could supply electrons for denitrification and provide precursors for the synthesis of extracellular polysaccharides and proteins.
Our research studied Thauera sp. JM12B12, an aerobic denitrifying bacterium that uniquely combines the capability to produce bioflocculants with efficient denitrification, even under low C/N ratio conditions. Consequently, we proposed that JM12B12 held significant promise as a candidate for wastewater treatment in RASs. However, it is important to acknowledge the limitations of our study, which can be addressed and supplemented in the future. Firstly, we will elucidate the chemical composition and mechanisms of EPS produced by JM12B12. To advance the practical application of Thauera sp. JM12B12 in N removal from aquaculture water, future research could focus on assessing its biosafety, validating its denitrification efficiency in real RASs aquaculture water. Additionally, employ the molecular biology, genetic biology, transcriptomics, and metabolomics techniques to elucidate the underlying mechanisms behind JM12B12’s flocculation and its exceptional denitrification performance at low C/N ratios.
4 Conclusion
Thauera sp. JM12B12, a novel denitrifying bacterium with bioflocculant-producing capability, demonstrated high NO3−-N and NO2−-N removal and bioflocculation efficiency under low C/N ratio conditions. Using sodium lactate as the sole carbon source, it achieved complete removal of NO2−-N and NO3−-N at a low C/N ratio of 5, with TN-N removal efficiencies reaching 93.8 and 93.1%, respectively. Optimal denitrification occurred across a broad pH range (7–9), salinity (0–1.5%), and temperature (25–40 °C) under static conditions. Additionally, the cell-free supernatant during NO3−-N removal exhibited a high flocculation activity of 91.4%. Genome analyses revealed a complete denitrification pathway and 80 potential functional genes associated with the production of bioflocculants, highlighting its dual functionalities for removing N and suspended particles from wastewater.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MZ: Validation, Data curation, Conceptualization, Investigation, Writing – original draft, Methodology, Funding acquisition, Visualization, Writing – review & editing. YZ: Formal analysis, Writing – review & editing, Data curation. QY: Visualization, Supervision, Project administration, Writing – review & editing. YH: Software, Investigation, Writing – original draft, Conceptualization. HZ: Data curation, Project administration, Writing – review & editing, Funding acquisition.
Funding
The author(s) declared that financial support was received for this work and/or its publication. This work was supported by the National Natural Science Foundation of China (32200086), the GDAS’ Project of Science and Technology Development (2022GDASZH-2022010101), the Guangzhou Science and Technology Program (2024A04J3662), the GDAS’s 2024 Young Talents Project (2024GDASQNRC-0216), and the Guangdong Special Support Program (2021JC06N628).
Conflict of interest
The author(s) declared that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declared that Generative AI was not used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1730924/full#supplementary-material
References
Alias, J., Abu Hasan, H., Sheikh Abdullah, S. R., and Othman, A. R. (2024). Flocculating activity and kinetic properties of bioflocculant produced by bacillus species for wastewater treatment. J Water Process Eng 68:106501. doi: 10.1016/j.jwpe.2024.106501
Baird, R. B., Eaton, A. D., and Rice, E. W. (2017). Standard methods for the examination of water and wastewater. Washington, DC: American Public Health Association.
Cai, X., Liu, R., Cheng, M., Liu, T., Zhang, Y., Li, X., et al. (2023). Characterization of a novel Pseudomonas mosselii 9-1 capable of nitrate reduction to ammonium, ammonium assimilation, and denitrification in aerobic conditions. J Water Process Eng 52:103531. doi: 10.1016/j.jwpe.2023.103531
Chalita, M., Kim, Y. O., Park, S., Oh, H. S., Cho, J. H., Moon, J., et al. (2024). EzBioCloud: a genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 74:006421. doi: 10.1099/ijsem.0.006421,
Chen, M., He, T., Wu, Q., Zhang, M., and He, K. (2023). Enhanced heterotrophic nitrification and aerobic denitrification performance of Glutamicibacter arilaitensis EM-H8 with different carbon sources. Chemosphere 323:138266. doi: 10.1016/j.chemosphere.2023.138266,
Chen, Z., Hu, Y., Qiu, G., Liang, D., Li, Y., Cheng, J., et al. (2024). Genomics and metabolic characteristics of simultaneous heterotrophic nitrification aerobic denitrification and aerobic phosphorus removal by Acinetobacter indicus CZH-5. Bioresour. Technol. 395:130322. doi: 10.1016/j.biortech.2024.130322,
Chen, W., Liang, J., Shen, L., Lv, X., Zhang, S., Zhang, X., et al. (2024). Production and characterization of efficient bioflocculant in high-turbidity drinking water treatment: identification of flocculation-related genes. Chemosphere 366:143452. doi: 10.1016/j.chemosphere.2024.143452,
Chen, J., Shen, L., Li, Y., Cao, H., Chen, C., Zhang, G., et al. (2024). Insights into the nitrogen transformation mechanism of pseudomonas sp. Y15 capable of heterotrophic nitrification and aerobic denitrification. Environ. Res. 240:117595. doi: 10.1016/j.envres.2023.117595,
Chen, L., Zhao, B., An, Q., Qiu Guo, Z., and Huang, C. (2024). The characteristics and flocculation mechanisms of SMP and B-EPS from a bioflocculant-producing bacterium pseudomonas sp. XD-3 and the application for sludge dewatering. Chem. Eng. J. 479:147584. doi: 10.1016/j.cej.2023.147584
Christiaens, A. S., Daenen, R., and Smets, I. (2023). Bioaugmentation of a structural extracellular polymeric substances (EPS) producer to improve activated sludge bioflocculation: lessons learned. Water Pract. Technol. 18, 1663–1680. doi: 10.2166/wpt.2023.103
Flemming, H. C., van Hullebusch, E. D., Little, B. J., Neu, T. R., Nielsen, P. H., Seviour, T., et al. (2025). Microbial extracellular polymeric substances in the environment, technology and medicine. Nat. Rev. Microbiol. 23, 87–105. doi: 10.1038/s41579-024-01098-y,
Gao, Y., Zhu, J., Wang, K., Ma, Y., Fang, J., and Liu, G. (2023). Discovery of a heterotrophic aerobic denitrification pseudomonas sp. G16 and its unconventional nitrogen metabolic pathway. Bioresour. Technol. 387:129670. doi: 10.1016/j.biortech.2023.129670,
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0,
Gu, X., Leng, J., Zhu, J., Zhang, K., Zhao, J., Wu, P., et al. (2022). Influence mechanism of C/N ratio on heterotrophic nitrification- aerobic denitrification process. Bioresour. Technol. 343:126116. doi: 10.1016/j.biortech.2021.126116,
Han, C., Shi, C., Liu, L., Han, J., Yang, Q., Wang, Y., et al. (2024). Majorbio cloud 2024: update single-cell and multiomics workflows. iMeta 3:e217. doi: 10.1002/imt2.217,
He, S., Zhao, L., Liu, Y., Feng, L., Hu, T., Gao, Z., et al. (2024). Multiple drivers and mechanisms of solid-water interfacial interactions in sludge dewatering: roles of polarity and molecular structure of extracellular polymeric substances. Water Res. 263:122180. doi: 10.1016/j.watres.2024.122180
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281,
Hu, B., Lu, J., Qin, Y., Zhou, M., Tan, Y., Wu, P., et al. (2023). A critical review of heterotrophic nitrification and aerobic denitrification process: influencing factors and mechanisms. J Water Process Eng 54:103995. doi: 10.1016/j.jwpe.2023.103995
Huang, M. Q., Cui, Y. W., Yang, H. J., Xu, M. J., Cui, Y., and Chen, Z. (2023). A halophilic aerobic-heterotrophic strain Halomonas venusta SND-01: nitrogen removal by ammonium assimilation and heterotrophic nitrification-aerobic denitrification. Bioresour. Technol. 374:128758. doi: 10.1016/j.biortech.2023.128758,
Huang, F., Liu, S., Cao, J., Yu, Y., Zhao, L., Zhao, S., et al. (2025). Nitrogen removal performance and antibiotic resistance of the heterotrophic nitrification-aerobic denitrification bacterium Stutzerimonas sp. X87. J. Environ. Chem. Eng. 13:118975. doi: 10.1016/j.jece.2025.118975
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9,
Kishi, K., Onodera, R., Matsumoto, A., Matsubayashi, Y., and Arakawa, H. (2025). Effects of particle concentration in river water on survival of juvenile chum salmon in hatcheries. Aquaculture 602:742320. doi: 10.1016/j.aquaculture.2025.742320
Kumar, M., Chadha, N. K., Prakash, S., Pavan-Kumar, A., Harikrishna, V., Gireesh-Babu, P., et al. (2023). Salinity, stocking density, and their interactive effects on growth performance and physiological parameters of white-leg shrimp, Penaeus vannamei (Boone, 1931), reared in inland ground saline water. Aquac. Int. 32, 675–690. doi: 10.1007/s10499-023-01181-w
Lan, M., Yin, Q., Wang, J., Li, M., Li, Y., and Li, B. (2023). Heterotrophic nitrification-aerobic denitrification performance of a novel strain, pseudomonas sp. B-1, isolated from membrane aerated biofilm reactor. Environ. Res. 220:115199. doi: 10.1016/j.envres.2022.115199,
Lauber, F., Deme, J. C., Liu, X., Kjær, A., Miller, H. L., Alcock, F., et al. (2024). Structural insights into the mechanism of protein transport by the type 9 secretion system translocon. Nat. Microbiol. 9, 1089–1102. doi: 10.1038/s41564-024-01644-7,
Li, H., Cui, Z., Cui, H., Bai, Y., Yin, Z., and Qu, K. (2023). Hazardous substances and their removal in recirculating aquaculture systems: a review. Aquaculture 569:739399. doi: 10.1016/j.aquaculture.2023.739399
Li, Q., He, Y., Wang, B., Weng, N., Zhang, L., Wang, K., et al. (2024). Heterotrophic nitrification–aerobic denitrification by bacillus sp. L2: mechanism of denitrification and strain immobilization. Water 16:416. doi: 10.3390/w16030416
Li, T., Liu, C., Xi, Z., Li, N., Liao, L., Zhou, Y., et al. (2025). Screening and characterization of an Alcaligenes sp. HHVWA23 and the efficiency of nitrogen removal by biochar immobilization. J. Environ. Manag. 390:126253. doi: 10.1016/j.jenvman.2025.126253,
Li, W., Peng, Y., Gao, R., Zhang, Q., Li, X., Kao, C., et al. (2022). Effect of low salinity on nitrogen removal from municipal wastewater via a double-anammox process coupled with nitritation and denitratation: performance and microbial structure. Bioresour. Technol. 346:126633. doi: 10.1016/j.biortech.2021.126633,
Liang, X., Gan, L., He, T., Chen, M., Zhang, M., and Wu, Q. (2023). The coexisted nitrate and nitrite as a driving force for the aerobic denitrification of Peribacillus sp. EM-C3. Environ. Technol. Innov. 32:103299. doi: 10.1016/j.eti.2023.103299
Liao, Y., Li, S., Zhu, X., Dang, Z., Tang, S., and Ji, G. (2021). The promotion and inhibition effect of graphene oxide on the process of microbial denitrification at low temperature. Bioresour. Technol. 340:125636. doi: 10.1016/j.biortech.2021.125636,
Liu, S., Dai, J., Wei, H., Li, S., Wang, P., Zhu, T., et al. (2021). Dissimilatory nitrate reduction to ammonium (DNRA) and denitrification pathways are leveraged by cyclic AMP receptor protein (CRP) paralogues based on electron donor/acceptor limitation in Shewanella loihica PV-4. Appl. Environ. Microbiol. 87, e01964–e01920. doi: 10.1128/AEM.01964-20,
Lou, L., Luo, H., Fang, J., and Liu, G. (2023). The advance of heterotrophic nitrification aerobic denitrification microorganisms in wastewater treatment. Bioresour. Technol. Rep. 22:101495. doi: 10.1016/j.biteb.2023.101495,
Lu, J., Tan, Y., Tian, S., Qin, Y., Zhou, M., Hu, H., et al. (2024). Effect of carbon source on carbon and nitrogen metabolism of common heterotrophic nitrification-aerobic denitrification pathway. Chemosphere 361:142525. doi: 10.1016/j.chemosphere.2024.142525,
Ma, T., He, Q., Cao, G., and Li, X. (2024). Enhanced nitrogen removal from a recirculating aquaculture system using a calcined FeS (x)-packed denitrification bioreactor. ACS Omega 9, 51089–51097. doi: 10.1021/acsomega.4c06374,
Mao, Q., Bao, J., Du, J., Zhang, Y., Zhou, Y., He, T., et al. (2025). Comprehensive revealing the destructive effect and inhibitory mechanism of oxytetracycline on aerobic denitrification bacteria Acinetobacter sp. AD1 based on cell state, electron behavior and intracellular environment. J. Hazard. Mater. 492:138058. doi: 10.1016/j.jhazmat.2025.138058,
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60,
Ming, Y., Abdullah Al, M., Zhang, D., Zhu, W., Liu, H., Cai, L., et al. (2024). Insights into the evolutionary and ecological adaption strategies of nirS- and nirK-type denitrifying communities. Mol. Ecol. 33:e17507. doi: 10.1111/mec.17507,
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37:2461. doi: 10.1093/molbev/msaa131,
Pacheco, P. J., Cabrera, J. J., Jiménez-Leiva, A., Bedmar, E. J., Mesa, S., Tortosa, G., et al. (2022). Effect of copper on expression of functional genes and proteins associated with Bradyrhizobium diazoefficiens denitrification. Int. J. Mol. Sci. 23:3386. doi: 10.3390/ijms23063386,
Pan, Y., She, D., Shi, Z., Cao, T., Xia, Y., and Shan, J. (2023). Salinity and high pH reduce denitrification rates by inhibiting denitrifying gene abundance in a saline-alkali soil. Sci. Rep. 13:2155. doi: 10.1038/s41598-023-29311-7,
Pan, C., Zhao, Y. G., Mupindu, P., and Zhao, S. (2024). The denitrification ability and nitrogen metabolism pathway of aerobic denitrifier Marinobacter alkaliphilus SBY-1 under low C/N ratios. Sci. Total Environ. 957:177651. doi: 10.1016/j.scitotenv.2024.177651,
Qin, S., Li, Q., Dou, J., Man, Y., Wu, L., Tian, H., et al. (2024). Isolation and characterization of Stenotrophomonas pavanii GXUN74707 with efficient flocculation performance and application in wastewater treatment. Front. Microbiol. 15:1367043. doi: 10.3389/fmicb.2024.1367043,
Ren, T., Jin, X., Deng, S., Guo, K., Gao, Y., Shi, X., et al. (2024). Oxygen sensing regulation mechanism of Thauera bacteria in simultaneous nitrogen and phosphorus removal process. J. Clean. Prod. 434:140332. doi: 10.1016/j.jclepro.2023.140332
Ren, J., Tang, J., Min, H., Tang, D., Jiang, R., Liu, Y., et al. (2024). Nitrogen removal characteristics of novel bacterium Klebsiella sp. TSH15 by assimilatory/dissimilatory nitrate reduction and ammonia assimilation. Bioresour. Technol. 394:130184. doi: 10.1016/j.biortech.2023.130184,
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131. doi: 10.1073/pnas.0906412106,
Riesco, R., and Trujillo, M. E. (2024). Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 74:006300. doi: 10.1099/ijsem.0.006300,
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M., and Katoh, K. (2019). MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47, W5–w10. doi: 10.1093/nar/gkz342,
Selepe, T. N., and Maliehe, T. S. (2024). Bioflocculation of pollutants in wastewater using flocculant derived from Providencia huaxiensis OR794369.1. BMC Microbiol. 24:39. doi: 10.1186/s12866-023-03144-w,
Show, S., Akhter, R., Paul, I., Das, P., Bal, M., Bhattacharya, R., et al. (2024). Efficacy of exopolysaccharide in dye-laden wastewater treatment: a comprehensive review. Chemosphere 355:141753. doi: 10.1016/j.chemosphere.2024.141753
Shu, H., Ma, Y., Lu, H., Sun, H., Zhao, J., Ruan, Z., et al. (2024). Simultaneous aerobic nitrogen and phosphate removal capability of novel salt-tolerant strain, Pseudomonas mendocina A4: characterization, mechanism and application potential. Bioresour. Technol. 393:130047. doi: 10.1016/j.biortech.2023.130047,
Stephens, Z., Wilson, L. F. L., and Zimmer, J. (2023). Diverse mechanisms of polysaccharide biosynthesis, assembly and secretion across kingdoms. Curr. Opin. Struct. Biol. 79:102564. doi: 10.1016/j.sbi.2023.102564,
Sun, W., Hu, C., Wu, J., Wei, M., Lin, J. G., and Hong, Y. (2024). Efficient nitrogen removal via simultaneous ammonium assimilation and heterotrophic denitrification of Paracoccus denitrificans R-1. iScience 27:110599. doi: 10.1016/j.isci.2024.110599,
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120,
Vandana, P. M., and Das, S. (2023). Bacterial extracellular polymeric substances: biosynthesis and interaction with environmental pollutants. Chemosphere 332:138876. doi: 10.1016/j.chemosphere.2023.138876,
Wang, F., Cui, Q., Liu, W., Jiang, W., Ai, S., Liu, W., et al. (2024). Synergistic denitrification mechanism of domesticated aerobic denitrifying bacteria in low-temperature municipal wastewater treatment. NPJ Clean Water 7:6. doi: 10.1038/s41545-024-00299-5
Wang, Q., and He, J. (2020). Complete nitrogen removal via simultaneous nitrification and denitrification by a novel phosphate accumulating Thauera sp. strain SND5. Water Res. 185:116300. doi: 10.1016/j.watres.2020.116300
Wang, F., Mei, X., Liu, B., Zhao, Y., Li, J., Li, W., et al. (2025). Recirculating aquaculture mode has high sustainability and environmental accommodation capacity via better energy-saving and water-saving effects in Cynoglossus semilaevis production. Aquaculture 597:741904. doi: 10.1016/j.aquaculture.2024.741904
Wang, H., Sun, Y., Zhou, X., Zhu, C., Wang, X., Abbasi, H. N., et al. (2025). Simultaneous removal of nitrogen and phosphorus by aerobic denitrifying Paracoccus versutus JUST-3. Bioresour. Technol. 428:132457. doi: 10.1016/j.biortech.2025.132457
Wu, Y., Cui, Y., Li, D., Yin, M., Pei, Y., Wang, X., et al. (2024). Fulvic acid mediated highly efficient heterotrophic nitrification-aerobic denitrification by Paracoccus denitrificans XW11 with reduced C/N ratio. Water Res. 267:122557. doi: 10.1016/j.watres.2024.122557,
Wu, T., Li, J., Cao, R., Chen, X., Wang, B., Huang, T., et al. (2024). Nitrate removal by a novel aerobic denitrifying Pelomonas puraquae WJ1 in oligotrophic condition: performance and carbon source metabolism. Sci. Total Environ. 954:176614. doi: 10.1016/j.scitotenv.2024.176614,
Xie, Y., Tian, X., He, Y., Dong, S., and Zhao, K. (2023). Nitrogen removal capability and mechanism of a novel heterotrophic nitrification-aerobic denitrification bacterium Halomonas sp. DN3. Bioresour. Technol. 387:129569. doi: 10.1016/j.biortech.2023.129569,
Yan, W., Wang, N., Wang, Z., Shi, J., Tang, T., and Liu, L. (2025). Nitrogen removal characteristics and mechanism of the aerobic denitrifying bacterium Stutzerimonas stutzeri os3 isolated from shrimp aquaculture sediment. Mar. Pollut. Bull. 214:117711. doi: 10.1016/j.marpolbul.2025.117711,
Yang, J., Feng, L., Pi, S., Cui, D., Ma, F., Zhao, H. P., et al. (2020). A critical review of aerobic denitrification: insights into the intracellular electron transfer. Sci. Total Environ. 731:139080. doi: 10.1016/j.scitotenv.2020.139080,
Yang, Q., Shi, Y., Xin, Y., Yang, T., Zhang, L., Gu, Z., et al. (2023). Insight into the cold adaptation mechanism of an aerobic denitrifying bacterium: Bacillus simplex H-b. Appl. Environ. Microbiol. 89:e0192822. doi: 10.1128/aem.01928-22,
Yu, P., Tu, W., Wu, M., Zhang, Z., Liu, X., and Wang, H. (2024). Evaluation of polyhydroxyalkanoate recovery from food waste by sequencing batch biofilm reactor with high mixed microbial biomass. Resour. Conserv. Recycl. 205:107536. doi: 10.1016/j.resconrec.2024.107536
Yuan, X., Cui, K., Chen, Y., Xu, W., Li, P., and He, Y. (2023). Response of microbial community and biological nitrogen removal to the accumulation of nonylphenol in sequencing batch reactor. Int. J. Environ. Sci. Technol. 20, 12669–12680. doi: 10.1007/s13762-023-04825-9,
Yue, X., Liu, H., Wei, H., Chang, L., Gong, Z., Zheng, L., et al. (2023). Reactive and microbial inhibitory mechanisms depicting the panoramic view of pH stress effect on common biological nitrification. Water Res. 231:119660. doi: 10.1016/j.watres.2023.119660,
Zhang, Q., Chen, W., Yuan, C., Liu, H., Liang, S., and Tan, S. (2024). A novel pure biofilm system based on aerobic denitrification for nitrate wastewater treatment: exploring the feasibility of high total nitrogen removal under low-carbon condition. Chem. Eng. J. 480:147978. doi: 10.1016/j.cej.2023.147978
Zhang, M., He, T., Wu, Q., and Chen, M. (2023a). Efficient detoxication of hydroxylamine and nitrite through heterotrophic nitrification and aerobic denitrification by Acinetobacter johnsonii EN-J1. Front. Microbiol. 14:1130512. doi: 10.3389/fmicb.2023.1130512,
Zhang, M., Li, A., Yao, Q., Xiao, B., and Zhu, H. (2022). Pseudomonas oligotrophica sp. nov., a novel denitrifying bacterium possessing nitrogen removal capability under low carbon-nitrogen ratio condition. Front. Microbiol. 13:882890. doi: 10.3389/fmicb.2022.882890,
Zhang, M., Lu, H., Cai, L., Sun, P., Ma, B., Li, J., et al. (2023b). C/N ratios inform sustainable aerobic denitrification for nitrogen pollution control: insights into the key parameter from a view of metabolic division. J. Clean. Prod. 414:137565. doi: 10.1016/j.jclepro.2023.137565
Zhang, Y., Wang, Q., Rogers, M. J., and He, J. (2025). Autotrophic denitrification under anoxic conditions by newly discovered mixotrophic sulfide-oxidizing bacterium. Bioresour. Technol. 430:132553. doi: 10.1016/j.biortech.2025.132553,
Keywords: aerobic denitrification, flocculation activity, genome analysis, low C/N ratio, Thauera
Citation: Zhang M, Zhang Y, Yao Q, Hu Y and Zhu H (2025) Dual-functionality of Thauera sp. JM12B12: aerobic denitrification and bioflocculation for nitrogen and suspended particles removal at low carbon-to-nitrogen ratios. Front. Microbiol. 16:1730924. doi: 10.3389/fmicb.2025.1730924
Edited by:
Bin Ji, Wuhan University of Science and Technology, ChinaReviewed by:
Xiaoxia Wang, Qingdao University, ChinaXiaoxu Zheng, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Zhang, Zhang, Yao, Hu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghui Zhu, emh1aGhfZ2RpbUAxNjMuY29t
 Mingxia Zhang
Mingxia Zhang Yulian Zhang1
Yulian Zhang1 Qing Yao
Qing Yao Honghui Zhu
Honghui Zhu