- 1Regenerative Medicine Cluster, Advanced Medical and Dental Institute (AMDI), Universiti Sains Malaysia, Kepala Batas, Penang, Malaysia
- 2College of Life Science and Technology, Xinxiang Medical University (XXMU), Xinxiang, China
- 3College of Biomedical Engineering, Xinxiang Medical University (XXMU), Xinxiang, China
Lung cancer is the most common cancer worldwide, accounting for 1.8 million new cases and 1.6 million deaths in 2012. Non-small cell lung cancer (NSCLC), which is one of two types of lung cancer, accounts for 85–90% of all lung cancers. Despite advances in therapy, lung cancer still remains a leading cause of death. Cancer relapse and dissemination after treatment indicates the existence of a niche of cancer cells that are not fully eradicated by current therapies. These chemoresistant populations of cancer cells are called cancer stem cells (CSCs) because they possess the self-renewal and differentiation capabilities similar to those of normal stem cells. Targeting the niche of CSCs in combination with chemotherapy might provide a promising strategy to eradicate these cells. Thus, understanding the characteristics of CSCs has become a focus of studies of NSCLC therapies.
Introduction
Lung cancer has remained as the leading cancer cases worldwide in term of the high incidence and mortality rate (1). Based on data published in GLOBOCAN, 2012, there are 1.8 million new cases of lung cancer (or 12.8% in total) and 1.6 million deaths (19.4%) in 2012 (1). In 2016, the number of new lung cancer diagnosed was increased to 14% and remained as the highest mortality rate accounting for about 1 in 4 cancer deaths (2). Based on pathological features, lung cancer has been classified into two major groups: small cell lung cancer (SCLC) that accounts for only 15% of all lung cancer cases and non-small cell lung cancer (NSCLC) that accounts for 85% of lung cancer cases. NSCLC is further classified into three subtypes: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (LCC). Due to very poor prognosis (only 15% of patients surviving 5 years after treatment), treatment for lung cancer remain challenging.
Researchers believe that there is a presence of small subpopulation of cells within the tumor cells that driven the aggressive behavior of cancer cells and chemotherapy resistance of the cancer cells. The subpopulation of cancer cells is known as cancer stem cells (CSCs) or cancer-initiating cells (CICs) (3). According to American Association for Cancer Research, CSC was defined as “a cell within a tumour that possesses the capacity to self-renew and to cause the heterogeneous lineages of cancer cells that comprise the tumour” (4). The theory of CSC suggests that this subpopulation of cells has the capacity to self-renew, initiate tumors, and undergo multipotent differentiation (3). Many of the concepts that arise in cancer research, such as self-renewal, heterogeneity, and relapse after treatment and resistance to conventional chemotherapies, can be explained by this theory.
The first solid evidence to support the CSC theory was the identification of relatively rare population (1:250,000 cells) of stem-like cells in acute myeloid leukemia (AML) (5). The cells were isolated based on the expression of surface protein markers and able to re-grown human AML when transplanted in immuno-compromised mice (5). Later, the analysis of stem-like cells from various AML subtypes has found that the cells were immature in terms of differentiation and closely related to the hematopoietic stem cells (HSCs) rather than mature, terminally differentiated blood cells (6). This breakthrough finding has fostered an intense effort to characterize and isolate CSCs in various solid tumor including breast, brain, prostate, colon, and pancreatic cancer (7–11) thus making the CSCs field at the rapidly evolving field that may play a pivotal role in changing how basic cancer researchers, clinical investigators, physicians, and cancer patients view cancer the CSCs study.
Biology of CSCs
CSCs is defined as a small population of cells within tumor with share characteristics of normal stem cells. The cells have the capacity to initiate tumor formation, extensive proliferation, and resistant to chemotherapy (Figure 1). Apart from that, CSCs also share other similar characteristics like normal stem cells including self-renewal, expression of specific markers and genes, and utilization of common signaling pathways (3). The source of CSCs can be either from somatic stem cells or differentiated progenitor cells (3, 12, 13). Those cells initiate tumorigenesis by undergoing self-renewal and differentiation and thus resulting in tumor relapse, therapy resistance, and metastasis (3, 14).
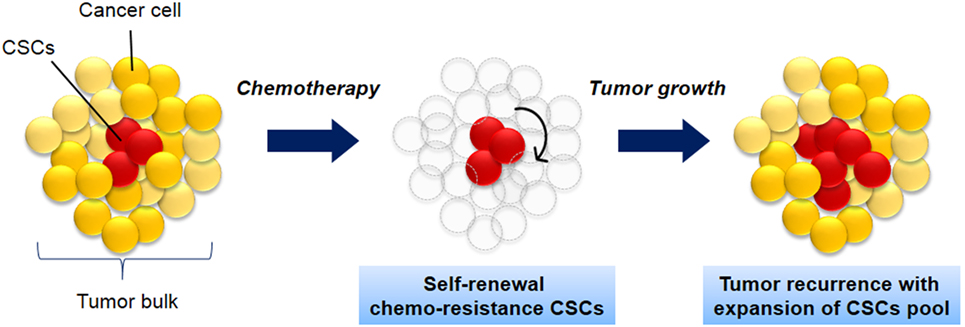
Figure 1. The biology of cancer stem cells (CSCs). CSCs (red) self-renew and differentiate within tumors to form CSCs pool and non-tumorigenic cancer cells (yellow), which have limited proliferative potential. As the tumor grows, these cells can either undergo limited benign growth or form disseminated malignancies. These cells are resistance to chemotherapy and leads to cancer recurrence.
CSCs are identified based on several approaches: (1) ability to self-renewal (sphere and colony forming assay), (2) ability to differentiate, and (3) ability to form secondary or tertiary tumors when transplanted into immunodeficient rodent (15). The ability of the CSCs to form tumor in immunodeficient mice was the commonest method to characterize the human CSCs. For instance, Dick et al. (16) had reported that the CD34+CD38− cells derived from human leukemia cells, which constituted less that 1% cells in 10,000 leukemia cells had the ability to form leukemia in non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice (16).
Similar to normal cells, tumors are composed of heterogeneous populations (17), which distinct in tumor subtypes. The heterogeneous tumor cells are different in terms of morphological and phenotypic profiles including proliferation and differentiation capacities. To illustrate, the progression of each cells within the tumor are different individually, in which there are some cells classified as cycling or non-cycling tumor cells, some may be in dormant state or reproductively dead (18). If the cells are in cycling mode, therefore, the cells may be at any stage of cell cycle which coincidentally may influence the cellular properties such as membrane properties (19, 20), antigen expression (21), sensitivity to immune killing (22), drug cytotoxicity (23), and ability to metastasize (24). Therefore, the characteristics of the tumor cells will be dependent on these properties. Back then, the heterogeneity of tumor cells may also eventually arise from genetic changes, environmental differences, and reversible changes in cells properties that resulted different in terms of morphology and biology of the tumor cells (25). Unfortunately, the diversity of tumor heterogeneity results in greater challenge in targeting specific CSCs. Thus, a number of specific markers are required in order to identify the CSCs population specifically lung CSCs (see Markers for Lung CSCs). Taken into consideration the tumor niches or microenvironment that control the stemness of the CSCs, the differences in the histological types and clinical presentations of lung cancer have contributed to the fact that the CSCs can produce morphologically and biologically different tumor within the same tissue.
Lung cancer has been considered as the most complex type of cancer due to the genotypic and histological varieties. This in turn, making the study of lung CSCs less explored as compared to other types of cancer (26). CSCs of lung cancer has been proposed to originate from several sources including from airway stem cells, bronchiole alveolar progenitor cells, basal/mucous secretary bronchial progenitor cells, or neuroendocrine progenitor cells. The different origin has resulting in the development of region-specific lung cancers (27). The study of CSCs has to be specific for respective subtypes of lung cancer and therefore making the CSCs study in lung cancer become more challenging.
The concept of CSCs in human lung cancer was initially coined 30 years ago (28). The study by Carney et al. (28) demonstrated that less than 1.5% of cells taken from patients with adenocarcinoma (SCLC) were able to form colonies when cultured in vitro. The selected colonies were able to generate the formation of tumor with similar features of the original tumor when transplanted into nude mice. Another study done by Kim et al. (29) has identified the bronchioalveolar stem cells from the mouse bronchioalveolar duct. They found out that the cells could initiate the development of lung adenocarcinoma (29). Recently, our group has identified the lung CSCs in NSCLC that was isolated from lung adenocarcinoma cells (A549 and H2170). Apart from showing characteristics similar to multipotent stem cells, the microarray analysis of the isolated lung CSCs also showed that the cells possess the biological characteristics associated with cancer and stem cells (30).
Methods for Identification of Lung CSCs
CSCs have been identify and isolated using functional assay such as side population (SP) assay and based on the expression of CSCs surface marker.
SP Assay
The SP assay method discriminate cells based on the differential potential of cells to efflux fluorescent Hoechst dye (a DNA-binding dye) via the ATP-binding cassette (ABC) transporters (31). The ABC transporter proteins are expressed within cell membrane, and they belong to the superfamily of membrane pumps that catalyze the adenosine triphosphate (ATP) and transport various endogenous compounds out of the cells (32). In human, it has been estimated between 500 and 1,200 genes encoded for drug transporter protein (33). Among them, the P-glycoprotein (ABCB1, MCR1), ABCG2 (breast cancer resistant protein, BRCP1), and ABCC1–5 (multidrug-resistant proteins, MRP1–5) are the drug transporter proteins involved in the establishment of SP phenotype (32). The SP assay was first described by Goodell et al. (34) for isolation of mouse bone marrow cells and shown to be enriched with HSCs (34). Moreover, the SP cells are overlapping with HSC phenotype CD177+Sca1+Lin−Thy1− (35, 36). The SP cells have been described in various tumor types as being enriched in stem-like properties (37, 38). The SP assay was the first approach used for the identification of lung CSCs (39, 40). The SP cells isolated from established lung cancer cell lines are more tumorigenic and display increased invasive capability compared to the non-SP cells (39). Moreover, the cells are resistance toward multiple chemotherapeutic drugs and display high expression of ABCG2 and other ABC transporters (39). The SP cells also exhibit the self-renewal characteristics display by the ability to generate floating spheres and poses high proliferative potential (41).
Surface Marker Expression
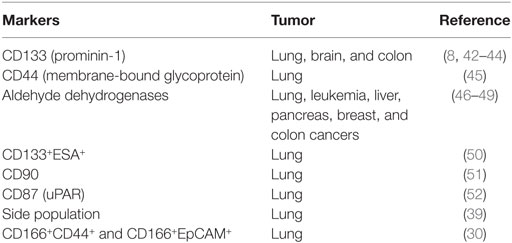
Another strategy used to identify lung CSCs is by expression of stem cell surface marker. Currently, there are relatively few lung CSC markers that have been validated. However, extensive studies have led to the identification of various CSCs that differ from other cells in the tumor. Most CSCs express multiple markers at the same time and using one marker to define CSC is not possible (Table 1). For example, in cell lines A549 and H446, CD133-positive and -negative populations contain the same amount of CSCs. Studying CSC markers may present new insight that will improve current lung cancer therapy and better patient prognosis.
CD133
CD133, also known as prominin-1, is one of the most commonly used markers for detecting CSCs. CD133 is a five-transmembrane glycoprotein that has been demonstrated to be highly expressed in various carcinomas of various origins including brain (8) and colon (42). The exact role of CD133 in lung cancer is still unclear. However the CD133+ cells are thought to be CICs on the basis of their ability to induce tumor development, invasion, and metastases. Previous study on NSCLC showed that CD133+ cells isolated from NSCLC tumor specimens have higher self-renewal capacity compared to CD133− cells with 40-fold higher number of colonies/spheres formed. Self-renewal capacity of CD133+ and CD133− cells were further confirmed by injecting the cells into NOD/SCID mice. The results had demonstrated that CD133+ cells possessed more tumourigenicity compared to CD133− cells (44). Moreover, in tumor specimens of NSCLC patients, significant increase in CD133+ cells and capillary structure have been detected, suggesting the involvement of this cell population in tumor growth and tumor vasculogenesis (44). However, another study on NSCLC patients’ exhibit opposite findings where CD133 cells were found to acts as resistant phenotype, not as a prognostic marker for survival like previously reported (53). Salnikov and colleagues reported that even though CD133 was found in various lung cancer cell lines, including A549, H157, H226, Calu-1, H292, and H446, the CD133 was only exhibited CSC characteristics such as self-renewal, differentiation, proliferation, and tumorigenic capacity in H446 cell line. In another study, Eramo and colleagues were able to isolate small niche of CD133+ from SCLC and NSCLC and found that both the CD133+/− populations have the ability to self-renew, but the CD133− population were non-tumorigenic (43).
CD44
Another marker that has been proposed as CSCs marker is the CD44. CD44 was initially proposed as CSCs marker for colorectal cancer (54–56). CD44 is a membrane-bound glycoprotein that plays several important roles including cell migration, cell adhesion, and modulation of cell–matrix interaction (57). Besides, CD44 has been reported to be associated with many cancer-mediated signaling pathways that explain the involvement of this marker in cancer initiation and enhancement. For example, CD44 acts as a coreceptor with epidermal growth factor receptor and as an indirect activator of the cell proliferation pathway through the ligand presentation (58). In addition, CD44 has been reported to enhance cancer cells invasiveness and multidrug resistance due to its association with mitogen-activated protein kinase-phosphatidyl inositol 3 kinase signaling pathway. This association led to the overexpression of urokinase plasminogen activator/urakinase plasminogen activator receptor that results in enhancing invasiveness and multidrug resistance of cancer cells (52). Profiling of the expression of several markers including CD44 from 10 NSCLC cell lines by using flow cytometry found that CD44+ from certain cancer cell lines acts as a tumor initiator marker in lung cancer cells when tested in both in vitro and in vivo (45).
CD166
CD166 is another CSCs that have been described as CSCs marker for lung cancer. While CD166 has been studied extensively in other solid tumor (59–61), little is known about CD166 expression and its function in lung cancer. The role of CD166 as marker for lung CSCs has been demonstrated by Zhang et al. (62) in their comprehensive study on the potential of different surface marker candidates (CD166, CD133, CD44, and EpCAM) for the identification of CSCs in NSCLC (62). The isolated CD166 shows higher self-renewal potential and initiates formation of in vivo xenograft. Moreover, CD166 population shows higher in vivo tumor initiating capacity in comparison to CD133+, CD44+, and EpCAM+ cells isolated from the same cells. Based on this evidence, CD166 marker is considered as the most robust CSCs marker for identification of lung cancer.
Aldehyde Dehydrogenase (ALDH)
Another marker for CSCs that recently caught the attention of researchers is ALDHs. ALDH superfamily is composed of 19 known functional genes that can be classified into 11 families and 4 subfamilies (63). ALDH superfamily is NADP+-dependent enzymes that play crucial role oxidation of aldehydes into carboxylic acid. In retinoic signaling, ALDHs are required to produce the active form of retinoic acid by oxidation of all-trans-retinal and 9-cis-retinal (64, 65). There are several isoforms of ALDH that play important role in retinoic acid signaling including ALDH1A1, ALDH1A2, ALDH1A3, and ALDH8A1 (66). Several ALDH isoforms has been reported to involve in stem cell and also CSCs populations including ALDH1A1, ALDH1A2, ALDH1A3, ALDH1L1, and ALDH1L2 (63). ALDH was found to be highly expressed in various cancers such as leukemia, liver, pancreas, breast, and colon cancers (46, 47, 67). Recently, ALDH have been recognized as common markers not only for normal but also for CSCs (63, 68, 69). Profiling of ALDH expression in 12 different human lung cancer cell lines by using flow cytometry-based assay exhibited high expression of the cytosolic form of ALDH1A1 and ALDH3A1 (70). The involvement of ALDH in CSCs was further clarified by Ucar and colleagues (49). In their study, they compare the proliferative potential between two subpopulations of CSCs: a population with bright ALDH activity and population with dim ALDH activity in H522, lung cancer cell line. The obtained results show that the cells’ population with bright ALDH activity has higher and long-term proliferative capability compared to cells population with dim ALDH activity (49). Consistent with this result, Jiang et al. (48) found that ALDH+ cells, isolated from human NCSLC cell lines had important properties of CSCs in both in vitro and in vivo studies (48). Further clarification of the specificity of ALDH in lung cancer development is needed, as ALDH expression in normal pneumocytes increases due to exposure to cigarette smoking (70). Both these ALDH isozymes have been reported to play crucial role in stem cells (63).
Other Markers
CSCs that were known to have a capability in self-renewal, differentiation, and tumor initiation might express multiple markers instead of only single marker. For example, in a study on lung cancer cell lines A549 and H446, it was proved that CD133 alone could not be used to isolate the CSCs since both CD133+ and CD133− cell populations contained similar number of cells that contains CSCs characteristic (71). In another study on lung cancer cells, CSCs have been isolated by using combination CD133 with other markers such as ABCG2, chemokine receptor 4, and epithelial-specific antigen (ESA) (50). Population of CD133+ESA+ population was higher in NSCLC compared with normal lung tissue. Comparison between positive and negative populations of CD133 found that CD133+ population exhibit higher tumorigenic capacity when injected in SCID. Recently, our group had found that CD166+CD44+ and CD166+EpCAM+ cells isolated from A549 and H2170 lung cancer cells showed the stemness characteristics including the sphere and colony-forming capabilities, expression of stem cells transcription factors (Sox2 and Oct 3/4), and tumorigenic capacity when transplanted in nude mice (30, 72, 73). Thus, it is clearly shown that cancer cells contain a heterogeneous population of CSCs that expresses various cell surface markers and the use of more than one marker for isolating CSCs might increase the stringency of the CSCs population. However, it is still not isolating the whole population of CSCs in any cancer cell populations. Therefore, more research should be done in order to explore more markers that can be used to identify lung CSCs.
Therapeutic Strategies of Lung CSCs
CSCs are believed to sustain the progenitor of cancer cells and regulate the progression of cancer development in human. Therefore, targeting CSCs may provide a strategic way in therapeutic of cancer. Signaling pathway such as Hedhgog (Hh) is function in development and regeneration or repairing of stem cells (74). However, due to mutation, the aberrant of Hh signaling pathway activated the oncogenic pathway and eventually lead to tumorigenesis including initiate the CSCs progression that is responsible in cancer relapse. Therefore, inhibit the Hh pathway would be a great strategic by aiming on (1) inhibition of ligand processing, (2) disruption of receptor ligand, and (3) inhibition of transcription factor, the glioma-associated oncogene transcription factors 1–3 (GLI1, GLI2, and GLI3) (74). In addition, Hh pathway is also associated with chemoresistance (75, 76), which is also a characteristic that defines the properties of CSCs and causes major failure in chemotherapy (77).
Understanding the biological properties of lung CSCs is a fundamental toward improvement of the therapeutic effectiveness in cancer therapy. M2 isoform of glycolytic enzyme pyruvate kinase (PKM2) is well known in regulating the tumorigenic of cancer and associated with therapeutic resistance (78) that eventually contributes to poor prognosis of cancer. A recent study has revealed that PKM2 expression was associated with the biological properties of lung CSCs in which when the function of PKM2 was silent in CD44+ cells, the capability of CSCs to form spheroid and colony were reduced, and the sensitivity toward cisplatin was increased (79). On the other hand, an activation of D2 dopamine (DA), a receptor in CD133+ adenocarcinoma NSCLC were significantly inhibited the proliferation, colonies formation, and invasiveness of this tumor cells (80). In a different study, the lysine demethylase 1 (LSD1) was discovered to play a role in maintaining SCLC stemness, thus could be a potential therapeutic candidate in targeting lung CSCs. Interestingly, a potent inhibitors for LSD1 known as GSK2879552 was discovered, which was found to be sensitive specifically to only SCLC and AML as compared to the other 165 cell lines that were tested (81). Therefore, it is essential to discover the biological properties of lung CSCs as a promising in selective targeted therapy.
Cellular surface markers are one of the common method uses in discriminating CSCs population. Markers such as CD166+/EpCAM+ and CD166+/CD44+ has been correlated to CSCs since it possesses the characteristics of CSCs including the ability to self-renewal, differentiated into adipogenic and osteogenic, and expressed transcriptomic profile of multipotent cells (30) that therefore could be a potential in targeting the CSCs population. Another strengthen markers that is CD44HighCD90+ which eventually represent a CSCs population in SCLC and LCC as it capable on forming a spheroid that is recognized as tumor model for in vitro study (82). Furthermore, the clinical study showed that dual expression of stem cells markers, the CD133+ABCG2+ has showed early relapse in stage 1 NSCLC as compare to non-CD133+ABCG2+ (83), which will be useful target in cancer chemotherapeutic. Perhaps such marker as CD133 (44), CD44 (45), CD24 (84, 85), and CD166 (62) are known to detect lung CSCs but are also used in other study to detect CSCs in head and neck region (86, 87), gastric (88), colorectal cancer (89–92), breast (93), and pancreatic (94). Nevertheless, functional assay including SP and Aldelflour assay also has been used as a strategy to distinct between CSCs and non-CSCs. Based on Hoechst 33342 dye efflux assay, the cells will be isolated as it develop the ability of stem cells to efficiently efflux the dye and consequently represent a Hoechst 33342 dye negative and will be characterize as SP (41). Likewise, Adelflour assay was used to isolate cells that possess high ALDH enzymatic activity which in the same way mimic the capacity of stem cells (95, 96) that encode for resistance potential.
Natural product has widely use in cancer treatment as it contain low or fewer side effect but exhibit strong antitumor activity. Curcumin, a yellow-pigmented polyphenol, derived from curcuma longa (commonly known as turmeric) has been used in traditional Indian Ayurvedic as anti-inflammatory agent. Finally, scientist has showed that curcumin has chemopreventive and therapeutic properties against many tumors and enable it to suppress cell proliferation and inflammation, induce apoptosis, suppress many cancer signaling and pathway, inhibit cells metastasis and angiogenesis, and sensitize tumor cells to cancer therapies (97, 98). Curcumin has effectively sensitizes NSCLC CSCs and enhances cisplatin effectiveness thus induce cells apoptosis and suppression of cell migration of CD166+/EpCAM+ population (99), which subsequently reverse cisplatin resistant (100) in CSCs. Another recent study also shown that curcumin effectively inhibit the lung CSCs traits as evidence of shrinking the tumoursphere formation and also decreasing the expression of lung CSC marker as well as suppressing cells proliferation and induce cells apoptosis through a specific signaling known as Wnt/β-catenin and sonic Hedgehog pathways (101). Perhaps, not only lung CSCs but also curcumin has shown to efficiently inhibit several others types of cancer including colon cancer stem-like cells (102), breast CSCs (103), SP of C6 glioma cell (104), and CSC of colorectal liver metastasis (105).
The tumor microenvironment offer a niche that can sustain the growth (106) of CSCs as well as a protective role by sheltering the CSCs from any genotoxic insults, which results in therapy resistant (107). The niche possesses a complex structure (107) composed of diverse of stroma cells including mesenchymal and immune cells, vascular network, soluble factor extracellular matrix components, and also homeostatic processes (108), which is important in order to retain the abilities of CSCs to self-renewal and give rise to progenitor cells through differentiation process (109). Thus, interrupting CSCs niche would be necessary and could be a promising strategy for eradicating CSCs. Current study on targeting CSCs niche found that integrin, which is the primary receptors that involved in cell–matrix adhesion have a profound impact on the ability of CSCs to survive in specific locations. Recent findings demonstrated that the integrins, which play important role in CSCs biology, are required for cancer progression and drug resistance. Previous study on K-ras mouse model of NSCLC demonstrated that CSCs expressing integrin β4 are enriched after treatment with cisplatin (110). In consistent with this study, study on breast cancer showed that after treatment of cancer cell with taxol, the integrin α6+ CSC population is upregulated (111). Also, study of β1 integrins on head and neck cancer cell also support that integrins can act as potential cancer targets (112). However, specific integrins that is responsible to promote stemness, drug resistance, and metastasis of CSCs are yet to be determined (113). Recently, there are a few integrin α2-binding agents that have been invented to reduce the proliferation capability of CSCs (114). Therefore, it is clear that identifying specific integrins that is responsible to maintain survival of CSCs might be a promising target for cancer therapy.
Current technique of targeting CSCs is using miRNA therapy. microRNAs or miRNA are short nucleotide (~18–25 nt) of non-coding RNA that is responsible in regulation of gene expression by binding to 3′-untranslated regions or open reading frame of target genes (115). Formerly, the important characteristics that determine the CSCs are self-renewal, and study has shown that miRNA regulates this important feature. Miao et al. (116) discover that downregulation of miR-27 enhanced the properties of stem-like SCLC, whereby downregulation of miR-27 dedifferentiate non-tumorigenic cells into highly tumorigenic cells and was consistently downregulated in spheroid cells even at passage 4 (116). In addition, miR-27 was also reported to be downregulated in A549 cell that derived from NSCLC type (117). There were many more miRNA that act as tumor suppressor such as miR-145 (118) and miRNA-34a (119) demonstrating the inhibition proliferation of lung cancer-initiating cells and epithelial–mesenchymal transition (118), the key step of metastasis of cells.
Challenges in Lung CSCs
Overcoming the CSCs could be one of the strategic ways in increasing the therapeutic of cancer diseases as it is associated with poor prognosis. However, the fact that CSCs should be eliminated is not as simple as the theory as it faces many challenges and limitations. There are several things need to be aware and considered in order to improve the method in targeting the CSCs populations. The heterogeneity in CSCs, which results in diverse phenotype, has become a great challenge in identifying the prominent CSCs subpopulation especially in lung. Moreover, lung structure consist of complex structure comprise of a large variety of morphologies that differently function and responsible in facilitating gas exchange, balancing fluid in the lung, detoxifying and clearing foreign agents, and the activation of inflammatory due to injury (120, 121). Therefore, to track the CSCs, a list of markers has been widely select but is not always reliable because certain marker may not be specific in targeting the CSC. Study by Meng et al. (71) revealed that single marker of CD133+ showed no differences with CD133− as both of these markers display similar CSCs features such as the ability of self-renewal, colony formation, differentiation, and invasion (71). Perhaps, during the selection of CSCs markers, there could be a contradiction in declaration of certain markers used, for example, Roudi et al. (122) mentioned CD44 and CD24 is not a potential CSCs used in A549 cell derived from NSCLC because both CD44+/CD24+ and CD44+CD24low are capable of forming the holoclone, meraclone, and paraclone colonies and develop spheroid formation (122). Furthermore, cellular markers are difficult to stand alone and have to be used in combination of other markers in order to improve the selection of CSCs. However, targeting CSCs in lung would be challenge due to heterogeneity (123) of the cells and various genomic pathways involved. Therefore, many studies are focusing on using combination of cellular markers as it increase the specificity of targeted population.
CSCs are known to be resistant to chemotherapy and therefore are accountable for cancer relapse. This would be the major challenge in targeting CSCs as it usually causes failure in chemotherapy. CSCs usually display high expression of multidrug-resistant gene whose role involved in drug-efflux pump and is regulated by ABC transporter family such as ABCB1 and ABCB2 (77). Liu described CD133+ lung cancer cells exhibited drug resistance when treated with cisplatin with both ABCG2 and ABCB1 was upregulated and therefore increased the cross resistant to doxorubicin and paclitaxel that usually used for second-line agent (124). In addition, the enrichment of CSCs population were also reported in recent study, whereby the subpopulation of A549 and H2170 with markers CD166+/EpCAM+ showed higher expression upon cisplatin treatment as comparing to basal expression (99). In another study of NSCLC, the CD166+ cells with strong CSCs characteristics also were resistant when exposed to cisplatin drug (125). Furthermore, CSCs can adopt in quiescent state where it resist any cytotoxic insult (126) due to high expression of Bcl-2/Bcl-XL that function in mitochondrial ATP/ADP exchange as to prevent apoptosis which probably contributes to resistant of CSCs upon chemotherapy (127). Thus, in future, it is important to break the CSCs resistance properties as this controlled countless cytotoxic agents from entering the plasma membrane, which afterward cause relapse in cancer.
Current research is struggling in finding a selective ways to inhibit the CSCs and its characteristics. Though certain things need to be considered as CSCs share a similar characteristics to stem cells, whereby targeting the CSCs could also give effect to normal stem cells and toxic to health tissue. Therefore, a selective method have to be done to specifically aiming on CSCs without affect any others normal cells.
Conclusion
As the second most common cancer in men and third most common cancer prevalence in Malaysians diagnosed each year, lung cancer is most complicated cancer diseases to treat due to chemodrug resistant and relapse. Therefore, the fundamental understanding on the basic science studies relating to CSCs development and maintenance are gaining momentum recently due to the fact that it has been known to be responsible to lead cancer progression, tumourigenicity, and therapeutic resistance. Thus, CSCs has become a novel and important clinical targets in cancer therapy in modern treatment. However, considering the fact that each type of tumor involves different epithelial/progenitor or stem cell types that controlled by various molecular pathways, variation in expression of the markers is even existent in the lung cancer subtypes making them all more difficult to be identified and targeted. Therefore, this review had discussed the importance and prospect of lung CSCs that would beneficial for future therapeutic of lung cancer.
Author Contributions
All authors have contributed in writing and editing the manuscript text.
Conflict of Interest Statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Acknowledgments
This manuscript was supported by grants from Universiti Sains Malaysia (USM) Research University Grant (RUI) (1001/CIPPT/811204) and Fundamental Research Grant Scheme (FRGS) (203/CIPPT/6711509), Ministry of Higher Education (MOHE) Malaysia, Universiti Sains Malaysia Fellowship scheme, and scholarship from MyPhD, MOHE.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin (2016) 66(1):7–30. doi:10.3322/caac.21332
3. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature (2001) 414(6859):105–11. doi:10.1038/35102167
4. Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells – perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res (2006) 66(19):9339–44. doi:10.1158/0008-5472.CAN-06-3126
5. Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature (1994) 367(6464):645–8. doi:10.1038/367645a0
6. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med (1997) 3(7):730–7. doi:10.1038/nm0797-730
7. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A (2003) 100(7):3983–8. doi:10.1073/pnas.0530291100
8. Singh S, Hawkins C, Clarke I, Squire J, Bayani J, Hide T, et al. Identification of human brain tumor initiating cells. Nature (2004) 19:396–401. doi:10.1038/nature03128
9. Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res (2005) 65(23):10946–51. doi:10.1158/0008-5472.can-05-2018
10. Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res (2007) 67(3):1030–7. doi:10.1158/0008-5472.can-06-2030
11. O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature (2007) 445(7123):106–10. doi:10.1038/nature05372
12. Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol (2007) 23:675–99. doi:10.1146/annurev.cellbio.22.010305.104154
13. Yang Y, Chang J. Current status and issues in cancer stem cell study. Cancer Invest (2008) 26(7):741–55. doi:10.1080/07357900801901856
14. Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea – a paradigm shift. Cancer Res (2006) 66(4):1883–90. doi:10.1158/0008-5472.CAN-05-3153
15. Kitamura H, Okudela K, Yazawa T, Sato H, Shimoyamada H. Cancer stem cell: implications in cancer biology and therapy with special reference to lung cancer. Lung Cancer (2009) 66(3):275–81. doi:10.1016/j.lungcan.2009.07.019
16. Dick JE, Bhatia M, Gan O, Kapp U, Wang JC. Assay of human stem cells by repopulation of NOD/SCID mice. Stem Cells (1997) 15(Suppl 1):199–203; discussion 204–7. doi:10.1002/stem.5530150826
17. Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev (1983) 2(1):5–23. doi:10.1007/BF00046903
18. Dethlefsen L. The growth dynamics of murine mammary tumor cells in situ. In: McGrath CM, Brennan MJ, Rich MA, editors. Cell Biology of Breast Cancer. New York: Academic Press (1980). p. 145–60.
19. Bosmann HB, Winston RA. Synthesis of glycoprotein, glycolipid, protein, and lipid in synchronized L5178Y cells. J Cell Biol (1970) 45(1):23–33. doi:10.1083/jcb.45.1.23
20. Pasternak C, Warmsley A, Thomas D. Structural alterations in the surface membrane during the cell cycle. J Cell Biol (1971) 50(2):562–4. doi:10.1083/jcb.50.2.562
21. Cikes M, Klein G. Quantitative studies of antigen expression in cultured murine lymphoma cells. I. Cell-surface antigens in “asynchronous” cultures. J Natl Cancer Inst (1972) 49(6):1599–606. doi:10.1093/jnci/49.6.1599
22. Shipley WU. Immune cytolysis in relation to the growth cycle of Chinese hamster cells. Cancer Res (1971) 31(7):925–9.
23. Valeriote F, Van Putten L. Proliferation-dependent cytotoxicity of anticancer agents: a review. Cancer Res (1975) 35(10):2619–30.
24. Weiss L. Cancer cells in primary tumors and their metastases. In: McGrath CM, Brennan MJ, Rich MA, editors. Cell Biology of Breast Cancer. New York: Academic Press (1980). p. 189–205.
25. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature (2013) 501(7467):328–37. doi:10.1038/nature12624
26. Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev (2010) 29(1):61–72. doi:10.1007/s10555-010-9216-5
27. Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? Am J Respir Crit Care Med (2007) 175(6):547–53. doi:10.1164/rccm.200607-984PP
28. Carney D, Gazdar A, Bunn P Jr, Guccion J. Demonstration of the stem cell nature of clonogenic tumor cells from lung cancer patients. Stem Cells (1982) 1(3):149.
29. Kim CFB, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell (2005) 121(6):823–35. doi:10.1016/j.cell.2005.03.032
30. Zakaria N, Yusoff NM, Zakaria Z, Lim MN, Baharuddin PJN, Fakiruddin KS, et al. Human non-small cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer (2015) 15(1):84. doi:10.1186/s12885-015-1086-3
31. Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell (2011) 8(2):136–47. doi:10.1016/j.stem.2011.01.007
32. Zhou S, Schuetz J, Bunting K, Colapietro A, Sampath J, Morris J, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med (2001) 7:1028–34. doi:10.1038/nm0901-1028
33. Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Hum Genomics (2009) 3(3):281. doi:10.1186/1479-7364-3-3-281
34. Goodell M, Brose K, Paradis G, Conner A, Mulligan R. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med (1996) 183:1797–806. doi:10.1084/jem.183.4.1797
35. Camargo FD, Chambers SM, Drew E, McNagny KM, Goodell MA. Hematopoietic stem cells do not engraft with absolute efficiencies. Blood (2006) 107(2):501–7. doi:10.1182/blood-2005-02-0655
36. Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGFβ1. Cell Stem Cell (2010) 6(3):265–78. doi:10.1016/j.stem.2010.02.002
37. Hirschmann-Jax C, Foster A, Wulf G, Nuchtern J, Jax T, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A (2004) 101(39):14228–33. doi:10.1073/pnas.0400067101
38. Wu C, Alman BA. Side population cells in human cancers. Cancer Lett (2008) 268(1):1–9. doi:10.1016/j.canlet.2008.03.048
39. Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res (2007) 67(10):4827–33. doi:10.1158/0008-5472.can-06-3557
40. Seo D-C, Sung J-M, Cho H-J, Yi H, Seo K-H, Choi I-S, et al. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer (2007) 6(1):75. doi:10.1186/1476-4598-6-75
41. Shi Y, Fu X, Hua Y, Han Y, Lu Y, Wang J. The side population in human lung cancer cell line NCI-H460 is enriched in stem-like cancer cells. PLoS One (2012) 7(3):e33358. doi:10.1371/journal.pone.0033358
42. Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature (2007) 445(7123):111–5. doi:10.1038/nature05384
43. Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ (2008) 15(3):504–14. doi:10.1038/sj.cdd.4402283
44. Tirino V, Camerlingo R, Franco R, Malanga D, La Rocca A, Viglietto G, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg (2009) 36(3):446–53. doi:10.1016/j.ejcts.2009.03.063
45. Leung EL-H, Fiscus RR, Tung JW, Tin VP-C, Cheng LC, Sihoe A, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One (2010) 5(11):e14062. doi:10.1371/journal.pone.0014062
46. Chang C, Hsu LC, Dave V, Yoshida A. Expression of human aldehyde dehydrogenase-3 associated with hepatocellular carcinoma: promoter regions and nuclear protein factors related to the expression. Int J Mol Med (1998) 2:333–8.
47. Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of Hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res (2007) 67(5):2187–96. doi:10.1158/0008-5472.CAN-06-3281
48. Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res (2009) 7(3):330–8. doi:10.1158/1541-7786.MCR-08-0393
49. Ucar D, Cogle CR, Zucali JR, Ostmark B, Scott EW, Zori R, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact (2009) 178(1):48–55. doi:10.1016/j.cbi.2008.09.029
50. Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A (2009) 106(38):16281–6. doi:10.1073/pnas.0905653106
51. Yan X, Luo H, Zhou X, Zhu B, Wang Y, Bian X. Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol Rep (2013) 30(6):2733–40. doi:10.3892/or.2013.2784
52. Gutova M, Najbauer J, Gevorgyan A, Metz MZ, Weng Y, Shih C-C, et al. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS One (2007) 2(2):e243. doi:10.1371/journal.pone.0000243
53. Salnikov AV, Gladkich J, Moldenhauer G, Volm M, Mattern J, Herr I. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. Int J Cancer (2010) 126(4):950–8. doi:10.1002/ijc.24822
54. Napier SL, Healy ZR, Schnaar RL, Konstantopoulos K. Selectin ligand expression regulates the initial vascular interactions of colon carcinoma cells: the roles of CD44v and alternative sialofucosylated selectin ligands. J Biol Chem (2007) 282(6):3433–41. doi:10.1074/jbc.M607219200
55. Du L, Wang H, He L, Zhang J, Ni B, Wang X, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res (2008) 14(21):6751–60. doi:10.1158/1078-0432.CCR-08-1034
56. Zeilstra J, Joosten SP, Dokter M, Verwiel E, Spaargaren M, Pals ST. Deletion of the WNT target and cancer stem cell marker CD44 in Apc (Min/+) mice attenuates intestinal tumorigenesis. Cancer Res (2008) 68(10):3655–61. doi:10.1158/0008-5472.CAN-07-2940
57. Fasano M, Sabatini MT, Wieczorek R, Sidhu G, Goswami S, Jagirdar J. CD44 and its v6 spliced variant in lung tumors: a role in histogenesis? Cancer (1997) 80(1):34–41. doi:10.1002/(SICI)1097-0142(19970701)80:1<34::AID-CNCR5>3.3.CO;2-Z
58. Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev (2002) 16(23):3074–86. doi:10.1101/gad.242602
59. Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One (2012) 7(8):e42564. doi:10.1371/journal.pone.0042564
60. Tachezy M, Effenberger K, Zander H, Minner S, Gebauer F, Vashist YK, et al. ALCAM (CD166) expression and serum levels are markers for poor survival of esophageal cancer patients. Int J Cancer (2012) 131(2):396–405. doi:10.1002/ijc.26377
61. Tachezy M, Zander H, Marx AH, Stahl PR, Gebauer F, Izbicki JR, et al. ALCAM (CD166) expression and serum levels in pancreatic cancer. PLoS One (2012) 7(6):e39018. doi:10.1371/journal.pone.0039018
62. Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell (2012) 148(1):259–72. doi:10.1016/j.cell.2011.11.050
63. Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev Rep (2011) 7(2):292–306. doi:10.1007/s12015-010-9208-4
64. Moreb JS, Gabr A, Vartikar GR, Gowda S, Zucali JR, Mohuczy D. Retinoic acid down-regulates aldehyde dehydrogenase and increases cytotoxicity of 4-hydroperoxycyclophosphamide and acetaldehyde. J Pharmacol Exp Ther (2005) 312(1):339–45. doi:10.1124/jpet.104.072496
65. Black W, Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum Genomics (2009) 4(2):136. doi:10.1186/1479-7364-4-2-136
66. Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle (2011) 10(9):1378–84. doi:10.4161/cc.10.9.15486
67. Ma S, Chan KW, Lee TK-W, Tang KH, Wo JY-H, Zheng B-J, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res (2008) 6(7):1146–53. doi:10.1158/1541-7786.MCR-08-0035
68. Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells (2005) 23(6):752–60. doi:10.1634/stemcells.2004-0292
69. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell (2007) 1(5):555–67. doi:10.1016/j.stem.2007.08.014
70. Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer (2008) 59(3):340–9. doi:10.1016/j.lungcan.2007.08.033
71. Meng X, Li M, Wang X, Wang Y, Ma D. Both CD133+ and CD133- subpopulations of A549 and H446 cells contain cancer-initiating cells. Cancer Sci (2009) 100(6):1040–6. doi:10.1111/j.1349-7006.2009.01144.x
72. Halim N, Zakaria N, Satar NA, Yahaya BH. Isolation and characterization of cancer stem cells of the non-small-cell lung cancer (A549) cell line. Methods Mol Biol (2016) 1516:371–88. doi:10.1007/7651_2016_326
73. Halim NHA, Zakaria N, Yahaya BH. Down-regulation of ALDH1A1 increased expression of carcinogenesis-related genes in non-small cell lung cancer cell line of A549. J Teknologi (Sci Eng) (2016) 78:11–3, 19–25.
74. Justilien V, Fields AP. Molecular pathways: novel approaches for improved therapeutic targeting of Hedgehog signaling in cancer stem cells. Clin Cancer Res (2015) 21(3):505–13. doi:10.1158/1078-0432.CCR-14-0507
75. Singh RR, Kunkalla K, Qu C, Schlette E, Neelapu SS, Samaniego F, et al. ABCG2 is a direct transcriptional target of Hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene (2011) 30(49):4874–86. doi:10.1038/onc.2011.195
76. Leprieur EG, Vieira T, Antoine M, Rozensztajn N, Rabbe N, Ruppert A-M, et al. Sonic Hedgehog pathway activation is associated with resistance to platinum-based chemotherapy in advanced non-small-cell lung carcinoma. Clin Lung Cancer (2015) 17(4):301–8. doi:10.1016/j.cllc.2015.12.007
77. Donnenberg VS, Donnenberg AD. Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J Clin Pharmacol (2005) 45(8):872–7. doi:10.1177/0091270005276905
78. Li SL, Ye F, Cai WJ, Hu HD, Hu P, Ren H, et al. Quantitative proteome analysis of multidrug resistance in human ovarian cancer cell line. J Cell Biochem (2010) 109(4):625–33. doi:10.1002/jcb.22413
79. Guo C-Y, Yan C, Luo L, Goto S, Urata Y, Xu J-J, et al. Enhanced expression of PKM2 associates with the biological properties of cancer stem cells from A549 human lung cancer cells. Oncol Rep (2017) 37(4):2161–6. doi:10.3892/or.2017.5438
80. Roy S, Lu K, Nayak MK, Bhuniya A, Ghosh T, Kundu S, et al. Activation of D2 dopamine receptors in CD133+ ve cancer stem cells in non-small cell lung carcinoma inhibits proliferation, clonogenic ability and invasiveness of these cells. J Biol Chem (2017) 292(2):435–45. doi:10.1074/jbc.M116.748970
81. Stewart CA, Byers LA. Altering the course of small cell lung cancer: targeting cancer stem cells via LSD1 inhibition. Cancer Cell (2015) 28(1):4–6. doi:10.1016/j.ccell.2015.06.011
82. Wang P, Gao Q, Suo Z, Munthe E, Solberg S, Ma L, et al. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS One (2013) 8(3):e57020. doi:10.1371/journal.pone.0057020
83. Li F, Zeng H, Ying K. The combination of stem cell markers CD133 and ABCG2 predicts relapse in stage I non-small cell lung carcinomas. Med Oncol (2011) 28(4):1458–62. doi:10.1007/s12032-010-9646-5
84. Kristiansen G, Schlüns K, Yongwei Y, Denkert C, Dietel M, Petersen I. CD24 is an independent prognostic marker of survival in nonsmall cell lung cancer patients. Br J Cancer (2003) 88(2):231–6. doi:10.1038/sj.bjc.6600702
85. Karimi-Busheri F, Rasouli-Nia A, Zadorozhny V, Fakhrai H. CD24+/CD38- as new prognostic marker for non-small cell lung cancer. Multidiscip Respir Med (2013) 8(1):65. doi:10.1186/2049-6958-8-65
86. Lun SW-M, Cheung ST, Cheung PFY, To K-F, Woo JK-S, Choy K-W, et al. CD44+ cancer stem-like cells in EBV-associated nasopharyngeal carcinoma. PLoS One (2012) 7(12):e52426. doi:10.1371/journal.pone.0052426
87. Yang C-H, Wang H-L, Lin Y-S, Kumar KS, Lin H-C, Chang C-J, et al. Identification of CD24 as a cancer stem cell marker in human nasopharyngeal carcinoma. PLoS One (2014) 9(6):e99412. doi:10.1371/journal.pone.0099412
88. Nosrati A, Naghshvar F, Khanari S. Cancer stem cell markers CD44, CD133 in primary gastric adenocarcinoma. Int J Mol Cell Med (2014) 3(4):279.
89. Horst D, Kriegl L, Engel J, Kirchner T, Jung A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest (2009) 27(8):844–50. doi:10.1080/07357900902744502
90. Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells (2009) 27(5):1006–20. doi:10.1002/stem.30
91. Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology (2010) 139(6):2072–2082.e5. doi:10.1053/j.gastro.2010.08.053
92. Lugli A, Iezzi G, Hostettler I, Muraro M, Mele V, Tornillo L, et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer (2010) 103(3):382–90. doi:10.1038/sj.bjc.6605762
93. Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res (2008) 10(1):R10. doi:10.1186/bcr1855
94. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell (2007) 1(3):313–23. doi:10.1016/j.stem.2007.06.002
95. Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res (2010) 16(1):45–55. doi:10.1158/1078-0432.CCR-09-1630
96. Liu X, Wang L, Cui W, Yuan X, Lin L, Cao Q, et al. Targeting ALDH1A1 by disulfiram/copper complex inhibits non-small cell lung cancer recurrence driven by ALDH-positive cancer stem cells. Oncotarget (2016) 7(36):58516–30. doi:10.18632/oncotarget.11305
97. Lin S-S, Lai K-C, Hsu S-C, Yang J-S, Kuo C-L, Lin J-P, et al. Curcumin inhibits the migration and invasion of human A549 lung cancer cells through the inhibition of matrix metalloproteinase-2 and-9 and vascular endothelial growth factor (VEGF). Cancer Lett (2009) 285(2):127–33. doi:10.1016/j.canlet.2009.04.037
98. Li Y, Zhang T. Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett (2014) 346(2):197–205. doi:10.1016/j.canlet.2014.01.012
99. Baharuddin P, Satar N, Fakiruddin KS, Zakaria N, Lim MN, Yusoff NM, et al. Curcumin improves the efficacy of cisplatin by targeting cancer stem-like cells through p21 and cyclin D1-mediated tumour cell inhibition in non-small cell lung cancer cell lines. Oncol Rep (2016) 35(1):13–25. doi:10.3892/or.2015.4371
100. Chen P, Li J, Jiang H-G, Lan T, Chen Y-C. Curcumin reverses cisplatin resistance in cisplatin-resistant lung caner cells by inhibiting FA/BRCA pathway. Tumor Biol (2015) 36(5):3591–9. doi:10.1007/s13277-014-2996-4
101. Zhu JY, Yang X, Chen Y, Jiang Y, Wang SJ, Li Y, et al. Curcumin suppresses lung cancer stem cells via inhibiting Wnt/β-catenin and Sonic Hedgehog pathways. Phytother Res (2017) 31(4):680–8. doi:10.1002/ptr.5791
102. Yu Y, Kanwar SS, Patel BB, Nautiyal J, Sarkar FH, Majumdar AP. Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Transl Oncol (2009) 2(4):321–8. doi:10.1593/tlo.09193
103. Kakarala M, Brenner DE, Korkaya H, Cheng C, Tazi K, Ginestier C, et al. Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat (2010) 122(3):777–85. doi:10.1007/s10549-009-0612-x
104. Fong D, Yeh A, Naftalovich R, Choi TH, Chan MM. Curcumin inhibits the side population (SP) phenotype of the rat C6 glioma cell line: towards targeting of cancer stem cells with phytochemicals. Cancer Lett (2010) 293(1):65–72. doi:10.1016/j.canlet.2009.12.018
105. James MI, Iwuji C, Irving G, Karmokar A, Higgins JA, Griffin-Teal N, et al. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett (2015) 364(2):135–41. doi:10.1016/j.canlet.2015.05.005
106. Ke SZ, Ni XY, Zhang YH, Wang YN, Wu B, Gao FG. Camptothecin and cisplatin upregulate ABCG2 and MRP2 expression by activating the ATM/NF-? B pathway in lung cancer cells. Int J Oncol (2013) 42(4):1289–96. doi:10.3892/ijo.2013.1805
107. Borovski T, Felipe De Sousa EM, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res (2011) 71(3):634–9. doi:10.1158/0008-5472.CAN-10-3220
108. Han L, Shi S, Gong T, Zhang Z, Sun X. Cancer stem cells: therapeutic implications and perspectives in cancer therapy. Acta Pharm Sin B (2013) 3(2):65–75. doi:10.1016/j.apsb.2013.02.006
109. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A perivascular niche for brain tumor stem cells. Cancer Cell (2007) 11(1):69–82. doi:10.1016/j.ccr.2006.11.020
110. Zheng Y, Cecile C, Sayles LC, Alleyne-Chin C, Vaka D, Knaak TD, et al. A rare population of CD24+ ITGB4+ Notch hi cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell (2013) 24(1):59–74. doi:10.1016/j.ccr.2013.05.021
111. Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, et al. Regulated splicing of the α6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep (2014) 7(3):747–61. doi:10.1016/j.celrep.2014.03.059
112. Dickreuter E, Eke I, Krause M, Borgmann K, van Vugt M, Cordes N. Targeting of β1 integrins impairs DNA repair for radiosensitization of head and neck cancer cells. Oncogene (2016) 35(11):1353–62. doi:10.1038/onc.2015.212
113. Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol (2015) 25(4):234–40. doi:10.1016/j.tcb.2014.12.006
114. Weiss SJ, Dudley DT. Integrin Alpha-2 Binding Agents and Use Thereof to Inhibit Cancer Cell Proliferation. (2015). Google patent US 8975029 B2 (2015).
115. Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev (2015) 81:53–61. doi:10.1016/j.addr.2014.11.014
116. Miao Y, Li J, Qiu X, Li Y, Wang Z, Luan Y. miR-27a regulates the self renewal of the H446 small cell lung cancer cell line in vitro. Oncol Rep (2013) 29(1):161–8. doi:10.3892/or.2012.2095
117. Hua S, Xiaotao X, Renhua G, Yongmei Y, Lianke L, Wen G, et al. Reduced miR-31 and let-7 maintain the balance between differentiation and quiescence in lung cancer stem-like side population cells. Biomed Pharmacother (2012) 66(2):89–97. doi:10.1016/j.biopha.2011.09.013
118. Hu J, Qiu M, Jiang F, Zhang S, Yang X, Wang J, et al. miR-145 regulates cancer stem-like properties and epithelial-to-mesenchymal transition in lung adenocarcinoma-initiating cells. Tumor Biol (2014) 35(9):8953–61. doi:10.1007/s13277-014-2158-8
119. Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One (2014) 9(3):e90022. doi:10.1371/journal.pone.0090022
120. Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology (2003) 8(4):432–46. doi:10.1046/j.1440-1843.2003.00493.x
121. Mercer BA, Lemaître V, Powell CA, D’Armiento J. The epithelial cell in lung health and emphysema pathogenesis. Curr Respir Med Rev (2006) 2(2):101. doi:10.2174/157339806776843085
122. Roudi R, Madjd Z, Ebrahimi M, Samani FS, Samadikuchaksaraei A. CD44 and CD24 cannot act as cancer stem cell markers in human lung adenocarcinoma cell line A549. Cell Mol Biol Lett (2014) 19(1):23–36. doi:10.2478/s11658-013-0112-1
123. Okudela K, Nagahara N, Katayama A, Kitamura H. Cancer stem cells in lung cancer: distinct differences between small cell and non-small cell lung carcinomas. In: Stanley S, editor. Cancer Stem Cells Theories and Practice. InTech (2011). Available from: http://www.intechopen.com/books/cancer-stem-cells-theories-and-practice/cancer-stem-cells-in-lung-cancerdistinct-differences-between-small-cell-and-non-small-cell-lung-car
124. Liu Y-P, Yang C-J, Huang M-S, Yeh C-T, Wu AT, Lee Y-C, et al. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signaling. Cancer Res (2013) 73(1):406–16. doi:10.1158/0008-5472.CAN-12-1733
125. Su J, Wu S, Tang W, Qian H, Zhou H, Guo T. Reduced SLC27A2 induces cisplatin resistance in lung cancer stem cells by negatively regulating Bmi1-ABCG2 signaling. Mol Carcinog (2016) 55(11):1822–32. doi:10.1002/mc.22430
126. Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature (2012) 488(7412):522–6. doi:10.1038/nature11287
Keywords: lung cancer, non-small cell lung cancer, cancer stem cells, neoplastic stem cells, surface markers, therapeutics
Citation: Zakaria N, Satar NA, Abu Halim NH, Ngalim SH, Yusoff NM, Lin J and Yahaya BH (2017) Targeting Lung Cancer Stem Cells: Research and Clinical Impacts. Front. Oncol. 7:80. doi: 10.3389/fonc.2017.00080
Received: 19 February 2017; Accepted: 11 April 2017;
Published: 05 May 2017
Edited by:
Ander Matheu, Biodonostia, SpainReviewed by:
Maria Buoncervello, Istituto Superiore di Sanità, ItalyShree Ram Singh, National Cancer Institute, USA
Debangshu Samanta, Johns Hopkins University, USA
Copyright: © 2017 Zakaria, Satar, Abu Halim, Ngalim, Yusoff, Lin and Yahaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Badrul Hisham Yahaya, YmFkcnVsQHVzbS5teQ==
†These authors have contributed equally as the first author.
 Norashikin Zakaria
Norashikin Zakaria Nazilah Abdul Satar
Nazilah Abdul Satar Noor Hanis Abu Halim
Noor Hanis Abu Halim Siti Hawa Ngalim1
Siti Hawa Ngalim1 Badrul Hisham Yahaya
Badrul Hisham Yahaya