- 1Gillies McIndoe Research Institute, Wellington, New Zealand
- 2Wellington Regional Plastic, Maxillofacial and Burns Unit, Hutt Hospital, Wellington, New Zealand
Cancer stem cells (CSCs) have been identified in oral cavity squamous cell carcinoma (OCSCC). CSCs possess the ability for perpetual self-renewal and proliferation, producing downstream progenitor cells and cancer cells that drive tumor growth. Studies of many cancer types including OCSCC have identified CSCs using specific markers, but it is still unclear as to where in the stem cell hierarchy these markers fall. This is compounded further by the presence of multiple CSC subtypes within OCSCC, making investigation reliant on the use of multiple markers. This review examines the current knowledge in CSC markers OCT4, SOX2, NANOG, ALDH1, phosphorylated STAT3, CD44, CD24, CD133, and Musashi-1, specifically focusing on their use and validity in OCSCC CSC research and how they may be organized into the CSC hierarchy. OCSCC CSCs also express components of the renin–angiotensin system (RAS), which suggests CSCs may be novel therapeutic targets by modulation of the RAS using existing medications.
Introduction
The overall 5-year survival of oral cavity squamous cell carcinoma (OCSCC) has remained at 50%, largely unchanged for 40 years (1), despite intensive research. This high mortality has been largely attributed to high rates of loco-regional recurrence (2, 3). An emerging hierarchical concept of carcinogenesis proposes that cancer stem cells (CSCs) sit atop a hierarchy of a heterogeneous population of cells within cancer and are defined functionally as a subset of cells that display stemness characteristics, including the ability to asymmetrically divide, resulting in self-renewal of CSCs and the production of heterogeneous populations of cancer cells that are further down the hierarchical ladder (4, 5). CSCs are highly tumorigenic compared to the other cancer cells and are believed to be largely responsible for the biological characteristics of cancer, namely, rapid growth, invasion, and metastasis (Figure 1). CSCs also show a greater capacity for migration, invasion, and proliferation in vitro (6, 7), and they generate far larger tumors in immunocompromised xenograft mice with fewer transplanted cells compared to large numbers of unsorted cancer cells (8, 9).
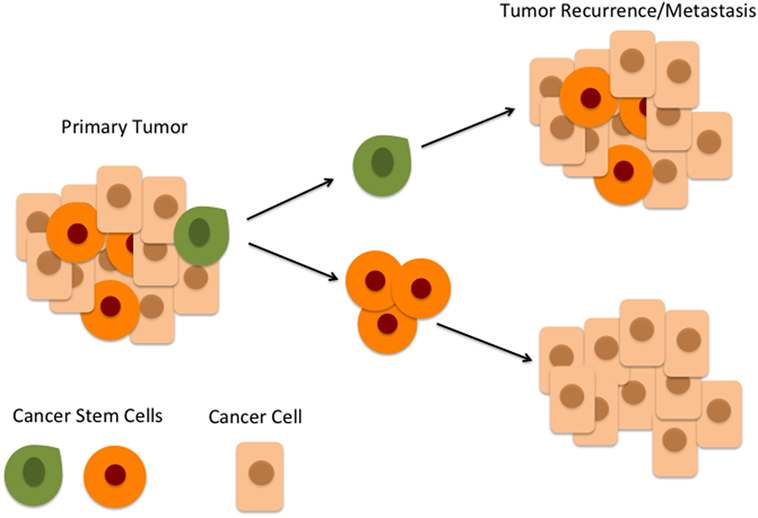
Figure 1. According to the hierarchical model of cancer, an oral cavity squamous cell carcinoma consists of a heterogeneous population of cells. At the top of the hierarchy is a small number of cancer stem cells (CSCs) within the peritumoral stroma (green) which differentiate and give rise to downstream CSCs (orange) which in turn give rise to cancer cells (beige). CSCs at the top of the hierarchy (green) are highly tumorigenic and are responsible for tumor recurrence and metastasis.
Surgery with adjuvant radiotherapy (RT) and occasionally chemotherapy (ChT) is the mainstay treatment for OCSCC, which effectively reduce total tumor size (10). However, CSCs are predominantly in the inactive G0 phase and thus avoid destruction by RT and ChT that target active cells (11). CSCs in OSCCC are resistant to both RT and ChT agents such as cisplatin (6, 8, 12, 13), carboplatin (6), doxetaxel (6), paclitaxel (6, 14), etoposide (15), gemcitabine (9), and 5-fluorouracil (6–8). Thus, treatment results in an enriching effect on CSCs within the post-treatment cancer cell population (16), providing a plausible rationale for loco-regional recurrence and distant metastasis from RT- and ChT-resistant cells, despite aggressive treatment.
The rapidly accumulating evidence supporting the existence and the role of CSCs in carcinogenesis in recent years is largely due to the advances in cell biology and the discovery of reliable markers of CSC (17). The expression profiles of a number of protein markers have been studied as putative CSC markers within OCSCC tumor samples and cell lines. No single marker has been shown to unequivocally identify CSCs, and it is likely that CSCs exist in an overlapping hierarchy of cell population subsets. Consequently, the majority of CSC characterization research relies on using combinations of these markers. This article reviews the common markers that have been used in CSC research into OCSCC and attempts to place them within the context of a hierarchical model of cancer.
Embryonic Stem Cell (ESC) Marker Master Regulators
Cancer stem cells in OCSCC express many of the same proteins involved in the core network that regulates ESCs. OCT4 and NANOG are two of the possible six major factors involved in reprogramming of somatic cells into ESC-like states (18, 19). OCT4, NANOG, and SOX2 are considered master regulators for self-renewal and maintenance of the stem cell population in the undifferentiated state (17, 20). Immunohistochemical staining for OCT4, SOX2, and NANOG in OCSCC demonstrates that OCT4 and SOX2 are expressed significantly higher in tumor-adjacent tissue compared to both normal tissue and the tumor (21). However, NANOG is highly expressed in both tumor tissue and peritumoral tissue, compared to normal tissue (21).
OCT4
The transcription factor OCT4 is a regulator of the POU domain and is critical in early embryogenesis and maintenance of ESC pluripotency (22). As such, OCT4 is commonly used as a marker of “stemness” of primitive cells. OCT4 has also been linked to oncogenic processes (17). It has been suggested that OCT4 plays a role in tumor transformation, tumorigenicity, invasion, and metastasis within OCSCC (23). It has also been proposed that OCT4 promotes tumor initiation by playing a role in the regulation of epithelial–mesenchymal transition (EMT) (13). Expression of OCT4 has been used to define the CSC population in OCSCC in conjunction with other primitive and CSC markers (24–26) and has been shown to contribute to tumorigenicity in murine models (27).
OCT4 has been hypothesized to play an important role in aberrant cell reprogramming resulting in carcinogenesis (28). In moderately differentiated buccal mucosal SCC (BMSCC), expression of OCT4 has been demonstrated in a distinct subpopulation of CSCs within the tumor nests, the peritumoral stroma, and the microvessels within the peritumoral stroma (29). Interestingly in moderately differentiated oral tongue SCC (OTSCC), the expression of OCT4 is restricted to a subpopulation of CSCs within the peritumoral stroma (30). Intriguingly, high levels of expression of OCT4 in OCSCC have been associated with early stage of disease, and better prognosis, and a molecular mechanism explaining this association has yet to be elucidated (21).
SOX2
The SOX2 protein is a high-mobility SRY-related HMG box transcription factor (31, 32). SOX2 is involved in multiple signal transduction pathways and has been shown to be involved in normal developmental and many pathological processes including cell proliferation, migration, invasion, stemness, tumorigenesis, anti-apoptosis, and chemoresistance (31, 33). SOX2 is known to complex with OCT4 (34), and in murine cell lines has been shown to control downstream embryonic genes including NANOG (20, 35). SOX2 overexpression has been used in combination with other markers, including ALDH1, CD44, OCT4, and NANOG, to identify the CSC population in SCC including OTSCC (26, 30, 31, 36). In BMSCC, SOX2 is expressed within the tumor nests, the peritumoral stroma and the endothelium of the microvessels within the peritumoral stroma (29). In OCSCC and oropharyngeal SCC cell lines, SOX2 is overexpressed in CSCs compared to the parental cell population (37). In OTSCC, SOX2 is expressed by cells that also express SALL4, NANOG, phosphorylated STAT3 (pSTAT3), and CD44 (30). In OCSCC, SOX2 expression is significantly higher in tumor tissue compared to normal tissue and is weakly correlated with OCT4 (21). In addition, SOX2 expression is correlated with small tumor size and early tumor stage, and better disease-free survival (21). SOX2 staining in OCSCC has been demonstrated in both a peripheral and diffuse staining pattern, and the diffuse staining pattern was significantly associated with lymph node metastasis (38). Chien et al. (39) demonstrate that regulation by the Lin28B-Let7 pathway, with the Lin28Bhigh-Let7low expression pattern highly correlated with high levels of expression of OCT4 and SOX2 in OCSCC specimens, and a high percentage of CD44+/ALDH1+ CSC in OCSCC. Overexpression of SOX2 has been demonstrated to enhance invasiveness, anchorage-independent growth, and xenotransplantation tumorigenicity in OCSCC cells. Conversely, silencing SOX2 effectively suppresses the expression of drug resistance and anti-apoptotic genes and increased the sensitivity of the cells to radiation combined cisplatin treatment (33).
NANOG
NANOG is a homeodomain transcription factor widely known as a marker for primitiveness or “stemness” (20). In murine cell lines, NANOG has been shown to be involved in functionally blocking differentiation and thus maintenance of ESC pluripotency (20, 35). NANOG has been shown to be upregulated in different types of cancers and plays a role in tumor transformation, tumorigenicity, and metastasis within OCSCC (23). Overexpression of NANOG is also correlated with poor differentiation status and chemoresistance (40). In OCSCC, increased expression of NANOG has been found to be associated with poor prognosis (41). NANOG expression has also been confirmed in OTSCC (30). In BMSCC, NANOG is expressed in cells within the tumor nests and the peritumoral stroma (29). In OCSCC and oropharyngeal SCC cell lines, NANOG is overexpressed in the CSC population compared to the parental population (37). Similarly in lip SCC, NANOG is expressed by three distinct putative CSC subpopulations, both within the tumor nests and the peritumoral stroma (42).
Signal Transducer and Activator of Transcription 3 (STAT3)
Signal transducer and activator of transcription 3 has long been recognized as an oncogene playing a key role in control of cell-cycle progression and anti-apoptosis (43). pSTAT3 plays a critical role in pluripotent stem cells, promoting cell proliferation and resistance to apoptosis, angiogenesis, invasion, and migration (44). In OCSCC, expression of STAT3 within a cell population is localized to the tumor nests that also express CD44, NANOG, and SOX2 (30). Constitutive activation of the STAT3 signaling pathway possesses confirmed oncogenic potential in OCSCC (45). Cross talk with other molecular pathways contributes to STAT3 regulation in cancer (45), and STAT3 is also aberrantly activated by the oversupply of growth factors from the tumor microenvironment (43). For example, Erk1/2 appears to promote serine-pSTAT3, but inhibit tyrosine-pSTAT3 resulting in an overall increased cell growth and varying roles for the different STAT3 phosphorylation sites in OCSCC (45). STAT3 has also been recently found to function co-operatively with SOX2 in the initiation of SCC (32). This further highlights the crucial role of those transcription factors in stem and/or cellular proliferation (44).
Signal transducer and activator of transcription has a dual role in tumor inflammation and immunity by promoting pro-oncogenic inflammatory pathways, including NF-κB and IL-6–GP130–JAK pathways, and by opposing STAT1- and NF-κB-mediated T(h)1 antitumor immune response (46). Continuous deregulation of these genes in tumor cells and the tumor microenvironment by persistently activated STAT3 and NF-κB, in contrast to their tightly controlled regulation in normal physiology, is considered crucial for inflammation and malignant progression (46). In a murine SCC model, forced constitutive activation of pSTAT3 shortens the latency period and increases the number of skin lesions progressing rapidly to SCC (47–54).
OCSCC Cancer Stem Markers
CD44
CD44 is a large cell surface hyaluronan receptor protein (36) involved with contrasting roles in both cell migration and adhesion (55). In OCSCC cell lines, CD44 is expressed significantly more highly in CSCs compared to parental cells (37). It has been widely used as a CSC marker in epithelial cancers including OCSCC. CD44 has been identified within normal oral epithelium, carcinoma in situ, and in some infiltrating lymphocytes, with the highest expression in carcinoma cells (56, 57). In OCSCC cell lines, Song et al. (15) have demonstrated increased expression of CD44 in side populations that also highly express ABC transporter proteins and Hoechst 33342 efflux, compared to the non-side population.
Overexpression of CD44 within OCSCC has been associated with decreased overall survival (58, 59), increased loco-regional recurrence, and increased resistance to RT, thus exhibiting many of the characteristics of CSCs. In one study of OCSCC, irregular staining of CD44 in tumor cells is shown to be associated with poor tumor differentiation and advanced stage (60). Conversely, another study finds no prognostic significance of CD44v6 expression in OTSCC (61). These differences may, in part, be explained by the expression of CD44v6 by CSCs, as well as inflammatory cells (62). Expression of the variant isoform CD44v6 has also been found to be significantly associated with regional nodal metastasis, pattern of invasion, depth of invasion, perineural invasion, and local recurrence in multiple solid tumors including OCSCC (63). OCSCC cell clones expressing stable levels of CD44 after transfection with CD44 expression vector increases proliferation and migration, inhibition of apoptosis, and cisplatin resistance resulting in a more aggressive tumor phenotype in vivo (64).
In addition, cleavage of CD44 regulated by ADAM17 has been found to be necessary for tumor sphere formation in OTSCC cells (65).
Tumors generated from CSC sorted OTSCC cell line (SCC9-CD44high) cells demonstrate increased tumorigenicity and increased expression of CK19, B-catenin, E-cadherin, and CD44 when compared with wild-type SCC9 cells. These same tumors show lower expression of CK19/4/15/13, and interestingly low levels of NANOG, Bmi-1, Snail, and Slug (66). However, the role of CD44 as a marker of CSCs is controversial, with many authors arguing that it is actually expressed by more differentiated cells (67). Lee et al. (41) find that increased CD44 expression has limited correlation with high histological grade and late clinical stage. However, Kokko et al. (68) demonstrate no association between expression of CD44 and poor prognosis in OCSCC. A recent study suggests that CD44 loses its expression during induced cellular reprogramming to the undifferentiated state and is actually a marker of partially differentiated cells (69). This may indicate a progressive gain of CD44 expression as CSCs progress to a more differentiated phenotype, and this implies that CD44 is in fact a relatively mature marker, likely downstream of the true CSC population. Interestingly, downregulation of CD44 also leads to reduced expression of OCT4, suggesting that CD44 has a functional role in maintaining stem cell properties (70). CD44+/CD133+ cells demonstrate higher clonogenic capacity than CD133− cells in vitro, while higher CD44 expression is demonstrated in nodal metastases, suggesting a role for CD44 in tumor progression (71).
CD24
CD24 is a small cell surface glycoprotein involved in cell adhesion and metastasis and has been identified in wide variety of cancer cells (72). A recent study using sorted OCSCC cells in a NOD/SCID murine model suggests that CD24+ cells may have angiogenic potential. Tumors generated from CD24+ cells isolated show a significantly higher functional capillary density, confirmed by the expression of CD31, than those seeded with CD24− cells (73).
CD44high/CD24low cells demonstrate CSC and EMT characteristics, and are able to give rise to all other tumor cell types upon differentiation (74). In OCSCC cell lines, CD44v3+/CD24− population demonstrated higher sphere forming capacity, higher drug resistance, and expressed higher mRNA levels of CSC-related genes. Expression of CD44v3 is found to be higher in lymph node metastases and in the invasive portion of tumors and is associated with poorer overall survival (75).
CD133
CD133 is a pentaspan transmembrane protein that plays a key role in the organization of plasma membrane topology (76, 77). Overexpression of CD133, first identified in hematopoietic stem cells and endothelial progenitor cells (57), is often used as a CSC marker in many solid tumors including OCSCC (23). There remain controversies surrounding the role of CD133 in tumorigenesis with reports regarding the utility of this protein as a CSC marker often being contradictory (77). These conflicting reports are based on the observation that both the CD133+ and CD133− cell fractions display similar stemness and differentiation capabilities, and that the CD133− population is in fact more tumorigenic (77).
However, CD133+ oral leukoplakia has been shown to be more than three times as likely to progress to OCSCC than CD133− lesions (78). Of all CSC phenotypes studied, OCSCC lesions displaying triple-positive expression of OCT4, NANOG, and CD133, are associated with the worst survival (23). CD133+ cells have also been found to co-express CD44, and the CD133+/CD44+ immunophenotype has been found to correlate significantly with poorer overall survival, supporting the idea that cells expressing these proteins have a more aggressive phenotype (58). The expression of CD133 in oral epithelium increases from normal epithelium, through dysplasia, to carcinoma (79). Pozzi et al. (37) demonstrate that along with multiple CSC and ESC markers, CD133 is more highly expressed in the CSC population compared to the parental normal population. In several cell lines, CD133+ cells have been found to overexpress ESC markers, including OCT4 and NANOG, and also display CSC characteristics such as tumor sphere formation, tumorigenicity and chemoresistance (14). In a head and neck SCC cell line, inhibition of CD133 expression significantly reduces proliferation, expression of ESC marker OCT4, but increases the expression of the epithelial differentiation marker CK18, suggesting its role in the maintenance of the CSC-phenotype (80, 81).
Musashi-1
Musashi-1 is a translational regulator that has been identified within OSCC (17). Musashi-1 expression has been associated with higher stage and poorly differentiated status of OCSCC, and is significantly correlated with CD133, suggesting a functional role for these two proteins in oral carcinogenesis (79).
ALDH1
Aldehyde dehydrogenase (ALDH) is a cytosolic enzyme responsible for catalyzing the pyridine nucleotide-dependent oxidation of aldehydes to carboxylic acids (82). ALDH has increasingly been used as a CSC marker in OCSCC, with ALDH+ cells demonstrating plasticity with the ability to form tumor spheres in serum-free media as well as having the ability to generate ALDH− cells in vitro (83). Although there are many isoforms of ALDH, ALDH1 appears to be of particular importance (84). ALDH1 is likely to play a role in malignant transformation of oral leukoplakia to OCSCC given that ALDH1+ leukoplakia is more than three times more likely to develop OCSCC (78). Overexpression of ALDH1 is also found to be correlated with nodal metastasis (38). A suppression subtractive hybridization assay shows that the ALDH+ subpopulation expresses many known CSC-related genes not seen in the ALDH− population (83). Furthermore, in HNSCC, ALDHhigh cells are seen to be more tumorigenic than ALDHlow cells when implanted into a NOD/SCID murine model (85). In one study of OCSCC, ALDH1+ cells display radioresistance and co-expressed Snail, providing evidence of EMT. Interestingly, knockdown of Snail significantly decreased ALDH1 expression and inhibited CSC properties, with resultant decreased tumorigenicity (86).
Renin–Angiotensin System (RAS)
Cancer stem cells within OCSCC have been found to express components of the RAS. (Pro)renin receptor (PRR), angiotensin II receptor 1 (ATIIR1), and angiotensin II receptor 2 (ATIIR2) are expressed by two CSC subpopulations within OTSCC: one within the tumor nests that express SALL4 and another within the peritumoral stroma that express OCT4 (87). PRR, ATIIR1, and ATIIR2 are localized to the CSC subpopulations within the tumor nests and the peritumoral stroma, while PRR and ACE are localized to the endothelium of the microvessels within the peritumoral stroma (88). These findings suggest CSC as a potential novel therapeutic target by modulating the RAS using commonly used medications such as the aliskiren, a direct renin blocker; β-blockers which reduce renin levels; ACE inhibitors which inhibit conversion of angiotensin I to angiotensin II; and angiotensin receptor blockers which prevent binding of angiotensin II to ATIIR1 and ATIIR2 (89).
Discussion
The origin of CSCs remains unclear, and many hypotheses have been advanced (90). One of the most accepted theories proposes that CSCs arise as a result of epigenetic or genetic alterations to these resident tissue stem cells (55, 91–93). The CSC concept of cancer is evolving as evidenced from increasingly sophisticated research accumulates (94). Rather than a single small population of CSCs and a large majority of bulk tumor cells, the presence of a complex hierarchy of distinct, genetically heterogeneous subpopulations of CSCs, each expressing an overlapping array of markers (Table 1) is appreciated (94) (Figure 2).
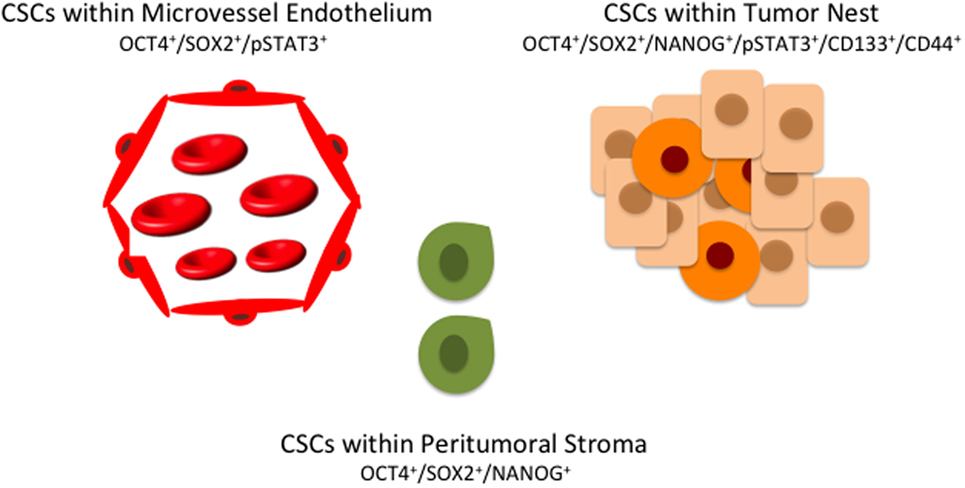
Figure 2. A hierarchy of cancer stem cells (CSCs) in oral cavity squamous cell carcinoma exists with multiple distinct subpopulations, each expressing overlapping markers. CSCs within the peritumoral stroma (green) co-express OCT4, SOX4, and NANOG. CSCs within the tumor nests (orange) co-express OCT, SOX2, NANOG, phosphorylated STAT3 (pSTAT3), CD133, and CD44. A further subpopulation of CSCs (red) co-expressing OCT4, SOX2, and pSTAT3 are present on the endothelium of the microvessels within the peritumoral stroma.
Cancer stem cells also display plasticity (95), and the complexity of these subpopulations increases as tumors progress to more advanced stages (94, 96). Increased density of CSCs, identified by high levels of expression of various primitive cell and CSC markers, has also been shown to be associated with poor prognosis, with much research focused on identifying CSC-related markers that have prognostic value (58, 59, 68).
It has also been shown that both moderately and poorly differentiated OSCC cells demonstrate higher expression of NANOG, SOX2, and OCT4 under hypoxic conditions, suggesting that CSCs share some similarities with induced pluripotent stem cells (97). It is increasingly recognized that the tumor microenvironment plays an important role in supporting tumor growth and metastasis, and contributes to tumor heterogeneity (98). Specialized niche CSC micro environments result from factors that stimulate CSC self-renewal, induce angiogenesis, and recruit cells that facilitate invasion and metastasis (95). It appears that CSCs in SCC switch between two distinct phenotypes that are preferentially migratory or proliferative (99). This plasticity presents an obvious challenge to the development of cancer therapeutics.
Nevertheless, CSC research has promising applications, and directly targeting CSCs has become increasingly appealing as it has the potential to be more effective than traditional approaches while having greater potential for organ preservation and reducing both immediate and long-term off-target toxicity. In addition, most stem cell markers currently used to identify CSCs in basic science research are not sufficiently specific and would be poor targets for direct therapy. Investigations into targeting CSCs vary, including targeting CSC markers and pathways, using epigenetic modulators, immunotherapy agents, and increasing CSC sensitivity to ChT and RT (100).
Oral cavity squamous cell carcinoma tumor spheres display higher expression of CSC and metastasis markers and are also more invasive and are resistant to cisplatin/RT. Increased fucosylation activity identified by upregulation of fucosyltransferases (FUT3 and FUT6) and increased expression of fucosylated polysaccharides such as Sialyl Lewis X are associated with invasion and metastasis (101). Conversely, inhibition of fucosylation negatively affected tumor sphere formation and invasiveness of OCSCC CSCs (101).
Expression of TNF receptor-associated factor 1 has been found to be higher in OCSCC than normal oral mucosa and oral epithelial dysplasia, and is associated with reduced overall survival, Moloney murine leukemia virus integration site 1, Lin28 homolog B, and most importantly ALDH1 in OSCC (102).
A single “silver bullet” that targets CSCs and effectively eliminates cancer remains elusive; however, alternative treatment using a combination of existing medications targeting critical steps of the RAS to control the CSCs may form a future approach to cancer treatment.
Author Contributions
TI and ST formulated the topic of the review. RB conducted the review. All authors were involved in the drafting, and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. RB, ST, and TI are inventors of the PCT patent application for Cancer Diagnosis and Therapy (No. PCT/NZ2015/050108), and ST and TI are inventors of the PCT patent application for Cancer Therapeutic (62/452479).
Funding
TI was supported, in part, by the Pacific Emerging Researcher First Grant, Health Research Council of New Zealand (Grant HRC16/434).
References
1. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol (2009) 45(4–5):309–16. doi: 10.1016/j.oraloncology.2008.06.002
2. Sano D, Myers J. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev (2007) 26(3–4):645–62. doi:10.1007/s10555-007-9082-y
3. Prince S, Bailey BM. Squamous carcinoma of the tongue: review. Br J Oral Maxillofac Surg (1999) 37(3):164–74. doi:10.1054/bjom.1999.0031
4. Tajbakhsh S. Stem cell: what’s in a name? Nat Rep Stem Cells (2009). doi:10.1038/stemcells.2009.90
5. da Silva SD, Ferlito A, Takes RP, Brakenhoff RH, Valentin MD, Woolgar JA, et al. Advances and applications of oral cancer basic research. Oral Oncol (2011) 47(9):783–91. doi:10.1016/j.oraloncology.2011.07.004
6. Okamoto A, Chikamatsu K, Sakakura K, Hatsushika K, Takahashi G, Masuyama K. Expansion and characterization of cancer stem-like cells in squamous cell carcinoma of the head and neck. Oral Oncol (2009) 45(7):633–9. doi:10.1016/j.oraloncology.2008.10.003
7. Yanamoto S, Kawasaki G, Yamada S, Yoshitomi I, Kawano T, Yonezawa H, et al. Isolation and characterization of cancer stem-like side population cells in human oral cancer cells. Oral Oncol (2011) 47(9):855–60. doi:10.1016/j.oraloncology.2011.06.501
8. Noto Z, Yoshida T, Okabe M, Koike C, Fathy M, Tsuno H, et al. CD44 and SSEA-4 positive cells in an oral cancer cell line HSC-4 possess cancer stem-like cell characteristics. Oral Oncol (2013) 49(8):787–95. doi:10.1016/j.oraloncology.2013.04.012
9. Han J, Fujisawa T, Husain SR, Puri RK. Identification and characterization of cancer stem cells in human head and neck squamous cell carcinoma. BMC Cancer (2014) 14(1):173. doi:10.1186/1471-2407-14-173
10. Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma – an update. CA Cancer J Clin (2015) 65(5):401–21. doi:10.3322/caac.21293
11. Chen X, Li X, Zhao B, Shang D, Zhong M, Deng C, et al. Dormancy activation mechanism of oral cavity cancer stem cells. Tumour Biol (2015) 36(7):5551–9. doi:10.1007/s13277-015-3225-5
12. Nör C, Zhang Z, Warner KA, Bernardi L, Visioli F, Helman JI, et al. Cisplatin induces bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia (2014) 16(2):137–46. doi:10.1593/neo.131744
13. Tsai LL, Hu FW, Lee SS, Yu CH, Yu CC, Chang YC. Oct4 mediates tumor initiating properties in oral squamous cell carcinomas through the regulation of epithelial-mesenchymal transition. PLoS One (2014) 9(1):e87207. doi:10.1371/journal.pone.0087207
14. Zhang Q, Shi S, Yen Y, Brown J, Ta JQ, Le AD. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett (2010) 289(2):151–60. doi:10.1016/j.canlet.2009.08.010
15. Song J, Chang I, Chen Z, Kang M, Wang CY. Characterization of side populations in HNSCC: highly invasive, chemoresistant and abnormal Wnt signaling. PLoS One (2010) 5(7):e11456. doi:10.1371/journal.pone.0011456
16. Rastogi P. Emergence of cancer stem cells in head and neck squamous cell carcinoma: a therapeutic insight with literature review. Dent Res J (Isfahan) (2012) 9(3):239–44.
17. Major AG, Pitty LP, Farah CS. Cancer stem cell markers in head and neck squamous cell carcinoma. Stem Cells Int (2013) 2013:13. doi:10.1155/2013/319489
18. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (2007) 318(5858):1917–20. doi:10.1126/science.1151526
19. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell (2006) 126(4):663–76. doi:10.1016/j.cell.2006.07.024
20. Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem (2005) 280(26):24731–7. doi:10.1074/jbc.M502573200
21. Fu TY, Hsieh IC, Cheng JT, Tsai MH, Hou YY, Lee JH, et al. Association of OCT4, SOX2, and NANOG expression with oral squamous cell carcinoma progression. J Oral Pathol Med (2016) 45(2):89–95. doi:10.1111/jop.12335
22. Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature (2006) 444(7117):364–8. doi:10.1038/nature05284
23. Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res (2008) 14(13):4085–95. doi:10.1158/1078-0432.ccr-07-4404
24. Zhang P, Zhang Y, Mao L, Zhang Z, Chen W. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotypes. Cancer Lett (2009) 277(2):227–34. doi:10.1016/j.canlet.2008.12.015
25. Chen YC, Chang CJ, Hsu HS, Chen YW, Tai LK, Tseng LM, et al. Inhibition of tumorigenicity and enhancement of radiochemosensitivity in head and neck squamous cell cancer-derived ALDH1-positive cells by knockdown of Bmi-1. Oral Oncol (2010) 46(3):158–65. doi:10.1016/j.oraloncology.2009.11.007
26. Huang CF, Xu XR, Wu TF, Sun ZJ, Zhang WF. Correlation of ALDH1, CD44, OCT4 and SOX2 in tongue squamous cell carcinoma and their association with disease progression and prognosis. J Oral Pathol Med (2014) 43(7):492–8. doi:10.1111/jop.12159
27. Koo BS, Lee SH, Kim JM, Huang S, Kim SH, Rho YS, et al. Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene (2015) 34(18):2317–24. doi:10.1038/onc.2014.174
28. Zeineddine D, Hammoud AA, Mortada M, Boeuf H. The Oct4 protein: more than a magic stemness marker. Am J Stem Cells (2014) 3(2):74–82.
29. Yu HH, Featherston T, Tan ST, Chibnall AM, Brasch HD, Davis PF, et al. Characterization of cancer stem cells in moderately differentiated buccal mucosal squamous cell carcinoma. Front Surg (2016) 3:46. doi:10.3389/fsurg.2016.00046
30. Baillie R, Itinteang T, Yu HH, Brasch HD, Davis PF, Tan ST, et al. Cancer stem cells in moderately differentiated oral tongue squamous cell carcinoma. J Clin Pathol (2016) 69(8):742–4. doi:10.1136/jclinpath-2015-203599
31. Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal (2013) 25(5):1264–71. doi:10.1016/j.cellsig.2013.02.013
32. Bass AJ, Wang TC. An inflammatory situation: SOX2 and STAT3 cooperate in squamous cell carcinoma initiation. Cell Stem Cell (2013) 12(3):266–8. doi:10.1016/j.stem.2013.02.004
33. Chou MY, Hu FW, Yu CH, Yu CC. Sox2 expression involvement in the oncogenicity and radiochemoresistance of oral cancer stem cells. Oral Oncol (2015) 51(1):31–9. doi:10.1016/j.oraloncology.2014.10.002
34. Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol (2005) 25(14):6031–46. doi:10.1128/MCB.25.14.6031-6046.2005
35. Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet (2006) 38(4):431–40. doi:10.1038/ng1760
36. Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem (2012) 287(39):32800–24. doi:10.1074/jbc.M111.308528
37. Pozzi V, Sartini D, Rocchetti R, Santarelli A, Rubini C, Morganti S, et al. Identification and characterization of cancer stem cells from head and neck squamous cell carcinoma cell lines. Cell Physiol Biochem (2015) 36(2):784–98. doi:10.1159/000430138
38. Michifuri Y, Hirohashi Y, Torigoe T, Miyazaki A, Kobayashi J, Sasaki T, et al. High expression of ALDH1 and SOX2 diffuse staining pattern of oral squamous cell carcinomas correlates to lymph node metastasis. Pathol Int (2012) 62(10):684–9. doi:10.1111/j.1440-1827.2012.02851.x
39. Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC, et al. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res (2015) 75(12):2553–65. doi:10.1158/0008-5472.can-14-2215
40. Tsai LL, Yu CC, Chang YC, Yu CH, Chou MY. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J Oral Pathol Med (2011) 40(8):621–8. doi:10.1111/j.1600-0714.2011.01015.x
41. Lee HJ, Kang YH, Lee JS, Byun JH, Kim UK, Jang SJ, et al. Positive expression of NANOG, mutant p53, and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma. BMC Oral Health (2015) 15(1):1–11. doi:10.1186/s12903-015-0120-9
42. Ram R, Brasch H, Dunne J, Davis PF, Tan ST, Itinteang T, et al. The identification of three cancer stem cell subpopulations within moderately differentiated lip squamous cell carcinoma. Front Surg (2017) 4:12. doi:10.3389/fsurg.2017.00012
43. Siveen KS, Sikka S, Surana R, Dai X, Zhang J, Kumar AP, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta (2014) 1845(2):136–54. doi:10.1016/j.bbcan.2013.12.005
44. Timofeeva OA, Tarasova NI, Zhang X, Chasovskikh S, Cheema AK, Wang H, et al. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci U S A (2013) 110(4):1267–72. doi:10.1073/pnas.1211805110
45. Gkouveris I, Nikitakis N, Karanikou M, Rassidakis G, Sklavounou A. Erk1/2 activation and modulation of STAT3 signaling in oral cancer. Oncol Rep (2014) 32(5):2175–82. doi:10.3892/or.2014.3440
46. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer (2009) 9(11):798–809. doi:10.1038/nrc2734
47. Chan KS, Sano S, Kataoka K, Abel E, Carbajal S, Beltran L, et al. Forced expression of a constitutively active form of Stat3 in mouse epidermis enhances malignant progression of skin tumors induced by two-stage carcinogenesis. Oncogene (2008) 27(8):1087–94. doi:10.1038/sj.onc.1210726
48. Abrigo M, Alvarez R, Paparella ML, Calb DE, Bal de Kier Joffe E, Gutkind JS, et al. Impairing squamous differentiation by Klf4 deletion is sufficient to initiate tongue carcinoma development upon K-Ras activation in mice. Carcinogenesis (2014) 35(3):662–9. doi:10.1093/carcin/bgt349
49. Li W, Liu M, Su Y, Zhou X, Liu Y, Zhang X. The Janus-faced roles of Krüppel-like factor 4 in oral squamous cell carcinoma cells. Oncotarget (2015) 6(42):44480–94. doi:10.18632/oncotarget.6256
50. He H, Li S, Hong Y, Zou H, Chen H, Ding F, et al. Kruppel-like factor 4 promotes esophageal squamous cell carcinoma differentiation by up-regulating keratin 13 expression. J Biol Chem (2015) 290(21):13567–77. doi:10.1074/jbc.M114.629717
51. Pai R, Pai S, Lalitha R, Kumaraswamy S, Lalitha N, Johnston R, et al. Over-expression of c-Myc oncoprotein in oral squamous cell carcinoma in the South Indian population. Ecancermedicalscience (2009) 3:128. doi:10.3332/ecancer.2008.128
52. Hu L, Liu J, Li Z, Wang C, Nawshad A. Transforming growth factor-β1 activates ΔNp63/c-Myc to promote oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol (2016) 122(4):460–82.e4. doi:10.1016/j.oooo.2016.05.018
53. Papakosta V, Vairaktaris E, Vylliotis A, Derka S, Nkenke E, Vassiliou S, et al. The co-expression of c-myc and p53 increases and reaches a plateau early in oral oncogenesis. Anticancer Res (2006) 26(4B):2957–62.
54. Sawant S, Gokulan R, Dongre H, Vaidya M, Chaukar D, Prabhash K, et al. Prognostic role of Oct4, CD44 and c-Myc in radio-chemo-resistant oral cancer patients and their tumourigenic potential in immunodeficient mice. Clin Oral Investig (2016) 20(1):43–56. doi:10.1007/s00784-015-1476-6
55. Monroe MM, Anderson EC, Clayburgh DR, Wong MH. Cancer stem cells in head and neck squamous cell carcinoma. J Oncol (2011) 2011:762780. doi:10.1155/2011/762780
56. Mack B, Gires O. CD44s and CD44v6 expression in head and neck epithelia. PLoS One (2008) 3(10):e3360. doi:10.1371/journal.pone.0003360
57. Mărgăritescu C, Pirici D, Simionescu C, Stepan A. The utility of CD44, CD117 and CD133 in identification of cancer stem cells (CSC) in oral squamous cell carcinomas (OSCC). Rom J Morphol Embryol (2011) 52(3 Suppl):985–93.
58. Oliveira LR, Castilho-Fernandes A, Oliveira-Costa JP, Soares FA, Zucoloto S, Ribeiro-Silva A. CD44+/CD133+ immunophenotype and matrix metalloproteinase-9: influence on prognosis in early-stage oral squamous cell carcinoma. Head Neck (2013) 36(12):1718–26. doi:10.1002/hed.23527
59. Koukourakis MI, Giatromanolaki A, Tsakmaki V, Danielidis V, Sivridis E. Cancer stem cell phenotype relates to radio-chemotherapy outcome in locally advanced squamous cell head-neck cancer. Br J Cancer (2012) 106(5):846–53. doi:10.1038/bjc.2012.33
60. Kosunen A, Pirinen R, Ropponen K, Pukkila M, Kellokoski J, Virtaniemi J, et al. CD44 expression and its relationship with MMP-9, clinicopathological factors and survival in oral squamous cell carcinoma. Oral Oncol (2007) 43(1):51–9. doi:10.1016/j.oraloncology.2006.01.003
61. Yu YH, Morales J, Feng L, Lee JJ, El-Naggar AK, Vigneswaran N. CD147 and Ki-67 overexpression confers poor prognosis in squamous cell carcinoma of oral tongue: a tissue microarray study. Oral Surg Oral Med Oral Pathol Oral Radiol (2015) 119(5):553–65. doi:10.1016/j.oooo.2014.12.022
62. Braumüller H, Gansauge S, Ramadani M, Gansauge F. CD44v6 cell surface expression is a common feature of macrophages and macrophage-like cells – implication for a natural macrophage extravasation mechanism mimicked by tumor cells. FEBS Lett (2000) 476(3):240–7. doi:10.1016/S0014-5793(00)01737-3
63. Yanamoto S, Yamada S, Takahashi H, Naruse T, Matsushita Y, Ikeda H, et al. Expression of the cancer stem cell markers CD44v6 and ABCG2 in tongue cancer: effect of neoadjuvant chemotherapy on local recurrence. Int J Oncol (2014) 44(4):1153–62. doi:10.3892/ijo.2014.2289
64. Perez A, Neskey DM, Wen J, Pereira L, Reategui EP, Goodwin WJ, et al. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol (2013) 49(4):306–13. doi:10.1016/j.oraloncology.2012.11.009
65. Kamarajan P, Shin JM, Qian X, Matte B, Zhu JY, Kapila YL. ADAM17-mediated CD44 cleavage promotes orasphere formation or stemness and tumorigenesis in HNSCC. Cancer Med (2013) 2(6):793–802. doi:10.1002/cam4.147
66. de Andrade N, Rodrigues M, Rodini C, Nunes FD. Cancer stem cell, cytokeratins and epithelial to mesenchymal transition markers expression in oral squamous cell carcinoma derived from orthtopic zenoimplantation of CD44high cells. Pathol Res Pract (2017) 213(3):235–44. doi:10.1016/j.prp.2016.12.009
67. Chen J, Zhou J, Lu J, Xiong H, Shi X, Gong L. Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer (2014) 14(1):15. doi:10.1186/1471-2407-14-15
68. Kokko LL, Hurme S, Maula SM, Alanen K, Grénman R, Kinnunen I, et al. Significance of site-specific prognosis of cancer stem cell marker CD44 in head and neck squamous-cell carcinoma. Oral Oncol (2011) 47(6):510–6. doi:10.1016/j.oraloncology.2011.03.026
69. Quintanilla RH Jr, Asprer JS, Vaz C, Tanavde V, Lakshmipathy U. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS One (2014) 9(1):e85419. doi:10.1371/journal.pone.0085419
70. Kidwai F, Costea DE, Hutchison I, Mackenzie I. The effects of CD44 down-regulation on stem cell properties of head and neck cancer cell lines. J Oral Pathol Med (2013) 42(9):682–90. doi:10.1111/jop.12076
71. Mannelli G, Magnelli L, Deganello A, Busoni M, Meccariello G, Parrinello G, et al. Detection of putative stem cell markers, CD44/CD133, in primary and lymph node metastases in head and neck squamous cell carcinomas. A preliminary immunohistochemical and in vitro study. Clin Otolaryngol (2015) 40(4):312–20. doi:10.1111/coa.12368
72. Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol (2012) 2012:11. doi:10.1155/2012/708036
73. Zimmerer R, Ludwig N, Kampmann A, Bittermann G, Spalthoff S, Jungheim M, et al. CD24+ tumor-initiating cells from oral squamous cell carcinoma induce initial angiogenesis in vivo. Microvasc Res (2017) 112:101–8. doi:10.1016/j.mvr.2017.03.006
74. Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, et al. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype on oral squamous cell carcinoma. Stem Cell Res (2016) 16(2):405–17. doi:10.1016/j.scr.2016.02.028
75. Todoroki K, Ogasawara S, Akiba J, Nakayama M, Naito Y, Seki N, et al. CD44v3+/CD24 cells possess cancer stem cell-like properties in human oral squamous cell carcinoma. Int J Oncol (2016) 48(1):99–109. doi:10.3892/ijo.2015.3261
76. Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med (2008) 86(9):1025–32. doi:10.1007/s00109-008-0357-8
77. Irollo E, Pirozzi G. CD133: to be or not to be, is this the real question? Am J Transl Res (2013) 5(6):563–81.
78. Liu W, Wu L, Shen XM, Shi LJ, Zhang CP, Xu LQ, et al. Expression patterns of cancer stem cell markers ALDH1 and CD133 correlate with a high risk of malignant transformation of oral leukoplakia. Int J Cancer (2013) 132(4):868–74. doi:10.1002/ijc.27720
79. Ravindran G, Devaraj H. Aberrant expression of CD133 and musashi-1 in preneoplastic and neoplastic human oral squamous epithelium and their correlation with clinicopathological factors. Head Neck (2012) 34(8):1129–35. doi:10.1002/hed.21896
80. Felthaus O, Ettl T, Gosau M, Driemel O, Brockhoff G, Reck A, et al. Cancer stem cell-like cells from a single cell of oral squamous carcinoma cell lines. Biochem Biophys Res Commun (2011) 407(1):28–33. doi:10.1016/j.bbrc.2011.02.084
81. Chen YS, Wu MJ, Huang CY, Lin SC, Chuang TH, Yu CC, et al. CD133/Src axis mediates tumor initiating property and epithelial-mesenchymal transition of head and neck cancer. PLoS One (2011) 6(11):e28053. doi:10.1371/journal.pone.0028053
82. Shen L-F, Zhou M-L, Zhou S-H, Bao Y-Y. Biomarkers of head and neck cancer stem cells and targeted therapeutic strategies. Int J Clin Exp Med (2016) 9(2):614–25.
83. Zou B, Sun S, Qi X, Ji P. Aldehyde dehydrogenase activity is a cancer stem cell marker of tongue squamous cell carcinoma. Mol Med Rep (2012) 5(4):1116–20. doi:10.3892/mmr.2012.781
84. Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle (2011) 10(9):1378–84. doi:10.4161/cc.10.9.15486
85. Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck (2010) 32(9):1195–201. doi:10.1002/hed.21315
86. Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun (2009) 385(3):307–13. doi:10.1016/j.bbrc.2009.05.048
87. Itinteang T, Dunne JC, Chibnall AM, Brasch HD, Davis PF, Tan ST, et al. Cancer stem cells in moderately differentiated oral tongue squamous cell carcinoma express components of the renin-angiotensin system. J Clin Pathol (2016) 69(10):942–5. doi:10.1136/jclinpath-2016-203736
88. Featherston T, Yu HH, Dunne JC, Chibnall AM, Brasch HD, Davis PF, et al. Cancer stem cells in moderately differentiated buccal mucosal squamous cell carcinoma express components of the renin-angiotensin system. Front Surg (2016) 3:52. doi:10.3389/fsurg.2016.00052
89. Cagnoni F, Njwe CA, Zaninelli A, Ricci AR, Daffra D, D’Ospina A, et al. Blocking the RAAS at different levels: an update on the use of the direct renin inhibitors alone and in combination. Vasc Health Risk Manag (2010) 6(1):539–59. doi:10.2147/VHRM.S11816
90. Sinha N, Mukhopadhyay S, Das DN, Panda PK, Bhutia SK. Relevance of cancer initiating/stem cells in carcinogenesis and therapy resistance in oral cancer. Oral Oncol (2013) 49(9):854–62. doi:10.1016/j.oraloncology.2013.06.010
91. Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer (2012) 12(2):133–43. doi:10.1038/nrc3184
92. Feller LL, Khammissa RR, Kramer BB, Lemmer JJ. Oral squamous cell carcinoma in relation to field precancerisation: pathobiology. Cancer Cell Int (2013) 13(1):31. doi:10.1186/1475-2867-13-31
93. Willis R. Targeted cancer therapy: vital oncogenes and a new molecular genetic paradigm for cancer initiation progression and treatment. Int J Mol Sci (2016) 17(9):1552. doi:10.3390/ijms17091552
94. Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol (2012) 198(3):281–93. doi:10.1083/jcb.201202014
95. Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell (2015) 16(3):225–38. doi:10.1016/j.stem.2015.02.015
96. Welch DR. Tumor heterogeneity – a ‘contemporary concept’ founded on historical insights and predictions. Cancer Res (2016) 76(1):4–6. doi:10.1158/0008-5472.can-15-3024
97. Takeda D, Hasegawa T, Ueha T, Iwata E, Harada R, Sakakibara A, et al. Induced pluripotent-stem-cell related genes contribute to de-differentiation in oral squamous cell carcinoma. Anticancer Res (2017) 37(3):1075–82. doi:10.21873/anticanres.11419
98. Kreso A, Dick John E. Evolution of the cancer stem cell model. Cell Stem Cell (2014) 14(3):275–91. doi:10.1016/j.stem.2014.02.006
99. Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res (2011) 71(15):5317–26. doi:10.1158/0008-5472.can-11-1059
100. Birkeland AC, Owen JH, Prince ME. Targeting head and neck cancer stem cells: current advances and future challenges. J Dent Res (2015) 94(11):1516–23. doi:10.1177/0022034515601960
101. Desiderio V, Papagerakis P, Tirino V, Zheng L, Matossian M, Prince ME, et al. Increased fucosylation has a pivotal role in invasive and metastatic properties of head and neck cancer stem cells. Oncotarget (2015) 1(6):71–84. doi:10.18632/oncotarget.2698
Keywords: oral cavity, squamous cell carcinoma, cancer, cancer stem cells, head and neck
Citation: Baillie R, Tan ST and Itinteang T (2017) Cancer Stem Cells in Oral Cavity Squamous Cell Carcinoma: A Review. Front. Oncol. 7:112. doi: 10.3389/fonc.2017.00112
Received: 19 March 2017; Accepted: 12 May 2017;
Published: 02 June 2017
Edited by:
Ander Matheu, Biodonostia, SpainReviewed by:
Shree Ram Singh, National Cancer Institute, United StatesVincenzo Desiderio, Università degli Studi della Campania “L. Vanvitelli” Naples, Italy
Copyright: © 2017 Baillie, Tan and Itinteang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tinte Itinteang, dGludGVAZ21yaS5vcmcubno=
†These authors have contributed equally to this work.
 Ranui Baillie
Ranui Baillie Swee T. Tan
Swee T. Tan Tinte Itinteang
Tinte Itinteang