- 1Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East, Kashiwa, Japan
- 2Department of Gastroenterology, Hokkaido University Hospital, Sapporo, Japan
Technologies for genomic analyses have revealed more details in cancer biology and have changed standard treatments for cancer, including the introduction of targeted gene-specific therapy. Currently, liquid biopsies are increasingly being utilized in clinical trials and research settings to analyze circulating tumor DNA (ctDNA) from peripheral blood. Several studies have shown the potential of ctDNA in the screening, prognostication, molecular profiling, and monitoring of gastrointestinal malignancies. Although limitations continue to exist in the use of ctDNA, such as method standardization, the sensitivity, concordance with tumor tissue, and regulatory issues, this field offers promising benefits for cancer treatment. A deeper understanding of tumor biology via ctDNA analyses and ctDNA-guided clinical trials will lead to the increasing use of ctDNA in clinical practice in the near future; this development will result in the improvement of outcomes among patients with gastrointestinal malignancies.
Introduction
Over several decades, the gold standard in the diagnosis and screening of tumors has been tissue biopsy (1). However, conventional tissue biopsies are invasive, painful, and carry a risk of complications such as bleeding, local infection, and damage to neighboring tissues. Moreover, a tissue biopsy cannot always reflect tumor dynamics or response to treatment. The recent era of precision medicine, which represents a paradigm shift in cancer, has challenged the gold standard in diagnosis by introducing a transition from tissue biopsy to liquid biopsy. Compared with tissue biopsies, liquid biopsies carry minimal potential risk and can be repeatedly performed in routine practice during cancer treatment by using peripheral blood. Furthermore, liquid biopsies have the potential to provide more complete information regarding the biology of whole tumors despite tumor heterogeneity. Liquid biopsies include the testing of soluble factors, such as circulating tumor DNA (ctDNA) and circulating cell-free DNA (cfDNA), as well as proteins and tumor markers (2). cfDNA is highly fragmented DNA that is released from necrotic or apoptotic cells into the bloodstream (3–5). cfDNA consists of DNA from healthy cells and tumor cells, whereas ctDNA is defined as DNA that is derived only from primary or metastatic tumor cells.
Since Mandel and Metais (6) reported fragmented DNA in blood for the first time in 1948, technologies for cfDNA quantification have changed over 70 years from quantitative polymerase chain reaction to complex BEAMing and deep next-generation sequencing (NGS), thus achieving improvements in the sensitivity and specificity of cfDNA detection. With the development of sensitive techniques that can detect rare mutations, the heterogeneous landscape of tumors can be determined using blood samples. In fact, National Comprehensive Cancer Network guideline for non-small cell lung cancer (version 4. 2018) states that plasma biopsy should be considered if repeat biopsy is not feasible (7).
Here, we review ctDNA in gastrointestinal malignancies by focusing on clinical utility and future perspectives.
ctDNA and Related Technologies
The presence of cfDNA in the blood is a well-established fact, and DNA fragments are released from dying cells because of cellular turnover or other types of cell death (2). In cancer patients, a fraction of cfDNA is tumor derived and is termed ctDNA. ctDNA originates from primary tumors, metastatic tumor cells, or circulating tumor cells. ctDNA molecules are shorter than non-mutant cfDNA molecules in plasma, as demonstrated by PCR (8, 9) and sequencing (10, 11).
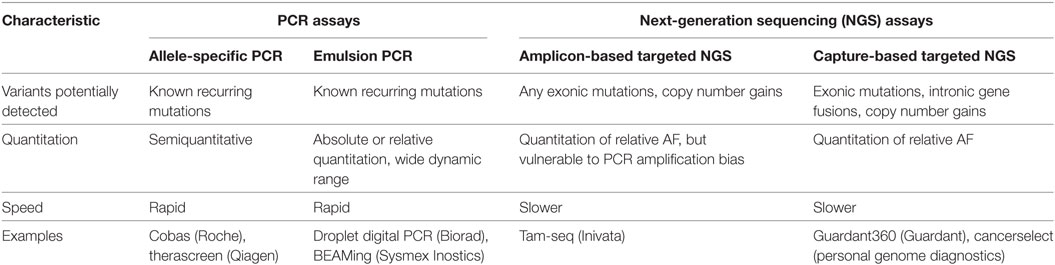
Representative approaches for analyzing ctDNA are summarized in Table 1 (12). Mutation-specific real-time or endpoint PCR has been used for the detection of point mutations in ctDNA (13–17). More recently, digital PCR methods such as BEAMing and droplet digital PCR have been developed to improve the identification of genomic alterations in ctDNA (18–20). The recent implementation of NGS has allowed the direct sequence-based detection of chromosomal alterations in plasma DNA (21–23); however, it is necessary to distinguish the relatively few somatic alterations in ctDNA from the larger numbers of structural variants present in the germline cells of all individuals. Bioinformatics-based filters that enrich high-confidence somatic structural alterations while eliminating germline and artifactual changes have been developed (12). In addition, importantly, amplification in ctDNA can be depend on both the amount of ctDNA in the plasma due to high tumor burden and high copy number of specific gene. Commercially available kits for the NGS assays of ctDNA are summarized in Table 2.
Early Detection of Cancer
The early detection of cancer is one of the most important issues in reducing cancer-related deaths. In many cases, gastrointestinal cancer is detected via endoscopy or CT scans conducted for symptoms such as anorexia, abdominal pain, or constipation. ctDNA may have a potential role in the noninvasive early diagnosis and screening of gastrointestinal cancer. Even localized cancers shed DNA into circulation; therefore, ctDNA can be detected in patients with localized cancers, in addition to patients with advanced or metastatic cancers.
In a study across several early and late-stage cancers, ctDNA was detected in 73, 57, and 48% of patients with colorectal cancer (CRC), gastroesophageal cancer, and pancreatic cancer, respectively (25). The use of several biomarkers in ctDNA including the levels of overall ctDNA, ALU247 fragment concentration (26), KRAS mutations (27, 28), TP53 mutations (29, 30), BRAF mutations (28), and septin 9 (SEPT9) methylation (31–34) have been demonstrated for the diagnosis of CRC. Also, detection of methylated SEPT9 DNA in plasma is US FDA approved as a blood test for CRC screening. Compared with biomarkers for CRC, biomarkers for the diagnosis of gastric cancer (GC) and esophageal cancer have been assessed in a relatively small number of cohorts (25, 35). For the early detection or screening of cancers including CRC, GC, and esophageal cancer, the sensitivity of ctDNA analysis needs to be improved. Analysis that can be performed using a few milliliters of blood would be suitable for cancer screening; however, increasing the analytical sensitivity beyond 0.1% may not provide clinical benefits because it also leads to difficulties in distinguishing oncological mutations and sampling noise. In fact, cancer-associated genomic alterations have been found in plasma from healthy individuals (36). In addition, because many cancers share common gene mutations such as TP53 mutations and KRAS mutations, ctDNA presents challenges in the detection of the specific organ sites of malignancies. To overcome these issues in ctDNA, the methylation profiling of cfDNA has been investigated in cancer diagnosis. Methylation haplotyping in plasma is a promising strategy for the early detection of a tumor and its primary growth site (37). Studies have reported the utility of methylation scores from over 9,000 CpG sites in cfDNA for cancer detection, with 76.3% accuracy for the prediction of cancer type (38).
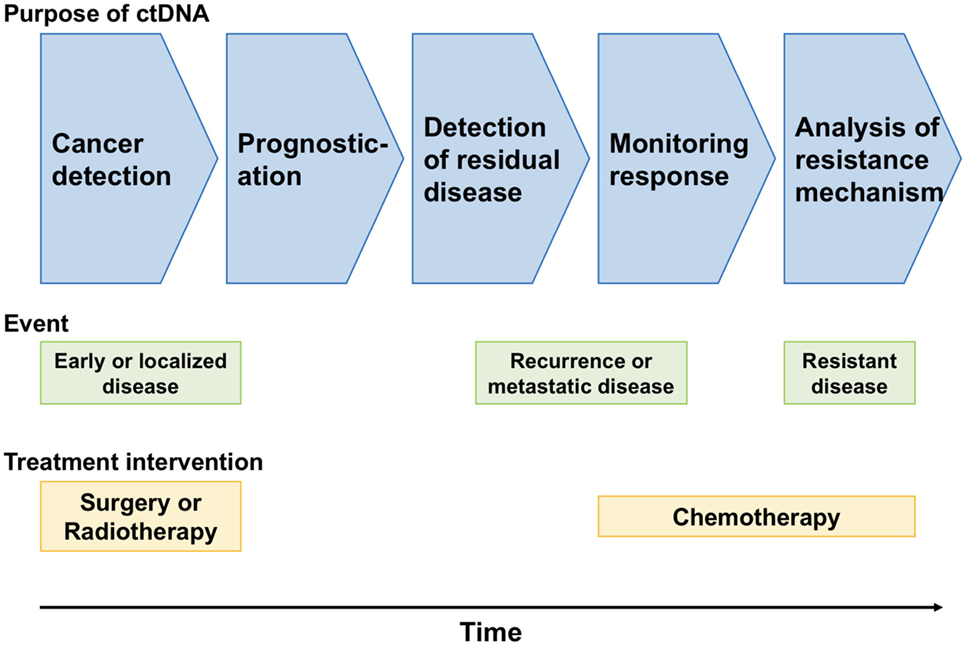
Despite the above hurdles to the use of ctDNA in cancer screening, it is expected that the clinical use of ctDNA is less than a decade away because of its utility and convenience in cancer screening (Figure 1).
Prognosis and Detection of Residual Disease
Following curative therapy for gastrointestinal malignancies, ctDNA may be a potential biomarker for minimal residual disease. The detection of ctDNA even in the absence of any other clinical evidence of disease may mean that the patient has higher risk of relapse. In a cohort of 230 patients with stage II CRC, the assessment of ctDNA using the Safe-SeqS NGS method at the first visit after surgery indicated that recurrence-free survival at 3 years was 0% in a ctDNA-positive group and 90% in a ctDNA-negative group (39). Other studies have also demonstrated that the persistent detection of ctDNA after local therapy (surgery or radical radiotherapy) predicts a high risk of relapse in patients with colon cancer (40, 41). In addition, methylated BCAT1/IKZF1 have been evaluated as biomarkers for CRC (42, 43). Of 397 patients with CRC who underwent primary tumor resection, odds ratio of a positive CEA test for recurrence was 6.9 (95% CI 2–22) compared to 14.4 (5–23, 25–40) for BCAT1/IKZF1.
In a meta-analysis of 16 studies including 1,193 patients with GC, the presence of ctDNA was significantly associated with the shorter disease-free survival (HR 4.36, 95% CI 3.08–6.16, p < 0.001) and overall survival (HR 1.77, 95% CI 1.38–2.28, p < 0.001) of GC patients, with high specificity (0.95, 95% CI 0.93–0.96) and relatively moderate sensitivity (0.62, 95% CI 0.59–0.65) (44). Another study demonstrated that the level of ctDNA was associated with tumor recurrence in patients who underwent curative surgery for GC (45). Similarly, several studies have reported the tumor-associated mutations in ctDNA and the prognosis of patients with esophageal cancer (46, 47); however, these studies included a limited number of patients, and further investigations are warranted. Almost all of these studies followed a retrospective design and provided limited validation for clinical use in gastrointestinal malignancies. One of the ideal applications of ctDNA is in the early detection of residual disease or recurrence compared with CT imaging and tumor markers. A more attractive idea is patient-specific ctDNA panels in patients who have undergone curative surgery (41, 48). Individual surgical tumor samples may provide a great opportunity to obtain tumor DNA from each patient to guide the design of patient-specific ctDNA panels from peripheral blood samples. Despite hurdles such as tumor heterogeneity, validation, regulatory issues, and quality control of individual panels, patient-specific ctDNA panels are potential biomarkers for postoperative monitoring.
Biomarkers of Chemotherapy Response and Resistance in Metastatic Disease
Another clinical potential of ctDNA is in the determination of systemic chemotherapy regimens, the prediction of response to chemotherapy, and the identification of resistance mechanisms. The short half-life of ctDNA enables the real-time monitoring of cancer burden, in contrast to radiological imaging or tumor markers. Indeed, a prospective study of 53 patients with metastatic CRC demonstrated that early changes in ctDNA during first-line chemotherapy predicted later radiologic response. Significant reductions in ctDNA levels were observed before cycle 2 and were correlated with CT response at 8–10 weeks (odds ratio 5.25 with a 10-fold ctDNA reduction; p = 0.016) (49). Blood-based monitoring is an ideal strategy during cancer treatment because of its minimal invasiveness and avoidance of radiation exposure.
RAS Mutations in Metastatic CRC
The assessment of RAS status has been mandatory in patients with metastatic CRC to predict the response of cetuximab and panitumumab to anti-EGFR antibodies (50–55). A double-blinded prospective study of 106 patients with mCRC has been performed to compare the KRAS mutation status assessed using tumor tissue via routine gold-standard methods to that assessed using plasma DNA via qPCR-based methods; the resultant specificity and sensitivity for the detection of KRAS point mutations were 98 and 92%, respectively, resulting in 96% concordance (28). In addition, the retrospective exploratory analysis in a biomarker subgroup of the CORRECT trial, which was a phase III trial investigating the efficacy and safety of regorafenib in patients with mCRC, confirmed the utility of detecting KRAS, BRAF, and PIK3CA mutations in ctDNA. Plasma DNA detected with BEAMing in 503 patients demonstrated that mutation status in ctDNA changed dynamically during chemotherapy and differed from that in pretreatment archival tissue (56). Currently, the OncoBEAM RAS CRC assay is the only European committee in vitro diagnostic test for RAS mutations in ctDNA. This assay is a qualitative PCR-based test and allows for the detection of 34 mutations within exons 2, 3, and 4 of KRAS and NRAS genes from a single blood sample. Four large cohort studies have been reported to achieve high concordance of approximately >90% (range, 89.7–93.3%) between OncoBEAM using plasma ctDNA and tumor tissue analysis in patients with CRC (57–60).
Acquired resistance to anti-EGFR antibody therapy has also been found by using ctDNA analyses. ctDNA from 28 patients receiving panitumumab monotherapy was assessed using qPCR, and 9 out of 24 (38%) patients whose tumors were initially KRAS wild-type showed KRAS mutations in ctDNA after panitumumab treatment (61). This study suggested that the emergence of KRAS mutations is a mechanism of resistance to anti-EGFR therapy and that these mutations may be detected in ctDNA as a more sensitive monitoring tool than radiological imaging. More recently, other studies have also demonstrated mutations associated with the resistance and decline of mutant KRAS clones after the withdrawal of anti-EGFR therapy (62, 63).
BRAF Mutations in Metastatic CRC
BRAF mutations have been confirmed to be associated with poor prognosis in patients with metastatic CRC; moreover, the limited efficacy of anti-EGFR therapy in patients with BRAF-mutant metastatic CRC has been shown in several studies (64–66). The analysis of BRAF mutations in ctDNA by using qPCR-based methods has shown specificity and sensitivity of 100% (28). Based on preclinical studies (67, 68), the clinical trials of dual EGFR and MAPK signaling pathway inhibition in patients with BRAF-mutant metastatic CRC is ongoing. In a phase Ib study of a combination therapy of dabrafenib, trametinib, and panitumumab, BRAF V600E mutant burden in ctDNA was more markedly reduced in responders than in nonresponders, and the emergence of RAS mutations was seen with disease progression in 9 of 22 patients (41%) (69). This exploratory analysis suggested that the monitoring of BRAF V600E mutant fraction in ctDNA could effectively predict response to combination therapy including a BRAF inhibitor and that overcoming the emergence of RAS-mutant subclones is important in combating resistance to this combination therapy.
Other Alterations in Metastatic CRC
HER2 or MET amplification is also known as a mechanism of resistance to anti-EGFR therapy in patients with metastatic CRC. The patient-derived xenograft models of HER2-amplified CRC showed resistance to anti-EGFR therapy (70, 71). In addition, the frequency of HER2 amplifications increased from approximately 3% in treatment-naïve patients to over 10% in patients who were administered anti-EGFR therapy (72). Although there are few studies regarding the concordance of HER2 status between ctDNA and tissue samples, 4 of 18 (22%) patients exhibited HER2 amplification in ctDNA by digital PCR after cetuximab therapy despite being negative for HER2 amplification prior to anti-EGFR therapy (73). The promising results of trastuzumab and T-DM1 combination therapy in the HERACLES trial (71) have encouraged clinical trials in patients with HER2-positive metastatic CRC such as the MyPathway trial (74) and the TRIUMPH trial (75); notably, the TRIUMPH trial includes patients with HER2 amplification detected using not only tissue samples but also ctDNA analysis using an NGS-based method.
Another important alteration that causes resistance to anti-EGFR therapy is MET amplification. A preclinical model of MET-amplified CRC also showed resistance to anti-EGFR therapy (76). In fact, MET amplification in ctDNA was detected using NGS in 12 of 53 (22.6%) patients who showed disease progression with anti-EGFR therapy; no such amplification was detected in patients before cetuximab therapy. Furthermore, MET amplification in ctDNA was not detected in patients with RAS mutations after cetuximab therapy, thus suggesting that MET amplification is one of the mechanisms (other than RAS mutations) that cause resistance to anti-EGFR therapy (77). In a phase Ib trial of cabozantinib and panitumumab combination therapy, the preliminary evidence of efficacy in patients with MET-amplified metastatic CRC was reported (78).
HER2 Amplification in GC
The amplification of the HER2 gene or overexpression of the HER2 protein, which contributes to cancer progression, has been reported in approximately 20% of patients with advanced GC (79, 80). According to the results of the ToGA trial, HER2 is a key biomarker of HER2-targeted therapy using trastuzumab for advanced GC (79). The gold-standard diagnostic method for detecting HER2 positivity and suitability for trastuzumab therapy is an immunohistochemistry score of 3+ or 2+ with a positive result in fluorescence in situ hybridization. A retrospective study of 52 patients with advanced GC and 40 healthy volunteers demonstrated that the plasma HER2–RPPH1 ratio (with RPPH1 as a reference gene) was significantly higher in patients with HER2-positive tumors than those with HER2-negative tumors (81). More recently, the droplet digital PCR of HER2 copy number in ctDNA has been reported. In a study of 60 patients with GC, including 17 patients who developed recurrence and 30 healthy volunteers, preoperative plasma HER2 ratio correlated with tumor HER2 status; postoperative plasma HER2 ratios were high during the recurrence of tumors, which were diagnosed as HER2-negative tumors in surgery samples (82). Considering that HER2 status may be altered after recurrence, the HER2 copy number analysis in ctDNA enables the real-time evaluation of HER2 status and leads to more effective treatment choices with HER2-targeted agents.
Future Perspectives
Overall, the data generated in all studies discussed above support the potential role of ctDNA in the diagnosis and treatment of patients with gastrointestinal malignancies. Despite a few limitations, including the standardization of detection and ctDNA quantification, the sensitivity, and concordance between ctDNA and tissue biopsies that currently hinder the routine use of ctDNA in clinical trials and clinical practice, its use would allow a deeper understanding of cancer biology and enable better cancer treatment, thus leading to improvements in patient survival.
In the context of clinical trials for metastatic disease treatment, several studies are ongoing or have been conducted using eligibility criteria based on gene alterations in ctDNA. A prospective study on the comprehensive ctDNA-guided treatment of advanced GC and lung cancers is ongoing in Korea (83). Another trial called the Targeted Agent and Profiling Utilization Registry, which is a large basket/umbrella trial sponsored by the American Society of Clinical Oncology, is accepting patient selection on the basis of ctDNA analysis (NCT02693535). In addition, an umbrella trial in patients with mCRC based on the molecular profiling of ctDNA, including the status of HER2, BRAF V600E, BRAF non-V600E, MET, or high tumor mutation burden, is ongoing in Japan. If promising results are obtained in these clinical trials, ctDNA will be used in routine clinical practice and in clinical trials in the near future.
Economic and regulatory issues still hinder the practical use of ctDNA. Although most guidelines recommend that comprehensive molecular profiling should be conducted, the substantial costs of NGS assays lead many community oncologists to rely on PCR tissue tests and do not understand the added benefit of a comprehensive genomic test. In addition, emerging ctDNA-guided clinical trials are essential to obtain approval for the use of ctDNA in clinical practice. These barriers need to be challenged, perhaps initially in patients with CRC, which is one of the most prevalent gastrointestinal malignancies worldwide. Simultaneously, further studies are needed on other gastrointestinal malignancies such as esophageal cancer and GC to identify the best gene biomarkers that are detectable in ctDNA for the diagnosis, prognosis, and prediction of therapy response.
Concluding Remarks
The potential role of ctDNA in gastrointestinal malignancies has been shown in basic studies, retrospective studies, and limited prospective studies. A paradigm shift in cancer diagnosis and treatment in ctDNA-based clinical trials and clinical practice will occur in the near future, thus leading to the availability of more DNA sequence information compared with that in the past decade. Although some limitations continue to exist on the use of ctDNA in clinical practice and clinical trials, ctDNA-based personalized therapy promises to improve patient outcomes and quality of life.
Author Contributions
This review was drafted by KS and DK and was revised by HB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors [DK].
References
1. Wright JR, Charles ES, Emile K. The first cancer biopsy. Int J Surg (2013) 11:106–7. doi:10.1016/j.ijsu.2012.11.017
2. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into management of cancer. Nat Rev Clin Oncol (2017) 14:531–48. doi:10.1038/nrclinonc.2017.14
3. Lecomte T, Ceze N, Dorval E, Laurent PP. Circulating free tumor DNA and colorectal cancer. Gastroenterol Clin Biol (2010) 34:662–81. doi:10.1016/j.gcb.2009.04.015
4. Alix PC, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med (2012) 63:199–215. doi:10.1146/annurev-med-062310-094219
5. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem (2015) 61:112–23. doi:10.1373/clinchem.2014.222679
6. Mandel P, Metais P. Les acides nucleiques du plasma sanguin chezl’homme. C R Seances Soc Biol Fil (1948) 142:241–3.
7. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Colon Cancer Version 2. Available from: https://www.nccn.org/professionals/physician_gls/recently_updated.aspx (Accessed: May 31, 2018).
8. Mouliere F, Robert B, Peyrotte EA, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One (2011) 6:e23418. doi:10.1371/journal.pone.0023418
9. Mouliere F, El Messaoudi S, Gongora C, Guedj AS, Robert B, Del Rio M, et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl Oncol (2013) 6:319–28. doi:10.1593/tlo.12445
10. Sun K, Jiang P, Chan KA, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genomewide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A (2015) 112:E5503–12. doi:10.1073/pnas.1508736112
11. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet (2016) 12:e1006162. doi:10.1371/journal.pgen.1006162
12. Haber DA, Valculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov (2014) 6:650–61. doi:10.1158/2159-8290.CD-13-1014
13. Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat (2010) 120:461–7. doi:10.1007/s10549-010-0747-9
14. Wang JY, Hsieh JS, Chang MY, Huang TJ, Chen FM, Cheng TL, et al. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg (2004) 28:721–6. doi:10.1007/s00268-004-7366-8
15. Yamada T, Nakamori S, Ohzato H, Oshima S, Aoki T, Higaki N, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res (1998) 4:1527–32.
16. Perkins G, Yap TA, Pope L, Cassidy AM, Dukes JP, Riisnaes R, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One (2012) 7:e47020. doi:10.1371/journal.pone.0047020
17. Shi J, Liu Q, Sommer SS. Detection of ultrarare somatic mutation in the human TP53 gene by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification. Hum Mutat (2007) 28:131–6. doi:10.1002/humu.20423
18. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variants. Proc Natl Acad Sci U S A (2003) 100:8817–22. doi:10.1073/pnas.1133470100
19. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewics AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem (2011) 83:8604–10. doi:10.1021/ac202028g
20. Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip (2011) 11:2156–66. doi:10.1039/c1lc20128j
21. Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med (2012) 4:162ra54. doi:10.1126/scitranslmed.3004742
22. Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem (2012) 59:211–24. doi:10.1373/clinchem.2012.196014
23. Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med (2013) 5:30. doi:10.1186/gm434
24. Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol (2017) 3:740–1. doi:10.1001/jamaoncol.2016.2835
25. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med (2014) 6:224ra24. doi:10.1126/scitranslmed.3007094
26. da Silva Filho BF, Gurgel AP, Neto MÁ, de Azevedo DA, de Freitas AC, Neto JD, et al. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol (2013) 66:775–8. doi:10.1136/jclinpath-2013-201521
27. Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer (2002) 100:542–8. doi:10.1002/ijc.10526
28. Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med (2014) 20:430–5. doi:10.1038/nm.3511
29. Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res (1990) 50:7717–22.
30. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell (1990) 61:759–67. doi:10.1016/0092-8674(90)90186-I
31. Tóth K, Sipos F, Kalmár A, Patai ÁV, Wichmann B, Stoehr R, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One (2012) 7:e46000. doi:10.1371/journal.pone.0046000
32. Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem (2009) 55:1337–46. doi:10.1373/clinchem.2008.115808
33. Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One (2008) 3:e3759. doi:10.1371/journal.pone.0003759
34. Lofton-Day C, Model F, DeVos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem (2008) 54:414–23. doi:10.1373/clinchem.2007.095992
35. Zhai R, Zhao Y, Su L, Cassidy L, Liu G, Christiani DC. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia (2012) 14:29–33. doi:10.1593/neo.111626
36. Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. High burden and perspective positive selection of somatic mutations in normal human skin. Science (2015) 348:880–6. doi:10.1126/science.aaa6806
37. Guo S, Diep D, Plongthongkum N, Fung HL, Zhang K, Zhan K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet (2017) 49(4):635–42. doi:10.1038/ng.3805
38. De la Cruz FF, Corcoran RB. Methylation in cell-free DNA for early cancer detection. Ann Oncol (2018) 29(6):1351–3. doi:10.1093/annonc/mdy134
39. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med (2016) 8:346ra92. doi:10.1126/scitranslmed.aaf6219
40. Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut (2016) 65:625–34. doi:10.1136/gutjnl-2014-308859
41. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med (2008) 14:985–90. doi:10.1038/nm.1789
42. Young GP, Pedersen SK, Mansfield S, Murray DH, Baker RT, Rabbitt P, et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med (2016) 5:2763–72. doi:10.1002/cam4.868
43. Symonds EL, Pedersen SK, Murray DH, Jedi M, Byrne SE, Rabbitt P, et al. Circulating tumour DNA for monitoring colorectal cancer-a prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin Epigenetics (2018) 10:63. doi:10.1186/s13148-018-0500-5
44. Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, et al. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget (2017) 8:6330–40. doi:10.18632/oncotarget.14064
45. Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of ctDNA after surgical resection. Ann Surg Treat Res (2014) 86:136–42. doi:10.4174/astr.2014.86.3.136
46. Ling ZQ, Zhao Y, Zhou SL, Mao WM. MSH2 promoter hypermethylation in circulating tumor DNA is a valuable predictor of disease-free survival for patients with esophageal squamous cell carcinoma. Eur J Surg Oncol (2012) 38:326–32. doi:10.1016/j.ejso.2012.01.008
47. Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, et al. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst (2000) 92:1805–11. doi:10.1093/jnci/92.22.1805
48. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A (2005) 102:16368–73. doi:10.1073/pnas.0507904102
49. Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol (2015) 26:1715–22. doi:10.1093/annonc/mdv177
50. Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med (2013) 369:1023–34. doi:10.1056/NEJMoa1305275
51. Patterson SD, Peeters M, Siena S, Van Cutsem E, Humblet Y, Van Laethem JL, et al. Comprehensive analysis of KRAS and NRAS mutations as predictive biomarkers for single agent panitumumab (pmab) response in a randomized, phase III metastatic colorectal cancer (mCRC) study (20020408). J Clin Oncol (2013) 31:3617. doi:10.1200/jco.2013.31.15_suppl.3617
52. Van Cutsem E, Lenz HJ, Köhne CH, Heinemann V, Tejpar S, Melezínek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol (2015) 33:692–700. doi:10.1200/JCO.2014.59.4812
53. Peeters M, Oliner K, Price TJ, Cervantes A, Sobrero AF, Ducreux M, et al. Analysis of KRAS/NRAS mutations in a phase III study of panitumumab with FOLFIRI compared with FOLFIRI alone as second-line treatment for metastatic colorectal cancer. Clin Cancer Res (2015) 21:5469–79. doi:10.1158/1078-0432.CCR-15-0526
54. Bokemeyer C, Köhne CH, Ciardiello F, Lenz HJ, Heinemann V, Klinkhardt U, et al. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. Eur J Cancer (2015) 51:1243–52. doi:10.1016/j.ejca.2015.04.007
55. Stintzing S, Modest DP, Rossius L, Lerch MM, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol (2016) 17:1426–34. doi:10.1016/S1470-2045(16)30269-8
56. Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol (2015) 16:937–48. doi:10.1016/S1470-2045(15)00138-2
57. Schmiegel W, Scott RJ, Dooley S, Lewis W, Meldrum CJ, Pockney P, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol (2017) 11:208–19. doi:10.1002/1878-0261.12023
58. Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol (2017) 28:1325–32. doi:10.1093/annonc/mdx125
59. Jones FS, Edelstein D, Wichner K, Ross C, Stieler K, Boehm V, et al. Performance of standardized BEAMing platform for detecting RAS mutations in the blood of metastatic colorectal cancer (mCRC) patients. J Clin Oncol (2016) 34:11538. doi:10.1200/JCO.2016.34.15_suppl.11538
60. Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol (2017) 28:1294–301. doi:10.1093/annonc/mdx112
61. Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature (2012) 486:537–40. doi:10.1038/nature11219
62. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med (2015) 21:795–801. doi:10.1038/nm.3870
63. Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, Auer M, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet (2014) 10:e1004271. doi:10.1371/journal.pgen.1004271
64. Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer (2011) 117:4623–32. doi:10.1002/cncr.26086
65. Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer (2015) 51:587–94. doi:10.1016/j.ejca.2015.01.054
66. Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer (2015) 112:1888–94. doi:10.1038/bjc.2015.173
67. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature (2012) 483:100–3. doi:10.1038/nature10868
68. Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov (2012) 2:227–35. doi:10.1158/2159-8290.CD-11-0341
69. Corcoran RB, Andre T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF V600E-mutant colorectal cancer. Cancer Discov (2018) 8:428–43. doi:10.1158/2159-8290.CD-17-1226
70. Leto SM, Sassi F, Catalano I, Torri V, Migliardi G, Zanella ER, et al. Sustained inhibition of HER3 and EGFR is necessary to induce regression of HER2-amplified gastrointestinal carcinomas. Clin Cancer Res (2015) 21:5519–31. doi:10.1158/1078-0432.CCR-14-3066
71. Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol (2016) 17:738–46. doi:10.1016/S1470-2045(16)00150-9
72. Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov (2011) 1:508–23. doi:10.1158/2159-8290.CD-11-0109
73. Takegawa N, Yonesaka K, Sakai K, Ueda H, Watanabe S, Nonagase Y, et al. HER2 genomic amplification in circulating tumor DNA from patients with cetuximab-resistant colorectal cancer. Oncotarget (2016) 7:3453–60. doi:10.18632/oncotarget.6498
74. Hainsworth JD, Bernstam FM, Swanton C, Hurwitz H, Spigel DR, Sweeney C, et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from MyPathway, an open-label, phase IIa multiple basket study. J Clin Oncol (2018) 36:536–42. doi:10.1200/JCO.2017.75.3780
75. Nakamura Y, Okamoto W, Sawada K, Komatsu Y, Kato K, Taniguchi H, et al. TRIUMPH Study: a multicenter Phase II study to evaluate efficacy and safety of combination therapy with trastuzumab and pertuzumab in patients with HER2-positive metastatic colorectal cancer (EPOC1602). Ann Oncol (2017) 28:612Ti. doi:10.1093/annonc/mdx393.137
76. Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov (2013) 3:658–73. doi:10.1158/2159-8290.CD-12-0558
77. Raghav K, Morris V, Tang C, Morelli P, Amin HM, Chen K, et al. MET amplification in metastatic colorectal cancer: an acquired response to EGFR inhibition, not a de novo phenomenon. Oncotarget (2016) 7:54627–31. doi:10.18632/oncotarget.10559
78. Strickler JH, Rushing CN, Uronis HE, Morse M, Blobe GC, Zafar Y, et al. Phase Ib study of cabozantinib plus panitumumab in KRAS wild-type (WT) metastatic colorectal cancer (mCRC). J Clin Oncol (2016) 34:3548. doi:10.1200/JCO.2016.34.15_suppl.3548
79. Bang YJ, Cutsem EV, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-esophageal junction cancer (ToGA): a phase 3, open-label, randomized controlled trial. Lancet (2010) 376:687–97. doi:10.1016/S0140-6736(10)61121-X
80. Gravalos C, Jimeno A. Her2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol (2008) 19:1523–9. doi:10.1093/annonc/mdn169
81. Shoda K, Masuda K, Ichikawa D, Arita T, Miyakami Y, Watanabe M, et al. HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study. Gastric Cancer (2015) 18:698–710. doi:10.1007/s10120-014-0432-5
82. Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer (2017) 20:126–35. doi:10.1007/s10120-016-0599-z
Keywords: circulating tumor DNA, colorectal cancer, gastric cancer, esophageal cancer, gastrointestinal malignancies
Citation: Sawada K, Kotani D and Bando H (2018) The Clinical Landscape of Circulating Tumor DNA in Gastrointestinal Malignancies. Front. Oncol. 8:263. doi: 10.3389/fonc.2018.00263
Received: 04 May 2018; Accepted: 25 June 2018;
Published: 16 July 2018
Edited by:
Pashtoon Murtaza Kasi, Mayo Clinic, United StatesReviewed by:
Ali Roberts, Guardant Health, Inc., United StatesPeter L. Molloy, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright: © 2018 Sawada, Kotani and Bando. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisuke Kotani, ZGtvdGFuaUBlYXN0Lm5jYy5nby5qcA==
 Kentaro Sawada1,2
Kentaro Sawada1,2 Daisuke Kotani
Daisuke Kotani