- Tumor Etiology and Screening Department of Cancer Institute and General Surgery, The First Hospital of China Medical University, Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang, China
Background: Accumulating studies have focused on the relationship between miRNAs polymorphisms and cancer prognosis. However, the results are conflicting and unconvincing. This systematic review and meta-analysis was conducted to explore the relationship between miRNAs polymorphisms and cancer prognosis, aiming to seek for markers with cancer prognostic function.
Methods: Hazard ratio of overall survival, disease-free survival (DFS) and recurrence-free survival were calculated to evaluate the association between miRNAs polymorphisms and cancer prognosis by using Stata software 11.0.
Results: We systematically reviewed the association of 17 miRNAs SNPs with cancer prognosis including 24,721 samples. It was shown that 6 miRNAs SNPs (miR-608 rs4919510, miR-492 rs2289030, miR-378 rs1076064, miR-499 rs4919510, miR-149 rs2292832, miR-196a2 rs11614913) were associated with better cancer overall survival (OS) while let-7i rs10877887 was associated with poor OS; the homozygous and heterozygote genotype of miR-423 were related to poor cancer relapse-free survival (RFS) when compared with the wild genotype; miR-146 rs2910164 was linked to favorable cancer DFS while miR-196a2 rs11614913 was associated with poor DFS.
Conclusions: In summary, let-7i rs10877887, miR-608 rs4919510, miR-492 rs2289030, miR-378 rs1076064, miR-423 rs6505162, miR-499 rs4919510, miR-149 rs2292832, miR-146 rs2910164, and miR-196a2 rs11614913 might serve as potential biomarkers for cancer prognosis.
Introduction
Despite emerging advances in the researches and understanding in tumor biology, cancer incidence remains rising and this global challenge further exacerbates by the increasing human life expectancy (1). It has been estimated that there will be ~19 million new cancer cases by 2025 (2). The high cancer-related morbidity and mortality contribute to urgent needs for novel biomarkers to help to evaluate the clinical outcome of cancer patients and enhance therapeutic effects to prolong their survival.
MicroRNAs (miRNAs), a class of non-coding RNAs with 19–25 nucleotides length, are small and regulatory RNAs binding to the 3′-UTR region of mRNA molecules. They have been regarded as key regulators in many diseases, particularly relevant in cancer (3, 4). It has been suggested that miRNAs may play oncogenic drivers or tumor suppressor roles in various cancer (3). The possible mechanism might be that the variation of miRNAs expression promote carcinogenesis, metastasis and many other characteristics of cancer by regulating the expression patterns of key genes involved in tumor growth and progression (5–7).
It has been widely recognized that the functional polymorphisms in miRNAs are the most common form of variation present in the human genome and could affect cancer susceptibility and prognosis (7–12). MiRNAs polymorphisms have been reported to influence the expression of mature miRNAs (13, 14). For instance, Chen et al. have reported that micRNA30c-1 polymorphisms could regulate the expression of mature miRNA 30c-1 and thus affect cancer prognosis (15, 16). A research by Yu et al. suggested the rs4938723 polymorphism could reduce micRNA-34b expression and increase the recurrence of early gastric cancer (17). Currently, accumulating studies have focused on the function of miRNAs polymorphisms and their relationship with cancer prognosis. However, the results were conflicting and unconvincing.
In the present study, a systematically review was conducted to investigate the association of miRNAs polymorphisms with overall survival (OS) time, recurrence-free survival (RFS) time as well as disease-free survival (DFS) time of cancer patients. Based on that, available data was collected to perform a meta-analysis to give a comprehensive assessment for the relationship between miRNAs polymorphisms and cancer prognosis. Data of this meta-analysis could expand our understanding of the role of miRNAs polymorphisms in human cancer prognosis, which may provide more credible evidences for future research in this field as well as find possible prognostic biomarkers and make an effort to assistant clinical decisions in the future.
Methods
Literature Mining
This study was carried out on the basis of Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) (18).
Studies published in English language up to 20 October 2018 reporting on the association between the miRNAs polymorphism and cancer prognosis were identified by entering the following search terms into PubMed and Web of Science: “miRNA/miRNAs”; “polymorphisms/variants/variation/single nucleotide polymorphism/SNP”; and “cancer/carcinoma/tumor/neoplasm” and “prognosis/prognostic/outcome/survival.” Two independent investigators (Hanxi Ding and Qian Xu) performed the literature search. Eligible studies met the following criteria: (1) Concerning the association between miRNAs SNPs and cancer prognosis; (2) Involving prognostic indicators such as OS, DFS or RFS; (3) Including available HR and 95% CI. Articles were excluded based on the followings: (1) Duplicated studies or data; (2) Not relevant with miRNAs SNPs and cancer prognosis; (3) Lacking of available data or figures.
Data Extraction
Two investigators (Hanxi Ding and Qian Xu) extracted the data independently and reached consensus regarding all the items. Study descriptions were obtained from each full text including author's name, year of publication, country of the origin, type of cancer, total number of the study population, the polymorphism site, the genotype, hazard ratio (HR), and corresponding 95% confidence interval (CI). In the absence of adequate information for estimation of HR or 95% CI, we have made all efforts to contact the authors to obtain sufficient information or extracted data from the Kaplan-Meier survival curves using a method suggested by Tierney et al. (19).
Methodology Quality Assessment
Two reviewers (Hanxi Ding and Qian Xu) independently evaluated the quality of selected studies according to Newcastle-Ottawa Scale (20). Eight items categorized of three dimensions were assessed, including selection, comparability and exposure were assessed. The quality scores ranged between 0 and 9 stars (Supplementary Table 2).
Statistical Analysis
The association of miRNAs polymorphisms with cancer OS, DFS and RFS was estimated through forest plots. Pooled HR and 95%CI were calculated by fixed-effects model or random-effects model. Pooled HR >1 suggested poor prognosis and was considered statistically significant if the 95%CI did not contain 1(21). In the absence of inter-study heterogeneity for Q-statistic with p > 0.05 and I2 < 50%, fixed-effect model was chosen to conserve statistical power, otherwise random-effect model was used (22, 23). Publication bias was assessed by Begger' test and if p > 0.05 was considered to be lack of publication bias (24). Sensitivity analysis was conducted by removing studies one by one. All analyses were performed using Stata software 11.0. All tests were two-sided and the results were considered to be statistically significant when the p < 0.05.
Results
Characteristics of the Eligible Studies
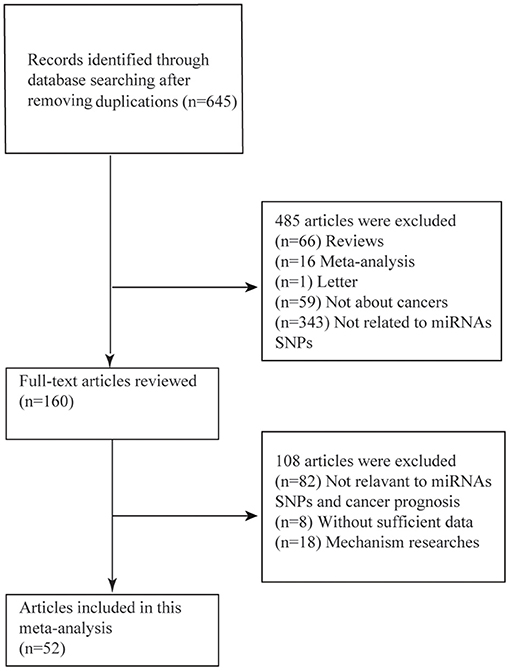
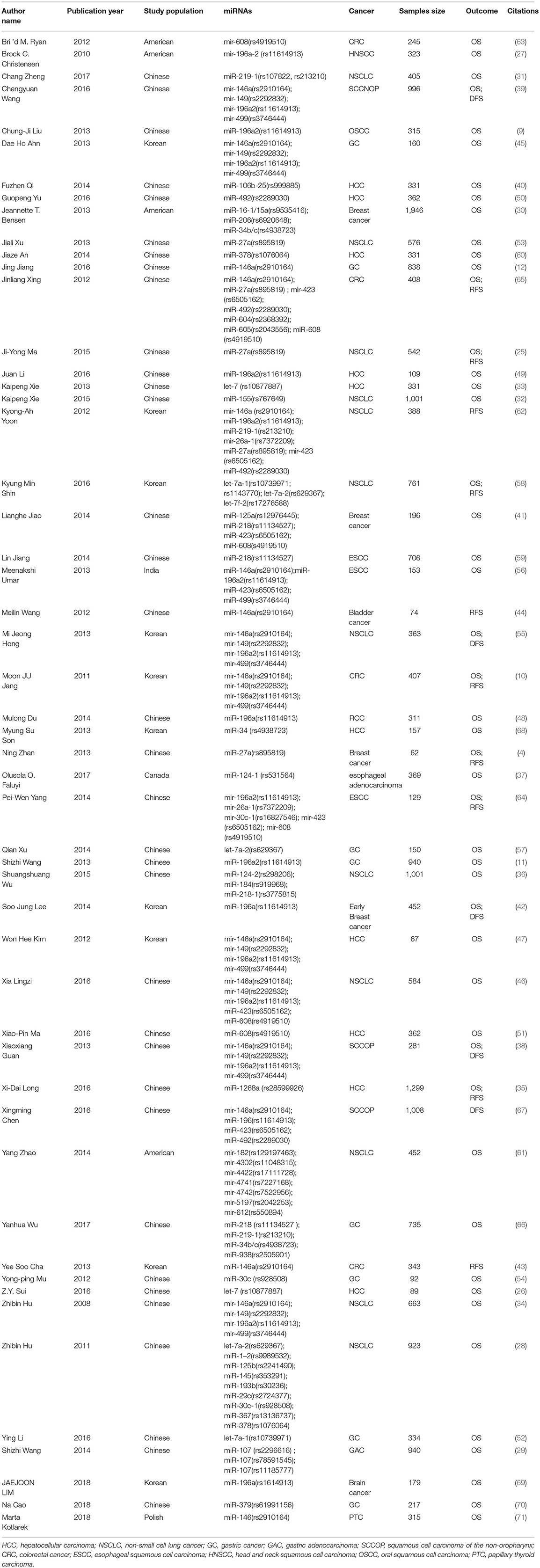
As was shown in the flow diagram, a total of 645 articles were enrolled in this systematic review. After multiple selections, 52 researches for 17 miRNAs SNPs including 24721 patients were involved in our meta-analysis (Figure 1) (4, 9–12, 25–68). Among the enrolled studies, 17 miRNAs were OS-related, 6 were RFS-related and 4 were DFS-related. The cancer type covered hepatocellular cancer (HCC), gastric cancer (GC), colorectal cancer (CRC), breast cancer, bladder cancer, non-small cell lung cancer (NSCLC), head and neck cancer, squamous cell carcinoma of the non-oropharynx (SCCOP). The original population came from Chinese, Korean, American, Canadian, Indian, and Polish. The characteristics of all the 52 studies were summarized in the Table 1 and the original data were shown in the Supplementary Tables 1, 4.
Quantitative Data Synthesis of miRNAs SNPs
Rs10877887 in let-7i
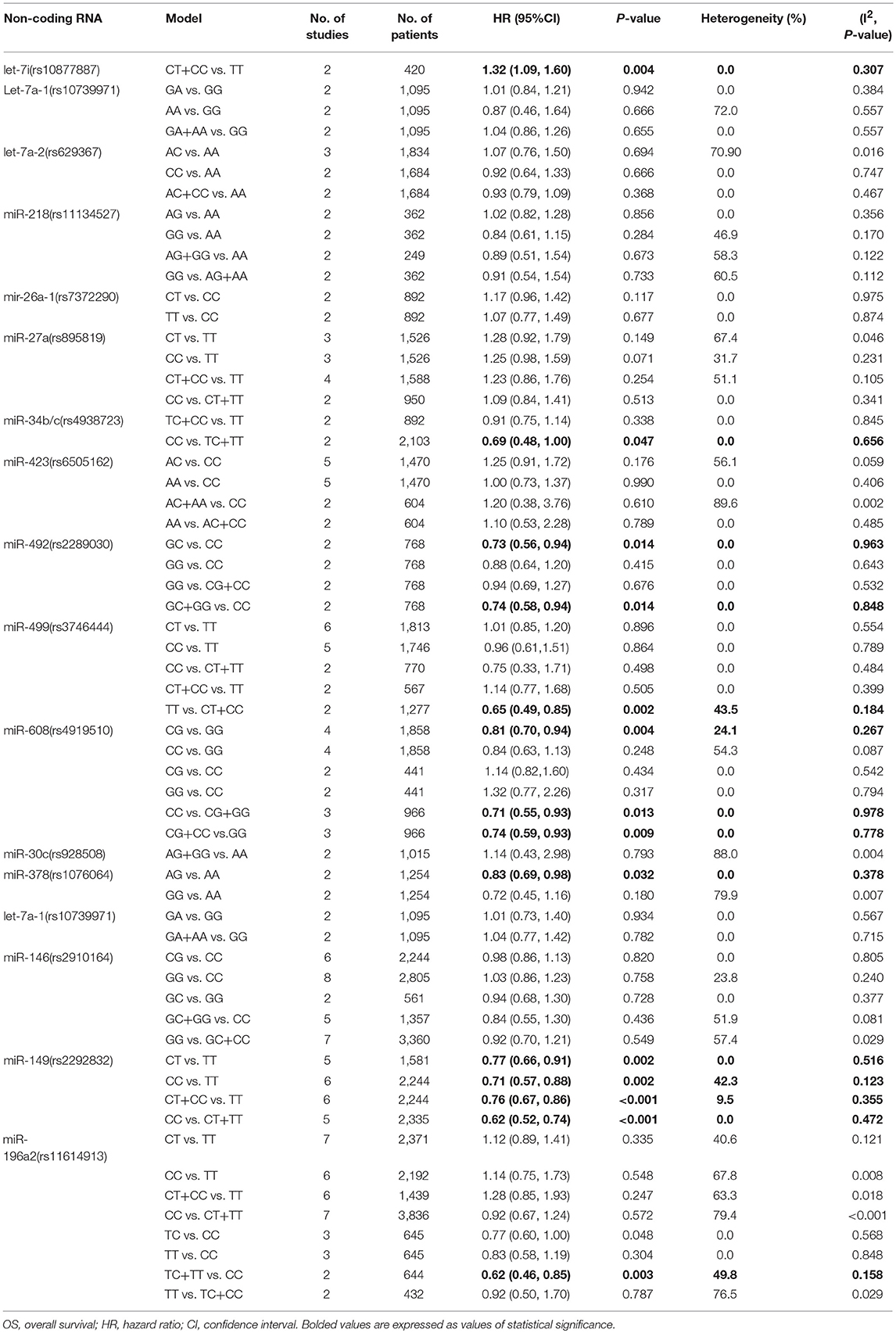
Two relevant studies were included into this analysis to investigate the possible association between rs10877887 and cancer prognosis and the results suggested poor OS in CT+CC vs. TT model (HR = 1.32, 95%CI 1.09–1.60, p = 0.004). No heterogeneity was found in the meta-analysis process (I2 = 0.0%, p = 0.307, Table 2).
Rs4919510 in miR-608
This polymorphism showed significant protective effects on cancer OS in CG vs. GG, CG+CC vs. GG and CC vs. CG+GG models (HR = 0.81, 95%CI 0.70–0.94, p = 0.004; HR = 0.74, 95%CI 0.59–0.93 p = 0.009; HR = 0.71, 95%CI 0.55–0.93, p = 0.013; respectively, Table 1). No heterogeneity was observed within the three models in the calculation processes (I2 = 24.1%, p = 0.267; I2 = 0.0%, p = 0.778; I2 = 0.0%, p = 0.978, respectively). Pooled data also suggested that heterozygote was associated with better RFS when compared with wild homozygote (HR = 0.73, 95%CI 0.60–0.88, p = 0.001). However, no significant relationship was indicated between rs4919510 and cancer DFS (Tables 2–4).
Rs2289030 in miR-492
For this polymorphism, only GC vs. CC and GC+GG vs. CC models were found to be associated with cancer OS (HR = 0.73, 95%CI 0.56–0.94, p = 0.014; HR = 0.74, 95%CI 0.58–0.94, p = 0.014, respectively, Table 2) but we failed to find any significant results about miR-429 SNP and RFS. And there was no meaningful heterogeneity among the molds in the meta-analysis.
Rs1076064 in miR-378
The heterozygote AG of rs1076064 in miR-378 was suggested to have a protective role in cancers OS when compared to the genotype AA (HR = 0.83, 95%CI 0.69–0.98, p = 0.032). No significant heterogeneity was found during calculation (I2 = 0.0%, p = 0.378, Table 2).
Rs6505162 in miR-423
The rs6505162 polymorphism was suggested to be associated with cancer RFS. The AC vs. CC and AC+AA vs. CC models indicated poor cancer prognosis (HR = 1.34, 95%CI 1.03–1.73, p = 0.026; HR = 1.37, 95%CI 1.01–1.86, p = 0.042, respectively) and we found no significant heterogeneity of these two SNPs (I2 = 24.1%, p = 0.268; I2 = 29.0%, p = 0.235, respectively Table 3). As for the relationship of rs6505162 with cancer OS and DFS, no related studies were involved in this meta-analysis.
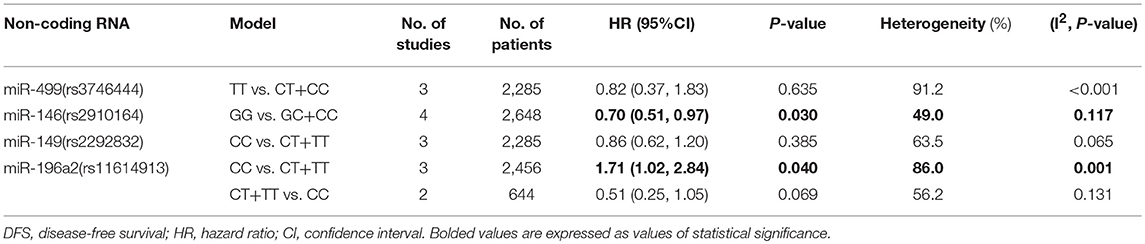
Rs3746444 in miR-499, rs2292832 in miR-149, rs2910164 in miR-146, and rs11614913 in miR-196a2
The TT vs. CT+CC model of rs3746444 polymorphism in miR-499 was associated with better OS (HR = 0.65, 95%CI 0.49–0.85, p = 0.002, Table 2). No association was suggested between miR-499 polymorphism and cancer RFS or DFS (Tables 3, 4). It was shown that miR-149 rs2292832 has significant protective effect on OS. Additionally, this effect was strengthened with the increasing variant C allele in each genetic model (CT vs. TT: HR = 0.77, 95%CI 0.66–0.91, p = 0.002; CC vs. TT: HR = 0.71, 95%CI 0.57–0.88, p = 0.002; CT+CC vs. TT: HR = 0.76, 95%CI 0.67–0.86, p < 0.001; CC vs. CT+TT: HR = 0.62, 95%CI 0.52–0.74, p < 0.001; respectively Table 2). No positive result was shown in it with cancer RFS or DFS. For the rs2910164 polymorphism in miR-146, only GG vs. GC+CC model suggested a protective role in DFS (HR = 0.70, 95%CI 0.51–0.97, p = 0.030; Table 4). No significant association was observed between rs2910164 and cancer OS or RFS in any genetic models. For miR-196a2 rs11614913 polymorphism, we also found that the TC+TT vs. CC model indicated better OS of cancer patients (HR = 0.62, 95%CI 0.46–0.85, p = 0.003; Table 2), and similar finding was indicated in cancer DFS (CC vs. CT+TT: HR = 1.71, 95%CI 1.02–2.84, p = 0.040; Table 4). However, it had no association with cancer RFS.
Other miRNAs SNPs and Cancers Prognosis
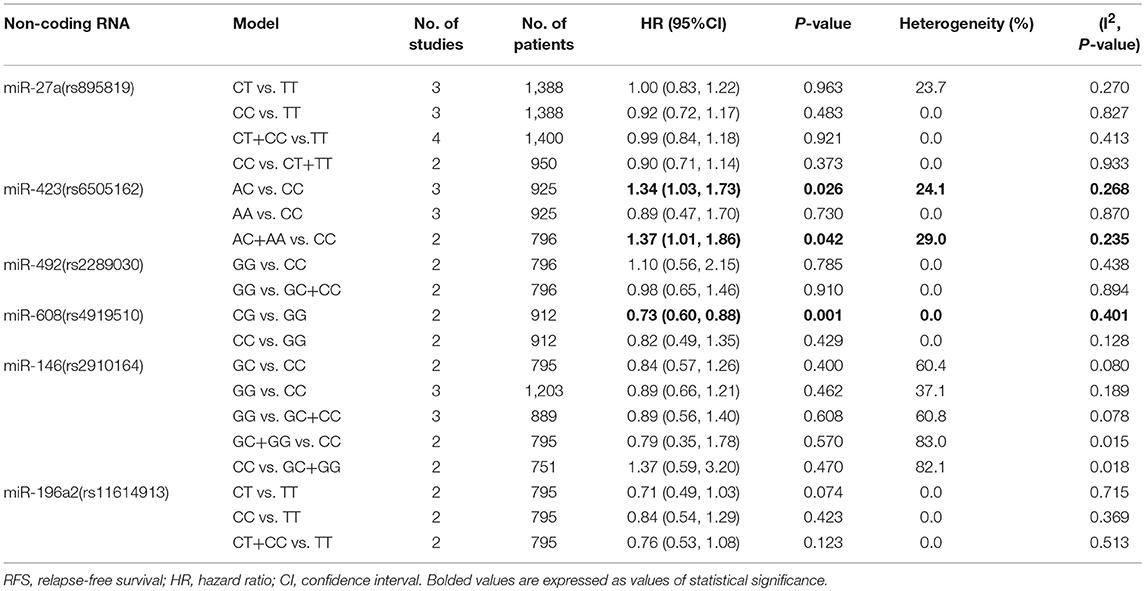
Other than the above-mentioned miRNAs SNPs, meta-analysis was also performed for the association of let-7a-2 (rs629367), miR-218 (rs11134527), miR-26a-1 (rs7372290), miR-27a (rs895819), miR-34b/c (rs4938723), miR-423 (rs6505162), miR-30c (rs928508), and let-7a-1 (rs10739971) with cancer OS and RFS. However, no statistical significance was demonstrated in them (Tables 2, 3).
Stratified Data and Cancers Prognosis
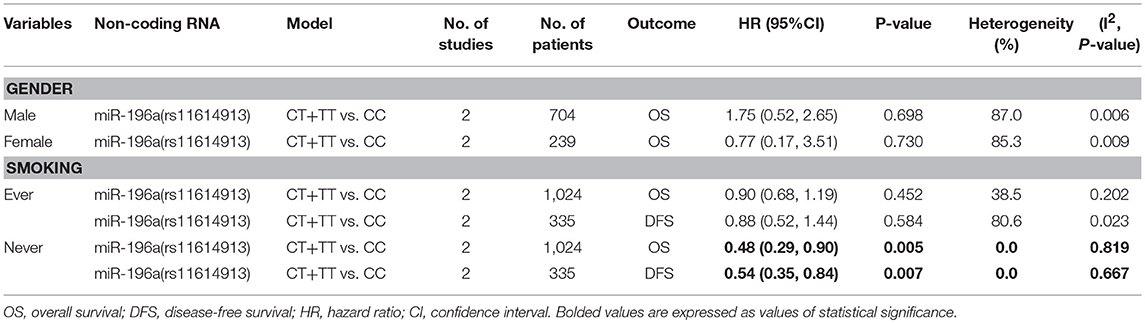
Meta-analysis of all the included stratified data was conducted and the results indicated that CT+TT vs. CC model of miR-196a (rs11614913) was related to better OS as well as DFS in the never smoking subgroup (HR = 0.48, 95%CI 0.29–0.90, p = 0.005 for OS; HR = 0.54, 95%CI 0.35–0.84, p = 0.007 for DFS; Table 5).
Heterogeneity
In the overall comparisons, no inter-study heterogeneity was associated with cancer prognosis (OS, RFS, and DFS, Tables 2–4). With respect to the heterogeneity in other overall comparisons, sensitivity analysis was performed subsequently.
Publication Bias
Begg's test was used to evaluate the potential publication bias of included studies. No statistically significant publication bias was indicated in any of the models for all involved miRNAs SNPs (Supplementary Table 3).
Discussion
In this study, we systematically reviewed 645 relevant published articles. Meanwhile, 52 researches for 17 SNPs including 24,721 patients were involved in this meta-analysis. The results indicated that except that the variant genotype of let-7i rs10877887 (OS), miR-423 rs6505162 (RFS), miR-196 rs11614913 (DFS) showed worse outcomes for cancer survival, others (including miR-608 rs4919510, miR-492 rs2289030, miR-378 rs1076064, miR-499 rs3746444, miR-149 rs29101644, miR-196 rs11614913, and miR-146 rs2292823) all played protective role in cancer survival. This study would provide theoretical and research clues for future exploration.
let-7i rs10877887: Association With Poor Cancer Prognosis
Let-7i rs10877887 is located in the promoter region of miRNA let-7, which was a well-known tumor suppressor of multiple cancers and incorporates a CpG island, TF biding sites, and DNase peak (72–74). Du et al. suggested miRNA let-7 acted as a tumor suppressor in RCC via down-regulating C-myc and C-myc's target gene (73). It has also been reported that low let-7i expression was independently associated with CRC distant metastasis and significantly linked to poor survival in CRC patients (75). Similar result was found in another research on lung cancer by Huang et al. (76).
On account of the location and incorporation of rs10877887, the T > C variant may influence the binding of transcription factors and let-7 expression (33, 77). Liu's research suggested that patients harboring rs10877887 CC genotype had a lower let-7i expression in CSCC tissues (78). In addition, the T > C variation of rs10877887 may have strong affinity with Myeloid zinc finger 1 (MAF1), a transcription factor which can promote the activity of Axl promoter, resulting in tumor cell migration, invasion and metastasis (33).
Our results suggested that the CT/CC genotype of rs10877887 plays a worse role in cancer prognosis when compared with wild homozygote. Probably it was because that deregulated let-7i expression associated with T > C mutation in cancer tissues. We could speculate that T > C variants may serve as biomarker of poor cancer prognosis as well as assistant clinical-decisions. Much more researches were needed to confirm this result as well as detect the potential mechanisms and functions in the future.
miR-608 rs4919510: Association With Better Cancer Prognosis
MiRNA 608 harbors one SNP, rs4919510 C > G, located in +22bp of its mature 25 bp sequence. It is at the joint of the stem with the canonical hairpin loop, which has been reported to play a key role in cancer progression and be associated with prognosis of several cancer recently (41, 46, 51, 63–65, 79–81). Zheng et al. have found that rs4919510 SNP might influence the expression of miR-608 target genes including cell growth-related genes, tumor invasion and metastasis-related genes and cancer death-related genes (82).
Among the studies included in our meta-analysis, CG vs. GG, CC vs. CG+GG, and CG+CC vs. GG models of miR-608 were associated with overall cancer prognosis, all having protective effects. The possible mechanism might be that the CG, CC, and CG+CC genotypes of rs4919510 could influence the expression level of miR-608 target genes by regulating the miR-608 expression and exert positive roles in cancer prognosis. The reason why no statistical significant results were found to be related to cancer RFS may result from the relatively small number of articles researching about the miR-608 rs4919510 polymorphisms and RFS. In conclusion, miR-608 rs4919510 is associated with cancer OS and could be used for specific prediction of cancer prognosis and may direct clinical-decisions in the future with abundant mechanisms as well as functions evidences.
miR-492 rs2289030: Association With Better Cancer Prognosis
As we know, mounting studies have shown that miR-492 plays an important role in cell tumorigenicity of multiple cancers (83–85). von Frowein et al found that up-regulation of miR-492 enhanced proliferation, anchorage-independent growth, migration, and invasion of hepatoblastoma by regulating CD44, which is a receptor for hyaluronan, the major component of the ECM and as well as a co-receptor for multiple cytokine signals and growth factors (83). Shen et al have revealed that ectopic expression of miR-492 contributed to deregulation of SOX7, leading to up-regulation of cyclin D1, c-Myc, and Rb phosphorylation, which could promote cell proliferation and cell cycle of breast cancer (84).
Single nucleotide polymorphisms of miRNAs have been identified to affect miRNA processing and alter miRNA expression (50). Some researchers have shown that miR-492 rs2289030 G > C was associated with various cancers (50, 62, 67, 86). Lee et al found CRC patients carried CG and GG genotype demonstrated worse RFS when compared with CC genotype. However, all enrolled studies in our meta-analysis showed that the GC and GC/GG genotypes played a positive role in cancers OS (87). Although our findings suggested that the GC and GG variation of miR-492 may improve cancers prognosis, more rounded investigations are needed to elucidate the association between miR-492 rs2289030 and cancer prognosis as well as the special mechanisms for the limited studies and inconsistent results.
miR-378 rs1076064: Association With Better Cancer Prognosis
It has been reported that the A to G base change of SNP rs1076064 at 222 bp upstream from miR-378 may alter the expression of miR-378 (60). Accumulating researches have indicated that miR-378 was down-regulated in CRC, GC, and oral cancer (88–90). MiR-378 may exert tumor suppressor roles by deregulating the expression of CDK6 and VEGF in GC. Wang et al suggested that miR-378 inhibited cell proliferation by targeting CDC40 (88, 90). However, other studies have demonstrated that the up-regulation of miR-378 was related to several kinds of cancer including breast cancer, RCC and AML (91–93). Lee et al found that miR-378 could promote cell survival, tumor growth and angiogenesis by targeting at SuFu and Fus-1 (94).
An's research revealed that the variant genotype of rs1076064 acted as a transcription regulator of miR-378 and the G allele of rs1076064 may exert higher promoter activity in many cancer cell lines and was associated with a better prognosis in HCC (60). The meta-analysis results of the two involved articles also showed that AG vs. AA model was associated with better cancer OS, which was consistent with previous study. However, more studies were needed to confirm this result and translate the mechanisms in the future.
miR-423 rs6505162: Association With Poor Cancer Prognosis
MiR-423, relevant to NSCLC, CRC, and breast cancer, is located in frequently amplified region of chromosome 17q11.2 (95–97). Zhao et al have revealed that miR-423 played a potentially oncogenic role in breast carcinogenesis by promoting cell proliferation of breast cancer cell lies (97). Similarly, Sun et al suggested the overexpression of miR-423 could decrease BRMS1 level obviously and promote cell invasion of HCC (98).
The study of Xing et al demonstrated that the variant-containing genotype AC/AA of rs6505162 in miR-423 was significantly associated with poor OS and RFS of CRC patients (65). However, another research by Lin et al suggested the A allele of miR-423 rs6505162 was associated with decreased RCC recurrence and better prognosis by weakening the capacity to target KLF2 mRNA, leading to inhibition of angiogenic pathways and cancer recurrence (99). The meta-analysis results of rs6505162 in our study showed that AC and AC/AA genotypes were associated with poor cancer RFS, which was consistent with Xing's research. However, only two related studies were enrolled in this meta-analysis, further relevant investigations are needed to obtain more reliable results.
miR-146 rs2910164, miR-196a2 rs11614193, miR-149 rs2292832, and miR-499 rs3746444: Association With Cancer Prognosis
It has been well acknowledged that miRNAs SNPs could influence cancer prognosis by affecting miRNAs maturation or ability to combine with mRNAs target gene (100, 101).
Researches showed the variant G allele of miR-146 rs2910164 G > C, located in the 3′ miRNA passenger strand, may enhance the expression of mature miR-146 and the miR-146 overexpression has been found to suppress breast cancer metastasis (102, 103). This meta-analysis results of miR-146 rs2910164 showed that only the GG vs. GC+CC model was associated with better cancer DFS, which was consistent with Xia's meta-analysis (14). It could be inferred that the variant G allele might lead to increased expression of miR-146a and thus better prognosis.
MiR-196a has been regarded as an oncogene in cancers pathogenesis such as proliferation, migration and invasion (55). MiR-196a2 rs11619413 T > C, located in the 3′ messenger strand of miR-196a2, could influence the combination with target gene and the CC genotype was related to increased expression of miR-196a2 (34, 49, 104). The Meta-analysis showed that TC+TT vs. CC model was associated with better cancer OS while CC vs. CT+TT model was associated with poor cancer DFS, suggesting that CC allele might have risk effects on cancer prognosis. These results were contrary to Wang et al but consistent with Hu's research about lung cancer (11, 34). The differences may result from the different types of cancers or the baseline characteristics and more large number samples researches are needed to clarify the relationship between miR-196a2 rs11614913 and cancer prognosis.
MiR-149 is reported to be a pro-apoptotic miRNA, which can inhibit the expression of Akt1 and E2F1 and thus induce cancer cell lines apoptosis (99, 105). A function study of Xia et al suggested that C allele could increase the expression of miR-149, therefore leading to better prognosis in NSCLC (46). In this meta-analysis, we found four models of miR-149 rs2292832 including CT vs. TT, CC vs. TT CT+CC vs. TT, and CC vs. CT+TT were associated with better cancer OS, which was consistent with previous meta-analysis (106). We could infer that variant C allele may enhance miR-149 expression which was associated with better cancer prognosis and may play role as specific biomarker of cancer prognosis.
MiR-499 rs3736444 is located in the stem region opposite to the mature miR-499 sequence genetic and the T to C variation may influence miR-499 expression (39, 107). Ma et al have reported that the C allele could suppress miR-499 expression, resulting in decreased expression of Ets1, which therefore promotes HCC development and cause poor cancer outcome (108). The present study showed that TT vs. CT+CC model was associated with better cancer OS which was consistent with the results of Qiu's study that rs3746444 C to T variant could contribute to unfavorable cancer prognosis by regulating the expression of cancer-related genes (109). Therefore, we could speculate that variants containing C allele may play the role of miR-499 expression inhibitor and results in poor cancer outcome. Although there were statistically significant relationships, it should be noticed that only two relevant studies were involved, thus more convincing results need more studies on this field.
Other miRNAs: Association With Cancer Prognosis
In addition to the above-mentioned 9 miRNAs polymorphisms, other miRNAs SNPs involved were also reviewed and processed in this meta-analysis. No significant association with cancer prognosis was discovered in our meta-analysis, but some of them were reported to influence the development and invasion of cancers and the corresponding mechanisms have been elaborated. For example, the transition from A to G of rs1113427 may alter the local secondary structure of miR-218 and then influence the expression of miR-218 which could inhibit the invasion and metastasis of GC by targeting the Robol receptor (66, 110, 111). Zhang et al. found rs895819 could affect the secondary structure of pre-miR-27a that subsequently influence the processing and maturation of miR-27a (4). Additionally, only two researches were available in the meta-analysis for multiple SNPs, so more relevant investigations should be included for updating the findings on the association between these polymorphisms and cancer prognosis in the future.
Stratified Meta-Analysis: miRNAs SNPs and Cancer Prognosis
Further, we have collected all the available stratified data of included researches. It is suggested only the CT+TT genotype of miR-196a rs11614913 was associated to better cancer prognosis in the never smoking subgroup. We may speculate that CT +TT genotype of miR-196a rs11614913 maybe a potential biomarker for cancer prognosis in the specific subgroup. More evidences were needed to strengthen this conclusion in the future.
Advantages and Limitations
Our study had some advantages. First, we collected all the published articles related to miRNAs SNPs and cancer prognosis, making the systematic review and meta-analysis comprehensive and complete. Second, this is the first meta-analysis concerning the association between miRNAs SNPs and cancer prognosis. Moreover, this study is reliable and stable due to the large number of enrolled patients (24,721) and the strict inclusion criteria.
Undoubtedly, some limitations should be acknowledged in our study. First, only English literature was searched, which may results in publication bias. Second, although the overall sample size were up to 24,721 patients enrolled, the number of some miRNAs SNP relevant studies was too small thus we could only preliminarily analyze the relationships between miRNAs polymorphisms and cancer prognosis with the currently published literatures. What's more, many of the included articles were without clear statistical power thus we could have no idea whether there were type I error and type II error in the original data. Finally, several original texts have no available data and we extracted data from figures which might lead to some bias.
Conclusion and Future Directions
We systematically reviewed the researches about the association between miRNAs SNPs and cancer prognosis. Meanwhile, available data was used to perform a meta-analysis for SNPs with OS, RFS, and DFS of solid cancer. The relationships between miRNAs polymorphisms and cancer prognosis could be categorized into four types: (1) a better association, SNPs were linked to a better cancer prognosis such as miR-608 rs4919510 and miR-378 rs1076064; (2) a worse association, SNPs were associated with poor cancer prognosis including miR-423 rs6505162 and miR-196a2 rs1161913; (3) no association, no significant association was found between miRNAs SNPs and cancer survival; (4) failed to be quantitatively synthesized due to limited studies. Our study suggested quite a few miRNAs SNPs were associated with cancer prognosis, which would provide clues for further exploration on prognostic biomarkers.
Following aspects should be focused on in the future investigations. First, more miRNAs SNPs that may be associated with cancer prognosis (OS, RFS, and DFS) should be screened out to provide more alternative prognostic biomarkers. Second, more functional studies are needed to explore the mechanisms of SNPs within miRNAs in caner prognosis. Thirdly, in addition to the association between SNPs and cancer prognosis, other aspects such as chemotherapeutic susceptibility and drug tolerance are also needed to be illuminated. Finally, whether the exosome could carry some specific miRNA SNPs to or leave neoplastic foci may be explored to find some new clinical targets as well as therapeutic targets. Therefore, the clinical application of miRNAs polymorphisms has extremely extensive prospects and requiring further exploration.
Author Contributions
HD and QX extracted and evaluated the data. HD calculated and wrote this paper. ZL revised this paper. QX and YY conceived the study. All authors read and approved the final manuscript.
Funding
This work is supported partly by grants from the Natural Science Foundation of Liaoning Province in China (Ref No. 20170540987).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2018.00596/full#supplementary-material
Abbreviations
OS, overall survival; DFS, disease-free survival; RFS, relapse-free survival; HCC, hepatocellular cancer; GC, gastric cancer; CRC, colorectal cancer; NSCLC, non-small cell lung cancer; SCCOP, squamous cell carcinoma of the non-oropharynx; PTC, papillary thyroid carcinoma.
References
1. Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature (2017) 552:194–9. doi: 10.1038/nature25016
2. Cupit-Link MC, Kirkland JL, Ness KK, Armstrong GT, Tchkonia T, LeBrasseur NK et al. Biology of premature ageing in survivors of cancer. ESMO Open (2017) 2:e000250. doi: 10.1136/esmoopen-2017-000250
3. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer (2018) 18:5–18. doi: 10.1038/nrc.2017.99
4. Zhang N, Huo Q, Wang X, Chen X, Long L, Jiang L, et al. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene (2013) 529:125–130. doi: 10.1016/j.gene.2013.07.041
5. Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. New Engl J Med. (2008) 359:2720–2. doi: 10.1056/NEJMe0808667
6. Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in cancer. Cell (2009) 137:586–586.e1. doi: 10.1016/j.cell.2009.04.040
7. Srivastava K, Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PLoS ONE (2012) 7:e50966. doi: 10.1371/journal.pone.0050966
8. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer (2010) 10:389–402. doi: 10.1038/nrc2867
9. Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, Lin SC. miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol. (2013) 20(Suppl. 3):S406–14. doi: 10.1245/s10434-012-2618-6
10. Jang MJ, Kim JW, Min KT, Jeon YJ, Oh D, Kim NK. Prognostic significance of microRNA gene polymorphisms in patients with surgically resected colorectal cancer. Exp Ther Med. (2011) 2:1127–32. doi: 10.3892/etm.2011.321
11. Wang S, Tao G, Wu D, Zhu H, Gao Y, Tan Y, et al. A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Mol Carcinog. (2013) 52(Suppl. 1):E87–95. doi: 10.1002/mc.22017
12. Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Fut Oncol. (2016) 12:2215–26. doi: 10.2217/fon-2016-0224
13. Mullany LE, Herrick JS, Wolff RK, Slattery ML. Single nucleotide polymorphisms within MicroRNAs, MicroRNA targets, and MicroRNA biogenesis genes and their impact on colorectal cancer survival. Genes Chromosomes Cancer (2017) 56:285–95. doi: 10.1002/gcc.22434
14. Xia L, Ren Y, Fang X, Yin Z, Li X, Wu W, et al. Prognostic role of common microRNA polymorphisms in cancers: evidence from a meta-analysis. PLoS ONE (2014) 9:e106799. doi: 10.1371/journal.pone.0106799
15. Xia L, Yin Z, Li X, Ren Y, Zhang H, Zhao Y, et al. Genetic polymorphisms in pre-miRNAs predict the survival of non-small-cell lung cancer in Chinese population: a cohort study and a meta-analysis. Oncotarget (2017) 8:77963–74. doi: 10.18632/oncotarget.20276
16. Chen JP, Liu Y, Hu ZB, Shen HB. [Single nucleotide polymorphism in flanking region of miR-30c influences the maturing process of miR-30c in lung carcinoma]. Chin J Oncol. (2012) 34:664–8. doi: 10.3760/cma.j.issn.0253-3766.2012.09.005
17. Yu K. The Rs4938723 polymorphism reduces expression of MicroRNA-34b and increases the risk of recurrence after endoscopic dissection in early gastric cancer. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. (2017) 43:1235–46. doi: 10.1159/000481764
18. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group: Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg (2010) 8:336–41. doi: 10.1016/j.ijsu.2010.02.007
19. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16
20. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
21. Yusuf S, Peto R, Lewis J, Collins R, Sleight P. Beta blockade during and after myocardial infarction: an overview of the randomized trials. Progr Cardiovasc Dis. (1985) 27:335–71. doi: 10.1016/S0033-0620(85)80003-7
22. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
23. Biggerstaff BJ, Tweedie RL. Incorporating variability in estimates of heterogeneity in the random effects model in meta-analysis. Stat Med. (1997) 16:753–68. doi: 10.1002/(SICI)1097-0258(19970415)16:7<753::AID-SIM494>3.0.CO;2-G
24. Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. (2001) 54:1046–55. doi: 10.1016/S0895-4356(01)00377-8
25. Ma JY, Yan HJ, Yang ZH, Gu W. Rs895819 within miR-27a might be involved in development of non small cell lung cancer in the Chinese Han population. Asian Pacific J Cancer Prevent. (2015) 16:1939–44. doi: 10.7314/APJCP.2015.16.5.1939
26. Sui ZY, Li J, Cheng GL, Wang SF. A single nucleotide polymorphism in the promoter region (rs10877887) of let-7 is associated with hepatocellular carcinoma in a Chinese population. Genet Mol Res. (2016) 15:gmr.15027661. doi: 10.4238/gmr.15027661
27. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, McClean MD, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. (2010) 16:3713–20. doi: 10.1158/1078-0432.CCR-10-0657
28. Hu Z, Shu Y, Chen Y, Chen J, Dong J, Liu Y, et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respirat Crit Care Med. (2011) 183:641–8. doi: 10.1164/rccm.201005-0717OC
29. Wang S, Lv C, Jin H, Xu M, Kang M, Chu H, et al. A common genetic variation in the promoter of miR-107 is associated with gastric adenocarcinoma susceptibility and survival. Mutat Res. (2014) 769:35–41. doi: 10.1016/j.mrfmmm.2014.07.002
30. Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Causes Control (2013) 24:1099–1109. doi: 10.1007/s10552-013-0187-z
31. Zheng C, Li X, Xia L, Fang X, Quan X, Yin Z, et al. Polymorphisms of pri-miR-219-1 are associated with the susceptibility and prognosis of non-small cell lung cancer in a Northeast Chinese population. Oncotarget (2017) 8:56533–41. doi: 10.18632/oncotarget.17035
32. Xie K, Ma H, Liang C, Wang C, Qin N, Shen W, et al. A functional variant in miR-155 regulation region contributes to lung cancer risk and survival. Oncotarget (2015) 6:42781–92. doi: 10.18632/oncotarget.5840
33. Xie K, Liu J, Zhu L, Liu Y, Pan Y, Wen J, et al. A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma. Cancer Epidemiol. (2013) 37:998–1002. doi: 10.1016/j.canep.2013.09.005
34. Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Investig. (2008) 118:2600–8. doi: 10.1172/JCI34934
35. Long XD, Huang XY, Yao JG, Liao P, Tang YJ, Ma Y, et al. Polymorphisms in the precursor microRNAs and aflatoxin B1-related hepatocellular carcinoma. Mol Carcinog. (2016) 55:1060–72. doi: 10.1002/mc.22350
36. Wu S, Shen W, Pan Y, Zhu M, Xie K, Geng L, et al. Genetic variations in key MicroRNAs are associated with the survival of nonsmall cell lung cancer. Medicine (2015) 94:e2084. doi: 10.1097/MD.0000000000002084
37. Faluyi OO, Eng L, Qiu X, Che J, Zhang Q, Cheng D, et al. Validation of microRNA pathway polymorphisms in esophageal adenocarcinoma survival. Cancer Med. (2017) 6:361–73. doi: 10.1002/cam4.989
38. Guan X, Sturgis EM, Song X, Liu Z, El-Naggar AK, Wei Q, et al. Pre-microRNA variants predict HPV16-positive tumors and survival in patients with squamous cell carcinoma of the oropharynx. Cancer Lett. (2013) 330:233–40. doi: 10.1016/j.canlet.2012.11.048
39. Wang C, Sturgis EM, Chen X, Zheng H, Wei Q, Li G. Pre-miRNA variants as predictors of clinical outcome in patients with squamous cell carcinomas of the nonoropharynx. Oncotarget (2016) 7:26444–53. doi: 10.18632/oncotarget.8512
40. Qi F, Huang M, Pan Y, Liu Y, Liu J, Wen J, et al. A genetic variant in the promoter region of miR-106b-25 cluster predict clinical outcome of HBV-related hepatocellular carcinoma in Chinese. PLoS ONE (2014) 9:e85394. doi: 10.1371/journal.pone.0085394
41. Jiao L, Zhang J, Dong Y, Duan B, Yu H, Sheng H, et al. Association between miR-125a rs12976445 and survival in breast cancer patients. Am J Transl Res. (2014) 6:869–75.
42. Lee SJ, Seo JW, Chae YS, Kim JG, Kang BW, Kim WW, et al. Genetic polymorphism of miR-196a as a prognostic biomarker for early breast cancer. Anticancer Res. (2014) 34:2943–9.
43. Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, Park JY, et al. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res. (2013) 33:3233–9.
44. Wang M, Chu H, Li P, Yuan L, Fu G, Ma L, et al. Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer Res. (2012) 72:6173–82. doi: 10.1158/0008-5472.CAN-12-0688
45. Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, Kwack K, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. (2013) 52(Suppl. 1):E39–51. doi: 10.1002/mc.21962
46. Lingzi X, Zhihua Y, Xuelian L, Yangwu R, Haibo Z, Yuxia Z, et al. Genetic variants in microRNAs predict non-small cell lung cancer prognosis in Chinese female population in a prospective cohort study. Oncotarget (2016) 7:83101–14. doi: 10.18632/oncotarget.13072
47. Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, et al. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene (2012) 504:92–7. doi: 10.1016/j.gene.2012.05.014
48. Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, et al. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis (2014) 35:1629–35. doi: 10.1093/carcin/bgu082
49. Li J, Cheng G, Wang S. A single-nucleotide polymorphism of miR-196a2T>C rs11614913 is associated with hepatocellular carcinoma in the Chinese population. Genet Test Mol Biomark. (2016) 20:213–5. doi: 10.1089/gtmb.2015.0271.
50. Yu G, Xiao Q, Ma XP, Chen X, Shi Z, Zhang LY, et al. miR-492G>C polymorphism (rs2289030) is associated with overall survival of hepatocellular carcinoma patients. Tumour Biol. (2016) 37:8961–72. doi: 10.1007/s13277-015-4752-9
51. Ma XP, Yu G, Chen X, Xiao Q, Shi Z, Zhang LY, et al. MiR-608 rs4919510 is associated with prognosis of hepatocellular carcinoma. Tumour Biol. (2016) 37:9931–42. doi: 10.1007/s13277-016-4897-1
52. Li Y, Xu Q, Liu J, He C, Yuan Q, Xing C, et al. Pri-let-7a-1 rs10739971 polymorphism is associated with gastric cancer prognosis and might affect mature let-7a expression. Onco Targets Ther. (2016) 9:3951–62. doi: 10.2147/OTT.S100481
53. Xu J, Yin Z, Shen H, Gao W, Qian Y, Pei D, et al. A genetic polymorphism in pre-miR-27a confers clinical outcome of non-small cell lung cancer in a Chinese population. PLoS ONE (2013) 8:e79135. doi: 10.1371/journal.pone.0079135
54. Mu YP, Su XL. Polymorphism in pre-miR-30c contributes to gastric cancer risk in a Chinese population. Med Oncol. (2012) 29:1723–32. doi: 10.1007/s12032-011-0115-6
55. Hong MJ, Choi YY, Jang JA, Jung HJ, Lee SY, Lee WK, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thor Oncol. (2013) 8:703–10. doi: 10.1097/JTO.0b013e318288dc0a
56. Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B. Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. (2013) 52(Suppl 1):E10–8. doi: 10.1002/mc.21931
57. Xu Q, Dong Q, He C, Liu W, Sun L, Liu J, et al. A new polymorphism biomarker rs629367 associated with increased risk and poor survival of gastric cancer in chinese by up-regulated miRNA-let-7a expression. PLoS ONE (2014) 9:e95249. doi: 10.1371/journal.pone.0095249
58. Shin KM, Jung DK, Hong MJ, Kang HJ, Lee WK, Yoo SS, et al. The pri-let-7a-2 rs1143770C>T is associated with prognosis of surgically resected non-small cell lung cancer. Gene (2016) 577:148–52. doi: 10.1016/j.gene.2015.11.036
59. Jiang L, Wang C, Sun C, Xu Y, Ding Z, Zhang X, et al. The impact of pri-miR-218 rs11134527 on the risk and prognosis of patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. (2014) 7:6206–12.
60. An J, Liu J, Liu L, Liu Y, Pan Y, Huang M, et al. A genetic variant in primary miR-378 is associated with risk and prognosis of hepatocellular carcinoma in a Chinese population. PLoS ONE (2014) 9:e93707. doi: 10.1371/journal.pone.0093707
61. Zhao Y, Wei Q, Hu L, Chen F, Hu Z, Heist RS, et al. Polymorphisms in MicroRNAs are associated with survival in non-small cell lung cancer. Cancer Epidemiol Biomark Prevent. (2014) 23:2503–11. doi: 10.1158/1055-9965.EPI-14-0389
62. Yoon KA, Yoon H, Park S, Jang HJ, Zo JI, Lee HS, et al. The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. J Thor Cardiovasc Surg. (2012) 144:794–807. doi: 10.1016/j.jtcvs.2012.06.030
63. Ryan BM, McClary AC, Valeri N, Robinson D, Paone A, Bowman ED, et al. rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer. PLoS ONE (2012) 7:e36306. doi: 10.1371/journal.pone.0036306
64. Yang PW, Huang YC, Hsieh CY, Hua KT, Huang YT, Chiang TH, et al. Association of miRNA-related genetic polymorphisms and prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. (2014) 21(Suppl. 4):S601–9. doi: 10.1245/s10434-014-3709-3
65. Xing J, Wan S, Zhou F, Qu F, Li B, Myers RE, et al. Genetic polymorphisms in pre-microRNA genes as prognostic markers of colorectal cancer. Cancer Epidemiol. (2012) 21:217–27. doi: 10.1158/1055-9965.EPI-11-0624
66. Wu Y, Jia Z, Cao D, Wang C, Wu X, You L, et al. Predictive value of MiR-219-1, MiR-938, MiR-34b/c, and MiR-218 polymorphisms for gastric cancer susceptibility and prognosis. Dis Mark. (2017) 2017:4731891. doi: 10.1155/2017/4731891
67. Chen X, Sturgis EM, Wang C, Cao X, Li Y, Wei Q, et al. Significance of microRNA-related variants in susceptibility to recurrence of oropharyngeal cancer patients after definitive radiotherapy. Oncotarget (2016) 7:35015–25. doi: 10.18632/oncotarget.9014
68. Son MS, Jang MJ, Jeon YJ, Kim WH, Kwon CI, Ko KH, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene (2013) 524:156–60. doi: 10.1016/j.gene.2013.04.042
69. Lim J, Kim JO, Park HS, Han IB, Kwack K, Kim NK, et al. Associations of miR146aC>G, miR149C>T, miR196a2C>T and miR499A>G polymorphisms with brain tumors. Oncol Rep. (2018) 40:1813–23. doi: 10.3892/or.2018.6557
70. Cao N, Li M, Han J, Wang Y, Wang X. rs61991156 in miR-379 is associated with low capability of glycolysis of gastric cancer by enhanced regulation of PKM2. Cancer Cell Int. (2018) 18:92. doi: 10.1186/s12935-018-0593-0
71. Kotlarek M, Kubiak A, Czetwertynska M, Swierniak M, Gierlikowski W, Kolanowska M, et al. The rs2910164 Genetic Variant of miR-146a-3p is associated with increased overall mortality in patients with follicular variant papillary thyroid carcinoma. Int J Mol Sci. (2018) 19:E655. doi: 10.3390/ijms19030655
72. Tvingsholm SA, Hansen MB, Clemmensen KKB, Brix DM, Rafn B, Frankel LB, et al. Let-7 microRNA controls invasion-promoting lysosomal changes via the oncogenic transcription factor myeloid zinc finger-1. Oncogenesis (2018) 7:14. doi: 10.1038/s41389-017-0014-6
73. Du M, Giridhar KV, Tian Y, Tschannen MR, Zhu J, Huang CC, et al. Plasma exosomal miRNAs-based prognosis in metastatic kidney cancer. Oncotarget (2017) 8:63703–14. doi: 10.18632/oncotarget.19476
74. Gowrishankar B, Ibragimova I, Zhou Y, Slifker MJ, Devarajan K, Al-Saleem T, et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. (2014) 15:329–41. doi: 10.4161/cbt.27314
75. Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Institute (2015) 107:dju492. doi: 10.1093/jnci/dju492
76. Huang J, Wu J, Li Y, Li X, Yang T, Yang Q, et al. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Bio Med Res Int. (2014) 2014:364316. doi: 10.1155/2014/364316
77. Mudduluru G, Vajkoczy P, Allgayer, H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol Cancer Res. (2010) 8:159–19. doi: 10.1158/1541-7786.mcr-09-0326
78. Liu J, Ni S. Association between genetic polymorphisms in the promoters of let-7 and risk of cervical squamous cell carcinoma. Gene (2018) 642:256–60. doi: 10.1016/j.gene.2017.11.038
80. Newby FD, Wilson LK, Thacker SV, DiGirolamo M. Adipocyte lactate production remains elevated during refeeding after fasting. Am J Physiol. (1990) 259(6 Pt 1):E865–71. doi: 10.1152/ajpendo.1990.259.6.E865
81. Lin M, Gu J, Eng C, Ellis LM, Hildebrandt MA, Lin J, et al. Genetic polymorphisms in MicroRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin Cancer Res. (2012) 18:3982–91. doi: 10.1158/1078-0432.CCR-11-2951
82. Zheng J, Deng J, Xiao M, Yang L, Zhang L, You Y, et al. A sequence polymorphism in miR-608 predicts recurrence after radiotherapy for nasopharyngeal carcinoma. Cancer Res. (2013) 73:5151–62. doi: 10.1158/0008-5472.CAN-13-0395
83. von Frowein J, Hauck SM, Kappler R, Pagel P, Fleischmann KK, Magg T, et al. MiR-492 regulates metastatic properties of hepatoblastoma via CD44. Liver Int. (2018) 38:1280–91. doi: 10.1111/liv.13687
84. Shen F, Cai WS, Feng Z, Li JL, Chen JW, Cao J, et al. MiR-492 contributes to cell proliferation and cell cycle of human breast cancer cells by suppressing SOX7 expression. Tumour Biol. (2015) 36:1913–21. doi: 10.1007/s13277-014-2794-z
85. Liu M, An J, Huang M, Wang L, Tu B, Song Y, et al. MicroRNA-492 overexpression involves in cell proliferation, migration, and radiotherapy response of cervical squamous cell carcinomas. Mol Carcinog. (2018) 57:32–43. doi: 10.1002/mc.22717
86. Iseki K, Kanayama T, Hayasi Y, Shibasaki M. Synthesis of a new chemically stable prostacyclin analogue with high and long-lasting activity. Chem Pharmac Bull. (1990) 38:1769–71. doi: 10.1248/cpb.38.1769
87. Lee HC, Kim JG, Chae YS, Sohn SK, Kang BW, Moon JH, et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. (2010) 136:1073–8. doi: 10.1007/s00432-009-0754-6
88. Wang KY, Ma J, Zhang FX, Yu MJ, Xue JS, Zhao JS. MicroRNA-378 inhibits cell growth and enhances L-OHP-induced apoptosis in human colorectal cancer. IUBMB Life (2014) 66:645–54. doi: 10.1002/iub.1317
89. Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, et al. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. (2010) 23:1229–34. doi: 10.1177/039463201002300427
90. Deng H, Guo Y, Song H, Xiao B, Sun W, Liu Z, et al. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene (2013) 518:351–9. doi: 10.1016/j.gene.2012.12.103
91. Grant D, Long WF, Moffat CF, Williamson FB. Infrared spectroscopy as a method for investigating the conformations of iduronate saccharide residues in glycosaminoglycans. Biochem Soc Transact. (1990) 18:1277–9. doi: 10.1042/bst0181277a
92. Samanek M, Hrobonova V, Bartakova H. Lorcainide treatment of Wolff-Parkinson-White syndrome in children and adolescents. Pediatr Cardiol. (1987) 8:3–9. doi: 10.1007/BF02308377
93. Qian J, Lin J, Qian W, Ma JC, Qian SX, Li Y, et al. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leuke Res. (2013) 37:765–8. doi: 10.1016/j.leukres.2013.03.014
94. Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. (2007) 104:20350–5. doi: 10.1073/pnas.0706901104
95. Chen X, Fan Z, McGee W, Chen M, Kong R, Wen P, et al. TDP-43 regulates cancer-associated microRNAs. Protein Cell (2017) 9:848–66. doi: 10.1007/s13238-017-0480-9
96. Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Polon. (2012) 59:467–74.
97. Zhao H, Gao A, Zhang Z, Tian R, Luo A, Li M, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. (2015) 36:4763–71. doi: 10.1007/s13277-015-3126-7
98. Sun X, Wang M, Liu H, Wang J. MicroRNA-423 enhances the invasiveness of hepatocellular carcinoma via regulation of BRMS1. Am J Transl Res. (2017) 9:5576–84.
99. Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis (2010) 31:1805–12. doi: 10.1093/carcin/bgq168
100. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature (2003) 425:415–9. doi: 10.1038/nature01957
101. Xiong XD, Cho M, Cai XP, Cheng J, Jing X, Cen JM, et al. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res. (2014) 761:15–20. doi: 10.1016/j.mrfmmm.2014.01.001
102. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. (2008) 105:7269–74. doi: 10.1073/pnas.0802682105
103. Ramkaran P, Khan S, Phulukdaree A, Moodley D, Chuturgoon AA. miR-146a polymorphism influences levels of miR-146a, IRAK-1, and TRAF-6 in young patients with coronary artery disease. Cell Biochem Biophys. (2014) 68:259–66. doi: 10.1007/s12013-013-9704-7
104. Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. (2010) 67:3535–48. doi: 10.1007/s00018-010-0394-7
105. Lin RJ, Lin YC, Yu AL. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog. (2010) 49:719–27. doi: 10.1002/mc.20647
106. Scheutz F: AIDS and the dentist: views and experiences among members of a group of patients with infection and AIDS. Tandlaegebladet (1989) 93:92–4.
107. Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. (2012) 39:7019–23. doi: 10.1007/s11033-012-1532-0
108. Ma Y, Wang R, Zhang J, Li W, Gao C, Liu J, et al. Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet Test Mol Biomark. (2014) 18:516–24. doi: 10.1089/gtmb.2013.0510
109. Qiu F, Yang L, Ling X, Yang R, Yang X, Zhang L, et al. Sequence variation in mature MicroRNA-499 confers unfavorable prognosis of lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. (2015) 21:1602–13. doi: 10.1158/1078-0432.CCR-14-1174
110. Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. (2010) 6:e1000879. doi: 10.1371/journal.pgen.1000879
Keywords: miRNAs, single nucleotide polymorphisms, cancers, systematic review, meta-analysis
Citation: Ding H, Lv Z, Yuan Y and Xu Q (2018) MiRNA Polymorphisms and Cancer Prognosis: A Systematic Review and Meta-Analysis. Front. Oncol. 8:596. doi: 10.3389/fonc.2018.00596
Received: 24 August 2018; Accepted: 26 November 2018;
Published: 13 December 2018.
Edited by:
Jianguang Ji, Lund University, SwedenCopyright © 2018 Ding, Lv, Yuan and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Xu, cXh1QGNtdS5lZHUuY24=
 Han-xi Ding
Han-xi Ding Zhi Lv
Zhi Lv Qian Xu
Qian Xu