- 1Institute for Infectious Diseases and Vaccine Development, Tongji University School of Medicine, Shanghai, China
- 2Department of Tropical Diseases, Second Military Medical University, Shanghai, China
MicroRNAs (miRNAs) play important roles in human diseases, such as cancer. Human miRNA-7-5p is a tumor suppressor miRNA that inhibits tumor growth by regulating multiple oncogenic signal pathways. Recently, studies revealed that plant miRNAs could regulate mammalian gene expression in a cross-kingdom manner. Schistosoma japonicum miRNA-7-5p (designated as sja-miR-7-5p) is conserved between the parasites and mammals. Thus, we investigated whether sja-miR-7-5p has similar antitumor activity to its mammalian counterpart. We first showed that sja-miR-7-5p was detected in host hepatocytes during S. japonicum infection. The sja-miR-7-5p mimics significantly inhibited the growth, migration, and colony formation of mouse and human hepatoma cell lines in vitro, and induced G1/G0 cell cycle arrest. In a xenograft animal model, the tumor volume and weight were significantly reduced in mice inoculated with hepatoma cells transfected with sja-miR-7-5p mimics compared with those transfected with NC miRNAs. Furthermore, the antitumor activity of sja-miR-7-5p was suggested by cross-species downregulation of the S-phase kinase-associated protein 2 gene in the host. Thus, sja-miR-7-5p is translocated into hepatocytes and exerts its anti-cancer activities in mammals, implying that sja-miR-7-5p might strengthen host resistance to hepatocellular carcinoma during schistosome infection.
Introduction
The primary pathology of schistosomiasis caused by S. japonicum is egg-induced granuloma and fibrosis. The female adult worms living in the host mesenteric veins lay numerous eggs, and most of them are trapped in the liver tissues via the portal venous system, causing a granulomatous reaction and fibrosis. The parasite eggs in the granuloma are surrounded by host cells, including immunocytes, hepatic mesenchymal cells, and hepatocytes (1). Our previous studies indicated that S. japonicum secretes many microRNAs (miRNAs), including Schistosoma-specific and conserved miRNAs (2), and parasite miRNA-containing exosomes (2).
MiRNAs are a class of highly conserved, small non-coding RNAs, with a length of about 20–24 nucleotides (nt) that post-transcriptionally regulate gene expression through complete or incomplete binding to their target mRNAs (3). MiRNAs have extensive effects on not only physiological processes, but also on the progression of many human diseases, such as cancers (4, 5). Aberrant miRNA expression promotes the occurrence and development of various cancers (6–8); however, some miRNAs can exert therapeutic effects on multiple cancers through regulation of tumor-related genes, including those that control tumor cell growth or apoptosis (9, 10). Interestingly, miRNAs derived from plants can regulate the expression of their target genes in mammals in a cross-kingdom manner (11–13). For example, miR-159 derived from plants was detectable in human sera and inhibited breast cancer growth by targeting the human transcription factor 7 (TCF7) gene (13). Accumulating evidence indicates that heterogeneous miRNAs can modulate cell functions in mammals. However, it remains unclear how the plant miRNAs can survive the passage through the gastrointestinal tract following ingestion.
Unlike plant miRNAs, which need to pass through the gastrointestinal tract before release into the host serum or entering host cells, schistosomal miRNAs from eggs trapped in liver tissue may be directly transferred to the neighboring host cells. Thus, we hypothesized that parasite miRNAs from the eggs might be translocated into neighboring hepatocytes to exert various biological effects, including some that are beneficial to the host, for example, strengthening the resistance of the host to diseases such as cancer, as do plant-derived miRNAs (13). Human miRNA-7-5p (designated as hsa-miR-7-5p) is a tumor suppressor miRNA that regulates multiple oncogenic signal pathways and reverses drug resistance in certain cancers (14–17). Our previous study identified a S. japonicum miRNA-7-5p (designated as sja-miR-7-5p) that is conserved between the parasite and mammals, i.e., there is an identical seed sequence (2–8 nt at the 5′ region) in both parasites and mammalian miRNA-7-5p, despite there being 6 nt differences in the rest of the sequence. Thus, it would be interesting to investigate if sja-miR-7-5 secreted by S. japonicum has a similar antitumor activity to hsa-miR-7-5p. In the present study, we demonstrated that sja-miR-7-5p is present in hepatocytes during the S. japonicum infection and the sja-miR-7-5p exerts anticancer effects on multiple hepatoma cells (assessed using in vitro and in vivo models) by targeting the S-phase kinase-associated protein 2(SKP2) gene, which is a component of the SCF (Skp1-Cullin 1-F-box) E3 ubiquitin-ligase complex. Previous studies have shown that overexpression of the SKP2 gene was observed in many cancers, such as in liver cancer (18), prostate cancer (19), lymphoma (20), melanoma (21), and breast cancer (22), which plays an important role in regulating cellular proliferation and cancer progression, mainly by targeting cell cycle regulators in an ubiquitin-dependent manner, followed by 26S proteasome degradation (23). In addition, the SKP2 overexpression also enhanced tumor cell invasion (24), metastasis (25), and resistance to apoptosis (26), and was associated with tumor aggressiveness (27) and poor prognosis (28).
Materials and Methods
Infection of Mice With S. japonicum Cercariae
Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and approved by the Internal Review Board of Tongji University School of Medicine. The animal surgeries were undertaken under sodium pentobarbital anesthesia. Cercariae of S. japonicum were provided by National Institute of Parasitic Disease, Chinese Center for Disease Control and Prevention (CDC). 36 six-week-old male C57BL/6J mice (18–20 g, 3 mice per group), purchased from experimental animal center of the Second Military Medical University and housed under specific pathogen-free conditions, were percutaneously infected with 50 or 100 cercariae of S. japonicum per mouse (50 for collection of infected hepatocytes and 100 for collection of early stage parasites). For collection of parasites, the hepatic schistosomula were isolated from the portal system and mesenteric veins of infected mice at 7, 14, and 42 days post-infection (dpi). In addition, 42 days male and female adult worms were manually separated under a light microscope. The eggs were isolated with a traditional method, as described by Cai et al. (29). All the freshly isolated parasites were washed three times with PBS (pH 7.4) and were immediately used for extraction of total RNA or frozen at −80°C until being subjected to further analysis.
Isolation of Primary Mouse Hepatocytes
The primary mouse hepatocytes were isolated by a two-step collagenase perfusion procedure, as described by He et al. (30) with minor modifications. Briefly, after infection, livers of the infected mice collected at various time points of 7, 9, 11, 14, 28, and 42 dpi (n = 5) along with the livers of uninfected mice were initially in situ digested with 0.03% collagenase type IV and then further digested with 0.08% collagenase type IV at 37°C in a shaking bath for 30 min. The single cell suspensions were harvested by filtration through 400-mesh sieves for removal of the remaining tissue debris and parasite eggs. Next, hepatocytes were isolated by centrifugation of the resulting cell suspensions at 50 × g for 4 min and further purified by centrifugation at 50 × g for 4 min. Purified hepatocytes were resuspend in DMEM containing 20 μg/ml Ribonuclease A (Sigma-Aldrich, USA) at 37°C for 30 min to eliminate any miRNA that might be released by schistosome eggs. After washing with PBS for three times, the cell pellet was immediately used for extraction of total RNA or frozen at −80°C until used.
Cell Proliferation Assay
Cells (2 × 105) were seeded in a 6-well plate overnight, respectively. Then cells were transfected with sja-miR-7-5p mimics or NC mimics, respectively, four replicates per group. And 24 h later, cells were digested and seeded in a 96-well plate (2 × 103) for 1, 2, 3, and 4 d. At each indicated time, 10 μL Cell Counting Kit-8(CCK-8, Dojindo, Japan) was added to each well and cells were incubated for 1 h at 37 °C, then, using the Microplate reader (Bio-Tek, USA) to measure the abosorbance at 450 nm.
Cell Cycle Analysis
Cells (1 × 105) were seeded in a 12-well plate overnight, respectively. Then, cells were transfected with sja-miR-7-5p mimics or NC mimics, respectively, three replicates per group. And 48 h later, cells were collected and fixed with ice-cold 75%(v/v) ethanol and stored at 4°C overnight, then, cells were washed and resuspended in 200 μL phosphate-buffered saline (PBS) contained with 0.05 mg/mL RNase A (Beyotime, China) and 25 mg/mL propidium iodide (PI) (Beyotime, China), cell cycle was determined by the FACSverse flow cytometer (BD Biosciences, USA).
Colony Formation Assay
Cells (2 × 105) were seeded in a 6-well plate overnight, then cells were transfected with sja-miR-7-5p mimics or NC mimics, respectively. And 24 h later, cells were digested and 200 cells in 500 μL complete medium were seeded in 24-well plate, three replicates per group. After incubation for 8 days, then cells were fixed in methanol for 30 min, followed by staining in crystal violet for 15 min. The number of colonies containing > 50 cells was counted under a light microscope.
Tumor Xenograft Animal Model
Male athymic nude mice were housed and manipulated according to the protocols approved by the Shanghai Medical Experimental Animal Care Commission. Hepa1-6 cells or HepG2 cells were transfected with sja-miR-7-5p mimics or NC mimics, respectively. And 24 h later, for each mouse, 1 × 106 cells in 100 μL PBS after treated with sja-miR-7-5p mimics were injected subcutaneously to the left scapula, while cells after treated with NC mimics were injected subcutaneously to the right scapula, respectively. Tumor volume was measured at 2, 4, 6, and 7 d after injection, At day 7, the mice were sacrificed and tumors were separated to measure their weight and volume. Tumor volume was measured using the formula: 0.5 × L × S2, where L is the longest diameter of tumor and S is the shortest diameter of tumor. The content of sja-miR-7-5p mimics transfected into the tumor cells measured by quantitative real-time reverse transcription PCR (qRT-PCR), the protein level of SKP2 was determined by Western blotting. And also the expression of Ki67 in the tumor was measured by immunohistochemistry (IHC) as described under this section.
Immunohistochemistry
To determine Ki67 expression in xenograft tumor tissues from the athymic nude mice, immunohistochemistry (IHC) was performed as described previously (31), Antibody against Ki67 was used (1:50 dilution).
Statistical Analysis
All experiments were performed in triplicate and the results were presented as mean ± standard deviation (mean ± SD). All data were analyzed by one-way ANOVA using the software GraphPad Prism 5.0(GraphPad Software, Inc. La Jolla, CA, USA). A value of P < 0.05 was considered statistically significant.
Results
Presence of sja-miR-7-5p in Infected Hepatocytes
We first investigated whether sja-miR-7-5p was present in the host liver cells during schistosome infection. For this purpose, we designed a set of two sets primers that could distinguish the sja-miR-7-5p from corresponding miRNA derived from mouse (mmu-miR-7-5p) and human (hsa-miR-7-5p). The sja-miR-7-5p has an identical seed sequence (2–8 nt at the 5' region), but there are 6 nt differences in the rest of the sequence among the species (Figure 1A, and Figure S1), which allowed us to design sets of specific primers for the mmu-miR-7a-5p (mmu-FP/RT) and the sja-miR-7-5(sja- FP/RT). We first tested specificity of the primers using the RNA samples derived from S. japonicum eggs (a), mouse Hepa1-6 cell line (c), and mixture of equal amount of a and c (b). As shown in Figures 1B,C, the sja-FP/RT pair primers successfully generated the sja-miR-7a-5p from the samples of a and b, but not c (Figure 1B), while the mmu-FP/RT pair primers generated the mmu-miR-7a-5p from the sample b and c, but not a (Figure 1C). These data indicated that the two sets of primers can effectively distinguish the sja-miR-7-5p and mmu-miR-7a-5p, and no cross reaction between the mmu-FP/RT and sja-FP/RT primers. All the primers are listed in Table S1.
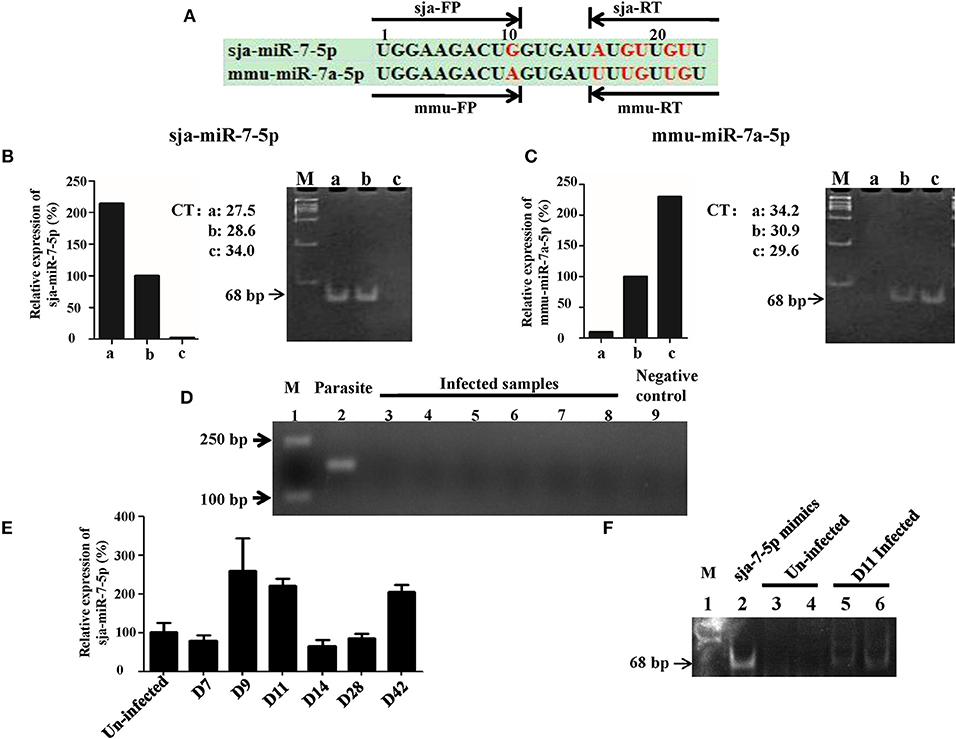
Figure 1. Detection of sja-miR-7-5p in infected hepatocytes. (A) A schematic diagram represents two sets of primers of reverse transcription stem-loop primer (RT) and forward primer (FP) for sja-miR-7-5p or mmu-miR-7a-5p, respectively. (B,C) Preparation of RNA samples: a. 200 ng Schistosoma japonicum egg RNA; b. mixture of equal amount of Schistosoma japonicum egg RNA (100 ng) and Hepa1-6 cell RNA (100 ng); c. 200 ng Hepa1-6 cell RNA. The three RNA templates were transcribed into cDNA using the corresponding reverse transcription stem-loop primer, respectively, which were used for qRT-PCR by the corresponding forward primer and common reverse primer, respectively. The PCR products were separated by polyacrylamide gel electrophoresis (PAGE). As shown in B and C, the two sets of primers can effectively distinguished the sja-miR-7-5p and mmu-miR-7a-5p, e.g., the set of sja-miR-7-5p RT and forward primer FP amplified the sja-miR-7-5p [(B), lane a and b] but not mmu-miR-7a-5p (lane c), while the set of mmu-miR-7a-5p RT and forward primer FP generated the mmu-miR-7a-5p but not the sja-miR-7-5p (C). (D) Analysis of the RNA samples to ensure no contamination with parasite RNA: the RNA samples used for the above analysis were detected as described in Method by PCR for presence of the NADH gene of S. japonicum. Lane 1: marker. Lane 2: parasite positive control: RNA samples of S. japonicum eggs as described above. Lane 3–8: six samples of infected hepatocytes with RNase pre-incubation. Lane 9: negative control without the template. (E) qRT-PCR analysis of sja-miR-7-5p in the infected hepatocytes at various days after infection; (F) 12% PAGE analysis showing sja-miR-7-5p PCR product (68 bp) from the infected hepatocytes: Lane 1: marker; Lane 2: sja-miR-7-5p mimics positive control; Lanes 3 and 4: two uninfected hepatocyte samples with pre-incubation with RNase; Lanes 5 and 6: two infected hepatocyte samples at day 11 post-infection with the pre-incubation. Data are presented as the mean ± SD, n = 3.
We next used the sja-FP/RT primers for detection of presence of sja-miR-7-5p in the liver cells of infected mice with S. japonicum. We prepared RNA samples from the infected liver cells, and carefully analyzed the samples to ensure no contamination with parasite RNA (Figure 1D). We showed that sja-miR-7-5p was detected by using qRT-PCR in the hepatocytes from infected mice at the early stage (i.e., days 9 and 11 post infection) and the late stage of infection (day 42) (Figure 1E). The presence of this parasite miRNA was further verified by PCR (Figure 1F) and cloning and sequencing of the PCR product showed identical sequence of sja-miR-7-5p (Figure S2A). In addition, we showed that sja-miR-7-5p was expressed at all these stages, and higher expression of sja-miR-7-5p was detected in adult males compared with that in adult females (Figure S2B). These findings indicated that this sja-miR-7-5p is present in the host liver cells during schistosome infection.
Inhibition of Proliferation and Migration of Hepatoma Cells by Sja-miR-7-5p
To investigated the effects of sja-miR-7-5p on the growth of hepatoma cells in vitro, both mouse and human hepatoma cells (e.g., Hepa1-6 cells and HepG2 cells) were transfected with the sja-miR-7-5p mimics, NC (a negative control mimics that has no target gene in mice and human) and Mock (transfection reagents only). As shown in Figure 2A, the sja-miR-7-5p mimics were effectively transfected into both cell lines. The schistosomal miRNA significantly suppressed the proliferation of both cell lines, as measured by the CCK-8 assay (Figure 2B), and substantially arrested the cell cycle at G1/G0 phase, as detected by flow cytometry (Figures 2C,D). We also showed that transfection of the sja-miR-7-5p mimics significantly suppressed cell migration, as assessed using the Transwell inserts without matrigel coating (Figure 2E) and by the wound-healing assay (Figures S3B,C) compared with the NC or Mock control cells. Colony formation assays showed that sja-miR-7-5p inhibited colony formation of hepatoma cells to a greater extent that those in the NC group or Mock group (Figure 2F). In addition, the Hepa1-6 cells transfected with sja-miR-7-5p mimics grew bigger and rounder compared with those in the NC or Mock control cells (Figure S3A). These data indicated that sja-miR-7-5p inhibited growth, migration, and colony formation of both mouse and human hepatoma cells and arrested their cell cycle at G1/G0 phase in vitro, indicating that the schistosomal miRNA is also a tumor suppressor.
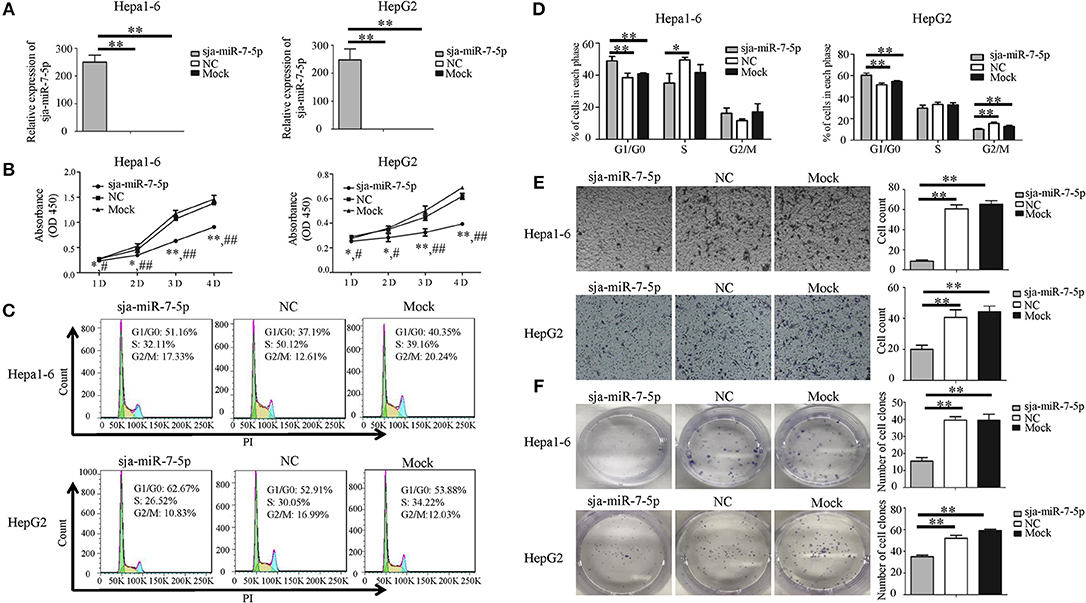
Figure 2. Sja-miR-7-5p inhibits cell proliferation and migration of Hepa1-6 and HepG2 cells in vitro. (A–F) Hepa1-6 and HepG2 cells were transfected with sja-miR-7-5p mimics and NC (negative control) mimics, respectively, and 48 h later [except for the cell counting kit-8(CCK-8) assay, which was 24 h later], the expression of sja-miR-7-5p was determined using qRT-PCR (A). Cell proliferation was evaluated by CCK-8 assay at 1, 2, 3, and 4 days (B), data are presented as the mean ± SD, n = 3, *p < 0.05 or **p < 0.01 indicates the comparison between the two groups of sja-miR-7-5p and NC; #p < 0.05 or ##p < 0.01 indicates the comparison between the groups of sja-miR-7-5p and Mock. Cell cycle was determined by flow cytometry analysis (C,D). Cell migration was evaluated using Transwell inserts without matrigel coating (E). The ability to form cell clones was determined using a colony formation assay (F). Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01.
Sja-miR-7-5p-Mediated Inhibition of Hepatoma Cell Growth in vivo
To further investigate whether sja-miR-7-5p inhibits growth of liver cancer cells in vivo, both Hepa1-6 and HepG2 cells were transfected with sja-miR-7-5p mimics or NC mimics, and then injected subcutaneously to the left and right scapula of athymic nude mice to generate subcutaneous tumors. The tumor volume was measured at days 2, 4, 6, and 7 after injection. At day 7, mice were sacrificed and tumors were excised to measure their weight and volume. The results showed that both the tumor volume and weight were significantly reduced in the mice inoculated with Hepa1-6 cells transfected with sja-miR-7-5p mimics compared with those in mice receiving cells transfected with NC miRNAs (Figures 3A–C). Similar results were obtained with the human cell line of HepG2 (Figures 3D–F). These data indicated that sja-miR-7-5p suppressed tumor growth in vivo.
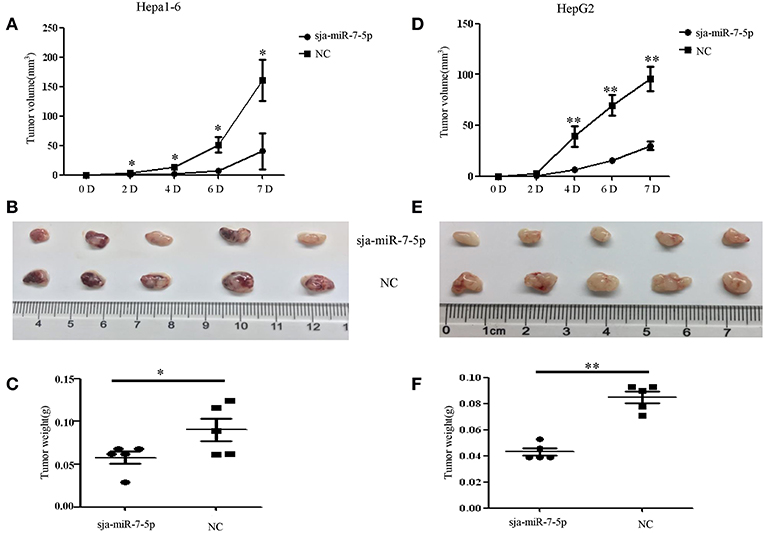
Figure 3. Sja-miR-7-5p inhibits hepatoma cell growth in vivo. (A–F) Hepa1-6 and HepG2 cells were transfected with sja-miR-7-5p mimics or NC mimics, respectively, and then the sja-miR-7-5p-treated cells (1 × 106) were injected subcutaneously to the left scapula of athymic nude mice, and the NC-treated cells were injected subcutaneously into the right scapula (n = 5), respectively. Tumor volumes were measured at days 2, 4, 6, and 7 after injection. At day 7, the mice were sacrificed and tumors were separated to measure their weight and volume, (A–C) for Hepa1-6 cells, (D–F) for HepG2 cells. Data are presented as the mean ± SD, n = 5, *p < 0.05, **p < 0.01.
SKP2 Is a Direct Target of Sja-miR-7-5p
To determine the molecular mechanisms by which sja-miR-7-5p inhibits hepatoma cell growth, we used the online software miRDB (32) (http://www.mirdb.org/miRDB/index.html), MR-microT (33) (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mrmicrot/index) and RNAhybrid (34) (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid?id=rnahybrid_view_submission).
To search for potential targets of sja-miR-7-5p. We identified the gene encoding S-phase kinase-associated protein 2(SKP2) as a potential target for sja-miR-7-5p, because a binding site was located at the 3' UTR of the both murine and human SKP2 gene that perfectly matched the seed sequence of sja-miR-7-5p. In addition, the SKP2 gene in human has been characterized as an oncogene during tumorigenesis (21, 35–38).
To investigate the relationship between sja-miR-7-5p and SKP2 gene in both human and mouse, first, we constructed two plasmids that contain the luciferase reporter gene: One was the pmirGLO-SKP2-WT construct in which the firefly luciferase gene is fused to the 3' UTR of SKP2 gene; the other was the pmirGLO-SKP2-MT in which the seven nucleotides in the miRNA binding site were mutated (Figure 4A). The constructs were simultaneously transfected with sja-miR-7-5p mimics or NC mimics into both Hepa1-6 cells and HepG2 cells. As shown in Figure 4B, the luciferase activity was significantly decreased in the cells transfected with the pmirGLO-SKP2-WT but not with the pmirGLO-SKP2-MT, indicating that sja-miR-7-5p mimics could directly bind to the site in the 3′ UTR of the SKP2 gene, while the mutations in the seed sequence abrogated the inhibitory effect.
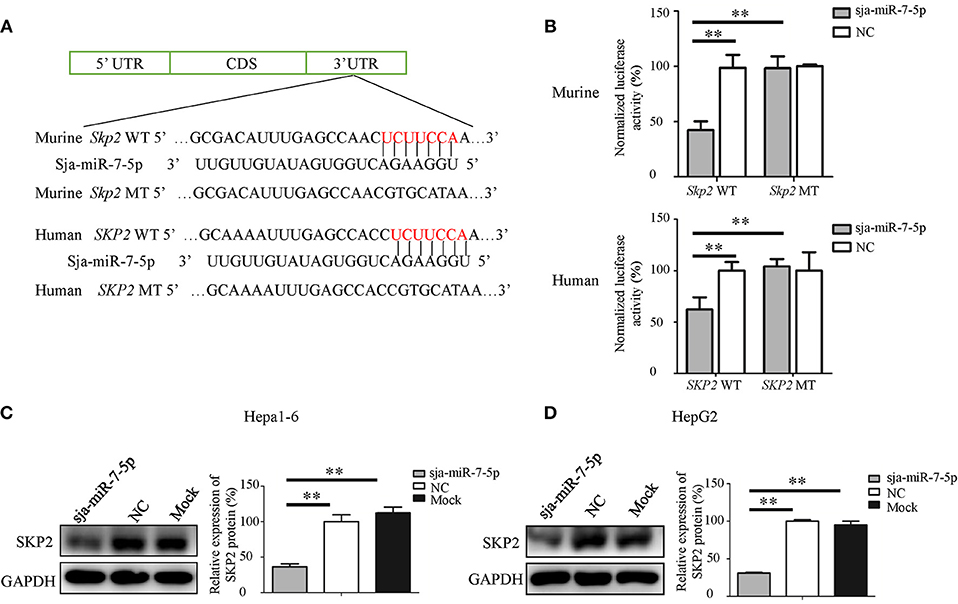
Figure 4. SKP2 (encoding S-phase kinase-associated protein 2) is a direct target of sja-miR-7-5p. (A) A schematic diagram representing the wild-type or mutant 3' untranslated region (UTR) sites of murine Skp2 and human SKP2 genes targeted by sja-miR-7-5p. (B) A dual-luciferase reporter assay was used to measure the activity of the reporter gene, and the firefly luciferase activity was normalized to renilla luciferase activity. (C,D) The protein levels of murine SKP2 (C) and human SKP2 (D) were measured using western blotting after transfection with sja-miR-7-5p mimics or NC mimics, respectively. Data are presented as the mean ± SD, n = 3, **p < 0.01.
We then detected the level of the SKP2 protein in both Hepa1-6 or HepG2 cells transfected with sja-miR-7-5p mimics using Western blotting. We found that sja-miR-7-5p downregulated the levels of SKP2 in both Hepa1-6 cells and HepG2 cells compared with that in cells transfected with NC or Mock controls (Figures 4C,D).
Sja-miR-7-5p-Mediated Suppression of the Hepatoma Cell Growth Through Downregulation of SKP2 Expression
To investigate whether sja-miR-7-5p inhibits the growth of hepatoma cells through inhibition of SKP2 expression, both Hepa1-6 cells and HepG2 cells were transfected with the SKP2 small interfering RNAs (siRNAs). We showed that both murine Skp2 siRNA (SKP2-786) and human SKP2 siRNA (SKP2-1291) significantly reduced the SKP2 expression in Hepa1-6 cells and HepG2 cells, respectively, at both transcriptional and translational levels detected by qRT-PCR and Western blotting (Figure 5A). Importantly, similar to the observations in the sja-miR-7-5p mimics-treated cells, the transfected Hepa1-6 cells and HepG2 cells with the siRNA showed cell cycle arrest at the G0/G1 phase (Figures 5C,D), and inhibition of cell proliferation (Figure 5B), cell migration (Figure 5E), and colony formation (Figure 5F), whereas these inhibitory effects were not observed in the cells treated with the NC siRNA. The phenotypes of the cells treated with SKP2 siRNA were similar to those of sja-miR-7-5p mimics-treated cells, which suggested that the inhibitory effects of the schistosome miRNA on hepatoma cells function by downregulating SKP2 expression.
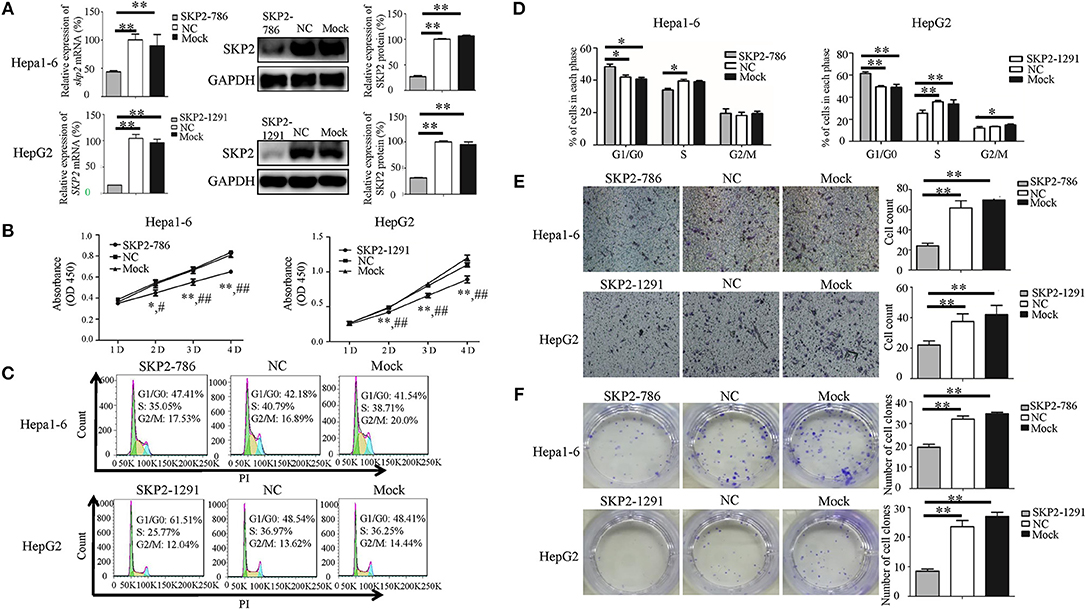
Figure 5. Knockdown of SKP2 inhibits cell proliferation and migration of Hepa1-6 and HepG2 cells in vitro. (A–F) Hepa1-6 and HepG2 cells were transfected with SKP2 siRNA and negative control (NC) siRNA, respectively, and 48 h later (except for the cell counting kit-8(CCK-8) assay, which was 24 h later), the expression of SKP2 was determined using qRT-PCR and western blotting (A). Cell proliferation was evaluated using the CCK-8 assay at 1, 2, 3, and 4 d (B), data are presented as the mean ± SD, n = 3, *p < 0.05 or **p < 0.01 indicates the comparison between the two groups of sja-miR-7-5p and NC; #p < 0.05 or ##p < 0.01 indicates the comparison between the two groups of sja-miR-7-5p and Mock. Cell cycle was determined using flow cytometry analysis (C,D). Cell migration was evaluated by using Transwell inserts without matrigel coating (E). The ability to form cell clones was determined using a colony formation assay (F). Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01.
We also detected the expression of SKP2 gene in the subcutaneous tumors generated by Hepa1-6 or HepG2 cells transfected with sja-miR-7-5p or NC mimics, respectively. As shown in Figures 6A,B, the transfected sja-miR-7-5p was detectable in the tumors on day 7 after injection. We then detected the SKP2 protein level using Western blotting, which showed that the level of SKP2 was significantly decreased in the tumors of both Hepa1-6 and HepG2 cells receiving sja-miR-7-5p compared with that in tumors from cells transfected with the NC control (Figures 6C,D). Meanwhile, we evaluated the proliferation of the tumor cells using immunohistochemistry (IHC) for Ki67, which showed that the protein level of Ki67 was also significantly decreased in tumor cells transfected with sja-miR-7-5p compared with that in cells transfected with the NC control (Figure 6E). These data further suggested that sja-miR-7-5p inhibited proliferation of both Hepa1-6 cells and HepG2 cells via downregulation of SKP2 expression.
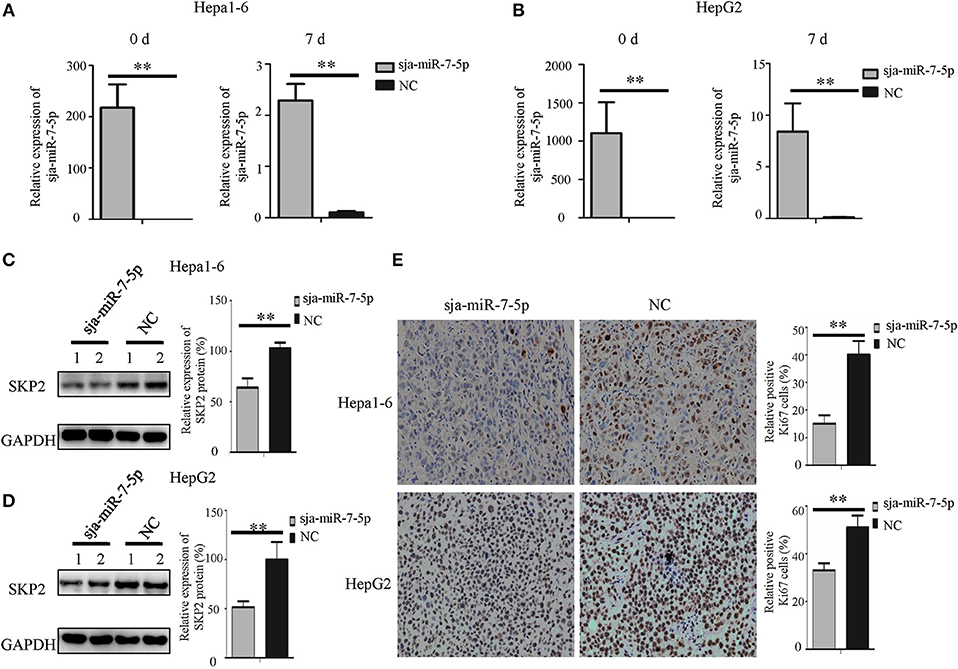
Figure 6. Sja-miR-7-5p inhibits the expression of SKP2 and Ki67 within hepatoma cell tumors. (A,B)The content of sja-miR-7-5p mimics after transfection into tumor cells was measured using qRT-PCR before inoculation (0 d) and after sacrifice (7 d), with U6 as the internal control, (A) for Hepa1-6 cells, (B) for HepG2 cells. Data are presented as the mean ± SD, n = 5, **p < 0.01. (C,D) The protein level of SKP2 was determined by Western blotting, with glyceraldehyde-3-phosphate (GAPDH) as the internal control, (C) for Hepa1-6 cells, (D) for HepG2 cells. (E) The level of Ki67 in tumors was determined using immunohistochemistry.
To further explore the molecular mechanism by which sja-miR-7-5p exerts its antitumor activities, we detected the expression of two downstream nodes of SKP2, e.g., P27 [also known as cyclin dependent kinase inhibitor 1B (CDKN1B)] and matrix metalloproteinase 9(MMP9), using Western blotting. We found that sja-miR-7-5p downregulated the level of SKP2, which led to significantly increased levels of P27 and reduced levels of MMP9 in the cells receiving sja-miR-7-5p mimics compared with those in the cells receiving the NC mimics (Figure 7A). In addition, transfection of the hepatoma cells with the murine Skp2 siRNA generated a similar outcome to that in cells transfected with sja-miR-7-5p mimics (Figure 7B). These data suggested that sja-miR-7-5p exerts its antitumor activity by targeting SKP2 to elevate P27 levels, which led to suppression of tumor cell growth, and reducing MMP9 levels, resulting in inhibition of cell migration.
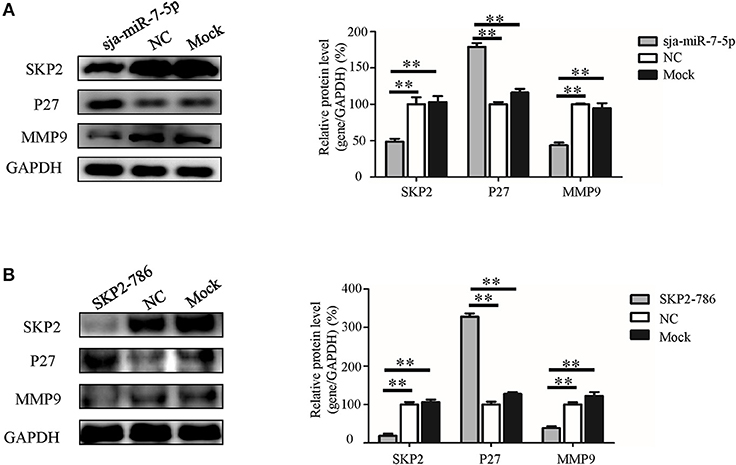
Figure 7. The molecular mechanism whereby sja-miR-7-5p exerts its antitumor activity in hepatoma cells. (A,B) In Hepa1-6 cells, the protein levels of SKP2, P27, and MMP9 were measured by Western blotting after transfection with sja-miR-7-5p mimics (A) or Skp2 siRNA (B) and their corresponding negative controls, with glyceraldehyde-3-phosphate (GAPDH) as the internal control. Data are presented as the mean ± SD, n = 3, **p < 0.01.
Discussion
Hsa-miR-7-5p is well-characterized as a tumor suppressor miRNA that suppresses survival, proliferation, invasion, and migration of multiple cancer cells, as well as increasing the sensitivity of resistant tumor cells to therapeutics. The molecular mechanism underpinning its anticancer activities involves regulation of multiple signaling related genes such as PI3K/Akt, FAK, KLF4, and REGγ (10, 39–41). This miRNA has therapeutic potential for human cancers (42). Our previous studies identified a conserved miR-7-5p from S. japonicum, sja-miR-7-5p, that has an identical seed sequence to those of hsa-miR-7-5p and mouse mmu-miR-7-5p, although there are 6 nt differences in the rest of the sequence of the miRNA among species. In this study, we have demonstrated that the schistosome miRNA, sja-miR-7-5p, is present in host hepatocytes during schistosome infection, and the in vitro transfection of sja-miR-7-5p mimics into hepatoma cells led to cell cycle arrest and inhibition of cell proliferation, colony formation, and cell migration. Furthermore, we showed that sja-miR-7-5p suppressed the growth of both human and mouse hepatoma cells in a xenograft animal model. Analysis of the molecular mechanisms revealed that sja-miR-7-5p exerts its activities by targeting the SKP2 gene, which is involved in regulation of cell viability and migration. Thus, the present data indicated that the schistosome sja-miR-7-5p is also a tumor suppressor miRNA that may have therapeutic potential for human cancers. In addition, both the presence of this miRNA in host hepatocytes and its antitumor effects on human hepatoma cells suggest that schistosome non-small RNA-mediated anticarcinogenic effects might exist in the host liver during schistosome infection.
Infection with several parasites, such as Opisthorchis viverrini and Clonorchis sinensis, has been reported to be associated with cancer (43, 44). Schistosomiasis is a neglected tropical parasitic disease, affecting approximately 210 million people worldwide. Infection with Schistosoma haematobium is associated with bladder cancer (43, 44). However, for infection with S. japonicum, the association with hepatocellular carcinoma (HCC) is less evident, although a potential association with colorectal cancer was reported (45). The large retrospective epidemiological surveys conducted in highly endemic areas for schistosomiasis in China showed no correlation between HCC and S. japonicum infection (46). Although several other epidemiological and case–control studies proposed a potential association between HCC and S. japonicum infection, the evidences for the association remain a matter of debate because the schistosomiasis patients are highly associated with HBV and HCV infections, which are hepatic carcinogens (47). However, accumulating evidence indicates that chronic inflammation plays an important role in carcinogenesis (48). For S. japonicum infection, the liver-trapped eggs induce severe hepatic chronic inflammation and fibrosis that could be risk factors for HCC (49). These factors derived from S. japonicum infection should contribute to HCC, but this does not seem to happen in S. japonicum schistosomiasis. Therefore, we speculated that the S. japonicum eggs trapped in the liver might play a dual role in the HCC occurrence and development, i.e., carcinogenic and anticancer activities, similar to those reported for the protozoan Trypanosoma cruzi, which has carcinogenic and anticancer activities during infection (50). This study demonstrated that a non-coding small RNA secreted by S. japonicum, sja-miR-7-5p, perhaps together with other miRNAs derived from the parasite, could be translocated into liver cells during parasitic infection, and exerts anticancer activity, implying that the S. japonicum-producing non-coding small RNAs may, in part, contribute to the anticancer activities in the infected host.
As described above, mammalian miR-7-5p exerts its anticancer activities through regulation of multiple target genes such as PI3K/Akt, FAK and KLF4. To identify the target gene of the parasite sja-miR-7-5p, we first used three online software to search for its potential target genes. We found 5 target gene candidates (Skp2, Psme3, Pik3cd, Klf4, and Hoxb5) that were consistently predicted by the three software and involved in tumor-related signaling pathway. Three of them (i.e., Pik3cd, Klf4, Hoxb5) were excluded through analysis of their expression in hepatoma cells transfected with the sja-miR-7-5p mimics. Although both Skp2 and Psme3 genes were validated as target gene by luciferase reporter assay, our experimental data with Skp2 and Psme3 siRNA showed that only the hepatoma cell transfected with the Skp2 siRNA produced similar phenotype to that of sja-miR-7-5p mimics-treated cells. Thus, Skp2 gene has been identified as the target gene of sja-miR-7-5p.
SKP2, also known as P45, FBL1, FLB1, and FBXL1, is a component of the SCF (Skp1-Cullin 1-F-box) E3 ubiquitin-ligase complex. Many studies have reported that SKP2 is overexpressed in various cancers of different organs, including the liver (18), colon (51), breast (52), prostate (53), and stomach (54). SKP2 is characterized as an oncogene, and is involved in modulation of the cell cycle, cell growth, and survival by regulation of its downstream node molecules, such as P27, P16, P21, P57, E2F-1, and c-MYC in an ubiquitin-dependent manner, followed by 26S proteasome degradation (23). Previous studies showed that loss of SKP2 reduced the migration and invasion abilities of oral squamous cell carcinoma cells by downregulating the expression of MMP2 and MMP9 (55). The best-known substrate of SKP2 is the cyclin dependent kinase (CDK) inhibitor, P27. Overexpression of SKP2 leads to reduction of P27, which is strongly associated with aggressive tumor behavior and poor clinical outcome (19, 36, 56), while knockdown of SKP2 resulted in the accumulation of P27, causing cell cycle arrest at G1/G0 phase (57). However, the relationship between miR-7-5p and SKP2 has not yet been reported in HCC. In the present study, we found that in liver cancer cells, including Hepa1-6 cells and HepG2 cells, sja-miR-7-5p inhibited the growth and migration of both mouse and human hepatoma cells by targeting SKP2 to elevate the expression of P27 and decrease the expression of MMP9. These data were consistent with the results of experiments using the SKP2 siRNA, and with the outcome of a study in which miRNA-7-5p could suppress cell proliferation of CHO cells partly by targeting skp2 (58). Therefore, our data demonstrated that sja-miR-7-5p suppresses hepatoma cell growth and migration by downregulating SKP2.
The present study demonstrated that sja-miR-7-5p is present in infected hepatocytes, selectively affects the growth and migration of human and mouse tumor cells by targeting the SKP2 gene, implying that sja-miR-7-5p might strengthen resistance of host to cancer during schistosome infection.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
CH and WP conceived and designed the study. CH, SZ, JW, YL, LM, LZ, PJ, and ZL performed the experiments. CH, SZ, and WP analyzed the data. CH and WP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81430051).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the National Institute of Parasitic Disease, Chinese Center for Disease Control and Prevention for their help with parasite infections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00175/full#supplementary-material
Abbreviations
S. japonicum, Schistosoma japonicum; HCC, hepatocellular cancer; SKP2, S-phase kinase associated protein 2; P27(also known as CDKN1B), cyclin dependent kinase inhibitor 1B; MMP9, matrix metallopeptidase 9; miRNA, microRNA; siRNA, small interfering RNA; NC, negative control; Mock, mock control.
References
1. Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. (2014) 383:2253–64. doi: 10.1016/S0140-6736(13)61949-2
2. Zhu SL, Wang S, Lin Y, Jiang PY, Cui XB, Wang XY, et al. Release of extracellular vesicles containing small RNAs from the eggs of Schistosoma japonicum. Parasit Vectors. (2016) 9:574. doi: 10.1186/s13071-016-1845-2
3. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
4. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. (2006) 6:857–66. doi: 10.1038/nrc1997
5. Berindan-Neagoe I, Monroig PD, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. Ca-a Cancer J Clini. (2014) 64:311–36. doi: 10.3322/caac.21244
6. Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. (2017) 389:11–22. doi: 10.1016/j.canlet.2016.12.033
7. Xue X, Fei X, Hou W, Zhang Y, Liu L, Hu R. miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancer. Cancer Lett. (2018) 412:170–8. doi: 10.1016/j.canlet.2017.10.024
8. Zhu Y, Gu J, Li Y, Peng C, Shi M, Wang X, et al. MiR-17-5p enhances pancreatic cancer proliferation by altering cell cycle profiles via disruption of RBL2/E2F4-repressing complexes. Cancer Lett. (2018) 412:59–68. doi: 10.1016/j.canlet.2017.09.044
9. Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4 alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. (2011) 147:1233–47. doi: 10.1016/j.cell.2011.10.043
10. Fang YX, Xue JL, Shen Q, Chen JZ, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. (2012) 55:1852–62. doi: 10.1002/hep.25576
11. Zhang L, Hou DX, Chen X, Li DH, Zhu LY, Zhang YJ, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. (2012) 22:107–26. doi: 10.1038/cr.2011.158
12. Zhou Z, Li XH, Liu JX, Dong L, Chen Q, Liu JL, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. (2015) 25:39–49. doi: 10.1038/cr.2014.130
13. Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu XW, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. (2016) 26:217–28. doi: 10.1038/cr.2016.13
14. Liu HX, Huang J, Peng J, Wu XX, Zhang Y, Zhu WL, et al. Upregulation of the inwardly rectifying potassium channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell lung cancer under the regulation of miR-7 and the Ras/MAPK pathway. Mol Cancer. (2015) 14:59. doi: 10.1186/s12943-015-0298-0
15. Liu HX, Wu XX, Huang J, Peng J, Guo LL. miR-7 modulates chemoresistance of small cell lung cancer by repressing MRP1/ABCC1. Int J Exp Pathol. (2015) 96:240–7. doi: 10.1111/iep.12131
16. Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. (2015) 36:338–45. doi: 10.1093/carcin/bgu242
17. Cheng MW, Shen ZT, Hu GY, Luo LG. Prognostic significance of microRNA-7 and its roles in the regulation of cisplatin resistance in lung adenocarcinoma. Cell Physiol Biochem. (2017) 42:660–72. doi: 10.1159/000477884
18. Lee SW, Li CF, Jin G, Cai Z, Han F, Chan CH, et al. Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress. Mol Cell. (2015) 57:1022–33. doi: 10.1016/j.molcel.2015.01.015
19. Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: a novel potential therapeutic target for prostate cancer. Biochim Biophys Acta. (2012) 1825:11–7. doi: 10.1016/j.bbcan.2011.09.002
20. Seki R, Okamura T, Koga H, Yakushiji K, Hashiguchi M, Yoshimoto K, et al. Prognostic significance of the F-box protein Skp2 expression in diffuse large B-cell lymphoma. Am J Hematol. (2003) 73:230–5. doi: 10.1002/ajh.10379
21. Rose AE, Wang G, Hanniford D, Monni S, Tu T, Shapiro RL, et al. Clinical relevance of SKP2 alterations in metastatic melanoma. Pigment Cell Melanoma Res. (2011) 24:197–206. doi: 10.1111/j.1755-148X.2010.00784.x
22. Radke S, Pirkmaier A, Germain D. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene. (2005) 24:3448–58. doi: 10.1038/sj.onc.1208328
23. Bochis OV, Irimie A, Pichler M, Berindan-Neagoe I. The Role of Skp2 and its Substrate CDKN1B (p27) in colorectal cancer. J Gastrointes Liver Dis. (2015) 24:225–34. doi: 10.15403/jgld.2014.1121.242.skp2
24. Hung WC, Tseng WL, Shiea J, Chang HC. Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett. (2010) 288:156–61. doi: 10.1016/j.canlet.2009.06.032
25. Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am J Pathol. (2004) 165:175–80. doi: 10.1016/S0002-9440(10)63286-5
26. Shi C, Pan BQ, Shi F, Xie ZH, Jiang YY, Shang L, et al. Sequestosome 1 protects esophageal squamous carcinoma cells from apoptosis via stabilizing SKP2 under serum starvation condition. Oncogene. (2018) 37:3260–74. doi: 10.1038/s41388-018-0217-0
27. Osoegawa A, Yoshino I, Tanaka S, Sugio K, Kameyama T, Yamaguchi M, et al. Regulation of p27 by S-phase kinase-associated protein 2 is associated with aggressiveness in non-small-cell lung cancer. J Clin Oncol. (2004) 22:4165–73. doi: 10.1200/JCO.2004.01.035
28. Zhu CQ, Blackhall FH, Pintilie M, Iyengar P, Liu N, Ho J, et al. Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker. Clin Cancer Res. (2004) 10:1984–91. doi: 10.1158/1078-0432.CCR-03-0470
29. Cai PF, Piao XY, Hao LL, Liu S, Hou N, Wang H, et al. A deep analysis of the small non-coding RNA population in Schistosoma japonicum eggs. PLoS ONE. (2013) 8:64003. doi: 10.1371/journal.pone.0064003
30. He X, Xie J, Zhang DM, Su Q, Sai X, Bai RP, et al. Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology. (2015) 61:2008–17. doi: 10.1002/hep.27671
31. Yin XD, Xiang TX, Li LL, Su XW, Shu XS, Luo XR, et al. DACT1, an antagonist to Wnt/beta-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. (2013) 15:R23. doi: 10.1186/bcr3399
32. Wong N, Wang XW. miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acids Res. (2015) 43:D146–52. doi: 10.1093/nar/gku1104
33. Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. (2012) 28:771–6. doi: 10.1093/bioinformatics/bts043
34. Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl Acids Res. (2006) 34:W451–4. doi: 10.1093/nar/gkl243
35. Lim MS, Adamson A, Lin ZS, Perez-Ordonez B, Jordan RCK, Tripp S, et al. Expression of Skp2, a p27(Kip1) ubliquitin ligase, in malignant lymphoma: correlation with p27(Kip1) and proliferation index. Blood. (2002) 100:2950–6. doi: 10.1182/blood.V100.8.2950
36. Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. (2005) 103:1336–46. doi: 10.1002/cncr.20917
37. Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. (2008) 112:1415–24. doi: 10.1002/cncr.23317
38. Schuler S, Diersch S, Hamacher R, Schmid RM, Saur D, Schneider G. SKP2 confers resistance of pancreatic cancer cells towards TRAIL-induced apoptosis. Int J Oncol. (2011) 38:219–25. doi: 10.3892/ijo_00000841
39. Kong XJ, Li GP, Yuan Y, He Y, Wu XL, Zhang WJ, et al. MicroRNA-7 Inhibits Epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE. (2012) 7:e41523. doi: 10.1371/journal.pone.0041523
40. Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. (2013) 73:1434–44. doi: 10.1158/0008-5472.CAN-12-2037
41. Shi Y, Luo XR, Li P, Tan JX, Wang XY, Xiang TXS, et al. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REG gamma. Cancer Lett. (2015) 358:27–36. doi: 10.1016/j.canlet.2014.12.014
42. Kalinowski FC, Brown RAM, Ganda C, Giles KM, Epis MR, Horsham J, et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. (2014) 54:312–7. doi: 10.1016/j.biocel.2014.05.040
43. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. (2009) 10:321–2. doi: 10.1016/S1470-2045(09)70096-8
44. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. (2012) 100:1–441.
45. Chen MG. Assessment of morbidity due to Schistosoma japonicum infection in China. Infect Dis Poverty. (2014) 3:6. doi: 10.1186/2049-9957-3-6
46. Liu BQ, Rong ZP, Sun XT, Wu YP, Gao RQ. Geographical correlation between colorectal cancer and schistosomiasis in China. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. (1983) 5:173–7.
47. Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. (2013) 13:123–35. doi: 10.1038/nrc3449
48. Zhang R, Takahashi S, Orita S, Yoshida A, Maruyama H, Shirai T, et al. p53 gene mutations in rectal cancer associated with schistosomiasis japonica in Chinese patients. Cancer Lett. (1998) 131:215–21. doi: 10.1016/S0304-3835(98)00154-2
49. Takemura Y, Kikuchi S, Inaba Y. Epidemiologic study of the relationship between schistosomiasis due to Schistosoma japonicum and liver cancer/cirrhosis. Am Journal Tropi Med Hygiene. (1998) 59:551–6. doi: 10.4269/ajtmh.1998.59.551
50. van Tong H, Brindley PJ, Meyer CG, Velavan TP. Parasite infection, carcinogenesis and human malignancy. EBioMedicine. (2017) 15:12–23. doi: 10.1016/j.ebiom.2016.11.034
51. Fujita T, Liu WJ, Doihara H, Wan Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am J Pathol. (2008) 173:217–28. doi: 10.2353/ajpath.2008.070957
52. Liu J, Wei XL, Huang WH, Chen CF, Bai JW, Zhang GJ. Cytoplasmic Skp2 expression is associated with p-Akt1 and predicts poor prognosis in human breast carcinomas. PLoS ONE. (2012) 7:e52675. doi: 10.1371/journal.pone.0052675
53. Moro L, Arbini AA, Marra E, Greco M. Up-regulation of Skp2 after prostate cancer cell adhesion to basement membranes results in BRCA2 degradation and cell proliferation. J Biol Chem. (2006) 281:22100–7. doi: 10.1074/jbc.M604636200
54. Ma XM, Liu Y, Guo JW, Liu JH, Zuo LF. Relation of overexpression of S phase kinase-associated protein 2 with reduced expression of p27 and PTEN in human gastric carcinoma. World J Gastroenterol. (2005) 11:6716–21. doi: 10.3748/wjg.v11.i42.6716
55. Yamada SI, Yanamoto S, Naruse T, Matsushita Y, Takahashi H, Umeda M, et al. Skp2 Regulates the Expression of MMP-2 and MMP-9, and enhances the invasion potential of oral squamous cell carcinoma. Pathol Oncol Res. (2016) 22:625–32. doi: 10.1007/s12253-016-0049-6
56. Wang Z, Fukushima H, Inuzuka H, Wan L, Liu P, Gao D, et al. Skp2 is a promising therapeutic target in breast cancer. Front Oncol. (2012) 1:18702. doi: 10.3389/fonc.2011.00057
57. Ding L, Li R, Han X, Zhou Y, Zhang H, Cui Y, et al. Inhibition of Skp2 suppresses the proliferation and invasion of osteosarcoma cells. Oncol Rep. (2017) 38:933–40. doi: 10.3892/or.2017.5713
Keywords: Schistosoma japonicum, microRNA, hepatoma cell, SKP2, cross-species regulation
Citation: Hu C, Zhu S, Wang J, Lin Y, Ma L, Zhu L, Jiang P, Li Z and Pan W (2019) Schistosoma japonicum MiRNA-7-5p Inhibits the Growth and Migration of Hepatoma Cells via Cross-Species Regulation of S-Phase Kinase-Associated Protein 2. Front. Oncol. 9:175. doi: 10.3389/fonc.2019.00175
Received: 24 October 2018; Accepted: 28 February 2019;
Published: 22 March 2019.
Edited by:
Yunkai Zhang, Vanderbilt University Medical Center, United StatesReviewed by:
Yang Li, University of Arizona, United StatesHui Wang, Vanderbilt University, United States
Copyright © 2019 Hu, Zhu, Wang, Lin, Ma, Zhu, Jiang, Li and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiqing Pan, d3FwYW4wOTEyQGFsaXl1bi5jb20=
 Chao Hu1
Chao Hu1 Li Ma
Li Ma Weiqing Pan
Weiqing Pan