- 1Department of Immunology, School of Basic Medical Science, Wuhan University, Wuhan, China
- 2Institute of Pathology, Hannover Medical School, Hanover, Germany
- 3Department of Hematology, Zhongnan Hospital of Wuhan University, Wuhan, China
- 4Hubei Provincial Key Laboratory of Developmentally Originated Disease, Wuhan University, Wuhan, China
Acute myeloid leukemia (AML) is a myeloid malignancy characterized by the proliferation of abnormal and immature myeloid blasts in the bone marrow. Circular RNA (circRNA) is a novel class of long non-coding RNA with a stable circular conformation that regulates various biological processes. The aberrant expression of circRNA and its impact on AML progression has been reported by a number of studies. Despite recent advances in circRNA research, our understanding of the leukemogenic mechanism of circRNA remains very limited, and translating the current circRNA-related research into clinical practice is challenging. This review provides an update on the functional roles of and research progress on circRNAs in AML with an emphasis on mechanistic insights. The challenges and opportunities associated with circRNA-based diagonostic and therapeutic development in AML are also outlined.
Introduction
Acute myeloid leukemia (AML) is the most common leukemia in adults, presenting great biological and clinical heterogeneity (1). In 2018, there were 19,520 new cases of AML in the United States (2), and, based on survey data, there were 14,100 reported cases in China in 2015 (3). Various genetic and epigenetic aberrations arrest hematopoietic cell differentiation and maturation events, leading to the accumulation of abnormal and immature hematopoietic progenitor cells in the bone marrow and peripheral blood. These abnormalities are often associated with lethal infection, organ infiltration, and cytopenias (4, 5).
Investigations on the genetic and molecular mechanisms of cancer have recently shifted from protein-coding genes to non-coding transcripts (6). About 98% of the human genome represents non-coding DNA sequences, and the majority of the human transcriptome is classified as non-coding RNAs (ncRNAs) (7). Circular RNA (circRNA) belongs to the family of endogenous ncRNA and has a non-polyadenylated closed single-stranded and continuous loop structure due to a covalent phosphodiester bond between the 3′ and 5′ ends (8). The size of circRNA ranges from a few hundred to thousands of nucleotides (9). Global circRNA expression profiling has revealed that they are abundant transcripts of well-regulated back-spliced RNA and that their biogenesis is regulated by cis-elements and/or trans-factors. The back-spliced RNA is produced as a result of back-splicing that ligates a splice donor site with an acceptor site present upstream and downstream, respectively, on the mRNA transcript, resulting in a covalently closed circRNA as well as an alternatively spliced linear RNA with skipped exon(s) (1). The circular and non-polyadenylated structure makes the circular RNA more stable. circRNAs exhibit evolutionary conservation across the eukaryotes (10), and circRNA expression is tissue- and developmental stage-specific (11–13). Studies have shown that circRNAs regulate gene expression (14, 15) and are involved in regulating vital cellular events such as differentiation, proliferation, growth, signaling, and aging (11–13, 16, 17). Recently, aberrant expression of circRNAs has been implicated in the progression and pathogenesis of hematopoietic malignancies (18–20) and solid tumors (21–23). Numerous circRNAs with altered expression have been reported to be involved in leukemogenesis (24–26). miRNAs, short stretches of RNA (23 nt), are associated with various biological processes (2). circRNAs are also implicated in tumorigenesis, metastasis, and drug resistance (22). The most established mechanism of action of circRNAs is its “sponge” function via binding to miRNAs (14, 27), proteins (21, 28, 29), or DNA (10, 30). The most documented role of circRNAs in AML is modulating mRNA stability and translation by sequestering the mRNA transcript and protein (31, 32), although circRNA-RNA binding protein (RBP)/DNA interactions remain obscure and need further exploration.
This review compiles the roles of recently reported circRNAs that show altered expression in AML. Moreover, we also present the potential molecular mechanisms of certain circRNAs. Finally, we discuss the main challenges related to research on circRNA.
Biogenesis and Functions of circRNA
The unique characteristic of circRNAs is associated with their production, which mostly occurs via the canonical and non-canonical splicing (3) of exons (4) and introns, non-coding antisense, 3′ and 5′ UTR or the intergenic region (4, 5). Non-canonical splicing is an unconventional mechanism of splicing characterized by the ligation of splice sites that are present at a distance from the currently annotated exons. The process aids in gene expression regulation and is important in evolution since it acts as a source of newly emerging transcripts (6).
Based on their origin of production and their determination by RNA-sequencing (RNA-seq), the circRNAs are classified into four types: exonic circRNA (ecircRNA) (4, 7), circular intronic RNA (ciRNA) (5), retained-intron or exon-intron circRNA (7, 8), and intergenic circRNA (4). Two models of circularization, illustrated in Figure 1, explain circRNA biogenesis via lariat-driven circularization (exon skipping) (Figure 1B) (9) or intron pairing-driven circularization (Figure 1C) (9). circRNA biogenesis is regulated by both cis- and trans-acting factors (33), with the latter also known as RNA binding proteins (RBP) (Figure 1D) (10). Some circRNAs, in which the introns are not trimmed and are retained in exons, are known as exon-intron circRNAs or EIciRNAs (Figure 1D) (8). Unique splicing mechanisms may also result in the generation of novel circRNAs known as ciRNA (intronic circRNA) (5) (Figure 1E) and tRNA intronic circular RNAs (tricRNAs) (Figure 1F) (11).
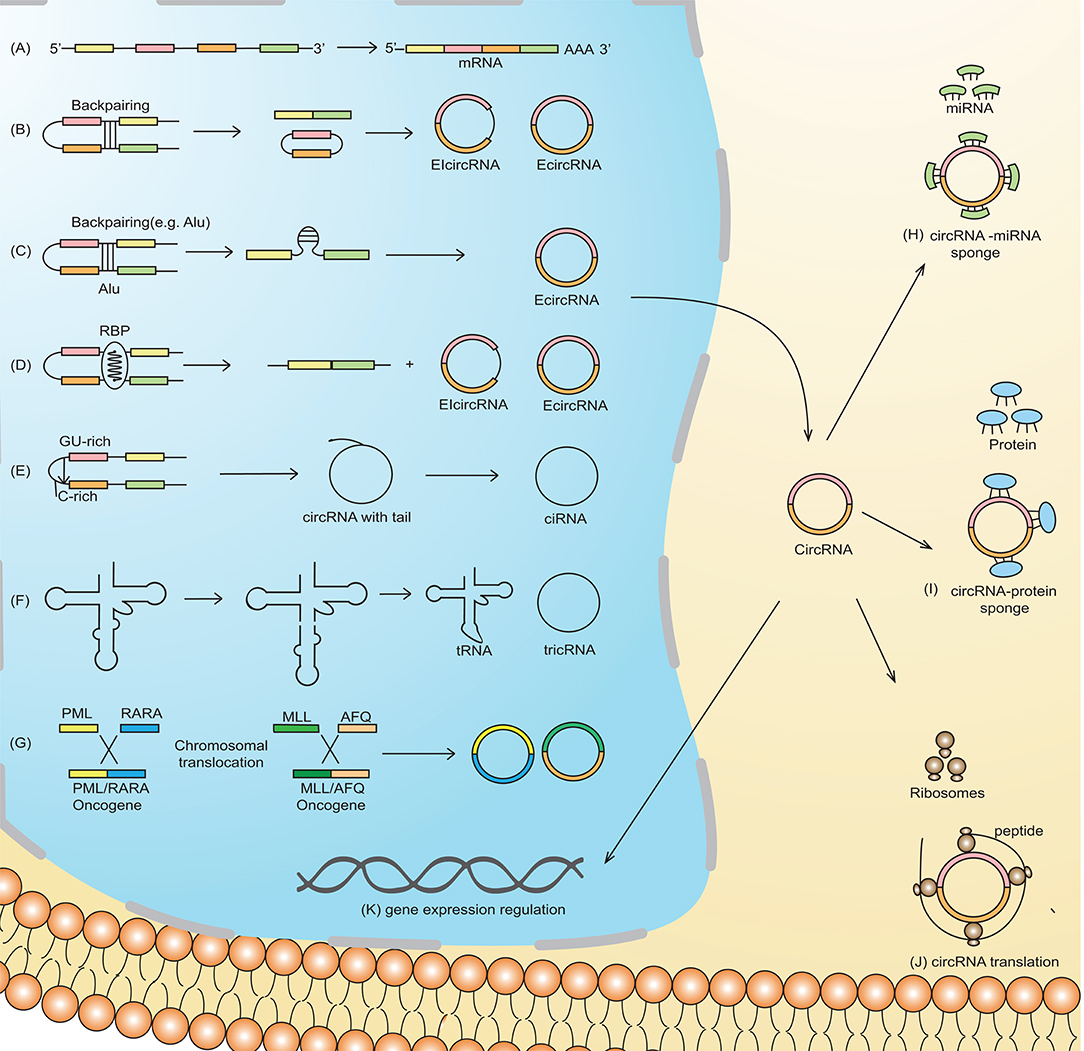
Figure 1. Schematic representation of circRNA biogenesis. (A) Canonical pre-mRNA producing a mature mRNA. (B) Lariat-driven circularization, also called exon-skipping, is a process in which the partial folding of pre-mRNA allows the covalent binding of the exonic 5′-splice donor site with the 3′-end acceptor site, thus resulting in the skipping of exons. (C) Intron-pairing-driven circularization is characterized by base-pairing between different repetitive elements such as Alu repeats across different introns. The intronic sequence is subsequently cleaved to produce exonic circRNA (ecircRNA). (D) RNA-binding proteins (RBP) working as trans-acting factors vitally regulate the production of circRNA. Quaking (QKI) and Muscleblind (MBL/MBNL1) proteins preferentially bind to a particular sequence motif on a linear RNA, thus bridging the two bracketing introns together and circularizing the linear RNA to generate circRNA. (E) A novel circRNA known as ciRNA is dependent on a 7-nt GU-rich motif and an 11-nt C-rich sequence near the 5′-splice site and branchpoint site. The lariat structure containing the excised intron and exons is trimmed by the spliceosome followed by the degradation of the 3′-end tail, ultimately generating ciRNA. (F) tRNA intronic circular RNAs (tricRNAs) are produced as a product of the splicing of tRNA independent of pre-mRNAs, which is catalyzed by an enzyme in a presence of conserved tRNA sequence motif. (G) Chromosomal translocation results in the fusion of two genes producing fusion circRNA (f-circRNA). Functions of circRNA: (H) circRNAs may act as miRNA sponges by competing for miRNA binding sites, diminishing the effect of miRNA-mediated regulatory activities. (I) cricRNA may acts as protein (RNA binding protein), which regulate gene expression and are thus involved in various biological processes (J) circRNAs can be translated to form functional proteins, e.g., ribosome-associated circRNAs (ribo-circRNAs) harbor start codon and evolutionarily conserved stop codon encoding at least in protein domain circRNAs. (K) circRNAs (e.g., EIciRNAs and ciRNAs) may interact with transcription complexes and enhance the expression of their parental genes. Moreover, some of the nuclear enriched circRNAs may bind with the genomic DNA, forming an RNA-DNA triplex that can regulate the DNA replication process.
The most established function of circRNAs is through sponge formation with miRNAs and proteins (Figure 1H). In the case of circRNA-miRNA sponge formation, the sequestration of miRNA by the circRNA allows the binding of translational machinery to the specific mRNA, thus causing gene derepression. Increased expression of genes that, for example, are associated with migration, differentiation, and proliferation, may contribute to leukemogenesis (10). Direct RBP sponging (circRNA-RBP) (Figure 1I) or indirect circRNA-miRNA-RBP interactions by circRNAs can also induce leukemogenesis, because RBPs are also implicated in cell cycle progression and the biogenesis of circRNA, ultimately thereby affecting cell differentiation, proliferation, and apoptosis (12).
Although circRNAs contain an open reading frame (ORF), they generally lack the essential components necessary for translation, such as a poly(A) tail and a 7-methylguanosine cap. Nevertheless, mounting evidence suggests the translation ability of circRNAs (Figure 1J) (13). For example, an RNA modification motif, N6-methyladenosine (m6A), enriched in circRNAs, facilitates the translation of circRNA in human cells (14). Overall, the translational mechanisms of circRNAs are not well-established and need further investigation.
circRNAs in AML
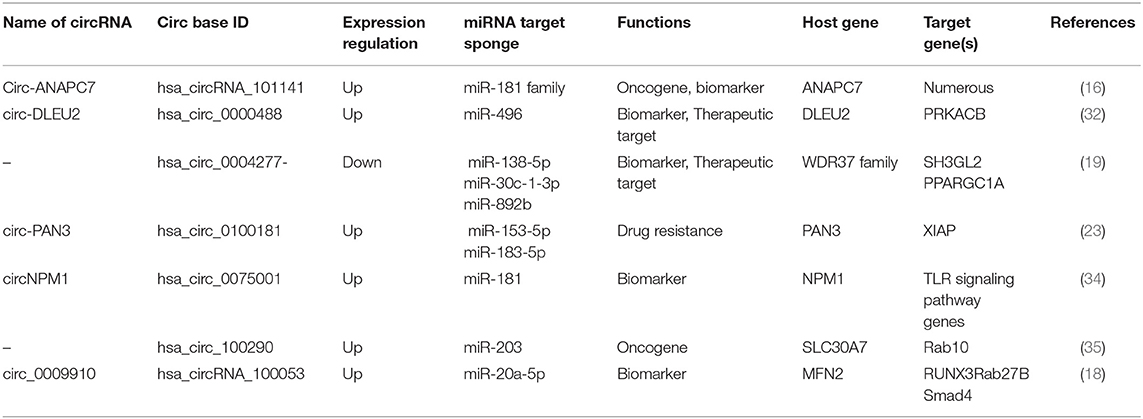
Circular RNAs as Potential Diagnostic and Prognostic Biomarkers (Table 1)
Various molecular-based biomarkers (e.g., cytogenetics, genetics, epigenetics, proteomics, and miRNA) have been reported in AML and have been comprehensively reviewed (15). circRNAs are emerging as potential biomarkers in the diagnosis and treatment of AML (Figure 2A) due to their stability against exonucleolytic degradation, tissue-specific expression patterns, and abundance in body fluids and exosomes. For example, microarray-based expression analysis of five bone marrow (BM) samples from AML patients revealed high levels of circ-ANAPC7 (16). Yi et al. examined the expression of circ-VIM (circRNA derived from Vitementin) in bone marrow mononuclear cells (BMNCs) derived from the bone marrow of 113 AML patients and 42 healthy donors and observed upregulation of circ-VIM in AML patients (17). Moreover, they found that overexpression of circ-VIM was an independent poor prognostic factor and was markedly associated with shorter overall survival in AML patients. Ping et al. recently profiled the expression of circRNAs in the bone marrow of three AML patients and three controls (iron deficiency anemia) via circRNA-microarray analysis and documented elevated expression of circ_0009910 (hsa_circRNA_100053, circBase). circ_009910-miR-20a-5p upregulation was found in AML bone marrow patients compared to the control, and this upregulation predicted a poor outcome for AML patients. Further, the silencing of this circRNA inhibited the proliferation of AML cells and initiated apoptosis, highlighting the diagnostic and therapeutic potential of circ_009910-miR-20a-5p AML (18). Hsa_circ_0004277 (present on chromosome 10: 1125950-1126416) is encoded by WDR37, and the WDR37 family is involved in cellular events such as the cell cycle, apoptosis, and signaling pathways (3). Hsa_circ_0004277 was found to have reduced expression in both newly diagnosed untreated and relapsed AML patients (19). Interestingly, expression of hsa_circ_0004277 and WDR37 was found to be positively correlated during different AML stages, implicating WDR37 as a positive regulator of hsa_circ_0004277. Taken together, these reports suggest that circRNAs might be used as potential diagnostic and prognostic markers in AML.
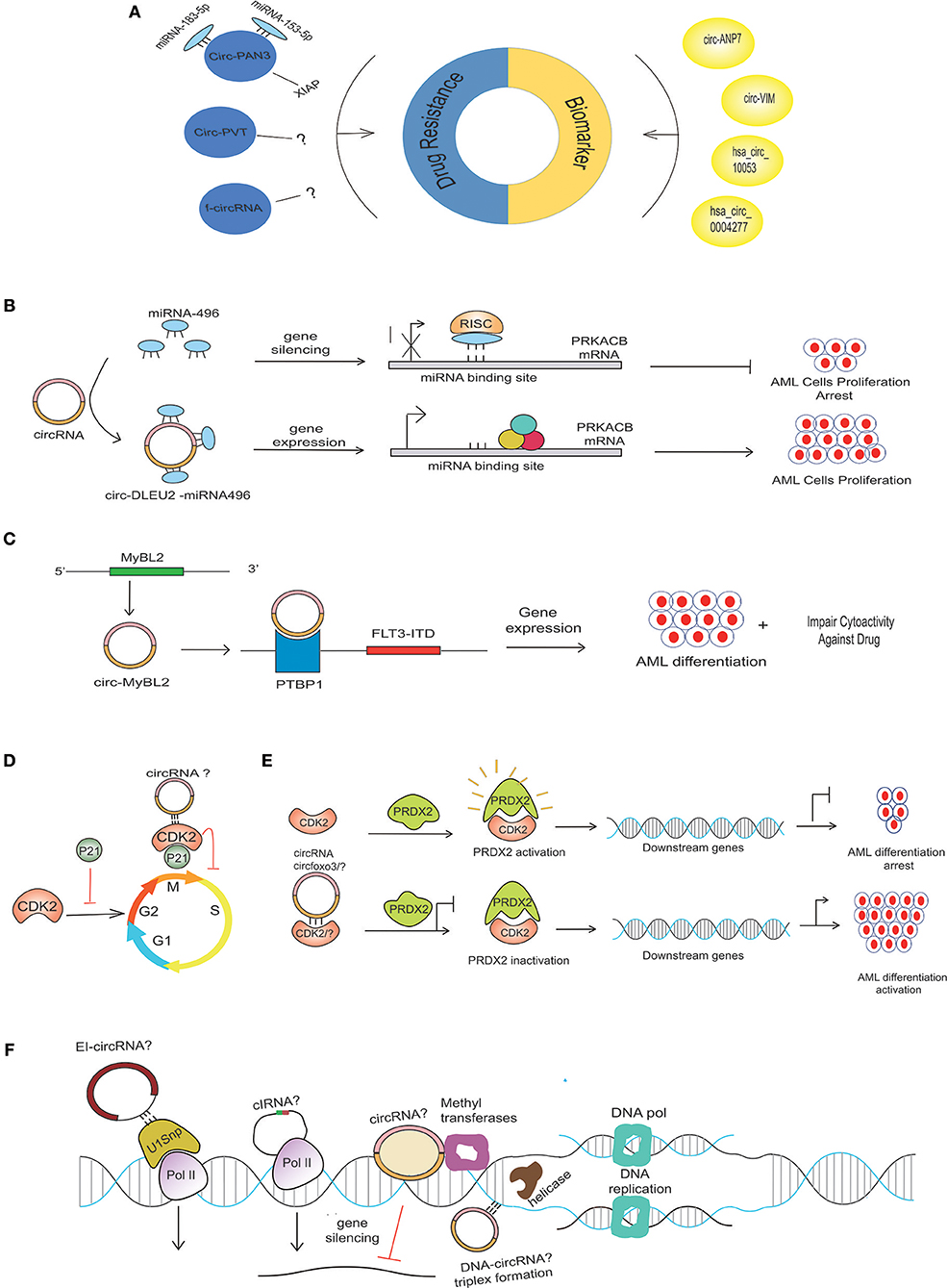
Figure 2. Possible mechanism of circRNA in AML pathogenesis. (A) Aberrant expression of various circRNAs detected in BM samples of AML patients can be implicated in the diagnosis and prognosis of AML. circRNAs use different mechanisms to contribute to drug resistance in AML. CircPAN3 binds with two different miRNAs (miRNA-183-5p and miRNA-153-5p) to increase the expression of XIAP, which is demonstrated to be a drug-resistance gene in AML. Several circRNAs generated from fused genes, known as fusion circRNA (f-circRNA), can also contribute to drug resistance in an unknown mechanism. CircPVT and f-circRNA (a product MLL/AF9) also potentiates drug resistance in an unknown mechanism. (B) miRNA-496 bind to their respective sites on mRNA, causing PRKACB repression, but circDLEU2-miRNA-496 sponge formation allows the binding of translational machinery to the mRNA, thus causing gene derepression. The increased expression of PRKACB is associated with the proliferation of AML cells. (C) CircMYBL2 produced from the MYBL2 gene positively regulates the expression of the FLT3-ITD mutant gene by recruiting PTBP1 to the mRNA of the FLT3-ITD gene. (D) CDK2, a cyclin-dependent kinase enzyme regulating the cell cycle, is negatively regulated by P21. Circ-Foxo3 binds to CDK2 inhibitor 21 (p21), forming circfoxo3-p21-CDK2 complexes, thus preventing cell cycle progression. (E) Interaction of PRDX2 with CDK2 causes PRDX2 activation that, in turn, regulates the downstream genes, thereby arresting AML cell differentiation; however, the depletion of CKD2 due to its possible binding with circRNA? or degradation could diminish its interaction with PRDX2, thus rendering PRDX2 to its inactivated stage. The inactivation of PRDX2 is associated with AML differentiation activation (36). (F) Possible circRNAs with various gene expression-regulating abilities in AML. CircRNAsX (e.g., EIciRNAs and ciRNAs) may interact with transcription complexes and enhance the expression of their parental genes; other circRNA (EcircRNA) may repress the gene expression. Certain circRNA, like the lncRNA, may interact with the DNA by forming a circRNA-DNA triplex to regulate DNA replication.
circRNAs in Drug Resistance
One of the hallmarks of AML relapse following transient remission upon conventional treatment is the development of chemo-resistance to therapy, resulting in insensitivity to further treatment (20, 21). Multiple genes and non-coding sequences are involved in the development of drug resistance in AML (22). Various non-coding RNAs are emerging as important players in mediating drug resistance, and their targeting offers avenues for the development of novel treatment options. Nonetheless, studies on the potential involvement of deregulated circRNAs in AML drug resistance are just emerging. For example, circRNA-microarray expression analysis of doxorubicin (ADM)-resistant THP1 cells revealed a set of 49 circRNAs with altered expression, and elevated circPAN3 (product PAN3 gene) in recurrent and refractory AML was defined as a key player in drug resistance in AML (23). Of interest, circPAN3 is predicted to interact with ten miRNAs, miR-153-5p, miR-183-5p, miR-338-3p, miR-346, miR-545-3p, miR-574-5p, miR-599, miR-653-5p, miR-766-3p, and miR-767-3p. Functional analysis via the KEGG and GO databases predicted that the target genes of these miRNAs are associated with various cellular processes and signaling pathways involved in cancer (24–27). Moreover, miR-153-5p and miR-183-5p were found to interact with X-linked inhibitor of apoptosis protein (XIAP), which has been demonstrated as a drug resistance gene in AML (28, 29). Consistent with the predicted circPAN3-miRNA interactions, inhibiting circPAN3 expression reduces the expression of XIAP, and this effect was reversed by miR-153-3p- or miR-183-5p-specific inhibitors. In view of circPAN3 interaction with miR-153-5p and miR-183-5p, which in turn bind with XIAP, it can be predicted that circPAN3 may contribute to doxorubicin resistance in THP1 cells (23).
Similarly, overexpression of circPVT1 (derived from Plasmacytoma Variant Translocation 1 or PVT1) has also been associated with resistance to vincristine in AML (30), and knockdown of f-circM9 (a fusion circRNA) showed increased sensitivity to anti-leukemic drugs (31). These findings suggest that circRNAs can potentially be manipulated to reverse drug resistance (Figure 2A).
circRNAs in Regulating Cell Proliferation and Apoptosis
Based on the circRNA microarray expression dataset GSE94591, it was previously shown that circ-DLEU2 (hsa_circ_0000488) was highly expressed in AML samples (37). circ-DLEU2 is the splicing product of the DLEU2 gene, which is located on chromosomes (32). Further studies demonstrated that circ-DLEU2 negatively regulated miRNA-496, whose downstream target gene is PRKACB (Figure 2B). The protein encoded by PRKACB is the catalytic subunit of cyclic AMP-dependent protein kinase, which regulates various signaling processes such as proliferation and differentiation through cAMP. The expression of PRKACB was negatively regulated by miRNA-496, while circ-DLEU2 promoted the expression of PRKACB by sponging miRNA-496 (32). Consequently, increased expression of circ-DLEU2 facilitated the in vitro proliferation of leukemic cells, blocked apoptosis, and promoted the formation of AML tumor in vivo. On the other hand, Morens et al. reported the deletion of the DLEU2 locus and embedded miRNA cluster miR-15a/16-1 in adult leukemia but its transcriptional repression due to hypermethylation in young AML patients (38). The DLEU2 gene contains the locus for miR-15a/16-1, the expression of which is negatively regulated by the binding of BSAP (a transcription factor) to the DLEU2 promoter. Increasing the expression of miR-15a/16-1 by inhibiting BASP facilitates apoptosis and leukemic cell cycle arrest (39). These findings suggest that circ-DLEU2 is crucial for in vitro proliferation and tumor formation in AML and, although the DLEU2 gene is identified as a tumor-suppressor gene, it can also be implicated in leukemogenesis.
Recently, Fan et al. reported that hsa_circ_100290, upregulated in AML, possesses oncogenic potential by enhancing cellular proliferation, inhibiting cell-cycle arrest, and suppressing cell apoptosis (35). They found that hsa_circ_100290 binds to miR-293, the expression of which is downregulated not only in AML but also in other cancers (40, 41). In many cancer types, miR-293 has been found to suppress the proliferation, invasion, and migration of cancer cells. Moreover, the target protein of miR-293 is Rab10, a member of the RAS superfamily of small GTPases (42). Furthermore, they found that the expression of miR-293 was decreased while Rab10 was elevated in AML patients, and hsa-circ_100290 expression was negatively and positively correlated with miR-293 and Rab10, respectively.
PVT1, on 8q24, is a non-protein coding region that produces numerous ncRNAs (43). The mechanistic role of the PVT1 region and its non-coding transcripts in cancers remains elusive. Apart from the role of PVT1 lncRNA in stabilizing MYC (44), several groups have reported the role of PVT1 as an oncogene in AML (45). Moreover, the circRNAs generated from exon 2 of the PVT1 region, termed circPVT1, possess oncogenic roles in AML by enhancing proliferation. Knocking down f-circRNAs9 (a product MLL/AF9) resulted in apoptosis, while its expression inhibited the drug-induced apoptosis of leukemic cells both in vitro and in vivo (31). To further corroborate these results, K562 cells were transduced with f-circRNA9 and treated with the standard drugs cytarabine (Arac-C) and ATO. Cells expressing f-circRNA9 inhibited the apoptosis induced by ATO and Arac-C (compared to empty vector-transfected cells) (31). These findings support the potential value of therapeutically targeting oncogenic circRNAs in AML (Figure 2B).
Fusion-circRNAs in Leukemogenic Properties
Leukemia is characterized by prevalent chromosomal translocation events (46). These chromosomal rearrangements often result in the formation of fusion genes, which may produce not only oncogenic chimeric proteins (47) but also circRNAs known as fusion circRNA (f-circRNA) (Figure 1G) (48). The fusion chimeric gene MLL/AF9 yields two fusion circRNA products, f-circM9_1 and f-circM9_2, which promote leukemogenesis by promoting cell proliferation (31, 49). Hypoxia-inducible protein kinase (HIPK)-2 is a protein that stabilizes PML oligomerization, a process that is important in nuclear body formation. The disruption of nuclear bodies by the fusion protein of PML and the retinoic acid receptor α (RARα) (PML-RARα) is often associated with APL and is thus implicated in leukemogenesis (50). In another study, the mutation in HIPK2 showed its impaired localization with AML1 transcription factor complex. This complex serves as a target for leukemia-associated chromosomal translocations. The mutant HIPK2 negatively regulated AML1- and P53-dependent transcription and thus possibly contributes to the pathogenesis of leukemia (51). PCR amplification and sequencing of the site flanking the PML-RARα junction revealed that the paradigmatic fusion gene PML-RARα also produced multiple isoforms of f-circRNA with oncogenic potential (31). In this scenario, the circ-HIPK2 produced from the host gene might serve as a transcription co-activator in the formation of nuclear bodies. Moreover, investigating the circular RNA transcript of the fusion PML-RARα in the development of AML would constitute interesting future research.
Nucleophosmin 1 (NPM1) encodes a multifunctional, highly conserved nuclear chaperone protein that is involved in ribosomal biogenesis, apoptosis, and cell proliferation (52). An insertion into the exon12 of NPM1 produces an abnormal protein that abnormally localizes in the cytoplasm and contributes to leukemia formation in 35% of AML cases (37). A study showed the differential expression of several linear isoforms of the NPM1 gene in a cohort of AML patients (53). Moreover, the dislocation and deletion and high expression of NPM1 also contribute to the AML phenotype (48, 54). Recently, RNA-seq analysis identified circNPM1 variant hsa_circ_0075001 as a signature for defining patient subgroups. Comparative gene expression profiling of high vs. low hsa_circ_0075001 revealed several genes with differential expression. Gene expression pathway analysis of low and high circNPM1 expression groups revealed a significant difference in the TLR gene expression groups, particularly TLR4, TLR5, TLR7, and TLR8 (34).
Web-Based Tools for circRNA Research Avenues
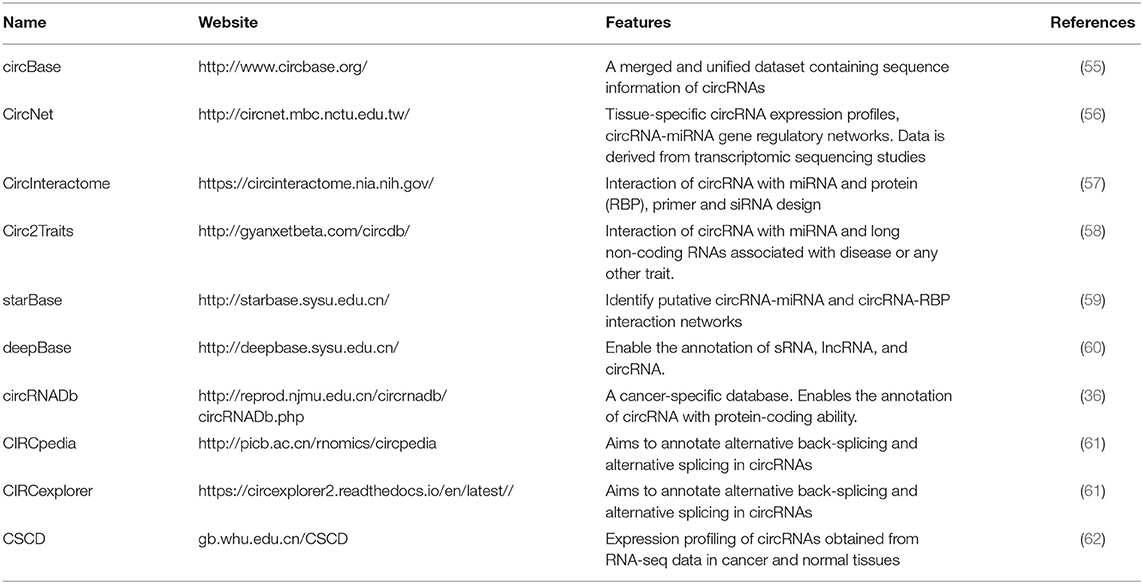
Because circRNA has multiple functions in various cancers, circRNA research is regarded as a hotspot in modern research. Various databases have been developed by researchers, mostly utilizing the data obtained from next-generation sequencing technology to shed light on the regulatory network and functional prediction of circRNA in various biological processes. The features of the most applied web-based tools are summarized in Table 2. The characteristics of the commonly employed databases are as follows. circBase, developed by Glazar et al., collects and integrates data related to circRNAs from public references and hosts information from various species such as model organisms. The database not only provides free access to the user to browse and download gene annotations but also allows the user to obtain evidence supporting the expression details of a particular circRNA (55). CircNet is a freely accessible database developed by Liu et al., which incorporates the transcriptomic datasets of novel and previously published circRNAs. The database provides tissue-specific circRNA expression profiling in terms of a heatmap. Furthermore, CircNet also depicts the regulatory network between circRNA, miRNA, and a particular gene (56). CircInteractome, developed by Dawood et al., is a freely accessible tool that enables the user to find the relevant miRNA- and RNA-binding protein sites on a particular circRNA. This database also facilitates circRNA-related research by allowing the researcher to design primers for qRT-PCR- and siRNA-based gene silencing. Moreover, CircInteractome can reveal the protein-coding abilities of a particular circRNA by deciphering its interaction with translational regulation factors and confirmation of IRES in circRNA (57). starBase depicts the interaction networks of circRNA with miRNA and RBP from 108 CLP-seq datasets generated by 37 different research studies. In addition, the other features of starBase v2.0 include miRNA-pseudogene interaction networks and miRNA-lncRNA interaction and competing endogenous RNA (ceRNA) functional networks (59). deepBase assembles NGS data from previous studies and enables users to annotate and discover small and long non-coding RNA and circRNAs. It also hosts features for decoding the evolution, expression profiles, and functions of non-coding RNAs across 19 species. Moreover, a spectrum of circRNA and lncRNA evolutionary patterns is also revealed by deepBase (60). Despite several worthwhile state-of-the-art toolkits for the identification, characterization, expression, and functional analysis of circRNA, problems such as a lack of a gold standard and overlap in predictions still need to be addressed. To this end, further innovation based on currently available methods with perfect precision and sensitivity should be pursued to overcome these challenges.
Future Perspective
Mechanism Exploration
One of the major functions of circRNA is sponging miRNA, thus regulating gene expression at an mRNA level. The dysregulation of miRNAs is implicated in the initiation and progression of leukemia (63, 64). miRNA sponging by circRNA can either transform into a leukemogenic property or its inhibition depending upon the type of miRNA inhibited by the circRNA. Numerous miRNAs with oncogenic potential have been reported in hematopoietic malignancies, e.g., miR22 causes the repression of the TET2 gene, thus causing defective differentiation of leukemic cells (65). We expect future research to explore circRNAs with binding ability to oncogenic miRNA(s). Such research will be challenging, since multiple miRNAs can regulate a particular gene. For instance, an extensive network of TET2-targeting miRNAs (≥30 miRNAs) has been documented to inhibit the expression of the TET2 gene (66). These findings pave the way for the possible development of therapeutic strategies based on circRNA targeting mechanisms to rescue tumor suppressor gene expression.
circRNA-RBP Sponge
Mounting evidence is revealing the interaction of circRNA with RNA binding proteins (RBPs) and its potential functional aspects (67). These RBPs include Argonaute (AGO) (4, 68), RNA polymerase (RNAPII) (5), Muscleblind protein (MBL) (69), Quanking I (QKI) (10), and Elongation Initiation Factor (EIF4A3) (57). These RBPs regulate gene expression and are thus involved in vital cellular processes. CRISPR-Cas9-based screening of RBP in AML has revealed a network of physically upregulated interacting RBPs that are crucial in regulating the splicing of RNA and maintaining the leukemic condition. The depletion of RBM39 (as the main regulator of this network) results in altered splicing of the mRNAs essential for AML, thereby causing the mortality of AML cells (70). Recently, the interaction of hsa_circ_0004870 with RBM39 has been shown to be associated with enzalutamide resistance in castration-resistant prostate cancer (71). This finding highlights the importance of investigating circRNA interactions with RBM in AML. In addition, mutational profiling of leukemic patients has uncovered somatic genetic mutations in RBP that are associated with splicing, as extensively reviewed by (72). Very recently, the high expression circMYBL2, a product of the MYBL2 gene, was reported in AML patients within internal tandem duplication (ITD) mutations in the FLT3 (FLT3-ITD) gene. A positive regulation was observed between circMYBL2 and FLT3-ITD mutant kinase. In terms of mechanism, circMYBL2 was found to enhance FLT3-ITD mutant protein expression by facilitating the binding of PTBP1 to the mRNA of mutant FLT3 kinase. The knockdown of circMYBL2 not only inhibits the proliferation and supports the differentiation of AML cells both in vitro and in vivo but also compromises the cytoactivity of cells with the FLT3-ITD mutation against quizartinib (Figure 2C) (73). Circ-Foxo3 binds to cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 21 (p21), forming the circfoxo3-p21-CDK2 complex, thus preventing the cell cycle progression by inhibiting CDK2 (74) (Figure 2D). In AML, CDK2 blocks myeloid differentiation (Figure 2E). The ubiquitin (KLHL6-E3 ubiquitin ligase)-mediated proteasome degradation of CDK2 is associated with the activation of AML cell differentiation (75). The deletion of FOXO-1/3/4 family members facilitates the maturation of myeloid cells, AML cell death, and a diminution of leukemia-initiating cell function, thus improving animal survival (76). In the scenario of circ-FOXO3-p21-CDK2 ternary complex formation, it will be worthwhile to investigate the affinity of circ-FOXO3 or any other relevant circRNAs with CDK2 or any key cell cycle-regulating proteins, thus ultimately controlling the differentiation of cells in AML (Figure 2D). Molecular insights into understanding how malignancies are produced due to perturbation in pathways downstream of these sites will help in the identification of drug target regions.
Generally, owing to their stable structure, circRNAs are present in the cytoplasm, but some circular isoforms (EIcircRNA) are also present in the nucleus. These circular isoforms interact with chromatin modifiers, thereby causing repression or activation of the gene (77, 78). EIcircRNAs such as circEIF3J and circPAIP2 are associated with RNA polymerase II and recruit U1 small nuclear ribonucleoprotein (snRNP), which promotes the transcription of genes (8) (Figure 2F). In addition, certain circRNAs positively regulate the expression of their parent gene, such as in the case of the down-regulation of the ciRNA known as ci-ankrd52, which reduces the expression of ankrd52 without affecting the surrounding genes (Figure 2F) (5). circRNA derived from the SEP3 gene regulated the expression of the linear transcript by binding to its cognate DNA. The linear counterpart of circRNA-SEP3 with the same sequence binds to the DNA with weak affinity. Thus, circRNA-DNA formation presumably results in transcriptional repression and the generation of a SEP3 linear transcript with exon skipping (79). In addition, promoter-associated RNA (pRNA) suppresses the expression of the rRNA genes by recruiting DNMT3b to the target site of TTF-I (a transcription factor) via complementarity with the rDNA promoter (Figure 2F) (80). Watanabe et al. found that long non-coding RNA near the MYC gene could facilitate the amplification of MYC by influencing the movement of the replication fork (Figure 2E) (81). Another lncRNA known as ANRASSF1, produced from the opposite strand of a tumor-suppressor gene RASSF1 gene, binds with the promoter region of the parental gene, thus forming an RNA/DNA hybrid, and recruits polycomb repressive complex 2 protein (PRC2) to regulate its expression (82, 83). Like other ncRNAs, the circRNA may possibly affect DNA replication by binding with the genomic DNA, thus forming a DNA-RNA triplex (80). These findings highlight that circular and linear non-coding RNA may have the same binding pattern with the DNA and manifest unique effects on gene expression and DNA replication.
Anti-Oncogenic Potential of circRNA
Some circRNAs, for instance, circ-ZEB1.19, circZEB-1.17, circZEB1.5, and circZEB1.33, by trapping miRNA-200, are involved in the suppression of lung cancer progression (56). The downregulation of circular RNA produced from mitochondrial translocation optimization 1 homolog (MTO1), known as circMTO1, was reported in hepatocellular carcinoma tissue. circMOT1 decoys miR-9 to allow the expression of tumor suppressor gene p21 (84). The transfection of synthesized small circular single-stranded DNA-9 (CSSD-9), which mimics the circRNA, resulted in increased expression of various tumor suppressor genes such as LF17, CDH1, and LASS2 by absorbing miR-9. The artificially synthesized CSSD-9, containing multiple binding sites for miR-9, impaired lung tumor progression and metastasis (85).
circRNAs were initially considered non-coding RNA due to their disability to recruit ribosome (9, 86). However, recent studies have documented the presence of an open reading frame (ORF) driven by the ribosomal machinery (13, 14). The circular transcript of the SNF2 histone linker PHD RING helicase (SHPRH) gene encodes a fully functional protein, 17kDa, termed circ-SHPRH, which suppresses the tumor transformation of glioma cells. In terms of mechanism, the circ-SHPRH protects SHPRH-146aa from ubiquitin-mediated proteasomal degradation, and subsequently, SHPRH ubiquitinates the proliferating cell nuclear antigen (PCNA) as an E3 ligase, thereby inhibiting cell proliferation (87). circRNA expression profiling of AML cell lines has identified that circKHLH8 is associated with clinical outcome, as its overexpression enhanced overall survival, with a higher level of platelets and reduced blast percentage in peripheral blood. Moreover, the inhibition of circFBXW7 facilitated the proliferation of AML cells, indicating the tumor-suppressive function of circFBXW7 in AML (88). Based on these findings, it is speculated that circRNA can work as an anti-oncogenic to promote the expression of tumor-suppressor genes by the derepression of miRNAs or by recruiting the transcription factor to the promoter of tumor suppressor genes.
Targeted Therapy
Of note, the circRNA-miRNA-mRNA axis is a widely accepted mechanism in the establishment of leukemia. Circularization-based approaches can be used as a treatment strategy. Generally, a circRNA contains multiple miRNA binding sites (MRE). Therefore, targeting the inhibition of circRNA expression rather than a single miRNA/gene offers treatment advantages. Inhibiting circRNA expression will promote the protective effect of the corresponding miRNA in suppressing oncogenes, for instance, XIAP (23), Rab10 (35), PRACKB (32), and TLR pathway-related genes (34). CRISPR-Cas9-based engineering is a robust tool for performing functional studies of circRNA in cancer (89). However, depleting the circRNAs without affecting the existing genes is a challenge. In order to achieve circRNA-specific knockdown and avoid the targeting of circRNA from its linear RNA, the guide sequence must be designed to target the back-splice junction (BSJ) site that is uniquely present in circRNA to avoid its binding with the linear transcript. The non-protein-coding property of circRNA makes it a tough choice for depletion by incorporating insertions/deletions (INDELs). A CRISPR-Cas-assisted homologous recombination approach may overcome the first challenge by replacing the circRNA gene with a marker gene (90). Moreover, strategies to avoid the off-target effects should be considered to circumvent undesirable mutations (91). Nevertheless, CRISPR-Cas9-mediated deletion of the cdr1 locus has been achieved by Piwecka et al. (92). Similarly, disruption of the formation of RNA pairing reduces the expression of circRNA, and, under certain conditions, the knock-out of a circRNA has been achieved, for example, circGCN1LI in human PA1 cells (93). In addition, the currently developed tool for RNA targeting, CRISPR-Cas13, represents a promising method for targeting a specific circRNA (94). Cas13, a principal enzyme of the type VI CRISPR-Cas system guided by a 20 to 30-nt-long CRISPR RNA (crRNA), could specifically bind to the BSJ site and thus, in principle, be able to distinguish between the linear transcript and circular RNA. CRISPR-Cas-based circRNA engineering approaches can be harnessed for high-throughput screening (89), and future research using well-characterized guide RNA (gRNA) libraries designed for circRNAs may facilitate global screening studies.
Challenges
Advancements in Next-Generation Sequencing (NGS) technologies and the availability of user-friendly bioinformatics tools have facilitated circRNA biology-related research in cancer. These advancements have helped in the transformation of circRNA research from considering it a redundant splicing byproduct to exploring its possible potential targeted therapeutic application in AML. However, the field is in its infancy and faces enormous challenges (Figure 3) and raises questions that need comprehensive consideration. First, the expense of NGS technologies limits the sample size, and there is a lack of gold-standard studies for data comparison. Second, despite extensive research related to the biogenesis of circRNA, knowledge on the metabolic processing of circRNA within cells is limited. Owing to the high stability of these molecules, the accumulation of circRNAs may have toxic effects on the cell. Studies have shown that surplus circRNAs are exported out of the cells in vesicles such as exosomes (95). Further molecular studies are needed to uncover the factors controlling the circulation, metabolism, and turnover of circRNA. Third, there is a lack of a gold-standard nomenclature system for circRNA. The naming of circRNA in future research should be based on the host gene along with the term “circ.” Fourth, lack of perfection in the web-based tools, characterized by the low correspondence of the circRNAs tested by these different tools, is not helpful. To this end, in the future, we expect the development of a database integrating more comprehensive information about function and molecular mechanism. Finally, our knowledge of circRNA in AML is limited. In the last couple of years, researchers have shown that the circRNA expression pattern is related of clinical outcome and a poor prognosis. The most common mode of action of circRNA in AML is miRNA sponging, underscoring the need for investigation of other biological functions of circRNA such as the circRNA-RBP and circRNA-DNA regulatory networks. Although current research is being focused on the validation of these findings, further studies should focus on questions such as how circRNAs contribute to AML and the correlation between altered expression of circRNA and the mutational analysis of candidate genes that are crucial in alternative splicing events.
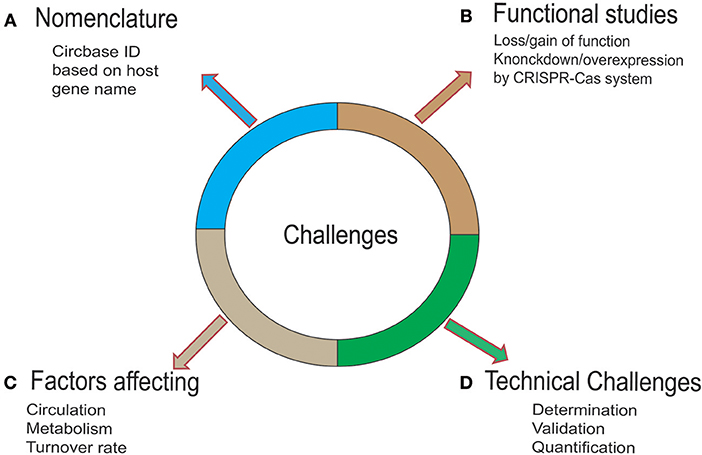
Figure 3. Challenges and strategies for circRNA. (A) To date, circRNAs have been named based on various parameters by various available databases, creating chaos in scientific research. Therefore, a standardized and uniform naming system is needed. (B) To gain mechanistic insights into the biological functions of circRNA, mechanism exploration is needed. To this end, an optimized CRISPR-Cas system would offer a robust and easy platform to carry out these studies. (C) The knowledge of the fate of circRNA in a cell is not fully known. Owing to the high stability of these molecules, the accumulation of circRNAs may exert toxic effects on the cell. Studies have shown that the surplus circRNAs are exported out of the cells in vesicles such as exosomes. Further molecular studies are needed to uncover the factors controlling the circulation, metabolism, and turnover of the circRNA. (D) Factors such as the peculiarity of the back-splice junction in circRNA, sequence similarity with the linear transcript, and low abundance compared to its linear counterpart make determination challenging. A multistep strategy for overcoming this challenge is promising. These steps include the discovery/identification of circRNA (by RNA-sequencing or microarray expression analysis), validation of circRNA by qRT-PCR, Sanger sequencing, Nothern Blotting, fluorescent in-situ hybridization (FISH), and the quantification of circRNA by targeted RT-qPCR assay.
Conclusion
In conclusion, the role of circRNA in various cancers, including AML, is a major focus in cancer research. Although the detailed mechanisms of its biogenesis, transportation, function, and metabolism have not been fully studied, yet the glimpses provided by modern research suggest that circRNA as a potential candidate for AML diagnosis and therapy.
Author Contributions
MJ, QZ, and LS designed and conceived the study. TS, BC, ZH, and TX helped with figure design and literature selection and drafted the manuscript. MF, LS, YW, and SP revised the language. QY helped in manuscript formatting and language revision. All of the authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81770180 and 81500151) and Hubei Provincial Natural Science Fund for Creative Research Groups (2018CFA018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Dr. Dean Tang (Professor and Chair in the Department of Pharmacology and Therapeutics at Roswell Park Comprehensive Cancer Center) for critically reviewing and improving the language of the manuscript.
Abbreviations
AML, Acute myeloid leukemia; circRNA, Circular RNA; miRNA, Micro RNA; CeRNA, Competing endogenousRNA; f-circRNA, Fusion circRNA; EIcircRNA, Exonic-intronic circRNA; CiRNA, Circular intronic RNA; EcircRNA, Exonic circRNA; TLR, Toll-like receptor; PML-RARα, Promyelocytic leukemia and the retinoic acid receptor α (RARα); IRES, Internal ribosome entry site; HIPK, Hypoxia-inducible protein kinase; XIAP, X-linked inhibitor of apoptosis protein; KEGG and GO, Kyoto encyclopedia of gene and genome and gene ontology; CLIP-seq, Crosslinking immunoprecipitation sequencing; qRT-PCR, Quantitative reverse transcriptase polymerase chain reaction; SiRNA, Small interfering RNA; CDK2, Cyclin-dependent kinase; AGO, Argonaute; QKI, Quanking I; EIF4A3, Elongation Initiation Factor; U1 snRNP, U1 small nuclear ribonucleoprotein; MRE, miRNA recognition element CRISPR-Cas9, Clustered regularly spaced palindromic repeats and CRISPR-associated gene/protein 9; NGS:, Next-Generation Sequencing.
References
1. Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
2. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
3. Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. (2017) 2:10. doi: 10.1186/s13045-016-0370-2
4. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
5. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
6. Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet. (2016) 17:407–21. doi: 10.1038/nrg.2016.46
7. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Correction: cell-type specific features of circular RNA expression. PLoS Genet. (2014) 9:e1003777. doi: 10.1371/journal.pgen.1003777
8. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
9. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
10. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
11. Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. (2017) 14:978–84. doi: 10.1080/15476286.2017.1317911
12. Balci Okcanoglu T, Gündüz C. Circular RNAs in leukemia. Biomed Rep. (2019) 10:87–91. doi: 10.3892/br.2018.1178
13. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
14. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
15. Prada-Arismendy J, Arroyave JC, Röthlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. (2017) 31:63–76. doi: 10.1016/j.blre.2016.08.005
16. Chen H, Liu T, Liu J, Feng Y, Wang B, Wang J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. (2018) 47:1998–2007. doi: 10.1159/000491468
17. Yi Y-Y, Yi J, Zhu X, Zhang J, Zhou J, Tang X, et al. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J Cell Physiol. (2019) 234:3711–9. doi: 10.1002/jcp.27145
18. Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. (2019) 75:41–7. doi: 10.1016/j.bcmd.2018.12.006
19. Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. (2017) 18:E597. doi: 10.3390/ijms18030597
20. Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. (2012) 481:506–10. doi: 10.1038/nature10738
21. Zahreddine H, Borden K. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. (2013) 4:28. doi: 10.3389/fphar.2013.00028
22. Bell CC, Fennell KA, Chan Y-C, Rambow F, Yeung MM, Vassiliadis D, et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat Commun. (2019) 10:2723. doi: 10.1038/s41467-019-10652-9
23. Shang J, Chen W-M, Wang Z-H, Wei T-N, Chen Z-Z, Wu W-B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p–XIAP axis. Exp Hematol. (2019) 70:42–54.e3. doi: 10.1016/j.exphem.2018.10.011
24. Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA Repair. (2016) 45:25–33. doi: 10.1016/j.dnarep.2016.06.003
25. Willems S, Kellner U, Midgley R, Simon I, Bernards R. TGF-β: an emerging player in drug resistance AU - Brunen, Diede. Cell Cycle. (2013) 12:2960–8. doi: 10.4161/cc.26034
26. Jiang B-H, Liu L-Z. Role of mTOR in anticancer drug resistance: Perspectives for improved drug treatment. Drug Resistance Updates. (2008) 11:63–76. doi: 10.1016/j.drup.2008.03.001
27. Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. (2014) 25:282–303. doi: 10.1016/j.ccr.2014.02.025
28. Shaffer BC, Gillet J-P, Patel C, Baer MR, Bates SE, Gottesman MM. Drug resistance: Still a daunting challenge to the successful treatment of AML. Drug Resistance Updates. (2012) 15:62–9. doi: 10.1016/j.drup.2012.02.001
29. Prabhu KS, Siveen KS, Kuttikrishnan S, Iskandarani A, Tsakou M, Achkar IW, et al. Targeting of X-linked inhibitor of apoptosis protein and PI3-kinase/AKT signaling by embelin suppresses growth of leukemic cells. PLoS ONE. (2017) 12:e0180895. doi: 10.1371/journal.pone.0180895
30. L′Abbate A, Tolomeo D, Cifola I, Severgnini M, Turchiano A, Augello B, et al. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia. (2018) 32:2152–66. doi: 10.1038/s41375-018-0033-0
31. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. (2016) 165:289–302. doi: 10.1016/j.cell.2016.03.020
32. Wu D-M, Wen X, Han X-R, Wang S, Wang Y-J, Shen M, et al. Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol. (2018) 38:e00259–18. doi: 10.1128/MCB.00259-18
33. Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. (2016) 17:205–11. doi: 10.1038/nrm.2015.32
34. Hirsch S, Blätte TJ, Grasedieck S, Cocciardi S, Rouhi A, Jongen-Lavrencic M, et al. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica. (2017) 102:2039–47. doi: 10.3324/haematol.2017.172866
35. Fan H, Li Y, Liu C, Liu Y, Bai J, Li W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. (2018) 507:178–84. doi: 10.1016/j.bbrc.2018.11.002
36. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. (2016) 6:34985. doi: 10.1038/srep34985
37. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. (2017) 129:424–47. doi: 10.1182/blood-2016-08-733196
38. Morenos L, Chatterton Z, Ng JL, Halemba MS, Parkinson-Bates M, Mechinaud F, et al. Hypermethylation and down-regulation of DLEU2 in paediatric acute myeloid leukaemia independent of embedded tumour suppressor miR-15a/16-1. Mol Cancer. (2014) 13:123. doi: 10.1186/1476-4598-13-123
39. Kasar S, Underbayev C, Yuan Y, Hanlon M, Aly S, Khan H, et al. Therapeutic implications of activation of the host gene (Dleu2) promoter for miR-15a/16-1 in chronic lymphocytic leukemia. Oncogene. (2013) 33:3307–15. doi: 10.1038/onc.2013.291
40. Chiu SH, Martínez MR, Komissarova EV, Llobet-Navas D, Bansal M, Paull EO, et al. The number of titrated microRNA species dictates ceRNA regulation. Nucleic Acids Res. (2018) 46:4354–69. doi: 10.1093/nar/gky286
41. Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, et al. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging. (2018) 10:775–88. doi: 10.18632/aging.101427
42. Wang W, Jia WD, Hu B, Pan YY. RAB10 overexpression promotes tumor growth and indicates poor prognosis of hepatocellular carcinoma. Oncotarget. (2017) 8:26434–47. doi: 10.18632/oncotarget.15507
43. Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. (2008) 6:212–21. doi: 10.1158/1541-7786.MCR-07-0105
44. Tseng Y-Y, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. (2014) 512:82–6. doi: 10.1038/nature13311
45. Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet. (2010) 42:1126–30. doi: 10.1038/ng.696
46. You X, Conrad TOF. Acfs: accurate circRNA identification and quantification from RNA-Seq data. Sci Rep. (2016) 6:38820. doi: 10.1038/srep38820
47. Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer. (2013) 13:559–71. doi: 10.1038/nrc3563
48. Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
49. Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. (2007) 7:823–33. doi: 10.1038/nrc2253
50. Shima Y, Honma Y, Kitabayashi I. PML-RARα and its phosphorylation regulate PML oligomerization and HIPK2 stability. Cancer Res. (2013) 73:4278–88. doi: 10.1158/0008-5472.CAN-12-3814
51. Li XL, Arai Y, Harada H, Shima Y, Yoshida H, Rokudai S, et al. Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene. (2007) 26:7231–9. doi: 10.1038/sj.onc.1210523
52. Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. (2007) 92:519–32. doi: 10.3324/haematol.11007
53. Zajac M, Dolnik A, Stasiak G, Zaleska J, Kielbus M, Czapinski J, et al. Analysis of NPM1 splice variants reveals differential expression patterns of prognostic value in acute myeloid leukemia. Oncotarget. (2017) 8:95163–75. doi: 10.18632/oncotarget.19871
54. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. (2013) 42:D68–73. doi: 10.1093/nar/gkt1181
55. GlaŽar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. (2014) 20:1666–70. doi: 10.1261/rna.043687.113
56. Liu Y-C, Li J-R, Sun C-H, Andrews E, Chao R-F, Lin F-M, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. (2016) 44:D209–15. doi: 10.1093/nar/gkv940
57. Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs AU - Dudekula, Dawood B. RNA Biol. (2016) 13:34–42. doi: 10.1080/15476286.2015.1128065
58. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. (2013). 4:283. doi: 10.3389/fgene.2013.00283
59. Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. (2014) 42:D92–7. doi: 10.1093/nar/gkt1248
60. Zheng L-L, Li J-H, Wu J, Sun W-J, Liu S, Wang Z-L, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. (2016) 44:D196–202. doi: 10.1093/nar/gkv1273
61. Zhang X-O, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. (2016) 26:1277–87. doi: 10.1101/gr.202895.115
62. Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. (2018) 46:D925–9. doi: 10.1093/nar/gkx863
63. Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. (2006) 6:259–69. doi: 10.1038/nrc1840
64. Wang X, Zhu B, Huang Z, Chen L, He Z, Zhang H. MicroRNAs as biomarkers in leukemia. Stem Cell Investig. (2014) 1:11. doi: 10.3978/j.issn.2306-9759.2014.04.01
65. Su Song J, Ito K, Ala U, Kats L, Webster K, Su Sun M, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. (2013) 13:87–101. doi: 10.1016/j.stem.2013.06.003
66. Cheng J, Guo S, Chen S, Stephen Mastriano J, Liu C, Ana D'Alessio C, et al. An extensive network of TET2-targeting microRNAs regulates malignant hematopoiesis. Cell Rep. (2013) 5:471–81. doi: 10.1016/j.celrep.2013.08.050
67. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
68. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
69. Ashwal-Fluss R, Meyer M, Nagarjuna Pamudurti R, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
70. Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell. (2019) 35:369–84.e7. doi: 10.1016/j.ccell.2019.01.010
71. Greene J, Baird A-M, Casey O, Brady L, Blackshields G, Lim M, et al. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep. (2019) 9:10739. doi: 10.1038/s41598-019-47189-2
72. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. (2011) 478:64–9. doi: 10.1038/nature10496
73. Sun Y-M, Wang W-T, Zeng Z-C, Chen T-Q, Han C, Pan Q, et al. CircMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. (2019). doi: 10.1182/blood.2019000802
74. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
75. Ying M, Shao X, Jing H, Liu Y, Qi X, Cao J, et al. Ubiquitin-dependent degradation of CDK2 drives the therapeutic differentiation of AML by targeting PRDX2. Blood. (2018) 131:2698–711. doi: 10.1182/blood-2017-10-813139
76. Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. (2011) 146:697–708. doi: 10.1016/j.cell.2011.07.032
77. Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. (2012) 19:1068–75. doi: 10.1038/nsmb.2428
78. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. (2013) 20:300–7. doi: 10.1038/nsmb.2480
79. Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. (2017) 3:17053. doi: 10.1038/nplants.2017.53
80. Schmitz K-M, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. (2010) 24:2264–9. doi: 10.1101/gad.590910
81. Watanabe T, Marotta M, Suzuki R, Diede SJ, Tapscott SJ, Niida A, et al. Impediment of replication forks by long non-coding RNA provokes chromosomal rearrangements by error-prone restart. Cell Rep. (2017) 21:2223–35. doi: 10.1016/j.celrep.2017.10.103
82. Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. (2013) 368:2059–74. doi: 10.1056/NEJMoa1301689
83. Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. (2013) 9:e1003705. doi: 10.1371/journal.pgen.1003705
84. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. (2017) 66:1151–64. doi: 10.1002/hep.29270
85. Meng J, Chen S, Han J-X, Tan Q, Wang X-R, Wang H-Z, et al. Derepression of co-silenced tumor suppressor genes by nanoparticle-loaded circular ssDNA reduces tumor malignancy. Sci Transl Med. (2018) 10:eaao6321. doi: 10.1126/scitranslmed.aao6321
86. Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev. (2018) 9:e1478. doi: 10.1002/wrna.1478
87. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. (2018) 37:1805–14. doi: 10.1038/s41388-017-0019-9
88. Papaioannou D, Volinia S, Nicolet D, Swierniak M, Petri A, Mrózek K, et al. Prognostic and biologic significance of circular rna profiling in younger adult patients with cytogenetically normal acute myeloid leukemia (CN-AML). EHA Learn Center. (2018) 214594.
89. Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR–Cas9. Nat Rev Genet. (2015) 16:299–311. doi: 10.1038/nrg3899
90. Ho T-T, Zhou N, Huang J, Koirala P, Xu M, Fung R, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. (2015) 43:e17. doi: 10.1093/nar/gku1198
91. Jamal M, Khan FA, Da L, Habib Z, Dai J, Cao G. Keeping CRISPR/Cas on-target. Curr Issues Mol Biol. (2015) 20:1–20. doi: 10.21775/cimb.020.001
92. Piwecka M, GlaŽar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. (2017) 35:eaam8526. doi: 10.1126/science.aam8526
93. Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J-L, et al. The biogenesis of nascent circular RNAs. Cell Rep. (2016) 15:611–24. doi: 10.1016/j.celrep.2016.03.058
94. Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR–Cas13. Nature. (2017) 550:280–4. doi: 10.1038/nature24049
Keywords: acute myeloid leukemia (AML), circular RNA (circRNA), biomarker, apoptosis, cell proliferation, drug resistance, targeted therapy
Citation: Jamal M, Song T, Chen B, Faisal M, Hong Z, Xie T, Wu Y, Pan S, Yin Q, Shao L and Zhang Q (2019) Recent Progress on Circular RNA Research in Acute Myeloid Leukemia. Front. Oncol. 9:1108. doi: 10.3389/fonc.2019.01108
Received: 22 February 2019; Accepted: 07 October 2019;
Published: 06 November 2019.
Edited by:
Cesar Wong, Hong Kong Polytechnic University, Hong KongReviewed by:
Kira Gritsman, Albert Einstein College of Medicine, United StatesAmaresh Chandra Panda, Institute of Life Sciences (ILS), India
Copyright © 2019 Jamal, Song, Chen, Faisal, Hong, Xie, Wu, Pan, Yin, Shao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Shao, bGlhbmdzbWRAMTYzLmNvbQ==; Qiuping Zhang, cXB6aGFuZ0B3aHUuZWR1LmNu
 Muhammad Jamal
Muhammad Jamal Tianbao Song
Tianbao Song Bei Chen
Bei Chen Muhammad Faisal
Muhammad Faisal Zixi Hong
Zixi Hong Tian Xie
Tian Xie Yingjie Wu
Yingjie Wu Shan Pan
Shan Pan Qian Yin1
Qian Yin1 Liang Shao
Liang Shao Qiuping Zhang
Qiuping Zhang