- 1Shenzhen People's Hospital, The First Affiliated Hospital of Southern University of Science and Technology, The Second Clinical Medical College of Jinan University, Shenzhen, China
- 2Shenzhen Nanshan District Shekou People's Hospital, Shenzhen, China
- 3Institute of Biomedicine, College of Life Science and Technology, Jinan University, Guangzhou, China
Breast cancer is the leading cause of cancer-related deaths in women; however, its underlying etiology remains largely unknown. In this study, we systematically analyzed breast cancer tissues using comprehensive iTRAQ labeled quantitative proteomics, identifying 841 differentially expressed proteins (474 and 367 significantly over- and under-expressed, respectively), which were annotated by protein domain analysis. All the heat shock proteins identified were upregulated in breast cancer tissues; Hsp90 upregulation was also validated by RT-qPCR and immunohistochemistry, and high Hsp90 protein levels correlated with poorer survival. Hsp90AA1 overexpression promoted MDA-MB-231 cell proliferation, whilst BJ-B11, an Hsp90 inhibitor, hampered their invasion, migration, and proliferation in a time and dose-dependent manner and induced cell cycle arrest and apoptosis. BJ-B11 inhibited the expression of epithelial-mesenchymal transition (EMT) marker in MDA-MB-231 cells, whereas Hsp90AA1 promoted its expression. Moreover, BJ-B11 inhibited tumor growth in xenograft model. Altogether, Hsp90 activation is a risk factor in breast cancer patients, and BJ-B11 could be used to treat breast cancer.
Introduction
Breast cancer is the most frequently diagnosed cancer in women (1), and its incidence has increased in most developing countries over the past few decades (1, 2). Although patients with breast cancer have a high 5-year survival rate following treatment, the survival rate decreases rapidly for patients with more advanced disease (3–5). Triple-negative breast cancers (TNBCs) account for 15% of all breast cancers and lack estrogen, progesterone, and ERBB2 receptor expression (6). TNBCs are poorly differentiated, and there are no specific treatment guidelines for this breast cancer subgroup (7, 8); therefore, biomarkers and more effective medical therapies are urgently required.
Breast cancer is coordinately controlled by regulatory networks; thus, understanding these networks could help identify candidates for the diagnosis, prediction, and therapy of breast cancer. Proteomics approaches are often used to acquire a comprehensive and quantitative profile of protein expression. Isobaric tags for relative and absolute quantitation (iTRAQ) is a novel and unbiased approach to simultaneously quantify relative protein abundance (9); in particular, the method enables protein quantification during various developmental stages (10).
Hsp90 is highly expressed in various cancers (11). It is responsible for the stability and function of client proteins, including Akt, IKKα, B-Raf, and GSK3β, which are critical for cell survival and proliferation (12). Therefore, Hsp90 is a potential therapeutic target and diagnostic marker for cancer (13, 14). BJ-B11 is a novel Hsp90 inhibitor that reportedly exhibits antitumor activity in myeloid leukemia and esophageal carcinoma (15, 16); however, its antitumor activity in breast cancer has not yet been investigated.
In this study, we investigated whether Hsp90 was associated with breast cancer and whether BJ-B11 affected the functions of breast cancer cells. Our findings suggested that Hsp90 could be a candidate for the early diagnosis, prognosis, and therapy of breast cancer and that BJ-B11 could be used to treat breast cancer.
Materials and Methods
Primary Breast Cancer Samples
Tumor tissue and adjacent normal tissue samples were collected at the Department of Thyroid and Breast Surgery, Shenzhen People's Hospital, with the informed consent of the patients. The study (LL-KT-2015002) was approved by the Ethics Committee of the hospital. The clinicopathological information regarding the samples is detailed in the Table S1. Tumor tissues and normal tissues (10 mg) were homogenized for proteomics and iTRAQ labeling followed by LC-MS/MS analysis. The protein with iTRAQ ratio (tumor tissue/normal tissue) <0.83 or > 1.2 (P < 0.05) was considered to be significantly differentially expressed.
Cell Culture and Reagents
The human breast cancer cell line MDA-MB-231 was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM/F12 supplemented with 10% FBS, 100 μg/mL streptomycin, and 100 unit/mL penicillin in a humidified incubator in a 5% CO2 atmosphere at 37°C.
BJ-B11 was prepared in our lab, as previously described (17), and the 10 mmol/L BJ-B11 stock solution in DMSO was stored at 4°C. Plasmids expressing wild-type Hsp90AA1 were provided by SAGENE (Guangzhou, Guangdong, China). Mouse anti-E-cadherin (cat: 14472), rabbit anti-vimentin (cat: 3932), and mouse anti-β-actin (cat: 3700) antibodies were purchased from CST (MA, USA).
Cell Viability and Apoptosis Assay
CCK-8 (Dojindo, Japan) was used to detect cell viability. Cell apoptosis induced by BJ-B11 was determined using AnnexinV/PI (KeyGEN, Nanjing, China) staining, followed by flow cytometry (Beckman Coulter, CA, USA) according to the manufacturer's instructions.
Cell Cycle Analysis
Cells were treated with BJ-B11 for 48 h, harvested in cold PBS, fixed in 70% ethanol, and stored overnight at 4°C. The cells were then washed twice with cold PBS, resuspended in 50 μg/mL PI staining reagent containing 100 μg/mL RNase and 0.1% Triton X-100 for 30 min in the dark, and analyzed by flow cytometry (Becton-Dickinson, CA, USA).
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted using TRIZOL (Thermo Fisher Scientific) and subjected to qRT-PCR using the primers shown in Table S2). Gene expression was normalized against GAPDH using the relative ΔΔCT method and is reported as relative expression compared to the control.
Cell Invasion Assay
A total of 2 × 104 MDA-MB-231 cells treated with or without BJ-B11 were added to Transwell inserts and cultured in an incubator for 16 h. Cells inside the insert were cleaned thoroughly with a cotton swab, while those on the underside were fixed in 4% paraformaldehyde for 5 min and stained with 0.5% crystal violet solution. At least five random fields were counted per insert, and each group consisted of three replicates.
Tissue Microarray
Human breast tissue (HBreD077Su01, Shanhai Xinchao, China) and breast cancer tissue (HBreD140Su05, Shanhai Xinchao, China) microarrays consisting of 77 adjacent non-malignant tissue samples and 140 breast cancer tissue samples, were stained with rabbit anti Hsp90 (4874, CST, USA). Immunohistochemical staining was carried out according to the manufacturer's instructions. Slides were evaluated for their positive staining rate (0, negative; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%) and the staining intensity of the positively stained cells (0, none; 1, weak; 2, moderate; and 3, strong). Samples were grouped according to the H score, which was the product of the “staining intensity” and “staining positive rate” scores: low expression group, <8; and high expression group, ≥ 8. Two investigators evaluated each tissue section independently.
The Cancer Genome Atlas (TCGA) Data Analysis
The expression level and survival of Hsp90AA1 and Hsp90AB1 in breast cancer were analyzed using the UALCAN platform.
Xenograft Model
Female BALB/c Nude Mice (6-week-old) were obtained from the Guangdong Medical Laboratory Animal Center. They were maintained in an air-conditioned room with controlled temperature of 21 ± 2°C, and humidity of 30–70% in a 12 h light/darkness cycle regulation and were fed laboratory chow and water ad libitum. All animal experiments were approved by the Animal Ethics Committee of Shenzhen People's Hospital (No. LL-KY-2019512). The athymic female nude mice were injected with about 5 × 106 MDA-MB-231 cells subcutaneously. When the tumors were measurable, the mice were randomly assigned into treatment group receiving oral BJ-B11 (20 mg/kg, daily for a total of 27 days) or the control group receiving oral vehicle alone. All mice were euthanized at day 28, and the tumors were excised and weighed to evaluate tumor growth inhibition.
Statistical Analysis
Data are presented as the mean ± SD, and Student's unpaired t-tests were used for the statistical analysis of differences between two groups. Differences between groups were analyzed using Prism 6 (GraphPad, Inc., San Diego, CA). P < 0.05 were considered statistically significant.
Results
Identification of Hsp90 as a Diagnostic Marker
We screened proteins that were differentially expressed between tumor tissues and adjacent normal tissues using iTRAQ with ratio (tumor:adjacent normal tissue) thresholds of >1.2 or <0.83, which indicated higher or lower protein expression in tumor tissue than in adjacent normal tissue, respectively. A total of 841 differentially expressed proteins were identified, among which 474 and 367 were up- and down-regulated in breast cancer tissues (Figure 1A).
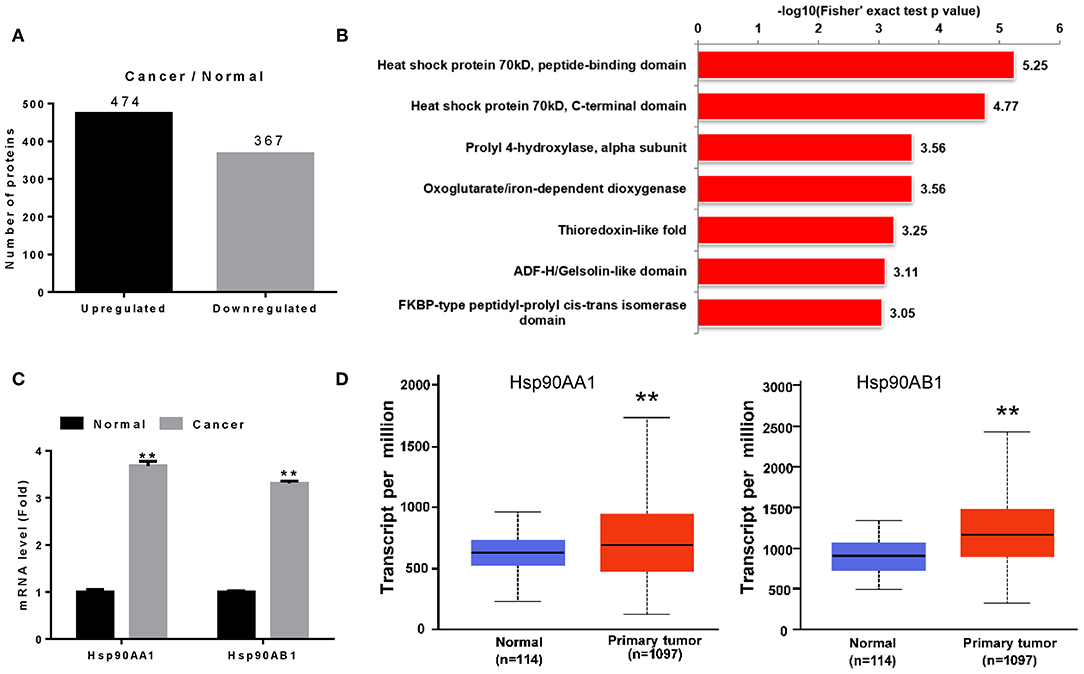
Figure 1. Identification of Hsp90 as a diagnostic marker. (A) Differentially expressed proteins in breast cancer. (B) Differentially expressed proteins were classified according to their protein domains (P < 0.001). (C) Validation of Hsp90AA1 and Hsp90AB1 mRNA levels in healthy and breast cancer tissues by RT-qPCR. (D) Hsp90AA1 and Hsp90AB1 mRNA levels in healthy and breast cancer tissues from TCGA database (**P < 0.01).
To understand the functions of these differentially expressed proteins, they were classified according to their protein domains (P < 0.001; Figure 1B). Heat shock proteins showed the most significant change of all the upregulated domains, with Hsp90AA1, Hsp90AB1, TRAP1, HspA5, HspB1, HspE1, HspD1, HspA1B, HspA8, HspA9, and HspA4 identified in breast cancer tissue (Table S3). RT-qPCR revealed that Hsp90AA1 and Hsp90AB1 mRNA levels increased in breast cancer (Figure 1C), consistent with TCGA data (Figure 1D). These results suggest that the highly expressed Hsp90 could act as a diagnostic marker for breast cancer.
Identification of Hsp90 as a Prognostic Marker
We then performed tissue microarray analysis, finding that Hsp90 protein levels increased in breast cancer tissue (P = 0.014; Figure 2A and Table S4). Moreover, Hsp90 expression was significantly correlated with clinical tumor grade (Table S5); therefore, we analyzed the survival rate of patients with breast cancer. High Hsp90 levels predicted poor survival (Figure 2B), consistent with TCGA data (Figure S1). Thus, the univariate and multivariate analyses suggested that Hsp90 could serve as a prognostic indicator for breast cancer (Table S6).
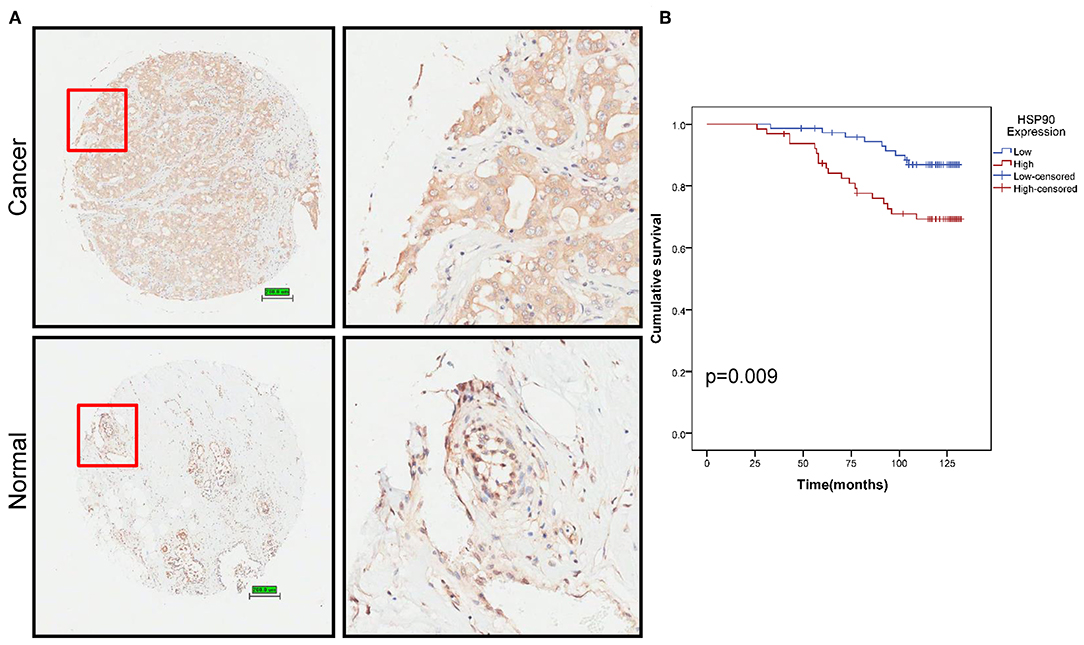
Figure 2. Identification of Hsp90 as a prognostic marker. (A) Immunohistochemical staining of Hsp90 in normal (n = 77) and breast cancer (n = 140) tissues. Representative images are shown. Scale bar, 200 μm. (B) Cumulative survival of breast cancer patients with low or high Hsp90 expression.
BJ-B11 Induces Apoptosis and Cell Cycle Arrest in Breast Cancer Cells
Since our findings suggested that Hsp90 was involved in breast cancer, we examined whether manipulating the Hsp90 gene affected the proliferation of breast cancer cells. Hsp90AA1 overexpression increased MDA-MB-231 cell proliferation (Figure 3A), whereas treating the cells with BJ-B11, an Hsp90 inhibitor, for 24, 48, or 72 h inhibited their growth in a time- and dose-dependent manner (Figure 3B) and induced dose-dependent apoptosis (Figure 3C) and G2/M cell cycle arrest (Figure 3D). Taken together, Hsp90AA1 promoted the proliferation of breast cancer cells, whilst BJ-B11 could be used to treat breast cancer.
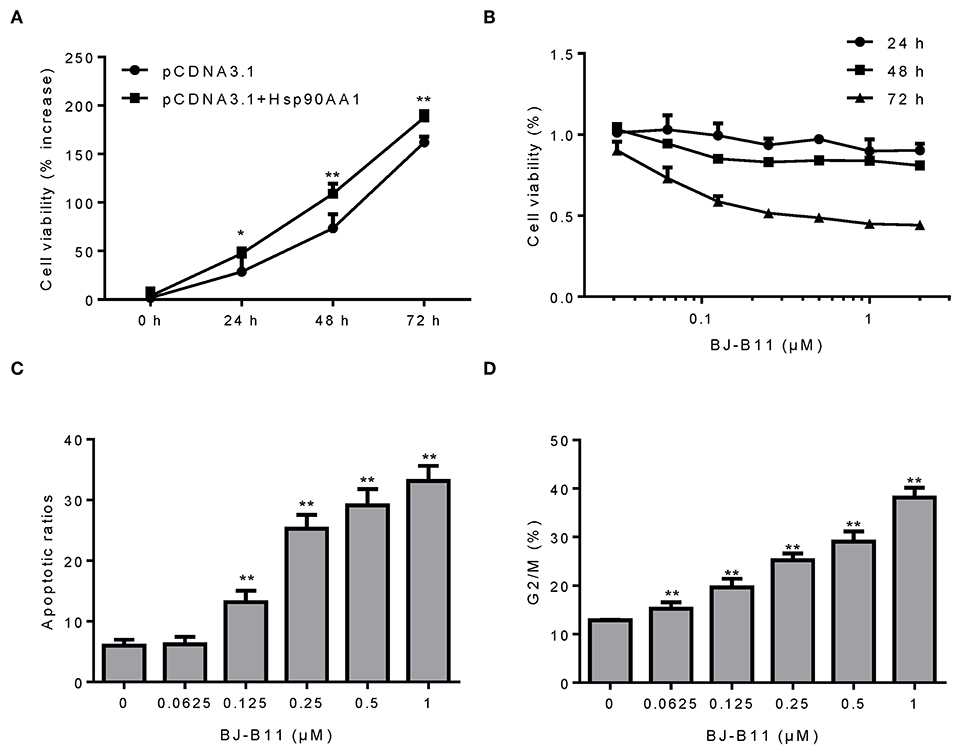
Figure 3. BJ-B11 induces apoptosis and cell cycle arrest in breast cancer cells. (A) The viability of MDA-MB-231 cells overexpressing Hsp90AA1 was tested by CCK8 after 24, 48, and 72 h (*P < 0.05, **P < 0.01). (B) The viability of cells treated with the indicated concentrations of BJ-B11 for 24, 48, or 72 h was tested by CCK8. (C) Cells were treated with BJ-B11 for 48 h, collected, and stained with AnnexinV/PI. Apoptotic cells were analyzed by flow cytometry (**P < 0.01). (D) Cells were treated with BJ-B11 for 48 h, collected, fixed, and stained with PI. The cell cycle stage was analyzed by flow cytometry (**P < 0.01).
BJ-B11 Inhibited Invasion and Migration of Breast Cancer Cells
MDA-MB-231 cells were cultured with or without BJ-B11 in Transwell inserts for 16 h. BJ-B11 inhibited the invasion of the MDA-MB-231 cells (Figure 4A) and inhibited Hsp90, thus significantly suppressing MDA-MB-231 cell migration (Figure 4B). We also analyzed epithelial-mesenchymal transition (EMT) markers related to invasion and migration, finding that BJ-B11 upregulated E-cadherin and downregulated vimentin (Figure 4C). Conversely, Hsp90AA1 overexpression upregulated vimentin and downregulated E-cadherin and occludin (Figure 4D), suggesting that Hsp90 plays a vital role in EMT in breast cancer. Taken together, these data strongly suggest that BJ-B11 inhibits cell invasion and migration by affecting EMT in breast cancer.
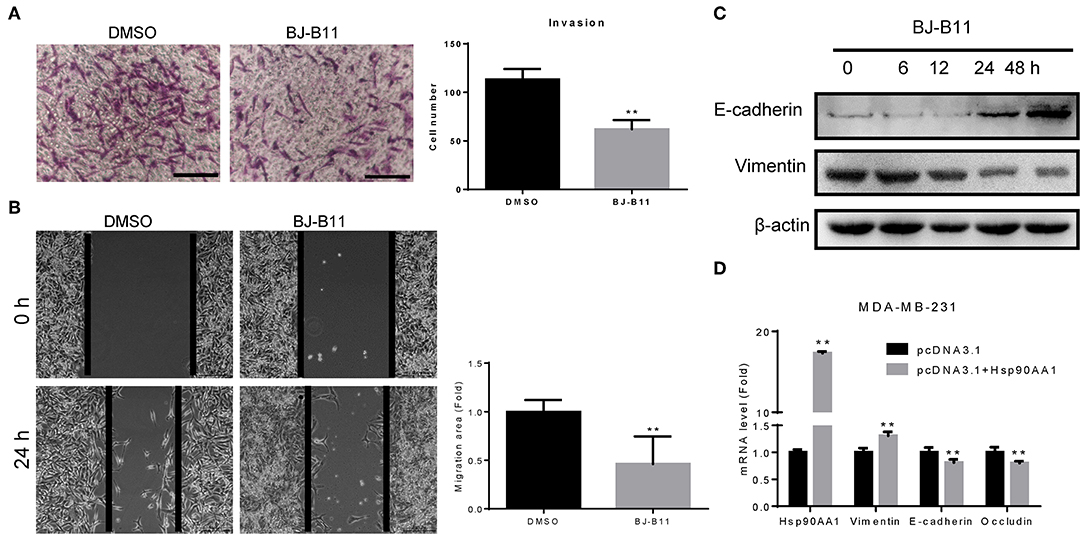
Figure 4. BJ-B11 inhibited the invasion and migration of breast cancer cells. (A) Cells were cultured in Transwell inserts for 16 h with or without 0.0625 μM BJ-B11 and then stained with crystal violet. Cell number was calculated and analyzed (**P < 0.01). Scale bar, 100 μm. (B) Wound healing assay of MDA-MB-231 cell treated with or without 0.0625 μM BJ-B11 for 24 h (**P < 0.01). Scale bar, 200 μm. (C) Western blot assay of MDA-MB-231 cells treated with 0.0625 μM BJ-B11 for the indicated length of time. (D) MDA-MB-231 cells overexpressing Hsp90AA1 were cultured for 48 h. Hsp90AA1, E-cadherin, vimentin, and occludin mRNA levels were tested by RT-qPCR (**P < 0.01).
BJ-B11 Inhibited Tumor Growth in vivo
We further tested whether BJ-B11 could suppress cell growth of breast cancer in vivo. First, we established xenograft models by subcutaneous injection of MDA-MB-231 cells into the right flanks of mice. We then tested the anti-tumor effects of BJ-B11 on MDA-MB-231 cancer cells. Nude mice bearing MDA-MB-231 tumor xenografts were treated with 20 mg/kg BJ-B11 (n = 7) or physiological saline (n = 7) for 27 days. Bodyweight was measured each day before the administration of BJ-B11. Results showed that BJ-B11 inhibited tumor growth significantly in vivo (Figures 5A,B).
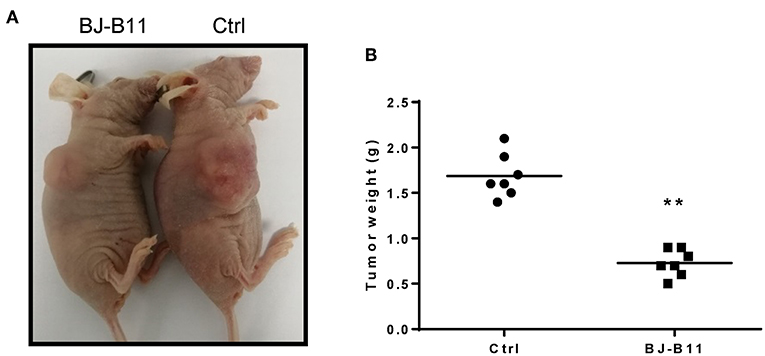
Figure 5. BJ-B11 inhibited tumor growth in vivo. (A) Representative picture of the xenograft model. (B) Analysis of tumor weight of BJ-B11 treated group and control group (n = 7, **P < 0.01).
Discussion
This study investigated breast cancer proteome using iTRAQ to obtain a global view and identify therapeutic targets for breast cancer. We identified 841 differentially expressed proteins and showed that heat shock proteins could be candidate biomarkers for the early diagnosis and therapy of breast cancer. Moreover, this is the first study to investigate the antitumor activity of BJ-B11 in TNBCs, showing that BJ-B11 could be used to treat breast cancer.
Multiple heat shock proteins, including Hsp90AA1, Hsp90AB1, TRAP1, HspA5, HspB1, HspE1, HspD1, HspA1B, HspA8, HspA9, and HspA4, were significantly upregulated in breast cancer tissue. Hsp70, HspA8, HspA9, HspA5, and Hsp110s may constitute up to 3% of the total protein in unstressed human cells (18). Moreover, Hsp70 is upregulated in various human cancers and associated with tumorigenesis (19, 20). HspA8 is overexpressed in cancer cells and it belongs to the Hsp70 family (21). Its depletion in RL-95-2 and HEC-1B cells was shown to suppress cell growth and promote apoptosis, suggesting that HspA8 could be a candidate biomarker for endometrial carcinoma (22).
Hsp90 is an ATP-dependent molecular chaperone (23). Reportedly, Hsp90 expression levels are associated with disease progression and survival in melanoma, gastrointestinal stromal tumors and non-small cell lung cancer (24, 25). Moreover, high Hsp90 expression has been associated with decreased survival in breast cancer (26) whilst its inhibition can suppress growth and promote apoptosis in breast cancer cells, suggesting that Hsp90 could act as both a biomarker and a therapeutic target for breast cancer (27). A phase II study of 17-AAG in breast cancer showed that Hsp90 inhibitors exhibit significant anticancer activity (28). Previously, we demonstrated that the Hsp90 inhibitor SNX-2112 suppressed MCF-7 cell proliferation and induced apoptosis (27), whilst the Hsp90 inhibitor PU-H71 has been shown to induce a complete response in TNBC models (29). BJ-B11 is a novel Hsp90 inhibitor that can inhibit cancer cell proliferation and exhibits anti-HSV activity (15, 16). To the best of our knowledge, this is the first study to investigate the antitumor activity of this molecule in TNBCs; BJ-B11 inhibited the proliferation, invasion, and migration of breast cancer cells, which may be associated with EMT. In addition, BJ-B11 showed significant antitumor activity in vivo. However, the underlying mechanism requires further clarification.
In summary, our study provides new insights into the molecular changes that occur in breast cancer. Hsp90AA1, Hsp90AB1, TRAP1, HspA5, HspB1, HspE1, HspD1, HspA1B, HspA8, HspA9, and HspA4 were validated as potential biomarkers for breast cancer tissue. In particular, Hsp90 plays important roles in breast cancer development and could be a candidate biomarker for the early diagnosis and therapy of breast cancer. Moreover, BJ-B11 could open new, potential therapeutic alternatives for breast cancer.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Shenzhen People's Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Ethics Committee of Shenzhen People's Hospital.
Author Contributions
KL conceived, designed the experiments, and wrote the manuscript. KL, YL, and YG performed the experiments and analyzed the data. KL, JC, FY, ZZ, HH, HG, HL, WZ, BQ, and YW contributed reagents, materials, and analysis tools.
Funding
This research was supported by the National Natural Science Foundation of China (81802749), the Science and Technology Foundation of Shenzhen (JCYJ20180301170047864, JCYJ20180228164300106), and the Cultivating Fund Project of Shenzhen People's Hospital (No. SYLY201704).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01447/full#supplementary-material
Figure S1. Cumulative survival analysis of breast cancer patients with low or high Hsp90AA1 (A) or Hsp90AB1 (B) expression.
Table S1. Sample information.
Table S2. Primers used in this study.
Table S3. Hsp family members upregulated in breast cancer tissue.
Table S4. Differential Hsp90 expression in healthy and breast cancer tissues.
Table S5. Correlation between Hsp90 expression and clinicopathological characteristics.
Table S6. Univariate and multivariate analyses of factors correlated with the overall survival of breast cancer patients.
Abbreviations
Hsp90, heat shock protein 90; TNBC, triple-negative breast cancer; iTRAQ, isobaric tags for relative and absolute quantitation; GO, Gene Ontology; RT-qPCR, real-time quantitative polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EMT, epithelial-mesenchymal transition.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. (2017) 67:439–48. doi: 10.3322/caac.21412
3. DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin. (2019) 69:211–33. doi: 10.3322/caac.21555
4. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. (2019) 69:363–85. doi: 10.3322/caac.21565
5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
6. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. (2010) 363:1938–48. doi: 10.1056/NEJMra1001389
7. Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. (2007) 8:235–44. doi: 10.1016/S1470-2045(07)70074-8
8. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. (2007) 13(15 Pt 1):4429–34. doi: 10.1158/1078-0432.CCR-06-3045
9. Chong PK, Gan CS, Pham TK, Wright PC. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J Proteome Res. (2006) 5:1232–40. doi: 10.1021/pr060018u
10. Palma JM, Corpas FJ, del Rio LA. Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J Proteomics. (2011) 74:1230–43. doi: 10.1016/j.jprot.2011.04.010
11. Liu KS, Liu H, Qi JH, Liu QY, Liu Z, Xia M, et al. SNX-2112, an Hsp90 inhibitor, induces apoptosis and autophagy via degradation of Hsp90 client proteins in human melanoma A-375 cells. Cancer Lett. (2012) 318:180–8. doi: 10.1016/j.canlet.2011.12.015
12. Liu KS, Ding WC, Wang SX, Liu Z, Xing GW, Wang Y, et al. The heat shock protein 90 inhibitor SNX-2112 inhibits B16 melanoma cell growth in vitro and in vivo. Oncol Rep. (2012) 27:1904–10. doi: 10.3892/or.2012.1738
13. Jin L, Xiao CL, Lu CH, Xia M, Xing GW, Xiong S, et al. Transcriptomic and proteomic approach to studying SNX-2112-induced K562 cells apoptosis and anti-leukemia activity in K562-NOD/SCID mice. FEBS Lett. (2009) 583:1859–66. doi: 10.1016/j.febslet.2009.04.046
14. Wang R, Shao F, Liu Z, Zhang J, Wang S, Liu J, et al. The Hsp90 inhibitor SNX-2112, induces apoptosis in multidrug resistant K562/ADR cells through suppression of Akt/NF-kappaB and disruption of mitochondria-dependent pathways. Chem Biol Interact. (2013) 205:1–10. doi: 10.1016/j.cbi.2013.06.007
15. Ju HQ, Wang SX, Xiang YF, Liu Z, Liu JY, Chen ZP, et al. BJ-B11, a novel Hsp90 inhibitor, induces apoptosis in human chronic myeloid leukemia K562 cells through the mitochondria-dependent pathway. Eur J Pharmacol. (2011) 666:26–34. doi: 10.1016/j.ejphar.2011.05.020
16. Liu KS, Zhang Y, Ding WC, Wang SX, Xiang YF, Yang P, et al. The selective Hsp90 inhibitor BJ-B11 exhibits potent antitumor activity via induction of cell cycle arrest, apoptosis and autophagy in Eca-109 human esophageal squamous carcinoma cells. Int J Oncol. (2012) 41:2276–84. doi: 10.3892/ijo.2012.1670
17. Ju HQ, Xiang YF, Xin BJ, Pei Y, Lu JX, Wang QL, et al. Synthesis and in vitro anti-HSV-1 activity of a novel Hsp90 inhibitor BJ-B11. Bioorg Med Chem Lett. (2011) 21:1675–7. doi: 10.1016/j.bmcl.2011.01.098
18. Finka A, Sharma SK, Goloubinoff P. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front Mol Biosci. (2015) 2:29. doi: 10.3389/fmolb.2015.00029
19. Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. (2006) 31:164–72. doi: 10.1016/j.tibs.2006.01.006
20. Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. (2016) 21:379–404. doi: 10.1007/s12192-016-0676-6
21. Stricher F, Macri C, Ruff M, Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. (2013) 9:1937–54. doi: 10.4161/auto.26448
22. Shan N, Zhou W, Zhang S, Zhang Y. Identification of HSPA8 as a candidate biomarker for endometrial carcinoma by using iTRAQ-based proteomic analysis. Onco Targets Ther. (2016) 9:2169–79. doi: 10.2147/OTT.S97983
23. Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. (2005) 5:761–72. doi: 10.1038/nrc1716
24. Workman P, Powers MV. Chaperoning cell death: a critical dual role for Hsp90 in small-cell lung cancer. Nat Chem Biol. (2007) 3:455–7. doi: 10.1038/nchembio0807-455
25. McCarthy MM, Pick E, Kluger Y, Gould-Rothberg B, Lazova R, Camp RL, et al. HSP90 as a marker of progression in melanoma. Ann Oncol. (2008) 19:590–4. doi: 10.1093/annonc/mdm545
26. Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, et al. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. (2007) 67:2932–7. doi: 10.1158/0008-5472.CAN-06-4511
27. Wang SX, Ju HQ, Liu KS, Zhang JX, Wang X, Xiang YF, et al. SNX-2112, a novel Hsp90 inhibitor, induces G2/M cell cycle arrest and apoptosis in MCF-7 cells. Biosci Biotechnol Biochem. (2011) 75:1540–5. doi: 10.1271/bbb.110225
28. Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, et al. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res. (2011) 17:5132–9. doi: 10.1158/1078-0432.CCR-11-0072
Keywords: breast cancer, Hsp90, BJ-B11, proliferation, invasion, migration, EMT
Citation: Liu K, Chen J, Yang F, Zhou Z, Liu Y, Guo Y, Hu H, Gao H, Li H, Zhou W, Qin B and Wang Y (2019) BJ-B11, an Hsp90 Inhibitor, Constrains the Proliferation and Invasion of Breast Cancer Cells. Front. Oncol. 9:1447. doi: 10.3389/fonc.2019.01447
Received: 03 September 2019; Accepted: 03 December 2019;
Published: 18 December 2019.
Edited by:
Dong-Hua Yang, St. John's University, United StatesReviewed by:
Haitao Mao, UT Southwestern Medical Center, United StatesTao Hu, University of Maryland, Baltimore, United States
Copyright © 2019 Liu, Chen, Yang, Zhou, Liu, Guo, Hu, Gao, Li, Zhou, Qin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbin Zhou, emhvdXdiMTAxNkAxNjMuY29t; Bo Qin, cWluYm96ZkAxNjMuY29t; Yifei Wang, dHdhbmcteWZAMTYzLmNvbQ==
 Kaisheng Liu
Kaisheng Liu Juan Chen2
Juan Chen2 Bo Qin
Bo Qin Yifei Wang
Yifei Wang