- 1Department of Molecular and Bio Physics, Moscow Institute of Physics and Technology, Dolgoprudny, Russia
- 2School of Biological Sciences, University of Auckland, Auckland, New Zealand
- 3Harefield Hospital, The Royal Brompton and Harefield Hospitals NHS Foundation Trust, Harefield, United Kingdom
- 4Department of Oncology and Reconstructive Surgery, Sechenov Medical University, Moscow, Russia
The tumor biomarkers already have proven clinical value and have become an integral part in cancer management and modern translational oncology. The tumor tissue microenvironment (TME), which includes extracellular matrix (ECM), signaling molecules, immune and stromal cells, and adjacent non-tumorous tissue, contributes to cancer pathogenesis. Thus, TME-derived biomarkers have many clinical applications. This review is predominately based on the most recent publications (manuscripts published in a last 5 years, or seminal publications published earlier) and fills a gap in the current literature on the cancer biomarkers derived from the TME, with particular attention given to the ECM and products of its processing and degradation, ECM-associated extracellular vesicles (EVs), biomechanical characteristics of ECM, and ECM-derived biomarkers predicting response to the immunotherapy. We discuss the clinical utility of the TME-incorporating three-dimensional in vitro and ex vivo cell culture models for personalized therapy. We conclude that ECM is a critical driver of malignancies and ECM-derived biomarkers should be included in diagnostics and prognostics panels of markers in the clinic.
Introduction
Cancer remains one of the leading causes of deaths globally with a strong tendency to become the “number one killer disease” in the 21st century (1). Despite the recent achievements in understanding how malignant tumor arise and develop there are many unique aspects of tumorigenesis which are not fully understood, one of them is how the tumor microenvironment (TME) orchestrates a wide array of events in the tumor initiation and progression. The studies focusing on the role of TME in cancer initiation and progression may identify novel therapeutic targets and biomarkers derived from within the TME, with clinical translational potential.
A biomarker is a biological characteristic that can be identified and objectively evaluated as an indicator of a normal or pathological biological process (2) and may serve for various clinical purposes (3, 4). The prognostic biomarkers can predict favorable or unfavorable outcome of disease irrespective of the therapy, predictive biomarkers may foresee favorable or unfavorable response to the therapy. It is apparent now that only genomic biomarkers are not clinically informative enough, and the set of available diagnostic tools should be expanded (5). The growing number of studies demonstrates that biomarkers identified not only in the tumor cell itself, but also within the TME are valuable diagnostic tools (6–8) (Supplementary Table 1). These biomarkers include bio-mechanical characteristics of ECM, structural components of ECM, products of ECM biosynthesis, processing, degradation, proteinase inhibitors, as well as activators, circulating EVs, cytokines, and others. There are a number of techniques allowing detection of such biomarkers in a clinical setting, such as ELISA (9), microscopy and imaging analysis (10), mass-spectrometry (11) including MALDI imaging mass-spectrometry applied to analysis of formalin-fixed paraffin-embedded tissues (12), immunohistochemistry (13), Western blotting (14), RT-qPCR (15), and others.
In the present review, we will provide a framework for understanding the role of TME-associated biomarkers in cancer pathogenesis and discuss their clinical utility in precision oncology, with special emphasis on biomarkers predicting response to immunotherapy. Particular focus is given to the ECM-derived protein markers, EVs and their molecular cargo, biomechanical characteristics of ECM, and ECM-incorporating 3D cell culture models for translational oncology.
ECM Components as Cancer Biomarkers
ECM is an extracellular three-dimensional (3D) maze-like structure formed by a variety of macromolecules such as proteins, proteoglycans, glycoproteins, polysaccharides, and others (16, 17). It also contains multitude of matrix-stored regulatory and signaling biomolecules, such as growth factors and cytokines, circular RNAs (circRNAs), and miRNAs within the TME-associated exosomes, and others (18, 19). Structurally, the ECM comprises the basal membrane and the interstitial tissue. The components of ECM, also referred to as “matrisome” (20), are produced by the cells of several types, predominantly fibroblasts (21). Interactions of cell surface receptors with the components of ECM enable cell-ECM adhesion, which is vital for many types of anchorage-dependent cells (22). ECM has a plethora of functions—it creates a niche for stem cells and regulates intercellular chemical and mechanical signaling networks, angiogenesis, innate and adaptive immune response, and migration and invasion of the cells (23–25). All this makes the ECM one of the key regulators of cancer progression and response to the therapy, capable of modulating fundamental hallmarks of cancer (26).
The molecular composition, mechanical properties of ECM, its infiltration by immune cells and stromal cells is heterogeneous and immensely diverse in different types of tumor tissues. To accommodate the specific needs of the tumor, both cancer cells and tumor-associated stromal cells modify ECM by producing and secreting ECM-modifying enzymes. For example, fibroblasts associated with tumor tissue (cancer-associated fibroblasts, CAFs) and tumor-associated macrophages (TAMs) modify ECM to create a metastasis-permissive environment (27, 28). Many components of ECM are deregulated in cancer, and some oncogenic macromolecules within the tumor ECM are upregulated whereas tumor-suppressors are downregulated (29) (Supplementary Table 1). The analysis of expression of 820 matrisome genes across a panel of 32 malignant tumors has identified universal pan-cancer gene signatures which supposedly might be used for diagnostics (30). Recent study of the changes in the matrisome during the cancer progression identified expression patterns of the 22 genes associated with shorter overall survival of patients with ovarian and several other solid tumors (31). Several independent attempts have also been made to characterize the profile of ECM-derived biomarkers for a particular type of cancer and identify cancer-specific markers for clinical application (Supplementary Table 1).
Some ECM-derived peptides, termed “matrikines” or “matricryptines”, have cytokine-like activity (32). The matricryptines are generated by the structural or enzymatic modification of ECM resulting in exposure of the biologically active and previously hidden (“cryptic”) sites. It has been suggested recently that cryptic collagen elements serve as signaling hubs regulating tumor metastasis and growth (33). ECM may also evolve releasing biologically active substances, including matrikines, which may be used as “protein fingerprint” of cancer. One of them is Tumstatin derived from collagen type IVα3 and described as a biomarker for non-small-cell lung cancer (NSCLC) (34).
Importantly for transnational oncology, ECM-derived biomarkers may reflect response to therapy, including immunotherapy. Whereas the role of stromal cells within TME in immune response is comprehensively studied and reviewed elsewhere (35), the role of ECM and products of its modification as biomarkers of tumor response to immunotherapy is not well known. Recent study demonstrated that tumor matrisome gene signatures are predictive biomarkers of resistance to ICT immunotherapy (36). Versican-derived matrikine versikine is a biomarker of tumor response to immunotherapy (37) and regulator of tumor infiltration by T-cells (38, 39) [notably, versican itself is upregulated in cervical cancer and leiomyosarcoma (40, 41)]. In patients with stage IV melanoma, collagen-derived biomolecules RO-C3, C1M, C3M, and C4M are biomarkers of poor response to the therapy with immune checkpoint inhibitor (ICI) ipilimumab (42). In patients with metastatic melanoma, blood-based biomarkers of type III collagen turnover are associated with worse overall survival and progress-free survival following PD-1 inhibition immunotherapy (43). In a clinical scenario, the ECM-turnover associated with the response of melanoma to immuno-therapy might be assessed in a “liquid biopsy” (44), and allows to stratify patients with metastatic melanoma according to their response to ICI therapy (45). Finally, many protein biomarkers of tumor invasiveness localized in ECM have been identified (comprehensively reviewed in (46).
Aforementioned, many soluble ECM-derived molecules arising from within a solid tumor can be found in a peripheral blood, are detectable using routine laboratory methods such as immunoassays (42, 43), and may therefore be used as a non-invasive “liquid biopsy” biomarkers. This makes them very attractive for use in clinics (47), the only limitation of their use being sensitivity and specificity of the immunoassay.
Mechanical and Physical Properties of ECM as Cancer Biomarkers
Mechanotransduction, also known as mechanosignaling, is a process through which cells initiate a biochemical process in response to mechanical signals. The stiffness, topology, and other mechanistic characteristics of the ECM are critical drivers and regulators of the tumor progression, affecting cancer cell biology via the mechanotransduction [comprehensively reviewed in (48–50)] and therefore can be used as biomarkers of malignancy (51, 52). The phenomenon of durotaxis (directed migration of the cells in response to the gradient of stiffness of the substrate) also plays an important role in tumorigenesis (53, 54).
The biomechanical properties of the ECM dynamically change over the course of the disease and differ between tumor and matched normal tissue. In many types of solid tumors, ECM within the tumor tissue is more rigid than ECM of matched non-tumorous tissue (55) mostly because of the elevated deposition and cross-linking of collagen type I, which can be detected by the imaging or manual examination. Such stiffness of the ECM induces epithelial-to-mesenchymal transition (EMT) of the cancer cells, thus resulting in a metastatic phenotype, for example, in pancreatic ductal adenocarcinoma (56) and in hepatocellular carcinoma (57). On the other hand, ovarian cancer cells undergo EMT on softer substrates (58), and soft matrices enhance cancer stem cell phenotype in hepatocellular carcinoma (59). This should be considered then developing therapeutic approaches aimed to modify softness/rigidity of the ECM and targeting its mechanical features (60, 61).
The overall role of the biomechanical properties of ECM in several types of cancer, for example, esophageal cancer (62), ovarian cancer (63), and colorectal cancer (64), has been comprehensively reviewed recently (Supplementary Table 1). The stiffness of the ECM can also be a biomarker predicting response to the chemotherapy; for example, it has been shown that in case of pancreatic ductal adenocarcinoma it induces chemoresistance to paclitaxel, but not to gemcitabine (56). Furthermore, the mechanical characteristics of ECM play a role in immune oncology and therefore might be a biomarker of response to immunotherapy. For example, stiffness of ECM modulates PD-L1 expression in lung cancer (65) and breast cancer cells (66) and regulates activity of T-cells within the tumor tissue (67).
There are several powerful tools and approaches available to assess mechanical and physical properties of TME, for example, high resolution Atomic Force Microscopy (AFM), Scanning Electron Microscopy (SEM), Spatial Light Interference Microscopy (SLIM), and others (64, 68–70). The Multiphoton Microscopy and Second Harmonic Generation (SHG) imaging can be applied to analyze “evolution” of collagen within the ECM during tumor progression (71). That said, non-invasive imaging techniques, such as Ultrasound Elastography, Magnetic resonance elastography, Magnetic Resonance Imaging (MRI), and others might still be a good option for assessing biomechanical characteristics of the tumor tissue in clinic (72–74).
Extracellular Vesicles as Cancer Biomarkers and Regulators of TME
Within the TME, cells communicate via different mechanisms including extracellular vesicles (EVs). EVs are carriers of a biologically active molecular cargo (lipids, nucleic acids, proteins, mRNA, miRNA, circRNA, lncRNA, and others). As some of the contents of EVs may modulate ECM [for example, matrix-remodeling enzymes (75)] or participate in a cross-talk of the cancer cell with stromal cells, thus contributing to chemotherapy resistance or metastasis, there is a possibility to use EVs within a TME as a therapeutic targets and therapeutic biomarkers. Moreover, the possibility to detect tumor-derived EVs in a bloodstream makes them attractive for use in a clinical setting (76).
In the context of ECM, there is a subset of matrix-bound nanovesicles (MBVs) (77, 78) present within the ECM rather than in biological fluids. They are embedded into the ECM, express surface antigens that are commonly found on exosomes, and can be isolated from the matrix only by methods of enzymatic digestion of ECM scaffolds (77). Their molecular cargo comprises miRNAs and is capable of changing the phenotype of the cells exposed to the contents of MBVs, for example, affecting the phenotype of macrophages (78). MBVs are integral and distinct components of ECM, and their content is unique to cellular origin (78). Recently, it has been demonstrated that MBVs can suppress pro-inflammatory signaling in microglia and astrocytes (79). So far, the literature exists only on MBVs found in non-timorous ECM, but we propose that tumor-specific MBVs may also be found. If molecular cargo of MBVs is cell type-dependent and unique to cellular origin, as demonstrated by Hussey et al., the MBVs derived from tumor ECM most likely will also have unique and tumor-specific characteristics. Further studies on this subject should be carried out on various types of malignancies to assess feasibility of using MBVs as potential biomarkers.
Finally, the promising avenue in translational oncology is a possibility to study EVs in vitro using cell culture models to identify and characterize novel cancer biomarkers. It has been demonstrated recently that there are cell culture-dependent differences in the content and production of EVs (80). The essential molecular cargo components of EVs secreted by cancer cells cultured in vitro in two-dimensional (2D) or 3D format are different, and EVs from 3D culture have much higher similarity to the EVs secreted in vivo by tumor tissue (81), and the spectrum of small RNAs in EVs derived from cells in 3D culture has approximately 96% similarity to EVs from cancer patient’s plasma (81). This provides a rationale for developing 3D cell culture-based in vitro model systems for cancer biomarkers identification.
3D Cell Culture Models Incorporating TME as a Testing System in Translational Oncology and Personalized Therapy
Over the past decades, significant progress has been made in developing ex vivo models that recapitulate in vivo tumor characteristics including response to the therapy. It is apparent now that in vitro 2D culture of cells on glass or plastic is not an accurate model of in vivo “biological reality”. Moreover, mono-culture of cancer cells is a less accurate model compared to the co-culture of cancer cells and stromal cells. Adding to this complexity, compared to the 2D culture, the in vitro 3D cell culture models, especially the models including ECM, more closely resemble in vivo TME, better reproduce a variety of conditions such as inter-tumor heterogeneity of hypoxia in vivo, and more closely resemble a patient’s response to the therapy compared to a 2D mono-culture, as have been demonstrated in many studies.
Currently, 3D systems with tunable ECM stiffness, bio-printed 3D cell culture systems incorporating TME, systems based on 3D culture of patient’s tumor tissue, and systems utilizing decellularized ECM from the patient’s tumor have been established (82–85). Such systems have a clear potential for use in translational oncology. For example, a 3D in vitro model of pancreatic ductal adenocarcinoma (PDAC) mimicking mechanical properties of the TME potentially allows more accurately distinguish between pancreatic cancer and pancreatitis (86). A host of technologies and tools have been developed to study the impact of ECM biomechanics on a cell behavior in a variety of 3D cell culture models [comprehensively reviewed in (87)].
Three-dimensional cell culture systems also have proven to be a “biomarkers goldmine”—a valuable tool for biomarker identification (88). For example, using 3D culture model with decellularized ECM scaffolds (dECM) allowed to identify full-length Collagen VI as a driver of breast cancer cell invasion in obesity and metastasis (89). In colorectal cancer, the patterns of expression of miRNA dependent on 3D microenvironment have been characterized, and one of them (miR-142-5p) was identified as a theranostic biomarker (90). As applied to clinical scenarios, the 3D cell culture models incorporating TME are referred to as a “patient’s avatar” (91) and “patient-on-a-chip” (92) models, and can allow to identify biomarkers of individual response to the therapy. For example, patient-derived 3D organoid culture of breast tumor was utilized to choose personalized chemotherapy (93). The feasibility of the automated real-time pharmacokinetic profiling in 3D tumor models has been demonstrated (94), and 3D micro-tumor platform comprising ECM-derived hydrogel and patient-derived colorectal tumor tissue has been created for high-throughput screening of the chemotherapies in a patient-specific format (95). Further development of ECM‐mimicking scaffolds and 3D bio-printing (comprehensively reviewed in (96)) can potentially assist in personalized therapy, although it has been suggested that some 3D cell culture models are rather too complex for routine implementation in clinics at this stage (97), and clinical use of such models would require a high level of methodological (as well as clinical) validation (98). In the next few years, we expect to see a growing number of publications in this emerging field of research.
Overall, the recognition that TME is one of the drivers of malignancy (Figure 1) changes the current approach to how malignant tumors will be diagnosed and treated. Here, we emphasize that all types of the TME components depicted in Figure 1 (such as ECM and its mechanical or biological characteristics, EVs, phenotype of stromal cells, and others) have a potential to serve as biomarkers.
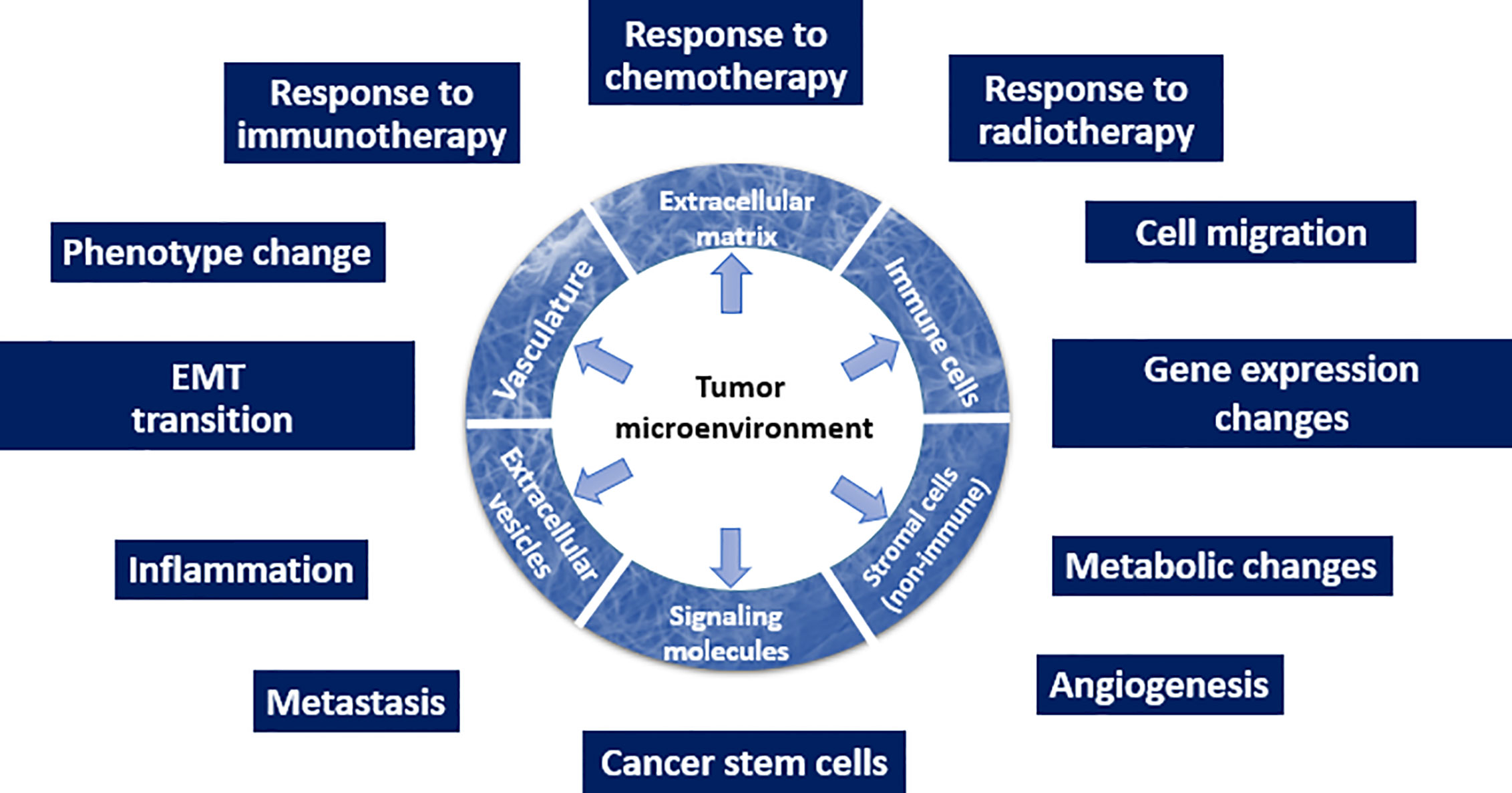
Figure 1 Schematic depiction of the TME and its multiple roles in the tumor initiation and progression.
Conclusion
ECM-derived biomarkers have a great potential in translational oncology and in clinical use. Hitherto, many novel biomarkers arising from within the ECM have been identified, although the clinical utility of many of them remains to be assessed. Based on a multitude of recent studies, we conclude that TME should be included into the in vitro and ex vivo models for cancer drug development and personalized therapy. In particular, 3D cell culture models incorporating TME and tumor-specific mechanistic characteristics of ECM, such as stiffness and topology, are more accurate and physiologically relevant models of the tumor compared to the traditional cell culture or animal xenograft models.
Author Contributions
All authors contributed to the article and approved the submitted version. Conception of the study and critical revision of the manuscript: DC, EP, IR, OM, and VA. Literature collection, analysis, and Supplementary Table preparation: ES. Literature collection, analysis, and manuscript writing: DC and EP.
Funding
This work was partially supported by the Russian Science Foundation under grant No.18-15-00391.
Conflict of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DC dedicates this mini-review to her uncle and his personal brave battle with cancer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.575569/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther (2001) 69:89–95. doi: 10.1067/mcp.2001.113989
3. Carlomagno N, Incollingo P, Tammaro V, Peluso G, Rupealta N, Chiacchio G, et al. Diagnostic, Predictive, Prognostic, and Therapeutic Molecular Biomarkers in Third Millennium: A Breakthrough in Gastric Cancer. BioMed Res Int (2017) 2017):7869802. doi: 10.1155/2017/7869802
4. Ballman KV. Biomarker: Predictive or Prognostic? J Clin Oncol: Off J Am Soc Clin Oncol (2015) 33(33):3968–71. doi: 10.1200/jco.2015.63.3651
5. Letai A. Functional precision cancer medicine-moving beyond pure genomics. Nat Med (2017) 23:1028–35. doi: 10.1038/nm.4389
6. Andriani F, Landoni E, Mensah M, Facchinetti F, Miceli R, Tagliabue E, et al. Diagnostic role of circulating extracellular matrix-related proteins in non-small cell lung cancer. BMC Cancer (2018) 18:899. doi: 10.1186/s12885-018-4772-0
7. Frezzetti D, De Luca A, Normanno N. Extracellular matrix proteins as circulating biomarkers for the diagnosis of non-small cell lung cancer patients. J Thorac Dis (2019) 11:S1252–6. doi: 10.21037/jtd.2019.02.46
8. Jia Z, Zhu J, Zhuo Y, Li R, Qu H, Wang S, et al. Offsetting expression profiles of prognostic markers in prostate tumour vs. ITS microenvironment. Front Oncol (2019) 9:539. doi: 10.3389/fonc.2019.00539
9. Attallah AM, El-Far M, El Sayes EA, Omran MM. Diagnostic role of collagen III in the diagnosis of breast cancer in Egyptian women. J Biosci Appl Res (2020) 6(1):20–9. doi: 10.21608/jbaar.2020.115764
10. Huang X, Wang W, Fu F, Kang D, Guo W, Wang C, et al. Monitoring collagen changes in tumor microenvironment using multiphoton microscopy. Proc SPIE 11553 Optics Health Care Biomed Optics X 1155321 (2020). doi: 10.1117/12.2573884
11. Macklin A, Khan S, Kislinger T. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin Proteom (2020) 17:17. doi: 10.1186/s12014-020-09283-w
12. Angel PM, Comte-Walters S, Ball LE, Talbot K, Mehta A, Brockbank KG, et al. Mapping extracellular matrix proteins in formalin-fixed, paraffin-embedded tissues by MALDI imaging mass spectrometry. J Proteome Res (2018) 17(1):635–46. doi: 10.1021/acs.jproteome.7b00713
13. Kujawa KA, Zembala-Nożyńska E, Cortez AJ, Kujawa T, Kupryjańczyk J, Lisowska KM. Fibronectin and Periostin as Prognostic Markers in Ovarian Cancer. Cells (2020) 9:149. doi: 10.3390/cells9010149
14. Wang Z, Zhou Q, Li A, Huang W, Cai Z, Chen W. Extracellular matrix protein 1 (ECM1) is associated with carcinogenesis potential of human bladder cancer. Onco Targets Ther (2019) 12:1423–32. doi: 10.2147/OTT.S191321
15. Pang X, Xie R, Zhang Z, Liu Q, Wu S, Cui Y. Identification of SPP1 as an Extracellular Matrix Signature for Metastatic Castration-Resistant Prostate Cancer. Front Oncol (2019) 9:924:924. doi: 10.3389/fonc.2019.00924
16. Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Adv Drug Deliv Rev (2016) 97:4–27. doi: 10.1016/j.addr.2015.11.001
17. Lanzi С, Yates EA, Cassinelli G. Editorial: Heparan Sulfate Proteoglycans and Their Endogenous Modifying Enzymes: Cancer Players, Biomarkers and Therapeutic Targets. Front Oncol (2020) 10:195. doi: 10.3389/fonc.2020.00195
18. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer (2020) 19(1):8. doi: 10.1186/s12943-019-1113-0
19. Shao Y, Lu B. The crosstalk between circular RNAs and the tumor microenvironment in cancer metastasis. Cancer Cell Int (2020) 20:448. doi: 10.1186/s12935-020-01532-0
20. Hynes RO, Naba A. Overview of the matrisome-An inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol (2012) 4(1):a004903. doi: 10.1101/cshperspect.a004903
21. Alkasalias T, Moyano-Galceran L, Arsenian-Henriksson M, Lehti K. Fibroblasts in the tumour microenvironment: Shield or spear? Int J Mol Sci (2018) 19(5):1532. doi: 10.3390/ijms19051532
22. Gkretsi V, Stylianopoulos T, Cell adheGkretsi V, Stylianopoulos T. Cell adhesion and matrix stiffness: Coordinating cancer cell invasion and metastasis. Front Oncol (2018) 8:145. doi: 10.3389/fonc.2018.00145
23. Hynes RO. The extracellular matrix: Not just pretty fibrils. Science (2009) 326(5957):1216–9. doi: 10.1126/science.1176009
24. Hastings JF, Skhinas JN, Fey D, Croucher DR, Cox TR. The extracellular matrix as a key regulator of intracellular signalling networks. Br J Pharmacol (2019) 176:82–92. doi: 10.1111/bph.14195
25. Nallanthighal S, Heiserman JP, Cheon D-J. The Role of the Extracellular Matrix in Cancer Stemness. Front Cell Dev Biol (2019) 7:86. doi: 10.3389/fcell.2019.00086
26. Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep (2014) 15:1243–53. doi: 10.15252/embr.201439246
27. Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumour metastasis. Biochem Soc Trans (2017) 45:229–36. doi: 10.1042/BST20160387
28. Paolillo M, Schinelli S. Extracellular Matrix Alterations in Metastatic Processes. Int J Mol Sci (2019) 20:4947. doi: 10.3390/ijms20194947
29. Venning FA, Wullkopf L, Erler JT. Targeting ECM disrupts cancer progression. Front Oncol (2015) 5:224. doi: 10.3389/fonc.2015.00224
30. Izzi V, Lakkala J, Devarajan R, Kääriäinen A, Koivunen J, Heljasvaara R, et al. Pan-Cancer analysis of the expression and regulation of matrisome genes across 32 tumour types. Matrix Biol Plus (2014) 1:100004. doi: 10.1016/j.mbplus.2019.04.001
31. Pearce OMT, Delaine-Smith R, Maniati E, Nichols S, Wang J, Böhm S, et al. Deconstruction of a metastatic tumour microenvironment reveals a common matrix response in human cancers. Cancer Discovery (2018) 8(3):304–19. doi: 10.1158/2159-8290.CD-17-0284
32. Ricard-Blum S, Salza R. Matricryptins and matrikines: Biologically active fragments of the extracellular matrix. Exp Dermatol (2014) 23:457–63. doi: 10.1111/exd.12435
33. Han XH, Caron JM, Brooks PC. Cryptic collagen elements as signaling hubs in the regulation of tumour growth and metastasis. J Cell Physiol (2020) 235(12):9005–20. doi: 10.1002/jcp.29752
34. Nielsen SH, Willumsen N, Brix S, Sun S, Manon-Jensen T, Karsdal M, et al. Tumstatin, a Matrikine Derived from Collagen Type IVα3, is Elevated in Serum from Patients with Non–Small Cell Lung Cancer. Transl Oncol (2018) 11:528–34. doi: 10.1016/j.tranon.2018.02.005
35. Zemek RM, Chin WL, Nowak AK, Millward MJ, Lake RA, Lesterhuis WJ. Sensitizing the Tumour Microenvironment to Immune Checkpoint Therapy. Front Immunol (2020) 11:223:223. doi: 10.3389/fimmu.2020.00223
36. Lim SB, Chua MLK, Yeong JPS, Tan SJ, Lim W-T, Lim CT. Pan-cancer analysis connects tumour matrisome to immune response. NPJ Precis Oncol (2019) 3:1–9. doi: 10.1038/s41698-019-0087-0
37. Mushtaq MU, Papadas A, Pagenkopf A, Flietner E, Morrow Z, Chaudhary SG, et al. Tumour matrix remodeling and novel immunotherapies: The promise of matrix-derived immune biomarkers. J Immunother Cancer (2018) 6:1–14. doi: 10.1186/s40425-018-0376-0
38. Hope C, Emmerich PB, Papadas A, Pagenkopf A, Matkowskyj KA, Van De Hey DR, et al. Versican-Derived Matrikines Regulate Batf3–Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J Immunol (2017) 199:1933–41. doi: 10.4049/jimmunol.1700529
39. Hope C, Foulcer S, Jagodinsky J, Chen SX, Jensen JL, Patel S, et al. Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood (2016) 128:680–5. doi: 10.1182/blood-2016-03-705780
40. Kodama J, Hasengaowa, Kusumoto T, Seki N, Matsuo T, Nakamura K, et al. Versican expression in human cervical cancer. Eur J Cancer (2007) 43:1460–6. doi: 10.1016/j.ejca.2007.02.007
41. Keire PA, Kang I, Wight TN. Versican: Role in cancer tumourigenesis. In: Brekken RA, Stupack DG, editors. Extracellular Matrix in Tumour Biology Cham: Springer (2017). p. 51–74. doi: 10.1007/978-3-319-60907-2_4
42. Jensen C, Madsen DH, Hansen M, Schmidt H, Svane IM, Karsdal MA, et al. Non-invasive biomarkers derived from the extracellular matrix associate with response to immune checkpoint blockade (anti-CTLA-4) in metastatic melanoma patients. J Immunother (2018) Cancer 6:152. doi: 10.1186/s40425-018-0474-z
43. Hurkmans DP, Jensen C, Koolen SLW, Aerts J, Karsdal MA, Mathijssen RHJ, et al. Blood-based extracellular matrix biomarkers are correlated with clinical outcome after PD-1 inhibition in patients with metastatic melanoma. J Immunother Cancer (2020) 8(2):e001193. doi: 10.1136/jitc-2020-001193
44. Willumsen N, Thomsen LB, Bager CL, Jensen C, Karsdal MA. Quantification of altered tissue turnover in a liquid biopsy: a proposed precision medicine tool to assess chronic inflammation and desmoplasia associated with a pro-cancerous niche and response to immuno-therapeutic anti-tumor modalities. Cancer Immunol Immunother (2018) 67(1):1–12. doi: 10.1007/s00262-017-2074-z
45. Willumsen N, Bager CL, Jensen C, Karsdal MA, Madsen DH, Hansen M, et al. 1250P Extracellular matrix and tissue derived metabolites in a liquid biopsy identifies endotypes of metastatic melanoma patients with differential response to immune checkpoint inhibitor treatment. Ann Oncol (2019) 30(Supplement_5):mdz253–075. doi: 10.1093/annonc/mdz253.075
46. Pouliquen DL, Boissard A, Coqueret O, Guette C. Biomarkers of tumor invasiveness in proteomics. Int J Oncol (2020) 57(2):409–32. doi: 10.3892/ijo.2020.5075
47. Giussani M, Triulzi T, Sozzi G, Tagliabue E. Tumor Extracellular Matrix Remodeling: New Perspectives as a Circulating Tool in the Diagnosis and Prognosis of Solid Tumors. Cells (2019) 8(2):81. doi: 10.3390/cells8020081
48. Deville SS, Cordes N. The Extracellular, Cellular, and Nuclear Stiffness, a Trinity in the Cancer Resistome—A Review. Front Oncol (2019) 9:1376. doi: 10.3389/fonc.2019.01376
49. Northcott JM, Dean IS, Mouw JK, Weaver VM. Feeling Stress: The Mechanics of Cancer Progression and Aggression. Front Cell Dev Biol (2018) 6:17:17. doi: 10.3389/fcell.2018.00017
50. Kalli M, Stylianopoulos T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front Oncol (2018) 8:55. doi: 10.3389/fonc.2018.00055
51. Stylianou А, Lekka М, Stylianopoulos T. AFM assessing of nanomechanical fingerprints for cancer early diagnosis and classification: from single cell to tissue level. Nanoscale (2018) 7;10(45):20930–45. doi: 10.1039/c8nr06146g
52. Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, et al. The nanomechanical signature of breast cancer. Nat Nanotechnol (2012) 7(11):757–65. doi: 10.1038/nnano.2012.167
53. DuChez BJ, Doyle AD, Dimitriadis EK, Yamada KM. Durotaxis by Human Cancer Cells. Biophys J (2019) 116(4):670–83. doi: 10.1016/j.bpj.2019.01.009
54. Zhang H, Lin F, Huang J, Xiong C. Anisotropic stiffness gradient-regulated mechanical guidance drives directional migration of cancer cells. Acta Biomater (2020) 106:181–92. doi: 10.1016/j.actbio.2020.02.004
55. Ondeck MG, Kumar A, Placone JK, Plunkett CM, Matte BF, Wong KC, et al. Dynamically stiffened matrix promotes malignant transformation of mammary epithelial cells via collective mechanical signaling. Proc Natl Acad Sci USA (2019) 116:3502–7. doi: 10.1073/pnas.1814204116
56. Rice AJ, Cortes E, Lachowski D, Cheung BCH, Karim SA, Morton JP, et al. Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis (2017) 6:e352. doi: 10.1038/oncsis.2017.54
57. Dong Y, Zheng Q, Wang Z, Lin X, You Y, Wu S, et al. Higher matrix stiffness as an independent initiator triggers epithelial-mesenchymal transition and facilitates HCC metastasis. J Hematol Oncol (2019) 12:112. doi: 10.1186/s13045-019-0795-5
58. McGrail DJ, Kieu QMN, Dawson MR. The malignancy of metastatic ovarian cancer cells is increased on soft matrices through a mechanosensitive Rho-ROCK pathway. J Cell Sci (2014) 127(Pt 12):2621–6. doi: 10.1242/jcs.144378
59. Tian B, Luo Q, Ju Y, Song GA. Soft Matrix Enhances the Cancer Stem Cell Phenotype of HCC Cells. Int J Mol Sci (2019) 20(11):2831. doi: 10.3390/ijms20112831
60. Vennin C, Murphy KJ, Morton JP, Cox TR, Pajic M, Timpson P. Reshaping the Tumor Stroma for Treatment of Pancreatic Cancer. Gastroenterology (2018) 154(4):820–38. doi: 10.1053/j.gastro.2017.11.280
61. Lampi MC, Reinhart-King CA. Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials. Sci Transl Med (2018) 10(422):eaao0475. doi: 10.1126/scitranslmed.aao0475
62. Palumbo А, Da Costa NM, Pontes B, de Oliveira L, Codeço ML, Pinto LFR, et al. Esophageal Cancer Development: Crucial Clues Arising from the Extracellular Matrix. Cells (2020) 9(2):455. doi: 10.3390/cells9020455
63. Novak C, Horst E, Mehta G. Review: Mechanotransduction in ovarian cancer: Shearing into the unknown. APL Bioeng (2018) 2(3):031701. doi: 10.1063/1.5024386
64. Liu C, Pei H, Tan F. Matrix Stiffness and Colorectal Cancer. Onco Targets Ther (2020) 13:2747–55. doi: 10.2147/OTT.S231010
65. Miyazawa A, Ito S, Asano S, Tanaka I, Sato M, Kondo M, et al. Regulation of PD-L1 expression by matrix stiffness in lung cancer cells. Biochem Biophys Res Commun (2017) 495(3):2344–9. doi: 10.1016/j.bbrc.2017.12.115
66. Azadi S, Aboulkheyr Es H, Bazaz SR, Thiery JP, Asadnia M, Warkiani ME. Upregulation of PD-L1 expression in breast cancer cells through the formation of 3D multicellular cancer aggregates under different chemical and mechanical conditions. Biochim Biophys Acta Mol Cell Res (2019) 1866(12):118526. doi: 10.1016/j.bbamcr.2019.118526
67. Kuczek DE, Larsen AMH, Thorseth M-L, Carretta M, Kalvisa A, Siersbæk MS, et al. Collagen density regulates the activity of tumour-infiltrating T cells. J Immunother Cancer (2019) 7:68. doi: 10.1186/s40425-019-0556-6
68. Li M, Xi N, Wang Y, Liu L. Atomic force microscopy for revealing micro/nanoscale mechanics in tumor metastasis: from single cells to microenvironmental cues. Acta Pharmacol Sin (2020). doi: 10.1038/s41401-020-0494-3
69. Li Y, Fanous MJ, Kilian KA, Popescu G. Quantitative phase imaging reveals matrix stiffness-dependent growth and migration of cancer cells. Sci Rep (2019) 9:248. doi: 10.1038/s41598-018-36551-5
70. Darling EM, Di Carlo D. High-Throughput assessment of cellular mechanical properties. Annu Rev Biomed Eng (2015) 17:35–62. doi: 10.1146/annurev-bioeng-071114-040545
71. Burke K, Brown E. The Use of Second Harmonic Generation to Image the Extracellular Matrix During Tumor Progression. Intravital (2014) 3(3):e984509. doi: 10.4161/21659087.2014.984509
72. Riegler J, Labyed Y, Rosenzweig S, Javinal V, Castiglioni A, Dominguez CX, et al. Tumor Elastography and Its Association with Collagen and the Tumor Microenvironment. Clin Cancer Res (2018) 24(18):4455–67. doi: 10.1158/1078-0432.CCR-17-3262
73. Manduca A, Oliphant TE, Dresner MA, Mahowald JL, Kruse SA, Amromin E, et al. Magnetic resonance elastography: non-invasive mapping of tissue elasticity. Med Image Anal (2001) 5(4):237–54. doi: 10.1016/s1361-8415(00)00039-6
74. Narunsky L, Oren R, Bochner F, Neeman M. Imaging aspects of the tumor stroma with therapeutic implications. Pharmacol Ther (2014) 141(2):192–208. doi: 10.1016/j.pharmthera.2013.10.003
75. Shimoda M, Khokh R. Metalloproteinases in extracellular vesicles. Biochim Biophys Acta (2017) 1864(11):Part A, 1989–2000. doi: 10.1016/j.bbamcr.2017.05.027
76. Kuhlmann JD, Chebouti I, Kimmig R, Buderath P, Reuter M, Puppel S-H, et al. Extracellular vesicle-associated miRNAs in ovarian cancer – design of an integrated NGS-based workflow for the identification of blood-based biomarkers for platinum-resistance. Clin Chem Lab Med (2019) 57:1053–62. doi: 10.1515/cclm-2018-1048
77. Huleihel L, Bartolacci JG, Dziki JL, Vorobyov T, Arnold B, Scarritt ME, et al. Matrix-Bound Nanovesicles Recapitulate Extracellular Matrix Effects on Macrophage Phenotype. Tissue Eng (2017) Part A 23:1283–94. doi: 10.1089/ten.tea.2017.0102
78. Huleihel L, Hussey GS, Naranjo JD, Zhang L, Dziki JL, Turner NJ, et al. Matrix-bound nanovesicles within ECM bioscaffolds. Sci Adv (2016) 2:e1600502. doi: 10.1126/sciadv.1600502
79. van der Merwe Y, Faust AE, Sakalli ET, Westrick CC, Hussey G, Chan KC, et al. Matrix-bound nanovesicles prevent ischemia-induced retinal ganglion cell axon degeneration and death and preserve visual function. Sci Rep (2019) 9(1):3482. doi: 10.1038/s41598-019-39861-4
80. Palviainen M, Saari H, Kärkkäinen O, Pekkinen J, Auriola S, Yliperttula M, et al. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J Extracell Vesicles (2019) 8:1596669. doi: 10.1080/20013078.2019.1596669
81. Thippabhotla S, Zhong C, He M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep (2019) 9:1–14. doi: 10.1038/s41598-019-49671-3
82. Bonnesoeur S, Morin-Grognet S, Thoumire O, Le Cerf D, Boyer O, Vannier J-P, et al. Hyaluronan-based hydrogels as versatile tumour-like models: Tunable ECM and stiffness with genipin-crosslinking. J Biomed Mater Res (2020) 108(5):1256–68. doi: 10.1002/jbm.a.36899
83. Wang C, Sinha S, Jiang X, Murphy L, Fitch S, Wilson C, et al. Matrix stiffness modulates patient-derived glioblastoma cell fates in 3D hydrogels. Tissue Eng Part A (2020). doi: 10.1089/ten.TEA.2020.0110
84. Monferrer E, Martín-Vañó S, Carretero A, García-Lizarribar A, Burgos-Panadero R, Navarro S, et al. A three-dimensional bioprinted model to evaluate the effect of stiffness on neuroblastoma cell cluster dynamics and behavior. Sci Rep (2020) 10(1):6370. doi: 10.1038/s41598-020-62986-w
85. Wishart AL, Conner SJ, Guarin JR, Fatherree JP, Peng Y, McGinn RA, et al. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci Adv (2019) 6:43, eabc3175. doi: 10.1126/sciadv.abc3175
86. Rubiano A, Delitto D, Han S, Gerber M, Galitz C, Trevino J, et al. Viscoelastic properties of human pancreatic tumors and in vitro constructs to mimic mechanical properties. Acta Biomater (2018) 67:331–40. doi: 10.1016/j.actbio.2017.11.037
87. Urbanczyk M, Layland SL, Schenke-Layland K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol (2020) 85–86:1–14. doi: 10.1016/j.matbio.2019.11.005
88. Rossetti S, Sacchi N. 3D Mammary Epithelial Cell Models: A Goldmine of DCIS Biomarkers and Morphogenetic Mechanisms. Cancers (Basel) (2019) 11:130. doi: 10.3390/cancers11020130
89. Wishart AL, Conner SJ, Guarin JR, Fatherree JP, Peng Y, McGinn RA, et al. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci Adv (2020) 6(43):eabc3175. doi: 10.1126/sciadv.abc3175
90. Kunigenas L, Stankevicius V, Dulskas A, Budginaite E, Alzbutas G, Stratilatovas E, et al. 3D Cell Culture-Based Global miRNA Expression Analysis Reveals miR-142-5p as a Theranostic Biomarker of Rectal Cancer Following Neoadjuvant Long-Course Treatment. Biomolecules (2020) 10(4):613. doi: 10.3390/biom10040613
91. Fong ELS, Toh TB, Yu H, Chow EK-H. 3D Culture as a Clinically Relevant Model for Personalized Medicine. SLAS Technol (2017) 22:245–53. doi: 10.1177/2472630317697251
92. Caballero D, Reis RL, Kundu SC. Engineering Patient-on-a-Chip Models for Personalized Cancer Medicine. In: . Biomaterials-and Microfluidics-Based Tissue Engineered 3D Models. Cham: Springer (2020). p. 43–64. doi: 10.1007/978-3-030-36588-2_4
93. Nayak B, Balachander GM, Manjunath S, Rangarajan A, Chatterjee K. Tissue mimetic 3D scaffold for breast tumour-derived organoid culture toward personalized chemotherapy. Colloids Surfaces B Biointerfaces (2019) 180:334–43. doi: 10.1016/j.colsurfb.2019.04.056
94. Joseph JF, Gronbach L, García-Miller J, Cruz LM, Wuest B, Keilholz U, et al. Automated Real-Time Tumour Pharmacokinetic Profiling in 3D Models: A Novel Approach for Personalized Medicine. Pharmaceutics (2020) 12(5):413. doi: 10.3390/pharmaceutics12050413
95. Forsythe S, Mehta N, Devarasetty M, Sivakumar H, Gmeiner W, Soker S, et al. Development of a Colorectal Cancer 3D Micro-tumour Construct Platform From Cell Lines and Patient Tumour Biospecimens for Standard-of-Care and Experimental Drug Screening. Ann Biomed Eng (2020) 48:940–52. doi: 10.1007/s10439-019-02269-2
96. Da Silva K, Kumar P, Choonara YE, du Toit LC, Pillay V. Three-dimensional printing of extracellular matrix (ECM)-mimicking scaffolds: A critical review of the current ECM materials. J BioMed Mater Res A (2020) 108(12):2324–50. doi: 10.1002/jbm.a.36981
97. Popova AA, Levkin PA. Precision Medicine in Oncology: In Vitro Drug Sensitivity and Resistance Test (DSRT) for Selection of Personalized Anticancer Therapy. Adv Ther (2020) 3(2):1900100. doi: 10.1002/adtp.201900100
98. Halfter K, Mayer B. Bringing 3D tumor models to the clinic - predictive value for personalized medicine. Biotechnol J (2017) 12(2). doi: 10.1002/biot.201600295
99. Wang X, Wang Q. Alpha-Fetoprotein and Hepatocellular Carcinoma Immunity(2018). Can J Gastroenterol Hepatol Article ID 9049252. doi: 10.1155/2018/9049252
100. Wang T-H, Hsia S-M, Shieh T-M. Lysyl Oxidase and the Tumour Microenvironment. Int J Mol Sci (2016) 18:62. doi: 10.3390/ijms18010062
101. Arcolia V, Journe F, Wattier A, Leteurtre E, Renaud F, Gabius HJ, et al. Galectin-1 is a diagnostic marker involved in thyroid cancer progression. Int J Oncol (2017) 51:760–70. doi: 10.3892/ijo.2017.4065
102. Dong R, Zhang M, Hu Q, Zheng S, Soh A, Zheng Y, et al. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy (Review). Int J Mol Med (2018) 41:599–614. doi: 10.3892/ijmm.2017.3311
103. Romagnoli M, Mineva ND, Polmear M, Conrad C, Srinivasan S, Loussouarn D, et al. ADAM 8 expression in invasive breast cancer promotes tumour dissemination and metastasis. EMBO Mol Med (2014) 6:278–94. doi: 10.1002/emmm.201303373
104. Conrad C, Benzel J, Dorzweiler K, Cook L, Schlomann U, Zarbock A, et al. ADAM8 in invasive cancers: Links to tumour progression, metastasis, and chemoresistance. Clin Sci (2019) 133:83–99. doi: 10.1042/CS20180906
105. Conrad C, Götte M, Schlomann U, Roessler M, Pagenstecher A, Anderson P, et al. ADAM8 expression in breast cancer derived brain metastases: Functional implications on MMP-9 expression and transendothelial migration in breast cancer cells. Int J Cancer (2018) 142:779–91. doi: 10.1002/ijc.31090
106. Xiao W, Wang X, Wang T, Xing J. Overexpression of BMP1 reflects poor prognosis in clear cell renal cell carcinoma. Cancer Gene Ther (2019) 27:330–40. doi: 10.1038/s41417-019-0107-9
107. Švajdler M, Mezencev R, Ondič O, Šašková B, Mukenšnábl P, Michal M. P16 is a useful supplemental diagnostic marker of pulmonary small cell carcinoma in small biopsies and cytology specimens. Ann Diagn Pathol (2018) 33:23–9. doi: 10.1016/j.anndiagpath.2017.11.008
108. Munari E, Cima L, Massari F, Bertoldo F, Porcaro AB, Caliò A, et al. Cathepsin K Expression in Castration-Resistant Prostate Carcinoma: A Therapeutical Target for Patients at Risk for Bone Metastases. Int J Biol Markers (2017) 32:243–7. doi: 10.5301/jbm.5000246
109. Banys-Paluchowski M, Loibl S, Witzel I, Mundhenke C, Lederer B, Solbach C, et al. Clinical Relevance of Collagen Protein Degradation Markers C3M and C4M in the Serum of Breast Cancer Patients Treated with Neoadjuvant Therapy in the GeparQuinto Trial. Cancers (Basel) (2019) 11:1186. doi: 10.3390/cancers11081186
110. Giussani M, Landoni E, Merlino G, Turdo F, Veneroni S, Paolini B, et al. Extracellular matrix proteins as diagnostic markers of breast carcinoma. J Cell Physiol (2018) 233:6280–90. doi: 10.1002/jcp.26513
111. Karaosmanoglu B, Kocaefe CY, Soylemezoglu F, Anlar B, Varan A, Vargel I, et al. Heightened CXCR4 and CXCL12 expression in NF1-associated neurofibromas. Child’s Nerv Syst (2018) 34:877–82. doi: 10.1007/s00381-018-3745-6
112. Järveläinen H, Sainio A, Koulu M, Wight TN, Penttinen R. Extracellular matrix molecules: Potential targets in pharmacotherapy. Pharmacol Rev (2009) 61:198–223. doi: 10.1124/pr.109.001289
113. Schaefer L, Schaefer RM. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res (2010) 339:237–46. doi: 10.1007/s00441-009-0821-y
114. Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol (2012) 196:395–406. doi: 10.1083/jcb.201102147
115. Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol (2014) 15:786–801. doi: 10.1038/nrm3904
116. Thorlacius-Ussing J, Kehlet SN, Rønnow SR, Karsdal MA, Willumsen N. Non-invasive profiling of protease-specific elastin turnover in lung cancer: biomarker potential. J Cancer Res Clin Oncol (2019) 145:383–92. doi: 10.1007/s00432-018-2799-x
117. Tada H, Hatoko M, Muramatsu T, Shirai T. Expression of E-Cadherin in Skin Carcinomas. J Dermatol (1996) 23:104–10. doi: 10.1111/j.1346-8138.1996.tb03979.x
118. Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, et al. Cathepsin D Is a Potential Serum Marker for Poor Prognosis in Glioma Patients. Cancer Res (2005) 15;65(12):5190–4. doi: 10.1158/0008-5472.CAN-04-4134
119. Zhou Q, Bauden M, Andersson R, Hu D, Marko-Varga G, Xu J, et al. YAP1 is an independent prognostic marker in pancreatic cancer and associated with extracellular matrix remodeling. J Trans Med (2020) 18:77. doi: 10.1186/s12967-020-02254-7
120. El-Khoury V, Béland M, Schritz A, Kim SY, Nazarov PV, Gaboury L, et al. Identification of beta-arrestin-1 as a diagnostic biomarker in lung cancer(2018). Br J Cancer 119:580–90. doi: 10.1038/s41416-018-0200-0
121. Morris LGT, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet (2013) 45:253–61. doi: 10.1038/ng.2538
122. Fanjul-Fernández M, Folgueras AR, Fueyo A, Balbín M, Suárez MF, Soledad Fernández-García M, et al. Erratum: Matrix metalloproteinase Mmp-1a is dispensable for normal growth and fertility in mice and promotes lung cancer progression by modulating inflammatory responses (Journal of Biological Chemistry (2018) 293:30 (11970–11970) DOI: 10.1074/jbc.AAC118.004704). J Biol Chem (2018) 293:11970. doi: 10.1074/jbc.AAC118.004704
123. Dumas V, Kanitakis J, Charvat S, Euvrard S, Faure M, Claudy A. Expression of Basement Membrane Antigens and Matrix Metalloproteinases 2 and 9 in Cutaneous Basal and Squamous Cell Carcinomas. Anticancer Res (1999) 19(4B):2929–38.
124. Vuoristo MS, Kellokumpu-Lehtinen P, Parvinen LM, Hahka-Kemppinen M, Korpela M, Kumpulainen E, et al. Serum matrix metalloproteinase-2 as a prognostic marker in advanced cutaneous melanoma. Acta Oncol (Acta Oncol) (2000), 877–9. doi: 10.1080/028418600750063659
125. Kawata R, Shinomiya T, Yasuda N, Takenaka H, Murakami Y. Matrix metalloproteinase-2 concentrations in squamous cell carcinoma of the head and neck and its clinical significance. J Otolaryngol (1996) 99:299–305. doi: 10.3950/jibiinkoka.99.299
126. Yuan X, Bian T, Liu J, Ke H, Feng J, Zhang Q, et al. Spondin2 is a new prognostic biomarker for lung adenocarcinoma. Oncotarget (2017) 8:59324–32. doi: 10.18632/oncotarget.19577
127. Duggan C, Maguire T, McDermott E, O’Higgins N, Fennelly JJ, Duffy MJ. Urokinase plasminogen activator and urokinase plasminogen activator receptor in breast cancer. Int J Cancer (1995) 61:597–600. doi: 10.1002/ijc.2910610502
128. Strojan P, Budihna M, Smid L, Svetic B, Vrhovec I, Kos J. Prognostic Significance of Cysteine Proteinases Cathepsins B and L and Their Endogenous Inhibitors Stefins A and B in Patients With Squamous Cell Carcinoma of the Head and Neck. Clin Cancer Res (2000) 6(3):1052–62.
129. Duffy MJ, Duggan C. The urokinase plasminogen activator system: A rich source of tumour markers for the individualised management of patients with cancer. Clin Biochem (2004) 37:541–8. doi: 10.1016/j.clinbiochem.2004.05.013
130. Schmidt M, Hoppe F. Increased levels of urokinase receptor in plasma of head and neck squamous cell carcinoma patients. Acta Otolaryngol (1999) 119:949–53. doi: 10.1080/00016489950180342
131. Langley RR, Carlisle R, Ma L, Specian RD, Gerritsen ME, Granger DN. Endothelial Expression of Vascular Cell Adhesion Molecule-1 Correlates with Metastatic Pattern in Spontaneous Melanoma. Microcirculation (2001) 8:335–45. doi: 10.1111/j.1549-8719.2001.tb00180.x
132. Liesche F, Griessmair M, Barz M, Gempt J, Schlegel J. ALDH1 - A new immunohistochemical diagnostic marker for Schwann cell-derived tumours. Clin Neuropathol (2019) 38:168–73. doi: 10.5414/NP301190
133. Azizidoost S, Ahmadzadeh A, Rahim F, Shahjahani M, Seghatoleslami M, Saki N. Hepatic metastatic niche: from normal to pre-metastatic and metastatic niche. Tumour Biol (2016) 37:1493–503. doi: 10.1007/s13277-015-4557-x
134. Shen Q, Nam SW. SF3B4 as an early-stage diagnostic marker and driver of hepatocellular carcinoma. BMB Rep (2018) 51:57–8. doi: 10.5483/BMBRep.2018.51.2.021
135. Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, et al. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer (2018) 18:116. doi: 10.1186/s12885-018-4006-5
136. Vicente-Munuera P, Burgos-Panadero R, Noguera I, Navarro S, Noguera R, Escudero LM. The topology of vitronectin: A complementary feature for neuroblastoma risk classification based on computer-aided detection. Tumour Markers Signatures Open Access (2020) 146(2):553–65. doi: 10.1002/ijc.32495
137. Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (United Kingdom) (2015) 7:1120–34. doi: 10.1039/c5ib00040h
138. Bozoky B, Savchenko A, Guven H, Ponten F, Klein G, Szekely L. Decreased Decorin Expression in the Tumour Microenvironment. Cancer Med (2014) 3(3):485–91. doi: 10.1002/cam4.231
139. Tada H, Hatoko M, Tanaka A, Kuwahara M, Muramatsu T. Expression of desmoglein I and plakoglobin in skin carcinomas. J Cutan Pathol (2000) 27:24–9. doi: 10.1034/j.1600-0560.2000.027001024.x
140. Jabbar KS, Arike L, Verbeke CS, Sadik R, Hansson GC. Highly accurate identification of cystic precursor lesions of pancreatic cancer through targeted mass spectrometry: A phase IIC diagnostic study. J Clin Oncol (American Soc Clin Oncol) (2018), 367–75. doi: 10.1200/JCO.2017.73.7288
141. Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, et al. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell (2004) 5:329–39. doi: 10.1016/S1535-6108(04)00081-9
142. Arnold SA, Brekken RA. SPARC: A matricellular regulator of tumourigenesis. J Cell Commun Signal (2009) 3:255–73. doi: 10.1007/s12079-009-0072-4
143. Gao D, Joshi N, Choi H, Ryu S, Hahn M, Catena R, et al. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res (2012) 72:1384–94. doi: 10.1158/0008-5472.CAN-11-2905
144. Johansen JS, Jensen BV, Roslind A, Nielsen D, Price PA. Serum YKL-40, a New Prognostic Biomarker in Cancer Patients? Cancer Epidemiol Biomarkers Prev (2006) 5(2):194–202. doi: 10.1158/1055-9965.EPI-05-0011
145. Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CMC, et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc Natl Acad Sci USA (2017) 114:E5625–34. doi: 10.1073/pnas.1707054114
146. O’Connell JT, Sugimoto H, Cooke VG, MacDonald BA, Mehta AI, LeBleu VS, et al. VEGF-A and Tenascin-C produced by S100A4 + stromal cells are important for metastatic colonization. Proc Natl Acad Sci USA (2011) 108:16002–7. doi: 10.1073/pnas.1109493108
147. Oskarsson T, Acharyya S, Zhang XHF, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med (2011) 17:867–74. doi: 10.1038/nm.2379
148. Nakayama K, Seike M, Noro R, Takeuchi S, Matsuda K, Kunugi S, et al. Tenascin XB is a novel diagnostic marker for malignant mesothelioma. Anticancer Res (2019) 39:627–33. doi: 10.21873/anticanres.13156
149. Yang Z, Zhang C, Qi W, Cui C, Cui Y, Xuan Y. Tenascin-C as a prognostic determinant of colorectal cancer through induction of epithelial-to-mesenchymal transition and proliferation. Exp Mol Pathol (2018) 105:216–22. doi: 10.1016/j.yexmp.2018.08.009
Keywords: biomarkers, cancer, tumor microenvironment, extracellular matrix, extracellular vesicles, 3D cell culture, personalized therapy
Citation: Petersen EV, Chudakova DA, Skorova EY, Anikin V, Reshetov IV and Mynbaev OA (2020) The Extracellular Matrix-Derived Biomarkers for Diagnosis, Prognosis, and Personalized Therapy of Malignant Tumors. Front. Oncol. 10:575569. doi: 10.3389/fonc.2020.575569
Received: 23 June 2020; Accepted: 10 November 2020;
Published: 18 December 2020.
Edited by:
Laura Rosanò, Italian National Research Council, ItalyReviewed by:
Etienne Becht, Fred Hutchinson Cancer Research Center, United StatesKexin Xu, The University of Texas Health Science Center at San Antonio, United States
Copyright © 2020 Petersen, Chudakova, Skorova, Anikin, Reshetov and Mynbaev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena V. Petersen, cGV0ZXJzZW4uZWxlbmEudkBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Elena V. Petersen
Elena V. Petersen Daria A. Chudakova
Daria A. Chudakova Ekaterina Yu. Skorova
Ekaterina Yu. Skorova Vladimir Anikin3,4
Vladimir Anikin3,4 Ospan A. Mynbaev
Ospan A. Mynbaev