- 1Laboratory of Molecular Medicine, Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of Russian Academy of Sciences, Novosibirsk, Russia
- 2Department of Mammology, Novosibirsk Regional Clinical Oncological Dispensary, Novosibirsk, Russia
- 3Probe and Electron Microscopy Resource Centre, National Research Center Kurchatov Institute, Moscow, Russia
- 4Institute of Nano-, Bio-, Information, Cognitive and Socio-humanistic Sciences and Technologies, Moscow Institute of Physics and Technology, Moscow, Russia
- 5Electron Microscopy Laboratory, Shubnikov Institute of Crystallography of Federal Scientific Research Centre “Crystallography and Photonics” of Russian Academy of Sciences, Moscow, Russia
- 6Laboratory of Protein Biosynthesis, Petersburg Nuclear Physics Institute Named by B.P. Konstantinov of National Research Center “Kurchatov Institute”, Gatchina, Russia
- 7Laboratory of Proteomics and Metabolomics, International Tomography Center, Siberian Branch of Russian Academy of Sciences, Novosibirsk, Russia
- 8Department of Molecular Biology and Biotechnology, Novosibirsk State University, Novosibirsk, Russia
Exosomes are directly involved in governing physiological and pathological processes of an organism by horizontal transfer of functional molecules (proteins, microRNA, etc.) from producing to receiving cells. We explored the relationship of proteins from plasma exosomes, and exosomes from the total blood of healthy females (HFs) and breast cancer patients (BCPs), with crucial steps of tumor progression: EMT, cell proliferation, invasion, cell migration, stimulation of angiogenesis, and immune response. A proteomic analysis of exosomes isolated from samples using ultrafiltration and ultracentrifugation was performed. Their nature has been verified using cryo-electron microscopy and flow cytometry. Bioinformatics analysis showed that 84% of common exosomal proteins were of cytoplasmic and vesicle origin. They perform functions of protein binding and signaling receptor binding, and facilitated the processes of the regulated exocytosis and vesicle-mediated transport. Half of the identified exosomal proteins from blood of HFs and BCPs are involved in crucial steps of the tumor progression: EMT, cell proliferation, invasion, cell migration stimulation of angiogenesis, and immune response. Moreover, we found that protein cargo of exosomes from HF total blood was enriched with proteins inhibiting EMT, cell migration, and invasion. Tumor diagnostic/prognostic protein markers accounted for 47% of the total composition of cell-surface-associated exosomes (calculated as the difference between the total blood exosomes and plasma exosomes) from BCP blood. Breast cancer-associated proteins were equally represented in the blood cell-surface-associated exosomes and in the plasma exosomes from BCPs. However, hyper-expressed proteins predominate in the blood cell-surface-associated exosomes as compared to the plasma exosomes (64 vs. 14%). Using breast cancer proteins data from the Human Protein Atlas (HPA) (www.proteinatlas.org/), three favorable (SERPINA1, KRT6B, and SOCS3), and one unfavorable (IGF2R) prognostic protein markers were found in the BCP total blood exosomes. Identified exosomal proteins from BCP blood can be recommended for further testing as breast cancer diagnostic/prognostic biomarkers or novel therapeutic targets.
Introduction
Cancer is a complex disease in which cells with tumorigenic traits (i.e., unregulated cell proliferation and resistance to cell death) have an ability to communicate with neighboring cells to initiate and facilitate the tumor progression. Although malignant cells themselves are the major source of tumorigenesis, their interactions with the tumor microenvironment are critical for the progression from a single tumor mass to distant metastases (1). It was shown that exosomes carry messages (proteins, microRNA, etc.) from transformed to healthy cells or to other cells in the tumor or they may signal in an autocrine manner back to the producing tumor cells causing changes in the recipient cells behavior and microenvironment alterations (2). Notably, exosomes may deliver signals to induce EMT, migration, invasion, angiogenesis, and metastasis processes (2–6). It has been shown blood circulating exosomes contain two fractions of free and cell-surface-associated vesicles (7, 8). Thus, the transport of vesicles is provided by both plasma exosomes and blood cell-surface-associated exosomes. It is known that plasma exosomes and cell-bound exosomes from the blood of cancer patients contain tumor-specific microRNAs (7) and proteins (9). Nonetheless, most studies of the involvement of blood exosomes in carcinogenesis focus on the study of plasma exosomes only. However, another fraction of exosomes—blood cell-surface-associated exosomes—is similarly noteworthy (7, 9).
The goal of the present study is to shed light on the differences between the proteomic cargos of free-circulating and total blood exosomes of healthy females (HFs) and breast cancer patients (BCPs), and to identify exosomal proteins involved in the crucial steps of the tumor progression: EMT, cell proliferation, invasion, cell migration stimulation of angiogenesis, and immune response.
Materials and Methods
Ethics Statement
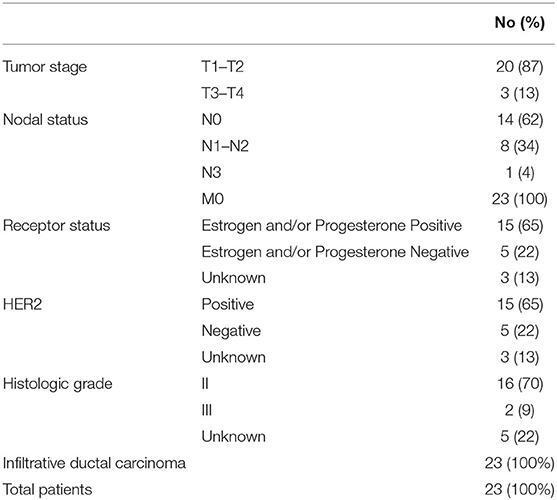
The study protocol was approved by the Ethics Committee of the Institute of Chemical Biology and Fundamental Medicine. Human samples were obtained according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from every female. Blood samples from HFs (n = 21, age 38–65 years, and median age 56) were obtained from Novosibirsk Central Clinical Hospital. HFs did not have any female-related disorders (mastopathy, endometriosis, etc.) or malignant diseases. Blood samples from previously untreated BCPs (n = 23, age 31–78 years, and median age 58) were obtained from Novosibirsk Regional Oncology Dispensary (Table 1).
Exosome Isolation
Venous blood (9 mL) was collected in K3EDTA vacutainers (Improvacuter, China, cat. no. 694091210), immediately mixed using a rotary mixer, placed at +4°C and processed within an hour after blood taking.
The blood sample was divided into two equal parts. One part was used for the isolation of plasma exosomes, and the second—for the isolation of total blood exosomes. To isolate plasma exosomes, blood cells were pelleted by centrifugation at 290 g for 20 min, blood plasma was transferred to a new tube and centrifuged a second time at 1,200 g for 20 min. To remove cell debris, plasma samples were centrifuged at 17,000 g at 4 °C for 20 min.
Plasma supernatants were diluted in PBS (P4417, Sigma, USA) with 5 mM EDTA in a 1:5 ratio, passed through a 100 nm pore-size filter (Minisart high flow, 16553-K, Sartorius, Germany), and the filtrates were centrifuged for 90 min at 100,000 g (4°C). Pellets were suspended in 12 mL PBS and again centrifuged for 90 min at 100,000 g (4°C). This washing stage was performed three times. Then supernatants were removed, and the pellets were resuspended in 150 μL PBS.
To isolate total blood exosomes, a previous protocol was used (9). Briefly, equal volumes of elution buffer (BioSilica Ltd., Russia) were added to blood samples, and incubated on a rotary mixer (10 rpm) for 4 min at room temperature. Total blood exosomes were isolated using the same procedures as for plasma exosome isolation.
For cryo-electron microscopy (cryo-EM) study 1/10 of the exosome samples from different individuals were mixed to generate two samples (plasma exosomes and total blood exosomes) from HFs and two samples (plasma exosomes and total blood exosomes) from BCPs. Individual and mixed samples were frozen in liquid nitrogen and stored in aliquots at −80°C until required.
Cryo-Electron Microscopy
Morphology of the isolated exosomes was assessed by cryo-EM using Titan Krios 60–300 TEM/STEM (Thermo Fisher Scientific, USA), equipped with TEM direct electron detector Falcon II (Thermo Fisher Scientific, USA) and Cs image corrector (CEOS, Germany) at accelerating voltage of 300 kV as described (10). The initial volumes of plasma or blood for the study of exosomes using the cryo-EM were 10 μL.
Protein Quantification
To evaluate the protein concentration of exosomes, a NanoOrange Protein Quantitation kit (NanoOrange® Protein Quantitation Kit, Molecular Probes, USA) was used in accordance with the manufacturer's recommendations.
Flow Cytometry Analysis
Flow cytometry was performed on the Cytoflex (Becman Coulter, USA), using CytExpert 2.0 Software. The MFI of stained exosomes was analyzed as described (11) and compared to the isotype control (BD bioscience, USA).
Mass Spectrometry Analysis
Individual exosome samples were separated according to their molecular weight using SDS disc electrophoresis. The gels were stained by Coomassie Brilliant Blue R250 (Sigma, United States). The PAAG fragments containing proteins under study were treated using the modified Rosenfeld method. Briefly, PAAG fragments with proteins were washed from Coomassie R250 and SDS with an aqueous solution containing 50% acetonitrile and 0.1% trifluoroacetic acid. Proteins absorbed in the gel were reduced with 45 mM DTT in 0.2 M ammonium bicarbonate at 60°C for 30 min, followed by protein alkylation with 100 mM iodoacetamide in 0.2 M ammonium bicarbonate, at room temperature for 30 min. The gel fragments were dehydrated in 100% acetonitrile. A 0.2 mM trypsin solution (modified by reductive methylation) (Sigma, T6567, USA), in a mixture with 0.1 M ammonium bicarbonate and 5 μM CaCl2, was added to each gel fragment and incubated for 30 min at room temperature. Then, peptide extraction buffer (60 μL) was added to the gel fragments, and samples were incubated for 16–18 h at 37°C. The peptide fragments of exosomal proteins extracted from the gel, concentrated, and desalted as described (9).
Mass-spectra were registered at the Center of Collective Use “Mass spectrometric investigations” SB RAS on an Ultraflex III MALDI-TOF/TOF mass spectrometer (BrukerDaltonics, Germany) as described (9). Exosomal proteins were identified by searching for appropriate candidates in annotated NCBI and SwissProt databases using Mascot software (Matrix Science Ltd., London, www.matrixscience.com/search_form_select.html).
Analysis of Exosomal Proteome Cargoes
Venn diagram illustrating common proteins in exosome samples from plasma and total blood was drawn using a FunRich version 3.1.3 software package using Vesiclepedia function and current versions of FunRich and Vesiclepedia databases (12). GO functional enrichment analysis of all exosomal fractions was performed using STRING software (www.string-db.org) with the base settings for multiple proteins analysis. The redundancy reduction of the obtained data was conducted using REVIGO software (https://www.irb.hr/) with the allowed similarity of <0.5.
Heatmapping of the enrichment results obtained through STRING was conducted via R (https://www.R-project.org), using packages gplots, dendextend, colorspace.
GO profiling of exosomal proteins involved in the cell migration and motility, immune response, vasculature development, and cell proliferation were performed using the QuickGO annotation terms (lists of obtained proteins were searched against GO terms: cell motility (GO:0048870), cell migration (GO:0016477), negative regulation of cell motility (GO:2000146), immune response (GO:0006955), negative regulation of immune response (GO:0050777), vasculature development (GO:0001944) negative regulation of vasculature development (GO:1901343), cell population proliferation (GO:0008283) and negative regulation of cell population proliferation (GO:0008285) (13). The involvement of exosomal proteins in cancer invasion and EMT were routinely analyzed by searching the PubMed database for relevant publications for each protein.
The relationship of exosomal proteins involved in key stages of tumor development was analyzed using Reactome (www.reactome.org) and NCI-Nature Pathway Interaction Database (www.ndexbio.org) by FunRich (http://www.funrich.org/).
Profiling of the exosomal proteins differently expressed during the development of various malignant diseases was performed using the dbDEPC 3.0 database (14). The search for cancer prognostic proteins in exosomal proteomes was conducted using Human Protein Atlas datasets (http://www.proteinatlas.org/) for breast, renal, thyroid, pancreatic, liver, endometric, head and neck, ovarian, stomach urothelial, cervical, lung and colorectal cancers as well as melanoma, and glioma datasets.
Statistical Analysis
Statistical calculations were performed using Statistica 6.0 software and GraphPad PRISM 5 software (GraphPad Software, La Jolla, CA, USA). All data were expressed either as the median with interquartile ranges or as means with standard errors. To evaluate the differences, the Mann–Whitney U-test was performed. P < 0.05 were considered statistically significant.
Results
Characterization of Exosomes Isolated From Plasma and Total Blood
The morphology of exosomes from plasma and total blood of HFs and BCPs was examined by cryo-EM. In total, the following sets of images have been analyzed in the present work: 100 images of vesicles from plasma and 116 images of vesicles from total blood of HFs (Figures 1A,B); 121 images of vesicles from plasma, and 117 images of vesicles from total blood of BCPs (Figures 1C,D). More than 90% of the extracellular vesicles were identified as exosome-like vesicles due to the clear presence of the lipid bilayer/membrane. Most of the vesicles were intact and had round or slightly elongated shapes. Single (Figures 1A–F), double (Figures 1A–D,G,H), double membrane (Figures 1I–L), and multilayer exosome-like vesicles (Figures 1A–D,I,J) were found in all samples of isolated vesicles from pooled plasma and pooled total blood of donors and patients. Protein aggregates outside EVs were also visualized. It should be noted that single vesicles were predominantly represented in the HF plasma, while in the total blood vesicles their content decreased (from 91 to 37%) with an increase of double vesicles (from 3 to 20%) and double-membrane vesicles (from 0 to 22%) fractions (Supplementary Table 1). The morphology of vesicles isolated from plasma and from the total blood of BCPs was comparable with the prevailing of single vesicles (81 and 70%, respectively; Supplementary Table 1).
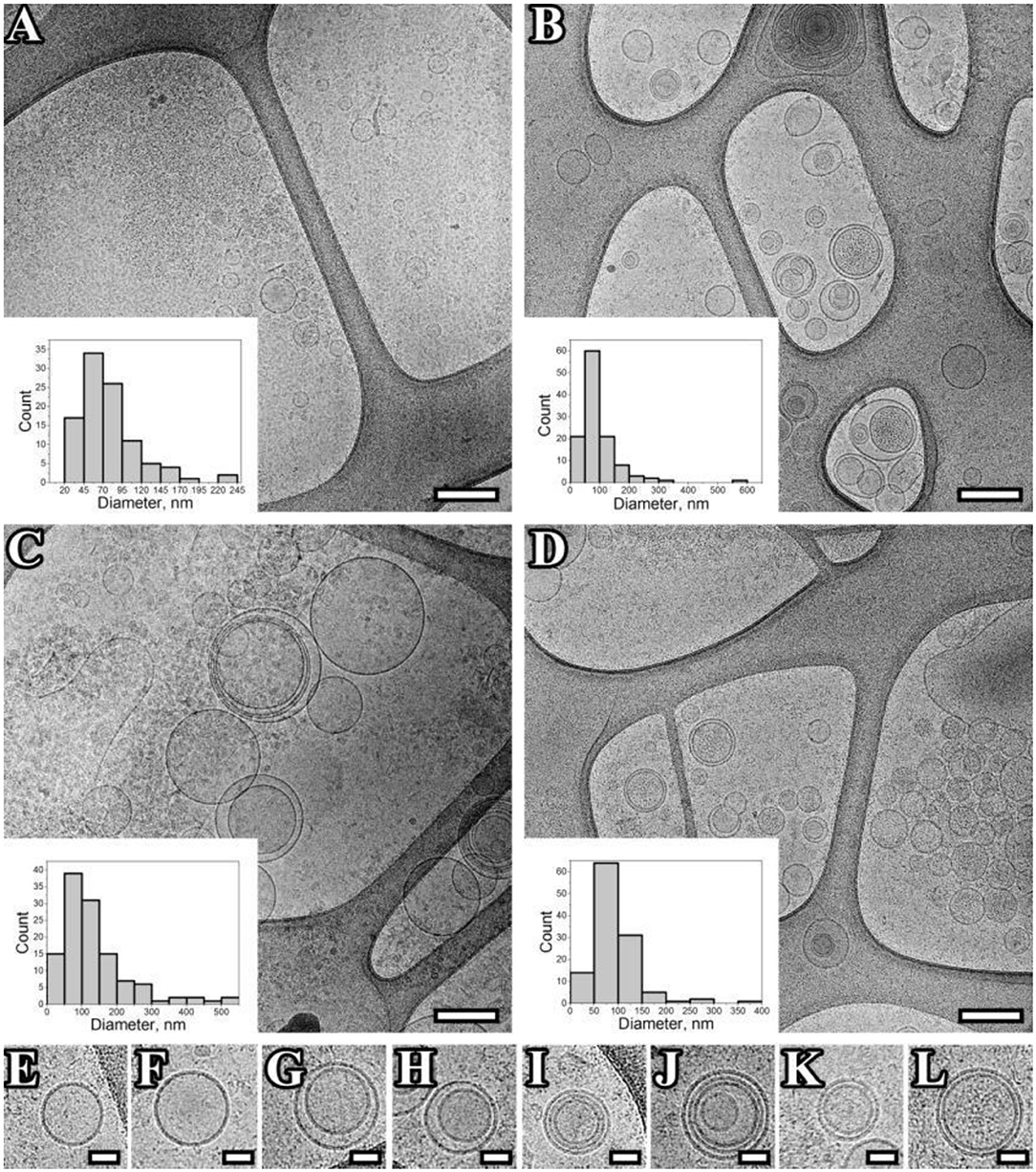
Figure 1. Representation of exosomes with various morphology. (A–L), Cryo-EM images of exosomes isolated from pooled samples of plasma of HFs (A), total blood of HFs (B), plasma of BCPs (C), total blood of BCPs (D), single vesicles (E,F), double vesicles (G,H), multilayer vesicles (I,J), double-membrane vesicles (I–K), vesicles with electron dense cargo in lumen (G,L). Scale bars are 500 nm for micrographs (A–D), and 100 nm for micrographs (E–L).
Exosome-like vesicles from plasma of BCPs were significantly larger compared to HFs (135 ± 9 and 78 ± 4 nm, respectively, p < 0.0001); however, the exosome-like vesicles from total blood had similar sizes (91 ± 5 and 92 ± 5 nm, respectively). It should be noted that the total blood exosomes consist of plasma exosomes and blood cell-surface-associated exosomes. Thus, the obtained data indirectly indicate that in HF blood, the blood-cell-surface associated exosome-like vesicles are larger than in plasma (p < 0.0322), and in BCP blood it is vice versa (p = 0.0007).
The isolated exosome-like vesicles were also inspected for the presence of exosomal markers (CD9, CD24, CD63, and CD81) using flow cytometry (Table 2). A subpopulation of CD9/CD24-positive exosome-like vesicles predominated in both plasma and total blood of HFs and BCPs. No significant differences in the MFI of populations of CD9/CD24-positive, CD9/CD63-positive, and CD9/CD81-positive exosome-like vesicles between plasma and total blood of HFs and BCPs were found.
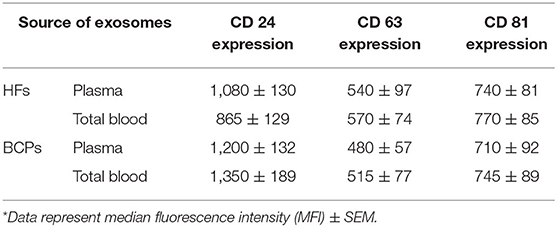
Table 2. Expression of CD 24, CD 63, and CD 81 on the surface of CD9-positive exosomes from plasma and total blood of HFs and BCPs*.
Collectively, obtained data reveal that the extracellular vesicles isolated from plasma and total blood have all properties of exosomes.
Annotation of Identified Exosomal Proteins
A total of 111 and 146 proteins were identified with high confidence (P < 0.05) using mass spectrometry in the blood-circulating exosomes from HFs and BCPs, respectively (Supplementary Tables 2–5). Of these, only 15% (34 proteins) were common for both groups (Figure 2). The percentage of proteins detected exclusively in the total blood exosomes of HFs and BCPs were 27 and 49%, respectively (Figure 2). To determine whether these proteins are associated with exosomes, the protein list was analyzed using Funrich to search against publicly available vesicle protein databases—Vesiclepedia and Exocarta. According to the analysis of the Vesiclepedia database (www.microvesicles.org), of the top 163 proteins associated with vesicles, a total of 80 and 114 proteins were identified in the HF and BCP exosomes, respectively (Figure 3A). Thus, about 27% of exosomal proteins are identified in our study for the first time as a part of extracellular vesicles; previously, they were not annotated in this database. Moreover, the analysis of the identified exosomal proteins by Exocarta database (www.exocarta.org) reveals that among 100 proteins that have been previously found to be associated with vesicles, a total of 59 and 72 proteins were identified in the HF and BCP exosomes, respectively. Thus, 55% of proteins identified in exosomes in this study were not previously annotated in the Exocarta database (Figure 3B).
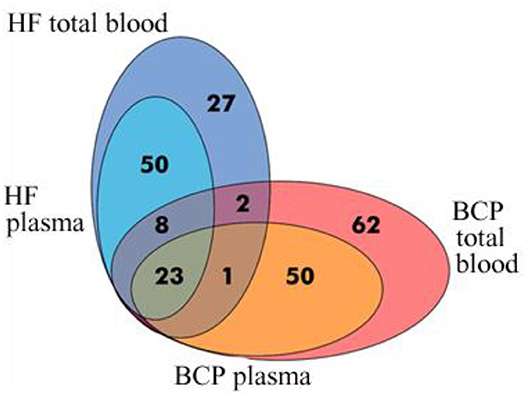
Figure 2. Venn-Euler diagram of proteins in exosomes from HF and BCP blood, composed using FunRich software.
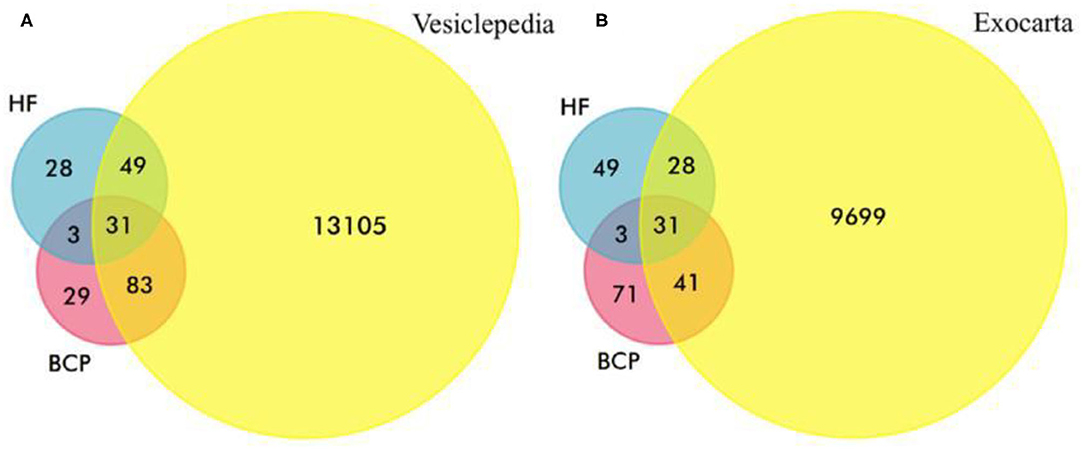
Figure 3. Venn-Euler diagrams of exosomal proteins from HFs and BCPs (A) with Vesiclepedia and (B) Exocarta databases.
Functional enrichment analysis of 32 common exosomal proteins was performed using STRING software (www.string-db.org), and the enriched GO terms for cellular components, molecular functions, and biological processes were highlighted (Figure 4). It was shown that 84% of exosomal proteins were of cytoplasmic and vesicle origin (Figure 4A). They perform functions of the protein binding and signaling receptor binding (Figure 4B), and facilitate processes of the regulated exocytosis and vesicle-mediated transport (Figure 4C). This highlights the vesicular nature of the majority of common proteins. The protein-protein interaction was mainly concentrated in the relevance among APOB, APOA1 and APOA4, FGA, TF, HPX, FGG, HP, C3, ALB, AHSG, FGB, TTR, HPR, and CLU (Figure 4).
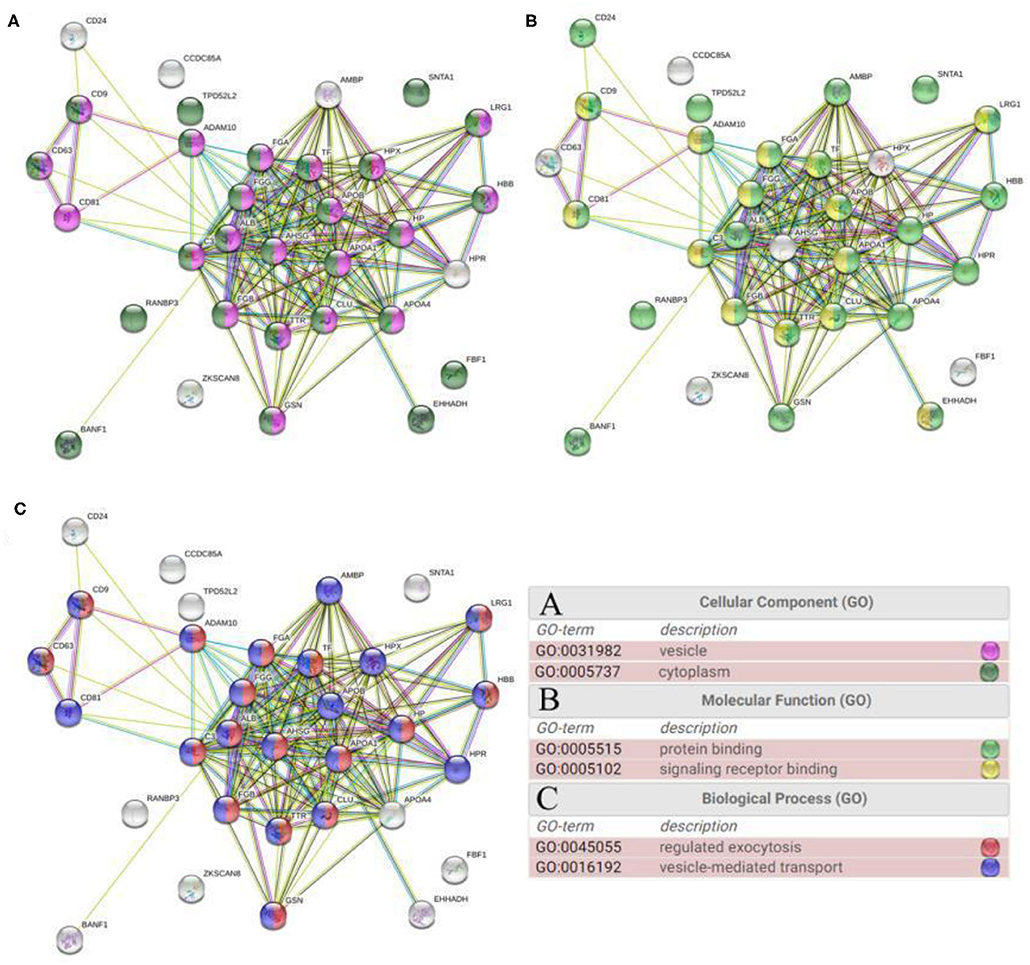
Figure 4. GO functional enrichment analysis of common proteins from total blood exosomes performed using STRING software: (A) cellular component, (B) molecular function, and (C) biological process.
Functional enrichment analysis was performed for all proteins from exosomes from plasma and total blood of HFs and BCPs using STRING software (www.string-db.org). The obtained lists of enriched 98 Cellular Components, 48 Molecular Functions, and 459 Biological Processes GO terms with corresponding p-values were clustered and visualized with heatmaps using R.
The universal terms of cellular components in exosomes included “vesicle” and “endocytic vesicle lumen,” “extracellular region part,” and “extracellular space.” It should be noted that exosomal proteins from HF blood exclusively have only five common GO with lg p > 5 (“fibrinogen complex,” “supramolecular fiber,” “platelet dence granule,” “endocytic vesicle membrane,” and “plasma membrane”). However, the total GO amount with lg p > 4 is 11 in these fractions. Only the exosomes from blood of BCPs were enriched for “cytoplasm,” “intracellular part,” and “perinuclear region of cytoplasm” (Figure 5). Moreover, the Molecular Function GO analysis revealed that the common exosomal proteins are involved in “protein binding,” “hemoglobin binding,” “cholesterol transporter activity,” “identical protein binding,” and “lipoprotein particle receptor binding.” The “heparin binding,” “cell adhesion molecule binding,” “peptidase regulator activity,” “macromolecular complex binding,” “chaperone binding,” and “cholesterol binding” were the most common terms for both plasma and total blood of HF exosomes, while “oxygen binding,” “oxygen transporter activity,” “tetrapyrrole binding,” “microtubule binding,” and “small molecule binding” were the most common for plasma and total blood of BCP exosomes (Figure 6).
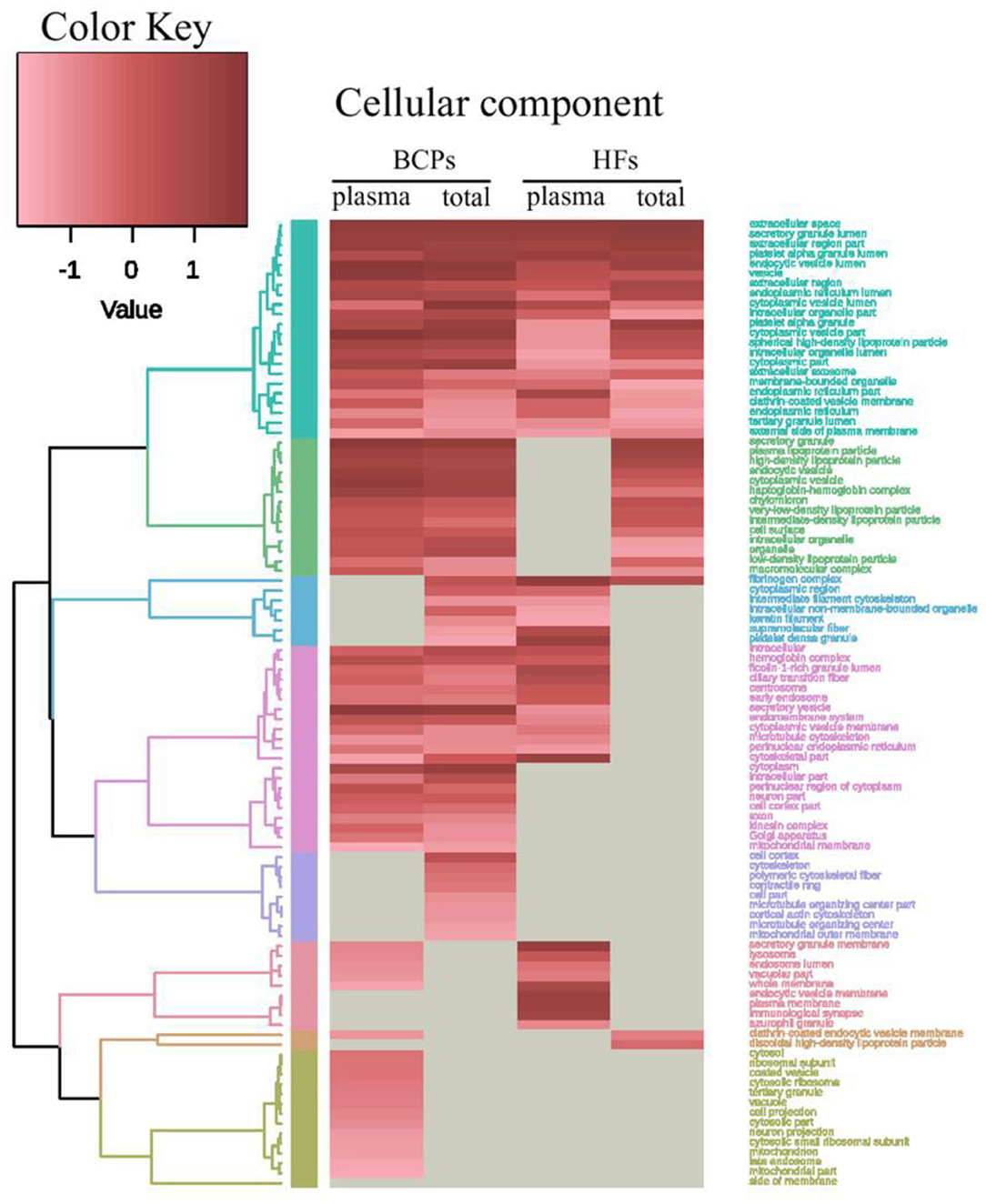
Figure 5. Heat map and dendrogram of the results of GO functional enrichment analysis of exosomal proteins from BCPs and HFs by Cellular Components obtained via STRING (https://string-db.org/) and visualized using R (https://www.r-project.org/).
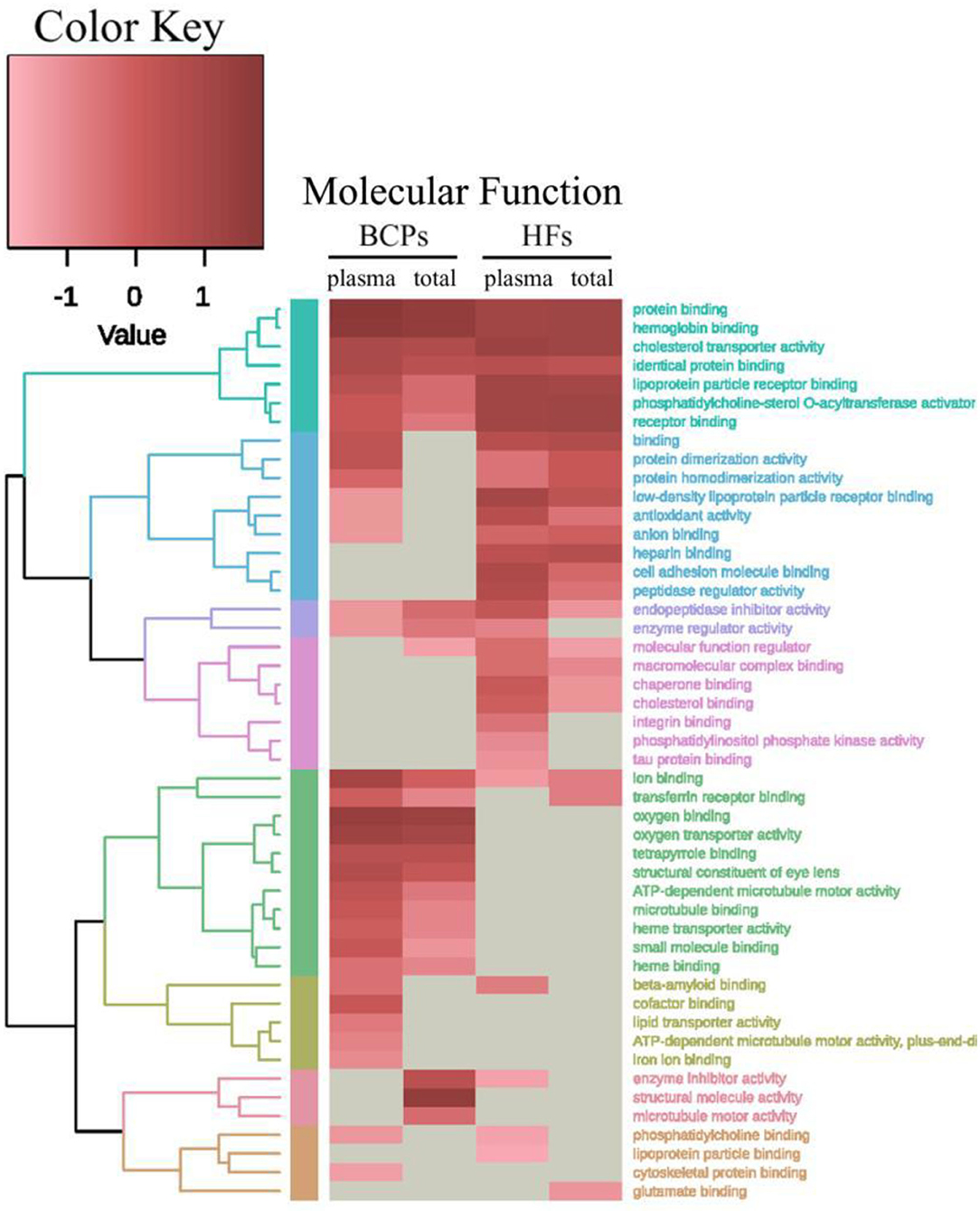
Figure 6. Heat map and dendrogram of the results of GO functional enrichment analysis of exosomal proteins from BCPs and HFs by Molecular Functions obtained via STRING (https://string-db.org/) and visualized using R (https://www.r-project.org/).
Although the results of the enrichment analysis regarding Biological Processes provided a detailed and broad picture, due to the number of resultant terms it was decided to put it through the REVIGO software (https://www.irb.hr/) before R visualization to summarize the GO list and to remove the redundant terms (Figure 7, Supplementary Figure 1). The common proteomic profiles of exosomes from blood of HFs and BCPs are enriched with “endocytosis,” “vesicle-mediated transport,” “cell activation,” “extracellular structure organization,” and “regulation of transport” terms. Some terms were enriched exclusively in total blood fractions (that is, only in blood cell-surface-associated exosomes) of both HF and BCP exosomal proteins: “regulation of blood vessel size” and “plasminogen activation.” Proteins from BCP exosomes are involved in biological processes such as “response to hypoxia,” “protein stabilization,” “membrane organization,” etc. HF exosomes, on the other hand, were enriched by “regulation of body fluid levels,” “protein metabolic process,” “transport,” “regulation of cell communication,” “secretion by cell,” etc.
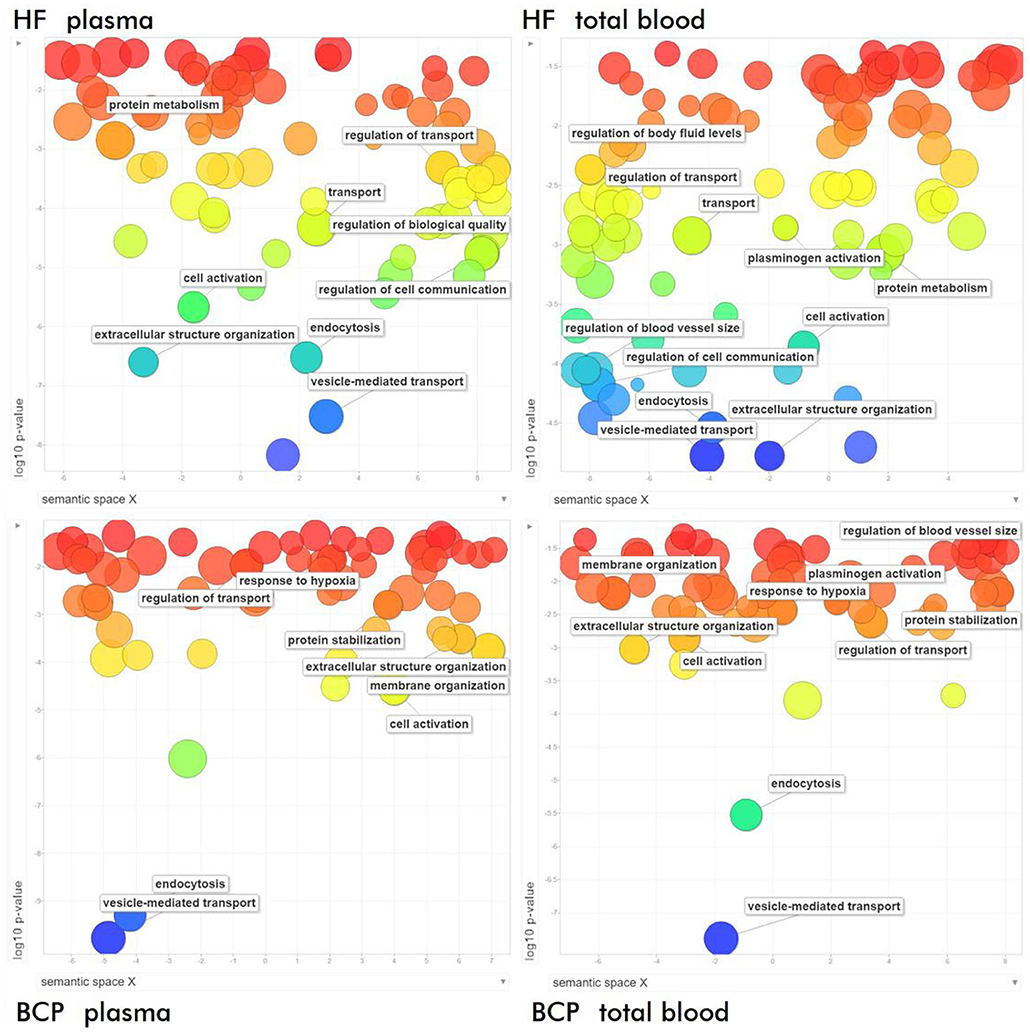
Figure 7. Semantic similarity-based scatterplots of the results of GO functional enrichment analysis of exosomal proteins from BCPs and HFs by Biological Processes obtained via STRING (https://string-db.org/) after redundancy reduction via REVIGO (https://www.irb.hr/).
Characterization of Exosomal Proteins Involved in Malignant Neoplasms Development
Literature analysis and QuickGO annotation of the exosomal cargo revealed that many proteins are involved in the crucial steps of the tumor progression: epithelial-to-mesenchymal transition (EMT), cell proliferation, invasion, cell migration, angiogenesis, and immune response (Table 3). In particular, a comparable amount of exosomal proteins from total blood of HFs and BCPs (32 (29%) and 35 (24%), respectively; 14 (29%) of which were common to both groups) are involved in EMT. However, the share of the EMT negative regulation proteins in control was two times higher than in disease (22 vs. 11%). Only 14 (13%) proteins from total blood HFs were involved in the cell proliferation, of which several proteins (APOE, CDC73, CHD5, FGF10, GDF2, and LMNA) inhibited this process. It is noteworthy that in the BCPs exosome proteome, only 6 (4%) of proteins regulating the cell proliferation were presented, of which four were the process inhibitors. Also, 51 (46%) proteins were involved in the invasion-associated functions in the total blood exosomes from HFs, and 54 (37%) such proteins were revealed in the total BCP blood exosomes, 18 (21%) of which were common for both groups. In the proteome cargo of HF exosomes, 6 (12%) proteins were engaged with the invasion suppression, while only IGF2R and KRT1 (4%) were detected in the exosomes from BCP blood. Moreover, 25 (23%) and 21 (14%) proteins involved in the regulation of the cell migration were revealed in HFs and BCPs total blood exosomes, respectively. GO analysis using QuickGO revealed that four (AKT1, APOE, BARD1, and GDF2) out of 16 unique proteins from exosomes from HF blood were associated with the negative migration regulation of endothelial and lymphocytic cells (15–17). In the vasculature development, 11 (10%) and 6 (4%) proteins of exosomal cargoes from total blood HFs and total blood BCPs were detected, of which only one inhibitor was identified in each group (GDF2 and BMP7, respectively). An equal amount of proteins (30 each) was involved in the regulation of the immune response in the total blood exosomes from HFs and BCPs, the number of the inhibitors in both cases was also equal (3 each) (Table 3). Thus, it turned out that the protein cargo of exosomes from HF blood was enriched by proteins for the negative regulation of EMT, cell proliferation, migration, and invasion. FunRich pathway analysis was used to visualize the interaction network of these proteins. The PPI was mainly concentrated in the relevance among TRAIL signaling pathway (Reactome: R-HSA-75158), Hemostasis (Reactome: R-HSA-109582), and Integrin family cell surface interactions (Reactome: R-HSA-216083) (Figure 8).
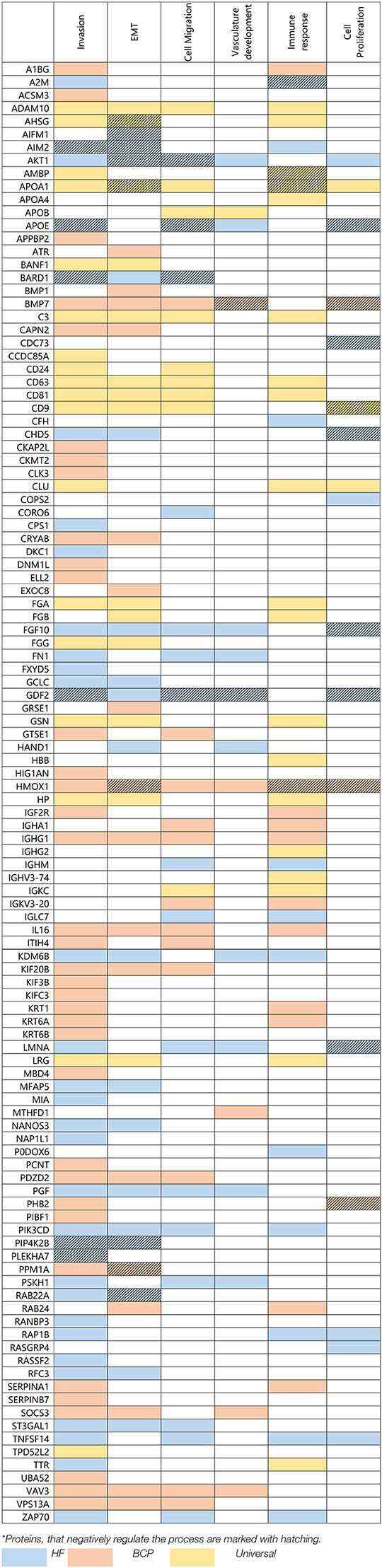
Table 3. Exosomal proteins from HFs and BCPs blood associated with crucial steps of tumor progression*.
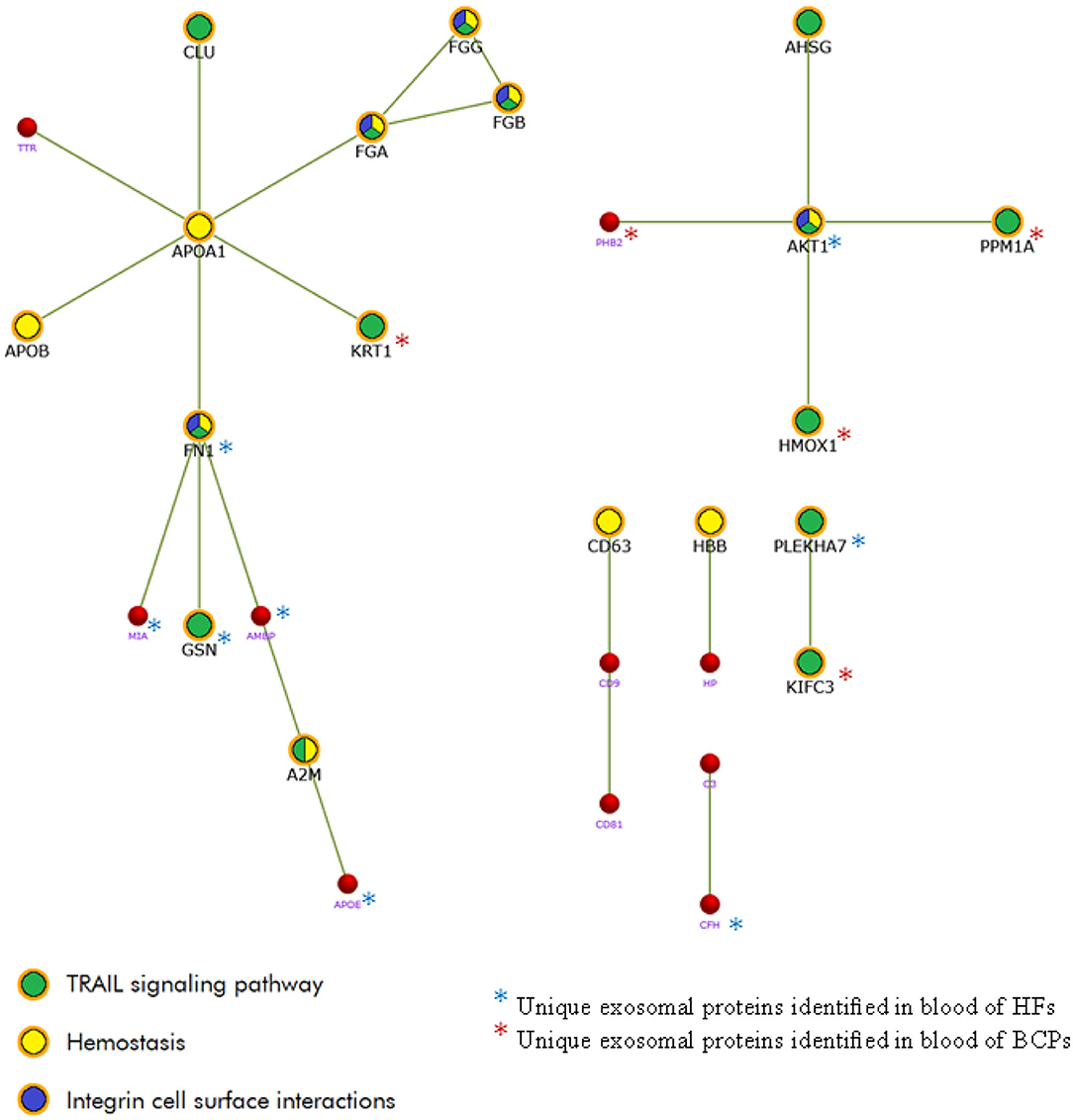
Figure 8. The PPI network of the exosomal proteins involved in key stages of tumor development was constructed using FunRich 3.1.3. (http://www.funrich.org/).
Using the dbDEPC 3.0 database (database of Differently Expressed Proteins in Human Cancer) (14) it was revealed that proteins associated with the breast cancer development were equally represented in the blood cell-surface-associated exosomes, calculated as the difference between the total blood exosomes and plasma exosomes, and in the plasma exosomes from BCPs [11 (15%) and 14 (19%) proteins, respectively] (Figure 9). However, the hyper-expressed proteins predominate in the blood cell-surface-associated exosomes as compared to the plasma exosomes (64 vs. 14%).
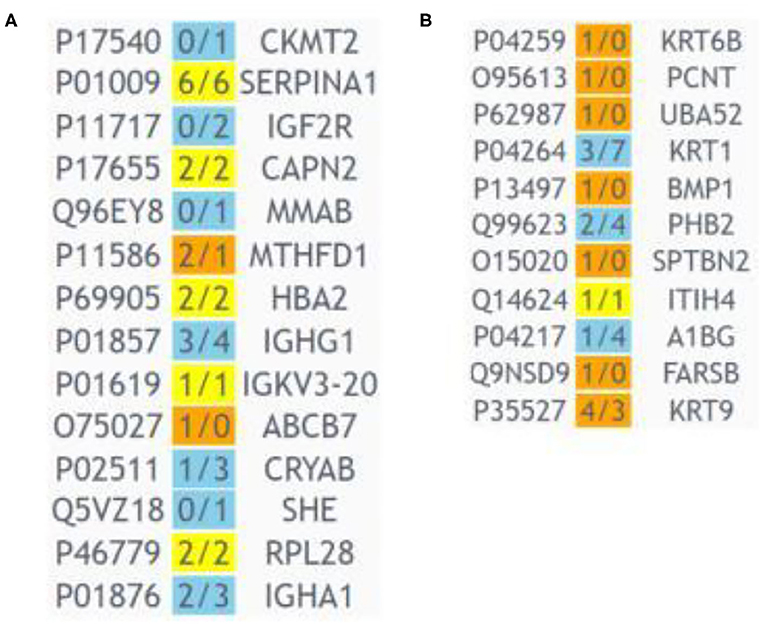
Figure 9. Heat map generated by dbDEPC 3.0 (14) from exosomal proteins from BCP blood, associated with breast cancer. (A) Proteins from plasma exosomes, (B) proteins from blood cell-surface-associated exosomes. Orange means that the number of the studies identified this protein as up-regulated is more than the number of the studies identified the protein as down-regulated; blue means that the number of the studies identified this protein as up-regulated is less than the number of the studies identified protein as down-regulated; yellow means that the number of the studies identified the protein as up-regulated is equal to the number of the studies identified protein as down-regulated.
Datasets of favorable/unfavorable prognostic proteins for all cancer types from the Human Protein Atlas (HPA) (www.proteinatlas.org/) were used to search for such proteins in the unique exosomal protein cargo of BCPs (Supplementary Table 6). In the BCP total blood exosomes, 64 prognostic proteins typical for different types of cancer were detected, and only 29 in the plasma exosomes. Thus, 35 of these 64 prognostic proteins found in the exosomes from total blood were detected solely through the analysis of exosomes associated with the surface of blood cells. Using the breast cancer proteomic data from HPA, we revealed three favorable (SERPINA1, KRT6B, and SOCS3) and one unfavorable (IGF2R) tumor prognostic protein markers. Notably, KRT6B and SOCS3 were revealed in the total blood exosomes from BCPs exclusively (i.e., in the composition of the blood cell-surface-associated exosomes). ROC analysis of revealed tumor markers (SERPINA1, KRT6B, SOCS3, and IGF2R) in total blood exosomes demonstrated that combination of these proteins enabled to distinguish BCPs at stage I and II of the disease with sensitivity of 73% and specificity of 100% (AUG 0.864), whereas the same analysis in the fraction of cell-free exosomes enabled to distinguish cancer individuals only with 45% sensitivity and 100% specificity.
Discussion
Exosomes are endosome-derived vesicles (30-120 nm) that are ubiquitous in biological fluids [tear (18), urine (19), cerebrospinal fluid (10), ascites (20), blood (21), and saliva (22)]. These vesicles are sufficiently small to penetrate from various tissues into biological fluids and then back, interacting with target cells, or tissues. There is a considerable research interest in understanding the physiological functions of exosomes, and in elucidating their role in diseases like cancer. Most studies are devoted to the study of the exosome proteome from cell cultures (1, 3, 23–27), which may be associated with various technical difficulties in analyzing exosomes from biological fluids (additional purification from concomitant protein impurities, heterogeneity of vesicles, etc.). Nevertheless, the study of exosomes isolated from clinical samples has greater significance and closer association with the real-life situation of patients. In the present, the studies of exosomes from plasma/serum in ovarian (21), breast (24), and colorectal cancers (28) have begun. A recent study has found that roughly two-thirds of the total blood extracellular vesicles of breast cancer patients exists in the blood cell-bound state (7, 8). A thorough investigation of blood cell-bound EVs as significant players of cell-to-cell communication and as potential cancer biomarkers is therefore entirely justified.
Our previously published data indicated that total blood exosomes morphologically resemble plasma exosomes, and that both pools of vesicles are positive for exosomal (CD24, CD63, CD81, and CD9) markers (9). We established with the use of cryo-EM a wide spectrum of the exosome morphologies with lipid bilayers and vesicular internal structures. To our knowledge, only two studies have examined several novel morphological EVs subcategories from human blood plasma by cryo-EM (29, 30) and none from human total blood. We revealed a variety of exosomes (single, double, double with two membrane bilayers and multilayer vesicles), which is in accordance with the previously obtained data (29, 30). Double and multilayered vesicles containing the electron-dense material were also visualized. These types of EVs were described earlier in the ejaculate and cerebrospinal fluid (10, 31). For the first time, we have found that single exosomes predominate in the plasma of HFs and the plasma and total blood of BCPs, while their proportion in the total blood of HFs is reduced by almost 2.5 times. Since the total blood exosomes contain two fractions (cell-free exosomes and blood cell-surface-associated exosomes), the revealed phenomenon indicates that exosomes of various morphologies predominate on the surface of blood cells. It allows suggesting that the subpopulations of exosomes with different and specific functions exist, and blood cells act as their transporters. Moreover, it seems that cancer can cause a redistribution of these subpopulations.
At the next step, using MALDI-TOF mass spectrometry we identified 223 proteins from plasma exosomes and total blood exosomes (containing plasma exosomes and blood-cell-surface associated exosomes) of HFs and BCPs. 27% of the identified proteins were absent in the Vesiclepedia database and 55% in the Exocarta database. This can be attributed to the difference in the sizes of the databases (13,105 Vesiclepedia proteins vs. 9,699 annotated in Exocarta), to the significant heterogeneity of extracellular vesicles, and to the presence of cell-bound exosomal proteins that elude researchers when only plasma exosomes are studied.
In according to previous studies, our obtained data shows that the main part of detected common exosomal proteins was of cytoplasmic and vesicle origin (3, 21, 28). The assignment of the subcellular localization revealed that a large proportion of unique exosomal proteins from plasma and total blood of BCPs is normally located in the cytoplasm of cells or vesicles; however, in the blood of HFs the unique exosomal proteins with GO term “fibrinogen complex” predominate. Unexpectedly, this GO term was also identified in a proteomic study of plasma exosomes in patients with prostate cancer (32). This finding is consistent the theory that biogenesis and selection of exosomal content is not a random procedure, but rather the result of a selective sorting process. Multilevel GO enrichment analysis of molecular functions revealed that the common exosomal proteins from plasma and total blood are involved in the protein binding, which is consistent with the earlier studies (21, 28, 33). Moreover, the presence of proteins associated with the receptor binding and cell adhesion molecule binding in the proteome cargo of exosomes from HF blood is consistent with the increased content of exosomes on the blood cell surface of healthy donors (7), while the share of blood-cell-associated exosomes is significantly reduced in tumor patients. The presence of proteins associated with the oxygen binding and oxygen transporter activity in the exosome composition is possibly associated with the adaptation of the tumor under conditions of hypoxia (34). The proteins involved in biological processes such as “endocytosis,” “vesicle-mediated transport,” “cell activation,” “extracellular structure organization,” and “regulation of transport” are common for the proteomic profiles of exosomes from blood of HFs and BCPs and are consistent with the previous studies (25, 35). An interesting finding was that the protein cargo from BCP exosomes is involved in the response to hypoxia; from the other hand, HF exosomes are enriched by proteins involved in the protein metabolic process, transport, regulation of cell communication, and secretion by cell (21, 28). Thus, the identified proteins from the total blood exosomes may play multiple roles in biological cell functions through the crucial mechanisms. These mechanisms include: (a) contact between surface molecules of exosomes and cells; (b) endocytosis and exocytosis of exosomes; (c) fusion of exosomes-cell membrane. It is possible that the production of exosomes allows neoplastic cells to exert various effects in accordance with possible acceptor targets. For example, exosomes can enhance the malignant properties of neighboring neoplastic cells and/or activate non-malignant adjacent cells through the transfer of multifunctional proteins, which play a key role in the cancerogenesis. To elucidate the molecular regulatory roles underlying blood exosome-mediated tumor progression, an analysis of protein cargoes in plasma and total blood exosomes was performed. Currently, there are few reports on the proteomic analysis of exosomes, especially for plasma exosomes in breast cancer in comparison with other types of cancer (24, 36) and a dearth of data on the proteomic profiles of blood cell-bound exosomes. Technical difficulties in the blood exosome proteomics are primarily associated with the heterogeneity of the composition of circulating vesicles. Nevertheless, we found that half of the identified exosomal proteins are involved in EMT, cell proliferation, invasion, cell migration, vasculature development, and immune response. Moreover, we revealed that the proteomic profiles of exosomes from HF total blood were enriched with the proteins inhibiting EMT, cell proliferation, migration, and invasion. Thus, the analysis of the proteomic composition of exosomes complements their biogenesis and functional roles in the cancer development.
Recently, liquid biopsy has gained much attention in cancer research, since it offers multiple advantages in the clinical settings, including their noninvasive nature, a suitable sample source for longitudinal disease monitoring, a better screenshot of tumor heterogeneity, and so on. Current liquid biopsies primarily focus on the detection and downstream analysis of circulating tumor cells, circulating tumor DNA, and EVs. The disadvantages of circulating tumor cells and circulating tumor DNA are heterogeneity and extreme rarity, while exosomes do not have such problems. Most of the studies on EVs focus on microRNAs and a small portion of exosomal proteins. Detection of functional proteins will provide real-time information about the body's physiological functions and disease progression, such as early detection and monitoring of cancer. In our study, we found that the breast cancer-associated proteins were equally represented in the blood cell-surface-associated exosomes (calculated as the difference between the total blood exosomes and plasma exosomes) and in the plasma exosomes from BCPs. However, the hyper-expressed proteins predominate in the blood cell-surface-associated exosomes as compared to the plasma exosomes (64 vs. 14%). Nevertheless, the use of the total blood exosomes without division into subfractions (plasma exosomes and blood cell-surface-associated exosomes) can reduce the time and labor required for the sample preparation before analysis, as well as increase the amount of the analyzed material, which is an extremely important aspect for further application of the developed methods to clinical practice. Using the data on favorable/unfavorable prognostic proteins in breast cancer from the Human Protein Atlas (HPA) (www.proteinatlas.org/), we found three favorable (SERPINA1, KRT6B, and SOCS3) and one unfavorable (IGF2R) prognostic protein markers in the total blood exosomes. Previously, these proteins were recognized as the prognostic markers (37, 38) in Human Pathology Atlas, and their expression in Breast tissue is linked to the overall higher/lower survival rates with significant (p < 0.001) association. Serpin family A member 1 (SERPINA1) is an inhibitor of serine proteases, namely elastase, trypsin, chymotrypsin, and plasminogen activator, which is also secreted to blood; it is involved in the invasion (39) and immune response, was found in vesicles, and it is a prognostic marker for the colorectal cancer (favorable prognosis). Keratin 6B (KRT6B) is a cytoskeleton protein involved in invasion (40) and prognostic in the urothelial cancer (unfavorable). Suppressor of the cytokine signaling 3 (SOCS3) is a suppressor of the cytokine signal transduction, localized in the cytosol and plasma membrane, involved in EMT (41), invasion (42), and vasculature development and prognostic for the renal cancer (unfavorable). Insulin-like growth factor 2 receptor (IGF2R) is a protein that transports phosphorylated lysosomal enzymes from the Golgi complex and the cell surface to lysosomes. It is situated in the Golgi apparatus and vesicles and acts as a positive regulator of the T-cell coactivation, shown to facilitate the immune response and invasion (43). It is also a prognostic marker for the cervical and urothelial cancers (unfavorable).
Thus, this study demonstrates that the proteomic analysis of exosomes from the total blood of cancer patients reveals more tumor markers. However, to facilitate the clinical use of such biomarker panels, a simple and reliable method for isolating exosomes from total blood should be developed.
In summary, the results of the present study indicate that the protein cargoes of both exosomes from plasma and blood cell-associated exosomes (calculated as the difference between the total blood exosomes and plasma exosomes) of HFs and cancer patients are enriched with proteins involving into the crucial steps of the tumor progression: EMT, cell proliferation, invasion, cell migration, stimulation of angiogenesis, and immune response. We found that the percentage of the tumor diagnostic/prognostic protein markers in the cell-surface-associated exosomes from BCP blood reaches 47%, with over-expressed proteins prevailing in this fraction (64 vs. 14% in the plasma exosomes). Further studies will be needed to determine whether identified new exosomal proteins from BCP blood are potential breast cancer diagnostic/prognostic markers or novel therapeutic targets.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committees of the Novosibirsk Regional Clinical Oncological Dispensary and the Institute of Chemical Biology and Fundamental Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ST: conceptualization. OT, KP, RK, TS, and ST: investigation. OT, RK, TS, YT, and ST: writing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.580891/full#supplementary-material
References
1. Sharma S, Alharbi M, Kobayashi M, Lai A, Guanzon D, Zuniga F, et al. Proteomic analysis of exosomes reveals an association between cell invasiveness and exosomal bioactivity on endothelial and mesenchymal cell migration in vitro. Clin Sci (Lond). (2018) 132:2029–44. doi: 10.1042/CS20180425
2. Conigliaro A, Cicchini C. Exosome-mediated signaling in epithelial to mesenchymal transition and tumor progression. J Clin Med. (2018) 8:26. doi: 10.3390/jcm8010026
3. Harris DA, Patel SH, Gucek M, Hendrix A, Westbroek W, Taraska JW. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS One. (2015) 10:e0117495. doi: 10.1371/journal.pone.0117495
4. Shtam T, Naryzhny S, Samsonov R, Karasik D, Mizgirev I, Kopylov A, et al. Plasma exosomes stimulate breast cancer metastasis through surface interactions and activation of FAK signaling. Breast Cancer Res Treat. (2019) 174:129–41. doi: 10.1007/s10549-018-5043-0
5. Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. (2019) 18:e18. doi: 10.1186/s12943-019-0948-8
6. Li Z, Tao Y, Wang X, Jiang P, Li J, Peng M, et al. Tumor-secreted exosomal miR-222 promotes tumor progression via regulating P27 expression and re-localization in pancreatic cancer. Cell Physiol Biochem. (2018) 51:610–29. doi: 10.1159/000495281
7. Tamkovich S, Tutanov O, Efimenko A, Grigor'eva A, Ryabchikova E, Kirushina N, et al. Blood circulating exosomes contain distinguishable fractions of free and cell-surface-associated vesicles. Curr Mol Med. (2019) 19:273–85. doi: 10.2174/1566524019666190314120532
8. Mamaeva SN, Kononova IV, Ruzhansky M, Nikiforov PV, Nikolaeva NA, Pavlov AN, et al. Using scanning electron microscopy and atomic force microscopy to study the formation of nanoparticles on red blood cell surface in cervical cancer patients. Int J Biomed. (2020) 10:70–5. doi: 10.21103/Article10(1)_OA12
9. Tutanov O, Orlova E, Proskura K, Grigor'eva A, Yunusova N, Tsentalovich Y, et al. Proteomic analysis of blood exosomes from healthy females and breast cancer patients reveals an association between different exosomal bioactivity on non-tumorigenic epithelial cell and breast cancer cell migration in vitro. Biomolecules. (2020) 10:495. doi: 10.3390/biom10040495
10. Emelyanov A, Shtam T, Kamyshinsky R, Garaeva L, Verlov N, Miliukhina I, et al. Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS One. (2020) 15:e0227949. doi: 10.1371/journal.pone.0227949
11. Tamkovich SN, Yunusova NV, Tugutova E, Somov AK, Proskura KV, Kolomiets LA, et al. Protease cargo in circulating exosomes of breast cancer and ovarian cancer patients. Asian Pac J Cancer Prev. (2019) 20:255–62. doi: 10.31557/APJCP.2019.20.1.255
12. Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang CS, Askenase P, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. (2017) 6:e1321455. doi: 10.1080/20013078.2017.1321455
13. Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. (2009) 25:3045–6. doi: 10.1093/bioinformatics/btp536
14. Yang Q, Zhang Y, Cui H, Chen L, Zhao Y, Lin Y, et al. dbDEPC 3.0: The database of differentially expressed proteins in human cancer with multi-level annotation and drug indication. Database (Oxford). (2018) 2018:bay015. doi: 10.1093/database/bay015
15. Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. (2012) 151:1068–82. doi: 10.1016/j.cell.2012.10.028
16. Luo C, Wang F, Ren X, Ke T, Xu C, Tang B, et al. Identification of a molecular signaling gene-gene regulatory network between GWAS susceptibility genes ADTRP and MIA3/TANGO1 for coronary artery disease. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:1640–53. doi: 10.1016/j.bbadis.2017.03.010
17. Suzuki Y, Ohga N, Morishita Y, Hida K, Miyazono K, Watabe T. BMP-9 induces proliferation of multiple types of endothelial cells in vitro and in vivo. J Cell Sci. (2010) 123(Pt 10):1684–92. doi: 10.1242/jcs.061556
18. Tamkovich S, Grigor'eva A, Eremina A, Tupikin A, Kabilov M, Chernykh V, et al. What information can be obtained from the tears of a patient with primary open angle glaucoma? Clin Chim Acta. (2019) 495:529–37. doi: 10.1016/j.cca.2019.05.028
19. Overbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. (2015) 6:30357–76. doi: 10.18632/oncotarget.4851
20. Yunusova NV, Patysheva MR, Molchanov SV, Zambalova EA, Grigor'eva AE, Kolomiets LA, et al. Metalloproteinases at the surface of small extrcellular vesicles in advanced ovarian cancer: relationships with ascites volume and peritoneal canceromatosis index. Clin Chim Acta. (2019) 494:116–22. doi: 10.1016/j.cca.2019.03.1621
21. Zhang W, Ou X, Wu X. Proteomics profiling of plasma exosomes in epithelial ovarian cancer: a potential role in the coagulation cascade, diagnosis and prognosis. Int J Oncol. (2019) 54:1719–33. doi: 10.3892/ijo.2019.4742
22. Xiao H, Wong DT. Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography-mass spectrometry. Anal Chim Acta. (2012) 723:61–7. doi: 10.1016/j.aca.2012.02.018
23. Lazar I, Clement E, Ducoux-Petit M, Denat L, Soldan V, Dauvillier S, et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. (2015) 28:464–75. doi: 10.1111/pcmr.12380
24. Vardaki I, Ceder S, Rutishauser D, Baltatzis G, Foukakis T, Panaretakis T. Periostin is identified as a putative metastatic marker in breast cancer-derived exosomes. Oncotarget. (2016) 7:74966–78. doi: 10.18632/oncotarget.11663
25. Wang S, Chen G, Lin X, Xing X, Cai Z, Liu X, et al. Role of exosomes in hepatocellular carcinoma cell mobility alteration. Oncol Lett. (2017) 14:8122–31. doi: 10.3892/ol.2017.7257
26. Gangoda L, Liem M, Ang CS, Keerthikumar S, Adda CG, Parker BS, et al. Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics. (2017) 17:1600370. doi: 10.1002/pmic.201600370
27. Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, et al. Breast cancer-derived exosomes reflect the cell-of-origin phenotype. Proteomics. (2019) 19:e1800180. doi: 10.1002/pmic.201800180
28. Chen Y, Xie Y, Xu L, Zhan S, Xiao Y, Gao Y, et al. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. (2017) 140:900–13. doi: 10.1002/ijc.30496
29. Arraud N, Linares R, Tan S, Gounou C, Pasquet J-M, Mornet S, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. (2014) 12:614–27. doi: 10.1111/jth.12554
30. Yuana Y, Koning RI, Kuil ME, Rensen PCN, Koster AJ, Bertina RM, et al. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J Extracell Vesicles. (2013) 2:10.3402/jev.v2i0.21494. doi: 10.3402/jev.v2i0.21494
31. Höög JL, Lötvall J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J Extracell Vesicles. (2015) 4:28680. doi: 10.3402/jev.v4.28680
32. Welton JL, Brennan P, Gurney M, Webber JP, Spary LK, Carton DG, et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array. J Extracell Vesicles. (2016) 5:31209. doi: 10.3402/jev.v5.31209
33. Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. (2013) 80:171–82. doi: 10.1016/j.jprot.2012.12.029
34. Kumar A, Deep G. Exosomes in hypoxia-induced remodeling of the tumor microenvironment. Cancer Lett. (2020) 488:1–8. doi: 10.1016/j.canlet.2020.05.018
35. Ge M, Ke R, Cai T, Yang J, Mu X. Identification and proteomic analysis of osteoblast-derived exosomes. Biochem Biophys Res Commun. (2015) 467:27–32. doi: 10.1016/j.bbrc.2015.09.135
36. Chen IH, Xue L, Hsu CC, Paez JSP, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. (2017) 114:3175–80. doi: 10.1073/pnas.1618088114
37. Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. (2017) 357:eaan2507. doi: 10.1126/science.aan2507
38. Pontén F, Schwenk JM, Asplund A, Edqvist PH. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med. (2011) 270:428–46. doi: 10.1111/j.1365-2796.2011.02427.x
39. Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK, Jo HJ, et al. Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget. (2015) 6:20312–26. doi: 10.18632/oncotarget.3964
40. Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. (2006) 19:264–71. doi: 10.1038/modpathol.3800528
41. Tang H, Long Q, Zhuang K, Yan Y, Han K, Guo H, et al. miR-665 promotes the progression of gastric adenocarcinoma via elevating FAK activation through targeting SOCS3 and is negatively regulated by lncRNA MEG3. J Cell Physiol. (2020) 235:4709–19. doi: 10.1002/jcp.29349
42. Chu Q, Shen D, He L, Wang H, Liu C, Zhang W. Prognostic significance of SOCS3 and its biological function in colorectal cancer. Gene. (2017) 627:114–22. doi: 10.1016/j.gene.2017.06.013
Keywords: exosomes, extracellular vesicles, mass-spectrometry, proteomics, breast cancer, blood
Citation: Tutanov O, Proskura K, Kamyshinsky R, Shtam T, Tsentalovich Y and Tamkovich S (2020) Proteomic Profiling of Plasma and Total Blood Exosomes in Breast Cancer: A Potential Role in Tumor Progression, Diagnosis, and Prognosis. Front. Oncol. 10:580891. doi: 10.3389/fonc.2020.580891
Received: 09 July 2020; Accepted: 16 September 2020;
Published: 28 October 2020.
Edited by:
Harsha Gowda, The University of Queensland, AustraliaReviewed by:
Maria Francesca Baietti, VIB KU Leuven Center for Cancer Biology, BelgiumRavi Manoharan, University of Madras, India
Copyright © 2020 Tutanov, Proskura, Kamyshinsky, Shtam, Tsentalovich and Tamkovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Svetlana Tamkovich, cy50YW1rb3ZpY2hAZy5uc3UucnU=
 Oleg Tutanov
Oleg Tutanov Ksenia Proskura
Ksenia Proskura Roman Kamyshinsky
Roman Kamyshinsky Tatiana Shtam
Tatiana Shtam Yuri Tsentalovich
Yuri Tsentalovich Svetlana Tamkovich
Svetlana Tamkovich