- 1Clinical Pharmacology and Pharmacogenetics Unit, Department of Clinical and Experimental Medicine, University of Pisa, Pisa, Italy
- 2Pneumology Unit, Cardiovascular and Thoracic Department, Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy
- 3Radiology Unit 2, Department of Diagnostics and Imaging, Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy
- 4Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa, Italy
- 5Section of Medical Oncology, Department of Surgical, Oncological and Stomatological Sciences, University of Palermo, Palermo, Italy
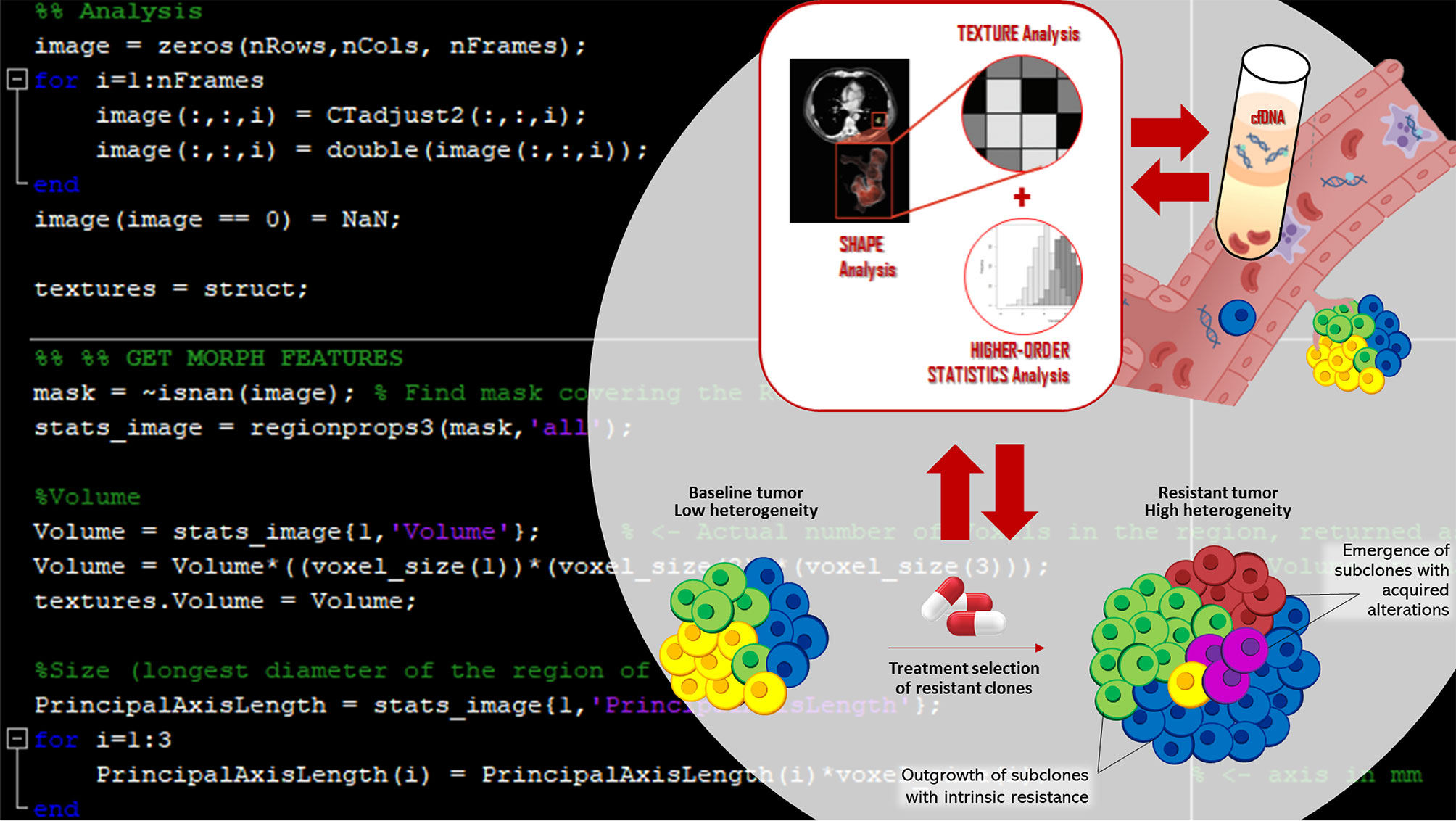
Background: EGFR-positive Non-small Cell Lung Cancer (NSCLC) is a dynamic entity and tumor progression and resistance to tyrosine kinase inhibitors (TKIs) arise from the accumulation, over time and across different disease sites, of subclonal genetic mutations. For instance, the occurrence of EGFR T790M is associated with resistance to gefitinib, erlotinib, and afatinib, while EGFR C797S causes osimertinib to lose activity. Sensitive technologies as radiomics and liquid biopsy have great potential to monitor tumor heterogeneity since they are both minimally invasive, easy to perform, and can be repeated over patient’s follow-up, enabling the extraction of valuable information. Yet, to date, there are no reported cases associating liquid biopsy and radiomics during treatment.
Case presentation: In this case series, seven patients with metastatic EGFR-positive NSCLC have been monitored during target therapy. Plasma-derived cell free DNA (cfDNA) was analyzed by a digital droplet PCR (ddPCR), while radiomic analyses were performed using the validated LifeX® software on computed tomography (CT)-images. The dynamics of EGFR mutations in cfDNA was compared with that of radiomic features. Then, for each EGFR mutation, a radiomic signature was defines as the sum of the most predictive features, weighted by their corresponding regression coefficients for the least absolute shrinkage and selection operator (LASSO) model. The receiver operating characteristic (ROC) curves were computed to estimate their diagnostic performance. The signatures achieved promising performance on predicting the presence of EGFR mutations (R2 = 0.447, p <0.001 EGFR activating mutations R2 = 0.301, p = 0.003 for T790M; and R2 = 0.354, p = 0.001 for activating plus resistance mutations), confirmed by ROC analysis.
Conclusion: To our knowledge, these are the first cases to highlight a potentially promising strategy to detect clonal heterogeneity and ultimately identify patients at risk of progression during treatment. Together, radiomics and liquid biopsy could detect the appearance of new mutations and therefore suggest new therapeutic management.
Introduction
Tyrosine kinases inhibitors (TKIs), such as gefitinib, erlotinib, afatinib or osimertinib, are the first-line treatments in patients with advanced NSCLC and activating EGFR mutation (1–4) since they improved progression-free survival (PFS) compared with conventional chemotherapy (5). Nevertheless, NSCLC is a dynamic entity, and tumor progression and resistance to treatment arise from the accumulation of independent genetic mutations in subclones, over time and across different disease sites, thereby resulting in temporal and spatial heterogeneity (6). Moreover, treatment exerts selective pressure on cancer cells, and only those bearing either primary or secondary resistance mutations will survive (7–9).
The concept of a single-site biopsy to monitor disease dynamics is practically unfeasible since it is invasive and may result in underestimation of heterogeneity (10). Instead, liquid biopsy—allowing the analysis of cell-free DNA (cfDNA) (11)—better reflects the mutational status from the overall sites of disease (12), being able to identify emerging sub-clones responsible for treatment resistance (13).
Besides, radiomics has emerged as a novel field of research (14), dealing with the extraction and analysis of specific features from diagnostic images (15), and potentially reflecting the pathophysiological processes and the heterogeneity of tumors genetics (16). Recent data has shown that also texture analysis of radiological images can identify NSCLCs bearing EGFR mutations (17, 18).
The combined approach of radiomics and liquid biopsy has the potential to understand the dynamics of molecular lesions, supporting clinical decision-making.
To date, no reports correlate the dynamics of EGFR mutations in cfDNA with that of radiomic features. The present study aimed to assess such correlation in a case series of seven patients with EGFR mutant NSCLC, and to build a multi-parametric signature of clonal heterogeneity.
Patients
The study retrospectively matched clinical, molecular and imaging databases of seven patients with histologically proven EGFR-positive NSCLC (exon 19 deletion [ex19del], exon 21 [L858R], or other mutations [i.e. L861Q]), and candidate to a first/second or third-generation EGFR-TKIs. Enrolled patients underwent blood sampling 1) before the first dose of TKI (baseline), 2) every two months, 3) and at each of the instrumental (i.e. imaging) disease re-evaluation throughout the follow-up. Complete (CR) and partial response (PR), disease stabilization (SD) and disease progression (PD) were defined following RECIST (v. 1.1) criteria. CT scans were collected at baseline and every 3–6 months as per clinical practice (19) and then used for radiomic analysis. The interval between follow-up medical visits (3 vs. 6 months) was based on clinical decision-making on an individual basis (19). Clinical data were collected from medical records. A written consent form was obtained from all patients. The study was approved by the institutional ethics committee of University Hospital, Pisa, Italy (protocol 5625/2015), and performed in accordance with the provisions established by the Helsinki Declaration.
Methods
cfDNA Extraction and Analysis
cfDNA was extracted from 3 ml of plasma using the QIAmp Circulating Nucleic Acid kit (Qiagen®, Hilden, Germany) and then eluted in 100 μl of the buffer, as previously described (20). EGFR mutations (ex19del, L858R, T790M, and C797S) were investigated by digital droplet polymerase chain reaction (ddPCR) using the ddPCR Mutation Assay (BioRad®, Hercules, CA). A fluorescence intensity threshold of 3,000 was set as a cut-off point; the sample was considered as mutant positive when at least one droplet was above the threshold level. The number of mutant alleles was reported as copies/ml.
CT Segmentation and Extraction of Radiomic Features
Images were extracted from multiple non-contrast material-enhanced thoraco-abdominal computed tomography (CT)-scans (SIEMENS CT Sensation 64®; kilovoltage = 120 KV and exposure = 165 mAs; CT slice = 1.5 mm) (21). All CT examinations were reconstructed using B30f kernel (22). The radiomic analysis was performed by one author, using the validated LifeX® software (LifeX®, IMIV, CEA, Inserm, CNRS, Orsay, France) (23–25), after appropriate manual segmentation of the volumes of interest (VOIs; i.e. the lesions). Thirty-six radiomic features including three shapes, two gray-level histogram, six gray-level co-occurrence matrix (GLCM), 11 gray-level run lengths matrix (GLRLM), three Neighborhood Grey-Level Different Matrix (NGLDM) and 11 Grey-Level Zone-Length Matrix (GLZLM) features, were computed.
Selection of Radiomic Features and Data Analysis
Each patient had a longitudinal dataset of several scans to match with respective temporally linked liquid biopsy data. To calculate to which extent the variation between the radiomic features is correlated to EGFR mutation status, a logistic least absolute shrinkage and selection operator (LASSO) regression model adopting a 27-fold Monte Carlo cross-validation was applied and executed in Matlab R2019a (MatLab® software, The Math Works Inc., Natick, MA) (26, 27). The LASSO logistic model was used to reduce the number of radiomic features and estimate the maximum-likelihood fitted regression coefficients for the remaining ones. The LASSO computation was performed to assess the radiomic features about the copies/ml of EGFR activating mutations (ex19del/L858R), as well as about the emergence of resistance mutations (T790M and C797S) and the total copies/ml (ex19del/L858R together with T790M and C797S) occurring in EGFR in patients progressed to TKI treatments. Selected radiomic features are reported in Supplemental Table 1 in the online version. Then, radiomic signatures were calculated as the sum of the selected features weighted by their corresponding regression coefficients for the LASSO models. The receiver operating characteristic (ROC) curve analysis was computed to estimate the diagnostic performance of such signatures and select the optimal thresholds.
Moreover, Kendall’s correlation coefficient (tau-b, τb) was calculated to determine the strength of the association between changes in radiomic features and changes in matched liquid biopsy-derived data, over time (28).
Differences were considered significant at p <0.05. Statistical analysis was performed using the open-source statistical language R (R Foundation for Statistical Computing, Vienna, Austria) through the free and open statistical software program JAMOVI® (Version 1.1.9; retrieved from https://www.jamovi.org).
Results
Five patients presented the ex19del activating mutation at diagnosis and two were carriers of the L858R. Three patients were treated with afatinib, two with erlotinib, and two with gefitinib as first-line TKI. Clinical characteristics are summarized in Supplemental Table 2 in the online version, while plasma monitoring for each of them is reported in Figure 1. Overall, at baseline, the median activating EGFR copies/ml was higher than T790M and was often related to disease control, whereas the T790M amount was not. At disease progression, T790M was detected in plasma and/or tissue in all patients; therefore, osimertinib treatment was started, except in one patient, since the drug was not yet available (Figure 1G). In two patients disease progression occurred due to C797S mutation, in addition to ex19del and T790M.
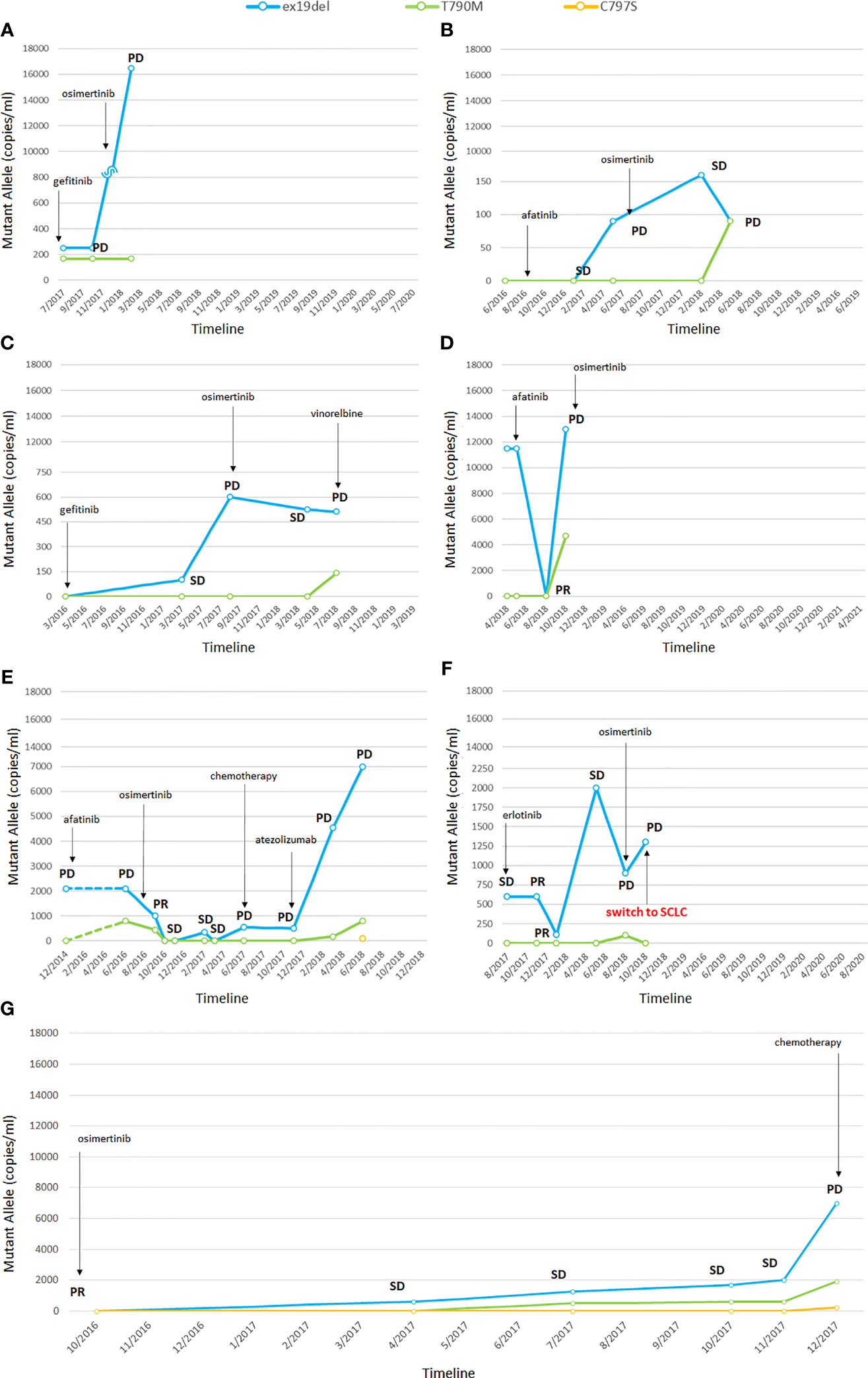
Figure 1 Changes in EGFR mutations detected by liquid biopsy (A–G). In one patient the tumor transformed into small cell lung cancer (SCLC) (F). PR, partial response; SD, stable disease; PD, progressive disease; SCLC, small cell lung cancer. The numbers before the year indicate the months while the letters (A–G) refer to single patients.
The dynamics of EGFR mutations were significantly associated to specific changes in radiomic features over time (p <0.05; Supplemental Table 3 in the online version). Radiomic features selected at least once in the LASSO models with respect to the number of copies/ml of mutant EGFR L858R/ex19del were combined in a signature (R).
The signature evidenced good capability—with acceptable representativeness—in predicting the number of copies/ml of the activating EGFR (R2 = 0.447, Akaike’s Information Criteria (AIC) = 515, p <0.001) (Figure 2A), and the optimal cut-point estimated from the ROC curve showed 88.9% accuracy, 90% sensitivity, and 85.7% specificity, with the area under the curve (AUC) of 0.90 (Figure 2B). Signatures and their predicting representativeness were also evaluated with respect to the copies/ml of T790M mutation and the total copies/ml of mutations in patients progressed to TKIs. The model predicting the T790M copies/ml showed R2 = 0.301 and AIC = 468 (p = 0.003), with 81.5% accuracy, 80% sensitivity and 83.3% specificity of the optimal threshold, and AUC of 0.84 (Figures 2C, D). The model predicting the total copies/ml of all mutations displayed R2 = 0.354 and AIC = 534 (p = 0.001), with 96.3% accuracy, 95% sensitivity, and 100% specificity of the optimal threshold, with AUC of 0.98 (Figures 2E, F); no correlation emerged for C797S.
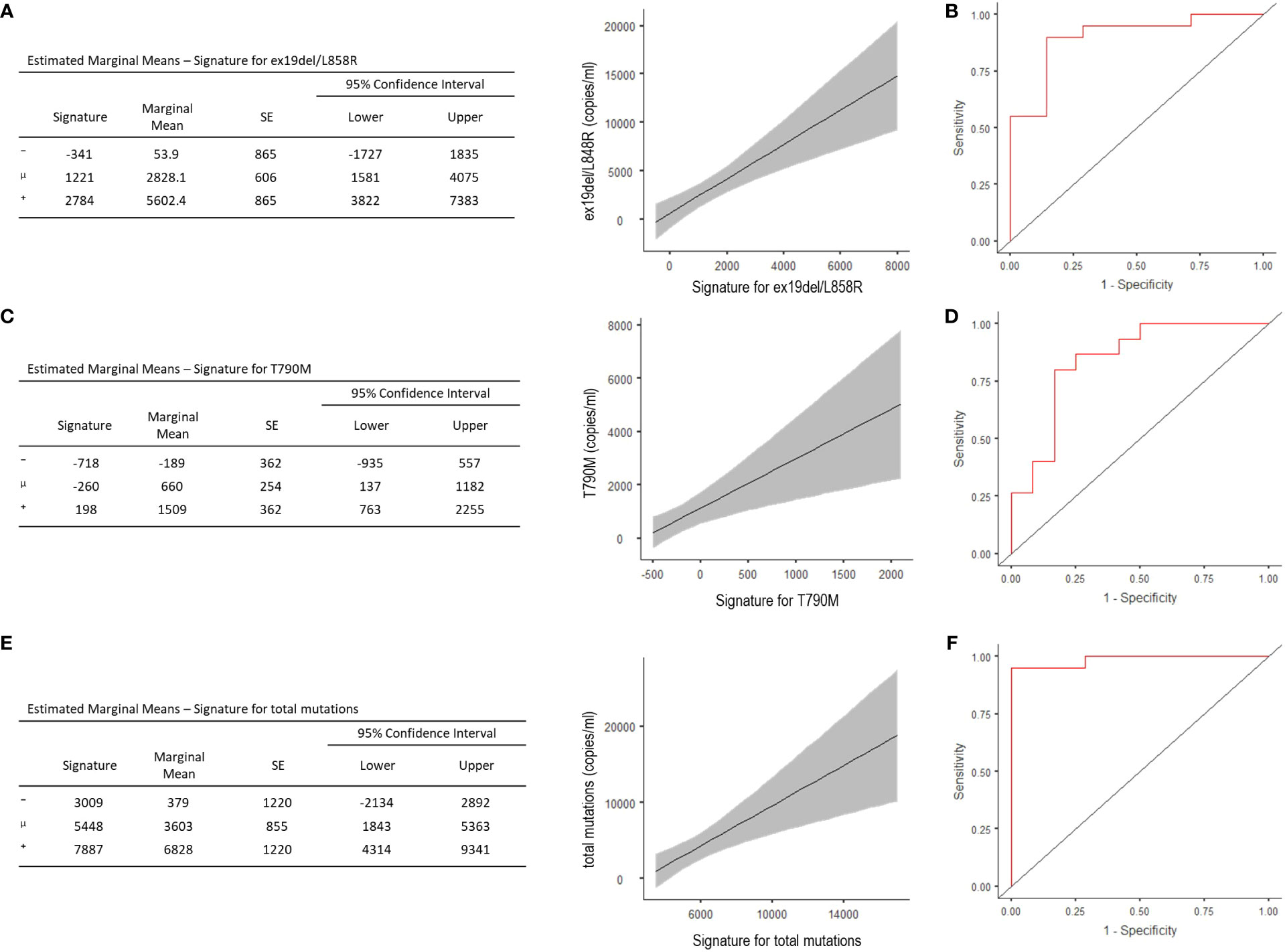
Figure 2 Estimated Marginal Means and ROC analysis for radiomic signature concerning ex19del/L858R (A, B), T790M (C, D), and total copies/ml (i.e. ex19del/L858R together with T790M and C797S) (E, F). ⁻, mean-1SD; μ, mean; ⁺, mean+1STD.
Discussion
To date, several works studied the potential of radiomics in the non-invasive prediction of EGFR mutational status and showed promising results (18, 24, 29–32). However, none of them addressed the subclonal heterogeneity that occurs asynchronously. In this study, we endeavored to highlight the great potential of integrating radiomics and liquid biopsy, as both are minimally invasive, easy to perform, and can be repeated over patients’ follow-up visits.
We did it by incorporating multiple radiomic functions into a signature (R) that could reliably predict the EGFR mutation status during treatment, and demonstrating significant correlations between radiomics and liquid biopsy data.
Of note, sphericity of lung lesions decreased with the increase of T790M copies/ml and, more generally, with the total copies/ml of mutant alleles, highlighting the association between spherical disproportion and neoplastic progression, aggressiveness and resistance to therapy (33). Besides, copies/ml of the T790M mutation were directly correlated with GLCM dissimilarity (a measure of local intensity variation of the voxel gray levels), and direct relationships also emerged between ex19del/L858R copies/ml, GLCM energy and contrast (Supplemental Table 3). The GLCM energy is a characteristic that describes the order status of the system and refers to the uniformity of the gray level between voxel pairs. The GLCM contrast highlights how many nearby sub-areas of heterogeneity differ within each lesion.
Interestingly, we found no significant correspondence between tumor volume (ml) and mutational status (Supplemental Table 3). Our results are consistent with those from Park et al. (34) and Lee et al. (35), testing the stability and reliability of radiomic features to evaluate tumor heterogeneity. Notably, Lee and collaborators (35), by studying the variability of radiomics features and their relationship with tumor size and shape upon 260 lung nodules, found that only a few features—including spherical disproportion and dissimilarity—showed high reproducibility in correlation with nodule status. This is probably because radiomics in lung cancer is different from in other oncology fields. Lung cancer resides in an environment rich with air, while other cancers primarily consist of soft tissue and reside in the interstitium (36). More than the usual volume changes, tumor progression is associated with shape and density changes from ground-glass opacity (GGO) to solid component (37–39). Thus, radiomics in the lung should jointly consider the tumor core geometry along with textural changes to properly model lung cancers. Nevertheless, reproducibility studies are lacking, and more evidences are needed to provide suggestions for future lung radiomics investigations.
There was also no significant correlation between changes in radiomic characteristics and C797S dynamics, probably due to the low sample size. However, the landscape of mechanisms of resistance dramatically changes considering osimertinib, and future studies to better investigate radiomic changes correlating with C797S dynamics will be needed (40). The appearance of the EGFR C797S mutation accounts for 6–10% after osimertinib as first line and 10–26% as second line (41). Furthermore, co-occurring with T790M has potential implications for treatment: when C797S and T790M occur on the same allele (cis), no response to EGFR TKIs alone or in combination can be expected, while the C797S in trans with the T790M mutation confers sensitivity to a combination of first/third-generation drugs (42–44).
Our results took advantage of consistently examining a few patients over a period, and of correlating changes in their radiomic features with the respective dynamics of EGFR mutations in cfDNA. The resulting signatures showed a good capability—with acceptable representativeness—in predicting the tumor mutational status. Unfortunately, given the low number of subjects, we found no radiomic signature that can be reliably associated with clinical outcomes, but we plan to look for it in the future. Future larger prospective clinical trials will also need to validate these findings and give us the chance to look for new resistance signatures, such as the one related to SCLC transformation, which is an important potential mechanism of resistance for to first/second and third-generation EGFR-TKIs (8), but to date, only a new tissue biopsy could allow to find it. Clinicians may consider using the signature as a new supporting tool, in accordance with their experience and judgment.
Liquid biopsy and radiomics have both advantages and drawbacks making them complementary methods. Although they are appealing options at progression, to track mechanisms of resistance (40, 45–48), there are still too few laboratory applications for liquid biopsy, and molecular protocols need to be standardized. Furthermore, there are difficulties in detection, and extremely sensitive and specific analytical methods are required to deal with small quantities of easily degradable materials. Lastly, it is still unclear whether liquid biopsy provides a representative sampling of all genetic clones or whether there is a propensity for specific subpopulations within the intra-tumor heterogeneity (49). Similarly, radiation, dearth of standardization for image acquisition, computational approaches and feature selection, as well as the black-box problem (i.e. non-interpretable advanced machine-learning algorithms that work like black boxes, hindering clinical translation), limit the use of radiomics, which should be considered as an indirect and non-detailed quantification of the underlying biological processes. Therefore, to strengthen the trustworthiness of the results, radiomics-based genotype predictions could be compared with information from liquid biopsy (16), over time. A combination of these two minimally invasive strategies, together with cutting-edge data analysis strategies, could be more valuable and reliable than their independent use and may help decode tumor information regarding the type, aggressiveness, progression, and response to treatment (29, 50). A study from the University of Oklahoma reported that while radiomics and genomics models were capable of predicting survival, accuracy significantly improved when both data were combined (51). Besides, while it is possible to avoid unnecessary radiation by using liquid biopsy, on the other hand, we can use radiomics to refine liquid biopsy results and provide a full-field analysis of patient’s lesions in virtually real-time response. Both techniques, providing a new instrumental and therefore objective diagnostic support, are able to reduce the need for invasive (and often difficult to perform) biopsies and favor an approach that promptly suggests a change in treatment strategy over the follow-up.
To the best of our knowledge, this is the first study investigating the longitudinal trajectory of NSCLC from both the radiomic and liquid biopsy points of view. As far as we know, the parallelism between the dynamics of EGFR mutation status and radiomic features is potentially dependent on the progressive enrichment of tumor tissue by treatment-resistant clones.
Conclusion
Radiomic signatures may represent a clinically relevant readout of EGFR mutational status and provide a non-invasive biomarker to monitor targeted drug therapies in NSCLC. Indeed, with the availability of big data and cutting-edge analysis strategies (such as machine learning), the information coming from tumor genotype and phenotype decoded via imaging (29), may predict treatment failures suggesting a change in treatment strategy earlier than with conventional methods. Nevertheless, it should be noted that such techniques will not substitute tissue biopsy in the near future, since they will require the aid of other parameters to be correctly interpreted and acted upon (52).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the institutional ethics committee of University Hospital, Pisa, Italy (protocol 5625/2015). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
Conception and design: RD, MDR, FC. Provision of study material or patients: IP, CR, SV, ML, AC, SC, ER, FC, AD, AR, RD. Collection and assembly of data: MDR, FC, SV, CR, IP, ML, SC, ER. Data analysis and interpretation: MDR, FC, CR, SC. Manuscript writing and final approval of manuscript: all authors. Financial support: RD. All authors contributed to the article and approved the submitted version.
Funding
The authors declare that this study received funding from Astra Zeneca. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
MDR received speaker honoraria from Astellas, Astra Zeneca, Celgene, Novartis, Pfizer, Bio-Rad Janssen-Cilag, Sanofi-Aventis; consulting fee from Ipsen and Janssen-Cilag; Speaker’s bureau: Celgene, Janssen, Sanofi; travel support from Janssen, Bio-Rad. RD received honoraria for scientific advisory board and consulting relationship from Ipsen, Novartis, Pfizer, Sanofi Genzyme, AstraZeneca, Janssen, Gilead, Lilly, Gilead, EUSA Pharma; travel support from Ipsen, Sanofi Genzyme. ML was a previous employee at Lilly and received travel support from M.S.D.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.593831/full#supplementary-material
Abbreviations
AIC, Akaike’s Information Criteria; AUC, area under the curve of the ROC; CT, computed tomography; cfDNA, cell-free DNA; ddPCR, digital droplet polymerase chain reaction; DNA, deoxyribonucleic acid; ECOG, Eastern Cooperative Oncology Group; EGFR, epidermal growth factor receptor; ex19del, exon 19 deletion; GLCM, gray level co-occurrence matrix; GLNUr, gray-level non-uniformity for run; GLNUz, gray-level non-uniformity for zone; GLRLM, gray-level run length matrix; GLZLM, gray-level zone length matrix; HGRE, high gray-level run emphasis; HGZE, high gray-level zone emphasis; KV, kilovolt; LASSO, least absolute shrinkage and selection operator; LGRE, low gray-level run emphasis; LGZE, low gray-level zone emphasis; LRE, long-run emphasis; LRHGE, long-run high gray-level emphasis; LRLGE, long-run low gray-level emphasis; LZE, long-zone emphasis; LZHGE, long-zone high gray-level emphasis; LZLGE, long-zone low gray-level emphasis; mAs, milliampere-seconds; min, minutes; ml, milliliter; mm, millimeter; n, number; NGLDM, neighborhood gray-level different matrix; NSCLC, non-small cell lung cancer; PD, progressive disease; PFS, progression-free survival; PR, partial response; rpm, revolutions per minute; RLNU, run length non-uniformity; ROC, receiver operating characteristic; RP, run percentage; RTKs, receptor tyrosine kinases; SCC, squamous cell cancer; SCLC, small cell lung cancer; SD, stable disease; SRE, short-run emphasis; SRHGE, short-run high gray-level emphasis; SRLGE, short-run low gray-level emphasis; STD, standard deviation; SZE, short-zone emphasis; SZHGE, short-zone high gray-level emphasis; SZLGE, short-zone low gray-level emphasis; TKI, tyrosine kinase inhibitor; ZLNU, zone length non-uniformity; ZP, zone percentage.
References
1. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol (2012) 13:239–46. doi: 10.1016/S1470-2045(11)70393-X
2. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med (2010) 362:2380–8. doi: 10.1056/NEJMoa0909530
3. Ricciuti B, Baglivo S, De Giglio A, Chiari R. Afatinib in the first-line treatment of patients with non-small cell lung cancer: clinical evidence and experience. Ther Adv Respir Dis (2018) 12:1–13. doi: 10.1177/1753466618808659
4. Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med (2018) 378:113–25. doi: 10.1056/NEJMoa1713137
5. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2017) 15:504–35. doi: 10.6004/jnccn.2017.0050
6. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol (2018) 15:81–94. doi: 10.1038/nrclinonc.2017.166
7. Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol Off J Eur Soc Med Oncol (2014) 25:423–8. doi: 10.1093/annonc/mdt573
8. Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med (2011) 3:75ra26. doi: 10.1126/scitranslmed.3002003
9. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res (2013) 19:2240–7. doi: 10.1158/1078-0432.CCR-12-2246
10. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol (2018) 13:1248–68. doi: 10.1016/j.jtho.2018.05.030
11. Alix-Panabières C, Pantel K. Clinical prospects of liquid biopsies. Nat BioMed Eng (2017) 1:65. doi: 10.1038/s41551-017-0065
12. Esposito A, Criscitiello C, Locatelli M, Milano M, Curigliano G. Liquid biopsies for solid tumors: Understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol Ther (2016) 157:120–4. doi: 10.1016/j.pharmthera.2015.11.007
13. Del Re M, Addeo A, Passaro A, Petrini I, van Schaik RH, Danesi R. Circulating tumor DNA and the future of EGFR-mutant lung cancer treatment. Pharmacogenomics (2019) 20:1255–7. doi: 10.2217/pgs-2019-0150
14. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer (2012) 48:441–6. doi: 10.1016/j.ejca.2011.11.036
15. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology (2016) 278:563–77. doi: 10.1148/radiol.2015151169
16. Neri E, Del Re M, Paiar F, Erba P, Cocuzza P, Regge D, et al. Radiomics and liquid biopsy in oncology: the holons of systems medicine. Insights Imaging (2018) 9:915–24. doi: 10.1007/s13244-018-0657-7
17. Nair JKR, Saeed UA, McDougall CC, Sabri A, Kovacina B, Raidu BVS, et al. Radiogenomic Models Using Machine Learning Techniques to Predict EGFR Mutations in Non-Small Cell Lung Cancer. Can Assoc Radiol J (2020) 17:1–11. doi: 10.1177/0846537119899526
18. Zhao W, Wu Y, Xu Y, Sun Y, Gao P, Tan M, et al. The Potential of Radiomics Nomogram in Non-invasively Prediction of Epidermal Growth Factor Receptor Mutation Status and Subtypes in Lung Adenocarcinoma. Front Oncol (2019) 9:1485. doi: 10.3389/fonc.2019.01485
19. Ng C, Pircher A, Augustin F, Kocher F. Evidence-based follow-up in lung cancer? memo - Mag Eur Med Oncol (2020) 13:73–7. doi: 10.1007/s12254-020-00575-3
20. Del Re M, Tiseo M, Bordi P, D’Incecco A, Camerini A, Petrini I, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget (2017) 8:13611–9. doi: 10.18632/oncotarget.6957
21. Berenguer R, Pastor-Juan MDR, Canales-Vazquez J, Castro-Garcia M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology (2018) 288:407–15. doi: 10.1148/radiol.2018172361
22. Choe J, Lee SM, Do K-H, Lee G, Lee J-G, Lee SM, et al. Deep Learning-based Image Conversion of CT Reconstruction Kernels Improves Radiomics Reproducibility for Pulmonary Nodules or Masses. Radiology (2019) 292:365–73. doi: 10.1148/radiol.2019181960
23. Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res (2018) 78(16):4786–9.
24. Jia T-Y, Xiong J-F, Li X-Y, Yu W, Xu Z-Y, Cai X-W, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol (2019) 29:4742–50. doi: 10.1007/s00330-019-06024-y
25. Li S, Ding C, Zhang H, Song J, Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys (2019) 46:4545–52. doi: 10.1002/mp.13747
26. de Jong EEC, Sanders KJC, Deist TM, van Elmpt W, Jochems A, van Timmeren JE, et al. Can radiomics help to predict skeletal muscle response to chemotherapy in stage IV non-small cell lung cancer? Eur J Cancer (2019) 120:107–13. doi: 10.1016/j.ejca.2019.07.023
27. Xu T, Sun J, Bi J. Longitudinal LASSO: Jointly Learning Features and Temporal Contingency for Outcome Prediction. KDD '15: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. (2015) 1345–54. doi: 10.1145/2783258.2783403
28. Ma Y. On Inference for Kendall’s τ within a Longitudinal Data Setting. J Appl Stat (2012) 39:2441–52. doi: 10.1080/02664763.2012.712954
29. Aerts HJWL, Grossmann P, Tan Y, Oxnard GR, Rizvi N, Schwartz LH, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep (2016) 6:33860. doi: 10.1038/srep33860
30. Yang X, Dong X, Wang J, Li W, Gu Z, Gao D, et al. Computed Tomography-Based Radiomics Signature: A Potential Indicator of Epidermal Growth Factor Receptor Mutation in Pulmonary Adenocarcinoma Appearing as a Subsolid Nodule. Oncologist (2019) 24:e1156–64. doi: 10.1634/theoncologist.2018-0706
31. Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer (2016) 17:441–8.e6. doi: 10.1016/j.cllc.2016.02.001
32. Digumarthy SR, Padole AM, Lo Gullo R, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Med (Baltimore) (2019) 98:e13963. doi: 10.1097/MD.0000000000013963
33. Limkin EJ, Reuzé S, Carré A, Sun R, Schernberg A, Alexis A, et al. The complexity of tumor shape, spiculatedness, correlates with tumor radiomic shape features. Sci Rep (2019) 9:4329. doi: 10.1038/s41598-019-40437-5
34. Park BW, Kim JK, Heo C, Park KJ. Reliability of CT radiomic features reflecting tumour heterogeneity according to image quality and image processing parameters. Sci Rep (2020) 10:3852. doi: 10.1038/s41598-020-60868-9
35. Lee S-H, Cho H-H, Lee HY, Park H. Clinical impact of variability on CT radiomics and suggestions for suitable feature selection: a focus on lung cancer. Cancer Imaging Off Publ Int Cancer Imaging Soc (2019) 19:54. doi: 10.1186/s40644-019-0239-z
36. Kang H, Lee HY, Lee KS, Kim J-H. Imaging-Based Tumor Treatment Response Evaluation: Review of Conventional, New, and Emerging Concepts. Korean J Radiol (2012) 13:371–90. doi: 10.3348/kjr.2012.13.4.371
37. Chong Y, Kim J-H, Lee HY, Ahn YC, Lee KS, Ahn M-J, et al. Quantitative CT variables enabling response prediction in neoadjuvant therapy with EGFR-TKIs: are they different from those in neoadjuvant concurrent chemoradiotherapy? PloS One (2014) 9:e88598. doi: 10.1371/journal.pone.0088598
38. Kobayashi Y, Fukui T, Ito S, Usami N, Hatooka S, Yatabe Y, et al. How long should small lung lesions of ground-glass opacity be followed? J Thorac Oncol (2013) 8:309–14. doi: 10.1097/JTO.0b013e31827e2435
39. Staring M, Pluim JPW, de Hoop B, Klein S, van Ginneken B, Gietema H, et al. Image Subtraction Facilitates Assessment of Volume and Density Change in Ground-Glass Opacities in Chest CT. Invest Radiol (2009) 44(2):61–6. doi: 10.1097/RLI.0b013e318197fcb7
40. Del Re M, Cucchiara F, Petrini I, Fogli S, Passaro A, Crucitta S, et al. erbB in NSCLC as a molecular target: current evidences and future directions. ESMO Open (2020) 5:e000724. doi: 10.1136/esmoopen-2020-000724
41. Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med (2017) 376:629–40. doi: 10.1056/NEJMoa1612674
42. Wang Z, Yang J-J, Huang J, Ye J-Y, Zhang X-C, Tu H-Y, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer (2017) 12:1723–7. doi: 10.1016/j.jtho.2017.06.017
43. Arulananda S, Do H, Musafer A, Mitchell P, Dobrovic A, John T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer (2017) 12:1728–32. doi: 10.1016/j.jtho.2017.08.006
44. Niederst MJ, Hu H, Mulvey HE, Lockerman EL, Garcia AR, Piotrowska Z, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res an Off J Am Assoc Cancer Res (2015) 21:3924–33. doi: 10.1158/1078-0432.CCR-15-0560
45. Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2009) 15:2630–6. doi: 10.1158/1078-0432.CCR-08-2592
46. Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Jänne PA, Koch WH, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res an Off J Am Assoc Cancer Res (2016) 22:1103–10. doi: 10.1158/1078-0432.CCR-15-1031
47. Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J Clin Oncol (2016) 34:3375–82. doi: 10.1200/JCO.2016.66.7162
48. Kim H, Park CM, Keam B, Park SJ, Kim M, Kim TM, et al. The prognostic value of CT radiomic features for patients with pulmonary adenocarcinoma treated with EGFR tyrosine kinase inhibitors. PloS One (2017) 12:e0187500. doi: 10.1371/journal.pone.0187500
49. Sisson BA, Uvalic J, Kelly K, Selvam P, Hesse AN, Ananda G, et al. Technical and Regulatory Considerations for Taking Liquid Biopsy to the Clinic: Validation of the JAX PlasmaMonitor(TM) Assay. Biomark Insights (2019) 14:1–11. doi: 10.1177/1177271919826545
50. Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife (2017) 6:e23421. doi: 10.7554/eLife.23421
51. Emaminejad N, Qian W, Guan Y, Tan M, Qiu Y, Liu H, et al. Fusion of Quantitative Image and Genomic Biomarkers to Improve Prognosis Assessment of Early Stage Lung Cancer Patients. IEEE Trans BioMed Eng (2016) 63:1034–43. doi: 10.1109/TBME.2015.2477688
Keywords: non-small cell lung cancer, EGFR, liquid biopsy, cell free DNA, radiomics, tyrosine kinase inhibitors, precision medicine
Citation: Cucchiara F, Del Re M, Valleggi S, Romei C, Petrini I, Lucchesi M, Crucitta S, Rofi E, De Liperi A, Chella A, Russo A and Danesi R (2020) Integrating Liquid Biopsy and Radiomics to Monitor Clonal Heterogeneity of EGFR-Positive Non-Small Cell Lung Cancer. Front. Oncol. 10:593831. doi: 10.3389/fonc.2020.593831
Received: 11 August 2020; Accepted: 30 October 2020;
Published: 16 December 2020.
Edited by:
Yunpeng Yang, Sun Yat-sen University Cancer Center (SYSUCC), ChinaReviewed by:
Dwight Hall Owen, The Ohio State University, United StatesAndrea Camerini, Azienda Usl Toscana nord ovest, Italy
Copyright © 2020 Cucchiara, Del Re, Valleggi, Romei, Petrini, Lucchesi, Crucitta, Rofi, De Liperi, Chella, Russo and Danesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Romano Danesi, cm9tYW5vLmRhbmVzaUB1bmlwaS5pdA==
†These authors have contributed equally to this work
 Federico Cucchiara
Federico Cucchiara Marzia Del Re
Marzia Del Re Simona Valleggi
Simona Valleggi Chiara Romei
Chiara Romei Iacopo Petrini
Iacopo Petrini Maurizio Lucchesi2
Maurizio Lucchesi2 Stefania Crucitta
Stefania Crucitta Eleonora Rofi
Eleonora Rofi Antonio Russo
Antonio Russo Romano Danesi
Romano Danesi