- 1Department of Thoracic Surgery, Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Hangzhou, China
- 2Institute of Cancer and Basic Medicine (IBMC), Chinese Academy of Sciences, Hangzhou, China
- 3Medical Department, Nanjing Geneseeq Technology Inc., Nanjing, China
- 4Department of Pathology, Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Hangzhou, China
- 5Department of Medical Oncology, Traditional Chinese Medical Hospital of Huzhou, Huzhou, China
- 6Department of Medical Oncology, Haining People’s Hospital, Haining Branch, The First Affiliated Hospital, Zhejiang University, Haining, China
Background: Invasive mucinous adenocarcinoma (IMA) of the lung is a distinct histological subtype with unique clinical and pathological features. Despite previous genomic studies on lung IMA, the genetic characteristics and the prognosis-related biomarkers in Chinese surgically resected lung IMA remain unclear.
Methods: We collected 76 surgically resected primary tumors of invasive lung adenocarcinoma, including 51 IMA and 25 non-mucinous adenocarcinomas (non-IMA). IMA was further divided into pure-IMA (mucinous features≥90%) and mixed-IMA subgroups. Comprehensive genomic profiling based on targeted next-generation sequencing (NGS) of 425 genes was explored and genomic characteristics were evaluated for the correlation with postoperative disease-free survival (DFS).
Results: IMA had a unique genetic profile, with more diverse driver mutations and more tumor drivers/suppressors co-occurrence than that of non-IMA. The frequency of EGFR (72.0% vs. 40.0% vs. 23.1%, p=0.002) and ALK (undetected vs. 20.0% vs. 26.9%, p=0.015) alterations showed a trend of gradual decrease and increase from non-IMA to mixed-IMA to pure-IMA, respectively. The frequency of KRAS mutations in pure-IMA was higher than that in mixed-IMA, albeit statistically insignificant (23.1% vs. 4.0%, p=0.10). TP53 mutation was significantly less in pure-IMA compared to mixed-IMA and non-IMA (23.1% vs. 52.0% vs. 56.0%, p=0.03). Besides, IMA exhibited less arm-level amplifications (p=0.04) and more arm-level deletions (p=0.004) than non-IMA, and the frequency of amplification and deletion also showed a trend of gradual decrease and increase from non-IMA to mixed-IMA to pure-IMA, respectively. Furthermore, prognosis analysis in stage III IMA patients showed that patients harboring alterations in EGFR (mDFS=30.3 vs. 16.0 months, HR=0.19, P=0.027) and PI3K pathway (mDFS=36.0 vs. 16.0 months, HR=0.12, P=0.023) achieved prolonged DFS, while patients with poorly differentiated tumors (mDFS=14.1 vs. 28.0 months, HR=3.75, p=0.037) or with KRAS mutations (mDFS=13.0 vs. 20.0 months, HR=6.95, p=0.027) had shorter DFS. Multivariate analysis showed that KRAS mutations, PI3K pathway alterations, and tumor differentiation status were independent factors that have statistically significant influences on clinical outcomes of IMA patients.
Conclusion: Our study provided genomic insights into Chinese surgically resected lung IMA. We also identified several genomic features that may serve as potential biomarkers on postoperative recurrence in IMA patients with stage III disease.
Introduction
Lung cancer is the leading cause of cancer-related mortality among various malignancies worldwide (1). In addition to surgery and chemoradiotherapy, targeted therapy has now become an effective treatment for lung cancer (2–5), which in turn emphasizes the importance of understanding tumors at gene levels.
According to the lung adenocarcinoma classification system proposed by the International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) in 2011 (6), invasive mucinous adenocarcinoma (IMA) is considered to be a distinct subtype of lung adenocarcinoma. IMA appears as translucent and grayish jelly-like lesions at gross observation, while the tumor cells are typified by a tall columnar or goblet cell morphology with abundant intracellular or extracellular mucus under the microscope (7). IMA has a low incidence, accounting for only 2%–5% of lung invasive adenocarcinoma, and IMA is associated with poor survival outcomes (6, 8). It has been shown that more than 70% of IMA tumors were associated with the spread-through-air-space pattern, which a poor prognostic predictor for various lung adenocarcinoma subtypes (9). In addition, some distinct clinic-pathological features were observed within the IMA subtype. Beck et al. reported spontaneous regression of airspace opacities in some patients with IMA, who tended to have worse overall survival than other IMA patients (10). Several pathological parameters of the MIA tumors, such as the mucin spread size, tumor cell spread size, and invasive size, were also negatively correlated with poor prognosis (11). Therefore, MIA is a clinically important and complex lung adenocarcinoma subtype that needs to be carefully investigated.
A number of studies have suggested that IMA differs from other adenocarcinomas in clinical manifestations and pathology (6, 8, 12, 13). Some studies indicated that certain genetic differences have been observed between IMA and other non-mucinous adenocarcinomas (non-IMA) particularly for driver genes. Early studies found that the mutation frequency of EGFR was lower than other adenocarcinomas (undetectable to ~28% in IMA vs. 47%–78% in non-IMA patients) (14, 15), while the frequency of KRAS mutations was higherin IMA than other adenocarcinomas (14%–86% in IMA vs. 1.5%–17% in non-IMA patients) (16, 17). These results implied that the mucinous differentiation might be partially relied on KRAS aberrations, whereas EGFR mutations tended to be involved in driving the development of other adenocarcinoma subtypes. However, these studies only detected a narrow range of genes and did not include other key oncogenic drivers, such as ALK fusions. In 2014, high rates of ALK fusions and KRAS mutations were observed in IMA from a study containing 13 samples, and a rare CD74-NRG1 fusion was also detected (18). In the same year, a study conducted on 44 pulmonary mucinous adenocarcinomas found that IMA had a lower frequency of EGFR mutations and a higher frequency of several other genetic alterations (e.g., KRAS mutations, HER2 mutations, and ALK fusions) than other invasive adenocarcinomas (19). These studies revealed that IMA is likely to have a unique genetic profile, especially for the driver genes. Although more and more genetic changes have been detected in IMA, the overall genetic spectrum is still unclear. In 2015, a study of 72 IMA patients from the USA and South Korea showed that KRAS mutations were the most common genetic aberrations (63%), whereas in KRAS-negative patients several rare gene fusions and mutations were observed, including CD74-NRG1, VAMP2-NRG1, TRIM4-BRAF, TPM3-NTRK1, and EML4-ALK fusions, as well as ERBB2, BRAF, and PIK3CA mutations (12). Among those 72 IMA patients, only 2 of them harbored TP53 mutations, which was much less than that observed in unselected lung adenocarcinomas (12). This study also employed next-generation sequencing (NGS) for gene detection, which increased the coverage and detecting sensitivity compared to previous studies. In 2016, another NGS-based study using a panel of 50 genes identified KRAS and TP53 as the most commonly mutated genes in IMA, as well as several rare EGFR gene mutations (20). Of note, this study categorized IMA into pure-IMA (≥ 90% invasive mucinous pattern) and mixed-IMA (> 10% of the non-mucinous invasive component) (20). Specifically, ALK rearrangements were found in two of mixed-IMA patients but none in pure-IMA patients, and patients with mixed-IMA had poorer prognosis (20), indicating that IMA patients with different proportions of mucinous components may have different molecular profiles and clinical prognosis, which is worthy of further investigations.
A review of existing studies suggested that the frequency of EGFR mutations was lower and the frequency of ALK fusions and KRAS mutations was markedly higher in IMA patients compared to the general lung invasive adenocarcinomas, whereas the frequency of rare mutations remains controversial. In addition, most of the previous studies were conducted on the Westerner population using small panel molecular assays, while the extensive genetic analysis in the Chinese population is still lacking. In this study, we retrospectively analyzed 76 surgically resected primary tumors from Chinese patients with invasive lung adenocarcinoma, including 51 IMA and 25 non-IMA. Furthermore, based on the presence of a non-mucinous component, IMA was further divided into pure-IMA and mixed-IMA subgroups. Targeted NGS of 425 cancer-relevant genes was performed on these tumors, and their genomic profile was comprehensively investigated and compared among 3 different histological subgroups. We also explored the association between molecular aberrations and postoperative disease-free survival (DFS) in stage III IMA patients. This study could enhance our understanding of the molecular characteristics and the potential prognostic biomarkers of East-Asian IMA patients.
Methods
Patients
A total of 79 surgically resected primary invasive lung adenocarcinomas diagnosed between October 2010 to July 2019 at the Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital). All cases were retrospectively collected and were centrally reviewed by two pathologists according to the 2015 WHO classification of lung adenocarcinoma. Specifically, during the screening process, patients with mucinous component higher than 10% were enrolled in the IMA group. Among the 79 patients, three did not pass the quality control of the sequencing analysis, resulting in 51 IMA and 25 non-mucinous adenocarcinomas (non-IMA). In addition, given that some IMAs have certain levels of the non-mucinous component to some extent, IMA can be further classified based on the percentage of mucinous pattern, that is, pure mucinous (≥90% of the invasive mucinous pattern) and mixed mucinous/non-mucinous (>10% of the non-mucinous invasive component). All patients provided informed consent forms in accordance with institutional regulations and study protocols were approved by the Ethical Review Community of Zhejiang Cancer Hospital.
DNA Extraction and Library Construction
NGS was performed in a centralized testing center (Nanjing Geneseeq Technology Inc.). DNA extraction, library preparation, and targeted-capture enrichment were performed as previously described (21). Briefly, genomic DNA from the white blood cells (WBCs) was extracted using the DNeasy Blood & Tissue Kit (Qiagen) and used as the normal control to distinguish germline mutations. Formalin-fixed and paraffin-embedded (FFPE) samples were de-paraffinized with xylene, and genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen). DNA was quantified by Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies), and the quality was evaluated by a Nanodrop 2000 (Thermo Fisher).
Libraries were prepared by the KAPA Hyper Prep kit (KAPA Biosystems) as previously described (22). Briefly, genomic DNA was sheared into fragments using a Covaris M220 instrument. End repair, A-tailing, and adaptor ligation of fragmented DNA were performed using the KAPA Hyper DNA Library Prep kit (Roche Diagnostics). DNA Libraries were then amplified by polymerase chain reaction (PCR) and purified using AgencourtAMPure XP beads.
Customized xGen lockdown probes panel (containing 425 predefined cancer-related genes) were used for selective enrichment. The capture reaction was performed with Dynabeads M-270 (Life Technologies) and the xGen Lockdown Hybridization and Wash kit (Integrated DNA Technologies). Captured libraries were PCR-amplified with KAPA HiFi HotStart ReadyMix (KAPA Biosystems). The purified library was quantified using the KAPA Library Quantification kit (KAPA Biosystems).
Sequencing and Bioinformatics Analysis
Target enriched libraries were sequenced on the HiSeq4000 platform (Illumina). Sequencing data were demultiplexed by bcl2fastq (v2.19), analyzed by Trimmomatic (23) to remove low-quality (quality<15) or N bases. Then the data were aligned to the hg19 reference human genome with the Burrows-Wheeler Aligner (bwa-mem) (24) and further processed using the Picard suite (available at https://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit (GATK) (25) SNPs and indels were called by VarScan2 (26) and HaplotypeCaller/UnifiedGenotyper in GATK, with the mutant allele frequency (MAF) cutoff as 0.5%. Common variants were removed using dbSNP and the 1000 Genome project databases. Germline mutations were filtered out by comparing to patient’s WBCs controls.
Gene fusions were identified by FACTERA (27) and copy number variations (CNVs) were analyzed with ADTEx (28). The log2 ratio cut-off for copy number gain was defined as 2.0 for tissue samples. A log2 ratio cut-off of 0.6 was used for copy number loss detection. Allele-specific somatic CNVs (SCNVs) were analyzed by FACETS (29) with a 0.2 drift cut-off for unstable joint segments. In pathway analysis, sets of genetic aberrations were included according to the ten previously defined canonical oncogenic signaling pathways (30).
Statistical Analysis
Quantitative data were presented as median (range) or the number of patients (percentage). Comparisons of proportion between groups were performed using Fisher’s exact test. Survival analysis was performed using Kaplan-Meier curves, and the p-value was determined with the log-rank test, and hazard ratios (HRs) were calculated by Cox proportional hazards model. A two-sided p-value of less than 0.05 was considered significant for all tests unless indicated otherwise. Univariate and multivariate analyses were used to study the associations between different variables and DFS, and the results were presented as HRs and their 95% confidence intervals (CIs). All analyses were performed with R 3.4.0.
Results
Patient Characteristics
Baseline demographics and clinical characteristics of 51 IMA and 25 non-IMA patients were summarized in Table 1. There were no significant differences between patients with IMA and those with non-IMA with respect to age, sex, stage, smoking status, intravascular tumor thrombus, perineural invasion, differentiation grading, and pleural invasion. The median age of IMA and non-IMA was 61 and 62 years, respectively. Twenty-eight females (55%) were enrolled in IMA cohort and 11 females (44%) were enrolled in non-IMA cohort. More than half of both IMA and non-IMA patients did not have a smoking history. The proportion of stage III patients in IMA cohort seemed more than that in non-IMA cohort, but no statistical significance was observed (p=0.11). The majority of the patients did not have intravascular tumor thrombus or perineural invasion, and slightly more patients had moderately differentiated tumors than those with poorly differentiated tumors. Nearly half of the IMA patients had pleural invasion, whereas there were a lower proportion of the non-IMA patients who had pleural invasion, although the difference was not statistically significant. According to the proportion of mucinous components, patients with IMA were divided into sub-groups of pure-IMA (n=25) and mixed-IMA (n=26).
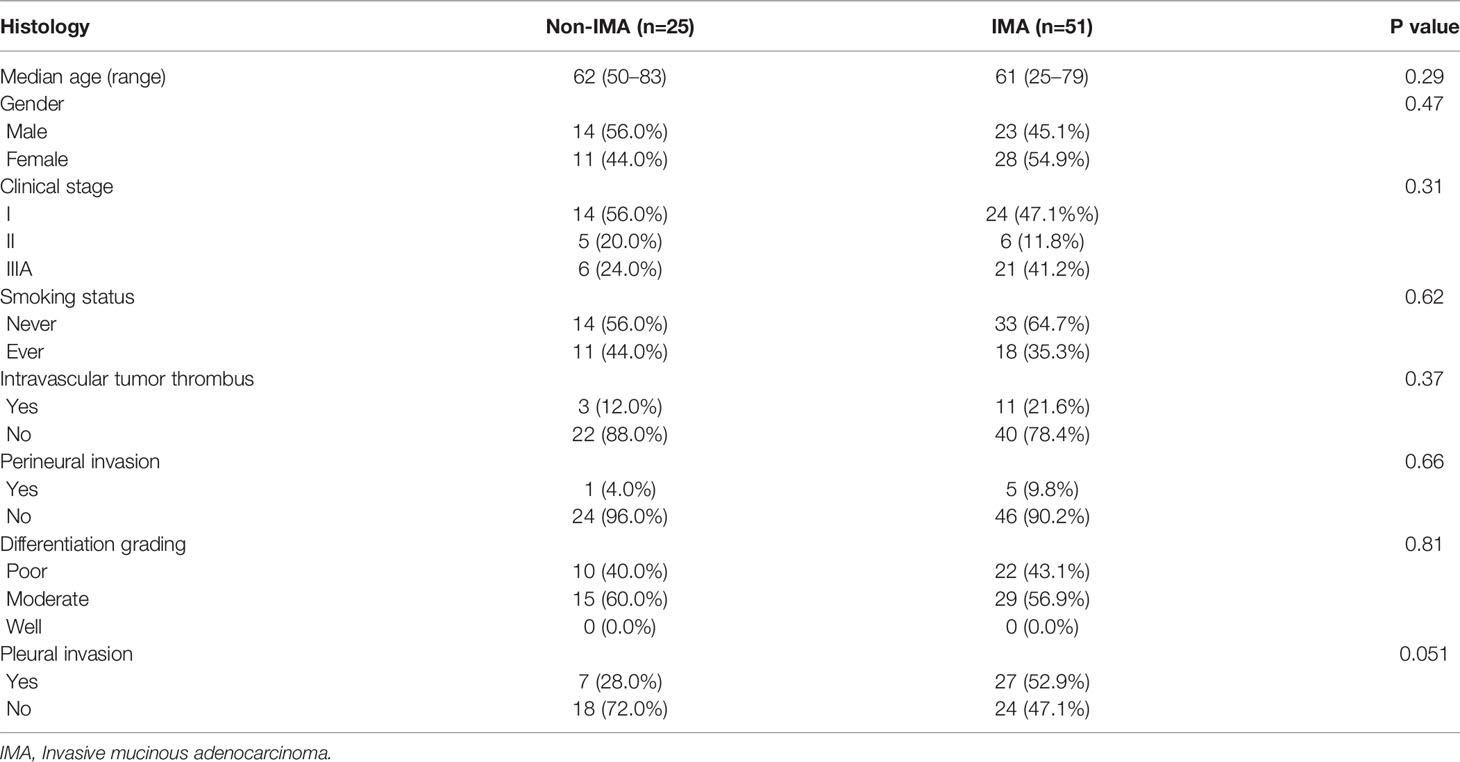
Table 1 Baseline demographics and clinical characteristics of non-IMA and IMA patients enrolled in this study.
Different Mutational Spectra of Driver Genes Between IMA and Non-IMA
Given that each tumor sample had the corresponding white blood cell sample as the normal control, we used these normal control samples to filter out the germline mutations, and all of our molecular analyses were based on tumor somatic alterations. In IMA patients, the most frequently altered driver genes were EGFR (33.3%), ALK (27.5%), KRAS (13.7%), ERBB2 (11.8%), and PIK3CA (11.8%) (Figure 1 and Figure S1). Among these alterations, the well-known EGFR driver mutations (i.e., L858R, exon 19 deletions, and G719S) and ALK rearrangements accounted for 31.4% and 23.5%, respectively (Figure S2). Specifically, EGFR G719S, L858R, and exon 19 deletion (E19 del) were detected in one (2.0%), 3 (5.9%), and 12 (23.5%) patients, respectively. In addition to these known driver mutations, EGFR A750P and E19 del coexisted in one patient, and a concomitant EGFR L747S mutation was also observed in a patient with EGFR G719S. Besides, an EGFR exon 20 insertion (20Ins) was detected in 1 (2.0%) patient. In terms of the ALK rearrangement, 10 (19.6%) patients harbored EML4-ALK fusions, one (2.0%) had an intergenic region (IGR)-ALK fusion (i.e., downstream TTC32-ALK exon 18 fusion), and one patient (2.0%) had IGR (downstream CTNNA2)-ALK exon 20 and ALK exon 18-LYN 5’ UTR co-fusions. ERBB2 20Ins were detected in four (7.8%) patients, which was much higher than that in unselected lung adenocarcinoma patients. In addition, the frequencies of RET and NRG1 alterations were both 5.9%, which includes two patients with RET fusions (KIF5B-RET and CCDC6-RET) and two patients with NRG1 fusions (SLC3A2-NRG1) (Figure 1 and Figure S2). Although MET aberrations were not detected in IMA patients, MET (12.0%), together with EGFR (72.0%) and KRAS (12.0%), was commonly mutated in non-IMA patients. The mutation frequency of ALK, PIK3CA, BRAF, and ROS1 was all 4.0% (Figure 1 and Figure S2), and MET exon 14 skipping was detected in two (8.0%) patients (Figure 1 and Figure S2). The frequency of EGFR (33.3% vs. 72.0%, p<0.01) and MET (undetected vs. 12.0%, p=0.03) mutations in IMA were significantly lower than that in non-IMA patients while the frequency of ALK rearrangements showed the opposite trend (27.5% vs. 4.0%, p=0.01). IMA group tended to have more ERBB2 mutations than non-IMA group, albeit no statistical difference (11.8% vs. 0%, p=0.17). Differences in frequencies of other driver genes were observed, although the results were not statistically significant (Figure 1 and Figure S2). In addition, all the gene mutations detected in three or more patients were shown in Figure S1. Among these genes, the mutation frequency of FBXW7 was significantly lower in IMA than in non-IMA (undetected vs. 12.0%, p=0.03) (Figure S1). In terms of gene-level copy number variations, there was no significant difference between non-IMA and IMA in our patient cohort (Data not shown).
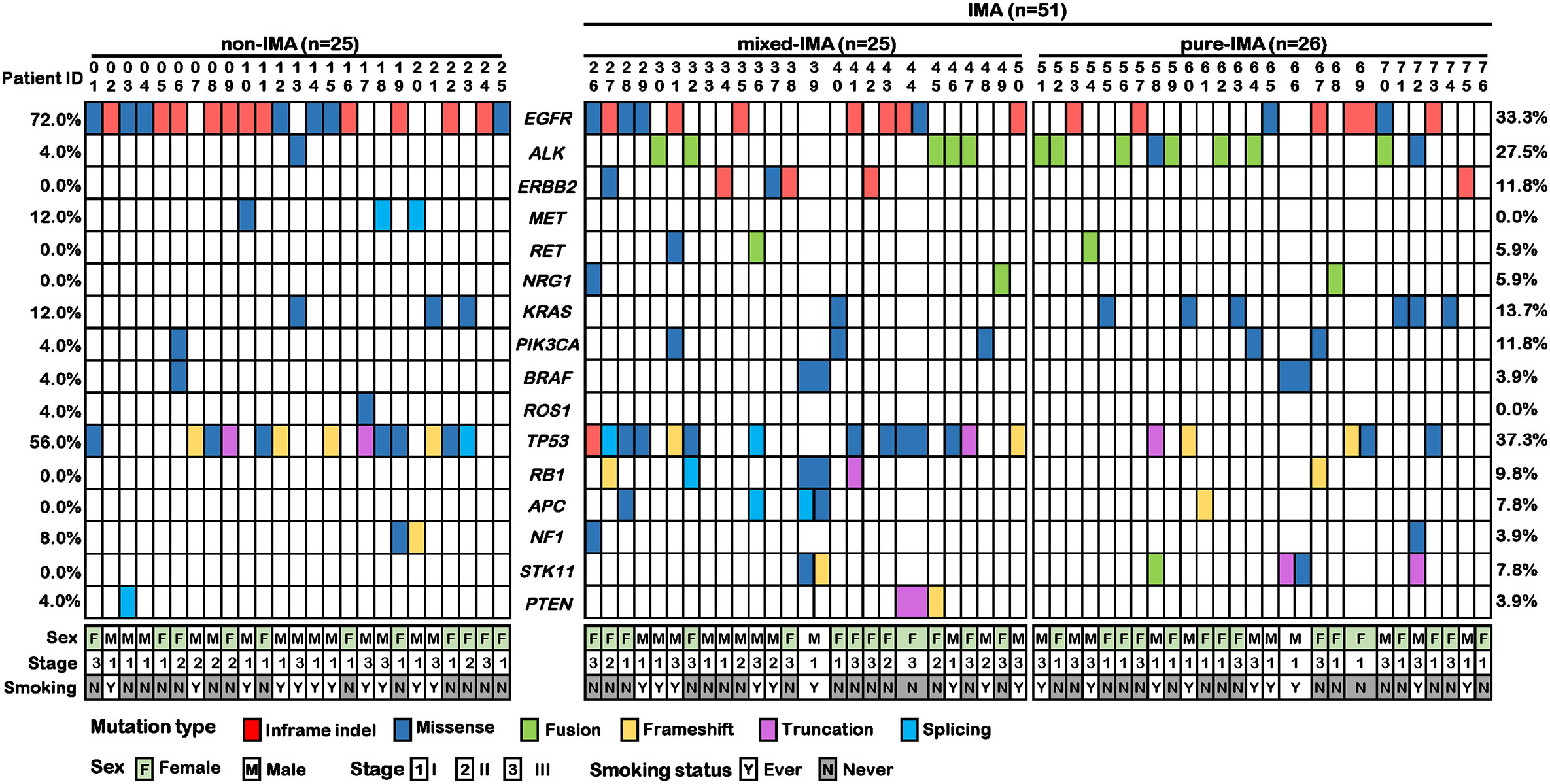
Figure 1 Co-mutation plot of common driver and suppressor genes in invasive mucinous adenocarcinoma (IMA) and non-IMA patients in our cohort. The mutation profile of 25 non-IMA patients and 51 IMA patients were shown. The IMA group could be further divided into mix-IMA (n=25) and pure IMA (n=26) based on the mucinous content of the tumor. Different colors indicate different mutational types. Clinical information on sex, TNM stage, and smoking history was shown at the bottom.
Frequency of Driver Mutations Related to the Proportion of Mucinous Component
Based on the presence of the non-mucinous component, IMA was further divided into pure-IMA and mixed-IMA subgroups, and comparative analysis of their driver genes was performed. Intriguingly, as the content of the mucinous component increased, the frequency of EGFR mutations gradually decreased, with significantly higher frequency in non-IMA comparing with mixed-IMA (72.0% vs. 40.0%, p=0.045) or pure-IMA (72.0% vs. 26.9%, p<0.001). The EGFR mutation frequency in mixed-IMA was higher than pure-IMA, but no significant difference was observed (p=0.24) (Figure 2A, Figure S2B). By contrast, as mucinous content increased, the frequency of ALK fusions gradually increased, with significantly higher frequencies in mixed-IMA (20.0% vs. undetected, p=0.05) and pure-IMA (26.9% vs. undetected, p=0.01) when compared with non-IMA (Figure S2B). The ALK fusion was slightly more frequent in pure-IMA than mixed-IMA, with no statistically significant difference (26.9% vs. 20.0%, p=0.74) (Figure 2B). On the other hand, the frequency of KRAS mutation in pure-IMA was higher than that in non-IMA (23.1% vs. 12.0%) and mixed-IMA ad less KRAS mutation than non-IMA (4.0% vs. 12.0%), although none of these results were statistically significant (pure-IMA vs. non-IMA, p=0.47; mixed-IMA vs. non-IMA, p=0.36) (Figure 2C and Figure S2B). Besides, mixed-IMA tended to have more ERBB2 20Ins than pure-IMA (11.5% vs. 4.0%, p=0.35). NRG1 fusion and RET fusion were individually observed in one patient from each of the two IMA cohorts of (Figure S2B).
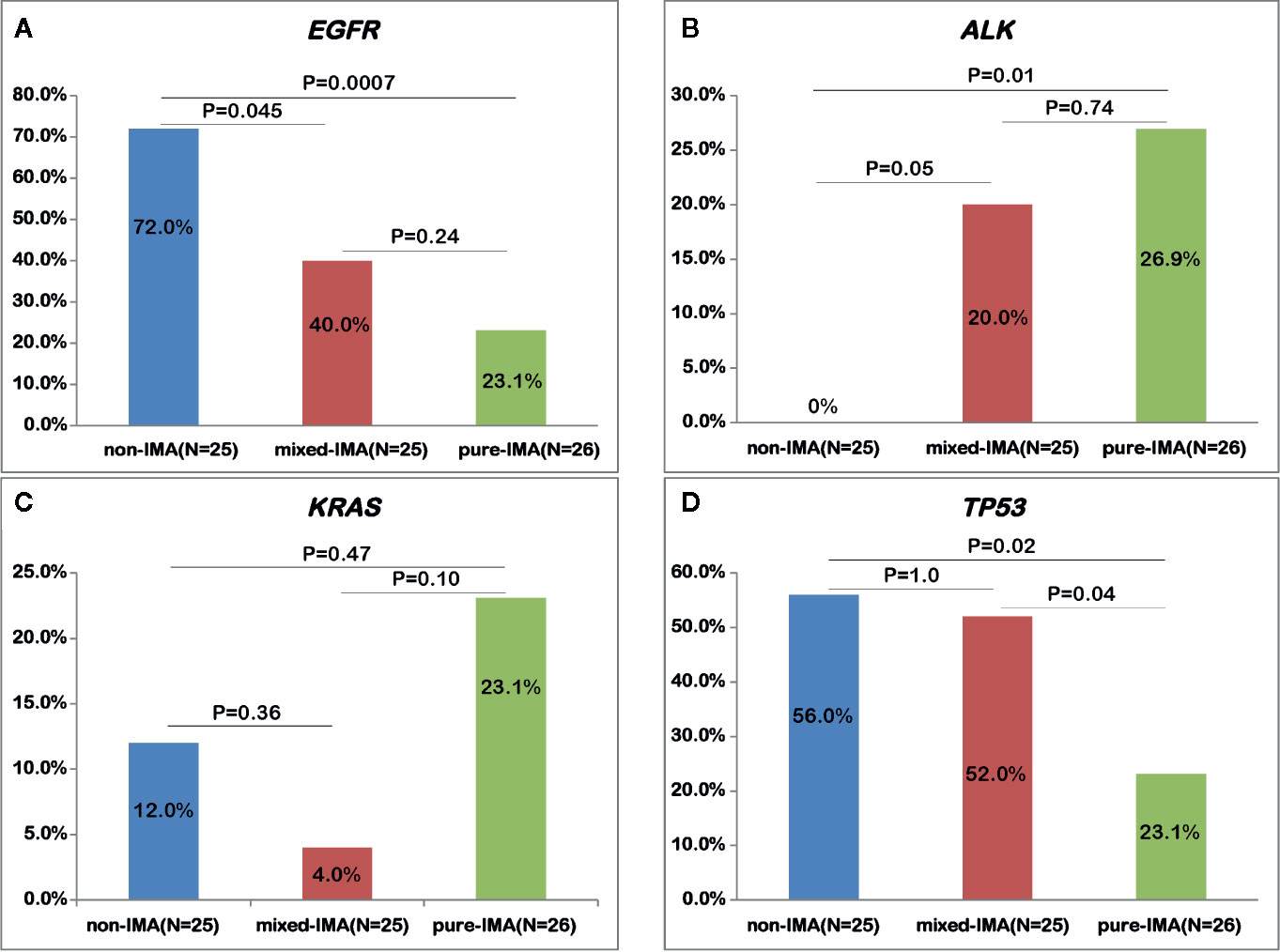
Figure 2 Comparing the alterations of common driver and suppressor genes among samples with different mucinous contents. The mutational frequency of EGFR mutation (A), ALK fusion (B), KRAS mutation (C), and TP53 mutation (D) was compared in patients with non-IMA, mixed-IMA and pure-IMA.
Different Profile of Tumor Suppressor Genes Between IMA and Non-IMA
Next, we investigated the mutational profile of tumor suppressors in patients with IMA and non-IMA. TP53 (37.3% vs. 56.0%, p=0.14) showed a lower frequency while RB1 (9.8% vs. 0%, p=0.17) showed a higher frequency in IMA than in non-IMA (Figure 1). In addition, the mutation frequency of other tumor suppressors, including NF1, APC, PTEN, and STK11, was higher in IMA than in non-IMA, albeit no significant difference (27.5% vs 12.0%, p=0.15). Interestingly, the frequency of co-mutations in these tumor suppressors was also higher in IMA than in non-IMA (19.6% vs 4.0%, p=0.09). In a specific subgroup of patients who harbored tumor suppressor mutations, the frequency of co-mutations in these suppressor genes was significantly higher in IMA patients than in non-IMA patients (40.0% vs 6.3%, p=0.003) (Figure 1). Similar to the results of EGFR mutations, as mucinous content increased, the frequency of TP53 mutations gradually decreased (Figure 1). TP53 mutations in pure-IMA were significantly more frequent than in non-IMA (23.1% vs. 56.0%, p=0.02) or mixed-IMA (23.1% vs. 52.0%, p=0.04), whereas no statistical difference was observed between non-IMA and mixed-IMA (p=1.0) (Figure 2D).
No Difference in the Number of Somatic Mutations Between IMA and Non-IMA
We then compared the number of somatic mutations between non-IMA and IMA patients, including missenses, insertions/deletions (indels), fusions, and splice-site mutations. No significant difference in the number of somatic mutations was observed between IMA and non-IMA (p=0.8), and the median number was four and five in IMA and non-IMA, respectively (Figure 3A). Similarly, there was no significant difference in the number of mutations among non-IMA, mixed-IMA, and pure-IMA (Figure 3B).
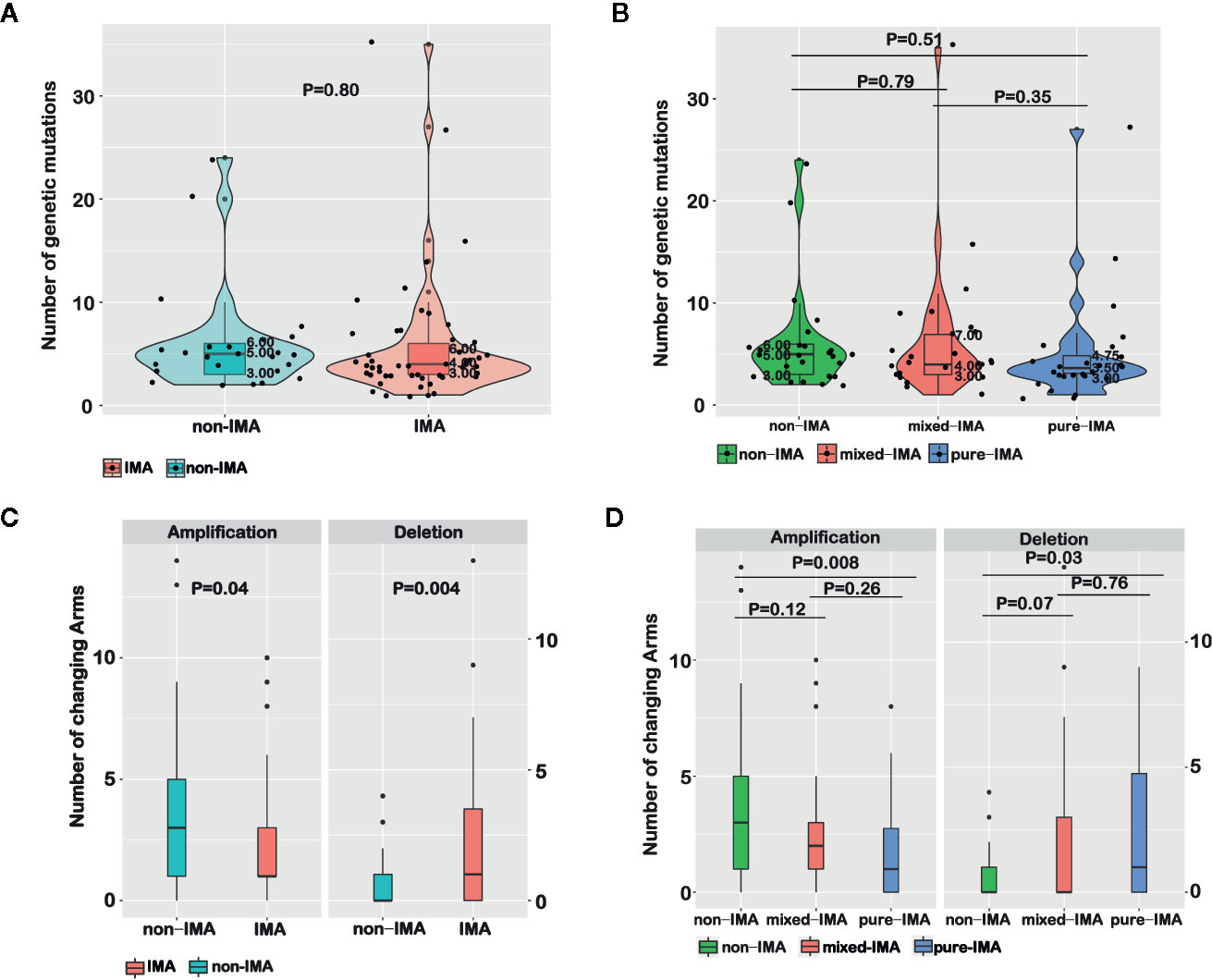
Figure 3 Comparing molecular features among samples with different mucinous contents. The number of genetic mutations for invasive mucinous adenocarcinoma (IMA) vs. non-IMA patients (A) or for patients that were divided based on the mucinous content (B) was shown in violin plots. The number of chromosome arm-level somatic CNVs (SCNVs) for IMA vs. non-IMA patients (C) or for patients that were divided based on the mucinous content (D) was shown in bar graphs.
The Different Pattern of Chromosome Arm-Level SCNVs in Non-IMA and IMA
The chromosome arm-level SCNVs in non-IMA and IMA were analyzed. In IMA, the number of amplifications (p=0.04) was significantly lower while the number of deletions (p=0.004) was significantly higher than in non-IMA (Figure 3C). In detail, the frequencies of 5p (OR=0.34, 95% CI, 0.11–1.04; P=0.04), 6q (OR=0.11, 95% CI, 0.00–1.18; P=0.04), 8q (OR=0.21, 95% CI, 0.05–0.74; P=0.01), 16p (OR=0.14, 95% CI, 0.02–0.65; P<0.01), 16q (OR=0.17, 95% CI, 0.01–1.13; P=0.04), and 20q (OR=0.21; 95% CI, 0.06–0.68; P<0.01) amplifications were significantly lower in IMA than in non-IMA (Table S1). The frequency of 18q deletion in IMA was higher than that in non-IMA (OR=Inf; 95% CI, 0.9–Inf; P=0.05). The frequencies of 12p (OR=Inf; 95% CI, 0.74–Inf; P=0.09) and 18p (OR=6.48; 95% CI, 0.84–295.3; P=0.09) deletions in IMA were higher than that in non-IMA (Table S1). In addition, the arm-level SCNVs in patients with different proportions of the mucinous component were also analyzed. As the proportion of mucinous component increased, the frequency of amplification gradually decreased while the frequency of deletions gradually increased. There were also trends in arm-level SCNV differences when comparing non-IMA and mixed-IMA (amplification, p=0.12; deletion, p=0.07). Significant differences in SCNVs were observed between non-IMA and pure-IMA (amplification, p=0.008; deletion, p=0.03), whereas no significant difference was found between mixed-IMA and pure-IMA (p>0.05) (Figure 3D).
Comparison of Mutation Profile Between East-Asian and Western IMA Patients
Herein, a study conducting on Caucasians reported by Righi et al. (20) was selected as the reference study, in which 49 out of 50 genes used in their panel were also analyzed in our sequencing panel. In addition, patients in this reference study were mainly Western patients who were also divided into pure-IMA and mixed-IMA subgroups. Several driver genes that we analyzed above were then compared between the two patient cohorts. The frequency of ALK fusions (23.5% vs. 4.2%, p=0.008) and ERBB2 mutations (11.8% vs. undetected, p=0.03) was significantly higher in our cohort while the frequency of EGFR mutations was comparable between the two cohorts (33.3% vs. 18.8%, P=0.12) (Figure 4). The frequency of KRAS mutations was significantly lower in our cohort (13.7% vs. 68.8%, p<0.001) (Figure 4). Furthermore, we compared the gene mutation profile between our cohort with the reference study in terms of the mucinous content. As shown in Figure S3, for mixed-IMA patients, there was a trend of higher, but not statistically significant frequency of ALK rearrangement (P=0.38) and EGFR mutation (P=0.33) in our study. ERBB2 mutation also had the trend to be more frequent in our patients (P=0.06). However, there was almost no difference in the frequency of TP53 and APC mutations (Figure S3). For pure-IMA patients, the frequency of ALK rearrangement was significantly higher in our study than in the reference study (26.9% vs. 3.3%, P=0.019), while the frequency of EGFR mutation was similar between the two studies (26.9% vs. 20.0%, p=0.52). Besides, there was almost no difference in mutation frequencies for ERBB2 and FLT3. Mutations in TP53 (23.1% vs. 76.5%, p=0.16) and APC (3.8% vs. 25.0%, p=0.11) were less in our cases, although no significant difference was observed. The frequency of KRAS mutations in both mixed-IMA and pure-IMA was significantly lower in our study than that in the reference study (p<0.001) (Figure S3).
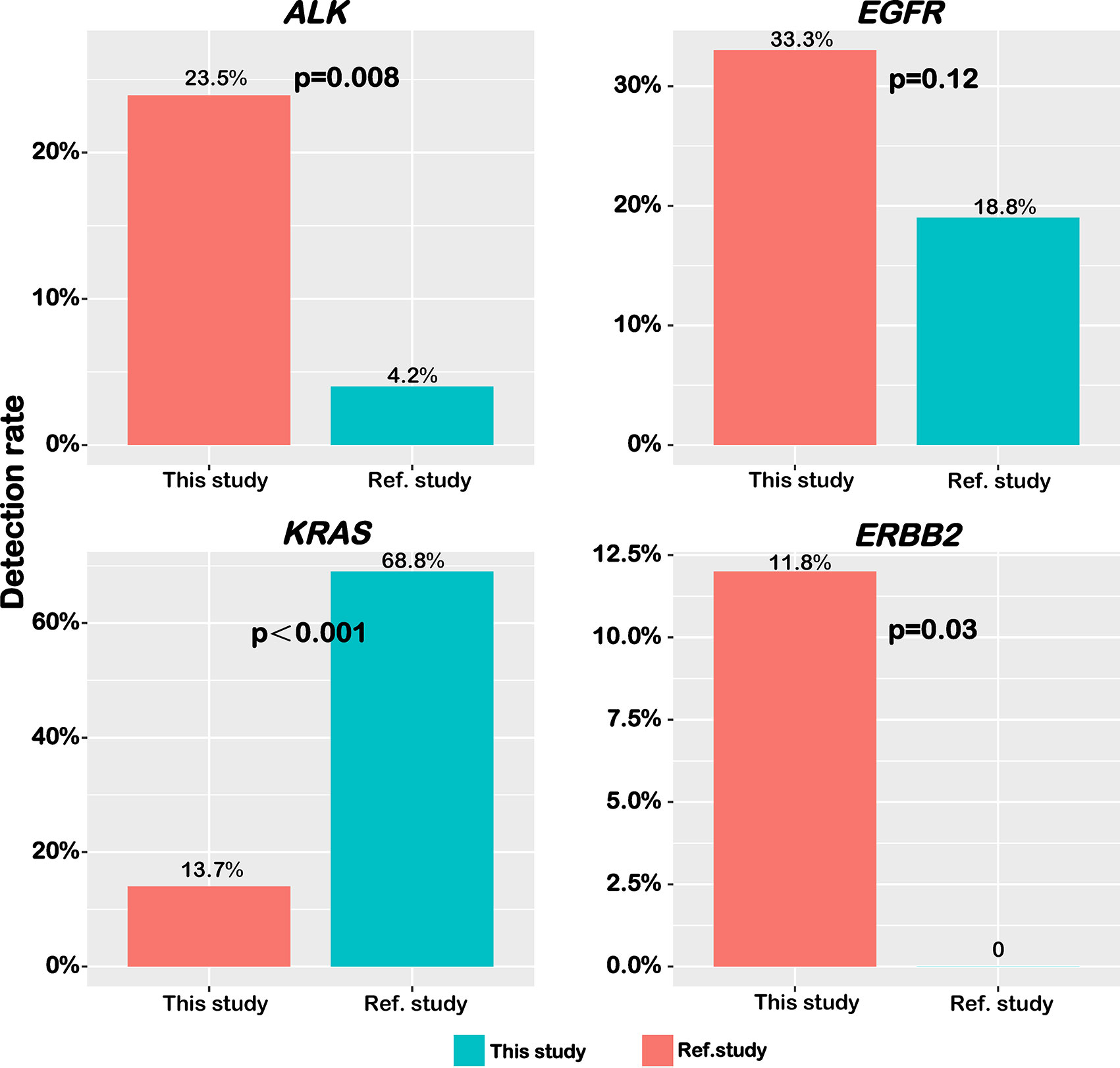
Figure 4 Distinct frequency of common driver genes between Chinese and Caucasian patients. The frequency of ALK fusion, EGFR mutation, KRAS mutation, and ERBB2 mutation was shown in bar graphs.
Individual Somatic Gene and Signaling Pathways Alterations Associated With Recurrence in Surgically Resected Patients With IMA
We then aimed to investigate whether the molecular profile in IMA patients affected their prognosis. We compared the postoperative disease-free survival (DFS) between patients with stage I/II tumors and stage III tumors. As shown in Figure 5A, the median DFS (mDFS) of patients with stage III disease was significantly worse than that of stage I/II (mDFS=17.0 months vs. Not reached; HR=6.58; 95% CI, 2.28–18.98; p<0.001). Due to the higher maturity of DFS in stage III patients, we further analyzed the correlations between molecular features and DFS in these patients. Survival data of 18 patients were available among the 21 stage III patients with IMA, and 14 of whom have relapsed after surgery. Our results showed patients with EGFR mutations had prolonged DFS than wildtype patients (Figure 5B). EGFR-mutated patients had an mDFS of 30.3 months, compared with an mDFS of 16.0 months in wildtype patients (HR=0.19; 95% CI, 0.04–0.96; P=0.027; Figure 5B). In contrast, patients with KRAS mutations showed shorter mDFS than those without KRAS mutations (mDFS=13.0 vs. 20.0 months; HR=6.95; 95% CI, 0.96–50.45; p=0.027) (Figure 5C). Previous studies reported that TP53 mutations may lead to a worse prognosis in TKI-treated NSCLC patients; however, we did not observe significant differences in mDFS between patients with TP53 mutations and those with wildtype TP53 (mDFS=16.0 vs. 20.0 months; HR=1.18; 95% CI, 0.39–3.57; p=0.771) (Figure S4A). Moreover, in EGFR-mutated patients, no significant difference of mDFS was found in cases who had concomitant TP53 mutations and those without TP53 co-mutations (mDFS=30.3 vs. 28.0 months; p=0.810) (Figure S4B). As for other known oncogene drivers, such as KRAS, ERBB2, RET, NRG1, and ALK, there were only a limited number of patients harboring these oncogenic mutations or having available clinical results, thus the clinical impact of concomitant TP53 mutations was still inconclusive in our patient cohort. Furthermore, the results of pathway analysis showed patients with genetic alterations in the PI3K pathway showed improved DFS (mDFS=36.0 vs. 16.0 months; HR=0.12; 95% CI, 0.02–0.99; P=0.023; Table S2 and Figure 5D).
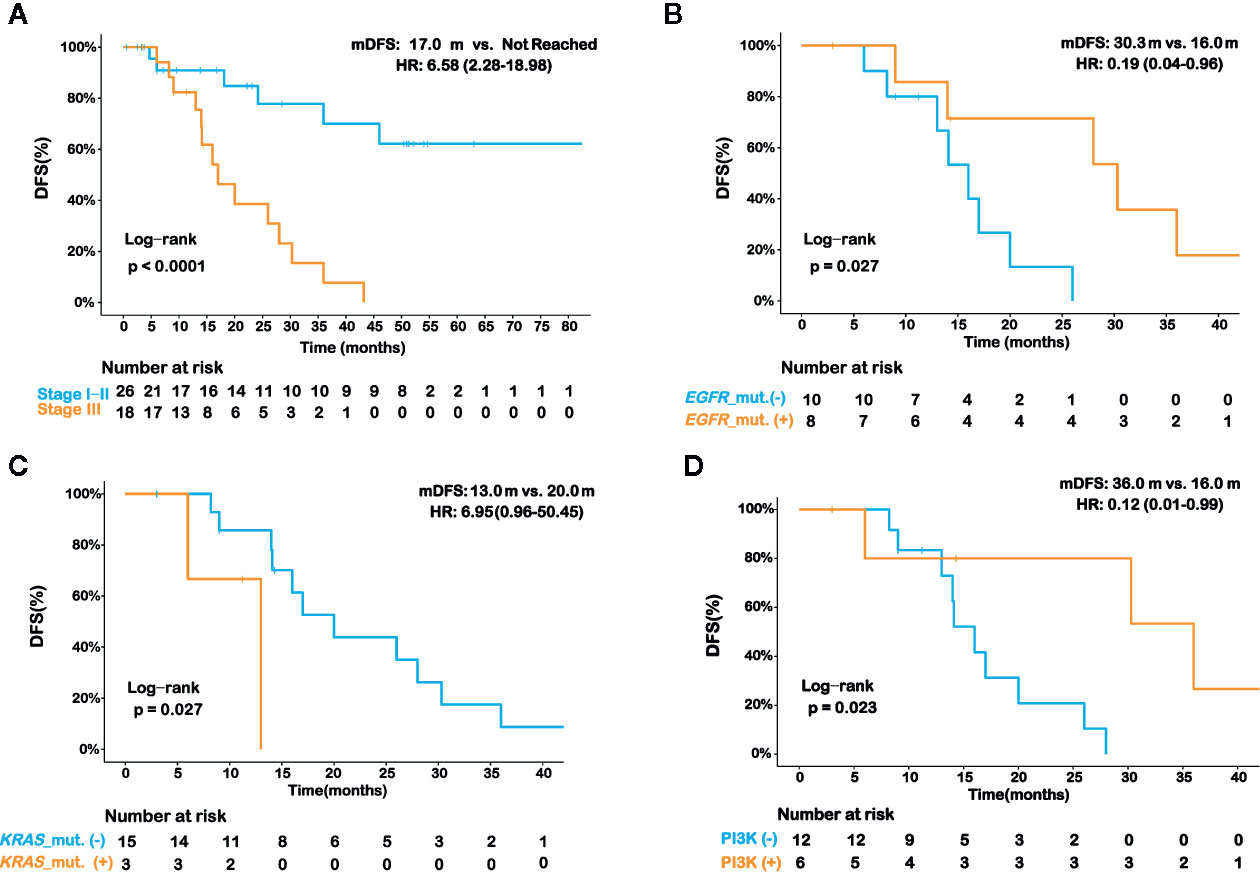
Figure 5 Prognostic analysis for various histopathologic and molecular features in invasive mucinous adenocarcinoma (IMA) patients. Kaplan-Meier curve of disease-free survival (DFS) in IMA patients in strata of different disease stages (A). Kaplan-Meier curve of DFS in stage III IMA patients in strata of EGFR mutation status (B), KRAS mutation status (C), and PI3K signaling pathway alteration status (D). The log-rank test was used to analyze the DFS for all of the survival analyses.
Lastly, we checked if some histopathologic features, such as intravascular tumor thrombus, perineural invasion, differentiation grading, and pleural invasion, were associated with prognosis in stage III IMA patients. By univariate analysis, we found that only the differentiation grading was significantly associated with DFS (Table S3). Specifically, IMA patients with moderately differentiated tumors had better DFS than those with poorly differentiated tumors (mDFS 28.0 vs. 14.1 mouths, p=0.037). In addition, we included both the differentiation grading and clinically-important molecular features that were identified in our study (Figures 5B–D) to perform a multivariate analysis. Intriguingly, DFS in these IMA patients remained to be significantly associated with KRAS mutation (p=0.016), PI3K pathway alteration (p=0.043), and differentiation grading (p=0.030), but not EGFR mutations (p=0.391) (Table 2), which might be due to the impact of EGFR mutations on lung adenocarcinomas differentiation, as previously described (31).
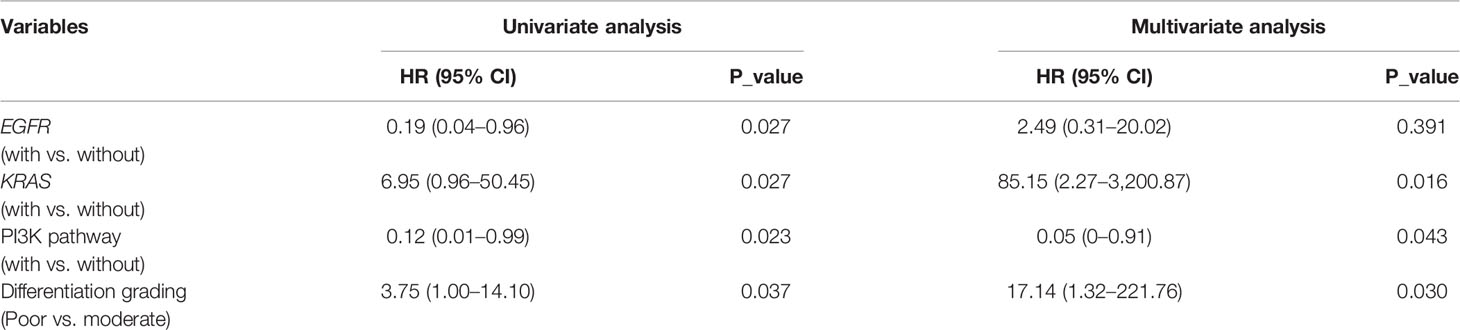
Table 2 Impact of genetic factors and differentiation grading on disease-free survival (DFS) in stage III patients with disease-free survival (IMA).
Discussion
In our study, the most commonly mutated driver gene in Chinese IMA patients was EGFR. The frequency of EGFR mutations was significantly lower while the frequency of ALK fusion was significantly higher in IMA compared to non-IMA, which was consistent with the previous studies (14–19, 32). The comparison of mixed-IMA and pure-IMA showed that there was no statistical difference in the frequency of EGFR or ALK variations, suggesting that these gene-level changes might happen in the entire tumor when lung adenocarcinoma harbors mucinous components. Interestingly, we observed a reverse trend in the frequency of EGFR and ALK alterations among patients with different proportions of the mucinous component, ranging from non-IMA, mixed-IMA, to pure-IMA. These results implied that adenocarcinoma with different proportion of mucinous component might be driven by distinct gene mutations.
In previous studies, KRAS mutation was more frequent in IMA, reaching as high as 63% (12, 17–19). In our study, the frequency of KRAS mutations was low in IMA (13.7%) and non-IMA (12.0%), which is likely due to the difference in KRAS mutation frequency between Caucasians and East-Asian populations (33). Of note, the frequency of KRAS mutations tended to be higher in pure-IMA compared to mixed-IMA (23.1% vs. 4.2%, p=0.1) in our East-Asian patient cohort, while almost no difference in KRAS mutation frequency was observed between pure-IMA and mixed-IMA in a study conducting on Caucasian patients (34). This suggested that, compared to Caucasians, there may be differences in the frequency of KRAS mutations in lung adenocarcinoma harbored different mucinous content in East-Asians. In a study performed on the Chinese population, reported by Chen et al., the frequency of KRAS mutations was 36%, which was higher than that of 13.7% in our study (19). As the mucinous content of IMA patients was not discussed in Chen et al.’s study, the detailed proportion of pure-IMA and mixed-IMA was unknown and, we speculated that the differences in KRAS mutation frequency may be partially due to the different proportion of pure-IMA patients.
In addition, multiple gene fusions, such as NRG1, RET, and ALK, were only detected in IMA patients in our cohort and the detecting rate (16/51, 31.4%) of gene fusions was significantly higher than the unselected lung adenocarcinoma patients (34). In addition, greater diversity in driver gene mutations in IMA compared to non-IMA was observed. For example, in a large cohort study of 10,966 Chinese NSCLC patients, the frequency of NRG1 fusions was only 0.16% in these unselected lung cancer patients, with CD74 being the most common fusion partner (35). In contrast, NRG1 fusions occurred in 2 out of 51 (3.9%) IMA patients in our cohort, both of which harbored the SLC3A2-NRG1 fusion that was previously reported by Jonna et al. (36) suggesting that NRG1 fusions were highly enriched in the IMA subtype. For NRG1 fusions, patients may benefit from the tyrosine kinase inhibitors (TKIs), e.g. afatinib (37). There are also several TKIs available for targeted therapy in advanced lung cancer patients with RET fusions, e.g. selpercatinib (38), pralsetinib (39), cabozantinib (40, 41), and vandetanib (42). In patients harboring ALK fusions, various ALK TKIs could be used in the clinical treatment of advanced NSCLC patients, such as crizotinib (43), alectinib (44–46), brigatinib (47, 48), ceritinib (49, 50), lorlatinib (51), ensartinib (52), and repotrectinib (53). Therefore, IMA patients may benefit from a broader option of TKIs compared to non-IMA patients.
Regarding tumor suppressors, TP53 mutation was less in IMA, especially in pure-IMA. However, the mutation frequency of other tumor suppressors was higher in IMA than that in non-IMA. Moreover, the frequency of tumor suppressor co-mutations was significantly higher in IMA than non-IMA. This indicates that in IMA patients, once a tumor suppressor gene is involved in tumorigenesis, mutating additional tumor suppressor genes may have a synergistic effect. This may be a potential molecular mechanism underlying the poor prognosis of IMA patients, but studies with larger sample sizes are required for further verification.
In terms of the number of somatic mutations, no difference between IMA and non-IMA was observed. However, there were significant differences in SCNVs between IMA and non-IMA at the chromosome arm-level. There were more arm-level deletions and fewer arm-level amplifications in IMA compared to non-IMA. In addition, gradual decreasing and increasing trends of arm-level amplifications and deletions were observed from non-IMA to mixed-IMA to pure-IMA, respectively. These findings suggest that differences in molecular characteristics do exist between IMA and non-IMA and they may belong to different diseases.
Lastly, the survival-related analysis showed that the TNM stage was still the main factor affecting DFS after surgery, which is consistent with previous reports (54–56). In stage III IMA patients, cases who harbored EGFR mutations showed significantly prolonged DFS. According to previous studies, the prognostic impact of EGFR mutation status in patients with NSCLC remains controversial. Several articles have reported that the presence of EGFR mutations is a favorable prognostic factor in patients with surgically resected NSCLC (56–60). However, some other studies did not show a significant difference in survival between patients with EGFR-mutated tumors and those with wild-type after surgical resection (55, 61–64). In our study, we found that EGFR mutation is associated with improved DFS in stage III IMA patients. Similarly, patients with abnormalities in the PI3K signaling pathway displayed improved DFS. A previous study had suggested that lung adenocarcinoma patients with positive PI3K expression had a favorable survival, although it failed to be an independent prognostic predictor (65). In addition, Shan et al. found that phosphorylated AKT expression in the PI3K signaling pathway was a significant independent favorable prognostic factor in stage I to IIIA NSCLC (66). These findings suggest that the PI3K signaling pathway may be a potential favorable prognostic factor in patients with NSCLC. Conversely, patients with KRAS mutations have a poorer prognosis than those without KRAS mutations, which is consistent with previous studies conducted on unselected lung adenocarcinoma (54, 62, 67, 68). This suggests that KRAS-activating mutation is still an unfavorable prognostic factor in particular cases with IMA. As we did not have a large enough sample size, studies with more samples are required to further verify the prognostic values of abnormalities in EGFR, KRAS, and the PI3K pathway in IMA patients.
In summary, firstly, our study confirmed that the pattern of driver gene mutations between IMA and non-IMA was different, and IMA had greater diversity in mutations of driver genes. In addition, significant differences in the mutation frequency of some driver genes were observed between East-Asian and Caucasian populations, i.e. ALK, KRAS, and ERBB2. Secondly, we found that there were differences in the frequency of EGFR, ALK, KRAS, TP53, and arm-level SCNVs in tumors with different mucinous content. EGFR and ALK showed gradual decreasing and increasing trends from non-IMA to mixed-IMA to pure-IMA, respectively. KRAS mutation was more frequent in pure-IMA than in mixed-IMA. Compared to mixed-IMA and non-IMA, mutations in TP53 were significantly enriched in pure-IMA. With regards to arm-level SCNVs, fewer amplifications but more deletions in IMA compared to non-IMA were observed, and these alterations also showed gradual decreasing and increasing trends from non-IMA to mixed-IMA to pure-IMA, respectively. Thirdly, co-mutations in tumor suppressors were more common in IMA compared to non-IMA, suggesting that this may be a potential molecular mechanism for poorer prognosis in IMA. Lastly, in stage III IMA patients, mutations in EGFR and the PI3K signaling pathway were associated with favorable postoperative DFS while patients with KRAS mutations or poorly differentiated tumors showed a shorter DFS. In conclusion, this study enhances our understanding of the genomic characteristics of IMA patients in an East-Asian population and helps us have a more comprehensive insight into this distinct histological subtype of lung adenocarcinoma.
Data Availability Statement
The data presented in the study are deposited in the Genome Sequence Archive for Human (GSA-Human) repository (https://bigd.big.ac.cn/gsa-human/), accession number (HRA000507).
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethics Committee of Zhejiang Cancer Hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
Study concept and design: YJ, LC, and JW. Analysis and interpretation of data: JY, YX, FW, YS, and QX. Clinical information collection: LC, JW, JZ, JL, CP, XH, GX. MX and XX. Pathological evaluation: LZ and JJ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the grants from the National Natural Science Foundation of China Grants (No. 81802995 and No. 81702851), and the Medical Science and Technology Project of Zhejiang Province (2018KY024).
Conflict of Interest
Authors JY, YX, FW, YS and QX were employed by Nanjing Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.603671/full#supplementary-material
Supplementary Figure 1 | The landscape of somatic mutations in all 76 patients in our cohort. The oncoprint only shows the mutated genes occurring in at least 3 patients of all the patients.
Supplementary Figure 2 | Pie chart showing the known driver mutations detected in IMA and non-IMA (A) or in mixed-IMA and pure-IMA (B) patients.
Supplementary Figure 3 | Comparison of gene mutations in pure-IMA or mixed-IMA between Chinese and Caucasian patients. EGFR and the other genes with p-value < 0.2 in any group of mixed-IMA or pure-IMA were shown here.
Supplementary Figure 4 | Prognostic analysis of TP53 mutations in IMA patients. Kaplan-Meier curve of DFS in strata of TP53 mutation status in all stage III patients (A), and concomitant TP53 mutation status in EGFR-mutated stage III patients (B). The log-rank test was used to analyze the DFS for the survival analyses.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or Platinum-Pemetrexed in Egfr T790m-Positive Lung Cancer. New Engl J Med (2017) 376(7):629–40. doi: 10.1056/NEJMoa1612674
3. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. New Engl J Med (2009) 361(10):947–57. doi: 10.1056/NEJMoa0810699
4. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib Versus Standard Chemotherapy as First-Line Treatment for European Patients with Advanced Egfr Mutation-Positive Non-Small-Cell Lung Cancer (Eurtac): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol (2012) 13(3):239–46. doi: 10.1016/S1470-2045(11)70393-X
5. Yang JCH, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib Versus Cisplatin-Based Chemotherapy for Egfr Mutation-Positive Lung Adenocarcinoma (Lux-Lung 3 and Lux-Lung 6): Analysis of Overall Survival Data from Two Randomised, Phase 3 Trials. Lancet Oncol (2015) 16(2):141–51. doi: 10.1016/S1470-2045(14)71173-8
6. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol (2011) 6(2):244–85. doi: 10.1097/JTO.0b013e318206a221
7. Nie K, Nie W, Zhang YX, Yu H. Comparing Clinicopathological Features and Prognosis of Primary Pulmonary Invasive Mucinous Adenocarcinoma Based on Computed Tomography Findings. Cancer Imaging (2019) 19:47. doi: 10.1186/s40644-019-0236-2
8. Casali C, Rossi G, Marchioni A, Sartori G, Maselli F, Longo L, et al. A Single Institution-Based Retrospective Study of Surgically Treated Bronchioloalveolar Adenocarcinoma of the Lung Clinicopathologic Analysis, Molecular Features, and Possible Pitfalls in Routine Practice. J Thorac Oncol (2010) 5(6):830–6. doi: 10.1097/JTO.0b013e3181d60ff5
9. Lee MA, Kang J, Lee HY, Kim W, Shon I, Hwang NY, et al. Spread through Air Spaces (Stas) in Invasive Mucinous Adenocarcinoma of the Lung: Incidence, Prognostic Impact, and Prediction Based on Clinicoradiologic Factors. Thorac Cancer (2020) 11(11):3145–54. doi: 10.1111/1759-7714.13632
10. Beck KS, Sung YE, Lee KY, Han DH. Invasive Mucinous Adenocarcinoma of the Lung: Serial Ct Findings, Clinical Features, and Treatment and Survival Outcomes. Thorac Cancer (2020) 11(12):3463–72. doi: 10.1111/1759-7714.13674
11. Saito T, Tsuta K, Honda O, Ishida M, Yamaka R, Tanaka N, et al. Prognostic Impact of Mucin Spread, Tumor Cell Spread, and Invasive Size in Invasive Mucinous Adenocarcinoma of the Lung. Lung Cancer (2020) 146:50–7. doi: 10.1016/j.lungcan.2020.05.030
12. Shim HS, Mari-Kenudson, Zheng ZL, Liebers M, Cha YJ, Ho QH, et al. Unique Genetic and Survival Characteristics of Invasive Mucinous Adenocarcinoma of the Lung. J Thorac Oncol (2015) 10(8):1156–62. doi: 10.1097/Jto.0000000000000579
13. Cha YJ, Shim HS. Biology of Invasive Mucinous Adenocarcinoma of the Lung. Transl Lung Cancer R (2017) 6(5):508–12. doi: 10.21037/tlcr.2017.06.10
14. Finberg KE, Sequist LV, Joshi VA, Muzikansky A, Miller JM, Han M, et al. Mucinous Differentiation Correlates with Absence of Egfr Mutation and Presence of Kras Mutation in Lung Adenocarcinomas with Bronchioloalveolar Features. J Mol Diagn (2007) 9(3):320–6. doi: 10.2353/jmoldx.2007.060182
15. Li H, Pan YJ, Li Y, Li CG, Wang R, Hu HC, et al. Frequency of Well-Identified Oncogenic Driver Mutations in Lung Adenocarcinoma of Smokers Varies with Histological Subtypes and Graduated Smoking Dose. Lung Cancer (2013) 79(1):8–13. doi: 10.1016/j.lungcan.2012.09.018
16. Zhang Y, Sun YH, Pan YJ, Li CG, Shen L, Li Y, et al. Frequency of Driver Mutations in Lung Adenocarcinoma from Female Never-Smokers Varies with Histologic Subtypes and Age at Diagnosis. Clin Cancer Res (2012) 18(7):1947–53. doi: 10.1158/1078-0432.Ccr-11-2511
17. Ichinokawa H, Ishii G, Nagai K, Kawase A, Yoshida J, Nishimura M, et al. Distinct Clinicopathologic Characteristics of Lung Mucinous Adenocarcinoma with Kras Mutation. Hum Pathol (2013) 44(12):2636–42. doi: 10.1016/j.humpath.2013.05.026
18. Gow CH, Wu SG, Chang YL, Shih JY. Multidriver Mutation Analysis in Pulmonary Mucinous Adenocarcinoma in Taiwan: Identification of a Rare Cd74-Nrg1 Translocation Case. Med Oncol (2014) 31(7):34. doi: 10.1007/s12032-014-0034-4
19. Hu HC, Pan YJ, Li Y, Wang L, Wang R, Zhang Y, et al. Oncogenic Mutations Are Associated with Histological Subtypes but Do Not Have an Independent Prognostic Value in Lung Adenocarcinoma. Oncotargets Ther (2014) 7:1423–37. doi: 10.2147/Ott.S58900
20. Righi L, Vatrano S, Di Nicolantonio F, Massa F, Rossi G, Cavazza A, et al. Retrospective Multicenter Study Investigating the Role of Targeted Next-Generation Sequencing of Selected Cancer Genes in Mucinous Adenocarcinoma of the Lung. J Thorac Oncol (2016) 11(4):504–15. doi: 10.1016/j.jtho.2016.01.004
21. Tong L, Ding N, Tong XL, Li JM, Zhang Y, Wang XD, et al. Tumor-Derived DNA from Pleural Effusion Supernatant as a Promising Alternative to Tumor Tissue in Genomic Profiling of Advanced Lung Cancer. Theranostics (2019) 9(19):5532–41. doi: 10.7150/thno.34070
22. Yang Z, Yang N, Ou QX, Xiang Y, Jiang T, Wu X, et al. Investigating Novel Resistance Mechanisms to Third-Generation Egfr Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res (2018) 24(13):3097–107. doi: 10.1158/1078-0432.Ccr-17-2310
23. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30(15):2114–20. doi: 10.1093/bioinformatics/btu170
24. Li H, Durbin R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
25. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat Genet (2011) 43(5):491–8. doi: 10.1038/ng.806
26. Koboldt DC, Zhang QY, Larson DE, Shen D, McLellan MD, Lin L, et al. Varscan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing. Genome Res (2012) 22(3):568–76. doi: 10.1101/gr.129684.111
27. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. Factera: A Practical Method for the Discovery of Genomic Rearrangements at Breakpoint Resolution. Bioinformatics (2014) 30(23):3390–3. doi: 10.1093/bioinformatics/btu549
28. Amarasinghe KC, Li J, Halgamuge SK. Convex: Copy Number Variation Estimation in Exome Sequencing Data Using Hmm. BMC Bioinf (2013) 14:S2. doi: 10.1186/1471-2105-14-S2-S2
29. Shen RL, Seshan VE. Facets: Allele-Specific Copy Number and Clonal Heterogeneity Analysis Tool for High-Throughput DNA Sequencing. Nucleic Acids Res (2016) 44(16):e131. doi: 10.1093/nar/gkw520
30. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in the Cancer Genome Atlas. Cell (2018) 173(2):321–37.e10. doi: 10.1016/j.cell.2018.03.035
31. Liu Y, Xu ML, Zhong HH, Heng WJ, Wu BQ. Egfr Mutations Are More Frequent in Well-Differentiated Than in Poor-Differentiated Lung Adenocarcinomas. Pathol Oncol Res (2008) 14(4):373–9. doi: 10.1007/s12253-008-9113-1
32. Liu B, Shi SS, Wang X, Xu Y, Zhang XH, Yu B, et al. [Relevance of Molecular Alterations in Histopathologic Subtyping of Lung Adenocarcinoma Based on 2011 International Multidisciplinary Lung Adenocarcinoma Classification]. Zhonghua Bing Li Xue Za Zhi (2012) 41(8):505–10. doi: 10.3760/cma.j.issn.0529-5807.2012.08.001
33. Tomizawa Y, Iijima H, Sunaga N, Sato K, Takise A, Otani Y, et al. Clinicopathologic Significance of the Mutations of the Epidermal Growth Factor Receptor Gene in Patients with Non-Small Cell Lung Cancer. Clin Cancer Res (2005) 11(19 Pt 1):6816–22. doi: 10.1158/1078-0432.CCR-05-0441
34. Kohno T, Nakaoku T, Tsuta K, Tsuchihara K, Matsumoto S, Yoh K, et al. Beyond Alk-Ret, Ros1 and Other Oncogene Fusions in Lung Cancer. Transl Lung Cancer Res (2015) 4(2):156–64. doi: 10.3978/j.issn.2218-6751.2014.11.11
35. Ke H, Shen W, Hu A, Ou Q, Lu S. Distribution of Nrg1 Gene Fusions in a Large Population of Chinese Patients with Nsclc. J Thorac Oncol (2019) 14(12):e263–6. doi: 10.1016/j.jtho.2019.07.012
36. Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of Nrg1 Gene Fusions in Solid Tumors. Clin Cancer Res (2019) 25(16):4966–72. doi: 10.1158/1078-0432.CCR-19-0160
37. Drilon A, Somwar R, Mangatt BP, Edgren H, Desmeules P, Ruusulehto A, et al. Response to Erbb3-Directed Targeted Therapy in Nrg1-Rearranged Cancers. Cancer Discovery (2018) 8(6):686–95. doi: 10.1158/2159-8290.Cd-17-1004
38. Drilon A, Oxnard G, Wirth L, Besse B, Gautschi O, Tan SWD, et al. Registrational Results of Libretto-001: A Phase 1/2 Trial of Loxo-292 in Patients with Ret Fusion-Positive Lung Cancers. J Thorac Oncol (2019) 14(10):S6–7. doi: 10.1016/j.jtho.2019.08.059
39. Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, et al. Precision Targeted Therapy with Blu-667 for Ret-Driven Cancers. Cancer Discovery (2018) 8(7):836–49. doi: 10.1158/2159-8290.CD-18-0338
40. Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, et al. Response to Cabozantinib in Patients with Ret Fusion-Positive Lung Adenocarcinomas. Cancer Discovery (2013) 3(6):630–5. doi: 10.1158/2159-8290.Cd-13-0035
41. Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, et al. Cabozantinib in Patients with Advanced Ret-Rearranged Non-Small-Cell Lung Cancer: An Open-Label, Single-Centre, Phase 2, Single-Arm Trial. Lancet Oncol (2016) 17(12):1653–60. doi: 10.1016/S1470-2045(16)30562-9
42. Lee SH, Lee JK, Ahn MJ, Kim DW, Sun JM, Keam B, et al. Vandetanib in Pretreated Patients with Advanced Non-Small Cell Lung Cancer-Harboring Ret Rearrangement: A Phase Ii Clinical Trial. Ann Oncol (2017) 28(2):292–7. doi: 10.1093/annonc/mdw559
43. Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-Line Crizotinib Versus Chemotherapy in Alk-Positive Lung Cancer. New Engl J Med (2014) 371(23):2167–77. doi: 10.1056/NEJMoa1408440
44. Shaw AT, Gandhi L, Gadgeel S, Riely GJ, Cetnar J, West H, et al. Alectinib in Alk-Positive, Crizotinib-Resistant, Non-Small-Cell Lung Cancer: A Single-Group, Multicentre, Phase 2 Trial. Lancet Oncol (2016) 17(2):234–42. doi: 10.1016/S1470-2045(15)00488-X
45. Hida T, Nokihara H, Kondo M, Kim YH, Azuma K, Seto T, et al. Alectinib Versus Crizotinib in Patients with Alk-Positive Non-Small-Cell Lung Cancer (J-Alex): An Open-Label, Randomised Phase 3 Trial. Lancet (2017) 390(10089):29–39. doi: 10.1016/S0140-6736(17)30565-2
46. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib Versus Crizotinib in Untreated Alk-Positive Non-Small-Cell Lung Cancer. New Engl J Med (2017) 377(9):829–38. doi: 10.1056/NEJMoa1704795
47. Kim DW, Tiseo M, Ahn MJ, Reckamp KL, Hansen KH, Kim SW, et al. Brigatinib in Patients with Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase Ii Trial. J Clin Oncol (2017) 35(22):2490–8. doi: 10.1200/Jco.2016.71.5904
48. Camidge DR, Kim HR, Ahn MJ, Yang JCH, Han JY, Lee JS, et al. Brigatinib Versus Crizotinib in Alk-Positive Non-Small-Cell Lung Cancer. New Engl J Med (2018) 379(21):2027–39. doi: 10.1056/NEJMoa1810171
49. Shaw AT, Kim TM, Crino L, Gridelli C, Kiura K, Liu G, et al. Ceritinib Versus Chemotherapy in Patients with Alk-Rearranged Non-Small-Cell Lung Cancer Previously Given Chemotherapy and Crizotinib (Ascend-5): A Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet Oncol (2017) 18(7):874–86. doi: 10.1016/S1470-2045(17)30339-X
50. Soria JC, Tan DSW, Chiari R. First-Line Ceritinib Versus Platinum-Based Chemotherapy in Advanced Alk-Rearranged Non-Small-Cell Lung Cancer (Ascend-4): A Randomised, Open-Label, Phase 3 Study (Vol 389, Pg 917, 2017). Lancet (2017) 389(10072):908–8. doi: 10.1016/S0140-6736(17)30123-X
51. Solomon BJ, Besse B, Bauer TM, Felip E, Soo RA, Camidge DR, et al. Lorlatinib in Patients with Alk-Positive Non-Small-Cell Lung Cancer: Results from a Global Phase 2 Study. Lancet Oncol (2018) 19(12):1654–67. doi: 10.1016/S1470-2045(18)30649-1
52. Yang YP, Zhou JY, Zhou JY, Feng JF, Zhuang W, Chen JH, et al. Efficacy, Safety, and Biomarker Analysis of Ensartinib in Crizotinib-Resistant, Alk-Positive Non-Small-Cell Lung Cancer: A Multicentre, Phase 2 Trial. Lancet Resp Med (2020) 8(1):45–53. doi: 10.1016/S2213-2600(19)30252-8
53. Cho BC, Drilon AE, Doebele RC, Kim DW, Lin JJ, Lee J, et al. Safety and Preliminary Clinical Activity of Repotrectinib in Patients with Advanced Ros1 Fusion-Positive Non-Small Cell Lung Cancer (Trident-1 Study). J Clin Oncol (2019) 37(suppl_15):9011. doi: 10.1200/JCO.2019.37.15_suppl.9011
54. Sonobe M, Kobayashi M, Ishikawa M, Kikuchi R, Nakayama E, Takahashi T, et al. Impact of Kras and Egfr Gene Mutations on Recurrence and Survival in Patients with Surgically Resected Lung Adenocarcinomas. Ann Surg Oncol (2012) 19:S347–54. doi: 10.1245/s10434-011-1799-8
55. Kim YT, Seong YW, Jung YJ, Jeon YK, Park IK, Kang CH, et al. The Presence of Mutations in Epidermal Growth Factor Receptor Gene Is Not a Prognostic Factor for Long-Term Outcome after Surgical Resection of Non-Small-Cell Lung Cancer. J Thorac Oncol (2013) 8(2):171–8. doi: 10.1097/JTO.0b013e318277a3bb
56. Takamochi K, Oh S, Matsunaga T, Suzuki K. Prognostic Impacts of Egfr Mutation Status and Subtype in Patients with Surgically Resected Lung Adenocarcinoma. J Thorac Cardiov Sur (2017) 154(5):1768–74.e1. doi: 10.1016/j.jtcvs.2017.06.062
57. Lee YJ, Park IK, Park MS, Choi HJ, Cho BC, Chung KY, et al. Activating Mutations within the Egfr Kinase Domain: A Molecular Predictor of Disease-Free Survival in Resected Pulmonary Adenocarcinoma. J Cancer Res Clin (2009) 135(12):1647–54. doi: 10.1007/s00432-009-0611-7
58. Izar B, Sequist L, Lee M, Muzikansky A, Heist R, Iafrate J, et al. The Impact of Egfr Mutation Status on Outcomes in Patients with Resected Stage I Non-Small Cell Lung Cancers. Ann Thorac Surg (2013) 96(3):962–8. doi: 10.1016/j.athoracsur.2013.05.091
59. Yotsukura M, Yasuda H, Shigenobu T, Kaseda K, Masai K, Hayashi Y, et al. Clinical and Pathological Characteristics of Egfr Mutation in Operable Early-Stage Lung Adenocarcinoma. Lung Cancer (2017) 109:45–51. doi: 10.1016/j.lungcan.2017.04.014
60. Isaka T, Nakayama H, Ito H, Yokose T, Yamada K, Masuda M. Impact of the Epidermal Growth Factor Receptor Mutation Status on the Prognosis of Recurrent Adenocarcinoma of the Lung after Curative Surgery. BMC Cancer (2018) 18:959. doi: 10.1186/s12885-018-4849-9
61. Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and Biological Features Associated with Epidermal Growth Factor Receptor Gene Mutations in Lung Cancers. J Natl Cancer Inst (2005) 97(5):339–46. doi: 10.1093/jnci/dji055
62. Kim YT, Kim TY, Lee DS, Park SJ, Park JY, Seo SJ, et al. Molecular Changes of Epidermal Growth Factor Receptor (Egfr) and Kras and Their Impact on the Clinical Outcomes in Surgically Resected Adenocarcinoma of the Lung. Lung Cancer (2008) 59(1):111–8. doi: 10.1016/j.lungcan.2007.08.008
63. Liu WS, Zhao LJ, Pang QS, Yuan ZY, Li B, Wang P. Prognostic Value of Epidermal Growth Factor Receptor Mutations in Resected Lung Adenocarcinomas. Med Oncol (2014) 31(1):771. doi: 10.1007/s12032-013-0771-9
64. Zhang ZX, Wang T, Zhang J, Cai XH, Pan CC, Long Y, et al. Prognostic Value of Epidermal Growth Factor Receptor Mutations in Resected Non-Small Cell Lung Cancer: A Systematic Review with Meta-Analysis. PloS One (2014) 9(8):e106053. doi: 10.1371/journal.pone.0106053
65. Li L, Liu D, Jin J, Zhu Y, Yang L, Song J, et al. Pi3k Expression Predicts Overall Survival in Lung Adenocarcinoma. Int J Clin Exp Pathol (2018) 11(10):4889–98.
66. Shah A, Swain WA, Richardson D, Edwards J, Stewart DJ, Richardson CM, et al. Phospho-Akt Expression Is Associated with a Favorable Outcome in Non-Small Cell Lung Cancer. Clin Cancer Res (2005) 11(8):2930–6. doi: 10.1158/1078-0432.CCR-04-1385
67. Marks JL, Broderick S, Zhou Q, Chitale D, Li AR, Zakowski MF, et al. Prognostic and Therapeutic Implications of Egfr and Kras Mutations in Resected Lung Adenocarcinoma. J Thorac Oncol (2008) 3(2):111–6. doi: 10.1097/JTO.0b013e318160c607
Keywords: invasive mucinous adenocarcinoma, surgical resection, next generation sequencing, genomic profiling, prognostic biomarker
Citation: Cai L, Wang J, Yan J, Zeng J, Zhu L, Liang J, Pan C, Huang X, Jin J, Xu Y, Wang F, Shao Y, Xu Q, Xia G, Xing M, Xu X and Jiang Y (2021) Genomic Profiling and Prognostic Value Analysis of Genetic Alterations in Chinese Resected Lung Cancer With Invasive Mucinous Adenocarcinoma. Front. Oncol. 10:603671. doi: 10.3389/fonc.2020.603671
Received: 22 September 2020; Accepted: 23 November 2020;
Published: 11 January 2021.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Alessandro Russo, A. O. Papardo, ItalyFrancesco Pepe, University of Naples Federico II, Italy
Copyright © 2021 Cai, Wang, Yan, Zeng, Zhu, Liang, Pan, Huang, Jin, Xu, Wang, Shao, Xu, Xia, Xing, Xu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youhua Jiang, amlhbmd5aEB6amNjLm9yZy5jbg==
†These authors have contributed equally to this work
 Lei Cai
Lei Cai Jiangfeng Wang1,2†
Jiangfeng Wang1,2† Junrong Yan
Junrong Yan