- 1Epigenome Research Center, China Medical University Hospital, Taichung, Taiwan
- 2Center for Precision Medicine, China Medical University Hospital, Taichung, Taiwan
- 3School of Medicine, China Medical University, Taichung, Taiwan
- 4Department of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan
- 5Department of Bioinformatics and Medical Engineering, Asia University, Taichung, Taiwan
- 6Department of Clinical Pharmacy, School of Pharmacy, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
- 7Division of Gastroenterology, Department of Internal Medicine, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
- 8Department of General Medicine, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
Background: Long noncoding RNA (lncRNA) mediates the pathogenesis of various diseases, including cancer and cardiovascular, infectious, and metabolic diseases. This study examined the role of lncRNA NTT in the development and progression of cancer.
Methods: The expression of NTT was determined using tissues containing complementary DNA (cDNA) from patients with liver, lung, kidney, oral, and colon cancers. The expression of cis-acting genes adjacent to the NTT locus (CTGF, STX7, MYB, BCLAF1, IFNGR1, TNFAIP3, and HIVEP2) was also assessed. We used knockdown and chromatin immunoprecipitation (ChIP) assays to identify the cis-acting genes that interact with NTT.
Results: NTT was most significantly downregulated in hepatocellular carcinoma (HCC), while a higher NTT level correlated with a shorter survival time of patients with HCC. Multivariate analysis indicated NTT was not an independent predictor for overall survival. MYB was significantly upregulated, and its increased expression was associated with dismal survival in HCC patients, similar to the results for NTT. NTT knockdown significantly decreased cellular migration. ChIP of HCC cell lines revealed that NTT is regulated by the transcription factor ATF3 and binds to the MYB promoter via the activated complex. Additionally, when NTT was knocked down, the expression of MYB target genes such as Bcl-xL, cyclinD1, and VEGF was also downregulated. NTT could play a positive or negative regulator for MYB with a context-dependent manner in both HCC tissues and animal model.
Conclusion: Our study suggests that NTT plays a key role in HCC progression via MYB-regulated target genes and may serve as a novel therapeutic target.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers and the third leading cause of cancer-related deaths worldwide (1). HCC accounts for more than 90% of liver cancers (2). Globally, approximately 600,000 patients with HCC die annually, and 78,200 new cases are diagnosed. Its incidence is highest in Asia and Africa and is higher among male than female individuals. The risk factors for developing HCC include infection with hepatitis B or C virus, alcohol abuse, nonalcoholic fatty liver disease, Budd–Chiari syndrome, aflatoxin B1 intake, and metabolic diseases (3, 4). Serum alpha-fetoprotein (AFP) and ultrasonography are used clinically for the early detection of HCC (5). However, the sensitivity and specificity of AFP for the diagnosis of HCC (cutoff 20 ng/ml) are 53% and 90%, respectively. Therefore, AFP has been removed from the HCC surveillance guidelines of the American Association for the Study of Liver Disease due to its low diagnostic accuracy (2, 6, 7). Cell-free nucleic acids could also contribute to the surveillance and diagnosis of HCC (8). Furthermore, because of the high postoperative tumor recurrence rate, which can reach 50%, surgical resection is effective for only about 20% of HCC patients (9). The overall 5-year survival is as low as 11–30% (10). A better understanding of the molecular mechanisms underlying the progression of HCC may contribute to the identification of therapeutic targets or diagnostic and prognostic markers.
Long noncoding RNAs (lncRNAs) are noncoding RNAs more than 200 nucleotides long. Because the expression of lncRNAs is lower than that of messenger RNAs (mRNAs), lncRNAs were considered transcriptional noise. However, lncRNAs play crucial roles in regulating various cellular processes and tumorigenesis, such as cell proliferation, metastasis, differentiation, and genomic instability via transcription, posttranscription, and epigenetic modifications of the related gene expression (11–15). Four typical molecular functions of lncRNAs are signal, decoy, manipulation, and scaffolding (16). The lncRNAs H19, HOTAIR, HULC, MALAT1, NEAT1, TUG1, UCA1, and ZFAS1 have been reported to be associated with HCC initiation, progression, and metastasis in humans (17).
Noncoding transcript in T cells (NTT) is located on chromosome 6q23-q24 and transcribes a 17-kb polyadenylated RNA, which is not spliced. It was first described in 1997 in activated CD4+ T cells (18). Subsequently, Amarante et al. detected NTT in peripheral blood mononuclear cells following stimulation with human immunodeficiency virus (HIV) peptide, suggesting it has a role in cellular immune responses (19). A recent study demonstrated that the NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis (20). NTT expression in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) has also been investigated (21). However, the expression pattern, biological function, and underlying mechanism of NTT in HCC are still unclear.
In this study, we analyzed NTT levels in HCC tissues and their correlation with the clinicopathological characteristics and prognosis of patients with HCC. Mechanistically, we suggest that lncRNA NTT interacts with MYB and epigenetically activates downstream target genes to facilitate cell migration in HCC. In vivo and in vitro assays indicated that NTT context-dependently regulated MYB.
Materials and Methods
Patient Samples
The samples examined in this study were from liver (n = 80), lung (n = 36), kidney (n = 34), oral (n = 28), and colon (n = 15) cancers. Tumor and adjacent normal tissues were collected from patients who underwent surgery at China Medical University Hospital. All samples were immediately snap-frozen in liquid nitrogen and stored at −80°C. The study was approved by the Research Ethics Committee of China Medical University Hospital (CMUH102-REC1-037).
RNA Extraction, Reverse Transcription, and Real-Time Quantitative Polymerase Chain Reaction
Total RNA was isolated using a NucleoSpin® RNA Kit (Macherey Nagel, Düren, Germany) and reverse transcribed into complementary DNA (cDNA) using a high-capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative PCR (qPCR) was performed using the IQ2 TaqMan Probe System (Biogenesis, Taiwan) on a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany) in accordance with the manufacturer’s instructions. The 2−ΔΔCt method was used to calculate the relative gene expression normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primer and probe sequences are listed in Supplementary Table 1.
Protein Extracts and Western Blotting
Proteins were extracted using a cell lysis solution [20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Nonidet P-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate, 1 mM Na3VO4, and 1 μg·ml−1 leupeptin] and separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE). After proteins were transferred onto polyvinylidene difluoride membranes (Millipore), the membranes were blocked with 5% bovine serum albumin (BSA) (Santa Cruz Biotechnology) and then exposed at 4°C overnight to the anti-c-MYB (Abcam, Cambridge, MA, USA) and anti-GAPDH (GeneTex, Inc., Taiwan), followed by horseradish peroxidase-conjugated secondary antibody for detection by an ECL chemiluminescence detection system (GE Healthcare, Pittsburgh, PA, USA).
Cell Culture and shRNA Transfection
The HCC cell lines (Huh7 and HepG2) were maintained in our laboratory and cultured in Dulbecco’s modified Eagle’s medium (GIBCO BRL, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (GIBCO BRL), 100 U/ml penicillin, and 100 mg/ml streptomycin in a humidified incubator containing 5% CO2 at 37°C. We used short tandem repeats to authenticate HCC cell lines that were used in this study. Primary human hepatocytes (HH) was from nearby normal parts of HCC tissues used as normal liver control.
For NTT knockdown, short hairpin RNA (shRNA) (5′-CAGAGCTGACTACACGGAGTGTT-3′) and negative control shRNA (5′-TTCTCCGAACGTGTCACGTTT-3′) were synthesized by MDBio (Taipei, Taiwan). The NTT and negative control shRNAs were constructed in pSUPER.neo (OligoEngine, Seattle, WA, USA). Huh7 and HepG2 cells were transiently transfected with shRNA or negative control using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and a stable clone was selected by G418. The transfection efficiency was determined by qPCR.
For ATF3 knockdown, shATF3 was obtained from the National RNAi Core Facility at Academia Sinica (Taiwan). The shRNA sequence was used for ATF3 knockdown (5′-GCTGAACTGAAGGCTCAGATT-3′).
Cytoplasmic and Nuclear RNA Extraction
Cytoplasmic and nuclear cell lysates were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions followed by TRIzol RNA extraction. The nuclear fraction was determined by qPCR.
Wound-Healing Assay
A wound-healing assay was used to assess cell migration. Huh7 and HepG2 cells were seeded and grown in 24-well plates until a monolayer formed. The cells were then scratched using a 200-μl plastic tip to create a straight wound. Wound gaps were photographed and analyzed using the free TScratch software.
Cell Proliferation Assay
Cell proliferation was assessed using a trypan blue exclusion assay. Cells were plated in 96-well plates at 1 × 105 cells per well and incubated for 24 or 48 h. After incubation, the number of cells was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
RNA-Binding Protein Immunoprecipitation
RNA-binding protein immunoprecipitation (RIP) was conducted using the RNA ChIP-IT kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, after cross-linking, cells were lysed, and the lysates were incubated with protein G agarose beads conjugated with specific antibodies (anti-EZH2, anti-EED, anti-MLL1, anti-PML, or anti-H3H4me3) or control immunoglobulin G (IgG). Then, the beads were incubated with proteinase K, and the purified RNA was subjected to qRT-PCR.
DNA ChIP
DNA ChIP was performed using the ChIP-IT kit (Active Motif). Cells were treated with formaldehyde and incubated for 10 min to generate DNA protein cross-links. The cell lysates were sonicated to obtain 200–500-bp chromatin fragments and immunoprecipitated with a specific antibody (anti-PML, anti-H3K4me3, anti-EZH2, or anti-ATF3) or IgG as the control. Precipitated chromatin DNA was recovered and analyzed by qPCR. The MYB promoter primer sequences were 5′-CCTAGCCAAACAGCCTATGAA-3′ (forward) and 5′-TGGAGACGGGGAAATTAGG-3′ (reverse). The NTT promoter primer sequences were 5′-CACCCACATGGTAGACAGGA-3′ (position 5350 forward) and 5′-CCCAGCTCCCAGAAGATACA-3′ (reverse).
Fluorescent In Situ Hybridization
Cells were harvested by directly culturing on cover slips. After fixation in 1% formaldehyde, the slides were inserted into solution A [80% formamide, 10% 1× saline sodium citrate (SSC) and 10% H2O] at room temperature for 2 min. Then, they were quenched in ice-cold 70% ethanol for 5 min to fix the slides. The RNA fluorescence in situ hybridization (FISH) locked nucleic acid (LNA) probe (corresponding to the sequence of U54776.1 from 10,024 to 10,044) was purchased from Exiqon Inc. (MA, USA) and labeled in orange. The slides were dehydrated, air dried, and preheated to 37°C for 2 min before probe addition. The slides were incubated overnight at 37°C in a humid chamber in an incubator. After RNA FISH washing, the slides were postfixed prior to DNA FISH in 4% paraformaldehyde for 15 min at room temperature and rinsed once with phosphate-buffered saline (PBS). The slides were then denatured in 70% formamide and 2× SSC at 80°C for 10 min. The slides were dehydrated before the denatured probes were added for overnight hybridization at 42°C in a dark humid environment. DNA FISH probe were purchased from Cytocell (Cat. No. LPH016). The red 183-kb MYB probe covers the entire MYB gene; the probe mix also contains a green control probe for the chromosome 6 centromere probe (D6Z1). Then, the slides were washed twice in 2× SSC at 45°C, and 4′,6-diamidino-2-phenylindole (DAPI) was added to this slides. Images were obtained using an E600 microscope (Nikon, Tokyo, Japan) with CytoVision software (Applied Imaging, Santa Clara, CA, USA).
Formalin-fixed, paraffin-embedded HCC and paired non-tumor tissue sections (1–2 μm) on poly-l-lysine-coated slides were deparaffinized. Before hybridization, the slides were incubated with 10% pepsin for 30 min at 37°C and then heated in denaturation solution at 72°C for 3 min. The RNA FISH probe (corresponding to the sequence of U54776.1 from 13,559 to 13,620) was labeled with Cy3 dyes using the Label IT labeling kit (Mirus, Madison, WI, USA). Hybridization with the denatured NTT probe was performed in a humid chamber at 37°C overnight. Then, the slides were washed at 45°C in washing buffer for 5 min. Finally, the slides were counterstained with DAPI and analyzed by fluorescence microscopy.
Animal Experiments
BALB/c athymic nude mice (male, 4–6 weeks old) were purchased from the National Laboratory Animal Breeding and Research Center, Taiwan. To establish a HCC xenograft model, 1 × 107 scramble or shNTT-Huh7 cells were suspended in 100 ml PBS and inoculated subcutaneously into the flanks of eight nude mice (left: shNTT; right: scramble). All animal experiments were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (CMU). All animals were housed in the Laboratory Animal Center of CMU under a 12 h light/dark (08:00/20:00) cycle with free access to food and water. The mice were sacrificed using CO2, and the tissues were subsequently harvested. All breeding and subsequent use of animals in this study, including sacrifice, was approved by the IACUC of CMU. The IACUC approval number was 102-203-N.
Luciferase Reporter Assay
Fragments of DNA from the NTT promoter region were obtained through PCR amplification and cloned into the pGL3-basic reporter vector (Promega Corporation). The constructs were transfected into Huh7 cells (1 × 105) using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The cells were lysed and assayed for luciferase activity using Steady-Glo Luciferase assay system (Promega Corporation) according to the manufacturer’s protocol at 24 h posttransfection.
Statistical Analysis
Student’s t-test was used to test for differences in mean values between two groups. Correlations between the NTT level and clinicopathological characteristics of the HCC patients were analyzed with the chi-square test. Survival analysis was performed using the Kaplan–Meier method, followed by the log-rank test. The level of statistical significance was set at p < 0.05. All statistical analyses were performed using GraphPad Prism software (ver. 8.0.2; GraphPad Software, Inc., La Jolla, CA, USA).
Results
NTT Expression in Various Cancer Types
We explored NTT expression in liver (HCC), lung (adenocarcinoma), kidney (renal cell carcinoma), oral (squamous cell carcinoma), and colon (adenocarcinoma) cancers. NTT expression was higher in cancer than in normal tissue in oral cancer but lower in cancer compared with normal tissues in the other types of cancer (Figure 1A). Our data showed that NTT was dysregulated in cancers. NTT showed the highest differences in HCC compared to its surrounding normal tissue, in respect to lung, kidney, oral, and colon cancers. Next, we investigated NTT expression in normal hepatocytes and HCC cell lines (Huh7 and HepG2). We found that the NTT expression in normal hepatocytes was higher than in Huh7 and HepG2 (Figure 1B), similar to the findings for HCC and nearby tissues. FISH confirmed that NTT expression was lower in tumor cells than in non-tumor cells (Figure 1C). Based on these results, we focused on NTT in HCC and the HCC cell lines Huh7 and HepG2 in the following experiments.
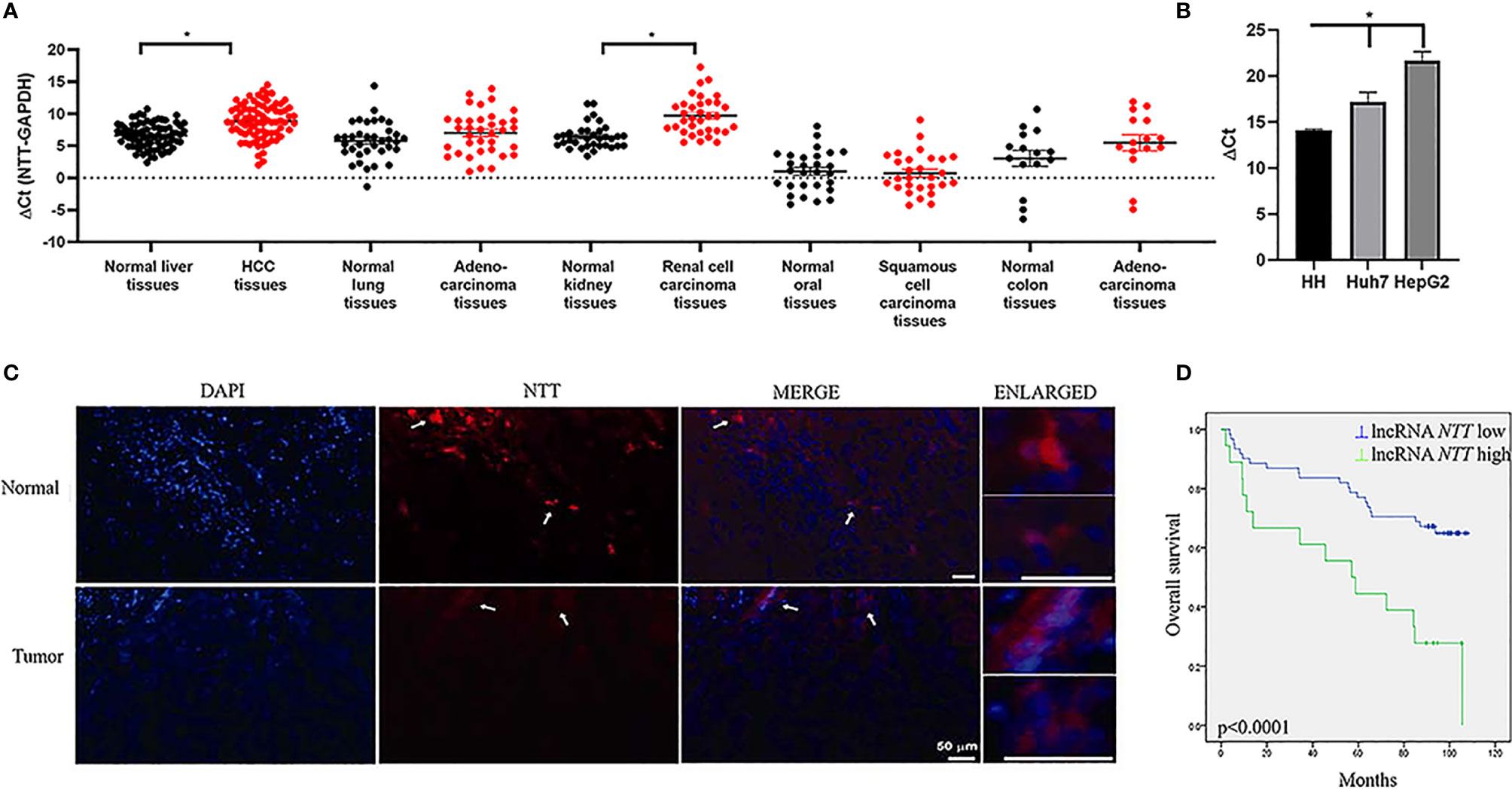
Figure 1 NTT is dysregulated in various cancers. (A) Expression of NTT was evaluated by qPCR in different cancer types. (B) Low expression of NTT in HCC cell lines, including Huh7 and HepG2 in comparison with that in normal primary human hepatocytes. (C) NTT expression was also evaluated in HCC by RNA FISH. (D) High NTT expression indicated a poor clinical outcome. *p < 0.05.
The NTT Level Is Correlated With the Clinical Characteristics of Patients With HCC
Using the mean NTT expression level in the 80 HCC patients, 62 patients (77.5%) were classified into a low-NTT expression group, and the remaining 18 patients (22.5%) were classified into a high-NTT expression group. Further survival analysis revealed that higher NTT expression was correlated with a shorter survival time in patients with HCC (p < 0.0001, Figure 1D). Moreover, there were significant correlations between NTT expression and age, tumor size, and tumor stage (Supplementary Table 2). In the multivariate analysis, which incorporated independent prognostic factors of gender, age, differentiation, tumor size, and stage, we found that NTT was not an independent predictor for overall survival (p = 0.059) (Supplementary Table 3).
Expression of Genes Near NTT in HCC: CTGF, STX7, MYB, BCLAF1, IFNGR1, TNFAIP3, and HIVEP2
lncRNA usually plays an important role in the regulation of genes nearby. We measured the expression of genes near NTT in HCC tissues. The genes were as follows: CTGF, STX7, MYB, BCLAF1, IFNGR1, TNFAIP3, and HIVEP2 (Supplementary Figure 1). The expression of CTGF and MYB was significantly higher in HCC tissues than that in the adjacent non-tumor tissues (p < 0.05, Figure 2A), while the expression of BCLAF1, IFNGR1, and HIVEP2 was significantly lower in HCC tissues than in adjacent non-tumor tissues (p < 0.05, Figure 2A). The high MYB levels in tumors predicted a poor clinical outcome (p = 0.014, Figure 2B), similar to the results for NTT expression. Based on these results, we explored the relationship between MYB and NTT, and the regulation mechanism.
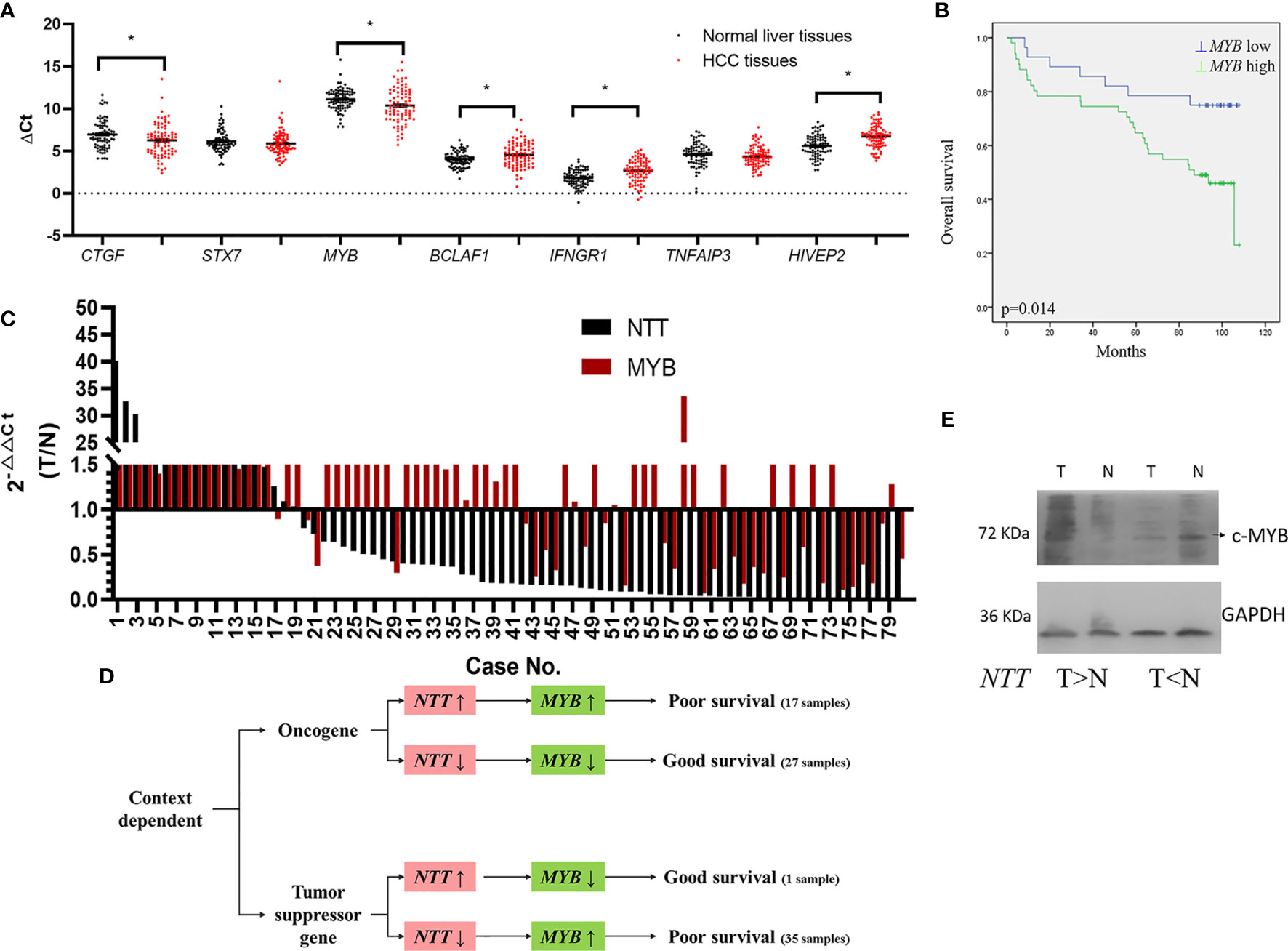
Figure 2 Genes near NTT are dysregulated in HCC. (A) Expression of CTGF, STX7, MYB, BCLAF1, IFNGR1, TNFAIP3, and HIVEP2 was evaluated by qPCR in HCC. (B) High MYB expression indicated a poor clinical outcome. *p < 0.05. (C) Log2 fold changes in NTT and MYB expressions (T/N) of each case are plotted. (D) The roles of NTT for MYB is context dependent. (E) MYB protein expression in HCC tissue samples. Left pair was MYB increased in HCC; right pair was MYB decreased in HCC.
First, we analyzed the correlation between the expressions of NTT and MYB in human tissues (Figure 2C). We found that NTT-positive regulation of MYB expression in 55% (44/80) of the HCC tissues, which including both NTT and MYB expressions were increased in 17 HCC tissues and both expressions were decreased in 27 HCC tissues (Figure 2D). From these results, we suggested that NTT plays like an oncogene.
However, NTT negative regulation of MYB expression in 45% (36/80) of the HCC tissues, which contained NTT expression, was decreased, and MYB expression was increased in 35 HCC tissues; NTT expression was increased and MYB expression was decreased in one HCC tissue (Figure 2D).
No differences were observed in the survival rates between patients with NTT positive and negative regulation of MYB expression (p = 0.796) (Supplementary Figure 2A). Further survival analysis revealed that there was significant correlation between higher and lower expression of both NTT and MYB (p = 0.000) (Supplementary Figure 2B). Survival is lower among both NTT- and MYB-upregulated patients compared with both NTT- and MYB-downregulated patients (Supplementary Figures 2B, D).
We also found that MYB play a determining role for HCC survival, overexpression of MYB resulted in poor survival, and downexpression of MYB correlated with good survival (Figure 2D). The expression of NTT could not determine patient’s survival directly. These data support the context-dependent role of NTT in liver tumorigenesis. Western bot analysis was also done to confirm the MYB protein expression in HCC tissue samples, the results of representative cases showed that NTT overexpression upregulated the expression of MYB, and NTT downexpression downregulated the expression of MYB in the paired normal and tumor tissues (Figure 2E).
NTT Promotes Cell Migration in HCC Cell Lines
We further explored the biological role of NTT in HCC cell lines. The shNTT plasmid was transfected into HCC Huh7 and HepG2 cells, which effectively knocked down the expression of NTT (Figure 3A). Knockdown of NTT significantly reduced the number of migratory cells in Huh7 and HepG2 cells compared with the sh-ctrl transfected cells (Figure 3B).
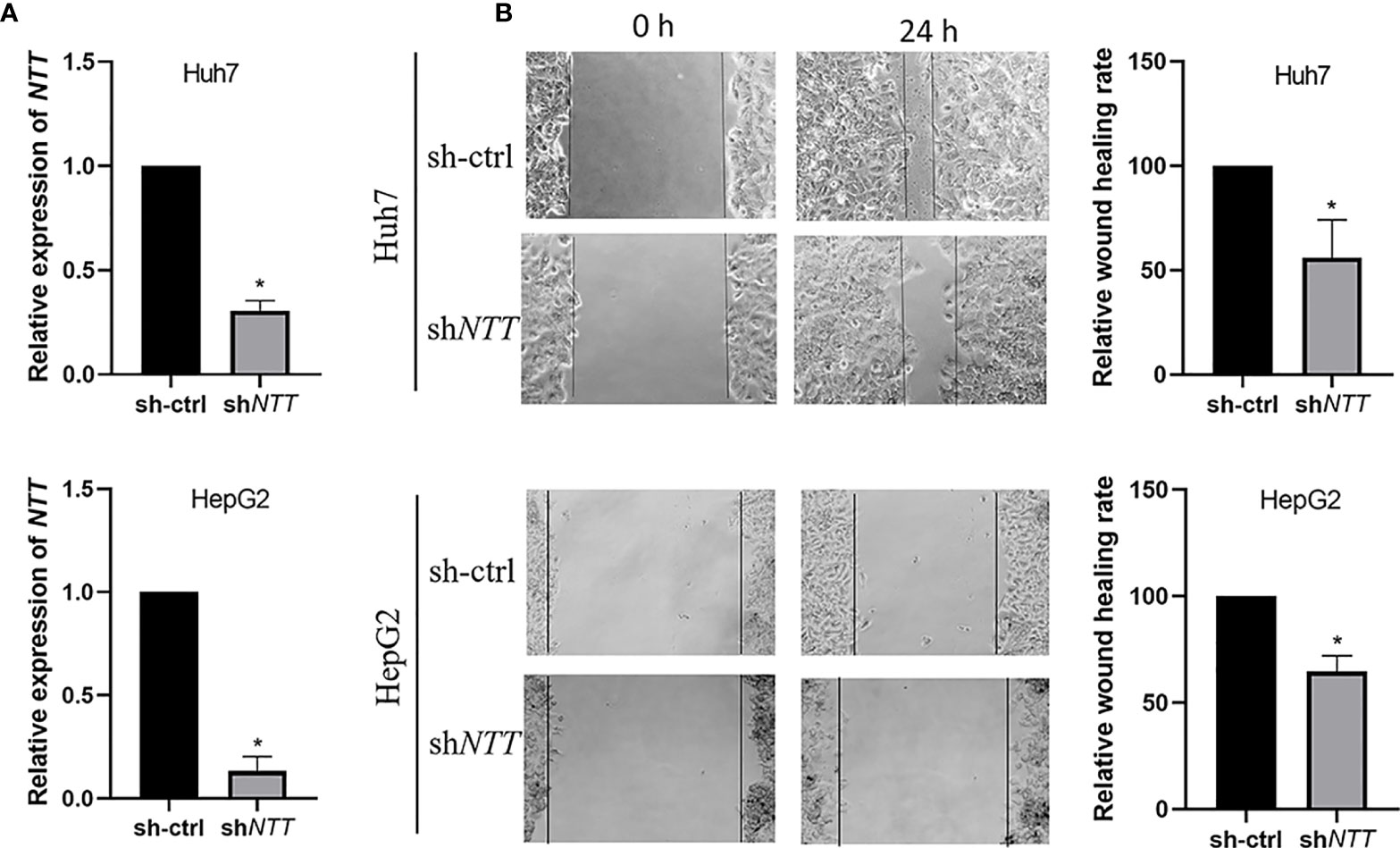
Figure 3 NTT knockdown reduced cell migration ability. (A) Expression of NTT was determined by qPCR. (B) Knockdown of NTT inhibited cell migration. Data are shown as the mean ± standard deviation (n = 3). *p < 0.05 for sh-ctrl vs. shNTT.
To test whether NTT levels affect cell proliferation, viable cells were counted using the MTT assay after Huh7 cells were transfected with shRNA for 48 h. We found no difference in cell proliferation between the Huh7 cells transiently transfected with sh-ctrl and shNTT (Supplementary Figure 3).
NTT Regulates the Nearby MYB Gene via Activated Complex Binding
Next, we investigated how NTT regulates the MYB gene. NTT expression in Huh7 and HepG2 were knocked down by shRNA, and the relative MYB mRNA level was analyzed (Figure 4A). Transfection of shNTT in Huh7 and HepG2 led to decreased MYB expression.
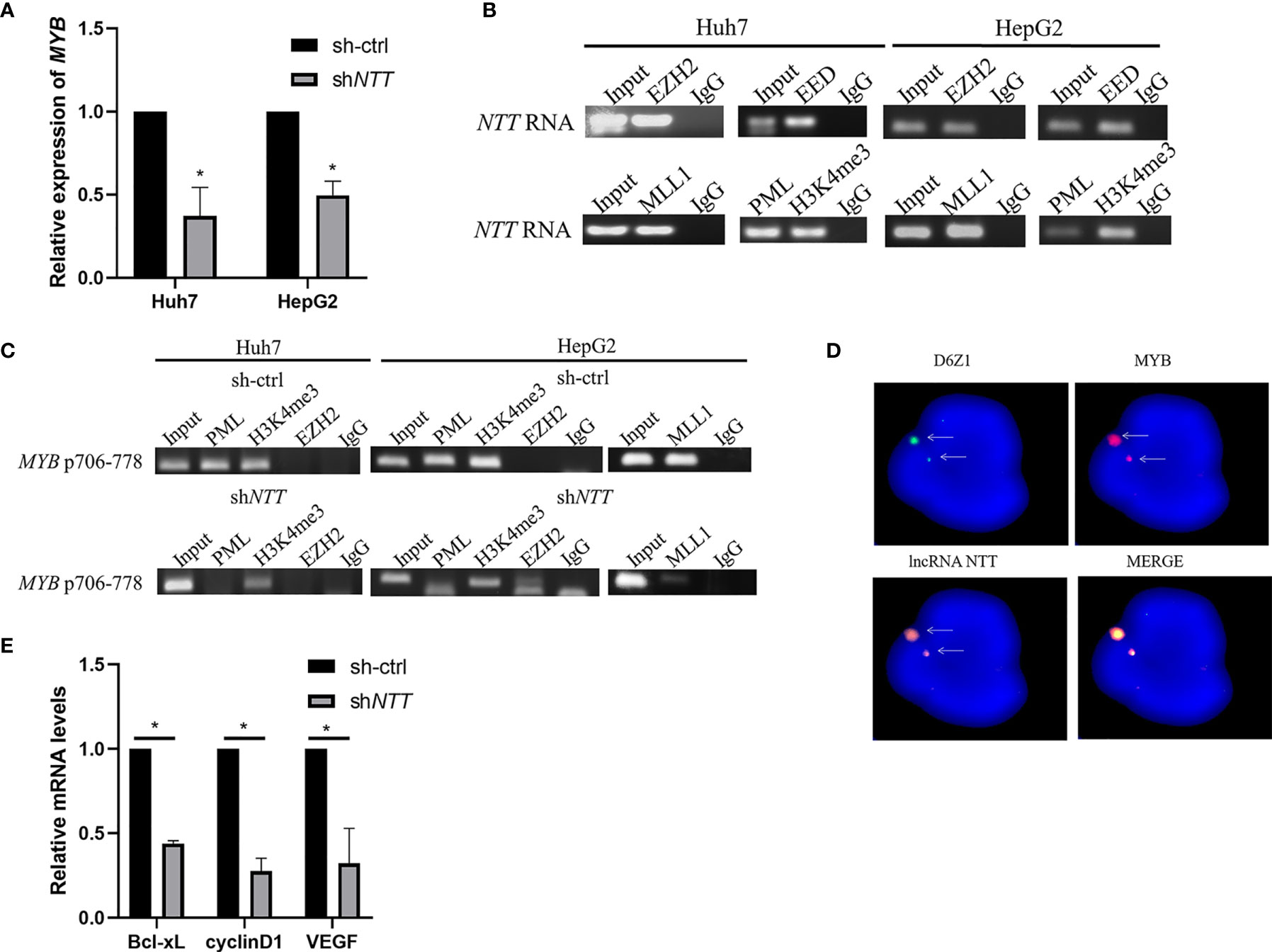
Figure 4 MYB expression may be regulated by NTT through its interaction with the activated complex. (A) MYB expression was examined in Huh7 and HepG2 cells transfected with NTT shRNA. (B) RIP-RT-qPCR analysis of NTT RNA binding to PRC2 (EZH2 and EED) and activated complex (MLL1, PML, and H3K4me3). (C) DNA ChIP-qPCR analysis of PML and H3K4me3 occupancy of the MYB promoter region. (D) RNA-DNA FISH conforming that NTT interacts with MYB. (E) RT-qPCR was used to measure expression of the selected genes in NTT knockdown cells. *p < 0.05.
Since it has been reported that lncRNAs may bind to polycomb-repressive complex 2 (PRC2), which consists of EZH2, EED, and SUZ12, to repress the expression of downstream genes, we examined whether NTT binds to the MYB promoter via interaction with PRC2. An RIP assay showed that NTT could bind EZH2 and EED (Figure 4B). Furthermore, we found that NTT not only bound to PRC2 but also bound to the activated complex (MLL1, PML, and H3K4me3) to activate gene transcription (Figure 4B). A similar result was observed with the HepG2 cells (Figure 4B).
DNA ChIP showed that the activated complex binds to positions 706–778 of the MYB promoter, and activated complex binding became undetectable or reduced following NTT knockdown (Figure 4C), suggesting that NTT enhances MYB expression by interacting with the activated complex binding to the MYB promoter. In agreement with the above results, we also found that NTT knockdown reduced the binding of PML and MLL1 to the MYB promoter (Figure 4C).
To further confirm the localization of NTT RNA transcript with MYB gene area, we used DNA (MYB genomic probe) FISH to detect the genomic location of MYB and RNA (NTT 21 base pair LNA probe) FISH to detect the NTT RNA transcript. Colocalization of NTT transcripts with the MYB gene was revealed (Figure 4D). These results suggest that NTT modulates nearby genes, such as MYB, via RNA transcripts and related protein complexes.
To explore the molecular mechanism underlying MYB-induced tumor promotion and metastasis, genes associated with tumor progression and metastasis were examined in sh-ctrl and shNTT cells by real-time quantitative PCR (RT-qPCR). Bcl-xL, cyclinD1, and VEGF are associated with cell proliferation, the cell cycle, and cell migration/invasion, respectively. All three were downregulated following NTT knockdown (Figure 4E).
NTT Is Regulated by ATF3
Examining the NTT promoter sequence, we identified the potential binding motif of ATF3, a key transcription factor in HCC cell lines. To verify if ATF3 binds to the NTT promoter and regulates NTT expression, we performed DNA ChIP and shRNA knockdown of ATF3. ATF3 binding was detected at position 5350 of the NTT promoter in Huh7 and HepG2 (Figure 5A), and ATF3 knockdown resulted in decreased NTT and its downstream genes expression (Figure 5B).
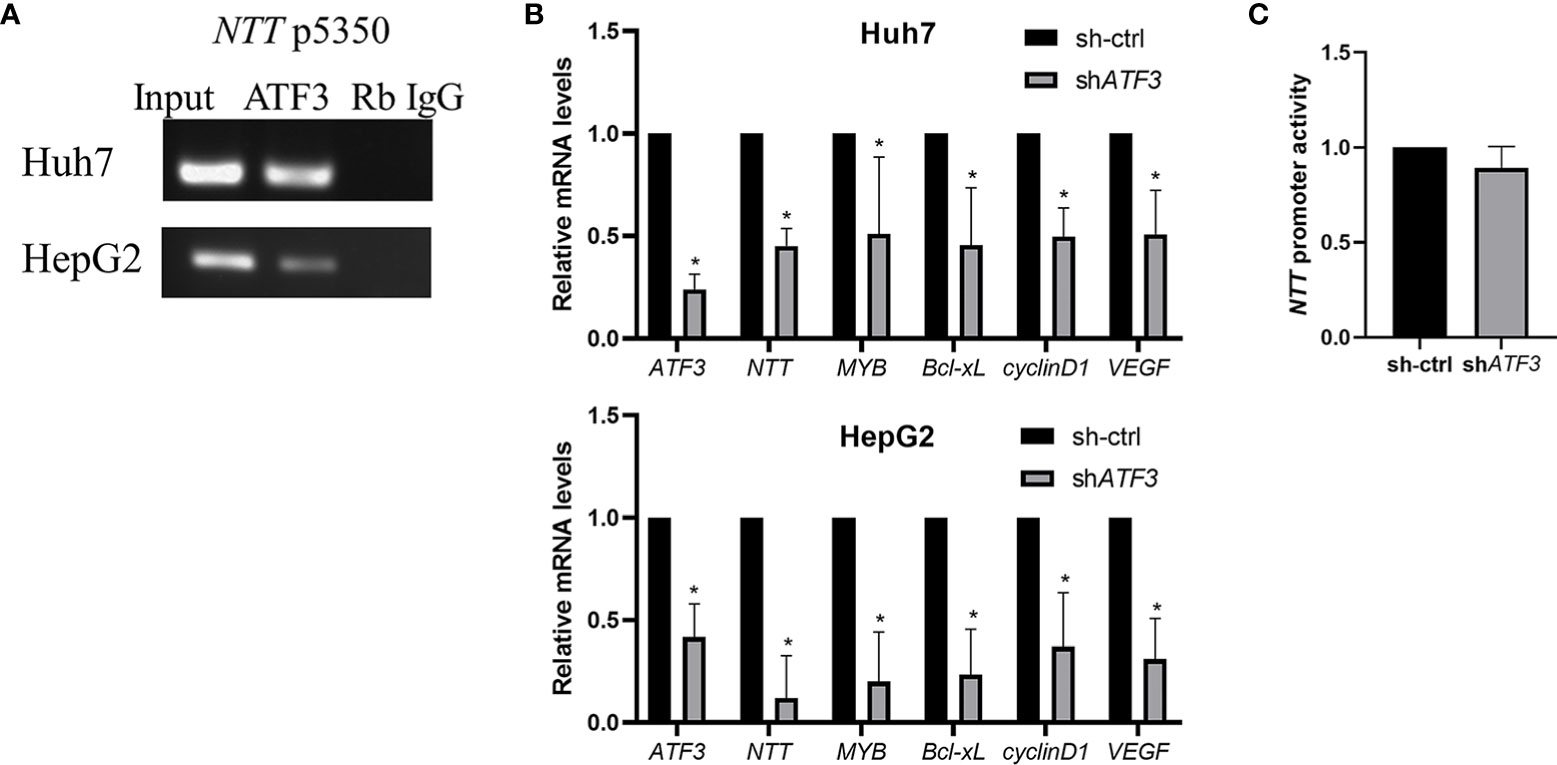
Figure 5 The functional effects of NTT are modulated by ATF3. (A) DNA ChIP-qPCR analysis of ATF3 occupancy of the NTT promoter region, shown by a representative graph of three independent experiments. (B) Knockdown of ATF3 by shRNA resulted in lower expression of NTT and its downstream genes. *p < 0.05. (C) Activity of promoter determined using a luciferase assay upstream of NTT.
We have performed the luciferase reporter assays to determine the NTT promoter activity in ATF3-knockdown cells, and the results showed that the luciferase activity was decreased after knockdown ATF3 (Figure 5C).
NTT Context-Dependently Regulating MYB In Vivo
We constructed stable cell lines that knocked down NTT or sh-ctrl control to verify the effect of NTT on tumorigenicity. From the in vivo experiments, we could not define NTT is an oncogene or tumor suppressor. Therefore, we examined NTT and MYB expression of eight mice tumor samples. The results showed both NTT and MYB overexpression result in larger tumor size than control in three-fourths of the mice, and NTT downexpression with MYB overexpression may result in either larger or smaller tumor or no change size than control (one-third with small, one-third with larger, and one-third with no definite change than control), and one mouse with both downexpression of NTT and MYB showed slight size change than the control (Supplementary Figure 4). These data are consistent with the findings of the human tissues experiments and suggested a context-dependent function of NTT for the regulation of MYB expression in in HCC development.
Discussion
Many studies have reported that lncRNAs are involved in the regulation of HCC differentiation and cell growth, including cell proliferation and apoptosis, invasion, and metastasis. For example, the lncRNA HOTAIR promotes cell invasion, proliferation, and metastasis and activates autophagy (22). HOTAIR also mediates hepatocarcinogenesis by suppressing miRNA-218 expression and activating P14 and P16 signaling (23). The lncRNA MALAT1 is associated with cell proliferation and migration by regulating Bax, bcl-2, bcl-xl, caspase-3, and caspase-8 (24). HULC promotes cell proliferation and triggers autophagy (25, 26). MEG3 expression in HCC samples is downregulated compared with normal controls. MEG3 interacts with the p53 DNA binding domain directly and regulates partial p53 target genes in hepatoma cells (27). As the role of NTT in HCC tumorigenesis is unknown, we explored it herein.
NTT was identified during the activation of T cells with phytohemagglutinin or with phorbol 12-myristate 13-acetate and ionomycin (18). NTT can be induced by synthetic peptide p9 of HIV-1 in human lymphocytes (19). Delgado et al. demonstrated that NTT plays a key role in the activation of lymphocytes mediated by RNA-dependent protein kinase through nuclear factor-kappa B activation (28). Recent studies have linked NTT overexpression to chronic inflammatory autoimmune disease, rheumatoid arthritis, and ME/CFS (20, 21). However, we did not find any correlations between NTT and hepatitis B or C infection in HCC patients. In addition, 17 primary/5 relapse HCC tumor samples and 22 adjacent normal liver tissue samples obtained from Gene Expression Omnibus (GEO) (GSE101432) were included in this study to validate the role of NTT in HCC. NTT is declined in HCC tissues compared with normal liver tissues (Supplementary Figure 5A). NTT in TCGA-HCC was also identified as compared to normal tissue samples, and decreased expression of NTT was also observed in HCC tissues (Supplementary Figure 5B). These results were consistent with our findings.
MYB is a transcription factor with three functional domains: DNA binding, transactivation, and negative regulatory domains (29). MYB is frequently overexpressed in human leukemias, breast cancers, and other solid tumors and is considered an oncogene. MYB overexpression promoted cell growth, cell-cycle progression, survival, and malignant behavior. Moreover, MYB is thought to be a potential therapeutic target in leukemia and β-hemoglobinopathies (30, 31). MYB has also been shown to interact with p300 protein (32). There are reports that MYB mRNA expression is higher in HCC tissues than in matched non-tumor tissues, and survival analysis revealed that strong MYB expression had lower disease-specific survival rates than in patients with negative MYB expression (33–35). Many studies have shown that MYB knockdown results in decreased cell migration (34, 36), similar to our results regarding NTT knockdown. We suggest that knockdown of either or both NTT and MYB produces similar results.
In this study, we confirmed that high MYB expression, determined by qRT-PCR, was associated with worse overall survival in a cohort of 80 patients with HCC. miR-424 plays a critical role in HCC tumorigenesis by targeting MYB mRNA (37). Liver cancer-associated lncRNAs participate in HCC processes by binding to the regulatory area of oncogenes, which is different from the translational regulation of microRNA (miRNA) (16). Our data suggest that the ability of NTT to target MYB may be one mechanism of the posttranscriptional control of MYB.
In conclusion, we found that NTT play a tumor-suppressor and tumor-promoting roles in human HCC tissues mostly from the ability to negatively or positively control MYB oncogene and suggested a context-dependent function of NTT in tumor development. Moreover, we report for the first time that the oncogenic activity of NTT is attributable to its activation of MYB by interacting with the activated complex in cellular experiments. Our results suggest that NTT may be a novel therapeutic target for the treatment of HCC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by China Medical University Hospital (CMUH102-REC1-037). The patients/participants provided their written informed consent to participate in this study. All animal experiments were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (CMU).
Author Contributions
Y-SC, Y-TL, and J-CY: investigation, methodology, and writing—original draft preparation. YC and L-LL: conceptualization and data curation. W-LC and W-CC: visualization, software, and supervision. S-YL and J-GC: writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the China Medical University (CMU103-BC-7) and China Medical University Hospital (DMR-110-238).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The data analyzed in this study were partially provided by the China Medical University Hospital Cancer Registry.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.592045/full#supplementary-material
References
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of Worldwide Burden of Cancer in 2008: GLOBOCAN 2008. Int J Cancer (2010) 127(12):2893–917. doi: 10.1002/ijc.25516
2. European Association For The Study Of The L, European Organisation For R, Treatment Of C. EASL-EORTC Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J Hepatol (2012) 56(4):908–43. doi: 10.1016/j.jhep.2011.12.001
3. Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, et al. Asia-Pacific Clinical Practice Guidelines on the Management of Hepatocellular Carcinoma: A 2017 Update. Hepatol Int (2017) 11(4):317–70. doi: 10.1007/s12072-017-9799-9
4. Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular Carcinoma: Epidemiology, Risk Factors and Pathogenesis. World J Gastroenterol (2008) 14(27):4300–8. doi: 10.3748/wjg.14.4300
5. Aghoram R, Cai P, Dickinson JA. Alpha-Foetoprotein and/or Liver Ultrasonography for Screening of Hepatocellular Carcinoma in Patients With Chronic Hepatitis B. Cochrane Database Syst Rev (2012) 9:CD002799. doi: 10.1002/14651858.CD002799.pub2
6. Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, et al. Alpha-Fetoprotein, Des-Gamma Carboxyprothrombin, and Lectin-Bound Alpha-Fetoprotein in Early Hepatocellular Carcinoma. Gastroenterology (2009) 137(1):110–8. doi: 10.1053/j.gastro.2009.04.005
7. Bruix J, Sherman M. American Association for the Study of Liver D. Management of Hepatocellular Carcinoma: An Update. Hepatology (2011) 53(3):1020–2. doi: 10.1002/hep.24199
8. Jiang P, Chan KCA, Lo YMD. Liver-Derived Cell-Free Nucleic Acids in Plasma: Biology and Applications in Liquid Biopsies. J Hepatol (2019) 71(2):409–21. doi: 10.1016/j.jhep.2019.04.003
9. Llovet JM, Burroughs A, Bruix J. Hepatocellular Carcinoma. Lancet (2003) 362(9399):1907–17. doi: 10.1016/S0140-6736(03)14964-1
10. Chapman BC, Paniccia A, Hosokawa PW, Henderson WG, Overbey DM, Messersmith W, et al. Impact of Facility Type and Surgical Volume on 10-Year Survival in Patients Undergoing Hepatic Resection for Hepatocellular Carcinoma. J Am Coll Surg (2017) 224(3):362–72. doi: 10.1016/j.jamcollsurg.2016.11.011
11. Geisler S, Coller J. RNA in Unexpected Places: Long Non-Coding RNA Functions in Diverse Cellular Contexts. Nat Rev Mol Cell Biol (2013) 14(11):699–712. doi: 10.1038/nrm3679
12. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev (2011) 25(18):1915–27. doi: 10.1101/gad.17446611
13. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of Somatic Tissue Differentiation by the Long Non-Coding RNA TINCR. Nature (2013) 493(7431):231–5. doi: 10.1038/nature11661
14. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
15. Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, et al. Long Non-Coding RNA ANRIL Is Upregulated in Hepatocellular Carcinoma and Regulates Cell Proliferation by Epigenetic Silencing of KLF2. J Hematol Oncol (2015) 8(1):57. doi: 10.1186/s13045-015-0153-1
16. Shi H, Xu Y, Yi X, Fang D, Hou X. Current Research Progress on Long Noncoding RNAs Associated With Hepatocellular Carcinoma. Anal Cell Pathol (Amst) (2019) 2019:1534607. doi: 10.1155/2019/1534607
17. DiStefano JK. Long Noncoding RNAs in the Initiation, Progression, and Metastasis of Hepatocellular Carcinoma. Noncoding RNA Res (2017) 2(3-4):129–36. doi: 10.1016/j.ncrna.2017.11.001
18. Liu AY, Torchia BS, Migeon BR, Siliciano RF. The Human NTT Gene: Identification of a Novel 17-Kb Noncoding Nuclear RNA Expressed in Activated CD4+ T Cells. Genomics (1997) 39(2):171–84. doi: 10.1006/geno.1996.4463
19. Amarante MK, De Lucca FL, de Oliveira CE, Pelegrinelli Fungaro MH, Reiche EM, Muxel SM, et al. Expression of Noncoding mRNA in Human Blood Cells Activated With Synthetic Peptide of HIV. Blood Cells Mol Dis (2005) 35(2):286–90. doi: 10.1016/j.bcmd.2005.06.004
20. Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, et al. lncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis. Int J Mol Sci (2018) 19(9):2806. doi: 10.3390/ijms19092806
21. Yang CA, Bauer S, Ho YC, Sotzny F, Chang JG, Scheibenbogen C. The Expression Signature of Very Long Non-Coding RNA in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. J Transl Med (2018) 16(1):231. doi: 10.1186/s12967-018-1600-x
22. Li E, Zhao Z, Ma B, Zhang J. Long Noncoding RNA HOTAIR Promotes the Proliferation and Metastasis of Osteosarcoma Cells Through the AKT/mTOR Signaling Pathway. Exp Ther Med (2017) 14(6):5321–8. doi: 10.3892/etm.2017.5248
23. Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, et al. Hotair Mediates Hepatocarcinogenesis Through Suppressing miRNA-218 Expression and Activating P14 and P16 Signaling. J Hepatol (2015) 63(4):886–95. doi: 10.1016/j.jhep.2015.05.016
24. Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long Non-Coding RNA MALAT-1 Overexpression Predicts Tumor Recurrence of Hepatocellular Carcinoma After Liver Transplantation. Med Oncol (2012) 29(3):1810–6. doi: 10.1007/s12032-011-0004-z
25. Xiong H, Li B, He J, Zeng Y, Zhang Y, He F. lncRNA HULC Promotes the Growth of Hepatocellular Carcinoma Cells via Stabilizing COX-2 Protein. Biochem Biophys Res Commun (2017) 490(3):693–9. doi: 10.1016/j.bbrc.2017.06.103
26. Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC Triggers Autophagy via Stabilizing Sirt1 and Attenuates the Chemosensitivity of HCC Cells. Oncogene (2017) 36(25):3528–40. doi: 10.1038/onc.2016.521
27. Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, et al. Long Noncoding RNA MEG3 Interacts With P53 Protein and Regulates Partial P53 Target Genes in Hepatoma Cells. PloS One (2015) 10(10):e0139790. doi: 10.1371/journal.pone.0139790
28. Delgado Andre N, De Lucca FL. Non-Coding Transcript in T Cells (NTT): Antisense Transcript Activates PKR and NF-kappaB in Human Lymphocytes. Blood Cells Mol Dis (2008) 40(2):227–32. doi: 10.1016/j.bcmd.2007.08.005
29. Wang X, Angelis N, Thein SL. MYB - A Regulatory Factor in Hematopoiesis. Gene (2018) 665:6–17. doi: 10.1016/j.gene.2018.04.065
30. Pattabiraman DR, Gonda TJ. Role and Potential for Therapeutic Targeting of MYB in Leukemia. Leukemia (2013) 27(2):269–77. doi: 10.1038/leu.2012.225
31. Thein SL. Molecular Basis of Beta Thalassemia and Potential Therapeutic Targets. Blood Cells Mol Dis (2018) 70:54–65. doi: 10.1016/j.bcmd.2017.06.001
32. Pattabiraman DR, McGirr C, Shakhbazov K, Barbier V, Krishnan K, Mukhopadhyay P, et al. Interaction of C-Myb With P300 Is Required for the Induction of Acute Myeloid Leukemia (AML) by Human AML Oncogenes. Blood (2014) 123(17):2682–90. doi: 10.1182/blood-2012-02-413187
33. Nakajima T, Yasui K, Zen K, Inagaki Y, Fujii H, Minami M, et al. Activation of B-Myb by E2F1 in Hepatocellular Carcinoma. Hepatol Res (2008) 38(9):886–95. doi: 10.1111/j.1872-034X.2008.00324.x
34. Chen RX, Xia YH, Xue TC, Ye SL. Transcription Factor C-Myb Promotes the Invasion of Hepatocellular Carcinoma Cells via Increasing Osteopontin Expression. J Exp Clin Cancer Res (2010) 29:172. doi: 10.1186/1756-9966-29-172
35. Yang H, Huang ZZ, Wang J, Lu SC. The Role of C-Myb and Sp1 in the Up-Regulation of Methionine Adenosyltransferase 2A Gene Expression in Human Hepatocellular Carcinoma. FASEB J (2001) 15(9):1507–16. doi: 10.1096/fj.01-0040com
36. Xu LH, Zhao F, Yang WW, Chen CW, Du ZH, Fu M, et al. MYB Promotes the Growth and Metastasis of Salivary Adenoid Cystic Carcinoma. Int J Oncol (2019) 54(5):1579–90. doi: 10.3892/ijo.2019.4754
Keywords: long noncoding RNA, NTT, MYB, hepatocellular carcinoma, context-dependent
Citation: Chang Y-S, Lee Y-T, Yen J-C, Chang YC, Lin L-L, Chan W-L, Chang W-C, Lin S-Y and Chang J-G (2021) Long Noncoding RNA NTT Context-Dependently Regulates MYB by Interacting With Activated Complex in Hepatocellular Carcinoma Cells. Front. Oncol. 11:592045. doi: 10.3389/fonc.2021.592045
Received: 06 August 2020; Accepted: 09 August 2021;
Published: 20 September 2021.
Edited by:
Yi Zhang, First Affiliated Hospital of Zhengzhou University, ChinaCopyright © 2021 Chang, Lee, Yen, Chang, Lin, Chan, Chang, Lin and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan-Gowth Chang, ZDY3ODFAbWFpbC5jbXVoLm9yZy50dw==; Shyr-Yi Lin, c3lsaW5AdG11LmVkdS50dw==
 Ya-Sian Chang1,2,3
Ya-Sian Chang1,2,3 Wen-Ling Chan
Wen-Ling Chan Wei-Chiao Chang
Wei-Chiao Chang Jan-Gowth Chang
Jan-Gowth Chang