- Queen Mary School, Nanchang University, Nanchang, China
Purpose: LRRC3B gene, as a tumor suppressor gene was involved in the development and progress of breast cancer (BC). However, the effect of LRRC3B polymorphisms on BC has rarely been reported. In the study, we aimed to evaluate the relation between LRRC3B variants and BC risk.
Methods: Among 563 BC patients and 552 healthy controls, ten single-nucleotide polymorphisms (SNPs) in LRRC3B were genotyped by Agena MassARRAY. Odds ratios (ORs) and 95% confidence interval (CI) were calculated using logistic regression model.
Results: Our study demonstrated that rs1907168 polymorphism (heterozygous: OR = 0.71, p = 0.017) was related to the reduced risk of BC in the overall. In stratified analyses by age, rs1907168 was associated with the decreased (heterozygous: OR = 0.53, p = 0.002) while rs78205284 (homozygote: OR = 2.83, p = 0.034) increased BC susceptibility among the population at age ≤51 years. Rs6551122 (recessive: OR = 0.51, p = 0.028) and rs12635768 (homozygote, OR = 0.36, p = 0.023) polymorphisms were related to the smaller BC tumor size (<2 cm). In addition, rs112276562 (heterozygote OR = 0.56, p = 0.002), rs6551122 (heterozygote OR = 0.63, p = 0.016), and rs73150416 (heterozygote OR = 0.57, p = 0.005) variants contributed to the lower incidence of PR-positive BC. Moreover, rs6788033 was associated with a lower expression level of Ki-67 (log-additive: OR = 0.68, p = 0.024). Furthermore, we found an association of ‘GATT’ haplotype with an increased risk for BC. In addition, LRRC3B gene was down-regulated in BC tumor and had a poor prognosis in BC in in silico analysis.
Conclusion: Our study firstly found LRRC3B SNPs contributed to the risk of BC, suggesting LRRC3B variants might help to predict BC progression.
Introduction
Breast cancer (BC) is the most commonly diagnosed malignant tumor and the primary cause of cancer death among women globally (1). In China, 268,600 new cases of BC were diagnosed and there were 69,500 deaths, and its morbidity and mortality rates have been increasing annually (2). BC is a multi-factorial, polygenic disease resulting from genetic, endocrine, and environmental factors (3). Age, serum hormone levels, family history, and environmental factors have been identified as potential contributors to BC susceptibility (4). Moreover, genetic factors were the major drivers in the genesis of BC (5–7).
Leucine-rich repeat-containing 3B (LRRC3B), also named LRP15, is one of the transmembrane proteins having an evolutionarily conserved LRR-domain (8). LRR-containing proteins participated in protein interaction, cell adherence, DNA repair, gene recombination, and so on (9). LRRC3B protein has been reported to regulate the DNA damage and repair pathways and cell cycle progression (10). Additionally, LRRC3B is down-regulated in non-small-cell lung, head and neck, gastric and breast cancers (11–14), suggesting LRRC3B involvement in carcinogenesis. Abnormal expression of LRRC3B in BC was associated with the development and prognosis of BC (13). In addition, LRRC3B also served an important role in BC recurrence and metastasis (15). Genetic polymorphisms may affect its gene expression, thus is associated with the occurrence of disease. In spite of many researchers’ reports on the role of LRRC3B expression in BC pathogenesis, the possible effects of LRRC3B polymorphisms on BC have not been studied.
Here, we investigated the frequency distribution of LRRC3B polymorphisms and the potential relationship of LRRC3B polymorphisms with BC susceptibility in Chinese Han females.
Materials and Methods
Study Subjects
A total of 1,115 unrelated Chinese Han female subjects (563 BC patients and 552 healthy controls) were enrolled from the Shaanxi Provincial Cancer Hospital. BC patients were newly diagnosed and were confirmed by histopathology based on the WHO BC classification. The blood samples of patients were collected before chemotherapy, radiotherapy, or endocrine therapy. Patients had no other cancer or other systemic inflammatory disease. In the meantime, age-matched healthy individuals who had a checkup at the hospital were recruited as controls. The controls had no mammary tumors by recent clinical and mammographic examinations, no cancer history, no BC family history, and no mammary diseases. The study was approved by the Nanchang University and in accordance with the 1964 Helsinki Declaration. Written informed consent was obtained.
In Silico Analysis
Prediction of LRRC3B SNPs’ possible function was performed with HaploReg v4.1 online tool (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). With the purpose to assess LRRC3B mRNA expression in BC, GEPIA database was used. Kaplan–Meier plotter was used to evaluate the prognosis of LRRC3B expression in BC.
Data Collection and DNA Extraction
Questionnaires and medical records were used to gather the demographic and clinical features. The following data were recorded: age, estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and Ki-67 status, clinical stage, tumor size, and tumor lymph node invasion. After interview, peripheral blood (5 ml) was gathered into EDTA-coated tubes from patients and healthy controls. Genomic DNA was isolated using the GoldMag genomic DNA purification kit (GoldMag Co. Ltd., Xi’an, China).
Genotyping of SNPs
We obtained the physical position of the LRRC3B gene on chromosome 3:26664297-26752267 through the e!GRCh37 (http://asia.ensembl.org/Homo_sapiens/Info/Index) database. In the VCF to PED Converter window (http://grch37.ensembl.org/Homo_sapiens/Tools/VcftoPed), we entered the gene location, selected the CHB and CHS population, and downloaded the “ped” file and “info” file for the variations of LRRC3B. Using Haploview software, we selected tagSNPs based on HWE >0.01, MAF >0.05, Min Genotype >75%, and Tagger r2 >0.8. Combining MassARRAY primer design software, HWE >0.05, MAF >0.05, and the call rate >95% in our study population, ten candidate SNPs (rs112276562, rs6790894, rs6551121, rs6551122, rs1907168, rs73150416, rs12635768, rs6551130, rs78205284 and rs6788033) in LRRC3B were randomly selected. LRRC3B SNPs were genotyped by MassARRAY platform (Agena Bioscience, San Diego, CA, USA) (16, 17). The primers were shown in Supplementary Table 1. About 10% of the samples were randomly selected and genotyped in duplicate for quality control, and the concordance was 100%.
Data Analysis
Student’s t test was performed to evaluate the age distribution. Hardy–Weinberg equilibrium (HWE) was tested by a goodness-of-fit χ2 test in controls. Genotype frequencies of LRRC3B SNPs between patients and controls were analyzed by χ2 test. Odds ratios (ORs) and 95% confidence interval (CI) adjusted age were calculated, and haplotype frequencies were estimated using PLINK software. False-positive report probability (FPRP) analysis was used to evaluate the noteworthy associations of the significant findings (18, 19). We set 0.2 as the FPRP threshold and assigned a prior probability of 0.1 for an association with genotypes under investigation. Pairwise linkage disequilibrium (LD) was analyzed using Haploview software (version 4.2). Multifactor dimensionality reduction (MDR) (version 3.0.2) was performed to evaluate the SNP–SNP interactions in the risk of BC. p-values <0.05 were considered significant, whereas a value of corrected p <0.05/10 was considered significant after Bonferroni correction.
Results
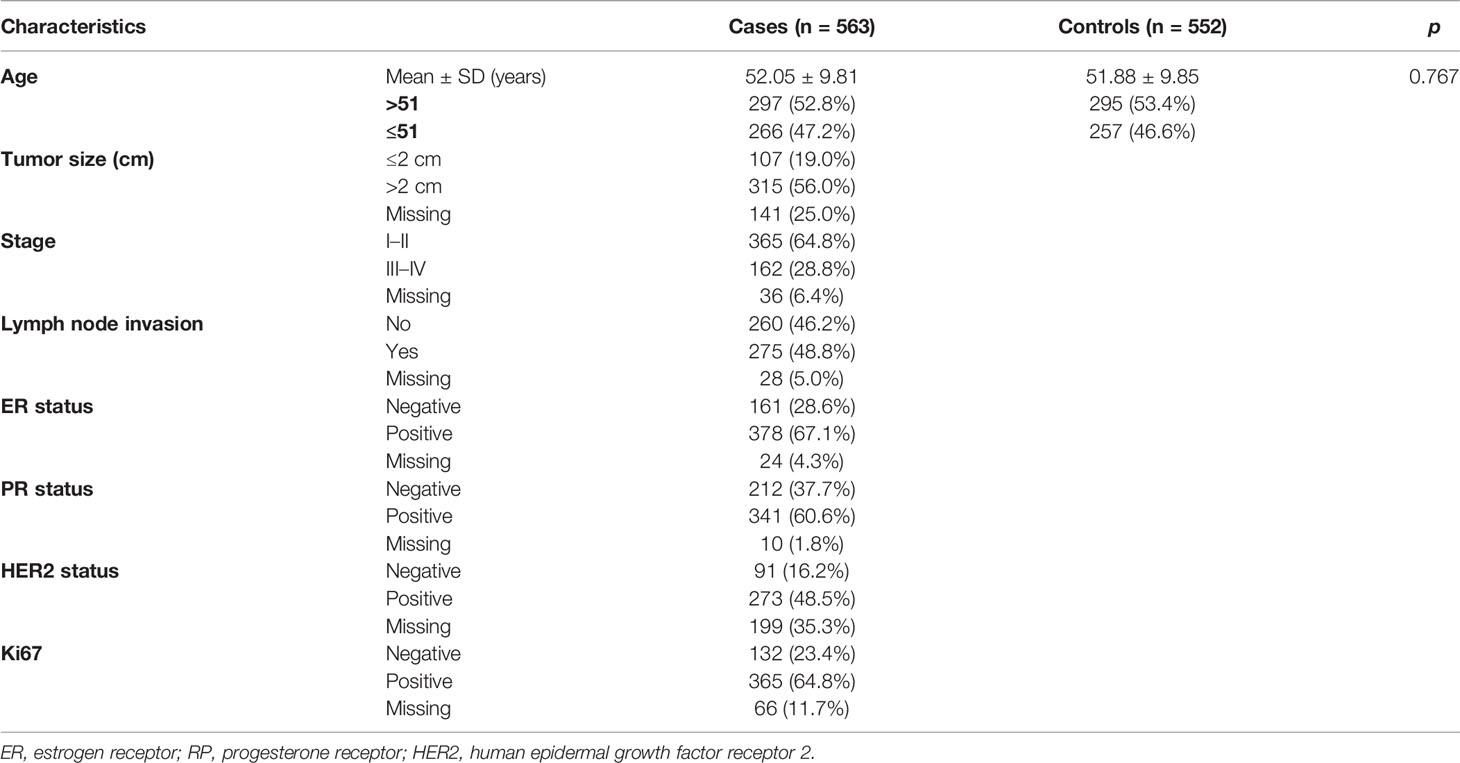
The features of all subjects were summarized in Table 1. 563 BC patients (52.05 ± 9.81 years) and 552 healthy controls (51.88 ± 9.85 years) participated in the present study. No significant difference of age distribution between cases and controls (p = 0.767) was found. Most patients had tumor size >2 cm (56.0%), stage I/II (64.8%), lymph node invasion (48.8%), positive hormone receptor status such as ER+ (67.1%), PR+ (60.6%), or HER2+ (48.5%) and positive Ki67 (64.8%).
According to the GEPIA database, LRRC3B gene was down-regulated in BC (p < 0.01, Supplementary Figure 1). Kaplan–Meier plotter showed high expression of LRRC3B was related to better BC overall survival (HR = 0.83, 95% CI: 0.71–0.97, p = 0.017, Supplementary Figure 2). We speculated that LRRC3B is a tumor suppressor gene, but more convincing studies are needed to validate the conclusions because these clinical data are from the database.
The MAFs of LRRC3B SNPs were more than 0.05 (Supplementary Table 2). No deviation from the HWE for the polymorphisms examined was observed in the genotype distribution of the controls (p > 0.05), suggesting these subjects could represent the general population. In addition, LRRC3B variants were related to enhancer histone, changed motifs, and GRASP QTL hits in in silico analysis.
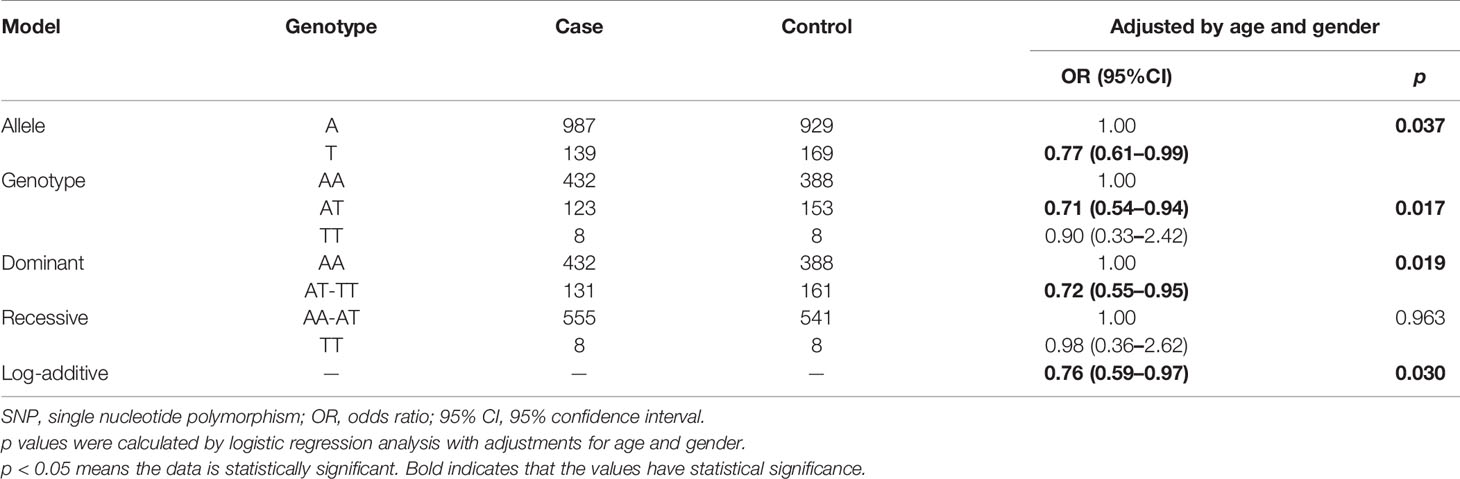
We performed allele and genetic model analyses of ten LRRC3B SNPs in the BC cases and control subjects. Genotype frequency distribution of SNPs was shown in Table 2 and Supplementary Table 3. The distribution of A and T alleles of LRRC3B rs1907168 polymorphism was 84.6 and 15.4% in controls and 87.7 and 12.3% in patients. An association between T allele and BC risk (OR = 0.77, 95% CI: 0.61–0.99, p = 0.037) was found. Similarly, rs1907168 had a decreased risk of BC (heterozygous: OR = 0.71, 95% CI: 0.54–0.94, p = 0.017; dominant: OR = 0.72, 95% CI: 0.55–0.95, p = 0.019; and log-additive: OR = 0.76, 95% CI: 0.59–0.97, p = 0.030). However, no significant association was found for other SNPs (Supplementary Table 3).
We further conducted subgroup analyses by age to examine the effects of LRRC3B SNPs on BC risk (Table 3). LRRC3B rs1907168 was related to the reduced risk of BC among young participants (age < 51 years) under the allele (OR = 0.69, 95% CI: 0.48–0.99, p = 0.043), heterozygote (OR = 0.53, 95% CI: 0.35–0.80, p = 0.002), dominant (OR = 0.58, 95% CI: 0.39–0.86, p = 0.008) and log-additive (OR = 0.68, 95% CI: 0.47–0.98, p = 0.038) models. The significance of the heterozygote model still existed after Bonferroni correction. In addition, rs78205284 polymorphism (TT vs. GG, OR = 2.83, 95% CI: 1.08–7.41, p = 0.034; and TT vs. GG-GT, OR = 2.72, 95% CI: 1.05–7.07, p = 0.040) contributed to the increased BC risk at age ≤51 years.
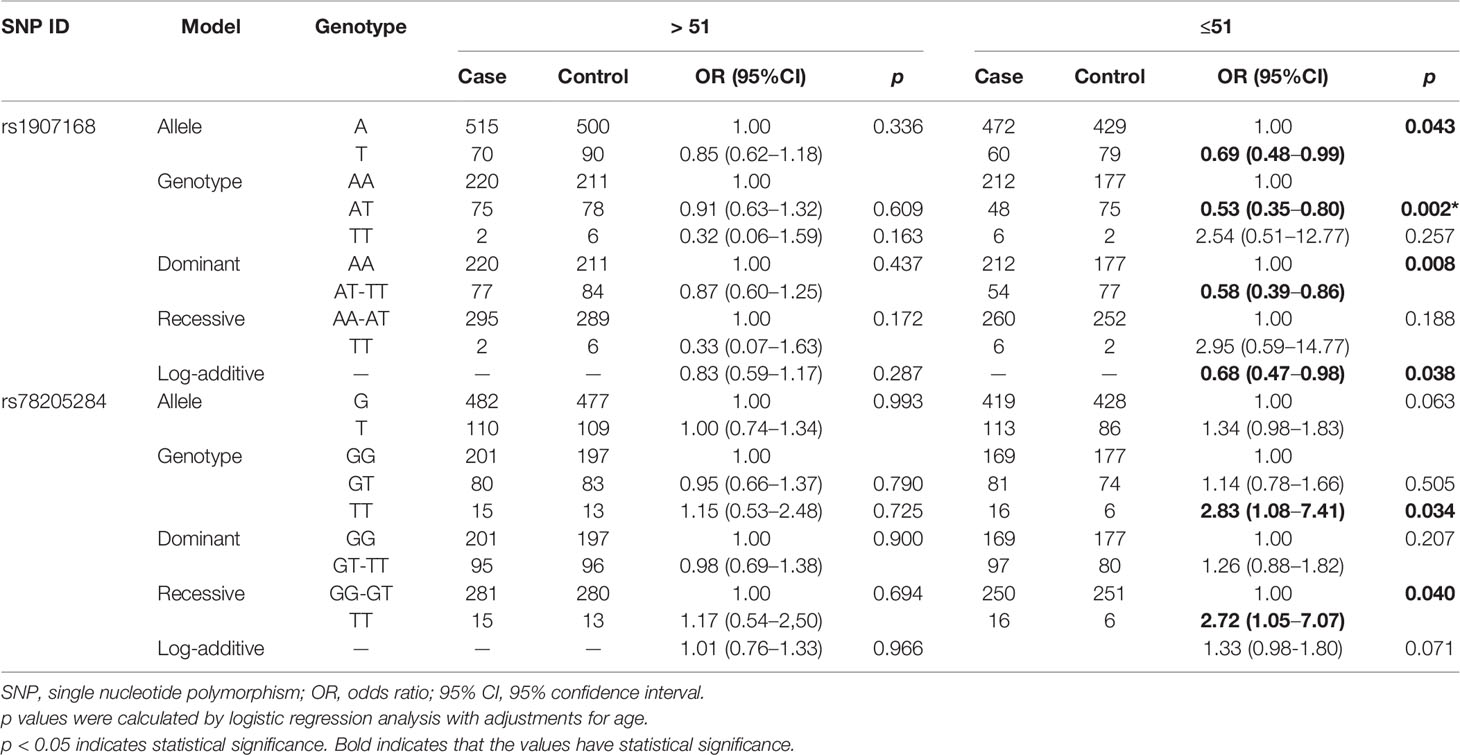
Table 3 Relationships between LRRC3B polymorphisms and breast cancer risk according to the stratification by age.
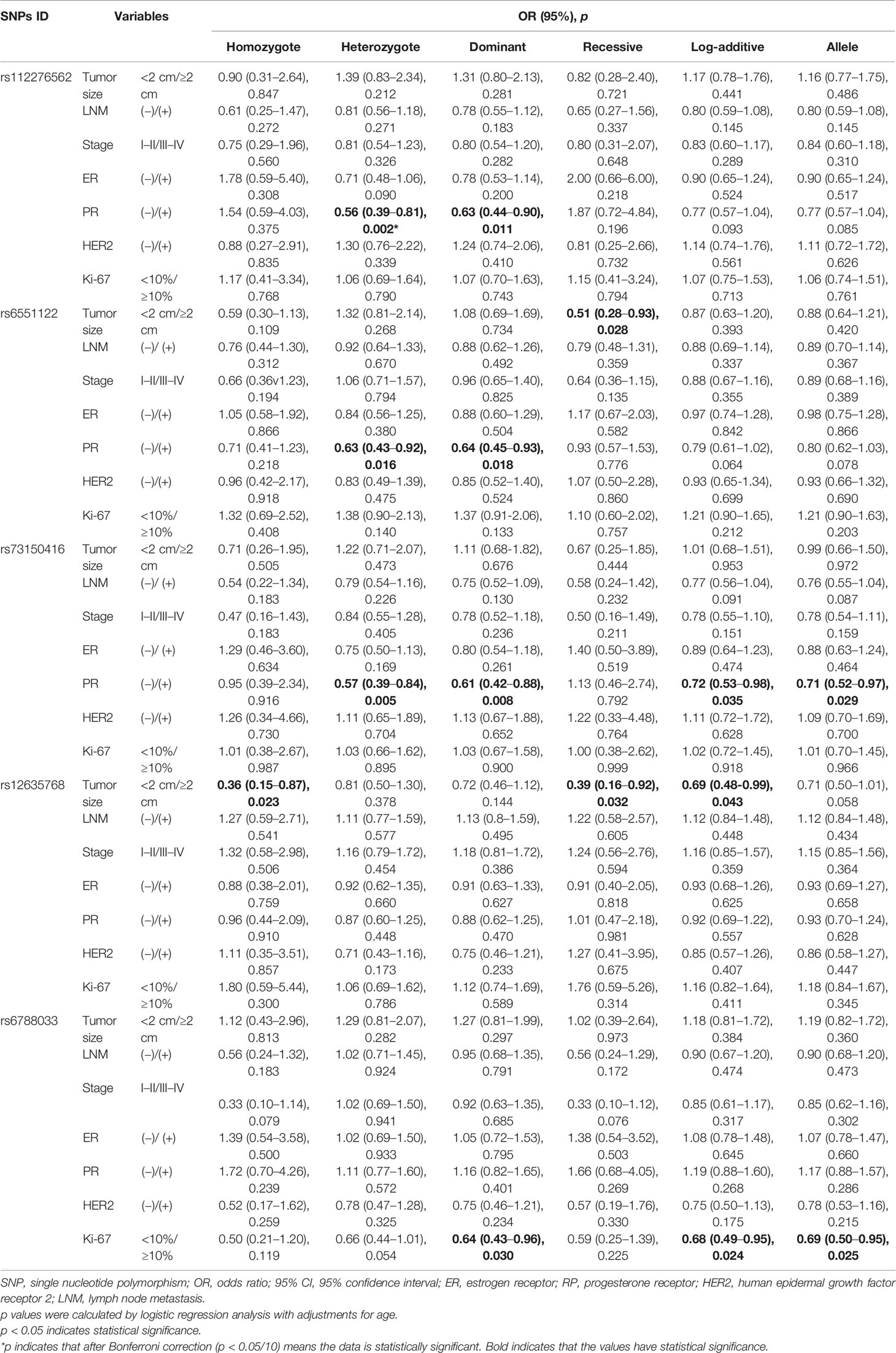
We also investigated the potential associations of LRRC3B polymorphisms with clinicopathological characteristics of BC, including tumor size, lymph node metastasis, clinical stage, the status of ER, PR, Her-2, and Ki67 (Table 4). We found that rs6551122 (OR = 0.51, p = 0.028) and rs12635768 (homozygote, OR = 0.36, p = 0.023; recessive, OR = 0.39, p = 0.032; log-additive, OR = 0.69, p = 0.043) polymorphisms were related to a smaller BC tumor size (<2 cm). In addition, rs112276562, rs6551122, and rs73150416 variants contributed to a lower incidence of PR-positive BC (for rs112276562, heterozygote OR = 0.56, p = 0.002 and dominant OR = 0.63, p = 0.011; for rs6551122, heterozygote OR = 0.63, p = 0.016 and dominant OR = 0.64, p = 0.018; and for rs73150416, heterozygote OR = 0.57, p = 0.005, dominant OR = 0.61, p = 0.008, log-additive OR = 0.72, p = 0.035, and allele OR = 0.71, p = 0.029). After Bonferroni correction, rs112276562 was associated with a lower incidence of PR-positive BC under the heterozygote model. Moreover, rs6788033 was associated with a lower the expression level of Ki-67 in the dominant (OR = 0.64, p = 0.030), log-additive (OR = 0.68, p = 0.024) and allele (OR = 0.69, p = 0.025) models. There was no significant association of LRRC3B polymorphisms with respect to the other variables like lymph node metastasis, clinical stage, and ER/HER2 status.
FPRP analysis was carried out to interrogate whether the significant findings were deserving attention (Table 5). At the prior probability level of 0.1, the significant association for rs1907168 (heterozygote model, FPRP = 0.184) remained noteworthy in the overall analysis. In the subgroup at age <51 years, significant findings remained noteworthy for rs1907168 under the heterozygote (FPRP = 0.141) and dominant (FPRP = 0.198) models. Moreover, the associations of rs112276562 (heterozygote model, FPRP = 0.095) and rs73150416 (heterozygote model, FPRP = 0.159; and dominant model FPRP = 0.189) with PR-positive BC were also positive at the prior probability level of 0.1.
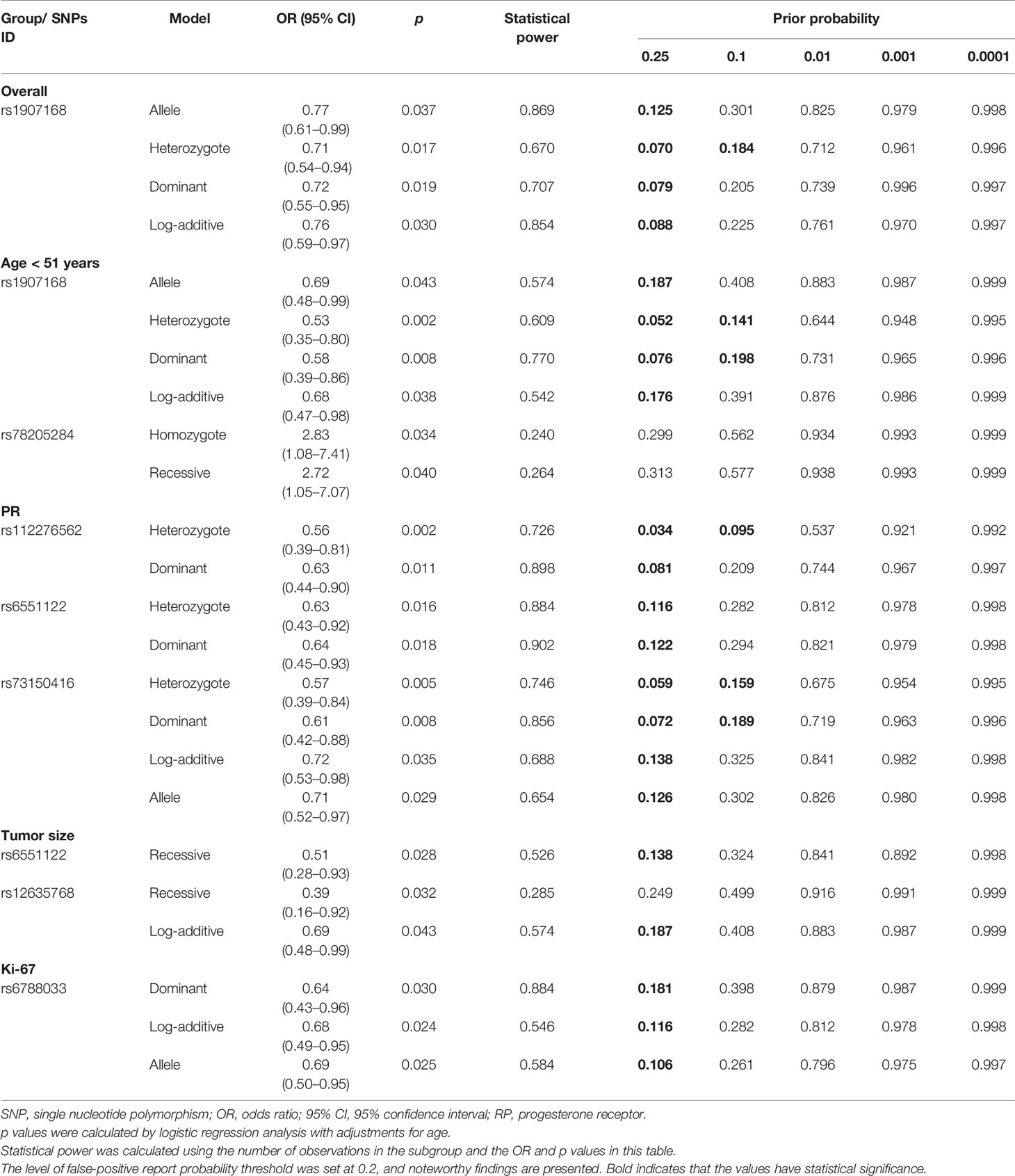
Table 5 False-positive report probability values for the associations between LRRC3B Polymorphisms and BC susceptibility.
Figure 1 represented the reconstructed LD plot and had three blocks in LRRC3B SNPs (Block 1, rs112276562 and rs6790894; Block 2, rs6551121, rs6551122, rs1907168, and rs73150416; and Block 3, rs12635768, rs6551130, rs78205284, and rs6788033). Moreover, ‘GATT’ haplotype in Block 2 had a higher risk for BC (OR = 1.29, 95% CI: 1.00–1.65, p = 0.048, Supplementary Table 4).
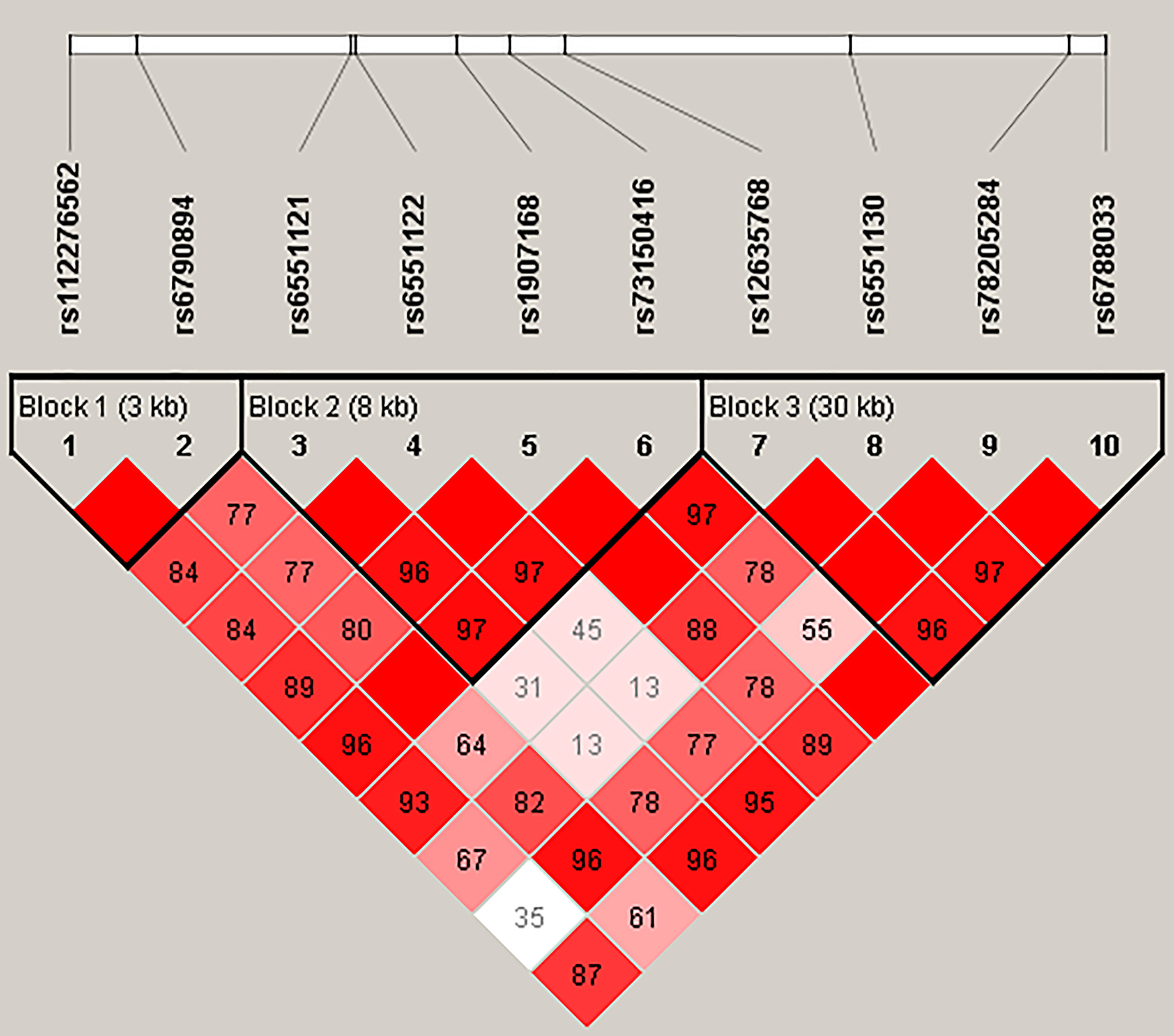
Figure 1 Haplotype block map for ten SNPs in the LRRC3B gene. The numbers inside the diamonds indicate the D′ for pairwise analyses.
Subsequently, MDR analysis was implemented to assess the impact of the SNP–SNP interactions. Dendrogram interaction analysis of our data showed that rs6551130, rs12635768, and rs6788033 exhibited a strong synergy effect, and rs1907168 and rs73150416 had a strong redundancy effect, as shown in Figure 2. Table 6 showed that rs1907168 was the best single-locus model to predict the risk of BC (testing accuracy = 0.5048, CVC = 8/10, p = 0.0298). The best multi-loci model included rs112276562, rs1907168, and rs6551130 (testing accuracy = 0.5205, CVC = 7/10, p <0.0001).
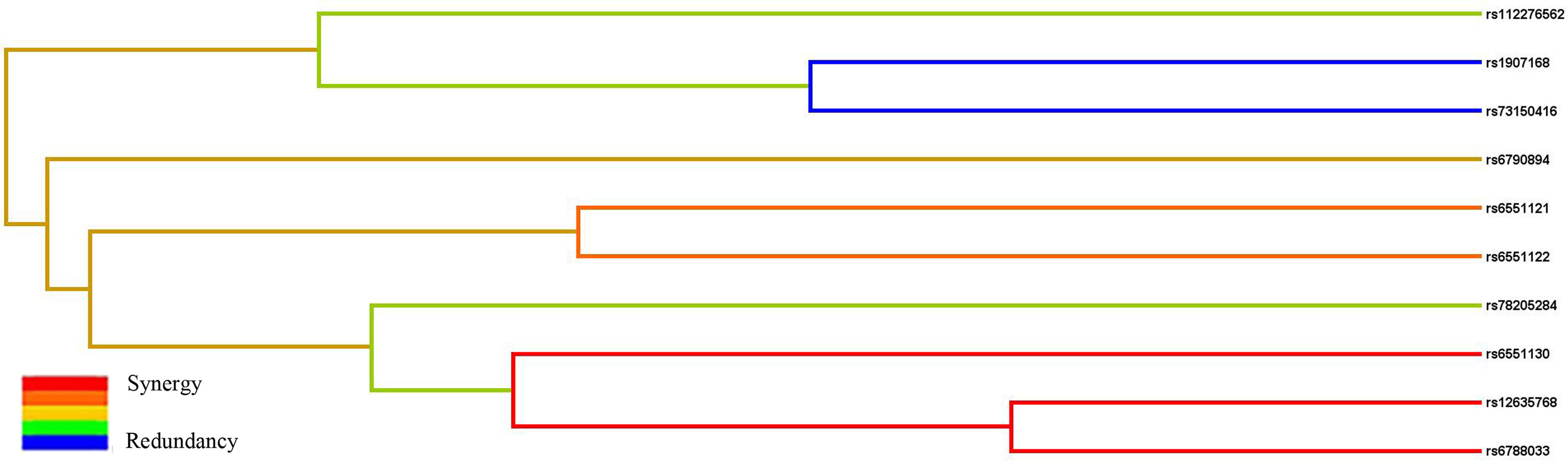
Figure 2 The interaction dendrogram derived from MDR for the SNP–SNP interaction in LRRC3B. Short connections among nodes represent stronger synergistic (red and orange) or redundant (green and blue) interactions.
Discussion
This study investigated the effect of LRRC3B genetic variation on BC risk in a Chinese Han population. The result revealed that rs1907168 conferred to the reduced BC risk overall. In additional, rs1907168 decreased the risk of BC while rs78205284 increased BC susceptibility among the population at age ≤51 years. Noticeably, clinical parameters such as the status of PR and Ki67 and tumor size were significantly associated with LRRC3B polymorphism. Furthermore, we found the existence of three blocks in LRRC3B SNPs and the association of ‘GATT’ haplotype with an increased BC risk. Our results suggested LRRC3B variants might be susceptibility markers for BC. Specially; this is firstly demonstrated by the effects of LRRC3B SNPs on BC susceptibility.
LRRC3B, located at 3p24, contained a leucine-rich repeat N-terminal domain (Leu >13%). The protein-conserved region of LRRC3B exists at cAMP- and cGMP-dependent protein kinase phosphorylation sites, thus, related to cell differentiation, cycle regulation, proliferation and invasion (20, 21). Several studies have revealed that LRRC3B could be a tumor suppressor in carcinogenesis (22, 23). Formerly, the expression of LRRC3B was reduced in BC tissues and associated with tumor grade of BC (13). Consistently, we also found that LRRC3B was down-regulated in BC, and low expression of LRRC3B was related to a poor overall survival for BC based on bioinformatics analysis. Moreover, LRRC3B might serve a protective role in preventing bupivacaine-induced BC recurrence and metastasis (15). These results hint us that LRRC3B has an important role in BC occurrence and progression.
Genetic variations in LRRC3B might be associated with the development and progression of disease. However, there is no finding of related research on LRRC3B polymorphisms. In this study, LRRC3B rs1907168 (A/T) variant had a protective effect in BC risk. Further, stratified analysis showed that rs1907168 decreased while rs78205284 increased BC susceptibility among the population at age ≤51 years. Especially, rs1907168 was associated with the decreased BC risk after Bonferroni correction. Notably, the peak age for BC is 40–50 years in the Asian countries, whereas 60–70 years in the Western countries (24). This finding suggested that age might affect the association of genetic variations and BC risk, and that rs78205284 was a risk factor to BC occurrence among younger women in the Chinese Han population.
As BC is a complex disease, the same variant might show a different response for BC risk across different tumor sizes, clinical stages and clinical features (25, 26). We further evaluated the association of LRRC3B polymorphisms with clinical features of BC, including tumor size, lymph node metastasis, clinical stage, the status of ER, PR, Her-2, and Ki67. We found that rs6551122 and rs12635768 polymorphisms were associated with a smaller BC tumor size (<2 cm). In addition, rs112276562, rs6551122, and rs73150416 variants contributed to a lower incidence of PR-positive BC. Especially, rs112276562 was associated with a lower incidence of PR-positive BC after Bonferroni correction. Our study suggested that LRRC3B polymorphisms might be involved in the pathogenesis of PR-positive BC. Moreover, rs6788033 was associated with a lower level of Ki-67. Our findings showed LRRC3B polymorphisms were associated with tumor size and PR/Ki-67 status, which might act as predictors for BC occurrence and development.
Our study provided evidence that LRRC3B variants were associated with BC susceptibility and related to clinical characteristic of BC patients. Inevitably, there were several intrinsic limitations in this study. First, data on LRRC3B SNPs’ potential function and LRRC3B expression/survival in BC were predicted in in silico analysis only; thus, further functional assay is necessary to explore the functions and the underlying mechanisms of these polymorphisms. Second, gene-to-environment interactions could not be further analyzed due to the unavailability of relevant information; therefore, additional studies will be required.
Conclusion
In conclusion, we firstly reported that LRRC3B polymorphisms might contribute to individual susceptibility and progression to BC among Chinese Han females. The study might help in understanding the possible effect of LRRC3B variants in the development of BC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Nanchang University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the individuals who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.657168/full#supplementary-material
Supplementary Figure 1 | LRRC3B gene expression is down-regulated in BC compared with that in normal lung tissues. Each bar represents the average level of LRRC3B expression. Error bars represent the standard deviation of the mean value. The data were extracted from the GEPIA database (http://gepia.cancer-pku.cn/). * indicates statistical significance (p < 0.01).
Supplementary Figure 2 | LRRC3B low expression is associated with poor survival in BC. Kaplan–Meier plots of overall survival: comparison of patients with high vs. low expression of LRRC3B in BC patients. The Kaplan–Meier plots were generated by the Kaplan–Meier Plotter (http://kmplot.com/analysis/).
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA: Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
3. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. New Engl J Med (2015) 373(21):2005–14. doi: 10.1056/NEJMoa1510764
4. Howell A, Anderson AS, Clarke RB, Duffy SW, Evans DG, Garcia-Closas M, et al. Risk Determination and Prevention of Breast Cancer. Breast Cancer Res BCR (2014) 16(5):446. doi: 10.1186/s13058-014-0446-2
5. Xia P, Li B, Geng T, Deng Z, Dang C, Chang D, et al. FGFR2 Gene Polymorphisms are Associated With Breast Cancer Risk in the Han Chinese Population. Am J Cancer Res (2015) 5(5):1854–61. eCollection 2015.
6. Dai ZJ, Liu XH, Ma YF, Kang HF, Jin TB, Dai ZM, et al. Association Between Single Nucleotide Polymorphisms in DNA Polymerase Kappa Gene and Breast Cancer Risk in Chinese Han Population: A Strobe-Compliant Observational Study. Medicine (2016) 95(2):e2466. doi: 10.1097/MD.0000000000002466
7. Ren HT, Li YM, Wang XJ, Kang HF, Jin TB, Ma XB, et al. PD-1 Rs2227982 Polymorphism is Associated With the Decreased Risk of Breast Cancer in Northwest Chinese Women: A Hospital-Based Observational Study. Medicine (2016) 95(21):e3760. doi: 10.1097/MD.0000000000003760
8. Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, et al. The Secreted Protein Discovery Initiative (SPDI), a Large-Scale Effort to Identify Novel Human Secreted and Transmembrane Proteins: A Bioinformatics Assessment. Genome Res (2003) 13(10):2265–70. doi: 10.1101/gr.1293003
9. Kobe B, Deisenhofer J. A Structural Basis of the Interactions Between Leucine-Rich Repeats and Protein Ligands. Nature (1995) 374(6518):183–6. doi: 10.1038/374183a0
10. Xu ZM, Gao WR, Mei Q, Chen J, Lu J. The Novel Gene LRP15 is Regulated by DNA Methylation and Confers Increased Efficiency of DNA Repair of Ultraviolet-Induced DNA Damage. BMB Rep (2008) 41(3):230–5. doi: 10.5483/BMBRep.2008.41.3.230
11. Guo Y, Zhang R, Shen S, Wei Y, Salama SM, Fleischer T, et al. Dna Methylation of LRRC3B: A Biomarker for Survival of Early-Stage Non-Small Cell Lung Cancer Patients. Cancer Epidemiol Biomarkers Prev (2018) 27: (12):1527–35. doi: 10.1158/1055-9965.EPI-18-0454
12. Kim M, Kim JH, Jang HR, Kim HM, Lee CW, Noh SM, et al. LRRC3B, Encoding a Leucine-Rich Repeat-Containing Protein, is a Putative Tumor Suppressor Gene in Gastric Cancer. Cancer Res (2008) 68(17):7147–55. doi: 10.1158/0008-5472.CAN-08-0667
13. Wang Y, Peng Y, Zhou Y, Zhou Y, Zhong H. The Clinical Value of LRRC3B Gene Expression and Promoter Hypermethylation in Breast Carcinomas. Cell Biochem Biophysics (2014) 70(2):1035–41. doi: 10.1007/s12013-014-0018-1
14. Demokan S, Chuang AY, Pattani KM, Sidransky D, Koch W, Califano JA. Validation of Nucleolar Protein 4 as a Novel Methylated Tumor Suppressor Gene in Head and Neck Cancer. Oncol Rep (2014) 31(2):1014–20. doi: 10.3892/or.2013.2927
15. Li GS, Kong GY, Zou Y. Protective Role of LRRC3B in Preventing Breast Cancer Metastasis and Recurrence Post-Bupivacaine. Oncol Lett (2017) 14(4):5013–7. doi: 10.3892/ol.2017.6773
16. Gabriel S, Ziaugra L, Tabbaa D. SNP Genotyping Using the Sequenom MassARRAY iPLEX Platform. Curr Protoc Hum Genet (2009). Chapter 2:Unit 2.12. doi: 10.1002/0471142905.hg0212s60
17. Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-Throughput Oncogene Mutation Profiling in Human Cancer. Nat Genet (2007) 39(3):347–51. doi: 10.1038/ng1975
18. Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the Probability That a Positive Report is False: An Approach for Molecular Epidemiology Studies. J Natl Cancer Institute (2004) 96(6):434–42. doi: 10.1093/jnci/djh075
19. Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z, et al. Mettl14 Gene Polymorphisms Confer Neuroblastoma Susceptibility: An Eight-Center Case-Control Study. Mol Ther Nucleic Acids (2020) 22:17–26. doi: 10.1016/j.omtn.2020.08.009
20. Bella J, Hindle KL, McEwan PA, Lovell SC. The Leucine-Rich Repeat Structure. Cell Mol Life Sci CMLS (2008) 65(15):2307–33. doi: 10.1007/s00018-008-8019-0
21. Kan L, Li H, Zhang Y, Wang J, Niu H, Jiang H, et al. LRRC3B is Downregulated in non-Small-Cell Lung Cancer and Inhibits Cancer Cell Proliferation and Invasion. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2016) 37(1):1113–20. doi: 10.1007/s13277-015-3833-0
22. Haraldson K, Kashuba VI, Dmitriev AA, Senchenko VN, Kudryavtseva AV, Pavlova TV, et al. LRRC3B Gene is Frequently Epigenetically Inactivated in Several Epithelial Malignancies and Inhibits Cell Growth and Replication. Biochimie (2012) 94(5):1151–7. doi: 10.1016/j.biochi.2012.01.019
23. Kondratov AG, Stoliar LA, Kvasha SM, Gordiyuk VV, Zgonnyk YM, Gerashchenko AV, et al. Methylation Pattern of the Putative Tumor-Suppressor Gene LRRC3B Promoter in Clear Cell Renal Cell Carcinomas. Mol Med Rep (2012) 5(2):509–12. doi: 10.3892/mmr.2011.681
24. Leong SP, Shen ZZ, Liu TJ, Agarwal G, Tajima T, Paik NS, et al. Is Breast Cancer the Same Disease in Asian and Western Countries? World J Surg (2010) 34(10):2308–24. doi: 10.1007/s00268-010-0683-1
25. Tian T, Wang M, Zheng Y, Yang T, Zhu W, Li H, et al. Association of Two FOXP3 Polymorphisms With Breast Cancer Susceptibility in Chinese Han Women. Cancer Manage Res (2018) 10:867–72. doi: 10.2147/CMAR.S158433
Keywords: breast cancer, LRRC3B, polymorphisms, susceptibility, case–control
Citation: Wang Y (2021) LRRC3B Polymorphisms Contributed to Breast Cancer Susceptibility in Chinese Han Population. Front. Oncol. 11:657168. doi: 10.3389/fonc.2021.657168
Received: 22 January 2021; Accepted: 24 May 2021;
Published: 10 June 2021.
Edited by:
Bairong Shen, Sichuan University, ChinaReviewed by:
Rahma Melki, Mohamed Premier University, MoroccoPrihantono Prihantono, Hasanuddin University, Indonesia
Copyright © 2021 Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxin Wang, aGVyYXdhbmd5eEBxcS5jb20=
 Yuxin Wang
Yuxin Wang