- 1Dr Panjwani Center for Molecular Medicine and Drug Research (PCMD), International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan
- 2Department of Human Genetics and Molecular Biology, University of Health Sciences, Lahore, Pakistan
- 3Atomic Energy Medical Centre (AEMC), Jinnah Postgraduate Medical Centre (JPMC), Karachi, Pakistan
- 4Centre for Human Genetics and Molecular Medicine, Sindh Institute of Urology and Transplantation (SIUT), Karachi, Pakistan
Purpose: Deletion of Glutathione S-Transferase Theta 1 (GSTT1) encoding gene is implicated in breast cancer susceptibility, clinical outcomes, and survival. Contradictory results have been reported in different studies. The present investigation based on a representative Pakistani population evaluated the GSTT1-absent genotype in breast cancer risk and prognosis.
Methods: A prospective study comprising case-control analysis and case series analysis components was designed. Peripheral blood samples were collected from enrolled participants. After DNA extraction, GSTT1 genotyping was carried out by a multiplex PCR with β-globin as an amplification control. Association evaluation of GSTT1 genotypes with breast cancer risk, specific tumor characteristics, and survival were the primary endpoints.
Results: A total of 264 participants were enrolled in the molecular investigation (3 institutions). The study included 121 primary breast cancer patients as cases and 143 age-matched female subjects, with no history of any cancer, as controls. A significant genetic association between GSTT1-absent genotype and breast cancer susceptibility (p-value: 0.03; OR: 2.13; 95% CI: 1.08-4.29) was reported. The case-series analysis showed lack of association of GSTT1 genotypes with menopause (p-value: 0.86), tumor stage (p-value: 0.12), grade (p-value: 0.32), and size (p-value: 0.07). The survival analysis revealed that GSTT1-absent genotype cases had a statistically significant shorter overall survival (OS) than those with the GSTT1-present genotype cases (mean OS: 23 months vs 33 months). The HR (95% CI) for OS in patients carrying GSTT1-absent genotype was 8.13 (2.91-22.96) when compared with the GSTT1-present genotype.
Conclusions: The present study is the first report of an independent significant genetic association between GSTT1-absent genotype and breast cancer susceptibility in a Pakistani population. It is also the foremost report of the association of this genotype with OS in breast cancer cases. Upon further validation, GSTT1 variation may serve as a marker for devising better population-specific strategies. The information may have translational implications in the screening and treatment of breast cancers.
Introduction
Glutathione is present in all living cells. Physiologically, it performs three important functions: protection of thiol groups in proteins from oxidation, intracellular redox buffering, storage for sulphur-containing cysteine. These functions are dependent upon the catalysis by Glutathione S-Transferases (GSTs), E.C. 2.5.1.18. Consequently, GSTs play a major role in the detoxification of potent endogenous and exogenous carcinogens (1). These enzymes constitute a superfamily of isoenzymes including GST- theta 1. GSTT1 gene is located on chromosome 22q11.2. It encodes the enzyme, which is involved in the conjugation of reduced glutathione to certain electrophiles and hydrophobic compounds (2). Ultimately, such toxic substrates may be removed from the body.
The absence of the GSTT1 gene, also known as homozygous deletion or null genotype and herein referred to as GSTT1-absent, has been reported with varying frequencies in different populations (3). The carriers of the GSTT1-absent genotype are unable to metabolize some mutagenic carcinogens (4). The deletion has been correlated with ovarian, bladder, colon, oral, lung, and pediatric cancers among different populations (5–10). It is a candidate genetic marker for cancer risk, prognosis, and treatment response. In the case of breast cancers, the independent contribution of GSTT1 null genotype to susceptibility, tumor characteristics, and response to prescribed regimens remains inconclusive in different populations across the world (11–13).
In Pakistan, the age-standardized rate (ASR) of the female breast cancer incidence is among the highest in Asia (34.4 per 100,000), whereas the mortality rate is one of the highest in the world (18.8 per 100,000) (14, 15). Therefore, it is essential to identify the underlying factors in breast cancer etiology and prognosis.
Two previous studies from Pakistan (16, 17) report no independent association between the absence of the GSTT1 gene and breast cancer susceptibility. Both the studies were published from the Punjab area. However, Pakistan shows ethnicity-specific genetic variation across its region (18). Furthermore, the small sample size, and conflicting frequencies in controls: 18.7% (16) vs 31.4% (erroneously reported as 16% in one of the studies) (17), limit the applicability of these conclusions.
This prospective observational molecular study was designed based on the biological plausibility of GSTT1 deletion in carcinogenesis. It addresses the paucity and contradiction in the available data from a region that has frequent and aggressive breast tumors. The first component of the study, the case-control analysis, evaluated the contribution of the GSTT1 gene in breast cancer risk. Simultaneously, the second part, comprising case series analysis, investigated the contribution of GSTT1 genotypes to specific tumor characteristics and breast cancer survival after standard treatment.
Materials and Methods
Study Design and Participant Enrollment
The overall study schema is shown in Figure 1.
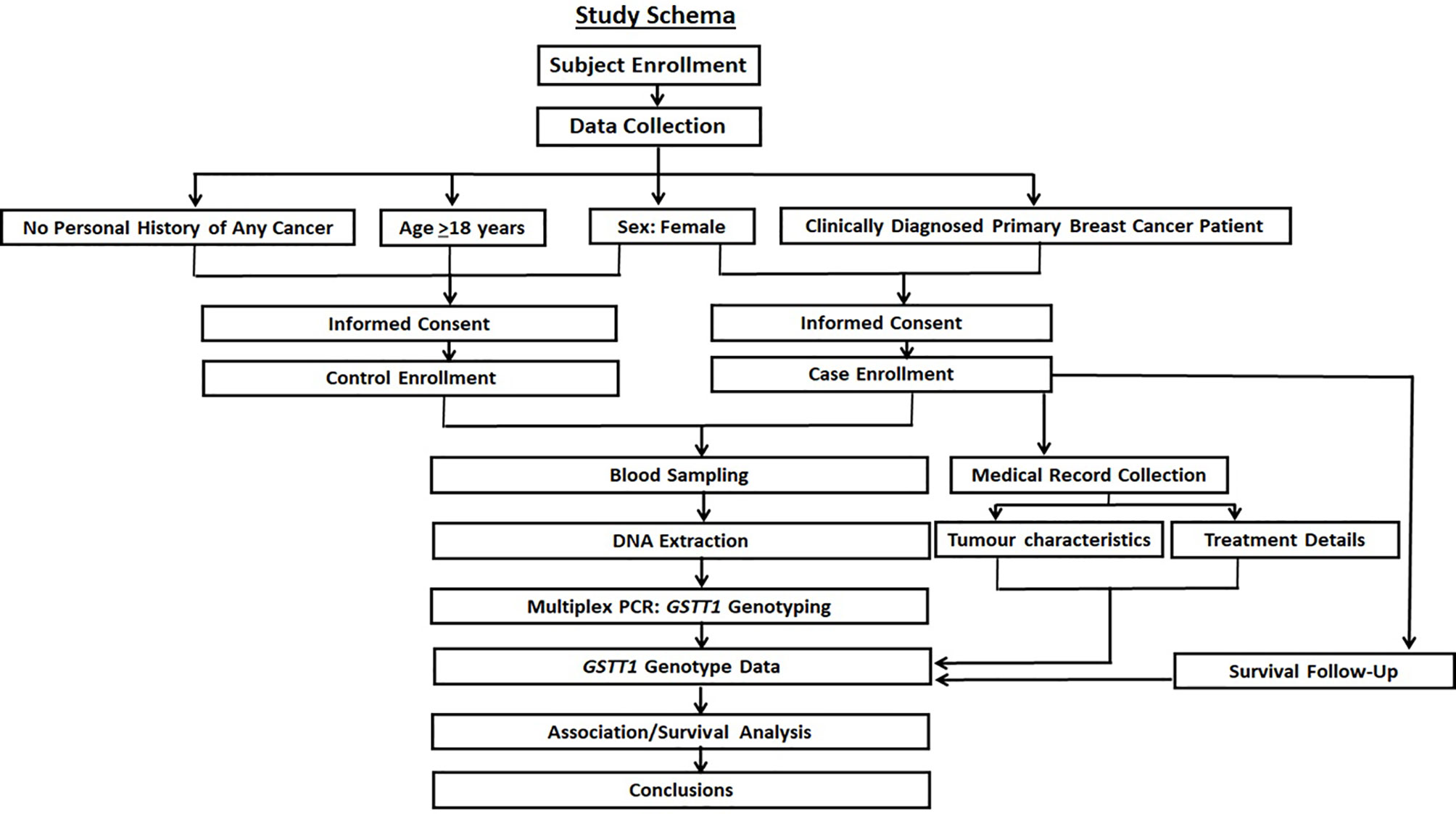
Figure 1 The study schema. The prospective recruitment of cases with age-matched female controls is shown along with the collection of specified molecular, clinical, and survival data. The objective was to investigate the contribution of GSTT1 variation in breast cancer risk, tumor characteristics, and survival after standard treatment.
The patients were recruited from the Atomic Energy Medical Centre (AEMC), Jinnah Postgraduate Medical Centre (JPMC), Karachi, Pakistan. The cases were clinically diagnosed primary breast cancer patients. The treatment included radiotherapy at the aforementioned participating institution following chemotherapy and surgery. The latter two components of treatment were carried out at hospitals other than AEMC. The details of control enrolment have been published elsewhere (19), with the modification that only age-matched (≥ 18 years), female participants’ data were included in the present study. All the subjects were recruited in Karachi, Pakistan and therefore, the distribution of ethnicities was the same in cases and controls. Sindhi, a self-defined Urdu-speaking ethnicity, Pathan, and Punjabi were the main ethnic groups. The research involved human participants and followed the provisions of the Declaration of Helsinki and its amendments. Research protocols were approved by the independent Ethics Review Committees of all the relevant institutions. The present study follows the reporting recommendations for tumor marker prognostic studies (REMARK) (20, 21) (Supplementary Table 1).
Data Collection
Information regarding age was recorded for all the participants. Patients’ family history, age at menarche, menopause (if applicable), and obstetrics and gynecology history were recorded in a questionnaire. In cases, tumor node metastasis (TNM) staging and histological grading were carried out according to the Union Internationale Contre le Cancer (UICC) recommendations (22). Data on tumor characteristics (tumor stage, grade, and size) and histology were obtained from the patients’ hospital medical files. Information relating to the parameters that were analyzed was documented in the majority of the hospital records. Three-year survival data were collected through telephonic follow-up. The missing information was due to: (i) the return of patients to their towns/villages after treatment at Karachi; (ii) erroneous contact information; and (iii) no response.
Sample Collection and DNA Extraction
All the participants volunteered 8-10ml of venous blood sample, which was collected in ACD-coated vacutainers (BD Vacutainer® BD Franklin Lakes NJ USA). Samples from the cases were collected at the time of radiotherapy, post-mastectomy, and chemotherapy treatment. The blood samples were either processed immediately or stored at 4°C until DNA extraction.
DNA was extracted from the white blood cells according to the standard phenol-chloroform method (23). It was quantified spectrophotometrically (Beckman Coulter™ DU® 530). The quality control cut-off for the 260/280 ratio was between 1.7-1.99. DNA quality was also analyzed by 0.7% agarose gel electrophoresis followed by UV visualization using a gel imaging system (Azure c300® biosystems). No fragmentation or smearing was observed in any of the samples (Figure 2).
Figure 2 A representative 0.7% agarose gel used for quality control of extracted DNA samples. Lane 1: 100bp DNA ladder; lanes 2-12: DNA samples.
The working dilutions for experiments were prepared at room temperature and stored at 4°C. The stock DNA samples were stored at -20°C.
Genotyping
Cases
GSTT1 genotyping was carried out by a multiplex polymerase chain reaction (PCR) with β-globin as an amplification control. The primer sequences have been published earlier (17).
PCR was carried out with a Taq DNA polymerase kit (Thermofisher Scientific Inc.). Total PCR reaction mix (10µl) consisted of 1X PCR buffer, 0.9mM MgCl2, 0.5mM dNTPs, 1.5U/µl Taq polymerase, 1.8µM primers each for GSTT1 and β-globin genes, and 70ng DNA. The PCR conditions were: initial denaturation at 94°C for 5 minutes, followed by 40 cycles of 94°C for 45 seconds, annealing at 60°C for 45 seconds, and extension at 72°C for 45 seconds. The final extension was carried at 72°C for 5 minutes. Amplicons were analyzed under UV on 2% agarose gel, which was stained with ethidium bromide. A fragment of 473bp indicated the GSTT1-present genotype. The GSTT1-absent genotype did not show this amplification. The amplification of the β-globin gene served as the control for successful PCR. A negative control was included in all the genotyping experiments (Figure 3).
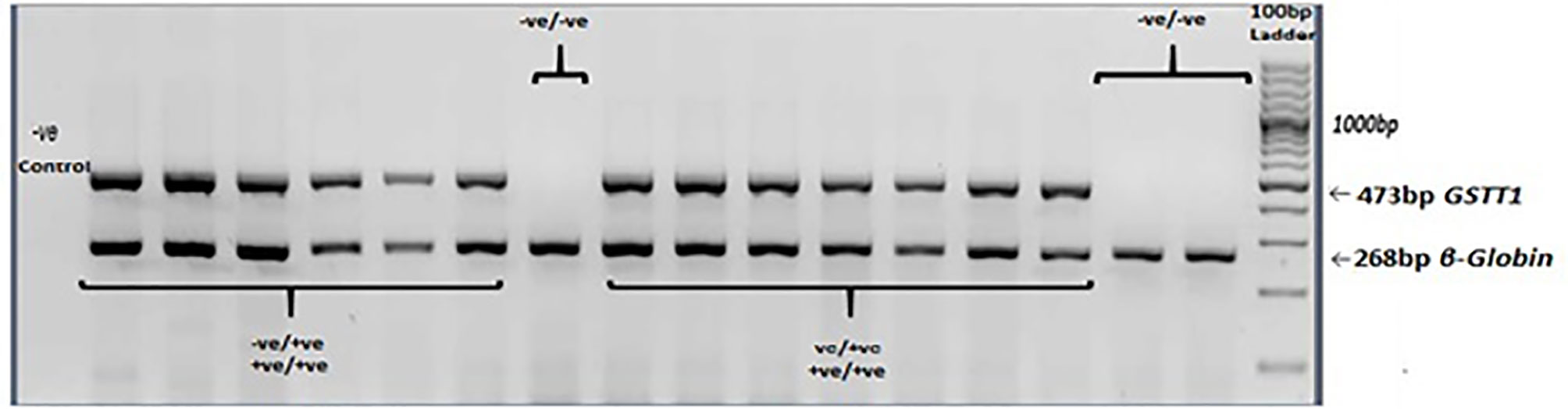
Figure 3 GSTT1 genotyping. Agarose gel electrophoresis (2%) of multiplex PCR amplified products: GSTT1-absent (–ve/-ve) genotype did not show amplification of a 473bp fragment. β-globin gene was included as an amplification control.
The results of genotyping were confirmed by double-blind evaluation, the inclusion of replicates, and negative controls.
Controls
The genotyping in controls has been described elsewhere (19). It identifies homozygous GSTT1-present, heterozygous GSTT1-present/absent, and homozygous GSTT1-absent genotypes. In the final observation, the GSTT1-present allele was determined by an amplicon of 466bp, while the GSTT1-absent allele was identified by an amplicon of 1,460bp.
Treatment of Breast Cancer Patients
All the participating cases underwent a mastectomy, adjuvant chemotherapy, and/or radiotherapy. Before the start of chemotherapy, echocardiography was done to assess cardiac function (ejection fraction cut-off for the start of doxorubicin based chemo was >55%). A test dose of docitaxel was given to rule out hypersensitivity before the first cycle. Neurological assessment was undertaken during taxane (paclitaxel) cycles. Chemotherapy-related toxicities were assessed after every cycle according to Common Toxicity Criteria of the National Cancer Institute (NCI-CTC, version 2.0) (24). “Severe toxicity” was defined as hematological or gastrointestinal toxicity of grades 3–4.
The chemotherapy regimen included: Adriamycin-Cyclophosphamide x4 followed by taxane x4 (docitaxel or paclitaxel): Doxorubicin 60mg/m2 on day 1, cyclophosphamide 600mg on day 1, paclitaxel 175mg/m2 on day 1 OR docitaxel 100mg/m2 on day 1, repeated every 3 weeks.
Complete blood count, liver function test, and renal function test were carried out to assess the treatment response.
Statistical Analysis
The allele distribution for GSTT1 polymorphism in the controls was assessed for Hardy-Weinberg equilibrium (25). The statistical tests for association analysis were carried out by using Statistical Package for Social Science (SPSS) for Windows v.19.0 (SPSS, Inc., Chicago, Illinois, USA) and online OpenEpi software (26).
In the case-control investigations, data for the GSTT1 genotype was obtained for all the participants, except two cases, where no amplification was recorded. The age-matching between cases and controls was analyzed by Student’s t-test for independent samples with the assumption of unequal variances. To achieve 80% power at a two-sided level of significance, various odds ratios (OR) of genetic risk due to GSTT1 polymorphism for breast cancers were calculated. The accrual of 260 participants (matched cases and controls) allows for the identification of OR≥2 for GSTT1 variation with the GSTT1-absent frequency of 0.24 [the median value of the reported prevalence in controls from Pakistan (16–18, 27–38) was used for the calculations (39)]:
Where,
n = number of subjects in each group
zα = Corresponding to α [level of significance (95%)] = 1.96
zβ= Corresponding to β (Probability of type II error. Power of study is 80%, 1- β = 0.2)
p0 = proportion of exposure among control groups (*prevalence of the polymorphism in general population without breast cancer).
p1 = proportion of exposure among cases based on the formula including odds ratios associated with exposure.
The missing information for case series analysis is itemized in the relevant tables in the results section.
The primary objective was the investigation of GSTT1 polymorphism association/s with breast cancer susceptibility, the selected clinical parameters, and survival. The data were assessed by Pearson χ2 test. The ORs were tabulated with a 95% confidence interval (95% CI) to evaluate the strength of the associations. Post hoc power analysis was carried out to assess the strength of the study (40, 41).
The overall survival (OS) and hazard ratios (HR) with 95% CI were assessed by the Kaplan-Meier method using MedCalc software v.19.2.6 (42, 43). In all the statistical tests, p-values <0.05 were considered to be significant.
Results
Participants’ Information and Clinical Data of Patients
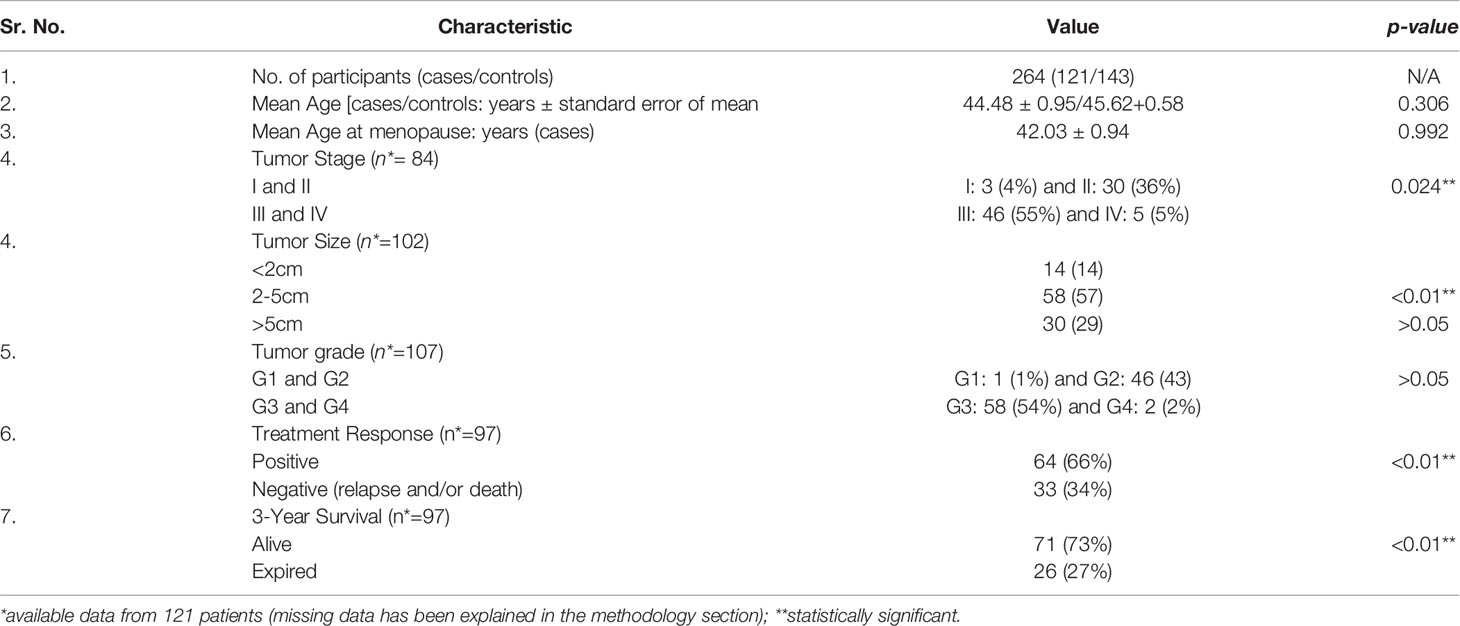
The total number of patients diagnosed with primary breast cancer disease was 121, whereas the total number of age- and gender-matched controls was 143. Characteristics of the 264 participants included in the study are presented in Table 1.
The mean age of the patients was 44.48 ± 0.95 years, whereas, for the controls, the mean age was 45.62 ± 0.58 years. All patients presented with invasive ductal carcinoma (IDC) of the breast. Majority of the patients had advanced tumor stage (stages III and IV; p-value: 0.024**), tumor size of >2cm (p-value: <0.01**), and high tumor grade (grades 3 and 4; p-value: >0.05).
Association Between GSTT1 Polymorphism and Breast Cancer Risk
The allelic and genotypic frequencies of GSTT1 polymorphism in controls are shown in Table 2. The proportions were in Hardy-Weinberg equilibrium.

Table 2 Distribution of GSTT1 genotypes and allele frequencies (with standard errors) in age- and gender-matched controls. Assessment of HWE test in controls.
Associations between the GSTT1 genotypes and breast cancer susceptibility are presented in Table 3.

Table 3 Distribution of GSTT1 genotypes in controls, breast cancer patients, and the association analysis with breast cancer risk*.
The comparison of GSTT1-present genotype with GSTT1-absent genotype in cases and controls revealed that GSTT1-absent genotype was significantly associated with risk for breast cancers (p-value: 0.03). The OR were 2.13 (95% CI: 1.08 - 4.29).
Lack of Association Between GSTT1 Polymorphism and Specific Parameters
In the case series analysis, the present study did not report any statistically significant association between the absence of the GSTT1 gene and the studied tumor characteristics, i.e., stage, grade, and size. The p-values were 0.12, 0.32, and 0.07, respectively. Similarly, the study did not find any correlation between the GSTT1 genotype and age at menopause (p-value: 0.86).
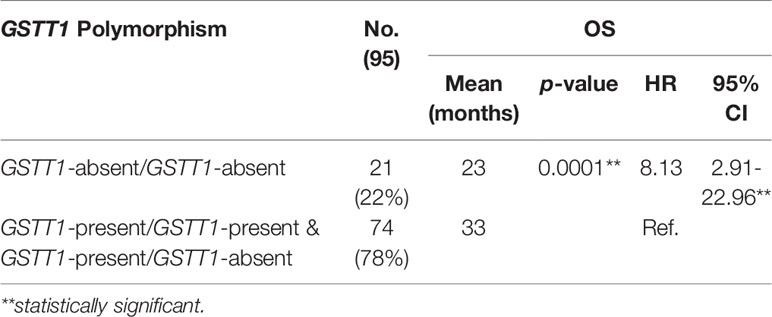
Association of the GSTT1 Polymorphism and OS in Breast Cancer Patients
As shown in Figure 4, GSTT1-present carriers had 10 months’ longer survival (mean OS: 33 months; 95% CI: 30.96-34.65) than those with GSTT1-absent genotype (mean OS: 23 months; 95% CI 17.90-28.59); p-value: 0.0001. The HR with 95% CI for OS in patients carrying GSTT1-absent genotype was 8.13 (2.91-22.96) with GSTT1-present genotype as the reference variable (Table 4).

Figure 4 Kaplan-Meier curve demonstrating the overall survival (OS) based on genotypes of GSTT1. The mean OS was 33 months (95% CI: 30.96-34.65) in GSTT1-present genotype carriers and 23 months (95% CI: 17.90-28.59) in GSTT1-absent genotype carriers; p-value: 0.0001.
Discussion
In the present study, the association/s of GSTT1 genotypes with breast cancer-related parameters were evaluated. In this study, we report a significant association of GSTT1-absent genotype with increased breast cancer risk in a representative sample from a Pakistani population. The OR were 2.13 (95% CI: 1.08 – 4.29). We also report a significant difference in the survival duration between GSTT1-present and GSTT1-absent carriers: mean OSGSTT1-present: 33 months (95% CI: 30.96-34.65) vs mean OSGSTT1-absent: 23 months (95% CI: 17.90-28.59). The present analysis is the first report of population-specific associations between GSTT1 genotypes and the specific factors associated with breast cancers.
The incidence of breast cancers varies across the globe. The highest estimated age-standardized incidence rates have been reported in Belgium (113.2 per 100,000), while the lowest was reported in Bhutan (5.0 per 100.000). Furthermore, the highest estimated age-standardized mortality rates are reported from Barbados (42.2 per 100,000), while the lowest is reported from Bhutan (2.6 per 100,000) (15). The known risk factors such as age, family history, different reproductive parameters, and obesity account for only one-third of the risk for breast cancers (11, 44). In addition, the reason(s) for high mortality rates across specific populations need to be determined (45–47).
A number of genes are likely involved in breast cancer characteristics, with the possibility of gene-environment interactions (48). The quantitative contributions of such genes remain to be delineated across different populations and regions.
A proposed mechanism of carcinogenesis due to the loss of function of GSTT1 isoenzyme is shown in Figure 5. Some of the exogenous and endogenous carcinogens are not metabolized to non-toxic components. Consequently, tumorigenesis and/or tumor progression are likely to occur (4). In addition, chemotherapeutic agents may also be metabolized by the pathways involving GSTT1, rendering the patients with GSTT1-present genotype irresponsive to either therapy or specific doses of therapy.
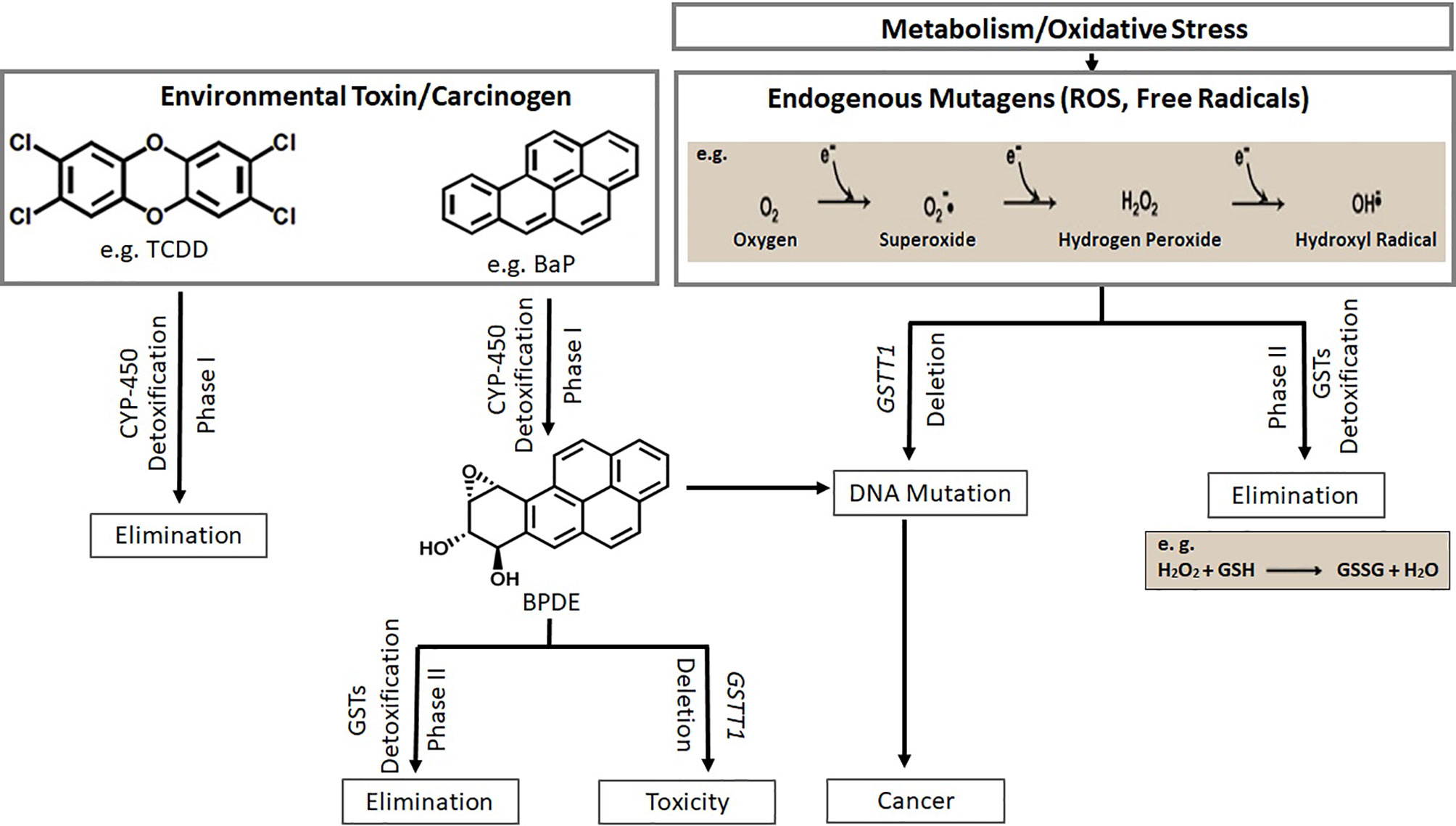
Figure 5 Xeno- and endo-biotic carcinogen metabolism through two pathway systems (phase I and phase II) is shown. Deletion of GSTT1 leads to toxicity and carcinogenesis (TCDD, 2,3,7,8-Tetrachlorodibenzodioxin; BaP, Benzo[a]pyrene; BPDE, Benzo(a)pyrene diolepoxide; ROS, Reactive Oxygen Species).
Among different populations, the loss-of-function polymorphism in the GSTT1 encoding gene occurs at varying frequencies (49). Null genotype is correlated with vulnerability to cancers, tumor characteristics, and differences in treatment response (1, 50).
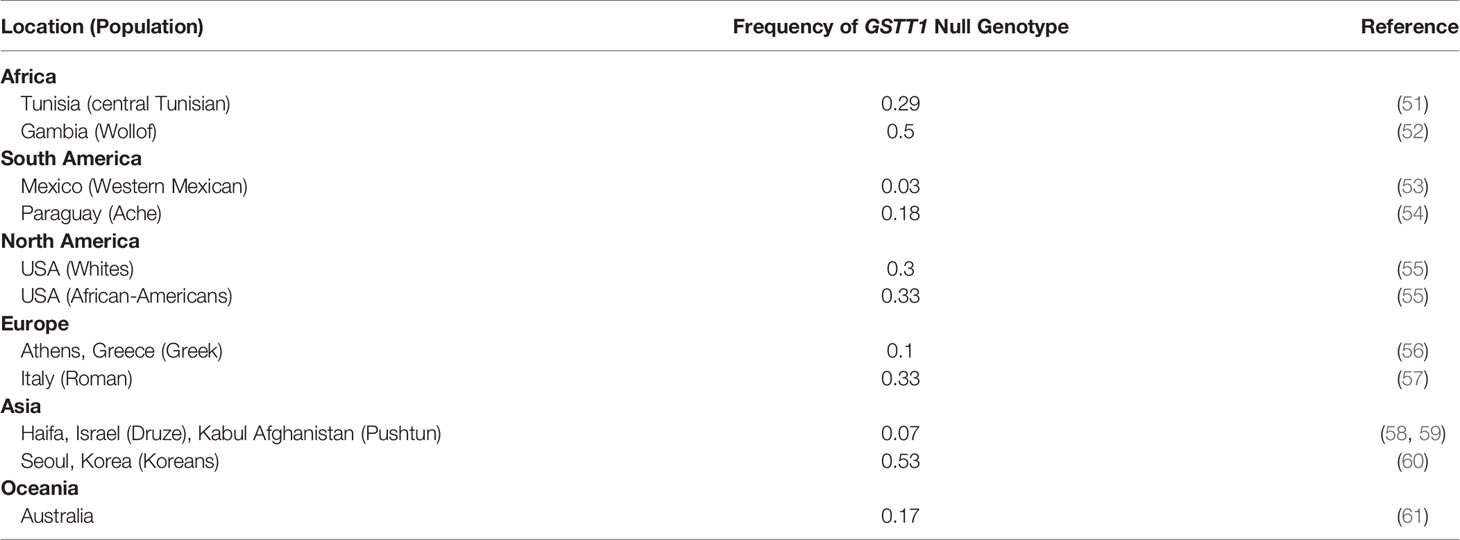
The examples of low and high frequencies of GSTT1-absent genotype across specified global regions according to WHO are listed in Table 5.
In Pakistan, the frequency of GSTT1 null genotype in healthy individuals has been reported in the range of 0.06 – 0.24 (16–18, 28–38). This wide range may be attributed to limited sample sizes, population admixture, and differences in methodologies. Similarly, variations in the frequency of this genotype in breast cancer patients from Pakistan have been reported as 8% (16) and 27% (erroneously reported as 49% in the text) (17). The present study reports a frequency of 21%. In contrast to the studies conducted in Punjab/Central Pakistan (16, 17), the present study was carried out in Southern-Pakistan, where the majority of the patients belong to Sindhi and other self-defined Urdu-Speaking ethnicities (25% each). Interestingly, Her2 +ve invasive ductal carcinomas were more frequent in GSTT1-absent genotype patients (although data is preliminary, which is available for only 59 samples). Our case-control analysis is in agreement with a number of other studies reported from different parts of the world (11, 62). However, it is the first report of significant association of GSTT1-absent genotype with decreased OS in primary breast cancer patients. An earlier study from China reported such an association with untreated metastatic breast cancers (63).
The strength of our study is the underlying unique population, for whom molecular data for breast cancer risk and clinical parameters are scarce. The limitations of the study are sample size and the paucity of information for known risk factors and clinical parameters, primarily due to the restriction of resources in healthcare. In terms of the limitation of the sample size, it should be emphasized that the results reported in the present study do not corroborate previously published reports from this region with the same limitation. The present study also notes that Chi-squared goodness-of-fit test was inappropriate for the association analysis as reported in earlier studies. The methodology results in a lack of data for heterozygous controls. This information is necessary to estimate HWE and subsequent association evaluation.
During the analysis, the present study takes into account missing information for risk factors and clinical parameters as illustrated in Table 1 and the results section. The post-hoc power analysis showed a value of 60.1%. However, it is pertinent to mention here that the calculated estimate does not capture the true power of the study (41). A three-year follow-up provides useful information for assessing the OS in breast cancers in Pakistan. The mortality rate (ASR: 18.8 per 100 000) attributed to breast cancer in the country is among the highest in Asia (15). As emphasized earlier, the reasons behind such a high mortality rate need to be investigated. The significant association of the investigated polymorphism with OS in breast cancers may aid in taking precautionary measures in treatment regimens early in the administration of therapy administration. This study highlights the importance of conducting rigorous molecular epidemiology studies to devise evidence-based better strategies in breast cancer management, particularly in resource-limited settings.
Conclusions
The present study reports significant contribution of the GSTT1-absent genotype to breast cancer risk in a Pakistani population for the first time. A unique finding of this study was the association of this genotype with significantly shorter OS in breast cancer patients post standard treatment, which has not been reported previously. These observations are biologically plausible. If validated further through multiple center studies and larger sample sizes, the absence of the GSTT1 gene could serve as a risk and survival marker in breast cancers, at least for a specific population.
Data Availability Statement
The raw data supporting the conclusions of this article is available as the Supplementary Material, without any reservations. The sample IDs are confidentially encoded and untraceable.
Ethics Statement
The studies involving human participants were reviewed and approved by the International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan [ICCBS/IEC-016-BS/HT-2016/Protocol/1.0]; the Atomic Energy Medical Centre (AEMC), Jinnah Postgraduate Medical Centre (JPMC), Karachi, Pakistan [Admin-3 (257)/2016]; and the Sindh Institute of Urology and Transplantation (SIUT), Karachi, Pakistan. All the participants signed a written informed-consent form prior to sampling. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: SA. Participant enrollment/Data collection: SA, S-E-ZZ, SMA, AS, MM, AA, SF, and SK. Benchwork: SA, S-E-ZZ, SMA, AS, AA, and SF. Analysis: SA. Original draft: SA, S-E-ZZ, and SMA. Reviewing and editing: SA. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the participants in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.678705/full#supplementary-material
References
1. Allocati N, Masulli M, Di Ilio C, Federici L. Glutathione Transferases: Substrates, Inihibitors and Pro-Drugs in Cancer and Neurodegenerative Diseases. Oncogenesis (2018) 7(1):8. doi: 10.1038/s41389-017-0025-3
2. Webb G, Vaska V, Coggan M, Board P. Chromosomal Localization of the Gene for the Human Theta Class Glutathione Transferase (GSTT1). Genomics (1996) 33(1):121–3. doi: 10.1006/geno.1996.0167
3. Kassogue Y, Diakite B, Kassogue O, Konate I, Tamboura K, Diarra Z, et al. Genetic Polymorphism of Drug Metabolism Enzymes (GSTM1, GSTT1 and GSTP1) in the Healthy Malian Population. Mol Biol Rep (2020) 47(1):393–400. doi: 10.1007/s11033-019-05143-5
4. Josephy PD. Genetic Variations in Human Glutathione Transferase Enzymes: Significance for Pharmacology and Toxicology. Hum Genomics Proteomics HGP (2010) 2010:876940. doi: 10.4061/2010/876940
5. Howells RE, Redman CW, Dhar KK, Sarhanis P, Musgrove C, Jones PW, et al. Association of Glutathione S-Transferase GSTM1 and GSTT1 Null Genotypes With Clinical Outcome in Epithelial Ovarian Cancer. Clin Cancer Res (1998) 4(10):2439–45.
6. Zhou T, Li HY, Xie WJ, Zhong Z, Zhong H, Lin ZJ. Association of Glutathione S-Transferase Gene Polymorphism With Bladder Cancer Susceptibility. BMC Cancer (2018) 18(1):1088. doi: 10.1186/s12885-018-5014-1
7. Qin XP, Zhou Y, Chen Y, Li NN, Chen B, Yang P, et al. Glutathione S-Transferase T1 Gene Polymorphism and Colorectal Cancer Risk: An Updated Analysis. Clinics Res Hepatol Gastroenterol (2013) 37(6):626–35. doi: 10.1016/j.clinre.2013.04.007
8. Zhang ZJ, Hao K, Shi R, Zhao G, Jiang GX, Song Y, et al. Glutathione S-Transferase M1 (GSTM1) and Glutathione S-Transferase T1 (GSTT1) Null Polymorphisms, Smoking, and Their Interaction in Oral Cancer: A HuGE Review and Meta-Analysis. Am J Epidemiol (2011) 173(8):847–57. doi: 10.1093/aje/kwq480
9. Yang X, Qiu MT, Hu JW, Wang XX, Jiang F, Yin R, et al. GSTT1 Null Genotype Contributes to Lung Cancer Risk in Asian Populations: A Meta-Analysis of 23 Studies. PLoS One (2013) 8(4):e62181. doi: 10.1371/journal.pone.0062181
10. Lopes BA, Emerenciano M, Goncalves BA, Vieira TM, Rossini A, Pombo-de-Oliveira MS. Polymorphisms in CYP1B1, CYP3A5, GSTT1, and SULT1A1 Are Associated With Early Age Acute Leukemia. PLoS One (2015) 10(5):e0127308. doi: 10.1371/journal.pone.0127308
11. Miao LF, Ye XH, He XF. Individual and Combined Effects of GSTM1, GSTT1, and GSTP1 Polymorphisms on Breast Cancer Risk: A Meta-Analysis and Re-Analysis of Systematic Meta-Analyses. PLoS One (2020) 15(3):e0216147. doi: 10.1371/journal.pone.0216147
12. Wang J, Wang T, Yin GY, Yang L, Wang ZG, Bu XB. Glutathione S-Transferase Polymorphisms Influence Chemotherapy Response and Treatment Outcome in Breast Cancer. Genet Mol Res GMR (2015) 14(3):11126–32. doi: 10.4238/2015.September.22.6
13. Bansal VK, Rajan K, Sharma A, Paliwal P, Chaubal G, Jindal V, et al. Prospective Case-Control Study to Evaluate the Role of Glutathione S Transferases (GSTT1 and GSTM1) Gene Deletion in Breast Carcinoma and Its Prognostic Significance. Indian J Surg (2015) 77(Suppl 3):1067–72. doi: 10.1007/s12262-014-1152-0
14. Badar F, Mahmood S, Yusuf MA, Sultan F. Epidemiology of Cancers in Lahore, Pakistan, 2010-2012: A Cross-Sectional Study. BMJ Open (2016) 6(6):e011828. doi: 10.1136/bmjopen-2016-011828
15. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
16. Nosheen M, Malik FA, Kayani MA. Lack of Influence of Glutathione S-Transferase Gene Deletions in Sporadic Breast Cancer in Pakistan. Asian Pacific J Cancer Prev APJCP (2011) 12(7):1749–52.
17. Sohail A, Kanwal N, Ali M, Sadia S, Masood AI, Ali F, et al. Effects of Glutathione-S-Transferase Polymorphisms on the Risk of Breast Cancer: A Population-Based Case-Control Study in Pakistan. Environ Toxicol Pharmacol (2017) 35(2):143–53. doi: 10.1016/j.etap.2012.11.014
18. Mohyuddin A, Ayub Q, Khaliq S, Mansoor A, Mazhar K, Rehman S, et al. HLA Polymorphism in Six Ethnic Groups From Pakistan. Tissue Antigens (2002) 59(6):492–501. doi: 10.1034/j.1399-0039.2002.590606.x
19. Abid A, Ajaz S, Khan AR, Zehra F, Hasan AS, Sultan G, et al. Analysis of the Glutathione S-Transferase Genes Polymorphisms in the Risk and Prognosis of Renal Cell Carcinomas. Case-Control and Meta-Analysis. Urol Oncol (2016) 34(9):419.e1–12. doi: 10.1016/j.urolonc.2016.04.005
20. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting Recommendations for Tumor MARKer Prognostic Studies (REMARK). Breast Cancer Res Treat (2006) 100(2):229–35. doi: 10.1007/s10549-006-9242-8
21. Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. J Natl Cancer Inst (2018) 110(8):803. doi: 10.1093/jnci/djy088
22. Brierley JD, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. Oxford UK, Hoboken NJ: John Wiley & Sons (2017).
23. Russell DW, Sambrook J. Molecular Cloning: A Laboratory Manual: Cold Spring Harbor Laboratory Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press (2001).
24. Health UNIo. National Cancer Institute Cancer Therapy Evaluation Program, Common Toxicity Criteria (CTC), Version 2.0. Medford, MA 02155, United States (1999). Available at: http://ctepinfonihgov.
25. Michael C. A Simple Calculator to Determine Whether Observed Genotype Frequencies Are Consistent With Hardy-Weinberg Equilibrium. Medford, MA, United States (2008). Available at: http://www.tufts.edu/~mcourt01/Documents/Courtlab-HWcalculator.xls.
26. Dean A, Sullivan K, Soe M. OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version 2.3. 1. Atlanta, GA, USA (2013). Available at: www.OpenEpi.com.
27. Dar A, Faryal R, Masood N. Possible Association of a Distinct Combined Glutathione-S-Transferase Members With Allergic Asthma Patients in Pakistan. Genes Dis (2017) 4(2):111–5. doi: 10.1016/j.gendis.2017.01.001
28. Hasan S, Hameed A, Saleem S, Shahid SM, Haider G, Azhar A. The Association of GSTM1 and GSTT1 Polymorphisms With Squamous Cell Carcinoma of Cervix in Pakistan. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2015) 36(7):5195–9. doi: 10.1007/s13277-015-3175-y
29. Khan MI, Micheal S, Akhtar F, Ahmed W, Ijaz B, Ahmed A, et al. The Association of Glutathione S-Transferase GSTT1 and GSTM1 Gene Polymorphism With Pseudoexfoliative Glaucoma in a Pakistani Population. Mol Vis (2010) 16:2146–52.
30. Malik SS, Masood N, Yasmin A. Prostate Cancer and Glutathione S-Transferase Deletions. EXCLI J (2015) 14:1049–54. doi: 10.17179/excli2015-192
31. Masood N, Kayani MA. Protection Against Laryngeal and Pharyngeal Carcinoma: Heterozygous vs. Homozygous Deletions of GSTM1 and GSTT1. Genet Mol Biol (2013) 36(1):1–6. doi: 10.1590/S1415-47572013005000006
32. Nabgha EA, Eqani S, Khuram F, Alamdar A, Tahir A, Shah STA, et al. Environmental Exposure Pathway Analysis of Trace Elements and Autism Risk in Pakistani Children Population. Sci Total Environ (2020) 712:136471. doi: 10.1016/j.scitotenv.2019.136471
33. Nosheen M, Ishrat M, Malik FA, Baig RM, Kayani MA. Association of GSTM1 and GSTT1 Gene Deletions With Risk of Head and Neck Cancer in Pakistan: A Case Control Study. Asian Pacific J Cancer Prev APJCP (2010) 11(4):881–5.
34. Rehman S, Ahmed P, Saba N, Munir S, Sajjad S, Satti TM, et al. Association of GSTM1 and GSTT1 Deletion Polymorphisms With Pakistani Aplastic Anemia Patients and Controls and Meta-Analysis. Ann Hematol (2015) 94(12):1965–. doi: 10.1007/s00277-015-2482-0
35. Shaikh RS, Amir M, Masood AI, Sohail A, Athar HU, Siraj S, et al. Frequency Distribution of GSTM1 and GSTT1 Null Allele in Pakistani Population and Risk of Disease Incidence. Environ Toxicol Pharmacol (2010) 30(1):76–9. doi: 10.1016/j.etap.2010.04.002
36. Zakiullah A, Khisroon M, Saeed M, Khan A, Khuda F, Ali S, et al. Genetic Susceptibility to Oral Cancer Due to Combined Effects of GSTT1, GSTM1 and CYP1A1 Gene Variants in Tobacco Addicted Patients of Pashtun Ethnicity of Khyber Pakhtunkhwa Province of Pakistan. Asian Pacific J Cancer Prev APJCP (2015) 16(3):1145–50. doi: 10.7314/APJCP.2015.16.3.1145
37. Zakiullah SM, Javed N, Khuda F, Ovais M. Association of Nasopharyngeal Cancer Risk With Genetic Polymorphisms of Drug-Metabolizing Enzyme Genes GSTM1, GSTT1 and CYP1A1 (Rs4646903 Variant), in Tobacco Addicted Patients of Pashtun Ethnicity of Khyber Pakhtunkhwa Province of Pakistan. Pakistan J Pharm Sci (2019) 32(5):2107–16.
38. Zehra A, Zehra S, Ismail M, Azhar A. Glutathione S-Transferase M1 and T1 Gene Deletions and Susceptibility to Acute Lymphoblastic Leukemia (ALL) in Adults. Pakistan J Med Sci (2018) 34(3):666–70. doi: 10.12669/pjms.343.14911
39. Schlesselman JJ. Case-Control Studies: Design, Conduct, Analysis. New York, Oxford: Oxford University Press (1982).
40. Rosner B. Fundamentals of Biostatistics: Cengage Learning. Boston, MA, USA: Cengage Learning (2015).
41. Levine M, Ensom MH. Post Hoc Power Analysis: An Idea Whose Time Has Passed? Pharmacotherapy (2001) 21(4):405–9. doi: 10.1592/phco.21.5.405.34503
42. Schoonjans F, Zalata A, Depuydt CE, Comhaire FH. MedCalc: A New Computer Program for Medical Statistics. Comput Methods Programs Biomed (1995) 48(3):257–62. doi: 10.1016/0169-2607(95)01703-8
43. MedCalc. Version 19.2.6 Software. Available at: https://www.medcalc.org/ (Accessed 11 May 2020).
44. Ng EH, Gao F, Ji CY, Ho GH, Soo KC. Risk Factors for Breast Carcinoma in Singaporean Chinese Women: The Role of Central Obesity. Cancer (1997) 80(4):725–31. doi: 10.1002/(SICI)1097-0142(19970815)80:4<725::AID-CNCR11>3.0.CO;2-V
45. De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, et al. Cancer Survival in Europe 1999-2007 by Country and Age: Results of EUROCARE–5-A Population-Based Study. Lancet Oncol (2014) 15(1):23–34. doi: 10.1016/S1470-2045(13)70546-1
46. Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, et al. Global Surveillance of Cancer Survival 1995-2009: Analysis of Individual Data for 25,676,887 Patients From 279 Population-Based Registries in 67 Countries (CONCORD-2). Lancet (Lond Engl) (2015) 385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9
47. Huang Z, Wen W, Zheng Y, Gao YT, Wu C, Bao P, et al. Breast Cancer Incidence and Mortality: Trends Over 40 Years Among Women in Shanghai, China. Ann Oncol (2016) 27(6):1129–34. doi: 10.1093/annonc/mdw069
48. Lauby-Secretan B, Scoccianti C, Loomis D, Benbrahim-Tallaa L, Bouvard V, Bianchini F, et al. Breast-Cancer Screening–Viewpoint of the IARC Working Group. N Engl J Med (2015) 372(24):2353–8. doi: 10.1056/NEJMsr1504363
49. Saitou M, Ishida T. Distributions of the GSTM1 and GSTT1 Null Genotypes Worldwide Are Characterized by Latitudinal Clines. Asian Pacific J Cancer Prev APJCP (2015) 16(1):355–61. doi: 10.7314/APJCP.2015.16.1.355
50. Ludovini V, Antognelli C, Rulli A, Foglietta J, Pistola L, Eliana R, et al. Influence of Chemotherapeutic Drug-Related Gene Polymorphisms on Toxicity and Survival of Early Breast Cancer Patients Receiving Adjuvant Chemotherapy. BMC Cancer (2017) 17(1):502. doi: 10.1186/s12885-017-3483-2
51. Lakhdar R, Denden S, Knani J, Leban N, Daimi H, Hassine M, et al. Association of GSTM1 and GSTT1 Polymorphisms With Chronic Obstructive Pulmonary Disease in a Tunisian Population. Biochem Genet (2010) 48(7-8):647–57. doi: 10.1007/s10528-010-9346-z
52. Kirk GD, Turner PC, Gong Y, Lesi OA, Mendy M, Goedert JJ, et al. Hepatocellular Carcinoma and Polymorphisms in Carcinogen-Metabolizing and DNA Repair Enzymes in a Population With Aflatoxin Exposure and Hepatitis B Virus Endemicity. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol (2005) 14(2):373–9. doi: 10.1158/1055-9965.EPI-04-0161
53. Gutierrez-Amavizca BE, Orozco-Castellanos R, Ortiz-Orozco R, Padilla-Gutierrez J, Valle Y, Gutierrez-Gutierrez N, et al. Contribution of GSTM1, GSTT1, and MTHFR Polymorphisms to End-Stage Renal Disease of Unknown Etiology in Mexicans. Indian J Nephrol (2013) 23(6):438–43. doi: 10.4103%2F0971-4065.120342
54. Gaspar PA, Hutz MH, Salzano FM, Hill K, Hurtado AM, Petzl-Erler ML, et al. Polymorphisms of CYP1a1, CYP2e1, GSTM1, GSTT1, and TP53 Genes in Amerindians. Am J Phys Anthropol (2002) 119(3):249–56. doi: 10.1002/ajpa.10128
55. Huang K, Sandler RS, Millikan RC, Schroeder JC, North KE, Hu J. GSTM1 and GSTT1 Polymorphisms, Cigarette Smoking, and Risk of Colon Cancer: A Population-Based Case-Control Study in North Carolina (United States). Cancer Causes Control CCC (2006) 17(4):385–94. doi: 10.1007/s10552-005-0424-1
56. Dialyna IA, Arvanitis DA, Spandidos DA. Genetic Polymorphisms and Transcriptional Pattern Analysis of CYP1A1, AhR, GSTM1, GSTP1 and GSTT1 Genes in Breast Cancer. Int J Mol Med (2001) 8(1):79–87. doi: 10.3892/ijmm.8.1.79
57. Piacentini S, Polimanti R, Porreca F, Martinez-Labarga C, De Stefano GF, Fuciarelli M. GSTT1 and GSTM1 Gene Polymorphisms in European and African Populations. Mol Biol Rep (2011) 38(2):1225–30. doi: 10.1007/s11033-010-0221-0
58. Karban A, Krivoy N, Elkin H, Adler L, Chowers Y, Eliakim R, et al. Non-Jewish Israeli IBD Patients Have Significantly Higher Glutathione S-Transferase GSTT1-Null Frequency. Digestive Dis Sci (2011) 56(7):2081–7. doi: 10.1007/s10620-010-1543-4
59. Saify K, Saadat I, Saadat M. Genetic Polymorphisms of Glutathione S-Transferase T1 (GSTT1) and M1 (GSTM1) in Selected Populations of Afghanistan. Mol Biol Rep (2012) 39(8):7855–9. doi: 10.1007/s11033-012-1628-6
60. Uhm YK, Yoon SH, Kang IJ, Chung JH, Yim SV, Lee MH. Association of Glutathione S-Transferase Gene Polymorphisms (GSTM1 and GSTT1) of Vitiligo in Korean Population. Life Sci (2007) 81(3):223–7. doi: 10.1016/j.lfs.2007.05.006
61. Spurdle AB, Chang JH, Byrnes GB, Chen X, Dite GS, McCredie MR, et al. A Systematic Approach to Analysing Gene-Gene Interactions: Polymorphisms at the Microsomal Epoxide Hydrolase EPHX and Glutathione S-Transferase GSTM1, GSTT1, and GSTP1 Loci and Breast Cancer Risk. Cancer Epidemiol Biomarkers Prev (2007) 16(4):769–74. doi: 10.1158/1055-9965.EPI-06-0776
62. Strange RC, Matharoo B, Faulder GC, Jones P, Cotton W, Elder JB, et al. The Human Glutathione S-Transferases: A Case-Control Study of the Incidence of the GST1 0 Phenotype in Patients With Adenocarcinoma. Carcinogenesis (1991) 12(1):25–8. doi: 10.1093/carcin/12.1.25
Keywords: breast cancer, molecular epidemiology, polymorphism, null genotype, GSTT1-absent, GSTT1-present
Citation: Ajaz S, Zaidi S-e-Z, Ali SM, Siddiqa A, Memon MA, Firasat S, Abid A and Khaliq S (2021) Absence of Glutathione S-Transferase Theta 1 Gene Is Significantly Associated With Breast Cancer Susceptibility in Pakistani Population and Poor Overall Survival in Breast Cancer Patients: A Case-Control and Case Series Analysis. Front. Oncol. 11:678705. doi: 10.3389/fonc.2021.678705
Received: 10 March 2021; Accepted: 01 November 2021;
Published: 06 December 2021.
Edited by:
Zhi-Ming Shao, Fudan University, ChinaReviewed by:
Isabel Aguilera, Institute of Biomedicine of Seville (IBIS), SpainCristian Scatena, University of Pisa, Italy
Copyright © 2021 Ajaz, Zaidi, Ali, Siddiqa, Memon, Firasat, Abid and Khaliq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadia Ajaz, c2FkaWFhamF6MUBnbWFpbC5jb20=
 Sadia Ajaz
Sadia Ajaz Sani-e-Zehra Zaidi
Sani-e-Zehra Zaidi Saleema Mehboob Ali
Saleema Mehboob Ali Aisha Siddiqa
Aisha Siddiqa Muhammad Ali Memon
Muhammad Ali Memon Sadaf Firasat4
Sadaf Firasat4 Aiysha Abid
Aiysha Abid