- Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha, China
The circulating tumor DNA (ctDNA), as a promising biomarker of liquid biopsy, has potential clinical relevance on the molecular diagnosis and monitoring of cancer. However, the trace concentration level of ctDNA in the peripheral blood restricts its extensive clinical application. Recently, high-throughput-based methodologies have been leveraged to improve the sensitivity and specificity of ctDNA detection, showing a promising avenue towards liquid biopsy. This review briefly summarizes the high-throughput data features concerned by current ctDNA detection strategies and the technical obstacles, potential solutions, and clinical relevance of current ctDNA profiling technologies. We also highlight future directions improving the limit of detection of ctDNA for better clinical application. This review may serve as a reference for the crosslinks between data science and ctDNA-based liquid biopsy, benefiting clinical translation in advanced cancer diagnosis.
Introduction
Liquid biopsy, a non-invasive real-time method, can provide diagnostic and prognostic information during cancer progression and treatment (1). Unlike tissue biopsy, liquid biopsy examines circulating tumor cells (2) or tumor-released molecules, such as DNAs (3) and RNAs (4), from the circulatory system. Circulating tumor DNA (ctDNA) is generated from tumor cells (5), which forms a small minority of the cell-free DNA (cfDNA) in circulation against a background of fragments mostly derived from normal cells in the event of cell death or exosome secretion (6, 7). Plasma ctDNA could originate from both the nuclei or the mitochondria of tumor cells (8). However, only nucleus ctDNA records sufficient information of tumor genome, revealing tumor generation, development, metastasis, and recurrence (9), while mitochondrial ctDNA often provides information noise due to its less genomic information and higher copy number (Supplementary Figure 1). Thus, the concentration and abnormal sequence features of nucleus ctDNA (hereinafter ctDNA for convenience) in patients’ blood are significantly correlated with the course of the disease and curative effect (10), rendering it an emerging tumor marker and an essential part of liquid biopsy (11). Although the trace concentration of ctDNA in the peripheral blood and intense background noises challenge the clinical application of ctDNA, a series of ctDNA capture methods based on data science aiming at its biological features improves the sensitivity and accuracy of ctDNA detection and gradually clears the obstacles in the potential clinical application of ctDNA detection (12, 13). This review briefly summarizes the recent development and application of data science for highly sensitive and robust ctDNA detection. We also discuss the current challenges of ctDNA detection technologies and provide insights into the potential development direction in their future application.
Data Features Utilized by Current ctDNA Detection Strategies
Current ctDNA detection strategies are developed mainly based on the fragment concentration and the sequence features, such as abnormal mutations and methylations. The dynamic concentration of ctDNA is significantly correlated with the progress of the cancer disease. Because of its short half-life of less than 2 hours (14) and its low concentration (5), ctDNA is almost undetectable in patients with primary tumors. However, along with the progression of the disease, the immune system is attenuated, and ctDNA is gradually accumulated, which could be discriminated from cfDNA under certain limits of detection (15). The increased concentration of ctDNA could identify patients with cancer from healthy cohorts and stratify patients in the early and advanced stages (16). Besides, the changes in ctDNA levels before and after drug treatment are related to the therapeutic effect for patients (17). Furthermore, for tumor-free patients after treatment, the concentration level of ctDNA indicates the risk of cancer recurrence (18).
Technically, concentration analysis of ctDNA is a challenging task because ctDNA makes up a small proportion of the total cfDNA extracted from serum (19, 20). For example, Diehl et al. found that the mean mutated allele frequency of APC gene of patients with colorectal cancer ranged from early stages’ 0.04% to late stages 11% (21). Extraction of ctDNA information from other cfDNA noise should be an initial step for ctDNA detection. Size selection–based data selection has been widely utilized in ctDNA detection to increase the signal-to-noise ratio. The size of cfDNA generated from the apoptosis of normal cells is about 167 bp, which is due to the structure of the histone octamer (22). However, studies show that the ctDNA is shorter than cfDNA in the meaning of statistics (23) and has a typical size of less than 142 bp in low molecular weight (24). The enriched mitochondrial ctDNA is about 100 bp in size, much smaller than that of nuclear ctDNA, further displaying the size variety of ctDNA fragments (8). Notably, there are long-size cfDNA fragments that exist, such as 2 kb and 20 kb fragments, which are probably generated from cancer cell necrosis (25) and blood cell surface (26), respectively. The accurate enrichment of ctDNA in a particular interval size eliminates some background noises to some extent and relatively enhances data processing efficiency. For example, Mouliere et al. mapped the distribution of ctDNA fragments and optimized the ctDNA capture by choosing to concentrate ctDNA fragments in the size of 90–150 bp from the blood samples (27). In parallel, the interference from mitochondrial ctDNA fragments can be significantly eliminated by reducing the interval size of captured DNA fragments (28, 29), and the size selection strategy not only greatly reduces the cost of sequencing but also considerably decreases the false-positive rates of results by data analysis (30).
The sequence information carried by ctDNA can reflect the mutation load and the methylation features of tumor cells. ctDNA profiling facilitates their delineation not only on a genome-wide scale but also in some specific genes or intervals of the tumor genome. Some studies showed that the mutational spectrum constructed by ctDNA is highly consistent with that of tissue biopsy (31, 32). Besides, ctDNA, which comes from a broader range of tumor cells, represents the heterogeneity of the tumor mutational spectrum better than the tissue biopsy (31). The spatial heterogeneity with the tumor’s continuous self-cloning and the temporal heterogeneity possibly resulting from drug resistance can be tracked by real-time monitoring in ctDNA fragments (33). An inspiring technology termed Cancer Personalized Profiling by deep sequencing (CAPP-Seq) preselected some specifically mutational exon regions by mining a large number of genetic mutations in silico. These exon-containing ctDNAs are subsequently extracted from serum cfDNAs using customized probes and then analyzed by high-throughput sequencing. This method could remarkably improve the detection sensitivity and specificity of ctDNAs by reducing the potential impact of stochastic noise and biological variability (34) (Supplementary Figure 2).
In addition, the methylation features of ctDNA reveal some epigenetic information of cancer patients. The methylation patterns can be maintained stably throughout the life span after de novo methylation (35), and the changes in the methylated patterns predict the risk of diseases (36). Because of the increased accuracy of high-throughput sequencing (HTS) technologies, the slight differences in methylation profiles between cancer patients and healthy cohorts shed light on the differential gene expression patterns at the epigenetic level and the relevance of epigenetic modification and tumor stage (37). For instance, the hypermethylation of tumor suppressor genes is strongly consistent with cancer occurrence, indicating that the methylation status of these modifications detected by ctDNA can play an essential role in the early detection of cancer and the determination of tissue of origin, and those patterns benefit machine learning for classification modeling (38).
Data mining has increasingly become a potential requirement for algorithm design of ctDNA detection. Given the rapid development in omics, the biological data in the open database online have increased exponentially, reforming the traditional data processing methods (39–41) (Supplementary Figure 3). By narrowing the scope of previous experimental data and using an appropriate workflow, the data mining system for ctDNA is simplified, accompanied by dramatically reduced costs of the whole research project. It provides the possibility for finding the new features of ctDNA hidden in the data structure to promote further development of ctDNA capture (Figure 1). For example, Misawa et al. mined the transcriptome data and filtered out the abnormal methylations as biomarkers in ctDNA, which assists in designing a mathematical model of ctDNA detection to identify patients with human papillomavirus–associated oropharyngeal cancer (42).
Technical Obstacles and Potential Solutions of Data Processing in ctDNA Profiling
With the deepening understanding of biological features of ctDNA, the prevalence of ctDNA detection in cancer diagnosis inspired researchers. However, several technical shortcomings limit the clinical application of ctDNA detection.
Firstly, the sensitivity and specificity of ctDNA profiling are remarkably influenced by poor experimental conditions when facing complex biological characteristics (43). The trace amount and inevitable degradation of plasma ctDNA jeopardize ctDNA detection, especially when the blood sample is isolated and collected by centrifugation. Recent related studies demonstrate that plasma ctDNA loses about 50% after centrifugation (44, 45). The current blood storage manners are always accompanied by hemagglutination and extravasation, which considerably hamper ctDNA detection. Moreover, several commercial kits have been developed but show different extraction efficiencies and fragment size preferences, thereby challenging the repeatability and comparability of ctDNA detection resulting from different studies (46–49). Thus, the development of a universal standard protocol used for ctDNA extraction is essential in the future clinic application of ctDNA detection strategies.
Furthermore, the low signal-to-noise ratio remains a major problem for data processing of ctDNA detection. In addition to the low proportion of ctDNA in the serum cfDNA pools, as mentioned above, somatic mutations deriving from clonal hematopoietic (CH) and mitochondrial ctDNA also bring significant background noises. CH variants are cumulated with age, which could be attributed to the cloning expansion of stem cells carrying somatic mutations (50). Due to the high false-positive rate in ctDNA detection results, CH variants also interfere with the construction of the ctDNA mutation spectrum (51). Although mitochondrial ctDNA could be roughly excluded by size selection manner, the leaking information still exists during ctDNA detection (52). Given the above, the development of data analysis algorithms to increase signal-to-noise rate will facilitate the reliability of ctDNA as a tumor biomarker applied in clinical diagnosis. For instance, Nassiri et al. have developed a machine learning–based model to analyze the data generated from methylated HTS in ctDNA detection, increasing the accuracy of subtyping intracranial tumors (53). Moreover, CH variances produced by white blood cells are recognized by the combination of computational algorithms, then the false-positive rates of ctDNA detection would be decreased (54). With the help of statistical analysis and machine learning models, the CH variances spectrum can be built quickly and will be removed effectively by comparing it to the mutational spectrum of tumors constructed by ctDNA assays (30).
Additionally, the technological bias of HTS platforms inevitably interferes with the high-throughput ctDNA detection. These sequencing errors can be partially reduced or corrected. For instance, increased sequencing depth dilutes the error information (55), and the introduction of appropriate barcodes and indexes could evaluate the sequence duplication bias produced through polymerase chain reaction (PCR) amplification (56). The erroneous sequencing on barcodes, which affects the deduplication of unique ctDNA molecules and results in errors in aligning molecules to unique ctDNA, can be optimized by increasing the hamming edit distance between different barcodes (57). However, these additional barcodes occupy some part of reads and then reduce the actual length of target ctDNA to be sequenced, attributing to the limitation of the reading sequence length of long-read assembly technology. Therefore, the choice of proper barcodes used for HTS of ctDNA detection is the critical factor in resolving the inevitable sequencing errors, calling for new methods for pre- and post-sequencing error correction based on a statistical landscape.
Clinical Relevance of Current ctDNA Profiling Technologies
The clinical relevance of the ctDNA biomarker shows substantial potential in non-invasive liquid biopsy, which may benefit millions of patients for early detection of tumor (58, 59), determination of tissue of origin (37), prediction of therapeutic effect, especially for immunotherapies (60–62), and monitoring (15, 30). The dynamic risk stratification correlated to the tumor genesis could also be facilitated by ctDNA detection such as occupation, age, living habits, and even mutational signatures (63). With the aid of classification or non-supervisor clustering, ctDNA detection technologies are conspicuously improved in terms of accuracy, sensitivity, specificity, operational convenience, and reasonable cost.
The patterns of mutational spectrums or epigenetic profiles recognized by data mining uncover the particular clinical relevance of ctDNA detection. The genome-wide mutational landscape is conducive to the evaluation of tumor mutation burden (13), neoplasm staging (64), genotyping (11), and the choice of therapies (65). Meanwhile, the methylation profiles of ctDNA contribute to discriminating patients from healthy cohorts (37), differentiating cancer types (53), and identifying the primary tumor location (66). These profiles can complement each other in many aspects, though those meaningful patterns below them require plenty of modeling theories for recognition accuracy. The combination of mutation and methylation spectrums makes the acquirement of detailed genomic landscapes possible, provides multiple insights into the tumor heterogeneity, and evaluates the impact of tumor heterogeneity on the selection of therapies, such as non-responders or drug resistance (67, 68).
In addition to its non-invasiveness, near real-time monitoring and prognosis prediction are additional advantages of ctDNA detection over tissue biopsy (69, 70). For example, the concentration of ctDNA was correlated with the prognosis of patients treated with pembrolizumab (10). Furthermore, ctDNA detection, as an auxiliary method for low-dose computed tomography, can track the molecular minimal residual disease and predict the risk of recurrence for tumor-free patients (18, 71). Finally, the real-time information of ctDNA detection reflects patients’ status and sheds light on the personalized profiling for each patient, which is essential for precision medicine (72).
Future Directions Improving the Limit of Detection of ctDNA
There is a definite clue that an evolution is happening in high-throughput ctDNA detection by introducing novel sequencing platforms, a combination of different biomarkers, and a development of new principles. New-generation sequencing technologies, such as nanopore sequencing, begin to be utilized in ctDNA detections (73). Compared with HTS technologies, nanopore sequencing exhibits real-time sequencing and long reads, resulting in its potentially broad application in the field of nucleic acid sequencing in the future (74). Moreover, nanopore sequencing is PCR-free, avoiding amplification bias and errors of PCR during the process of sequencing library preparation. Although nanopore sequencing remains to have some shortcomings in sequencing short DNA fragments, many efforts have been made to ameliorate these shortcomings (2, 75). For instance, Sun et al. have applied the solid-state nanopore to detect ctDNA originating from serum samples. This strategy cooperates with the hybridization chain reaction to amplify the target’s signals, improve data authenticity, and overcome the hurdles of nanopore application (76).
Combining multi-biomarkers in liquid biopsy, based on optimized models and algorithms, has a higher efficiency in tumor detections. The various biomarkers used as the inputs of the detection model have a complementary function because of their different sensitivity and specificity to patients. For example, combining exosome RNA and ctDNA in plasma, Krug et al. leveraged the threshold of a predefined model to detect EGFR mutations in non-small cell lung cancer, achieving a higher sensitivity than that of ctDNA detection alone (77). Cohen et al. utilized the protein biomarkers as a supplement to ctDNA detection, and a few patients with ctDNA undetectable were finally detected (78). Furthermore, the combination of multi-biomarkers provides the convenience of multi-parameters to machine learning in future liquid biopsy and promotes the development of detection tools in the diagnosis and prognosis for patients with cancer.
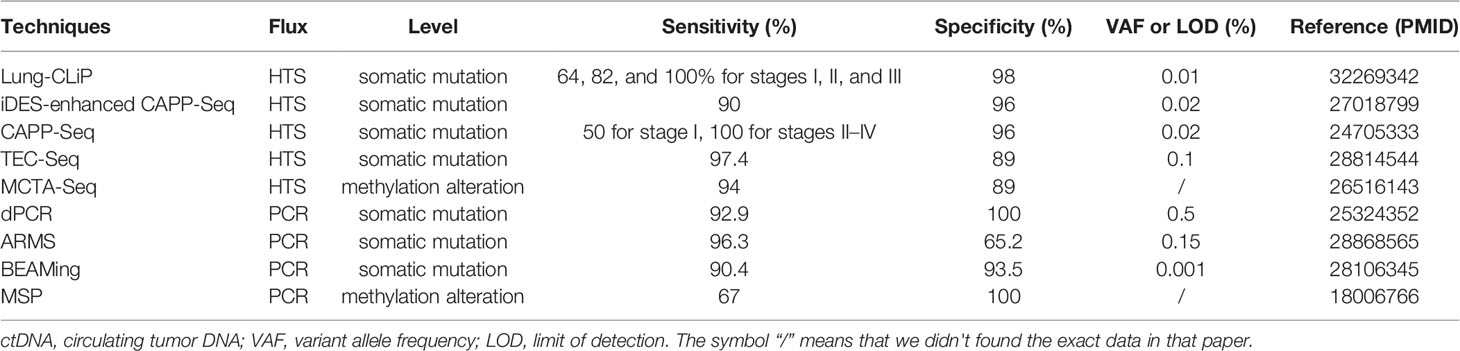
New principles, whether biological feature-based or data-driven, are the catalyzers of the ctDNA detection improvement. In the last decades, discovering new principles of ctDNA detection methods contributes to promoting the clinical application of this intriguing biomarker of liquid biopsy (Table 1). For example, the definition of recurrence index, an index equal to total unique patients with mutations covered per kb of an exon, has been introduced into CAPP-Seq as a selection principle that obviously improved the limit of detection of ctDNA (34). The continuous evolution of new technical principles of data analysis provides the substantial potential to ctDNA as a promising biomarker for its future clinical utility (79).
Conclusion
Embracing data science, ctDNA is a promising biomarker in cancer detection. ctDNA has several exciting characteristics, which could be handled to raise strategies to improve ctDNA detection performance. Herein, we reviewed data science that played an essential role in current strategies, such as data selection, data mining, and data correction, to overcome the technical obstacles in ctDNA detection. The recognition of the value of directing data processing indicates a possible trend to exploit ctDNA assays further. With the rapid development of data acquisition methodologies, modeling, and data processing algorithms, ctDNA detection enhanced its prevalent advantages in monitoring intrinsic tumor information. Novel high-throughput technology platforms and the combination of diverse biomarkers in liquid biopsy were also essential for this technology advancement. ctDNA-based liquid biopsies, as an alternative or even a substitutive choice of tissue biopsy, have significant clinical relevance in cancer diagnosis and prognosis. Subsequent efforts should be continued to promote the advancement of the detection technologies, theories, and principles accelerated by prosperously developed data science.
Author’s Note
The data of Supplementary Figure 3 coming from the GEOdatabase, including the R script and Rdata file.
Author Contributions
ML: writing—original draft, figure design, and visualization. SX: assistance with writing review and editing. CL: assistance with figure design and visualization. LiZ: necessary advice provision. LvZ: supervision, funding acquisition, writing review, and editing. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by grants from the National Natural Science Foundation of China (31500686, 31870855), the “Huxiang Young Talents Plan” Project of Hunan Province (2019RS2030), the Natural Science Foundation of Hunan Province (2020JJ5657), Fund for NUDT Young Innovator Awards (20190104), and Postgraduate Scientific Research Innovation Project of Hunan Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank ShineWrite for its linguistic assistance during this manuscript’s preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.692322/full#supplementary-material
Supplementary Figure 1 | Origination of ctDNA. ctDNA, circulating tumor DNA; cfDNA, cell-free DNA.
Supplementary Figure 2 | The workflow of CAPP-Seq. CAPP-Seq, the Cancer Personalized Profiling by deep sequencing; ctDNA, circulating tumor DNA; cfDNA, cell-free DNA.
Supplementary Figure 3 | Rapid growth in the number of samples uploaded in the GEO (Gene Expression Omnibus) database.
Abbreviations
CAPP-Seq, the Cancer Personalized Profiling by deep sequencing; CH, clonal hematopoietic; cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; HTS, high-throughput sequencing; PCR, polymerase chain reaction.
References
1. Mohan S, Foy V, Ayub M, Leong HS, Schofield P, Sahoo S, et al. Profiling of Circulating Free DNA Using Targeted and Genome-Wide Sequencing in Patients With SCLC. J Thorac Oncol (2020) 15(2):216–30. doi: 10.1016/j.jtho.2019.10.007
2. Li X, Zhang P, Dou L, Wang Y, Sun K, Zhang X, et al. Detection of Circulating Tumor Cells in Breast Cancer Patients by Nanopore Sensing With Aptamer-Mediated Amplification. ACS Sens (2020) 5(8):2359–66. doi: 10.1021/acssensors.9b02537
3. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med (2013) 368(13):1199–209. doi: 10.1056/NEJMoa1213261
4. Allegretti M, Casini B, Mandoj C, Benini S, Alberti L, Novello M, et al. Precision Diagnostics of Ewing's Sarcoma by Liquid Biopsy: Circulating EWS-FLI1 Fusion Transcripts. Ther Adv Med Oncol (2018) 10:1758835918774337. doi: 10.1177/1758835918774337
5. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin From Apoptotic and Necrotic Cells. Cancer Res (2020) 61(4):1659–65. doi: cancerres.aacrjournals.org/content/61/4/1659
6. Bronkhorst AJ, Wentzel JF, Aucamp J, van Dyk E, du Plessis L, Pretorius PJ. Characterization of the Cell-Free DNA Released by Cultured Cancer Cells. Biochim Biophys Acta (2016) 1863(1):157–65. doi: 10.1016/j.bbamcr.2015.10.022
7. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of Exosome Composition. Cell (2019) 177(2):428–45.e18. doi: 10.1016/j.cell.2019.02.029
8. Mair R, Mouliere F, Smith CG, Chandrananda D, Gale D, Marass F, et al. Measurement of Plasma Cell-Free Mitochondrial Tumor DNA Improves Detection of Glioblastoma in Patient-Derived Orthotopic Xenograft Models. Cancer Res (2019) 79(1):220–30. doi: 10.1158/0008-5472.CAN-18-0074
9. Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell (2017) 168(4):571–4. doi: 10.1016/j.cell.2017.01.030
10. Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized Circulating Tumor DNA Analysis as a Predictive Biomarker in Solid Tumor Patients Treated With Pembrolizumab. Nat Cancer (2020) 1(9):873–81. doi: 10.1038/s43018-020-0096-5
11. Jayaram A, Wetterskog D, Attard G. Plasma DNA and Metastatic Castration-Resistant Prostate Cancer: The Odyssey to a Clinical Biomarker Test. Cancer Discov (2018) 8(4):392–4. doi: 10.1158/2159-8290.CD-18-0124
12. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov (2017) 7(12):1394–403. doi: 10.1158/2159-8290.CD-17-0716
13. Ishii H, Azuma K, Sakai K, Naito Y, Matsuo N, Tokito T, et al. Determination of Somatic Mutations and Tumor Mutation Burden in Plasma by CAPP-Seq During Afatinib Treatment in NSCLC Patients Resistance to Osimertinib. Sci Rep (2020) 10(1):691. doi: 10.1038/s41598-020-57624-4
14. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat Med (2008) 14(9):985–90. doi: 10.1038/nm.1789
15. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct Detection of Early-Stage Cancers Using Circulating Tumor DNA. Sci Transl Med (2017) 9(403):eaan2415. doi: 10.1126/scitranslmed.aan2415
16. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med (2014) 6(224):224ra24. doi: 10.1126/scitranslmed.3007094
17. Blumendeller C, Boehme J, Frick M, Schulze M, Rinckleb A, Kyzirakos C, et al. Use of Plasma ctDNA as a Potential Biomarker for Longitudinal Monitoring of a Patient With Metastatic High-Risk Upper Tract Urothelial Carcinoma Receiving Pembrolizumab and Personalized Neoepitope-Derived Multipeptide Vaccinations: A Case Report. J Immunother Cancer (2021) 9(1):e001406. doi: 10.1136/jitc-2020-001406
18. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating Tumor DNA Analysis Detects Minimal Residual Disease and Predicts Recurrence in Patients With Stage II Colon Cancer. Sci Transl Med (2016) 8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219
19. Yang N, Li Y, Liu Z, Qin H, Du D, Cao X, et al. The Characteristics of ctDNA Reveal the High Complexity in Matching the Corresponding Tumor Tissues. BMC Cancer (2018) 18(1):319. doi: 10.1186/s12885-018-4199-7
20. Yong E. Cancer Biomarkers: Written in Blood. Nature (2014) 511(7511):524–6. doi: 10.1038/511524a
21. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and Quantification of Mutations in the Plasma of Patients With Colorectal Tumors. Proc Natl Acad Sci U S A (2005) 102(45):16368–73. doi: 10.1073/pnas.0507904102
22. Ivanov M, Baranova A, Butler T, Spellman P, Mileyko V. Non-Random Fragmentation Patterns in Circulating Cell-Free DNA Reflect Epigenetic Regulation. BMC Genomics (2015) 16 Suppl 13:S1. doi: 10.1186/1471-2164-16-S13-S1
23. Tamkovich SN, Kirushina NA, Voytsitskiy VE, Tkachuk VA, Laktionov PP. Features of Circulating DNA Fragmentation in Blood of Healthy Females and Breast Cancer Patients. Adv Exp Med Biol (2016) 924:47–51. doi: 10.1007/978-3-319-42044-8_10
24. Hellwig S, Nix DA, Gligorich KM, O'Shea JM, Thomas A, Fuertes CL, et al. Automated Size Selection for Short Cell-Free DNA Fragments Enriches for Circulating Tumor DNA and Improves Error Correction During Next Generation Sequencing. PLoS One (2018) 13(7):e0197333. doi: 10.1371/journal.pone.0197333
25. Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS One (2011) 6(9):e23418. doi: 10.1371/journal.pone.0023418
26. Tamkovich S, Laktionov P. Cell-Surface-Bound Circulating DNA in the Blood: Biology and Clinical Application. IUBMB Life (2019) 71(9):1201–10. doi: 10.1002/iub.2070
27. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced Detection of Circulating Tumor DNA by Fragment Size Analysis. Sci Transl Med (2018) 10(466):eaat4921. doi: 10.1126/scitranslmed.aat4921
28. Zhang R, Nakahira K, Guo X, Choi AM, Gu Z. Very Short Mitochondrial DNA Fragments and Heteroplasmy in Human Plasma. Sci Rep (2016) 6:36097. doi: 10.1038/srep36097
29. Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and Shortening of Plasma DNA in Hepatocellular Carcinoma Patients. Proc Natl Acad Sci U S A (2015) 112(11):E1317–25. doi: 10.1073/pnas.1500076112
30. Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating Genomic Features for Non-Invasive Early Lung Cancer Detection. Nature (2020) 580(7802):245–51. doi: 10.1038/s41586-020-2140-0
31. Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic Landscape of Cell-Free DNA in Patients With Colorectal Cancer. Cancer Discov (2018) 8(2):164–73. doi: 10.1158/2159-8290.CD-17-1009
32. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable Whole-Exome Sequencing of Cell-Free DNA Reveals High Concordance With Metastatic Tumors. Nat Commun (2017) 8(1):1324. doi: 10.1038/s41467-017-00965-y
33. De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CKY, Nuciforo P, et al. Capturing Intra-Tumor Genetic Heterogeneity by De Novo Mutation Profiling of Circulating Cell-Free Tumor DNA: A Proof-of-Principle. Ann Oncol (2014) 25(9):1729–35. doi: 10.1093/annonc/mdu239
34. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA With Broad Patient Coverage. Nat Med (2014) 20(5):548–54. doi: 10.1038/nm.3519
35. Dor Y, Cedar H. Principles of DNA Methylation and Their Implications for Biology and Medicine. Lancet (2018) 392(10149):777–86. doi: 10.1016/s0140-6736(18)31268-6
36. Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, et al. A DNA Hypermethylation Module for the Stem/Progenitor Cell Signature of Cancer. Genome Res (2012) 22(5):837–49. doi: 10.1101/gr.131169.111
37. Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat Commun (2018) 9(1):5068. doi: 10.1038/s41467-018-07466-6
38. Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, et al. Noninvasive Detection of Cancer-Associated Genome-Wide Hypomethylation and Copy Number Aberrations by Plasma DNA Bisulfite Sequencing. Proc Natl Acad Sci U S A (2013) 110(47):18761–8. doi: 10.1073/pnas.1313995110
39. Resteghini C, Trama A, Borgonovi E, Hosni H, Corrao G, Orlandi E, et al. Big Data in Head and Neck Cancer. Curr Treat Options Oncol (2018) 19(12):62. doi: 10.1007/s11864-018-0585-2
40. Ngiam KY, Khor IW. Big Data and Machine Learning Algorithms for Health-Care Delivery. Lancet Oncol (2019) 20(5):e262–e73. doi: 10.1016/S1470-2045(19)30149-4
41. Kantarjian H, Yu PP. Artificial Intelligence, Big Data, and Cancer. JAMA Oncol (2015) 1(5):573–4. doi: 10.1001/jamaoncol.2015.1203
42. Misawa K, Imai A, Matsui H, Kanai A, Misawa Y, Mochizuki D, et al. Identification of Novel Methylation Markers in HPV-Associated Oropharyngeal Cancer: Genome-Wide Discovery, Tissue Verification and Validation Testing in ctDNA. Oncogene (2020) 39(24):4741–55. doi: 10.1038/s41388-020-1327-z
43. Markus H, Contente-Cuomo T, Farooq M, Liang WS, Borad MJ, Sivakumar S, et al. Evaluation of Pre-Analytical Factors Affecting Plasma DNA Analysis. Sci Rep (2018) 8(1):7375. doi: 10.1038/s41598-018-25810-0
44. Rikkert LG, van der Pol E, van Leeuwen TG, Nieuwland R, Coumans FAW. Centrifugation Affects the Purity of Liquid Biopsy-Based Tumor Biomarkers. Cytomet A (2018) 93(12):1207–12. doi: 10.1002/cyto.a.23641
45. van der Pol Y, Mouliere F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell (2019) 36(4):350–68. doi: 10.1016/j.ccell.2019.09.003
46. Sorber L, Zwaenepoel K, Deschoolmeester V, Roeyen G, Lardon F, Rolfo C, et al. A Comparison of Cell-Free DNA Isolation Kits: Isolation and Quantification of Cell-Free DNA in Plasma. J Mol Diagn (2017) 19(1):162–8. doi: 10.1016/j.jmoldx.2016.09.009
47. Worm Orntoft MB, Jensen SO, Hansen TB, Bramsen JB, Andersen CL. Comparative Analysis of 12 Different Kits for Bisulfite Conversion of Circulating Cell-Free DNA. Epigenetics (2017) 12(8):626–36. doi: 10.1080/15592294.2017.1334024
48. Torga G, Pienta KJ. Patient-Paired Sample Congruence Between 2 Commercial Liquid Biopsy Tests. JAMA Oncol (2018) 4(6):868–70. doi: 10.1001/jamaoncol.2017.4027
49. Kuderer NM, Burton KA, Blau S, Rose AL, Parker S, Lyman GH, et al. Comparison of 2 Commercially Available Next-Generation Sequencing Platforms in Oncology. JAMA Oncol (2017) 3(7):996–8. doi: 10.1001/jamaoncol.2016.4983
50. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred From Blood DNA Sequence. N Engl J Med (2014) 371(26):2477–87. doi: 10.1056/NEJMoa1409405
51. Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res (2018) 24(18):4437–43. doi: 10.1158/1078-0432.CCR-18-0143
52. Weerts MJA, Timmermans EC, van de Stolpe A, Vossen R, Anvar SY, Foekens JA, et al. Tumor-Specific Mitochondrial DNA Variants Are Rarely Detected in Cell-Free DNA. Neoplasia (2018) 20(7):687–96. doi: 10.1016/j.neo.2018.05.003
53. Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, et al. Detection and Discrimination of Intracranial Tumors Using Plasma Cell-Free DNA Methylomes. Nat Med (2020) 26(7):1044–7. doi: 10.1038/s41591-020-0932-2
54. Zhang Y, Yao Y, Xu Y, Li L, Gong Y, Zhang K, et al. Pan-Cancer Circulating Tumor DNA Detection in Over 10,000 Chinese Patients. Nat Commun (2021) 12(1):11. doi: 10.1038/s41467-020-20162-8
55. Goodwin S, McPherson JD, McCombie WR. Coming of Age: Ten Years of Next-Generation Sequencing Technologies. Nat Rev Genet (2016) 17(6):333–51. doi: 10.1038/nrg.2016.49
57. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated Digital Error Suppression for Improved Detection of Circulating Tumor DNA. Nat Biotechnol (2016) 34(5):547–55. doi: 10.1038/nbt.3520
58. Yokoi K, Yamashita K, Watanabe M. Analysis of DNA Methylation Status in Bodily Fluids for Early Detection of Cancer. Int J Mol Sci (2017) 18(4):735. doi: 10.3390/ijms18040735
59. Wan N, Weinberg D, Liu TY, Niehaus K, Ariazi EA, Delubac D, et al. Machine Learning Enables Detection of Early-Stage Colorectal Cancer by Whole-Genome Sequencing of Plasma Cell-Free DNA. BMC Cancer (2019) 19(1):832. doi: 10.1186/s12885-019-6003-8
60. Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res (2018) 24(8):1872–80. doi: 10.1158/1078-0432.CCR-17-1341
61. Lee JH, Long GV, Menzies AM, Lo S, Guminski A, Whitbourne K, et al. Association Between Circulating Tumor DNA and Pseudoprogression in Patients With Metastatic Melanoma Treated With Anti-Programmed Cell Death 1 Antibodies. JAMA Oncol (2018) 4(5):717–21. doi: 10.1001/jamaoncol.2017.5332
62. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol (2019) 5(5):696–702. doi: 10.1001/jamaoncol.2018.7098
63. Carroll PR, Parsons JK, Andriole G, Bahnson RR, Castle EP, Catalona WJ, et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J Natl Compr Canc Netw (2016) 14(5):509–19. doi: 10.6004/jnccn.2016.0060
64. Song CX, Yin S, Ma L, Wheeler A, Chen Y, Zhang Y, et al. 5-Hydroxymethylcytosine Signatures in Cell-Free DNA Provide Information About Tumor Types and Stages. Cell Res (2017) 27(10):1231–42. doi: 10.1038/cr.2017.106
65. Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda S, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment With PARP Inhibition. Cancer Discov (2017) 7(9):1006–17. doi: 10.1158/2159-8290.CD-17-0261
66. Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA Tissue Mapping by Genome-Wide Methylation Sequencing for Noninvasive Prenatal, Cancer, and Transplantation Assessments. Proc Natl Acad Sci U S A (2015) 112(40):E5503–12. doi: 10.1073/pnas.1508736112
67. Kim MS, Yamashita K, Chae YK, Tokumaru Y, Chang X, Zahurak M, et al. A Promoter Methylation Pattern in the N-Methyl-D-Aspartate Receptor 2B Gene Predicts Poor Prognosis in Esophageal Squamous Cell Carcinoma. Clin Cancer Res (2007) 13(22 Pt 1):6658–65. doi: 10.1158/1078-0432.CCR-07-1178
68. Claus R, Lucas DM, Stilgenbauer S, Ruppert AS, Yu L, Zucknick M, et al. Quantitative DNA Methylation Analysis Identifies a Single CpG Dinucleotide Important for ZAP-70 Expression and Predictive of Prognosis in Chronic Lymphocytic Leukemia. J Clin Oncol (2012) 30(20):2483–91. doi: 10.1200/JCO.2011.39.3090
69. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci Transl Med (2012) 4(136):136ra68. doi: 10.1126/scitranslmed.3003726
70. Burnham P, Dadhania D, Heyang M, Chen F, Westblade LF, Suthanthiran M, et al. Urinary Cell-Free DNA is a Versatile Analyte for Monitoring Infections of the Urinary Tract. Nat Commun (2018) 9(1):2412. doi: 10.1038/s41467-018-04745-0
71. Comino-Mendez I, Turner N. Predicting Relapse With Circulating Tumor DNA Analysis in Lung Cancer. Cancer Discov (2017) 7(12):1368–70. doi: 10.1158/2159-8290.CD-17-1086
72. Girotti MR, Gremel G, Lee R, Galvani E, Rothwell D, Viros A, et al. Application of Sequencing, Liquid Biopsies, and Patient-Derived Xenografts for Personalized Medicine in Melanoma. Cancer Discov (2016) 6(3):286–99. doi: 10.1158/2159-8290.CD-15-1336
73. Burck N, Gilboa T, Gadi A, Patkin Nehrer M, Schneider RJ, Meller A. Nanopore Identification of Single Nucleotide Mutations in Circulating Tumor DNA by Multiplexed Ligation. Clin Chem (2021) 67(5):753–62. doi: 10.1093/clinchem/hvaa328
74. Deamer D, Akeson M, Branton D. Three Decades of Nanopore Sequencing. Nat Biotechnol (2016) 34(5):518–24. doi: 10.1038/nbt.3423
75. Martignano F, Munagala U, Crucitta S, Mingrino A, Semeraro R, Del Re M, et al. Nanopore Sequencing From Liquid Biopsy: Analysis of Copy Number Variations From Cell-Free DNA of Lung Cancer Patients. Mol Cancer (2021) 20(1):32. doi: 10.1186/s12943-021-01327-5
76. Sun H, Yao F, Su Z, Kang XF. Hybridization Chain Reaction (HCR) for Amplifying Nanopore Signals. Biosens Bioelectron (2020) 150:111906. doi: 10.1016/j.bios.2019.111906
77. Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann Oncol (2018) 29(3):700–6. doi: 10.1093/annonc/mdx765
78. Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined Circulating Tumor DNA and Protein Biomarker-Based Liquid Biopsy for the Earlier Detection of Pancreatic Cancers. Proc Natl Acad Sci U S A (2017) 114(38):10202–7. doi: 10.1073/pnas.1704961114
Keywords: cancer diagnosis, ctDNA detection, data science, liquid biopsy, technological advancement
Citation: Li M, Xie S, Lu C, Zhu L and Zhu L (2021) Application of Data Science in Circulating Tumor DNA Detection: A Promising Avenue Towards Liquid Biopsy. Front. Oncol. 11:692322. doi: 10.3389/fonc.2021.692322
Received: 08 April 2021; Accepted: 05 July 2021;
Published: 21 July 2021.
Edited by:
George Calin, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Svetlana Tamkovich, Novosibirsk State University, RussiaElizabeth A. Proctor, The Pennsylvania State University, United States
Copyright © 2021 Li, Xie, Lu, Zhu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lvyun Zhu, emh1bHZ5dW5AbnVkdC5lZHUuY24=; Lingyun Zhu, bGluZ3l1bnpodUBudWR0LmVkdS5jbg==
†These authors have contributed equally to this work
 Ming Li
Ming Li Sisi Xie†
Sisi Xie† Lingyun Zhu
Lingyun Zhu Lvyun Zhu
Lvyun Zhu