- 1Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, Qingdao Medical College, Qingdao University, Qingdao, China
- 2School of Basic Medical Sciences, Qingdao Medical College, Qingdao University, Qingdao, China
- 3Clinical Laboratory, Linqu People’s Hospital, Linqu, China
Gastric cancer (GC) is one of the most common malignant tumors of digestive systems worldwide, with high recurrence and mortality. Chemotherapy is still the standard treatment option for GC and can effectively improve the survival and life quality of GC patients. However, with the emergence of drug resistance, the clinical application of chemotherapeutic agents has been seriously restricted in GC patients. Although the mechanisms of drug resistance have been broadly investigated, they are still largely unknown. MicroRNAs (miRNAs) are a large group of small non-coding RNAs (ncRNAs) widely involved in the occurrence and progression of many cancer types, including GC. An increasing amount of evidence suggests that miRNAs may play crucial roles in the development of drug resistance by regulating some drug resistance-related proteins as well as gene expression. Some also exhibit great potential as novel biomarkers for predicting drug response to chemotherapy and therapeutic targets for GC patients. In this review, we systematically summarize recent advances in miRNAs and focus on their molecular mechanisms in the development of drug resistance in GC progression. We also highlight the potential of drug resistance-related miRNAs as biomarkers and therapeutic targets for GC patients.
Introduction
Gastric cancer (GC) is one of the most common malignant diseases of the digestive tract and the third leading cause of cancer-related deaths worldwide (1). According to global cancer statistics, GC ranks fifth for incidence and fourth for mortality among all types of cancer, with more than one million new cases and an estimated 769,000 deaths occurring in 2020 (2). Although the incidence of GC is declining gradually, it is still a major public health problem that seriously threatens patients’ health and lives (3). Currently, common treatment approaches to GC include chemotherapy, radiation, surgery, and targeted therapies. Depending on the resectability, stage, and status of GC patients, these therapies can be used in combination to improve their survival and life quality. However, for metastatic GC, chemotherapy is the main treatment method since most advanced patients fail to benefit from surgical resection or radiotherapy (4).
Chemotherapy is the first-line standard treatment option for all stages of cancer and can effectively delay or avoid cancer recurrence in the short term. However, the long-term role of chemotherapy in extending patient survival is very limited (5). One of the main reasons is the development of drug resistance, which results in chemotherapy failure, cancer recurrence, and finally patient death. It has been reported that drug resistance is correlated with more than 90% of cancer-related death (6, 7). Overall, drug resistance can be classified into two categories: intrinsic and acquired. Its underlying mechanisms are very complicated, mainly including changes in drug efflux, the inhibition of cell apoptosis, alterations of the cell cycle, enhancement of DNA damage repair, mutations of drug target genes, and the dysregulation of epithelial mesenchymal transformation (EMT) as well as the acquisition of cancer stem cell (CSC) properties (8). However, the detailed mechanisms involved in drug resistance are still unclear.
MicroRNAs (miRNAs) are a large group of small ncRNAs involved in practically all major biological processes via the direct post-transcriptional inhibition of target mRNAs. It has been reported that approximately 2600 miRNAs molecules have been identified in the human genome, and more than 60% of human protein coding genes are regulated by miRNAs (9, 10). MiRNAs can simultaneously regulate multiple target genes involved in different cellular processes, such as signal transduction, cell differentiation, apoptosis and proliferation (11). Therefore, the dysregulation of miRNAs contributes to many pathological processes, including GC. Moreover, the aberrant expression of miRNAs has been observed in GC (12). A growing amount of evidence suggests that miRNAs may play crucial roles in the drug resistance of GC. In this review, we provide a brief description of recent findings regarding the biogenesis and functions of miRNAs and highlight their underlying mechanisms in the drug resistance of GC.
Overview: Biogenesis and Functions of MiRNAs
Biogenesis of MiRNAs
MiRNAs are endogenous RNA-type molecules transcribed by RNA polymerase II (Pol II) with 19–25 nucleotides in length (13). The biogenesis mechanism of miRNAs has been well-studied. The majority of miRNAs are generated by the canonical biogenesis pathway (Figure 1). In this pathway, primary miRNAs (pri-miRNAs) transcribed from original miRNA genes are processed into precursor miRNAs (pre-miRNAs) in the nucleus by a microprocessor complex consisting of DiGeorge syndrome critical region 8 (DGCR8) and ribonuclease III enzyme Drosha. Subsequently, pre-miRNAs are exported to the cytoplasm through exportin 5 (EXP5) and then further recognized and processed into double-stranded miRNAs by the Dicer/TRBP/PACT complex. Next, the double-stranded miRNAs are unwound into a guide strand and a passenger strand via helicase. The passenger strand (with a higher stability) is subsequently degraded, whereas the guide strand (with a lower stability) is incorporated into the RNA-induced silencing complex (RISC) to form a mature miRNA (6). In addition, multiple non-canonical miRNA biogenesis pathways have been clarified. These non-canonical biogenesis pathways are group into Drosha/DGCR8-independent and Dicer-independent. For instance, the precursor stem lengths of some miRNAs are shorter than canonical pri-miRNAs, which can’t be recognized and processed by Drosha/DGCR8. These miRNA precursors are encoded in short introns that are named mirtrons. Mirtrons possess a hairpin structure and undergo splicing. The resulting products form a lariat structure and are processed into pre-miRNAs by DBR1 (lariat debranching enzyme), which are followed by further processing via Dicer (14). MiR-451 is a typical miRNA produced in a Dicer-independent manner. Its maturation does not require Dicer catalysis. The pre-miRNA of miR-451 is cleaved by Ago2 to generate an intermediate 3’ end, which is then further trimmed (15).
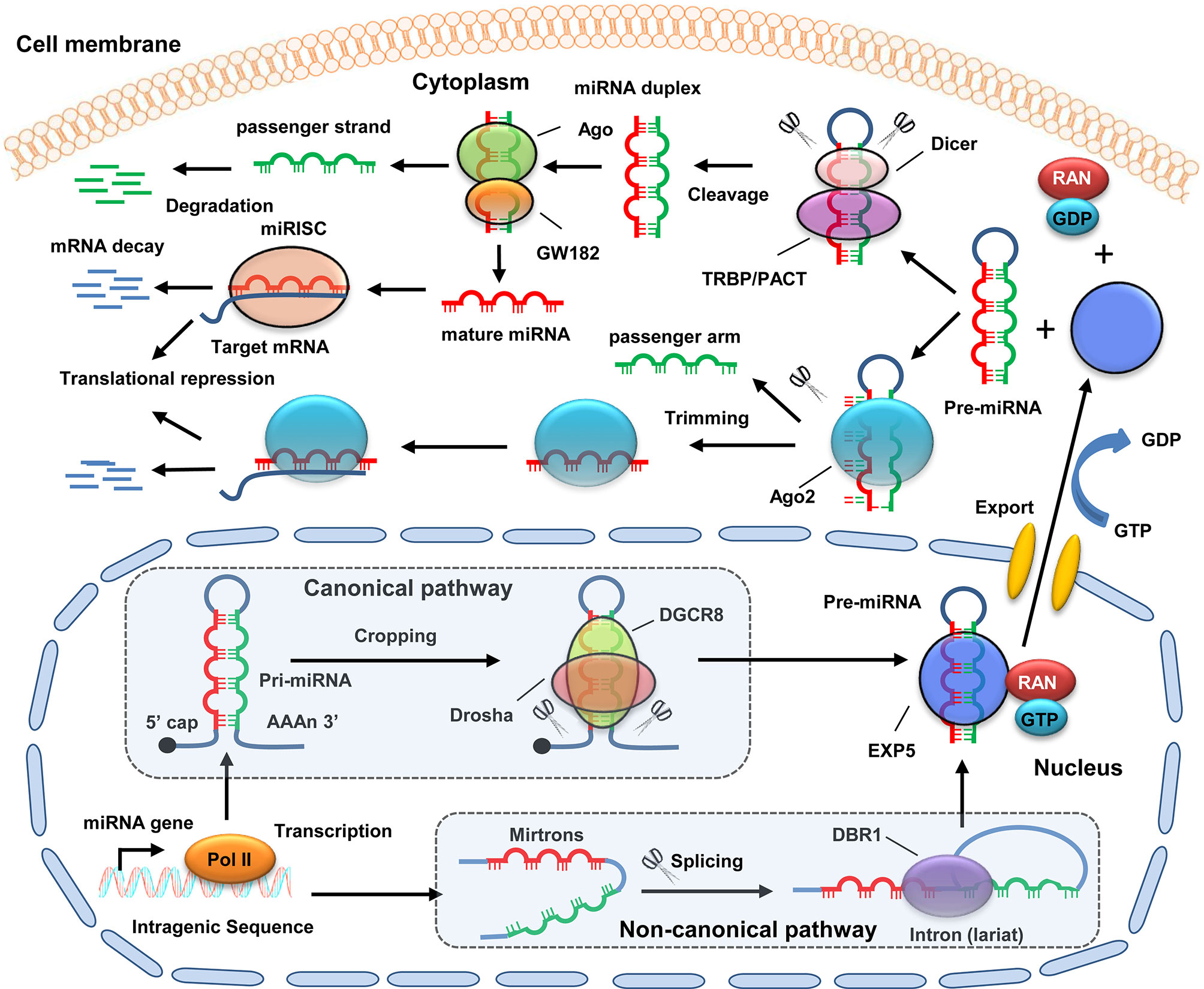
Figure 1 Schematic diagram of miRNA biogenesis. For canonical pathway, pri-miRNA is transcribed by Poll II from original miRNA genes in the nucleus. Then, pri-miRNA is processed into pre-miRNAs by a microprocessor complex consisting of Drosha and DGCR8. For non-canonical pathway, mirtron is transcribed from original miRNA genes. Then, mirtrons undergo splicing to form a lariat structure, which is further processed into pre-miRNAs by DBR1. Next, the pre-miRNAs are exported to the cytoplasm with the help of EXP5, GTP, and RAN. In the cytoplasm, the pre-miRNA is further recognized and processed into double-stranded miRNAs by the Dicer/TRBP/PACT complex. Subsequently, the double-stranded miRNAs are unwound into a guide strand and a passenger strand via Ago protein. The passenger strand is then degraded, whereas the guide strand is incorporated into RISC to form a mature miRNA. MiRNA binds to the 3’-UTR of mRNAs, and ten promotes its degradation. In addition, some pre-miRNAs (e.g., miR-451) can be cleaved by Ago2 to generate an intermediate 3’ end, which is then further trimmed.
The biogenesis of miRNAs is modulated by various factors, including post-translational modifications, target mRNAs, RNA binding proteins (RBPs), and long non-coding RNAs (lncRNAs). For instance, DGCR8 can be modified by SUMO1. The SUMOylation of DGCR8 regulates its affinity with pri-miRNAs, leading to an alteration in the pri-miRNA functions of the recognition and repression of the target mRNAs (16). Bose et al. found that target mRNA promotes the biogenesis of its cognate miR-122 by enhancing the activity of AGO2-associated DICER1 (17). Moreover, Treiber et al. screened approximately 180 RBPs that interact specifically with different pre-miRNAs. Functional analysis showed that a large number of these RBPs, including splicing factors and other mRNA processing proteins, play a role in regulating miRNA processing (18). In addition, lncRNA NEAT1 is reported to modulate global pri-miRNA biogenesis by broadly interacting with the NONO–PSF heterodimer and many other RBPs (19). Collectively, the biogenesis of miRNAs is tightly regulated by diverse mechanisms. The dysregulation of miRNA biogenesis would contribute to many pathological progresses, particularly cancer progression.
Functions of MiRNAs
MiRNAs exert their biological functions by negatively regulating the expression of their target mRNAs via directly binding to 3′ untranslated regions (UTR), leading to the inhibition of their translation (20). The precise regulation of miRNA expression and activity is crucial for maintaining common physiological conditions. MiRNAs are involved in the regulation of almost all major physiological and pathological processes, such as DNA damage, encompassing metabolism, apoptosis, differentiation, proliferation, and cell cycle as well as drug resistance (21–23). Therefore, any dysregulation of miRNA function and aberrant expression may lead to the occurrence of pathological events, particularly cancer. In fact, the abnormal expression of miRNAs has been observed in a number of cancer types, including GC. MiRNAs act as oncogenes or tumor suppressors to play crucial roles in cancer progression (24). Moreover, increasing evidence suggests that miRNAs are closely associated with drug resistance of GC, and some of miRNAs possess great potential as novel biomarkers and therapeutic targets in reversing drug resistance in GC (25). However, further studies are still required to elucidate the detailed mechanism of miRNAs in the drug resistance of GC.
Implication of MiRNAs in GC Drug Resistance
Drug resistance is the most critical obstacle in GC effective treatment, blocking novel therapies and bringing about huge financial burden to patients and their families (26). The underlying mechanisms of drug resistance are very complicated and have not been fully elucidated. MiRNAs play crucial roles in the development of drug resistance in GC progression (Figure 2). For instance, miRNA expression profiles are associated with drug resistance (27), suggesting the potential of miRNA analysis as a valuable tool in precisely assessing the sensitivity of cancer cells to chemotherapy in GC treatment. MiRNAs participate in drug resistance by modulating the drug targets of multiple cellular pathways involved in the response to chemotherapy (28, 29). However, the detailed mechanisms of miRNA in drug resistance in GC are still inconclusive and are required to be elucidated.
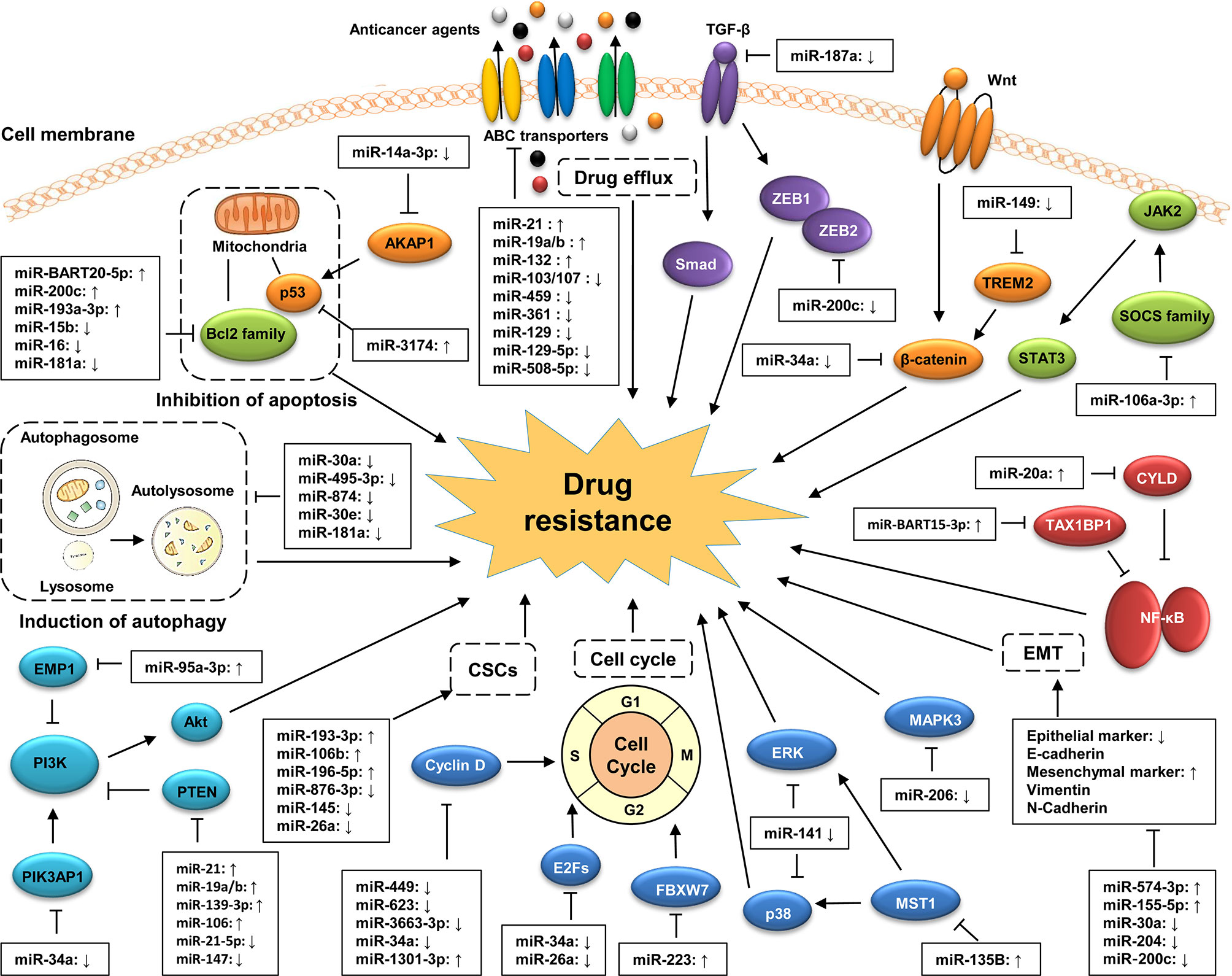
Figure 2 Classic mechanisms of miRNA involved in the drug resistance of GC. MiRNAs participate in the development of GC drug resistance by regulating multiple biological processes of GC cells, including drug efflux, cell apoptosis, cell cycle, and EMT as well as the acquisition of cancer stem cell (CSC) properties. MiRNAs also involves in the regulation of GC drug resistance by targeting cancer-related signaling pathways, such as the PI3K/Akt, Wnt/β-catenin, MAPK, TGF-β/Smad, and NF-κB signaling pathways.
MiRNAs and Drug Resistance
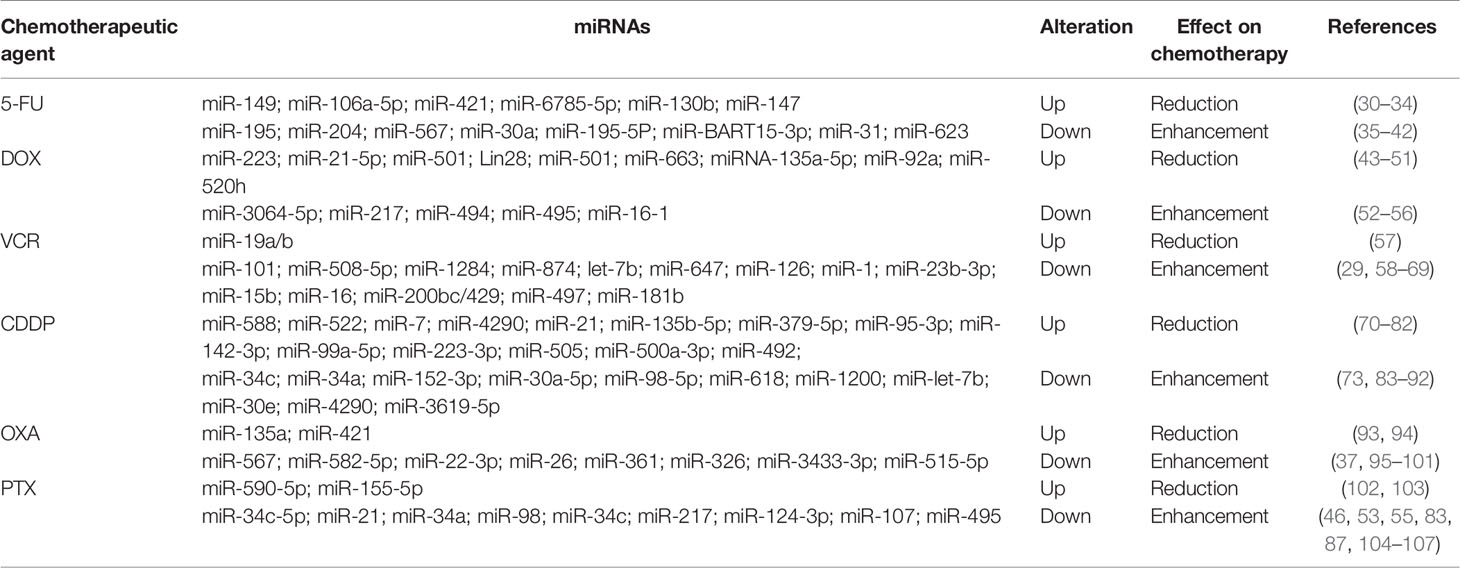
Currently, the first-line chemotherapeutic agents for GC mainly include 5-fluorouracil (5-FU), doxorubicin (DOX), vincristine (VCR), platinum drugs, and paclitaxel (PTX). The involvement of miRNAs in GC resistance to these drugs is summarized in Table 1.
MiRNAs and 5-FU Resistance
5-FU is a synthetic fluorinated pyrimidine analog with a fluorine atom at the C5 position in place of hydrogen. It shares a common structure with pyrimidine and can replace uracil to incorporate into RNA, leading to the disruption of RNA synthesis (108). Moreover 5-FU can also disrupt the intracellular deoxynucleotide pools by inhibiting thymidylate synthase (TS) required for DNA replication, thereby resulting in apoptosis and cell cycle arrest (109). 5-FU has been widely used to treat GC in clinic. However, the 5‐year survival rate of patients is still low due to the development of resistance to this chemotherapeutic agent. Multiple miRNAs are reported to be involved in the development of 5-FU resistance in GC. Several oncogenic miRNAs, such as miR-149, miR-130b, and miR-147, have been found to promote 5-FU resistance in gastric cancer (30, 33, 34). For instance, Wang et al. revealed that miR-149 confers 5-FU resistance by inhibiting TREM2 expression and modulating β-catenin pathway in GC cells (30). Similarly, miR-130b enhances the resistance of GC cells to 5-FU through targeting CMPK1 (33). By contrast, multiple tumor suppressor miRNAs, such as miR-195 and miR-204, can reverse 5-FU resistance of GC (35, 36). For instance, the overexpression of miR-195 enhanced the sensitivity of GC cells to 5-FU by downregulating HMGA1 expression (35). Exogenous expression of miR-204 sensitized GC cells to 5-FU by inhibiting EMT process via targeting TGFBR2 (36).
MiRNAs and DOX Resistance
DOX, also known as adriamycin (ADR), is an anthracycline and one of the more effective chemotherapy agents applied for GC treatment. Clinically, it is commonly used in combination with other chemotherapy agents, such as 5-FU, VCR, and PTX (27). DOX could exert strong cytotoxic effects on tumor cells through different molecular mechanisms including inhibition of DNA topoisomerase II, intercalation of DNA, and generation of free radicals (110). Multiple miRNAs have been shown to be responsible for the development of DOX resistance in GC. Several oncogenic miRNAs can promote DOX resistance in GC. For instance, miR-21-5p was highly upregulated in GC cell lines, and its overexpression increased DOX resistance in GC cells by inhibiting gene expression of phosphatase and tensin homologue (PTEN) and TIMP3 (44)(X21). Exosomal miR-501 could also significantly enhance DOX resistance by silencing cell death inducer (BLID), and, thus, suppressing caspase-9/-3 and p-AKT (45). Conversely, several tumor suppressor miRNAs can reverse DOX resistance of GC. MiR-16-1, for instance, has been found to increase the sensitivity of GC cells to DOX via inhibiting FUBP1 expression (56). The overexpression of miR-494 could also enhance the sensitivity of GC cells to DOX by directly targeting phosphodiesterases 4D (PDE4D) expression (54).
MiRNAs and VCR Resistance
VCR is an alkaloid extracted from vinca, which suppresses tubulin polymerization and disable spindles formation, thereby causing mitosis arrest of tumor cells (111). VCR is one of the first-line chemotherapy agents for GC treatment and often used in combination with other chemotherapeutic drugs. Multiple miRNAs have been reported to be involved in VCR resistance of GC. For instance, oncogenic miR-19a/b was highly expressed in VCR-resistant GC cells. The overexpression of miR-19a/b significantly enhanced the resistance of GC cells to VCR by directly targeting PTEN expression (57). On the contrary, some tumor suppressor miRNAs, such as miR-101, miR-126, and miR-181b, have been found to reverse the resistance of GC cells to VCR. Bao et al. showed that miR-101 was downregulated in GC tissues and cell lines. Forced expression of miR-101 could promote the sensitivity of GC cells to VCR by downregulating the expression of P-gp via targeting ANXA2 (58). Wang et al. revealed that miR-126 overexpression increased the sensitivity of GC cells to VCR via suppressing EZH2 expression (63). In addition, Zhu et al. found that ectopic expression of miR-181b could sensitize GC cells to VCR-induced apoptosis by downregulating BCL2expression (69).
MiRNAs and Resistance to Platinum Drugs
Platinum drugs are a class of cell cycle non-specific chemotherapeutic agent that widely used in clinical treatment of cancer patients. Currently, five platinum chemotherapy analogues have been approved for use in clinic, including cisplatin (CDDP), oxaliplatin (OXA), carboplatin, nedaplatin, and lobaplatin. These drugs directly insert platinum into DNA to form cross-links, which is either removed by specific DNA repair processes or it triggers a signaling cascade resulting in apoptosis of cancer cells (112). It has been reported that multiple miRNAs are involved in resistance to platinum drugs in GC. Several oncogenic miRNAs can facilitate the resistance of GC cells to platinum drugs. For instance, exosomal miR-588 from M2 macrophages increased the resistance of GC cells to CDDP via inhibiting the expression of cylindromatosis (CYLD) (71). In another study, exosomal miR-522 derived from cancer-associated fibroblasts (CAFs) was found to promote acquired resistance of GC cells to CDDP by inhibiting ALOX15 expression (70). Additionally, miR-135a was highly expressed in GC samples. The overexpression of miR-135a could promote the resistance of GC cells to OXA by inhibiting E2F1 and DAPK2 expression (93). By contrast, multiple tumor suppressor miRNAs can reverse platinum drugs resistance of GC. For instance, miR-34c overexpression was found to promote the sensitivity of drug-resistant GC cells to CDDP combined with paclitaxel by increasing the expression of apoptosis-related proteins. Consistent with this, inhibition of miR-34c significantly decreased the sensitization of drug resistant GC cells to CDDP combined with paclitaxel (83). Furthermore, the overexpression of miR-7 was able to enhance the sensitivity of GC cells to CDDP by inhibiting LDH-A expression (72). In addition, Qian et al. showed that miR-4290 was significantly downregulated in both GC samples and cell lines. Overexpression of miR-4290 could increase the sensitivity of GC cells to CDDP by suppressing PDK1-mediated glycolysis (73).
MiRNAs and PTX Resistance
PTX, a class of taxanes, is the most successful and widely used chemotherapeutic agent. As a natural anticancer drug, PTX promotes the assembly of tubulin into microtubules and resists the dissociation of microtubules, blocking mitosis progression, which resulting in programmed cell death (113). PTX is frequently used as the first-line treatment drug in GC. However, the resistance of GC to PTX is a great obstacle in its clinical applications. It has been reported that multiple miRNAs are involved in PTX resistance. Oncogenic miRNAs can promote PTX resistance. For instance, miR-590-5p was highly expressed in GC tissues and cells. Overexpression of miR-590-5p significantly decreased the sensitivity of GC cells to PTX by inhibiting RECK expression. Opposite result was obtained in miR-590-5p knockout GC cells (102). In another study, exosomal miR-155-5p derived from PTX-resistant GC cells has been found to promote drug resistance in PTX-sensitive GC cells by inhibiting the expression of GATA3 and TP53INP1 (103). On the contrary, several tumor suppressor miRNAs are able to reverse the resistance of GC cells to PTX. For instance, low expression of miR-34c-5p was observed in PTX-resistant GC tissues. Overexpression of miR-34c-5p significantly promoted the chemosensitivity of PTX-resistant GC cells (104). Tumor suppressor miR-21 has also been found to enhance the sensitivity of GC cells to PTX, at least in part, by modulating P-glycoprotein expression (105). In addition, overexpression of miR-34a and miR-98 could sensitize GC cells to PTX by suppressing the expression of E2F5 and BCAT1, respectively (87, 106).
Expression Profiles of Drug Resistance-Related MiRNAs in GC
With the rapid development of detecting techniques and bioinformatics, a large number of miRNAs have been identified in GC. Many of them have aberrant expression levels and are closely correlated with the drug resistance of GC, indicating their great potential in predicting survival and response of patients to therapy (26). For instance, Sun et al. performed a microarray analysis to detect the miRNA expression profiles of GC cell-derived exosomes. They found that miR-106a-5p, miR-421, miR-19b-3p, miR-133a, and miR-214 are upregulated, whereas miR-144, miR-16-5p, miR-100, miR-30a-5p, and miR-361-5p are downregulated in MGC-803/5-FU exosomes compared with those in MGC-803 exosomes. High levels of miR-106a-5p and miR-421 in exosomes indicate 5-FU resistance in GC (31). Zhang et al. showed that 68 miRNAs were differentially expressed in SGC-7901/CDDP cells compared to SGC-7901 cells, including 41 upregulated miRNAs and 27 downregulated miRNAs. In BGC-823 and BGC-823/CDDP cells, 94 differently expressed miRNAs were observed, including 40 upregulated miRNAs and 54 downregulated miRNAs in BGC-823/CDDP cells. Among these differently expressed miRNAs, high levels of miR-99a and miR-491 indicate CDDP resistance in GC (114). In another study, using the Gene Expression Omnibus (GEO) database and GEO2R analysis, Zhou et al. confirmed 244 differentially expressed miRNAs in drug-resistant GC patients compared with in GC patients, among which 1120 were upregulated, and 124 were downregulated (115). Furthermore, Wei et al. reported 48 differentially expressed miRNAs (more than two-fold) in SGC7901/DDP cells when compared with SGC7901 cells, including 19 upregulated miRNAs and 29 downregulated miRNAs (116). Due to several factors, such as the heterogeneity of cancer cells, differences in research strategies, and differences in criteria for selecting significant miRNA profile data, the expression profiles of drug resistance-related miRNAs in GC may vary among studies. However, these studies still provide researchers with a new direction in showing that these altered miRNAs may be used to predict drug response to chemotherapy for GC patients.
MiRNAs Regulate the Drug Resistance of GC by Targeting Signaling Pathways
An accumulating amount of evidence has shown that miRNAs are involved in the regulation of GC drug resistance by targeting cancer-related signaling pathways, such as the phosphatidylinositol 3-kinase (PI3K)/AKT, mitogen-activated protein kinase (MAPK), Wnt/β−catenin, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and signal transducers and activators of transcription (STAT) signaling pathways. MiRNAs can change the chemotherapeutic sensitivity of GC via modulating the function or expression of some components of these pathways (117). Understanding the mechanisms of miRNAs in signaling pathway regulation may facilitate the development of new therapeutic strategies against GC drug resistance.
The PI3K/AKT signaling pathway is involved in the regulation of various cellular functions during cancer progression, such as proliferation, apoptosis, and metastasis (118, 119). This pathway also confers drug resistance to various types of cancer, including GC (120). Some miRNAs have been shown to mediate the drug resistance of GC by targeting the PI3K/AKT signaling pathway. For instance, Zhang et al. revealed that miR-567 enhances the sensitivity of GC cells to 5-FU and oxaliplatin. Mechanistically, miR-567 inhibits PI3K/AKT/c-Myc signaling pathway by blocking PIK3AP1 activity. Interestingly, c-Myc inversely regulates the expression of miR-567, leading to the formation of a miR-567-PIK3AP1- PI3K/AKT-c-Myc feedback loop (37). Ni et al. found that miR-95-3p activates the PI3K/AKT signaling pathway by upregulating the expression of p-PI3K and p-AKT, leading to the enhancement of CDDP resistance of GC (76). PTEN is a key negative regulator of the PI3K/AKT signaling pathway (121). Shen et al. showed that miR-147 is upregulated in GC tissues and cell lines. MiR-147 decreases the sensitivity of GC cells to 5-FU by targeting PTEN to activate the PI3K/AKT signaling pathway (34). In addition, high levels of exosomal miR-21 derived from M2 macrophages have also been demonstrated to facilitate CDDP resistance in GC cells by enhancing the activation of the PI3K/AKT signaling pathway via the downregulation of PTEN (74).
The MAPK signaling pathway is a classical carcinogenic pathway, and its aberrant activation has been shown to promote the initiation and progression of many cancer types, including GC (122, 123). It has been reported that some miRNAs contribute to the drug resistance of GC by regulating the expression of key components in the MAPK signaling pathway. For instance, Chen et al. found that low expression of miR-206 is closely correlated with the CDDP resistance of GC cells. The overexpression of miR-206 inhibits the proliferation of drug-resistant GC cells and decreases CDDP resistance by downregulating the expression of MAPK3 and p-MAPK3 (124). Another study showed that the downregulation of miR-135b enhances the CDDP sensitivity of GC cells. Mechanistically, miR-135b increases the expression of p-p38MAPK p38MAPK, p-ERK1/2, and ERK1/2 by targeting mammalian ste20-like kinase 1 (MST1), leading to the activation of the MAPK signaling pathway (125). Moreover, miR-20a is reported to promote the multidrug resistance (MDR) of GC cells by activating the epidermal growth factor receptor-mediated PI3K/AKT and MAPK/ERK signaling pathways by targeting LRIG1 (126). In addition, miR-27a-5p has been shown to enhance the sensitivity of GC cells to DOX by inhibiting the MAPK and AKT signaling pathways by targeting apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1) (127).
The Wnt/β−catenin signaling pathway plays crucial roles in maintaining the fundamental function of cells. Dysregulation of the Wnt/β−catenin signaling pathway has been widely observed in multiple types of cancer, including GC. Wang et al. showed that the overexpression of miR-149 enhances the 5-FU resistance of GC cells by activating the Wnt/β−catenin signaling pathway by targeting TREM2 (30). In another study, Chen et al. revealed that miR-34a mediates the enhancement of lncRNA HOTAIR on the CDDP resistance of GC cell lines by inactivating the Wnt/β−catenin signaling pathway (128). MiRNAs can also modulate the drug resistance of GC by targeting the transforming growth factor (TGF)-β signaling pathway. For instance, the expression of miR-187 is negatively associated with the CDDP-resistance of GC cells. The overexpression of miR-187 inhibits CDDP resistance in GC cells by downregulating the expression of TGF-β1 and p-Smad4 to inactivate the TGF-β/Smad signaling pathway (129). Additionally, miR-200c overexpression decreases the resistance of GC cells to trastuzumab by suppressing the TGF-β/Smad signaling pathway via downregulating zinc finger E-box-binding homeobox 1 (ZEB1) and ZEB2 expression (130). In addition, miR-362 and miR-20a have been reported to activate the NF-κB signaling pathway and upregulate the expression of NF-κB-regulated genes by targeting CYLD, leading to the enhancement of the CDDP resistance of GC cells (131, 132). In a study by Guo et al., miR-106a-3p was found to increase the apatinib resistance of GC cells by targeting SOCS genes (SOCS2, SOCS4, and SOCS5) to activate the JAK2/STAT3 signaling pathway (133). Taken together, these studies suggest that targeting the cancer-related signaling pathways is a common miRNA regulation mechanism for miRNAs in GC drug resistance. Understanding the mechanisms of miRNAs in the regulation of the GC signaling pathway may thus provide new insights on therapeutic strategies against GC drug resistance.
MiRNAs and Drug Efflux in GC
Excessive drug efflux is one of the classical mechanisms of the generation of drug resistance during cancer treatment. Human ATP-binding cassette (ABC) transporters belong to the P-type membrane ATPase superfamily and are closely associated with excessive drug efflux (26). It has been reported that ABC transporters are usually highly expressed in drug-resistant cancer cells, and their overexpression promotes the efflux of excessive intracellular drugs, leading to the impairment of chemotherapeutic effects (134, 135). An increasing number of studies have suggested that miRNAs are involved in the regulation of excessive drug efflux in GC cells by modulating ABC transporters.
Permeability glycoprotein (P-gp), also known as ABCB1 and MDR1, was the first identified ABC transporter closely associated with MDR. Currently, a series of miRNAs, such as miR-103/107, miR-459, miR-361, miR-21, miR-19a/b and mir-129, have been shown to regulate the resistance of GC cells to chemotherapeutic drugs by directly or indirectly targeting P-gp (55, 57, 98, 105, 136, 137). Furthermore, Zhang et al. found that miR-132 is upregulated in Lgr5+ GC cells with stem cell-like features, and the overexpression of miR-132 promotes the CDDP resistance of these Lgr5+ GC cells. Mechanistically, miR-132 increases the expression of ABCG2 by targeting SIRT1 to downregulate the deacetylation of CREB (138). In addition, Wu et al. reported that the overexpression of miR-129-5p decreases the drug resistance of GC cells by targeting MDR-related ABC transporters, including ABCB1, ABCC5, and ABCG1, whereas the silencing of miR-129-5p shows the opposite effect (139). Shang et al. showed that miR-508-5p overexpression sufficiently reverses the resistance of GC cells to multiple chemotherapeutics and enhances the sensitivity of tumors to chemotherapy by targeting ABCB1 and Zinc ribbon domain-containing 1 (29). Collectively, these findings suggest that miRNAs play a key role in regulating the drug efflux of GC cells, which may provide new insight in the investigation of the roles of miRNA in the drug resistance of GC cells.
MiRNAs Regulation of Apoptosis in GC Drug Resistance
Inducing the apoptosis of cancer cells is one of the main roles of chemotherapeutic drugs. Therefore, the dysregulation of apoptosis (or its evasion) is commonly characterized as a crucial hallmark of GC drug resistance (140). This may be the result of the aberrant expression of crucial apoptotic proteins or the dysregulation of apoptotic pathways. A large number of studies have shown that miRNAs are involved in the regulation of apoptosis in drug-resistant GC cells by targeting apoptotic proteins or pathways (26, 27, 141).
B-cell lymphoma-2 (BCL-2) family proteins are well-known proteins that regulate mitochondrial apoptosis and are closely associated with chemotherapy resistance (142, 143). Due to their distinct effects on apoptosis, BCL-2 proteins are classified into anti-apoptotic (e.g., Bcl-2, Bcl‐XL, and Mcl-1) and pro-apoptotic categories (e.g., Bax, Bad, and Bak) (134, 144). Xia et al. found that miR-15b and miR-16 are downregulated in GC cells. The overexpression of these two miRNAs enhances the sensitivity of GC cells to VCR-induced apoptosis by directly targeting Bcl-2 (66). A study of miR-BART20-5p in Epstein-Barr virus-associated gastric carcinoma showed that miR-BART20-5p promotes the resistance of gastric carcinoma cell line AGS to 5-FU and docetaxel by downregulating the BAD expression (145). Chen et al. showed that miRNA-200c significantly decreases Bax expression and increases Bcl-2 expression by targeting E-cadherin, leading to the inhibition of the resistance of GC cells to 5-FU, PTX, and ADR (146). In our previous work, miR-185 was found to be downregulated in GC tissues and cell lines. The overexpression of miR-185 enhances the sensitivity of GC cells to low-dose CDDP or DOX by targeting apoptosis repressors with a caspase recruitment domain. Conversely, silencing miR-185 inhibits high-dose chemotherapy-induced apoptosis (147). In another study, we showed that miR-633 promotes DOX/CDDP resistance in GC cells by decreasing the expression of Fas-associated proteins with the death domain. Forkhead box O 3 is an upstream regulator of miR-633 that can directly inhibit miR-633 transcription by binding to its promoter region (48). In addition, we also revealed that miR-422a mediates the facilitation effect of lncR-D63785 on the sensitivity of GC cells to DOX (148).
P53 is a well-studied tumor suppressor that mediates major apoptotic pathways to protect cells from malignant transformation. The dysregulation or mutation of p53 contributes to the development of chemotherapeutic drug resistance in cancer treatment (149). Li et al. showed that miR-148a-3p enhances the sensitivity of GC cells to CDDP by promoting mitochondrial fission-induced apoptosis. Mechanistically, miR-148a-3p promotes the activation of P53 to induce DRP1 dephosphorylation by targeting AKAP1, leading to mitochondrial fission and apoptosis in GC cells (150). They also found that the high expression of miR-3174 is closely associated with CDDP resistance in GC cells. MiR-3174 decreases the expression of p53 to inhibit Bax trans-activation by targeting ARHGAP10, thereby suppressing mitochondria-dependent apoptosis (151). In addition, Lee et al. revealed that miR‐193a‐3p triggers the resistance of CD44‐positive GC stem cells against CDDC by regulating the mitochondrial apoptosis pathway. Mechanistically, the high expression of miR‐193a‐3p decreases the expression of Bax, cytochrome C, cleaved caspase 3, and cleaved caspase 9 and increases the expression of Bcl‐XL and Bcl‐2 via targeting SRSF2, leading to the enhancement of CDDC resistance in CD44‐positive GC stem cells (152). Furthermore, miR-20a is reported to inhibit apoptosis in CDDC-resistant GC cells by activating the NF-κB pathway. The overexpression of miR-20a upregulates the expression of p65, livin, and survivin by targeting NFKBIB (also known as IκBβ) (132).
MiRNAs Are Involved in GC Drug Resistance by Modulating Autophagy
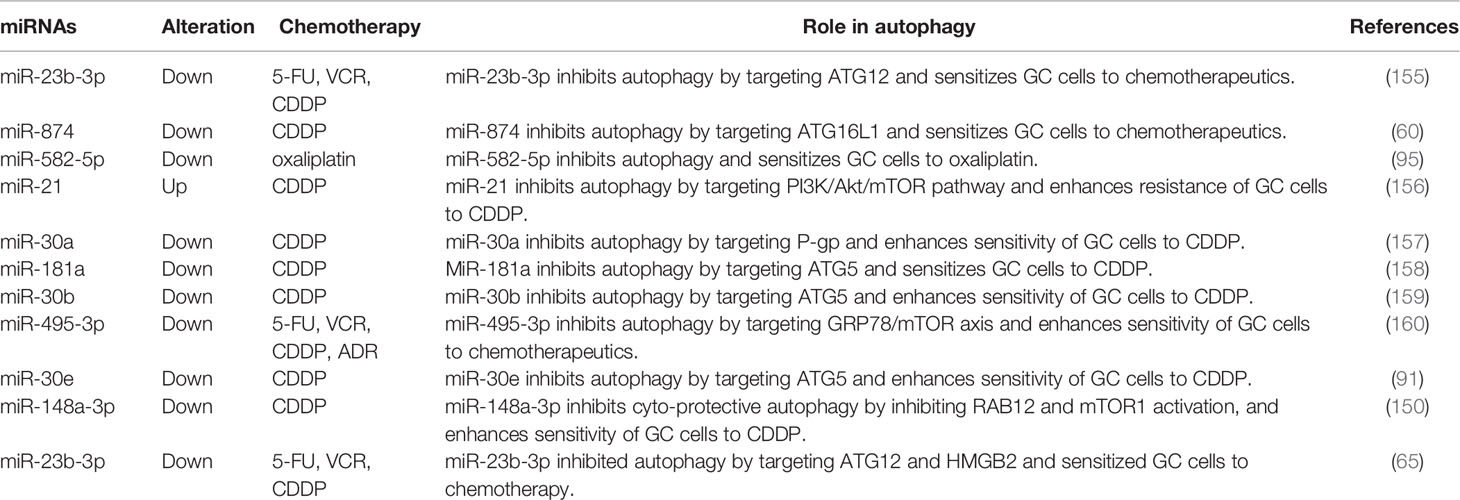
Autophagy is a crucial intracellular degradation system that protects cells from the damage of stressors, such as hypoxia and nutrient deprivation (153). It has been reported that chemotherapy-induced autophagy contributes to the acquired drug resistance of cancer cells by helping them escape from deadly cell damage (154). Increasing evidence shows that miRNAs regulate the drug resistance of GC cells by targeting autophagy-related genes (Table 2). For instance, the overexpression of miR-30a is found to inhibit chemoresistance-associated autophagy in GC cells by downregulating the expression of light chain (LC)3-II (157). Another study revealed that miR-495-3p overexpression enhances the sensitivity of GC MDR cells to chemotherapy by inhibiting autophagy via activating mTOR (a key upstream mediator of autophagy) and targeting GRP78 (160). Moreover, Huang et al. showed that the overexpression of miR-874 enhances the sensitivity of GC cells to chemotherapy by inhibiting the targeting of autophagy occurrence by targeting autophagy-related 16-like 1 (60). Additionally, miR-30e and miR-181a have been shown to enhance the sensitivity of GC cells to chemotherapeutic agents by inhibiting chemo-induced autophagy via targeting ATG5 (91, 158). Research on the function of miRNAs in regulating chemotherapy-induced autophagy is currently limited. Further studies are thus required to elucidate its exact mechanisms.
MiRNAs Control Drug Resistance in GC by Modulating Cancer Stem Cell Features
CSCs are recognized as the main cause of chemotherapeutic drug resistance due to their unique characteristics, such as their high DNA repair ability, genomic instability, and overexpression of ABC transporters (161, 162). CSCs are closely associated with the proliferation, metastasis, and recurrence of cancer. An increasing amount of evidence has shown that miRNAs are involved in the regulation of GC drug resistance by affecting CSC properties. For instance, Peng et al. showed that miR-876-3p is downregulated in CDDP-resistant GC cells and its low expression is closely associated with the CDDP resistance of GC. MiR-876-3p confers sensitivity to CDDP-resistant GC cells. Mechanistically, the overexpression of miR-876-3p downregulates the expression of Sox-2, Oct-4, CD133, and CD44 by targeting TMED3, thereby inhibiting the stem cell-like features of GC cells (163). Zeng et al. revealed that the expression of miR-145 is decreased in GC cells. The overexpression of miR-145 enhances the sensitivity of GC cells to chemotherapeutic drugs by inhibiting the stem-like properties of GC via directly targeting CD44 (164). Lee et al. reported that miR‐193a‐3p overexpression promotes the resistance of CD44‐positive gastric CSCs to CDDC by modulating the mitochondrial apoptosis pathway via targeting SRSF2 (152). In addition, some miRNAs, such as miRNA-106b, miR‐196a‐5p, and miR-26a, are also found to regulate GC stem-like cell properties (165–167), indicating their great potential in regulating the drug resistance of GC.
MiRNAs and EMT in GC Drug Resistance
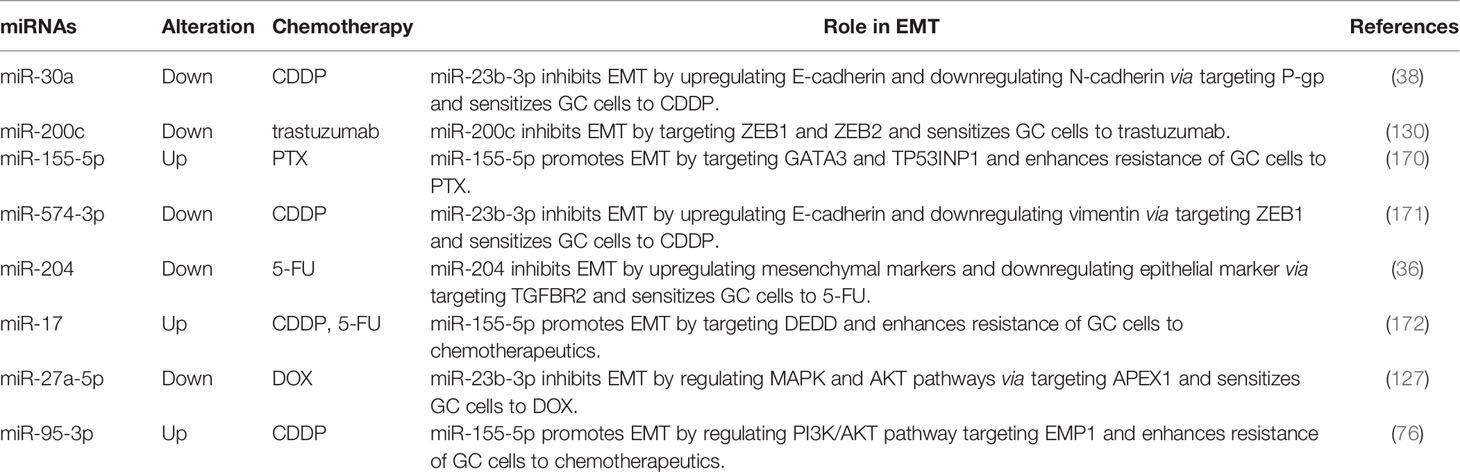
EMT is a morphogenetic process that changes epithelial cells from a pebble-like phenotype to a fibroblast-like phenotype, which endows cells with migratory and invasive properties. During the EMT process, the mesenchymal markers (e.g., Vimentin, N-Cadherin) are upregulated, whereas the epithelial markers (e.g., E-cadherin) are downregulated (168). It has been reported that the aberrant activation of EMT contributes to the development of drug resistance in cancer by enabling the conversion of non-CSCs into CSCs (169). An increasing number of studies have suggested that miRNAs play crucial roles in drug resistance by directly targeting the EMT process (Table 3). For instance, Wang et al. showed that miR-30a is downregulated in CDDP-resistant GC cells. The overexpression of miR-30a increases the CDDP sensitivity of GC cells by inhibiting EMT via downregulating the Snail and Vimentin levels. GC cells with miR-30a knockdown show decreased sensitivity to CDDP (173). Li et al. revealed that miR-204 is downregulated in 5-FU-resistant GC cells, with the epithelial markers (E-cadherin) decreased and the mesenchymal markers (N-cadherin, Fibronectin, Twist, and Snail) increased. The overexpression of miR-204 sensitized GC to 5-FU by inhibiting TGF-β-induced EMT via targeting TGFBR2 (36). ZEB1 is a crucial EMT-inducing transcription factor. Wang et al. found that miR-574-3p overexpression inhibits the CDDP resistance of GC cells. Mechanistically, the overexpression of miR-574-3p increases E-cadherin expression and decreases vimentin expression by targeting ZEB1 via binding to its 3’-UTR, thereby enhancing the sensitivity of GC cells to CDDP (171). Moreover, miR‐200c overexpression is found to enhance the trastuzumab sensitivity of GC cells by inhibiting ZEB1 and ZEB2 (130). Additionally, Wang et al. showed that the overexpression of exosomal miR-155-5p derived from PTX-resistant GC cells induces EMT progress and enhances drug resistance in PTX-sensitive GC cells by targeting GATA binding protein 3 (GATA3) and tumor protein p53-inducible nuclear protein 1 (TP53INP1) (103). Collectively, these findings suggest that miRNAs are crucial regulators determining the fate of drug-resistant GC cells by targeting the EMT process.
MiRNAs Influence Cell Cycle Progression in GC Drug Resistance
Cell cycle alteration is one of the main processes involved in drug resistance. The dysregulation of the cell cycle may lead to drug resistance (28, 174). Therefore, targeting cell cycle progression factors may provide new insight into therapeutic strategies for cancer. Many miRNAs have been proven to be involved in GC drug resistance by regulating cell cycle progression. Cyclin D1 is a major regulator of cell cycle progression that governs the entrance of a cell from the G1 phase into S (175). Hu et al. revealed that miR-449a is downregulated in both GC tissues and cell lines. The overexpression of miR-449a suppresses proliferation and promotes CDDP-mediated apoptosis in GC cells. They further showed that miR-449a reduced the percent of S phase cells and increased the percent of G1/G0 phase cells by targeting BCL2 and cyclin D1, leading to the enhancement of the CDDP sensitivity of GC cells (176). In another study, Jiang et al. showed that miR-623 is downregulated in both GC tissues and cell lines. The overexpression of miR-623 enhances the sensitivity of GC cells to 5-FU by targeting Cyclin D1 (42). In addition, some miRNAs, such as miR-1301-3p, miR-3663-3p, and miR-34a, are also found to regulate cell cycle progression by targeting cyclin D1, indicating their potential role in the drug resistance of GC (177–179). F-box and WD repeat domain-containing 7 (FBXW7) is a classical tumor suppressor that promotes the ubiquitination and degradation of several oncoproteins, such as Cyclin E, c-MYC, and c-JUN (180). Zhou et al. demonstrated that miR-223 is up-regulated in CDDP-resistant GC cells. The knockdown of miR-223 enhances the sensitivity of resistant GC cells to CDDP by inducing cell arrest in the G0/G1 phase. Mechanistically, miR-223 modulates the cell cycle of GC cells by targeting FBXW7 via binding to its 3’-UTR, thus affecting the sensitivity of the GC cells to CDDP (181). The early region 2 binding factor (E2F) family of transcription factors are well-studied major transcriptional regulators of cell cycle-dependent gene expression (182). Wen et al. showed that miR-26a is downregulated in CDDP-resistant GC cells. Function analysis demonstrated that miR-26a improves the sensitivity of GC cells to CDDP by targeting E2F2 and NRAS (183). Another study revealed that the overexpression of miR-34a enhances the sensitivity of GC cells to PTX by targeting E2F5 (106). Taken together, these findings provide new insights into the mechanisms of GC drug resistance mediated by miRNAs involved in cell cycle regulation, which may improve chemotherapy effectiveness.
MiRNAs and T Cells in GC Drug Resistance
T cells are the major effector cells in tumor immunity and produce cytokines in immune responses to mediate inflammation and regulate other types of immune cells (184, 185). It has been reported that the immune escape and immune tolerance induced by the dysregulation of cytotoxic T cell activity are closely associated with drug resistance in cancer (186). Some studies suggested that miRNAs are involved in drug resistance by regulating T cell activity in multiple cancer types. Xu et al. demonstrated that the restoration of miR-424 (322) expression in chemoresistant ovarian cancer cells could activate the T-cell immune response by regulating the production of CD8+ T, MDSC, and Treg cells via targeting PD-L1, resulting in the reversal of drug resistance (187). Qian et al. showed that the overexpression of miR-101 inhibits cell proliferation and invasion and induces apoptosis by targeting Notch1 in T-cell acute lymphoblastic leukemia (T-ALL) cells. Further analysis revealed that miR-101 enhances the sensitivity of T-ALL cells to the chemotherapeutic agent ADR (188). MiRNAs can also regulate GC progression by influencing T cells. For instance, miR−140 plays an anti-tumoral effect in GC by increasing cytotoxic CD8+ T cell and reducing myeloid-derived suppressive and regulatory T cell infiltration (189). Exosomal miRNA-16-5p derived from M1 macrophages inhibits GC development through activation of T cell immune response via PD-L1. These findings strongly indicate that miRNAs may participate in the development of GC drug resistance by regulating T cells. Thus, in-depth studies are required to elucidate the mechanism of miRNAs in regulating T cells in GC drug resistance, which may provide new insights into the development of miRNA-based therapeutics strategies in GC.
Clinical Applications of Drug Resistance-Related MiRNAs in GC
MiRNAs as Diagnostic and Prognostic Biomarkers
Currently, most GC patients are still diagnosed at an advanced stage with poor prognosis due to the lack of an effective approach for early detection and prognostic evaluation in clinical practice. Some protein biomarkers, such as CEA, uPA, and CA 19-9, have been applied in clinic, but the low specificity and sensitivity of these biomarkers limit their further utilization (190–192). Therefore, it is important to screen and identify novel biomarkers for the early detection and prognostic evaluation of GC patients, particularly those demonstrating a poor response to chemotherapy.
An increasing number of studies suggest that miRNAs possess great potential to be biomarkers for the diagnosis and prognosis of GC patients. In a latest clinical trial consist of 5248 GC and control subjects, So et al. developed a clinical diagnostic assay for GC from a high-risk population based on a serum 12-miRNA biomarker panel. In the discovery cohorts, the 12-miRNA panel has an area under the curve (AUC) of 0.93 in discriminating early GC patients from normal controls. Excitingly, the AUC value also reached 0.92 in the verification cohorts. Further prospective study revealed that the AUC value for the 12-miRNA panel was 0.848, which is higher than HP serology (0.635), PG 1/2 ratio (0.641), PG index (0.576), ABC method (0.647), CEA (0.576), and CA19-9 (0.595). Moreover, the overall sensitivity of the 12-miRNA assay was 87.0% at specificity of 68.4% (193). These data strongly suggest that serum 12-miRNA panel is a promising biomarker with higher GC diagnostic accuracy than traditional serum-based biomarkers. In another study consist of 354 GC patients, Shimura et al. identified a miRNA-based signature (including miR-30a-5p, -134-5p, -337-3p, -659-3p, and -3917), which can be used as a biomarker to identify peritoneal metastasis (PM) in GC patients. The AUC value for the combination miRNA signature was 0.82 in distinguishing GC patients with versus without PM. In an independent validation cohort, the AUC value reached 0.74 (194).
It has been reported that miRNAs are involved in drug resistance and may serve as promising biomarkers for predicting drug resistance and prognosis in GC (26) (Table 4). For instance, Jin et al. found that miR-3180-3p is significantly upregulated and miR-124-3p is downregulated in both drug-resistant GC patients and cell lines. The combination of the two miRNAs can effectively distinguish drug-resistant GC patients from drug-sensitive GC patients (AUC = 0.946 ± 0.023, p < 0.001), indicating their potential as serum-based biomarkers in predicting the therapeutic benefit of CDDP in GC (204). Ji et al. showed that miR-374a-5p is upregulated in GC serum, and its upregulation predicts poor prognosis of GC patients. The overexpression of miR-374a-5p promotes the drug resistance of GC cells by targeting Neurod1, indicating the great value of miR-374a-5p as a biomarker for GC diagnosis and prognosis (195). Besides, miR-1229-3p is significantly upregulated in the plasma of GC patients, and its upregulation is an independent poor prognostic factor for recurrence-free survival. The overexpression of miR-1229-3p promotes the significant drug resistance of GC cells to 5-FU both in vitro and in vivo. This data indicate that plasma miR-1229-3p can serve as a clinically useful biomarker for predicting drug resistance to 5-FU in GC patients (200).
In addition, Huang’s group found that partial response rates of GC patients with high miRNA27a expression and low miRNA27a expression to fluoropyrimidine‐based chemotherapy to be 7.7% and 25.9%, respectively (P = 0.018). GC patients with high miRNA27a expression have a significantly worse overall survival (OS) than those with lower miRNA27a expression (P = 0.024). These results suggest that miRNA27a is a promising biomarker for predicting resistance to fluoropyrimidine‐based chemotherapy and a novel prognostic biomarker for metastatic or recurrent GC (201). MiR-21 in both tumor tissue and plasma is significantly upregulated in drug-resistant GC patients compared to the drug-sensitive GC patients (p < 0.001). ROC analysis showed that the expression of miR-21 in tissue distinguished drug-resistant GC patients from drug-sensitive GC patients, with an AUC of 0.830, 88.0% sensitivity, and 68.7% specificity; moreover, the expression of miR-21 in tissue plasma distinguished drug-resistant GC patients from drug-sensitive GC patients, with an AUC of 0.759, 52.0% sensitivity, and 88.1% specificity. Moreover, GC patients with high miR-21 expression exhibit shorter OS time than patients with low miR-21 expression, indicating that miR-21 might be a promising biomarker for identifying metastatic GC with drug resistance (202).
In a recent study of Jin et al., they found that miR-9-3p, miR-9-5p, miR-146a-5p, and miR-433-3p were closely associated with chemotherapy responses in GC patients and cells. MiR-9-5p distinguished drug-resistant GC patients from drug-sensitive GC patients with an AUC of 0.856 and p < 0.0001; moreover, the combination of miR-9-3p, miR-146a-5p, and miR-433-3p distinguished drug-resistant GC patients from drug-sensitive GC patients, with an AUC of 0.915, and p < 0.0001, suggesting the great potential of miR-9-5p and the combined group (miR-9-3p, miR-146a-5p, and miR-433-3p) as serum-based biomarkers distinguishing drug-resistant GC (199). These findings strongly suggest that miRNAs possess great potential as valuable biomarkers for predicting the drug response and prognosis of GC. However, more in-depth studies are required to overcome their limitations in clinical application using high-quality samples and larger patient cohorts.
Therapeutic Potential of MiRNAs in GC Drug Resistance
A single miRNA can simultaneously modulate several genes by targeting different mRNAs, and one gene can also be regulated by multiple miRNAs, indicating that miRNAs possess great potential as effective therapeutic targets or therapeutic agents in cancer treatment. Aberrantly expressed miRNAs have been shown to play crucial roles in the development of GC drug resistance. Therefore, correcting these miRNA deficiencies by either antagonizing or restoring miRNA functions may provide new insights into the development of therapeutic strategies for reversing GC drug resistance. Currently, the main miRNA-based therapies contain nanoparticles, miRNA mimics, miRNA inhibitors, locked nucleic acid, decoy vectors, DNA sponge, antagomiRs, and small compounds (205–208). Wang et al. found that exosomes can act as nanoparticles to deliver anti-miR-214 to reverse the resistance of GC cells to CDDP. Exosome-delivered anti-miR-214 is maintained at a stable level in the blood and downregulated the expression of miR-214 in the tumor tissues of mice, thereby enhancing the sensitivity of refractory GC to CDDP (209). Ghasabi et al. showed that miR-200c mimics significantly reduced the resistance of GC cells to CDDP and increased CDDP-induced apoptosis by targeting RhoE (210). Ji et al. revealed that the knockdown of miR-374a-5p using miR-374a-5p inhibitor repressed the resistance of GC cells to oxaliplatin and promoted cell apoptosis induced by oxaliplatin (195). In addition, miR-21 has been found to confer CDDP resistance in GC. The miR-21 inhibitor sensitized CDDP-resistant GC cells by inducing autophagy via the PI3K/Akt/mTOR pathway (156). These findings support that miRNA could become effective therapeutic target or therapeutic agent reversing GC drug resistance. However, miRNA-based therapies have not been translated into GC clinical treatment. More detailed and in-depth studies are still required to investigate the roles of miRNAs in GC drug resistance.
Conclusion and Perspective
Chemotherapy is still the first-line standard treatment option for cancer patients. However, the development of drug resistance results in chemotherapy failure and cancer recurrence, both of which seriously threaten patients’ health and lives. In recent years, increasing numbers of miRNAs have been found to be aberrantly expressed in drug-resistant GC tissues. Most have been shown to be involved in the regulation of GC sensitivity or resistance to chemotherapeutic agents by influencing various aspect of GC cell function, including drug efflux, apoptosis, autophagy, EMT, CSCs, and cell cycle. Moreover, a complex cross-talk network has been observed between miRNAs and several drug resistance-related signaling pathways in GC (Figure 2).
MiRNAs have been shown to be crucial regulators of several oncogenic pathways during GC progression. Moreover, miRNAs are small in size. Thus, it is easier to design specific drugs targeting them or deliver them to target tissues. These features strongly suggest that miRNAs are ideal therapeutic targets for GC patients. In the other hand, miRNAs in plasma/serum are well protected from RNases, they remain stable under harsh conditions (211), which endows them with great potential as biomarkers for the early diagnosis and prognostic evaluation of GC patients. Therefore, the identification of drug resistance-related miRNAs and the investigation of their mechanisms in GC progression are crucial for designing novel effective therapeutic strategies for GC patients, especially those exhibiting a poor response to chemotherapy. In addition, the combination of miRNAs with existing chemotherapeutic agents may provide a new option for maximizing therapeutic effects and improving clinical outcomes in GC patients (Figure 3). However, there are still some challenges in translating the findings on miRNA-mediated drug resistance in GC into clinical utilization, such as side effects, minimized off-target effects, and the modes of targeted delivery. Nevertheless, miRNA-based therapies may provide promising therapeutics for GC patients to overcome drug resistance and increase their survival in the future. Further studies are needed to clarify the exact mechanism of miRNAs in the regulation of GC drug resistance using large-scale clinical trials.
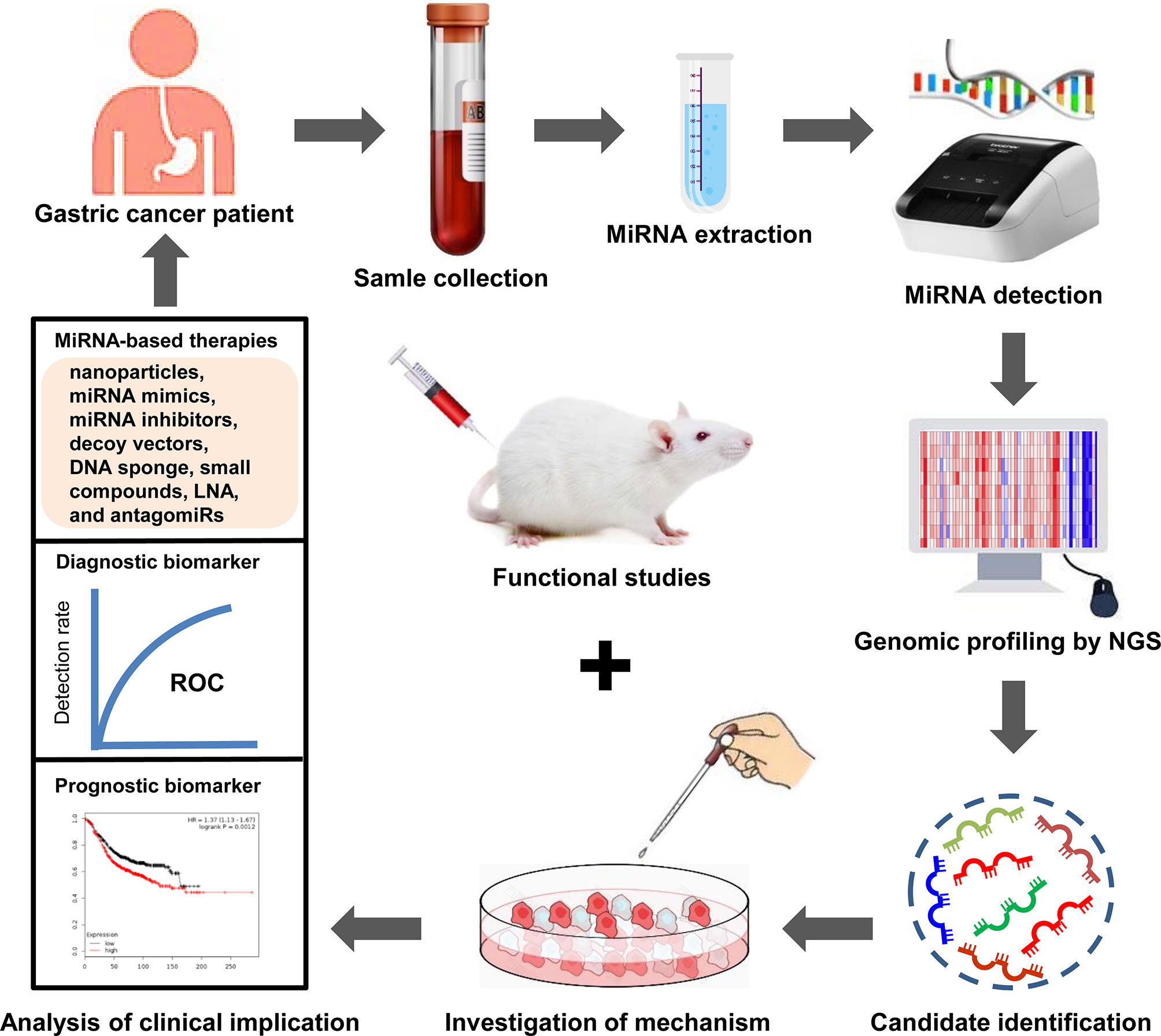
Figure 3 Clinical implications of miRNAs in GC drug resistance. MiRNAs are enriched in blood samples from drug resistant-GC patients. Dysregulated miRNAs are identified through high-throughput sequencing. Subsequently, the mechanisms of dysregulated miRNA in the drug resistance of GC are investigated using cell models. The functions of dysregulated miRNA in GC drug resistance are investigated using animal models. Next, the dysregulated miRNAs that can act as diagnostic or prognostic biomarkers for drug resistant-GC patients are selected. Moreover, targeting specific miRNAs may produce reliable therapeutic effect. Finally, GC patients with drug resistance receive individualized precision treatment based on these effective strategies.
In summary, recent studies have shown that miRNAs possess the great potential to be effective therapeutic targets and promising biomarkers for predicting drug resistance and prognosis in GC treatment. In-depth understanding of their mechanism in GC progression may facilitate the design of potential therapeutic strategies that can be used to reverse drug resistance in GC patients. However, overcoming resistance to chemotherapy still remains a big challenge. Continuous efforts are required to develop miRNA-based therapies that can provide novel therapeutic options and thus improve the clinical outcomes of GC patients in the future.
Author Contributions
YL: Writing- Conceptualization, Original draft preparation, Writing - Review and Editing, Funding acquisition. XA: Data Curation, Writing - Review and Editing. GJ: Data Curation. YZ: Data Curation. WY: Data Curation. JW: Writing - Review and Editing. All authors contributed to the article and approved the submitted version.
Funding
All authors are supported by Qingdao Medical College, Qingdao University. This work was funded by the National Natural Science Foundation of China (81802822), the China Postdoctoral Science Foundation (2018M642607), the Natural Science Foundation of Shandong Province (ZR2021MC189), and the Qingdao Applied Basic Research Project (19-6-2-50-cg).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, Functions and Clinical Significance of circRNAs in Gastric Cancer. Mol Cancer (2019) 18(1):136. doi: 10.1186/s12943-019-1069-0
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
4. Charalampakis N, Economopoulou P, Kotsantis I, Tolia M, Schizas D, Liakakos T, et al. Medical Management of Gastric Cancer: A 2017 Update. Cancer Med (2018) 7(1):123–33. doi: 10.1002/cam4.1274
5. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer Treatment and Survivorship Statistics, 2019. CA Cancer J Clin (2019) 69(5):363–85. doi: 10.3322/caac.21565
6. Si W, Shen J, Zheng H, Fan W. The Role and Mechanisms of Action of microRNAs in Cancer Drug Resistance. Clin Epigenet (2019) 11(1):25. doi: 10.1186/s13148-018-0587-8
7. Zhang Y, Jia DD, Zhang YF, Cheng MD, Zhu WX, Li PF, et al. The Emerging Function and Clinical Significance of circRNAs in Thyroid Cancer and Autoimmune Thyroid Diseases. Int J Biol Sci (2021) 17(7):1731–41. doi: 10.7150/ijbs.55381
8. Min HY, Lee HY. Mechanisms of Resistance to Chemotherapy in non-Small Cell Lung Cancer. Arch Pharm Res (2021) 44(2):146–64. doi: 10.1007/s12272-021-01312-y
9. Plotnikova O, Baranova A, Skoblov M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front Genet (2019) 10:933. doi: 10.3389/fgene.2019.00933
10. Gorski SA, Vogel J, Doudna JA. RNA-Based Recognition and Targeting: Sowing the Seeds of Specificity. Nat Rev Mol Cell Biol (2017) 18(4):215–28. doi: 10.1038/nrm.2016.174
11. Konoshenko MY, Bryzgunova OE, Laktionov PP. miRNAs and Radiotherapy Response in Prostate Cancer. Andrology (2021) 9(2):529–45. doi: 10.1111/andr.12921
12. Liu X, Ma R, Yi B, Riker AI, Xi Y. MicroRNAs Are Involved in the Development and Progression of Gastric Cancer. Acta Pharmacol Sin (2021) 42(7):1018–26. doi: 10.1038/s41401-020-00540-0
13. Yu X, Wang M, Li L, Zhang L, Chan MTV, Wu WKK. MicroRNAs in Atopic Dermatitis: A Systematic Review. J Cell Mol Med (2020) 24(11):5966–72. doi: 10.1111/jcmm.15208
14. Westholm JO, Lai EC. Mirtrons: microRNA Biogenesis via Splicing. Biochimie (2011) 93(11):1897–904. doi: 10.1016/j.biochi.2011.06.017
15. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A Dicer-Independent miRNA Biogenesis Pathway That Requires Ago Catalysis. Nature (2010) 465(7298):584–9. doi: 10.1038/nature09092
16. Zhu C, Chen C, Huang J, Zhang H, Zhao X, Deng R, et al. SUMOylation at K707 of DGCR8 Controls Direct Function of Primary microRNA. Nucleic Acids Res (2015) 43(16):7945–60. doi: 10.1093/nar/gkv741
17. Bose M, Bhattacharyya SN. Target-Dependent Biogenesis of Cognate microRNAs in Human Cells. Nat Commun (2016) 7:12200. doi: 10.1038/ncomms12200
18. Treiber T, Treiber N, Plessmann U, Harlander S, Daiss JL, Eichner N, et al. A Compendium of RNA-Binding Proteins That Regulate MicroRNA Biogenesis. Mol Cell (2017) 66(2):270–84 e13. doi: 10.1016/j.molcel.2017.03.014
19. Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, et al. NEAT1 Scaffolds RNA-Binding Proteins and the Microprocessor to Globally Enhance pri-miRNA Processing. Nat Struct Mol Biol (2017) 24(10):816–24. doi: 10.1038/nsmb.3455
20. Zhou C, Zhao X, Duan S. The Role of miR-543 in Human Cancerous and Noncancerous Diseases. J Cell Physiol (2021) 236(1):15–26. doi: 10.1002/jcp.29860
21. Goodall GJ, Wickramasinghe VO. RNA in Cancer. Nat Rev Cancer (2021) 21(1):22–36. doi: 10.1038/s41568-020-00306-0
22. Abedi F, Rezaee R, Hayes AW, Nasiripour S, Karimi G. MicroRNAs and SARS-CoV-2 Life Cycle, Pathogenesis, and Mutations: Biomarkers or Therapeutic Agents? Cell Cycle (2021) 20(2):143–53. doi: 10.1080/15384101.2020.1867792
23. Liu Y, Ao X, Wang Q, Wang J, Ge H. PiViewer: An Open-Source Tool for Automated Detection and Display of Pi-Pi Interactions. Chem Biol Drug Des (2018) 92(4):1809–14. doi: 10.1111/cbdd.13340
24. Ashrafizadeh M, Zarrabi A, Hushmandi K, Hashemi F, Hashemi F, Samarghandian S, et al. MicroRNAs in Cancer Therapy: Their Involvement in Oxaliplatin Sensitivity/Resistance of Cancer Cells With a Focus on Colorectal Cancer. Life Sci (2020) 256:117973. doi: 10.1016/j.lfs.2020.117973
25. Riquelme I, Letelier P, Riffo-Campos AL, Brebi P, Roa JC. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int J Mol Sci (2016) 17(3):424. doi: 10.3390/ijms17030424
26. Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang Z, et al. Molecular Mechanisms and Theranostic Potential of miRNAs in Drug Resistance of Gastric Cancer. Expert Opin Ther Targets (2017) 21(11):1063–75. doi: 10.1080/14728222.2017.1389900
27. Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in Gastric Cancer: Implications for Drug Resistance. Mol Cancer (2020) 19(1):62. doi: 10.1186/s12943-020-01185-7
28. Poursheikhani A, Bahmanpour Z, Razmara E, Mashouri L, Taheri M, Morshedi Rad D, et al. Non-Coding RNAs Underlying Chemoresistance in Gastric Cancer. Cell Oncol (Dordr) (2020) 43(6):961–88. doi: 10.1007/s13402-020-00528-2
29. Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li K, et al. miR-508-5p Regulates Multidrug Resistance of Gastric Cancer by Targeting ABCB1 and ZNRD1. Oncogene (2014) 33(25):3267–76. doi: 10.1038/onc.2013.297
30. Wang XY, Zhou YC, Wang Y, Liu YY, Wang YX, Chen DD, et al. miR-149 Contributes to Resistance of 5-FU in Gastric Cancer via Targeting TREM2 and Regulating Beta-Catenin Pathway. Biochem Biophys Res Commun (2020) 532(3):329–35. doi: 10.1016/j.bbrc.2020.05.135
31. Jingyue S, Xiao W, Juanmin Z, Wei L, Daoming L, Hong X. TFAP2E Methylation Promotes 5fluorouracil Resistance via Exosomal Mir106a5p and Mir421 in Gastric Cancer MGC803 Cells. Mol Med Rep (2019) 20(1):323–31. doi: 10.3892/mmr.2019.10237
32. Yu C, Chen DQ, Liu HX, Li WB, Lu JW, Feng JF. Rosmarinic Acid Reduces the Resistance of Gastric Carcinoma Cells to 5-Fluorouracil by Downregulating FOXO4-Targeting miR-6785-5p. BioMed Pharmacother (2019) 109:2327–34. doi: 10.1016/j.biopha.2018.10.061
33. Chu H, Han N, Xu J. CMPK1 Regulated by miR-130b Attenuates Response to 5-FU Treatment in Gastric Cancer. Front Oncol (2021) 11:637470. doi: 10.3389/fonc.2021.637470
34. Shen J, Niu W, Zhang H, Jun M, Zhang H. Downregulation of MicroRNA-147 Inhibits Cell Proliferation and Increases the Chemosensitivity of Gastric Cancer Cells to 5-Fluorouracil by Directly Targeting PTEN. Oncol Res (2018) 26(6):901–11. doi: 10.3727/096504017X15061902533715
35. Wang CQ. MiR-195 Reverses 5-FU Resistance Through Targeting HMGA1 in Gastric Cancer Cells. Eur Rev Med Pharmacol Sci (2019) 23(9):3771–8. doi: 10.26355/eurrev_201905_17803
36. Li LQ, Pan D, Chen Q, Zhang SW, Xie DY, Zheng XL, et al. Sensitization of Gastric Cancer Cells to 5-FU by MicroRNA-204 Through Targeting the TGFBR2-Mediated Epithelial to Mesenchymal Transition. Cell Physiol Biochem (2018) 47(4):1533–45. doi: 10.1159/000490871
37. Zhang F, Li K, Yao X, Wang H, Li W, Wu J, et al. A miR-567-PIK3AP1-PI3K/AKT-C-Myc Feedback Loop Regulates Tumour Growth and Chemoresistance in Gastric Cancer. EBioMedicine (2019) 44:311–21. doi: 10.1016/j.ebiom.2019.05.003
38. Li C, Zou J, Zheng G, Chu J. MiR-30a Decreases Multidrug Resistance (MDR) of Gastric Cancer Cells. Med Sci Monit (2016) 0. doi: 10.12659/MSM.898415
39. Mu L, Yang F, Guo D, Li P, Zhang M. Overexpression of Secretory Clusterin (sCLU) Induces Chemotherapy Resistance in Human Gastric Cancer Cells by Targeting miR-195-5p. Bioengineered (2020) 11(1):472–83. doi: 10.1080/21655979.2020.1747825
40. Choi H, Lee SK. TAX1BP1 Downregulation by EBV-miR-BART15-3p Enhances Chemosensitivity of Gastric Cancer Cells to 5-FU. Arch Virol (2017) 162(2):369–77. doi: 10.1007/s00705-016-3109-z
41. Korourian A, Madjd Z, Roudi R, Shariftabrizi A, Soleimani M. Induction of miR-31 Causes Increased Sensitivity to 5-FU and Decreased Migration and Cell Invasion in Gastric Adenocarcinoma. Bratisl Lek Listy (2019) 120(1):35–9. doi: 10.4149/BLL_2019_005
42. Jiang L, Yang W, Bian W, Yang H, Wu X, Li Y, et al. MicroRNA-623 Targets Cyclin D1 to Inhibit Cell Proliferation and Enhance the Chemosensitivity of Cells to 5-Fluorouracil in Gastric Cancer. Oncol Res (2018) 27(1):19–27. doi: 10.3727/096504018X15193469240508
43. Gao H, Ma J, Cheng Y, Zheng P. Exosomal Transfer of Macrophage-Derived miR-223 Confers Doxorubicin Resistance in Gastric Cancer. Onco Targets Ther (2020) 13:12169–79. doi: 10.2147/OTT.S283542
44. Chen J, Zhou C, Li J, Xiang X, Zhang L, Deng J, et al. Mir215p Confers Doxorubicin Resistance in Gastric Cancer Cells by Targeting PTEN and TIMP3. Int J Mol Med (2018) 41(4):1855–66. doi: 10.3892/ijmm.2018.3405
45. Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B, et al. Exosomal Transfer of miR-501 Confers Doxorubicin Resistance and Tumorigenesis via Targeting of BLID in Gastric Cancer. Cancer Lett (2019) 459:122–34. doi: 10.1016/j.canlet.2019.05.035
46. Teng R, Hu Y, Zhou J, Seifer B, Chen Y, Shen J, et al. Overexpression of Lin28 Decreases the Chemosensitivity of Gastric Cancer Cells to Oxaliplatin, Paclitaxel, Doxorubicin, and Fluorouracil in Part via microRNA-107. PloS One (2015) 10(12):e0143716. doi: 10.1371/journal.pone.0143716
47. Xu YC, Liu X, Li M, Li Y, Li CY, Lu Y, et al. A Novel Mechanism of Doxorubicin Resistance and Tumorigenesis Mediated by MicroRNA-501-5p-Suppressed BLID. Mol Ther Nucleic Acids (2018) 12:578–90. doi: 10.1016/j.omtn.2018.06.011
48. Pang X, Zhou Z, Yu Z, Han L, Lin Z, Ao X, et al. Foxo3a-Dependent miR-633 Regulates Chemotherapeutic Sensitivity in Gastric Cancer by Targeting Fas-Associated Death Domain. RNA Biol (2019) 16(2):233–48. doi: 10.1080/15476286.2019.1565665
49. Pan Y, Ren F, Zhang W, Liu G, Yang D, Hu J, et al. Regulation of BGC-823 Cell Sensitivity to Adriamycin via miRNA-135a-5p. Oncol Rep (2014) 32(6):2549–56. doi: 10.3892/or.2014.3546
50. Tao XC, Zhang XY, Sun SB, Wu DQ. Mir92a Contributes to Cell Proliferation, Apoptosis and Doxorubicin Chemosensitivity in Gastric Carcinoma Cells. Oncol Rep (2019) 42(1):313–20. doi: 10.3892/or.2019.7168
51. Shen Q, Yao Q, Sun J, Feng L, Lu H, Ma Y, et al. Downregulation of Histone Deacetylase 1 by microRNA-520h Contributes to the Chemotherapeutic Effect of Doxorubicin. FEBS Lett (2014) 588(1):184–91. doi: 10.1016/j.febslet.2013.11.034
52. Wang S, Ping M, Song B, Guo Y, Li Y, Jia J. Exosomal CircPRRX1 Enhances Doxorubicin Resistance in Gastric Cancer by Regulating MiR-3064-5p/PTPN14 Signaling. Yonsei Med J (2020) 61(9):750–61. doi: 10.3349/ymj.2020.61.9.750
53. Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia H, et al. HOTAIR Enhanced Paclitaxel and Doxorubicin Resistance in Gastric Cancer Cells Partly Through Inhibiting miR-217 Expression. J Cell Biochem (2018) 119(9):7226–34. doi: 10.1002/jcb.26901
54. Peng QP, Du DB, Ming Q, Hu F, Wu ZB, Qiu S. MicroRNA 494 Increases Chemosensitivity to Doxorubicin in Gastric Cancer Cells by Targeting Phosphodiesterases 4D. Cell Mol Biol (Noisy-le-grand) (2018) 64(15):62–6. doi: 10.14715/cmb/2017.64.15.10
55. Zou Z, Zou R, Zong D, Shi Y, Chen J, Huang J, et al. miR-495 Sensitizes MDR Cancer Cells to the Combination of Doxorubicin and Taxol by Inhibiting MDR1 Expression. J Cell Mol Med (2017) 21(9):1929–43. doi: 10.1111/jcmm.13114
56. Zhao D, Zhang Y, Song L. MiR-16-1 Targeted Silences Far Upstream Element Binding Protein 1 to Advance the Chemosensitivity to Adriamycin in Gastric Cancer. Pathol Oncol Res (2018) 24(3):483–8. doi: 10.1007/s12253-017-0263-x
57. Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, et al. MicroRNA-19a/B Regulates Multidrug Resistance in Human Gastric Cancer Cells by Targeting PTEN. Biochem Biophys Res Commun (2013) 434(3):688–94. doi: 10.1016/j.bbrc.2013.04.010
58. Bao J, Xu Y, Wang Q, Zhang J, Li Z, Li D, et al. miR-101 Alleviates Chemoresistance of Gastric Cancer Cells by Targeting ANXA2. BioMed Pharmacother (2017) 92:1030–7. doi: 10.1016/j.biopha.2017.06.011
59. Cao W, Wei W, Zhan Z, Xie Y, Xiao Q. MiR-1284 Modulates Multidrug Resistance of Gastric Cancer Cells by Targeting EIF4A1. Oncol Rep (2016) 35(5):2583–91. doi: 10.3892/or.2016.4643
60. Huang H, Tang J, Zhang L, Bu Y, Zhang X. miR-874 Regulates Multiple-Drug Resistance in Gastric Cancer by Targeting ATG16L1. Int J Oncol (2018) 53(6):2769–79. doi: 10.3892/ijo.2018.4593
61. Yang X, Cai H, Liang Y, Chen L, Wang X, Si R, et al. Inhibition of C-Myc by Let-7b Mimic Reverses Mutidrug Resistance in Gastric Cancer Cells. Oncol Rep (2015) 33(4):1723–30. doi: 10.3892/or.2015.3757
62. Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Regulation of Drug Resistance and Metastasis of Gastric Cancer Cells via the Microrna647-ANK2 Axis. Int J Mol Med (2018) 41(4):1958–66. doi: 10.3892/ijmm.2018.3381
63. Wang P, Li Z, Liu H, Zhou D, Fu A, Zhang E. MicroRNA-126 Increases Chemosensitivity in Drug-Resistant Gastric Cancer Cells by Targeting EZH2. Biochem Biophys Res Commun (2016) 479(1):91–6. doi: 10.1016/j.bbrc.2016.09.040
64. Deng LM, Tan T, Zhang TY, Xiao XF, Gu H. Mir1 Reverses Multidrug Resistance in Gastric Cancer Cells via Downregulation of Sorcin Through Promoting the Accumulation of Intracellular Drugs and Apoptosis of Cells. Int J Oncol (2019) 55(2):451–61. doi: 10.3892/ijo.2019.4831
65. An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, et al. miR-23b-3p Regulates the Chemoresistance of Gastric Cancer Cells by Targeting ATG12 and HMGB2. Cell Death Dis (2015) 6:e1766. doi: 10.1038/cddis.2015.123
66. Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 Modulate Multidrug Resistance by Targeting BCL2 in Human Gastric Cancer Cells. Int J Cancer (2008) 123(2):372–9. doi: 10.1002/ijc.23501
67. Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, et al. miR-200bc/429 Cluster Modulates Multidrug Resistance of Human Cancer Cell Lines by Targeting BCL2 and XIAP. Cancer Chemother Pharmacol (2012) 69(3):723–31. doi: 10.1007/s00280-011-1752-3
68. Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, et al. miR-497 Modulates Multidrug Resistance of Human Cancer Cell Lines by Targeting BCL2. Med Oncol (2012) 29(1):384–91. doi: 10.1007/s12032-010-9797-4
69. Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b Modulates Multidrug Resistance by Targeting BCL2 in Human Cancer Cell Lines. Int J Cancer (2010) 127(11):2520–9. doi: 10.1002/ijc.25260
70. Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, et al. CAF Secreted miR-522 Suppresses Ferroptosis and Promotes Acquired Chemo-Resistance in Gastric Cancer. Mol Cancer (2020) 19(1):43. doi: 10.1186/s12943-020-01168-8
71. Cui HY, Rong JS, Chen J, Guo J, Zhu JQ, Ruan M, et al. Exosomal microRNA-588 From M2 Polarized Macrophages Contributes to Cisplatin Resistance of Gastric Cancer Cells. World J Gastroenterol (2021) 27(36):6079–92. doi: 10.3748/wjg.v27.i36.6079
72. Jin HF, Wang JF, Shao M, Zhou K, Ma X, Lv XP. Down-Regulation of miR-7 in Gastric Cancer Is Associated With Elevated LDH-A Expression and Chemoresistance to Cisplatin. Front Cell Dev Biol (2020) 8:555937. doi: 10.3389/fcell.2020.555937
73. Qian Y, Wu X, Wang H, Hou G, Han X, Song W. MicroRNA-4290 Suppresses PDK1-Mediated Glycolysis to Enhance the Sensitivity of Gastric Cancer Cell to Cisplatin. Braz J Med Biol Res (2020) 53(5):e9330. doi: 10.1590/1414-431X20209330
74. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal Transfer of Tumor-Associated Macrophage-Derived miR-21 Confers Cisplatin Resistance in Gastric Cancer Cells. J Exp Clin Cancer Res (2017) 36(1):53. doi: 10.1186/s13046-017-0528-y
75. Shao L, Chen Z, Soutto M, Zhu S, Lu H, Romero-Gallo J, et al. Helicobacter Pylori-Induced miR-135b-5p Promotes Cisplatin Resistance in Gastric Cancer. FASEB J (2019) 33(1):264–74. doi: 10.1096/fj.201701456RR
76. Ni Q, Zhang Y, Tao R, Li X, Zhu J. MicroRNA-95-3p Serves as a Contributor to Cisplatin Resistance in Human Gastric Cancer Cells by Targeting EMP1/PI3K/AKT Signaling. Aging (Albany NY) (2021) 13(6):8665–87. doi: 10.18632/aging.202679
77. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. Circcul2 Regulates Gastric Cancer Malignant Transformation and Cisplatin Resistance by Modulating Autophagy Activation via miR-142-3p/ROCK2. Mol Cancer (2020) 19(1):156. doi: 10.1186/s12943-020-01270-x
78. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 Inhibits Cisplatin Resistance in Gastric Cancer by miR-99a-5p-Mediated Induction of MTMR3 Expression. J Exp Clin Cancer Res (2020) 39(1):246. doi: 10.1186/s13046-020-01758-w
79. Ren N, Jiang T, Wang C, Xie S, Xing Y, Piao D, et al. LncRNA ADAMTS9-AS2 Inhibits Gastric Cancer (GC) Development and Sensitizes Chemoresistant GC Cells to Cisplatin by Regulating miR-223-3p/NLRP3 Axis. Aging (Albany NY) (2020) 12(11):11025–41. doi: 10.18632/aging.103314
80. Wang Z, Wang Q, Xu G, Meng N, Huang X, Jiang Z, et al. The Long Noncoding RNA CRAL Reverses Cisplatin Resistance via the miR-505/CYLD/AKT Axis in Human Gastric Cancer Cells. RNA Biol (2020) 17(11):1576–89. doi: 10.1080/15476286.2019.1709296
81. Lin H, Zhang L, Zhang C, Liu P. Exosomal MiR-500a-3p Promotes Cisplatin Resistance and Stemness via Negatively Regulating FBXW7 in Gastric Cancer. J Cell Mol Med (2020) 24(16):8930–41. doi: 10.1111/jcmm.15524
82. Wu S, Xie J, Shi H, Wang ZW. miR-492 Promotes Chemoresistance to CDDP and Metastasis by Targeting Inhibiting DNMT3B and Induces Stemness in Gastric Cancer. Biosci Rep (2020) 40(3):BSR20194342. doi: 10.1042/BSR20194342
83. Zheng H, Wang JJ, Yang XR, Yu YL. Upregulation of miR-34c After Silencing E2F Transcription Factor 1 Inhibits Paclitaxel Combined With Cisplatin Resistance in Gastric Cancer Cells. World J Gastroenterol (2020) 26(5):499–513. doi: 10.3748/wjg.v26.i5.499
84. Pei LJ, Sun PJ, Ma K, Guo YY, Wang LY, Liu FD. LncRNA-SNHG7 Interferes With miR-34a to De-Sensitize Gastric Cancer Cells to Cisplatin. Cancer Biomark (2021) 30(1):127–37. doi: 10.3233/CBM-201621
85. Wang X, Zhang Y, Li W, Liu X. Knockdown of Cir_RNA PVT1 Elevates Gastric Cancer Cisplatin Sensitivity via Sponging miR-152-3p. J Surg Res (2021) 261:185–95. doi: 10.1016/j.jss.2020.12.013
86. Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis Via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother Radiopharm (2021) 36(4):347–59. doi: 10.1089/cbr.2020.3578
87. Zhan P, Shu X, Chen M, Sun L, Yu L, Liu J, et al. miR-98-5p Inhibits Gastric Cancer Cell Stemness and Chemoresistance by Targeting Branched-Chain Aminotransferases 1. Life Sci (2021) 276:119405. doi: 10.1016/j.lfs.2021.119405
88. Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng A, et al. CircRNACCDC66 Regulates Cisplatin Resistance in Gastric Cancer via the miR-618/BCL2 Axis. Biochem Biophys Res Commun (2020) 526(3):713–20. doi: 10.1016/j.bbrc.2020.03.156
89. Zhang Z, Yu X, Zhou B, Zhang J, Chang J. Circular RNA Circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. BioMed Res Int (2020) 2020:5103272. doi: 10.1155/2020/5103272
90. Han X, Zhang HB, Li XD, Wang ZA. Long non-Coding RNA X-Inactive-Specific Transcript Contributes to Cisplatin Resistance in Gastric Cancer by Sponging miR-Let-7b. Anticancer Drugs (2020) 31(10):1018–25. doi: 10.1097/CAD.0000000000000942
91. Zhang YF, Li CS, Zhou Y, Lu XH. Propofol Facilitates Cisplatin Sensitivity via lncRNA MALAT1/miR-30e/ATG5 Axis Through Suppressing Autophagy in Gastric Cancer. Life Sci (2020) 244:117280. doi: 10.1016/j.lfs.2020.117280
92. Wu C, Hu Y, Ning Y, Zhao A, Zhang G, Yan L. Long Noncoding RNA Plasmacytoma Variant Translocation 1 Regulates Cisplatin Resistance via miR-3619-5p/TBL1XR1 Axis in Gastric Cancer. Cancer Biother Radiopharm (2020) 35(10):741–52. doi: 10.1089/cbr.2019.3342
93. Yan LH, Chen ZN, Li L, Chen J, Wei WE, Mo XW, et al. miR-135a Promotes Gastric Cancer Progression and Resistance to Oxaliplatin. Oncotarget (2016) 7(43):70699–714. doi: 10.18632/oncotarget.12208
94. Wu Q, Wang H, Liu L, Zhu K, Yu W, Guo J. Hsa_circ_0001546 Acts as a miRNA-421 Sponge to Inhibit the Chemoresistance of Gastric Cancer Cells via ATM/Chk2/p53-Dependent Pathway. Biochem Biophys Res Commun (2020) 521(2):303–9. doi: 10.1016/j.bbrc.2019.10.117
95. Hu Y, Su Y, Lei X, Zhao H, Wang L, Xu T, et al. LINC00641/miR-582-5p Mediate Oxaliplatin Resistance by Activating Autophagy in Gastric Adenocarcinoma. Sci Rep (2020) 10(1):14981. doi: 10.1038/s41598-020-70913-2
96. Zhang Z, Li M, Zhang Z. lncRNA MALAT1 Modulates Oxaliplatin Resistance of Gastric Cancer via Sponging miR-22-3p. Onco Targets Ther (2020) 13:1343–54. doi: 10.2147/OTT.S196619
97. Li Y, Peng C, Fang C, Huang K. Upregulation of Nuclear-Enriched Abundant Transcript 1 Confers Oxaliplatin Resistance to Gastric Cancer. Cell Biol Int (2020) 44(2):446–55. doi: 10.1002/cbin.11245
98. Wu X, Zheng Y, Han B, Dong X. Long Noncoding RNA BLACAT1 Modulates ABCB1 to Promote Oxaliplatin Resistance of Gastric Cancer via Sponging miR-361. BioMed Pharmacother (2018) 99:832–8. doi: 10.1016/j.biopha.2018.01.130
99. Song W, Qian Y, Zhang MH, Wang H, Wen X, Yang XZ, et al. The Long Non-Coding RNA DDX11-AS1 Facilitates Cell Progression and Oxaliplatin Resistance via Regulating miR-326/IRS1 Axis in Gastric Cancer. Eur Rev Med Pharmacol Sci (2020) 24(6):3049–61. doi: 10.26355/eurrev_202003_20669
100. Wang J, Sun Y, Zhang X, Cai H, Zhang C, Qu H, et al. Oxidative Stress Activates NORAD Expression by H3K27ac and Promotes Oxaliplatin Resistance in Gastric Cancer by Enhancing Autophagy Flux via Targeting the miR-433-3p. Cell Death Dis (2021) 12(1):90. doi: 10.1038/s41419-020-03368-y
101. Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA Circ_0032821 Contributes to Oxaliplatin (OXA) Resistance of Gastric Cancer Cells by Regulating SOX9 via miR-515-5p. Biotechnol Lett (2021) 43(2):339–51. doi: 10.1007/s10529-020-03036-3
102. Shen B, Yu S, Zhang Y, Yuan Y, Li X, Zhong J, et al. miR-590-5p Regulates Gastric Cancer Cell Growth and Chemosensitivity Through RECK and the AKT/ERK Pathway. Onco Targets Ther (2016) 9:6009–19. doi: 10.2147/OTT.S110923
103. Wang M, Qiu R, Yu S, Xu X, Li G, Gu R, et al. Paclitaxelresistant Gastric Cancer MGC803 Cells Promote Epithelialtomesenchymal Transition and Chemoresistance in Paclitaxelsensitive Cells via Exosomal Delivery of Mir1555p. Int J Oncol (2019) 54(1):326–38. doi: 10.3892/ijo.2018.4601
104. Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P, et al. Regulation of Microtubule-Associated Protein Tau (MAPT) by miR-34c-5p Determines the Chemosensitivity of Gastric Cancer to Paclitaxel. Cancer Chemother Pharmacol (2013) 71(5):1159–71. doi: 10.1007/s00280-013-2108-y
105. Jin B, Liu Y, Wang H. Antagonism of miRNA-21 Sensitizes Human Gastric Cancer Cells to Paclitaxel. Cell Biochem Biophys (2015) 72(1):275–82. doi: 10.1007/s12013-014-0450-2
106. Li L, Wu C, Zhao Y. miRNA-34a Enhances the Sensitivity of Gastric Cancer Cells to Treatment With Paclitaxel by Targeting E2F5. Oncol Lett (2017) 13(6):4837–42. doi: 10.3892/ol.2017.6041
107. Liu YY, Zhang LY, Du WZ. Circular RNA Circ-PVT1 Contributes to Paclitaxel Resistance of Gastric Cancer Cells Through the Regulation of ZEB1 Expression by Sponging miR-124-3p. Biosci Rep (2019) 39(12):BSR20193045. doi: 10.1042/BSR20193045
108. Blondy S, David V, Verdier M, Mathonnet M, Perraud A, Christou N. 5-Fluorouracil Resistance Mechanisms in Colorectal Cancer: From Classical Pathways to Promising Processes. Cancer Sci (2020) 111(9):3142–54. doi: 10.1111/cas.14532
109. Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 5-Fluorouracil and Other Fluoropyrimidines in Colorectal Cancer: Past, Present and Future. Pharmacol Ther (2020) 206:107447. doi: 10.1016/j.pharmthera.2019.107447
110. Prathumsap N, Shinlapawittayatorn K, Chattipakorn SC, Chattipakorn N. Effects of Doxorubicin on the Heart: From Molecular Mechanisms to Intervention Strategies. Eur J Pharmacol (2020) 866:172818. doi: 10.1016/j.ejphar.2019.172818
111. Li GZ, Hu YH, Li DY, Zhang Y, Guo HL, Li YM, et al. Vincristine-Induced Peripheral Neuropathy: A Mini-Review. Neurotoxicology (2020) 81:161–71. doi: 10.1016/j.neuro.2020.10.004
112. van Zyl B, Tang D, Bowden NA. Biomarkers of Platinum Resistance in Ovarian Cancer: What can We Use to Improve Treatment. Endocr Relat Cancer (2018) 25(5):R303–R18. doi: 10.1530/ERC-17-0336
113. Abu Samaan TM, Samec M, Liskova A, Kubatka P, Busselberg D. Paclitaxel's Mechanistic and Clinical Effects on Breast Cancer. Biomolecules (2019) 9(12):789. doi: 10.3390/biom9120789
114. Zhang Y, Xu W, Ni P, Li A, Zhou J, Xu S. MiR-99a and MiR-491 Regulate Cisplatin Resistance in Human Gastric Cancer Cells by Targeting Capns1. Int J Biol Sci (2016) 12(12):1437–47. doi: 10.7150/ijbs.16529
115. Zhou D, Li X, Zhao H, Sun B, Liu A, Han X, et al. Combining Multi-Dimensional Data to Identify a Key Signature (Gene and miRNA) of Cisplatin-Resistant Gastric Cancer. J Cell Biochem (2018) 119(8):6997–7008. doi: 10.1002/jcb.26908
116. Wei X, Gao M, Ahmed Y, Gao M, Liu W, Zhang Y, et al. MicroRNA-362-5p Enhances the Cisplatin Sensitivity of Gastric Cancer Cells by Targeting Suppressor of Zeste 12 Protein. Oncol Lett (2019) 18(2):1607–16. doi: 10.3892/ol.2019.10496
117. Chen C, Tang X, Liu Y, Zhu J, Liu J. Induction/reversal of Drug Resistance in Gastric Cancer by non-Coding RNAs (Review). Int J Oncol (2019) 54(5):1511–24. doi: 10.3892/ijo.2019.4751
118. Ao X, Ding W, Zhang Y, Ding D, Liu Y. TCF21: A Critical Transcription Factor in Health and Cancer. J Mol Med (Berl) (2020) 98(8):1055–68. doi: 10.1007/s00109-020-01934-7
119. Liu Y, Chen DD, Xing YH, Ge N, Zhang Y, Liu J, et al. A New Oxovanadium Complex Enhances Renal Function by Improving Insulin Signaling Pathway in Diabetic Mice. J Diabetes Complic (2014) 28(3):265–72. doi: 10.1016/j.jdiacomp.2014.02.001
120. Ma L, Zong X. Metabolic Symbiosis in Chemoresistance: Refocusing the Role of Aerobic Glycolysis. Front Oncol (2020) 10:5. doi: 10.3389/fonc.2020.00005
121. Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, et al. Critical Role of FOXO3a in Carcinogenesis. Mol Cancer (2018) 17(1):104. doi: 10.1186/s12943-018-0856-3
122. Magnelli L, Schiavone N, Staderini F, Biagioni A, Papucci L. MAP Kinases Pathways in Gastric Cancer. Int J Mol Sci (2020) 21(8):2893. doi: 10.3390/ijms21082893
123. Barbosa R, Acevedo LA, Marmorstein R. The MEK/ERK Network as a Therapeutic Target in Human Cancer. Mol Cancer Res (2021) 19(3):361–74. doi: 10.1158/1541-7786.MCR-20-0687
124. Chen Z, Gao YJ, Hou RZ, Ding DY, Song DF, Wang DY, et al. MicroRNA-206 Facilitates Gastric Cancer Cell Apoptosis and Suppresses Cisplatin Resistance by Targeting MAPK2 Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(1):171–80. doi: 10.26355/eurrev_201901_16761
125. Zhou J, Chen Q. Poor Expression of microRNA-135b Results in the Inhibition of Cisplatin Resistance and Proliferation and Induces the Apoptosis of Gastric Cancer Cells Through MST1-Mediated MAPK Signaling Pathway. FASEB J (2019) 33(3):3420–36. doi: 10.1096/fj.201800618RRR
126. Zhou L, Li X, Zhou F, Jin Z, Chen D, Wang P, et al. Downregulation of Leucine-Rich Repeats and Immunoglobulin-Like Domains 1 by microRNA-20a Modulates Gastric Cancer Multidrug Resistance. Cancer Sci (2018) 109(4):1044–54. doi: 10.1111/cas.13538
127. He H, Song F, Gao Q, Lu Z, Yuan Y, Li X, et al. The APEX1/miRNA-27a-5p Axis Plays Key Roles in Progression, Metastasis and Targeted Chemotherapy of Gastric Cancer. Int J Pharm (2021) 599:120446. doi: 10.1016/j.ijpharm.2021.120446
128. Jiang D, Li H, Xiang H, Gao M, Yin C, Wang H, et al. Long Chain Non-Coding RNA (lncRNA) HOTAIR Knockdown Increases miR-454-3p to Suppress Gastric Cancer Growth by Targeting STAT3/Cyclin D1. Med Sci Monit (2019) 25:1537–48. doi: 10.12659/MSM.913087
129. Zhu QL, Li Z, Lv CM, Wang W. MiR-187 Influences Cisplatin-Resistance of Gastric Cancer Cells Through Regulating the TGF-Beta/Smad Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(22):9907–14. doi: 10.26355/eurrev_201911_19556
130. Zhou X, Men X, Zhao R, Han J, Fan Z, Wang Y, et al. miR-200c Inhibits TGF-Beta-Induced-EMT to Restore Trastuzumab Sensitivity by Targeting ZEB1 and ZEB2 in Gastric Cancer. Cancer Gene Ther (2018) 25(3-4):68–76. doi: 10.1038/s41417-017-0005-y
131. Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, et al. MicroRNA-362 Induces Cell Proliferation and Apoptosis Resistance in Gastric Cancer by Activation of NF-kappaB Signaling. J Transl Med (2014) 12:33. doi: 10.1186/1479-5876-12-33
132. Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J, et al. miR-20a Enhances Cisplatin Resistance of Human Gastric Cancer Cell Line by Targeting NFKBIB. Tumour Biol (2016) 37(1):1261–9. doi: 10.1007/s13277-015-3921-1
133. Guo W, Li W, Yuan L, Mei X, Hu W. MicroRNA-106a-3p Induces Apatinib Resistance and Activates Janus-Activated Kinase 2 (JAK2)/Signal Transducer and Activator of Transcription 3 (STAT3) by Targeting the SOCS System in Gastric Cancer. Med Sci Monit (2019) 25:10122–8. doi: 10.12659/MSM.919610
134. Duan L, Yang W, Feng W, Cao L, Wang X, Niu L, et al. Molecular Mechanisms and Clinical Implications of miRNAs in Drug Resistance of Colorectal Cancer. Ther Adv Med Oncol (2020) 12:1758835920947342. doi: 10.1177/1758835920947342
135. Kathawala RJ, Gupta P, Ashby CR Jr., Chen ZS. The Modulation of ABC Transporter-Mediated Multidrug Resistance in Cancer: A Review of the Past Decade. Drug Resist Update (2015) 18:1–17. doi: 10.1016/j.drup.2014.11.002
136. Zhang Y, Qu X, Li C, Fan Y, Che X, Wang X, et al. miR-103/107 Modulates Multidrug Resistance in Human Gastric Carcinoma by Downregulating Cav-1. Tumour Biol (2015) 36(4):2277–85. doi: 10.1007/s13277-014-2835-7
137. Lu C, Shan Z, Li C, Yang L. MiR-129 Regulates Cisplatin-Resistance in Human Gastric Cancer Cells by Targeting P-Gp. BioMed Pharmacother (2017) 86:450–6. doi: 10.1016/j.biopha.2016.11.139
138. Zhang L, Guo X, Zhang D, Fan Y, Qin L, Dong S, et al. Upregulated miR-132 in Lgr5(+) Gastric Cancer Stem Cell-Like Cells Contributes to Cisplatin-Resistance via SIRT1/CREB/ABCG2 Signaling Pathway. Mol Carcinog (2017) 56(9):2022–34. doi: 10.1002/mc.22656
139. Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y, et al. Methylation of miR-129-5p CpG Island Modulates Multi-Drug Resistance in Gastric Cancer by Targeting ABC Transporters. Oncotarget (2014) 5(22):11552–63. doi: 10.18632/oncotarget.2594
140. Marin JJ, Al-Abdulla R, Lozano E, Briz O, Bujanda L, Banales JM, et al. Mechanisms of Resistance to Chemotherapy in Gastric Cancer. Anticancer Agents Med Chem (2016) 16(3):318–34. doi: 10.2174/1871520615666150803125121
141. Shi WJ, Gao JB. Molecular Mechanisms of Chemoresistance in Gastric Cancer. World J Gastrointest Oncol (2016) 8(9):673–81. doi: 10.4251/wjgo.v8.i9.673
142. Liang Y, Liang Q, Qiao L, Xiao F. MicroRNAs Modulate Drug Resistance-Related Mechanisms in Hepatocellular Carcinoma. Front Oncol (2020) 10:920. doi: 10.3389/fonc.2020.00920
143. Marin W, Marin D, Ao X, Liu Y. Mitochondria as a Therapeutic Target for Cardiac Ischemiareperfusion Injury (Review). Int J Mol Med (2021) 47(2):485–99. doi: 10.3892/ijmm.2020.4823
144. Leibowitz B, Yu J. Mitochondrial Signaling in Cell Death via the Bcl-2 Family. Cancer Biol Ther (2010) 9(6):417–22. doi: 10.4161/cbt.9.6.11392
145. Kim H, Choi H, Lee SK. Epstein-Barr Virus miR-BART20-5p Regulates Cell Proliferation and Apoptosis by Targeting BAD. Cancer Lett (2015) 356(2 Pt B):733–42. doi: 10.1016/j.canlet.2014.10.023
146. Chen Y, Zuo J, Liu Y, Gao H, Liu W. Inhibitory Effects of miRNA-200c on Chemotherapy-Resistance and Cell Proliferation of Gastric Cancer SGC7901/DDP Cells. Chin J Cancer (2010) 29(12):1006–11. doi: 10.5732/cjc.010.10236
147. Li Q, Wang JX, He YQ, Feng C, Zhang XJ, Sheng JQ, et al. MicroRNA-185 Regulates Chemotherapeutic Sensitivity in Gastric Cancer by Targeting Apoptosis Repressor With Caspase Recruitment Domain. Cell Death Dis (2014) 5:e1197. doi: 10.1038/cddis.2014.148
148. Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M, et al. The Long Noncoding RNA D63785 Regulates Chemotherapy Sensitivity in Human Gastric Cancer by Targeting miR-422a. Mol Ther Nucleic Acids (2018) 12:405–19. doi: 10.1016/j.omtn.2018.05.024
149. Moxley AH, Reisman D. Context Is Key: Understanding the Regulation, Functional Control, and Activities of the P53 Tumour Suppressor. Cell Biochem Funct (2021) 39(2):235–47. doi: 10.1002/cbf.3590
150. Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J, et al. MicroRNA-148a-3p Enhances Cisplatin Cytotoxicity in Gastric Cancer Through Mitochondrial Fission Induction and Cyto-Protective Autophagy Suppression. Cancer Lett (2017) 410:212–27. doi: 10.1016/j.canlet.2017.09.035
151. Li B, Wang L, Li Z, Wang W, Zhi X, Huang X, et al. miR-3174 Contributes to Apoptosis and Autophagic Cell Death Defects in Gastric Cancer Cells by Targeting Arhgap10. Mol Ther Nucleic Acids (2017) 9:294–311. doi: 10.1016/j.omtn.2017.10.008
152. Lee SD, Yu D, Lee DY, Shin HS, Jo JH, Lee YC. Upregulated microRNA-193a-3p Is Responsible for Cisplatin Resistance in CD44(+) Gastric Cancer Cells. Cancer Sci (2019) 110(2):662–73. doi: 10.1111/cas.13894
153. Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab (2017) 25(5):1037–43. doi: 10.1016/j.cmet.2017.04.004
154. Huang Z, Zhou L, Chen Z, Nice EC, Huang C. Stress Management by Autophagy: Implications for Chemoresistance. Int J Cancer (2016) 139(1):23–32. doi: 10.1002/ijc.29990
155. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long Noncoding RNA MALAT1 Regulates Autophagy Associated Chemoresistance via miR-23b-3p Sequestration in Gastric Cancer. Mol Cancer (2017) 16(1):174. doi: 10.1186/s12943-017-0743-3
156. Gu Y, Fei Z, Zhu R. miR-21 Modulates Cisplatin Resistance of Gastric Cancer Cells by Inhibiting Autophagy via the PI3K/Akt/mTOR Pathway. Anticancer Drugs (2020) 31(4):385–93. doi: 10.1097/CAD.0000000000000886
157. Du X, Liu B, Luan X, Cui Q, Li L. miR-30 Decreases Multidrug Resistance in Human Gastric Cancer Cells by Modulating Cell Autophagy. Exp Ther Med (2018) 15(1):599–605. doi: 10.3892/etm.2017.5354
158. Zhao J, Nie Y, Wang H, Lin Y. MiR-181a Suppresses Autophagy and Sensitizes Gastric Cancer Cells to Cisplatin. Gene (2016) 576(2 Pt 2):828–33. doi: 10.1016/j.gene.2015.11.013
159. Xi Z, Si J, Nan J. LncRNA MALAT1 Potentiates Autophagyassociated Cisplatin Resistance by Regulating the Microrna30b/Autophagyrelated Gene 5 Axis in Gastric Cancer. Int J Oncol (2019) 54(1):239–48. doi: 10.3892/ijo.2018.4609
160. Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D, et al. MicroRNA-495-3p Inhibits Multidrug Resistance by Modulating Autophagy Through GRP78/mTOR Axis in Gastric Cancer. Cell Death Dis (2018) 9(11):1070. doi: 10.1038/s41419-018-0950-x
161. Vaghari-Tabari M, Majidinia M, Moein S, Qujeq D, Asemi Z, Alemi F, et al. MicroRNAs and Colorectal Cancer Chemoresistance: New Solution for Old Problem. Life Sci (2020) 259:118255. doi: 10.1016/j.lfs.2020.118255
162. Liu Y. Targeting the non-Canonical AKT-FOXO3a Axis: A Potential Therapeutic Strategy for Oral Squamous Cell Carcinoma. EBioMedicine (2019) 49:6–8. doi: 10.1016/j.ebiom.2019.10.020
163. Peng C, Huang K, Liu G, Li Y, Yu C. MiR-876-3p Regulates Cisplatin Resistance and Stem Cell-Like Properties of Gastric Cancer Cells by Targeting TMED3. J Gastroenterol Hepatol (2019) 34(10):1711–9. doi: 10.1111/jgh.14649
164. Zeng JF, Ma XQ, Wang LP, Wang W. MicroRNA-145 Exerts Tumor-Suppressive and Chemo-Resistance Lowering Effects by Targeting CD44 in Gastric Cancer. World J Gastroenterol (2017) 23(13):2337–45. doi: 10.3748/wjg.v23.i13.2337
165. Pan Y, Shu X, Sun L, Yu L, Sun L, Yang Z, et al. Mir196a5p Modulates Gastric Cancer Stem Cell Characteristics by Targeting Smad4. Int J Oncol (2017) 50(6):1965–76. doi: 10.3892/ijo.2017.3965
166. Peng X, Kang Q, Wan R, Wang Z. miR-26a/HOXC9 Dysregulation Promotes Metastasis and Stem Cell-Like Phenotype of Gastric Cancer. Cell Physiol Biochem (2018) 49(4):1659–76. doi: 10.1159/000493502
167. Yu D, Shin HS, Lee YS, Lee YC. miR-106b Modulates Cancer Stem Cell Characteristics Through TGF-Beta/Smad Signaling in CD44-Positive Gastric Cancer Cells. Lab Invest (2014) 94(12):1370–81. doi: 10.1038/labinvest.2014.125
168. Liu Y, Ding W, Ge H, Ponnusamy M, Wang Q, Hao X, et al. FOXK Transcription Factors: Regulation and Critical Role in Cancer. Cancer Lett (2019) 458:1–12. doi: 10.1016/j.canlet.2019.05.030
169. Shibue T, Weinberg RA. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat Rev Clin Oncol (2017) 14(10):611–29. doi: 10.1038/nrclinonc.2017.44
170. Wei J, Zhang L, Li J, Zhu S, Tai M, Mason CW, et al. MicroRNA-205 Promotes Cell Invasion by Repressing TCF21 in Human Ovarian Cancer. J Ovarian Res (2017) 10(1):33. doi: 10.1186/s13048-017-0328-1
171. Wang M, Zhang R, Zhang S, Xu R, Yang Q. MicroRNA-574-3p Regulates Epithelial Mesenchymal Transition and Cisplatin Resistance via Targeting ZEB1 in Human Gastric Carcinoma Cells. Gene (2019) 700:110–9. doi: 10.1016/j.gene.2019.03.043
172. Wu DM, Hong XW, Wang LL, Cui XF, Lu J, Chen GQ, et al. MicroRNA-17 Inhibition Overcomes Chemoresistance and Suppresses Epithelial-Mesenchymal Transition Through a DEDD-Dependent Mechanism in Gastric Cancer. Int J Biochem Cell Biol (2018) 102:59–70. doi: 10.1016/j.biocel.2018.06.007
173. Wang LL, Zhang XH, Zhang X, Chu JK. MiR-30a Increases Cisplatin Sensitivity of Gastric Cancer Cells Through Suppressing Epithelial-to-Mesenchymal Transition (EMT). Eur Rev Med Pharmacol Sci (2016) 20(9):1733–9.
174. Ao X, Ding W, Ge H, Zhang Y, Ding D, Liu Y. PBX1 Is a Valuable Prognostic Biomarker for Patients With Breast Cancer. Exp Ther Med (2020) 20(1):385–94. doi: 10.3892/etm.2020.8705
175. Liu Y, Ao X, Jia Z, Bai XY, Xu Z, Hu G, et al. FOXK2 Transcription Factor Suppresses ERalpha-Positive Breast Cancer Cell Growth Through Down-Regulating the Stability of ERalpha via Mechanism Involving BRCA1/BARD1. Sci Rep (2015) 5:8796. doi: 10.1038/srep08796
176. Hu J, Fang Y, Cao Y, Qin R, Chen Q. miR-449a Regulates Proliferation and Chemosensitivity to Cisplatin by Targeting Cyclin D1 and BCL2 in SGC7901 Cells. Dig Dis Sci (2014) 59(2):336–45. doi: 10.1007/s10620-013-2923-3
177. Luo D, Fan H, Ma X, Yang C, He Y, Ge Y, et al. miR-1301-3p Promotes Cell Proliferation and Facilitates Cell Cycle Progression via Targeting SIRT1 in Gastric Cancer. Front Oncol (2021) 11:664242. doi: 10.3389/fonc.2021.664242
178. Sun C, Zhang S, Liu C, Liu X. Curcumin Promoted miR-34a Expression and Suppressed Proliferation of Gastric Cancer Cells. Cancer Biother Radiopharm (2019) 34(10):634–41. doi: 10.1089/cbr.2019.2874
179. Takao K, Konishi H, Matsubara D, Shoda K, Arita T, Shimizu H, et al. MiR-3663-3p Inhibits the Progression of Gastric Cancer Through the CCND1 Pathway. Anticancer Res (2021) 41(5):2441–9. doi: 10.21873/anticanres.15019
180. Cui D, Xiong X, Shu J, Dai X, Sun Y, Zhao Y. FBXW7 Confers Radiation Survival by Targeting P53 for Degradation. Cell Rep (2020) 30(2):497–509 e4. doi: 10.1016/j.celrep.2019.12.032
181. Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 Promotes the Cisplatin Resistance of Human Gastric Cancer Cells via Regulating Cell Cycle by Targeting FBXW7. J Exp Clin Cancer Res (2015) 34:28. doi: 10.1186/s13046-015-0145-6
182. Kent LN, Leone G. The Broken Cycle: E2F Dysfunction in Cancer. Nat Rev Cancer (2019) 19(6):326–38. doi: 10.1038/s41568-019-0143-7
183. Wen L, Cheng F, Zhou Y, Yin C. MiR-26a Enhances the Sensitivity of Gastric Cancer Cells to Cisplatin by Targeting NRAS and E2F2. Saudi J Gastroenterol (2015) 21(5):313–9. doi: 10.4103/1319-3767.166206
184. Dong C. Cytokine Regulation and Function in T Cells. Annu Rev Immunol (2021) 39:51–76. doi: 10.1146/annurev-immunol-061020-053702
185. Raskov H, Orhan A, Christensen JP, Gogenur I. Cytotoxic CD8(+) T Cells in Cancer and Cancer Immunotherapy. Br J Cancer (2021) 124(2):359–67. doi: 10.1038/s41416-020-01048-4
186. Lee DY, Im E, Yoon D, Lee YS, Kim GS, Kim D, et al. Pivotal Role of PD-1/PD-L1 Immune Checkpoints in Immune Escape and Cancer Progression: Their Interplay With Platelets and FOXP3+Tregs Related Molecules, Clinical Implications and Combinational Potential With Phytochemicals. Semin Cancer Biol (2020) S1044-579X(20)30258–3. doi: 10.1016/j.semcancer.2020.12.001
187. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) Reverses Chemoresistance via T-Cell Immune Response Activation by Blocking the PD-L1 Immune Checkpoint. Nat Commun (2016) 7:11406. doi: 10.1038/ncomms11406
188. Qian L, Zhang W, Lei B, He A, Ye L, Li X, et al. MicroRNA-101 Regulates T-Cell Acute Lymphoblastic Leukemia Progression and Chemotherapeutic Sensitivity by Targeting Notch1. Oncol Rep (2016) 36(5):2511–6. doi: 10.3892/or.2016.5117
189. Zhao M, Liu Q, Liu W, Zhou H, Zang X, Lu J. MicroRNA140 Suppresses Helicobacter Pyloripositive Gastric Cancer Growth by Enhancing the Antitumor Immune Response. Mol Med Rep (2019) 20(3):2484–92. doi: 10.3892/mmr.2019.10475
190. Lu H, Shi C, Liu X, Liang C, Yang C, Wan X, et al. Identification of ZG16B as a Prognostic Biomarker in Breast Cancer. Open Med (Wars) (2021) 16(1):1–13. doi: 10.1515/med-2021-0004
191. Liu X, Wang Q, Song S, Feng M, Wang X, Li L, et al. Epithelial Splicing Regulatory Protein 1 Is Overexpressed in Breast Cancer and Predicts Poor Prognosis for Breast Cancer Patients. Med Sci Monit (2021) 27:e931102. doi: 10.12659/MSM.931102
192. Liu X, Liu Y, Wang Q, Song S, Feng L, Shi C. The Alterations and Potential Roles of MCMs in Breast Cancer. J Oncol (2021) 2021:7928937. doi: 10.1155/2021/7928937
193. So JBY, Kapoor R, Zhu F, Koh C, Zhou L, Zou R, et al. Development and Validation of a Serum microRNA Biomarker Panel for Detecting Gastric Cancer in a High-Risk Population. Gut (2021) 70(5):829–37. doi: 10.1136/gutjnl-2020-322065
194. Shimura T, Toden S, Kandimalla R, Toiyama Y, Okugawa Y, Kanda M, et al. Genomewide Expression Profiling Identifies a Novel miRNA-Based Signature for the Detection of Peritoneal Metastasis in Patients With Gastric Cancer. Ann Surg (2021) 274(5):e425–e34. doi: 10.1097/SLA.0000000000003647
195. Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J, et al. miR-374a-5p: A New Target for Diagnosis and Drug Resistance Therapy in Gastric Cancer. Mol Ther Nucleic Acids (2019) 18:320–31. doi: 10.1016/j.omtn.2019.07.025
196. Pang K, Song J, Bai Z, Zhang Z. miR-15a-5p Targets PHLPP2 in Gastric Cancer Cells to Modulate Platinum Resistance and Is a Suitable Serum Biomarker for Oxaliplatin Resistance. Neoplasma (2020) 67(5):1114–21. doi: 10.4149/neo_2020_190904N861
197. Shekari N, Asghari F, Haghnavaz N, Shanehbandi D, Khaze V, Baradaran B, et al. Let-7a Could Serve as A Biomarker for Chemo-Responsiveness to Docetaxel in Gastric Cancer. Anticancer Agents Med Chem (2019) 19(3):304–9. doi: 10.2174/1871520619666181213110258
198. Zhang PF, Sheng LL, Wang G, Tian M, Zhu LY, Zhang R, et al. miR-363 Promotes Proliferation and Chemo-Resistance of Human Gastric Cancer via Targeting of FBW7 Ubiquitin Ligase Expression. Oncotarget (2016) 7(23):35284–92. doi: 10.18632/oncotarget.9169
199. Jin L, Zhang N, Zhang Q, Ding G, Yang Z, Zhang Z. Serum microRNAs as Potential New Biomarkers for Cisplatin Resistance in Gastric Cancer Patients. PeerJ (2020) 8:e8943. doi: 10.7717/peerj.8943
200. Nishibeppu K, Komatsu S, Imamura T, Kiuchi J, Kishimoto T, Arita T, et al. Plasma microRNA Profiles: Identification of miR-1229-3p as a Novel Chemoresistant and Prognostic Biomarker in Gastric Cancer. Sci Rep (2020) 10(1):3161. doi: 10.1038/s41598-020-59939-8
201. Huang D, Wang H, Liu R, Li H, Ge S, Bai M, et al. Mirna27a Is a Biomarker for Predicting Chemosensitivity and Prognosis in Metastatic or Recurrent Gastric Cancer. J Cell Biochem (2014) 115(3):549–56. doi: 10.1002/jcb.24689
202. Qi M, Liu D, Zhang S. MicroRNA-21 Contributes to the Discrimination of Chemoresistance in Metastatic Gastric Cancer. Cancer Biomark (2017) 18(4):451–8. doi: 10.3233/CBM-161732
203. Venturutti L, Cordo Russo RI, Rivas MA, Mercogliano MF, Izzo F, Oakley RH, et al. MiR-16 Mediates Trastuzumab and Lapatinib Response in ErbB-2-Positive Breast and Gastric Cancer via its Novel Targets CCNJ and FUBP1. Oncogene (2016) 35(48):6189–202. doi: 10.1038/onc.2016.151
204. Jin L, Zhang Z. Serum miR-3180-3p and miR-124-3p may Function as Noninvasive Biomarkers of Cisplatin Resistance in Gastric Cancer. Clin Lab (2020) 66(12). doi: 10.7754/Clin.Lab.2020.200302
205. He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, et al. miRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int J Biol Sci (2020) 16(14):2628–47. doi: 10.7150/ijbs.47203
206. Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-Based Therapeutic Approaches for Cancer Patients. Cancer Sci (2019) 110(4):1140–7. doi: 10.1111/cas.13965
207. Tian L, Wang S, Jiang S, Liu Z, Wan X, Yang C, et al. Luteolin as an Adjuvant Effectively Enhances CTL Anti-Tumor Response in B16F10 Mouse Model. Int Immunopharmacol (2021) 94:107441. doi: 10.1016/j.intimp.2021.107441
208. Zhang YF, Shan C, Wang Y, Qian LL, Jia DD, Zhang YF, et al. Cardiovascular Toxicity and Mechanism of Bisphenol A and Emerging Risk of Bisphenol S. Sci Total Environ (2020) 723:137952. doi: 10.1016/j.scitotenv.2020.137952
209. Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, et al. Exosomes Serve as Nanoparticles to Deliver Anti-miR-214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer. Mol Ther (2018) 26(3):774–83. doi: 10.1016/j.ymthe.2018.01.001
210. Ghasabi M, Majidi J, Mansoori B, Mohammadi A, Shomali N, Shirafkan N, et al. The Effect of Combined miR-200c Replacement and Cisplatin on Apoptosis Induction and Inhibition of Gastric Cancer Cell Line Migration. J Cell Physiol (2019) 234(12):22581–92. doi: 10.1002/jcp.28823
Keywords: microRNA, gastric cancer, drug resistance, biomarker, therapeutic target
Citation: Liu Y, Ao X, Ji G, Zhang Y, Yu W and Wang J (2021) Mechanisms of Action And Clinical Implications of MicroRNAs in the Drug Resistance of Gastric Cancer. Front. Oncol. 11:768918. doi: 10.3389/fonc.2021.768918
Received: 01 September 2021; Accepted: 15 November 2021;
Published: 29 November 2021.
Edited by:
Farhad Jadidi-Niaragh, Tabriz University of Medical Sciences, IranReviewed by:
Xiao Zheng, First People’s Hospital of Changzhou, ChinaHuilin Zheng, Zhejiang University of Science and Technology, China
Copyright © 2021 Liu, Ao, Ji, Zhang, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Liu, bGl1eWluZ19oZXJvQDE2My5jb20=
 Ying Liu
Ying Liu Xiang Ao2
Xiang Ao2 Yuan Zhang
Yuan Zhang Wanpeng Yu
Wanpeng Yu