- 1Department of Gastroenterology, Institute of Liver and Gastrointestinal Diseases, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Hubei Key Laboratory of Hepato-Pancreato-Biliary Diseases, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 3Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
The homeobox (HOX) genes encoding an evolutionarily highly conserved family of homeodomain-containing transcriptional factors are essential for embryogenesis and tumorigenesis. HOX genes are involved in cell identity determination during early embryonic development and postnatal processes. The deregulation of HOX genes is closely associated with numerous human malignancies, highlighting the indispensable involvement in mortal cancer development. Since most HOX genes behave as oncogenes or tumor suppressors in human cancer, a better comprehension of their upstream regulators and downstream targets contributes to elucidating the function of HOX genes in cancer development. In addition, targeting HOX genes may imply therapeutic potential. Recently, novel therapies such as monoclonal antibodies targeting tyrosine receptor kinases, small molecular chemical inhibitors, and small interfering RNA strategies, are difficult to implement for targeting transcriptional factors on account of the dual function and pleiotropic nature of HOX genes-related molecular networks. This paper summarizes the current state of knowledge on the roles of HOX genes in human cancer and emphasizes the emerging importance of HOX genes as potential therapeutic targets to overcome the limitations of present cancer therapy.
1 Introduction
1.1 General Overview
The homeobox (HOX) genes were initially discovered in 1992 with the roles in embryogenesis in Drosophila melanogaster, where mutations in antennapedia and bithorax clusters were observed to lead to abnormal body development (1). These mutations lead one body segment to emerge similar to another segment and is originally called the term “homeotic” mutations since the 1990s (2).
The HOX genes encode a family of transcription factors that regulate embryogenesis and morphogenesis during the embryonic period and adulthood. These genes are highly conserved and contain homologous domains in almost all eukaryotic cells (3). The HOX genes comprise a highly conserved sequence of 180-183 base pairs encoding a homeodomain of 60 or 61 amino acids helix-turn-helix motif (2). The specific structure was initially characterized by magnetic resonance spectroscopy imaging.
In the human genome, according to the sequence similarity and position correlation in the chromosome, 39 HOX family genes can be divided into 4 clusters, namely HOXA, HOXB, HOXC, HOXD. The number after the subgroup HOXA, HOXB, HOXC, HOXD increases orderly in a 3’ to 5’ orientation, which is shown in Figure 1. Each cluster has between 9 and 11 genes in a row on a homologous strand of DNA, which contains duplication and divergence of ancestral HOX genes.
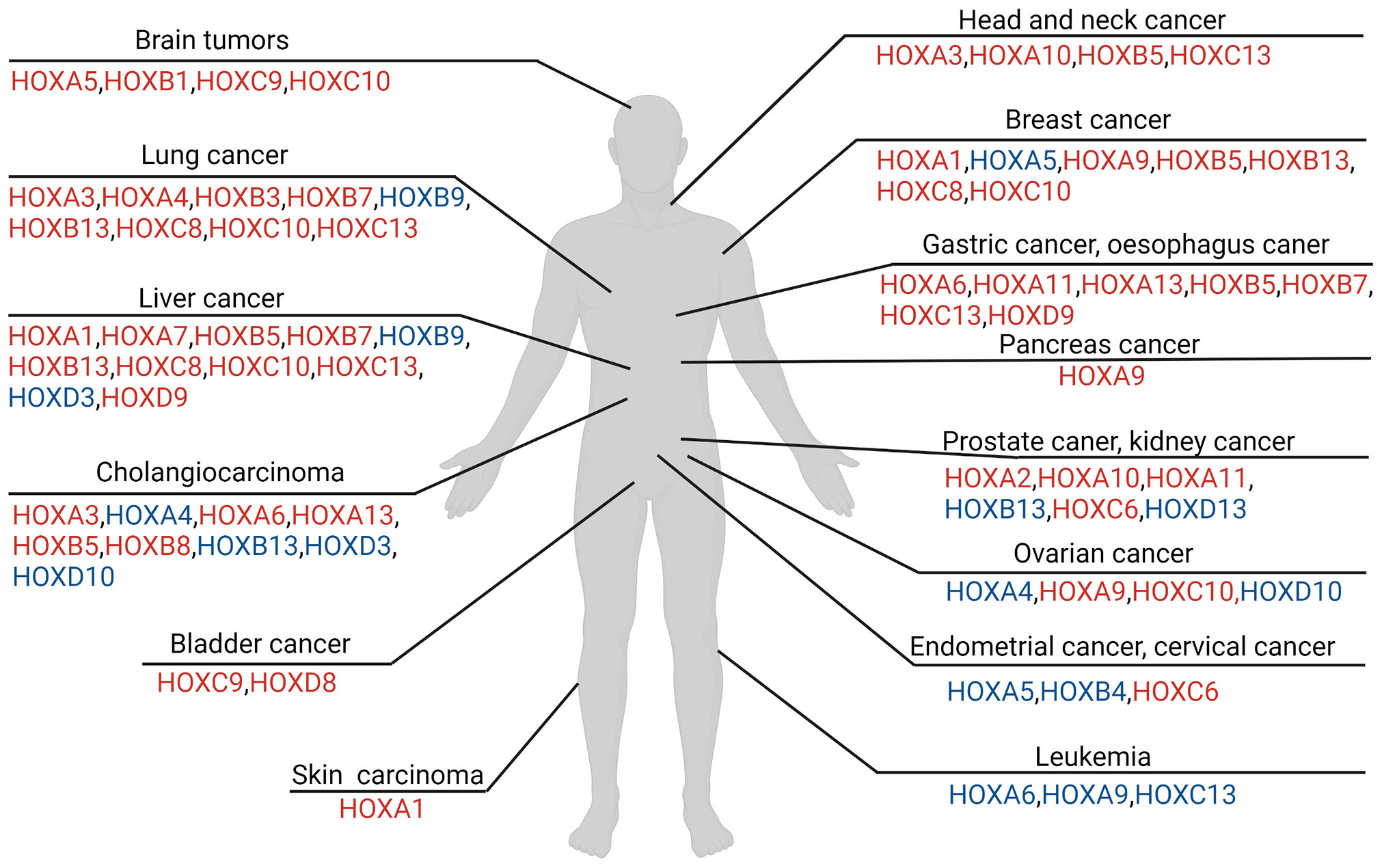
Figure 1 Overview of HOX involvement in different tumors in human. The colors of HOX molecules indicate regulations in relevant tumor cell. The red font means the corresponding HOX genes is up-regulated in tumor, while the blue font means the corresponding HOX genes is down-regulated in comparison. Regulation of HOX factors in various tumor types described in this figure legend. [brain cancer mainly includes the glioma and neuroblastoma (4–11), lung cancer (12–24), hepatocellular carcinoma (25–31), bladder cancer (32, 33), cholangiocarcinoma (34), head and neck squamous cell carcinoma (35, 36), breast cancer (21–23, 37–54), gastric carcinoma and esophagus cancer (55–64), prostate cancer and renal carcinoma (43, 65–72), ovarian cancer (73–79), endometrial cancer and cervical cancer (80–84), leukemia (85–95), skin carcinoma (96–98)].
1.2 Physiological Function of HOX Genes
HOX genes encode a family of master transcriptional regulators throughout growth and development in human tissues and organs. This period monitoring elicits distinct and temporospatial limb and organ developmental programs along the anterior-posterior axis (99). Researchers found the chromosomal arrangement of HOX genes was closely related to their localization order of genetic expression (100).
Plenty of scientific evidence suggests that the specific functions of individual HOX genes largely reflect their regional and restricted expression patterns. The disruption of the chromosome region related expression pattern may lead to developmental defects and diseases, especially human cancer (101).
HOX proteins encoded by HOX genes are key components of substantial metabolic processes such as lipidic metabolism, and their roles in organogenesis and tumorigenesis have been studied in detail over the past decade (12, 37, 73, 102–105). Pluripotent embryonic carcinoma cells have been associated with the differentiation of a broad spectrum of tissues early in development in mice embryos (106). This process appears to be related to retinoic acid reactions, the chromosomal region determining the aforementioned relation was regulated by HOX gene (96, 107).
In conclusion, HOX genes act as primary regulators to regulate downstream target molecules (103). HOX genes play an essential role in embryonic development, including morphogenesis, organogenesis, and differentiation of a variety of tissues and organs (108).
2 Deregulation of HOX Genes in Human Cancer
Over the past several decades, we have come to understand that a great many genes and proteins controlling embryogenesis usually partake indispensable effects in carcinogenesis likewise. Many genes identified as pivotal genes in carcinogenesis, for example, oncogenes or tumor suppressor genes, have been discovered to paly primary roles in embryogenesis correspondingly. It indicates both processes are tightly linked (109). The familiar components of signal transduction pathways, such as the Sonic Hedgehog pathway, Wingless-Type MMTV Integration Site Family, Notch pathway, Paxillin pathway, and Sry-box transcription factors family members, closely act in accordance with the above rules.
This close relationship between embryogenesis and carcinogenesis supports the lineage-dependency theory. The theory proposes that the cellular mechanisms participating in lineage heredity and adaptability during development, potentially underlies tumorigenic mechanisms in humans (110). The HOX genes might be applicable to the lineage-dependency theory. Apart from the indispensable involvement of HOX genes in embryonic development in physiological status, their abnormal expression, has long been linked to tumors. Initially, these genes were thought to be related to human cancers in the hematologic system and embryo. Later, many other kinds of tumors were also noticed to be associated with deregulated HOX genes (85, 101, 111). In the context of cancer development, the abnormal expression of HOX gene may affect cell proliferation, differentiation, apoptosis, motility, angiogenesis, autophagy, and cell receptor signaling (112). HOX genes in the hallmarks of cancer are shown in Figure 2.
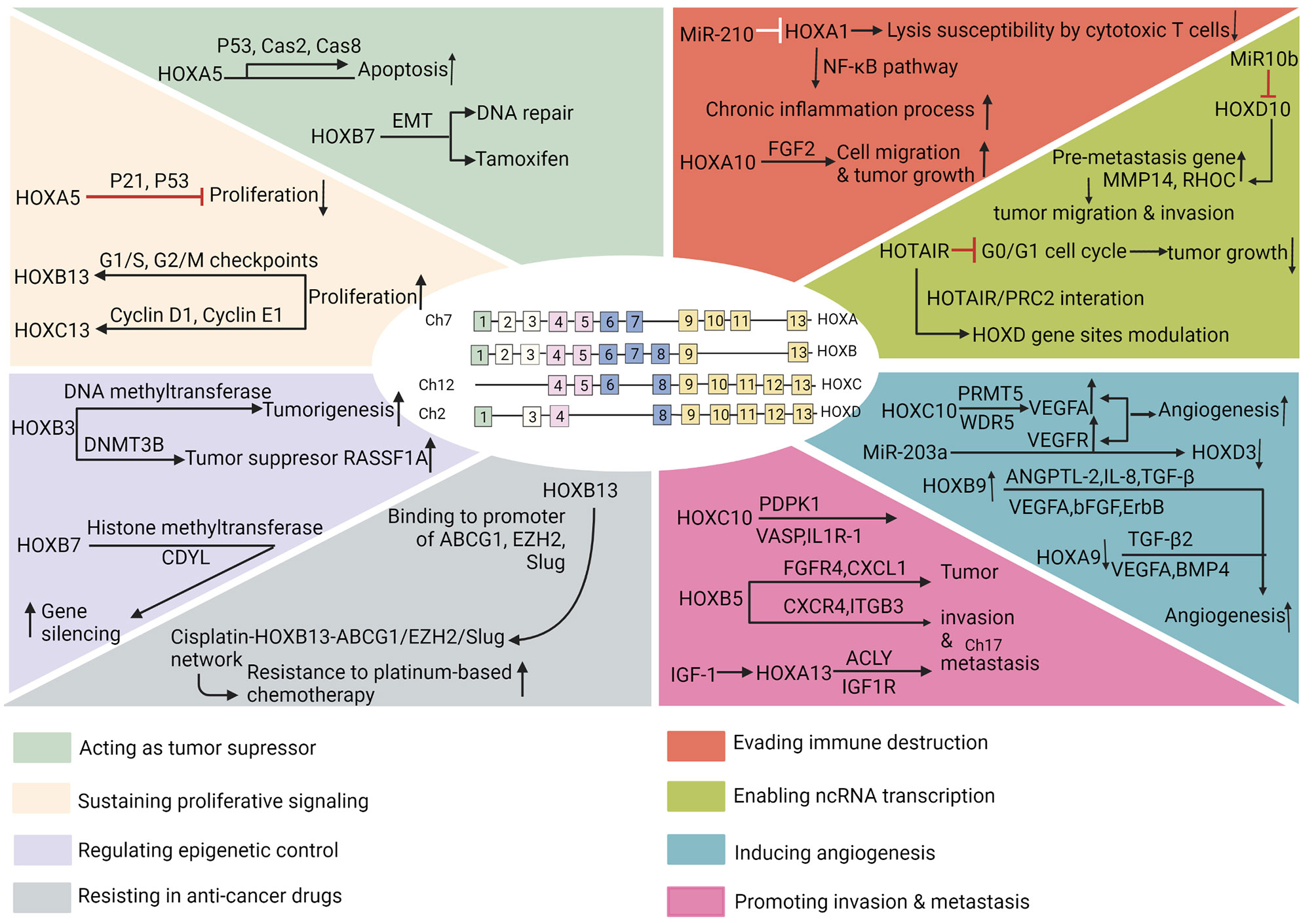
Figure 2 HOX genes in the hallmarks of cancer. The hallmarks of cancer comprise eight biological capabilities acquired during multistep development of human tumors. The hallmarks constitute an organizing principle for rationalizing the complexities of neoplastic disease. They include acting as tumor suppressor, sustaining proliferative signaling, regulating epigenetic control, resisting in anti-cancer drugs, evading immune destruction, enabling ncRNA transcription, inducing angiogenesis, and promoting invasion and metastasis.
The protein products of HOX genes are initially thought to be transcription factors that accelerate cancerization since HOX proteins are deregulated in carcinogenesis. In the above process, HOX proteins govern the intricate balance of multiple signaling pathways on and off, and affect downstream targets of these pathways (113), so as to determine different cancer outcomes.
Subsequent studies have shown that HOX genes act not only as transcriptional activators but also as transcriptional repressors in cancer. This abnormal regulation of HOX genes in cancer suggests that HOX genes expression is an integral part of the regulatory network (103, 114, 115).
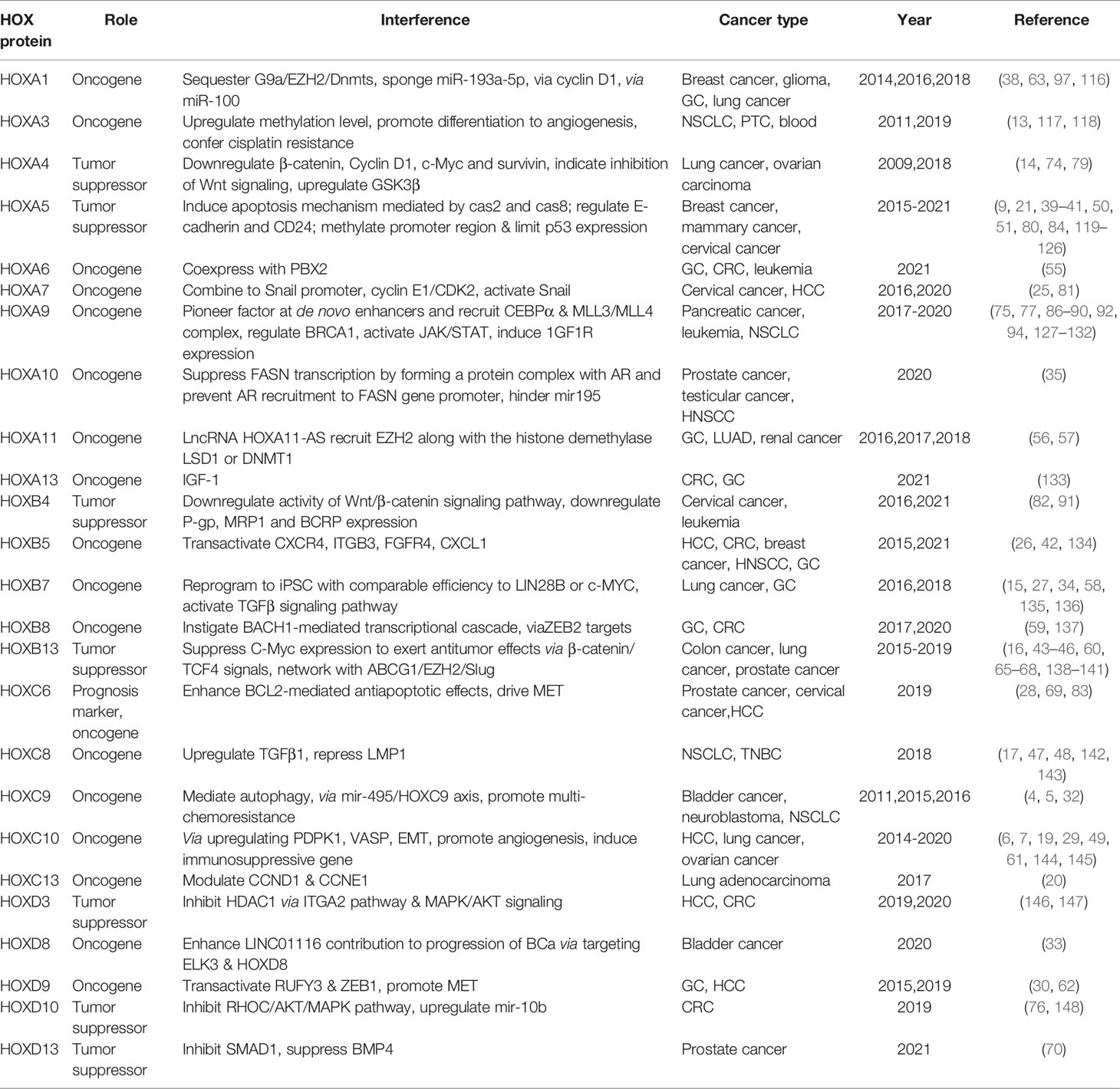
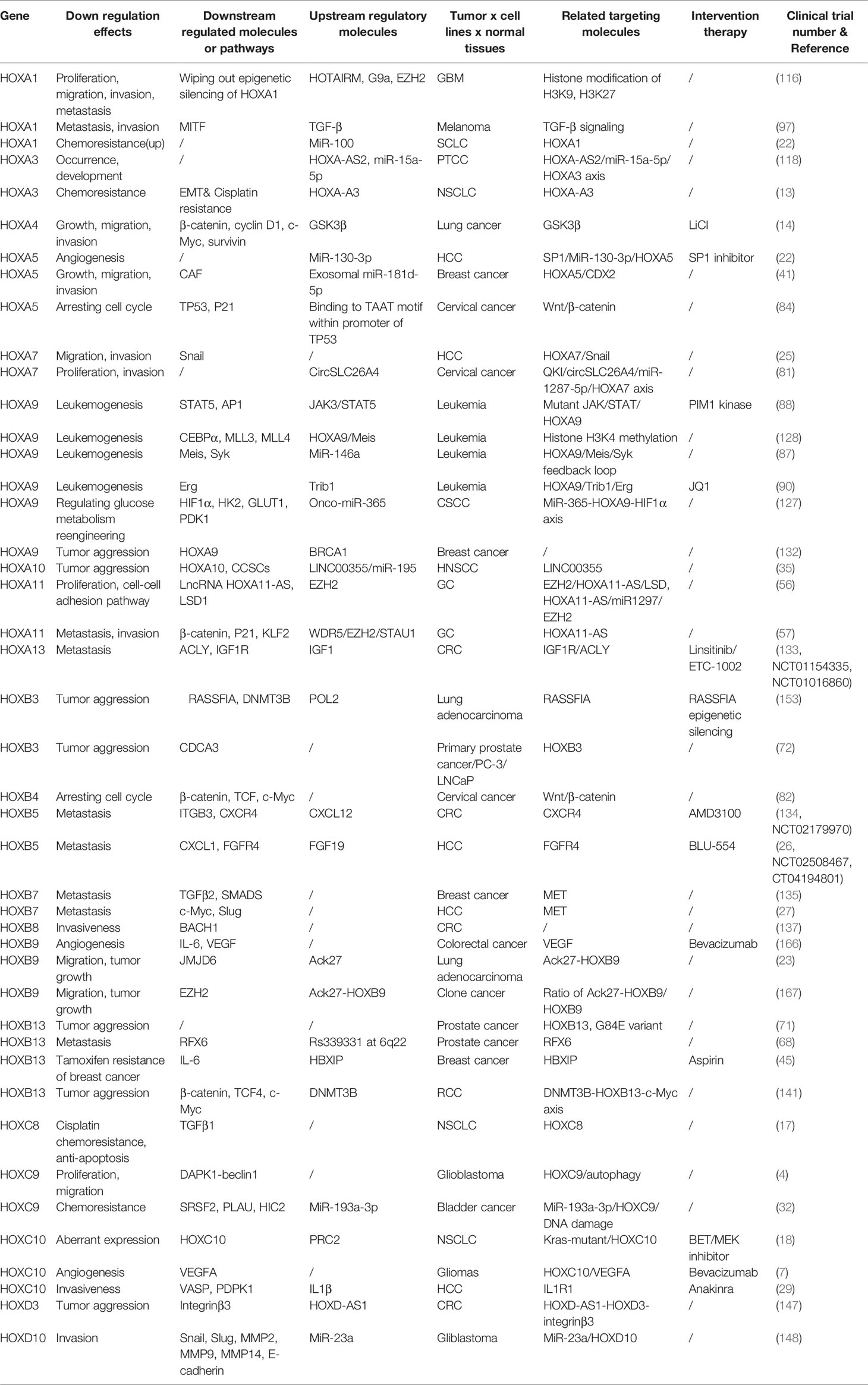
HOX genes in tumor show temporospatial deregulation pattern, different from that in normal tissues and organs. Besides, the gene dominance in expression level, that is, the aberrantly increased expression level of HOX genes in specific tissue types, the mechanism is also proposed to explain HOX genes relevant to cancer. The targets identified for HOX factors in human cancer are shown in Table 1 (4–7, 13–20, 25–30, 32–35, 38–49, 55–62, 65–70, 74–76, 80–83, 86–91, 116–121, 127–130, 133–149).
2.1 Direct Role: As Oncogenes
The HOX genes have been reported to act as oncogenes and contribute to tumor progression in several types of cancer. For example, simultaneous overexpression of HOXA9 and MEIS1A induces acute myeloid leukemia in rats (92, 131). In breast cancer, HOXB7 has been reported as an oncogene because its upregulation appears to promote the expression of bFGF and induce the epithelial-mesenchymal transitions (EMT) (15, 27, 34). In colorectal cancer, HOXB5 overexpression mediated by CXC chemokine ligand 12 facilitates metastasis through transactivating downstream protein CXCR4 and ITGB3 (134). HOXB13 seems to be an oncogene for ovarian and prostate cancer. Its knockout in ovarian cancer cells results in reduced tumor invasion (45, 65, 71, 138, 141). On the contrary, its ectopic expression promotes cell proliferation and non-anchoring. Mutations and growing resistance to tamoxifen-mediated apoptosis in the cell tumor antigens P53, Myc, and Ras are included in mouse ovarian cancer cells (43–46). In addition, HOXB13 overexpression appears to promote invasion of these types of cancers. In prostate cancer, HOXB13 regulates the prostate-derived ETS family members and also facilitates cell invasion (140). As HOXB3 in glioblastoma, its knockout leads to cancer cell suppression (150). In squamous carcinoma of the cervix, HOXC10, which is overexpressed and thought to be an oncogene, is knocked out to reduce invasiveness (6, 19, 29, 61). In colorectal cancer, HOXA13 overexpression mediated by insulin-like growth factor 1 promotes metastasis through upregulating downstream targets ATP-citrate lyase and insulin-like growth factor 1 receptor (133). Ectopic overexpression of ATP-citrate lyase and insulin-like growth factor 1 receptor rescues the decreased colorectal cancer metastasis induced by HOXA13 knockdown (133). HOXC10 overexpression mediated by Interleukin 1β facilitates hepatocellular carcinoma (HCC) metastasis (29). The above studies implicate HOX genes as oncogenes and prognostic biomarkers. Hence, targeting HOX genes’ relevant pathways is likely to be the promising therapeutic option for clinical cancer prevention (29).
2.2 Direct Role: As Tumor-Suppressor Genes
Basing on our review of the literature, we found the expression of specific HOX genes in cancers tends to vary with tissue type and tumor sites. Besides, the HOX genes have been found to behave as tumor-suppressor genes and dedicate to tumor suppression in several cancers.
HOXC8 in nasopharyngeal cancer is silent and its ectopic expression causes the inhibition of tumor growth (143). In vitro and in vivo, in addition, HOXA5 is often down-regulated in breast cancer and appears to mediate apoptosis through p53 or caspases 2 and 8 in normal cells, thus having a tumor-like effect inhibition (50, 121, 122). Downregulation of HOXC9 in infant neuroblastoma appears to lead to increased cancer cell survival and tumor growth (5). HOXB1 is also significantly down-regulated in glioma cells, its knockdown promotes proliferation and invasion, inhibits apoptosis of cancer cells in vitro, and leads to poor survival (5, 8, 151).
The expression changes of HOXA4, HOXA9, and HOXD10 cause abnormal proliferation and differentiation of colorectal carcinoma cells and contribute to tumor development (14, 74, 87, 127). In addition, HOXB3 overexpression promotes the proliferation and invasion of glioblastoma cells, acute myeloid leukemia, pancreas, prostate, ovarian, and lung cancer (74, 93, 150, 152, 153). Excessive HOXB7 inducing MET in breast cancer cells also interferes with DNA repair and tamoxifen (136). HOXA5 down-regulation is discovered in multiple tumors, including liposarcoma, cervical cancer, breast cancer, which suggests that HOXA5 may be an important tumor suppressor (21, 51, 84, 123). The promoter methylation and downregulation of HOXA5 during the epigenetic deregulation decrease RARβ-driven apoptosis mediated by caspase 2 and caspase 8. Therefore, HOXA5 consumption in breast cells causes a lack of epithelial cell characteristics as well as an increase of stem cell property and cellular plasticity, eventually leads to a more aggressive phenotype (119).
Therefore, the abnormal expression of the HOX genes in various cancers seems to indicate that random alterations are not necessary for the health maintenance and survival of cancer cells. Instead, abnormal expression of specific HOX genes in cancer seems to play a large part in the development of cancer.
However, HOX genes may show a predisposition. It is up-regulated or down-regulated in certain tumors, promoting cancer progression or inhibition in certain cases. Thus, they can be regarded as oncogenes or tumor suppressor genes, usually depending on the corresponding tumor microenvironment.
2.3 Indirect Role: Epigenetic Control via HOX Genes
HOX proteins are also involved in chromatin posttranslational modifications, epigenetically affect the expression of crucial cancer progression genes. For example, HOXB3 regulates the expression of DNA methyltransferase, which can add methyl to the 5 positions of cytosine ring carbon in CpG dinucleotide (93, 152, 153). Therefore, HOX proteins can indirectly regulate the expression of other genes by controlling the expression of methyltransferase. Thus, the HOX genes may have an indirect effect on cancer by controlling the expression of DNA methyltransferase, which seems to be of considerable relevance to understanding the mechanisms involved in tumorigenesis.
In fact, all cancer progression has common abnormalities, such as the massive epigenetic silencing of tumor suppressor genes. The mechanisms behind this phenomenon are remained to be fully understood. The DNA methyltransferase protein family consists of DNA methyltransferase (DNMT)-1, DNMT3A, DNMT3B, and DNMT3L (154). The last is associated with de novo methylation of DNMT3A/B, which is required during embryonic development, imprinting, and X chromosome inactivation. DNMT1 maintains the DNA methylation profile of the genome during each cell division. DNMT3A and DNMT3B can actively add de novo methyl to DNA sequences to control gene expression and play a key role in cancer progression.
Palakurthy has shown that DNMT3B is the target of HOXB3 protein and is involved in the epigenetic regulation of tumor suppressor gene RASSF1A in lung adenocarcinoma and other cancers (153). HOXB3 also directly interacts with DNMT3B to promote the occurrence of leukemia in acute myeloid leukemia.
In addition, HOX genes may also play a role in post-translational modification of chromatin and affect gene expression. For instance, Heinonen has shown that HOXB7 can directly bind to chromodomain protein Y-like and enhance the histone methyltransferase activity of Polycomb Complex 2 and induce gene silencing as typical for trimethylate lysine 27 of histone H3 (155).
2.4 Tumor Proliferation, Invasion, and Metastasis
Proliferation phenotype has been found in various HOX-related researches, especially in leukemia, where HOX upregulated expression is often resulted from translocation mutations or altered regulation in the trithorax homologue myeloid-lymphoid leukemia (112). Invasion and metastasis phenotypes caused by abnormal HOX gene expression have been mostly studied in solid tumors, where HOX gene deregulation is usually due to loss-of-function mutations or gain-of-function mutations, or undefined mutations in upstream regulators.
In recent work, HOXC13 promotes cell proliferation by modulating the expression of cyclin D1 and cyclin E1 in lung adenocarcinoma. HOXA5 inhibits cell proliferation by regulating p53 expression in liposarcomas and p21 in non-small lung cancers (20, 63, 84, 120). Although HOXB13 overexpression promotes prostate cancer metastasis by downregulating intracellular zinc and upregulating NF-kappaB signaling pathways, HOXB13 consumption promotes proliferation of PC-3 and LNCaP cells by controlling G1/S and G2/M checkpoints (16, 60, 67, 138, 141).
HOX proteins affect cell cycle process by regulating cell cycle-related proteins (156), thus also affect proliferation and apoptosis in cancer progression. Growing evidence is noticeable that many HOX transcription factors are abnormally expressed in cancer, and their dysregulation significantly promotes tumor invasion and metastasis.
HOXD9 interacts with the promoter region of zinc-finger E-box binding homeobox (ZEB)-1, inhibition of ZEB1 induced by HOXD9 suppresses HCC cell migration, invasion as well as EMT (30). In addition, according to microarray analysis, ZEB2 may be a downstream cofactor of HOXB8 (59). Patients enrolled in studies have shown that high HOXB13 expression promotes the progression of lung adenocarcinoma and predicts poor prognosis (16).
In epithelial ovarian cancer cells, HOXA9 not only promotes the growth of epithelial ovarian cancer cells in vivo by activating transcriptional activity of the gene encoding transforming growth factor β (TGFβ)-2, but also binds to the promoter of the cadherin3 gene that encodes P-cadherin to induce intraperitoneal dissemination (77, 94). Many reports suggest the temporospatial deregulation of HOXA9 is associated with primary tumors and specific histological subtypes. HOXA9 is of therapeutic potency, while the potency is limited by the low membrane permeability.
2.5 Angiogenesis
Angiogenesis is an important link during tumor progression. HOX proteins affect angiogenesis mainly by regulating VEGF expression (7). For example, HOXC10 level is statistically correlated with VEGFA expression in gliomas. HOXC10 upregulates VEGFA expression transcriptionally by binding to its promoter, and the post-translational modification of histones mediated by protein arginine methyltransferase 5 and WD repeat domain 5 is required in angiogenesis (7).MiR-203a negatively targets HOXD3 directly by targeting the VEGFR promoter region and increases VEGFR expression.
HOX protein expression also plays an important role in endothelial cells (EC) (146). For instance, HOXA5 is expressed in static EC but not in activated angiogenic EC. HOXA5 continuously increases the TSP-2 expression and decreases the VEGF expression, thereby inhibiting histopathological angiogenesis (124, 125). In addition, HOXA5 is absent in EC of proliferative hemangioma (9). HOXA5 increases the mRNA and protein expression of Akt1, further enhances Akt activity by coordinating the down-regulation of PTEN, thereby increasing the stability of the capsular patellar junction (126).
2.6 Resistance in Anti-Cancer Drugs
HOX proteins are involved in resistance to the anti-cancer drugs in cancers, especially in myeloid leukemia, HCC, breast cancer, and lung cancer. HOX proteins regulate various non-coding RNAs (ncRNAs) which influence cancer cells chemotherapy resistance. In particular, we take the example of the role of HOXB13 in mediating chemotherapy resistance in lung adenocarcinoma. HOXB13 upregulates a series of drug-transfer and drug-resistance-related genes by directly binding to their promoters and forming the cisplatin-HOXB13-ABCG1/EZH2/Slug network, including ATP Binding Cassette Subfamily G Member 1, Enhancer Of Zeste 2 Polycomb Repressive Complex 2, and Slug (16, 139). Levels of HOXB13 and its target genes may help predict sensitivity of platinum-based chemotherapy in lung adenocarcinoma patients (16).
2.7 HOX-Mediated Molecular Crosstalk During Tumorigenesis
2.7.1 Post-Translational Modifications
A variety of signaling pathways that regulate proliferation, apoptosis, differentiation, movement, and angiogenesis, interact with HOX transcription factors family. Post-translational modifications of HOX protein also play a key role in tumorigenesis. HOX post-translational modifications are an under-valued project. In most eukaryotic proteins, the turn-over, intracellular localization, molecular interactions and activity are modulated by post-translational modifications. The post-translational modifications of HOX proteins in cancer mainly contributes to the modulation of protein stability, DNA binding, transcriptional activator-like effectors interaction, transcriptional activation capacity, unidentified cellular impact (157).
2.7.2 MicroRNA and Non-Coding RNA
Of the 39 HOX genes, 30 HOX genes contain more than one conserved nucleotide sequences that are expected to be targets of miRNAs in vertebrate (2). We found that the HOX targeting genes are mainly located on the 3’ side of each HOX miRNA site. This study shows that HOX miRNAs help inhibiting abundant pre-gene expression, hence enhancing the prevalence of post-genes. In patients with gastrointestinal stromal tumors, the level of miRNA196a is positively related to higher tumor histologic grade, higher recurrence rate, and lower survival rate (64).
In addition, HOX transcribed antisense RNA (HOTAIR) also contributes to HOX expression regulation. HOTAIR is a 2.2-kilobase long non-coding RNA that is transcribed from the antisense strand of HOXC clusters, and its function is to inhibit the transcription of 40 kilobases of HOXD clusters (158, 159). The mechanism of action of these long non-coding RNAs has not been fully determined. However, HOTAIR has been shown to regulate chromatin state and kinetics by binding to specific chromatin modification complexes. The trimethylation of histone H3 lysine 27 at the HOXD site requires HOTAIR/PRC2 interactions. Knocking out HOTAIR activates HOXD gene transcriptional activity on human chromosome 2 (158, 160, 161). Thus, it is possible to modulate the HOXD gene by modulating HOTAIR levels, which has implications in cancer therapy. For example, the expression of HOTAIR is closely related to the neoplasm staging and poor prognosis of glioma. Reducing HOTAIR expression induces inhibition of colony formation, G0/G1 cell cycle arrest, and inhibition of tumor growth in situ (10). Thus, HOTAIR seems to serve as a prognostic factor for survival, as well as a biomarker for identifying molecular subtypes in cancer (162).
Recent evidence underscored the function of long noncoding RNAs (lncRNAs)-driven hepatocarcinogenesis. The expression level of lncRNA HOXA transcript at the distal tip (HOTTIP) and HOXA13 is associated with metastasis and survival in HCC patients. It indicates the prospective potency of HOTTIP and HOXA13 to be the predictive biomarker in HCC (163).
Summing up the above, we highlight the specific roles of miRNAs and lncRNAs in human cancer. They perform not only as the intermediary between DNA and protein, including chromosome remodeling, transcription, and post-transcriptional processing, but as leading characters in body balance adjustment during congenic malformation, oncogenesis, metabolic processes, and deregulation of cell cycle (164).
3 HOX Transcription Factors as Therapeutic Targets
3.1 HOX Genes Act as a Key Intermediate Point in the Anti-Cancer Progression
HOX proteins interact with specific cofactors to select their downstream binding sites in the genome. In vertebrates, those including pre-B cell leukemia transcription factor (PBX) and mouse heterotopic integration, belong to the tricarboxylic acid cycle family of homologous domain protein. In addition, these HOX cofactors can increase the nuclear translocation of HOX protein from the cytoplasm to the nucleus. Nuclear translocation of HOX protein is inhibited by suppressing the formation of HOX/PBX dimer, which impairs the function of HOX transcript factors (165). Therefore, considering the use of HOX protein as cancer therapeutic targets, its interaction with cofactors needs to be determined. Therapeutic values of HOX factors in human cancer are seen in Table 2 (4, 7, 13, 14, 17, 18, 22, 23, 25–27, 29, 31, 32, 35, 41, 45, 56, 57, 68, 71, 72, 81, 82, 84, 87, 88, 90, 97, 116, 117, 127, 128, 132–135, 137, 141, 147, 148, 153, 166, 167).
Morgan et al. have found HXR9 peptides specifically target the interaction between HOX and PBX (78, 168). HXR9 inhibits HOX function by preventing its binding to PBX, which leads to apoptosis in multiple mouse breast cancer derived cell lines (168). In addition, the interaction between HOX and HXR9 has been shown to cause apoptosis in numerous cancers, including melanoma, mesothelioma, myeloma, renal cancer, prostate cancer, lung cancer, ovarian cancer, pancreatic cancer, squamous cell cancer of the head and neck, and oral cancer (58, 78, 95, 98, 168). Kaspar and Reichert have already used HOX/HXR9-based treatments in cancer treatments, and Morgan and others are exploring a variant of HXR9 for intratumoral injection in clinical trials (168). The HOX gene targeting therapy is also being explored through RNA interference approaches (158). For example, MicroRNAs transcribed in HOX clusters, namely miR10A32 and miR196B33, regulate HOX expression through RNA interference (158). These miRNAs cleave or inhibit the translation of HOX mRNA. The use of these miRNAs in cancer cells in vitro seems to modulate HOX gene expression and its effect on cancer progression.
In fact, HOTAIR has the potential to be an effective therapeutic target for many types of cancer, where abnormal HOXD expression has been found to be associated with cancer progressions, such as cancers in breast, stomach, colon, cervix, lung, and liver (11, 12, 24, 36, 52–54, 158, 159, 169, 170). For example, HOTAIR is highly expressed in cervical cancer compared to normal cells (158, 159, 169). However, its knockout in cervical carcinoma cells induces apoptosis and inhibits tumor proliferation, migration, and invasion.
3.2 HOX Genes as Therapeutic Targets
Previous studies have shown that HOX protein exerts carcinogenic activity not only through its reversed transcription ability but through its protein interaction network.
When designing domain-specific HOX inhibitors, gene targeting must be taken into account. The problem in bringing monoclonal antibodies into clinical therapies is their nuclear localization. To address the above issue, the focus of next-generation immunotherapies is to develop smaller monoclonal antibody fragments or totally new entities to improve tissue permeability and subcellular localization. These crucial immunotherapeutic strategies are also focused on addressing the treatment of hematologic tumors and solid tumors.
Since HOX protein is closely linked to brain malignancies where the blood brain barrier is a major challenge for drug infiltration, this provides instructive thinking of treatment. Another strategy for targeting oncogenes in cancer cells is to use small interfering RNAs (siRNAs) (171). Many studies have shown inhibition of HOX expression by siRNAs can distinctly retard tumor growth and aggressiveness. Similarly, the use of siRNA strategies may be an effective therapeutic approach for targeting HOX genes in vivo.
Although the strategy is not as progressive as small molecular chemical compound inhibitors or monoclonal antibodies, efforts have been made over the past decade to implement it in cancer treatment. At present, poor uptake of cells, side effects of packaging-related methods are major obstacles to HOX targeting clinical application. As a result, current efforts are made to address a range of novel strategies for delivering siRNAs in vivo.
The small-molecule chemical inhibitors are of anticancer potential. We highlight the recent therapies targeting HOX genes-downstream proteins especially in HCC and colorectal cancer. Overexpression of HOXB5 transactivates downstream protein expression of FGFR4 and CXCL1, hence promoting HCC metastasis. The small chemical compounds application of FGFR4 inhibitor BLU-554 and CXCR2 inhibitor SB265610 sharply inhibits HCC metastasis mediated by HOXB5 (26). Integrins are also involved in HOX-induced cancer metastasis. In CAOV-3 cells, HOXA4 overexpression inhibited migration and increased the protein level of β1 integrin, suggesting that the β1 integrin may be involved in HOXA4’s inhibitory effect on cell motility (79). In HOXC10-VASP/IL-1R1 mediated HCC metastasis, daily administration of IL-1R1 antagonist anakinra dramatically prolongs survival time (29). In colorectal cancer, the molecule network IGF1-HOXA13-ACLY/IGF1R and CXCL12-HOXB5-CXCR4/ITGB3, targeted blocking the downstream protein with small molecular compounds serves as a promising anticancer therapy (133, 134).
Furthermore, natural and synthetic drugs regulating HOX genes network are associated with anticancer and antibacterial activities. For instance, researchers synthesized the new molecule 5H-pyrido[3,2-a] phenoxazine-5-one in the laboratory, evaluated its ability to regulate the activity of lncRNA HOTAIR and HOXC locus genes from HOXC9 to HOXC13 in MCF-7 human breast cancer cell lines, and confirmed that 5H-pyrido[3,2-a] phenoxazine-5-one was able to inhibit the relevant HOX gene expression and counteract the pathogenesis of breast cancer (172).
The discovery of targeting downstream molecules with chemical compounds has enhanced the potential to specifically target HOX genes intermediated network and eliminate the cancer development, which may improve clinical outcomes in cancers.
4 Conclusion
Although it is difficult to intercept various transcription factors, alternative strategies based on the exploitation of the associated molecular networks are emerging. The HOX genes play an important role in cancer progression, showing great plasticity and interfering with many molecular mechanisms. Abnormal HOX expression by altering its homologous box methylation profile, is often associated with cancer. In addition, different HOX gene regulatory features describe different cancer types and are increasingly being used as cancer biomarkers. Their role in cancer progression is based on the ability to control gene expression either directly as transcriptional regulators or indirectly through epigenetic control. In fact, HOX proteins have been shown to be involved in different epigenetic mechanisms involved in DNA and histone methylation, which may be related to the epigenetic regulation of multiple cancer related genes. Therefore, it is very important to seek pharmacological agents that are synthetically lethal in conjunction with HOX overexpression. Therapies targeting HOX molecules should intake the functional redundancy among the different HOX family members, so that appropriate therapeutic combination can be created. There is an urgent need for a more comprehensive understanding of their biology, and identifying their gene targets and molecular networks involved in tumor progression.
Author Contributions
Conceptualization, YF, and LX. Writing—original draft preparation, YF. Writing—review and editing, YF, TZ, YW, MX, XJ, WH, and LX. Supervision, TZ, MX, XJ, XL, and LX. Project administration, WH and LX. Funding acquisition, WH and LX. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by grants from the National Natural Science Foundation of China No. 81871911 (WH), No. 81972237 (LX), and No.81772623 (LX), and the National Key Research and Development Program of China 2018YFC1312103 (LX).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
NSCLC, non-small-cell lung carcinoma; CRC, colorectal cancer; PTC, papillary thyroid cancer; LSCC, laryngeal squamous cell cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; FASN, fatty acid synthase; LUAD, lung adenocarcinoma; HNSCC, head and neck squamous cell carcinoma; TNBC, triple negative breast cancer; CMM, cutaneous malignant melanoma; ESCC, esophageal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; CHOL, cholangiocarcinoma.
References
1. Mallo M, Alonso CR. The Regulation of Hox Gene Expression During Animal Development. Dev (Cambridge England) (2013) 140:3951–63. doi: 10.1242/dev.068346
2. Holland PW. Evolution of Homeobox Genes. Wiley Interdiscip Rev Dev Biol (2013) 2:31–45. doi: 10.1002/wdev.78
3. Montavon T, Soshnikova N. Hox Gene Regulation and Timing in Embryogenesis. Semin Cell Dev Biol (2014) 34:76–84. doi: 10.1016/j.semcdb.2014.06.005
4. Xuan F, Huang M, Liu W, Ding H, Yang L, Cui H. Homeobox C9 Suppresses Beclin1-Mediated Autophagy in Glioblastoma by Directly Inhibiting the Transcription of Death-Associated Protein Kinase 1. Neuro-Oncology (2016) 18:819–29. doi: 10.1093/neuonc/nov281
5. Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W, et al. HOXC9 Links Cell-Cycle Exit and Neuronal Differentiation and Is a Prognostic Marker in Neuroblastoma. Cancer Res (2011) 71:4314–24. doi: 10.1158/0008-5472.Can-11-0051
6. Li S, Zhang W, Wu C, Gao H, Yu J, Wang X, et al. HOXC10 Promotes Proliferation and Invasion and Induces Immunosuppressive Gene Expression in Glioma. FEBS J (2018) 285:2278–91. doi: 10.1111/febs.14476
7. Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G, et al. Overexpression of HOXC10 Promotes Angiogenesis in Human Glioma via Interaction With PRMT5 and Upregulation of VEGFA Expression. Theranostics (2018) 8:5143–58. doi: 10.7150/thno.27310
8. Han L, Liu D, Li Z, Tian N, Han Z, Wang G, et al. HOXB1 Is a Tumor Suppressor Gene Regulated by miR-3175 in Glioma. PloS One (2015) 10:e0142387. doi: 10.1371/journal.pone.0142387
9. Zhu Y, Cuevas IC, Gabriel RA, Su H, Nishimura S, Gao P, et al. Restoring Transcription Factor HoxA5 Expression Inhibits the Growth of Experimental Hemangiomas in the Brain. J Neuropathol Exp Neurol (2009) 68:626–32. doi: 10.1097/NEN.0b013e3181a491ce
10. Zhang L, He A, Chen B, Bi J, Chen J, Guo D, et al. A HOTAIR Regulatory Element Modulates Glioma Cell Sensitivity to Temozolomide Through Long-Range Regulation of Multiple Target Genes. Genome Res (2020) 30:155–63. doi: 10.1101/gr.251058.119
11. Angelopoulou E, Paudel YN, Piperi C. Critical Role of HOX Transcript Antisense Intergenic RNA (HOTAIR) in Gliomas. J Mol Med (Berlin Germany) (2020) 98:1525–46. doi: 10.1007/s00109-020-01984-x
12. Li L, Wang Y, Song G, Zhang X, Gao S, Liu H. HOX Cluster-Embedded Antisense Long Non-Coding RNAs in Lung Cancer. Cancer Lett (2019) 450:14–21. doi: 10.1016/j.canlet.2019.02.036
13. Lin S, Zhang R, An X, Li Z, Fang C, Pan B, et al. LncRNA HOXA-AS3 Confers Cisplatin Resistance by Interacting With HOXA3 in Non-Small-Cell Lung Carcinoma Cells. Oncogenesis (2019) 8:60. doi: 10.1038/s41389-019-0170-y
14. Cheng S, Qian F, Huang Q, Wei L, Fu Y, Du Y. HOXA4, Down-Regulated in Lung Cancer, Inhibits the Growth, Motility and Invasion of Lung Cancer Cells. Cell Death Dis (2018) 9:465. doi: 10.1038/s41419-018-0497-x
15. Monterisi S, Lo Riso P, Russo K, Bertalot G, Vecchi M, Testa G, et al. HOXB7 Overexpression in Lung Cancer Is a Hallmark of Acquired Stem-Like Phenotype. Oncogene (2018) 37:3575–88. doi: 10.1038/s41388-018-0229-9
16. Zhan J, Wang P, Li S, Song J, He H, Wang Y, et al. HOXB13 Networking With ABCG1/EZH2/Slug Mediates Metastasis and Confers Resistance to Cisplatin in Lung Adenocarcinoma Patients. Theranostics (2019) 9:2084–99. doi: 10.7150/thno.29463
17. Liu H, Zhang M, Xu S, Zhang J, Zou J, Yang C, et al. HOXC8 Promotes Proliferation and Migration Through Transcriptional Up-Regulation of Tgfβ1 in Non-Small Cell Lung Cancer. Oncogenesis (2018) 7:1. doi: 10.1038/s41389-017-0016-4
18. Guerra SL, Maertens O, Kuzmickas R, De Raedt T, Adeyemi RO, Guild CJ, et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell (2020) 37:705–719.e6. doi: 10.1016/j.ccell.2020.03.004
19. Tang XL, Ding BX, Hua Y, Chen H, Wu T, Chen ZQ, et al. HOXC10 Promotes the Metastasis of Human Lung Adenocarcinoma and Indicates Poor Survival Outcome. Front Physiol (2017) 8:557. doi: 10.3389/fphys.2017.00557
20. Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M, et al. HOXC13 Promotes Proliferation of Lung Adenocarcinoma via Modulation of CCND1 and CCNE1. Am J Cancer Res (2017) 7:1820–34.
21. Duriseti S, Winnard PT Jr., Mironchik Y, Vesuna F, Raman A, Raman V. HOXA5 Regulates Hmlh1 Expression in Breast Cancer Cells. Neoplasia (New York N.Y.) (2006) 8:250–8. doi: 10.1593/neo.05766
22. Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang J, et al. Downregulation of HOXA1 Gene Affects Small Cell Lung Cancer Cell Survival and Chemoresistance Under the Regulation of miR-100. Eur J Cancer (Oxford England: 1990) (2014) 50:1541–54. doi: 10.1016/j.ejca.2014.01.024
23. Wan J, Xu W, Zhan J, Ma J, Li X, Xie Y, et al. PCAF-Mediated Acetylation of Transcriptional Factor HOXB9 Suppresses Lung Adenocarcinoma Progression by Targeting Oncogenic Protein JMJD6. Nucleic Acids Res (2016) 44:10662–75. doi: 10.1093/nar/gkw808
24. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in Lung Cancer. J Hematol Oncol (2014) 7:90. doi: 10.1186/s13045-014-0090-4
25. Tang B, Qi G, Sun X, Tang F, Yuan S, Wang Z, et al. HOXA7 Plays a Critical Role in Metastasis of Liver Cancer Associated With Activation of Snail. Mol Cancer (2016) 15:57. doi: 10.1186/s12943-016-0540-4
26. He Q, Huang W, Liu D, Zhang T, Wang Y, Ji X, et al. Homeobox B5 Promotes Metastasis and Poor Prognosis in Hepatocellular Carcinoma, via FGFR4 and CXCL1 Upregulation. Theranostics (2021) 11:5759–77. doi: 10.7150/thno.57659
27. Huan HB, Yang DP, Wen XD, Chen XJ, Zhang L, Wu LL, et al. HOXB7 Accelerates the Malignant Progression of Hepatocellular Carcinoma by Promoting Stemness and Epithelial-Mesenchymal Transition. J Exp Clin Cancer Res: CR (2017) 36:86. doi: 10.1186/s13046-017-0559-4
28. Li PD, Chen P, Peng X, Ma C, Zhang WJ, Dai XF. HOXC6 Predicts Invasion and Poor Survival in Hepatocellular Carcinoma by Driving Epithelial-Mesenchymal Transition. Aging (2018) 10:115–30. doi: 10.18632/aging.101363
29. Dang Y, Chen J, Feng W, Qiao C, Han W, Nie Y, et al. Interleukin 1β-Mediated HOXC10 Overexpression Promotes Hepatocellular Carcinoma Metastasis by Upregulating PDPK1 and VASP. Theranostics (2020) 10:3833–48. doi: 10.7150/thno.41712
30. Lv X, Li L, Lv L, Qu X, Jin S, Li K, et al. HOXD9 Promotes Epithelial-Mesenchymal Transition and Cancer Metastasis by ZEB1 Regulation in Hepatocellular Carcinoma. J Exp Clin Cancer Res: CR (2015) 34:133. doi: 10.1186/s13046-015-0245-3
31. Liao Y, Wang C, Yang Z, Liu W, Yuan Y, Li K, et al. Dysregulated Sp1/miR-130b-3p/HOXA5 Axis Contributes to Tumor Angiogenesis and Progression of Hepatocellular Carcinoma. Theranostics (2020) 10:5209–24. doi: 10.7150/thno.43640
32. Lv L, Li Y, Deng H, Zhang C, Pu Y, Qian L, et al. MiR-193a-3p Promotes the Multi-Chemoresistance of Bladder Cancer by Targeting the HOXC9 Gene. Cancer Lett (2015) 357:105–13. doi: 10.1016/j.canlet.2014.11.002
33. Meng L, Xing Z, Guo Z, Liu Z. LINC01106 Post-Transcriptionally Regulates ELK3 and HOXD8 to Promote Bladder Cancer Progression. Cell Death Dis (2020) 11:1063. doi: 10.1038/s41419-020-03236-9
34. Dai L, Hu W, Yang Z, Chen D, He B, Chen Y, et al. Upregulated Expression of HOXB7 in Intrahepatic Cholangiocarcinoma Is Associated With Tumor Cell Metastasis and Poor Prognosis. Lab investigation; J Tech Methods Pathol (2019) 99:736–48. doi: 10.1038/s41374-018-0150-4
35. Lu S, Sun Z, Tang L, Chen L. LINC00355 Promotes Tumor Progression in HNSCC by Hindering MicroRNA-195-Mediated Suppression of HOXA10 Expression. Mol Ther Nucleic Acids (2020) 19:61–71. doi: 10.1016/j.omtn.2019.11.002
36. Sun S, Wu Y, Guo W, Yu F, Kong L, Ren Y, et al. STAT3/HOTAIR Signaling Axis Regulates HNSCC Growth in an EZH2-Dependent Manner. Clin Cancer Res: an Off J Am Assoc Cancer Res (2018) 24:2665–77. doi: 10.1158/1078-0432.Ccr-16-2248
37. de Bessa Garcia SA, Araújo M, Pereira T, Mouta J, Freitas R. HOX Genes Function in Breast Cancer Development. Biochim Biophys Acta Rev Cancer (2020) 1873:188358. doi: 10.1016/j.bbcan.2020.188358
38. Li J, Zeng T, Li W, Wu H, Sun C, Yang F, et al. Long Non-Coding RNA SNHG1 Activates HOXA1 Expression via Sponging miR-193a-5p in Breast Cancer Progression. Aging (2020) 12:10223–34. doi: 10.18632/aging.103123
39. Teo WW, Merino VF, Cho S, Korangath P, Liang X, Wu RC, et al. HOXA5 Determines Cell Fate Transition and Impedes Tumor Initiation and Progression in Breast Cancer Through Regulation of E-Cadherin and CD24. Oncogene (2016) 35:5539–51. doi: 10.1038/onc.2016.95
40. Hussain I, Deb P, Chini A, Obaid M, Bhan A, Ansari KI, et al. HOXA5 Expression Is Elevated in Breast Cancer and Is Transcriptionally Regulated by Estradiol. Front Genet (2020) 11:592436. doi: 10.3389/fgene.2020.592436
41. Wang H, Wei H, Wang J, Li L, Chen A, Li Z. MicroRNA-181d-5p-Containing Exosomes Derived From CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol Ther Nucleic Acids (2020) 19:654–67. doi: 10.1016/j.omtn.2019.11.024
42. Lee JY, Hur H, Yun HJ, Kim Y, Yang S, Kim SI, et al. HOXB5 Promotes the Proliferation and Invasion of Breast Cancer Cells. Int J Biol Sci (2015) 11:701–11. doi: 10.7150/ijbs.11431
43. Shah N, Jin K, Cruz LA, Park S, Sadik H, Cho S, et al. HOXB13 Mediates Tamoxifen Resistance and Invasiveness in Human Breast Cancer by Suppressing Erα and Inducing IL-6 Expression. Cancer Res (2013) 73:5449–58. doi: 10.1158/0008-5472.Can-13-1178
44. Jansen MP, Sieuwerts AM, Look MP, Ritstier K, Meijer-van Gelder ME, van Staveren IL, et al. HOXB13-To-IL17BR Expression Ratio Is Related With Tumor Aggressiveness and Response to Tamoxifen of Recurrent Breast Cancer: A Retrospective Study. J Clin Oncology: Off J Am Soc Clin Oncol (2007) 25:662–8. doi: 10.1200/jco.2006.07.3676
45. Liu B, Wang T, Wang H, Zhang L, Xu F, Fang R, et al. Oncoprotein HBXIP Enhances HOXB13 Acetylation and Co-Activates HOXB13 to Confer Tamoxifen Resistance in Breast Cancer. J Hematol Oncol (2018) 11:26. doi: 10.1186/s13045-018-0577-5
46. Jerevall PL, Jansson A, Fornander T, Skoog L, Nordenskjöld B, Stål O. Predictive Relevance of HOXB13 Protein Expression for Tamoxifen Benefit in Breast Cancer. Breast Cancer Res: BCR (2010) 12:R53. doi: 10.1186/bcr2612
47. Shah M, Cardenas R, Wang B, Persson J, Mongan NP, Grabowska A, et al. HOXC8 Regulates Self-Renewal, Differentiation and Transformation of Breast Cancer Stem Cells. Mol Cancer (2017) 16:38. doi: 10.1186/s12943-017-0605-z
48. Zhang Y, Yang C, Zhang M, Liu H, Gong C, Zhang J, et al. Interleukin Enhancer-Binding Factor 3 and HOXC8 Co-Activate Cadherin 11 Transcription to Promote Breast Cancer Cells Proliferation and Migration. Oncotarget (2017) 8:107477–91. doi: 10.18632/oncotarget.22491
49. Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, et al. Epigenetic Reprogramming of HOXC10 in Endocrine-Resistant Breast Cancer. Sci Trans Med (2014) 6:229ra41. doi: 10.1126/scitranslmed.3008326
50. Chen H, Chung S, Sukumar S. HOXA5-Induced Apoptosis in Breast Cancer Cells Is Mediated by Caspases 2 and 8. Mol Cell Biol (2004) 24:924–35. doi: 10.1128/mcb.24.2.924-935.2004
51. Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P Jr., Raman V. HOXA5-Twist Interaction Alters P53 Homeostasis in Breast Cancer Cells. J Biol Chem (2005) 280:2294–9. doi: 10.1074/jbc.M411018200
52. Cantile M, Di Bonito M, Cerrone M, Collina F, De Laurentiis M, Botti G. Long Non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers (2020) 12. doi: 10.3390/cancers12051197
53. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR Enhances ER Signaling and Confers Tamoxifen Resistance in Breast Cancer. Oncogene (2016) 35:2746–55. doi: 10.1038/onc.2015.340
54. Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR Influences Cell Growth, Migration, Invasion, and Apoptosis via the miR-20a-5p/HMGA2 Axis in Breast Cancer. Cancer Med (2018) 7:842–55. doi: 10.1002/cam4.1353
55. Lin J, Zhu H, Hong L, Tang W, Wang J, Hu H, et al. Coexpression of HOXA6 and PBX2 Promotes Metastasis in Gastric Cancer. Aging (2021) 13:6606–24. doi: 10.18632/aging.202426
56. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res (2016) 76:6299–310. doi: 10.1158/0008-5472.Can-16-0356
57. Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, et al. Over-Expressed Long Noncoding RNA HOXA11-AS Promotes Cell Cycle Progression and Metastasis in Gastric Cancer. Mol Cancer (2017) 16:82. doi: 10.1186/s12943-017-0651-6
58. Shen LY, Zhou T, Du YB, Shi Q, Chen KN. Targeting HOX/PBX Dimer Formation as a Potential Therapeutic Option in Esophageal Squamous Cell Carcinoma. Cancer Sci (2019) 110:1735–45. doi: 10.1111/cas.13993
59. Ding WJ, Zhou M, Chen MM, Qu CY. HOXB8 Promotes Tumor Metastasis and the Epithelial-Mesenchymal Transition via ZEB2 Targets in Gastric Cancer. J Cancer Res Clin Oncol (2017) 143:385–97. doi: 10.1007/s00432-016-2283-4
60. Zhang E, Han L, Yin D, He X, Hong L, Si X, et al. H3K27 Acetylation Activated-Long Non-Coding RNA CCAT1 Affects Cell Proliferation and Migration by Regulating SPRY4 and HOXB13 Expression in Esophageal Squamous Cell Carcinoma. Nucleic Acids Res (2017) 45:3086–101. doi: 10.1093/nar/gkw1247
61. Li J, Tong G, Huang C, Luo Y, Wang S, Zhang Y, et al. HOXC10 Promotes Cell Migration, Invasion, and Tumor Growth in Gastric Carcinoma Cells Through Upregulating Proinflammatory Cytokines. J Cell Physiol (2020) 235:3579–91. doi: 10.1002/jcp.29246
62. Zhu H, Dai W, Li J, Xiang L, Wu X, Tang W, et al. HOXD9 Promotes the Growth, Invasion and Metastasis of Gastric Cancer Cells by Transcriptional Activation of RUFY3. J Exp Clin Cancer Res: CR (2019) 38:412. doi: 10.1186/s13046-019-1399-1
63. Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, et al. Elevated HOXA1 Expression Correlates With Accelerated Tumor Cell Proliferation and Poor Prognosis in Gastric Cancer Partly via Cyclin D1. J Exp Clin Cancer Res: CR (2016) 35:15. doi: 10.1186/s13046-016-0294-2
64. Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, et al. Upregulation of miR-196a and HOTAIR Drive Malignant Character in Gastrointestinal Stromal Tumors. Cancer Res (2012) 72:1126–36. doi: 10.1158/0008-5472.Can-11-1803
65. Chen Z, Wu D, Thomas-Ahner JM, Lu C, Zhao P, Zhang Q, et al. Diverse AR-V7 Cistromes in Castration-Resistant Prostate Cancer Are Governed by Hoxb13. Proc Natl Acad Sci USA (2018) 115:6810–5. doi: 10.1073/pnas.1718811115
66. Kim YR, Kim IJ, Kang TW, Choi C, Kim KK, Kim MS, et al. HOXB13 Downregulates Intracellular Zinc and Increases NF-κb Signaling to Promote Prostate Cancer Metastasis. Oncogene (2014) 33:4558–67. doi: 10.1038/onc.2013.404
67. VanOpstall C, Perike S, Brechka H, Gillard M, Lamperis S, Zhu B, et al. MEIS-Mediated Suppression of Human Prostate Cancer Growth and Metastasis Through HOXB13-Dependent Regulation of Proteoglycans. eLife (2020) 9. doi: 10.7554/eLife.53600
68. Huang Q, Whitington T, Gao P, Lindberg JF, Yang Y, Sun J, et al. A Prostate Cancer Susceptibility Allele at 6q22 Increases RFX6 Expression by Modulating HOXB13 Chromatin Binding. Nat Genet (2014) 46:126–35. doi: 10.1038/ng.2862
69. Zhou J, Yang X, Song P, Wang H, Wang X. HOXC6 in the Prognosis of Prostate Cancer. Artif Cells Nanomedicine Biotechnol (2019) 47:2715–20. doi: 10.1080/21691401.2019.1635136
70. Xu F, Shangguan X, Pan J, Yue Z, Shen K, Ji Y, et al. HOXD13 Suppresses Prostate Cancer Metastasis and BMP4-Induced Epithelial-Mesenchymal Transition by Inhibiting SMAD1. Int J Cancer (2021) 148:3060–70. doi: 10.1002/ijc.33494
71. Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline Mutations in HOXB13 and Prostate-Cancer Risk. N Engl J Med (2012) 366:141–9. doi: 10.1056/NEJMoa1110000
72. Chen J, Zhu S, Jiang N, Shang Z, Quan C, Niu Y. HoxB3 Promotes Prostate Cancer Cell Progression by Transactivating CDCA3. Cancer Lett (2013) 330:217–24. doi: 10.1016/j.canlet.2012.11.051
73. Idaikkadar P, Morgan R, Michael A. HOX Genes in High Grade Ovarian Cancer. Cancers (2019) 11:1107. doi: 10.3390/cancers11081107
74. Miller KR, Patel JN, Zhang Q, Norris EJ, Symanowski J, Michener C, et al. HOXA4/HOXB3 Gene Expression Signature as a Biomarker of Recurrence in Patients With High-Grade Serous Ovarian Cancer Following Primary Cytoreductive Surgery and First-Line Adjuvant Chemotherapy. Gynecologic Oncol (2018) 149:155–62. doi: 10.1016/j.ygyno.2018.01.022
75. Ko SY, Naora H. HOXA9 Promotes Homotypic and Heterotypic Cell Interactions That Facilitate Ovarian Cancer Dissemination via Its Induction of P-Cadherin. Mol Cancer (2014) 13:170. doi: 10.1186/1476-4598-13-170
76. Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, et al. Loss of HOXD10 Expression Induced by Upregulation of miR-10b Accelerates the Migration and Invasion Activities of Ovarian Cancer Cells. Int J Oncol (2013) 43:63–71. doi: 10.3892/ijo.2013.1935
77. Ko SY, Barengo N, Ladanyi A, Lee JS, Marini F, Lengyel E, et al. HOXA9 Promotes Ovarian Cancer Growth by Stimulating Cancer-Associated Fibroblasts. J Clin Invest (2012) 122:3603–17. doi: 10.1172/jci62229
78. Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS, Targeting HOX. And PBX Transcription Factors in Ovarian Cancer. BMC Cancer (2010) 10:89. doi: 10.1186/1471-2407-10-89
79. Klausen C, Leung PC, Auersperg N. Cell Motility and Spreading are Suppressed by HOXA4 in Ovarian Cancer Cells: Possible Involvement of Beta1 Integrin. Mol Cancer research: MCR (2009) 7:1425–37. doi: 10.1158/1541-7786.Mcr-08-0466
80. Wang Z, Yu C, Wang H. HOXA5 Inhibits the Proliferation and Induces the Apoptosis of Cervical Cancer Cells via Regulation of Protein Kinase B and P27. Oncol Rep (2019) 41:1122–30. doi: 10.3892/or.2018.6874
81. Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X, et al. Circular RNA Circslc26a4 Accelerates Cervical Cancer Progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids (2020) 19:413–20. doi: 10.1016/j.omtn.2019.11.032
82. Lei D, Yang WT, Zheng PS. HOXB4 Inhibits the Proliferation and Tumorigenesis of Cervical Cancer Cells by Downregulating the Activity of Wnt/β-Catenin Signaling Pathway. Cell Death Dis (2021) 12:105. doi: 10.1038/s41419-021-03411-6
83. Wang Y, Wang C, Liu N, Hou J, Xiao W, Zhao L, et al. HOXC6 Promotes Cervical Cancer Progression via Regulation of Bcl-2. FASEB Journal: Off Publ Fed Am Societies Exp Biol (2019) 33:3901–11. doi: 10.1096/fj.201801099RR
84. Ma HM, Cui N, Zheng PS. HOXA5 Inhibits the Proliferation and Neoplasia of Cervical Cancer Cells via Downregulating the Activity of the Wnt/β-Catenin Pathway and Transactivating TP53. Cell Death Dis (2020) 11:420. doi: 10.1038/s41419-020-2629-3
85. Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-Mutated Acute Myeloid Leukemia: From Bench to Bedside. Blood (2020) 136:1707–21. doi: 10.1182/blood.2019004226
86. Zhang H, Zhang Y, Zhou X, Wright S, Hyle J, Zhao L, et al. Functional Interrogation of HOXA9 Regulome in MLLr Leukemia via Reporter-Based CRISPR/Cas9 Screen. eLife (2020) 9. doi: 10.7554/eLife.57858
87. Mohr S, Doebele C, Comoglio F, Berg T, Beck J, Bohnenberger H, et al. Hoxa9 and Meis1 Cooperatively Induce Addiction to Syk Signaling by Suppressing miR-146a in Acute Myeloid Leukemia. Cancer Cell (2017) 31:549–562.e11. doi: 10.1016/j.ccell.2017.03.001
88. de Bock CE, Demeyer S, Degryse S, Verbeke D, Sweron B, Gielen O, et al. HOXA9 Cooperates With Activated JAK/STAT Signaling to Drive Leukemia Development. Cancer Discov (2018) 8:616–31. doi: 10.1158/2159-8290.Cd-17-0583
89. Whelan JT, Ludwig DL, Bertrand FE. HoxA9 Induces Insulin-Like Growth Factor-1 Receptor Expression in B-Lineage Acute Lymphoblastic Leukemia. Leukemia (2008) 22:1161–9. doi: 10.1038/leu.2008.57
90. Yoshino S, Yokoyama T, Sunami Y, Takahara T, Nakamura A, Yamazaki Y, et al. Trib1 Promotes Acute Myeloid Leukemia Progression by Modulating the Transcriptional Programs of Hoxa9. Blood (2021) 137:75–88. doi: 10.1182/blood.2019004586
91. Wang H, Jia XH, Chen JR, Yi YJ, Wang JY, Li YJ, et al. HOXB4 Knockdown Reverses Multidrug Resistance of Human Myelogenous Leukemia K562/ADM Cells by Downregulating P-Gp, MRP1 and BCRP Expression via PI3K/Akt Signaling Pathway. Int J Oncol (2016) 49:2529–37. doi: 10.3892/ijo.2016.3738
92. Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a Suppresses Differentiation by G-CSF and Promotes Proliferation by SCF: Potential Mechanisms of Cooperativity With Hoxa9 in Myeloid Leukemia. Proc Natl Acad Sci USA (2001) 98:13120–5. doi: 10.1073/pnas.231115398
93. Bi L, Zhou B, Li H, He L, Wang C, Wang Z, et al. A Novel miR-375-HOXB3-CDCA3/DNMT3B Regulatory Circuitry Contributes to Leukemogenesis in Acute Myeloid Leukemia. BMC Cancer (2018) 18:182. doi: 10.1186/s12885-018-4097-z
94. Quéré R, Karlsson G, Hertwig F, Rissler M, Lindqvist B, Fioretos T, et al. Smad4 Binds Hoxa9 in the Cytoplasm and Protects Primitive Hematopoietic Cells Against Nuclear Activation by Hoxa9 and Leukemia Transformation. Blood (2011) 117:5918–30. doi: 10.1182/blood-2010-08-301879
95. Daniels TR, Neacato II, Rodríguez JA, Pandha HS, Morgan R, Penichet ML. Disruption of HOX Activity Leads to Cell Death That Can be Enhanced by the Interference of Iron Uptake in Malignant B Cells. Leukemia (2010) 24:1555–65. doi: 10.1038/leu.2010.142
96. Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic Regulation in Human Melanoma: Past and Future. Epigenetics (2015) 10:103–21. doi: 10.1080/15592294.2014.1003746
97. Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, et al. HOXA1 Drives Melanoma Tumor Growth and Metastasis and Elicits an Invasion Gene Expression Signature That Prognosticates Clinical Outcome. Oncogene (2014) 33:1017–26. doi: 10.1038/onc.2013.30
98. Morgan R, Simpson G, Gray S, Gillett C, Tabi Z, Spicer J, et al. HOX Transcription Factors Are Potential Targets and Markers in Malignant Mesothelioma. BMC Cancer (2016) 16:85. doi: 10.1186/s12885-016-2106-7
99. Gaunt SJ. Hox Cluster Genes and Collinearities Throughout the Tree of Animal Life. Int J Dev Biol (2018) 62:673–83. doi: 10.1387/ijdb.180162sg
100. Lewis EB. A Gene Complex Controlling Segmentation in Drosophila. Nature (1978) 276:565–70. doi: 10.1038/276565a0
101. Quinonez SC, Innis JW. Human HOX Gene Disorders. Mol Genet Metab (2014) 111:4–15. doi: 10.1016/j.ymgme.2013.10.012
102. Paço A, Aparecida de Bessa Garcia S, Leitão Castro J, Costa-Pinto AR, Freitas R. Roles of the HOX Proteins in Cancer Invasion and Metastasis. Cancers (2020) 13:10. doi: 10.3390/cancers13010010
103. Li B, Huang Q, Wei GH. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers (2019) 11:528. doi: 10.3390/cancers11040528
104. Kuo TL, Cheng KH, Chen LT, Hung WC. Deciphering The Potential Role of Hox Genes in Pancreatic Cancer. Cancers (2019) 11:734. doi: 10.3390/cancers11050734
105. Procino A, Cillo C. The HOX Genes Network in Metabolic Diseases. Cell Biol Int (2013) 37:1145–8. doi: 10.1002/cbin.10145
106. Jambhekar A, Dhall A, Shi Y. Roles and Regulation of Histone Methylation in Animal Development. Nat Rev Mol Cell Biol (2019) 20:625–41. doi: 10.1038/s41580-019-0151-1
107. Cunningham TJ, Duester G. Mechanisms of Retinoic Acid Signalling and Its Roles in Organ and Limb Development. Nat Rev Mol Cell Biol (2015) 16:110–23. doi: 10.1038/nrm3932
108. Gaunt SJ. Hox Cluster Genes and Collinearities Throughout the Tree of Animal Life. Cell Mol Life Sci: CMLS (2018) 62:673–83. doi: 10.1387/ijdb.180162sg
109. Peters JM, Gonzalez FJ. The Evolution of Carcinogenesis. Toxicological Sci: an Off J Soc Toxicol (2018) 165:272–6. doi: 10.1093/toxsci/kfy184
110. Cairns RA, Harris IS, Mak TW. Regulation of Cancer Cell Metabolism. Nat Rev Cancer (2011) 11:85–95. doi: 10.1038/nrc2981
111. Magli MC, Largman C, Lawrence HJ. Effects of HOX Homeobox Genes in Blood Cell Differentiation. J Cell Physiol (1997) 173:168–77. doi: 10.1002/(sici)1097-4652(199711)173:2<168::Aid-jcp16>3.0.Co;2-c
112. Shah N, Sukumar S. The Hox Genes and Their Roles in Oncogenesis. Nat Rev Cancer (2010) 10:361–71. doi: 10.1038/nrc2826
113. Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The Pathophysiology of HOX Genes and Their Role in Cancer. J Pathol (2005) 205:154–71. doi: 10.1002/path.1710
114. Kamkar F, Xaymardan M, Asli NS. Hox-Mediated Spatial and Temporal Coding of Stem Cells in Homeostasis and Neoplasia. Stem Cells Dev (2016) 25:1282–9. doi: 10.1089/scd.2015.0352
115. Luo F, Yu S, Jin LH. The Posterior Signaling Center Is an Important Microenvironment for Homeostasis of the Drosophila Lymph Gland. Front Cell Dev Biol (2020) 8:382. doi: 10.3389/fcell.2020.00382
116. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-Expressed lncRNA HOTAIRM1 Promotes Tumor Growth and Invasion Through Up-Regulating HOXA1 and Sequestering G9a/EZH2/Dnmts Away From the HOXA1 Gene in Glioblastoma Multiforme. J Exp Clin Cancer Res: CR (2018) 37:265. doi: 10.1186/s13046-018-0941-x
117. Jiang L, Wu Z, Meng X, Chu X, Huang H, Xu C. LncRNA HOXA-AS2 Facilitates Tumorigenesis and Progression of Papillary Thyroid Cancer by Modulating the miR-15a-5p/HOXA3 Axis. Hum Gene Ther (2019) 30:618–31. doi: 10.1089/hum.2018.109
118. Mahdipour E, Charnock JC, Mace KA. Hoxa3 Promotes the Differentiation of Hematopoietic Progenitor Cells Into Proangiogenic Gr-1+CD11b+ Myeloid Cells. Blood (2011) 117:815–26. doi: 10.1182/blood-2009-12-259549
119. Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell (2015) 28:815–29. doi: 10.1016/j.ccell.2015.11.001
120. Liang Y, Zhou R, Fu X, Wang C, Wang D. HOXA5 Counteracts the Function of Pathological Scar-Derived Fibroblasts by Partially Activating P53 Signaling. Cell Death Dis (2021) 12:40. doi: 10.1038/s41419-020-03323-x
121. Chen YQ, Yang TQ, Zhou B, Yang MX, Feng HJ, Wang YL. HOXA5 Overexpression Promotes Osteosarcoma Cell Apoptosis Through the P53 and P38α MAPK Pathway. Gene (2019) 689:18–23. doi: 10.1016/j.gene.2018.11.081
122. Lee DH, Forscher C, Di Vizio D, Koeffler HP. Induction of P53-Independent Apoptosis by Ectopic Expression of HOXA5 in Human Liposarcomas. Sci Rep (2015) 5:12580. doi: 10.1038/srep12580
123. Strathdee G, Sim A, Soutar R, Holyoake TL, Brown R. HOXA5 is Targeted by Cell-Type-Specific CpG Island Methylation in Normal Cells and During the Development of Acute Myeloid Leukaemia. Carcinogenesis (2007) 28:299–309. doi: 10.1093/carcin/bgl133
124. Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ. HoxA5 Stabilizes Adherens Junctions via Increased Akt1. Cell Adhesion Migration (2007) 1:185–95. doi: 10.4161/cam.1.4.5448
125. Cuevas I, Layman H, Coussens L, Boudreau N. Sustained Endothelial Expression of HoxA5 In Vivo Impairs Pathological Angiogenesis and Tumor Progression. PloS One (2015) 10:e0121720. doi: 10.1371/journal.pone.0121720
126. Feng F, Ren Q, Wu S, Saeed M, Sun C. Hoxa5 Increases Mitochondrial Apoptosis by Inhibiting Akt/mTORC1/S6K1 Pathway in Mice White Adipocytes. Oncotarget (2017) 8:95332–45. doi: 10.18632/oncotarget.20521
127. Zhou L, Wang Y, Zhou M, Zhang Y, Wang P, Li X, et al. HOXA9 Inhibits HIF-1α-Mediated Glycolysis Through Interacting With CRIP2 to Repress Cutaneous Squamous Cell Carcinoma Development. Nat Commun (2018) 9:1480. doi: 10.1038/s41467-018-03914-5
128. Sun Y, Zhou B, Mao F, Xu J, Miao H, Zou Z, et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell (2018) 34:643–58.e5. doi: 10.1016/j.ccell.2018.08.018
129. Gonçalves CS, Xavier-Magalhães A, Martins EP, Pinto AA, Pires MM, Pinheiro C, et al. A Novel Molecular Link Between HOXA9 and WNT6 in Glioblastoma Identifies a Subgroup of Patients With Particular Poor Prognosis. Mol Oncol (2020) 14:1224–41. doi: 10.1002/1878-0261.12633
130. Yu SL, Koo H, Lee HY, Yeom YI, Lee DC, Kang J. Recombinant Cell-Permeable HOXA9 Protein Inhibits NSCLC Cell Migration and Invasion. Cell Oncol (Dordrecht) (2019) 42:275–85. doi: 10.1007/s13402-019-00424-4
131. Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 Transforms Primary Bone Marrow Cells Through Specific Collaboration With Meis1a But Not Pbx1b. EMBO J (1998) 17:3714–25. doi: 10.1093/emboj/17.13.3714
132. Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, et al. HOXA9 Regulates BRCA1 Expression to Modulate Human Breast Tumor Phenotype. J Clin Invest (2010) 120:1535–50. doi: 10.1172/jci39534
133. Qiao C, Huang W, Chen J, Feng W, Zhang T, Wang Y, et al. IGF1-Mediated HOXA13 Overexpression Promotes Colorectal Cancer Metastasis Through Upregulating ACLY and IGF1R. Cell Death Dis (2021) 12:564. doi: 10.1038/s41419-021-03833-2
134. Feng W, Huang W, Chen J, Qiao C, Liu D, Ji X, et al. CXCL12-Mediated HOXB5 Overexpression Facilitates Colorectal Cancer Metastasis Through Transactivating CXCR4 and ITGB3. Theranostics (2021) 11:2612–33. doi: 10.7150/thno.52199
135. Liu S, Jin K, Hui Y, Fu J, Jie C, Feng S, et al. HOXB7 Promotes Malignant Progression by Activating the Tgfβ Signaling Pathway. Cancer Res (2015) 75:709–19. doi: 10.1158/0008-5472.Can-14-3100
136. Errico MC, Jin K, Sukumar S, Carè A. The Widening Sphere of Influence of HOXB7 in Solid Tumors. Cancer Res (2016) 76:2857–62. doi: 10.1158/0008-5472.Can-15-3444
137. Ying Y, Wang Y, Huang X, Sun Y, Zhang J, Li M, et al. Oncogenic HOXB8 is Driven by MYC-Regulated Super-Enhancer and Potentiates Colorectal Cancer Invasiveness via BACH1. Oncogene (2020) 39:1004–17. doi: 10.1038/s41388-019-1013-1
138. Nguyen NUN, Canseco DC, Xiao F, Nakada Y, Li S, Lam NT, et al. A Calcineurin-Hoxb13 Axis Regulates Growth Mode of Mammalian Cardiomyocytes. Nature (2020) 582:271–6. doi: 10.1038/s41586-020-2228-6
139. Barresi V, Ieni A, Cardia R, Licata L, Vitarelli E, Reggiani Bonetti L, et al. HOXB13 as an Immunohistochemical Marker of Prostatic Origin in Metastatic Tumors. APMIS: Acta Pathologica Microbiologica Immunologica Scandinavica (2016) 124:188–93. doi: 10.1111/apm.12483
140. Kim IJ, Kang TW, Jeong T, Kim YR, Jung C. HOXB13 Regulates the Prostate-Derived Ets Factor: Implications for Prostate Cancer Cell Invasion. Int J Oncol (2014) 45:869–76. doi: 10.3892/ijo.2014.2485
141. Xie B, Bai B, Xu Y, Liu Y, Lv Y, Gao X, et al. Tumor-Suppressive Function and Mechanism of HOXB13 in Right-Sided Colon Cancer. Signal Transduction Targeted Ther (2019) 4:51. doi: 10.1038/s41392-019-0086-1
142. Adwan H, Zhivkova-Galunska M, Georges R, Eyol E, Kleeff J, Giese NA, et al. Expression of HOXC8 Is Inversely Related to the Progression and Metastasis of Pancreatic Ductal Adenocarcinoma. Br J Cancer (2011) 105:288–95. doi: 10.1038/bjc.2011.217
143. Jiang Y, Yan B, Lai W, Shi Y, Xiao D, Jia J, et al. Repression of Hox Genes by LMP1 in Nasopharyngeal Carcinoma and Modulation of Glycolytic Pathway Genes by Hoxc8. Oncogene (2015) 34:6079–91. doi: 10.1038/onc.2015.53
144. Suo D, Wang Z, Li L, Chen Q, Zeng T, Liu R, et al. HOXC10 Upregulation Confers Resistance to Chemoradiotherapy in ESCC Tumor Cells and Predicts Poor Prognosis. Oncogene (2020) 39:5441–54. doi: 10.1038/s41388-020-1375-4
145. Guan Y, He Y, Lv S, Hou X, Li L, Song J. Overexpression of HOXC10 Promotes Glioblastoma Cell Progression to a Poor Prognosis via the PI3K/AKT Signalling Pathway. J Drug Targeting (2019) 27:60–6. doi: 10.1080/1061186x.2018.1473408
146. Wang L, Yao J, Yu T, Zhang D, Qiao X, Yao Z, et al. Homeobox D3, A Novel Link Between Bone Morphogenetic Protein 9 and Transforming Growth Factor Beta 1 Signaling. J Mol Biol (2020) 432:2030–41. doi: 10.1016/j.jmb.2020.01.043
147. Yang MH, Zhao L, Wang L, Ou-Yang W, Hu SS, Li WL, et al. Nuclear lncRNA HOXD-AS1 Suppresses Colorectal Carcinoma Growth and Metastasis via Inhibiting HOXD3-Induced Integrin β3 Transcriptional Activating and MAPK/AKT Signalling. Mol Cancer (2019) 18:31. doi: 10.1186/s12943-019-0955-9
148. Yachi K, Tsuda M, Kohsaka S, Wang L, Oda Y, Tanikawa S, et al. miR-23a Promotes Invasion of Glioblastoma via HOXD10-Regulated Glial-Mesenchymal Transition. Signal transduction targeted Ther (2018) 3:33. doi: 10.1038/s41392-018-0033-6
149. Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S. MicroRNA-10b Pleiotropically Regulates Invasion, Angiogenicity and Apoptosis of Tumor Cells Resembling Mesenchymal Subtype of Glioblastoma Multiforme. Cell Death Dis (2012) 3:e398. doi: 10.1038/cddis.2012.134
150. Bi Y, Mao Y, Su Z, Du J, Ye L, Xu F. HOXB-AS1 Accelerates the Tumorigenesis of Glioblastoma via Modulation of HOBX2 and HOBX3 at Transcriptional and Posttranscriptional Levels. J Cell Physiol (2021) 236:93–106. doi: 10.1002/jcp.29499
151. Petrini M, Felicetti F, Bottero L, Errico MC, Morsilli O, Boe A, et al. HOXB1 Restored Expression Promotes Apoptosis and Differentiation in the HL60 Leukemic Cell Line. Cancer Cell Int (2013) 13:101. doi: 10.1186/1475-2867-13-101
152. Diehl F, Rössig L, Zeiher AM, Dimmeler S, Urbich C. The Histone Methyltransferase MLL Is an Upstream Regulator of Endothelial-Cell Sprout Formation. Blood (2007) 109:1472–8. doi: 10.1182/blood-2006-08-039651
153. Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, et al. Epigenetic Silencing of the RASSF1A Tumor Suppressor Gene Through HOXB3-Mediated Induction of DNMT3B Expression. Mol Cell (2009) 36:219–30. doi: 10.1016/j.molcel.2009.10.009
154. Lorton BM, Shechter D. Cellular Consequences of Arginine Methylation. Cell Mol Life Sci: CMLS (2019) 76:2933–56. doi: 10.1007/s00018-019-03140-2
155. Boudadi E, Stower H, Halsall JA, Rutledge CE, Leeb M, Wutz A, et al. The Histone Deacetylase Inhibitor Sodium Valproate Causes Limited Transcriptional Change in Mouse Embryonic Stem Cells But Selectively Overrides Polycomb-Mediated Hoxb Silencing. Epigenet Chromatin (2013) 6:11. doi: 10.1186/1756-8935-6-11
156. Del Bene F, Wittbrodt J. Cell Cycle Control by Homeobox Genes in Development and Disease. Semin Cell Dev Biol (2005) 16:449–60. doi: 10.1016/j.semcdb.2005.02.001
157. Yu M, Zhan J, Zhang H. HOX Family Transcription Factors: Related Signaling Pathways and Post-Translational Modifications in Cancer. Cell Signal (2020) 66:109469. doi: 10.1016/j.cellsig.2019.109469
158. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464:1071–6. doi: 10.1038/nature08975
159. Qu X, Alsager S, Zhuo Y, Shan B. HOX Transcript Antisense RNA (HOTAIR) in Cancer. Cancer Lett (2019) 454:90–7. doi: 10.1016/j.canlet.2019.04.016
160. Li Y, Ren Y, Wang Y, Tan Y, Wang Q, Cai J, et al. A Compound AC1Q3QWB Selectively Disrupts HOTAIR-Mediated Recruitment of PRC2 and Enhances Cancer Therapy of DZNep. Theranostics (2019) 9:4608–23. doi: 10.7150/thno.35188
161. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Sci (New York N.Y.) (2010) 329:689–93. doi: 10.1126/science.1192002
162. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum Long Noncoding RNA HOTAIR as a Novel Diagnostic and Prognostic Biomarker in Glioblastoma Multiforme. Mol Cancer (2018) 17:74. doi: 10.1186/s12943-018-0822-0
163. Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long Noncoding RNA HOTTIP/HOXA13 Expression Is Associated With Disease Progression and Predicts Outcome in Hepatocellular Carcinoma Patients. Hepatol (Baltimore Md.) (2014) 59:911–23. doi: 10.1002/hep.26740
164. Procino A. Class I Homeobox Genes, “The Rosetta Stone of the Cell Biology”, in the Regulation of Cardiovascular Development. Curr medicinal Chem (2016) 23:265–75. doi: 10.2174/0929867323666151207111302
165. Selleri L, Zappavigna V, Ferretti E. Building a Perfect Body’: Control of Vertebrate Organogenesis by PBX-Dependent Regulatory Networks. Genes Dev (2019) 33:258–75. doi: 10.1101/gad.318774.118
166. Hoshino Y, Hayashida T, Hirata A, Takahashi H, Chiba N, Ohmura M, et al. Bevacizumab Terminates Homeobox B9-Induced Tumor Proliferation by Silencing Microenvironmental Communication. Mol Cancer (2014) 13:102. doi: 10.1186/1476-4598-13-102
167. Song J, Wang T, Xu W, Wang P, Wan J, Wang Y, et al. HOXB9 Acetylation at K27 Is Responsible for Its Suppression of Colon Cancer Progression. Cancer Lett (2018) 426:63–72. doi: 10.1016/j.canlet.2018.04.002
168. Morgan R, Boxall A, Harrington KJ, Simpson GR, Gillett C, Michael A, et al. Targeting the HOX/PBX Dimer in Breast Cancer. Breast Cancer Res Treat (2012) 136:389–98. doi: 10.1007/s10549-012-2259-2
169. Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: A Novel Oncogenic Propellant in Human Cancer. Clinica Chimica Acta; Int J Clin Chem (2020) 503:1–18. doi: 10.1016/j.cca.2019.12.028
170. Tatangelo F, Di Mauro A, Scognamiglio G, Aquino G, Lettiero A, Delrio P, et al. Posterior HOX Genes and HOTAIR Expression in the Proximal and Distal Colon Cancer Pathogenesis. J Trans Med (2018) 16:350. doi: 10.1186/s12967-018-1725-y
171. Singh A, Trivedi P, Jain NK. Advances in siRNA Delivery in Cancer Therapy. Artif Cells Nanomedicine Biotechnol (2018) 46:274–83. doi: 10.1080/21691401.2017.1307210
Keywords: homeobox genes, transcription factors, therapy, biomarker, cancer progression
Citation: Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo X, Huang W and Xia L (2021) Homeobox Genes in Cancers: From Carcinogenesis to Recent Therapeutic Intervention. Front. Oncol. 11:770428. doi: 10.3389/fonc.2021.770428
Received: 03 September 2021; Accepted: 28 September 2021;
Published: 14 October 2021.
Edited by:
Alfredo Procino, Rudjer Boskovic Institute, CroatiaReviewed by:
Manfra Michele, University of Basilicata, ItalyMohammad Hojjat-Farsangi, Karolinska Institutet (KI), Sweden
Copyright © 2021 Feng, Zhang, Wang, Xie, Ji, Luo, Huang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limin Xia, eGlhbGltaW5AdGpoLnRqbXUuZWR1LmNu; Wenjie Huang, aHVhbmd3ZW5qaWVAdGpoLnRqbXUuZWR1LmNu
 Yangyang Feng
Yangyang Feng Tongyue Zhang
Tongyue Zhang Yijun Wang
Yijun Wang Meng Xie1,2
Meng Xie1,2 Wenjie Huang
Wenjie Huang Limin Xia
Limin Xia