- 1Department of Radiology, Peking University First Hospital, Beijing, China
- 2Department of Radiology, Peking University Shenzhen Hospital, Shenzhen, China
Objective: To establish and evaluate the 3D U-Net model for automated segmentation and detection of pelvic bone metastases in patients with prostate cancer (PCa) using diffusion-weighted imaging (DWI) and T1 weighted imaging (T1WI) images.
Methods: The model consisted of two 3D U-Net algorithms. A total of 859 patients with clinically suspected or confirmed PCa between January 2017 and December 2020 were enrolled for the first 3D U-Net development of pelvic bony structure segmentation. Then, 334 PCa patients were selected for the model development of bone metastases segmentation. Additionally, 63 patients from January to May 2021 were recruited for the external evaluation of the network. The network was developed using DWI and T1WI images as input. Dice similarity coefficient (DSC), volumetric similarity (VS), and Hausdorff distance (HD) were used to evaluate the segmentation performance. Sensitivity, specificity, and area under the curve (AUC) were used to evaluate the detection performance at the patient level; recall, precision, and F1-score were assessed at the lesion level.
Results: The pelvic bony structures segmentation on DWI and T1WI images had mean DSC and VS values above 0.85, and the HD values were <15 mm. In the testing set, the AUC of the metastases detection at the patient level were 0.85 and 0.80 on DWI and T1WI images. At the lesion level, the F1-score achieved 87.6% and 87.8% concerning metastases detection on DWI and T1WI images, respectively. In the external dataset, the AUC of the model for M-staging was 0.94 and 0.89 on DWI and T1WI images.
Conclusion: The deep learning-based 3D U-Net network yields accurate detection and segmentation of pelvic bone metastases for PCa patients on DWI and T1WI images, which lays a foundation for the whole-body skeletal metastases assessment.
Introduction
The nature of bone marrow makes it a favorite fertile soil into which prostate tumors incline to colonize and grow (1, 2); up to 84% of patients with advanced prostate cancer (PCa) experience bone metastases (3), and more than 80% PCa patients developed relapse in the bone following treatment of the primary site (4). The mortality of PCa is 6.6-fold for those with bone metastases compared to those without bone metastases (5). Accurate detection and assessment of metastatic burden in bone are of fundamental importance for radiologists.
Bone scintigraphy (BS) and computed tomography (CT) scans were endorsed as the standard imaging method in the staging and follow-up of metastatic PCa (6), while it is gradually clear that the reduced accuracy of BS and CT in the detection and therapeutic response evaluation of bone metastases reduces their effectiveness in therapy management (7). Multiparametric magnetic resonance imaging (mpMRI) is emerging as a powerful alternative for metastatic PCa. One of the main strengths of mpMRI is to achieve a precise evaluation of bone metastasis via the incorporation of anatomic [e.g., T1 weighted imaging (T1WI)] and functional imaging sequences [e.g., diffusion-weighted imaging (DWI)] (7, 8). The value of volumetric measurements for assessing treatment response has been increasingly discussed, and the measurements of lesion volume on mpMRI should be undertaken on high-quality T1WI images according to the METastasis Reporting and Data System (MET-RADS) for PCa (9). Additionally, the volume of bone metastasis assessed with DWI was reported to show a correlation with established prognostic biomarkers and is associated with overall survival in metastatic castration-resistant PCa (10). In short, the detection and delineation of metastases and evaluation of volume change concerning disease progression or therapy on DWI and T1WI images are key tasks as part of optimal patient management.
Heavy workload of mpMRI images evaluation can be tiresome for radiologists, hence bearing the risk of missed diagnosis for lesions and leading to decreased sensitivity. The measurements of all the metastatic lesions are time consuming, in particular, if multiple metastases are present. In this context, automated and accurate segmentation of bone metastases would be highly beneficial.
Driven by the rapid growth in computer science, the performance of deep learning is on par with or even outperforms radiologists in visual identification, which can perform automated data-oriented feature extraction and thus learning directly the most relevant feature representation from the input images (11, 12). The U-Net algorithm is one of the most commonly used deep learning-based convolutional neural networks (CNNs) (13), which shows potential in detection, segmentation, and classification of metastatic lesions on MRI images such as brain metastases (14, 15) and liver metastases (16). Concerning the automated bone metastasis analysis using the deep learning technique, the research trend is mainly on BS (17, 18) and single-photon emission computerized tomography (SPECT) images (19, 20); less attention has been paid to the diagnosis of mpMRI (21, 22). To this end, we intend to apply the 3D U-Net (23) algorithm for the segmentation of bone metastases on mpMRI images. For proof-of-concept, we focused on the detection and segmentation of bone metastases in the pelvic area.
Materials and Methods
This retrospective single-center study was approved by the institutional review board, and written informed consent was waived.
Patient Cohort
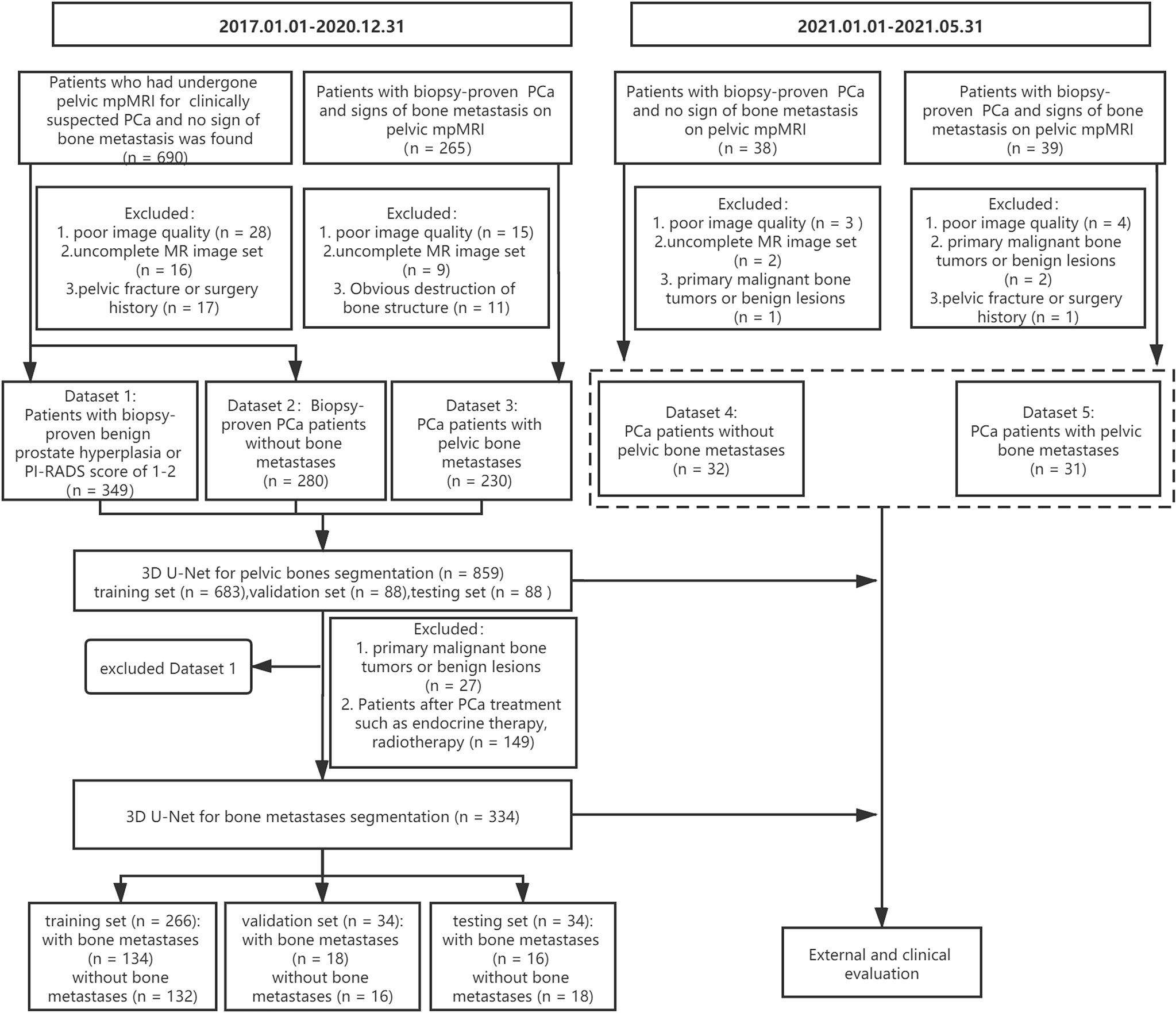
A cohort of 955 consecutive patients who had undergone pelvic mpMRI for either clinically suspected or confirmed PCa between January 2017 and December 2020 was reviewed using our institutional image archiving system. The exclusion criteria were as follows: (1) poor image quality (significant motion artifact or chemical shift artifact), (2) uncomplete MR image set, (3) obvious destruction of bone structure, and (4) patients with a history of pelvic fracture or surgery. Finally, the images from 859 patients were included for the 3D U-Net model development of pelvic bony structures segmentation, including a dataset of patients with PI-RADS score of 1–2 or biopsy-proven benign prostate hyperplasia (dataset 1, n = 349), a dataset of biopsy-proven PCa patients without bone metastases (dataset 2, n = 280), and a dataset of biopsy-proven PCa patients with bone metastases (dataset 3, n = 230).
All three datasets were used to develop a pelvic bony structure segmentation model. Then, a 3D U-Net model for bone metastases segmentation was developed using datasets 2 and 3. The patients with primary malignant bone tumors (such as osteosarcoma and myeloma) or definite benign findings (hemangiomas, bone island) on pelvic bones (n = 27) and patients who underwent PCa treatment (endocrine therapy, chemotherapy, or radiotherapy, n = 149) were excluded. In total, 334 patients were enrolled for the model development, including 168 PCa patients with bone metastases and 166 PCa patients without bone metastases.
Additionally, 77 patients with biopsy-proven PCa who performed pelvic mpMRI scanning from January 2021 and May 2021 were acquired; according to the above excluding criteria, 63 patients were finally recruited for the external evaluation of the 3D U-Net model including a dataset of 31 PCa patients with bone metastases (dataset 4) and a dataset of 32 PCa patients without bone metastases (dataset 5). The workflow of data enrollment is shown in Figure 1.
Image Acquisition
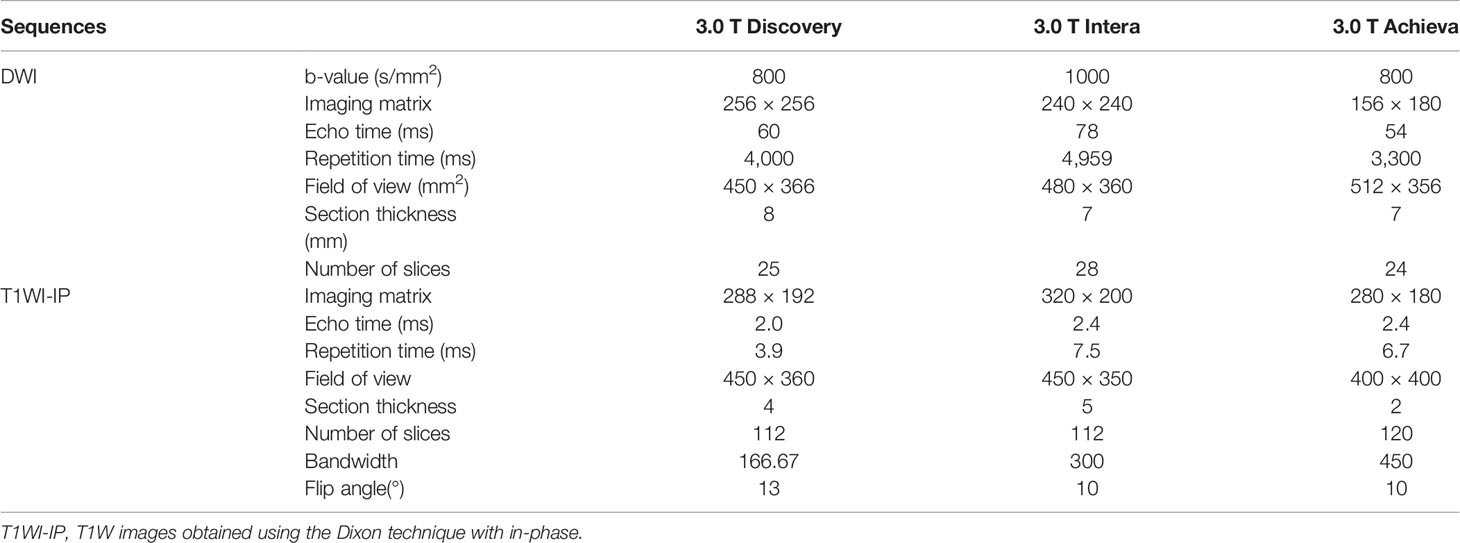
The pelvic mpMRI acquisitions were performed on three 3.0 T MR units (Achieva, Philips Healthcare; Discovery, GE Healthcare; Interia, Philips Healthcare). The standard pelvic mpMRI protocol at our institution included a T1/T2-weighted sequence, DWI with b-values of 0, 800, or 1,000 s/mm2 along with reconstructed ADC images, T1W images obtained using the 2-point Dixon technique with in-phase (T1WI-IP) and out-phase (T1WI-OP), and dynamic contrast-enhanced imaging. DWI images with high b-values (b = 800 or 1,000 s/mm2) and T1WI-IP images were selected for PCa bone metastases analyses in this study. Detailed MR imaging parameters of DWI and T1WI-IP sequence are shown in Table 1.
Manual Annotation
The manual annotations were performed with an image segmentation software (ITK-SNAP 3.6; Penn Image Computing and Science Laboratory, Philadelphia, PA). Under the supervision of a board-certified radiology expert (with more than 15 years of reading experience), a radiology resident with 3 years of reading experience evaluated all mpMRI examinations and, section by section, manually annotated eight pelvic bony structures (lumbar vertebra, sacrococcyx, ilium, acetabulum, femoral head, femoral neck, ischium, and pubis) on DWI images and T1WI-IP images. The bone metastases were included in the annotations, which were recognized as bone tissue in this bony structure segmentation model. The manual annotations of the pelvic bony structures were regarded as the reference standard for the 3D U-Net model evaluation.
To establish the reference standard of bone metastases, the radiology resident and expert radiologist conducted a review of the original radiology report and double reviewed the included MR imaging scans and prior/follow-up imaging before annotation. A bone lesion was considered as a metastasis if it showed an MR imaging correlated with adequate image contrast (positive image contrast on DWI images and negative image contrast on those obtained with the T1WI-IP images). The radiology resident performed manual annotations of the metastatic lesions on DWI and T1WI-IP images in a voxel-wise manner (indicated as A1.1). Then, the expert radiologist modified the annotations of A1.1 and the annotations after modification were indicated as A2.1. Both the resident and expert radiologist repeated the annotations and modifications at least 3 weeks later (indicated as A1.2 and A2.2, respectively). The inter- and intraobserver agreement between the manual annotations (A1.1 vs. A2.1; A1.1 vs. A1.2; A2.1 vs. A2.2; and A1.2 vs. A2.2) were estimated using Dice similarity coefficient (DSC).
The bony metastatic lesions in the 31 PCa patients of the external dataset were manually annotated by the resident radiology under the supervision of the expert radiologist, which was taken as the reference standard for external evaluation of the model.
Model Development
A two-step method for the bone metastases segmentation was proposed using the 3D U-Net model: the first step with a 3D U-Net algorithm for pelvic bone segmentation followed by a second step with a 3D U-Net for bone lesion segmentation within the segmented pelvic bony structures. Both the CNNs were coded by Python3.6, Pytorch 0.4.1, Opencv, Numpy, and SimpleITK, and trained on the GPU NVIDIA Tesla P100 16G.
Model Development for Pelvic Bones Segmentation
The model of the pelvic bony structure segmentation takes the combination of DWI images and T1WI-IP images as input, and each image sequence is used as an independent input data (Figure 2). The 859 patients were randomly divided into either training (n = 683), validation (n = 88), or testing (n = 88) sets with a ratio of 8:1:1. During the image preprocessing, the pixel values in images were scaled between 0 and 65,535. Then, the images were resized to 64 × 224 × 224 (z, y, x) by resampling to maintain the optimal image features, and z-score intensity normalization was applied to all images. Skewing (angel: 0–5), shearing (angel: 0–5), and translation (scale: −0.1,0.1) of the images were applied for data augmentation. To remove small spurious segmentation, the two largest connected components of each bone were selected as the final segmentation. A total of 300 epochs of training were performed until validation loss failed to rise. The Adam optimizer was employed to minimize loss with a learning rate of 0.0001, a batch size of 2, and a Dice loss function. Other hyperparameters (such as weight initialization and dropout for regularization) were randomly searched and automatically executed in the validation set during model development.
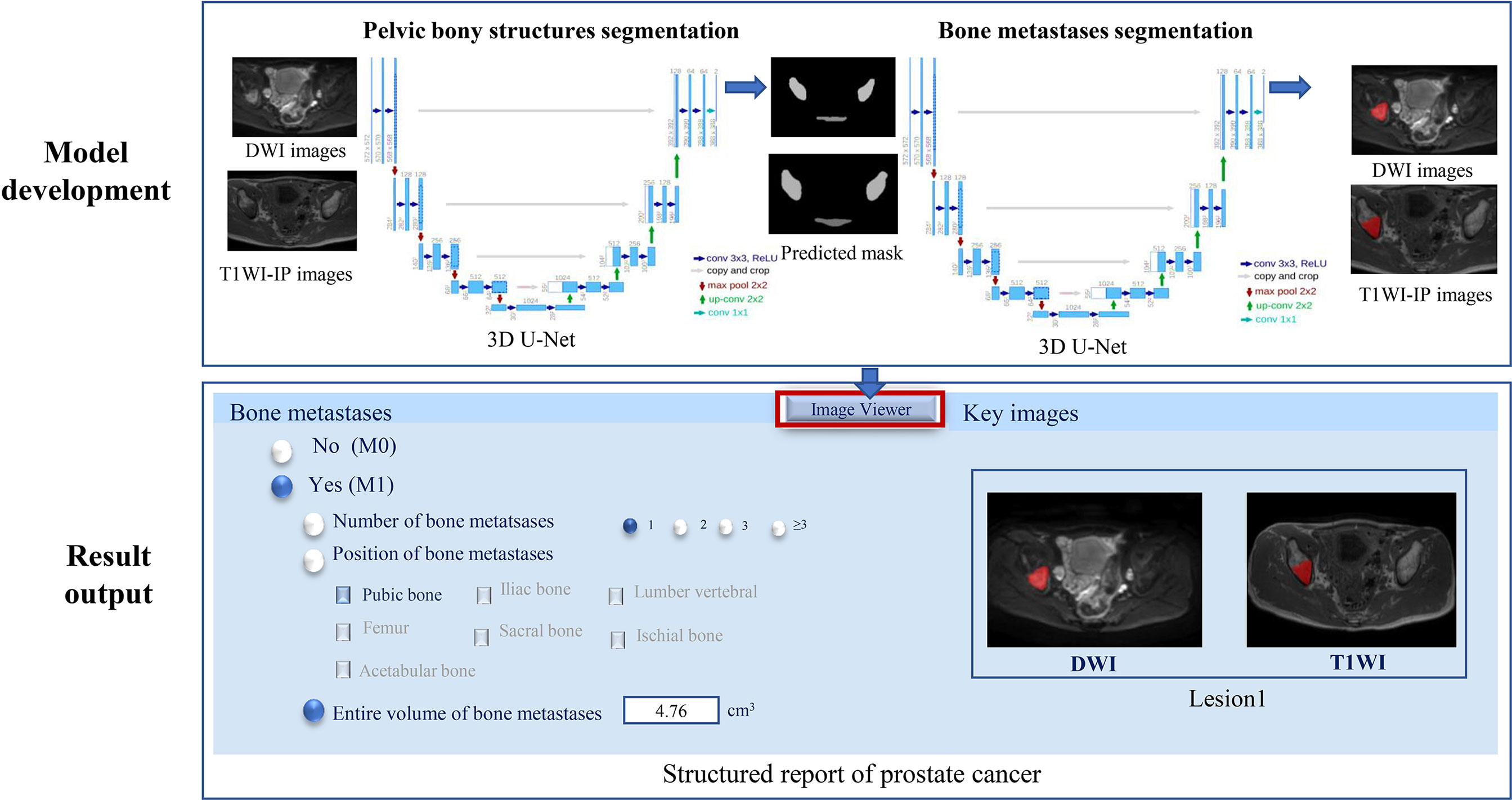
Figure 2 The two-step 3D U-Net for bone metastases segmentation on DWI and T1WI-IP images. T1WI-IP, T1W images obtained using the Dixon technique with in-phase.
Model Development for Bone Metastases Segmentation
The volume of interest predicted by the model of pelvic bony structure segmentation was used as the mask for the bone metastases segmentation (Figure 2). The network configurations were set as follows: training epoch, 250; learning rate, 0.01; batch size, 5; optimizer, Adam optimizer; and loss function, Dice loss.
For post-processing, automatically detected metastases of <0.2 cm3 during inference of testing set were regarded as image noise and discarded. The threshold was based on the resolution of T1WI-IP sequences and is determined by referring to the smallest annotated metastases (0.356 cm3).
Model Evaluation
Model Evaluation for Pelvic Bony Structure Segmentation
The performance of the network was evaluated by comparing the segmentations generated by the 3D U-Net based on image data from the testing set to the corresponding reference standard represented by the manual segmentations on DWI and T1WI-IP images quantitatively. The evaluation metrics used for the bony structures segmentation include the overlap-based metric (DSC), the volume-based metric [volumetric similarity (VS)], and the spatial distance-based metric [Hausdorff distance (HD)] (24).
Model Evaluation for Bone Metastases Segmentation
The performance of the bone metastases segmentation model was evaluated both on detection and segmentation. Detection is defined as the network’s ability to detect a metastasis annotated by the radiologist. One bone metastasis was considered detected when the manual annotation and the predicted segmentation had an overlap >0. Segmentation is defined as its ability to provide a contour identical to that of the radiologist.
The detection performance of the network was quantified at the patient and lesion levels. The sensitivity, specificity, accuracy, positive predictive value (PPV), negative predictive value (NPV), and area under the receiver operating characteristic curve (AUC) were used to assess the performance of the model to discriminate between patients with bone metastases and patients without bone metastases. To determine the detection accuracy of the metastases at the lesion level, we compared the lesions obtained with model predictions and manual annotations to determine the true-positive (TP), false-negative (FN), and false-positive (FP) findings. The recall (correctly detected metastases divided by all metastases contained in reference standard), precision (correctly detected metastases divided by all the detected metastases), and F1 score (harmonic mean of precision and recall) were calculated to assess the detection performance of the model on a lesion-by-lesion basis. In addition, we determined the number of distinct metastatic lesions in each case in the testing set and then divided the data into groups with (a) 1, (b) 2–3, (c) 4–5, and (d) >5 lesions to facilitate subgroup analysis of metastases detection at lesion level.
The metastases segmentation performance of the network was assessed using the metrics of DSC, VS, and HD by comparing the CNN-predicted segmentation and manual segmentation. Besides, the volume of the bone metastases in manual annotations and automated segmentations was calculated to further quantitatively estimate the segmentation efficacy of the U-Net algorithm.
Model Evaluation on an External Dataset
The external dataset was used to further assess the efficiency of the model on bone metastases evaluation in the clinical setting. Given the new mpMRI data of PCa patients, the 3D U-Net was supposed to determine the existence of bone metastases (M0 or M1) and output the number, location, and volume of the bone metastases with corresponding segmented masks (Figure 2). A bone lesion was considered as being detected if it was segmented on at least one of the two MR imaging sequences (DWI/T1WI-IP). The accuracy of the M-staging of the model was assessed using the receiver operating characteristic curve analysis, and the segmentation performance (DSC, VS, HD) and quantitative measurements (volume) were assessed by comparison with manual annotations.
Statistical Analysis
MedCalc (version 14.8; MedCalc Software, Ostend, Belgium) and SPSS (version 22.0, IBM Corp., Armonk, NY, USA) were used for the statistical analyses. Numerical data of patients’ age were reported as the mean ± SD (standard deviation), and prostate-specific antigen (PSA) levels were reported as (median, quartile). One-way analysis of variance (ANOVA) was used to compare the characteristics of patients (age, PSA level) among training, validation, and testing sets. The segmentation performance of the algorithm (DSC, VS, and HD) between DWI and T1WI-IP images were compared by paired t-test. The McNemar’s test was used to compare the detection performance (sensitivity, specificity, PPV, NPV, recall, and precision) between the two sequences. Bland–Altman analyses were performed to compare manual versus automated bone metastases volume. p < 0.05 was considered indicative of a statistically significant difference.
Results
Characteristics of Patients
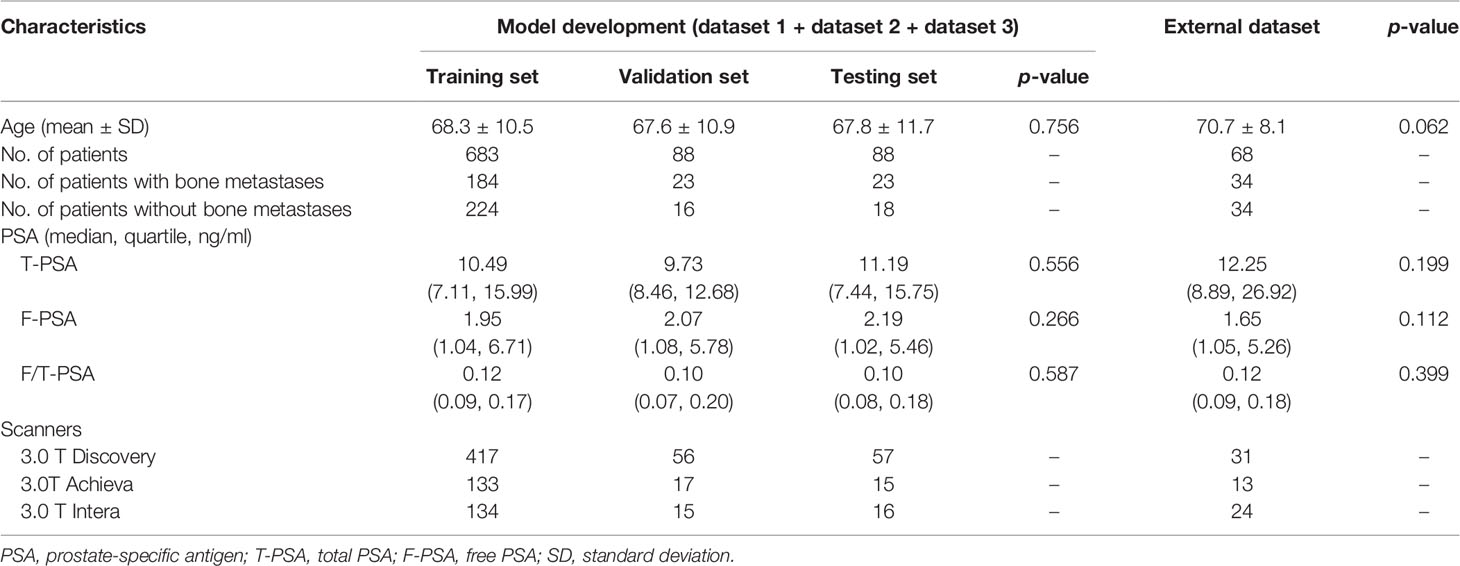
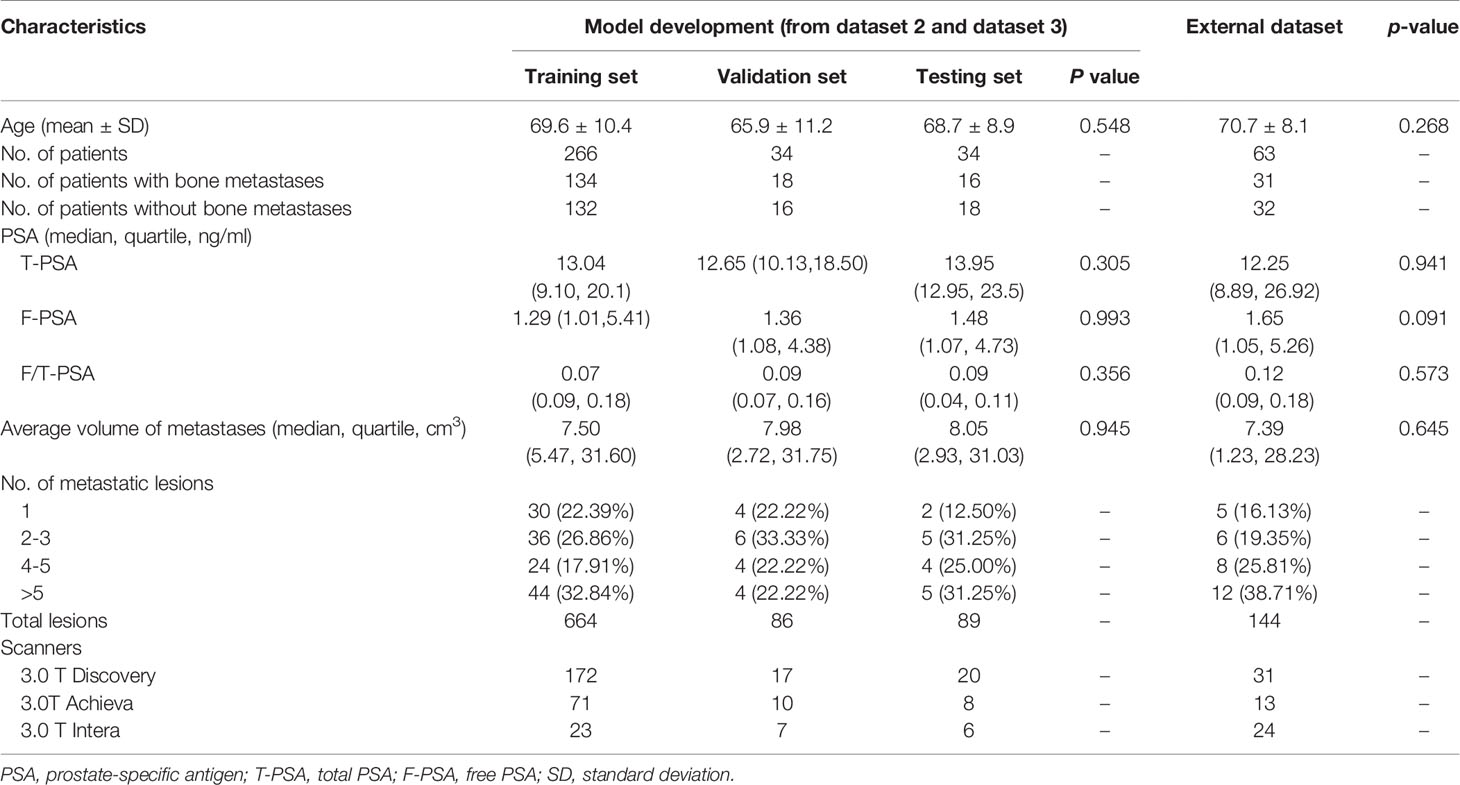
The characteristics of patients are shown in Tables 2, 3. The age and PSA level showed no significant difference among the training, validation, and testing sets on both models (all with p > 0.05). The average volume of metastases in the external dataset was 7.39 cm3, and no difference was found between the external dataset and model development dataset (p = 0.645). Of the 16 PCa patients with bone metastases in the testing set, 2 patients (12.50%) had one metastasis, 5 patients (31.25%) had two to three metastases, 4 patients (25.00%) had four to five metastases, and 5 patients (31.25%) had more than five metastases.
Assessment of Pelvic Bony Structures Segmentation
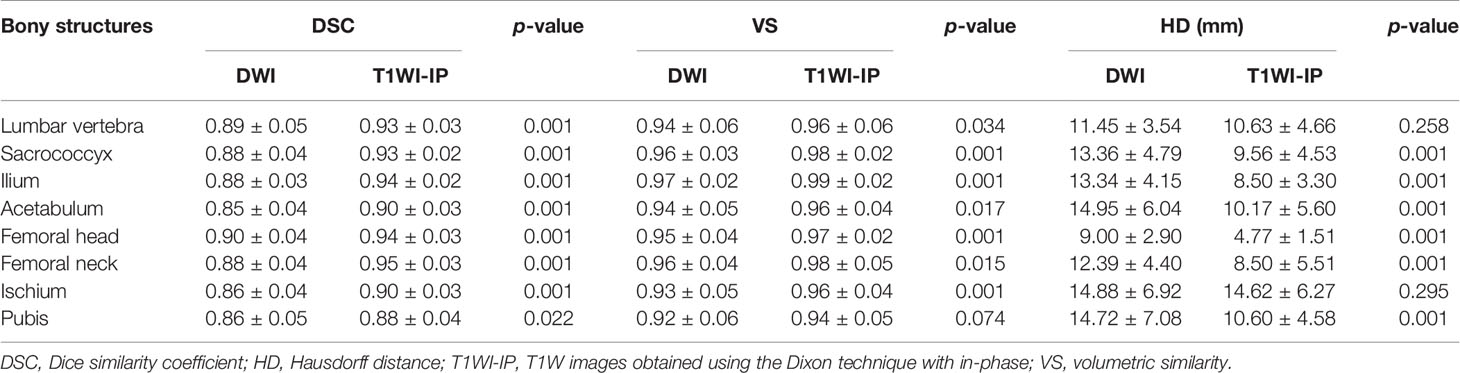
As shown in Table 4 and Figure 3, in the testing set of pelvic bone segmentation model, the DSC and VS values of eight pelvic bony structures between model prediction and manual annotation are all above 0.85 on both DWI and T1WI-IP images, while the mean DSC and VS values on T1WI-IP images are significantly higher than those on DWI images (all with p < 0.05), and the HD is significantly lower. This may be explained by the higher spatial resolution of the T1WI-IP images. Additionally, as detailed in the Supplementary materials (Supplementary Tables S1–S4), no significant difference was found among the patients from different datasets (dataset 1 vs. dataset 2 vs. dataset 3) and different scanners (3.0 T Discovery vs. 3.0 T Achieva vs. 3.0 T Intera) on both DWI and T1WI-IP images.
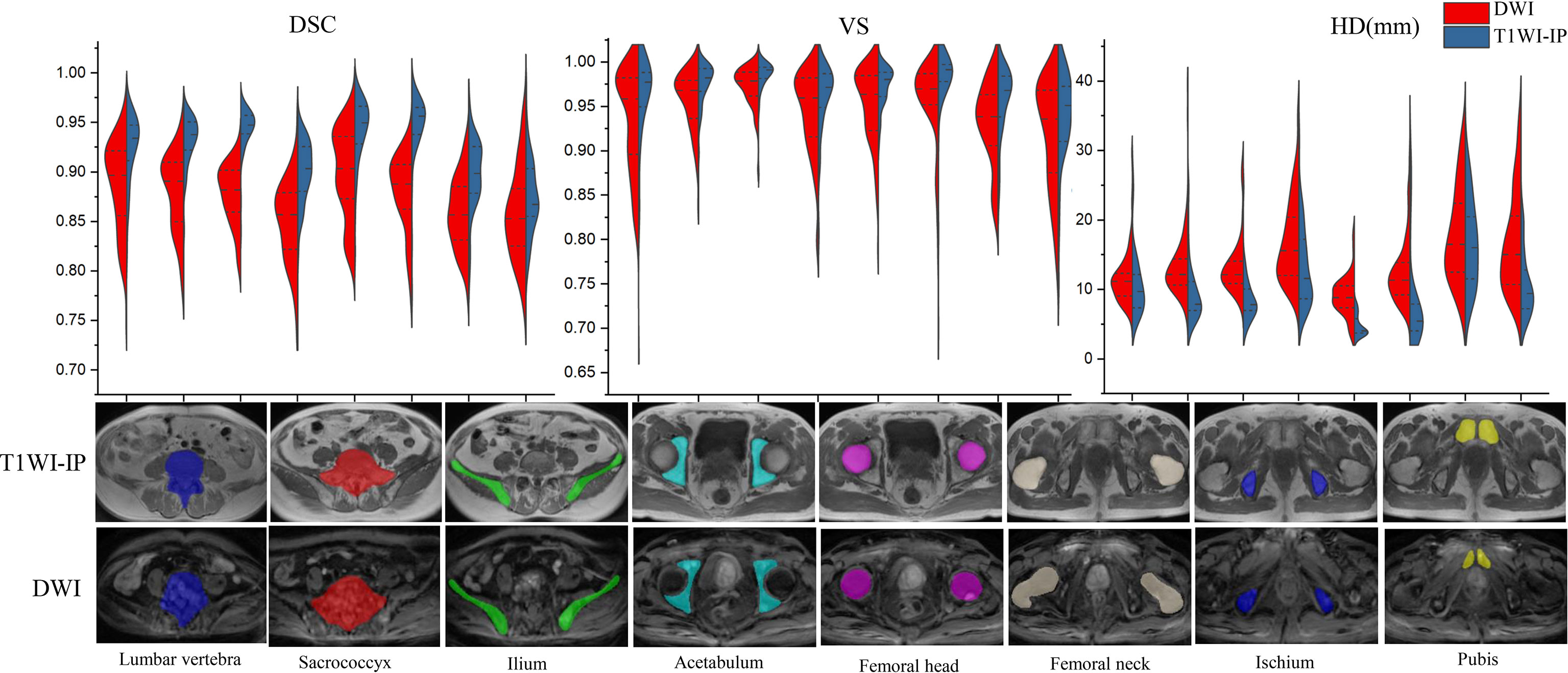
Figure 3 Split violin plots of DSC, VS, and HD (mm) for pelvic bony structures segmentation. DSC, Dice similarity coefficient; HD, Hausdorff distance; T1WI-IP, T1W images obtained using the Dixon technique with in-phase; VS, volumetric similarity.
The Inter- and Intraobserver Agreement of Bone Metastases Annotations
The interobserver agreement of the manual annotations of bone metastases was assessed by calculating the DSC values between A1.1 and A2.1, and A1.2 and A2.2. The intraobserver agreement was assessed by A1.1 vs. A1.2 and A2.1 vs. A2.2. The DSC values on DWI images were as follows: A1.1 vs. A2.1, 0.90 ± 0.08; A1.1 vs. A2.1, 0.91 ± 0.09; A2.1 vs. A2.2, 0.94 ± 0.05; A1.2 vs. A2.2, 0.91 ± 0.08. In T1WI-IP images, the DCS values were as follows: A1.1 vs. A2.1, 0.89 ± 0.09; A1.1 vs. A2.1, 0.90 ± 0.09; A2.1 vs. A2.2, 0.97 ± 0.04; and A1.2 vs. A2.2, 0.92 ± 0.08. The high DSC values between A2.1 vs. A2.2 confirmed the reliability of the manual annotations. A2.2 was regarded as the reference standard for the lesion segmentation model evaluation.
The Detection Accuracy of Bone Metastases
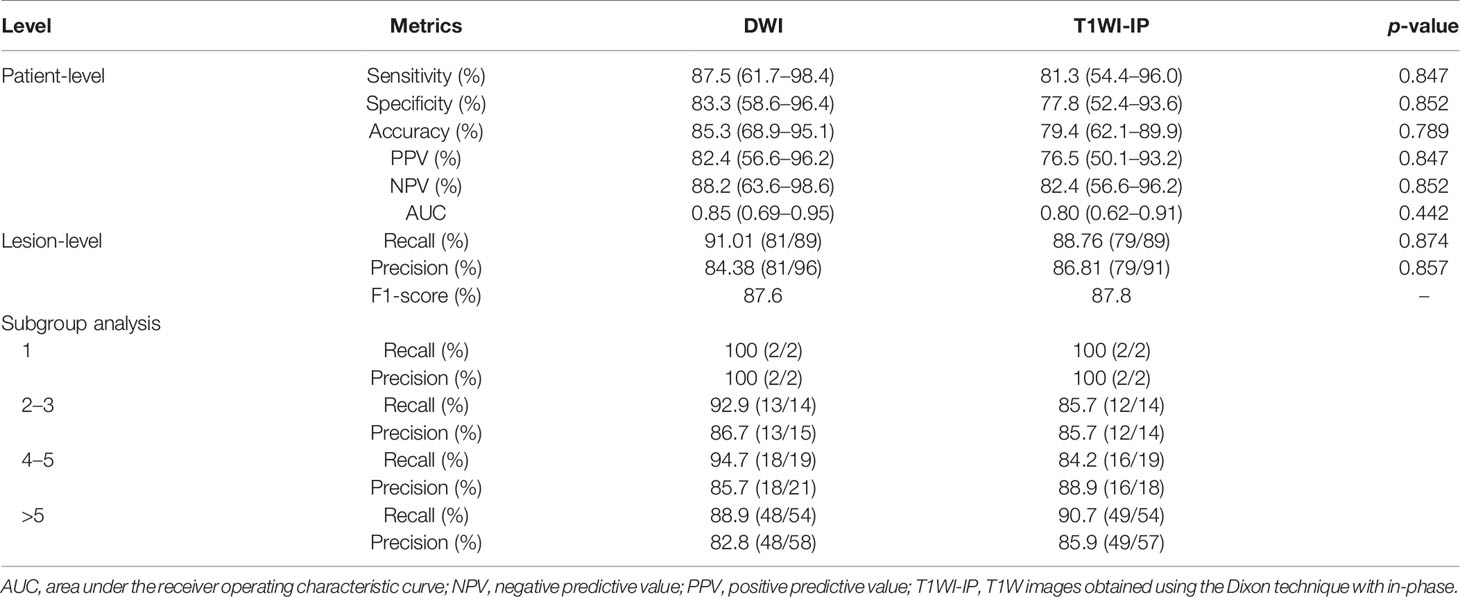
The detection performance of the CNN on DWI and T1WI-IP images at the patient and lesion levels are shown in Table 5. The detection performance of the model on DWI images was better than on T1WI-IP images concerning the values of the evaluation metrics, while no significant difference was found between the two sequences (all with p > 0.05). The results of the subgroup analysis of detection accuracy at lesion level in the testing set showed the highest recall and precision values in patients with single metastases, and both the recall and precision were above 80% for few metastases (≤5 metastases) and multiple metastases (>5 metastases).
The Segmentation Accuracy of Bone Metastases
The mean DSC, VS, and HD for the automatic metastases segmentation are 0.79 ± 0.05, 0.84 ± 0.09, and 15.05 ± 3.61 mm on DWI images and 0.80 ± 0.06, 0.85 ± 0.08, and 13.39 ± 3.20 mm on T1WI images (Figure 4A), which showed no significant difference between the two sequences (p = 0.627, 0.741, and 0.175, respectively).
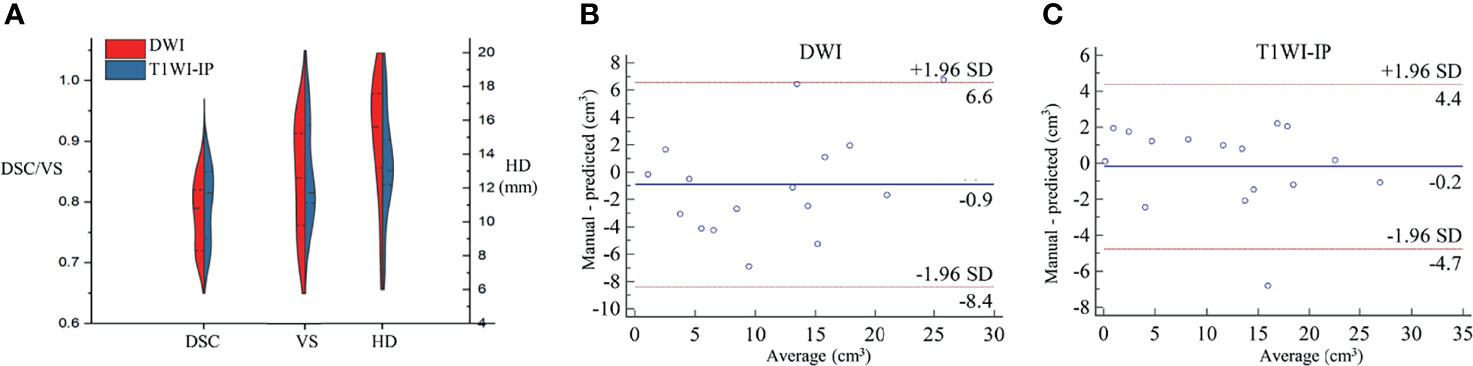
Figure 4 The segmentation accuracy of bone metastases in the testing set. (A) Split violin plot of DSC, VS, and HD of the bone metastases on DWI and T1WI-IP images. (B) The Bland–Altman plot of the volume difference between manual annotation and model prediction on DWI images. (C) The Bland–Altman plot of the volume difference between manual annotation and model prediction on T1WI-IP images. DSC, Dice similarity coefficient; HD, Hausdorff distance; T1WI-IP, T1W images obtained using the Dixon technique with in-phase; VS, volumetric similarity.
The volume differences between manual annotation and model prediction of bone metastases on DWI and T1WI-IP images are shown in Figures 4B, C. The limit of agreement (LOA) between the automated and manual segmentation on DWI images was −8.4–6.6 cm3 and −4.4–4.4 cm3 on T1WI-IP images. Most of the difference values were within the LOA, which showed that the volume of overall metastatic lesions in each patient between manual and automated segmentations agreed closely. Example results of the automatic bone metastases segmentation are shown in Figure 5.
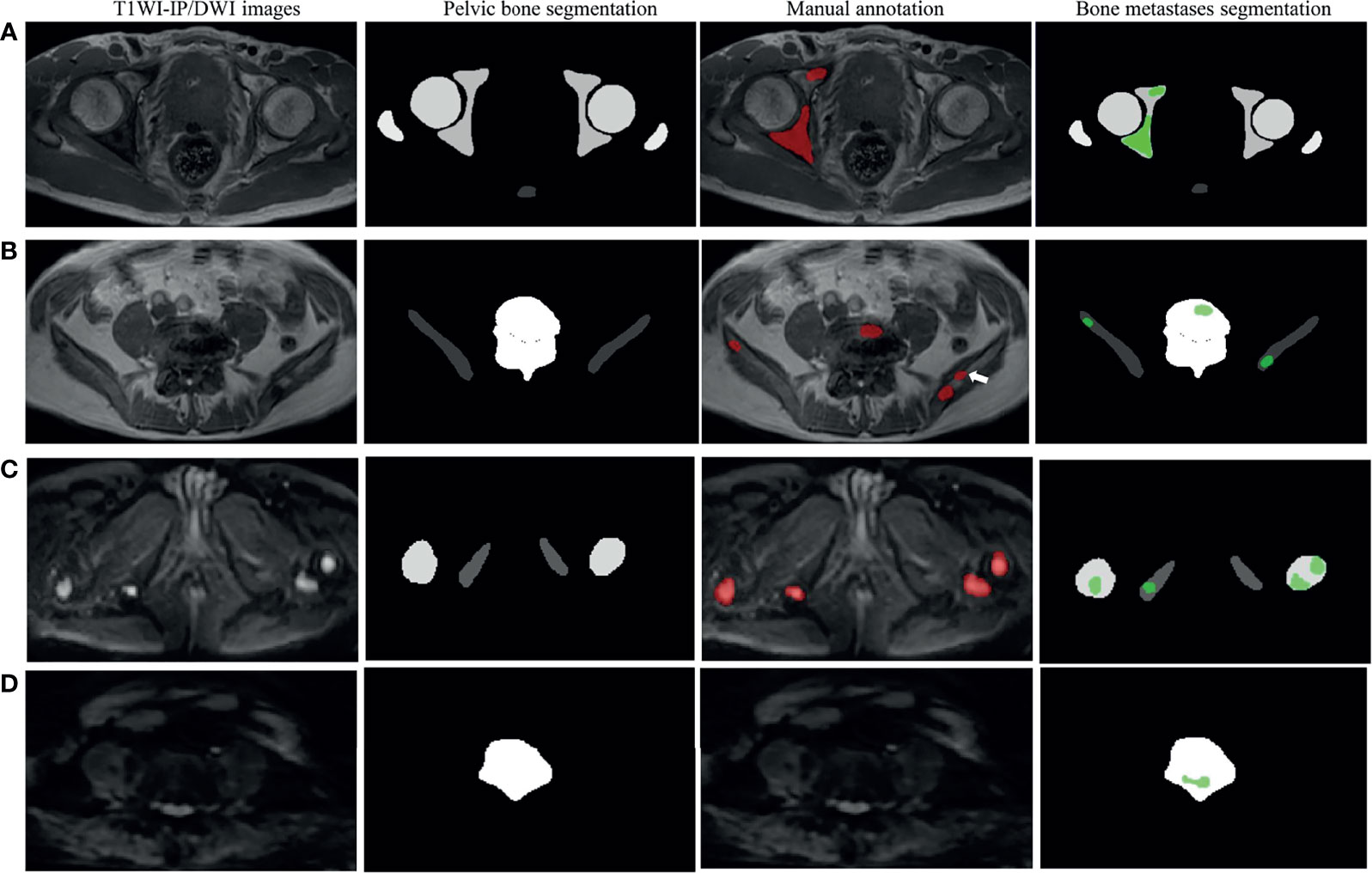
Figure 5 Examples of pelvic bony structure and bone metastases segmentations. (A) Two metastases of acetabulum annotated by radiologists were corrected segmented by the model on T1WI-IP images (true positive). (B) Four of five metastases annotated by the radiologists were corrected segmented by model on T1WI-IP images; one metastasis on the right ilium was missed (the white arrow pointed, false negative). (C) All the four metastases of femoral head and ischium annotated by radiologists were correctly segmented by the model on DWI images (true positive). (D) One metastasis of lumbar vertebra was segmented by the model by error, which was not annotated by the radiologists (false positive). T1WI-IP, T1W images obtained using the Dixon technique with in-phase.
Detection and Segmentation Accuracy on the External Dataset
The sensitivity, specificity, and AUC values of the model in determining the M-staging (M0 or M1) were 93.6% (29/31; 95%CI, 78.6%–99.2%), 93.8% (30/32; 95%CI, 79.6%–99.2%), and 0.94 (95%CI, 0.85–0.98) on DWI images and 87.1% (27/31; 95%CI, 70.2%–96.4%), 90.6% (29/32; 95%CI, 75.0%–98.0%), and 0.89 (95%CI, 0.85–0.98) on T1WI-IP images. The AUC values between the two sequences showed no significant difference (p = 0.368).
At lesion level, the segmentation accuracy of the model for bone metastases achieved average DSC, VS, and HD values of 0.79 ± 0.06, 0.83 ± 0.08, and 16.03 ± 9.74 mm on DWI images, 0.81 ± 0.06, 0.82 ± 0.07, and 17.20 ± 6.73 mm on T1WI-IP images (Figure 6A). The mean volumes of manual annotation and model prediction were 15.35 and 14.10 cm3 on DWI images and 15.68 and 14.40 cm3 on T1WI-IP images. The volume difference is shown in Figures 6B, C.
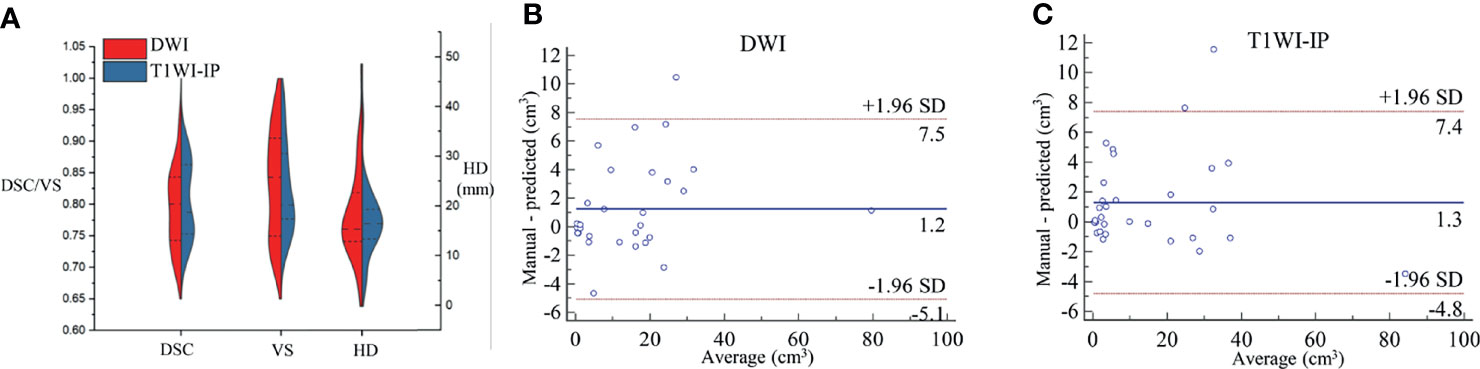
Figure 6 The segmentation accuracy of bone metastases on an external dataset. (A) Split violin plot of DSC, VS, and HD of the bone metastases on DWI and T1WI-IP images. (B) The Bland–Altman plot of the volume difference between manual annotation and model prediction on DWI images. (C) The Bland–Altman plot of the volume difference between manual annotation and model prediction on T1WI-IP images. DSC, Dice similarity coefficient; HD, Hausdorff distance; T1WI-IP, T1W images obtained using the Dixon technique with in-phase; VS, volumetric similarity.
Discussion
In this work, we developed a two-step deep learning-based 3D CNN for automated detection and segmentation of bone metastases in PCa patients using whole 3D MR images (DWI and T1WI-IP images), in which the first 3D U-Net focuses on the segmentation of pelvic bony structures and the second one on bone metastases segmentation. On heterogeneous scanner data, the first CNN performed excellent segmentation of pelvic bony structures on both DWI and T1WI-IP images (all with DSC > 0.85), which provides a reliable foundation for the subsequent bone metastases segmentation. Furthermore, our result showed that the proposed CNN provided an AUC of 0.854 and 0.795 on DWI and T1WI-IP images for bone metastases detection at the patient level, and high overlap between automated and manual metastases segmentations was observed (DSC = 0.79 and 0.80 on DWI and T1WI-IP images, respectively). Additionally, by testing on an external dataset, this work demonstrates the CNN’s potential ability of M-staging in clinical practice (with AUC of 0.936 and 0.889 on DWI and T1WI-IP images).
mpMRI has been identified as an essential and crucial imaging modality in PCa diagnosis and metastases evaluation (25, 26). The importance of DWI and T1WI in the detection and quantification of osseous metastasis in patients with PCa has been widely recognized (9, 27). In this study, to avoid the limitation of the application of the CNN if one of these sequences is unavailable, we trained the two-step 3D U-Net CNN using DWI and T1WI-IP images as independent input data. The enrolled participants performed the mpMRI examinations on one of the three different 3.0-T MR scanners with different protocols, and the b-values of the DWI images were different (b = 0, 800 or 0, 1,000 s/mm2). In a previous publication (28), we proposed a deep learning-based approach for the segmentation of normal pelvic bony structures. It was the proof-of-concept study for the possibility to detect skeletal metastases located on the pelvic bones. In this study, we used two 3D U-Nets in cascade. The first model was trained to segment the pelvic bony structures. Taking the areas predicted by the first model as the mask, the second model was trained to segment the metastatic lesions on the pelvic bones. The combination of the two 3D U-Nets offers the potential for efficient bone metastases location and quantification. It is important to note that the two-step deep learning model has been widely used to improve the accuracy and stability of the system, such as lymph node detection (29) and PCa segmentation (30).
The high number of FP lesions poses a common drawback in automated detection of metastatic lesions, which has been reported to be approximately seven to eight per scan for brain metastases (31, 32). In the present study, by providing high-quality pelvic bone segmentation masks on DWI and T1WI-IP images, the FP interference from other tissues within the pelvic region (such as metastatic lymph nodes, colon, bladder, etc.) can be effectively eliminated. Moreover, a simple post-processing step was added to avoid FP findings by rejecting all structures with a volume <0.2 cm3, which was smaller than the smallest annotated metastases.
Our CNN not only detects almost all metastases but also incorrectly marks other objects as metastases. Most of these FPs were caused by objects that showed a similar radiological appearance to metastatic lesions on DWI and T1WI-IP images. As shown in Figure 5D, the high-intensity spinal cord on DWI images within the mask of the lumbar vertebra was detected as metastases by mistake. In addition, the objects that were not or scarcely represented in the training set and thus had an appearance unknown to the network could result in FP as well. These unknown appearances could be other lesions or conditions such as incidental cysts. An inspection of the 15 FP findings on DWI images showed that nine of the FPs were the spinal cord and nerve root structure, and six of the FPs were benign lesions: four cysts and two hemangiomas. The 12 FP objects on T1WI-IP images included eight spinal cord and nerve root structures, three cysts, and one blood vessel structure.
The FN metastases missed by the CNN networks were the small ones, as can be seen in Figure 5A, which might be due to the few occupied voxels compared with large metastases. Additionally, on a subgroup analysis, our results suggest that the networks perform well on patients with few metastases (≤5 metastases) and multiple metastases (>5 metastases) in terms of recall and precision, which boosts the clinical utility of the CNN.
Automated segmentation can help radiologists in dealing with an increased number of image interpretations while maintaining high diagnostic accuracy and, simultaneously, may also assist in evaluating treatment response during oncological follow-up. Volumetric assessment proves to be a promising tool for quantification of tumor burden and treatment response evaluation, which is superior to user-dependent conventional linear measurements because metastatic lesions are irregular (33). Compared with manual segmentation, our proposed CNN achieved a high volumetric correlation on both the testing set and the external dataset, which is crucial to help treatment decision-making and potentially improve patient care.
TNM is considered to be one of the most pivotal factors in evaluating the prognosis of PCa, and the existence of bone metastases is a decisive index for the M-staging (34). Concerning M-staging, on the external dataset, our model achieved an AUC of 0.936 (95%CI, 0.845–0.982) on DWI images and 0.889 (95%CI, 0.845–0.982) on T1WI-IP images, which demonstrated that the two-step 3D U-Net algorithm could be used in a clinical context. Besides, the output of the automated segmentation result to the structure report essentially combines visualization, quantification, and segmentation into one step, producing results that can be directly displayed to the radiologists.
U-Net has been proven to possess the potential for bone metastases segmentation. Lin et al. (19) built two deep learning networks based on U-Net and Mask R-CNN to segment hotspots in bone SPECT images for automatic assessment of metastasis. Their results showed that the U-Net-based model achieved better segmentation performance with a precision and recall value of 0.76 and 0.67 than the Mask R-CNN model (precision, 0.72; recall, 0.65). In addition, Chang et al. (35) demonstrated the capability of U-Net in segmenting spinal sclerotic bone metastases on CT images with a Dice score of 0.83. In this study, we explored the feasibility of the 3D U-Net network for pelvic bone metastases segmentation on DWI and T1WI-IP images, and our results further confirmed the segmentation accuracy of the U-Net for bone metastases. However, the comparisons among a couple of other architectures may be helpful to choose an optimal model for metastases segmentation and detection. In the future, we should further explore the performance of other models.
While this study shows high accuracy and performance using CNNs for bone metastases segmentation, several potential study limitations exist. First, the study has a typical drawback of retrospective setting. Testing of the network performance on prospective multicenter data remains a key step towards understanding its clinical value. Second, the relatively small number of patients needs to be noted. Only patients with PCa were included here, which potentially limits the transferability of our CNN to a broad range of bone metastases of other primary tumors (rectal cancer, bladder cancer, etc.). In this context, future studies are needed to evaluate the feasibility of the CNN for bone metastases segmentation of other tumors. Third, in clinical practice, the detection of the lesion by the radiologist is usually done by simultaneous review of anatomical and functional MR images. Besides the Dixon T1WI-IP and DWI images, the Fat or Water images from the Dixon sequence and the short time inversion recovery sequence may also be helpful for the bone metastases evaluation (36, 37). Last, the choice of pelvic examinations as the anatomic target to detect bone metastases and assess the positive–negative status of the patients in terms of metastases is insufficient in clinical practice. The axial and probably whole skeleton, at least from skull to thighs, is necessary, as metastases affect the red marrow-containing areas. Future research is needed to allow for the whole-body bone metastases assessment.
Conclusion
In summary, our study shows that the deep learning-based 3D U-Net network can automatically detect and segment bone metastases on DWI and T1WI-IP images with high accuracy and thus illustrates the potential use of this technique in a clinically relevant setting.
Data Availability Statement
The datasets presented in this article are not readily available because the datasets are privately owned by Peking University First Hospital and are not made public. Requests to access the datasets should be directed to wangxiaoying@bjmu.edu.cn.
Ethics Statement
The studies involving human participants were reviewed and approved by Peking University First Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
XL and XW contributed to the study concept and design. TX and CH contributed to acquisition of data. XL and XW annotated the images data. YC and XZ designed the model and implemented the main algorithm. XL and CH contributed to drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Capital’s Funds for Health Improvement and Research (2020-2-40710).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors gratefully acknowledge the technical support of Yaofeng Zhang, Xiangpeng Wang, and Jiahao Huang from Beijing Smart Tree Medical Technology Co. Ltd.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.773299/full#supplementary-material
References
1. Buenrostro D, Mulcrone PL, Owens P, Sterling JA. The Bone Microenvironment: A Fertile Soil for Tumor Growth. Curr Osteoporos Rep (2016) 14:151–8. doi: 10.1007/s11914-016-0315-2
2. Park SH, Eber MR, Shiozawa Y. Models of Prostate Cancer Bone Metastasis. Methods Mol Biol (2019) 1914:295–308. doi: 10.1007/978-1-4939-8997-3_16
3. Hensel J, Thalmann GN. Biology of Bone Metastases in Prostate Cancer. Urology (2016) 92:6–13. doi: 10.1016/j.urology.2015.12.039
4. Clamp A, Danson S, Nguyen H, Cole D, Clemons M. Assessment of Therapeutic Response in Patients With Metastatic Bone Disease. Lancet Oncol (2004) 5:607–16. doi: 10.1016/s1470-2045(04)01596-7
5. Sathiakumar N, Delzell E, Morrisey MA, Falkson C, Yong M, Chia V, et al. Mortality Following Bone Metastasis and Skeletal-Related Events Among Men With Prostate Cancer: A Population-Based Analysis of US Medicare Beneficiaries, 1999-2006. Prostate Cancer Prostatic Dis (2011) 14:177–83. doi: 10.1038/pcan.2011.7
6. Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M, et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment With Curative Intent. Eur Urol (2017) 71:618–29. doi: 10.1016/j.eururo.2016.08.003
7. Padhani AR, Lecouvet FE, Tunariu N, Koh DM, De Keyzer F, Collins DJ, et al. Rationale for Modernising Imaging in Advanced Prostate Cancer. Eur Urol Focus (2017) 3:223–39. doi: 10.1016/j.euf.2016.06.018
8. Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol (2016) 34:1402–18. doi: 10.1200/jco.2015.64.2702
9. Padhani AR, Tunariu N. Metastasis Reporting and Data System for Prostate Cancer in Practice. Magn Reson Imaging Clin N Am (2018) 26:527–42. doi: 10.1016/j.mric.2018.06.004
10. Perez-Lopez R, Lorente D, Blackledge MD, Collins DJ, Mateo J, Bianchini D, et al. Volume of Bone Metastasis Assessed With Whole-Body Diffusion-Weighted Imaging Is Associated With Overall Survival in Metastatic Castration-Resistant Prostate Cancer. Radiology (2016) 280:151–60. doi: 10.1148/radiol.2015150799
11. Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep Learning in Medical Image Analysis. Adv Exp Med Biol (2020) 1213:3–21. doi: 10.1007/978-3-030-33128-3_1
12. Kriegeskorte N, Golan T. Neural Network Models and Deep Learning. Curr Biol (2019) 29:R231–r236. doi: 10.1016/j.cub.2019.02.034
13. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation, 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) Munich, GERMANY (2015) 9351:234–41. doi: 10.1007/978-3-319-24574-4_28
14. Bousabarah K, Ruge M, Brand JS, Hoevels M, Rueß D, Borggrefe J, et al. Deep Convolutional Neural Networks for Automated Segmentation of Brain Metastases Trained on Clinical Data. Radiat Oncol (2020) 15:87. doi: 10.1186/s13014-020-01514-6
15. Park YW, Jun Y, Lee Y, Han K, An C, Ahn SS, et al. Robust Performance of Deep Learning for Automatic Detection and Segmentation of Brain Metastases Using Three-Dimensional Black-Blood and Three-Dimensional Gradient Echo Imaging. Eur Radiol (2021) 31:6686–95. doi: 10.1007/s00330-021-07783-3
16. Goehler A, Harry Hsu TM, Lacson R, Gujrathi I, Hashemi R, Chlebus G, et al. Three-Dimensional Neural Network to Automatically Assess Liver Tumor Burden Change on Consecutive Liver MRIs. J Am Coll Radiol (2020) 17:1475–84. doi: 10.1016/j.jacr.2020.06.033
17. Aoki Y, Nakayama M, Nomura K, Tomita Y, Nakajima K, Yamashina M, et al. The Utility of a Deep Learning-Based Algorithm for Bone Scintigraphy in Patient With Prostate Cancer. Ann Nucl Med (2020) 34:926–31. doi: 10.1007/s12149-020-01524-0
18. Wuestemann J, Hupfeld S, Kupitz D, Genseke P, Schenke S, Pech M, et al. Analysis of Bone Scans in Various Tumor Entities Using a Deep-Learning-Based Artificial Neural Network Algorithm-Evaluation of Diagnostic Performance. Cancers (Basel) (2020) 12:2654. doi: 10.3390/cancers12092654
19. Lin Q, Luo M, Gao R, Li T, Man Z, Cao Y, et al. Deep Learning Based Automatic Segmentation of Metastasis Hotspots in Thorax Bone SPECT Images. PloS One (2020) 15:e0243253. doi: 10.1371/journal.pone.0243253
20. Lin Q, Li T, Cao C, Cao Y, Man Z, Wang H. Deep Learning Based Automated Diagnosis of Bone Metastases With SPECT Thoracic Bone Images. Sci Rep (2021) 11:4223. doi: 10.1038/s41598-021-83083-6
21. Colombo A, Saia G, Azzena AA, Rossi A, Zugni F, Pricolo P, et al. Semi-Automated Segmentation of Bone Metastases From Whole-Body MRI: Reproducibility of Apparent Diffusion Coefficient Measurements. Diagnostics (Basel) (2021) 11:499. doi: 10.3390/diagnostics11030499
22. Heredia V, Gonzalez CA, Azevedo RM, Semelka RC. Bone Metastases: Evaluation of Acuity of Lesions using Dynamic Gadolinium-chelate Enhancement, Preliminary Results. J Magn Reson Imaging (2011) 34:120–7. doi: 10.1002/jmri.22495
23. Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3d U-Net:Learning Dense Volumetric Segmentation From Sparse Annotation. 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) Munich, GERMANY (2016) 9901:24–32. doi: 10.1007/978-3-319-46723-8_49
24. Taha AA, Hanbury A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med Imaging (2015) 15:29. doi: 10.1186/s12880-015-0068-x
25. Stabile A, Giganti F, Rosenkrantz AB, Taneja SS, Villeirs G, Gill IS, et al. Multiparametric MRI for Prostate Cancer Diagnosis: Current Status and Future Directions. Nat Rev Urol (2020) 17:41–61. doi: 10.1038/s41585-019-0212-4
26. Patel P, Wang S, Siddiqui MM. The Use of Multiparametric Magnetic Resonance Imaging (mpMRI) in the Detection, Evaluation, and Surveillance of Clinically Significant Prostate Cancer (csPCa). Curr Urol Rep (2019) 20:60. doi: 10.1007/s11934-019-0926-0
27. Padhani AR, Lecouvet FE, Tunariu N, Koh DM, De Keyzer F, Collins DJ, et al. METastasis Reporting and Data System for Prostate Cancer: Practical Guidelines for Acquisition, Interpretation, and Reporting of Whole-Body Magnetic Resonance Imaging-Based Evaluations of Multiorgan Involvement in Advanced Prostate Cancer. Eur Urol (2017) 71:81–92. doi: 10.1016/j.eururo.2016.05.033
28. Liu X, Han C, Wang H, Wu J, Cui Y, Zhang X, et al. Fully Automated Pelvic Bone Segmentation in Multiparameteric MRI Using a 3D Convolutional Neural Network. Insights Imaging (2021) 12:93. doi: 10.1186/s13244-021-01044-z
29. Hu Y, Su F, Dong K, Wang X, Zhao X, Jiang Y, et al. Deep Learning System for Lymph Node Quantification and Metastatic Cancer Identification From Whole-Slide Pathology Images. Gastric Cancer (2021) 24:868–77. doi: 10.1007/s10120-021-01158-9
30. Zhu Y, Wei R, Gao G, Ding L, Zhang X, Wang X, et al. Fully Automatic Segmentation on Prostate MR Images Based on Cascaded Fully Convolution Network. J Magn Reson Imaging (2019) 49:1149–56. doi: 10.1002/jmri.26337
31. Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic Detection and Segmentation of Brain Metastases on Multimodal MR Images With a Deep Convolutional Neural Network. Comput Biol Med (2018) 95:43–54. doi: 10.1016/j.compbiomed.2018.02.004
32. Grøvik E, Yi D, Iv M, Tong E, Rubin D, Zaharchuk G. Deep Learning Enables Automatic Detection and Segmentation of Brain Metastases on Multisequence MRI. J Magn Reson Imaging (2020) 51:175–82. doi: 10.1002/jmri.26766
33. Chang V, Narang J, Schultz L, Issawi A, Jain R, Rock J, et al. Computer-Aided Volumetric Analysis as a Sensitive Tool for the Management of Incidental Meningiomas. Acta Neurochir (Wien) (2012) 154:589–97. doi: 10.1007/s00701-012-1273-9
34. Paner GP, Stadler WM, Hansel DE, Montironi R, Lin DW, Amin MB. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur Urol (2018) 73(4):560–9. doi: 10.1016/j.eururo.2017.12.018
35. Chang CY, Buckless C, Yeh KJ, Torriani M. Automated Detection and Segmentation of Sclerotic Spinal Lesions on Body CTs Using a Deep Convolutional Neural Network. Skeletal Radiol (2021). doi: 10.1007/s00256-021-03873-x
36. Costelloe CM, Kundra V, Ma J, Chasen BA, Rohren EM, Bassett RL, et al. Fast Dixon Whole-Body MRI for Detecting Distant Cancer Metastasis: A Preliminary Clinical Study. J Magn Reson Imaging (2012) 35(2):399–408. doi: 10.1002/jmri.22815
Keywords: pelvic bones, metastases, prostate cancer, deep learning, magnetic resonance imaging
Citation: Liu X, Han C, Cui Y, Xie T, Zhang X and Wang X (2021) Detection and Segmentation of Pelvic Bones Metastases in MRI Images for Patients With Prostate Cancer Based on Deep Learning. Front. Oncol. 11:773299. doi: 10.3389/fonc.2021.773299
Received: 09 September 2021; Accepted: 08 November 2021;
Published: 29 November 2021.
Edited by:
Oliver Diaz, University of Barcelona, SpainReviewed by:
Kavita Singh, Massachusetts General Hospital and Harvard Medical School, United StatesWei Wei, Xi’an Polytechnic University, China
Copyright © 2021 Liu, Han, Cui, Xie, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoying Wang, d2FuZ3hpYW95aW5nQGJqbXUuZWR1LmNu
 Xiang Liu
Xiang Liu Chao Han
Chao Han Yingpu Cui
Yingpu Cui Tingting Xie
Tingting Xie Xiaodong Zhang
Xiaodong Zhang Xiaoying Wang
Xiaoying Wang