- 1Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China
- 2Department of Hepatobiliary Tumour Surgery, Chenzhou No.1 People’s Hospital, Chenzhou, China
- 3Department of Medicine, Geneplus-Beijing, Beijing, China
- 4Department of Liver Disease Center, The Affiliated Hospital of Qingdao University, Qingdao, China
Majority of patients with resected early- and intermediate-stage liver cancer will experience postoperative recurrence. This study aimed to investigate the application of ctDNA sequencing in the postoperative period of hepatocellular carcinoma. A total of 96 patients with liver cancer were enrolled in this study. Postoperative peripheral blood samples were collected from all patients after surgery and analyzed using hybridization capture-based next-generation sequencing. Identification of at least one somatic mutation in the peripheral blood was defined as ctDNA+. Five genetic features in tumor tissues were associated with disease-free survival (DFS) using Lasso-Cox model. The area under the receiver operating characteristic curve was 0.813 and 0.882 in training and validation cohorts, respectively. The recurrence rate in ctDNA+ and ctDNA- groups was 60.9% and 27.8%, respectively. Multivariate Cox regression analysis showed that the postoperative ctDNA was an independent prognostic predictor of DFS (HR [hazard ratio]: 6.074, 95% Cl [confidence interval]: 2.648-13.929, P<0.001) and overall survival (OS) (HR: 4.829, 95% CI: 1.508-15.466, P=0.008). Combined ctDNA with AFP improved prediction performance. The median DFS was 2.0, and 8.0 months in ctDNA+/AFP-H and ctDNA+/AFP-L groups, respectively; while ctDNA-/AFP-H and ctDNA-/AFP-L groups had not reached the median time statistically (Log-rank test, P < 0.0001). Furthermore, ctDNA- patients had better prognosis than ctDNA+ patients irrespective of tumor stage. Postoperative ctDNA sequencing has great prognostic value in patients with liver cancer. Patients with positive ctDNA should receive more intensive disease monitoring and more aggressive treatment strategies to improve the survival time.
Background
The incidence and mortality of hepatocellular carcinoma (HCC) are among the top five in all tumors in China (1). Evidence suggests that surgical resection, liver transplantation and other curative therapies can significantly improve the survival for those HCC patients with tumor diagnosed at early stage (2). However, the overall survival of HCC patients remains unsatisfactory due to frequent tumor recurrence and metastasis after curative treatments (3). Since the recurrence rate of HCC within 5 years following liver resection has been reported to exceed 70% (4), it is of vital importance to effectively identify those patients who were at a high risk of recurrence after surgery.
Various prognostic tools which interpret surrogate clinicopathologic features such as liver cirrhosis, tumor size, vascular invasion and serological markers have been proposed to predict HCC tumor recurrence with varying degree of reliability (5, 6). Although these prognostic markers are related to tumor recurrence and patient survival, it is difficult to accurately identify those patients who are at a high risk of recurrence immediately after surgery. Furthermore, there is a lack of effective markers to identify earlier staged patients who require more aggressive adjuvant therapy.
Circulating tumor DNA (ctDNA) had been reported as a potent biomarker to reflect tumor load and treatment efficacy in varieties of cancers irrespective of tumor stage (7–10). Previous study has shown that among patients with colorectal cancer, the recurrence rate of ctDNA-positive patients was 77% and ctDNA-negative patients were 0% during the median follow-up time (11). In patients with liver cancer, ctDNA has been proved to be associated with clinical characteristics and clinicopathologic parameters, and could be used as a non-invasive biomarker to monitor tumor progression in real-time (12–14). However, at present, alpha-fetoprotein (AFP) and computed tomography (CT) are routinely used methods for postoperative monitoring (15, 16).
In this study, we aimed to verify the clinical value of postoperative ctDNA in patients with early- or intermediate-stage HCC. Mutations in cancer-related genes were detected through next-generation sequencing of resected tumor tissues and blood samples after resection. We identified five genes whose mutations in tissue samples were associated with survival outcomes, and confirmed that postoperative ctDNA (combined with AFP or not) could be used to stratify the recurrence risk and evaluate prognosis in patients with resected HCC.
Method and Materials
Patients and Samples
A total of 96 patients diagnosed with primary HCC were enrolled in this study. All patients provided written informed consent and received radical resection. The study was approved by the Ethics Committee of Xiangya Hospital Central South University (No. 201703377). After surgery, all patients were monitored regularly in the outpatient clinic with examinations of tumor markers, liver function and abdominal ultrasound every 3-6 months for two years, and then every 6 months. Further CT or magnetic resonance imaging scans were performed as needed if recurrence was suspected. Disease-free survival (DFS) was calculated from the date of liver resection to the date of diagnosis of tumor recurrence. Tumor burden was measured according to the modified Response Evaluation Criteria in Solid Tumors (mRECIST) (17). Tumor size was evaluated as the longest axial diameter of lesions.
Resected tumor tissue and peripheral blood samples (10 mL) within 7-10 days after surgery were collected from each patient. Peripheral blood was collected in Streck tubes (Streck, Omaha, NE, USA) and processed within 72 h to separate plasma and buffy coat (used to filter germline variants).
DNA Extraction
Circulating free DNA (cfDNA) was isolated from plasma using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Buffy coat and tumor tissue DNA were extracted using the DNeasy Blood & Tissue Kit (Qiagen). DNA concentration was measured using a Qubit fluorometer and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen, Carlsbad, CA, USA). The size distribution of the cfDNA was assessed using an Agilent 2100 BioAnalyzer and a DNA HS kit (Agilent Technologies, Santa Clara, CA, USA).
Targeted Capture Sequencing
Before library construction, 1 μg of tissue or buffy coat DNA was sheared to 300 bp fragments with a Covaris S2 Ultrasonicator (Covaris, Woburn, MA, USA). Indexed libraries were prepared from tissue, buffy coat and cfDNA using the KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) as previously described (18). Libraries were then hybridized to custom-designed biotinylated oligonucleotide probes (Integrated DNA Technologies, Iowa, IA, USA). Capture probe for tissue samples was designed to cover whole coding regions or partial exons with mutations frequently detected (hot exons) of 1,021 genes (Supplementary Table 1). Capture probe for postoperative ctDNA samples was designed to cover coding sequences or hot exons of 293 genes, including frequently mutated genes in liver cancer or driver mutations in other cancer types (Supplementary Table 1). Matched tumor-normal sequencing was performed using Illumina 2×100 bp paired-end reads on an Illumina HiSeq 3000 instrument according to the manufacturer’s recommendations using a TruSeq PE Cluster Generation Kit v3 and a TruSeq SBS Kit v3 (Illumina, San Diego, CA, USA). Hybridization capture sequencing revealed a median value of the mean effective depth of coverage of 829× in resected tissue samples and 3470× in postoperative plasma samples (Supplementary Table 2).
Sequencing Data Analysis
Terminal adaptor sequences and low-quality reads were removed from raw data of paired samples. Burrows-Wheeler Aligner (BWA, version 0.7.12-r1039) tool was used to align clean reads to the reference human genome (hg19). Somatic mutations were detected in tissue and ctDNA. Non-synonymous mutations including SNVs and InDels were detected using MuTect (version 1.1.4) and GATK, respectively, and hotspot variants were reviewed by NChot software. Mutations related to clonal hematopoiesis were filtered as previously described, including those in DNMT3A, IDH1, and IDH2 and specific alterations within ATM, GNAS or JAK2 (19, 20). The final candidate variants were all manually verified in the Integrative Genomics Viewer. Whether a tumor specific mutant was detected in the peripheral blood or not was defined as ctDNA (+) and ctDNA (-), respectively.
Statistical Analysis
Nonparametric comparisons were preformed using Wilcoxon t test. Kaplan-Meier survival, univariate and multivariate Cox regression analyses were used to analyze associations between prognostic factors and survival. Univariate comparisons of proportion were analyzed using Fisher’s exact test. All statistical analyses were performed with SPSS (v.21.0; STATA, College Station, TX, USA) or GraphPad Prism (GraphPad Software 7.0, La Jolla, CA, USA) software. Statistical significance was defined as a two-sided P value of smaller than 0.05. LASSO analysis was used to identify prognostic features.
Results
Patient Characteristics
In this study, 96 patients with early- or intermediate-stage HCC were enrolled eligibly. Baseline characteristics were summarized in Table 1. All patients had pathologically confirmed primary HCC before surgical operation, with 78.1% (75/96) patients diagnosed as Barcelona Clinic Liver Cancer (BCLC) 0/A/B stage, and the remaining diagnosed as BCLC C stage. Enrolled patients including 85 males and 11 females. The median age at diagnosis was 50 years, ranging from 18 to 75 years. More than 95% (92/96) patients were infected with hepatitis B virus (HBV). The median diameter of the largest tumor was 5 cm, ranging from 1-18.5 cm. Microvascular invasion was seen in 50% (48/96) of cases, and multifocal lesions were occurred in 32.2% (31/96) of patients.
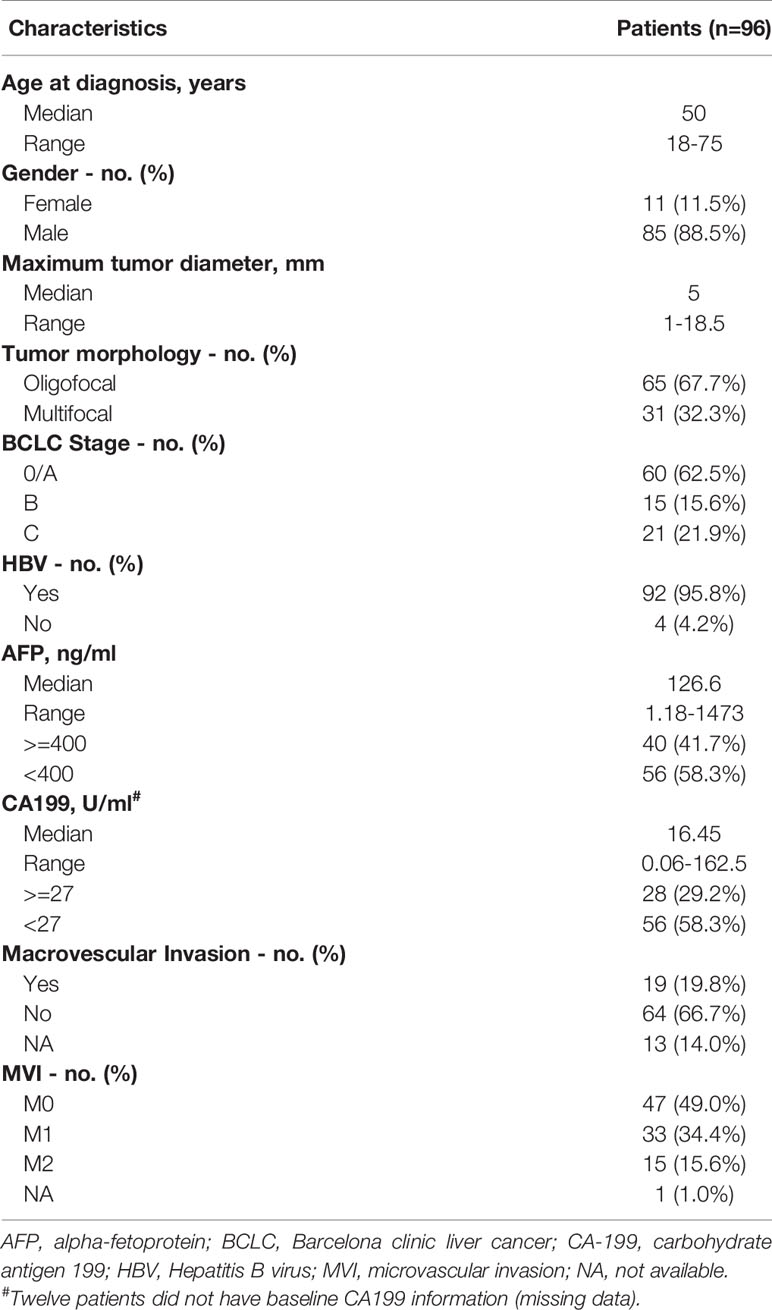
Table 1 Clinical characteristics of patients with primary hepatocellular carcinoma enrolled in this study.
Mutational Profiling of Tumor Tissue Samples
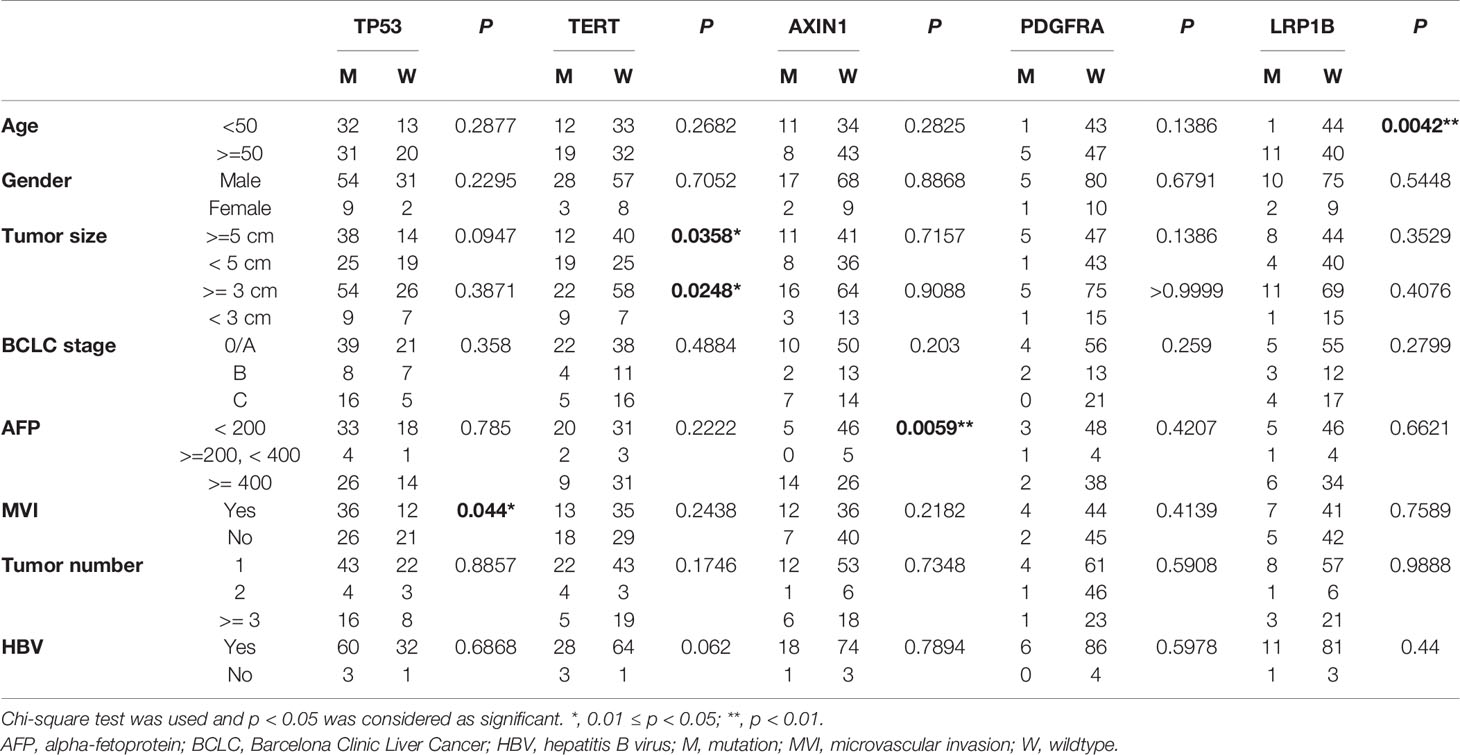
Based on the targeted capture sequencing, a total of 1184 (including 292 CNVs) somatic mutations were detected in 96 tissue samples, with a positive detection rate of 100% (96/96, Figure 1A and Supplementary Table 3). The top three frequently mutated genes were TP53 (65.6%, 63/96), TERT (32.3%, 31/96) and AXIN1 (19.8%, 19/96) (Figures 1A, B). TP53 mutations detected in 28.6% (18/63) of patients were concentrated at the R249S site, which was shown in Figure 1C. All the TERT mutations occurred in promoter regions (100%, 31/31). The mutational landscape of tumor tissue was further compared with data downloaded from Memorial Sloan Kettering Cancer Center (MSKCC) and the cancer genome atlas (TCGA) Cohort. As a result, the mutation frequencies of TP53 (65.6% vs 27.2% and 30.3%, respectively), AXIN1 (18.5% vs 3.2% and 6.9%, respectively), TERT (32.3% vs 14.5% and 0.6%, respectively) in our cohort were higher than that in MSKCC and TCGA database; while a relatively low detection rate of CTNNB1 (14.6% vs 12.4% and 26.2%, respectively) was observed in our cohort, possibly implying the disparate molecular mechanisms of tumorigenesis between Eastern Asia population and Western or/and Caucasian population.
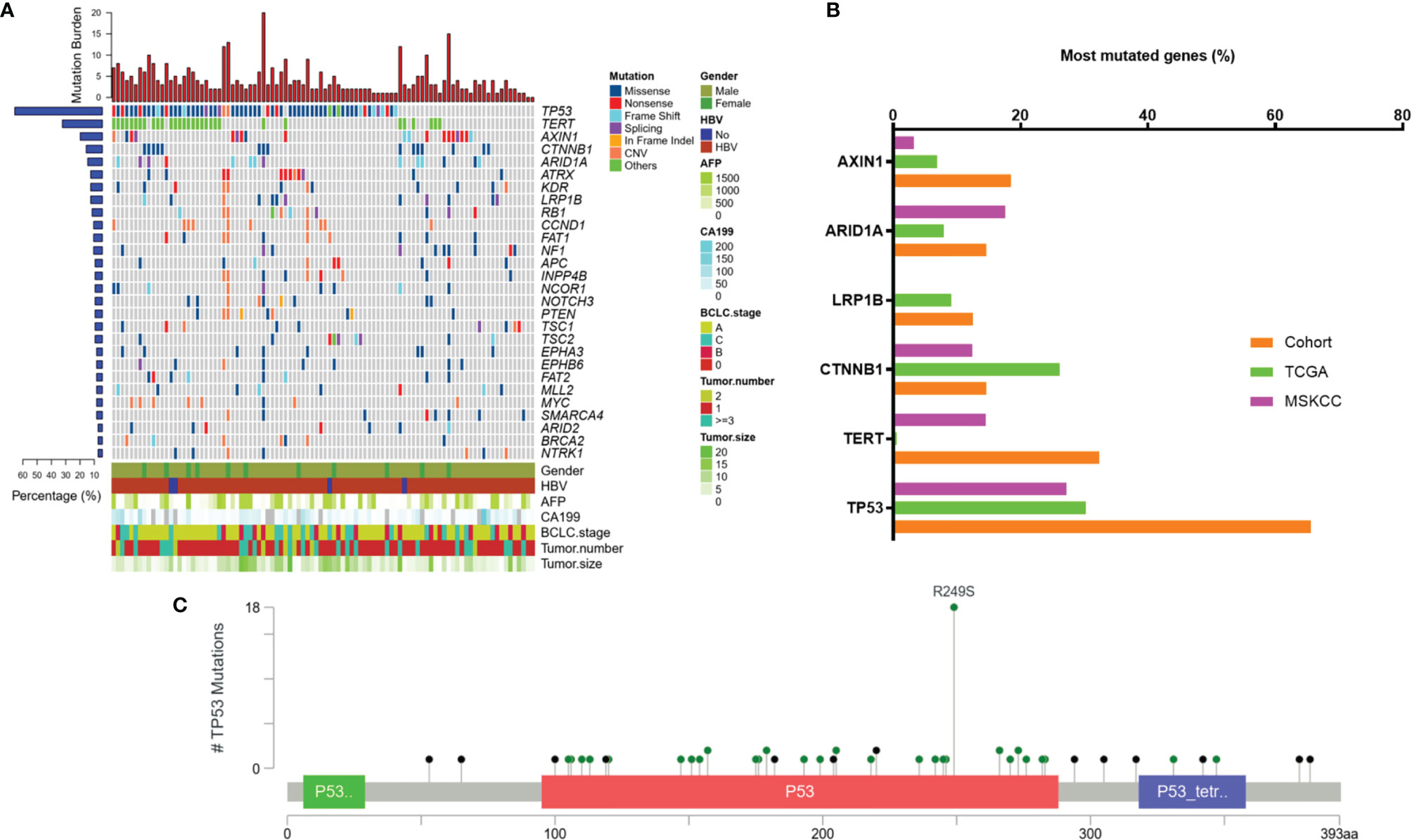
Figure 1 Mutation characteristics of tumor tissue samples. (A) Mutation profiles of tissue samples of 96 enrolled patients. (B) Frequently mutated genes in this cohort and the incidence of mutations in these genes in our cohort compared to that in public databases. (C) Lollipop diagram showing mutations in TP53. TCGA, The Cancer Genome Atlas; MSKCC, Memorial Sloan Kettering Cancer Center; AFP, alpha-fetoprotein; MVI, microvascular invasion; BCLC, Barcelona Clinic Liver Cancer; CA 19-9, carbohydrate antigen 19-9; HBV, hepatitis B virus.
Mutational Features of Tumor Tissue Related to Prognosis
To select prognostic genetic features, we screened out 123 common genes in recurrence and non-recurrence group from 1021 cancer-related genes and performed the least absolute shrinkage and selection operator (LASSO) regression model on the basis of recurrence and DFS (Figures 2A, B). Five genetic features were identified in the training cohort: AXIN1, CTNNB1, LRP1B, PDGFRA and TP53. These five genes were also the frequently mutated genes in liver cancer. The area under the ROC curve was 0.813 and 0.882 in training and validation cohorts, respectively (Figures 2C, D).
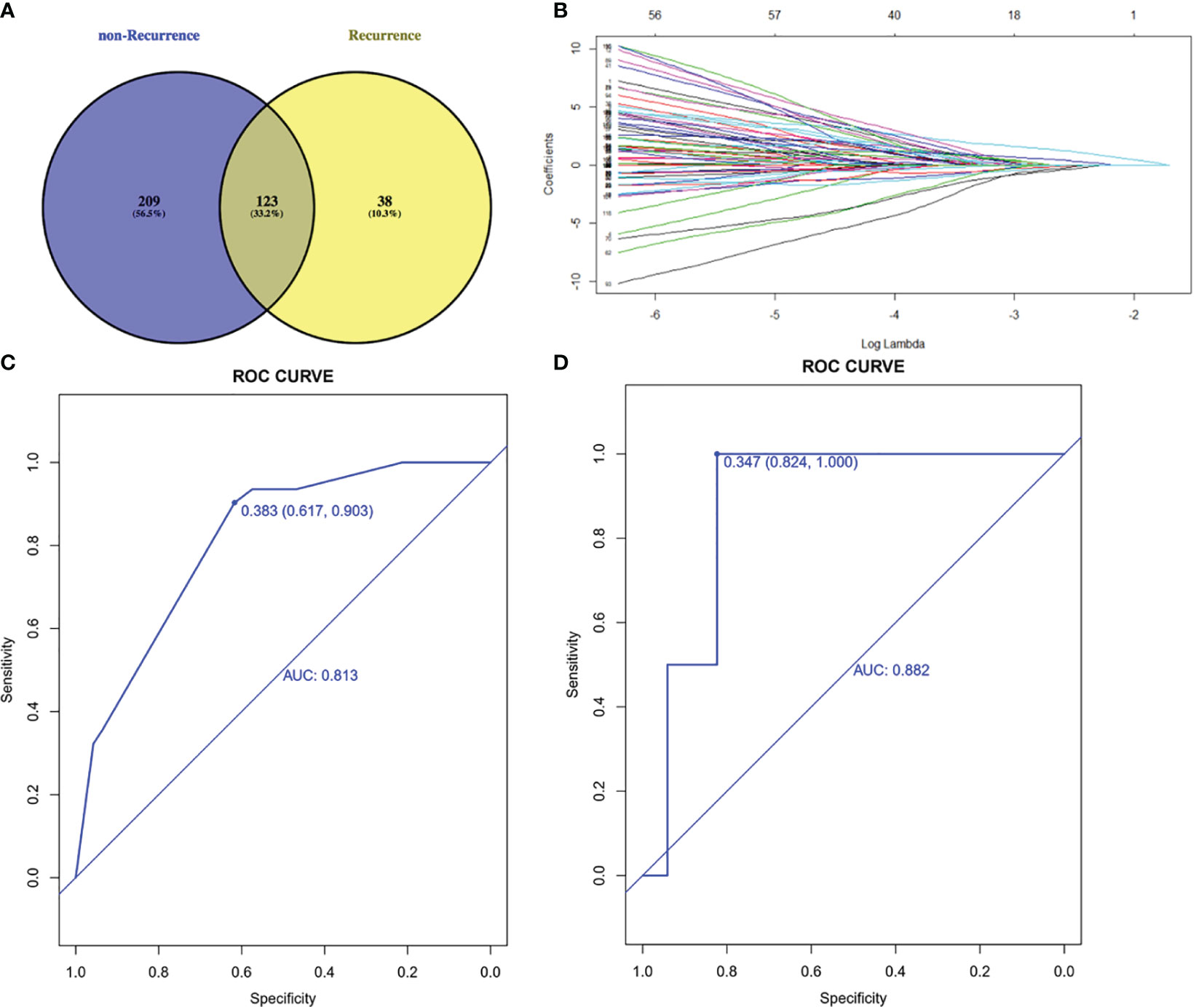
Figure 2 Lasso-Cox regression model. (A) 123 common genes mutated in both recurrence and non-recurrence group. (B) Least absolute shrinkage and selection operator (LASSO) regression model. (C) ROC curve of training cohort. (D) ROC curve of validation cohort. AUC, area under curve; ROC, receiver operating characteristic.
Furthermore, as shown in Table 2, specific gene mutations were related with patients’ clinical features. TP53 mutations were associated with relapse (p=0.0106). LRP1B mutations were more likely to occur in older patients. The mutation rate in patients <50 years and ≥50 years was 2% and 21.6%, respectively (p=0.0042). Surprisingly, the difference in tumor size between TERT-wildtype and mutation group was also significant. The mutation rate was 23.1% in patients with tumor size ≥5 cm and 43.2% in patients with tumor size <5 cm (p=0.0358), which may suggest that TERT promoter mutations were more likely to be detected in tumor tissues in the early stage. TP53 R249S mutation frequency was extremely high in liver cancer, which was considered to be closely associated with aflatoxin exposure and HBV infection (21, 22). In our cohort, TP53 R249S mutation was detected in 18 patients and 94.4% (17/18) of them were identified with positive HBs-Ag. 55.5% (10/18) R249S mutation patients had recurred within 8 months, and 29.5% (23/78) patients without TP53 R249S mutation had recurred in 19 months (P=0.0003, Supplementary Table 4 and Figure 3A). Patients with R249S point mutation showed a worse prognosis. The median DFS time was 7.0 months for R249S mutated group (n =18) and not reached for R249S wildtype group (n= 78), respectively (Log-rank test, P = 0.0124) (Figure 3B).
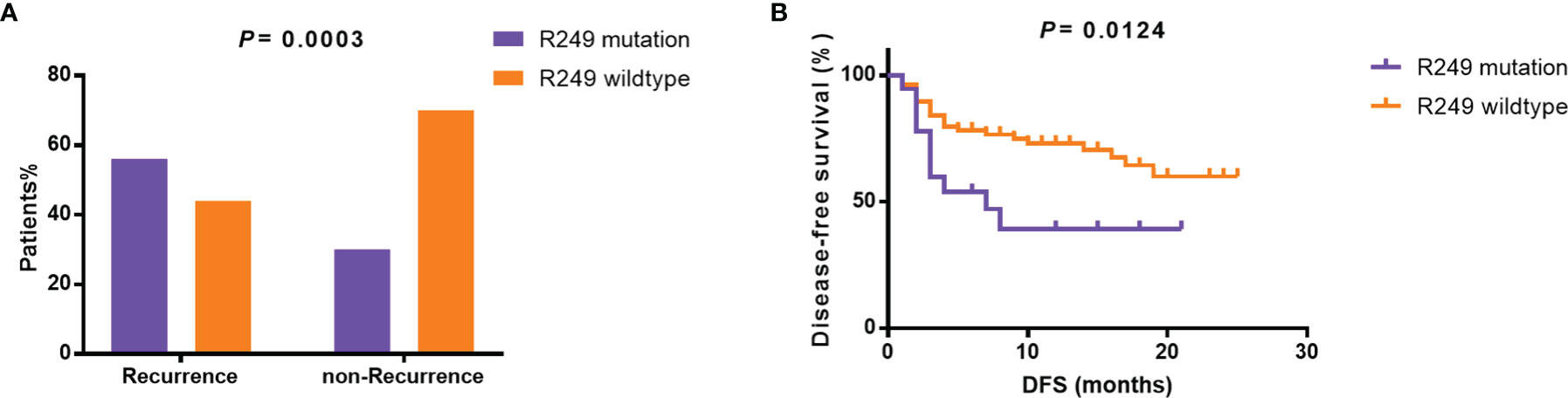
Figure 3 Relationship between TP53 R249S mutation and prognosis. (A) Incidence of R249 mutation in the recurrent and non-recurrent groups. (B) Disease-free survival in the R249 mutation group and the R249 wildtype group. DFS, disease-free survival.
ctDNA Positive Patients Had the Worse Prognosis
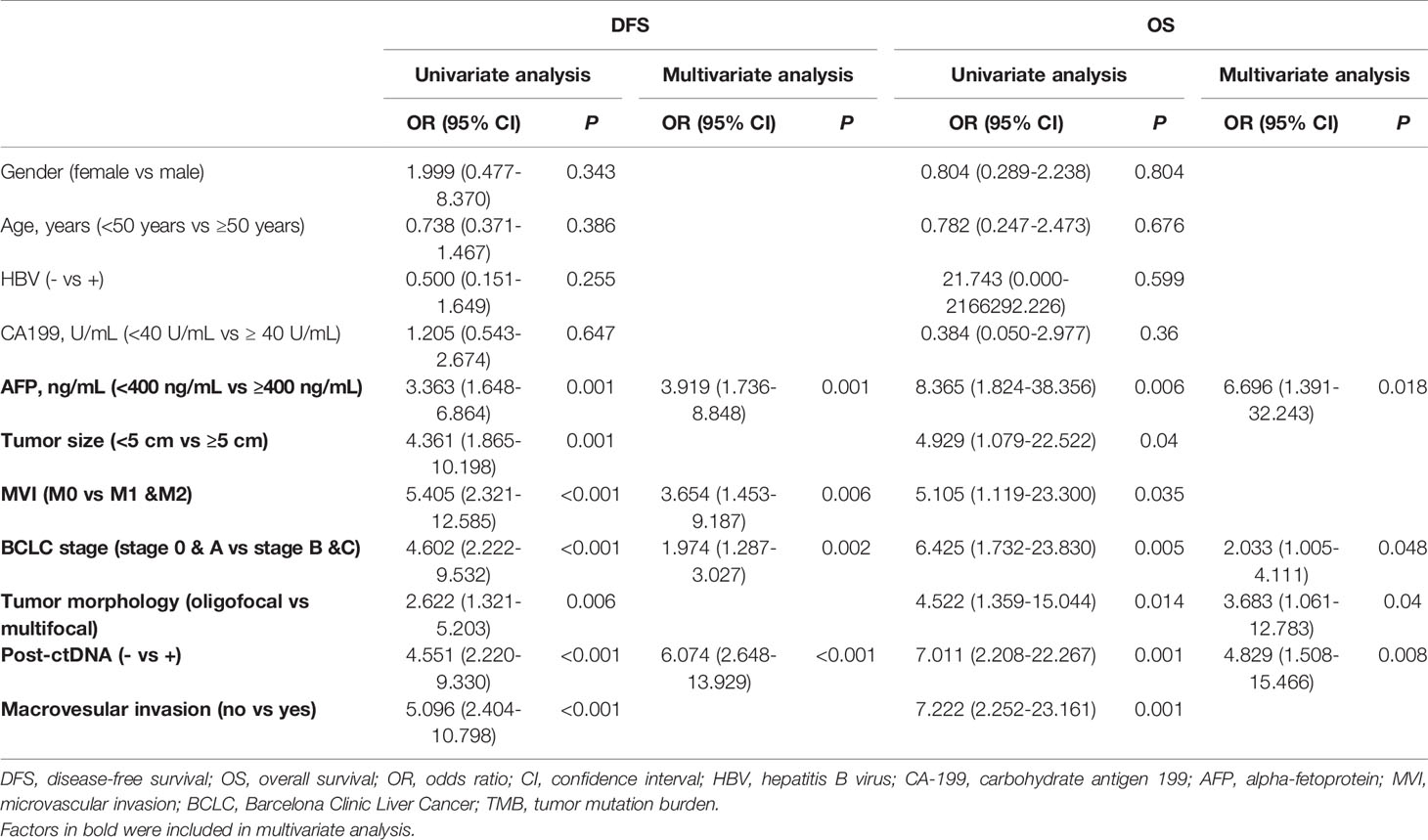
Postoperative blood samples were collected within 7 days after surgery from all of the 96 patients. The sequencing results were classified based on whether mutations were detected in blood samples or not. Twenty-three patients were considered as ctDNA (+), and 60.9% (14/23) of them had recurrence; while 72 patients were ctDNA (-), and only 27.8% (20/72) patients had recurrence (P = 0.0059) (Supplementary Figure 1). The recurrence in 27 patients was in the liver only and 7 patients had distant metastases, and ctDNA positivity rate in these patients was 96.3% (26/27) and 100% (7/7), respectively. The median DFS time was 4.0 months in ctDNA (+) group, and the ctDNA (-) group had not reached the median time (Log-rank test, P < 0.0001) (Figure 4A). Multivariate Cox regression analysis showed that the postoperative ctDNA was an independent prognostic predictor of DFS (HR [hazard ratio]: 6.074, 95% Cl [confidence interval]: 2.648-13.929, P<0.001) and OS (HR: 4.829, 95% CI: 1.508-15.466, P=0.008 (Table 3).
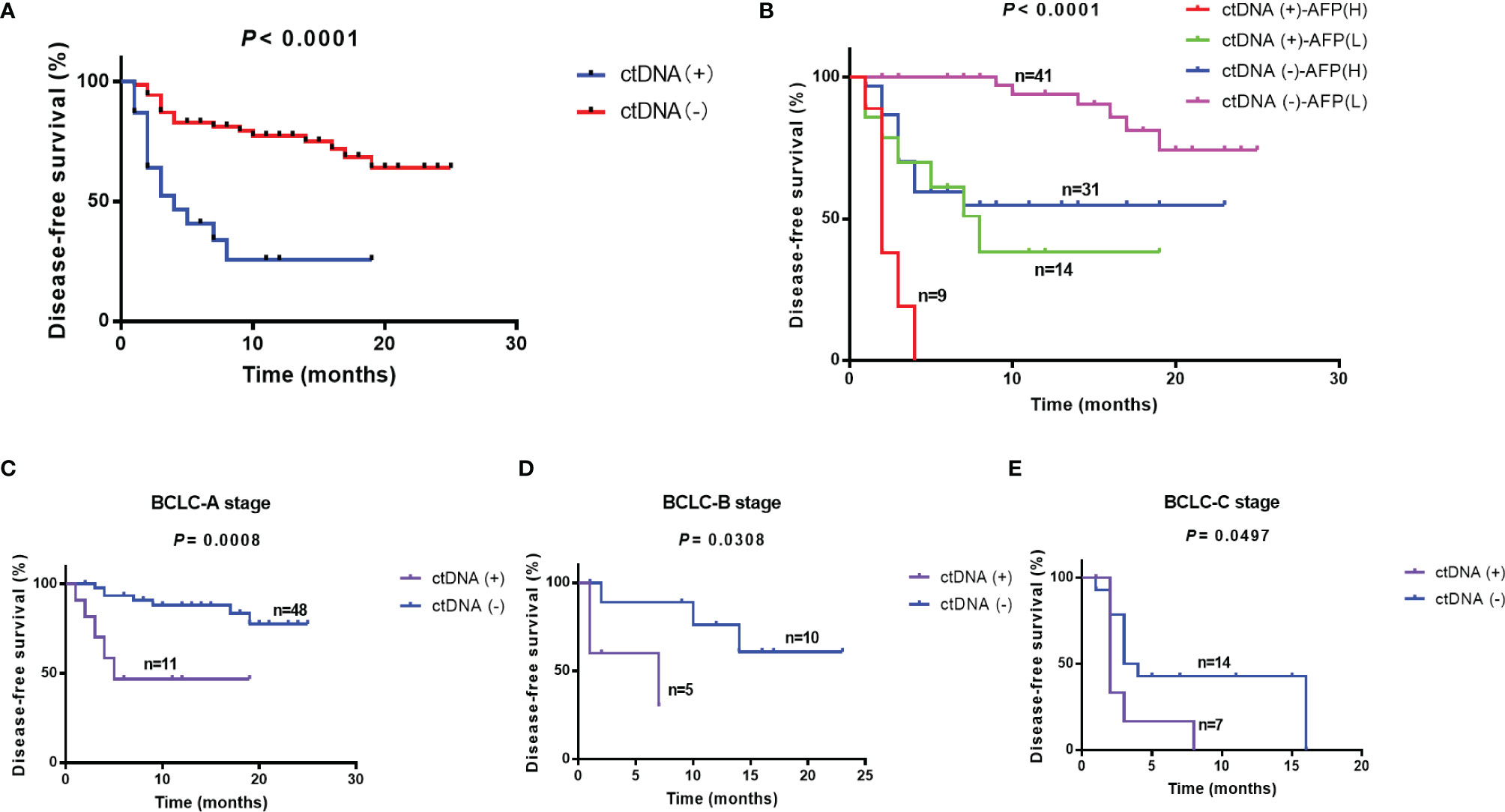
Figure 4 Relationship between ctDNA positivity and other clinical factors and prognosis. (A) Disease-free survival in ctDNA (-) and ctDNA (+) groups. (B) Prediction of prognosis using ctDNA combined with AFP. Prediction of prognosis using ctDNA in patients with different BCLC stages: (C) BCLC-A stage, (D) BCLC-B stage, (E) BCLC-C stage. AFP, alpha-fetoprotein; BCLC, Barcelona Clinic Liver Cancer.
Combined ctDNA With AFP and BCLC Stage Improved Prediction Performance
BCLC stage and baseline AFP level were indispensable clinical indicators to assess prognosis of liver cancer. In our study, BCLC stage and baseline AFP were independent prognostic predictors (Table 3). We classified patients based on postoperative ctDNA combined with baseline AFP or BCLC stage. The results showed that ctDNA combined with AFP would effectively predict the prognosis of patients after surgery. CtDNA (+)-AFP (H) (>= 400 ng/mL) patients had the worst prognosis and 77.8% (7/9) of them had relapsed; while ctDNA (-)-AFP (L) (<400 ng/mL) patients had the best prognosis, with less than 15% (6/41) had relapsed (Log-rank test, P < 0.0001). The median DFS time was 2.0, and 8.0 months in ctDNA (+)-AFP (H) (n=9) and ctDNA (+)-AFP (L) (n=14) groups, respectively; while ctDNA (-)-AFP (H) (n=31) and ctDNA (-)-AFP (L) groups (n=41) had not reached the median time statistically (Log-rank test, P < 0.0001) (Figure 4B).
In the previous study, BCLC stage was related to baseline ctDNA abundance. BCLC C patients had a higher ctDNA abundance than A and B stage patients (23). Combined ctDNA and BCLC stage may effectively predict the prognosis of patients after surgery. In BCLC A group, 11 patients were ctDNA positive and 48 patients were ctDNA negative, the ctDNA (-)-A patients had the better prognosis (P=0.0008) and the median DFS was 5 months in ctDNA(+)-A (n=11) and ctDNA(-)-A (n=48) had not reached the median DFS time. Similarly, the ctDNA (-)-B (n=10) and ctDNA (-)-C (n=14) patients had the better prognosis than ctDNA (+)-B(n=5) (P=0.0308) and ctDNA (+)-C (n=7) patients (P=0.0497). The median DFS was 7 months in ctDNA (+)-B and ctDNA (-)-B had not reach the median DFS time statistically. The median DFS was 2 and 3.5 months in ctDNA (+)-C and ctDNA (-)-C groups (Figures 4C–E).
Discussion
The prognosis of patients with resectable hepatocellular carcinoma is affected by various factors. We portrayed the mutational spectrum of the surgical tumor tissues of HCC patients in our cohort, and finally determined 5 genes related to prognosis after resection through Lasso-Cox regression model. According to previous study, TP53 is the most frequently mutated gene in HCC among all somatic mutated genes and its mutations were significantly associated with poorer overall survival in HCC patients (24). In various cancer types, TP53 has a broad-spectrum mutation, but only in liver cancer, TP53 mutations are highly concentrated at the R249 point, which was considered to be related to aflatoxin infection and positive HBs-Ag (21, 22). This is also observed in our research. Furthermore, we found that DFS and OS of patients harboring TP53 R249 mutation were significantly shorter than those of wild-type patients. This finding may provide a clue for subsequent targeted treatment of liver cancer. Unexpectedly, TERT promoter mutations were related with smaller tumor sizes in our study. Previous studies revealed the inconsistent relationship between TERT mutations and tumor sizes. TERT promoter mutations in tissue samples did not correlated with tumor size (25–27). However, TERT mutations in ctDNA samples were correlated with large intrahepatic tumor size and increased mortality (28). One study compared the presence of TERT mutation in ctDNA and corresponding tumor tissue, and found that non-concordance (ctDNA-, tissue+) was associated with an early TNM stage. The TERT mutations were detected in 84.6% (11/13), 42.9% (3/7), 80% (4/5) and 55.6% (5/9) of patients with stage I, II, III and IV, respectively (27). These results were consistent with our findings to a certain degree.
In addition to the tumor mutational features and clinical factors, postoperative minimal residual disease (MRD) is considered as a high risk factor for tumor recurrence, which refers to postoperative tumor burden that cannot be identified with traditional diagnostic methods (29). In recent years, ctDNA-based sequencing has gradually been explored as a possible tool to identify MRD. Many previous studies demonstrated that detection of ctDNA after surgery was strongly associated with increased risk of disease recurrence (30–32). A few studies in HCC also revealed that the dynamic change of ctDNA after surgery could accurately and better evaluate patients’ prognosis and detect tumor occurrence prior to traditional strategies (13, 33). However, several studies generated conflicting data about the prognostic value of ctDNA (34). Indeed, low ctDNA concentration in early-stage cancers is a nonnegligible limitation of ctDNA analysis in early detection of relapse (35). Combination with other protein markers has been proved to improve the sensitivity and specificity of ctDNA for predicting prognosis (36, 37).
This prospective study evaluated the clinical utility of ctDNA for detection of MRD in HCC. We found that ctDNA positivity in the immediate post-operative period was significantly associated with worse DFS and OS independently, suggesting the clinical significance of ctDNA-based MRD detection. We further combined ctDNA with clinical indicator AFP and BCLC stage and found that patients with positive postoperative ctDNA-AFP (H) subgroups and patients with positive ctDNA and BCLC staging C subgroups had the worst prognosis. It is not enough for us to evaluate the possibility of postoperative recurrence by relying solely on traditional clinical indicators. Especially for patients with earlier stage tumors, postoperative ctDNA has unique advantages in identifying high-risk recurrence patients.
In summary, postoperative ctDNA detection has great potential value in clinic, and patients with positive ctDNA after surgery should receive individualized medication to improve the survival time.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://ngdc.cncb.ac.cn/gsa-human, HRA001983.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Xiangya Hospital Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
KY and LZ contributed to the conceptualization and design. KY, QF, MY, TF and ZL contributed to the data curation and writing. DW, LX, and GL analyzed and interpreted the data. RC contributed to the conceptualization. All authors contributed to the article and approved the submitted version.
Conflict of Interest
MY and RC are employees of Geneplus-Beijing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.834992/full#supplementary-material
Supplementary Figure 1 | The proportion of ctDNA positivity in the recurrence and non-recurrence groups.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
2. Chen L, Ho DW, Lee NP, Sun S, Lam B, Wong KF, et al. Enhanced Detection of Early Hepatocellular Carcinoma by Serum SELDI-TOF Proteomic Signature Combined With Alpha-Fetoprotein Marker. Ann Surg Oncol (2010) 17(9):2518–25. doi: 10.1245/s10434-010-1038-8
3. Lee J, Long G, Menzies A, Lo S, Guminski A, Whitbourne K, et al. Association Between Circulating Tumor DNA and Pseudoprogression in Patients With Metastatic Melanoma Treated With Anti-Programmed Cell Death 1 Antibodies. JAMA Oncol (2018) 4(5):717–21. doi: 10.1001/jamaoncol.2017.5332
4. Bruix J, Sherman M. Management of Hepatocellular Carcinoma. Hepatology (Baltimore Md) (2005) 42(5):1208–36. doi: 10.1002/hep.20933
5. Wang Y, Tan PY, Handoko YA, Sekar K, Shi M, Xie C, et al. NUF2 Is a Valuable Prognostic Biomarker to Predict Early Recurrence of Hepatocellular Carcinoma After Surgical Resection. Int J Cancer (2019) 145(3):662–70. doi: 10.1002/ijc.32134
6. Ueno M, Hayami S, Shigekawa Y, Kawai M, Hirono S, Okada K, et al. Prognostic Impact of Surgery and Radiofrequency Ablation on Single Nodular HCC 5 Cm: Cohort Study Based on Serum HCC Markers. J Hepatol (2015) 63(6):1352–9. doi: 10.1016/j.jhep.2015.07.013
7. Parikh AR, Mojtahed A, Schneider JL, Kanter K, Van Seventer EE, Fetter IJ, et al. Serial ctDNA Monitoring to Predict Response to Systemic Therapy in Metastatic Gastrointestinal Cancers. Clin Cancer Res (2020) 26(8):1877–85. doi: 10.1158/1078-0432.ccr-19-3467
8. Horn L, Whisenant JG, Wakelee H, Reckamp KL, Qiao H, Leal TA, et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J Thorac Oncol (2019) 14(11):1901–11. doi: 10.1016/j.jtho.2019.08.003
9. Garcia-Murillas I, Chopra N, Comino-Méndez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol (2019) 5(10):1473–8. doi: 10.1001/jamaoncol.2019.1838
10. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov (2017) 7(12):1394–403. doi: 10.1158/2159-8290.cd-17-0716
11. Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-Small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front Immunol (2019) 10:1113. doi: 10.3389/fimmu.2019.01113
12. Ng CKY, Di Costanzo GG, Tosti N, Paradiso V, Coto-Llerena M, Roscigno G, et al. Genetic Profiling Using Plasma-Derived Cell-Free DNA in Therapy-Naive Hepatocellular Carcinoma Patients: A Pilot Study. Ann Oncol (2018) 29(5):1286–91. doi: 10.1093/annonc/mdy083
13. Cai Z, Chen G, Zeng Y, Dong X, Li Z, Huang Y, et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients With Hepatocellular Carcinoma. Clin Cancer Res (2019) 25(17):5284–94. doi: 10.1158/1078-0432.CCR-18-3477
14. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and Localization of Surgically Resectable Cancers With a Multi-Analyte Blood Test. Science (New York NY) (2018) 359(6378):926–30. doi: 10.1126/science.aar3247
15. Ferris JV, Baron RL, Marsh JW Jr., Oliver JH 3rd, Carr BI, Dodd GD 3rd. Recurrent Hepatocellular Carcinoma After Liver Transplantation: Spectrum of CT Findings and Recurrence Patterns. Radiology (1996) 198(1):233–8. doi: 10.1148/radiology.198.1.8539385
16. Tateishi R, Shiina S, Yoshida H, Teratani T, Obi S, Yamashiki N, et al. Prediction of Recurrence of Hepatocellular Carcinoma After Curative Ablation Using Three Tumor Markers. Hepatology (Baltimore Md) (2006) 44(6):1518–27. doi: 10.1002/hep.21408
17. Lencioni R, Llovet JM. Modified RECIST (mRECIST) Assessment for Hepatocellular Carcinoma. Semin Liver Dis (2010) 30(1):52–60. doi: 10.1055/s-0030-1247132
18. Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, et al. Circulating Tumor DNA Analysis Depicts Subclonal Architecture and Genomic Evolution of Small Cell Lung Cancer. Nat Commun (2018) 9(1):3114. doi: 10.1038/s41467-018-05327-w
19. Hu ZY, Xie N, Tian C, Yang X, Liu L, Li J, et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients With Multiline Resistance. EBioMedicine (2018) 32:111–8. doi: 10.1016/j.ebiom.2018.05.015
20. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct Detection of Early-Stage Cancers Using Circulating Tumor DNA. Sci Trans Med (2017) 9(403):eaan2415. doi: 10.1126/scitranslmed.aan2415
21. Ortiz-Cuaran S, Villar S, Gouas D, Ferro G, Plymoth A, Khuhaprema T, et al. Association Between HBX Status, Aflatoxin-Induced R249S TP53 Mutation and Risk of Hepatocellular Carcinoma in a Case-Control Study From Thailand. Cancer Lett (2013) 331(1):46–51. doi: 10.1016/j.canlet.2012.11.012
22. Villar S, Le Roux-Goglin E, Gouas DA, Plymoth A, Ferro G, Boniol M, et al. Seasonal Variation in TP53 R249S-Mutated Serum DNA With Aflatoxin Exposure and Hepatitis B Virus Infection. Environ Health Perspect (2011) 119(11):1635–40. doi: 10.1289/ehp.1103539
23. Zhao W, Qiu L, Liu H, Xu Y, Zhan M, Zhang W, et al. Circulating Tumor DNA as a Potential Prognostic and Predictive Biomarker During Interventional Therapy of Unresectable Primary Liver Cancer. J Gastrointest Oncol (2020) 11(5):1065–77. doi: 10.21037/jgo-20-409
24. Li X, Xu W, Kang W, Wong SH, Wang M, Zhou Y, et al. Genomic Analysis of Liver Cancer Unveils Novel Driver Genes and Distinct Prognostic Features. Theranostics (2018) 8(6):1740–51. doi: 10.7150/thno.22010
25. Chen YL, Jeng YM, Chang CN, Lee HJ, Hsu HC, Lai PL, et al. TERT Promoter Mutation in Resectable Hepatocellular Carcinomas: A Strong Association With Hepatitis C Infection and Absence of Hepatitis B Infection. Int J Surg (Lond Engl) (2014) 12(7):659–65. doi: 10.1016/j.ijsu.2014.05.066
26. Lee HW, Park TI, Jang SY, Park SY, Park WJ, Jung SJ, et al. Clinicopathological Characteristics of TERT Promoter Mutation and Telomere Length in Hepatocellular Carcinoma. Medicine (2017) 96(5):e5766. doi: 10.1097/md.0000000000005766
27. Oversoe SK, Clement MS, Pedersen MH, Weber B, Aagaard NK, Villadsen GE, et al. TERT Promoter Mutated Circulating Tumor DNA as a Biomarker for Prognosis in Hepatocellular Carcinoma. Scand J Gastroenterol (2020) 55(12):1433–40. doi: 10.1080/00365521.2020.1837928
28. Hirai M, Kinugasa H, Nouso K, Yamamoto S, Terasawa H, Onishi Y, et al. Prediction of the Prognosis of Advanced Hepatocellular Carcinoma by TERT Promoter Mutations in Circulating Tumor DNA. J Gastroenterol Hepatol (2021) 36(4):1118–25. doi: 10.1111/jgh.15227
29. Chae YK, Oh MS. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J Thorac Oncol (2019) 14(1):16–24. doi: 10.1016/j.jtho.2018.09.022
30. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA Analysis Depicts Early-Stage Lung Cancer Evolution. Nature (2017) 545(7655):446–51. doi: 10.1038/nature22364
31. Bogani G, Ricci MT, Vitellaro M, Ditto A, Chiappa V, Raspagliesi F. Impact of Gene-Specific Germline Pathogenic Variants on Presentation of Endometrial Cancer in Lynch Syndrome. Int J Gynecol Cancer (2019) 29(4):705–10. doi: 10.1136/ijgc-2019-000277
32. Coenegrachts L, Garcia-Dios DA, Depreeuw J, Santacana M, Gatius S, Zikan M, et al. Mutation Profile and Clinical Outcome of Mixed Endometrioid-Serous Endometrial Carcinomas Are Different From That of Pure Endometrioid or Serous Carcinomas. Virchows Arch (2015) 466(4):415–22. doi: 10.1007/s00428-015-1728-5
33. Wang J, Huang A, Wang YP, Yin Y, Fu PY, Zhang X, et al. Circulating Tumor DNA Correlates With Microvascular Invasion and Predicts Tumor Recurrence of Hepatocellular Carcinoma. Ann Transl Med (2020) 8(5):237. doi: 10.21037/atm.2019.12.154
34. Tuaeva NO, Falzone L, Porozov YB, Nosyrev AE, Trukhan VM, Kovatsi L, et al. Translational Application of Circulating DNA in Oncology: Review of the Last Decades Achievements. Cells (2019) 8(10):1251. doi: 10.3390/cells8101251
35. Heidrich I, Ačkar L, Mossahebi Mohammadi P, Pantel K. Liquid Biopsies: Potential and Challenges. Int J Cancer (2021) 148(3):528–45. doi: 10.1002/ijc.33217
36. Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined Circulating Tumor DNA and Protein Biomarker-Based Liquid Biopsy for the Earlier Detection of Pancreatic Cancers. Proc Natl Acad Sci USA (2017) 114(38):10202–7. doi: 10.1073/pnas.1704961114
Keywords: circulating tumor DNA, liver cancer, alpha-fetoprotein, next-generation sequencing, postoperative recurrence
Citation: Ye K, Fan Q, Yuan M, Wang D, Xiao L, Long G, Chen R, Fang T, Li Z and Zhou L (2022) Prognostic Value of Postoperative Circulating Tumor DNA in Patients With Early- and Intermediate-Stage Hepatocellular Carcinoma. Front. Oncol. 12:834992. doi: 10.3389/fonc.2022.834992
Received: 14 December 2021; Accepted: 31 January 2022;
Published: 04 March 2022.
Edited by:
Guoliang Qiao, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Simon Chang-Hao Tsao, The University of Melbourne, AustraliaLuca Falzone, G. Pascale National Cancer Institute Foundation (IRCCS), Italy
Copyright © 2022 Ye, Fan, Yuan, Wang, Xiao, Long, Chen, Fang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ledu Zhou , emhvdWxkQGNzdS5lZHUuY24=
 Ke Ye1
Ke Ye1 Mingming Yuan
Mingming Yuan Rongrong Chen
Rongrong Chen Ledu Zhou
Ledu Zhou