- 1Xi’an Key Laboratory of Basic and Translation of Cardiovascular Metabolic Disease, Shaanxi Key Laboratory of Ischemic Cardiovascular Disease, Institute of Basic & Translational Medicine, Xi’an Medical University, Xi’an, China
- 2College of Bioresources Chemical and Materials Engineering, Shaanxi University of Science & Technology, Xi’an, China
- 3College of Pharmacy, Shaanxi University of Chinese Medicine, Xianyang, China
- 4Shanghai Synchrotron Radiation Facility, Shanghai Advanced Research Institute, Chinese Academy of Sciences, Shanghai, China
- 5Institute for Advanced Studies, Wuhan University, Wuhan, China
New targeted chemotherapy agents greatly improved five-year survival in NSCLC patients, but which were susceptible to drug resistance. NVP-AUY922, terminated in phase II clinical trials, exhibited promising anti-NSCLC (non-small-cell lung cancer) activity targeting to Hsp90N (heat shock protein), which demonstrated advantages in overcoming drug resistance as a broad-spectrum anti-cancer target. It was expected to develop novel anti-NSCLC drugs to overcome drug resistance by the structural optimization of NVP-AUY922. However, the absence of high-resolution complex crystal structure of Hsp90N-NVP-AUY922 blocked the way. Herein, 1.59 Å-resolution complex crystal structure of Hsp90N-NVP-AUY922 (PDB ID 6LTI) was successfully determined by X-ray diffraction. Meanwhile, there was a strong binding capability between NVP-AUY922 and its target Hsp90N verified by TSA (ΔTm, −15.56 ± 1.78°C) and ITC (Kd, 5.10 ± 2.10 nM). Results by the complex crystal structure, TSA and ITC verified that NVP-AUY922 well accommodated in the ATP-binding pocket of Hsp90N to disable the molecular chaperone activity of Hsp90. Therefore, NVP-AUY922 exhibited approving inhibitory activity on NSCLC cell line H1299 (IC50, 2.85 ± 0.06 μM) by inhibiting cell proliferation, inducing cell cycle arrest and promoting cell apoptosis. At the basis of the complex crystal structure and molecular interaction analysis, thirty-two new NVP-AUY922 derivatives were further designed, and among which twenty-eight new ones display enhanced binding force with Hsp90N by molecular docking evaluation. The results would promote anti-NSCLC new drug development to overcome drug resistance based on the lead compound NVP-AUY922.
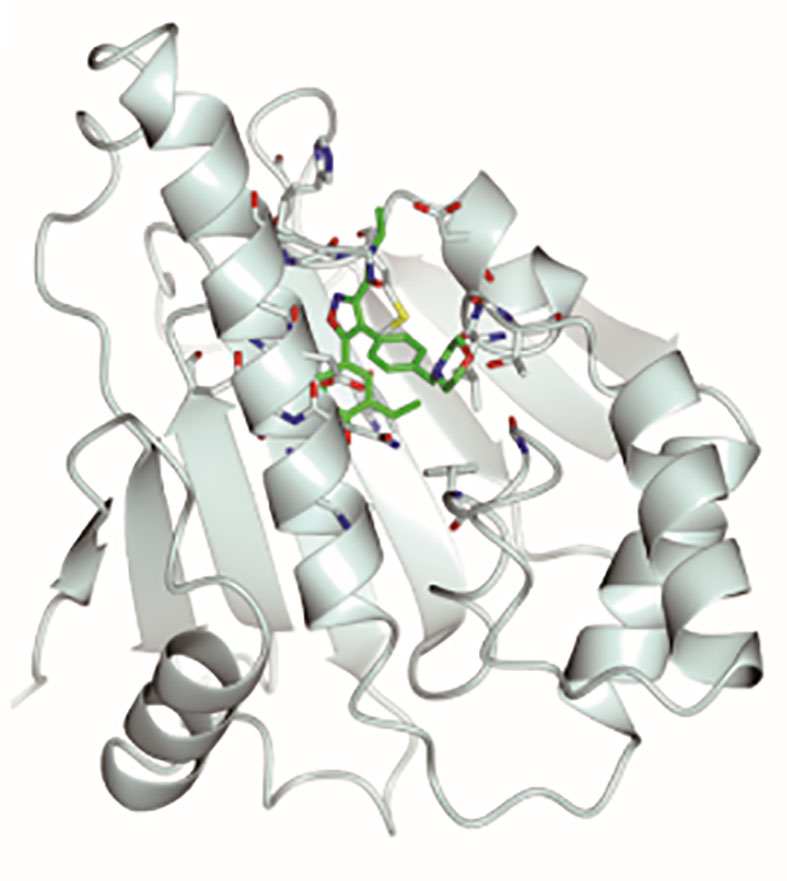
Graphical Abstract Stereoscopic images of complex crystal structure of Hsp90N-NVP-AUY922. A high-resolution complex crystal structure of Hsp90N-NVP-AUY922 was determined by X-ray diffraction (resolution limit 1.59 Å, PDB ID 6LTI), which suggested that NVP-AUY922 perfectly bound in the N-terminal ATP-binding pocket of Hsp90 to disable its molecular chaperone function, therefore suppressed or killed cancer cells.
Highlights
1. A high-resolution complex crystal structure of Hsp90N-NVP-AUY922 was successfully determined by X-ray diffraction.
2. NVP-AUY922 well accommodated in the ATP-binding pocket to disable the molecular chaperone function of Hsp90N to inhibit cancer cells verified by the complex crystal structure, thermal shift assay (TSA) and isothermal titration calorimetry (ITC).
3. Thirty-two new NVP-AUY922 derivatives were designed and twenty-eight new ones exhibited increased binding force with the target Hsp90N, which was proven a feasible scheme by molecular docking evaluation.
4. NVP-AUY922 exhibited favourable in vitro anti-NSCLC activity.
Introduction
Lung cancer remained the top mortality cancer causing 1.8 million deaths (18%) among 10.0 million deaths for cancers worldwide in 2020 (1). Lung cancer is therapeutically divided into non-small-cell lung carcinoma (NSCLC) and small cell lung cancer (SCLC), in which NSCLC covers 80% of patients (2). Recently, targeted therapies for lung cancer have been made remarkable progress based on the genomic studies on the subtypes of NSCLC, leading to the improvement of clinical outcomes for NSCLC, but there is still a low 5-year survival rate of less than 20% and susceptible to drug resistance (3). As is well known, cancer cells carry mutant genes and proteins to survive from therapeutic toxins (4). Thus, heat shock protein 90 (Hsp90) inhibitors with non-targeted therapies with broad-spectrum effects demonstrated advantages in overcoming drug resistance, which would be another choice for NSCLC patients with drug resistance.
Hsp90 inhibitors resulted in unfolded and unmatured multiple important client proteins to be captured and degraded by the proteasome by inhibiting molecular chaperone function of Hsp90, therefore suppressed cancer cells (5–7). Hsp90 inhibitors were designed targeting to three domains of Hsp90 monomer that consequently typed as Hsp90N inhibitors, Hsp90M inhibitors, and Hsp90C inhibitors. Hsp90N inhibitors have been a hot spot against tumor cells in vitro and in vivo (8–12) and many papers and patents focused on Hsp90N inhibitors (13–15). NVP-AUY922, a small molecule inhibitor for Hsp90N, is a resorcinol derivative based on the structure of 4,5-diarylisoxazole scaffold, and displayed favorable anti-cancer activity in vitro and in vivo (16). NVP-AUY922 was evaluated in a phase I clinical trial concerning solid-organ malignancies (primarily breast, ovarian, and colon cancer), and a phase II clinical trial evaluation in NSCLC patients was also carried out by grouping to epidermal growth factor receptor (EGFR) mutations, KRAS proto-oncogene GTPase (KRAS) mutations, anaplastic lymphoma kinase (ALK) rearrangement, and wild-type NSCLC. In phase I clinical trial, major side effects were dose-limiting toxicities, namely, visual symptoms and diarrhea. In phase II clinical trial, NVP-AUY922 exhibited high efficacy in the ALK rearrangement and EGFR mutations. Unfortunately, further development of AUY922 was stopped by Novartis in December of 2014 on account of the failure in meeting its endpoint of complete and partial response of phase II clinical trial in NSCLC patients (7, 17–21).
In brief, NVP-AUY922 was a promising inhibitor for anti-NSCLC new drug development and it was expected to develop novel anti-NSCLC drugs by structural improvement of NVP-AUY922. The absence of high-resolution complex crystal structure of Hsp90N-NVP-AUY922 blocked the way. Herein, we successfully determined a high-resolution complex crystal structure (1.59 Å) of Hsp90N-NVP-AUY922 by X-ray diffraction (XRD). The molecular interaction mechanism of Hsp90N-AUY922 was explored based on the complex crystal structure, thermal shift assay (TSA) and isothermal titration calorimetry (ITC). The findings would promote novel anti-NSCLC drug development to overcome drug resistance based on the lead compound NVP-AUY922.
Materials and Methods
Materials and Methods
Human NSCLC cell line H1299 was acquired from the AOLUKEJI (Shanghai, China). The medium RPMI 1640 and fetal bovine serum were purchased from Gibco-BRL (Gaithersburg, MD, USA).
Tris, Tris–HCl, sodium acetate (NaAc), magnesium chloride (MgCl2), dimethylsulfoxide (DMSO) and PEG4000 were all from Sigma-Aldrich Corp. (St. Louis, MO, USA). Crystal loop was supplied by Hampton Research Corp. (Aliso Viejo, CA, USA). Sodium chloride (NaCl), glycerol, hydrochloric acid (HCl), sodium hydroxide (NaOH) and ethyl alcohol were obtained from the Xi’an Chemical Reagent Company (Xi’an, China). NVP-AUY922 was from Target Molecule Corp. (Boston, MA, USA). DMSO was used to dissolve NVP-AUY922 and double distilled water (ddH2O) was used for other chemicals.
Cell Viability Assay
First of all, quantitative IC50 value was determined. RPMI 1640 with 10% (v/v) fetal bovine serum was used to cultured NSCLC cell line H1299 cells at 37°C in a humidified 5% CO2 atmosphere incubator (Thermo Fisher Scientific Inc., Waltham, MA, USA). Briefly, cells were seeded in 96-well plates at 4 × 103 per well in triplicates and were treated with NVP-AUY922 (100, 10, 1, 0.1, 0.01 μM) for 72 h after incubating for 24 h. OD450 was then detected after 2.5 h adding CCK-8 (7Sea, Shanghai, China) by ELISA plate reader (BioTek Instruments, USA).
Cell viability assay was followed and performed. H1299 cells were placed in 96-well plates as described above. Afterwards, cells were treated with 4 × 2.85 μM NVP-AUY922 for 24, 48, and 72 h. After incubating with CCK-8 for 2.5 h, OD450 was valued to analyze the cell viability.
Cell Cycle Analysis
To monitor effects of NVP-AUY922 on cell cycle progression, cells were seeded in 6-well plates at 3 × 105 per well. After treating with 4 × 2.85 μM NVP-AUY922, or the same amount of DMSO for 72 h at 37°C, cells were harvested and resuspended with 3 ml PBS/ethanol mixture (v:v = 1:2) and then preserved at 4°C. The samples were resuspended with 400 μl buffer after 5 min-centrifugation at 1,000g and washing in 2 ml PBS once. Lastly, 50 μl propidium iodide (PI) and the same volume of RNase were then added into the cells to be incubated for 30 min in the dark at 37°C (22). Flow cytometry (Becton Dickinson, NJ, USA) was used to analyze the distribution of cell cycle phases.
Apoptosis Assay
The apoptosis rate was detected by the Annexin V-FITC/PI Apoptosis Detection Kit (556547, Becton Dickinson, USA) according to the manufacturer’s instructions. Briefly, cells were seeded and incubated with NVP-AUY922 as is mentioned in Cell Cycle Analysis. Subsequently, cells were resuspended in 100 μl binding buffer after washing with 2 ml PBS. Ultimately, 5 μl Annexin V-FITC and 10 μl PI were added and incubated at 4°C for 30 min in the dark. Also, flow cytometry was adopted for cell apoptosis analysis.
Protein Purification and Crystallization
The recombination plasmid, pET28a with the synthesized Hsp90N gene (residues 9–236) was transferred into E. coli BL21 (DE3) (TIANGEN Biotech Corp., Beijing, China) for the target Hsp90N expression. The protein was purified by Nickel affinity chromatography and molecular sieve chromatography, as previously reported (23).
For crystallization, NVP-AUY922 was mixed with Hsp90N protein (20 mg/ml) with 5:1 molar ratio to incubate for 30 min at 4°C. The mixture was then centrifuged for 10 min at 3,000g to collect the supernate. Mixing with the same volume of precipitant (pH 8.5, 0.1 M Tris–HCl, 0.2 M MgCl2, 25% PEG4000) (24) was conducted for 3–7 d at 4°C. Hanging drop vapor diffusion method was used. Co-crystallization was accomplished in a bath circulator controlled incubator (PolyScience 9712, PolyScience, USA).
Data Collection, Structure Determination and Refinement
Hsp90N-NVP-AUY922 complex crystal image of was obtained by a stereomicroscope (M165, Leica Microsystems, Germany). Mounted with cryo-loop (Hampton Research Corp., Aliso Viejo, CA, USA), the complex crystals were flash-frozen in liquid nitrogen for XRD after soaking briefly in the cryoprotectant solution (pH8.5, 0.1 M Tris–HCl, 0.2 M MgCl2, 25% PEG4000, and 20–25% glycerol). All data sets were acquired at 100 K on Macromolecular Crystallography Beamline17U1 (BL17U1) using an ADSC Quantum 315r CCD detector at Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) (25) and the diffraction data were auto-processed by aquarium pipeline (26). The complex crystal structure was analyzed by molecular replacement method using the PHENIX software (27) according to the reported Hsp90N-FS23 (PDB ID, 5CF0) (28). The program Coot was used to rebuild the initial model (29). CCP4MG software was used for graphic reconstruction and molecular interaction analysis (30).
Thermal Shift Assay (TSA)
The ligand NVP-AUY922, dissolved in DMSO, and the target Hsp90N were mixed at a molar ratio of 1:5 in a 20 μl TSA dye reaction buffer with 20 mM Tris–HCl, 150 mM NaCl and 10% glycerol (pH 7.5). The melting-curve (25 to 95°C with a ramp rate of 1°C/min) was determined on ABI 7500 system (7500, ABI Corp., USA). Fluorescence was stimulated by environmentally-sensitive TSA dye binding to the exposed hydrophobic regions during protein unfolding as it is heated. Binding capacity of protein–ligand can be closely evaluated by the melting temperature (Tm) and melting temperature shift (ΔTm) which was calculated based on the melt curves (31).
Isothermal Titration Calorimetry (ITC)
ITC was performed to further illustrate molecular interaction mechanism of Hsp90N-NVP-AUY922 using an ITC-200 calorimeter (Malvern Instrument Ltd., UK). All measurements were carried out in PBS (pH 7.5) at 25°C. Briefly, fresh-purified Hsp90N protein was concentrated to 50 μM. The ligand NVP-AUY922 was prepared in a concentration of 500 μM. Sample was degassed before use. For each titration in the cell, 2 μl aliquot of NVP-AUY922 was added into Hsp90N solution at a 200 s-interval and stirring speed of 750 rpm. The experimental data was fitted to a bimolecular binding model by adjusting three parameters, namely, stoichiometry (n), enthalpy (ΔH°) and association constant (Ka) using Microcal Origin software embedded in the equipment. The thermodynamic parameters ΔG° (free energy) and ΔS° (entropy) were obtained according to the formula, −RT ln Ka = ΔG° = ΔH° − TΔS° (32).
New NVP-AUY922 Derivatives Design and Molecular Docking Evaluation
Based on the analysis of the Hsp90N-NVP-AUY922 crystal structure, TSA and ITC, we designed a series of novel NVP-AUY922 derivatives. Evaluation of the derivatives was performed through molecular docking on software SYBYL-X 2.0 (Tripos Associates Inc., St. Louis, MO, USA).
First, a library of novel NVP-AUY922 derivatives was built and a geometric and force field optimization was carried out over 10,000 times. Second, the protein optimization was performed, such as adding hydrogenation and charge. Lastly, the molecular docking evaluation was performed by Surflex-Dock module of SYBYL software. Further, complex 3D structure reconstruction was simulated by CCP4MG software and molecular interaction mechanism was also analyzed (30).
Statistical Analysis
All data were exhibited as the mean ± standard deviation (SD). The difference significance between two variables was evaluated by Unpaired Student’s t-test, and P <0.05 was considered having statistical significance. Data analysis was performed using GraphPad Prism 5.01 (GraphPad Software, San Diego, USA) and SPSS 13.0 (International Business Machines Corporation, USA). Flow cytometry (Becton Dickinson, NJ, USA) was adopted for cell apoptosis analysis.
Results
Anti-NSCLC Activity In Vitro
AUY-922, as a positive prodrug control, performed favorable anti-NSCLC activity on NSCLC cell lines H460, A549 and H1975 (33). Herein, the effects of NVP-AUY922 on cell viability, cell cycle progression and cell apoptosis of another NSCLC cell line H1299 were analyzed.
NVP-AUY922 exhibited favorable inhibitory activity against H1299 cells (IC50, 2.85 ± 0.063 μM). As shown in Figure 1A, NVP-AUY922 significantly inhibited H1299 cell viability at 24 h (P <0.01), 48 h (P <0.001) and 72 h (P <0.0001), and its inhibitory activity was increased with extended treat time.
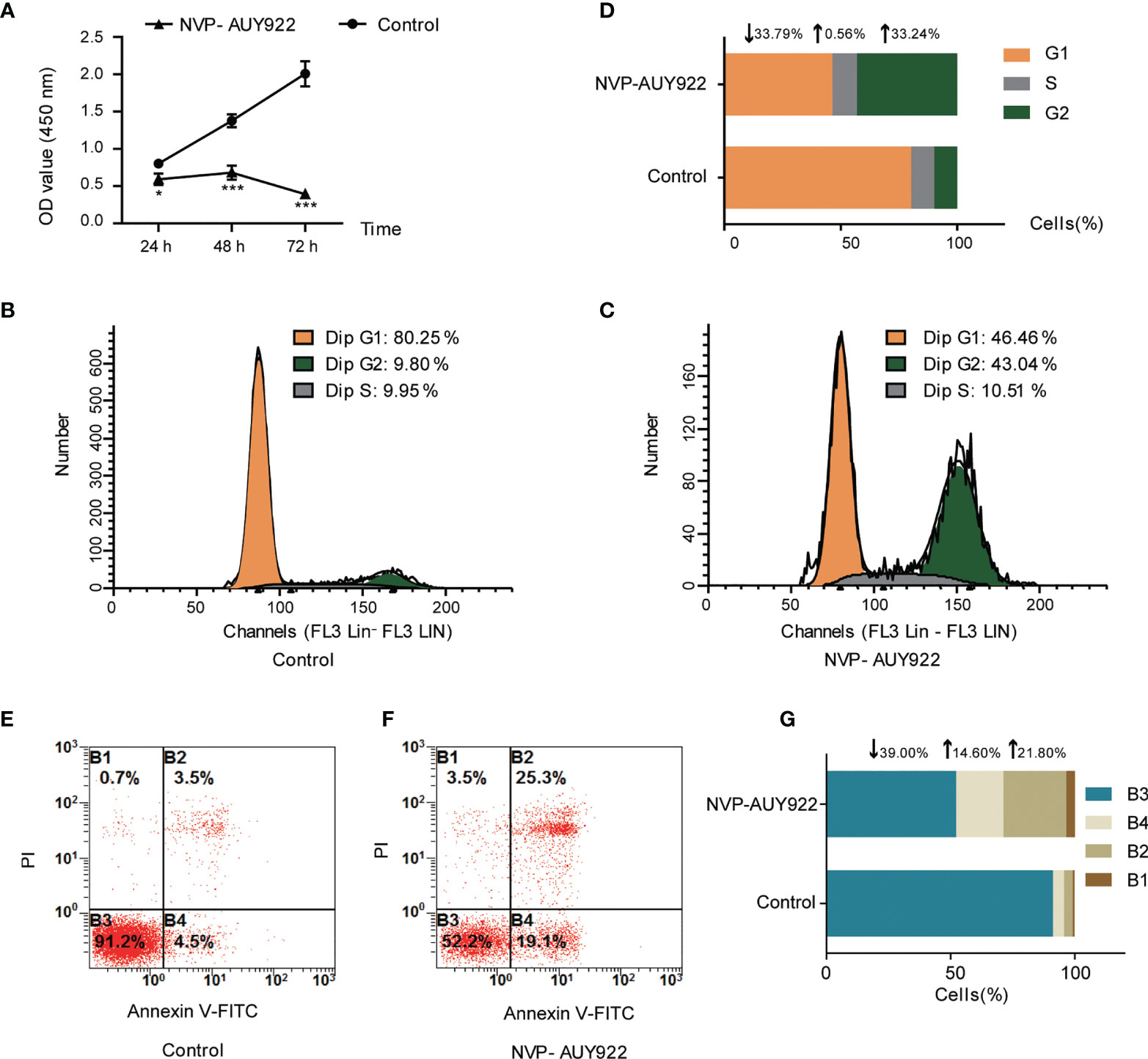
Figure 1 Cell inhibition activity of NVP-AUY922 on NSCLC cell line H1299. (A) NVP-AUY922 inhibited H1299 cell viability. NVP-AUY922 significantly suppressed cell viability against H1299 and its inhibition activity increased with elongated treat time. The data was presented as Mean ± SD, *P <0.05, ***P <0.001 (vs Control, n = 3) indicated a significant difference versus the control. (B) Cell cycle distribution for H1299 treated with DMSO for 72 h. (C) Cell cycle distribution for H1299 treated with NVP-AUY922 for 72 h. (D) Bar graph for cell percentage in G1, S and G2 phase of H1299 treated with NVP-AUY922 or DMSO for 72 h NVP-AUY922 induced H1299 cell cycle arrest in G2 phase. (E) Cell distribution in four quadrants of H1299 treated with DMSO. (F) Cell distribution in four quadrants of H1299 treated with NVP-AUY922. (G) Bar graph for cell percentage in four quadrants of H1299 treated with NVP-AUY922 or DMSO. Control, H1299 treated with DMSO; NVP-AUY922, H1299 treated with NVP-AUY922. The four quadrants represented as: B1, necrotic cell rate; B2, late apoptotic rate; B3, Normal cell rate; B4, early apoptotic rate. NVP-AUY922 promoted H1299 cell apoptosis. The results indicated that NVP-AUY922 exhibited favorable inhibition activity against H1299 cells by suppressing cell viability, inducing cell cycle arrest and promoting cell apoptosis.
As can be seen from Figures 1B–D, after treated with NVP-AUY922 for 72 h, the percentage of H1299 cells in G1, S and G2 phases was obviously changed (NVP-AUY922, G1,↓33.79%; S,↑0.56%; G2,↑33.24% vs Control), indicating that when treated with NVP-AUY922, H1299 cells were notably arrested at G2 phase.
Effects of NVP-AUY922 on H1299 cell apoptosis was also analyzed by the annexin V and PI double staining kit by flow cytometry. Briefly, during early apoptosis, Annexin V can bind to phosphatidylserine (PS) that exists on the external leaflet of the plasma membrane. While, early apoptotic cells exclude PI, late apoptotic cells and necrotic cells can be stained positively (34). Cell percentage at different apoptosis stages are shown in Figures 1E–G. Compared with the control, NVP-AUY922 elevated both the early apoptosis rate and later apoptosis rate of H1299 cells obviously (NVP-AUY922, normal cell rate,↓39.00%; early apoptosis rate,↑14.60%; late apoptosis rate,↑21.80% vs Control).
Molecular Interaction Mechanism Analysis
The thermal stability of proteins with or without ligands was detected by TSA according to the Tm and ΔTm calculated by the protein melting curve, which closely reflect the protein–ligand binding force. The affinity between target Hsp90N and ligand can be indicated by the absolute value of ΔTm, while |ΔTm| >3 is usually considered as a favorable ligand. The results showed that after binding with NVP-AUY922, the thermal stability of Hsp90N was shifted by approximately 15.56°C (ΔTm, 15.56 ± 1.78°C), suggesting a strong binding force between the Hsp90N and NVP-AUY922 (Figures 2A–C).
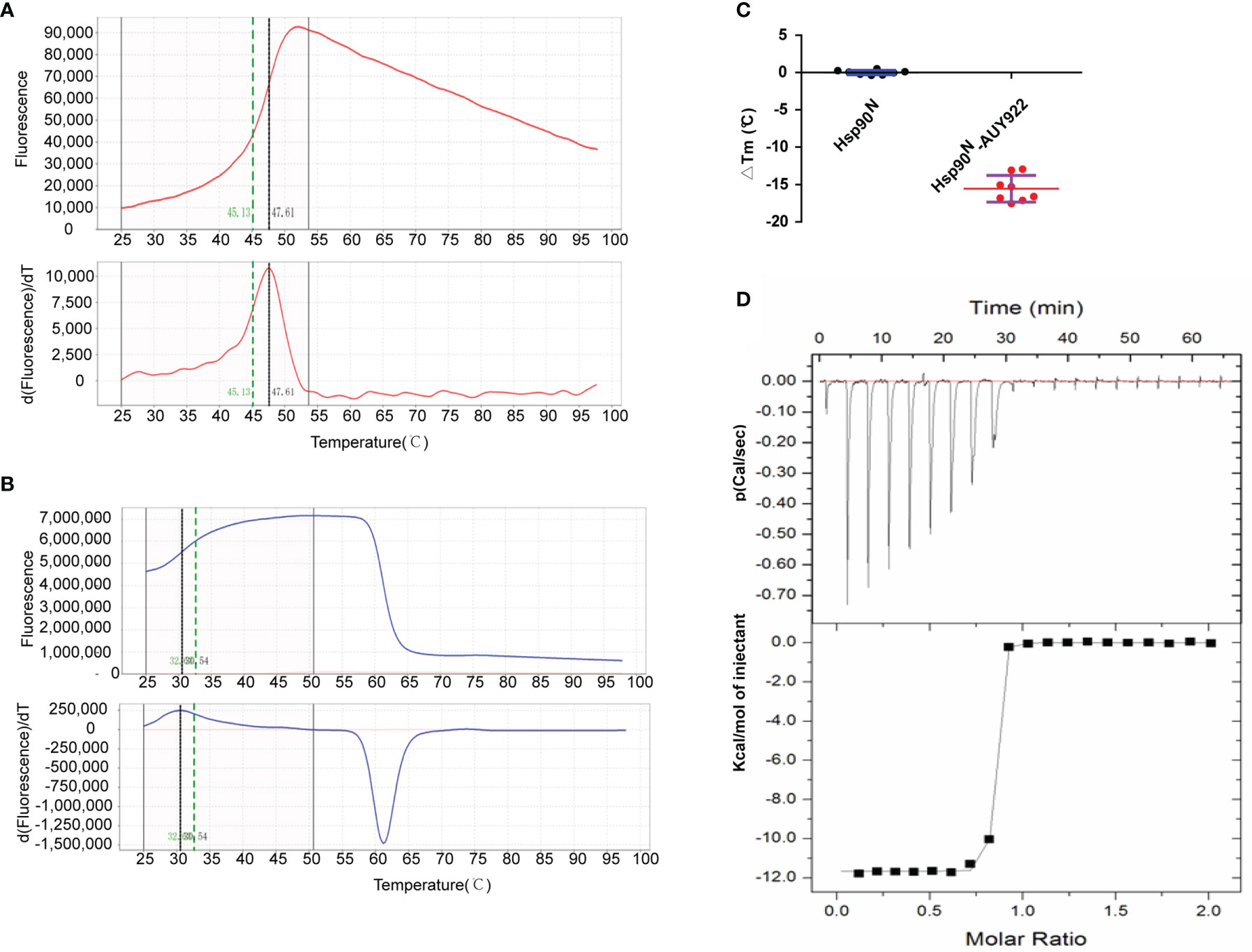
Figure 2 New NVP-AUY922 derivatives design according to complex crystal structure and molecular docking evaluation. (A) Design scheme for new NVP-AUY922 derivatives. (B) Top ten new derivatives with the highest Total scores evaluated by molecular docking using SYBYL-X 2.0 software. (C) Simulated overall 3D structure of Hsp90N-A15. Cartoon for Hsp90N and stick for A15. (D) Simulated intermolecular force between Hsp90N and A15. For A15, the carbon, nitrogen, oxygen and sulfur atoms of were marked in green, blue, red and yellow, respectively. For Hsp90N, the carbon, nitrogen, oxygen and sulfur atoms were marked in cyan, blue, red and yellow, respectively. The red dashed line represents the hydrogen bond. (E) Connolly surface of simulated complex 3D structure of Hsp90N-A15. (F) Molecular structure of A15.
The dissociation constant of Hsp90N-NVP-AUY922 was further determined by ITC at 25°C in PBS. As can be seen in Figure 2D, a perfect S-shaped curve was established by nonlinear data fitting. The negative peaks in the raw ITC titration data revealed that the binding process of Hsp90N-NVP-AUY922 was exothermic at 25°C. The stoichiometric ratio was 0.78 ± 0.04, suggesting a 1:1 binding mode between Hsp90N and NVP-AUY922. The Kd values derived here was 5.10 ± 2.10 nM, further indicating a strong binding force of Hsp90N-NVP-AUY922, as shown in Table 1. The thermodynamic signature was dominated by a large enthalpy change, indicating favorable thermodynamic interactions established in the Hsp90N-NVP-AUY922 binding process.

Table 1 Dissociation constant and thermodynamic data for the target Hsp90N binding with its ligand NVP-AUY922 at 25°C.
As is shown in Figure 3, the empty ATP-binding site of Hsp90N showed continuous and strong electron density, which was well matched with NVP-AUY922 molecule structure (Figures 3A, C, D). A major part of NVP-AUY922 was buried in the ATP-binding pocket of Hsp90N, while the morpholine ring was extending out of the pocket. As shown in Figure 3B, the complex hydrogen bond networks were responsible for strong binding force of Hsp90N-NVP-AUY922. Three direct hydrogen bonds were formed between NVP-AUY922 and residue D93 (2.7 Å), residue K58 (3.0 Å) and residue G97 (3.0 Å) of Hsp90N, respectively. Furthermore, one hydroxy group of the isoxazole ring of NVP-AUY922 formed four water-mediated hydrogen bonds with residues D93, T184, and G97. The other hydroxy group of phenyl ring formed three water-mediated hydrogen bonds with residues L48 and S52. The carbanyl group of carboxamide moiety formed three water-mediated hydrogen bonds with residues K58 and D54. In addition to hydrogen bonds, hydrophobic effects, derived from isopropyl group of the benzene ring in NVP-AUY922 with surrounding residues of Hsp90N, also played an important role for their powerful binding force. The results suggested a competitive ATP-binding inhibition of NVP-AUY922 against Hsp90N, which inactivated the molecular chaperone function of Hsp90N, resulting in inhibiting or killing cancer cells.
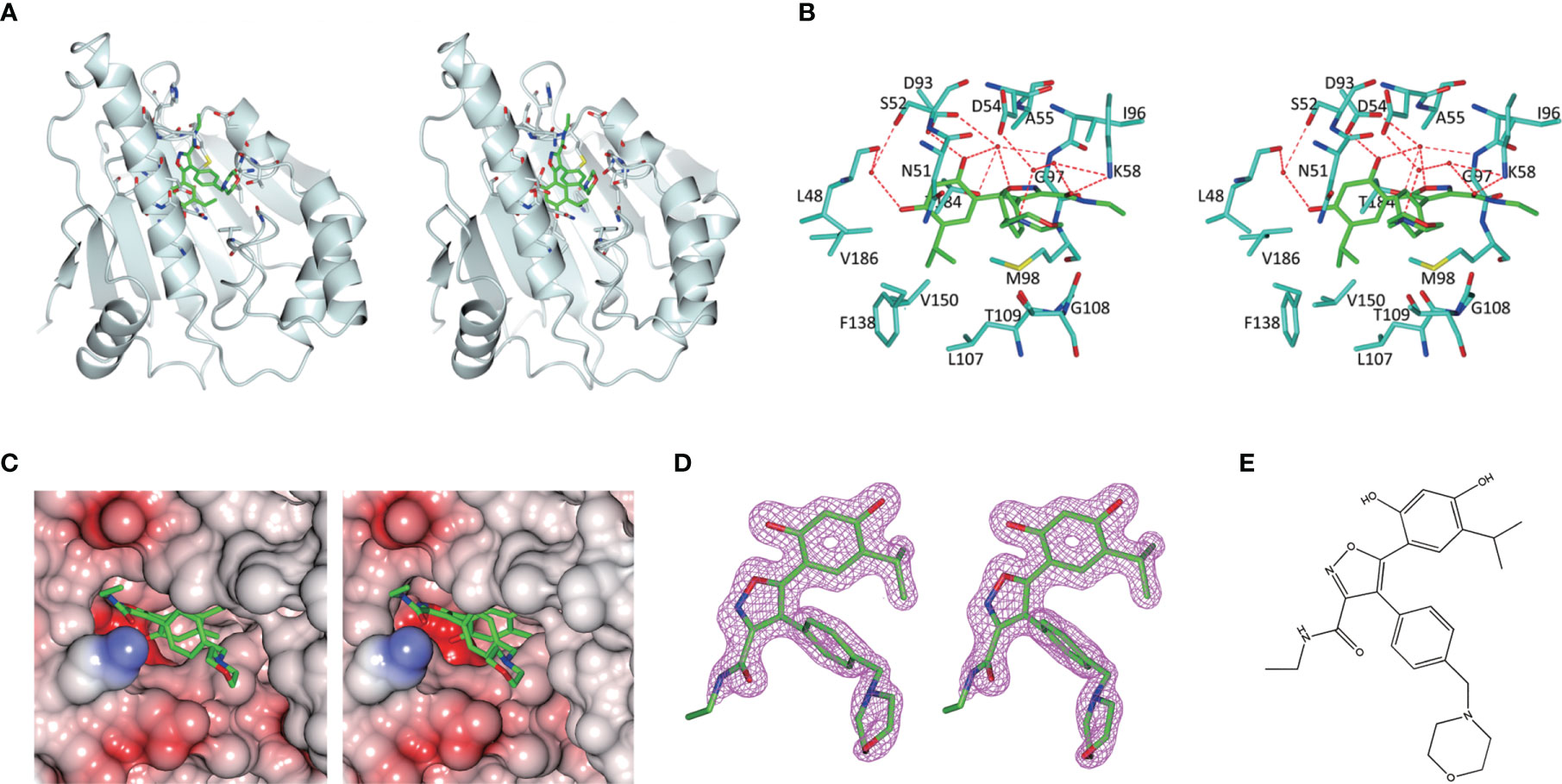
Figure 3 Molecular interaction analysis for NVP-AUY922 and Hsp90N by thermal shift assay (TSA) and isothermal titration calorimetry (ITC). (A) Melting curve of protein Hsp90N. (B) Melting curve of Hsp90N binding with NVP-AUY922. (C) ΔTm. (°C) The melting temperature differences of Hsp90N binding with or without NVP-AUY922 (−15.56 ± 1.78°C). Values were presented as Mean ± SD, ***P < 0.001, n = 8. (D) ITC plots for Hsp90N binding with its ligand NVP-AUY922. Upper panel: the raw titration data. A constant temperature against time was maintained, interaction heat for injection was given by the area of each peak. Lower panel: the bimolecular data fitting of normalized heats against the molar concentration during Hsp90N-NVP-AUY922 binding process. Titrations were performed in PBS (pH 7.5) at 25°C.
Complex Crystal Structure Determination
Hsp90N protein, expressed in E. coli BL21 DE3, was purified by immobilized Ni+ affinity chromatography, and followed by gel filtration chromatography. Hsp90N was co-crystallized with NVP-AUY922 by hanging-drop vapor diffusion method. The complex crystals of Hsp90N-NVP-AUY922 were obtained at 4 °C after growing for 3–5 d. Crystals were rhombus, with the average dimension of approximately 120 μm × 70 μm × 50 μm (Supplementary 1, Figure S1).
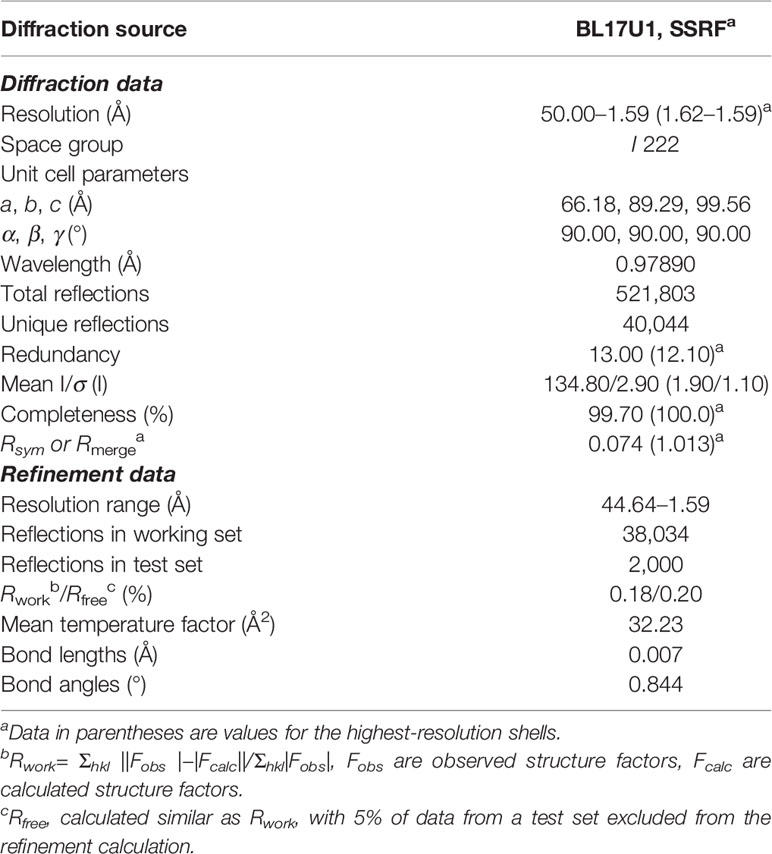
Meanwhile, the complex crystal structure of Hsp90N-NVP-AUY922 was determined by molecular replacement method, and Hsp90N-FS23 (PDB ID 5CF0) (25) was taken as the analytic model. The coordinates and structure factors have been deposited in PDB (PDB ID 6LTI). Data for collection and refinement statistics are described in Table 2. A 1.59 Å resolution limit diffraction data was collected with a final Rfree of 0.20 and Rwork of 0.18, co-crystal space group of I 222, and unit cell parameters of a = 66.18 Å, b = 89.29 Å, c = 99.56 Å; α = β = γ = 90.00°. Residues Glu16–Lys224 were observed in the refined model structure with no electron density for N-terminal residues Asp9–Glu15 and C-terminal residues Glu225–Glu236. The missing residues were conceivable for the N- and C-termini disordered electron density.
New NVP-AUY922 Derivatives Design and Molecular Docking Evaluation
A series of novel NVP-AUY922 derivatives was designed according to the complex crystal structure and molecular interaction analysis, as is shown in Figure 4A. For the new derivative design, it was crucial to maintain the special molecular conformation of NVP-AUY922. First, hydrogen bonds played a key role and introduction of hydrophilic groups could be a favorable strategy. For instance, the 2,4-Dihydroxy-5-isopropyl-phenyl ring moiety was varied to an amino group or sulfydryl group, the isoxazole ring was modified to a 1H-pyrazole ring and the carbanyl group of formamide group was changed by a sulfonyl group. Second, hydrophobic effects were another specific factor contributed to their affinity. Therefore, transformation of hydrophobic functional groups was considered as appropriate structural modification scheme, followed by isopropyl group on the 2,4-Dihydroxy-5-isopropyl-phenyl ring moiety was replaced by a dimethylamino, trifluoromethyl, ethane or tert-butyl group, respectively. Last, considering the further drug metabolism, a thiomorpholine, piperazine or 1-methyl-piperazine was used to substitute the morpholine ring.
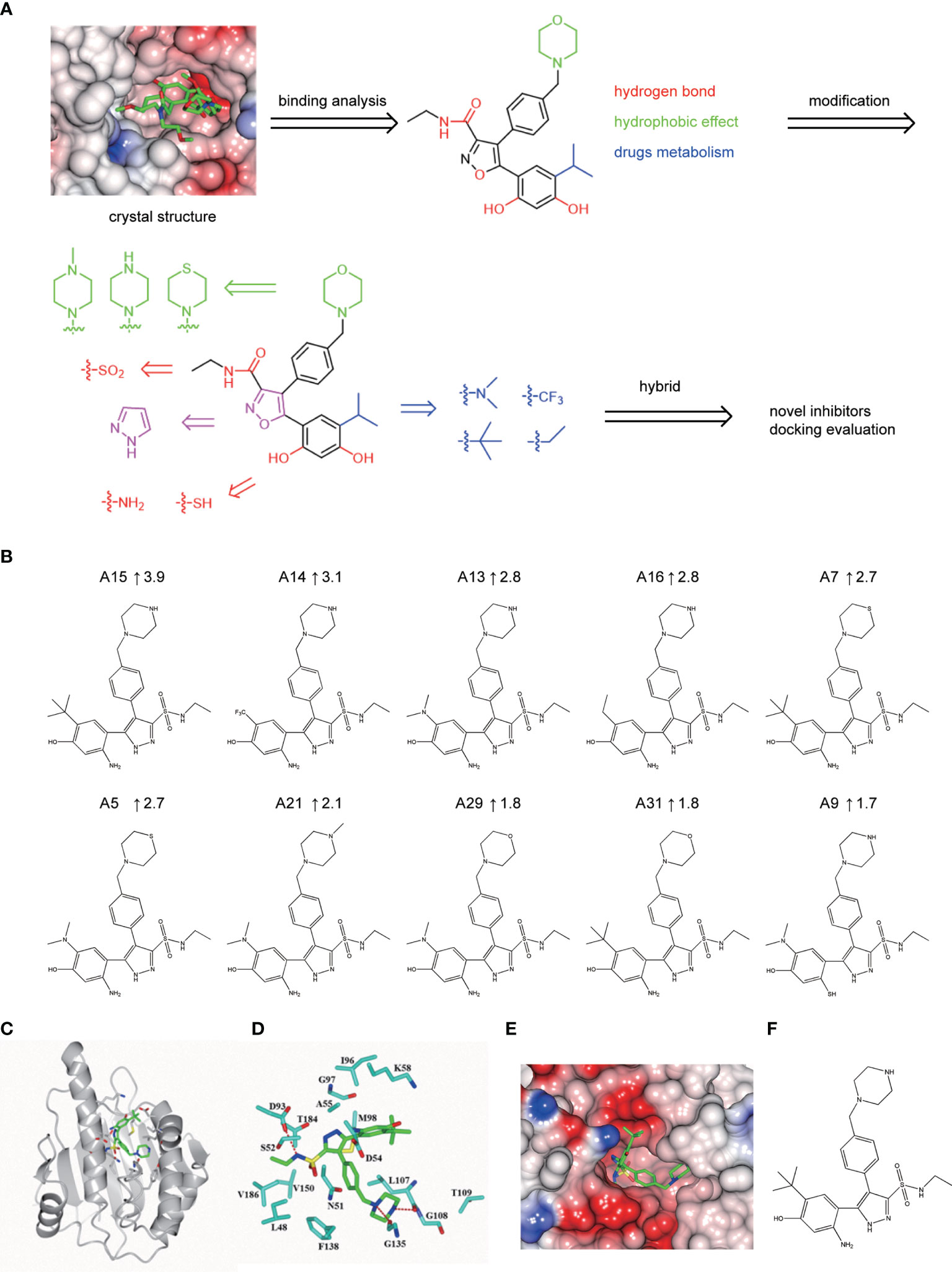
Figure 4 Hsp90N-NVP-AUY922 complex crystal structure. (A) Overall structural view of Hsp90N-NVP-AUY922. Hsp90N and NVP-AUY922 were indicated by cartoon and stick, respectively. (B) Intermolecular forces between Hsp90N and NVP-AUY922. For NVP-AUY922, the carbon, nitrogen and oxygen atoms were colored in green, blue and red, respectively. For Hsp90N, the carbon, nitrogen, oxygen and sulfur atoms were shown in cyan, blue, red and yellow, respectively. The red dashed line represents the hydrogen bond. The red spheres showed water molecules. (C) Distribution of electrostatic potential on Hsp90N-NVP-AUY922 crystal structure surface. Red colored surface reflects the negative charge of electrostatic potential, and blue for positive charge. (D) Electron density map (2Fo-Fc) of NVP-AUY922. The map was calculated in the vicinity of NVP-AUY922. (E) Molecular structure of NVP-AUY922.
Based on the scheme, thirty-two novel NVP-AUY922 derivatives were successfully designed and evaluated by molecular docking with Total score and Cscore. The molecular structures, Total scores and Total score increments compared with NVP-AUY922 for new derivatives are presented in Supplementary 2 Table S1. Total score, evaluates the binding force between target and ligand, is the negative logarithmic value (−log) of the dissociation constant (Kd) of target-ligand, with effects of hydrophobic interaction, polarity, repulsion, entropy change, hydrogen bonds, collision, and so on, to be considered. Higher values indicate stronger binding force of target-ligand. Cscore is a comprehensive score of D-score, PMF-score, G-score, Chem-score, and Total score. The max score is five points. Usually, derivatives with high scores (Total score ≥6, Csore ≥4) were recognized as favorable ligands. Twenty-eight novel NVP-AUY922 derivatives showed increased binding force with the target Hsp90N shown as high Total scores, which manifested a feasible design scheme by molecular docking evaluation. The molecular structures and the Total score increment of top ten new derivatives are shown in Figure 4B.
A15 perfomed the top optimal representative (Total score, 11.9). Thereby, simulated complex 3D structure of Hsp90N-A15 was built, and analyzed their molecular interaction mechanism in detail. A15 was accommodated appropriately in the ATP-binding pocket of Hsp90N (Figure 4C). Compared with NVP-AUY922, the 5-amino-2-tert-butyl-phenol group of A15 was obviously shifted from the interior of cavity to the exterior. Meanwhile, the branched chain of pyrazole ring was moved oppositely from the external space into the internal pocket. Besides, the piperazine ring of A15 was shrunk gently into the pocket (Figures 4C–E), while the morpholine ring of NVP-AUY922 was extended out of the cavity (Figure 4A).
Three hydrogen bonds between A15 and residues D93 (2.9 Å), L107 (2.7 Å) and G135 (3.0 Å) of Hsp90N were formed, respectively. Meanwhile, the piperazine ring together with adjacent aromatic ring formed hydrophobic interactions with residue G135, the pyrazole ring with branched chain arranged hydrophobic contacts with residues S52, M98, and V186, the 5-amino-2-tert-butyl-phenol group formed hydrophobic contacts with residues D54 and A55 (Figure 4D). Therefore, the hydrogen bonds and hydrophobic interactions played an important role for the intense binding of Hsp90N-A15. The results would provide promising lead compounds for anti-NSCLC new drug development based on NVP-AUY922.
Discussion
Lung cancer is the top one mortality cancer and NSCLC covers 80% of patients (2). Recently, targeted therapies for lung cancer have been made remarkable progress based on the genomic studies on the subtypes of NSCLC, leading to the improvement of clinical outcomes for NSCLC, but there is still a low 5-year survival rate of less than 20% and susceptible to drug resistance (3). As is well known, cancer cells carry mutant genes and proteins to survive from therapeutic toxins (4). Hsp90, a well-known molecular chaperone, played a crucial role in tumorigenesis and progression. Hsp90 can assist over 280 client proteins in correct folding and conformational maturation, which are widely involved in signal transduction, cell cycle and apoptosis regulation in cancer cells. Therefore, Hsp90 demonstrated advantages in overcoming drug resistance as a broad-spectrum anti-cancer target, which would be another choice for NSCLC patients with drug resistance. Because the activity of chaperone molecule must obtain energy from ATP hydrolysis, Hsp90N inhibitors disabled molecular chaperone function of Hsp90 by competitively suppressing ATP-binding to result in multiple misfolded or immature client proteins that were captured and degraded by the proteasome, cancer cells were thereby suppressed or killed. Therefore, the development of Hsp90 inhibitors has always been a significant tactic for anti-NSCLC drug development (5–9).
NVP-AUY922, a promising second generation Hsp90N inhibitor, exhibited high objective response rate in NSCLC patients with the ALK rearrangement and EGFR mutations. Though NVP-AUY922 was terminated in a phase II clinical trial by Novartis for unsatisfied endpoint of complete and partial response (7, 17–20), NVP-AUY922 was still a promising lead compound for anti-NSCLC new drug development by the structural optimization of NVP-AUY922.
Herein, a 1.59 Å-high-resolution complex crystal structure of Hsp90N-NVP-AUY922 was successfully determined by XRD, which made it possible to clearly exhibit the binding mode and molecular interaction in detail between NVP-AUY922 and its target Hsp90N. The continuous and clear electron density was presented in the empty ATP-binding pocket of Hsp90N and well matched with NVP-AUY922 molecule structure. Except for hydrophobic interactions, NVP-AUY922 exhibited strong affinity to Hsp90N for complex hydrogen bond networks, namely, direct hydrogen bonds and water-mediated hydrogen bonds. There was still a vast room around NVP-AUY922 in the ATP-binding pocket for further structural optimization.
Crystal structure analysis, TSA and ITC were comprehensively assessed to study the molecular interaction mechanism of Hsp90N-NVP-AUY922. Melting temperature (Tm) is a definite parameter for protein under certain conditions which is changed while binding ligands (ΔTm). TSA was applied to detect the protein thermal stability with or without ligands according to the Tm and ΔTm calculated by protein melting curve. Greater affinity was indicated by Greater ΔTm and |ΔTm| >3 is usually deemed as a favorable ligand. There was a strong affinity between the Hsp90N and its ligand NVP-AUY922 (ΔTm, −15.56 ± 1.78°C), suggesting that NVP-AUY922 binding to Hsp90N enhanced the protein thermostability. This was further verified by ITC (Kd, 5.10 ± 2.10 nM). ITC evaluated target-ligand binding force by examining the endothermic and exothermic amount when ligand binding to its target. Smaller the Kd is, stronger the binding force is. The large enthalpy change indicated favorable thermodynamic interactions established in the Hsp90N-NVP-AUY922 binding process. The molecular interaction analysis suggested that NVP-AUY922 can inactivate the molecular chaperone function of Hsp90 by accommodating in the ATP-binding pocket, which took the responsibility for the favorable anti-NSCLC activity of NVP-AUY922. In practice, NVP-AUY922 suppressed cell proliferation (IC50, 2.85 ± 0.06 μM), induced cell cycle arresting at G2 phase and accelerated cell apoptosis of the NSCLC cell line H1299.
For further structural optimization, it was crucial to keep the special molecular conformation of NVP-AUY922, which was kept by hydrogen bonds and hydrophobic interactions. The design scheme devoted to promote these beneficial interactions by functional group replacements. Under the precise reference of molecular interaction analysis by complex crystal structure, TSA and ITC of Hsp90N-NVP-AUY922, thirty-two new NVP-AUY922 derivatives were designed and molecular docking evaluation showed twenty-eight of them displayed enhanced binding force to the target Hsp90N. This also verified a practicable design scheme. A15 as the optimal representative, simulated Hsp90N-A15 3D structure was built and detailed molecular interaction mechanism was analyzed. Compared with NVP-AUY922, A15 exhibited increased binding force with Hsp90N and hydrophobic interactions contributed more than hydrogen bonds for Hsp90N-A15 binding. The results would promote novel anti-NSCLC drug development to overcome drug resistance based on the lead compound NVP-AUY922.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
H-LC designed the research study. C-XH, YL, MG, H-JL, LX, XZ, and P-QL performed the research. FY, HZ and J-HH provided help and advice on study design. C-XH, WQ, DZ, and P-QL analyzed the data. C-XH, YL, and MG wrote the manuscript. H-LC, FY, and J-HH revised the paper. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was funded by the National Natural Science Foundation of China (U1932130), the Program of Shaanxi Provincial Science and Technology Department (2021JQ-786), the Key Program of Shaanxi Provincial Education Department (17JS117), the Program of Shaanxi Administration of Traditional Chinese Medicine (2019-ZZ-ZY009), the Key Program of Weiyang District Bureau of Science, Technology and Industry Information Technology (201928), the Program of Shaanxi Provincial Research Center for the Project of Prevention and Treatment of Respiratory Diseases (2016HXKF05), and the Talent Program of Xi’an Medical University (2016DOC13, 2017GJFY03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.847556/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 0:1–41. doi: 10.3322/caac.21660
2. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin Proc (2008) 83(5):584–94. doi: 10.1016/S0025-6196(11)60735-0
3. O smani L, Askin F, Gabrielson E, Li QK. Current WHO Guidelines and the Critical Role of Immunohistochemical Markers in the Subclassification of Non-Small Cell Lung Carcinoma (NSCLC): Moving From Targeted Therapy to Immunotherapy. Semin Cancer Biol (2018) 52(Pt 1):103–9. doi: 10.1016/j.semcancer
4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2015. CA Cancer J Clin (2015) 65(1):5–29. doi: 10.3322/caac.21254
5. Vahid S, Thaper D, Zoubeidi A. Chaperoning the Cancer: The Proteostatic Functions of the Heat Shock Proteins in Cancer. Recent Pat Anticancer Drug Discov (2017) 12(1):35–47. doi: 10.2174/1574892811666161102125252
6. Taldone T, Ochiana SO, Patel PD, Chiosis G. Selective Targeting of the Stress Chaperome as a Therapeutic Strategy. Trends Pharmacol Sci (2014) 35(11):592–603. doi: 10.1016/j.tips.2014.09.001
7. Chatterjee S, Bhattacharya S, Socinski MA, Burns TF. HSP90 Inhibitors in Lung Cancer: Promise Still Unfulfilled. Clin Adv Hematol Oncol (2016) 14(5):346–56.
8. Jiang F, Guo AP, Xu JC, Wang HJ, Mo XF, You QD, et al. Identification and Optimization of Novel 6-Acylamino-2-Aminoquinolines as Potent Hsp90 C-Terminal Inhibitors. Eur J Med Chem (2017) 141:1–14. doi: 10.1016/j.ejmech.2017.07.080
9. Akram A, Khalil S, Halim SA, Younas H, Iqbal S, Mehar S. Therapeutic Uses of HSP90 Inhibitors in Non-Small Cell Lung Carcinoma (NSCLC). Curr Drug Metab (2018) 19(4):335–41. doi: 10.2174/1389200219666180307122441
10. Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, et al. NVP-AUY922: A Novel Heat Shock Protein 90 Inhibitor Active Against Xenograft Tumor Growth, Angiogenesis, and Metastasis. Cancer Res (2008) 68(8):2850–60. doi: 10.1158/0008-5472.CAN-07-5256
11. Moser C, Lang SA, Hackl C, Wagner C, Scheiffert E, Schlitt HJ, et al. Targeting HSP90 by the Novel Inhibitor NVP-AUY922 Reduces Growth and Angiogenesis of Pancreatic Cancer. Anticancer Res (2012) 32(7):2551–61.
12. Best OG, Mulligan SP. Heat Shock Protein-90 Inhibitor, NVP-AUY922, Is Effective in Combination With Fludarabine Against Chronic Lymphocytic Leukemia Cells Cultured on CD40L-Stromal Layer and Inhibits Their Activated/Proliferative Phenotype. Leuk Lymphoma (2012) 53(11):2314–20. doi: 10.3109/10428194.2012.698278
13. Fernandes JC, Alves P. Recent Patents on Heat Shock Proteins Targeting Antibodies. Recent Pat Anticancer Drug Discov (2017) 12(1):48–54. doi: 10.2174/1574892812666161123141516
15. David P. Combination Therapy of Hsp90 Inhibitors With Platinum-Containing Agents. (2018). US20180221345.
16. Sidera K, Patsavoudi E. HSP90 Inhibitors: Current Development and Potential in Cancer Therapy. Recent Pat Anticancer Drug Discov (2014) 9(1):1–20. doi: 10.2174/15748928113089990031
17. Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M, et al. First-In-Human Phase I Dose-Escalation Study of the HSP90 Inhibitor AUY922 in Patients With Advanced Solid Tumors. Clin Cancer Res (2013) 19:3671–80. doi: 10.1158/1078-0432.CCR-12-3404
18. Felip E, Barlesi F, Besse B, Chu Q, Gandhi L, Kim SW, et al. Phase 2 Study of the HSP-90 Inhibitor AUY922 in Previously Treated and Molecularly Defined Patients With Advanced Non-Small Cell Lung Cancer. J Thorac Oncol (2018) 13(4):576–84. doi: 10.1016/j.jtho.2017.11.131
19. Johnson ML, Yu HA, Hart EM, Weitner BB, Rademaker AW, Patel JD, et al. Phase I/II Study of HSP90 Inhibitor AUY922 and Erlotinib for EGFR-Mutant Lung Cancer With Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J Clin Oncol (2015) 33(15):1666–73. doi: 10.1200/JCO.2014.59.7328
20. Vermes K. Vernalis PLC Stumbles As Novartis AG Abandons Work On Auy922. Available at: http://www.biospace.com/News/vernalisplc-stumbles-as-novartis-ag-abandons-/358835.
21. Brough PA, Aherne W, Barril X, Borgognoni J, Boxall K, Cansfield JE, et al. 4,5-Diarylisoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer. J Med Chem (2008) 51(2):196–218. doi: 10.1021/jm701018h
22. Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal Structure of an Hsp90–geldanamycin Complex: Targeting of a Protein Chaperone by an Antitumor Agent. Cell (1997) 89:239–50. doi: 10.1016/S0092-8674(00)80203-2
23. Wang Q, Zhang K, Cui Y, Wang ZJ, Pan QY, Liu K, et al. Upgrade of Macromolecular Crystallography Beamline BL17U1 at SSRF. Nucl Sci Tech (2018) 29(5):68. doi: 10.1007/s41365-018-0398-9
24. Yu F, Wang Q, Li M, Zhou H, Liu K, Zhang KH, et al. Aquarium: An Automatic Data-Processing and Experiment Information Management System for Biological Macromolecular Crystallography Beamlines. J Appl Crystallogr (2019) 52:472–7. doi: 10.1107/S1600576719001183
25. Li J, Shi F, Chen DQ, Cao HL, Xiong B, Shen JK, et al. FS23 Binds to the N-Terminal Domain of Human Hsp90: A Novel Small Inhibitor for Hsp90. Nucl Sci Tech (2015) 26:060503. doi: 10.13538/j.1001-8042/nst.26.060503
26. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, et al. Macromolecular Structure Determination Using X-Rays, Neutrons and Electrons: Recent Developments in Phenix. Acta Crystallogr D Struct Biol (2019) 75(Pt 10):861–77. doi: 10.1107/S2059798319011471
27. Cao H, Lyu KK, Liu B, Li J, He JH. Discovery of a Novel Small Inhibitor RJ19 Targeting to Human Hsp90. Nucl Sci Tech (2017) 28:148. doi: 10.1007/s41365-017-0300-1
28. Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr D (2004) 60:2126–32. doi: 10.1107/S0907444904019158
29. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr D (2011) 67:235–42. doi: 10.1107/S0907444910045749
30. Andreotti G, Monticelli M, Cubellis MV. Looking for Protein Stabilizing Drugs With Thermal Shift Assay. Drug Test Anal (2015) 7:831–4. doi: 10.1002/dta.1798
31. He Y, Roth C, Turkenburg JP, Davies GJ. Three-Dimensional Structure of a Streptomyces Sviceus GNAT Acetyltransferase With Similarity to the C-Terminal Domain of the Human GH84 O-GlcNAcase. Acta Crystallogr D (2014) 70:186–95. doi: 10.1107/S1399004713029155
32. Olszewska P, Cal D, Zagórski P, Mikiciuk-Olasik E. A Novel Trifluoromethyl 2-Phosphonopyrr- Ole Analogue Inhibits Human Cancer Cell Migration and Growth by Cell Cycle Arrest at G1 Phase and Apoptosis. Eur J Pharmacol (2020) 871:172943. doi: 10.1016/j.ejphar.2020.172943
33. Wu WC, Liu YM, Liao YH, Hsu KC, Lien ST, Chen IC, et al. Fluoropyrimidin-2,4-Dihydroxy-5-Isopropylbenzamides as Antitumor Agents Against CRC and NSCLC Cancer Cells. Eur J Med Chem (2020) 203(1):12540. doi: 10.1016/j.ejmech.2020.112540
Keywords: NVP-AUY922, Hsp90N, non-small-cell lung cancer (NSCLC), complex crystal structure, molecular interaction, drug design
Citation: He C-X, Lv Y, Guo M, Zhou H, Qin W, Zhao D, Li H-J, Xing L, Zhou X, Li P-Q, Yu F, He J-H and Cao H-L (2022) Complex Crystal Structure Determination of Hsp90N-NVP-AUY922 and In Vitro Anti-NSCLC Activity of NVP-AUY922. Front. Oncol. 12:847556. doi: 10.3389/fonc.2022.847556
Received: 02 January 2022; Accepted: 31 January 2022;
Published: 24 February 2022.
Edited by:
Qingbin Cui, University of Toledo, United StatesCopyright © 2022 He, Lv, Guo, Zhou, Qin, Zhao, Li, Xing, Zhou, Li, Yu, He and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Yu, eXVmZW5nQHNhcmkuYWMuY24=; Jian-Hua He, aGVqaWFuaHVhQHdodS5lZHUuY24=; Hui-Ling Cao, Y2FvaHVpbGluZ19qenNAeGl5aS5lZHUuY24=
†These authors have contributed equally to this work
 Chun-Xia He
Chun-Xia He You Lv2†
You Lv2† Wei Qin
Wei Qin Dong Zhao
Dong Zhao Hui-Jin Li
Hui-Jin Li Lu Xing
Lu Xing Xin Zhou
Xin Zhou Peng-Quan Li
Peng-Quan Li Jian-Hua He
Jian-Hua He Hui-Ling Cao
Hui-Ling Cao