- Department of Oncology, Yongchuan Hospital of Chongqing Medical University, Chongqing, China
Background: Genetic variability in DNA double-strand break repair genes such as RAD51 gene and its paralogs XRCC2、XRCC3 may contribute to the occurrence and progression of breast cancer. To obtain a complete evaluation of the above association, we performed a meta-analysis of published studies.
Methods: Electronic databases, including PubMed, EMBASE, Web of Science, and Cochrane Library, were comprehensively searched from inception to September 2022. The Newcastle-Ottawa Scale (NOS) checklist was used to assess all included non-randomized studies. Odds ratios (OR) with 95% confidence intervals (CI) were calculated by STATA 16.0 to assess the strength of the association between single nucleotide polymorphisms (SNPs) in these genes and breast cancer risk. Subsequently, the heterogeneity between studies, sensitivity, and publication bias were performed. We downloaded data from The Cancer Genome Atlas (TCGA) and used univariate and multivariate Cox proportional hazard regression (CPH) models to validate the prognostic value of these related genes in the R software.
Results: The combined results showed that there was a significant correlation between the G172T polymorphism and the susceptibility to breast cancer in the homozygote model (OR= 1.841, 95% CI=1.06–3.21, P=0.03). Furthermore, ethnic analysis showed that SNP was associated with the risk of breast cancer in Arab populations in homozygous models (OR=3.52, 95% CI=1.13-11.0, P= 0.003). For the XRCC2 R188H polymorphism, no significant association was observed. Regarding polymorphism in XRCC3 T241M, a significantly increased cancer risk was only observed in the allelic genetic model (OR=1.05, 95% CI= 1.00–1.11, P=0.04).
Conclusions: In conclusion, this meta-analysis suggests that Rad51 G172T polymorphism is likely associated with an increased risk of breast cancer, significantly in the Arab population. The relationship between the XRCC2 R188H polymorphism and breast cancer was not obvious. And T241M in XRCC3 may be associated with breast cancer risk, especially in the Asian population.
Introduction
In all countries around the world, cancer is the leading cause of death and an important obstacle to improving life expectancy. Female breast cancer (BC) has overtaken lung cancer as the leading cause of global cancer incidence in 2020, with an estimated 2.3 million new cases, representing 11.7% of all cancer cases (1). The mechanism of breast carcinogenesis is not yet fully understood. It is considered a polygenic disease and has a component of inheritance due to low-penetrant and common genetic variants. The steady repair of DNA damage is very important for the survival of cells and the maintenance of genetic stability (2).
Over the years, it has been increasingly recognized that variations in the genetic background of individuals combined with environmental exposure can ultimately lead to the occurrence and progression of cancer. DNA repair genes have been considered considerable factors in the prevention of genomic damage and continuously monitor chromosomes to correct injuries caused by exogenous agents such as ultraviolet light or endogenous mutagens (3, 4). Aberrant double-stranded break (DSB) repair leads to genomic instability, a hallmark of malignant cells. Double-stranded breaks are repaired by two pathways: homologous recombination (HR) and non-homologous end joining (NHEJ). Previous analysis has revealed several important features of DSB repair in breast cancer cells: (i) HR is evidently increased in breast cancer cells compared with normal cells; (ii) Non-homologous end joining(NHEJ)repair is the major DSB repair route in both normal and malignant breast epithelial cells; (iii) NHEJ efficiency does not differ significantly between normal and cancerous cells (5). The two pathways of DSB repair are independently controlled, and only HR is increased in breast cancer cells compared with normal breast epithelial cells. RAD51 is a homolog of the E. coli RecA protein, which is essential for maintainability such as meiotic and mitotic recombination, and also plays a critical role in homologous recombination repair (HR) of DNA double-strand breaks (DSB) (6–8).
Researchers recently discovered that the Rad51 promoter in cancer cells is on average 840-fold more active in cancer cells than in normal cells and the fusion of RAD51 promoter and diphtheria toxin gene selectively kills cancer cells. Transcriptional targeting therapy using up-regulated HR gene expression can effectively eliminate cancer cells without toxicity to normal tissues. The human RAD51 gene, located on chromosome 15q15.1, is considered to participate in a common DSB repair pathway and is involved in the development of breast cancer development (9). RAD51 functions by assembling on a single-stranded DNA, inducing homologous pairing, and in turn mediates strand invasion and exchange between homologous DNA and damaged site (10). In recent years, the RAD51 gene polymorphism has attracted a great deal of attention. The RAD51 family of genes, including RAD51 and the five RAD51-like genes, are known to have crucial non-redundant roles in this pathway. Recently, researchers have revealed that RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3) could serve as central proteins during the HRR process. The function of RAD51-like genes is to transduce DNA damage signals to effector kinases that promote break repair. A central player in homologous recombination is the RAD51 recombinase that binds to single-stranded DNA at break sites, the XRCC2 and XRCC3 genes are structurally and functionally related to the RAD51 genes (11). Two commonly studied polymorphisms of the RAD51 gene are G135C (rs1801320), a G to C transversion at position +135, and G172T (rs1801321), a G to T transversion at position +172, both of which are located in the 5 Untranslated region (5’UTR) and appear to be related to functional polymorphisms. Two variants of 135G/C and 172G/T would affect mRNA stability or translational efficiency, resulting in altered levels of polypeptide products, altering the function of encoding the RAD51 protein, and in some way influencing DNA repair capacity and malignancies (12). RAD51 interacts with BRCA1 and BRCA2, acting through HR and NHEJ. For example, down-regulation or mutation of DNA DSB repair proteins involved in the NHEJ pathway was shown to be associated with both BC risk and increased chromosomal radiosensitivity (CRS) (13–15). In addition, RAD51 overexpression is acknowledged to be associated with therapeutic antagonism, aggressiveness, metastatic behavior, and poor prognosis.
X-ray repair cross complementing group 2(XRCC2)gene, located in 7q36.1, is an essential part of the homologous recombination repair pathway and a functional candidate for involvement in cancer progression. Its XRCC2 protein product, together with other proteins encoded by the XRCC2 gene such as RAD51L3, forms a complex that plays a critical role in chromosome segregation and the apoptotic response to DSBs (16, 17). As a member of the RAD51 family of proteins, it is widely acknowledged to mediate HRR (18). However, the exact function of SNPs in the XRCC2 gene in response to different DNA-damaging agents still remains unclear. There is a G-to-A polymorphism located in exon 3 of the XRCC2 gene resulting in a substitution of histidine (His) for arginine (Arg). Known as Arg188His (R188H, rs3218536), this polymorphism has been widely investigated to explore its potential impact on cancer susceptibility. Furthermore, DNA damage caused by anticancer drugs and radiation have been documented to require XRCC2 for repair in mammalian cells (19–22). Several pieces of evidence stress that high levels of expression of The X-ray repair cross complementing group 3 (XRCC3), another member of the RAD51 family of proteins, are correlated with radioresistance and cytotoxic resistance in human tumor cell lines, suggesting that XRCC2 could also play a relevant role in the effects of oncotherapy (23–25). XRCC3, as we know, is localized on human chromosomes 14q32.325. A coding SNP (T241M, rs861539) has been reported at the 18,067th nucleotide in exon 7 of the XRCC3 gene, resulting in a substitution of methionine (Met) for threonine(Thr) (25). The XRCC3 protein is involved in the joining of single-strand DNA breaks and the joining of double-strand DNA breaks (26). As a member of the Rad51 DNA repair gene family. It functions in the HRR pathway by repairing double-strand breaks. XRCC3 helps the assembly of the nucleofilament protein and its selection and interaction with the appropriate recombination substrates (12). Likewise, XRCC3 controls HR fidelity and is essential to stabilize heteroduplex DNA in HRR. Furthermore, a mutation in XRCC3 generates severe chromosomal instability. The XRCC2 and XRCC3 genes are necessary for HRR and are required for the formation of RAD51 focus (27, 28). In recent studies, common variants of XRCC2, particularly the encoding SNP of exon 3 (Arg188His), have been identified as potential cancer susceptibility sites, although in this case, the association with breast cancer susceptibility remains unclear. Earlier studies have shown that the XRCC3 Thr241Met polymorphism has long been regarded as a risk factor for many cancers.
We examined whether polymorphisms in these three genes involving homologous recombination with DSB were associated with the risk of breast cancer.
Materials and methods
Search strategy and data extraction
All studies investigating the association between polymorphisms in the RAD51 gene and paralog genes, such as the XRCC2 & XRCC3, and the risk of breast cancer, were identified by comprehensive computer-based searches of the PubMed, Embase, Web of Knowledge, and Cochrane Library databases(the last search update on September 2022). The search was carried out using various combinations of keywords such as (‘RAD51 gene’ OR ‘RAD51 recombinase gene’ OR ‘XRCC3 polymorphism’ OR ‘XRCC3 Thr241Met polymorphism’ OR ‘XRCC2’ OR ‘XRCC2 Arg188His polymorphism’) AND (‘polymorphism’ OR ‘variant’ OR ‘variants’).
Eligibility criteria and selection process
Inclusion criteria
Studies included in our meta-analysis needed to have met the following criteria: 1)published in public, full text only; 2) case-control study; 3) sufficient data (genotype distributions for cases and controls) to calculate an odds ratio (OR) with its 95%CI; 4) studies published in English; 5) genotype distribution of the control population consistent with the Hardy-Weinberg Equilibrium (HWE).
Data extraction
Two authors independently extracted information from all eligible publications according to the inclusion criteria listed above. Disagreement was resolved by evaluating a third reviewer and discussing until a consensus was reached. The following characteristics were collected from each study: first author, year of publication, country, ethnicity, methods in experiments, source of control groups and genotype frequencies in case and control groups, and the value of HWE. Duplicated primary studies were deleted and only one version of duplicated documents was kept.
Data collection
The transcriptome data and clinical information of BC patients were obtained from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). In total, 903 patients with BC were selected from the TCGA cohort. For the transcriptome data from TCGA-BRCA, we download their series files. Some important clinical characteristics including age, pathologic stage (I, II, III, IV, V, and NA), and pathology stage (T, N, M) are available. The datasets listed in Table 6 are used to discover and verify prognostic factors of BC patients. We assessed the association of each gene with overall survival by univariate and multivariable Cox proportional hazard regression analysis. All statistical tests were two-sided. The Cox proportional hazard model, including several important factors, was employed to estimate the hazard ratio (HR) and 95% CI for each gene for breast cancer survival. We use normalized P values of <0.05 to define statistical significance. This part of statistical tests was performed using the R software.
Statistical analysis
We first analyze HWE in the controls for each study using a goodness-of-fit test (chi-square or Fisher’s exact test) and the departure of HWE genotype frequency among control subjects was determined by P <0.05. Crude odds ratios (OR) with 95% confidence intervals (CI) were used to assess the strength of the association between the RAD51 gene and its paralog polymorphisms and breast cancer susceptibility. The pooled ORs for the RAD51 G172T polymorphism were performed under the dominant model (GG vs. TT+GT), recessive model (TT vs. GG+GT), homozygote model (TT vs. GG), and allelic genetic model (T vs. G). T and G represent the minor and the major alleles, respectively. The same methods were applied to the analysis of other polymorphisms. Stratified analyzes were performed on ethnicity and source of control. A Q-test was performed to assess statistical heterogeneity among studies. The pooled OR was calculated using a fixed effect model if the result of the p-value of the Q test<0.1 indicated significant heterogeneity according to the previous study(Davey and Egger,1997) (29, 30). If the result of the Q test was P>0.1, which indicated that the heterogeneity between studies was not significant. Otherwise, a random-effects model was used. Given the potential heterogeneity among studies with different ethnicities and sources of control, the random-effects model was adopted (30). Sensitivity analysis was carried out by removing each study at a time to evaluate the stability of the results under either genotypic models or the allelic model. In addition, the Begg test and Egger’s linear regression test by visual inspection of the funnel plot were carried out to address the potential publication bias, and P <0.05 was considered an indicator of significant publication bias (30, 31). Cox regression was used to analyze the impact of genes on the prognosis of BC patients and its value in prognostic diagnosis
The Newcastle-Ottawa Scale (NOS) was applied to assess the quality of all studies. The NOS checklist includes three parameters of quality: (i) selected population, (ii) comparability of groups, and (iii) assessment of either the exposure or outcome of interest for case-control studies. The studies scored greater than or equal to 7 were considered to be high quality articles.
Results
Studies included in the meta-analysis
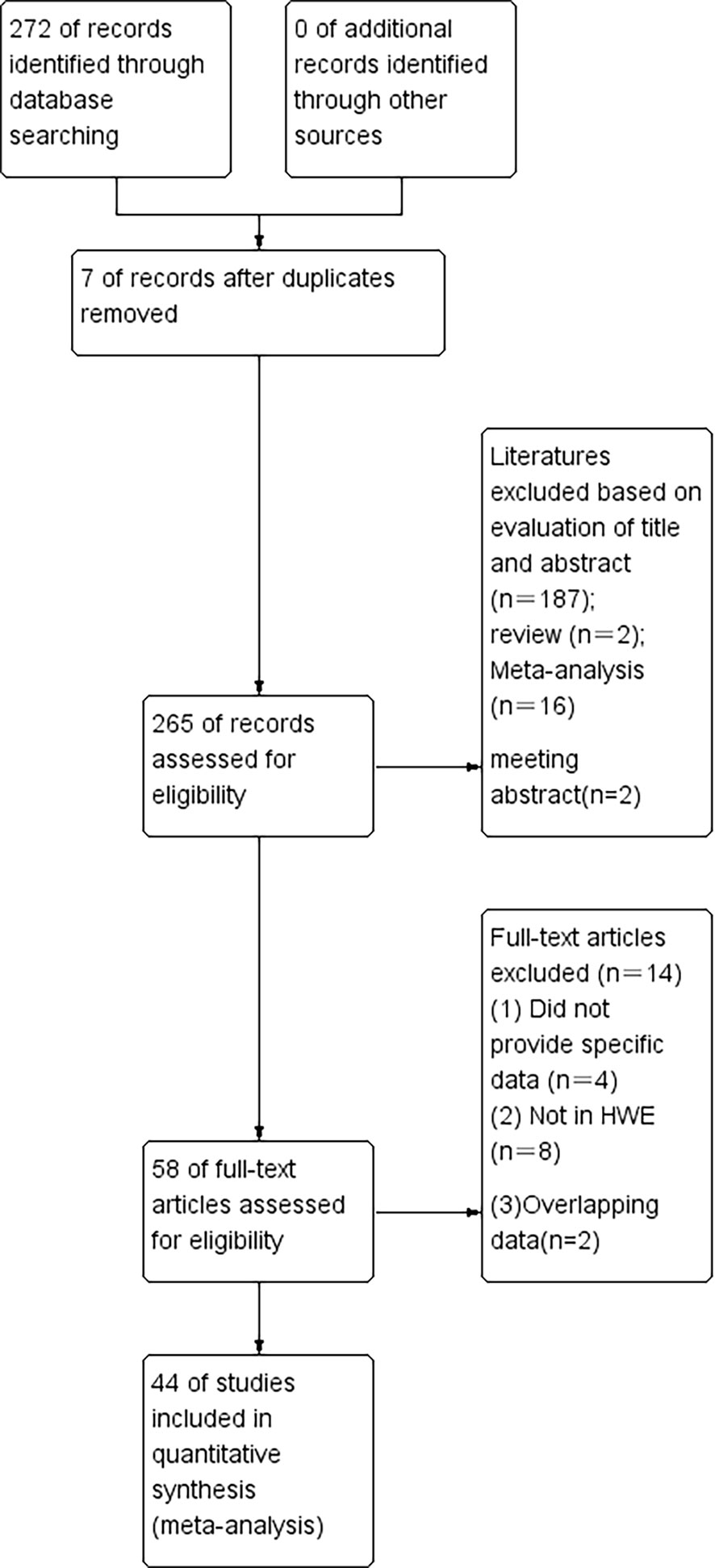
According to our first database search, 272 items were identified (Figure 1). An initial literature search through the PubMed, Embase, Web of Science, and Cochrane database databases yielded 265 published articles after duplicates were removed. When reviewed by titles or abstracts,187 records did not meet the inclusion criteria, leaving 88 potentially relevant studies that were reviewed in full text. Among the remaining 88 articles, 2 were reviews, 16 were meta-analyzes and 2 were meeting conferences; these publications were also excluded. Left 58 publications were left, 2 were insufficient data, 4 were overlapping data, and 8 were not in HWE (Tables 1–3). Finally, a total of 44 publications were included in the meta-analysis, among which 9 case-control studies from 9 publications with 4111 cases and 2669 controls for the RAD51 G172T polymorphism, and 20 case-control studies from 11 publications with 20183 cases and 20321 controls for the XRCC2 R188H polymorphism and a total of 47 studies from 38 publications with 26667 cases and 27912 controls for the XRCC3 T241M polymorphism were eventually included in our meta-analysis. We checked the symmetry of the Begg funnel plot and the results of Egger’s test to assess publication bias. All statistical analyzes were performed with STATA version 16.0.
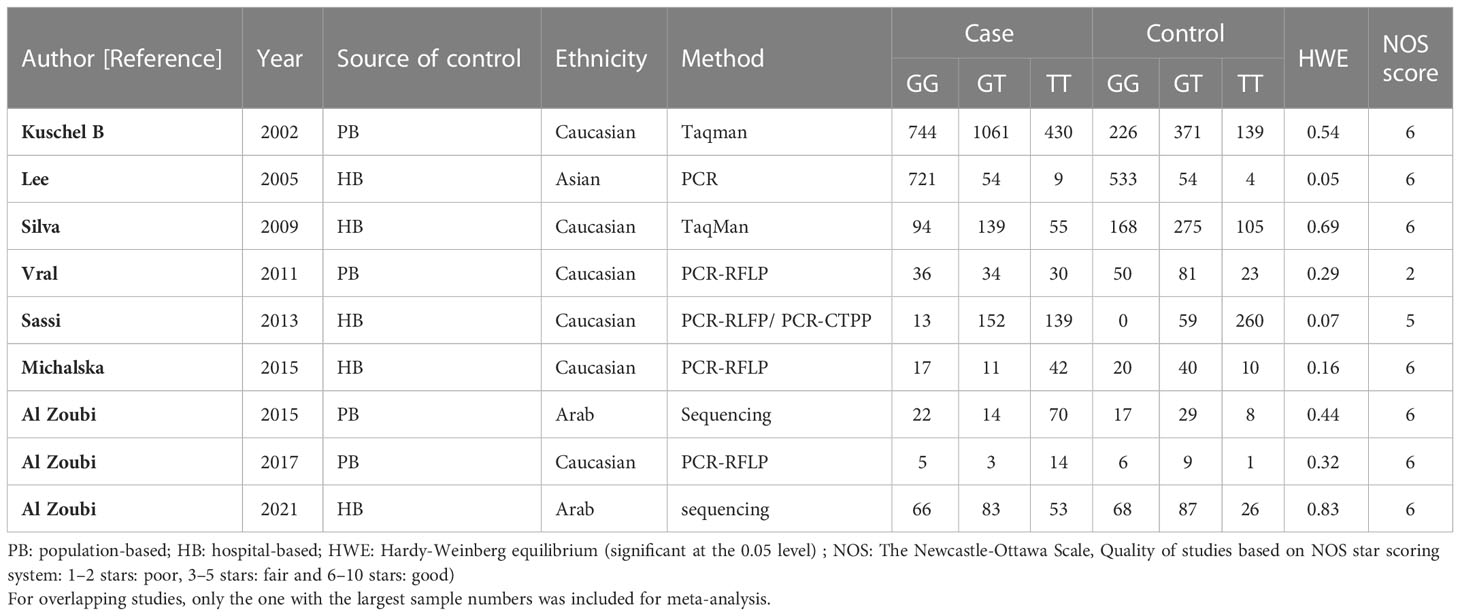
Table 1 Main characteristics of all studies included in the meta-analysis of the RAD51 G172T polymorphism.
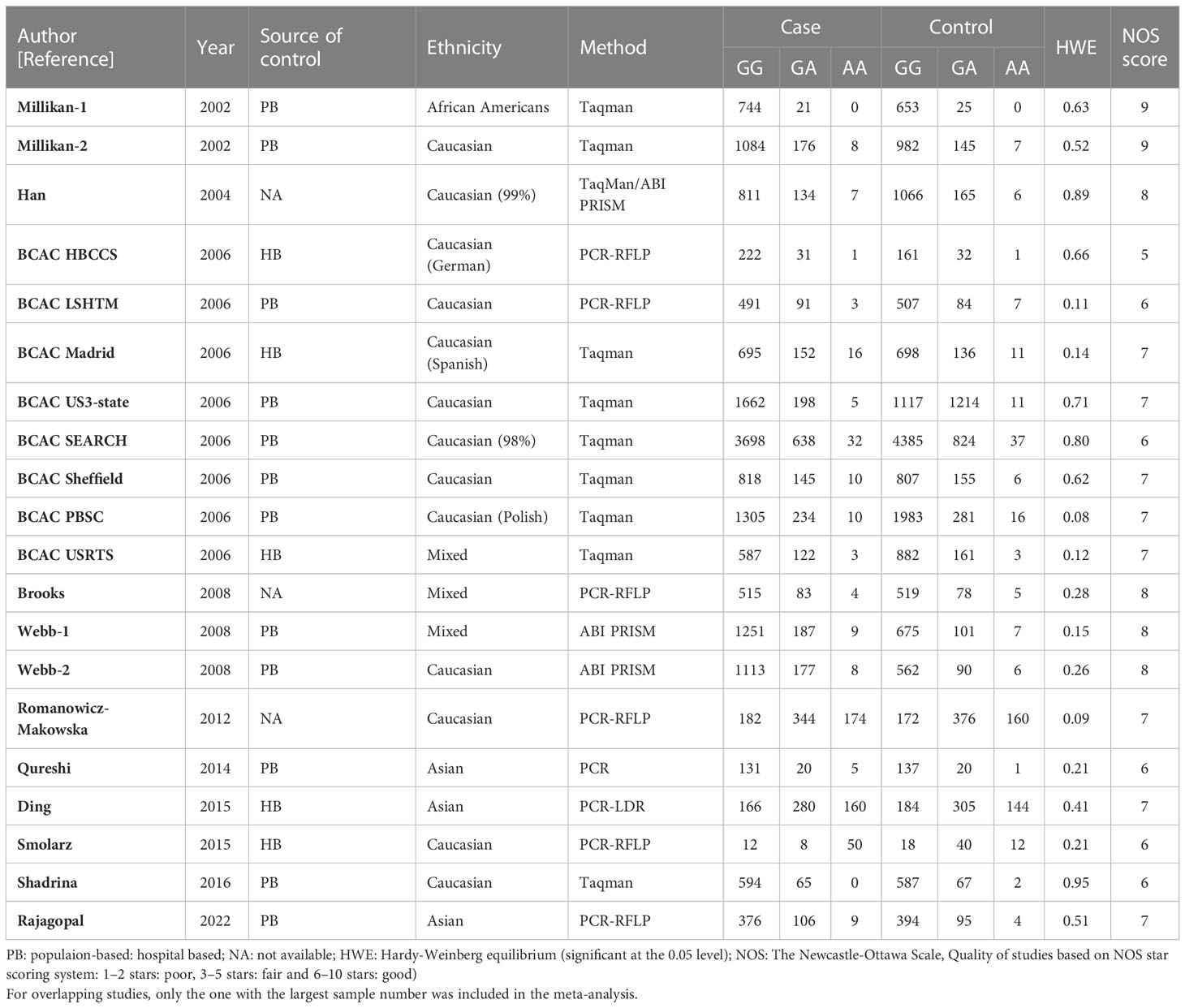
Table 2 Main characteristics of all studies included in the meta-analysis of the XRCC2 R188H polymorphism.
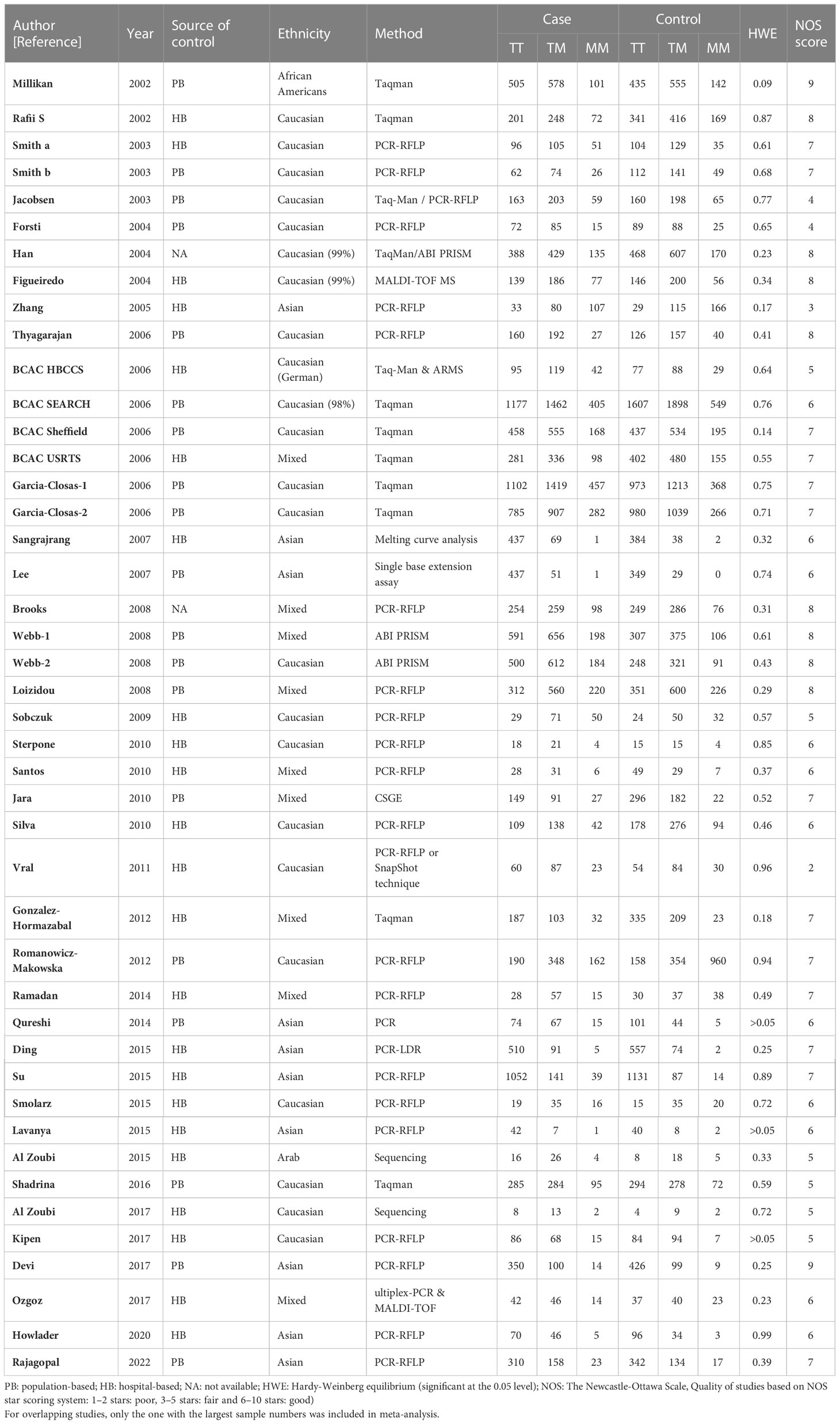
Table 3 Main characteristics of all studies included in the meta-analysis of the XRCC3 T241M polymorphism.
Meta-analysis result
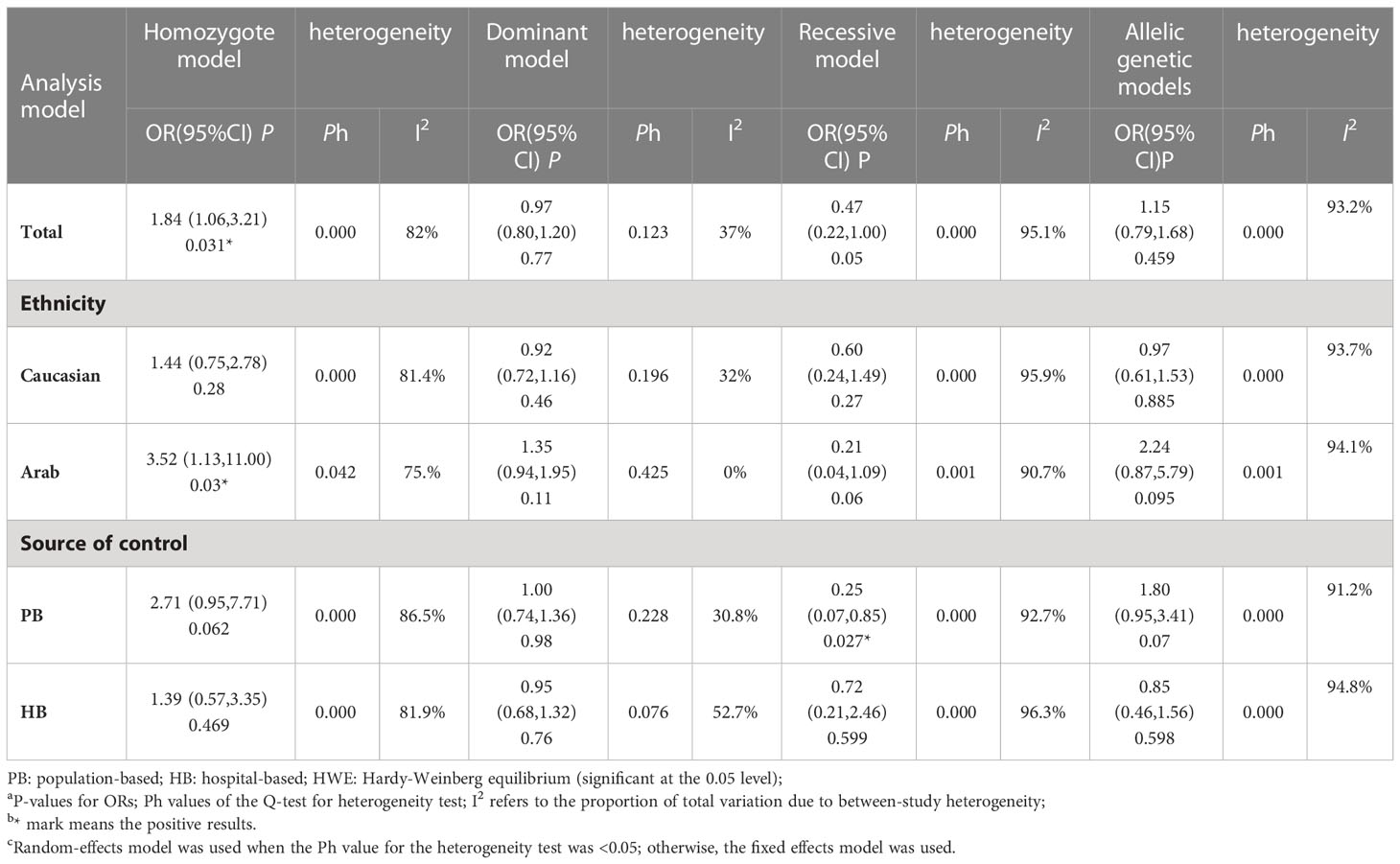
Among these 9 case-control studies from 9 publications with 4111 cases and 2669 controls for the RAD51 G172T polymorphism (32–40). The combined results showed that there was no significant correlation between the G172T polymorphism and breast cancer susceptibility in all genetic models except the homozygote model (homozygote model: OR = 1.84, 95% CI = 1.06-3.21, Figure 2; dominant model: OR = 0.97, 95% CI = 0.80–1.18; recessive model: OR = 0.47, 95% CI = 0.22–1.00; allelic genetic model: OR = 1.15, 95% CI = 0.79–1.68). Additionally, ethnic-based analysis showed that SNP was associated with breast cancer risk in Arab populations in homozygous models (OR=3.52, 95% CI=1.13-11.0, P= 0.003) (Figure 3). It suggests that the G172T polymorphism may be associated with an increased risk of breast cancer in the Arab population in some cases. When stratified by the source of controls, our results found evidence of an association between cancer risk and the G172T polymorphism in population-based controls in the recessive model (OR=0.25, 95% CI=0.07-0.85, P= 0.027), suggesting that it is marginally related to the population-based group.
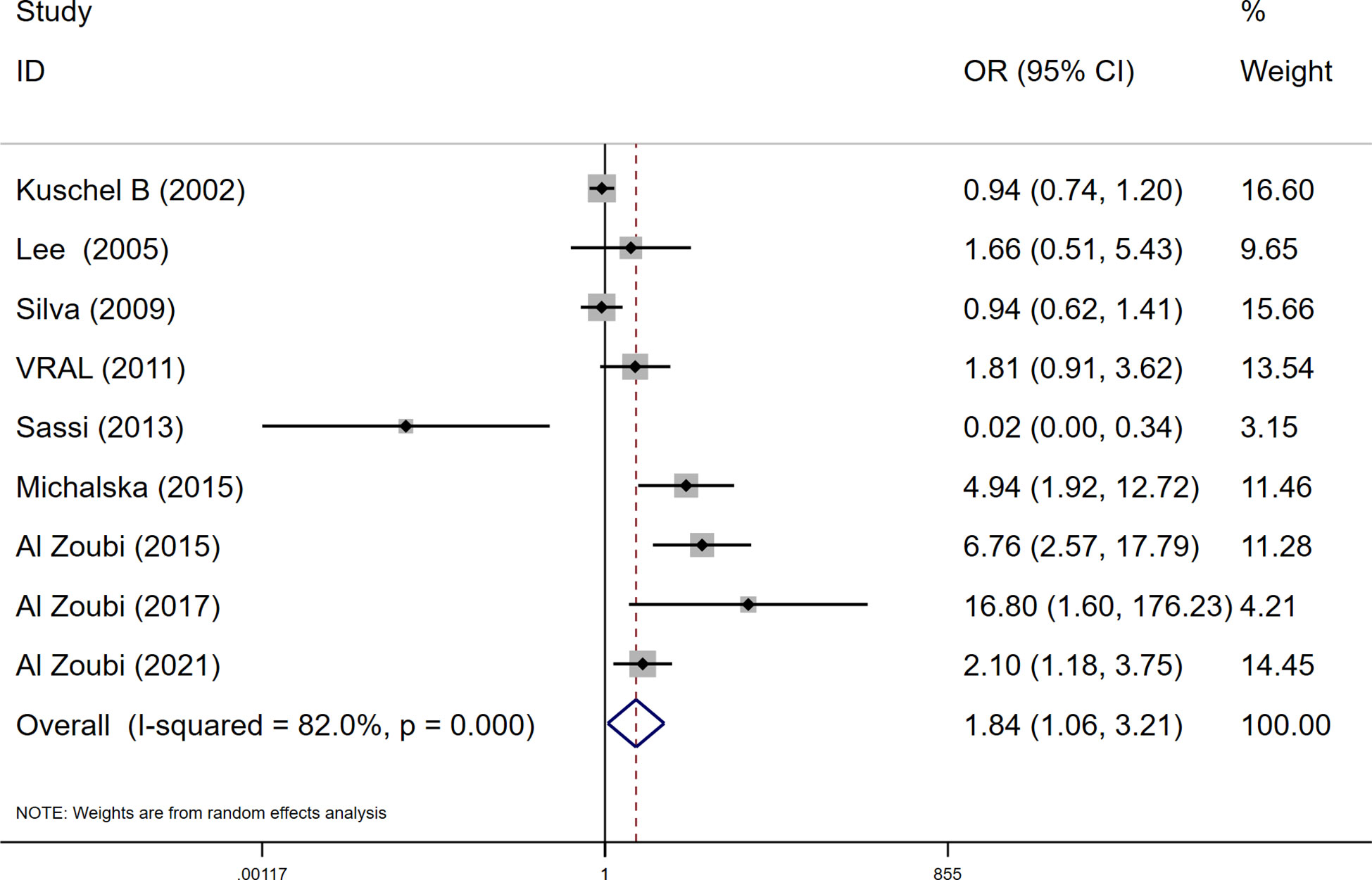
Figure 2 Meta-analysis of the RAD51 G172T polymorphism and risk in breast cancer (homozygote model, TT vs. GG).
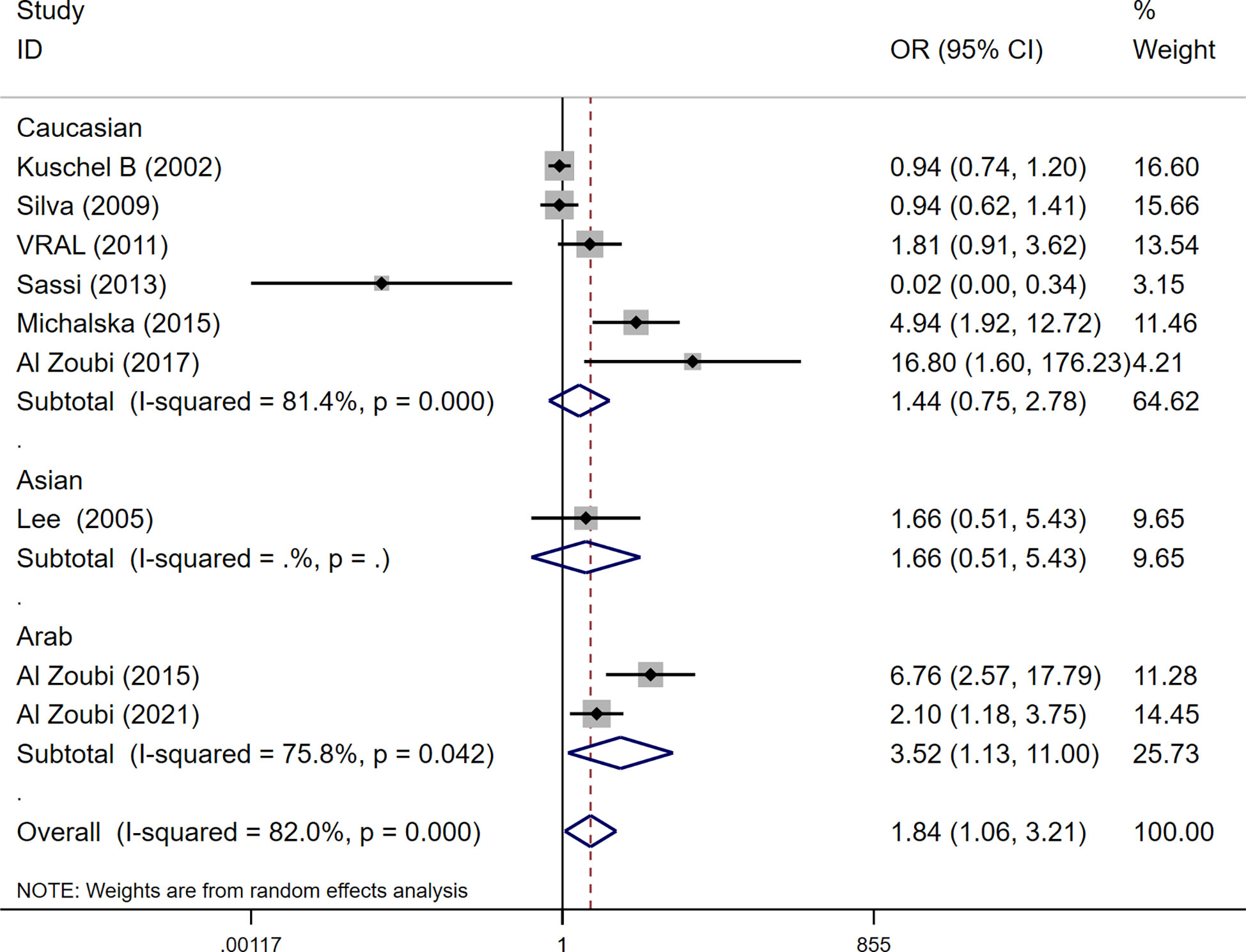
Figure 3 Subgroup analysis according to the ethnicity of the RAD51 G172T polymorphism (homozygote model, TT vs. GG).
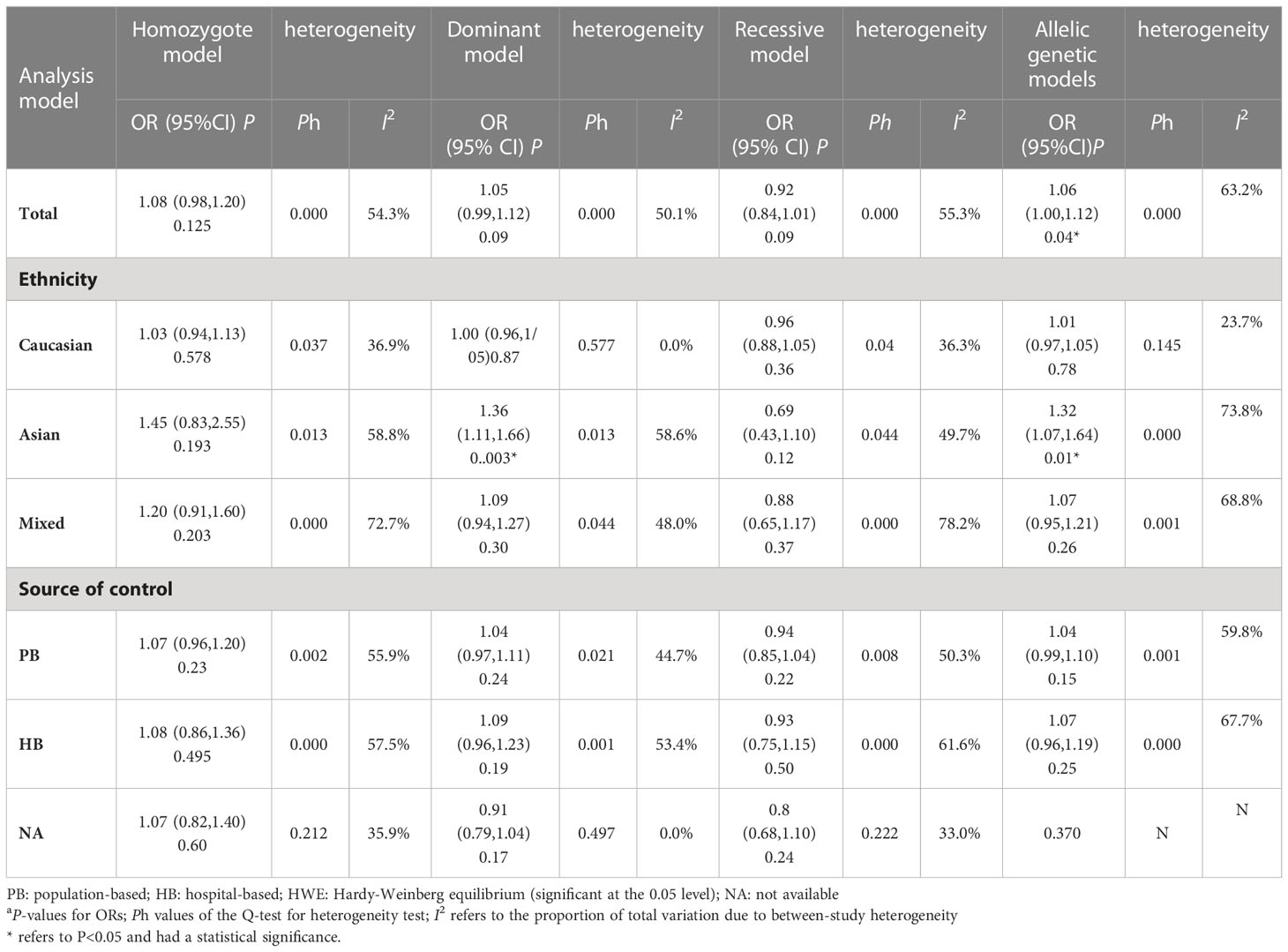
For the R188H polymorphism XRCC2, 20 case-control studies from 11 articles with 20183 cases and 20321 controls for the XRCC2 R188H polymorphism (41–51). No significant association was observed between this polymorphism and breast cancer susceptibility (homozygote model: OR = 1.13, 95% CI = 0.88–1.46; dominant model: OR = 1.01, 95% CI = 0.92-1.11, recessive model: OR = 0.83, 95% CI = 0.61-1.12; allelic genetic model: OR = 1.05, 95% CI = 0.95-1.17.).
For the polymorphism in XRCC3 Thr241Met, a total of 47 studies of 38 articles with 26667 cases and 27912 controls were eventually included in our meta-analysis (32, 37, 38, 44, 45, 47, 48, 52–80). A significant increase in cancer risk was observed only in the allelic genetic model (homozygote model: OR = 1.08, 95% CI = 0.98–1.20; dominant model: OR = 1.05, 95% CI = 0.99–1.12; recessive model: OR = 0.92, 95% CI = 0.84–1.01; allelic genetic model: OR = 1.05, 95% CI = 1.00–1.11) (Figure 4). In addition, ethnic-based analysis showed that SNP was associated with breast cancer risk in Asian populations in dominant genetic (OR = 1.36,95% CI= 1.11–1.66, P = 0.003) and allelic genetic models (OR = 1.32,95% CI 1.07–1.64, P = 0.01) (Tables 4, 5).
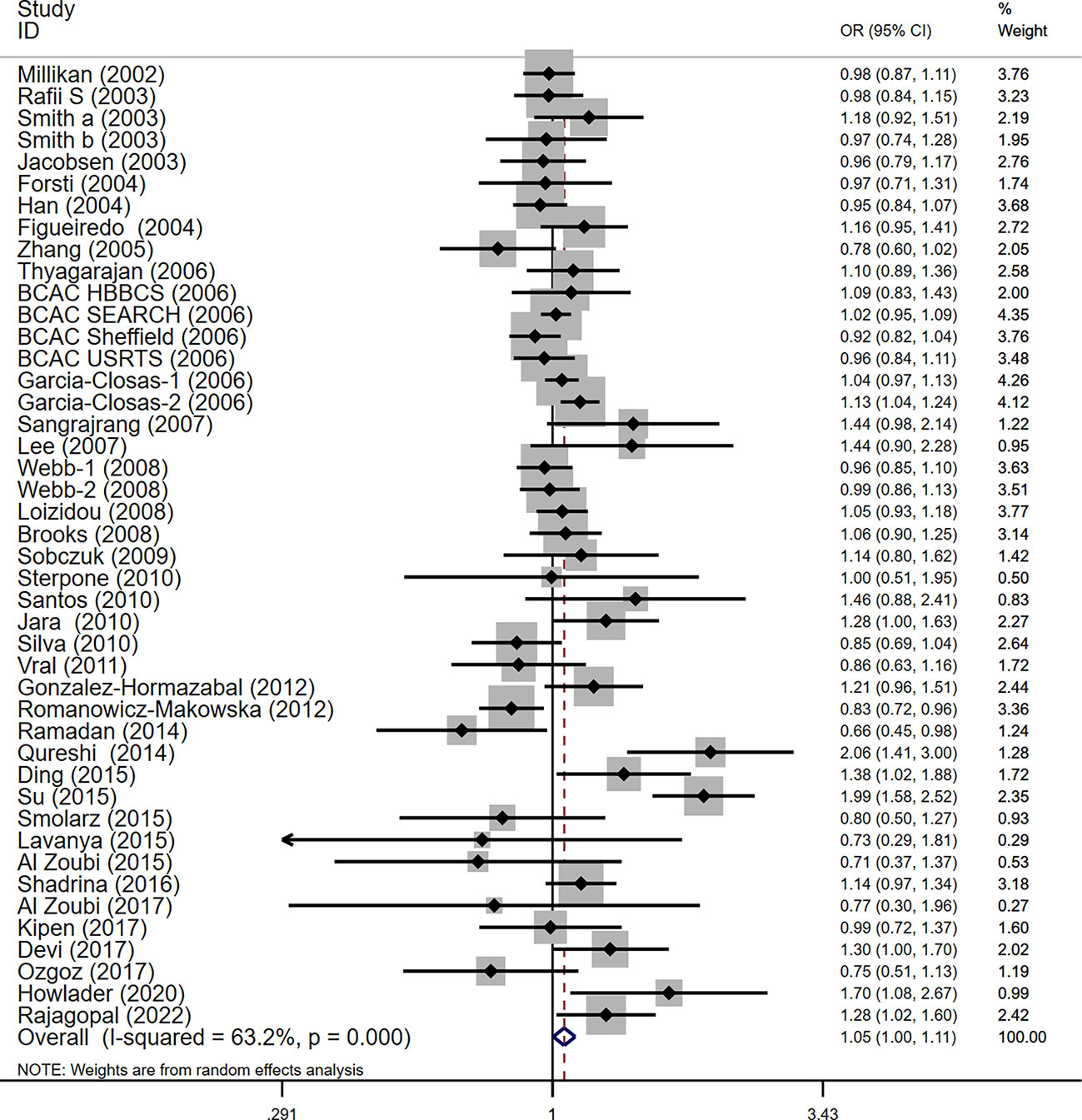
Figure 4 Forest plots of the XRCC3 T241M polymorphism and risk of breast cancer (allelic genetic model, M vs. T).
Prognostic factors
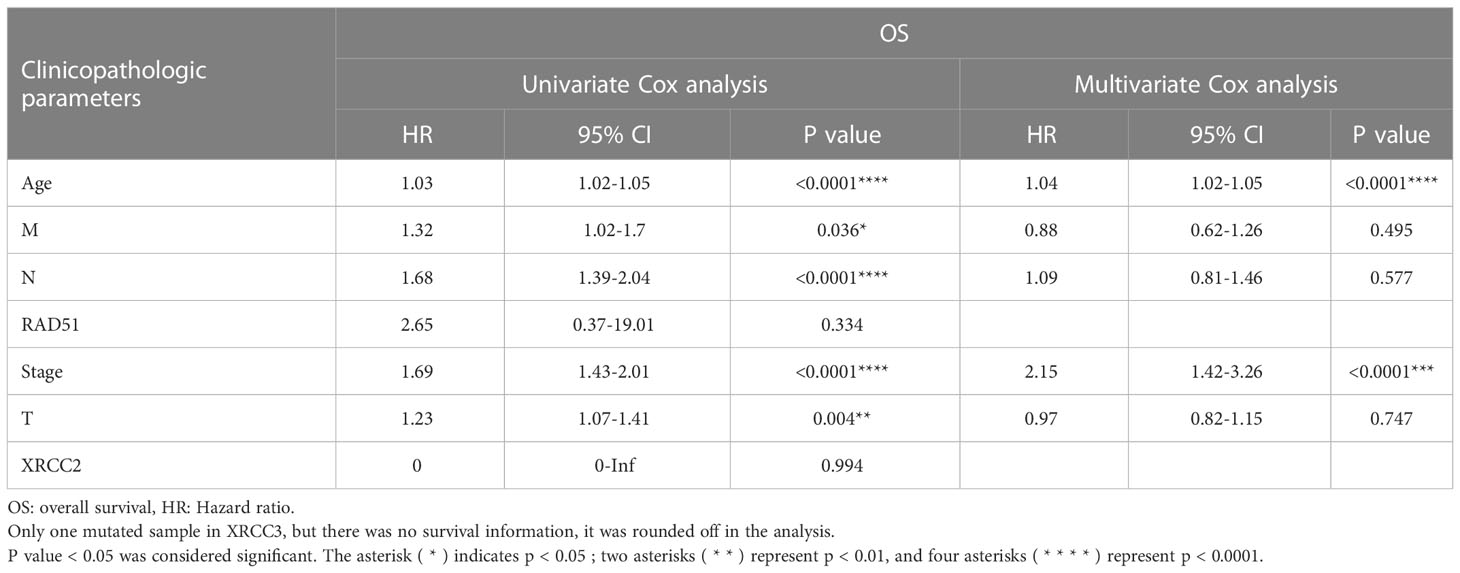
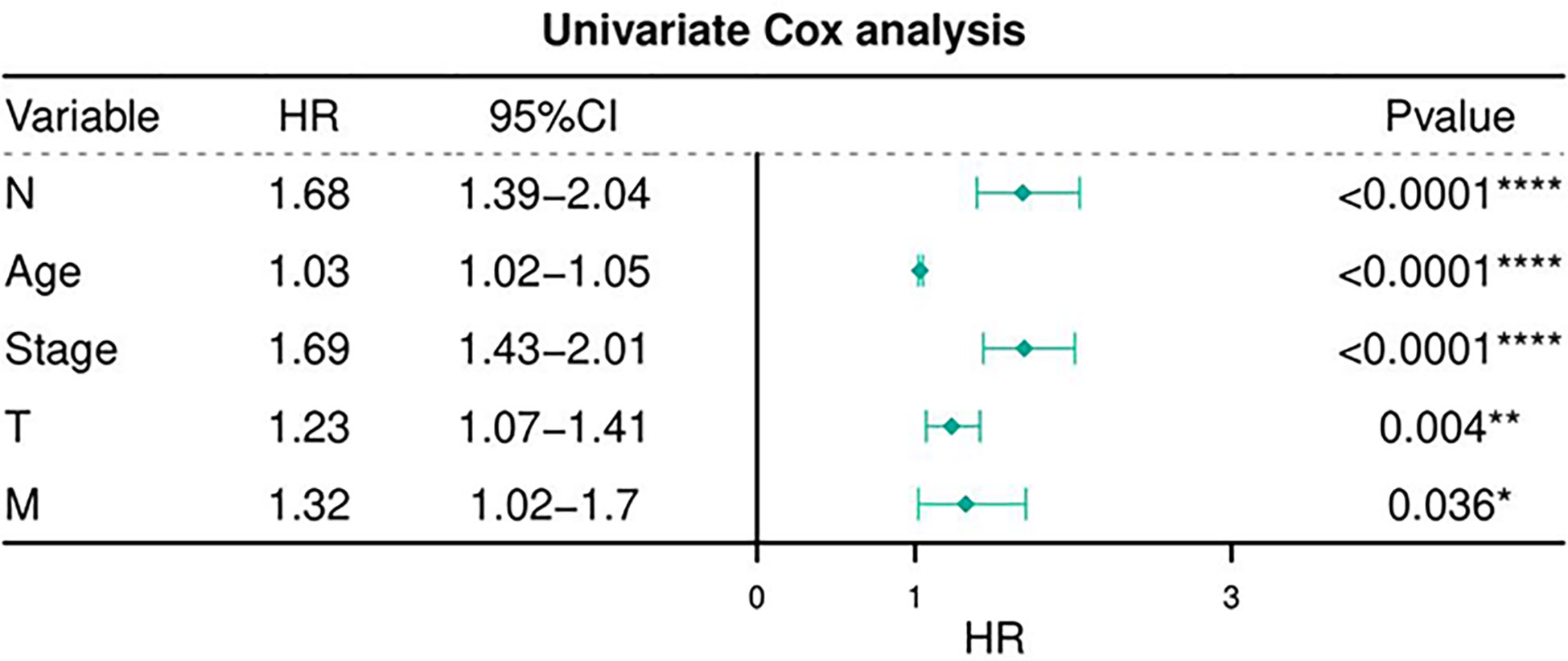
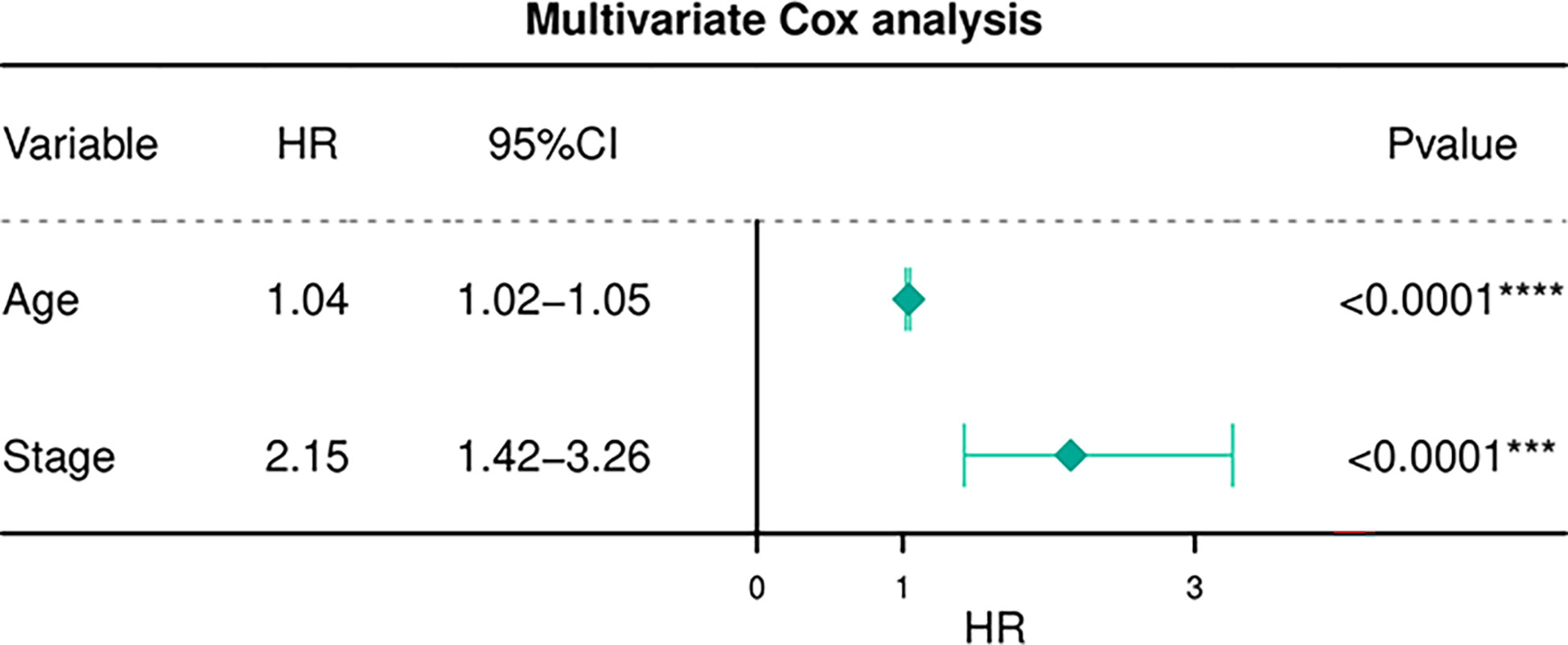
Table 6 depicts the pooled results from the univariable and the multivariable analyses of OS in BC patients (HR). Univariate and multivariate Cox regression analysis was performed to determine whether gene expression is an independent prognostic model of OS in breast cancer patients. As shown in Figure 5, the p values of T, N, M, Stage, and Age were less than 0.05. The results of the univariate Cox regression analysis of OS showed that pathology stage, age, and stage could effectively predict survival in BC patients. Then, we took these factors into the multivariate Cox regression analysis. Furthermore, after the multivariate analyses (Figure 6), the results showed that stage (HR =2.15; 95% CI, 1.42−3.26), age (HR = 1.04; 95% CI, 1.02-1.05) remained independent prognostic factors with an adjusted P value < 0.0001.
Sensitive analysis
Given the significant heterogeneity between studies for the polymorphisms, the random-effect model was used to calculate the pooled results if the heterogeneity was significant. Meanwhile, we also performed a sensitivity analysis to assess the effects of each study on the pooled ORs by omission of individual studies. The sensitivity analysis showed that, for each polymorphism, no single study qualitatively changed the pooled ORs, suggesting that the results of this meta-analysis were statistically stable and reliable.
Publication bias diagnostics
We further identify potential publication biases of the literature using the Egger test and funnel plot. In all studies, no funnel plot asymmetry was found. The results of Egger’s test for the RAD51 G172T polymorphism did not show any evidence of publication bias. For the homozygote model, the funnel plot p-value was 0.47, and Egger’s test p-value was 0.185. In the dominant model, Begg’s test results of the R188H P value were 0.67, and Egger’s test P value was 0.319. Begg’s test result of the allelic genetic model in XRCC3 T241M P = 0.65 and Egger’s test result showed P = 0.52, suggesting no publication bias. All P-values> 0.05, suggesting that there was no publication bias.
Discussion
Screening for some frequent polymorphisms has improved our understanding of the critical roles that inheritance plays in BC susceptibility. To date, associations between genetic variants in HRR genes and BC development have been investigated, but the results remain unexplained to the best of our knowledge. However, new discoveries in drug research aimed at these gene mutations are always innovative. Some experiments suggest that the inhibition of HR will be selective against breast tumor cells. Inhibitors of HR proteins can be used in combination with radiotherapy or chemotherapy to sensitize the cells [5]. A more intriguing possibility would be to use anti-HR agents alone, avoiding the toxicity of DNA-damaging agents. Such a strategy has been applied to selectively kill BRCA2-deficient cells using poly-ADP-ribose-polymerase inhibitors (PARP). The first phase III clinical study of PARP inhibitor for adjuvant treatment of early breast cancer, OlympiA study, aims to evaluate the efficacy and safety of olaparib compared with placebo in the adjuvant treatment of early breast cancer with clinically and pathologically high-risk, HER2-negative, BRCA1/2 mutation. And randomized phase II GeparOLA study showed olaparib plus paclitaxel (PO) in early HER2-negative homologous recombination deficiency (HRD) breast cancer. In conclusion, germline BRCA 1/2 status and HRD predict a higher pathological complete response (pCR) rate in the neoadjuvant treatment (81). The molecular mechanism of breast cancer is very complex. Therefore, in the post-PARP inhibitor era, there is a great clinical need to find therapeutic targets and analyze prognostic factors to benefit patients, which is conducive to drug development and expansion of new indications and provides the possibility of individualized treatment for breast cancer.
Our analysis demonstrated the importance of recombination repair processes for the fidelity of chromosome segregation and reinforce the functional connection between genes involved in HRR and those that predispose to breast cancer. We also found that patients in our prediction models tended to be older, have an advanced-stage disease, and have a poorer prognosis. Current literature varies widely in experimental methods, stage of disease, family history of cancer, patients with the type of tumor therapy, and the duration since cancer diagnosis, all of which can lead to inconsistent results in case-control studies. Additionally, most of the studies did not specify the immunohistochemical indicators of breast cancer that are relevant to determine which factors can exert a dominating effect. Some research data indicate that double-strand break damage is the most fatal lesion observed in eukaryotic cells because it can cause cell death or create a serious threat to cell viability and genome stability. It has the potential to permanently arrest cell cycle progression and endanger cell survival [10]. Due to the fact that DNA repair mechanisms are crucial to preserving genomic stability and functionality, DNA repair defects can result in the development of chromosomal aberrations that can lead to increased susceptibility to cancer (4, 82, 83). A Japanese study showed that Rad51 gene polymorphisms were found in two patients with bilateral breast cancer (10). It proves that germline mutations in the RAD51 gene may modulate the risk of breast cancer. Previous meta-analyses evaluated the effect of the Rad51 G135C polymorphism on the risk of breast cancer and other cancers. Some experts performed relevant meta-analyses of the analysis and concluded that the Rad51 G172T polymorphism may play a protective role in the development of head and neck cancer, but no significant correlation was found between the Rad51 G172T polymorphism and breast and ovarian cancer (84). It is inconsistent with our conclusion and hypothesized that it was related to inadequate inclusion of the sample size, neglecting gene-gene and gene-environment interactions for some reason. However, there were some approvals on the connection of polymorphism in XRCC2 R188H and the risk of breast cancer before, which has not been confirmed in two population studies in the United States and Poland and several case-control experimental studies (39, 42–44, 50, 51, 68). Moreover, an experiment conducted by RafiiS was hardly replicated in the latest BCAC study (41). Several studies describe a marginally protective effect for rare allele carriers (188His) (64, 85). Interestingly, Silva suggested that the potential protective role of the variant allele of XRCC2, in women who have never breastfed, could be related to a more efficient DNA repair activity (37). On the other hand, Han described a protective effect for women with high plasma α-carotene levels. However, current evidence shows that in most studies the XRCC2 R188H polymorphism is considered to have little relationship with the risk of breast cancer. According to our meta-analysis of breast cancer, we did not find a significant association between this polymorphism and breast cancer susceptibility, which is consistent with the previous meta-analysis. In previous studies, a relevant study reported their results with significant unexplained heterogeneity (Ph = 0.014) (86). Furthermore, studies that depart from the Hardy-Weinberg equilibrium (HWE) were included in the meta-analysis, which may lead to potential bias. Current evidence suggests that XRCC2 R188H polymorphism is considered to have a weak protective effect against breast cancer development in most studies, but the association did not reach statistical significance. As we mentioned above, since this effect is very weak and R188H may serve as a positional marker for other potentially functional SNPs or haplotypes, it is not surprising that this SNP is not associated with breast cancer, or even in an inverse relationship. Therefore, limited by the above factors, the interpretation of the results of previous research should be cautious. A common polymorphism in the XRCC3 gene is at nucleotide 1,8607C/T which results in the substitution of the amino acid threonine for methionine at codon 241 (Thr241Met) of exon 7 of the XRCC3 gene, which may affect the function of the encoding enzyme or/and its interaction with other proteins involved in DNA repair. Inheritance of functional polymorphisms in DNA repair genes may influence the capacity of the DNA repair process, thus leading to increased cancer risk. Due to a C18607T transition at exon 7 of the XRCC3 gene, the substitution of amino acids Thr241Met is functionally active, as it is associated with an increase in the number of micronuclei in human lymphocytes exposed to ionizing radiation (59, 67, 72, 87, 88). The variant allele (241Met) is associated with high levels of DNA adducts in lymphocyte DNA, which could be associated with reduced DNA repair capacity (88). A case-control study in Pakistan found that homozygous (TT) and heterozygous (TM) genotypes of the T241M polymorphism were associated with an increased risk of breast cancer compared to controls (47). Similar results have previously been observed in different studies, suggesting an association between Met allele variants and breast cancer in Caucasian and Asian populations (63, 65). Interestingly, Rajagopal found that heterozygous genotype (TM) and homozygous mutant genotype (MM) were not significantly associated with breast cancer risk when it comes to the role of the T241M variation in XRCC3 (48). Chai performed a meta-analysis of 23 case-control studies on the association of XRCC3 SNPs with the risk of breast cancer in the above SNPs and the general population and the Asian population in both recessive and homozygous models (89). Our results based on racial stratification analysis are consistent with their observed correlations in Asian populations, but not the same with their associated models. Although they found an association between this SNP and the risk of sporadic breast cancer, based on the conclusive results obtained, we believe that this association is not accurate enough. Although other studies have not shown an association between T241M polymorphism and the risk of breast cancer (52, 54). Therefore, more studies are needed to confirm these associations.
Compared with studies before, our study has some improvements. First, Our study had the advantage of including higher numbers of cases and controls. Second, these polymorphisms in RAD51 and paralog genes were analyzed and associated with the risk of specific cancer, breast cancer. Third, we provided a more comprehensive analysis of the data by calculating four different genetic models and performing a subgroup analysis by ethnicity, and source of controls (population or hospital-based). Finally, we excluded studies in which the distribution of genotypes in the control group was inconsistent with HWE because they might influence the results. The results of this study further revealed the correlation between the polymorphism in these genes and the occurrence and development of breast cancer, providing a direction for the study of molecular mechanisms of cancer in the future.
The main limitations of our meta-analysis are: 1) This meta-analysis only searched published studies in English, ignoring some unpublished studies or studies in other languages that may also meet the inclusion criteria. 2) Some studies did not provide enough clinical data such as patient family history, ER/PR, HER-2 hormone receptor status, tissue type, and tumor grade, leading to failure to conduct a comprehensive subgroup analysis to explore the source of heterogeneity. 3) Gene-gene and gene-environment interactions were not considered in current meta-analyses. Possible gene-gene and gene-environment interactions between Rad51 gene polymorphism and cancer susceptibility need to be further studied. 4) some patients were chosen from hospital-based groups, and these women may have benign breast disease, corresponding to an increased potential risk of breast cancer. 5) Most of the patients in our study were Caucasian, which may limit the general application of our results.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by JY and C-GW. Manuscript drafting and reviewing: All authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Science Foundation of Chongqing Yongchuan science and Technology Commission (No. Ycstc2018nb0208).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1047336/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Mohindra A, Hays LE, Phillips EN, Preston BD, Helleday T, Meuth M., et al. Defects in homologous recombination repair in mismatch-repair-deficient tumour cell lines. Hum Mol Genet (2002) 11(18):2189–200. doi: 10.1093/hmg/11.18.2189
3. Dixon K, Kopras E. Genetic alterations and DNA repair in human carcinogenesis. Semin Cancer Biol (2004) 14(6):441–8. doi: 10.1016/j.semcancer.2004.06.007
4. Yu Z, Chen J, Ford BN, Glickman BW. Human DNA repair systems: an overview. Environ Mol Mutagen (1999) 33(1):3–20. doi: 10.1002/(SICI)1098-2280(1999)33:1<3::AID-EM2>3.0.CO;2-L
5. Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA Repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia (New York NY). (2009) 11(7):683–91. doi: 10.1593/neo.09312
6. Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res (2009) 69(3):863–72. doi: 10.1158/0008-5472.CAN-08-3057
7. Richardson C. RAD51, genomic stability, and tumorigenesis. Cancer Lett (2005) 218(2):127–39. doi: 10.1016/j.canlet.2004.08.009
8. Khanna KK, Jackson SP. DNA Double-strand breaks: signaling, repair and the cancer connection. Nat Genet (2001) 27(3):247–54. doi: 10.1038/85798
9. Kato M, Yano K, Matsuo F, Saito H, Katagiri T, Kurumizaka H, et al. Identification of Rad51 alteration in patients with bilateral breast cancer. J Hum Genet (2000) 45(3):133–7. doi: 10.1007/s100380050199
10. Vispé S, Defais M. Mammalian Rad51 protein: a RecA homologue with pleiotropic functions. Biochimie (1997) 79(9-10):587–92. doi: 10.1016/S0300-9084(97)82007-X
11. Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol (2011) 22(8):898–905. doi: 10.1016/j.semcdb.2011.07.019
12. Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett (2005) 219(2):125–35. doi: 10.1016/j.canlet.2004.08.018
13. Willems P, Claes K, Baeyens A, Vandersickel V, Werbrouck J, De Ruyck K, et al. Polymorphisms in nonhomologous end-joining genes associated with breast cancer risk and chromosomal radiosensitivity. Genes Chromosomes Cancer (2008) 47(2):137–48. doi: 10.1002/gcc.20515
14. Vandersickel V, Depuydt J, Van Bockstaele B, Perletti G, Philippe J, Thierens H, et al. Early increase of radiation-induced γH2AX foci in a human Ku70/80 knockdown cell line characterized by an enhanced radiosensitivity. J Radiat Res (2010) 51(6):633–41. doi: 10.1269/jrr.10033
15. Willems P, De Ruyck K, Van Den Broecke R, Makar A, Perletti G, Thierens H, et al. A polymorphism in the promoter region of Ku70/XRCC6, associated with breast cancer risk and oestrogen exposure. J Cancer Res Clin Oncol (2009) 135(9):1159–68. doi: 10.1007/s00432-009-0556-x
16. Braybrooke JP, Spink KG, Thacker J, Hickson ID. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J Biol Chem (2000) 275(37):29100–6. doi: 10.1074/jbc.M002075200
17. Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol (2000) 2(10):757–61. doi: 10.1038/35036399
18. Rajesh C, Gruver AM, Basrur V, Pittman DL. The interaction profile of homologous recombination repair proteins RAD51C, RAD51D and XRCC2 as determined by proteomic analysis. Proteomics (2009) 9(16):4071–86. doi: 10.1002/pmic.200800977
19. Tsaryk R, Fabian K, Thacker J, Kaina B. Xrcc2 deficiency sensitizes cells to apoptosis by MNNG and the alkylating anticancer drugs temozolomide, fotemustine and mafosfamide. Cancer Lett (2006) 239(2):305–13. doi: 10.1016/j.canlet.2005.08.036
20. Sprong D, Janssen HL, Vens C, Begg AC. Resistance of hypoxic cells to ionizing radiation is influenced by homologous recombination status. Int J Radiat Oncol Biol Phys (2006) 64(2):562–72. doi: 10.1016/j.ijrobp.2005.09.031
21. Evans JW, Chernikova SB, Kachnic LA, Banath JP, Sordet O, Delahoussaye YM, et al. Homologous recombination is the principal pathway for the repair of DNA damage induced by tirapazamine in mammalian cells. Cancer Res (2008) 68(1):257–65. doi: 10.1158/0008-5472.CAN-06-4497
22. De Silva IU, Mchugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol (2000) 20(21):7980–90. doi: 10.1128/MCB.20.21.7980-7990.2000
23. Wang ZM, Chen ZP, Xu ZY, Christodoulopoulos G, Bello V, Mohr G, et al. In vitro evidence for homologous recombinational repair in resistance to melphalan. J Natl Cancer Inst (2001) 93(19):1473–8. doi: 10.1093/jnci/93.19.1473
24. Bello VE, Aloyz RS, Christodoulopoulos G, Panasci LC. Homologous recombinational repair vis-à-vis chlorambucil resistance in chronic lymphocytic leukemia. Biochem Pharmacol (2002) 63(9):1585–8. doi: 10.1016/S0006-2952(02)00954-1
25. Yanagisawa T, Urade M, Yamamoto Y, Furuyama J. Increased expression of human DNA repair genes, XRCC1, XRCC3 and RAD51, in radioresistant human KB carcinoma cell line N10. Oral Oncol (1998) 34(6):524–8. doi: 10.1016/S1368-8375(98)00045-1
26. Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, et al. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology (2003) 193(1-2):43–65. doi: 10.1016/S0300-483X(03)00289-0
27. Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, et al. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem (1998) 273(34):21482–8. doi: 10.1074/jbc.273.34.21482
28. O'regan P, Wilson C, Townsend S, Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem (2001) 276(25):22148–53. doi: 10.1074/jbc.M102396200
29. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst (1959) 22(4):719–48.
30. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629
31. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics (1994) 50(4):1088–101. doi: 10.2307/2533446
32. Al Zoubi MS, Zavaglia K, Mazanti C, Al Hamad M, Al Batayneh K, Aljabali AAA, et al. Polymorphisms and mutations in GSTP1, RAD51, XRCC1 and XRCC3 genes in breast cancer patients. Int J Biol Markers (2017) 32(3):e337–43. doi: 10.5301/ijbm.5000258
33. Al-Zoubi MS, Mazzanti CM, Zavaglia K, Al Hamad M, Armogida I, Lisanti MP, et al. Homozygous T172T and heterozygous G135C variants of homologous recombination repairing protein RAD51 are related to sporadic breast cancer susceptibility. Biochem Genet (2016) 54(1):83–94. doi: 10.1007/s10528-015-9703-z
34. Michalska MM, Samulak D, Romanowicz H, Smolarz B. Single nucleotide polymorphisms (SNPs) of RAD51-G172T and XRCC2-41657C/T homologous recombination repair genes and the risk of triple- negative breast cancer in polish women. Pathol Oncol Res (2015) 21(4):935–40. doi: 10.1007/s12253-015-9922-y
35. Salim Al Zoubi MS, Al-Eitan LN, Rababa'h DM, Al-Batayneh K, Farzand R, Quinn GA, et al. RAD51-UTR haplotype genetic polymorphisms and susceptibility to breast cancer in women from Jordanian population. Exp Oncol (2021) 43(2):149–54. doi: 10.32471/exp-oncology.2312-8852.vol-43-no-2.16338
36. Sassi A, Popielarski M, Synowiec E, Wozniak K. BLM and RAD51 genes polymorphism and susceptibility to breast cancer. Pathol Oncol Res (2013) 19(3):451–9. doi: 10.1007/s12253-013-9602-8
37. Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, et al. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol (2010) 34(1):85–92. doi: 10.1016/j.canep.2009.11.002
38. Vral A, Willems P, Claes K, Poppe B, Perletti G, Thierens H. Combined effect of polymorphisms in Rad51 and Xrcc3 on breast cancer risk and chromosomal radiosensitivity. Mol Med Rep (2011) 4(5):901–12. doi: 10.3892/mmr2011523
39. Kuschel B, Auranen A, Mcbride S, Novik KL, Antoniou A, Lipscombe JM, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet (2002) 11(12):1399–407. doi: 10.1093/hmg/11.12.1399
40. Lee KM, Choi JY, Kang C, Kang CP, Park SK, Cho H, et al. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res (2005) 11(12):4620–6. doi: 10.1158/1078-0432.CCR-04-2534
41. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the breast cancer association consortium. J Natl Cancer Inst (2006) 98(19):1382–96.
42. Brooks J, Shore RE, Zeleniuch-Jacquotte A, Currie D, Afanasyeva Y, Koenig KL, et al. Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer Epidemiol Biomarkers Prev (2008) 17(4):1016–9. doi: 10.1158/1055-9965.EPI-08-0065
43. Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet (2006) 119(4):376–88. doi: 10.1007/s00439-006-0135-z
44. Han J, Hankinson SE, Ranu H, De Vivo I, Hunter DJ. Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the nurses' health study. Carcinogenesis (2004) 25(2):189–95. doi: 10.1093/carcin/bgh00244
45. Millikan RC, Player JS, Decotret AR, Tse CK, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev (2005) 14(10):2326–34. doi: 10.1158/1055-9965.EPI-05-0186
46. Pelttari LM, Kiiski JI, Ranta S, Vilske S, Blomqvist C, Aittomäki K, et al. RAD51, XRCC3, and XRCC2 mutation screening in Finnish breast cancer families. Springerplus (2015) 4:92. doi: 10.1186/s40064-015-0880-3
47. Qureshi Z, Mahjabeen I, Baig R, Kayani M. Correlation between selected XRCC2, XRCC3 and RAD51 gene polymorphisms and primary breast cancer in women in Pakistan. Asian Pac J Cancer Prev (2014) 15(23):10225–9. doi: 10.7314/apjcp.2014.15.23.10225
48. Rajagopal T, Seshachalam A, Rathnam KK, Talluri S, Venkatabalasubramanian S, Dunna NR, et al. Homologous recombination DNA repair gene RAD51, XRCC2 & XRCC3 polymorphisms and breast cancer risk in south Indian women. PloS One (2022) 17(1):e0259761. doi: 10.1371/journal.pone.0259761
49. Ramadan RA, Desouky LM, Elnaggar MA, Moaaz M, Elsherif AM. Association of DNA repair genes XRCC1 (Arg399Gln), (Arg194Trp) and XRCC3 (Thr241Met) polymorphisms with the risk of breast cancer: a case-control study in Egypt. Genet Test Mol Biomarkers (2014) 18(11):754–60. doi: 10.1089/gtmb.2014.0191
50. Romanowicz-Makowska H, Smolarz B, Zadrozny M, Westfal B, Baszczynski J, Polac I, et al. Single nucleotide polymorphisms in the homologous recombination repair genes and breast cancer risk in polish women. Tohoku J Exp Med (2011) 224(3):201–8. doi: 10.1620/tjem.224.201
51. Webb PM, Hopper JL, Newman B, Chen X, Kelemen L, Giles GG, et al. Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer Epidemiol Biomarkers Prev (2005) 14(2):319–23. doi: 10.1158/1055-9965.EPI-04-0335
52. Jacobsen NR, Nexø BA, Olsen A, Overvad K, Wallin H, Tjønneland A, et al. No association between the DNA repair gene XRCC3 T241M polymorphism and risk of skin cancer and breast cancer. Cancer Epidemiol Biomarkers Prev (2003) 12(6):584–5.
53. Rafii S. A naturally occurring mutation in an ATP-binding domain of the recombination repair gene XRCC3 ablates its function without causing cancer susceptibility. Hum Mol Genet (2003) 12(8):915–23. doi: 10.1093/hmg/ddg102
54. Smith TR, Miller MS, Lohman K, Lange EM, Case LD, Mohrenweiser HW, et al. Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer. Cancer Lett (2003) 190(2):183–90. doi: 10.1016/S0304-3835(02)00595-5
55. Su CH, Chang WS, Hu PS, Hsiao CL, Ji HX, Liao CH, et al. Contribution of DNA double-strand break repair gene XRCC3 genotypes to triple-negative breast cancer risk. Cancer Genomics Proteomics (2015) 12(6):359–67.
56. Al Zoubi MS. X-Ray repair cross-complementing protein 1 and 3 polymorphisms and susceptibility of breast cancer in a Jordanian population. Saudi Med J (2015) 36(10):1163–7. doi: 10.15537/smj.2015.10.12659
57. Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, et al. DNA Repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: a study on a Portuguese population. Breast Cancer Res Treat (2007) 103(2):209–17. doi: 10.1007/s10549-006-9364-z
58. Devi KR, Ahmed J, Narain K, Mukherjee K, Majumdar G, Chenkual S, et al. DNA Repair mechanism gene, XRCC1A ( Arg194Trp) but not XRCC3 ( Thr241Met) polymorphism increased the risk of breast cancer in premenopausal females: A case-control study in northeastern region of India. Technol Cancer Res Treat (2017) 16(6):1150–9. doi: 10.1177/1533034617736162
59. Ding P, Yang Y, Cheng L, Zhang X, Cheng L, Li C, et al. The relationship between seven common polymorphisms from five DNA repair genes and the risk for breast cancer in northern Chinese women. PloS One (2014) 9(3):e92083. doi: 10.1371/journal.pone.0092083
60. Gonzalez-Hormazabal P, Reyes JM, Blanco R, Bravo T, Carrera I, Peralta O, et al. The BARD1 Cys557Ser variant and risk of familial breast cancer in a south-American population. Mol Biol Rep (2012) 39(8):8091–8. doi: 10.1007/s11033-012-1656-2
61. Jara L, Dubois K, Gaete D, de Mayo T, Ratkevicius N, Bravo T, et al. Variants in DNA double-strand break repair genes and risk of familial breast cancer in a south American population. Breast Cancer Res Treat (2010) 122(3):813–22. doi: 10.1007/s10549-009-0709-2
62. Krupa R, Synowiec E, Pawlowska E, Morawiec Z, Sobczuk A, Zadrozny M, et al. Polymorphism of the homologous recombination repair genes RAD51 and XRCC3 in breast cancer. Exp Mol Pathol (2009) 87(1):32–5. doi: 10.1016/j.yexmp.2009.04.005
63. Lee SA, Lee KM, Park SK, Choi JY, Kim B, Nam J, et al. Genetic polymorphism of XRCC3 Thr241Met and breast cancer risk: case-control study in Korean women and meta-analysis of 12 studies. Breast Cancer Res Treat (2007) 103(1):71–6. doi: 10.1007/s10549-006-9348-z
64. Loizidou MA, Michael T, Neuhausen SL, Newbold RF, Marcou Y, Kakouri E, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat (2008) 112(3):575–9. doi: 10.1007/s10549-007-9881-4
65. Romanowicz-Makowska H, Smolarz B, Polac I, Sporny S. Single nucleotide polymorphisms of RAD51 G135C, XRCC2 Arg188His and XRCC3 Thr241Met homologous recombination repair genes and the risk of sporadic endometrial cancer in polish women. J Obstet Gynaecol Res (2012) 38(6):918–24. doi: 10.1111/j.1447-0756.2011.01811.x
66. Sangrajrang S, Schmezer P, Burkholder I, Boffetta P, Brennan P, Woelfelschneider A, et al. The XRCC3 Thr241Met polymorphism and breast cancer risk: a case-control study in a Thai population. Biomarkers (2007) 12(5):523–32. doi: 10.1080/13547500701395602
67. Shadrina AS, Ermolenko NA, Boyarskikh UA, Sinkina TV, Lazarev AF, Petrova VD, et al. Polymorphisms in DNA repair genes and breast cancer risk in Russian population: a case-control study. Clin Exp Med (2016) 16(1):21–8. doi: 10.1007/s10238-014-0329-y
68. Smolarz B, Makowska M, Samulak D, Michalska MM, Mojs E, Wilczak M, et al. Association between single nucleotide polymorphisms (SNPs) of XRCC2 and XRCC3 homologous recombination repair genes and triple-negative breast cancer in polish women. Clin Exp Med (2015) 15(2):151–7. doi: 10.1007/s10238-014-0284-7
69. Sterpone S, Cornetta T, Padua L, Mastellone V, Giammarino D, Testa A, et al. DNA Repair capacity and acute radiotherapy adverse effects in Italian breast cancer patients. Mutat Res (2010) 684(1-2):43–8. doi: 10.1016/j.mrfmmm.2009.11.009
70. Thyagarajan B, Anderson KE, Folsom AR, Jacobs DR Jr, Lynch CF, Bargaje A, et al. No association between XRCC1 and XRCC3 gene polymorphisms and breast cancer risk: Iowa women's health study. Cancer Detect Prev (2006) 30(4):313–21. doi: 10.1016/j.cdp.2006.07.002
71. Zhang L, Ruan Z, Hong Q, Gong X, Hu Z, Huang Y, et al. Single nucleotide polymorphisms in DNA repair genes and risk of cervical cancer: A case-control study. Oncol Lett (2012) 3(2):351–62. doi: 10.3892/ol.2011.463
72. Aka P, Mateuca R, Buchet JP, Thierens H, Kirsch-Volders M. Are genetic polymorphisms in OGG1, XRCC1 and XRCC3 genes predictive for the DNA strand break repair phenotype and genotoxicity in workers exposed to low dose ionising radiations? Mutat Res (2004) 556(1-2):169–81. doi: 10.1016/j.mrfmmm.2004.08.002.
73. Figueiredo JC, Knight JA, Briollais L, Andrulis IL, Ozcelik H. Polymorphisms XRCC1-R399Q and XRCC3-T241M and the risk of breast cancer at the Ontario site of the breast cancer family registry. Cancer Epidemiol Biomarkers Prev (2004) 13(4):583–91.
74. Försti A, Angelini S, Festa F, Sanyal S, Zhang Z, Grzybowska E, et al. Single nucleotide polymorphisms in breast cancer. Oncol Rep (2004) 11(4):917–22.
75. Santos RA, Teixeira AC, Mayorano MB, Carrara HH, Andrade JM, Takahashi CS, et al. DNA Repair genes XRCC1 and XRCC3 polymorphisms and their relationship with the level of micronuclei in breast cancer patients. Genet Mol Biol (2010) 33(4):637–40. doi: 10.1590/S1415-47572010005000082
76. Dashti S, Taherian-Esfahani Z, Keshtkar A, Ghafouri-Fard S. Associations between XRCC3 Thr241Met polymorphisms and breast cancer risk: systematic-review and meta-analysis of 55 case-control studies. BMC Med Genet (2019) 20(1):79. doi: 10.1186/s12881-019-0809-8
77. Dufloth RM, Costa S, Schmitt F, Zeferino LC. DNA Repair gene polymorphisms and susceptibility to familial breast cancer in a group of patients from campinas, Brazil. Genet Mol Res (2005) 4(4):771–82.
78. Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta (2005) 359(1-2):150–5. doi: 10.1016/j.cccn.2005.03.047
79. Howlader NR, Rahman MM, Hossain MA, Sultana R, Hossain SM, Mazid MA, et al. Genetic polymorphisms in DNA repair genes XRCC1 and 3 are associated with increased risk of breast cancer in Bangladeshi population. Breast Cancer Res Treat (2020) 182(3):739–50. doi: 10.1007/s10549-020-05738-8
80. Sobczuk A, Romanowicz-Makowska H, Fiks T, Baszczyński J, Smolarz B. XRCC1 and XRCC3 DNA repair gene polymorphisms in breast cancer women from the lodz region of Poland. Pol J Pathol (2009) 60(2):76–80.
81. Fasching PA, Link T, Hauke J, Seither F, Jackisch C, Klare P, Seither F, Jackisch C, Klare P, et al. Neoadjuvant paclitaxel/olaparib in comparison to paclitaxel/carboplatinum in patients with HER2-negative breast cancer and homologous recombination deficiency (GeparOLA study). Ann Oncol Off J Eur Soc Med Oncol (2021) 32:49–57. doi: 10.1016/j.annonc.2020.10.471
82. Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst (2000) 92(11):874–97. doi: 10.1093/jnci/92.11.874
83. Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res (2005) 577(1-2):275–83. doi: 10.1016/j.mrfmmm.2005.03.007
84. Zhao M, Chen P, Dong Y, Zhu X, Zhang X. Relationship between Rad51 G135C and G172T variants and the susceptibility to cancer: a meta-analysis involving 54 case-control studies. PloS One (2014) 9(1):e87259. doi: 10.1371/journal.pone.0087259
85. Pooley KA, Baynes C, Driver KE, Tyrer J, Azzato EM, Pharoah PD, et al. Common single-nucleotide polymorphisms in DNA double-strand break repair genes and breast cancer risk. Cancer Epidemiol Biomarkers Prev (2008) 17(12):3482–9. doi: 10.1158/1055-9965.EPI-08-0594
86. Yu KD, Chen AX, Qiu LX, Fan L, Yang C, Shao ZM.. XRCC2 Arg188His polymorphism is not directly associated with breast cancer risk: evidence from 37,369 subjects. Breast Cancer Res Treat (2010) 123(1):219–25. doi: 10.1007/s10549-010-0753-y
87. Kipen VN, Melnov SB, Smolyakova RM. The role of the XRCC1, XRCC3, and PALB2 genes in the genesis of sporadic breast cancer. Russian J Genetics: Appl Res (2017) 7(6):705–11. doi: 10.1134/S2079059717060090
88. Angelini S, Kumar R, Carbone F, Maffei F, Forti GC, Violante FS, et al. Micronuclei in humans induced by exposure to low level of ionizing radiation: influence of polymorphisms in DNA repair genes. Mutat Res (2005) 570(1):105–17. doi: 10.1016/j.mrfmmm.2004.10.007
Keywords: breast neoplasms, Rad51 recombinase, single nucleotide polymorphism, DNA repair mechanism mutations, meta-analysis
Citation: Yu J and Wang C-G (2023) Relationship between polymorphisms in homologous recombination repair genes RAD51 G172T、XRCC2 & XRCC3 and risk of breast cancer: A meta-analysis. Front. Oncol. 13:1047336. doi: 10.3389/fonc.2023.1047336
Received: 18 September 2022; Accepted: 09 January 2023;
Published: 24 January 2023.
Edited by:
Anna Diana, Ospedale del Mare, ItalyReviewed by:
Juan Zhang, Zhuzhou Central Hospital, ChinaEwa Grzybowska, Maria Skoldowska-Curie National Research Institute of Oncology, Poland
Copyright © 2023 Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Guang Wang, MTc4MzA4NDIzNkBxcS5jb20=
 Jiayang Yu
Jiayang Yu Chun-Guang Wang*
Chun-Guang Wang*