- Department of Respiratory and Critical Care Medicine, the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
Background: Our study aimed to explore the potential association of CYP4F2 gene polymorphisms with lung cancer (LC) risk.
Methods: The five variants in CYP4F2 were genotyped using Agena MassARRAY in 507 cases and 505 controls. Genetic models and haplotypes based on logistic regression analysis were used to evaluate the potential association between CYP4F2 polymorphisms and LC susceptibility.
Results: This study observed that rs12459936 was linked to an increased risk of LC in no-smoking participants (allele: OR = 1.38, p = 0.035; homozygote: OR = 2.00, p = 0.035; additive: OR = 1.40, p = 0.034) and females (allele: OR = 1.64, p = 0.002; homozygote: OR = 2.57, p = 0.006; heterozygous: OR = 2.56, p = 0.001; dominant: OR = 2.56, p < 0.002; additive: OR = 1.67, p = 0.002). Adversely, there was a significantly decreased LC risk for rs3093110 in no-smoking participants (heterozygous: OR = 0.56, p = 0.027; dominant: OR = 0.58, p = 0.035), rs3093193 (allele: OR = 0.66, p = 0.016; homozygote: OR = 0.33, p = 0.011; recessive: OR = 0.38, p = 0.021; additive: OR = 0.64, p = 0.014), rs3093144 (recessive: OR = 0.20, p = 0.045), and rs3093110 (allele: OR = 0.54, p = 0.010; heterozygous: OR = 0.50, p = 0.014; dominant: OR = 0.49, p = 0.010; additive: OR = 0.54, p = 0.011) in females.
Conclusions: The study demonstrated that CYP4F2 variants were associated with LC susceptibility, with evidence suggesting that this connection may be affected by gender and smoking status.
Introduction
Lung cancer (LC) has been regarded as one of the most common causes of cancer-related death worldwide over the past few decades, with an estimated 2.1 million new diagnoses of LC in 2018, accounting for 12% of the total increase in cancer cases (1). In recent years, the incidence of LC in China has been consistent with the global trend, showing a rapid increase, and LC has since become the main cause of cancer-related deaths in China (2). It is predicted that the mortality of LC in China is likely to increase by about 40% between 2015 and 2030 (3). Despite advances in early detection, the majority of LC patients are often diagnosed at a later stage, resulting in a 5-year overall survival rate of only 10% to 15%, according to statistics (4). The burden of LC on our society is increasing day by day and cannot be ignored. Various factors can predispose people to LC, with smoking being the most prevalent factor. In addition, other potential risk factors include gender, age, race, ethnicity, and especially single nucleotide polymorphisms (SNPs) (5, 6).
Cytochrome P450s (CYP), phase I drug metabolizing enzymes, encode 57 CYP proteins in the human genome and are responsible for the metabolism of numerous endogenous and xenobiotic compounds (7). The CYP4F2 gene, a member of the CYP450 superfamily, is an ω-hydroxylase that catalyzes the first step of the vitamin E metabolic pathway (8), as well as the metabolism of arachidonic acid (AA) to generate 20-hydroxyethyl hexadecanoic acid (20-HETE) through ω-hydroxylation (9). 20-HETE is known to promote tumorigenesis by increasing a variety of pro-inflammatory mediators, cytokines, and chemokines. Previous studies have demonstrated that the elevated expression of CYP4F2 enzymes and 20-HETE is closely related to ovarian cancer (10). We hypothesized that CYP4F2 might be involved in tumor genesis and development by accelerating the production of 20-HETE. Additionally, Geng et al. have proved that rs1558139 and rs2108622 of CYP4F2 are associated with hypertension, and the association between rs1558139 and hypertension is particularly strong in men (11). Despite this, there is a lack of studies investigating the association between CYP4F2 polymorphisms and LC risk.
In this case–control study, five SNPs (rs3093203, rs3093144, rs12459936, rs3093110, and rs3093193) in CYP4F2 were genotyped by the Agena MassARRAY platform. The gender- and smoking-stratified analyses on the correlation between CYP4F2 variants and LC risk were performed.
Materials and methods
Study subjects
A total of 507 newly diagnosed LC patients (353 males and 154 females) were randomly recruited from the Second Affiliated Hospital of Xi’an Jiaotong University in the case–control association analysis between CYP4F2 polymorphisms and the risk of LC. All patients had no history of any other cancers and had not received chemotherapy before acquiring blood samples. Further, the control group comprised 505 unrelated healthy controls (354 males and 151 females) from the physical examination center of the hospital. Information about all subjects, including age, gender, height (cm), weight (kg), smoking status, drinking status, tumor stage, and lymph node metastasis, was collected from questionnaires and clinical data. Peripheral blood samples were collected from all study subjects into vacutainer tubes containing EDTA, and genomic DNA was then isolated from the collected blood samples using the GoldMag-Mini Purification Kit (GoldMag Co. Ltd., Xi’an, China) and stored at −80°C. DNA concentration and purity were determined by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
SNP selection and genotyping
In this study, five SNPs (rs3093203, rs3093144, rs12459936, rs3093110, and rs3093193) in CYP4F2 were selected according to previously published studies on the association between CYP4F2 polymorphisms and disease susceptibility (12–14). The genotype distributions of the candidate SNPs in controls met Hardy–Weinberg equilibrium (HWE) (p >0.05). All the candidate SNPs had a minor allele frequency (MAF) of >5% in the Han Chinese in Beijing (CHB) population from the 1,000 Genomes Project (http://www.internationalgenome.org/). The primers for five SNPs were designed by Agena Bioscience Assay Design Suite version 2.0 software. The polymorphisms were genotyped using the Agena MassARRAY platform (Agena Bioscience, San Diego, CA, USA) with iPLEX gold chemistry. Ultimately, Agena Bioscience TYPER version 4.0 software was used for data management and genotyping result analysis.
Expression analysis
We extracted the data for CYP4F2 expression in normal lung tissues and lung squamous cell carcinoma (LUSC) tissues under different subgroups from the TCGA database and analyzed them via UALCAN (http://ualcan.path.uab.edu/index.html), which is an interactive web resource for tumor subgroup gene expression analysis and survival analysis.
Statistical analysis
SPASS version 22.0 software was applied for statistical analysis. HWE was calculated for the control group by the chi-square test. Differences in the continuous characteristic (age) and categorical variable (gender) between patients with LC and controls were measured by the student’s t-test and Pearson Chi-Square test, respectively. The correlation between CYP4F2 variants and LC susceptibility was evaluated by logistic regression analysis adjusted for age and gender using PLINK software (version 1.07) under multiple genetic models (allele, genotype, dominant, recessive, and additive). Odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the relationship between CYP4F2 SNPs and LC risk (OR = 1: no impact; OR <1: protective factor; OR >1: risk factor). Finally, PLINK (version 1.07) and Haploview (version 4.2) softwares were used to analyze the pairwise linkage disequilibrium (LD) among five SNPs and generate an LD map to observe the linkage degree among them based on D’ and r-squared values. The SNPStats software (https://www.snpstats.net/start.htm) was used to estimate the correlation between CYP4F2 haplotypes and LC risk. In our study, the p-values of all tests were two-sided, and p <0.05 was considered statistically significant.
Results
Participant characteristics
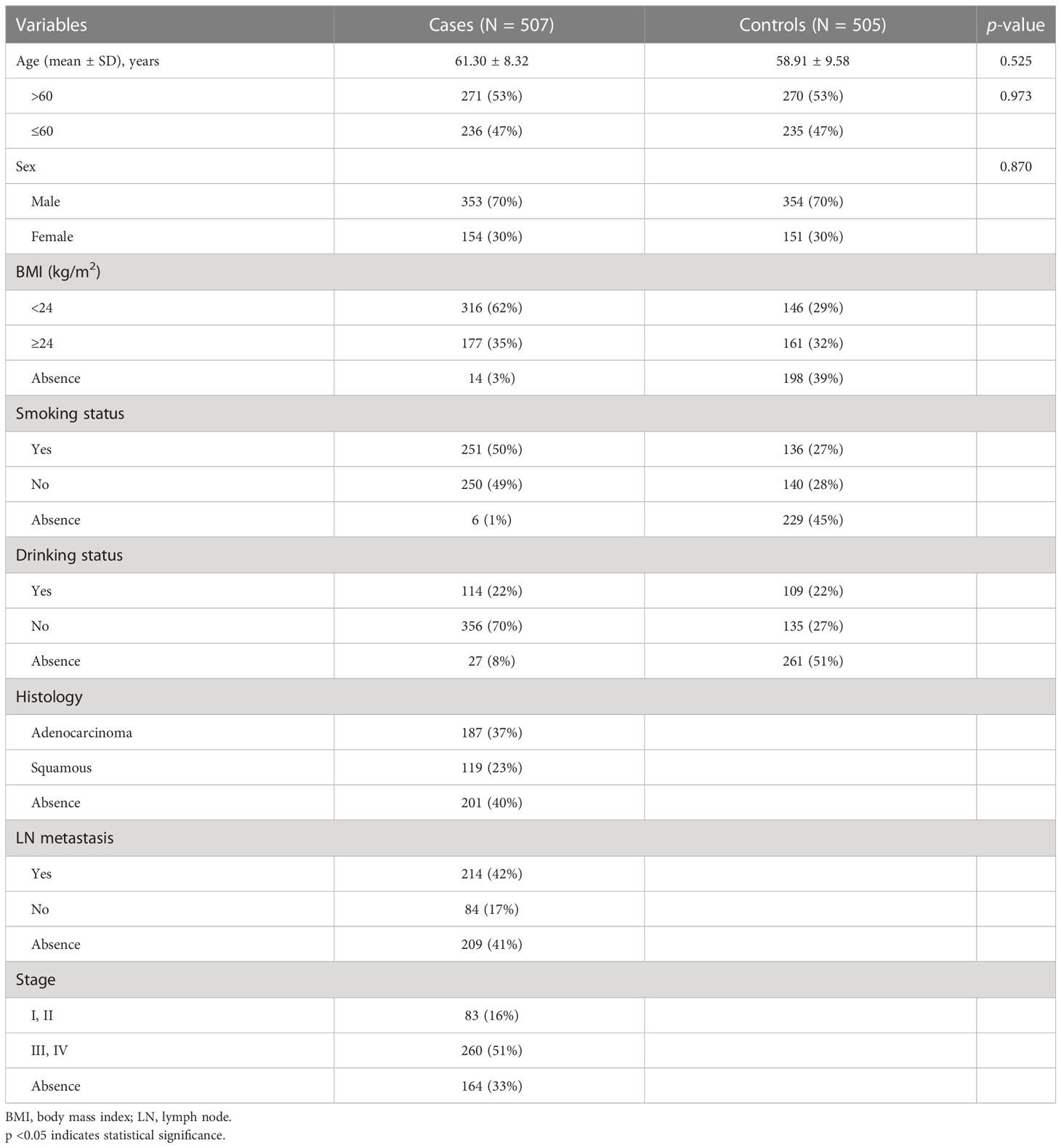
The mean ages of 507 LC patients and 505 unrelated healthy controls were 61.30 ± 8.32 years and 58.91 ± 9.58 years, respectively (Table 1). In our study, there were no statistically significant differences in age (p = 0.525) and gender (p = 0.870) distribution between cases and controls.
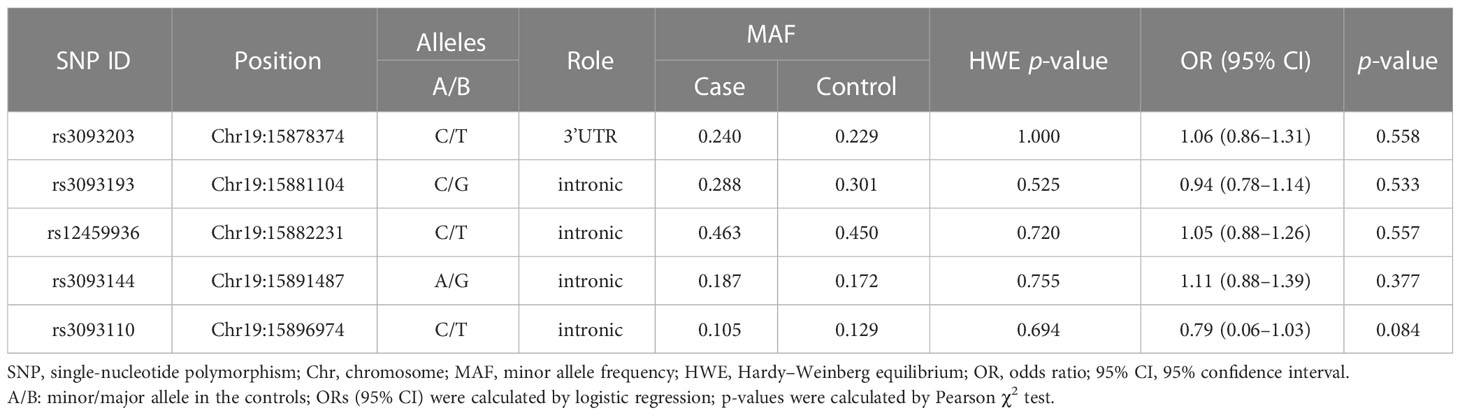
Basic information about the selected SNPs in CYP4F2
The basic information about the five SNPs in CYP4F2 (rs3093203, rs3093144, rs12459936, rs3093110, and rs3093193) among cases and controls was displayed (Table 2), including gene, SNP ID, position, alleles, HWE, and OR (95% CI). The five SNPs in controls were in accordance with HWE (p >0.05). We further evaluated the association between the five SNPs and LC susceptibility by logistic regression (Table 2). The four genetic models (genotype, dominant, recessive, and additive) were also applied to analyze the association by logistic regression adjusted for age and gender (Table S1). Unfortunately, there was no significant association between these five SNPs in CYP4F2 and LC susceptibility under the allelic and genetic models.
Stratification analysis by smoking status
The smoking-stratified analysis (Table 3) was performed to examine the relationship between CYP4F2 variants and LC risk. Our results showed that rs12459936 in CYP4F2 was associated with an increased risk of LC in no-smoking individuals under the allele (T vs. C: OR = 1.38, 95% CI: 1.02–1.85, p = 0.035), genotype (TT vs. CC: OR = 2.00, 95% CI: 1.05–3.82, p = 0.035), and additive (OR = 1.40, 95% CI: 1.03–1.92, p = 0.034) models. On the contrary, rs3093110 was found to have a protective effect against LC risk in no-smoking individuals under the genotype (GA vs. AA: OR = 0.56, 95% CI: 0.33–0.94, p = 0.027) and dominant (GG + GA vs. AA: OR = 0.58, 95% CI: 0.35–0.96, p = 0.035) models.
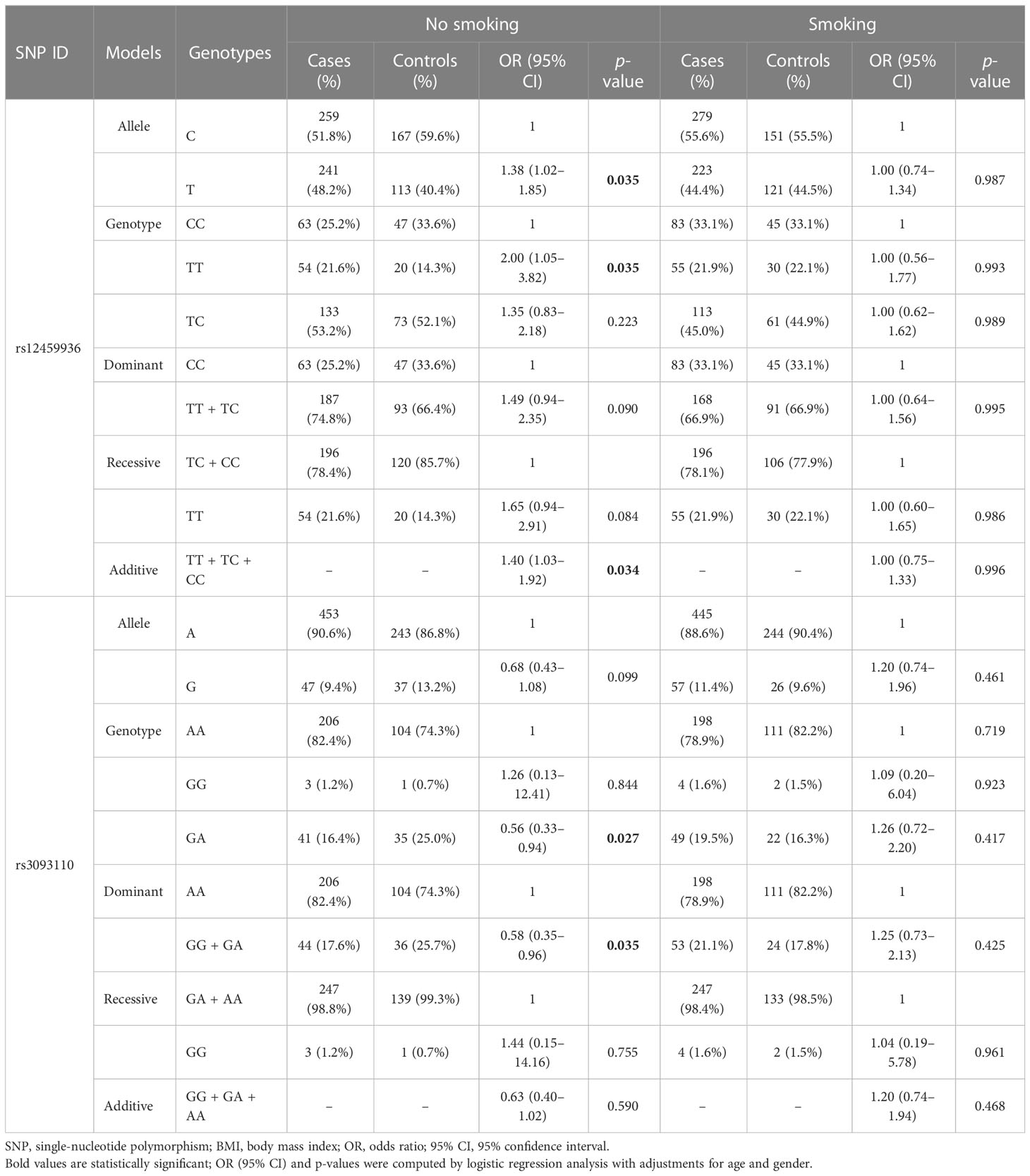
Table 3 The association of variants in CYP4F2 with lung cancer susceptibility stratified by smoking status.
Stratification analysis by gender
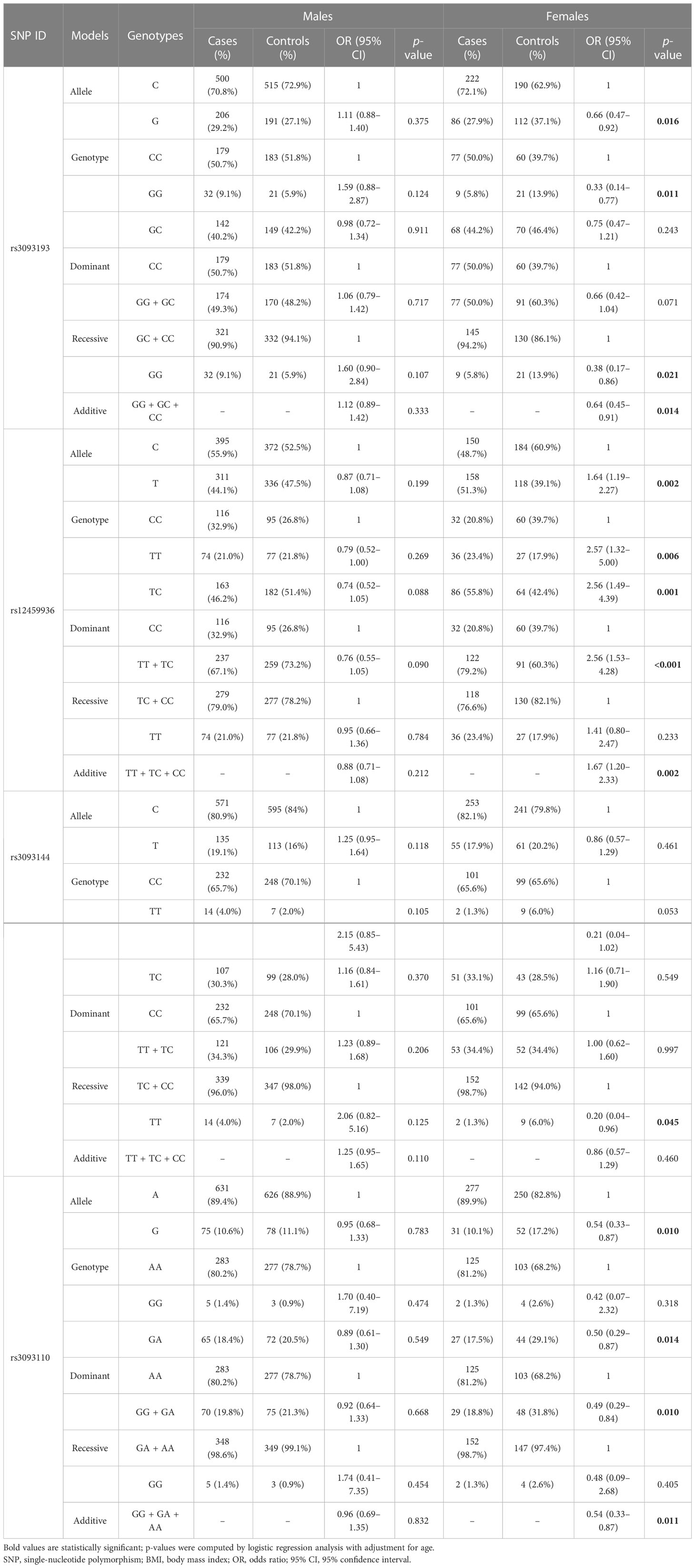
In addition, the analysis stratified by gender (Table 4) demonstrated that rs3093193 (G vs. C: OR = 0.66, 95% CI: 0.47–0.92, p = 0.016; GG vs. CC: OR = 0.33, 95% CI: 0.14–0.77, p = 0.011; GG vs. GC + CC: OR = 0.38, 95% CI: 0.17–0.86, p = 0.021; additive: OR = 0.64, 95% CI: 0.45–0.91, p = 0.014) was related to a decreased risk of LC in females. Rs3093144 in the recessive model (TT vs. TC + CC: OR = 0.20, 95% CI: 0.04–0.96, p = 0.045) and rs3093110 in the allele, genotype, dominant, and additive models (G vs. A: OR = 0.54, 95% CI: 0.33–0.87, p = 0.010; GA vs. AA: OR = 0.50, 95% CI: 0.29–0.87, p = 0.014; GG + GA vs. AA: OR = 0.49, 95% CI: 0.29–0.84, p = 0.010; additive: OR = 0.54, 95% CI: 0.33–0.87, p = 0.011) showed a protective effect on LC in females. However, the CYP4F2 rs12459936 was associated with an increased risk of LC in females under the allele, genotype, dominant, and additive models (T vs. C: OR = 1.64, 95% CI: 1.19–2.27, p = 0.002; TT vs.CC: OR = 2.57, 95% CI: 1.49–4.39, p = 0.006; TC vs.CC: OR = 2.56, 95% CI: 1.49–4.39, p = 0.001; TT + TC vs.CC: OR = 2.56, 95% CI: 1.53–4.28, p <0.001; additive: OR = 1.67, 95% CI: 1.20–2.33, p = 0.002).
Haplotype analysis
Finally, the results of haplotype analysis indicated a strong 18-kb LD block among the five SNPs (rs3093203, rs3093193, rs12459936, rs3093144, and rs3093110) (Figure 1 and Table S2). Compared with haplotype “GCTCA,” haplotypes “GGCTA” (OR = 0.63, 95% CI: 0.40–1.00, p = 0.048) and “GGCCG” (OR = 0.46, 95% CI: 0.27–0.78, p = 0.004) were associated with a decreased risk of LC in females (Table 5). For non-smokers, the haplotype “GGCCG” (OR = 0.54, 95% CI: 0.32–0.90, p = 0.046) was also associated with decreased susceptibility to LC (Table 5).
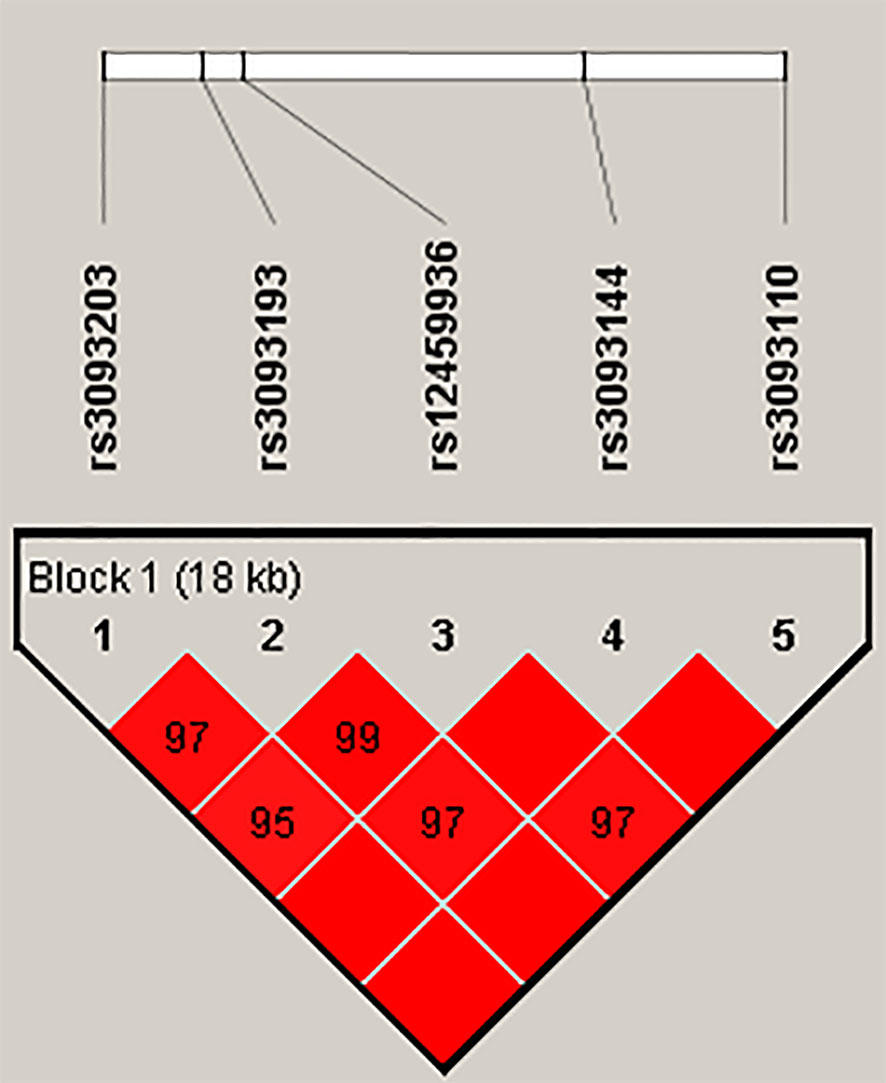
Figure 1 Haplotype block map for SNPs in the CYP4F2 gene. The numbers inside the diamonds indicate the D′ value × 100 for pairwise analyses.
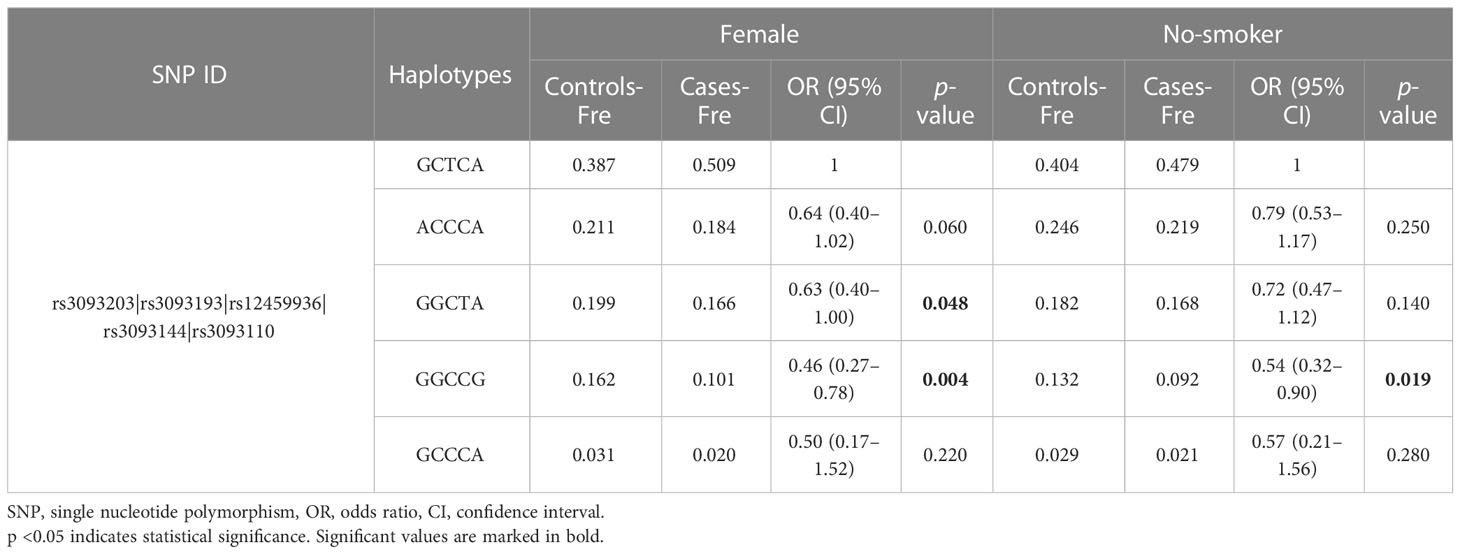
Table 5 The frequency of CYP4F2 haplotypes and their association with the risk of lung cancer in subgroups.
Bioinformatics analysis of CYP4F2 expression in LC
The analysis of the expression level of CYF4F2 in normal and LUSC tissues and its effect on the survival of these patients was conducted using UALCAN online analysis software based on the TCGA database, as shown in Figure 2. We observed that the expression level of CYP4F2 was significantly different between normal and LUSC tissues (p <0.001). In addition, the expression level of CYP4F2 was higher in non-smoking LUSC patients than in normal and smoking ones (p <0.001). The expression level was higher in males than in females (p <0.001). Moreover, a high expression level of CYP4F2 was found to be significantly related to the poor prognosis of non-smoking LUSC patients (p = 0.033).
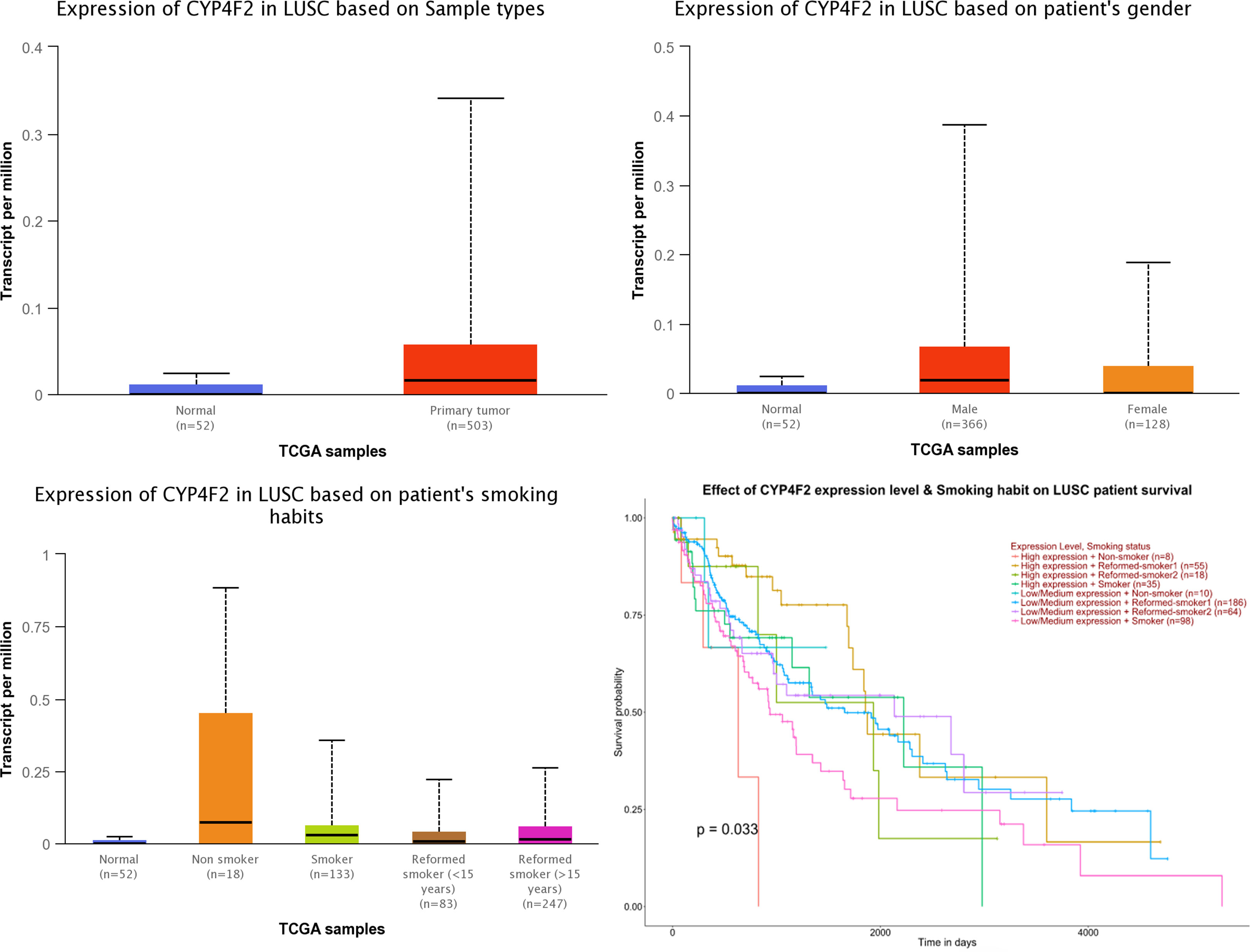
Figure 2 The expression of CYP4F2 in normal lung squamous cell carcinoma tissues based on different types.
Discussion
In our study, the connection between five variants in CYP4F2 and LC risk in the Chinese Han population was detected. Association analyses revealed that CYP4F2 rs12459936 increased susceptibility to LC in non-smoking individuals and females. In contrast, rs3093110 showed a protective effect on LC susceptibility in non-smoking groups and females. The two SNPs (rs3093193 and rs3093144) were also associated with a decreased risk of LC in females.
The CYP4F2 gene, a member of the CYP450 superfamily, located on chromosome 19p13.12, has been shown to be expressed at higher levels in certain types of cancerous tissues, such as the thyroid, ovarian, breast, and colon (10). Eun et al. have confirmed that low expression of CYP4F2 may contribute to the progression of hepatocellular carcinoma (HCC) and decrease survival rates due to its involvement in various metabolic pathways (15). A similar study showed that CYP4F2 expression was higher in pancreatic ductal adenocarcinoma (PDA) patients than in normal ones and negatively correlated with age (16). Database prediction found that CYP4F2 was highly expressed in lung cancer tissues. The expression of CYP4F2 was higher in men than women and higher in non-smokers than smokers. Additionally, Xu et al. have reported that CYP4F generates 20-HETE by catalyzing ω-hydroxylation of arachidonic acid (17). According to previous studies, 20-HETE plays a significant role in tumor progression. Colombero et al. have demonstrated that HET0016, a selective inhibitor of 20-HETE synthesis, can reduce the proliferation of prostate cancer (18), while another study has revealed that the antagonist of 20-HETE, WIT002, is able to inhibit tumor growth in a renal cell carcinoma cell line (19). This suggests that CYP4F2 polymorphisms may be related to susceptibility to LC by affecting the metabolism of 20-HETE, although further verification is required. Studies have also indicated a significant association between CYP4F2 polymorphisms and a variety of diseases, including ischemic stroke and various other cardiovascular and cerebrovascular diseases (12, 20).
Our study focused on the association between CYP4F2 polymorphisms and susceptibility to LC. Five sites were selected for statistical analyses: rs3093203, rs3093193, rs12459936, rs3093144, and rs3093110. However, none of these loci were found to be significantly associated with LC susceptibility under the allelic model or any of the five genetic models. The actual increase in LC risk may be underestimated due to the limited sample size. To further examine the potential influence of LC, we conducted a stratified analysis. Tobacco has long been recognized as an independent risk factor for tumorigenesis, as it contains many carcinogens, such as nitrosamines, polycyclic aromatic hydrocarbons, and volatile organic compounds (21). However, our analysis stratified by smoking revealed that the rs12459936 and rs3093110 loci were significantly associated with increased susceptibility to LC in the non-smoking population but not in the smoking population.
In addition, gender has been found to have a notable impact on the toxicity of therapeutic treatments and the response to them in many types of cancer. The underlying cause of this difference is likely related to a complex interplay of several factors, including sex hormones, which have been shown to affect the self-renewal of tumor stem cells, the tumor microenvironment, the immune system, and metabolism (22). It is well established that there are considerable differences in the immune system between men and women. In general, women have a stronger immune system than men, leading to distinct sex-based differences in both innate and adaptive immune responses. These disparities in immune systems likely play a role in cancer susceptibility between males and females (23). In our study, analysis stratified by gender was performed, and we found that rs309319, rs12459936, and rs3093110 all had a protective role against LC in females.
Taken together, our study observed that variants in CYP4F2 were associated with LC susceptibility. However, our research had some limitations. First, the potential functional implications of CYP4F2 polymorphisms were not addressed in this study. The expression data for CYP4F2 in LC cases were sourced from the database. To properly elucidate the genetic mechanism of CYP4F2 in LC, expression analysis of CYP4F2 mRNA and annotation of the functional significance of variants are necessary. Second, the sample size was relatively small. In the following steps, we will perfect this information and expand the sample size to explore the molecular mechanism of CYP4F2 polymorphisms affecting the development of LC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
Our study complied with the Declaration of Helsinki, and the protocol in our experience was approved by the Second Affiliated Hospital of Xi’an Jiaotong University. All participants have been informed and provided written informed consent for the study.
Author contributions
HS designed this study and drafted the manuscript. YZ performed the DNA extraction and genotyping. YW revised the manuscript and performed the data analysis. PF and YL performed the samples collection and information recording. HS conceived and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Basic Research Plan in Shaanxi Province of China (2020JM-410) and the hospital fund of the Second Affiliated Hospital of Xi’an Jiaotong University (RC(GG)201809).
Acknowledgments
We thank all volunteers in the study. We would also like to thank the clinicians and hospital staff who contributed to the collection of samples and data for our study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1114218/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA: Cancer J Clin (2016) 66:115–32. doi: 10.3322/caac.21338
3. Martín-Sánchez JC, Lunet N, González-Marrón A, Lidón-Moyano C, Matilla-Santander N, Clèries R, et al. Projections in breast and lung cancer mortality among women: A Bayesian analysis of 52 countries worldwide. Cancer Res (2018) 78:4436–42. doi: 10.1158/0008-5472.Can-18-0187
4. Cagle PT, Allen TC and Olsen RJ. Lung cancer biomarkers: Present status and future developments. Arch Pathol Lab Med (2013) 137:1191–8. doi: 10.5858/arpa.2013-0319-CR
5. O'Keeffe LM, Taylor G, Huxley RR, Mitchell P, Woodward M, Peters SAE. Smoking as a risk factor for lung cancer in women and men: A systematic review and meta-analysis. BMJ Open (2018) 8:e021611. doi: 10.1136/bmjopen-2018-021611
6. Siegel RL, Miller KD and Jemal A. Cancer statistics, 2019. CA: Cancer J Clin (2019) 69:7–34. doi: 10.3322/caac.21551
7. Drolet B, Pilote S, Gélinas C, Kamaliza AD, Blais-Boilard A, Virgili J, et al. Altered protein expression of cardiac CYP2J and hepatic CYP2C, CYP4A, and CYP4F in a mouse model of type II diabetes-a link in the onset and development of cardiovascular disease? Pharmaceutics (2017) 9:44. doi: 10.3390/pharmaceutics9040044
8. Sontag TJ, Parker RS. Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. novel mechanism of regulation of vitamin e status. J Biol Chem (2002) 277:25290–6. doi: 10.1074/jbc.M201466200
9. Zhang C, Booz GW, Yu Q, He X, Wang S, Fan F. Conflicting roles of 20-HETE in hypertension and renal end organ damage. Eur J Pharmacol (2018) 833:190–200. doi: 10.1016/j.ejphar.2018.06.010
10. Alexanian A, Miller B, Roman RJ, Sorokin A. 20-HETE-producing enzymes are up-regulated in human cancers. Cancer Genomics Proteomics (2012) 9:163–9.
11. Geng H, Li B, Wang Y, Wang L. Association between the CYP4F2 gene rs1558139 and rs2108622 polymorphisms and hypertension: A meta-analysis. Genet Test Mol Biomarkers (2019) 23:342–7. doi: 10.1089/gtmb.2018.0202
12. Wu Y, Zhao J, Zhao Y, Huang T, Ma X, Pang H, et al. Genetic variants in CYP4F2 were significantly correlated with susceptibility to ischemic stroke. BMC Med Genet (2019) 20:155. doi: 10.1186/s12881-019-0888-6
13. Ding Y, Yang Y, Li Q, Feng Q, Xu D, Wu C, et al. The correlation between CYP4F2 variants and chronic obstructive pulmonary disease risk in hainan han population. Respir Res (2020) 21:86. doi: 10.1186/s12931-020-01348-6
14. Si L, Wang H, Wang R, Tsering L, Long Q, Jiang Y, et al. Suggestive evidence of CYP4F2 gene polymorphisms with HAPE susceptibility in the Chinese han population. PloS One (2023) 18:e0280136. doi: 10.1371/journal.pone.0280136
15. Eun HS, Cho SY, Lee BS, Seong IO, Kim KH Profiling cytochrome P450 family 4 gene expression in human hepatocellular carcinoma. Mol Med Rep (2018) 18:4865–76. doi: 10.3892/mmr.2018.9526
16. Gandhi AV, Saxena S, Relles D, Sarosiek K, Kang CY, Chipitsyna G, et al. Differential expression of cytochrome P450 omega-hydroxylase isoforms and their association with clinicopathological features in pancreatic ductal adenocarcinoma. Ann Surg Oncol (2013) 20(Suppl 3):S636–643. doi: 10.1245/s10434-013-3128-x
17. Kroetz DL, Xu F. Regulation and inhibition of arachidonic acid omega-hydroxylases and 20-HETE formation. Annu Rev Pharmacol Toxicol (2005) 45:413–38. doi: 10.1146/annurev.pharmtox.45.120403.100045
18. Colombero C, Papademetrio D, Sacca P, Mormandi E, Alvarez E, Nowicki S. Role of 20-hydroxyeicosatetraenoic acid (20-HETE) in androgen-mediated cell viability in prostate cancer cells. Horm Cancer (2017) 8:243–56. doi: 10.1007/s12672-017-0299-0
19. Alexanian A, Rufanova VA, Miller B, Flasch A, Roman RJ, Sorokin A. Down-regulation of 20-HETE synthesis and signaling inhibits renal adenocarcinoma cell proliferation and tumor growth. Anticancer Res (2009) 29:3819–24.
20. Zhang T, Yu K and Li X. Cytochrome P450 family 4 subfamily f member 2 (CYP4F2) rs1558139, rs2108622 polymorphisms and susceptibility to several cardiovascular and cerebrovascular diseases. BMC Cardiovasc Disord (2018) 18:29. doi: 10.1186/s12872-018-0763-y
21. Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, et al. A review of human carcinogens–part e: Tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol (2009) 10:1033–4. doi: 10.1016/s1470-2045(09)70326-2
22. Dart A. Sexual dimorphism in cancer. Nat Rev Cancer (2020) 20:627. doi: 10.1038/s41568-020-00304-2
Keywords: CYP4F2, lung cancer, single nucleotide polymorphisms, smoking, gender
Citation: Shi H, Zhang Y, Wang Y, Fang P and Liu Y (2023) Genetic polymorphisms in CYP4F2 may be associated with lung cancer risk among females and no-smoking Chinese population. Front. Oncol. 13:1114218. doi: 10.3389/fonc.2023.1114218
Received: 02 December 2022; Accepted: 21 February 2023;
Published: 14 March 2023.
Edited by:
Dana Kristjansson, Norwegian Institute of Public Health (NIPH), NorwayReviewed by:
Guangzhao Qi, First Affiliated Hospital of Zhengzhou University, ChinaLucia Guadalupe Taja Chayeb, National Institute of Cancerology (INCAN), Mexico
Copyright © 2023 Shi, Zhang, Wang, Fang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyang Shi, c2hpaHkyMDAzQDEyNi5jb20=
 Hongyang Shi
Hongyang Shi Yonghong Zhang
Yonghong Zhang