- 1School of Medicine, University of Electronic Science and Technology of China, Chengdu, China
- 2Department of Radiation Oncology, Radiation Oncology Key Laboratory of Sichuan Province, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Center, Sichuan Cancer Hospital & Institute, Affiliated Cancer Hospital of University of Electronic Science and Technology of China, Chengdu, China
- 3School of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, China
Background: The intratumoral microbiota has attracted considerable interest in carcinogenesis, progression, and treatment owing to advancements in sequencing technology. This systematic review provides a comprehensive overview of the current literature regarding the diversity and compositional characteristics of the intratumoral microbiota in women’s cancers. Additionally, it also explores potential associations among intratumoral microbiota, estrogen, and anti-tumor therapies.
Methods: A comprehensive literature search was conducted using PubMed, Embase, Web of Science, and the Cochrane Library from their inception to May 1, 2024. The review protocol was pre-registered in PROSPERO (CRD 42024601213). Articles were assessed utilizing the Newcastle-Ottawa Scale (NOS). To estimate the effect size and variability in microbial diversity changes, the standardized mean difference (SMD) and 95% confidence intervals (CIs) were employed. The systematic review adhered to PRISMA reporting guidelines, and meta-analyses were performed using Review Manager version 5.4.
Results: This systematic review included 29 of 8,291 studies after a thorough screening process. Of the 22 studies investigating α-diversity in women’s cancers, disease-free controls, and those with benign conditions, notable changes in diversity indices were observed. Compared to adjacent normal tissues, the Simpson index significantly decreased in breast cancer (SMD = -0.75, 95% CI: [-0.94, -0.55]) and endometrial cancer (SMD = -0.83, 95% CI: [-1.37, -0.28]). The Chao1 index was reduced in endometrial cancer tumor tissues relative to normal tissues (SMD = -2.25, 95% CI: [-3.13, -1.36]), while the Shannon index decreased in ovarian cancer tumor tissues (SMD = -0.61, 95% CI: [-1.18, -0.04]). In comparisons between tumor and benign tissues, the Chao1 index was decreased (SMD = -0.64, 95% CI: [-1.20, -0.08], I² = 0%), while the Simpson index was increased (SMD = 0.36, 95% CI: [0.01, 0.71], I² = 0%) in patients with ovarian cancer. Other microbial diversity indices showed no significant differences between tumor and non-tumor tissues. At the phylum level, Fusobacteriota were enriched in tumor tissues, while Firmicutes and Actinobacteria predominated in non-tumor tissues. At the genus level, Pseudomonas, Porphyromonas, Atopobium, Peptoniphilus, and Acinetobacter were consistently more abundant in cancerous tissues. Microbial alterations were also linked to estrogen receptor (ER) status, with Alkanindiges negatively correlated with ER status in two studies. Furthermore, one study on the effect of antineoplastic therapy indicated that neoadjuvant chemotherapy reduced microbial diversity in breast cancer patients (n = 15 vs. n = 18) (Shannon index: SMD = -0.95, 95% CI: [-1.68, -0.22]).
Conclusion: This study highlights significant differences in microbiota composition between tumor and non-tumor tissues in women’s cancers, emphasizing changes in intratumoral microbiota influenced by estrogen and antineoplastic treatments. Further research is needed to explore the potential for developing targeted therapies based on estrogen-driven microbiota alterations. Investigations may yield insights into the enhancement of female reproductive health and the improvement of treatment efficacy for female cancers.
Systematic review registration: https://www.crd.york.ac.uk/PROSPERO/view/CRD42024601213, identifier CRD 42024601213.
1 Introduction
The human microbiota, comprising a diverse community of microorganisms—including bacteria, fungi, viruses, and archaea—resides across various body sites and plays a crucial role in human health and disease. In particular, accumulating evidence has highlighted its significant involvement in cancer initiation, progression, and prognosis (1). Currently, considerable attention has been devoted to the gut, skin, oral, and vaginal microbiota due to their interactions with the host and therapeutic implications. With the discovery of the microbiota within tumor tissues, researchers are increasingly investing in the interaction between the intratumoral microbiota and cancer. Discoveries have identified microbiota within tumor tissues, which has sparked increasing interest in elucidating the interactions between the intratumoral microbiota and cancer. Mounting evidence indicates that the intratumoral microbiota may influence tumor development through several mechanisms, including DNA mutations, activation of oncogenic pathways, and promotion of chronic inflammation (2, 3). Additionally, intratumoral bacteria may modulate antitumor immunity by activating immune cells, triggering the STING signaling pathway, promoting the maturation of tertiary lymphoid structures (TLS), and presenting microbiota-derived antigens, thereby influencing cancer progression (4, 5).
Among women cancers—namely, breast, ovarian, cervical, and endometrial cancers—the role of the microbiota in tumor development has garnered increasing recognition. Certain bacterial taxa, such as Firmicutes and Proteobacteria, have been associated with immune escape and chemoresistance in breast cancer (BC) (6, 7). In ovarian cancer (OC), the intratumoral microbiota may also play a role in tumorigenesis by modulating hormone metabolism. In endometrial cancer (EC), the relative abundance of Micrococcus has been positively correlated with elevated levels of inflammatory cytokines, including IL-6 and IL-17 (8, 9). Furthermore, in cervical cancer (CC), Lactobacillus iners has been shown to induce resistance to chemotherapy and radiotherapy, potentially through lactic acid-mediated metabolic alterations (10).
Estrogen, a key sex hormone in women, plays a central role in regulating cell proliferation, differentiation, and apoptosis through its binding to intracellular estrogen receptors. Dysregulations in estrogen levels, metabolism, and receptor expression are implicated in cancer development. Notably, complex interactions between estrogen and the microbiota have emerged as critical factors in the pathophysiology of hormone-dependent women cancers. The microbiota can directly or indirectly modulate estrogen metabolism by altering systemic estrogen concentrations and the profile of its bioactive metabolites through mechanisms such as enterohepatic circulation and microbial enzymatic activity (e.g., β-glucuronidase). These alterations can, in turn, impact tumor development. Conversely, estrogen can shape the composition of the microbiota in gynecologic tumors, promoting the growth of Lactobacillus species and potentially affecting the tumor microenvironment and immune modulation. (11, 12).
Despite increasing recognition of the interconnected roles of women cancers, intratumoral microbiota, and estrogen, systematic evaluations of the intratumoral microbiota across the four major types of women cancers—breast, ovarian, endometrial, and cervical—remain scarce. To address this gap, we conducted a systematic review to evaluate the composition of the intratumoral microbiota in women cancers. This review aims to explore the diversity, taxonomic abundance, and estrogen-mediated alterations in the intratumoral microbiota, as well as its potential role in tumor progression and therapeutic responses. To visually summarize the conceptual framework of this study, Figure 1 illustrates the dynamic interplay between the intratumoral microbiota, estrogen, and the tumor microenvironment, highlighting our hypothesis that microbial–estrogen crosstalk plays a pivotal role in shaping tumor progression. Specifically, the diagram depicts (1) microbial modulation of estrogen metabolism and antitumor immunity, and (2) estrogen-driven feedback on microbial composition.
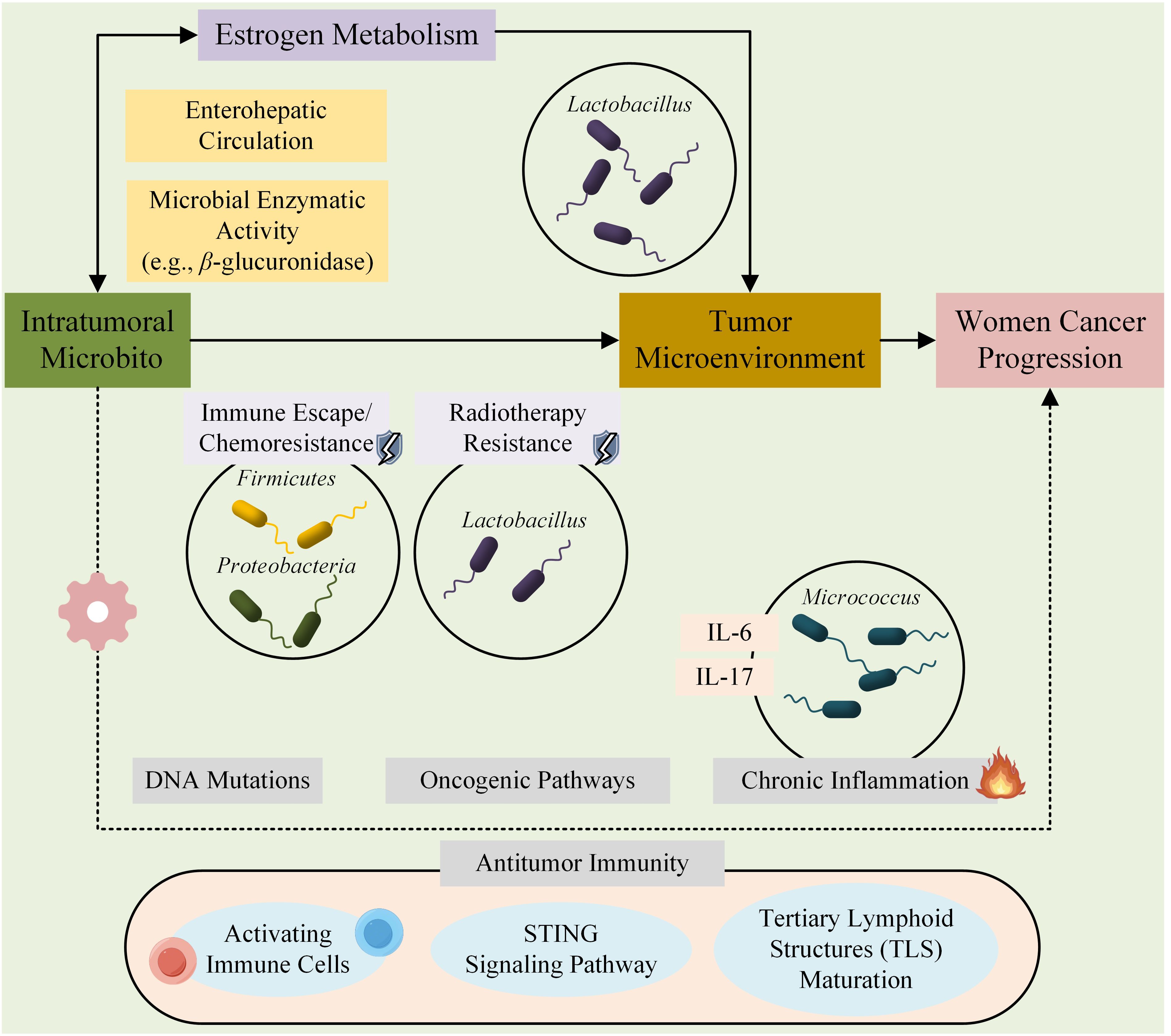
Figure 1. Proposed conceptual framework of the microbiota-estrogen-tumor microenvironment (TME) crosstalk in cancer progression. This schematic illustrates the complex and bidirectional interactions between the intratumoral microbiota, estrogen signaling, and components of the tumor microenvironment (TME). The microbiota can influence tumor development through multiple mechanisms, including modulation of estrogen metabolism via microbial enzymes (e.g., β-glucuronidase), induction of DNA damage, activation of oncogenic signaling pathways, and shaping of the immune landscape (e.g., through STING pathway activation, antigen presentation, and tertiary lymphoid structure formation). Conversely, estrogen may regulate the composition and function of the tumor-associated microbiota by promoting or inhibiting the growth of specific taxa (e.g., Lactobacillus spp.), thereby further influencing immune responses and tumor behavior. This tripartite interaction contributes to cancer initiation, progression, and treatment response in women’s cancers such as breast, ovarian, endometrial, and cervical cancers.
2 Methods
2.1 Search strategy
PubMed, Embase, Web of Science, and the Cochrane Library were searched from their inception to May 1, 2024, to identify the literature included in this systematic review. The search strategy is outlined in Supplementary Table S1. Search criteria were restricted to English-language publications, human studies, and those focused on women’s cancers. The systematic review was conducted in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) guidelines (13), and the review protocol was pre-registered with PROSPERO (CRD 42024601213).
2.2 Selection of articles and data extraction
Studies included in this review adhered to the following inclusion criteria: 1) adult women (over 18 years old) who have undergone tissue sampling for gynecological cancer or breast cancer; 2) use of high-throughput sequencing to evaluate the diversity and composition of intratumoral microbiota; 3) analysis of microbial alterations across different disease states or treatment periods. Studies were excluded based on the following criteria: 1) articles related to in vitro studies, case reports, review articles, letters to the editor, comments, protocols, conference abstracts, or guidelines; 2) duplicate studies or overlapping study populations; 3) absence of a healthy or benign control group, or failure to evaluate microbiota composition.
Data from the selected studies were extracted as follows: study characteristics (first author’s name, publication year, study duration, country, participants, nationality, age, BMI, sample type, and estrogen receptor status or estrogen levels), sequence characteristics (sequencing technology, amplification region, and sequencing platform), and outcomes. This review primarily examines alterations in α-diversity, β-diversity, and relative abundance across various disease states, as well as the impact of antineoplastic therapy on the tumor microbiota.
2.3 Quality assessment
The Newcastle-Ottawa Scale (NOS) was employed to assess the quality of the included studies (14). The scale evaluates three key domains: selection, comparability, and exposure of cases and controls, yielding a maximum score of 9. Studies with scores below 6 were excluded from this systematic review. All included studies were independently assessed by two researchers, and any discrepancies were resolved through group consensus.
2.4 Data analysis
Estimated data from images were extracted using WebPlotDigitizer (15), and the estimated analysis of means (M) and standard deviations (SD) was calculated using an online calculator (http://www.math.hkbu.edu.hk/~tongt/papers/median2mean.html) (16). Data analysis was performed using Review Manager version 5.4. For continuous variables, standardized mean difference (SMD) and 95% confidence intervals (CI) were used to calculate effect sizes and represent variation in microbial diversity across studies. Heterogeneity was assessed using the I2 statistic, except in cases where only a single study was included, in which heterogeneity was not evaluated. An I2 value greater than 75% was considered to indicate substantial heterogeneity. In the presence of moderate to high heterogeneity (I² > 50%), a random-effects model was employed to obtain a more conservative and generalizable pooled estimate. When heterogeneity was low (I²< 50%), a fixed-effects model was considered. Given the overall heterogeneity observed among studies, a random-effects model was consistently applied throughout the analysis. Subgroup analyses stratified by tumor type were performed to explore potential sources of heterogeneity. Publication bias was assessed using funnel plots.
3 Results
3.1 Characteristics of included studies
In total, 8,291 records were retrieved from the following databases: Web of Science (4,010), PubMed (4,190), Embase (51), and Cochrane Library (40). After a thorough examination of the titles, abstracts, and full texts, duplicates, irrelevant publications, and those containing inappropriate research content were excluded, resulting in the inclusion of 29 articles in the systematic review (Figure 2). Nineteen articles focused on breast cancer, three on ovarian cancer, six on endometrial cancer, and one on cervical cancer.
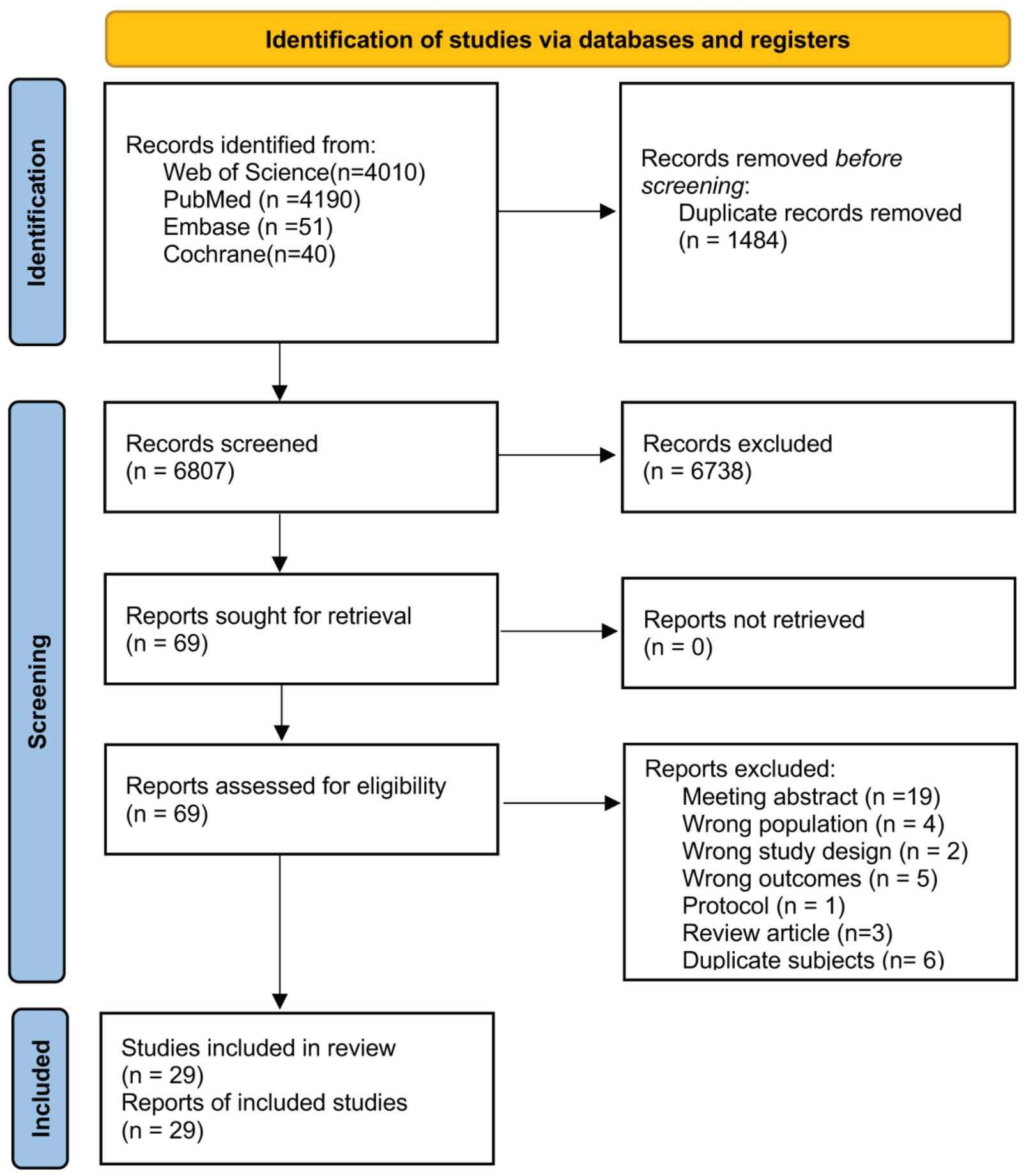
Figure 2. Flow diagram of the study selection process following the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines.
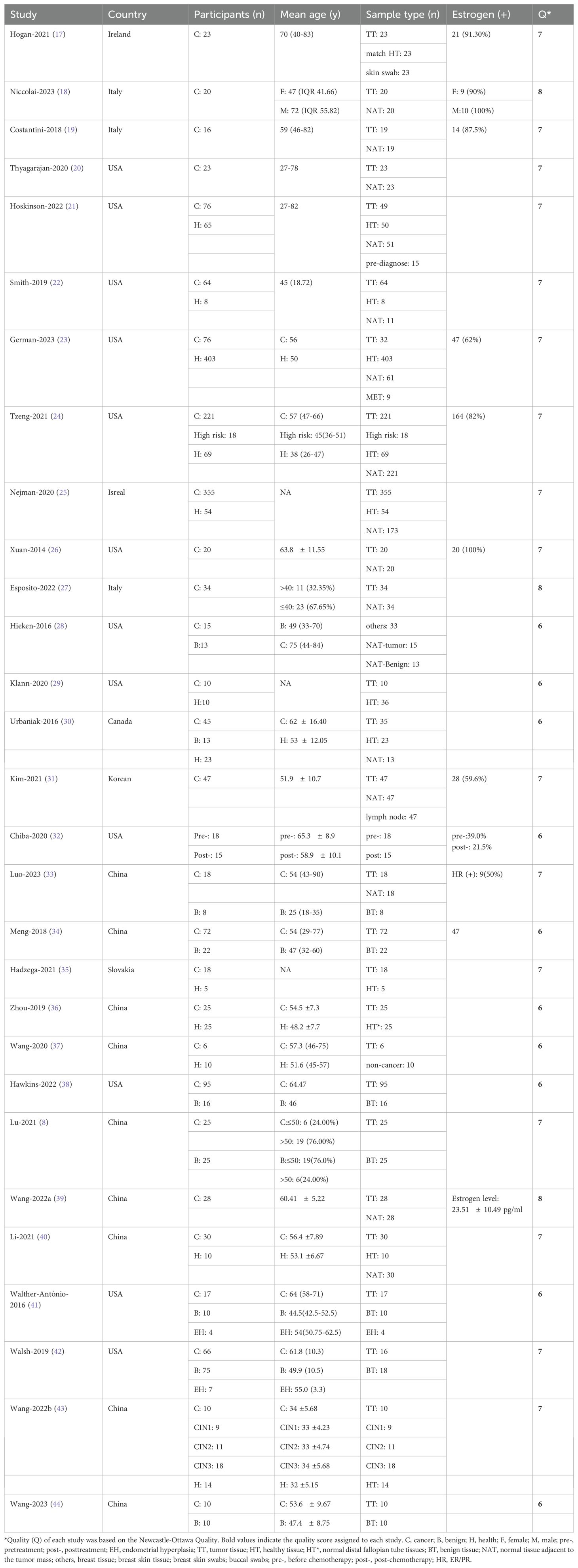
The characteristics of the included studies are summarized in Table 1. Thirteen studies were conducted in North America (12 in the USA and 1 in Canada), 11 in Asia (9 in China, 1 in Korea, and 1 in Israel), and 5 in Europe (3 in Italy, 1 in Ireland, and 1 in Slovakia). A total of 2,448 participants were enrolled in the research, comprising 99.41% females (n=2,007) and 0.59% males (n=12). In two studies, the sex of 429 participants was unspecified, and these participants were classified as female for subsequent analysis. The average age was 52.73 years, with a standard deviation of 12.36 years. The sample categories primarily included tumor tissue (n=1,322), normal tissue adjacent to the tumor mass (NAT, n=792), normal tissue (n=697), and benign disease tissue (n=157). NAT was defined as normal tissues located approximately 5 cm from the tumor. Regarding sequencing characteristics (Supplementary Table S2), 16S rRNA sequencing was the predominant method utilized (79.31%, n=23), followed by 16S rDNA (6.9%, n=2). The V4 region was the most frequently sequenced, appearing in 21 out of 29 studies, followed by V3 region in 20 out of 29 studies, and V1, V2, and V5 regions, each appearing in 7 out of 29 studies.
3.2 α-diversity
3.2.1 α-diversity alterations in different disease states
Twenty-two studies evaluated changes in α-diversity among women with cancer, disease-free controls, and individuals with benign disease. The sequencing reads originated mainly from tumor tissue, NAT, healthy control tissue (HC), and benign tissue. Species richness indexes, specifically the Chao1 index and observed species, along with metrics for richness and evenness such as the Shannon index and Simpson index, were the most commonly used measures to assess differences among disease states. The articles on breast cancer and endometrial cancer analyzed the distinctions between tumor tissue and adjacent normal tissue (Figure 3). In both cancer studies, species richness exhibited no significant alteration, with the Chao1 index values indicating SMD= -0.23, (95% CI: [-2.05,1.59], I2 = 80%) and SMD= 0.42, (95% CI: [-0.11, 0.95]), respectively (Figure 3A). The assessment of both richness and evenness using the Shannon index, evaluated in eight studies, revealed no significant changes (BC: SMD= -0.37, 95% CI: [-0.99, 0.25], I2 = 95%; EC: SMD= 1.04, 95% CI: [0.48, 1.60]) (Figure 3B). The Simpson index, although examined in only two studies, indicated a declining trend in both cancers (BC: SMD= -0.75, 95% CI: [-0.94, -0.55]; EC: SMD= -0.83, 95% CI: [-1.37, -0.28]) (Figure 3C).
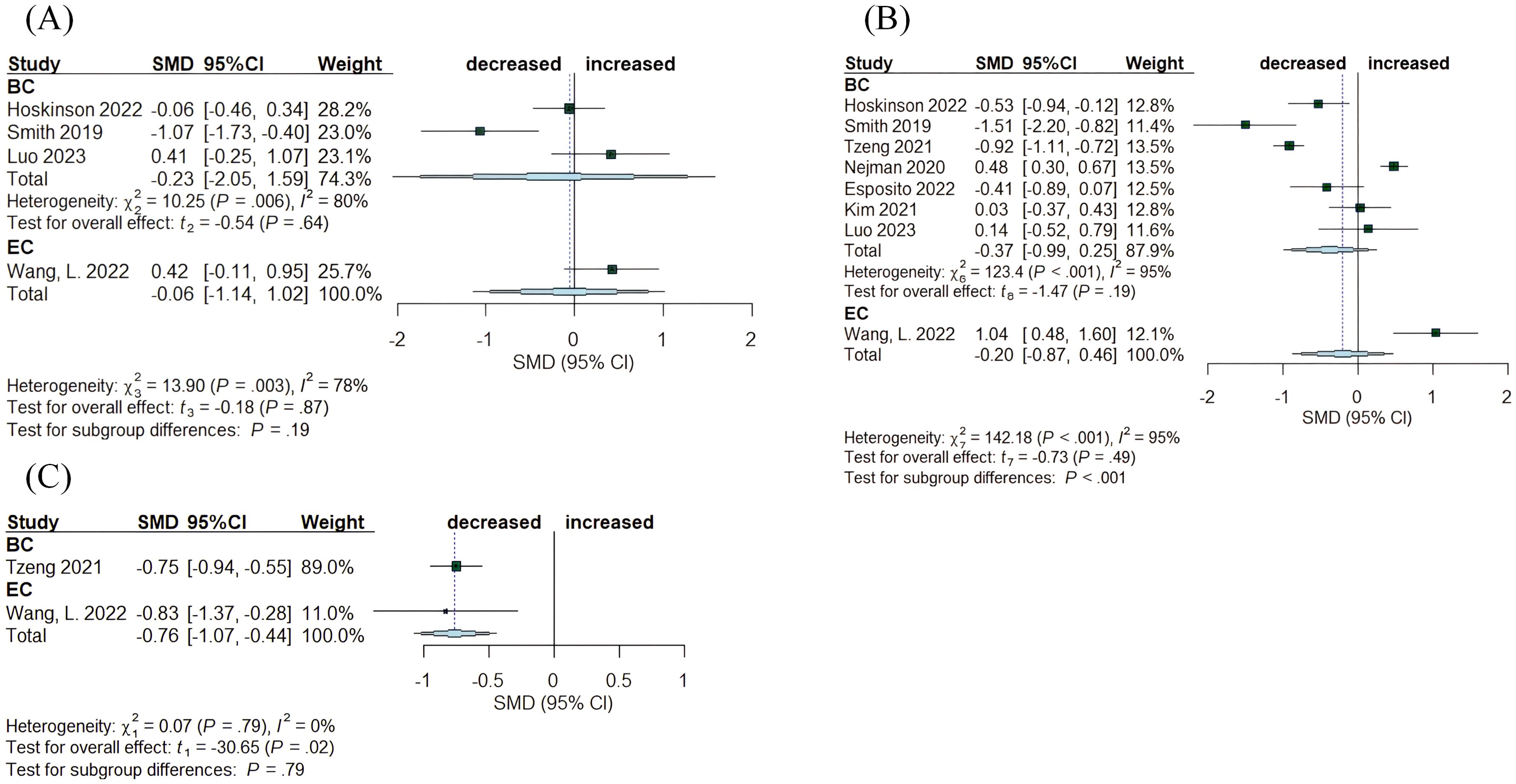
Figure 3. Forest plots illustrating alpha diversity indices comparing intratumoral microbiota with matched normal adjacent tissues (NATs) across female cancers. (A) Chao1 index; (B) Shannon index; (C) Simpson index. Each plot presents standardized mean differences (SMDs) with 95% confidence intervals (CIs), calculated using a random-effects model. Squares indicate individual study effect sizes (with sizes proportional to study weight); horizontal lines represent 95% CIs; diamonds denote pooled estimates. Positive SMD values indicate higher diversity in tumor tissues, while negative values indicate lower diversity compared to NATs. Statistical significance was defined as p < 0.05. BC, breast cancer; EC, endometrial cancer; CC, cervical cancer.
A total of ten articles compared tumor tissue with healthy normal tissue, encompassing breast, endometrial, and ovarian cancer (Figure 4). In BC, the Chao1 index exhibited no significant changes (SMD= 0.16, 95% CI: [-1.03,1.36], I2 = 83%), while in EC, the index demonstrated a reduction (SMD= -2.25, 95% CI: [-3.13, -1.36]) (Figure 4A). The Simpson and Shannon indices were computed for the three tumor types, revealing a decrease in the Shannon index for OC (SMD= -0.61, 95% CI: [-1.18, -0.04]). The other indices exhibited no significant changes: for BC, the Simpson index had an SMD of -0.45 (95% CI: [-2.96, 2.06], I2 = 30%) and the Shannon index had an SMD of -0.12, (95% CI: [-0.56, 0.31], I²= 82%); for OC, Simpson SMD= 0.18, (95% CI: [-0.38, 0.73]); for EC, Shannon SMD= -0.40, (95% CI: [-1.12, 0.32]) (Figures 4C, B).
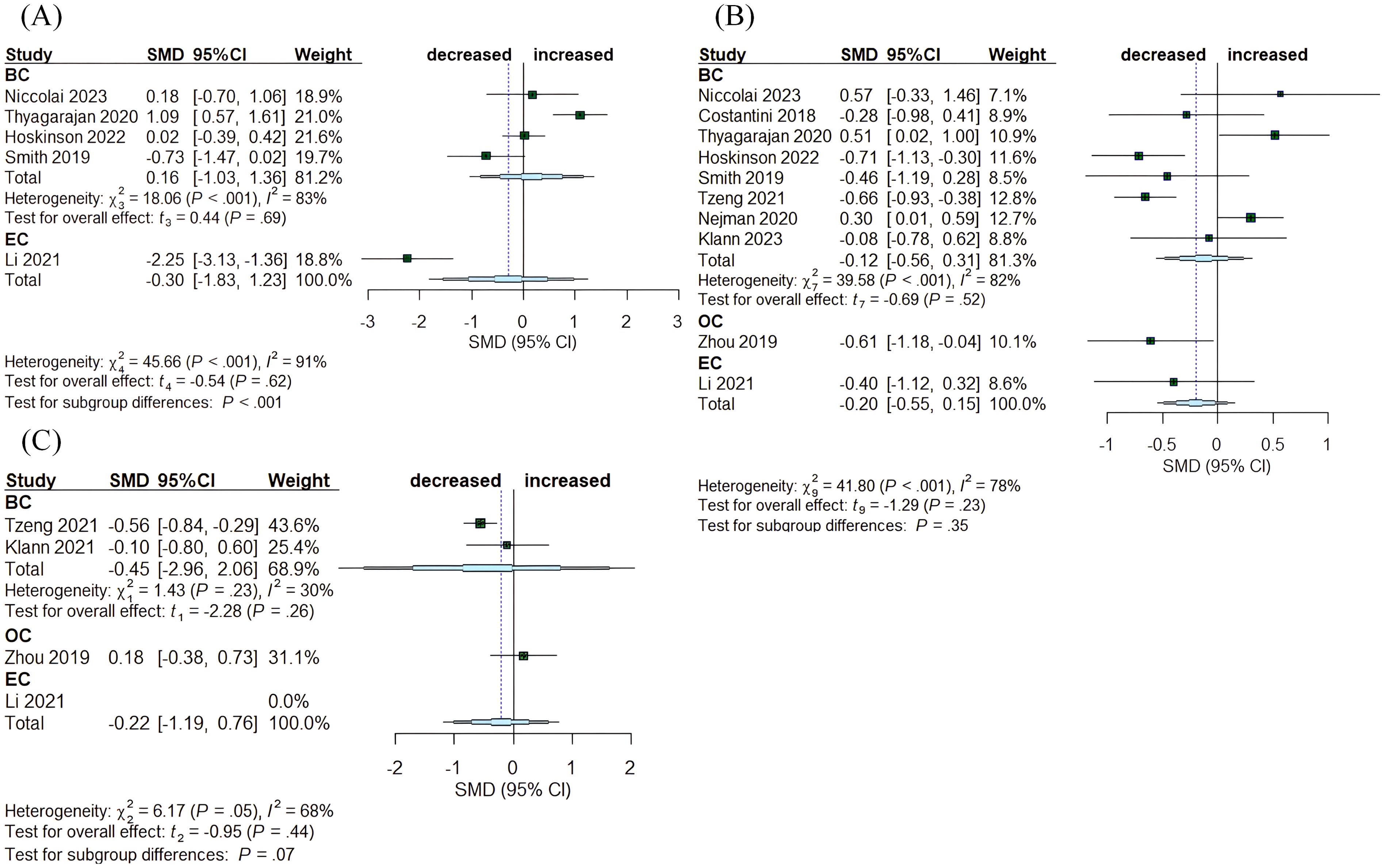
Figure 4. Forest plots illustrating alpha diversity indices comparing intratumoral microbiota with healthy controls (HC) across female cancers. (A) Chao1 index; (B) Shannon index; (C) Simpson index. Each plot presents standardized mean differences (SMDs) with 95% confidence intervals (CIs), calculated using a random-effects model. Squares indicate individual study effect sizes (with sizes proportional to study weight); horizontal lines represent 95% CIs; diamonds denote pooled estimates. Positive SMD values indicate higher diversity in tumor tissues, while negative values indicate lower diversity compared to HCs. Statistical significance was defined as p < 0.05. BC, breast cancer; EC, endometrial cancer; OC, ovarian cancer.
Benign diseases are frequently considered precancerous. Eight articles assessed the differences between benign and tumor tissue in the breast, ovary, and endometrium (Figure 5). The Chao1 index showed a decrease in OC (SMD= -0.64, 95% CI: [-1.20, -0.08], I2 = 0%), while no significant alterations were observed in BC (SMD= 0.44, 95% CI: [-0.40, 1.28]) (Figure 5A). Simpson index indicated an increasing trend in OC (SMD= 0.36, 95% CI: [0.01, 0.71], I2 = 0%) (Figure 5C). In contrast, Shannon index exhibited no significant changes across the three cancers: BC (SMD= -0.15, 95% CI: [-2.01, 1.72], I2 = 76%), OC (SMD= -0.02, 95% CI: [-3.75, 3.71], I2 = 0%), and EC (SMD= 0.18, 95% CI: [-3.23, 3.59], I2 = 91%) (Figure 5B).
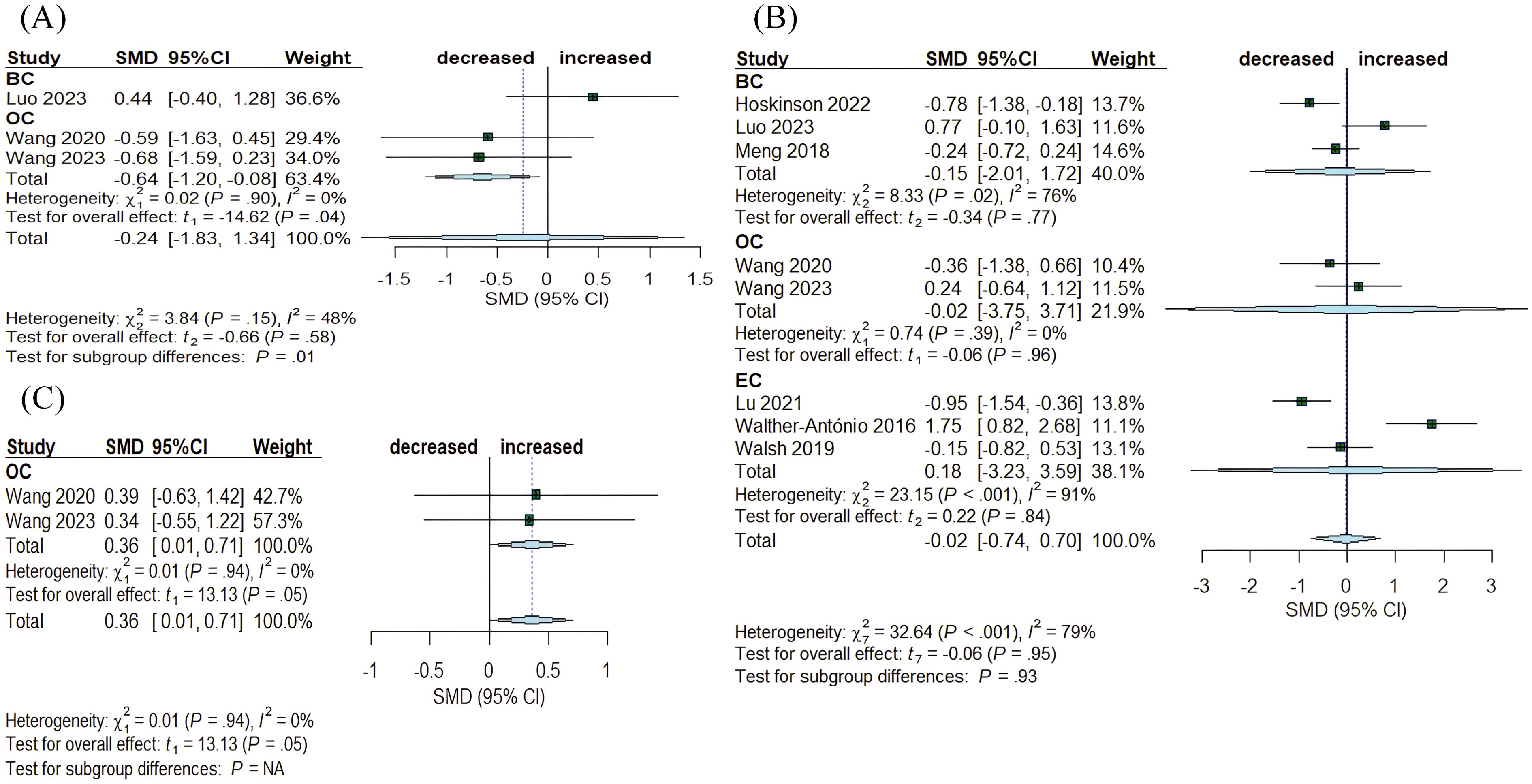
Figure 5. Forest plots illustrating alpha diversity indices comparing intratumoral microbiota with benign disease tissues across female cancers. (A) Chao1 index; (B) Shannon index; (C) Simpson index. Each plot presents standardized mean differences (SMDs) with 95% confidence intervals (CIs), calculated using a random-effects model. Squares indicate individual study effect sizes (with sizes proportional to study weight); horizontal lines represent 95% CIs; diamonds denote pooled estimates. Positive SMD values indicate higher diversity in tumor tissues, while negative values indicate lower diversity compared to benign disease tissues. Statistical significance was defined as p < 0.05. BC, breast cancer; EC, endometrial cancer; OC, ovarian cancer.
3.2.2 Analysis of publication bias
Given the limited sample size, the analysis for publication bias was restricted to the Shannon indices, employing funnel plots and Egger’s test. Funnel plots indicated an absence of significant publication bias (Supplementary Figure S1). Quantitative analysis utilizing Egger’s test indicated an absence of significant publication bias in comparisons of tumor tissue to normal tissue (t= 0.2146, p= 0.8345), normal tissue adjacent to tumor (t= -0.1101, p= 0.91), and benign tissue (t= 1.8306, p= 0.1169).
3.3 Microbial taxa abundance
Microbiota referenced in a minimum of three articles were incorporated into the analysis (Figure 6). Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes represented the four predominant phyla across all tissues in BC, OC, EC, and CC. The microbiota abundance varied between tumor and non-tumor tissues. At the phylum level, four publications indicated that Fusobacteriota were enriched in tumor tissues, while five publications noted that Firmicutes and four publications highlighted that Actinobacteria were more abundant in non-tumor tissues. Proteobacteria and Bacteroidetes exhibited controversy, with four studies indicating a greater abundance of Proteobacteria in tumor tissues, while three reported the contrary. In contrast, two studies suggested a higher presence of Bacteroidetes in tumor tissue, with one presenting opposing evidence. At the genus level, Pseudomonas (n= 4), Porphyromonas (n= 3), Atopobium (n= 3), Peptoniphilus (n= 3) and Acinetobacter (n= 4) were significantly overrepresented in cancer tissues. Nonetheless, the prevalence of certain genera across various tissues continues to be a subject of debate, despite examination in multiple studies. Streptococcus shows 2 instances of increase compared to 3 instances of decrease; Staphylococcus exhibits 3 instances of increase versus 2 instances of decrease; Lactococcus has 1 instance of increase against 2 instances of decrease; Lactobacillus presents 1 instance of increase relative to 2 instances of decrease; Bacteroides indicates 1 instance of increase compared to 2 instances of decrease; Prevotella reflects 3 instances of increase alongside 3 instances of decrease; Micrococcus demonstrates 2 instances of increase in contrast to 1 instance of decrease.
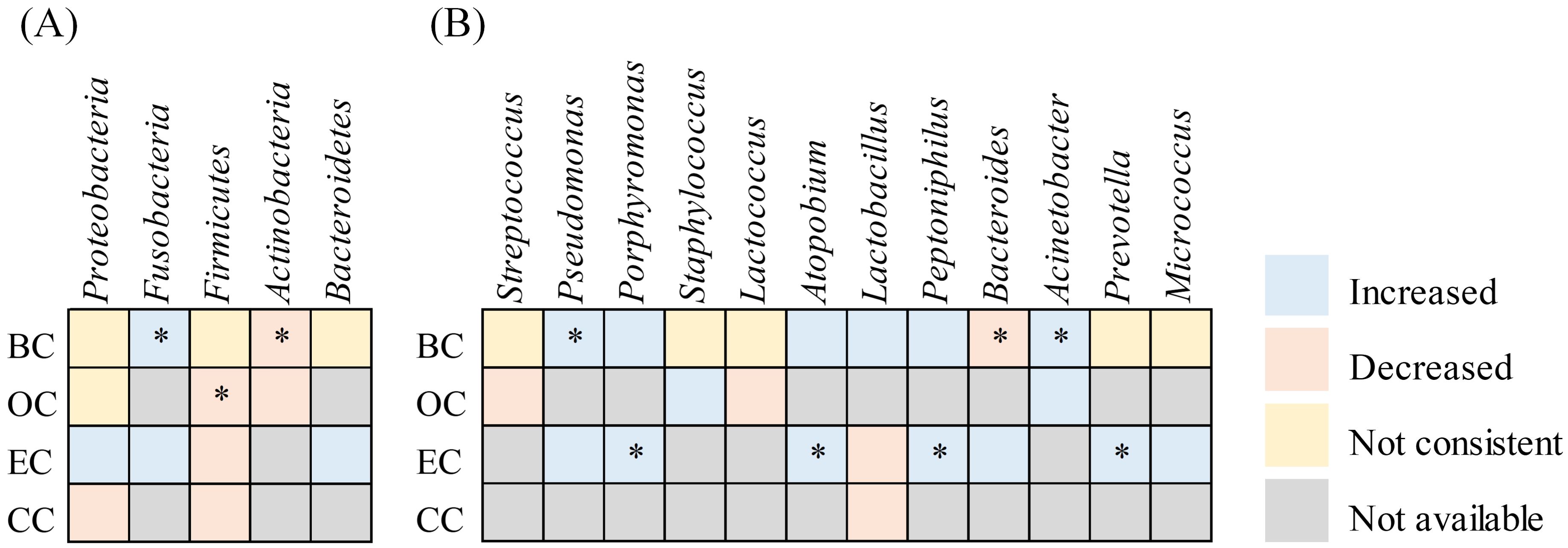
Figure 6. Heatmap summarizing changes in the relative abundance of microbial taxa in tumor tissues compared to non-tumor tissues across female cancer types. This heatmap illustrates reported increases or decreases in specific microbial taxa across endometrial cancer (EC), ovarian cancer (OC), breast cancer (BC), and cervical cancer (CC). An asterisk (*) denotes consistent findings reported in two or more independent studies.
3.4 Microbial alterations with estrogen
In BC, six studies assessed microbial changes associated with varying estrogen levels: three focused on estrogen receptor (ER) status, two on hormone receptor (HR) status (including ER or progesterone receptor (PR)), and one on menopause status. The genus Alkanindiges was negatively correlated with ER in two studies. Additionally, Micrococcus, Caulobacter, Proteus, Brevibacillus, Kocuria, Parasediminibacterium, Comamonas, and Pseudoxanthomonas were identified in one study. Corynebacterium was the sole genus positively associated with ER. Various HR statuses were also correlated with bacterial abundance. α-diversity and β-diversity did not exhibit significant differences across various HR statuses. In HR (-), Acinetobacter, Priestia, Streptomyces, Rhodobacter, Bradyrhizobium, Pseudolysobacter, Gammaretrovirus, Adidovorax exhibited higher abundance, while Lawsonella, Spirosoma, Paracoccus, Actinomyces, Clostridium, Bacillus, Hydrogenophaga, and Staphylococcus, along with Halomonas, were positively correlated with HR (+). A significantly increased relative abundance of Acinetobacter was noted in HR (+) breast tissue. Premenopause showed a higher abundance of Ralstonia, Acetobacter aceti, Lactobacillus vini, Lactobacillus paracasei, and Xanthomonas sp. in comparison to the postmenopausal period.
Correlation analysis for EC indicated that the genera Dialister, Rhodococcus, Delftia, and Parvimonas exhibited a positive correlation with estrogen levels (p< 0.05). Additionally, A. tetradius, A. lactolyticus, P. coxii, and C. ureolyticus were found to be associated with EC and postmenopause.
3.5 Microbial alterations with treatment
Only one study examined the effect of antineoplastic therapy on intratumoral microbiota (32). Chiba et al. reported that the neoadjuvant chemotherapy significantly decreased the diversity of intratumor microbiota by comparing breast cancer patients who underwent neoadjuvant chemotherapy (n= 15) with those who did not (n= 18) (Shannon index: SMD= -0.95, 95% CI: [- 1.68, -0.22]). Additionally, neoadjuvant chemotherapy-induced alterations in the composition of intratumoral microbiota, were notably characterized by a significant increase in the abundance of the genus Pseudomonas and a decrease in the genus Prevotella (P< 0.05).
4 Discussion
Previous studies have identified distinct microbiota profiles in various tumor tissues, which are associated with clinical characteristics such as tumor stage and estrogen levels (25). This systematic review aims to evaluate prevalent changes in the intratumoral microbiota associated with estrogen in female cancers. Our findings revealed alterations in the α-diversity and composition of the intratumoral microbiota, which were linked to estrogen and antitumor therapy.
α-diversity was assessed primarily using the Chao1, Shannon, and Simpson indices. Variations in α-diversity have not been consistently reported in previous studies, with literature indicating increases, decreases, and instances of no significant changes in diversity. Our study indicated that the Simpson index was the only measure exhibiting a decreasing trend in both BC (SMD= -0.75, 95% CI: [-0.94, -0.55]) and EC (SMD= -0.83, 95% CI: [-1.37, -0.28]) when comparing tumor tissues to adjacent normal tissues. In comparisons of tumor tissues with normal tissues, the Chao1 index showed a reduction in EC (SMD= -2.25, 95% CI: [-3.13, -1.36]) while the Shannon index revealed a decrease in OC (SMD= -0.61, 95% CI: [-1.18, -0.04]). When comparing tumor tissues to benign tissues, the Chao1 index revealed a decrease in OC (SMD= -0.64, 95% CI: [-1.20, -0.08], I2 = 0%), while the Simpson index demonstrated an increase in OC (SMD= 0.36, 95% CI: [0.01, 0.71], I2 = 0%). Other indices exhibited no significant differences between tumor and non-tumor tissues. The reduction in α-diversity in tumor tissues compared to non-tumor tissues suggests that dysbiosis may play a role in tumor development. Given the limited number of included studies and the potential influence of various factors on the intratumoral microbiota, a larger sample size may be required to confirm changes in α-diversity. Moreover, despite performing subgroup analyses based on tumor type, significant heterogeneity remained, which may be attributed to the inclusion of study populations with varying stages, grades, races, and hormonal statuses.
Additionally, the composition of intratumoral microbiota displayed statistically significant variations across different tissue types. At the phylum level, Fusobacteriota were consistently reported to be enriched in tumor tissues across four studies, whose critical role were previously highlighted in the tumor microenvironment of various cancer types, particularly colorectal cancer. The presence of Fusobacterium in intratumor colonization influences the immune response, tumor cell proliferation, and drug resistance. Although research on Fusobacterium in women’s cancers is limited, a negative correlation has been observed between the abundance of Fusobacterium nucleatum in cervical cancer tissues and prognosis (45). Furthermore, an animal model of breast cancer demonstrated that Fusobacterium nucleatum binds to breast cancer tissues via its leptin Fap2, inhibiting the accumulation of tumor-infiltrating T cells and thereby promoting tumor growth and metastatic progression (46).
At the genus level, Pseudomonas, Porphyromonas, Atopobium, Peptoniphilus, and Acinetobacter were consistently found to be enriched in tumor tissues across multiple studies. These microbial signatures may hold functional relevance for tumor behavior and therapeutic response. For example, Pseudomonas aeruginosa has demonstrated anti-proliferative properties, with its ExoT effector protein shown to impede tumor cell division. A randomized, double-blind, placebo-controlled trial is currently assessing Pseudomonas aeruginosa-mannose sensitive hemagglutinin (PA-MSHA) as a neoadjuvant agent in HER2-negative breast cancer, with preliminary findings suggesting clinical benefit (47). These observations raise the possibility that the intratumoral enrichment of Pseudomonas may serve as a favorable prognostic indicator in specific contexts. In contrast, other taxa appear to promote oncogenic processes. Porphyromonas gingivalis, for instance, can enhance tumor invasiveness by stimulating IL-8 secretion and promoting IL-8-dependent matrix metalloproteinase (MMP) activity (48). Furthermore, Atopobium vaginae and Porphyromonas somerae have been shown to induce proinflammatory cytokines—such as IL-1α, IL-1β, IL-17α, and TNFα—when co-cultured with endometrial cells, potentially contributing to a pro-tumorigenic inflammatory microenvironment (49). Collectively, these findings indicate that bacterial enrichment in tumor tissues may influence carcinogenesis through distinct molecular pathways, including immune modulation, cytokine-driven inflammation, and matrix remodeling. These microbe-host interactions may also intersect with hormonal signaling and therapeutic exposures, particularly in hormone-sensitive malignancies such as breast and endometrial cancers. Future studies should aim to delineate these pathway-specific effects and evaluate their potential utility as biomarkers or therapeutic targets.
In addition to microbial composition and diversity, recent studies have highlighted the critical role of microbial metabolic functions and immune modulation in tumor biology. Of particular interest is the estrobolome - a collection of microbial genes involved in estrogen metabolism - which has been implicated in the regulation of systemic estrogen levels and the pathogenesis of hormone-driven malignancies such as breast and endometrial cancer (50). Dysbiosis within the estrobolome may disrupt estrogen homeostasis, potentially contributing to oncogenesis or resistance to therapy. Estrogen itself, a pivotal hormone in female reproductive physiology, is known to drive the development of hormone-sensitive cancers when present at dysregulated or excessive levels. Increasing evidence suggests that estrogen homeostasis is partially governed by the gut microbiota through mechanisms such as deconjugation and enterohepatic recirculation, thereby influencing systemic estrogen exposure (51, 52). However, the relationship between intratumoral microbiota and estrogen signaling remains underexplored. Limited but notable studies have reported tumor-specific microbial shifts associated with estrogenic states. In BC, the genera Alkanindiges, Micrococcus, Caulobacter, Proteus, Brevibacillus, Kocuria, Parasediminibacterium, Comamonas, and Pseudoxanthomonas exhibited a decrease in ER (+) tumors, while Corynebacterium showed increased abundance. In EC, genera such as Dialister, Rhodococcus, Delftia, and Parvimonas were enriched in patients with elevated estrogen levels, whereas A. tetradius, A. lactolyticus, P. coxii, and C. ureolyticus were more prevalent in postmenopausal women. These findings hint at a context-specific microbial modulation potentially linked to estrogen availability, yet inconsistencies across studies limit the ability to draw definitive conclusions. Further research is warranted to delineate the functional pathways through which intratumoral microbiota may influence tumor behavior in estrogen-dependent cancers. Potential mechanisms may include microbial regulation of local estrogen metabolism, modulation of hormone receptor expression, or interactions with estrogen-responsive immune pathways.
While numerous studies have investigated the role of microbiota in antitumor efficacy (53), relatively few have explored the bidirectional interactions between intratumor microbiota and antitumor therapy. Intratumoral microbiota may influence the effectiveness of antineoplastic treatment by modulating antitumor immunity through shaping the tumor microenvironment. Previous murine models have demonstrated that the antitumor efficacy of gemcitabine was diminished in Mycoplasma hyorhinis-infected murine mammary tumors compared to uninfected murine mammary tumors (54). Conversely, antitumor therapy can reshape the tumor microenvironment, thereby affecting the composition and diversity of the intratumoral microbiota. Chiba et al. observed that breast cancer patients undergoing neoadjuvant chemotherapy exhibited decreased microbial diversity within tumors compared to untreated patients. Moreover, emerging evidence suggests that microbiota can influence immune checkpoint activity, including PD-1/PD-L1 signaling, potentially shaping responsiveness to immunotherapy (55). These therapy-induced microbial changes may be associated with adverse treatment effects or tumor recurrence (56, 57). Notably, such interactions may be subtype-specific, as microbial compositions have been shown to differ between luminal and triple-negative breast cancer, and between endometrioid and serous subtypes of endometrial cancer (9).
Several limitations exist in this study. First, the small sample sizes in the included studies undermine the robustness and generalizability of the findings. Few studies have investigated the effects of estrogen levels and antitumor therapy on intratumoral microbiota, highlighting the need for further research with larger sample sizes. Second, the inclusion of only English-language literature may introduce publication bias, which may have limited the comprehensiveness of the findings. Third, there was substantial heterogeneity observed in the diversity analyses, which may be attributable to clinical factors such as ethnicity, cancer stage, and treatment regimen. However, due to the lack of consistently reported data on these variables across the included studies, subgroup meta-analyses could not be performed to explore their potential contributions to the heterogeneity. An additional important limitation is that several included studies did not clearly report the sex of their participants. In these cases, the sex of participants was inferred as female based on contextual cues, which may have introduced classification bias. This limitation may reduce the generalizability of the findings and underscores the need for future studies to provide explicit sex-disaggregated data.
Overall, our study revealed notable alterations in the composition of the intratumoral microbiota in women’s cancers, particularly in relation to estrogenic status and exposure to antitumor therapies. These findings suggest that hormonal regulation and therapeutic interventions may reshape the tumor-associated microbial landscape, potentially influencing tumor progression and treatment response. Further investigations are warranted to elucidate the microbial metabolic pathways, host-microbe interactions, and immune modulatory roles involved, which could help identify novel biomarkers or therapeutic targets for precision oncology.
5 Conclusion
In summary, this systematic review highlights distinct alterations in the intratumoral microbiota associated with female malignancies. Notably, a consistent enrichment of Fusobacteriota was observed in tumor tissues, while Firmicutes and Actinobacteria were more abundant in adjacent non-tumor tissues. Reductions in α-diversity were frequently reported in tumor samples, suggesting a less diverse microbial community within the tumor microenvironment. In addition, estrogen-related microbial shifts - such as the increased prevalence of Dialister, Rhodococcus, and Parvimonas under elevated estrogenic states - and treatment-induced changes, including reduced microbial diversity following chemotherapy, were observed. These findings point to a potential role of the intratumoral microbiota in modulating tumor progression and treatment response. Future mechanistic studies are warranted to elucidate the functional contributions of specific microbial taxa, which may ultimately aid in the development of targeted microbiome-based strategies for cancer therapy and women’s health.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
QW: Methodology, Writing – review & editing. SW: Data curation, Software, Writing – original draft. SF: Data curation, Writing – original draft. XZ: Data curation, Writing – original draft. YM: Data curation, Writing – original draft. JL: Methodology, Supervision, Writing – original draft. MC: Data curation, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. MC was supported by the Health Commission of Sichuan Province Medical Science and Technology Program (24QNMP038). JL was supported by the Chengdu Technology Bureau (2024-YF05-02230-SN) and the Health Commission of Sichuan Province (2024–803).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1544786/full#supplementary-material
References
1. Elinav E, Garrett WS, Trinchieri G, and Wargo J. The cancer microbiome. Nat Rev Cancer. (2019) 19:371–6. doi: 10.1038/s41568-019-0155-3
2. Li ZR, Li J, Cai W, Lai JYH, McKinnie SMK, Zhang WP, et al. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat Chem. (2019) 11:880–9. doi: 10.1038/s41557-019-0317-7
3. Yu F, Yu C, Li F, Zuo Y, Wang Y, Yao L, et al. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther. (2021) 6:307. doi: 10.1038/s41392-021-00701-5
4. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806.e712. doi: 10.1016/j.cell.2019.07.008
5. Zhu G, Su H, Johnson CH, Khan SA, Kluger H, and Lu L. Intratumour microbiome associated with the infiltration of cytotoxic CD8+ T cells and patient survival in cutaneous melanoma. Eur J Cancer. (2021) 151:25–34. doi: 10.1016/j.ejca.2021.03.053
6. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–1372.e1326. doi: 10.1016/j.cell.2022.02.027
7. Ma W, Zhang L, Chen W, Chang Z, Tu J, Qin Y, et al. Microbiota enterotoxigenic Bacteroides fragilis-secreted BFT-1 promotes breast cancer cell stemness and chemoresistance through its functional receptor NOD1. Protein Cell. (2024) 15:419–40. doi: 10.1093/procel/pwae005
8. Lu W, He F, Lin Z, Liu S, Tang L, Huang Y, et al. Dysbiosis of the endometrial microbiota and its association with inflammatory cytokines in endometrial cancer. Int J Cancer. (2021) 148:1708–16. doi: 10.1002/ijc.33428
9. Semertzidou A, Whelan E, Smith A, Ng S, Roberts L, Brosens JJ, et al. Microbial signatures and continuum in endometrial cancer and benign patients. Microbiome. (2024) 12:118. doi: 10.1186/s40168-024-01821-0
10. Colbert LE, El Alam MB, Wang R, Karpinets T, Lo D, Lynn EJ, et al. Tumor-resident Lactobacillus iners confer chemoradiation resistance through lactate-induced metabolic rewiring. Cancer Cell. (2023) 41:1945–1962 e1911. doi: 10.1016/j.ccell.2023.09.012
11. Samavat H and Kurzer MS. Estrogen metabolism and breast cancer. Cancer Lett. (2015) 356:231–43. doi: 10.1016/j.canlet.2014.04.018
12. Łaniewski P, Ilhan ZE, and Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. (2020) 17:232–50. doi: 10.1038/s41585-020-0286-z
13. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. (2021) 372:n71. doi: 10.1136/bmj.n71
14. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
15. Rohatgi A. WebPlotDigitizer (2020). Available online at: https://automeris.io/WebPlotDigitizer (Accessed October 8, 2024).
16. Shi J, Luo D, Weng H, Zeng XT, Lin L, Chu H, et al. Optimally estimating the sample standard deviation from the five-number summary. Res Synth Methods. (2020) 11:641–54. doi: 10.1002/jrsm.1429
17. Hogan G, Eckenberger J, Narayanen N, Walker SP, Claesson MJ, Corrigan M, et al. Biopsy bacterial signature can predict patient tissue Malignancy. Sci Rep. (2021) 11:18535. doi: 10.1038/s41598-021-98089-3
18. Niccolai E, Baldi S, Nannini G, Gensini F, Papi L, Vezzosi V, et al. Breast cancer: the first comparative evaluation of oncobiome composition between males and females. Biol Sex Differ. (2023) 14:37. doi: 10.1186/s13293-023-00523-w
19. Costantini L, Magno S, Albanese D, Donati C, Molinari R, Filippone A, et al. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci Rep. (2018) 8:16893. doi: 10.1038/s41598-018-35329-z
20. Thyagarajan S, Zhang Y, Thapa S, Allen MS, Phillips N, Chaudhary P, et al. Comparative analysis of racial differences in breast tumor microbiome. Sci Rep. (2020) 10:14116. doi: 10.1038/s41598-020-71102-x
21. Hoskinson C, Zheng K, Gabel J, Kump A, German R, Podicheti R, et al. Composition and functional potential of the human mammary microbiota prior to and following breast tumor diagnosis. mSystems. (2022) 7(3):e0148921. doi: 10.1128/msystems.01489-21
22. Smith A, Pierre JF, Makowski L, Tolley E, Lyn-Cook B, Lu L, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci Rep. (2019) 9:11940. doi: 10.1038/s41598-019-48348-1
23. German R, Marino N, Hemmerich C, Podicheti R, Rusch DB, Stiemsma LT, et al. Exploring breast tissue microbial composition and the association with breast cancer risk factors. Breast Cancer Res. (2023) 25(1):82. doi: 10.1186/s13058-023-01677-6
24. Tzeng A, Sangwan N, Jia M, Liu CC, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. (2021) 13(1):60. doi: 10.1186/s13073-021-00874-2
25. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
26. Xuan C, Shamonki JM, Chung A, DiNome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PloS One. (2014) 9(1):e83744. doi: 10.1371/journal.pone.0083744
27. Esposito MV, Fosso B, Nunziato M, Casaburi G, D’Argenio V, Calabrese A, et al. Microbiome composition indicate dysbiosis and lower richness in tumor breast tissues compared to healthy adjacent paired tissue, within the same women. BMC Cancer. (2022) 22:30. doi: 10.1186/s12885-021-09074-y
28. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and Malignant disease. Sci Rep. (2016) 6:30751. doi: 10.1038/srep30751
29. Klann E, Williamson JM, Tagliamonte MS, Ukhanova M, Asirvatham JR, Chim H, et al. Microbiota composition in bilateral healthy breast tissue and breast tumors. Cancer Causes Control. (2020) 31:1027–38. doi: 10.1007/s10552-020-01338-5
30. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, and Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. (2016) 82:5039–48. doi: 10.1128/aem.01235-16
31. Kim H-E, Kim J, Maeng S, Oh B, Hwang K-T, and Kim B-S. Microbiota of breast tissue and its potential association with regional recurrence of breast cancer in korean women. J Microbiol Biotechnol. (2021) 31:1643–55. doi: 10.4014/jmb.2106.06039
32. Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Mol Cancer Res. (2020) 18:130–9. doi: 10.1158/1541-7786.Mcr-19-0451
33. Luo L, Fu A, Shi M, Hu J, Kong D, Liu T, et al. Species-level characterization of the microbiome in breast tissues with different Malignancy and hormone-receptor statuses using nanopore sequencing. J Personalized Med. (2023) 13(2):174. doi: 10.3390/jpm13020174
34. Meng S, Chen B, Yang J, Wang J, Zhu D, Meng Q, et al. Study of microbiomes in aseptically collected samples of human breast tissue using needle biopsy and the potential role of in situ tissue microbiomes for promoting Malignancy. Front Oncol. (2018) 8:318. doi: 10.3389/fonc.2018.00318
35. Hadzega D, Minarik G, Karaba M, Kalavska K, Benca J, Ciernikova S, et al. Uncovering microbial composition in human breast cancer primary tumour tissue using transcriptomic RNA-seq. Int J Mol Sci. (2021) 22(16):9058. doi: 10.3390/ijms22169058
36. Zhou B, Sun C, Huang J, Xia M, Guo E, Li N, et al. The biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. (2019) 9:1691. doi: 10.1038/s41598-018-38031-2
37. Wang Q, Zhao L, Han L, Fu G, Tuo X, Ma S, et al. The differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J Ovarian Res. (2020) 13(1):8. doi: 10.1186/s13048-019-0603-4
38. Hawkins GM, Burkett WC, McCoy AN, Nichols HB, Olshan AF, Broaddus R, et al. Differences in the microbial profiles of early stage endometrial cancers between Black and White women. Gynecol Oncol. (2022) 165:248–56. doi: 10.1016/j.ygyno.2022.02.021
39. Wang L, Yang J, Su H, Shi L, Chen B, and Zhang S. Endometrial microbiota from endometrial cancer and paired pericancer tissues in postmenopausal women: differences and clinical relevance. Menopause. (2022) 29:1168–75. doi: 10.1097/gme.0000000000002053
40. Li C, Gu Y, He Q, Huang J, Song Y, Wan X, et al. Integrated analysis of microbiome and transcriptome data reveals the interplay between commensal bacteria and fibrin degradation in endometrial cancer. Front Cell Infection Microbiol. (2021) 11:748558. doi: 10.3389/fcimb.2021.748558
41. Walther-António MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. (2016) 8:122. doi: 10.1186/s13073-016-0368-y
42. Walsh DM, Hokenstad AN, Chen J, Sung J, Jenkins GD, Chia N, et al. Postmenopause as a key factor in the composition of the Endometrial Cancer Microbiome (ECbiome). Sci Rep. (2019) 9(1):19213. doi: 10.1038/s41598-019-55720-8
43. Wang H, Jiang Y, Liang Y, Wei L, Zhang W, and Li L. Observation of the cervical microbiome in the progression of cervical intraepithelial neoplasia. BMC Cancer. (2022) 22:362. doi: 10.1186/s12885-022-09452-0
44. Wang X, Zheng Y, Chen X, Peng C, Zhou S, Shen S, et al. 2bRAD-M reveals the difference in microbial distribution between cancerous and benign ovarian tissues. Front Microbiol. (2023) 14:1231354. doi: 10.3389/fmicb.2023.1231354
45. Huang ST, Chen J, Lian LY, Cai HH, Zeng HS, Zheng M, et al. Intratumoral levels and prognostic significance of Fusobacterium nucleatum in cervical carcinoma. Aging (Albany NY). (2020) 12:23337–50. doi: 10.18632/aging.104188
46. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. (2020) 11:3259. doi: 10.1038/s41467-020-16967-2
47. Gong Y, Zuo H, Zhou Y, Yu KD, Liu GY, Di GH, et al. Neoadjuvant Pseudomonas aeruginosa mannose-sensitive hemagglutinin (PA-MSHA) and chemotherapy versus placebo plus chemotherapy in patients with HER2-negative breast cancer: a randomized, controlled, double-blind trial. Ann Transl Med. (2023) 11:243. doi: 10.21037/atm-22-4093
48. Ha NH, Park DG, Woo BH, Kim DJ, Choi JI, Park BS, et al. Porphyromonas gingivalis increases the invasiveness of oral cancer cells by upregulating IL-8 and MMPs. Cytokine. (2016) 86:64–72. doi: 10.1016/j.cyto.2016.07.013
49. Caselli E, Soffritti I, D’Accolti M, Piva I, Greco P, and Bonaccorsi G. Atopobium vaginae And Porphyromonas somerae Induce Proinflammatory Cytokines Expression In Endometrial Cells: A Possible Implication For Endometrial Cancer? Cancer Manag Res. (2019) 11:8571–5. doi: 10.2147/cmar.S217362
50. Kovacs T, Miko E, Ujlaki G, Yousef H, Csontos V, Uray K, et al. The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer. Cancer Metastasis Rev. (2021) 40:1223–49. doi: 10.1007/s10555-021-10013-3
51. Dabek M, McCrae SI, Stevens VJ, Duncan SH, and Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. (2008) 66:487–95. doi: 10.1111/j.1574-6941.2008.00520.x
52. Parida S and Sharma D. The microbiome-estrogen connection and breast cancer risk. Cells. (2019) 8(12):1642. doi: 10.3390/cells8121642
53. Herrera-Quintana L, Vazquez-Lorente H, Silva R, Olivares-Arancibia J, Reyes-Amigo T, Pires BRB, et al. The role of the microbiome and of radiotherapy-derived metabolites in breast cancer. Cancers (Basel). (2024) 16(21):3671. doi: 10.3390/cancers16213671
54. Vande Voorde J, Sabuncuoğlu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, et al. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem. (2014) 289:13054–65. doi: 10.1074/jbc.M114.558924
55. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
56. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, and Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. (2017) 14:356–65. doi: 10.1038/nrgastro.2017.20
Keywords: intratumoral microbiota, 16s rrna gene sequencing, breast cancer, gynecologic cancer, estrogen, meta-analysis
Citation: Wen Q, Wang S, Fu S, Zhou X, Min Y, Lang J and Chen M (2025) Intratumoral microbiota composition in women’s cancers: a systematic review and meta-analysis. Front. Oncol. 15:1544786. doi: 10.3389/fonc.2025.1544786
Received: 17 December 2024; Accepted: 30 May 2025;
Published: 12 June 2025.
Edited by:
Louis Dubeau, University of Southern California, United StatesReviewed by:
Amit Kumar, Virginia Commonwealth University, United StatesAleksander Szymczak, Oklahoma Medical Research Foundation, United States
Copyright © 2025 Wen, Wang, Fu, Zhou, Min, Lang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinyi Lang, bGFuZ2p5NjEwQDE2My5jb20=; Meihua Chen, Y2hlbm1laWh1YUBzY3N6bHl5Lm9yZy5jbg==
 Qin Wen
Qin Wen Shubin Wang
Shubin Wang Shunlian Fu
Shunlian Fu Xinxiang Zhou1,2
Xinxiang Zhou1,2 Jinyi Lang
Jinyi Lang