- 1Department of Maxillofacial Surgery, Faculty of Medicine, Medical University of Gdansk, Gdańsk, Poland
- 2Department of Maxillofacial Surgery, University Clinical Centre in Gdańsk, Gdańsk, Poland
- 3Department of Pathomorphology, Faculty of Medicine, Medical University of Gdansk, Gdańsk, Poland
- 4Department of Clinical Pathomorphology, University Clinical Centre in Gdańsk, Gdańsk, Poland
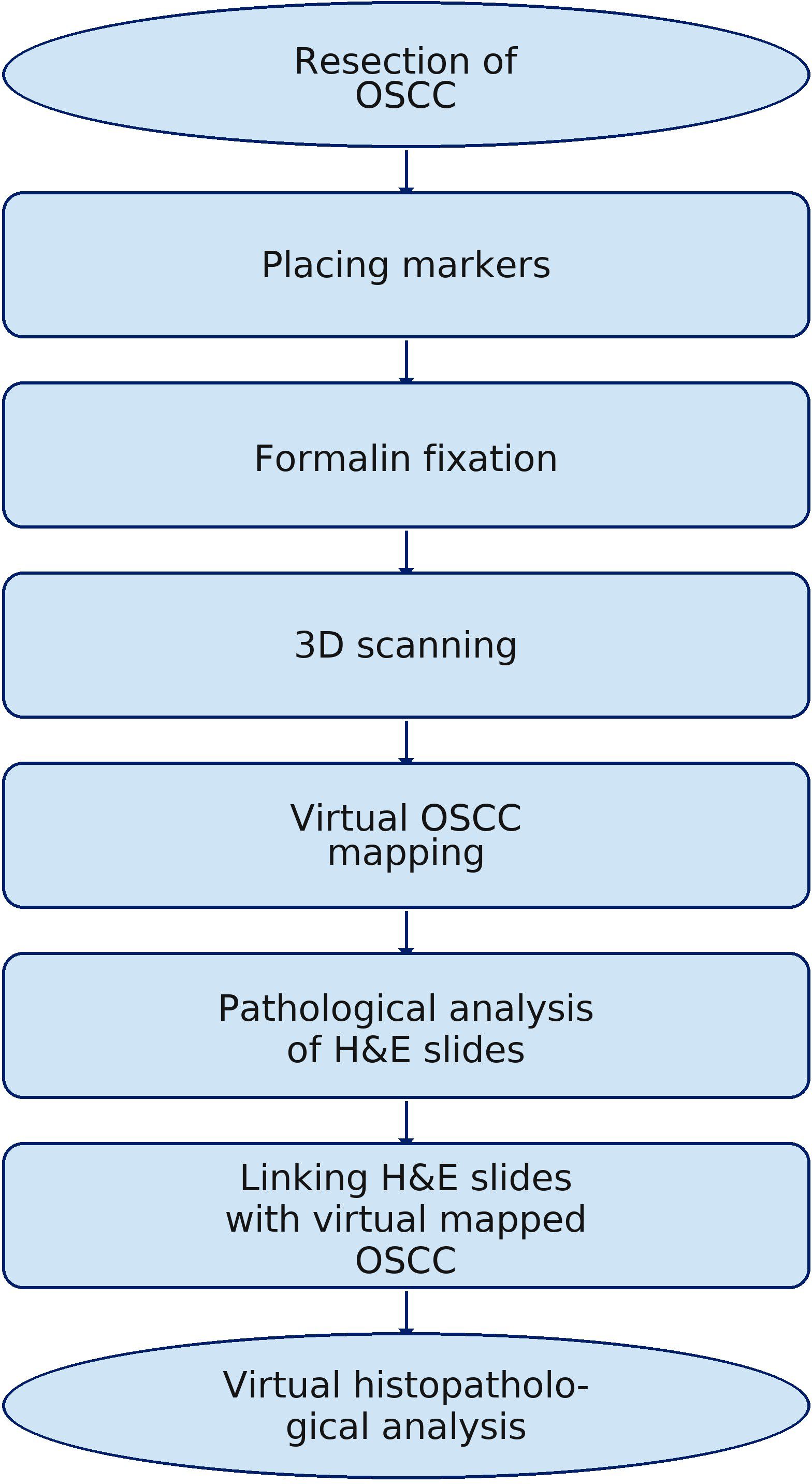
One factor that enhances the survival of patients with oral squamous cell carcinoma (OSCC) is improved local control. Many prognostic factors (PF) influence a patient’s prognosis, one of which is the correct histopathological excision margins. Standard histopathological analysis does not allow for the precise determination of the locations of individual hematoxylin and eosin (H&E) slides, which is clinically very important. In extensive tumors, the ability to precisely identify the sites with the narrowest microscopic excision is essential for accurately interpreting postoperative results and making proper clinical decisions. Therefore, a protocol has been developed to precisely localize individual H&E-stained tissue slides on virtual three-dimensional (3D) OSCC images. This approach provides a virtual map of the entire excised tumor, allowing accurate identification of areas with the narrowest histopathological margins (HM). As a result, if reoperation is necessary, it is possible to precisely determine where to widen the OSCC excision or plan adjuvant radiotherapy with greater accuracy. The proposed pilot study is based on the virtual three-dimensional OSCC model of the floor of the mouth cancer. Using 3D scanning technology, a user-friendly protocol has been created.
Introduction
Among malignant tumors, oral squamous cell carcinoma ranks 16th worldwide (1) and is the most common malignant tumor of the oral cavity (2, 3). Persistent unfavorable outcomes, including a 5-year survival rate of 50% (4), remain unsatisfactory. The literature describes numerous prognostic factors (PF) that influence patient prognosis (5). Some of these are determined by the biological characteristics of oral squamous cell carcinoma (OSCC) (6–8) and nodal metastases (9, 10), which increase the risk of treatment failure. The depth of invasion (DOI), lymphovascular invasion (LVI), perineural invasion (PNI), and extranodal extension (ENE) (11–13) are of significant importance and directly influence the risk of recurrence. The worst pattern of invasion (WPOI) describes tissue infiltration by cancer cells at the tumor interface and is considered a key factor in histological grading systems, especially for OSCC (14). It is well known that when nodal metastases are present, regional control of the tumor is challenging, and the likelihood of nodal recurrence is increased (9). According to existing research, locoregional spread is the most common cause of treatment failure in patients with OSCC (15, 16). However, tumor local control is equally essential. To increase it, procedures should be performed with a safe macroscopic excision margin, defined as 1 cm of macroscopically unchanged tissue around the tumor (17, 18). The UK Royal College of Pathologists issued guidance on pathological margins for OSCC (19). A margin less than 1 mm is considered involved, less than 5 mm as close, and greater than 5 mm as clear. In available studies, we observe various cutoffs that have been identified as sufficient (20–22) and did not impair local tumor control. When planning OSCC excision with an appropriate margin, it is also necessary to consider how the processes involved in specimen preparation for histopathological analysis might affect the outcome. The influence of formalin fixation (FF) is important for the final measurement of the width of the microscopic margin width. Available publications confirm shrinkage occurring during FF (23–25).
Standard histopathological analysis of the resected OSCC includes cover-up and subsequent slicing according to a scheme recognized by the pathologist. During this process, the technician records the pathologist’s notes to aid in the later interpretation of the results. Then, the histopathological slides are prepared and stained using the standard hematoxylin and eosin (H&E) stain. Once the specimen is dissected, the ability to visualize the tumor and to connect the histopathological findings to it is lost, as well as the capacity to accurately identify which section of a given margin is affected by a specific H&E slide. This is especially critical in cases of irregular tumor shape and long margins. In such situations, the ability to retrospectively locate a specific area on the sample is essential. This underscores the need to explore new solutions to improve the accuracy of H&E slide locations and enable a more precise assessment of OSCC margins. The proposed pilot study assumed that three-dimensional (3D) scanning technology would create a virtual, interactive histopathological examination result that accurately and precisely determined each point of the excised and mapped cancer. This may improve the accuracy of histopathological slide analysis and enable archiving as well as interactive retrospective study analysis. The use of structured light in the histological analysis of OSCC is a novelty and opens up new, previously unattainable possibilities for digitizing this stage of patient oral cancer treatment.
Methods of the pilot study
The presented methodology is based on over 100 scans of resected OSCCs and the virtual mapping method. This helped eliminate errors that occurred during the initial phase of the process.
Scanning protocol
The excised OSCC with a margin of macroscopically unchanged tissue was marked in the operating room with beads to facilitate orientation in the subsequent 3D virtual object. Following the procedure, it was placed in formalin. After 24 to 48 h post-FF, the tumor was scanned according to our protocol in the pathology laboratory (26). After formalin fixation and drying, the tumor was placed on a specially constructed tripod (Figure 1).
The tumor was then contrasted with a matte spray designed for Aesub Blue 3D scanners (composition: propane, ethanol, tricyclodecane, hydrocarbons, C6–C7, iso-alkanes, cyclics, < 5% n-hexane, n-hexane) (27). Next, scanning was performed using a 3D scanner: Revopoint MINI—Dual-Axis Turntable Package (Shenzhen, China), adapted to scan small objects (minimum 10 mm × 10 mm × 10 mm) with an accuracy of 0.02 mm at 10 frames per second and a point distance of up to 0.05 mm. The scanner was selected for its exceptional resolution and resistance to ambient lighting. Finally, the acquired virtual models in STL format (triangulated surface geometry in three-dimensional space) were processed using Revo Scan 5 (https://global.revopoint3d.com), which included removing environmental contaminants and enhancing the tumor surface detail. The processed 3D images were saved as OBJ files, and the models’ textures were saved as JPG files. The total time to complete the described process was approximately 10 min (Figure 2).
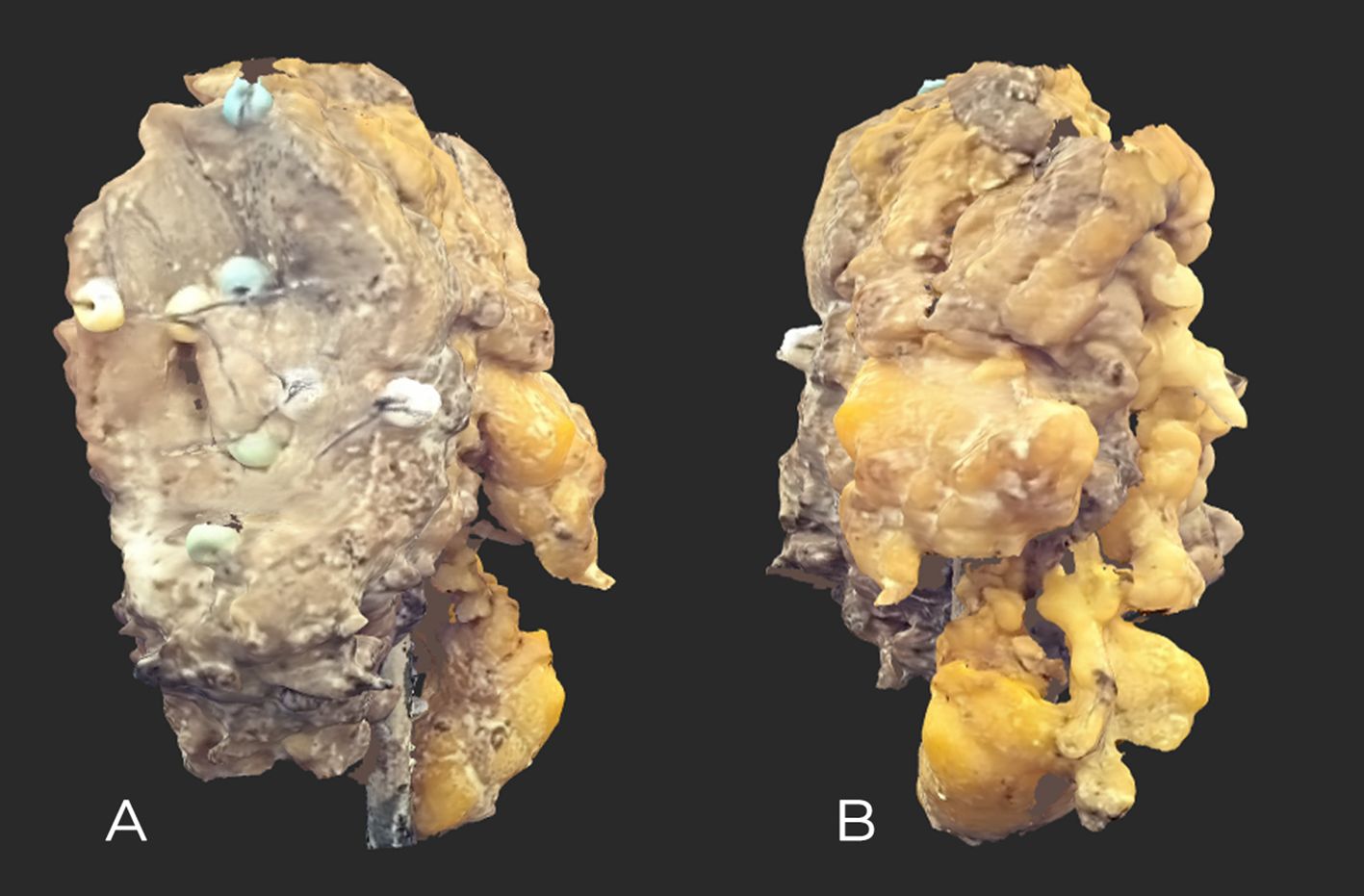
Figure 2. High-resolution maxilla tumor scan after FF. (A) Front view showing slight ulceration and sewn-on markers. (B) Rear view; Bichat’s buccal fat is visible.
Image annotation
The image labeling process was conducted using Blender 4.1.1, an open-source application (www.blender.org, accessed on 25 September 2023). This software was chosen for its accessibility, free availability, and user-friendly interface. Originally developed as a 3D modeling tool for digital creators, its capabilities have expanded over time to include film editing and motion capture. The application allowed users to add annotations directly onto the surfaces of 3D objects and create markers with notes.
Histopathological processing of the floor of the mouth cancer specimen with parallel annotation on the virtual OSCC object
Stage I: Covering up the specimen
In the first stage of the pathological protocol, the specimen was inked (Figure 3).
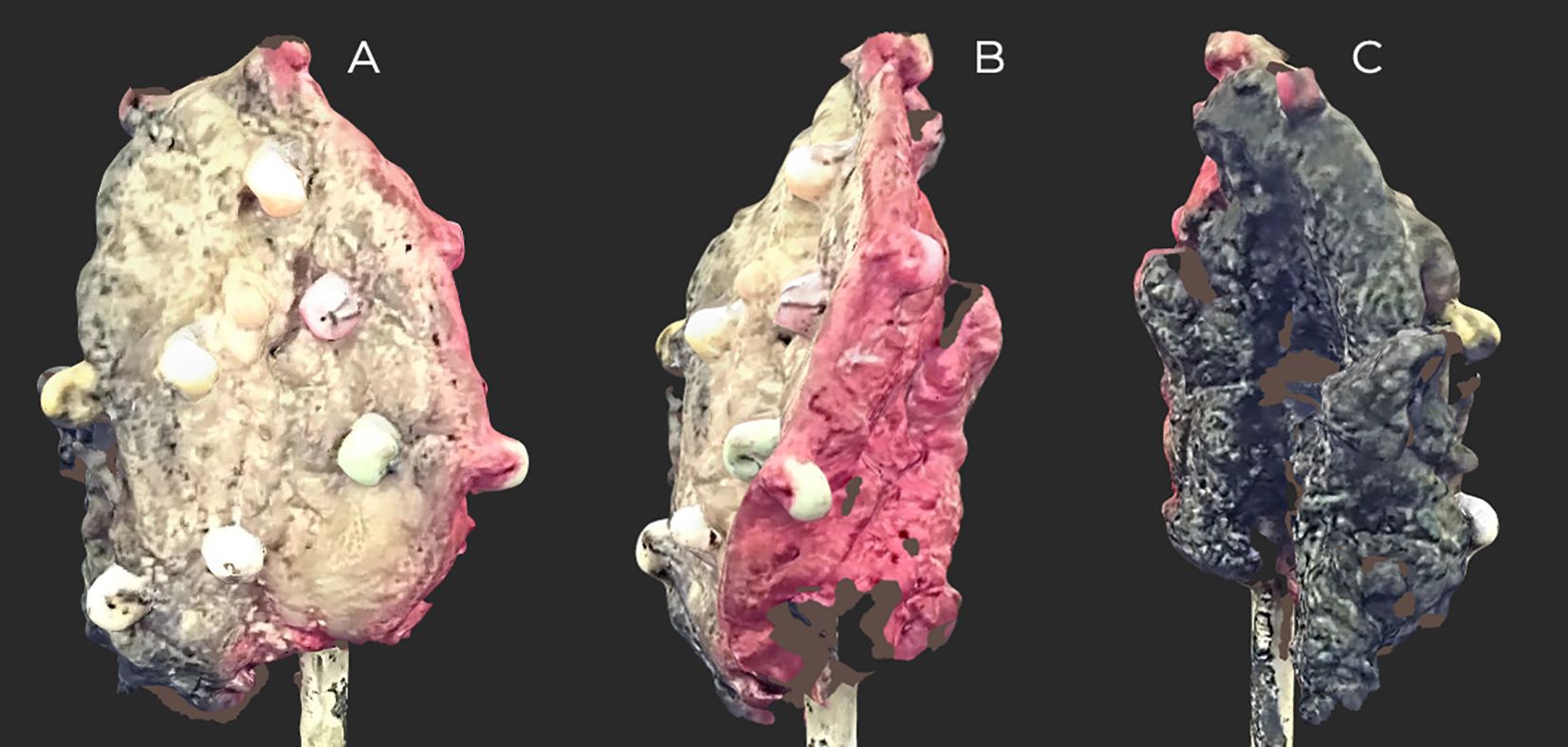
Figure 3. Scan of the inked floor of the mouth (FOM) specimen before slicing. On the black-inscribed specimen, small areas are invisible to the scanner. Inked: side and back, red; front and middle, black. (A) Front view. (B, C) Side views.
Stage II
After FF, the specimen was sliced while simultaneously mapping the tumor on the scan. The visible lines correspond to the actual lines cut on the specimen by the pathologist (Figures 4, 5).
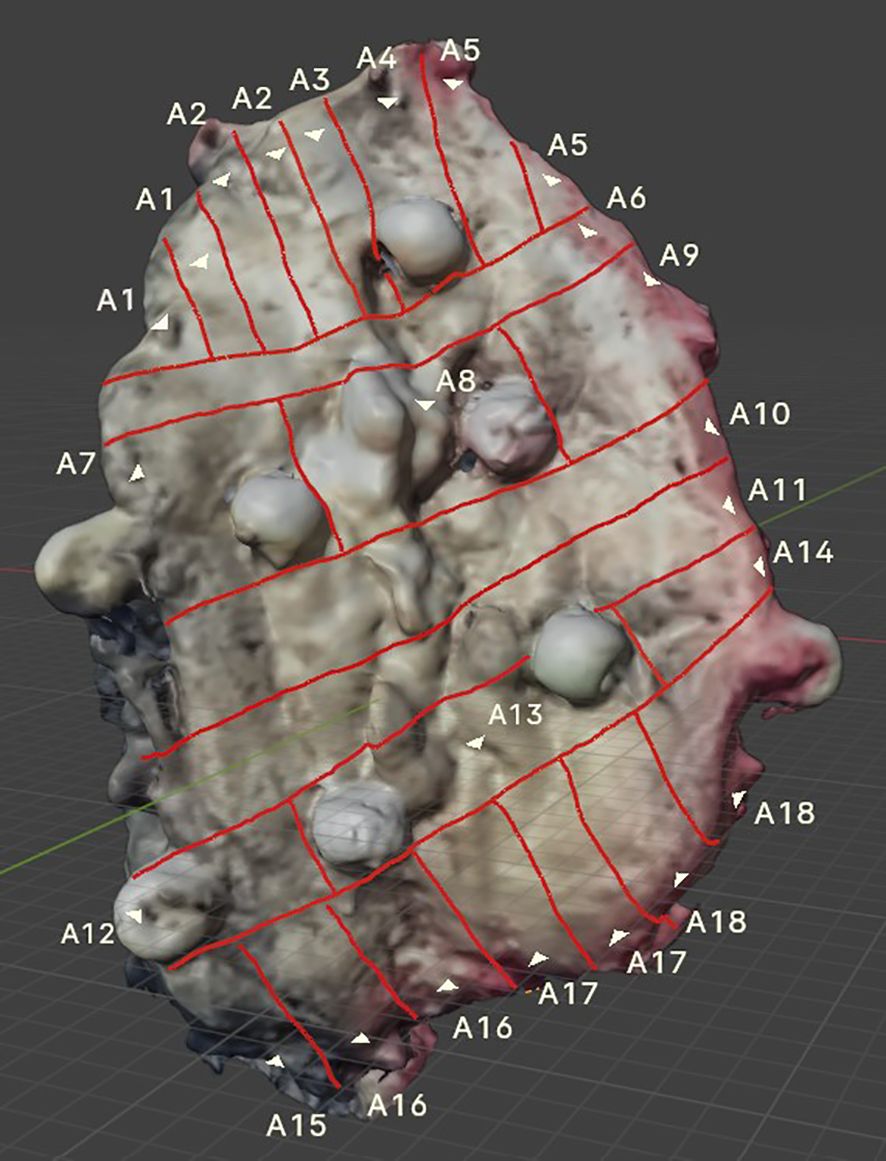
Figure 4. 3D FOM cancer scan showing visible incision lines applied to the tumor surface, created in parallel with the incisions made on the actual specimen. Sewn-on markers enhance the accuracy of tumor mapping.
Histopathological analysis
According to the presented numbering (A1–A18), H&E-stained tissue slides were collected (Figure 6).
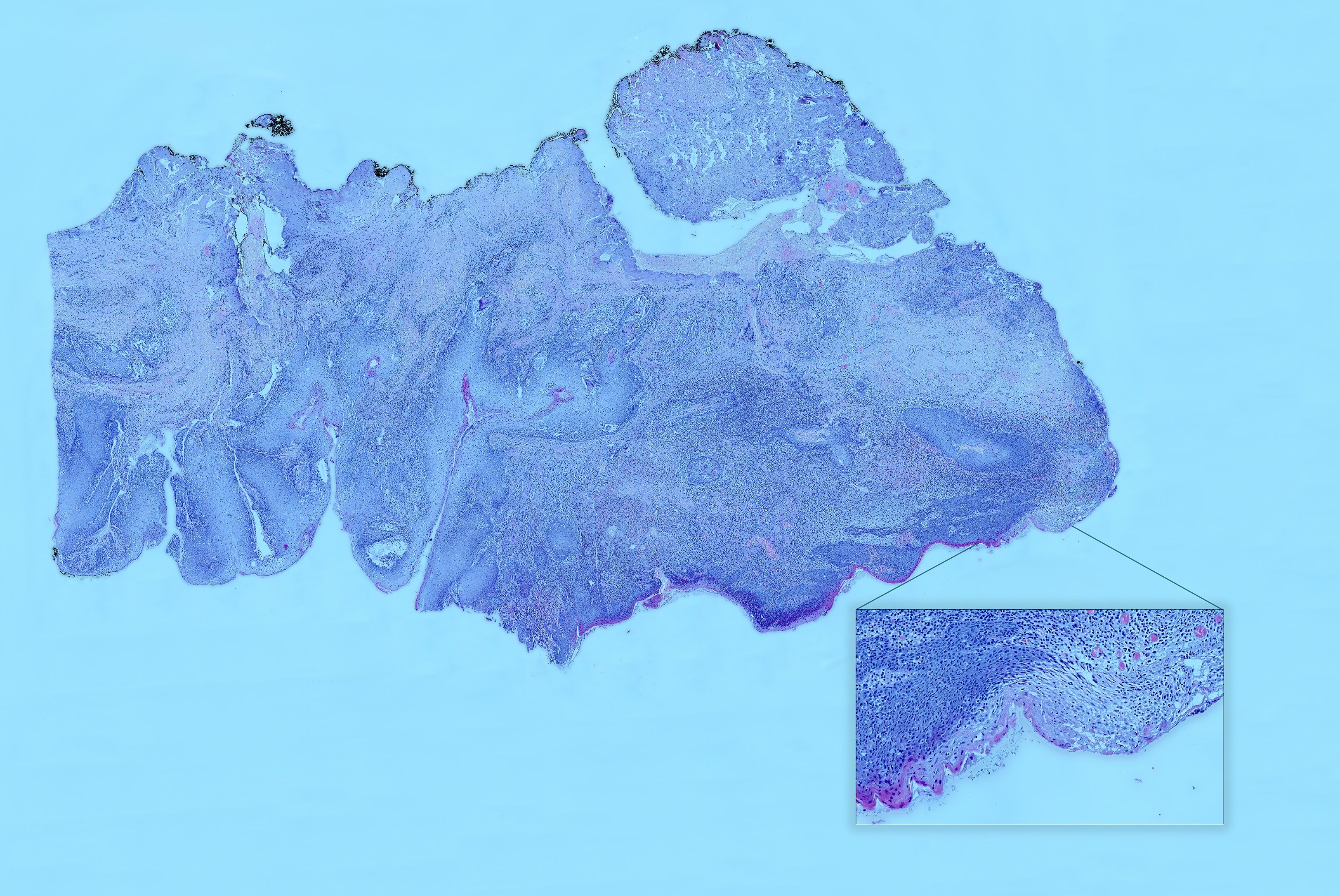
Figure 6. Hematoxylin and eosin (H&E) slice scan No. A3. A high-resolution scan showing the entire margin. A magnified view of a specimen fragment displays detailed images in an interactive histopathological report. In the interactive version, users can view H&E slide scans of specific areas selected on a virtual OSCC, providing easy and unlimited access to archived H&E images.
Results
After the pathologist evaluated the received H&E slides, the results were recorded in a description of the virtually mapped tumor, numbered accordingly. Additionally, to assist with orientation, the tumor’s margins are described, and annotations have been added to support retrospective analysis (Figure 7).

Figure 7. The mapped FOM scan includes a description of all evaluated H&E slides, enabling precise identification of areas with the narrowest margin. In the draft version, details about individual H&E slides can be provided as a legend (as shown earlier) or directly attached to a virtual object (expanded through cursor selection). Additionally, in the interactive version, users can upload H&E slide scans after selecting a specific area on a particular slide.
For simplicity and accessibility, the archived OSCC with histopathological margin (HM) results, initially saved as a.obj file (Blender 4.1.1), has been converted to a Portable Document Format (PDF) that allows interactive highlighting of specific areas corresponding to H&E slide numbers. While support for virtual objects in Blender 4.1.1 offers great editing options, it requires additional learning, which can be challenging. To make the file easier and more shareable, it was converted into a PDF that can be viewed on any computer with basic software. This significantly enhances the accessibility of virtual results, which is especially important during oncology team meetings or when sharing images with other centers. Consequently, there is greater potential for information flow among the surgeon, the pathologist, and, subsequently, the oncologist. This improved communication fosters a more integrated approach to patient care, ultimately contributing to more informed clinical decision-making (Figure 8, Video).
Discussion
Using 3D scanning technology allowed modifications to the OSCC histopathological examination process. By archiving spatial, mapped 3D objects and creating an interactive map, we have gained previously unavailable opportunities for more precise interpretation of histopathological results. Currently, the locations corresponding to a specific H&E slide can be identified with high precision, even along long margins or within complex tumor shapes. This can help guide postoperative decisions, such as planning reoperations or adjuvant radiotherapy. The proposed method has the potential to improve local control of OSCC by more precisely identifying the narrowest areas of the microscopic margin. A postoperative clinical challenge faced by all surgeons treating OSCC is accurately interpreting histopathological results, including precisely determining the width of the HM along its entire length. In standard histopathological analysis, the result obtained is the H&E slide with the narrowest margin. In this case, it is not possible to identify which part of the margin the result pertains to. Furthermore, after cutting the specimen, the pathologist permanently loses the overall view and the ability to accurately identify the location of a specific slice later on. This makes it impossible to track the actual changes in the HM’s width along its entire length. Consequently, in some clinical cases, especially those involving primary extensive tumors, histopathological results are often difficult to interpret and can lead to incorrect conclusions. Making further therapeutic decisions based on standard histopathological analysis is often challenging and depends primarily on clinical experience.
It is well known that histological differentiation (28), DOI, LVI, PNI, a diffuse pattern of invasion (29), and pT stage (30) affect the local recurrence (LR). However, patients with close HM generally have a poor prognosis (31). Therefore, the postoperative therapeutic decision on eligibility for reoperation, observation, or adjuvant radiotherapy must be made based on an analysis of all the above factors. Studies indicate that margin status is one of the most important factors influencing local cancer control (31–33). Regardless of the factors mentioned, the margin status determines the radicalness (34). Therefore, increasing the detail of histopathological results, including identifying the narrowest points of the HM or detecting epithelial dysplasia, will improve the accuracy of interpretation. In the absence of the remaining PFs, the decision to reoperate will be based on the width of the HM. Precisely locating the site with the narrowest HM enables a more accurate, targeted reoperation. This is especially important in cases of extensive primary resections or near critical organs, where limiting the extent of surgical excision is necessary. Additionally, in radiotherapy planning, the oncologist can more precisely identify where to deliver the highest dose of radiation using a virtual map of the area. This approach will help reduce radiation-related complications, including osteoradionecrosis. The primary premise behind the proposed methodology is its potential to improve local control of OSCC through virtual tumor mapping.
A personalized approach to patients, digitalization of medicine, and the use of modern imaging techniques present new challenges and opportunities for improving the effectiveness of cancer treatment. The continuous advancement of precision in various areas of oncological surgery, including robotic surgery and virtual radiotherapy planning, requires updating the standard histopathological procedure. This procedure should align with and reflect the modern technologies used at other stages of patient care in OSCC.
The latest research on OSCC histopathological evaluation focuses on artificial intelligence (AI), which, through the use of specific algorithms, is expected to assist in histopathological diagnosis of OSCC (35–37). Using algorithms to quantify lesions, such as on the HALO platform, enables tissue segmentation with AI assistance (38). Despite the potential of AI in OSCC histopathological assessment, this technique does not allow the determination of the location of histopathological slides on the resected tumor. AI assists the pathologist in accurately evaluating the specimen. Further work is needed to integrate the proposed method with AI’s evolving capabilities. Currently, the technique requires manual processing and mapping on a virtual OSCC. In the future, by leveraging AI capabilities, specific stages of the proposed method can be automated, significantly speeding up and simplifying the procedure.
The ability to virtually combine individual patient treatment stages, including the surgical phase, with oncological therapy creates new opportunities for personalized treatment. Transferring a virtual, annotated 3D tumor model for planning adjuvant radiotherapy—allowing for precise identification of the narrowest or involved margins (HM)—will enhance the accuracy and likely the effectiveness of treatment.
However, the proposed methodology has limitations. It requires proper equipment and training for surgical and pathological teams, including scanning procedures and the use of appropriate software. Additionally, large tumors with highly irregular surfaces and multiple arcades may pose challenges for virtual imaging. Nevertheless, the advantages clearly outweigh the procedure’s inconveniences, which take no more than 10 min. We hope the proposed pilot study will establish a standard for OSCC histopathological analysis and contribute to improved local tumor control.
Conclusion
In summary, the protocol for the virtual presentation of histopathological results of resected OSCC, proposed in our pilot study, significantly improves the accuracy of interpretation and the planning of further treatment steps. Using modern, noninvasive virtual imaging technologies, these methods can be easily and consistently integrated into our daily clinical practice. Additionally, the capacity to archive data that would otherwise be permanently lost in traditional histopathological assessments allows for retrospective analysis of specimens during oncology team meetings or in cases of late local OSCC recurrence. Due to its simplicity and reproducibility, the protocol has the potential to become a routine part of OSCC treatment protocols and training programs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Medical University of Gdansk (Gdansk, Poland (KB/352/2023), approved date 7 August 2023). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AM: Validation, Conceptualization, Resources, Visualization, Investigation, Supervision, Methodology, Software, Data curation, Writing – original draft. RP: Methodology, Writing – review & editing. ŁG: Writing – review & editing, Visualization, Software, Data curation. AP: Data curation, Validation, Writing – review & editing. PC: Formal Analysis, Writing – original draft. BW: Writing – original draft, Formal Analysis. BD: Project administration, Writing – review & editing, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The publication costs were covered by the Medical University of Gdansk, Project number: 01-62025/0008403/643, Medical University of Gdansk, 3a, M. Skłodowskiej-Curie Street, 80–210 Gdansk, Poland.
Acknowledgments
The authors wish to thank Karolina Lis for graphic correction and Klaudia Stawarska, M.A., for performing H&E slide scans.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1617778/full#supplementary-material
Video | Interactive PDF with margin widths and the ability to view H&E slides.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. (2009) 45:309–16. doi: 10.1016/j.oraloncology.2008.06.002
3. Sarode G, Maniyar N, Sarode SC, Jafer M, Patil S, and Awan KH. Epidemiologic aspects of oral cancer. Disease-a-Month. (2020) 66:100988. doi: 10.1016/j.disamonth.2020.100988
4. Kumar M, Nanavati R, Modi T, and Dobariya C. Oral cancer: Etiology and risk factors: A review. J Cancer Res Ther. (2016) 12:458–63. doi: 10.4103/0973-1482.186696
5. Kanatas A, Walshaw EG, Wu J, Fabbroni G, and Chengot P. Prognostic factors in oral cancer surgery - results from a UK tertiary centre. Eur J Surg Oncol. (2023) 49:755–9. doi: 10.1016/j.ejso.2022.11.595
6. Dewenter I, Kumbrink J, Poxleitner P, Smolka W, Liokatis P, Fliefel R, et al. New insights into redox-related risk factors and therapeutic targets in oral squamous cell carcinoma. Oral Oncol. (2023) 147:106573. doi: 10.1016/j.oraloncology.2023.106573
7. Michcik A, Polcyn A, Sikora M, Wach T, Garbacewicz Ł, Drogoszewska B, et al. Oral squamous cell carcinoma-do we always need elective neck dissection? evaluation of clinicopathological factors of greatest prognostic significance: a cross-sectional observational study open access edited by. Front Oncol. (2023) 13:1203439. doi: 10.3389/fonc.2023.1203439
8. Nokovitch L, Maquet C, Crampon F, Taihi I, Roussel LM, Obongo R, et al. Oral cavity squamous cell carcinoma risk factors: state of the art. J Clin Med. (2023) 12(9):3264. doi: 10.3390/jcm12093264
9. D’Cruz AK, Vaish R, Kapre N, Dandekar M, Gupta S, Hawaldar R, et al. Elective versus therapeutic neck dissection in node-negative oral cancer. N Engl J Med. (2015) 373:521–9. doi: 10.1056/nejmoa1506007
10. Alqutub S, Alqutub A, Bakhshwin A, Mofti Z, Alqutub S, Alkhamesi AA, et al. Histopathological predictors of lymph node metastasis in oral cavity squamous cell carcinoma: a systematic review and meta-analysis. Front Oncol. (2024) 14:1401211. doi: 10.3389/fonc.2024.1401211
11. Sahoo A, Panda S, Mohanty N, Jena D, Mishra N, and Surabhi BM. Perinerural, lymphovascular and depths of invasion in extrapolating nodal metastasis in oral cancer. Clin Oral Investig. (2020) 24:747–55. doi: 10.1007/s00784-019-02921-0
12. Huang S, Zhu Y, Cai H, Zhang Y, and Hou J. Impact of lymphovascular invasion in oral squamous cell carcinoma: A meta-analysis. Oral Surg Oral Med Oral Pathol Oral Radiol. (2021) 131:319–328.e1. doi: 10.1016/j.oooo.2020.10.026
13. Wahab A, Onkamo O, Pirinen M, Almangush A, and Salo T. The budding and depth of invasion model in oral cancer: A systematic review and meta-analysis. Oral Dis. (2022) 28:275–83. doi: 10.1111/odi.13671
14. Binmadi NO and Mohamed YA. Impact of worst pattern of invasion on prognosis of oral squamous cell carcinoma: a systematic review and meta-analysis. J Int Med Res. (2023) 51(10):3000605231206260. doi: 10.1177/03000605231206260
15. Ferlito A, Rinaldo A, Robbins KT, Leemans CR, Shah JP, Shaha AR, et al. Changing concepts in the surgical management of the cervical node metastasis. Oral Oncol. (2003) 39:429–35. doi: 10.1016/S1368-8375(03)00010-1
16. Valero C, Zanoni DK, Pillai A, Xu B, Katabi N, Ghossein RA, et al. Nodal characteristics associated with adverse prognosis in oral cavity cancer are linked to host immune status. J Surg Oncol. (2021) 123:141–8. doi: 10.1002/jso.26235
17. Varvares MA, Poti S, Kenyon B, Christopher K, and Walker RJ. Surgical margins and primary site resection in achieving local control in oral cancer resections. Laryngoscope. (2015) 125:2298–307. doi: 10.1002/lary.25397
18. Ooms M, Ponke L, Winnand P, Heitzer M, Peters F, Steiner T, et al. Predictive factors and repetition numbers for intraoperative additional resection of initially involved soft tissue resection margins in oral squamous cell carcinoma: a retrospective study. World J Surg Oncol. (2023) 21:1–9. doi: 10.1186/s12957-023-03192-6
19. Bullock MJ, Beitler JJ, Carlson DL, Fonseca I, Hunt JL, Katabi N, et al. Data set for the reporting of nodal excisions and neck dissection specimens for head and neck tumors explanations and recommendations of the guidelines from the international collaboration on Cancer reporting. Arch Pathol Lab Med. (2019) 143:452–62. doi: 10.5858/arpa.2018-0421-SA
20. Lin MC, Leu YS, Chiang CJ, Ko JY, Wang CP, Yang TL, et al. Adequate surgical margins for oral cancer: A Taiwan cancer registry national database analysis. Oral Oncol. (2021) 119:105358. doi: 10.1016/j.oraloncology.2021.105358
21. Anderson CR, Sisson K, and Moncrieff M. A meta-analysis of margin size and local recurrence in oral squamous cell carcinoma. Oral Oncol. (2015) 51:464–9. doi: 10.1016/j.oraloncology.2015.01.015
22. Liao CT, Chang JT, Wang HM, Ng SH, Hsueh C, Lee LY, et al. Analysis of risk factors of predictive local tumor control in oral cavity cancer. Ann Surg Oncol. (2008) 15:915–22. doi: 10.1245/s10434-007-9761-5
23. El-Fol HA, Noman SA, Beheiri MG, Khalil AM, and Kamel MM. Significance of post-resection tissue shrinkage on surgical margins of oral squamous cell carcinoma. J Cranio-Maxillofacial Surg. (2015) 43:475–82. doi: 10.1016/j.jcms.2015.01.009
24. Chen CH, Hsu MY, Jiang RS, Wu SH, Chen FJ, and Liu SA. Shrinkage of head and neck cancer specimens after formalin fixation. J Chin Med Assoc. (2012) 75:109–13. doi: 10.1016/j.jcma.2012.02.006
25. Michcik A, Jopek M, Pęksa R, Choma P, Garbacewicz Ł, Polcyn A, et al. Oral squamous cell carcinoma and what we lose during formalin fixation: an evaluation of changes in macroscopic resection margins utilizing virtual three-dimensional imaging techniques with analysis based on 947 measurements. Biomedicines. (2024) 12(12):2805. doi: 10.3390/biomedicines12122805
26. Michcik A, Jopek M, Rafał P, Choma P, Garbacewicz Ł, and Polcyn A. Virtual tumor mapping: a new standard for surgeon – pathologist collaboration in treating oral squamous cell carcinoma. Cancers (Basel). (2024) 16(22):3761. doi: 10.3390/cancers16223761
27. Franke J, Koutecký T, and Koutný D. Comparison of sublimation 3D scanning sprays in terms of their effect on the resulting 3D scan, thickness, and sublimation time. Materials. (2023) 16(18):6165. doi: 10.3390/ma16186165
28. Brandwein-Gensler M, Teixeira MS, Lewis CM, Lee B, Rolnitzky L, Hille JJ, et al. Oral squamous cell carcinoma: histologic risk assessment, but not margin status, is strongly predictive of local disease-free and overall survival. Am J Surg Pathol. (2005) 29:167–78. doi: 10.1097/01.pas.0000149
29. Wong LS, McMahon J, Devine J, McLellan D, Thompson E, Farrow A, et al. Influence of close resection margins on local recurrence and disease-specific survival in oral and oropharyngeal carcinoma. Br J Oral Maxillofac Surg. (2012) 50:102–8. doi: 10.1016/j.bjoms.2011.05.008
30. Jerjes W, Upile T, Petrie A, Riskalla A, Hamdoon Z, Vourvachis M, et al. Clinicopathological parameters, recurrence, locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Head Neck Oncol. (2010) 2:1–21. doi: 10.1186/1758-3284-2-9
31. Mannelli G and Comini LV PC. Surgical margins in oral squamous cell cancer: intraoperative evaluation and prognostic impact. Curr Opin Otolaryngol Head Neck Surg. (2019) 27:98–103. doi: 10.1097/MOO.0000000000
32. Hakim SG, von Bialy R, Falougy M, Steller D, Tharun L, Rades D, et al. Impact of stratified resection margin classification on local tumor control and survival in patients with oral squamous cell carcinoma. J Surg Oncol. (2021) 124:1284–95. doi: 10.1002/jso.26655
33. Binahmed A, Nason RW, and Abdoh AA. The clinical significance of the positive surgical margin in oral cancer. Oral Oncol. (2007) 43:780–4. doi: 10.1016/j.oraloncology.2006.10.001
34. Fang FM, Chuang HC, Chou SY, Huang TL, Wang CJ, Lin YT, et al. The therapeutic benefit of radical resection for T4b oral cavity squamous cell carcinoma with partial or complete response after radical chemo-intensity-modulated radiotherapy (IMRT). Ann Surg Oncol. (2016) 23:866–73. doi: 10.1245/s10434-016-5
35. Ahmad M, Irfan MA, Sadique U, Haq IU, Jan A, Khattak MI, et al. Multi-method analysis of histopathological image for early diagnosis of oral squamous cell carcinoma using deep learning and hybrid techniques. Cancers (Basel). (2023) 15(21):5247. doi: 10.3390/cancers15215247
36. Abdul NS, Shivakumar GC, Sangappa SB, Di Blasio M, Crimi S, Cicciù M, et al. Applications of artificial intelligence in the field of oral and maxillofacial pathology: a systematic review and meta-analysis. BMC Oral Health. (2024) 24:1–12. doi: 10.1186/s12903-023-03533-7
37. Pirayesh Z, Mohammad-Rahimi H, Ghasemi N, Motamedian SR, Sadeghi TS, Koohi H, et al. Deep learning-based image classification and segmentation on digital histopathology for oral squamous cell carcinoma: A systematic review and meta-analysis. J Oral Pathol Med. (2024) 53(9):551–66. doi: 10.1111/jop.13578
Keywords: oral squamous cell carcinoma, histopathological analysis, three-dimensional imaging, local control, 3D objects, virtual medicine
Citation: Michcik A, Pęksa R, Garbacewicz Ł, Polcyn A, Choma P, Wojciechowska B and Drogoszewska B (2025) A novel method for histopathological analysis of oral squamous cell carcinoma: a pilot study using three-dimensional imaging to enhance local control. Front. Oncol. 15:1617778. doi: 10.3389/fonc.2025.1617778
Received: 24 April 2025; Accepted: 17 October 2025;
Published: 04 November 2025.
Edited by:
Manuel Tousidonis, Gregorio Marañón Hospital, SpainReviewed by:
Michał Gontarz, Jagiellonian University Medical College, PolandRené Andrés Martinez-Flores, Universidad Andrés Bello, Chile
Copyright © 2025 Michcik, Pęksa, Garbacewicz, Polcyn, Choma, Wojciechowska and Drogoszewska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam Michcik, YWRhbW1pY2hjaWtAZ3VtZWQuZWR1LnBs
 Adam Michcik
Adam Michcik Rafał Pęksa
Rafał Pęksa Łukasz Garbacewicz
Łukasz Garbacewicz Adam Polcyn
Adam Polcyn Piotr Choma
Piotr Choma Barbara Wojciechowska
Barbara Wojciechowska Barbara Drogoszewska1,2
Barbara Drogoszewska1,2