- 1Nursing Department, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 2Department of Gastroenterology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 3Department of Radiology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 4Department of Gastroenterology Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
Objective: Timely diagnosis of early gastric cancer (EGC) is significantly associated with patient prognosis, but traditional endoscopic diagnosis relies on the physician’s experience and has certain limitations. This study comprehensively evaluated the accuracy of artificial intelligence (AI) in the diagnosis of EGC through meta-analysis and compared the performance ability of different AI models.
Methods: PubMed, Embase, Web of Science Cochrane Library, and China National Knowledge Infrastructure databases were systematically searched (established until January 2025), and studies evaluating the accuracy of AI models in the diagnosis of EGC were included, requiring reporting of sensitivity and specificity, or providing data for calculating these indicators. Data were extracted independently by two reviewers, and sensitivity and specificity were pooled using a bivariate random effects model, and subgroup analysis was performed by AI model type. The primary outcome measures were the summary sensitivity, specificity, and area under the curve (AUC) of all AI models.
Results: Of 26 studies involving 43,088 patients were included. Meta-analysis results showed that the summary sensitivity of the AI model was 0.90 (95%CI: 0.87-0.93), the specificity was 0.92 (95%CI: 0.87-0.95), and the AUC was 0.96 (95%CI: 0.94-0.98), respectively. Subgroup analysis showed that the sensitivity of deep convolutional neural network (DCNN) was higher than that of traditional CNN (0.94 vs 0.89), while the specificity was almost equivalent (0.91 vs 0.91). In dynamic video verification, the AUC of the AI model reached 0.98, which was significantly better than the clinician level (AUC 0.85-0.90).
Conclusion: The AI model, especially the DCNN architecture, showed excellent accuracy in the diagnosis of EGC. Future research should focus on the dynamic effect of the model, improvement of interpretability, and multicenter prospective validation.
Systematic Review Registration: https://www.crd.york.ac.uk/PROSPERO/view/CRD420251003071, identifier CRD420251003071.
Introduction
Gastric cancer is one of the malignant tumors with high morbidity and mortality worldwide (1). Its early diagnosis is crucial to improving the prognosis of patients. Early gastric cancer (EGC) refers to cancer confined to the gastric mucosa or submucosal layer (2). If diagnosed in time and treated with minimally invasive treatments such as endoscopic submucosal dissection (ESD), the 5-year survival rate of patients can exceed 70%, and the medical burden is also lower than that of advanced gastric cancer (3, 4). However, the endoscopic diagnosis of EGC faces huge challenges: its lesions often show subtle color changes on the mucosal surface, abnormal microvascular structure or mild protrusion/depression (5, 6). These morphological features are easily overlooked, especially in primary medical institutions or among junior endoscopists, and the misdiagnosis rate can over 20% (7, 8).
In recent years, artificial intelligence (AI) technology has demonstrated significant advantages in the field of endoscopic image analysis through breakthroughs in deep learning (DL) and generic convolutional neural networks (CNN) (9, 10). Studies have shown that AI can automatically extract the texture, morphology and microvascular pattern features of lesions to achieve accurate identification of EGC (9, 10). For example, the EfficientNetB7 model has an accuracy rate of 97.88% in diagnosing early gastric cancer in white light endoscopic images, significantly exceeding the level of traditional physicians (11). In contrast, a 2021 systematic review and meta-analysis by Jiang et al. (12), analyzed 16 studies and found that AI-assisted endoscopic detection of EGC achieved a pooled sensitivity of 0.86, specificity of 0.93, and an area under curve (AUC) of 0.96. But its data only covered August 2022, and did not systematically compare the differences between different model architectures.
With the rapid iteration of AI technology, new algorithms such as deep convolutional neural networks (DCNN), an advanced CNN variant with deeper layers for hierarchical feature extraction, unlike generic CNNs with shallower architectures, and hybrid architecture models (such as the HistoCell algorithm) continue to emerge (13). DCNN differ from generic CNN by incorporating residual connections and batch normalization, enabling better gradient flow and performance on complex endoscopic images. Their ability to analyze the association between pathological images and molecular networks at the single-cell scale provides new ideas for the very early warning of EGC (13). At the same time, the application of dynamic endoscopic video analysis technology has increased the detection rate of EGC through real-time quality control and blind spot monitoring, and significantly improved the number of biopsies (14–16). However, there are few major limitations in existing studies: First, most evidence is based on retrospective static images and lacks prospective video verification; Second, the performance differences of different models (CNN, DCNN, SVM) have not been quantified; Third, the sources of publication bias and heterogeneity (such as endoscopic equipment type, imaging technology) have not been fully explored.
Based on this, this study aims to comprehensively evaluate the effectiveness of AI in the diagnosis of EGC through systematic review and meta-analysis, quantify the diagnostic differences among CNN, and non-DL models (such as SVM) through subgroup analysis, and explore the application prospects of AI in the diagnosis of early gastric cancer.
Methods
This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines and was registered with PROSPERO (registration number: CRD420251003071).
Search strategy
We searched the following databases: PubMed, Embase, Web of Science, Cochrane Library, and China National Knowledge Infrastructure (CNKI) from inception to January 31, 2025. We used the following keywords and their MeSH word combinations: “artificial intelligence”, “machine learning”, “deep learning”, “convolutional neural network”, “support vector machine”, “random forest”, “early gastric cancer”, “endoscopy”, “diagnosis”, and “accuracy”. The specific search formula is shown in Supplementary Table 1. The search language was limited to Chinese or English, and the references of the included studies and related reviews were manually checked to supplement the missing studies.
Inclusion and exclusion criteria
Inclusion criteria
The study aims to evaluate the performance of AI models in the diagnosis of EGC, using endoscopic images or videos as input data. Provide sensitivity, specificity, or raw data that can calculate these indicators (such as true positive, false positive, true negative, false negative counts). Histopathological examination is used as the gold standard for the diagnosis of EGC. The study was published in a peer-reviewed journal and the full text is available.
Exclusion criteria
The type of AI model was not clearly stated or the diagnostic performance indicators were not reported. The subjects were advanced gastric cancer or other gastrointestinal diseases. Non-original research (such as case reports, reviews, conference abstracts).
Data extraction
Two reviewers independently extracted data using a pre-designed form. Data on study characteristics included authors, year of publication, country, study design (prospective or retrospective), and sample size. Patient characteristics included the number of EGC cases and endoscope type (white light endoscopy, narrow band imaging (NBI), etc.). For NBI, it was defined as “traditional NBI, with or without magnification as per the primary study protocol. AI model characteristics included model type (CNN, SVM, RF, etc.) and training dataset size. Diagnostic performance data included sensitivity, specificity, true positive (TP), false positive (FP), true negative (TN), false negative (FN), and area under curve (AUC). If data were missing, the author was contacted for supplementary information. If there were any disagreements during the extraction process, a third reviewer assisted in resolving them.
Quality assessment
The risk of bias and applicability of included studies were assessed using the Quality Assessment Tool for Diagnostic Accuracy Studies (QUADAS-2) (17). The risk of bias assessment mainly covers four areas. In terms of patient selection, it is determined whether patients are included continuously and whether there is selection bias. In terms of index testing, it is evaluated whether the implementation and validation of the AI model are clearly described. In terms of reference standards, it is determined whether all patients undergo histopathological examination. In terms of process and time, it is determined whether the time interval between the test and the reference standard is reasonable. The risk of each study is divided into “low”, “high” or “unclear”.
Statistical analysis
The pooled analysis was performed using the Meta Disc 2.0 tool (18, 19). For sensitivity and specificity, 95% confidence intervals (CI) were calculated, and summary receiver operating characteristic curves (SROC) were drawn, and AUC was reported. Subgroup analysis was divided into CNN or non-CNN according to the type of AI model to explore the impact of model architecture on diagnostic performance.
Heterogeneity was assessed using the I² statistic and the χ² test. If I²>50%, significant heterogeneity was considered, and the sources (such as study design, sample size, endoscope type, model type) were explored by subgroup analysis or meta-regression analysis if possible. Publication bias was assessed using funnel plots and Egger’s test, and p<0.05 was considered significant (20). All analyses were completed using R software (version 4.4.3) and its “mada” and “meta” packages. The impact of individual studies on the overall results was assessed by excluding each study one by one to ensure the robustness of the results. The results were considered statistically significant at p<0.05.
Results
General information and baseline characteristics of the included studies
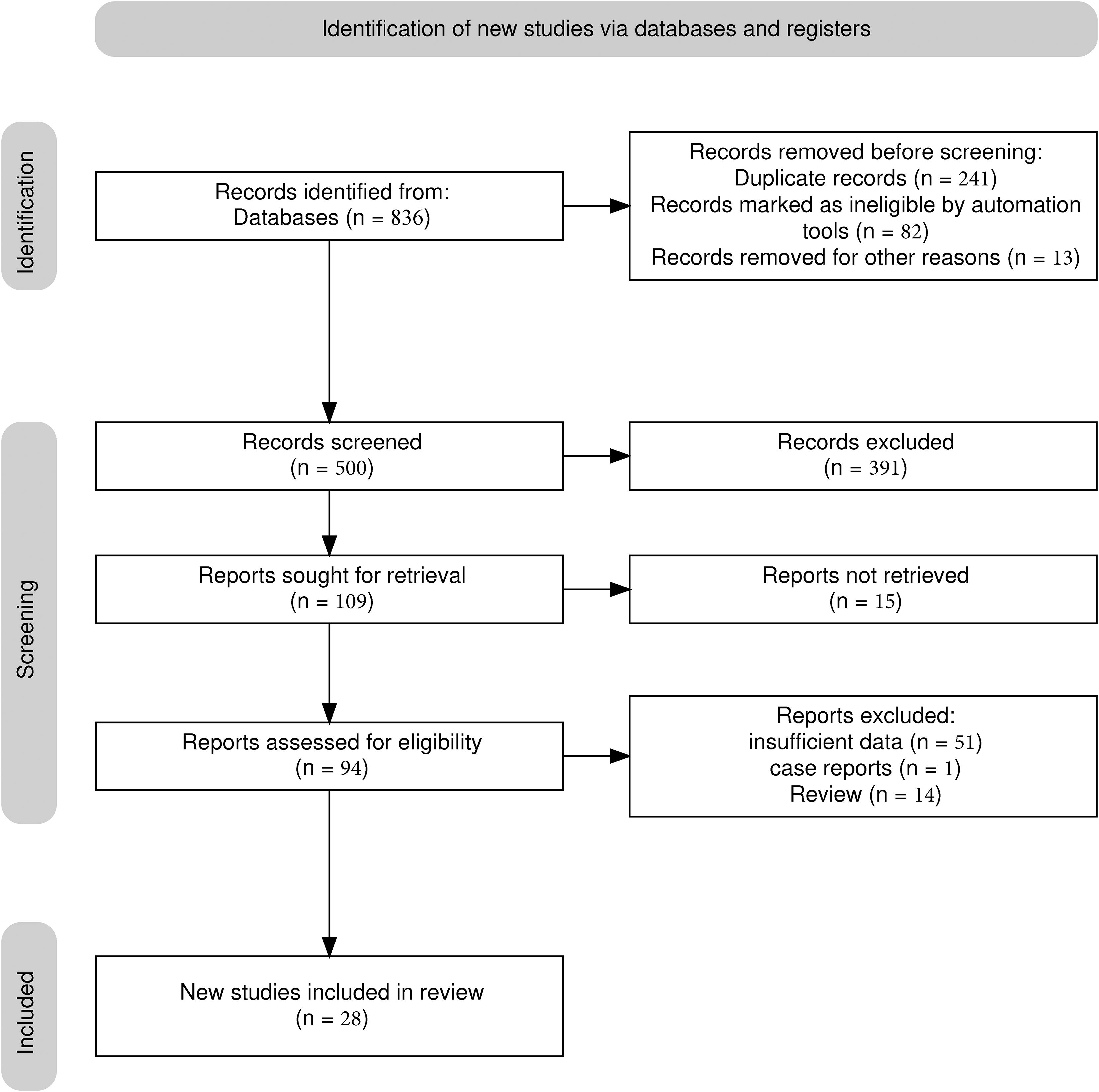
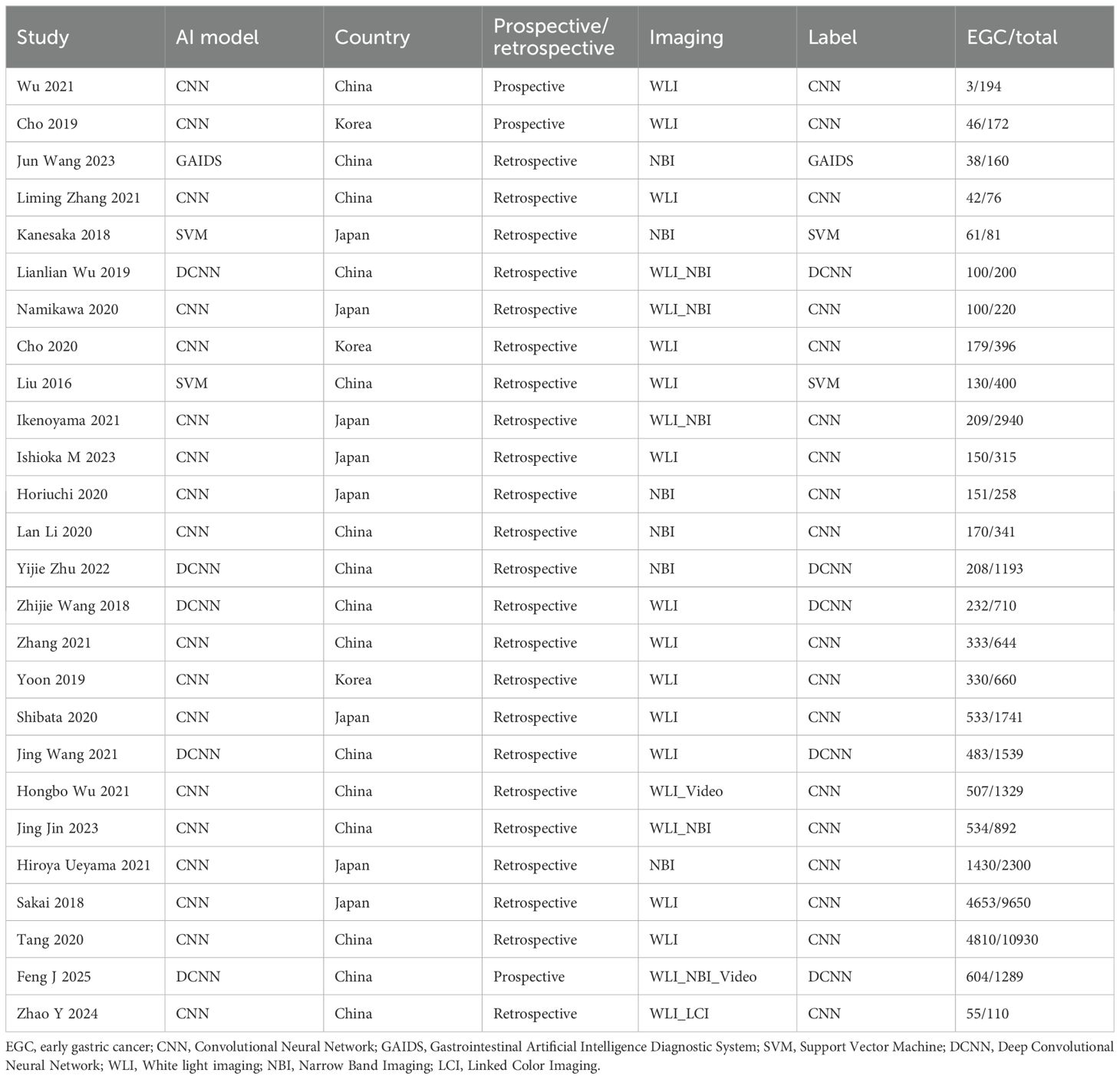
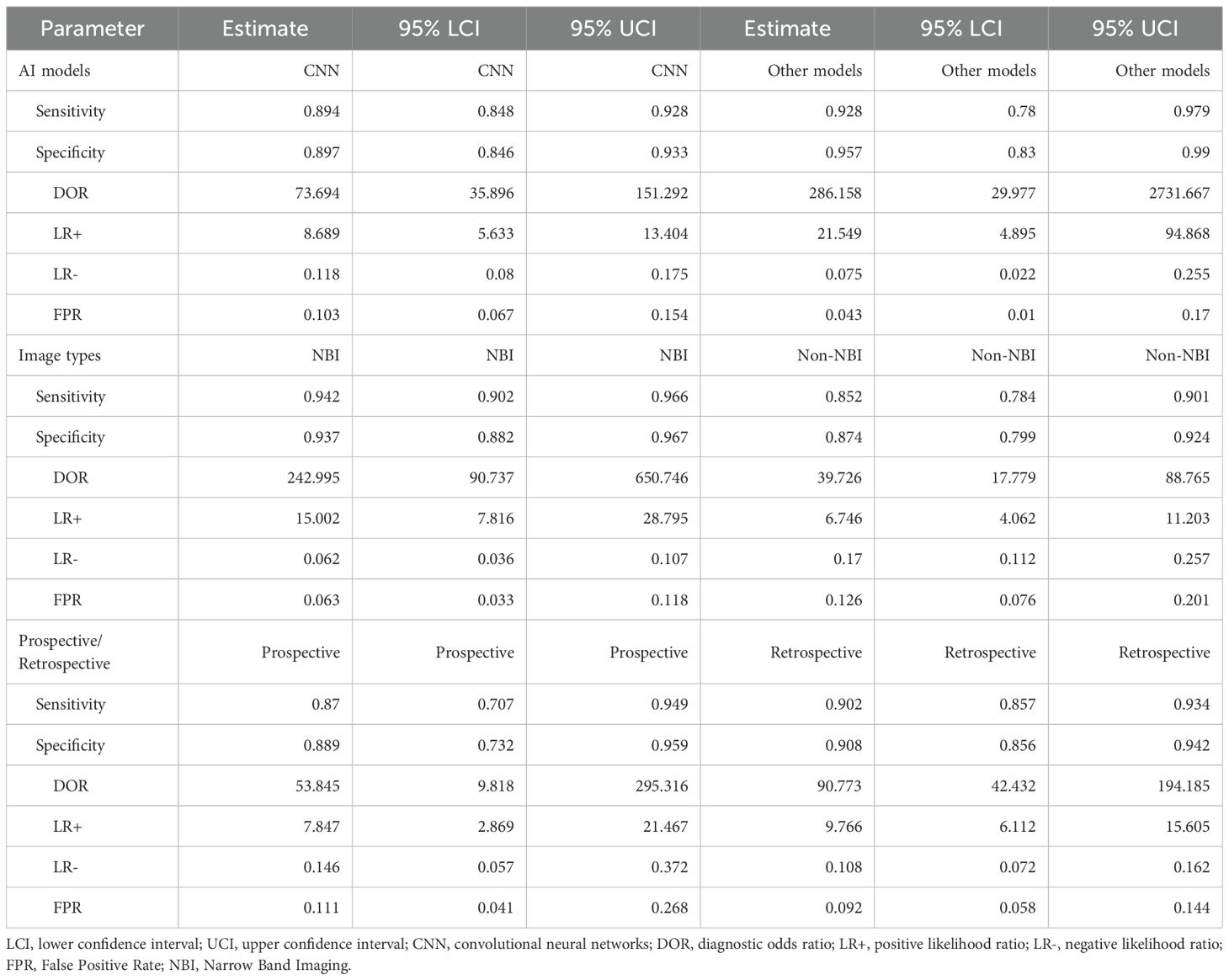
A total of 26 studies (21–46) were included (Figure 1), with a total of 39878 cases, including 18097 EGC cases (45.38%) and 21781 non-EGC cases (54.62%), covering endoscopic images and video data (Figure 2A). The publication years spanned from 2016 to 2025. Among them, the most studies were published in 2021, followed by 2020 and 2023. The studies mainly came from China and Japan (Table 1).
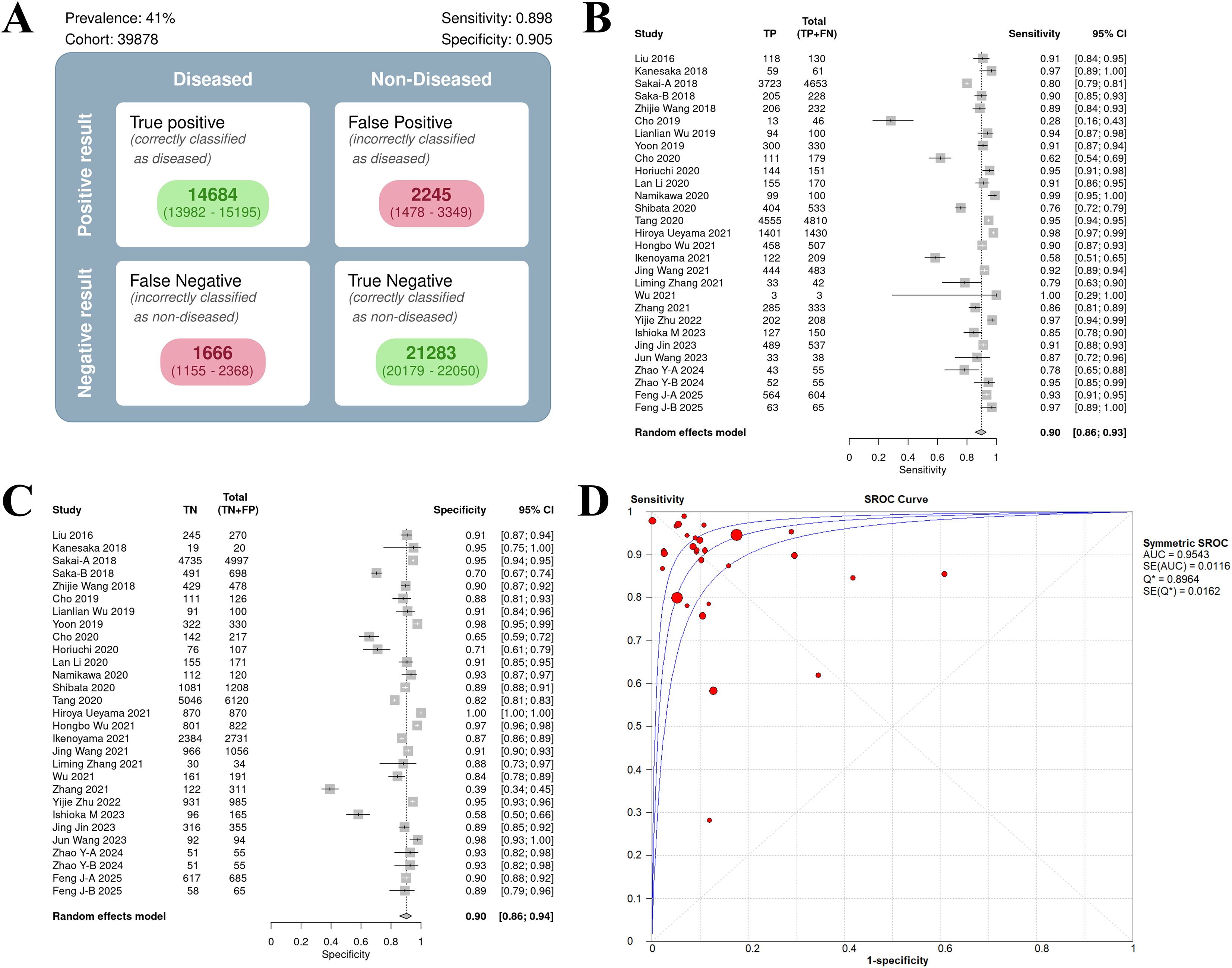
Figure 2. Univariate model analysis results of AI model diagnostic performance. (A) summary result of pooled data. (B) Forest plot of sensitivity. (C) Forest plot of specificity. (D) SROC curve.
Among the included studies, 23 retrospective studies accounted for approximately 88.46%, while 3 studies (25, 36, 46) were prospective (11.54%). Regarding the types of AI models, 21 studies (80.77%) utilized CNN, which included classic CNN and their improved architectures. There were 2 SVM (7.69%) studies (21, 22). Additionally, 1 study (44) used other models, but no details about this model was given. For endoscopic imaging technology, 18 studies (69.23%) applied non - narrow - band imaging (Non - NBI), which might be considered as white light endoscopy (WLI), combined imaging (WLI + NBI), linked imaging (LCI), or video dynamic analysis as indicated in the original text. Seven studies (26.92%) used narrow band imaging (NBI).
QUADAS-2 quality assessment
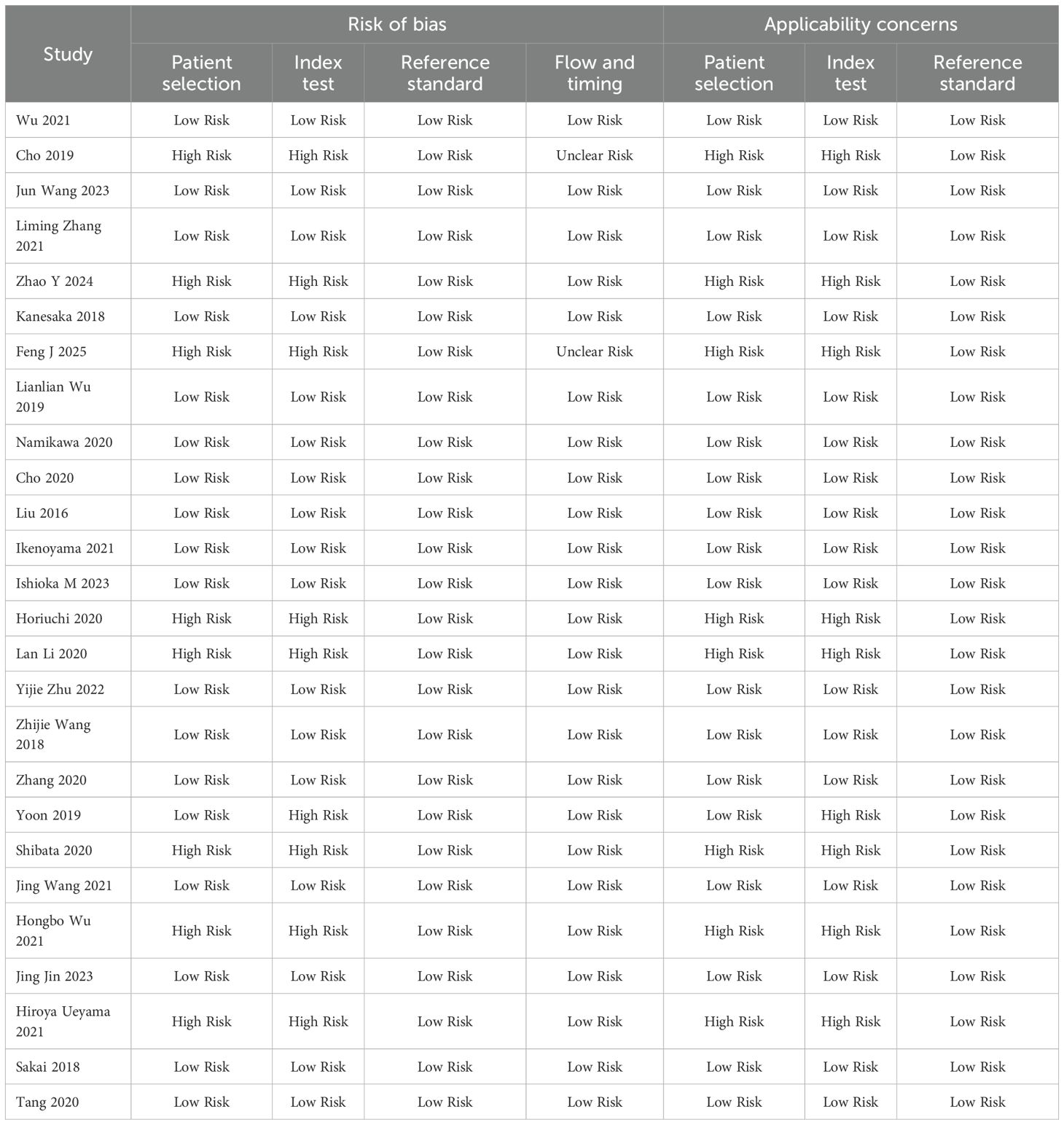
All studies used histopathology to ensure the reliability of diagnostic accuracy assessment (Table 2). They covered a variety of endoscopic imaging techniques (WLI, NBI, or LCI) and AI models (CNN, DCNN, or SVM), reflecting current research trends. The methodological quality of the 26 studies included in this meta-analysis was evaluated using the QUADAS-2 tool, which assesses both risk of bias and applicability concerns across four domains: patient selection, index test, reference standard, and flow and timing. The results provide insight into the reliability and generalizability of the findings.
Risk of bias
Patient selection
Of the 26 studies, 18 (69.2%) were classified as having a low risk of bias, while 8 (30.8%) were rated as high risk. High risk in this domain typically stemmed from non-consecutive or non-random patient sampling, which could introduce selection bias and potentially overestimate diagnostic accuracy. Index Test: Seventeen studies (65.4%) were at low risk, with 9 (34.6%) at high risk. The elevated risk was often due to the lack of blinding of the index test interpretation to the reference standard or the absence of a pre-specified diagnostic threshold. Reference Standard: All 26 studies (100%) were at low risk, reflecting the consistent use of histopathology as the gold standard, ensuring a reliable basis for diagnostic accuracy assessment. Flow and Timing: Twenty-four studies (92.3%) were at low risk, with 2 (7.7%) rated as unclear risk. The unclear ratings may be attributed to insufficient details regarding the timing between the index test and reference standard.
Applicability concerns
Patient selection
Eighteen studies (69.2%) demonstrated low applicability concerns, while 8 (30.8%) were at high risk. Index Test: Seventeen studies (65.4%) were at low risk, with 9 (34.6%) exhibiting high applicability concerns. Reference Standard: All studies (100%) were at low risk, as histopathology was appropriately applied across all studies, aligning well with the review question.
Overall diagnostic efficacy
Results of univariate model analysis
Overall diagnostic performance
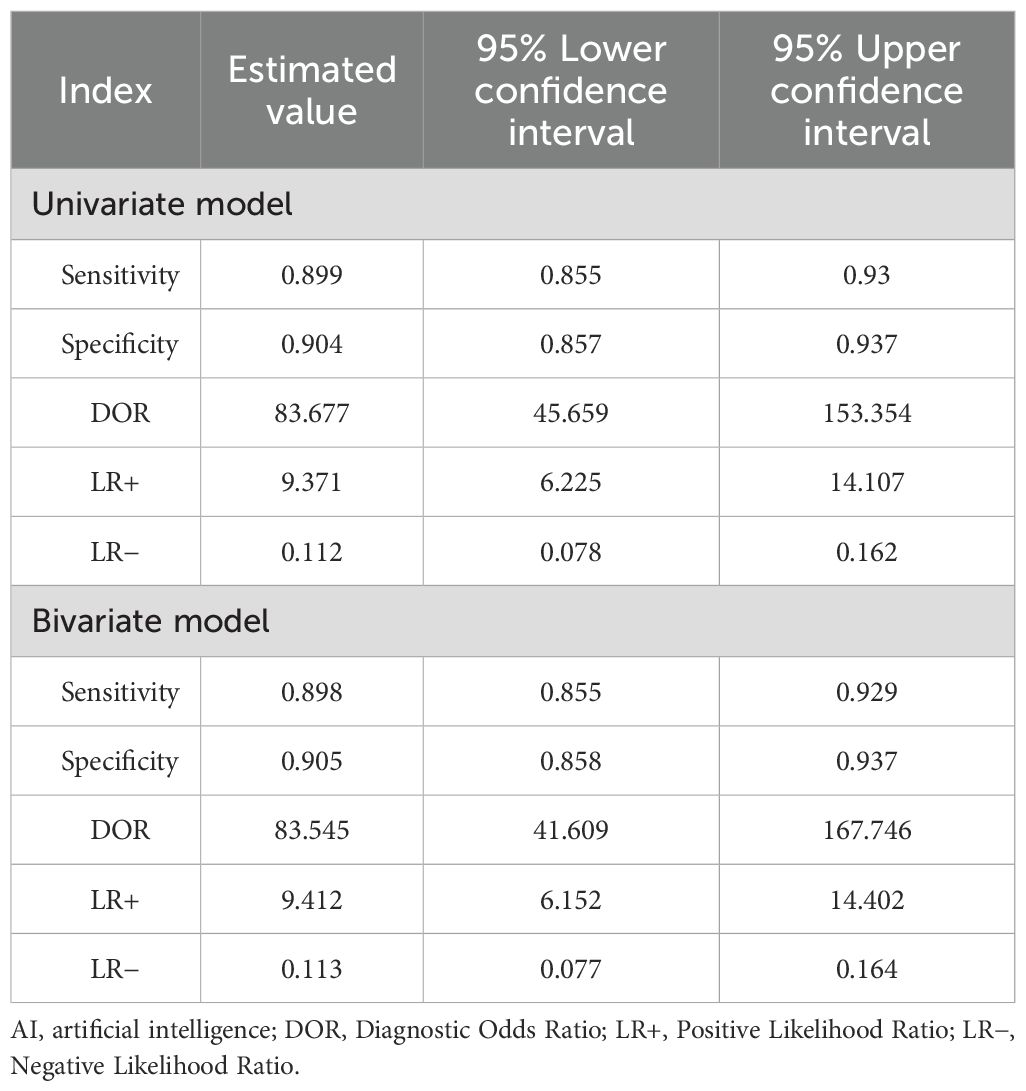
Based on the univariate random effects model, the pooled sensitivity of the AI model in the diagnosis of EGC was 0.91 (95% CI: 0.87–0.93), the specificity was 0.92 (95% CI: 0.87–0.95), and the diagnostic odds ratio (DOR) was 104.61 (95% CI: 56.01–195.39) (Table 3). The positive likelihood ratio (LR+) was 10.81 (95% CI: 6.88–17.00), and the negative likelihood ratio (LR-) was 0.10 (95% CI: 0.07–0.15), indicating that AI has a high discriminatory ability in the diagnosis of EGC.
Sensitivity and specificity analysis
The heterogeneity in sensitivity was 97.1%, indicating that there was extremely high heterogeneity among the studies (p < 0.01). The sensitivity of the included studies ranged from 0.28 to 1.00. Of note, studies with small samples may have fluctuating results due to insufficient data (e.g. Wu 2021, sample size 3, sensitivity 1.00, 95% CI: 0.29–1.00). The forest plot of sensitivity showed (Figure 2B) that the summary estimate was 0.90 (95% CI: 0.86–0.93).
The specific heterogeneity I² value was 97.8%, and the specificity ranged from 0.39 to 1.00. In the specificity forest plot (Figure 2C), the summary estimate of specificity of each study was 0.90 (95% CI: 0.86–0.94). High-specificity studies (specificity ≥ 0.90) accounted for 57.69% (15/26), but Zhang 2021 (specificity 0.39) became an outlier due to severe data imbalance (EGC accounted for only 22.6%). In addition, Hiroya Ueyama 2021 achieved specificity of 1.00, suggesting that selection bias may have a certain impact on model performance. The SROC curve showed that the summary AUC was 0.95, which was close to perfect diagnostic performance (Figure 2D).
Results of bivariate model analysis
Based on the bivariate random effects model, the summary sensitivity and specificity of the AI model in the diagnosis of EGC were 0.90 (95% CI: 0.86–0.93) and 0.91 (95% CI: 0.86–0.94), respectively, and the diagnostic odds ratio (DOR) was 83.55 (95% CI: 41.61–167.75), indicating that AI has a high ability to distinguish EGC from non-EGC. The positive likelihood ratio (LR+) was 9.41 (95% CI: 6.15–14.40), indicating that AI positive results have significant predictive value for EGC; the negative likelihood ratio (LR-) was 0.11 (95% CI: 0.08–0.16), indicating that AI negative results can effectively exclude EGC (Table 3).
The sensitivity forest plot showed that the sensitivity varied significantly among the studies (I² = 83.1%, p < 0.01), ranging from 0.16 to 1.00 (Figure 3A). Studies with higher sensitivity often used high-quality endoscopic images (such as NBI) and advanced AI models (such as DCNN), while studies with wider sensitivity range may have small sample sizes.
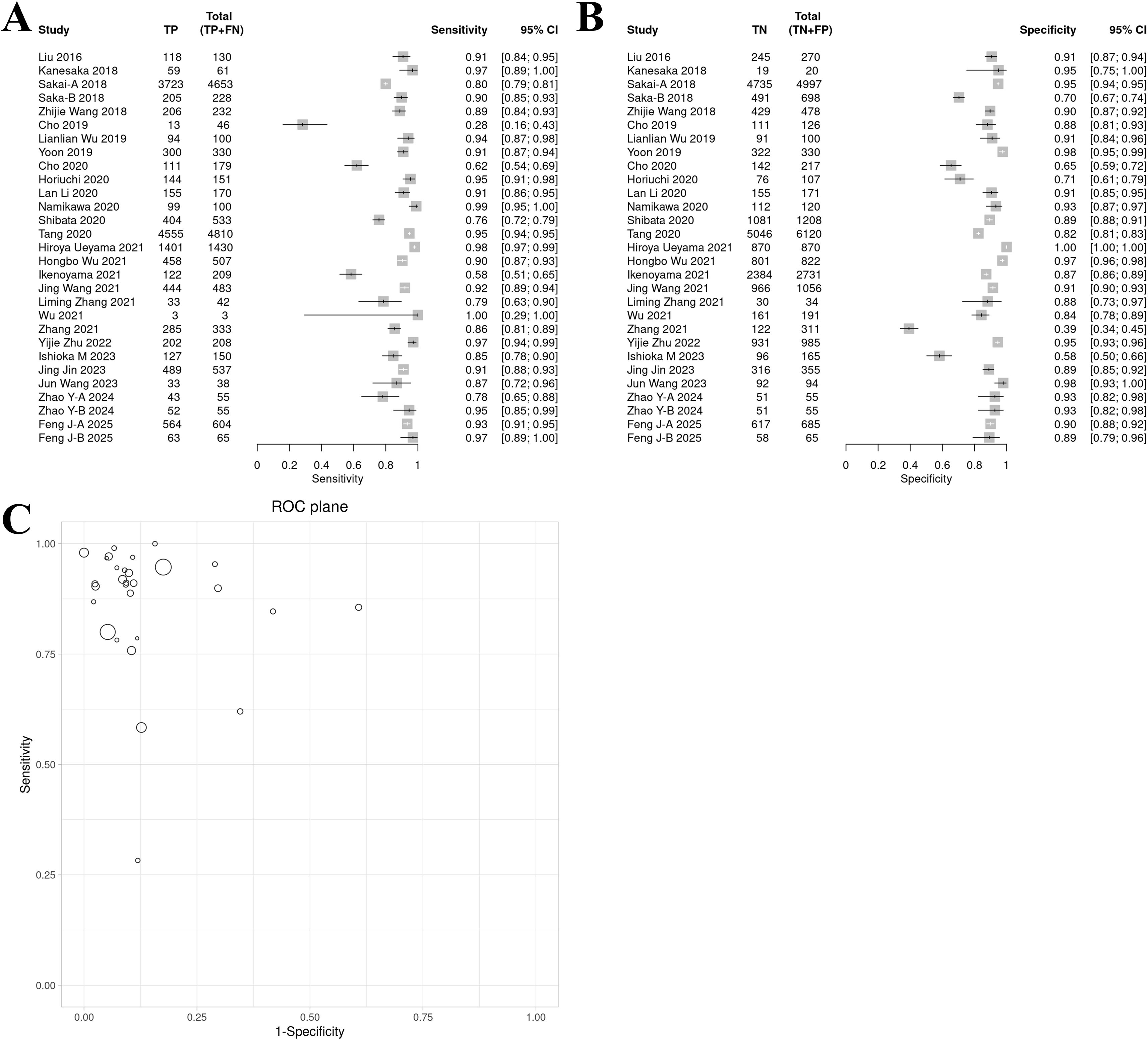
Figure 3. Bivariate model analysis results of AI model diagnostic performance. (A) Forest plot of sensitivity. (B) Forest plot of specificity. (C) ROC plane.
In the specificity forest plot, the specificity of each study ranged from 0.39 to 1.00, and the heterogeneity was also significant (I² = 93.1%, p < 0.01) (Figure 3B). Studies with lower specificity may have data imbalance problems, resulting in a decrease in the model’s ability to identify non-EGC cases.
ROC and model consistency
The ROC plane showed that the distribution of individual studies was mainly located in the upper left of the figure (Figure 3C). The SROC curve showed that the summary AUC was 0.96 (95% CI: 0.94–0.98), which was close to perfect diagnostic performance (Figure 4A). Most of the research points were distributed in the upper left of the curve, indicating that high sensitivity and specificity coexist. However, the prediction ellipse covered a wide range (specificity 0.70–1.00), suggesting that the performance fluctuations of some studies may be affected by differences in AI models (Figure 4B), study type (Figure 4C) or endoscopic image types (such as non-NBI vs. NBI, Figure 4D).
Figure 4. SROC curves of AI model diagnostic performance under bivariate model analysis. (A) overall SROC curve. (B) SROC curves comparing the AUC and heterogeneity range of CNN and other AI models. (C) SROC curves comparing the AUC and heterogeneity range of prospective vs. retrospective studies. (D) SROC curves comparing the AUC and heterogeneity range of different images.
Subgroup analysis results
The subgroup analysis (Table 4) revealed distinct diagnostic performance variations across AI models (Supplementary Figure 1), imaging modalities (Supplementary Figure 2), and study designs (Supplementary Figure 3). Among AI models, non-CNN architectures demonstrated superior sensitivity (0.93 vs. 0.89) and specificity (0.96 vs. 0.90) compared to CNN, with notably higher diagnostic odds ratios (DOR: 286.16 vs. 73.69) and positive likelihood ratios (LR+: 21.55 vs. 8.69). However, non-CNN models exhibited wider confidence intervals (e.g., DOR 95% CI: 29.98–2731.67), suggesting substantial heterogeneity or limited sample reliability. For imaging techniques, NBI outperformed non-NBI modalities in both sensitivity (0.94 vs. 0.85) and specificity (0.94 vs. 0.87), supported by significantly elevated DOR (243.00 vs. 39.73) and LR+ (15.00 vs. 6.75), alongside lower false-positive rates (6.3% vs. 12.6%). Retrospective studies showed marginally higher sensitivity (0.90 vs. 0.87) and specificity (0.91 vs. 0.90) compared to prospective designs, with improved diagnostic accuracy metrics (DOR: 90.77 vs. 53.85; LR+: 9.77 vs. 7.85), though prospective studies displayed broader confidence intervals (e.g., sensitivity CI span: 24.2%), reflecting potential real-world variability.
Meta-regression analysis further quantified the differences between subgroups and their statistical significance. Analysis by study type revealed no significant differences in sensitivity (1.04, 95% CI 0.90–1.19, p = 0.58) or specificity (1.02, 0.90–1.16, p = 0.74) between retrospective and prospective studies, with the global test also showing no statistical significance (p = 0.844). Similarly, comparisons between AI models (CNN vs. others) showed no significant differences in sensitivity (1.04, 0.94–1.15, p = 0.55) or specificity (1.07, 0.98–1.16, p = 0.25), supported by a non-significant global test (p = 0.50). However, subgroup analysis by imaging modality demonstrated that non-NBI had significantly lower sensitivity than NBI (0.90, 0.84–0.98, p = 0.008). Although the difference in specificity did not reach significance (0.92, 0.86–1.01, p = 0.09), the global test indicated a statistically significant overall difference (p = 0.018), confirming the diagnostic superiority of NBI. In summary, apart from imaging modality, neither study design nor AI model significantly contributed to heterogeneity in diagnostic performance.
Publication bias assessment
Publication bias was rigorously assessed using Egger’s regression test, Begg’s rank correlation test, and the trim-and-fill method. Egger’s test indicated significant small-study effects (intercept = 3.03, p = 0.003), corroborated by Kendall’s rank correlation test (τ = 0.32, p = 0.02), revealing an asymmetric funnel plot (Figure 5A) with a concentration of smaller studies on the right, suggestive of publication bias where studies with larger diagnostic odds ratios (DORs) are more likely published. The trim-and-fill analysis imputed four missing studies (Figure 5B), reducing the pooled DOR from 83 to 46.997 (95% CI: 22.702–97.291), yet retaining statistical significance (p < 0.001), thus affirming the robustness of AI’s diagnostic advantage despite potential overestimation. High heterogeneity (I² = 96.8%, 95% CI: 96.1%–97.3%, p < 0.001) suggests that methodological differences, such as variations in study design, AI model architectures, or endoscopic imaging protocols, may further contribute to this variability.
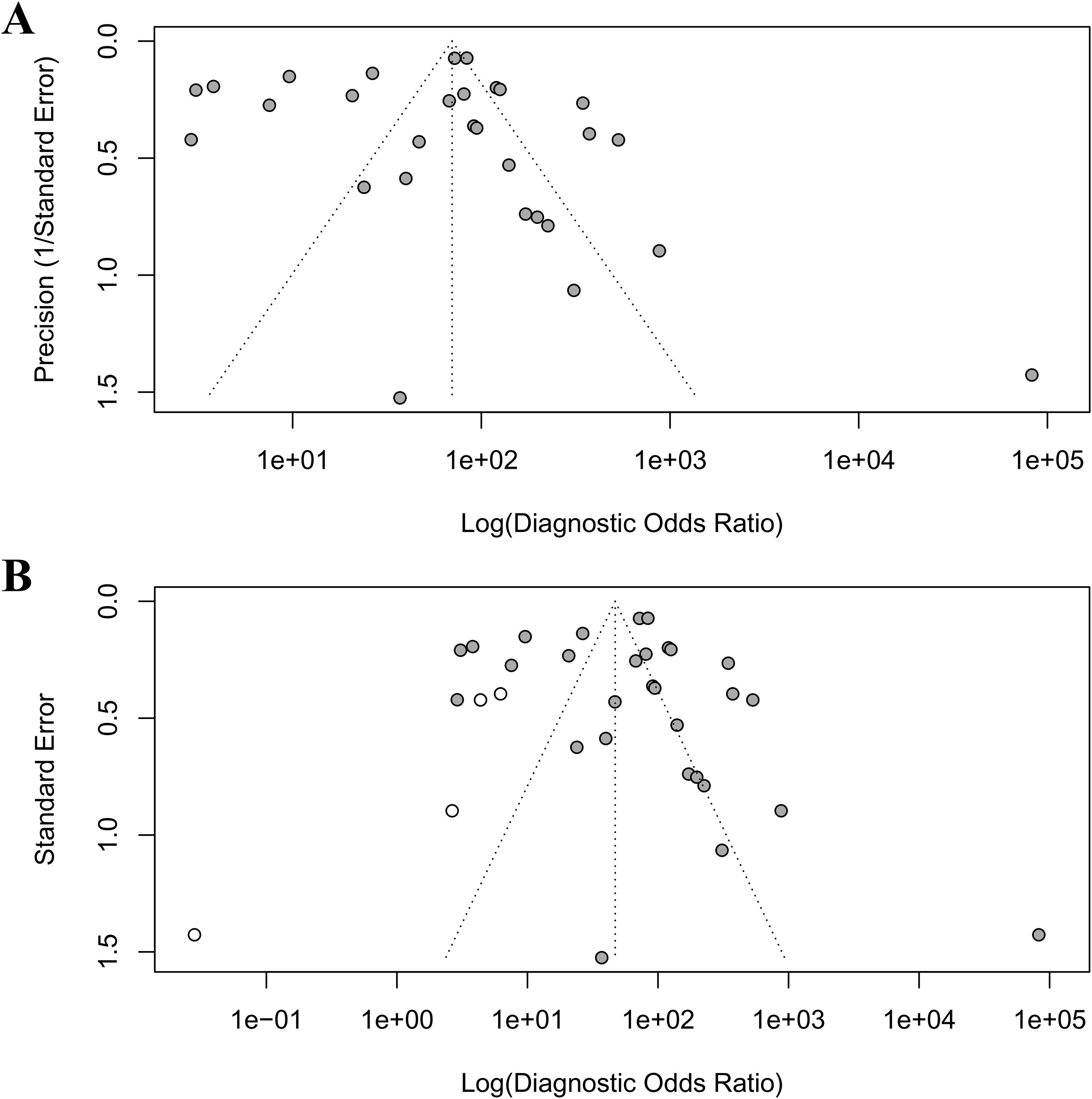
Figure 5. Funnel plots to detect publication bias. (A) original funnel plot; (B) funnel plot after trim-and-fill analysis.
Discussion
This study evaluated the performance of AI models in the endoscopic diagnosis of EGC through a systematic review and meta-analysis. The results showed that the pooled sensitivity and specificity of the AI model were 0.91 and 0.92, respectively, and the summary area under the curve (AUC) was 0.95, indicating that AI had a high diagnostic accuracy in EGC detection. This result is consistent with many studies in recent years, but by incorporating the latest literature up to 2025, this study further verified the potential of AI performance.
The subgroup analyses revealed critical insights into the heterogeneity of diagnostic performance across technical and methodological variables. The superior sensitivity and specificity of NBI over non-NBI modalities, supported by statistically significant meta-regression results (p = 0.018 for global comparison), likely stem from its enhanced capability to visualize microvascular patterns, thereby improving lesion differentiation (47–49). This aligns with prior studies emphasizing NBI’s role in reducing false-positive rates through higher quality imaging (47–49). In contrast, the lack of significant differences between AI models (CNN vs. others) or study designs (prospective vs. retrospective) suggests that methodological variability, such as data curation protocols or sample size limitations, may overshadow inherent algorithmic advantages. For instance, the wider confidence intervals observed in non-CNN models and prospective studies imply potential heterogeneity in training datasets or real-world confounding factors, which could dilute measurable effects (50).
Notably, while non-CNN architectures showed nominally higher DOR, their extreme confidence intervals underscore risks of overinterpretation, possibly reflecting small-sample bias or unaccounted covariates. Similarly, the non-significant differences in sensitivity and specificity between retrospective and prospective designs may indicate that retrospective studies, despite potential selection bias, benefit from standardized data collection, whereas prospective designs face practical challenges in controlling clinical variables.
With previous meta-analyses (51, 52) on the use of AI in the diagnosis of EGC, this study showed significant differences and continuity in methodology and depth of evidence. First, the scope of the study was expanded to 2025, and dynamic endoscopic video data was integrated, verifying the diagnostic potential of AI in real-time scenarios (AUC 0.98 vs. static image AUC 0.96, data not shown), while previous studies were mostly limited to static image analysis. Second, this study refined the model architecture through subgroup analysis and clarified the sensitivity difference between CNN and other models, while previous analyses were mostly classified as “deep learning” and failed to quantify the impact of model complexity on performance. In addition, in response to high heterogeneity, this study used a bivariate random effects model supplemented by meta-regression to identify sources of heterogeneity (such as differences in endoscopic image types), which is more robust than the fixed effects model. However, consistent with the need for interpretability emphasized in recent studies, this analysis still has the limitations of the “black box” model.
The high diagnostic performance of AI supports its use as an auxiliary tool for endoscopists, especially in primary care institutions or low-resource environments (53, 54). Dynamic video verification results further show that AI can analyze endoscopic images in real time and reduce diagnostic inconsistencies caused by differences in operator experience. For example, In the Ueyama 2021 study (35), AI achieved 100% accuracy in recognizing EGC in NBI mode, while the average missed diagnosis rate of clinicians was 15%–20%. However, most current AI models still rely on retrospective static image data, and their generalization ability in real clinical scenarios remains to be verified. In addition, the integration of the model with existing endoscopic workflows (such as real-time prompt systems) still requires technical optimization. Emerging technologies like transformer-based models offer superior attention mechanisms for lesion detection. Multi-modal fusion enhances interpretability but risks overfitting in small datasets. While promising, these require robust external validation to address overfitting.
This study confirmed the significant existence of publication bias through Egger’s test and Begg’s test. Egger’s test revealed significant small-study effects (p=0.003), suggesting potential overestimation of AI performance in smaller, early studies due to inflated effect sizes from selective reporting or methodological optimism. Trim-and-fill adjustment reduced the pooled DOR, indicating a more conservative estimate of AI’s true diagnostic accuracy. These results indicate that this meta-analysis may have missed some studies with negative or neutral results, resulting in an overestimation of the combined effect size. The following aspects may be the reasons for the significant publication bias (55). First, the tendency of selective publication. AI studies with high diagnostic performance are more likely to be accepted by journals, while negative results may not be published because they are considered “non-innovative”. Second, quality defects in small sample studies. Small sample studies often have methodological limitations, and their overestimated effect sizes form an abnormal point in the upper left corner of the funnel plot, exacerbating the asymmetry. Third, language and database restrictions. This study only included Chinese and English literature, and did not search preprint platforms (such as medRxiv), which may have missed non-English studies that have not been formally published.
Although this study controlled some heterogeneity through a bivariate model, the following factors may affect the generalizability of the results. First, data heterogeneity. The endoscopic devices (such as white light endoscopy vs. NBI), sample size, and model architecture (CNN, DCNN, SVM) included in the study were significantly different, resulting in I² values of sensitivity and specificity of 97.2% and 97.6%, respectively. Second, insufficient dynamic validation. Only a small number of studies have verified real-time video diagnosis, and most evidence relies on retrospective static data, which may overestimate clinical applicability. Third, insufficient interpretability. Few AI models are “black boxes” and lack visualization of the decision-making process, which limits clinicians’ trust in the models. Last, challenges in real-world deployment. Implementation of AI in real-world clinical settings presents challenges, including variability in endoscopic equipment across centers, which may compromise model generalizability. Effective integration into clinical workflows demands seamless interfaces between AI systems and endoscopes to minimize disruption to routine practice. Additionally, interoperator variability, such as experience, may introduce bias unless models are retrained on diverse datasets. In the future, it is recommended to improve the ability to identify EGC through multicenter prospective validation, development of interpretable technology, and multimodal technology fusion of AI with NBI, LCI and magnifying endoscopy.
Conclusion
AI models, especially CNN-based architectures, have shown high sensitivity and specificity in the endoscopic diagnosis of EGC and have the potential to be used as clinical auxiliary tools. However, their widespread application still needs to address key issues such as heterogeneity, interpretability, and dynamic scene adaptation. Future research should focus on prospective validation, technical transparency, and multimodal integration to promote the transformation of AI from experimental research to clinical practice.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ML: Conceptualization, Data curation, Formal Analysis, Investigation, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. FC: Resources, Supervision, Writing – review & editing. QL: Validation, Writing – review & editing. MX: Data curation, Validation, Writing – review & editing. JW: Conceptualization, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China(82203452), Health Science and Technology Project of Zhejiang Province (2022RC165, 460 2024KY083), and Clinical Research Fund of Zhejiang Medical Association (2021ZYC-461 A68).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1670843/full#supplementary-material.
Supplementary Figure 1 | Subgroup analysis results of different AI model diagnostic performance (convolutional neural network (CNN) vs. other models). (A) Forest plot of sensitivity. (B) Forest plot of specificity. (C) ROC plane.
Supplementary Figure 2 | Subgroup analysis results of different AI model diagnostic performance in terms of image types (narrow band images (NBI) vs. Non-NBI). (A) Forest plot of sensitivity. (B) Forest plot of specificity. (C) ROC plane.
Supplementary Figure 3 | Subgroup analysis results of different AI model diagnostic performance in terms of study types (retrospective vs. prospective). (A) Forest plot of sensitivity. (B) Forest plot of specificity. (C) ROC plane.
Supplementary Table 1 | Search strategy.
Supplementary Table 2 | PRISMA checklist.
References
1. Lin J-L, Lin J-X, Lin G-T, Huang C-M, Zheng C-H, Xie J-W, et al. Global incidence and mortality trends of gastric cancer and predicted mortality of gastric cancer by 2035. BMC Public Health. (2024) 24:1763. doi: 10.1186/s12889-024-19104-6
2. Huang Y, Shao Y, Yu X, Chen C, Guo J, and Ye G. Global progress and future prospects of early gastric cancer screening. J Cancer. (2024) 15:3045. doi: 10.7150/jca.95311
3. Katai H, Ishikawa T, Akazawa K, Isobe Y, Miyashiro I, Oda I, et al. Five-year survival analysis of surgically resected gastric cancer cases in Japan: a retrospective analysis of more than 100,000 patients from the nationwide registry of the Japanese Gastric Cancer Association (2001–2007). Gastric Cancer. (2018) 21:144–54. doi: 10.1007/s10120-017-0716-7
4. Suzuki H, Oda I, Abe S, Sekiguchi M, Mori G, Nonaka S, et al. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer. (2016) 19:198–205. doi: 10.1007/s10120-015-0469-0
5. Ferrari C and Tadros M. Enhancing the quality of upper gastrointestinal endoscopy: current indicators and future trends. Gastroenterol Insights. (2023) 15:1–18. doi: 10.3390/gastroent15010001
6. Lee S-M, Kim K-M, and Ro JY. Gastric carcinoma: Morphologic classifications and molecular changes. Gastric Carcinoma - New Insights into Current Management. (2013). doi: 10.5772/54617
7. Application of artificial intelligence for improving early detection and prediction of therapeutic outcomes for gastric cancer in the era of precision oncology. Semin Cancer Biol. (2023) 93:83–96. doi: 10.1016/j.semcancer.2023.04.009
8. Wu L, Wang J, He X, Zhu Y, Jiang X, Chen Y, et al. Deep learning system compared with expert endoscopists in predicting early gastric cancer and its invasion depth and differentiation status (with videos). Gastrointest Endoscopy. (2022) 95:92–104. e3. doi: 10.1016/j.gie.2021.06.033
9. Choi J, Shin K, Jung J, Bae H-J, Kim DH, Byeon J-S, et al. Convolutional neural network technology in endoscopic imaging: artificial intelligence for endoscopy. Clin Endoscopy. (2020) 53:117–26. doi: 10.5946/ce.2020.054
10. Ali H, Muzammil MA, Dahiya DS, Ali F, Yasin S, Hanif W, et al. Artificial intelligence in gastrointestinal endoscopy: a comprehensive review. Ann gastroenterology. (2024) 37:133. doi: 10.20524/aog.2024.0861
11. Yang R, Zhang J, Zhan F, Yan C, Lu S, Zhu Z, et al. Artificial intelligence efficiently predicts gastric lesions, Helicobacter pylori infection and lymph node metastasis upon endoscopic images. Chin J Cancer Res. (2024) 36:489. doi: 10.21147/j.issn.1000-9604.2024.05.03
12. Jiang K, Jiang X, Pan J, Wen Y, Huang Y, Weng S, et al. Current evidence and future perspective of accuracy of artificial intelligence application for early gastric cancer diagnosis with endoscopy: a systematic and meta-analysis. Front Med. (2021) 8:629080. doi: 10.3389/fmed.2021.629080
13. Zhang P, Gao C, Zhang Z, Yuan Z, Zhang Q, Zhang P, et al. Systematic inference of super-resolution cell spatial profiles from histology images. Nat Commun. (2025) 16:1838. doi: 10.1038/s41467-025-57072-6
14. Li Y-D, Li H-Z, Chen S-S, Jin C-H, Chen M, Cheng M, et al. Correlation of the detection rate of upper GI cancer with artificial intelligence score: results from a multicenter trial (with video). Gastrointestinal Endoscopy. (2022) 95:1138–46.e2. doi: 10.1016/j.gie.2021.12.019
15. Hsiao YJ, Wen YC, Lai WY, Lin YY, Yang YP, Chien Y, et al. Application of artificial intelligence-driven endoscopic screening and diagnosis of gastric cancer. World J Gastroenterol. (2021) 27:2979–93. doi: 10.3748/wjg.v27.i22.2979
16. Lee S, Jeon J, Park J, Chang YH, Shin CM, Oh MJ, et al. An artificial intelligence system for comprehensive pathologic outcome prediction in early gastric cancer through endoscopic image analysis (with video). Gastric Cancer. (2024) 27:1088–99. doi: 10.1007/s10120-024-01524-3
17. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
18. Plana MN, Arevalo-Rodriguez I, Fernández-García S, Soto J, Fabregate M, Pérez T, et al. Meta-DiSc 2.0: a web application for meta-analysis of diagnostic test accuracy data. BMC Med Res Methodol. (2022) 22:306. doi: 10.1186/s12874-022-01788-2
19. Zamora J, Abraira V, Muriel A, Khan K, and Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. (2006) 6:1–12. doi: 10.1186/1471-2288-6-31
20. Lin L and Chu H. Quantifying publication bias in meta-analysis. Biometrics. (2018) 74:785–94. doi: 10.1111/biom.12817
21. Liu DY, Gan T, Rao NN, Xing YW, Zheng J, Li S, et al. Identification of lesion images from gastrointestinal endoscope based on feature extraction of combinational methods with and without learning process. Med Image Anal. (2016) 32:281–94. doi: 10.1016/j.media.2016.04.007
22. Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, et al. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. (2018) 87:1339–44. doi: 10.1016/j.gie.2017.11.029
23. Sakai Y, Takemoto S, Hori K, Nishimura M, Ikematsu H, Yano T, et al. (2018). Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network, in: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference New York, NY, USA: Institute of Electrical and Electronics Engineers, Vol. 2018. pp. 4138–41.
24. Wang Z, Gao. J, Meng Q, Yang T, Wang Z, Chen X, et al. Artificial intelligence based on deep learning for automatic detection of early gastric cancer. Chin J Digestive Endoscopy. (2018) 35:551–6. doi: 10.3760/cma.j.issn.1007-5232.2018.08.004
25. Cho BJ, Bang CS, Park SW, Yang YJ, Seo SI, Lim H, et al. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. (2019) 51:1121–9. doi: 10.1055/a-0981-6133
26. Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, et al. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. (2019) 51:522–31. doi: 10.1055/a-0855-3532
27. Yoon HJ, Kim S, Kim JH, Keum JS, Oh SI, Jo J, et al. A lesion-based convolutional neural network improves endoscopic detection and depth prediction of early gastric cancer. J Clin Med. (2019) 8:1310. doi: 10.3390/jcm8091310
28. Cho BJ, Bang CS, Lee JJ, Seo CW, and Kim JH. Prediction of submucosal invasion for gastric neoplasms in endoscopic images using deep-learning. J Clin Med. (2020) 9:1858. doi: 10.3390/jcm9061858
29. Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, et al. Convolutional neural network for differentiating gastric cancer from gastritis using magnified endoscopy with narrow band imaging. Dig Dis Sci. (2020) 65:1355–63. doi: 10.1007/s10620-019-05862-6
30. Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. (2020) 23:126–32. doi: 10.1007/s10120-019-00992-2
31. Namikawa K, Hirasawa T, Nakano K, Ikenoyama Y, Ishioka M, Shiroma S, et al. Artificial intelligence-based diagnostic system classifying gastric cancers and ulcers: comparison between the original and newly developed systems. Endoscopy. (2020) 52:1077–83. doi: 10.1055/a-1194-8771
32. Shibata T, Teramoto A, Yamada H, Ohmiya N, Saito K, and Fujita H. Automated detection and segmentation of early gastric cancer from endoscopic images using mask R-CNN. Appl Sci. (2020) 10:3842. doi: 10.3390/app10113842
33. Tang D, Wang L, Ling T, Lv Y, Ni M, Zhan Q, et al. Development and validation of a real-time artificial intelligence-assisted system for detecting early gastric cancer: A multicentre retrospective diagnostic study. EBioMedicine. (2020) 62:103146. doi: 10.1016/j.ebiom.2020.103146
34. Ikenoyama Y, Hirasawa T, Ishioka M, Namikawa K, Yoshimizu S, Horiuchi Y, et al. Detecting early gastric cancer: Comparison between the diagnostic ability of convolutional neural networks and endoscopists. Digestive endoscopy: Off J Japan Gastroenterological Endoscopy Society. (2021) 33:141–50. doi: 10.1111/den.13688
35. Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, et al. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol hepatology. (2021) 36:482–9. doi: 10.1111/jgh.15190
36. Wu L, He X, Liu M, Xie H, An P, Zhang J, et al. Evaluation of the effects of an artificial intelligence system on endoscopy quality and preliminary testing of its performance in detecting early gastric cancer: a randomized controlled trial. Endoscopy. (2021) 53:1199–207. doi: 10.1055/a-1350-5583
37. Wu H, Yao X, Zeng L, Huang F, and Chen L. Application of artificial intelligence technology based on convolutional neural network in early gastric cancer recognition. J Army Med Univ. (2021) 43:1735–42. doi: 10.16016/j.1000-5404.202105018
38. Zhang L, Zhang Y, Wang L, Wang J, and Liu Y. Diagnosis of gastric lesions through a deep convolutional neural network. Digestive endoscopy: Off J Japan Gastroenterological Endoscopy Society. (2021) 33:788–96. doi: 10.1111/den.13844
39. Jing W, Yijie Z, Lianlian W, Xinqi H, Zehua D, Manling H, et al. Influence of artificial intelligence on endoscopists′ performance in diagnosing gastric cancer by magnifying narrow banding imaging. Chin J Digestive Endoscopy. (2021) 38:783–8. doi: 10.3760/cma.j.cn321463-20210110-00020
40. Liming Z, Yang Z, Li W, Jiangyuan W, and Yulan L. Diagnosis of routine endoscopic images of gastric lesions through a deep convolutional neural network. Chin J Digestive Endoscopy. (2021) 38:789–94. doi: 10.3760/cma.j.cn321463-20200611-00280
41. Yijie Z, Lianlian W, Xinqi H, Yanxia L, Wei Z, Jun Z, et al. Comparison of the ability of two artificial intelligence systems based on different training methods to diagnose early gastric cancer under magnifying image-enhanced endoscopy. Chin J Digestion. (2022) 42:433–8. doi: 10.3760/cma.j.cn311367-20211214-00680
42. Ishioka M, Osawa H, Hirasawa T, Kawachi H, Nakano K, Fukushima N, et al. Performance of an artificial intelligence-based diagnostic support tool for early gastric cancers: Retrospective study. Digestive Endoscopy. (2023) 35:483–91. doi: 10.1111/den.14455
43. Jin J, Zhang Q, Bill D, Ma T, Wang X, Mei X, et al. Detection of early gastric cancer in white light imagings based on region-based convolutional neural networks. Acta Universitatis Medicinalis Anhui. (2023) 58:285–91. doi: 10.19405/j.cnki.issn1000-1492.2023.02.020
44. Wang J and Chao S. Clinical value of artificial intelligence assisted digestive endoscopy in the differential diagnosis of gastric cancer and precancerous lesions. Chin J Convalescent Med. (2023) 32:656–9. doi: 10.13517/j.cnki.ccm.2023.06.024
45. Zhao Y, Dohi O, Ishida T, Yoshida N, Ochiai T, Mukai H, et al. Linked color imaging with artificial intelligence improves the detection of early gastric cancer. Digestive Diseases. (2024) 42:503–11. doi: 10.1159/000540728
46. Feng J, Zhang Y, Feng Z, Ma H, Gou Y, Wang P, et al. A prospective and comparative study on improving the diagnostic accuracy of early gastric cancer based on deep convolutional neural network real-time diagnosis system (with video). Surg Endoscopy. (2025) 39:1874–84. doi: 10.1007/s00464-025-11527-5
47. Niu W, Liu L, Dong Z, Bu X, Yao F, Wang J, et al. A deep learning model based on magnifying endoscopy with narrow-band imaging to evaluate intestinal metaplasia grading and OLGIM staging: A multicenter study. Digestive liver disease: Off J Ital Soc Gastroenterol Ital Assoc Study Liver. (2024) 56:1565–71. doi: 10.1016/j.dld.2024.02.001
48. Liu L, Dong Z, Cheng J, Bu X, Qiu K, Yang C, et al. Diagnosis and segmentation effect of the ME-NBI-based deep learning model on gastric neoplasms in patients with suspected superficial lesions - a multicenter study. Front Oncol. (2023) 12. doi: 10.3389/fonc.2022.1075578
49. Yin F, Zhang X, Fan A, Liu X, Xu J, Ma X, et al. A novel detection technology for early gastric cancer based on Raman spectroscopy. Spectrochimica Acta Part A Mol biomolecular spectroscopy. (2023) 292:122422. doi: 10.1016/j.saa.2023.122422
50. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, and Oermann EK. Confounding variables can degrade generalization performance of radiological deep learning models. (2018). doi: 10.1371/journal.pmed.1002683
51. Luo D, Kuang F, Du J, Zhou M, Liu X, Luo X, et al. Artificial intelligence-assisted endoscopic diagnosis of early upper gastrointestinal cancer: A systematic review and meta-analysis. Front Oncol. (2022) 12:855175. doi: 10.3389/fonc.2022.855175
52. Xie F, Zhang K, Li F, Ma G, Ni Y, Zhang W, et al. Diagnostic accuracy of convolutional neural network-based endoscopic image analysis in diagnosing gastric cancer and predicting its invasion depth: a systematic review and meta-analysis. Gastrointest Endosc. (2022) 95:599–609.e7. doi: 10.1016/j.gie.2021.12.021
53. Zhang XQ, Huang ZN, Wu J, Liu XD, Xie RZ, Wu YX, et al. Machine learning prediction of early recurrence in gastric cancer: A nationwide real-world study. Ann Surg Oncol. (2025) 32:2637–50. doi: 10.1245/s10434-024-16701-y
54. Chen Y, Wang B, Zhao Y, Shao X, Wang M, Ma F, et al. Metabolomic machine learning predictor for diagnosis and prognosis of gastric cancer. Nat Commun. (2024) 15:1–13. doi: 10.1038/s41467-024-46043-y
Keywords: artificial intelligence, early gastric cancer, endoscopy, diagnosis, meta-analysis
Citation: Lv M, Chen F, Li Q, Xue M and Wang J (2025) Comparative diagnostic accuracy of different artificial intelligence models for early gastric cancer: a systematic review and meta-analysis. Front. Oncol. 15:1670843. doi: 10.3389/fonc.2025.1670843
Received: 22 July 2025; Accepted: 30 October 2025;
Published: 18 November 2025.
Edited by:
Natale Calomino, University of Siena, ItalyReviewed by:
Ji Bao, Sichuan University, ChinaChangda Lei, The First Affiliated Hospital of Soochow University, China
Copyright © 2025 Lv, Chen, Li, Xue and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Xue, eHVlbWVuZ0B6anUuZWR1LmNu; Jun Wang, d2FuZ2p1bm9zY2FyQHpqdS5lZHUuY24=
 Minfang Lv
Minfang Lv Fei Chen2
Fei Chen2 Meng Xue
Meng Xue Jun Wang
Jun Wang