- 1Department of Paediatrics, College of Medicine, University of Ibadan, Ibadan, Nigeria
- 2Institute of Child Health, College of Medicine, University of Ibadan, Ibadan, Nigeria
- 3Department of Virology, College of Medicine, University of Ibadan, Ibadan, Nigeria
Objective: The pattern and timing of development of intestinal microflora in Nigerian infants have been scarcely researched. This study was carried out to investigate the bacteria flora in the rectum of healthy neonates in Ibadan, Nigeria.
Patients and methods: In this hospital-based longitudinal study, rectal swabs of 70 neonates were taken within 6–12 h of birth (day 1) and subsequently on days 3, 9, and 14. Information collected included maternal sociodemographic characteristics, antibiotic use for the neonates, and type of feeding during the first 14 days of life. Identification and speciation of gram-negative isolates were done using the Analytical Profile Index 20E® and 20NE® as appropriate. Gram-positive bacteria were identified biochemically using the catalase and coagulase tests. Data were analyzed using descriptive statistics and Chi-square at p = 0.05.
Results: Majority (92.9%) of the neonates were delivered vaginally with a median gestational age of 38 weeks (range = 34–42). On the first day of life, Escherichia coli was isolated more frequently from the rectal swabs of preterm (50.0%) than term (23.1%) neonates (p = 0.031). On day 3 of life, coagulase-negative staphylococcus was the most frequently isolated bacteria from the rectal swabs of nonasphyxiated (64.4%) compared with asphyxiated (27.3%) neonates’ rectal swabs (p = 0.042). Staphylococcus aureus was the most frequently isolated bacteria from the rectal swabs of nonexclusively breastfed (66.7%) than exclusively breastfed (21.3%) neonates on day 14 (p = 0.004).
Conclusion: Staphylococcus aureus and Escherichia coli were the predominant isolates from the rectum of Nigerian neonates, and these isolates were influenced by breastfeeding and mild–moderate asphyxia. In all, bacterial diversity in the rectum increased as the neonates got older.
Introduction
The composition of intestinal bacterial flora is critical because of its role in nutrition, immune modulation, and biosynthesis of vitamins among other benefits (1). Currently, attention is on the health benefits and consequences of modification of bacterial composition of the intestine in early life. The introduction of pre- and probiotics, use of antibiotics, and experimental fecal flora transplantation to modify bacterial composition of intestine have improved health outcomes in necrotizing enterocolitis and antibiotic-associated diarrhea in children (2, 3). Studies have also associated diseases such as kwashiorkor, obesity, asthma, inflammatory bowel disease, and type 2 diabetes mellitus with pattern of bacterial microflora in early life (2, 4–6).
Consequently, the knowledge of the composition of the intestinal bacteria flora and its developmental pattern as well as possible alterations in different populations has become important in recent times. Notably, the development of intestinal bacterial population starts early in neonatal life and it is, to an extent, determined by the exposure of the baby to bacteria in the immediate environment (7, 8). Common environmental exposures include bacterial flora in maternal feces, vagina, and skin. Therefore, the types of bacterial flora acquired by a neonate are influenced by the mode of delivery, infant care practices such as type of feeding, the cleanness of the nursing environment, hospitalization, administration of antibiotics to the neonate, and maturity at the time of delivery (7, 9).
Currently, there are many techniques that can be used to identify bacterial composition of samples from the intestine including the traditional microbial culture, polymerase chain reaction, microarrays, and high-throughput sequencing (HTS). Of these, microbial culture is still considered the gold standard for clinical diagnostic purposes, and it is widely used in clinical laboratories (10). The HTS technologies that give better insights into the gut microbiota, because it can identify noncultivable organisms, are very specialized, expensive, and time consuming. These techniques are not available in resource-limited countries like Nigeria and they are not routinely used. However, a recent review by called attention to the fact that results of these technologies give correlate closely with those from culture-based approaches, showing the ability to corroborate and supplement the culture-based results (11).
Moreover, it is worth noting that to date, the successes recorded in the understanding of intestinal bacterial composition among infants are largely based on findings from studies conducted in developed countries (12), while data from developing countries like Nigeria remain scarce with available reports being over 30 years old (13, 14). With changes in lifestyle and the dynamic of factors that may influence intestinal colonization as well as development of better microbiological identification methods over time, it is probable that the intestinal microflora of Nigerian children might have changed. This study was, therefore, carried out to investigate the intestinal bacterial flora of neonates in Ibadan, Nigeria.
Patients and Methods
Study Design and Setting
This longitudinal study was carried out between August and December 2013. The neonates were recruited from the labor and postnatal wards of the University College Hospital (UCH), Ibadan within 6–12 h of birth (day 1). The neonates were subsequently seen on days 3, 9, and 14 after birth. The UCH provides primary and specialist care to Ibadan and other parts of the South-West region of Nigeria. The labor and postnatal wards have a total of 75 beds, where healthy neonates and those who do not require intensive care are nursed besides their mothers after delivery. In the absence of maternal and neonatal complications, neonates are usually discharged within 48–72 h after delivery.
Study Population
All healthy neonates were considered eligible to participate in the study. A neonate was regarded as “healthy or needing no intensive care” if there were no clinical features indicating the need for admission into the special care baby unit, but well enough to be nursed by the mother’s bedside. Seventy-two neonates were enrolled at birth but two mothers declined further participation of their babies after the first day, giving a dropout rate of 2.8%.
Sample Size Calculation
The required sample size was determined using the colonization rate from the previous study by Rotimi et al. (14) in 1985 which identified Escherichia coli as the commonest organism in 18 out of 23 babies by the first day of life, giving 78.2% colonization rate. It was hypothesized that the percentage of neonates with Escherichia coli colonization rate would be ±10% than the previously reported. At 95% level of confidence, 65 babies were required. Considering the fact that each neonate was sampled four times, 264 rectal swabs samples were expected.
Sampling Technique
Systematic random sampling method was used to recruit every fifth live-born neonate in the UCH who met the inclusion criteria during the study period. Neonates who had anorectal anomalies were not eligible to participate in the study.
Data Collection and Follow-Up
Two trained research assistants collected data using a structured interviewer-administered questionnaire. All rectal swab samples were collected by two of the investigators (AK and AEO). Information recorded on questionnaire included mothers’ demographic characteristics, pregnancy and delivery history; infant’s characteristics including APGAR scores, mode of resuscitation, feeding methods, and antibiotic/antimycotic use. The 5-min APGAR scores of ≤3, 4–6, and 7–10 were regarded as indicative of low (severe asphyxia), mild–moderate abnormal, and reassuring (no asphyxia), respectively (15). We determined the gestational ages of the neonates using the date of the last menstrual period (LMP) reported by mothers corroborated with ultrasound report. In the event the mother’s LMP was unknown, the gestational age of the baby was estimated by the pediatrician using the New Ballard score system (16). Preterm neonates were those whose gestational ages were less than 37 weeks.
Rectal swabs were taken from the neonates on the first day within 6–12 h after birth while on the ward. Subsequent samples were collected on days 3, 9, and 14 when mothers brought their babies for follow-up. Samples were collected in an aseptic manner using sterile gel coated soft cotton tipped swab sticks (Transwab® Amies; Medical Wire & Equipment, Corsham, Wiltshire, England). The Transwab® is a sterile pack containing a gel-coated soft cotton tipped swab and a container containing Amies transport medium and recommended for recovery of aerobes, anaerobes, and fastidious organisms. All specimens were collected following the procedures described in Centre for Diseases Control manual (17). Each tube containing the swab was placed in a transport pack and in a cold box and transported to the laboratory within 3 h after collection by a laboratory motorcycle dispatcher. Five mothers who failed to present their babies for follow-up at different times were located through their addresses and phone numbers with the rectal swabs samples collected through home visits.
Microbiological Methods
The rectal swabs on arrival in the laboratory were plated onto MacConkey agar and MacConkey Crystal Violet agar alongside Salmonella Shigella agar plates. Each sample was also inoculated onto a Selenite F broth and further processed in accordance with the standard practice for bacteria isolation. Other media used were Columbia blood agar and Amikacin blood agar, which were both incubated anaerobically at 37°C for 2 days. Inoculation of the plates was carried out using standard methods such that a semiquantitative assessment of the density of growth could be made on the solid media. All primary plates were reincubated after 18 h, regardless of the culture results.
Upon isolation of organisms, gram staining was done, which divided isolates into gram-negative and -positive bacteria. The complete identification and speciation of all the gram-negative isolates were done using the Analytical Profile Index (API) 20E® and API 20NE® (API® bioMérieux Clinical Diagnostics Kits, USA) identification system in accordance with the standard procedures for bacteria classification. The API® is a miniaturized identification system that contains dehydrated sugars that gives a characteristic color change upon addition and incubation of substrates. Miniaturized reagents are also incorporated in the system. API 20E® identified members of the Enterobacteriaceae while API 20NE® identified the nonfermenters and biochemically inert gram-negative rods. Gram-positive cocci were identified biochemically using the catalase test and coagulase test; Pastorex for Staphylococci (18). Anaerobes were conclusively identified employing the use of AnaeroGen a miniaturized GasPak™ gas regenerating kit system (19). All plates were kept for 40 h before being read. Indicator strips were added into the anaerobic jars to ascertain the presence of anaerobiosis.
Data Analysis
The main outcome was the frequency of the types of isolates cultured on days 1, 3, 9, and 14 while independent variables included characteristics of neonates and mothers. Chi-square test was used to test associations between categorical variables, while comparisons between continuous variables were performed using Student’s t-test. The patterns of organisms identified on the different days were displayed in charts. All p < 0.05 were considered statistically significant. The data were analyzed using Statistical Package for Social Sciences (SPSS) for Windows 21 (IBM SPSS statistics 2012, Chicago, IL, USA).
Ethical Considerations
Approval for the study was obtained from the University of Ibadan/UCH Ethics Review Committee (approval number: UI/EC/13/0161), and written informed consent was obtained from each mother.
Results
Characteristics of the Mothers and Neonates
Of the 68 mothers whose neonates participated in the study, two had twin deliveries. The mothers’ age ranged from 17 to 40 years (mean = 28.5 ± 5.1 years). Twenty-six (38.2%) of the mothers were primipara, 65 (95.6%) booked and received antenatal care at the study center. Pregnancy-related medical conditions recorded in the hospital notes and corroborated by mothers’ reports were hypertension (n = 3; 4.4%), antepartum hemorrhage (n = 1; 1.5%), postpartum hemorrhage (n = 1; 1.5%), and polyhydramnios (n = 1; 1.5%). Only two (2.9%) mothers had PROMs both lasting >24 h and they were given intravenous Augmentin (amoxicillin and clavulanate) prophylactically for 48 h, then orally to complete a 7–10-day course. Three mothers (4.4%) had prolonged labor lasting 19 to 24 h. Sixty-six (97.1%) of the mothers reported no illness or complications during pregnancy, and only two (2.9%) were “HIV positive.”
Table 1 shows the characteristics of the neonates. There were 32 (45.7%) males and 38 (54.3%) female neonates. There were 66 singletons and two sets of twins. Sixty-one neonates (87.1%) were exclusively breastfed, and 9 (12.9%) were given water and/or formula in addition to breast milk. Two neonates required hospital admission, which occurred during the second week of life, with aspiration pneumonitis and acyanotic congenital cardiac defect, respectively. One mother reported administering oral antibiotics to her neonate (1.4%). Five (7.1%) neonates were delivered by elective cesarean section. One female term neonate had pustular skin rash on the third day of life was given a course of oral cephalexin, which resolved by the ninth day of life.
Bacterial Isolates on the First Day of Life and Associated Factors
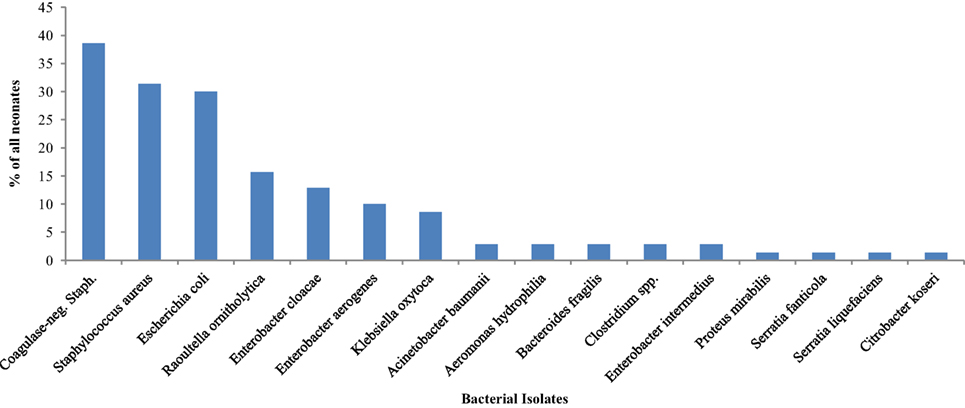
Bacteria were isolated from the rectal swab samples of 65 (92.9%) on day 1 mainly facultative anaerobes (15 of 16 different species). Figure 1 shows the species of isolates. The most frequently isolated bacteria was coagulase-negative Staphylococcus (n = 27; 38.6%); followed by Staphylococcus aureus (n = 22; 31.4%) and Escherichia coli (n = 21; 30.0%). Serratia fanticola, Serratia liquefaciens, Citrobacter koseri, and Proteus mirabilis were found in samples from one neonate each. With the exception of Escherichia coli in preterm neonates (n = 9; 50.0%) compared with term neonates 12 (23.1) (p = 0.031), there was no significant differences in the bacterial isolates in preterm and term neonates (Table 1).
Table 2 also shows comparisons of frequency of bacterial isolates from the neonates between neonates who were preterm versus term, had mild to moderate asphyxia versus no asphyxia, and delivered per vaginal versus cesarean section. On the first day of life, the odds of isolating Escherichia coli (OR = 3.3; 95% CI: 1.1, 10.2) from the rectal swabs of preterm neonates was significantly higher than that of term neonates.
Bacterial Isolates the Third Day of Life and Associated Factors
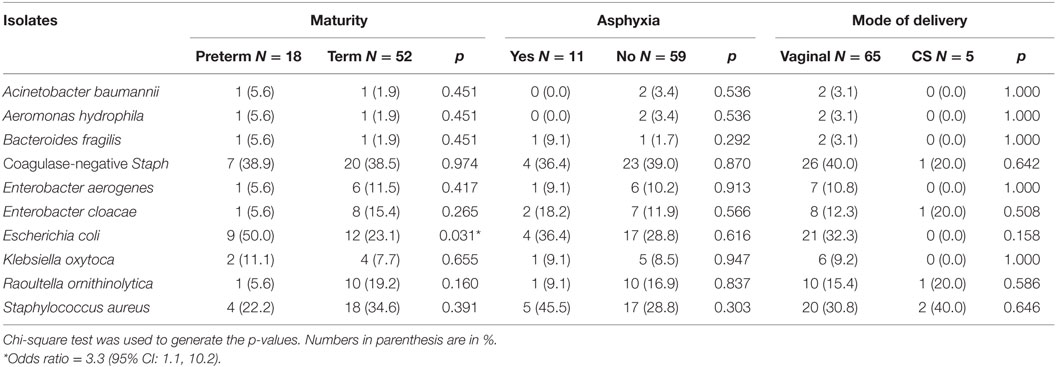
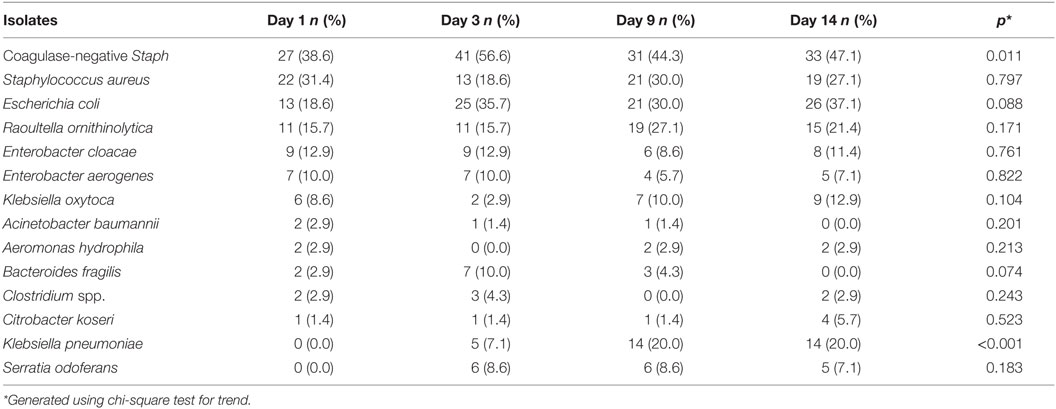
Different species of isolates were obtained from all 70 (100.0%) rectal samples on day 3 of life (Figure 2). Coagulase-negative Staphylococcus (58.6%) was the most frequently isolated bacteria on day 3. Table 3 shows the bacteria isolates from neonates by maturity, presence or absence of asphyxia at birth, mode of delivery, and whether or not neonate was exclusively breastfed for the first 3 days of life. The coagulase-negative Staphylococcus was more frequently (p = 0.042) isolated in nonasphyxiated neonates (64.6%) than asphyxiated neonates (27.3%). Asphyxiated neonates had significantly higher colonization by Enterobacter cloacae (36.4%) than nonasphyxiated neonates (8.5%) (p = 0.029). On day 3, the odds of isolating coagulase-negative Staphylococcus from the rectal swabs of nonasphyxiated neonates were significantly higher than that of asphyxiated neonates (OR = 4.83; 95% CI: 1.15, 20.16). Asphyxiated neonates had significantly higher odds of colonization by Enterobacter cloacae (OR = 6.1; 95% CI: 1.33, 28.57) than nonasphyxiated neonates.
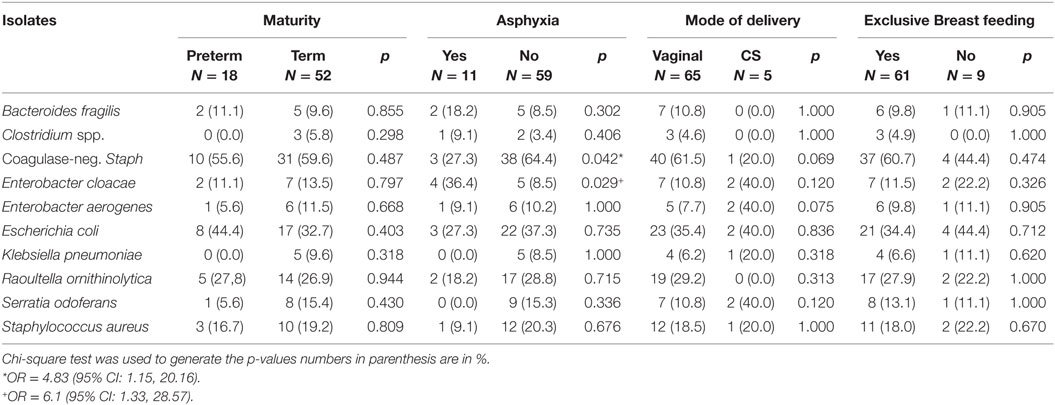
Table 3. Frequency of 10 common bacterial isolates from the rectum of neonates on the third day of life.
Bacterial Isolates on the Ninth Day of Life and Associated Factors
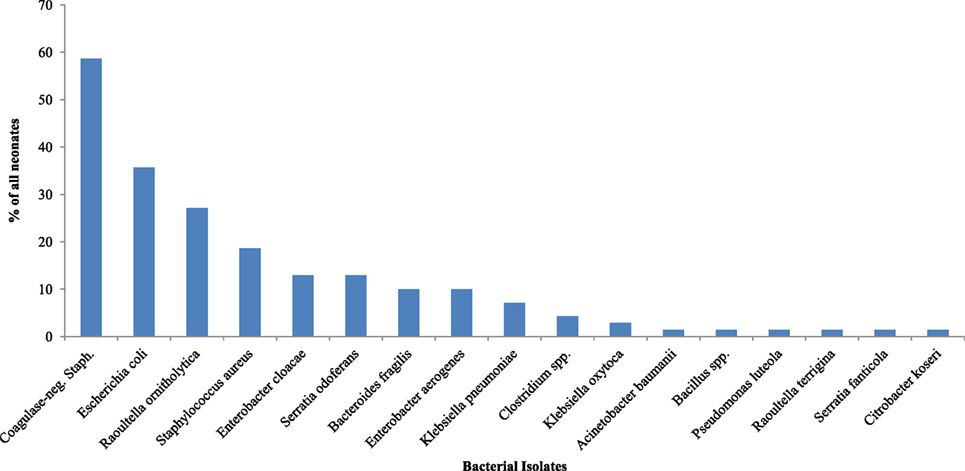
The isolates obtained from all 70 (100.0%) neonates on the ninth day of life were as displayed in Figure 3. The most frequently isolated bacteria were coagulase-negative Staphylococcus (44.3%). Table 4 shows comparisons of bacterial isolates from the neonates by maturity, presence or absence of asphyxia at birth, mode of delivery, and whether or not exclusively breastfed for the first 9 days of life. Remarkably, the odds of isolating coagulase-negative Staphylococcus (OR = 4.17; 95% CI: 1.21, 7.38) was significantly higher in nonasphyxiated than asphyxiated neonates as shown in Table 4.
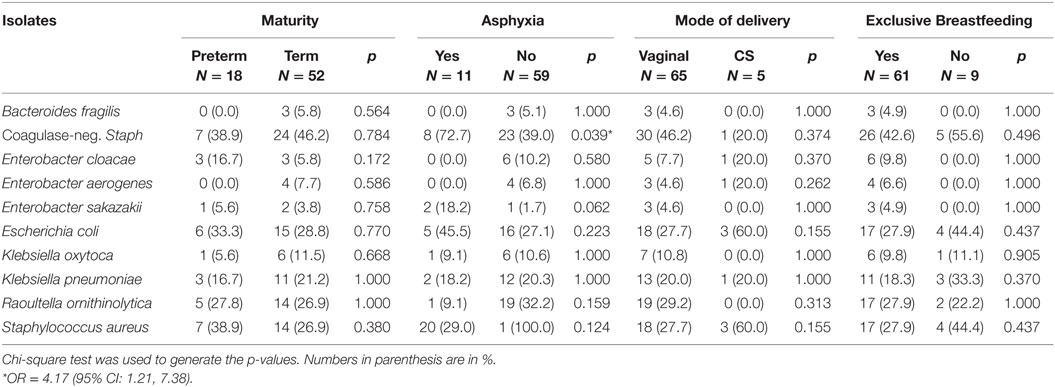
Table 4. Frequency of 10 common bacterial isolates from the rectum of neonates on the ninth day of life.
Bacterial Isolates on the 14th Day of Life and Associated Factors
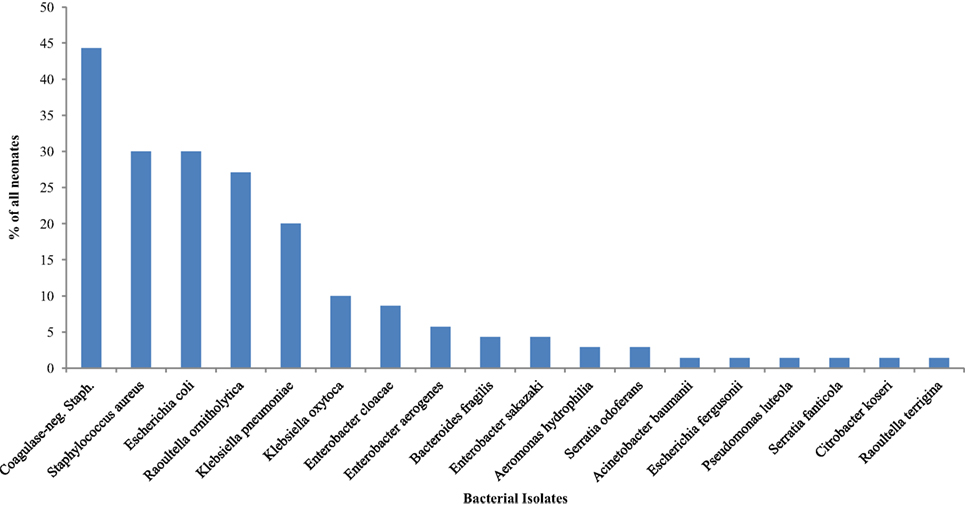
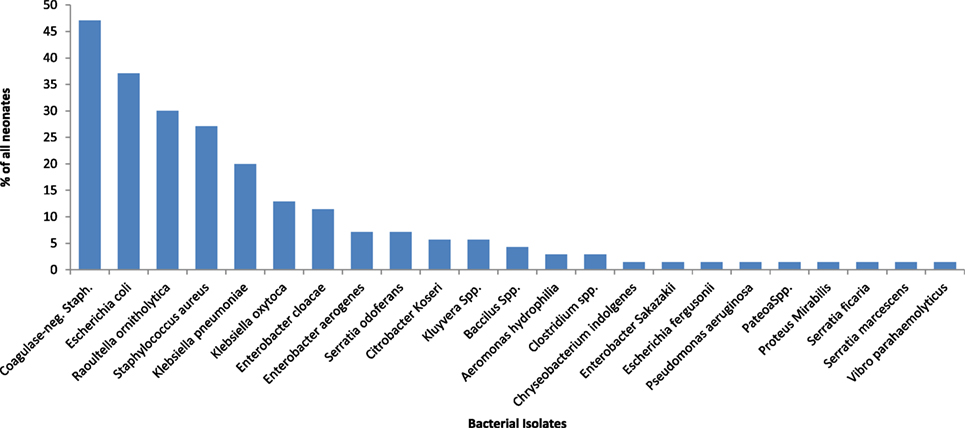
Figure 4 and Table 5 show the bacteria isolates obtained from the samples of all 70 (100.0%) neonates on day 14. Coagulase-negative Staphylococcus (47.1%) remains the most frequently isolated organism, it was isolated more from those who were not exclusively breastfed than those who had exclusively breastfeeding indicating significantly higher odds of Staphylococcus aureus (OR = 7.38; 95% CI: 1.62, 33.61) from nonexclusively breastfed than that of exclusively breastfed neonates.
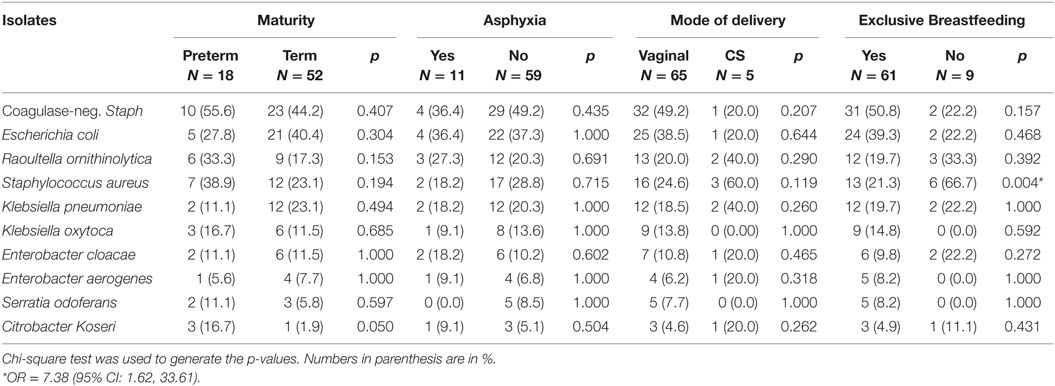
Table 5. Frequency of 10 common bacterial isolates from the rectum of neonates on the 14th day of life.
Trend of Acquisition of the Leading Bacteria in the First 2 Weeks of Life
Table 6 shows the trend and comparisons of the difference in colonization patterns between days 1, 3, 9, and 14. It appears that bacteria colonization in the neonates became established at slightly different times. On day 1, bacteria isolates were predominantly facultative anaerobes comprising coagulase-negative Staphylococcus, Staphylococcus aureus, Escherichia coli, and Enterobacteria other than Escherichia coli forming a large contingent (Enterobacter cloacae, Enterobacter aerogenes, Raoultella ornithinolytica, and Klebsiella oxytoca). On days 3 and 9, the bacteria isolates were heterogeneous but still dominated by coagulase-negative Staphylococcus and the Enterobacteria, Escherichia coli, and Raoultella ornithinolytica. On day 14, there was more diversity in the isolates with 23 different species but the dominance of coagulase-negative Staphylococcus and Staphylococcus aureus was maintained. Most of the remaining constituent bacteria were other members of the Enterobacteriaceae family. Statistically significant increasing trends were observed for Staphylococcus aureus and Klebsiella pneumoniae, respectively, as the neonates got older.
Discussion
This study showed that almost all (92.9%) neonates had rectal colonization by various bacteria on the first day of life and all subsequently became colonized by day 3. The bacteria isolates from rectal swabs were dominated by the phyla Firmicutes, Proteobacteria, and Bacteroidetes. However, the diversity of the bacteria composition was limited to 18 genera comprising Aeromonas, Acinetobacter, Bacillus, Bacteroides, Chryseobacteria, Citrobacter, Clostridium, Enterobacter, Escherichia, Klebsiella, Kluyvera, Pantoea, Proteus, Pseudomonas, Raoultella, Serratia, Staphylococcus, and Vibrio. The clinically important species amongst these bacteria were Bacteroides fragilis, Clostridium spp., coagulase-negative Staphylococcus, Enterobacter cloacae, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Raoultella ornithinolytica, and Staphylococcus aureus. These bacteria have the propensity for translocation into the blood stream and the resultant septicemia.
That the neonates had bacteria isolates on the first day of life is consistent with the report by Rotimi and colleagues, in Lagos Nigeria, about 30 years earlier (14) and Pakistan (20, 21). Conversely, reports from Europe showed lower bacterial colonization rates on the first day of life (20–22). These reports revealed the marked differences in the timing of bacterial colonization of rectum of neonates between developed and developing countries probably a reflection of the level of hygiene of the delivery environment (21).
The pattern of bacterial colonization, as shown by bacterial isolation in the rectal swabs on days 1, 3, 9, and 14, falls into the expected pattern of aerobes-facultative groups. The anaerobes were the initial colonizers of the neonates’ rectum, followed by sequential establishment of the expected anaerobic bacteria as earlier reported by Walker et al. (23) and Fanaro et al. (24). However, the composition of the individual bacteria in the present study differ from those found in European neonates (21, 23, 24). Notably, the trend observed in the first 2 weeks of life was that coagulase-negative Staphylococcus and Staphylococcus aureus were acquired from the first day of life, peaking on day 3 and remained dominant throughout the first 2 weeks of life.
The Enterobacteriaceae maintained the expected trend of colonization after the first day of life, but at a moderate level and the expected change to an anaerobic pattern had not occurred by the end of the second week. The findings in the present study are consistent with studies in Sweden (22) and France (25) that consistently reported rising rates of colonization by Staphylococcus aureus and coagulase-negative Staphylococcus but at variance with earlier reports from Nigeria (14), United Kingdom (26), and Sweden (27), which reported high rates of colonization with Escherichia coli, Enterococcus, and Streptococcus and moderate rates with Staphylococcus spp. A possible explanation for this trend is the practice of short hospital stay for newly delivered mothers, rooming-in, and improved hygiene may all have reduced exposure of infants to typical fecal bacteria such as Escherichia coli in the hospital (21). With low Escherichia coli colonization, the traditional skin bacteria including Staphylococcus aureus and coagulase-negative Staphylococcus might have expanded and become dominant due to lack of competition from other commensals.
The high predominance of Staphylococcus spp. in the present study has some clinical implications. The neonatal rectal carriage of Staphylococcus spp. may be the possible source of the blood infection, which may explain the dominance of Staphylococcus aureus as the cause of neonatal septicemia in our hospitals (28–30). However, despite the high colonization rate by Staphylococcus spp. in this study, none of the neonates with this isolate had clinical features suggestive of septicemia. The present study found no statistically significant association between maternal factors such as prolonged rupture of membrane (PROM), HIV status, and bacterial colonization of the rectum of the neonates. Nonetheless, a gross examination of the spectrum of isolates from these mothers with PROM and HIV infection showed that the pattern of isolates were similar to those of other mothers.
The higher rate of Escherichia coli colonization among preterm than term neonates may be as a result of acquisition from hospital staff, visitors, and the mothers. In general, preterm neonates cared for in specialized units, hygienically controlled, and more likely to be on antibiotics usually show delayed colonization and a limited number and diversity of bacteria (31). However, the preterm neonates in this study were neither admitted nor given antibiotics, they were nursed in cots besides their mothers in the postnatal wards. Also, the fact that nonexclusively breastfed neonates had higher colonization with Staphylococcus aureus than those exclusively breastfed corroborates the finding by Harmsen et al. in the Netherlands (32). Another likely source of Staphylococcus aureus was the vaginal canal during the delivery period and the skin of caregivers.
An important, but unexpected, finding in this study was the absence of bacteria from the genus Actinobacteria, specifically Bifidobacteria spp. which is consistent with the finding by Ekwempu et al. (13) who found neither Bifidobacteria nor Lactobacillus spp. among a population of children in Northern Nigeria. More recent studies have also questioned the dominance of Bifidobacteria, with many reporting very low to negligible quantity (33–35), though an earlier study in the same environment (14), albeit more than 25 years ago, reported high rates of colonization by Bifidobacteria. One possible explanation for the absence of Bifidobacteria in our data is that suppression of Bifidobacteria could occur in some instances of predominant growth of gram-negative bacteria and Staphylococcus spp. in neonates (22, 33) or as suggested by Brandt et al. (36) and Leke et al. (37) that the level of Bifidobacteria was probably under the cultural detection threshold. Another notable negative finding was the absence of Lactobacillus spp. The extent to which Lactobacilli colonize the intestines of neonates is controversial. Most studies that utilized traditional biochemical methods similar to the present study also reported low Lactobacillus colonization rates in infants (21).
The findings in our study need to be viewed and interpreted with caution bearing in mind the limitations of the methods used for bacteria identification. The microbiological culture methods used for bacterial flora identification could only detect cultivable organisms. Recent studies on the intestinal microbiome have used nonculture techniques that allow the identification of a large number of bacteria that may not grow on standard culture media. This limits the ability to compare the results of the present study with other recent studies performed by analyzing 16S RNA or by metagenomics (38). Also, the fact that the population of neonates who participated in the study were recruited from a tertiary hospital limits the generalizability of the findings. The differences that may exist between neonates delivered elsewhere such as home, private, secondary, and primary health facilities compared with the sampled population remain largely undetermined.
Conclusion
In all, using standard culture media, 12 species of bacterial flora were isolated from the stool of neonates delivered in Ibadan, Nigeria. These were Bacillus, Bacteroides, Candida, Clostridium, Staphylococcus, Enterobacter, Enterobacteriaceae, Klebsiella, Proteus, Pseudomonas, Raoultella, and Serratia. Staphylococcus aureus and Escherichia coli were the predominant isolates from the rectum of Nigerian neonates, and these isolates were influenced by breastfeeding and mild–moderate asphyxia. In all, bacterial diversity in the rectum increased as the neonates got older.
Author Contributions
AO, AK, and OA conceptualised the study. OT, GO, and DO contributed to the study design. AO analyzed the data and wrote the first draft of the manuscript. All the authors critically review and contributed to the final draft of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was partly funded by the Fogarty International Center (NIH) TW008878, Medical Education Partnership in Nigeria. The contents are solely the responsibility of the authors and do not represent the official views of any institution. The authors appreciate the contributions of the Medical Microbiologist at PATHCARE Nigeria limited (a private laboratory in Ibadan, Nigeria) for providing technical support.
References
1. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet (2003) 361:512–9. doi: 10.1016/S0140-6736(03)12489-0
2. Horvath A, Dziechciarz P, Szajewska H. Meta-analysis: Lactobacillus rhamnosus GG for abdominal pain-related functional gastrointestinal disorders in childhood. Aliment Pharmacol Ther (2011) 33:1302–10. doi:10.1111/j.1365-2036.2011.04665.x
3. Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res (2010) 20:1411–9. doi:10.1101/gr.107987.110
4. Garrett WS. Kwashiorkor and the Gut Microbiota. N Engl J Med (2013) 368:1746–7. doi:10.1056/NEJMcibr1301297
5. Cox MJ, Huang YJ, Fujimura KE, Liu JT, McKean M, Boushey HA, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One (2010) 5:e8745. doi:10.1371/journal.pone.0008745
6. Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science (2013) 339:548–54. doi:10.1126/science.1229000
7. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol (2007) 5:e177. doi:10.1371/journal.pbio.0050177
8. Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A (2006) 103:732–7. doi:10.1073/pnas.0506655103
9. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics (2006) 118:511–21. doi:10.1542/peds.2005-2824
10. Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet (2012) 13:601–12. doi:10.1038/nrg3226
11. Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes (2012) 3:203–20. doi:10.4161/gmic.20169
12. Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA, et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One (2013) 8:e53838. doi:10.1371/journal.pone.0053838
13. Ekwempu CC, Lawande RV, Egler LJ. Bacterial colonisation of various sites at birth of babies born in Zaria. J Infect (1982) 5:177–81. doi:10.1016/S0163-4453(82)91859-X
14. Rotimi VO, Olowe SA, Ahmed I. The development of bacterial flora of premature neonates. J Hyg (1985) 94:309–18. doi:10.1017/S0022172400061532
15. ACOG and AAP Task Force on Neonatal Encephalopathy. Neonatal Encephalopathy and Neurologic Outcome. Washington, DC: American College of Obstetrics and Gynecology (ACOG) and American Academy of Pediatrics (AAP) (2014). p. 1–236.
16. Ballard JL, Khoury JC, Wedig K, Wang L, Eilers-Walsman BL, Lipp R. New Ballard Score, expanded to include extremely premature infants. J Pediatr (1991) 119:417–23. doi:10.1016/S0022-3476(05)82056-6
17. Ministry of Health of the United Republic of Tanzania. Disease Outbreak Management: A Field Manual for Council Health Management Teams Integrated Disease Surveillance and Response. Tanzania: The CDSC Change Project for the Ministry of Health of the United Republic of Tanzania (2004). p. 1–45.
18. Compernolle V, Verschraegen G, Claeys G. Combined use of Pastorex Staph-Plus and either of two new chromogenic agars, MRSA ID and CHROMagar MRSA, for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol (2007) 45:154–8. doi:10.1128/JCM.01115-06
19. Ruangrungrote S, Intasorn A, Sindermsuk J. Gas analyses of anaerobic or microaerophillic generating systems using gas chromatography. J Met Miner (2008) 18:13–6.
20. Adlerberth I, Carlsson B, de Man P, Jalil F, Khan SR, Larsson P, et al. Intestinal colonization with Enterobacteriaceae in Pakistani and Swedish hospital-delivered infants. Acta Paediatr Scand (1991) 80:602–10. doi:10.1111/j.1651-2227.1991.tb11917.x
21. Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr (2009) 98:229–38. doi:10.1111/j.1651-2227.2008.01060.x
22. Adlerberth I, Lindberg E, Åberg N, Hesselmar B, Saalman R, Strannegård IL, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel – an effect of hygienic lifestyle? Pediatr Res (2006) 59:96–101. doi:10.1203/01.pdr.0000191137.12774.b2
23. Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab (2013) 63:8–15. doi:10.1159/000354907
24. Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl (2003) 441:48–55. doi:10.1111/j.1651-2227.2003.tb00646.x
25. Borderon JC, Lionnet C, Rondeau C, Suc AL, Laugier J, Gold F. Current aspects of faecal flora of the newborn without antibiotic therapy during the first 7 days of life: Enterobacteriaceae, enterococci, staphylococci. Pathol Biol (1996) 44:416–22.
26. Rotimi V, Duerden BI. The development of the bacterial flora in normal neonates. J Med Microbiol (1981) 14:15–61.
27. Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora of breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand (1985) 74:45–51. doi:10.1111/j.1651-2227.1985.tb10919.x
28. Anah MU, Udo JJ, Ochigbo SO, Abia-Bassesy LN. Neonatal septicaemia in Calabar, Nigeria. Trop Doct (2008) 38:126–8. doi:10.1258/td.2006.006037
29. Awoniyi DO, Udo SJ, Oguntibeju OO. An epidemiological survey of neonatal sepsis in a hospital in western Nigeria Afr. J Microbiol Res (2009) 3:385–9.
30. Ogunlesi TA, Ogunfowora OB, Osinupebi O, Olanrewaju DM. Changing trends in newborn sepsis in Sagamu, Nigeria: bacterial aetiology, risk factors and antibiotic susceptibility. J Paediatr Child Health (2011) 47:5–11. doi:10.1111/j.1440-1754.2010.01882.x
31. Bennet R, Erriksson M, Nord CE, Zetterstrom R. Fecal bacterial microflora of newborn infants during intensive care management and treatment with five antibiotic regimens. Pediatr Infect Dis J (1986) 5:533–9. doi:10.1097/00006454-198609000-00009
32. Harmsen JM, Wildeboer-Veloo CM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr (2000) 30:61–7. doi:10.1097/00005176-200001000-00019
33. Bennet R, Eriksson M, Tafari N, Nord CE. Intestinal bacteria of newborn Ethiopian infants in relation to antibiotic treatment and colonisation by potentially pathogenic gram-negative bacteria. Scand J Infect Dis (1991) 23:63–9. doi:10.3109/00365549109023376
34. Langhendries JP, Detry J, Van-Hees J, Lamboray JM, Darimont J, Mozin MJ, et al. Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full term infants. J Pediatr Gastroenterol Nutr (1995) 21:177–81. doi:10.1097/00005176-199508000-00009
35. Rubaltelli FF, Biadaioli R, Pecile P, Nicoletti P. Intestinal flora in breast- and bottle-fed infants. J Perinat Med (1998) 26:186191. doi:10.1515/jpme.1998.26.3.186
36. Brandt K, Taddei CR, Takagi EH, Oliveira FF, Duarte RTD, Irino I, et al. Establishment of the bacterial fecal community during the first month of life in Brazilian newborns. Clinics (2012) 67:113–23. doi:10.6061/clinics/2012(02)05
37. Léké A, Romond MB, Mullié C. Insights in the human bifidobacterial flora through culture dependent and independent techniques. In: Méndez-Vilas A, editor. Communicating Current Research and Educational Topics and Trends in Applied Microbiology. Badajoz: FORMATEX (2007). p. 758–65.
Keywords: bacterial colonization, early life, newborns, preterm infants, rectal swabs
Citation: Kigbu A, Orimadegun AE, Tongo OO, Odaibo GN, Olaleye DO and Akinyinka OO (2016) Intestinal Bacterial Colonization in the First 2 Weeks of Life of Nigerian Neonates Using Standard Culture Methods. Front. Pediatr. 4:139. doi: 10.3389/fped.2016.00139
Received: 28 August 2016; Accepted: 09 December 2016;
Published: 27 December 2016
Edited by:
Christoph Bührer, Charité – Universitätsmedizin Berlin, GermanyReviewed by:
Eric Giannoni, Centre Hospitalier Universitaire Vaudois, SwitzerlandJanet Elizabeth Berrington, Newcastle Upon Tyne Hospitals NHS Foundation Trust, UK
Copyright: © 2016 Kigbu, Orimadegun, Tongo, Odaibo, Olaleye and Akinyinka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adebola E. Orimadegun, YmVvcmltYWRlZ3VuQHlhaG9vLmNvbQ==
†These are joint first authors on this paper.
 Allan Kigbu
Allan Kigbu Adebola E. Orimadegun
Adebola E. Orimadegun Olukemi O. Tongo
Olukemi O. Tongo Georgina N. Odaibo
Georgina N. Odaibo David O. Olaleye3
David O. Olaleye3 Olusegun O. Akinyinka
Olusegun O. Akinyinka