- 1AESKU. KIPP Institute, Wendelsheim, Germany
- 2B. Rappaport School of Medicine, Technion-Israel Institute of Technology, Haifa, Israel
The enzyme microbial transglutaminase is heavily used in the food processing industries to ameliorate food qualities and elongate the products' shelf life. As a protein's glue, it cross-links gliadin peptides, creating neo-complexes that are immunogenic and potentially pathogenic to celiac disease communities. Even lacking sequence identity, it imitates functionally the endogenous tissue transglutaminase, known to be the autoantigen of celiac disease and representing an undisputable key player in celiac disease initiation and progress. The present review expend on the enzyme characteristics, exogenous intestinal sources, its cross-linking avidity to gluten or gliadin, turning naïve protein to immunogenic ones. Several observation on microbial transglutaminase cross linked complexes immunogenicity in celiac patients are reviewed and its pathogenicity is summarized. Warnings on its potential risks for the gluten dependent conditions are highlighted. When substantiated, it might represent a new environmental factor of celiac disease genesis. It is hoped that the presented knowledge will encourage further research to explore the mechanism and the pathogenic pathways taken by the gliadin cross linked enzyme in driving celiac disease.
Introduction
Celiac disease (CD) is an autoimmune disease, thus dependent on genetic background and on environmental factors. The list of the environmental components that might impact CD initiation or development is continuously expanding, but the precipitating events causing CD remain enigmatic. Except for the prolamins, none of them reached cause and effect relationship. The introduction is divided to 4 sub-heading in order to set the stage for microbial transglutaminase (mTg)-CD cross-talks.
Environmental Factors Associated With Celiac Disease
A plethora of environmental components were suggested to influence CD, spanning infections, food, drugs, vaccination, toxins and metals, abdominal or gynecological surgery, level of hygiene, socio-economic status, life style stress and processed food additives (1–5). Microbes like Helicobacter pylori, Campylobacter jejuni, pneumococcus, bacteroides species and tuberculosis were associated. In the virus's domain: CMV, HBV, HCV, Rotavirus, Adenovirus, Reovirus, and enteroviruses were implicated. To complicate the topic, same bacteria and viruses might play a protective role in CD (4). Pharmacological factors are early antibiotic therapy and proton pump inhibitors. Influenza and HPV vaccines, Heavy metals, Aflatoxin, smoking, alcohol, Cesarean section and abdominal surgery, Western higher hygiene and higher socioeconomic level, stressful life and Westernization of the diet, microbiome dysbalance characterized by an abundance of Proteobacteria and a decrease in Lactobacillus were associated with CD development or increased incidence (6, 7). Most recently the processed food additive were suggested to influence the intestinal microbiome and to increase intestinal permeability, thus contributing to luminal eco-events that drive autoimmunity (8). One of them is the mTg, the topic of the present review. In 2015, the hypothesis that mTg might play a role in CD genesis was forwarded and since then more studies were conducted to substantiate this hypothesis (9).
MTg: Characteristics, Functions, Food Applications
Prokaryotic mTg is a part of the global transglutaminases family (10), isolated and characterized from multiple bacterial strains, Streptomyces mobaraense being the first one (11, 12). The S. mobaraense mTg harbors 331 amino acids, its molecular weight is 37.9 kDa. Notably, list of additional microbes that secrete the enzyme and their enzymatic yield capacities is continuously expanding (13), a recent one is a novel Transglutaminase from Streptococcus suis (14).
Being a member of the Tg family, mTg catalyzes the formation of an isopeptide bond, cross-linking an amine group (containing the acyl acceptor lysine) and the acyl group (containing the acyl donor glutamine). As gluten is abundant in glutamine (~30%) and contains lysine (<2%) (15), representing an acyl donor and acceptor, respectively, it is an ideal substrate for post-translational modification of gluten, by transamidation, or deamidation. Based on its peptide modifying capacity it imitates functionally the endogenous tTg, the ultimate autoantigen in CD (9, 13, 16, 17). It should be emphasized that bonds formed by the mTg resist proteolytic degradation, it exhibits a higher reaction rate, substrate specificity, higher transamidation/ deamidation ratio resulting in improved cross-linking capacity. Due to its broad enzymatic activity, it is heavily used by the food processing industries (8, 9, 12, 18–23). In fact, the enzyme is consumed by most of the processed food industries, spanning the meat, dairy, sea food and fish, surimi, casein and gelatin, myosin and actin, confection, and convenience ones and many more (8, 9, 12, 18–22). The net % increase per year of enzyme usage in the processed food industries is estimated to 21.9%, mTg being a major one (8). In the food processed industries, mTg improves gelation and changes emulsification, foaming, viscosity and water-holding capacity. It is considered as the “glue of proteins” and polymerization agent, thus improving food palatability, texture and life time on the supermarkets' shelves.
Zooming on CD and the increased affinity of the mTg to gluten, the enzyme in increasingly used in the bakery industries (8, 9), where it lowers the products calories, improves texture, elasticity and dough characteristics (9, 12, 24).
Summarizing the literature, an estimated daily intake of mTg used in the processed food products can range up to 15 mg where every kg of mTg processed product contains around 50–100 mg of mTg (18, 25, 26). Finally, a direct positive correlation is found between the increased annual usage of industrial enzymes added to processed bakery products and the increase in CD frequency, in the last four decades (7, 13). It should be stressed that we are dealing with an associative correlation and no causality was determined.
MTg Is Structurally Different but Functionally Imitates the tTg
In contrast to endogenous human tTg, the microbial one is a calcium and nucleotide independent enzyme. It consists of a single domain, compared to the four domains of the tTg and has around half the molecular weight. MTg operates at a larger range of pH, buffers, and temperatures and has a much larger list of substrates. So, in opposition to the tTg, mTg less substrate specific. Those features are advantageous for many industrial applications and it is clear why the industries preferred mTg over its member, tTg. MTg lacks sequence homology to tTg, but, due to their active site performances, mTg has substantial functional similarity (9, 13, 18). Not surprisingly, the two enzymes can transamidate or deamidate proteins, based on the enzymatic reaction's conditions (23).
The Luminal Intestinal Sources of mTg
MTg can originate from intra luminal sources and from external sources. The luminal enteric sources come from the gut microbiome. Our lumen is over-crowded by bacteria but also by archaea, viruses and protozoa, reaching roughly 1014−16, and the ratio of microbes/ host cells ratio averages 1:1. Its dimensions, composition, diversity, mobile products and activities have led to its description as a “superorganism” (27). A component of the microbial Metabolome represented as a mobilome is the secreted mTg. It appears that by using sequence search programs, hundreds to thousands of mTgs-encoding bacteria can be detected, the majority belong to the Firmicutes phylum (13, 16). As a survival factor mTgs are secreted by the microbes in order to survive their overpopulated luminal compartment. When intestinal fluid was analyzed, the fluid was positive for transglutaminase activity, however, the authors did not distinguish between the endogenous tTg and the bacterial one, but to our knowledge, no tTg activity was reported in the duodenal lumen, so far, in contrast to the intestinal mucosa and the lamina propria (28).
The extra intestinal sources of mTg are much wider. Mtg as a processed food additive was extensively discussed above (8, 9, 12, 13, 16, 18–22). Belonging to the prokaryotes, the probiotics represent a reservoir of mTg, representing an active cargo that affects luminal events. It is well-known that microbes can transfer virulent factors to their mates by horizontal gene transfer (29). Major concern on the transfer of virulent genes, like antibiotic resistant genes, via the nutrients and food chain exists and many of those deleterious genes are carried by probiotics (30–32). The virulent genes were described to end up in the human gastrointestinal tract microbiota (33). The factors causing the transformation from benign inhabitant of the gut microbiome or ingested probiotics to virulent pathogen are not clear, but a combination of horizontal gene exchange of virulence factors and differential transcription of endogenous genes are clearly involved (34).
An additional enzymatic cargo of mTg delivered to the enteric lumen comes from the pathobionts. In addition to their capacity to post-translate and modify protein and breach tight-junction integrity (8, 13, 16) they add pathogenic burden, represented by the mTg, as was recently described (14). The environmental pathogenic microbes can exchange mobile elements with the luminal inhabitants. Surface water, sewage treatment plant effluents, soils, animal wastes, contaminated biofilms and urban rivers are increasingly reported, thus threatening global human health (35–37). Not surprising, commonly consumed plants and vegetables contain transglutaminase. Even if their sequence homology to mTg is not high, they are capably to cross-link peptides, including gluten. They were even implicated as a possible player in CD pathogenesis, starting the process in the intestinal lumen (38). MTg carrying examples are apple, soybean, bean sprouts, fodder beet, rosemary leaves, Jerusalem artichoke, spinach leaves and green peas, routinely consumed/ingested plants, fruits and vegetables, all have transglutaminase activities (38). Polyols, heavily used in the processed food industry in protein-based coating, biofilm formation, gelatination of products and bio-packaging, were shown to improve thermal stability and half-life of the mTg, thus potentiating the enzyme cross linking capacity (39).
A totally new topic of potential Tg delivery to the gut lumen is coming from the microbiota yeast's domain. Significantly higher fecal counts of candida and saccharomyces species were identified in CD patients (40) and CD patients have higher frequency and levels of anti-Saccharomyces cerevisiae antibodies (41). Both, Candida albicans and Saccharomyces cerevisiae synthesize the Tg enzyme (42, 43) to the point that Candida albicans was suggested to trigger CD (44, 45).
Summarizing the present paragraph, a plethora of extra intestinal and luminal sources of mTg exist that potentially can interact with gluten containing compounds. This enzymatic cargo can cross link those glutamine rich peptides, resulting in post-translated, modified immunogenic epitopes, potentially driving CD autoimmunogenesis (9, 13, 16). After setting the introductory stage, the following part will expend on the mTg cross linked gliadin complex immunogenicity and potential pathogenicity in CD.
Gliadin Docked mTg Complexes are Immunogenic in Pediatric CD
When mTg transamidates gliadin peptides a neo cross-linked complex is created, where the physico-chemical, electrical and three dimensional features are changed (46). This is a typical enzymatic post-translational modification of gliadin, resulting in a non-naïve, non-tolerogenic and potentially immunogenic protein (13, 16). To investigate its immunogenicity, CD associated antibodies, including the anti-neo-epitope mTg were investigated, back to back, in a pediatric CD population, compared to controls. The neo-epitope mTg IgG had a sensitivity of 94.9% and specificity of 93.9% and reflected the patient's intestinal pathology (46). Performing competition assays between the isolated antigens (gliadin, tTg and deamidated gliadin peptide) and the gliadin tTg/mTg neo complexes, mTg and tTg-neo epitopes display comparable immunopotent epitopes (46).
After describing the mTg-gliadin complex antigenicity in CD, the stage is ready to evaluate mTg potential pathogenicity in driving CD.
Pathogenic Aspects of mTg
Several aspects of the mTg hint at its potential pathogenicity and suggest involvement in CD initiation/progression. Sharing functional aspects with the autoantigen and the mucosal step limiting phase of the tTg enzyme in CD development puts mTg as a primary candidate as a partner for CD development. Following are several observations that support mTg involvement in the CD autoimmunogenesis.
MTg Suppresses Intestinal Luminal Protective Barriers
The human intestine possess multiple protective mechanisms to keep the microbes at bay. Several of those mechanisms are contradicted by the mTg, when bugs are fighting to survive inside us:
a. The highly physiological important isopeptide bonds created by the mTg are resistant to any known human enzyme, thus escaping the enzymatic hydrolysis, reducing or chaotrophic agents and detergents. Those mTg formed protective structures improve the microorganism's survival in the enteric hostile compartment. Even bile acids, antimicrobial peptides and immunoglobulins are ineffective facing those structures (13, 47, 48).
b. mTg was found to suppress human immunity by its anti-phagocytic property, thus suppressing a major immune protective system (14, 49, 50).
MTg Effects on Epithelial Gliadin Uptake and Transportation
a. MTg may enhances gliadin uptake through human intestinal cell-line, as was demonstrated for tTg. TTg was shown to facilitate apical-basal passage of gliadin peptides, cooperating with the apical transferrin receptor and secretory IgA (9, 13, 51). Imitating tTg functions, mTg potentially can facilitate this epithelial gliadin uptake pathway, thus enhancing CD.
b. MTg and gliadin share comparable trans-enterocytic transport pathway. A major indispensable step in the pathophysiology of CD is the transcytosis of gliadin peptide to the sub-epithelial space to be deamidated\ transamidated by the tTg. It appears that mTg and gliadin peptides use this port of entry and pathway. Most Lately, Stricker et al. (52) placed tagged mTg and gliadin to the enterocyte upper membrane of CD and control human intestinal specimen or human cell line. A simultaneous uptake of mTg and gliadin into the endoplasmic reticulum of the RACE-cells and co-localization of the mTg and gliadin at the basolateral membrane and the lamina propria of CD duodenal samples were detected. If their results are substantiated it might reinforce the potential pathogenic role of the exogenous mTg or mTg-gliadin complex in CD evolution, when the complexes are exposed to the local immune systems. In this case, one can envision that the sub-epithelial deposits of tTg-gliadin, described as an early histological finding in CD intestinal samples, contain mTg-gliadin complexes.
MTg can Potentially Enhance Gliadin Induced Intestinal Permeability
Several mechanisms can explain how mTg functions and products can increase enteric permeability:
a. Protecting intestinal dysbiota and pathobionts, the mTg can enhance their luminal survival. Intestinal infections are important breachers of tight junction functional integrity (53).
b. Gliadin is an ideal substrate for mTg cross-linking. Being a major disruptor of intestinal permeability (54), its cross-linked complexes might further augment gut permeability.
c. Some tight junction proteins and cytoskeleton elements like actin, e-cadherin or adherens junctions can be modified by Tgs (55, 56). By cross-linking those proteins the mTg can disrupt tight junction functionality (9).
Additional Facts Related to Potential mTg Pathogenicity in CD
In order that the industrial processed food additive will impact human health, it has to be present in the consumed products. Notably, when 60 meat and meat products from the supermarket shelves were checked by two sensitive (around 25 mg pure enzyme in 1 kg of product) analytical methods, many contained the enzyme mTg (57). A literature survey disclosed that around 50–100 mg of mTg is used to process 1 kg of food product and the average intake of mTg can amount to 15 mg per day (12, 18, 25, 26). Although not causative, epidemiological data show an associative correlation between the surges of CD incidence and the consumption of enzymes in the bakeries, mTg being a major one (8, 9). Finally, wheat or gluten containing products enzymatically treated by mTg were shown to be immunogenic, inducing antibodies when consumed by humans (9, 46, 58–67).
Academical and Authoritative Warnings on the Potential Harmful Functions of mTg Usage by the Processed Food Industries
The mTg is not labeled, since it is considered as a processing aid, thus escaping the definition of a food additive. Due to the potential detrimental public health aspects, several scientists and organizations issue warnings, trying to increase the awareness of the regulatory authorities, academical communities and the general public on the subject. Following are some citations: “The usage of transglutaminase as a food additive is permitted in some countries. However, its utilization has to be declared to ensure transparency for consumers” Kaufmann et al. (57), “Therefore mTg can enhance the immunogenicity of gluten and should not be used in food products intended for consumption by CD patients” Dekking et al. (66). Not surprisingly, the worries and warnings on mTg nutritional industrial usage safety appear in numerous publications (26, 38, 57, 62, 66–68). More so, at least, in Switzerland and Germany, public warning were issued concerning mTg food safety, recommending labeling of the enzyme (69)1. The warnings mentioned the risk of mTg consumption by CD patients, “Suitable labeling of foods produced using mTg would enable these patients to avoid the uncertainties that the scientific community has yet to clarify”1.
Conclusions
Microbial transglutaminase is a food additive, heavily used in a plethora of processed food industries. It is unlabeled and hidden from the public knowledge. Being functionally similar to the tTg, it can post-translate and modify gliadin peptides by cross-linking them, thus, inducing loss of tolerance. Figure 1 describe schematically the chain of events starting from the mTg sources ending in CD development.
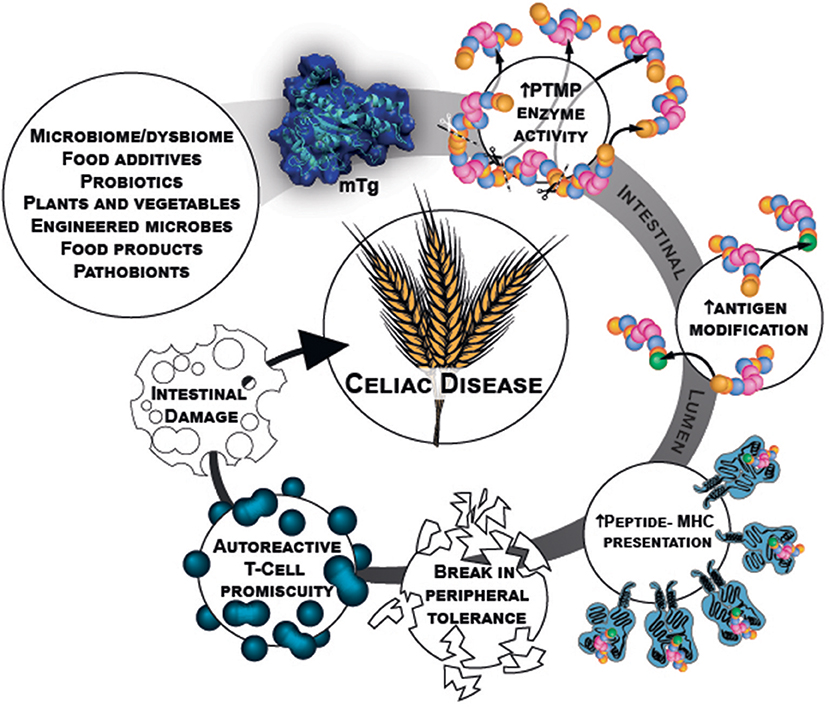
Figure 1. mTg sources and chain of events that potentially initiate gluten break of tolerance resulting in CD.
There are published warnings, alarming the public on the potential danger of using or consuming this enzyme. Recent publications found mTg to be immunogenic in CD patients and its pathogenicity is continuously unraveled. The logical theoretical basis for the mTg to be a new environmental factor in CD induction exist, however, causality should further be explored.
Author Contributions
MT designed, overviewed, searched, and analyzed the literature and edited the manuscript. LA designed and wrote the manuscript.
Conflict of Interest Statement
MT is the head of the Aesku. KIPP institute.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Mr. Neu Alf for the figure design and to Dr. Neidhöfer Sandra and Dr. Ramesh Ajay for the editing and reviewing the manuscript.
Abbreviations
CD, celiac disease; mTg, microbial transglutaminase; tTg, tissue transglutaminase.
Footnotes
References
1. Hardy MY, Tye-Din JA. Coeliac disease: a unique model for investigating broken tolerance in autoimmunity. Clin Transl Immunol. (2016) 5:e112. doi: 10.1038/cti.2016.58
2. Parzanese I, Qehajaj D, Patrinicola F, Aralica M, Chiriva-Internati M, Stifter S, et al. Celiac disease: from pathophysiology to treatment. World J Gastrointest Pathophysiol. (2017) 8:27–38. doi: 10.4291/wjgp.v8.i2.27
3. Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet (2018) 391:70–81. doi: 10.1016/S0140-6736(17)31796-8
4. Lerner A, Arleevskaya M, Schmiedl A, Matthias T. Microbes and viruses are bugging the gut in celiac disease. Are they friends or foes? Front Microbiol. (2017) 8:1392. doi: 10.3389/fmicb.2017.01392
5. Lionetti E, Catassi C. The role of environmental factors in the development of celiac disease: what is new? Diseases (2015) 3:282–93. doi: 10.3390/diseases3040282
6. Lerner A, Jeremias P, Matthias T. The world incidence of celiac disease is increasing: a review. Int J Recent Sci Res. (2015) 7:5491–6.
7. Lerner A, Jeremias P, Matthias T. The world incidence and prevalence of autoimmune diseases is increasing: a review. Int J Celiac Dis. (2015) 3:151–5. doi: 10.12691/ijcd-3-4-8
8. Lerner A, Matthias T. Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun Rev. (2015) 14:479–89. doi: 10.1016/j.autrev.2015.01.009
9. Lerner A, Matthias T. Possible association between celiac disease and bacterial transglutaminase in food processing: a hypothesis. Nutr Rev. (2015) 73:544–52. doi: 10.1093/nutrit/nuv011
10. Lerner A, Neidhöfer S, Matthias T. Transglutaminase 2 and anti-transglutaminase 2 autoantibodies in celiac disease and beyond: Part A: TG2 double-edged sword: gut and extraintestinal involvement. Immun Res. (2015) 11:101–5. doi: 10.4172/1745-7580.10000101
11. Ando H, Adachi M, Umeda K, Matsuura A, Nonaka M, Uchio R, et al. Purification and characteristics of a novel transglutaminase derived from microorganisms. Agric Biol Chem. (1989) 53:2613–7.
12. Martins IM, Matos M, Costa R, Silva F, Pascoal A, Estevinho LM, et al. Transglutaminases: recent achievements and new sources. Appl Microbiol Biotechnol. (2014) 98:6957–64. doi: 10.1007/s00253-014-5894-1
13. Lerner A, Aminov R, Matthias T. Transglutaminases in dysbiosis as potential environmental drivers of autoimmunity. Front Microbiol. (2017) 8:article 66. doi: 10.3389/fmicb.2017.00066
14. Yu J, Pian Y, Ge J, Guo J, Zheng Y, Jiang H, et al. Functional and structural characterization of the antiphagocytic properties of a novel transglutaminase from Streptococcus suis. J Biol Chem. (2015) 290:19081–92. doi: 10.1074/jbc.M115.643338
15. Rombouts I, Lamberts L, Celus I, Lagrain B, Brijs K, Delcour JA. Wheat gluten amino acid composition analysis by high-performance anion-exchange chromatography with integrated pulsed amperometric detection. J Chromatogr A (2009) 1216:5557–62. doi: 10.1016/j.chroma.2009.05.066
16. Lerner A, Aminov R, Matthias T. Dysbiosis may trigger autoimmune diseases via inappropriate posttranslational modification of host proteins. Front Microbiol. (2016) 7: 84. doi: 10.3389/fmicb.2016.00084
17. Reif S, Lerner A. Tissue transglutaminase – the key player in celiac disease: a review. Autoimm Rev. (2004) 3:40–5. doi: 10.1016/S1568-9972(03)00065-X
18. Kieliszek M, Misiewicz A. Microbial transglutaminase and its application in the food industry. Rev Folia Microbiol. (2014) 59:241–50. doi: 10.1007/s12223-013-0287-x
19. Rachel NM, Pelletier JN. Biotechnological applications of transglutaminases. Biomolecules (2013) 3:870–88. doi: 10.3390/biom3040870
20. Santhi D, Kalaikannan A, Malairaj P, Arun Prabhu S. Application of microbial transglutaminase in meat foods: a review. Crit Rev Food Sci Nutr. (2017) 57:2071–6. doi: 10.1080/10408398.2014.945990
21. Gaspar AL, de Góes-Favoni SP. Action of microbial transglutaminase (MTGase) in the modification of food proteins: a review. Food Chem. (2015) 171:315–22. doi: 10.1016/j.foodchem.2014.09.019
22. Motoki M, Seguro K. Transglutaminase and its use for food processing. Trends Food Sci Tech. (1998) 9:204–10. doi: 10.1016/S0924-2244(98)00038-7
23. Heil A, Ohsam J, Büchold C, Pasternack R, Yokoyama K, Kumazawa Y, et al. Microbial transglutaminase has a lower deamidation preference than human tissue transglutaminase on a celiac disease relevant wheat gliadin T-cell epitope. J Cereal Sci. (2016) 70:47–56. doi: 10.1016/j.jcs.2016.05.022
24. Gerrard JA, Fayle SE, Brown PA, Sutton KH, Simmons L, Rasiah I. Effects of microbial transglutaminase on the wheat proteins of bread and croissant dough. J Food Sci. (2001) 66:782–6. doi: 10.1111/j.1365-2621.2001.tb15172.x
25. Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol. (2004) 64:447–54. doi: 10.1007/s00253-003-1539-5
26. Malandain H. Transglutaminases: a meeting point for wheat allergy, celiac disease, and food safety Europ. Ann Aller Clin Immun. (2005) 37:397–403.
27. Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. (2005) 3:431–8. doi: 10.1038/nrmicro1152
28. Bruce SE, Bjarnason I, Peters TJ. Human jejunal transglutaminase: demonstration of activity, enzyme kinetics and substrate specificity with special relation to gliadin and coeliac disease. Clin Sci. (1985) 68:573–9. doi: 10.1042/cs0680573
29. Lerner A, Aminov R, Matthias T. Horizontal gene transfer in the human gut. Front Immunol. (2017) 8:1630. doi: 10.3389/fimmu.2017.01630
30. Devirgiliis C, Zinno P, Perozzi G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front Microbiol. (2013) 4:301. doi: 10.3389/fmicb.2013.00301
31. Flórez AB, Campedelli I, Delgado S, Alegría Á, Salvetti E, Felis GE, et al. Antibiotic susceptibility profiles of dairy leuconostoc, analysis of the genetic basis of atypical resistances and transfer of genes in vitro and in a food matrix. PLoS ONE (2016) 11:e0145203. doi: 10.1371/journal.pone.0145203
32. Rosander A, Connolly E, Roos S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl Environ Microbiol. (2008) 74:6032–40. doi: 10.1128/AEM.00991-08
33. Egervärn M, Lindmark H, Olsson J, Roos S. Transferability of a tetracycline resistance gene from probiotic Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie Van Leeuwenhoek (2010) 97:189–200. doi: 10.1007/s10482-009-9401-0
34. Tajkarimi M, Wexler HM. CRISPR-Cas systems in Bacteroides fragilis, an important pathobiont in the human gut microbiome. Front Microbiol. (2017) 8:2234. doi: 10.3389/fmicb.2017.02234
35. Zhou ZC, Zheng J, Wei YY, Chen T, Dahlgren RA, Shang X, et al. Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ Sci Pollut Res Int. (2017) 24:23753–62. doi: 10.1007/s11356-017-0032-0
36. Qiao M, Ying GG, Singer AC, Zhu YG. Review of antibiotic resistance in China and its environment. Environ Int. (2018) 110:160–72. doi: 10.1016/j.envint.2017.10.016
37. Mokracka J, Koczura R, Jabłonska L, Kaznowski A. Phylogenetic groups, virulence genes and quinolone resistance of integron-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Van Leeuwenhoek (2011) 99:817–24. doi: 10.1007/s10482-011-9555-4
38. Skovbjerg H, Norén O, Anthonsen D, Moller J, Sjöström H. Gliadin is a good substrate of several transglutaminases: possible implication in the pathogenesis of coeliac disease. Scand J Gastroenterol. (2002) 37:812–7.
39. Cui Li Du G, Zhang D, Chen J. Thermal stability and conformational changes of transglutaminase from a newly isolated Streptomyces hygroscopicus. Bioresour Technol. (2008) 99:3794–800. doi: 10.1016/j.biortech.2007.07.017
40. Harnett J, Myers SP, Rolfe M. Significantly higher faecal counts of the yeasts candida and saccharomyces identified in people with coeliac disease. Gut Pathog. (2017) 9:26. doi: 10.1186/s13099-017-0173-1
41. Ashorn S, Raukola H, Välineva T, Ashorn M, Wei B, Braun J, et al. Elevated serum anti-Saccharomyces cerevisiae, anti-I2 and anti-OmpW antibody levels in patients with suspicion of celiac disease. J Clin Immunol. (2008) 28:486–94. doi: 10.1007/s10875-008-9200-9
42. Iranzo M, Aguado C, Pallotti C, Cañizares JV, Mormeneo S. Transglutaminase activity is involved in Saccharomyces cerevisiae wall construction. Microbiol. (2002) 148:1329–34. doi: 10.1099/00221287-148-5-1329
43. Ruiz-Herrera J, Iranzo M, Elorza MV, Sentandreu R, Mormeneo S. Involvement of transglutaminase in the formation of covalent cross-links in the cell wall of Candida albicans. Arch Microbiol. (1995) 164:186–93. doi: 10.1007/BF02529970
44. Nieuwenhuizen WF, Pieters RH, Knippels LM, Jansen MC, Koppelman SJ. Is Candida albicans a trigger in the onset of coeliac disease? Lancet (2003) 361:2152–4. doi: 10.1016/S0140-6736(03)13695-1
45. Corouge M, Loridant S, Fradin C, Salleron J, Damiens S, Moragues MD, Souplet V, et al. Humoral immunity links Candida albicans infection and celiac disease. PLoS ONE (2015) 10:e0121776. doi: 10.1371/journal.pone.0121776
46. Matthias T, Jeremias P, Neidhöfer S, Lerner A. The industrial food additive microbial transglutaminase, mimics the tissue transglutaminase and is immunogenic in celiac disease patients. Autoimm Rev. (2016) 15:1111–9. doi: 10.1016/j.autrev.2016.09.011
47. Rao RU, Mehta K. Transglutaminases, thioredoxins and protein disulphide isomerase: diverse enzymes with a common goal of cross-linking proteins in lower organisms. Indian J Exp Biol. (2004) 42:235–43.
48. Tagami U, Shimba N, Nakaamura M, Yokoyama K, Suzuki E, Hirokawa T. Substrate specificity of microbial transglutaminase as revealed by three-dimentional docking simulation and mutagenesis. Protein Eng Des Sel. (2009) 22:747–52. doi: 10.1093/protein/gzp061
49. Fittipaldi N, Segura M, Grenier D, Gottschalk M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Fut Microbiol. (2012) 7:259–79. doi: 10.2217/fmb.11.149
50. Xu B, Zhang P, Li W, Liu R, Tang J, Fan H. hsdS, Belonging to the type i restriction-modification system, contributes to the Streptococcus suis serotype 2 survival ability in phagocytes. Front Microbiol. (2017) 8:1524. doi: 10.3389/fmicb.2017.01524
51. Lebreton C, Menard S, Abed J, Moura IC, Coppo R, Dugave C, et al. Interactions among secretory immunoglobulin A, CD71, and transglutaminase-2 affect permeability of intestinal epithelial cells to gliadin peptides. Gastroenterol (2012) 143:698–707. doi: 10.1053/j.gastro.2012.05.051
52. Stricker S, de Laffolie J, Rudloff S, Komorowski L, Zimmer KP. Intracellular localization of microbial transglutaminase and its influence on the transport of gliadin in enterocytes. J Pediatr Gastroenterol Nutr. doi: 10.1097/MPG.0000000000002171
53. El Asmar R, Panigrahi P, Bamford P, Berti I, Not T, Coppa GV, et al. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterol (2002) 123:1607–15. doi: 10.1053/gast.2002.36578
54. Hollon J, Puppa EL, Greenwald B, Goldberg E, Guerrerio A, Fasano A. Effect of gliadin on permeability of intestinal biopsy explants from celiac disease patients and patients with non-celiac gluten sensitivity. Nutrients (2015) 7:1565–76. doi: 10.3390/nu7031565
55. Terman JR, Kashina A. Post-translational modification and regulation of actin. Curr Opin Cell Biol. (2013) 25:30–8. doi: 10.1016/j.ceb.2012.10.009
56. Hiiragi T, Sasaki H, Nagafuchi A, Sabe H, Shen SC, Matsuki M, et al. Transglutaminase type 1 and its cross-linking activity are concentrated at adherens junctions in simple epithelial cells. J Biol Chem. (1999) 274:34148–54. doi: 10.1074/jbc.274.48.34148
57. Kaufmann A, Koppel R, Widmer M. Determination of microbial transglutaminase in meat and meat products. Food Addit Contam Part A Chem Anal Control, Exp Risk Assess. (2012) 29:1364–73. doi: 10.1080/19440049.2012.691557
58. Lerner A, Matthias T. Food industrial microbial transglutaminase in celiac disease: treat or trick. Int J Celiac Dis. (2015) 3:1–6. doi: 10.12691/ijcd-3-1-10
59. Cabrera-Cha'vez F, Rouzaud-Sa'ndez O, Sotelo-Cruz N, Calderón de la Barca AM. Bovine milk caseins and transglutaminase-treated cereal prolamines are differentially recognized by IgA of celiac disease patients according to age. J Agric Food Chem. (2009) 57:3754–9. doi: 10.1021/jf802596g
60. Elli L, Roncoroni L, Hils M, Pasternack R, Barisani D, Terrani C, et al. Immunological effects of transglutaminase-treated gluten in coeliac disease. Hum Immunol. (2012) 73:992–7. doi: 10.1016/j.humimm.2012.07.318
61. Falini ML, Elli L, Caramanico R, Bardella MT, Terrani C, Roncoroni L, et al. Immunoreactivity of antibodies against transglutaminase-deamidated gliadins in adult celiac disease. Dig Dis Sci. (2008) 53:2697–701. doi: 10.1007/s10620-007-0191-9
62. Cabrera-Cha'vez F, Rouzaud-Sa'ndez O, Sotelo-Cruz N, Calderón de la Barca AM. Transglutaminase treatment of wheat and maize prolamines of bread increase the serum IgA reactivity of celiac disease patients. J Agric Food Chem. (2008) 56:1387–91. doi: 10.1021/jf0724163
63. Berti C, Roncoroni L, Falini ML, Caramanico R, Dolfini E, Bardella MT, et al. Celiac-related properties of chemically and enzymatically modified gluten proteins. J Agric Food Chem. (2007) 55:2482–8. doi: 10.1021/jf062623n
64. Heredia-Sandoval NG, Islas-Rubio AR, Cabrera-Chávez F, Calderón de la Barca AM. Transamidation of gluten proteins during the bread-making process of wheat flour to produce breads with less immunoreactive gluten. Food Funct. (2014) 5:1813–8. doi: 10.1039/c4fo00118d
65. Ruh T, Ohsam J, Pasternack R, Yokoyama K, Kumazawa Y, Hils M. Microbial transglutaminase treatment in pasta-production does not affect the immunoreactivity of gliadin with celiac disease patients' sera. J Agric Food Chem. (2014) 62:7604–11. doi: 10.1021/jf501275c
66. Dekking EHA, Van Veelen PA, de Ru A, Kooy-Winkelaara EMC, Gröneveld T, Nieuwenhuizen WF, Koning F. Microbial transglutaminase generate Tcell stimulatory epitopes involved in celiac disease. J Cereal Sci. (2008) 47:339–46. doi: 10.1016/j.jcs.2007.05.004
67. Gerrard JA, Sutton KH. Addition of transglutaminase to cereal products may generate the epitope responsible for coeliac disease. Trend Food Sci Technol. (2005) 16:510–2. doi: 10.1016/j.tifs.2005.07.002
68. Cottam JRA, Gerrard JA (2012). Protein cross-linking in food-structure, applications, implications for health and food safety. In: Hui YH, editors. Food Biochemistry and Food Processing, 2nd ed. (Ames, IA: Blackwell), p. 207–23.
69. Eidgeno ssische Departement des Innern (EDI). Verordnung des EDI u ber Lebensmittel tierischer Herkunft [in German]. [Regulation of the EDI about foods of animal origin. SR 817.022.108. Approved November 23 2005]. Available online at: https://www.admin.ch/opc/de/classified-compilation/20050164/index.html. (Accessed November 23, 2005).
Keywords: celiac disease, microbiome, food additive, gluten, cross linking, microbial transglutaminase
Citation: Torsten M and Aaron L (2018) Microbial Transglutaminase Is Immunogenic and Potentially Pathogenic in Pediatric Celiac Disease. Front. Pediatr. 6:389. doi: 10.3389/fped.2018.00389
Received: 04 October 2018; Accepted: 27 November 2018;
Published: 11 December 2018.
Edited by:
Andrew S. Day, University of Otago, New ZealandReviewed by:
Marek Kieliszek, Warsaw University of Life Sciences, PolandVictor Manuel Navas-López, Hospital Materno-Infantil, Spain
Copyright © 2018 Torsten and Aaron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lerner Aaron, YWFyb25sZXJuZXIxOTQ4QGdtYWlsLmNvbQ==
 Matthias Torsten1
Matthias Torsten1 Lerner Aaron
Lerner Aaron