- 1Department of Biological Sciences, Indian Institute of Science Education and Research Bhopal, Bhopal, India
- 2School of Physical Sciences, Jawaharlal Nehru University, New Delhi, India
- 3Department of Physics, TDPG College, VBS Purvanchal University, Jaunpur, India
Silver nanoparticles (AgNPs) have many applications in various fields, including biomedical applications. Due to the broad range of applications, they are considered as the leading fraction of manufactured nanoparticles. AgNPs are synthesized by different types of chemical and biological (green) methods. Previously, biologically synthesized AgNPs were considered safe for the environment and humans. However, new toxicity evidence have initiated a more careful assessment to delineate the toxicity mechanisms associated with these nanoparticles. This study demonstrates the use of aqueous gooseberry extract for AgNP preparation in a time- and cost-effective way. Ultraviolet-visible spectroscopy, X-ray diffraction, transmission electron microscopy, and dynamic light scattering confirm the formation of AgNPs, with an average size between 50 and 100 nm. Untargeted 1H-nuclear magnetic resonance-based metabolomics revealed manyfold up- and down-regulation in the concentration of 55 different classes of annotated metabolites in AgNP-exposed yeast Saccharomyces cerevisiae cells. Based on their chemical nature and cellular functions, these metabolites are classified into amino acids, glycolysis and the tricarboxylic acid (TCA) cycle, organic acids, nucleotide metabolism, urea cycle, and lipid metabolism. Transcriptome analysis revealed that the genes involved in oxidative stress mitigation maintain their expression levels, whereas the genes of the TCA cycle and lipid metabolism show drastic down-regulation upon AgNP exposure. Moreover, they can induce alteration in histone epigenetic marks by altering the methylation and acetylation of selected histone H3 and H4 proteins. Altogether, we conclude that the selected dose of biologically synthesized AgNPs impose toxicity by modulating the transcriptome, epigenome, and metabolome of eukaryotic cells, which eventually cause disequilibrium in cellular metabolism leading to toxicity.
Introduction
The incorporation of engineered nanomaterials (ENMs) in several commercial applications and consumer products has been increasing rapidly. The accelerating and unregulated release of ENMs into the environment constitutes an emerging and widespread form of pollution (Oberdörster et al., 2005; Musee, 2011). Among the wide range of ENMs, silver nanoparticles (AgNPs), in particular, are considered as a major fraction of such nanoparticles. They exhibit antimicrobial and antitumor properties, leading to their application in several aspects of biomedical fields. They are often used in wound dressings, surgical instruments, medical catheters, and bone prostheses as well as in nanotheranostics and personalized medicine (Smolkova et al., 2017; Marassi et al., 2018). Earlier, biologically (using plant and microbial cell extracts) synthesized AgNPs were considered to be nontoxic for humans and its exposure with overdose was the only known parameter reported to impose toxicity (McShan et al., 2014; Singh et al., 2014; Vazquez-Muñoz et al., 2017; Siddiqi et al., 2018). However, in the recent past, several new toxicity evidence have initiated a more careful assessment to outline the toxicity mechanism(s) of AgNPs on various living systems as reviewed elsewhere (McShan et al., 2014; Vazquez-Muñoz et al., 2017). The literature describes that these mechanism(s) involve mechanical and physical damages to cell membranes by direct attachment resulting in the disruption of membrane potential. Among them, the release of silver ions (Ag+) and the generation of reactive oxygen species (ROS; oxidative stress) are assumed as the main factors closely associated with inflammatory cell responses, immunotoxicity, DNA damage, and global gene and/or protein expression and thus AgNP-induced toxicity (Dubey et al., 2015; Mytych et al., 2017).
Concrete information on AgNP-induced toxicity biomarkers is still lacking. Therefore, researchers are attempting to employ different “-omics”-based tools to gain a better understanding of the toxicity mechanism and precise biomarkers (Bouhifd et al., 2013). In this context, untargeted metabolomics offers a powerful tool for the quantification of small metabolites (Schnackenberg et al., 2012). Compared to transcriptomic and proteomic data, assessing the cellular metabolic alteration with nuclear magnetic resonance (NMR)-based metabolomics provides more valuable information for enlightening unknown biological effects and helps in better understanding the mechanisms of nanomaterial-induced toxicity (Lv et al., 2015). Researchers recently used this method to identify and characterize deregulated metabolites in various model systems under different physical and chemical stressors, including nanomaterials, but none has been used to assess the changes in most commonly used AgNP exposure in yeast Saccharomyces cerevisiae yet (Carrola et al., 2016; Carrola et al., 2018). Moreover, previous studies on nanomaterials revealed that, apart from cytotoxicity, genotoxicity, and immunotoxicity, nanomaterials used in personalized medicine could alter the cellular epigenome and induce epigenetic toxicity. These alterations were found to be associated with many pathophysiological diseases, including cancer (Smolkova et al., 2015; Sierra et al., 2016; Smolkova et al., 2017). Post-translational modifications (PTMs) of histone proteins, in particular, attained much attention as they are linked to a variety of biological processes, e.g., gene/protein regulation and cell signaling, as well as to disease states (Jenuwein and Allis, 2001). In case of AgNPs, most of these studies were focused on the DNA methylation mechanism of epigenetics, but the study that specifically deals with histone PTMs has not been reported. The availability of the whole-genome sequence, mutant strains, and metabolome database (YMDB) of S. cerevisiae makes it an excellent model organism to study the metabolomics and other “-omic” (e.g., epigenomic) approaches under diverse external stimuli, including nanotoxicity (Pereira et al., 2018).
The present study proceeds with the green synthesis (using the aqueous extract of gooseberry) and characterization of AgNPs. Thereafter, 1H-NMR analysis of the metabolic alterations induced by biologically synthesized AgNPs in the yeast S. cerevisiae has been done. Further, transcriptomics studies have been performed to validate the down-regulation in the expression of genes associated with the deregulated metabolites of different metabolic pathways. Finally, it is aimed to examine the potential effects on histone PTMs and its relevance in gene expression. The combined results of the present study demonstrate that biologically synthesized AgNPs significantly alter the metabolic/epigenetic markers in yeast and also illustrate a novel finding on the mechanism of nanotoxicity.
Materials and Methods
Chemicals and Synthesis of AgNPs
Silver nitrate (AgNO3) procured from Alfa Aesar was used without any further purification. Dried gooseberry was purchased from a local market (New Delhi, India). Gooseberry extract was prepared by rehydrating 25 g dried powder of gooseberry in 500 mL sterile deionized (DI) water and kept for 24 h. Further, the solution was filtered (by 0.5 µm membrane filter) and tested for contamination, if any, and stored in a cool, dark place under sterile conditions. AgNPs were synthesized by mixing gooseberry extract and AgNO3 (0.1 M) with constant stirring at 250 rpm at room temperature. To study the effect of the amount of gooseberry extract on the synthesis of AgNPs, two samples with different volume ratios of AgNO3 solution and gooseberry extract (1:1 and 1:0.5) were prepared. The instant change in color depends on the shape and size of the particles, which primarily indicate the successful formation of AgNPs. Reaction mixtures thus obtained were further centrifuged at 2000× g for 15 min. AgNPs thus settled at the bottom of tubes and were collected and suspended in sterile DI water.
Characterization of Synthesized AgNPs
X-ray diffraction (XRD) analysis: To confirm the formation of AgNPs using gooseberry extract, XRD analysis was carried out on a tabletop diffractometer (Rigaku Miniflex 600) using CuKα radiation in a wide range of Bragg angle (20° ≤ 2θ ≤ 80°).
Transmission electron microscopy (TEM) analysis: TEM investigations were carried out to determine the size and shape of biologically synthesized AgNPs. The samples for TEM analysis were prepared by placing a drop (∼6 µL) of dispersed AgNP suspension on 300-mesh carbon-coated copper grid and allowed to dry. Studies were done on JEOL-2100F (JEOL, USA) operated at an accelerating voltage 200 kV.
Ultraviolet (UV)-visible (UV-vis) spectroscopy: The formation of AgNPs was observed by evaluating UV-vis spectra of synthesized AgNP nanoparticles. The synthesized samples were analyzed using Perkin-Elmer Lambda 750S UV-vis spectrometer with a resolution of 1 nm. Then, 500 µL AgNP solution, diluted to 2 mL with water, was analyzed by a UV-vis spectrometer. Two different samples of AgNPs prepared with varying AgNO3 solution/gooseberry extract ratios (1:1 and 1:0.5) resulted in distinct colors due to the different shapes and sizes of the synthesized nanoparticles.
Yeast Growth and AgNP Treatment
Overnight-grown yeast S. cerevisiae BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) cells inoculated into 250 mL YPD medium (OD600 nm = 0.2) were used for cytotoxicity and metabolite extraction. Yeast cells were grown until mid-exponential growth phase (OD600 nm = 1.3–1.5), subsequently collected by centrifugation (1000× g; 5 min), and thoroughly washed three times with autoclaved Milli Q water. Water-suspended yeast cells were equally partitioned (10 mL each) into five culture tubes under sterile conditions; one tube was left untreated (control), whereas the others were used for AgNP treatment. AgNP stock solution (0.5 mg mL-1) was prepared in sterile DI water, further ultrasonicated for 1 h, and stored in the dark at room temperature.
Cytotoxicity, Cell Death, and ROS Measurements
Four concentrations (0.5, 1.0, 1.5, and 2.0 mg L-1) of AgNPs were prepared to be used for cytotoxicity, cell death, and ROS assays. For these assays, DI water-suspended yeast cells with different doses of AgNPs were incubated for 3 h (200 rpm, 30°C). Each experiment was performed in triplicate with similar culture/treatment conditions. The cytotoxicity and cell death measurements in yeast cells under AgNP exposure were analyzed by colony-forming units (CFU) and spot sensitivity assay methods, whereas cell death assay and ROS measurements were performed in accordance with our previously published research article (Babele et al., 2018). CFU and spot assay plates were scanned using an HP scanner and the respective colonies were counted and presented as percent survival (Babele et al., 2018). Propidium iodide (PI) and 2′,7′-dichlorofluorescin diacetate (DCFDA) fluorogenic probes were used for the detection of dead cells and ROS, respectively, using previously described methods (Babele et al., 2018). The percentage of PI-positive (dead) cells was calculated as the number of PI-positive cells divided by the number of total cells × 100. Approximately 300 cells from 10 different panels (under a fluorescence microscope) were counted from three independent repeats. For ROS measurement, fluorescence intensity (FLU; excitation λ = 488 nm and emission λ = 520 nm) was used (Babele et al., 2018). All fluorescence microscopic observations were done on a Zeiss Apotome 2 using 63× oil lens in RFP (PI stain)-GFP (DCF stain) filter sets. Quantitative estimation of ROS was done in a fluorescence microplate reader (BioTek FLx800, BioTek Instruments, Inc., USA) with FLU (excitation λ = 488 nm and emission λ = 520 nm).
Metabolite Extraction and Sample Preparation for NMR Analysis
For metabolite extraction, sterile DI water-suspended yeast cells were equally divided (100 mL each) into two flasks under sterile conditions: one was left untreated (control) and the other was treated with AgNPs (1.0 mg L-1). The respective experiments were performed in triplicate in similar culture/treatment conditions. Metabolites were extracted according to a previously described method (Airoldi et al., 2015; Babele, 2019). Briefly, the control and treated cells were pelleted through centrifugation and washed gently with 100 mL ice-cold water and instantaneously placed in a refrigerator (-80°C). Frozen cells were thoroughly washed (three times) and rinsed with 10 mL ice-cold water. Further, cell pellets were resuspended in 4 mL of 75%:25% ethanol/DI water and an equal volume of autoclaved glass beads was subsequently added. Tubes were allowed to be vigorously vortex mixed (10 times) for 35 s with a time interval of 40 s in the ice. Further, samples were centrifuged (5 min, 2000× g) to remove the glass beads. Cell debris from the extract and equal volumes of the supernatant (with metabolites) were vacuum evaporated. Finally, dried samples were properly dissolved in 600 μL deuterated phosphate buffer (Na2DPO4 100 mM, pH 7.5) in D2O. For chemical shift and the calculation of concentration, 0.1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS-d6) was used as an internal standard.
1H-NMR Experiments and Spectra Preprocessing
An Advance-III 700 MHz FT NMR spectrometer (AscendTM, Bruker) in digital quadrature detection mode at a frequency of 4323.96 Hz 5 mm QCI cryoprobe was used to assess NMR spectra (Babele, 2019). The deconvolution of the overlapping regions of the spectra and subsequently the absolute quantification for metabolites with resonance in populated spectral areas were performed (Puig-Castellví et al., 2016).
Metabolite Identification and Quantification
A targeted metabolite profiling analysis of 1H-NMR signals using both YMDB and HMDB was done for metabolite assignment. During analysis, each spectral intensity data set was normalized to the assigned chemical compounds in accordance with the total sum of the spectral regions. It was further converted to tab-delimited text (.txt) and comma-separated values (.csv) and fetched directly into Web-based MetaboAnalyst version 3.5 software to carry out data normalization, scaling, and multivariate analysis. p < 0.05 was used to provide the respective statistical significance. For the normalization of metabolite levels, log2 function and mean centering Pare to scaling were incorporated to all principal component analyses (PCA) and heatmaps by MetaboAnalyst version 3.5 (Xia et al., 2015).
Isolation of Total RNA and Quantitative Real-Time PCR (qRT-PCR)
To carry out qRT-PCR experiments, different concentrations (0.5, 1.0, and 2.0 mg L-1) of AgNPs were prepared. DI water-suspended yeast cells with varying doses of AgNPs were incubated for 3 h (200 rpm, 30°C). Each experiment was performed in triplicate under similar culture/treatment conditions. RNA extraction, cDNA preparation, and corresponding qRT-PCR experiments were carried out according to a previously described method (Babele, 2019). qRT-PCR was performed in an ABI-7300 with Sequence Detection System version 1.4 software (Applied Biosystems, CA) using 2× SYBR Green Master Mix (ABI). Housekeeping gene actin (ACT1) was used as an internal control to calculate the relative expression of genes. Relative changes (control vs. AgNPs) were calculated using the 2-ΔΔCT method and results are expressed as fold change. Data are the mean ± standard error of three independent experiments. The details of the genes and primers used in the present study for real-time PCR experiments are provided in Table S1.
Preparation of Protein Extracts and Immunoblotting Analysis
To carry out Western blotting experiments, different concentrations (0.5, 1.0, and 2.0 mg L-1) of AgNPs were used. DI water-suspended yeast cells with different doses of as-prepared AgNPs were incubated for 3 h (200 rpm, 30°C). Each experiment was performed in triplicate under similar culture/treatment conditions. Protein cell extraction from 20% trichloroacetic acid precipitation and further immunoblotting analysis were carried out in accordance to a procedure as described in our previous article (Babele et al., 2018). Primary antibodies general H3 (Sigma, H0164), H4 (Sigma, SAB4500312) H3K4me2 (Abcam, 8895), H3K4me3 (Abcam, 32356), H3K56Ac (Sigma, SAB4200328), H4K8Ac (Abcam, 46982), and H4K16Ac (Abcam, 61240) were used to investigate global histone modifications. The same blots were re-probed with anti-Tbp, -H3, and -H4 protein and used as loading control; purified H3 and H4 proteins were used as marker to represent the correct bands (Figure S4). The detection of proteins was carried out using a relevant secondary antibody, IRDye® 800CW goat anti-rabbit IgG or anti-mouse IgG (1:15,000; LI-COR Biosciences). An Odyssey infrared imager (LI-COR Biosciences) was used to scan the blots, and as-received images from three independent performances for each condition are shown.
Statistical Analysis
The analyzed data are the mean ± SD of three independent experiments. It favors the analysis of variance (ANOVA) assumptions as received from Shapiro-Wilk normality and Levene test for the equality of variances. Statistical significance (p < 0.05) was assessed using one-way ANOVA using GraphPad Prism 5 with Dunnett’s multiple comparison test.
Results
Synthesis and Characteristics of AgNPs
To test whether gooseberry extract has potential to synthesize AgNPs, we performed routine and confirmatory experiments. Initially, XRD analysis was performed. Figure S1A shows a typical XRD pattern of AgNPs synthesized with different concentrations of AgNO3 and gooseberry extract. The positions of the diffraction peaks of both samples were found to be in accordance with their reference of AgNPs and indexed well with JCPDS number 04-0783. The formation of AgNPs was further confirmed by a careful observation of change in color for AgNO3 solution by adding gooseberry extract. UV-vis absorption spectra of AgNPs with the above-mentioned variation of gooseberry extract exhibit prominent peaks at 442 and 457 nm (Figure S1B). It also reveals that the absorption spectrum with a 1:1 concentration ratio possesses two distinct maxima at 342 and 457 nm. The maxima peak of the longer-wavelength side shifted to 442 nm with a varied concentration of 1:0.5 of AgNO3 and gooseberry extract. The morphology and microstructure of the synthesized AgNPs were examined by TEM. Figure S2A depicts the TEM micrographs of AgNPs obtained from 1:0.5 reaction mixture of AgNO3 and gooseberry extract and the respective selected area electron diffraction (SAED) pattern was indexed well with the known F.C.C. structure of AgNPs. Further, it was found that, as we increase the ratio of AgNO3 to gooseberry extract from 1:0.5 to 1:1, the size of the synthesized particles increased and was found in the range of 80 to 90 nm. This change in the size of the nanoparticles was likely assumed as the primary cause for the red shift in the absorption spectra of AgNPs (Figure S1B). The SAED patterns associated with Figure S2B display four diffraction rings that correspond to the (111), (200), (220), and (311) reflection planes and were in good agreement with the XRD results. Figures S2C, D reveal an approximate statistical distribution of the particle size.
AgNPs Cause Dose-Dependent Toxicity, Cell Membrane Damage, and ROS Generation
Do biologically synthesized nanoparticles impose toxicity to biological systems? To answer this question, the effect of AgNPs on the growth of S. cerevisiae (BY4741) was studied. First, yeast cells were grown under different concentrations of AgNPs (0.5, 1.0, 1.5, and 2.0 mg L-1, respectively) to select the lethal concentration (LC50). A growth inhibition (CFU and spot assay) study indicated no apparent inhibition activity against cell growth with 0.5 mg L-1 AgNPs; however, 1.0 mg L-1 AgNPs led to a substantial decrease in cell numbers with 50% survival. Further, with the increase in AgNP concentration, i.e., 1.5 and 2.0 mg L-1, a drastic reduction in the cell numbers resulting in 20% cell survival was observed (Figures S3A– C). Furthermore, AgNP-exposed cells after PI staining were visualized under a microscope to investigate if the growth inhibition results in cell membrane damage. Fluorescence microscopy images (Figure 1A) depict that the selected doses (0.5–2.0 mg L-1) of AgNPs result in a gradual increase in PI-positive cells than the control cells (Figure 1B). Moreover, a dose-dependent gradual increase in ROS compared to control cells upon AgNP treatment was observed (Figure 1C). A similar trend was followed in a plate reader experiment (Figure 1D). These results helped us to select 1.0 mg L-1 as a LC50.
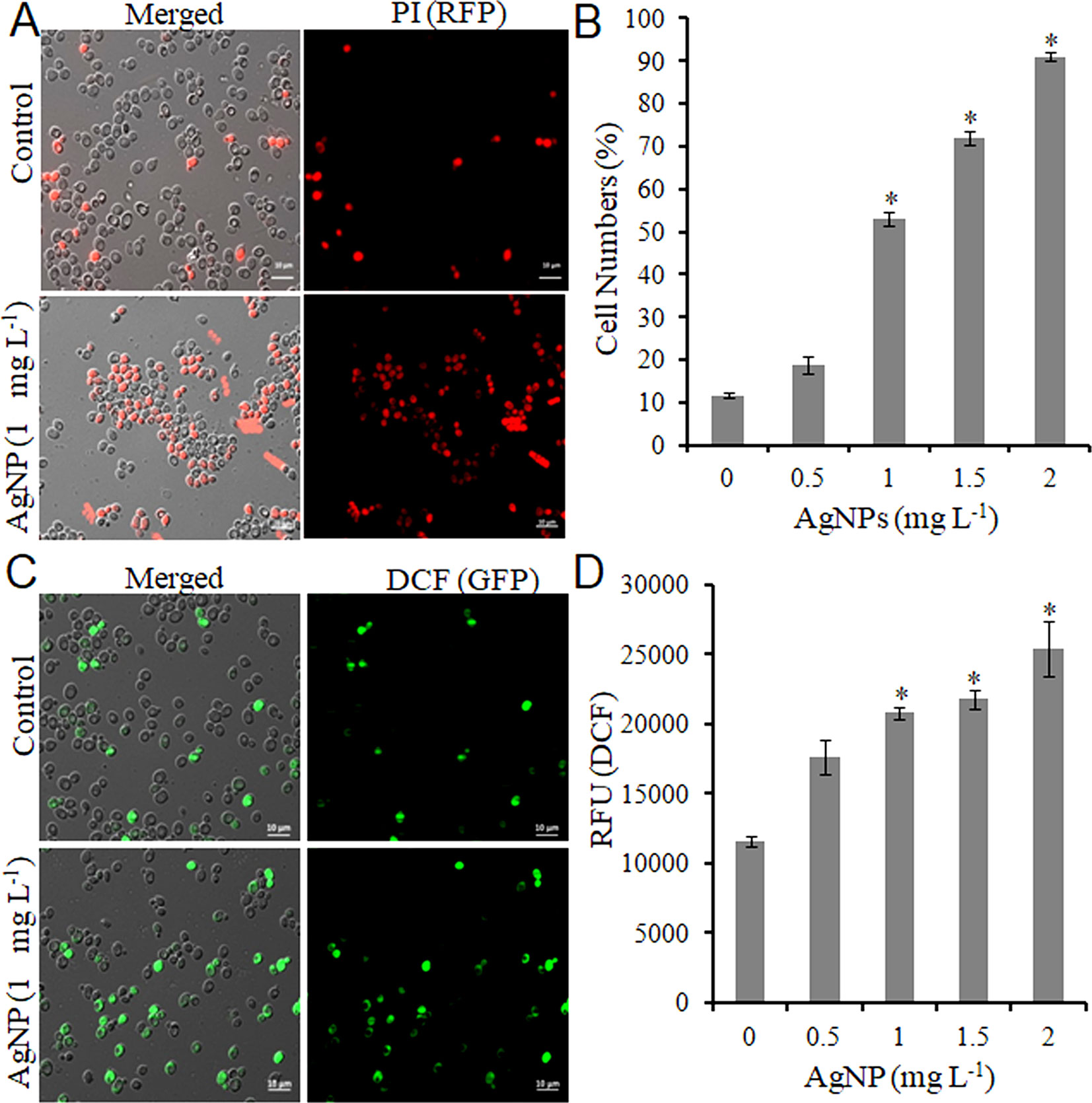
Figure 1 AgNPs causes cell death and oxidative stress by ROS accumulation. (A) Representative microscopic images of PI-stained yeast cells of control and AgNP-treated groups after 3 h. (B) Percentage of PI-positive (dead) cells was calculated as the number of PI-positive cells divided by the number of total cells × 100. (C) Representative microscopic images of DCFDA-stained yeast cells of control and AgNP-treated groups after 3 h. (D) ROS level in control and AgNP-treated cells was measured by a fluorescence plate reader after staining with DCFDA. Error bars, SD (n = 3). *p < 0.05, significant difference between the control and treatment groups. Bar, 10 μm.
Do AgNPs Cause Deregulation in the Metabolome and Transcriptome?
The evaluation of deregulation in metabolites was done by adopting an untargeted 1H-NMR analysis method. The complex experimental set-up limits the experimental conditions to a single dose of AgNPs (1.0 mg L-1; LC50). To partially overcome the complexity of metabolomic data analysis and to get insights into the molecular pathways from identified metabolite annotations, integrated metabolomic approaches were used. The respective chemical shifts resulting from 1H-NMR analysis have been submitted to the public database search to gain insights from the metabolite annotations and identification as described previously (Puig-Castellví et al., 2016). Based on the chemical shift data and coupling patterns, 55 metabolites were identified from S. cerevisiae whole-cell extracts (Figure 2). These annotated metabolites were classified into six different classes based on their chemical nature and metabolic functions and fold changes (control vs. AgNPs); their HMDB and KEGG IDs and cellular functions are summarized in Table 1. Deregulation in the metabolites were plotted in the form of fold changes (control vs. AgNP) as shown in Figure 3. The molecular pathway analysis conceded the fact that AgNPs exert deregulation of a wide range of key metabolites involved in amino acid metabolism, glycolysis and tricarboxylic acid (TCA) cycle (citric acid cycle), organic acids, nucleotide metabolism, urea cycle, and lipids metabolism. Results showed that AgNPs have a significant impact on the metabolic profiling of yeast cells as metabolites as shown by a significant deregulation pattern (p < 0.05). The variations in their regulation were shown in form of heatmaps (Figure 4). To visualize the differences in control and AgNP-treated S. cerevisiae cells, 55 identified metabolites were studied by PCA using MetaboAnalyst software known as a supervised clustering method to maximize the separation between groups. It is evident from the score plots that the cells divulged to AgNPs and detached to the first principal axis (PC1) and the separations between PC1 and PC2 revealed that AgNP exposure considerably altered the metabolic profiles of yeast cells (Figure 5). To further validate these results, some of the genes associated with these metabolic pathways have been selected. The functions of these genes are summarized in Table S1. Transcript-level analyses by qRT-PCR revealed that the genes of glycolysis and TCA cycle and lipid metabolism were significantly down-regulated; however, enzymes linked to oxidative stress mitigation showed the similar expression pattern in both control and treated cells (Figure 6).
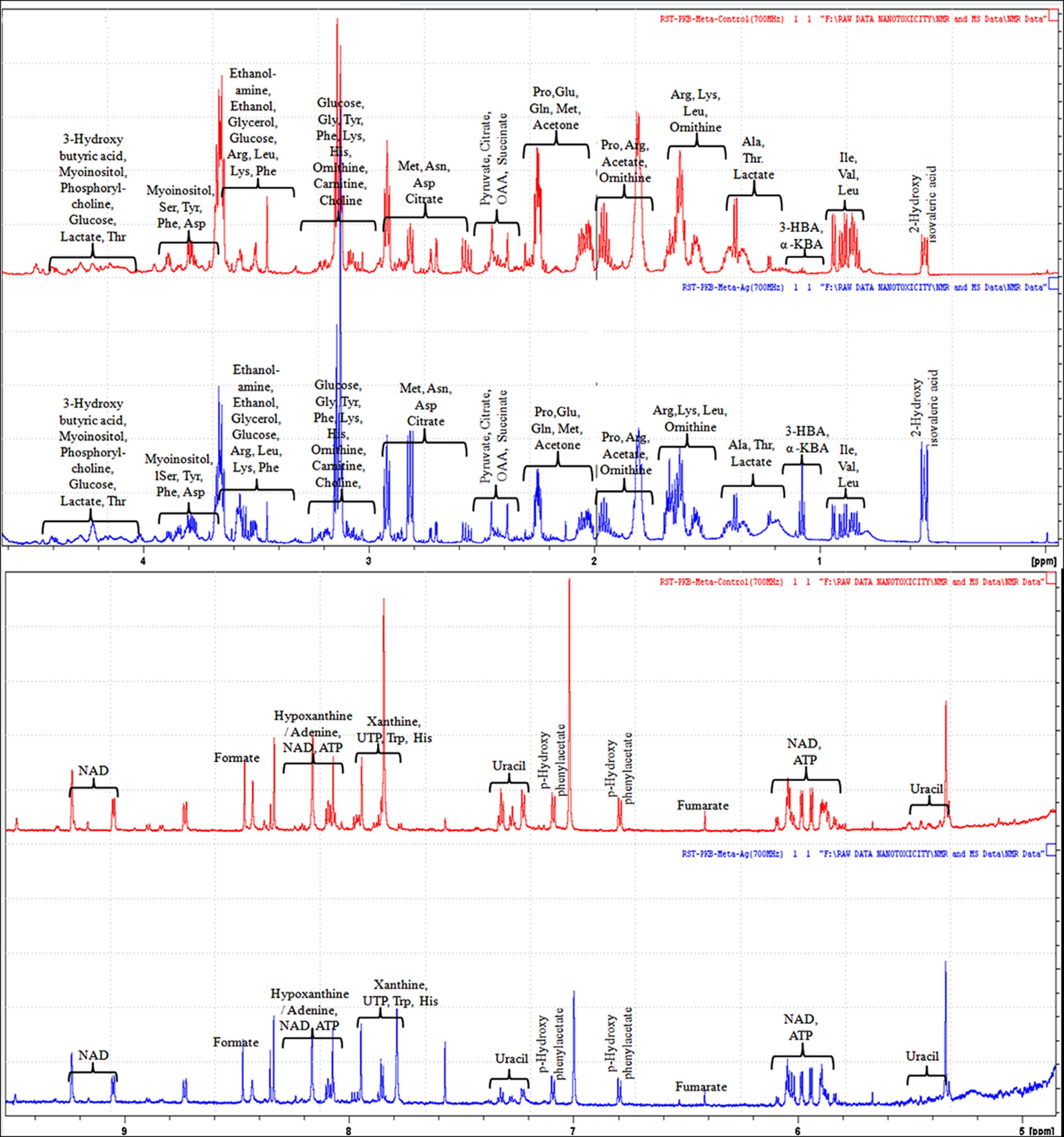
Figure 2 1H-NMR (700 MHz) spectra showing different types of metabolites, annotated based on their chemical shift (ppm) patterns. Spectra are obtained from whole-cell extracts of yeast S. cerevisiae cells incubated for 3 h in the absence of AgNPs (controls; red spectra) and exposed to AgNPs (1 mg L-1; blue spectra).
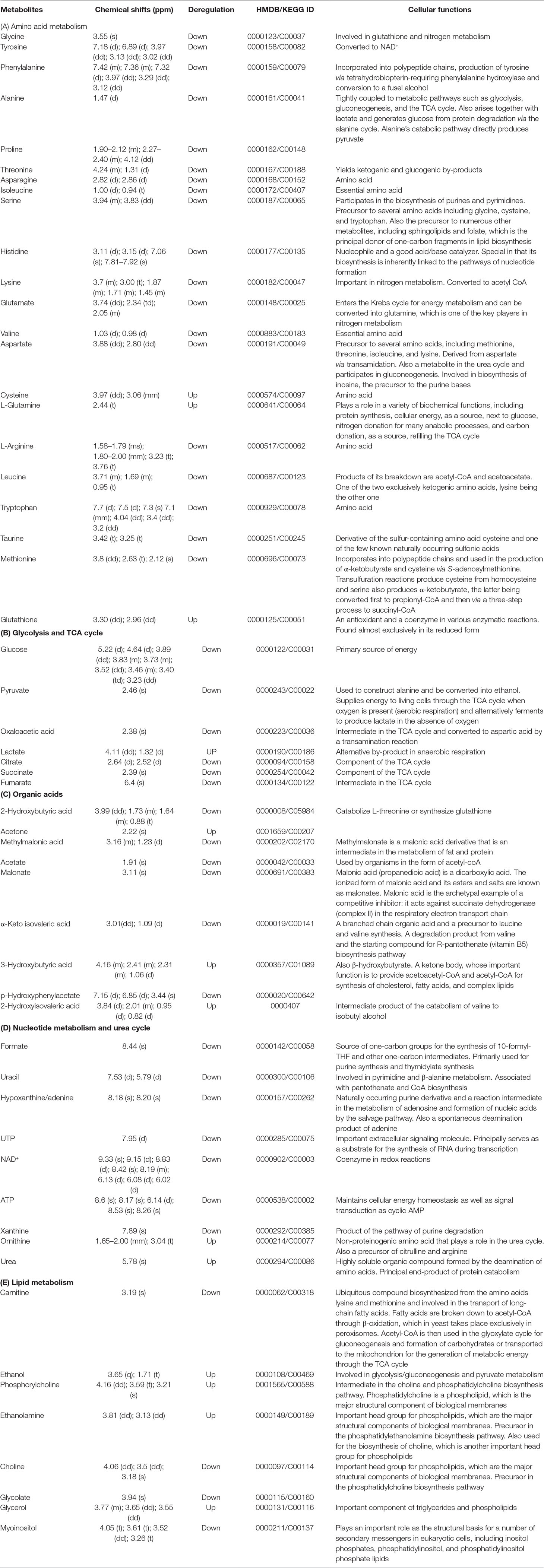
Table 1 1H-NMR spectroscopy (chemical shift) data and the regulation of the assigned metabolites with their respective HMDB/KEGG IDs and cellular functions.
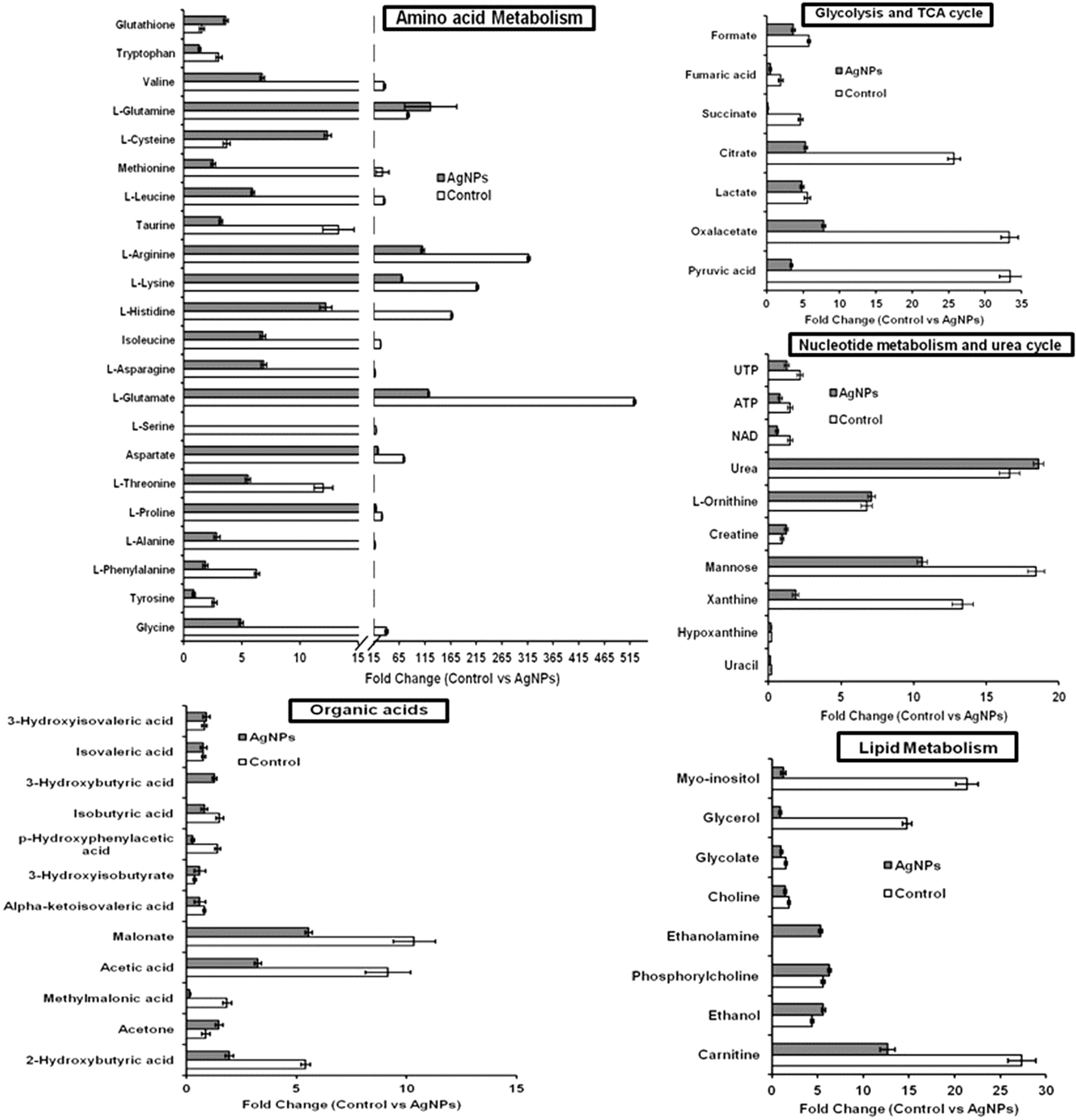
Figure 3 Deregulation in the metabolites involved in key metabolic pathways upon AgNP (1 mg L-1) exposure to S. cerevisiae BY4741 for 3 h. Relative changes in metabolite levels are plotted as fold change (control vs. AgNPs). Data are mean ± SE of three independent biological replicates. Values are normalized with 0.1 mM DSS-d6 (internal standard).
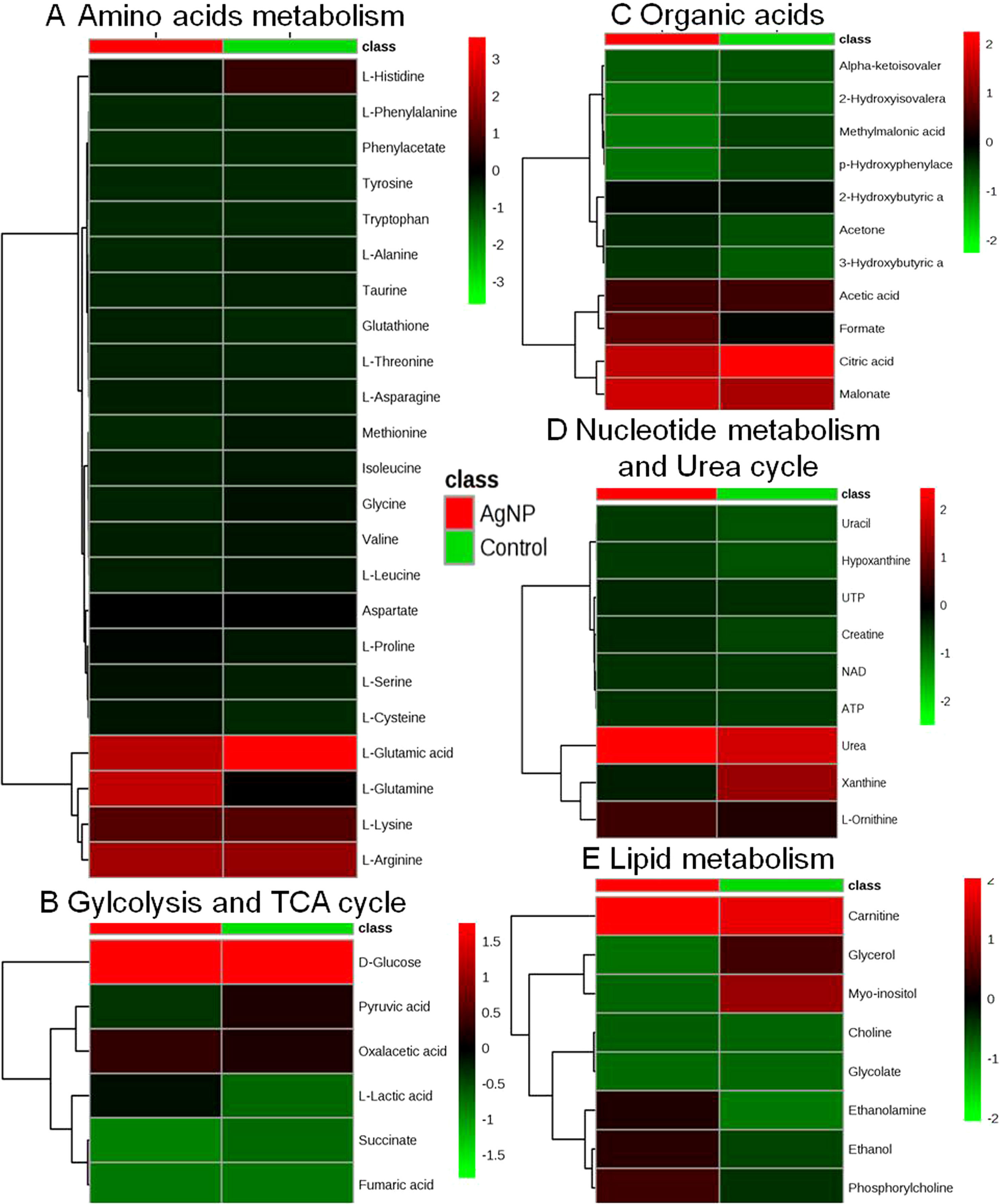
Figure 4 Heatmaps showing alteration in the metabolites of control vs. AgNP-treated yeast cells. The metabolites based on their chemical nature and metabolic functions are classified into (A) amino acid metabolism, (B) glycolysis and TCA cycle, (C) organic acids, (D) nucleotide metabolism and urea cycle, (E) and lipid metabolism.
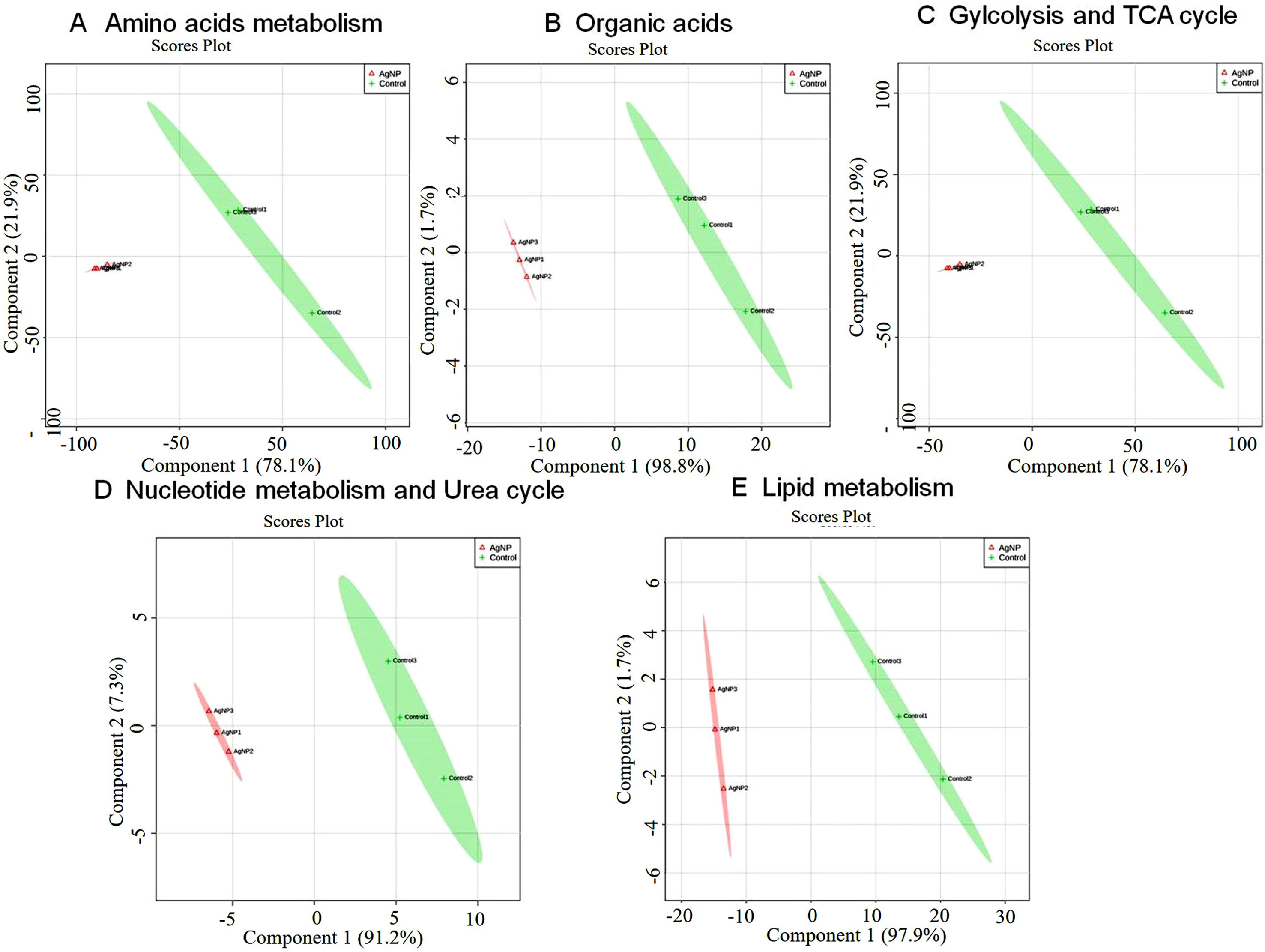
Figure 5 Principal component scores plot (PC1 vs. PC2) of partial least-squares discriminant analysis showing alteration in the different classes of metabolites in control vs. AgNP-treated yeast cells. The metabolites based on their chemical nature and metabolic functions are classified into (A) amino acid metabolism, (B) glycolysis and TCA cycle, (C) organic acids, (D) nucleotide metabolism, and (E) urea cycle and lipid metabolism.
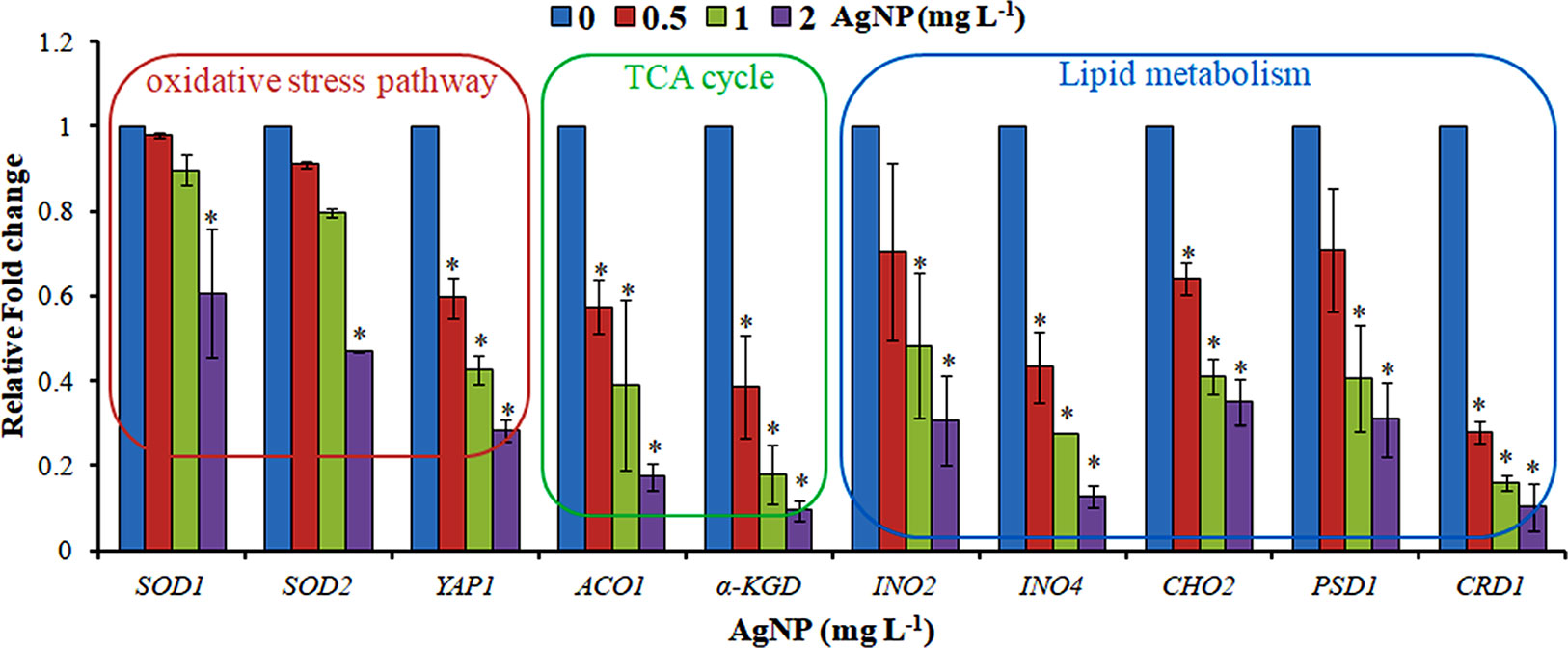
Figure 6 qRT-PCR showing the relative fold change in the expression profile of the genes selected from the oxidative stress pathway, TCA cycle, and lipid metabolism. Data are mean ± SE of three independent experiments. *p ≤ 0.05.
Do AgNPs Cause Alteration in Histone Epigenetic Marks?
To investigate the role of histone modification in cellular responses upon AgNP treatments, histone PTMs was analyzed by immunoblotting. A significant down-regulation in histone methylation (H3K4Me2 and H3K36Me3) was observed, whereas selected acetylation (H3K56Ac, H4K8Ac, and H4K16Ac) was up-regulated upon AgNP exposure (Figure 7).
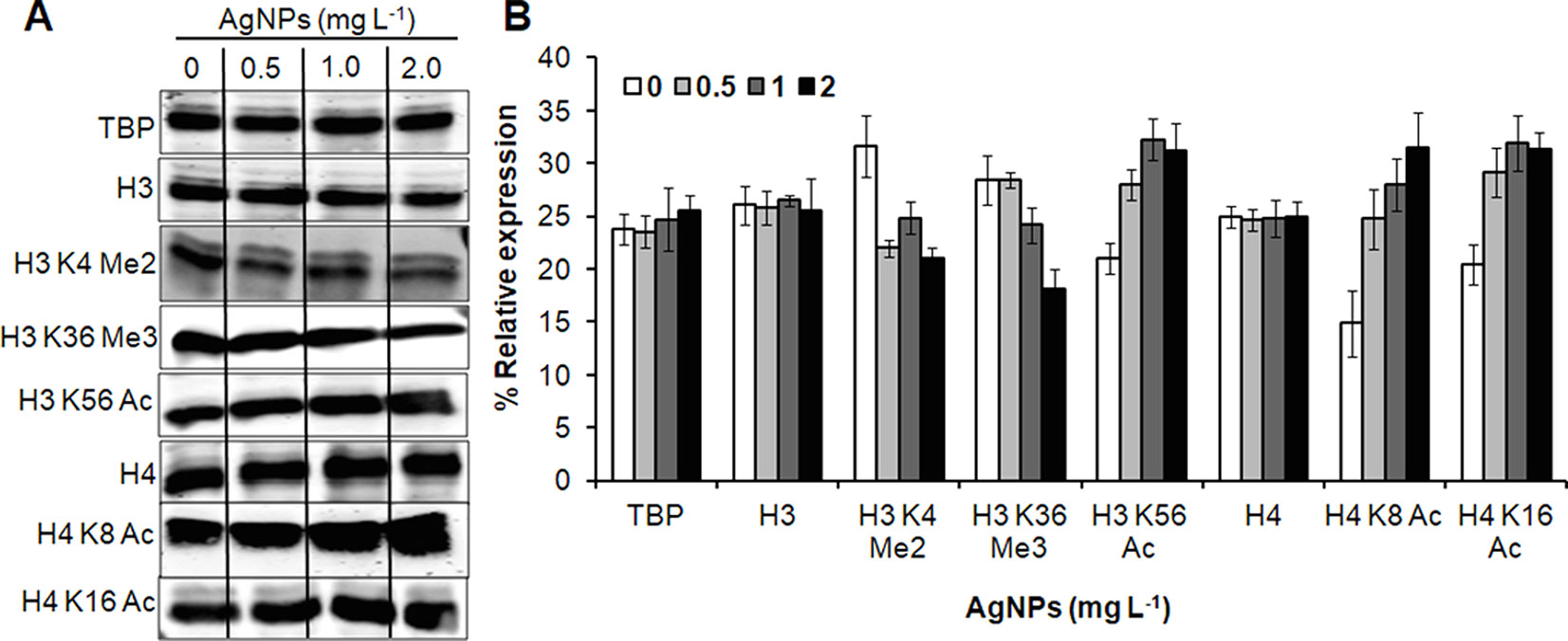
Figure 7 AgNPs cause alteration in histone PTMs. (A) Whole-cell protein extracts of untreated (0) and AgNP-treated yeast cells for 3 h at 30°C were analyzed for histone methylation (Me) and acetylation (Ac) marks by immunoblotting using the indicated antibodies. Anti-tata binding protein (Tbp), anti-histone H3, and anti histone H4 antibodies were used as loading control. (B) Densitometric analysis was performed by measuring the density of bands using the Alpha-Imager© software and the percent relative expression (control vs. AgNPs) of the indicated histone modification was quantified. Data are mean ± SEM of three independent experiments.
Discussion
A number of studies were reported previously to describe the green/biological methods of AgNP synthesis using a variety of plant and microbial cell extracts, including their toxic effects on different microbes, tumor cell lines, and mice models (McShan et al., 2014; Singh et al., 2014; Vazquez-Muñoz et al., 2017; Siddiqi et al., 2018). Here, we devised a new green (biological) method using gooseberry extract. Gooseberry was employed due to its very high amounts of antioxidants and polyphenolic compounds, thus acting as a strong reducing agent for AgNO3 and eventually providing stability to the particles (Rosarin et al., 2013; El-Sheekh and El-Kassas, 2014). It can also be seen that the absorption peaks were sharp, indicating the narrow size distribution of the nanoparticles. The recorded XRD pattern of synthesized AgNPs exhibits peaks at angles (2θ) 37.9°, 44.1°, 64.3°, and 77.3° corresponding to (111), (200), (220), and (311) reflection planes of silver lattice, respectively. In UV-vis spectra, it is quite obvious that there is a blue shift in absorption maxima, when gooseberry extract is reduced from 1 to 0.5 in ratio. The enhanced intensity and observed blue shift in the absorption peak suggest the decrease in the particle size of AgNPs. There are no extra peaks related to the by-products, which indicate the effectiveness of our synthesis method to fabricate the desired nanoparticles. TEM and SAED patterns further confirm the formation of AgNPs with supporting data of particle size distribution and dynamic light scattering (DLS). Our observations clearly suggest that the particles have no specific shapes, and average particle size ranges from approximately 50 to 70 nm.
After the successful synthesis and characterization of AgNPs, we investigated the effects of these AgNPs on the yeast S. cerevisiae to elucidate the mechanism(s) of toxicity in eukaryotes. Instead of rich growing media, sterilized DI water was used for AgNP treatment toward yeast cells. It was used to avoid the aggregation and accumulation of AgNPs as discussed previously (Babele et al., 2018; Babele, 2019). Preliminary results showed the inhibitory effect of AgNPs on the growth and survival of yeast cells as evidenced by CFU and spot assay in a dose-dependent manner. Furthermore, it was investigated whether the inhibition of cell growth was associated with cell membrane damage. PI and DCFDA staining revealed that AgNPs cause a remarkable increase in PI-positive cells and ROS accumulation, which indicate damage to the cell membrane and consequently cell death. Our results are in accordance to earlier reports, establishing that AgNPs cause several cellular responses, including ROS generation and cell death (McShan et al., 2014; Márquez et al., 2018).
Microorganisms that have evolved adaptive/stress responses were studied by adjusting metabolism through the modulation of gene expression, protein/enzyme level activity, and cellular concentration of metabolites, allowing them to survive under stressful and in ever-fluctuating environmental conditions (Pereira et al., 2018). Deregulation in the complete cellular metabolome and investigation of transcriptome and histone PTMs in response to AgNPs are the major objectives of this study. A significant deregulation in the key metabolites of the cells involved in various metabolic pathways has been observed. It has also been observed that almost all amino acids annotated in this study are down-regulated, except glutathione, glutamine, and cysteine. These observations indicate that AgNP exposure severely affects amino acid metabolism in yeast cells. The levels of reduced glutathione are increased, which is considered as a natural defense system of cells to cope with oxidative stress, as it plays an essential role in maintaining the intracellular redox environment for metal stress-generated inflammation and oxidative stress (Hatem et al., 2014; Ilyas and Rehman, 2015). The observed diminished level of glycine, a well-known precursor amino acid of glutathione biosynthesis, might be due to its utilization to replenish glutathione consumed by the cells (Ribas et al., 2014).
It is well established that the mitochondria (powerhouse of the cell) are fundamental bioenergetic organelles and fulfill the energy requirements of the cell. They participate in central carbon metabolism (CCM) and various other metabolic reactions of the cell, including stress mitigation, by providing precursors and intermediates for many other primary and secondary metabolites, including amino acids (Edirisinghe et al., 2016). The CCM and other metabolic pathways can be altered effectively in S. cerevisiae under stressed conditions (Wiebe et al., 2007; Guo et al., 2016). Metabolites identified in the present work show that metabolites linked to glycolysis, tricarboxylic acid (TCA) cycle, and pentose phosphate pathway, such as glucose, pyruvic acid, succinate, fumaric acid, citrate, and oxaloacetic acid, are drastically down-regulated, indicating that AgNPs affect the CCM. Earlier, an NMR based metabolomic study suggested that Ag+ and AgNPs significantly down-regulate the metabolites (citrate, succinate, and α-ketoglutarate) of glycolysis and Kreb’s (TCA) cycle in human skin keratinocytes (HaCaT cells; Carrola et al., 2016). Also, in an in vivo metabolic study, it appeared that the TCA cycle was down-regulated in the lungs of AgNP-injected mice (Jarak et al., 2017). Succinic acid, an indispensable metabolite in the TCA cycle, participates in the GABA pathway involved in the mitigation of ROS-mediated oxidative stress (Cao et al., 2013). The up-regulation of lactate, but down-regulation in the levels of glucose, manifests as a change into lactate or acetate to provide bounteous energy for survival. These results might encompass the imbalance in the aerobic metabolism and the TCA cycle. Moreover, acetate is also regarded as an expedient source of energy for stressed cells (Jiang et al., 2013). These observations are in consistent with the earlier reports in which nanoparticles cause oxidative stress and defects in mitochondrial and endoplasmic reticulum (ER) enzymes (Teodoro et al., 2011; Jarak et al., 2017). In aerobic metabolism, the formation of ROS is a natural by-product, but in excess they can chemically modify proteins and lipids by lipid peroxidation and oxidative stress, thus leading to a damage to vital organelles such as mitochondria, ER, and lysosomes (McShan et al., 2014; Márquez et al., 2018).
A drastic down-regulation in a number of organic acids such as malonate, α-keto isovaleric acid, p-hydroxyphenyl acetate, 2-hydroxybutyric acid, and methyl-malonic acid was also observed, whereas the concentrations of 2-hydroxyisovaleric acid, 3-hydroxybutyric acid, and acetone were up-regulated. These observations suggest that yeast cells modulate their metabolism to improve their viability under AgNP stress, as 3-hydroxybutyrate, one of the end products of fatty acid β-oxidation, functions as an alternative energy source (Newman and Verdin, 2014). Similar to our finding, there are several other metal oxide toxicity studies that have reported the increase in 3-hydroxybutyrate in the serum or urine, whereas the glucose level is found to be limited or the TCA cycle is impaired (Lei et al., 2008; Bu et al., 2010; Yan et al., 2012). In addition, it was proposed that the increased level of 3-hydroxybutyrate uses as an auxiliary energy source to replenish the energetic demands during tissue damage (Fabisiak et al., 2011; Lee et al., 2016).
AgNP exposure also affects the levels of many metabolites associated with nucleotide metabolism and urea cycle. These metabolites known to be involved in energy generation/transfer mechanisms, particularly uracil, UTP, hypoxanthine, NAD, and ATP, are down-regulated after treatment compared to control cells. Nucleotides are considered to play a vital role in diverse cellular processes. Moreover, nucleotides are assumed to be assimilated in various cofactors (e.g., NAD and coenzyme A) and also serve as precursors (e.g., UDP-glucose and GDP-mannose). A down-regulation in the level of formate was also observed, it is the only non-tetrahydrofolate (THF)-linked intermediate in one-carbon metabolism and produced from various cellular metabolic reactions. Formate is regarded as an important substitute to single-carbon source for N5, N10-methylene-THF generation; these are primarily used for purine and thymidylate biosynthesis and required for epigenetic methylation reactions of methyl groups for synthetic, regulatory mechanism (Brosnan and Brosnan, 2016). Recently, it was observed that this acid also has potential to combat against oxidative stress (Thomas et al., 2016).
Increment in the metabolites involved in lipid metabolism, i.e., glycerol, choline, ethanol, myo-inositol, and carnitine, are found in their levels. Glycerol is a well-known protective agent synthesized by yeast cells for environmental stress and maintains redox homeostasis (Paget et al., 2014). The increment in glycerol biosynthesis rate primarily signifies the alteration in stress defense of cells for AgNP toxicity. Choline and phosphorylcholine are also important metabolites that are found up-regulated. Glycerophosphocholine is also an important ingredient in eukaryotic cellular membranes. These are primarily convoluted in the membrane choline phospholipid metabolism pathway, in which phosphatidylcholine may be converted to glycerophosphocholine and further reconverted to phosphatidylcholine (Hu et al., 2008; Van Meer et al., 2008). As a result of nanotoxicity, cells consume glycerophosphocholine to generate phosphatidylcholine, which helps in maintaining the integrity of the cell membrane. This provides a path to propose a plausible mechanism of membrane component maintenance upon AgNP exposure. The results/findings of the present study are analogous and parallel to the results of a research conducted on human hepatoma (HepG2) cells exposed to citrate-coated AgNPs (Carrola et al., 2018).
Histone PTMs regulate the transcription and other DNA-dependent functions of eukaryotic cells and these PTMs are driven by specific histone-modifying enzymes. Cellular metabolites serve as co-substrates or enzyme inhibitors essentially required for proper enzyme activities. Therefore, cell metabolism can be influenced through histone modification dealing with local concentrations of metabolites (Fan et al., 2015). The epigenetic changes in response to cellular metabolism are vital for stress adaption and cell survival physiologically. There are numerous environmental factors that alter gene expression through covalent histone modifications and fundamental genome functions (Kouzarides, 2007; Fang et al., 2014). These modifications have not been much studied in response to biologically synthesized AgNPs. Some reports described that the sublethal dose of AgNP induces a drastic fall in hemoglobin levels in mouse erythroleukemia cells, imprinting diminished levels of H3K4me3 and H3K79me1 methylation. These results have been attributed to the direct binding of AgNPs to histone (H3/H4) proteins (Qian et al., 2015). A study on human cells reported the phosphorylation of histone H3 at serine 10 (p-H3S10) and its usefulness as a marker to scale the toxicity induced by AgNPs (Zhao and Ibuki, 2015). Here, we found significant changes in the methylation and acetylation marks of histone H3 and H4 proteins upon AgNP exposure, suggesting that either AgNPs induced metabolic alteration or their direct binding to nucleosomes may be involved in the histone modification.
Conclusion
The present study describes the potential of 1H-NMR-based metabolomics in nanotoxicology. This approach is successfully used for the rapid and untargeted screening of biochemical modulations in S. cerevisiae upon AgNP exposure. The combined metabolomic and epigenetic alterations demonstrate that a very low concentration of biologically synthesized AgNPs has potential to induce nanotoxicity by modulating the metabolic machinery of yeast cells.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
PB conceived the whole idea, designed and performed the metabolic toxicity experiments, ASi performed and wrote the nanoparticle synthesis part. ASr helped in writing and correction of the MS.
Funding
This work was supported by a project grant sanctioned to PB in the form of a national postdoctoral fellowship by SERB-DST, New Delhi, India (File No. PDF/000200).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
ASi is very much thankful to UGC, New Delhi, India, for providing financial support in the form of a Dr. D.S. Kothari postdoctoral fellowship. ASr acknowledges the UGC, New Delhi, India and TD College for providing opportunity in the form of a Raman fellowship. The authors thank the Advanced NMR and other facilities at IISER Bhopal.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01016/full#supplementary-material
References
Airoldi, C., Tripodi, F., Guzzi, C., Nicastro, R., Coccetti, P. (2015). NMR analysis of budding yeast metabolomics: a rapid method for sample preparation. Mol. Biosyst. 11 (2), 379–383. doi: 10.1039/C4MB00452C
Babele, P. K. (2019). Zinc oxide nanoparticles impose metabolic toxicity by de-regulating proteome and metabolome in Saccharomyces cerevisiae. Toxicol. Rep. 6, 64–73. doi: 10.1016/j.toxrep.2018.12.001
Babele, P. K., Thakre, P. K., Kumawat, R., Tomar, R. S. (2018). Zinc oxide nanoparticles induce toxicity by affecting cell wall integrity pathway, mitochondrial function and lipid homeostasis in Saccharomyces cerevisiae. Chemosphere 213, 65–75. doi: 10.1016/j.chemosphere.2018.09.028
Bouhifd, M., Hartung, T., Hogberg, H. T., Kleensang, A., Zhao, L. (2013). toxicometabolomics. J. Appl. Toxicol. 33 (12), 1365–1383. doi: 10.1002/jat.2874
Bu, Q., Yan, G., Deng, P., Peng, F., Lin, H., Xu, Y., et al. (2010). NMR-based metabonomic study of the sub-acute toxicity of titanium dioxide nanoparticles in rats after oral administration. Nanotechnology 21 (12), 125105. doi: 10.1088/0957-4484/21/12/125105
Brosnan, M. E., Brosnan, J. T. (2016). Formate: the neglected member of one-carbon metabolism. Annu. Rev. Nutr. 36, 369–388. doi: 10.1146/annurev-nutr-071715-050738
Cao, J., Barbosa, J. M., Singh, N. K., Locy, R. D. (2013). GABA shunt mediates thermotolerance in Saccharomyces cerevisiae by reducing reactive oxygen production. Yeast 30 (4), 129–144. doi: 10.1002/yea.2948
Carrola, J., Bastos, V., Jarak, I., Oliveira-Silva, R., Malheiro, E., Daniel-da-Silva, A. L., et al. (2016). Metabolomics of silver nanoparticles toxicity in HaCaT cells: structure-activity relationships and role of ionic silver and oxidative stress. Nanotoxicology 10 (8), 1105–1117. doi: 10.1080/17435390.2016.1177744
Carrola, J., Pinto, R. J., Nasirpour, M., Freire, C. S., Gil, A. M., Santos, C., et al. (2018). NMR metabolomics reveals metabolism-mediated protective effects in liver (HepG2) cells exposed to subtoxic levels of silver nanoparticles. J. Proteome Res. 17 (4), 1636–1646. doi: 10.1021/acs.jproteome.7b00905
Dubey, P., Matai, I., Kumar, S. U., Sachdev, A., Bhushan, B., Gopinath, P. (2015). Perturbation of cellular mechanistic system by silver nanoparticle toxicity: cytotoxic, genotoxic and epigenetic potentials. Adv. Colloid. Interface. Sci. 221, 4–21. doi: 10.1016/j.cis.2015.02.007
Edirisinghe, J. N., Weisenhorn, P., Conrad, N., Xia, F., Overbeek, R., Stevens, R. L., et al. (2016). Modeling central metabolism and energy biosynthesis across microbial life. BMC Genomics 17 (1), 568. doi: 10.1186/s12864-016-2887-8
El-Sheekh, M. M., El-Kassas, H. Y. (2014). Application of biosynthesized silver nanoparticles against a cancer promoter cyanobacterium, Microcystis aeruginosa. Asian Pac. J. Cancer Prev. 15 (16), 6773–6779. doi: 10.7314/APJCP.2014.15.16.6773
Fabisiak, J. P., Medvedovic, M., Alexander, D. C., McDunn, J. E., Concel, V. J., Bein, K., et al. (2011). Integrative metabolome and transcriptome profiling reveals discordant energetic stress between mouse strains with differential sensitivity to acrolein induced acute lung injury. Mol. Nutr. Food Res. 55 (9), 1423–1434. doi: 10.1002/mnfr.201100291
Fan, J., Krautkramer, K. A., Feldman, J. L., Denu, J. M. (2015). Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 10 (1), 95–108. doi: 10.1021/cb500846u
Fang, L., Wuptra, K., Chen, D., Li, H., Huang, S.-K., Jin, C., et al. (2014). Environmental-stress-induced chromatin regulation and its heritability. J. Carcinog. Mutagen. 5 (1). doi: 10.4172/2157-2518.1000156
Guo, W., Chen, Y., Wei, N., Feng, X. (2016). Investigate the metabolic reprogramming of Saccharomyces cerevisiae for enhanced resistance to mixed fermentation inhibitors via 13C metabolic flux analysis. PLoS One 11 (8), e0161448. doi: 10.1371/journal.pone.0161448
Hatem, E., Berthonaud, V., Dardalhon, M., Lagniel, G., Baudouin-Cornu, P., Huang, M.-E., et al. (2014). Glutathione is essential to preserve nuclear function and cell survival under oxidative stress. Free Radic. Biol. Med. 67, 103–114. doi: 10.1016/j.freeradbiomed.2013.10.807
Hu, J. Z., Rommereim, D. N., Minard, K. R., Woodstock, A., Harrer, B. J., Wind, R. A., et al. (2008). Metabolomics in lung inflammation: a high-resolution 1H NMR study of mice exposed to silica dust. Toxicol. Mech. Methods 18 (5), 385–398. doi: 10.1080/15376510701611032
Ilyas, S., Rehman, A. (2015). Oxidative stress, glutathione level and antioxidant response to heavy metals in multi-resistant pathogen, Candida tropicalis. Environ. Monit. Assess. 187 (1), 4115. doi: 10.1007/s10661-014-4115-9
Jarak, I., Carrola, J., Barros, A. S., Gil, A. M., Pereira, M. L., Corvo, M. L., et al. (2017). From the cover: metabolism modulation in different organs by silver nanoparticles: an NMR metabolomics study of a mouse model. Toxicol. Sci. 159 (2), 422–435. doi: 10.1093/toxsci/kfx142
Jenuwein, T., Allis, C. D. (2001). Translating the histone code. Science 293 (5532), 1074–1080. doi: 10.1126/science.1063127
Jiang, L., Gulanski, B. I., De Feyter, H. M., Weinzimer, S. A., Pittman, B., Guidone, E., et al. (2013). Increased brain uptake and oxidation of acetate in heavy drinkers. J. Clin. Investig. 123 (4), 1605–1614. doi: 10.1172/JCI65153
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 (4), 693–705. doi: 10.1016/j.cell.2007.02.005
Lee, S.-H., Wang, T.-Y., Hong, J.-H., Cheng, T.-J., Lin, C.-Y. (2016). NMR-based metabolomics to determine acute inhalation effects of nano- and fine-sized ZnO particles in the rat lung. Nanotoxicology 10 (7), 924–934. doi: 10.3109/17435390.2016.1144825
Lei, R., Wu, C., Yang, B., Ma, H., Shi, C., Wang, Q., et al. (2008). Integrated metabolomic analysis of the nano-sized copper particle-induced hepatotoxicity and nephrotoxicity in rats: a rapid in vivo screening method for nanotoxicity. Toxicol. Appl. Pharmacol. 232 (2), 292–301. doi: 10.1016/j.taap.2008.06.026
Lv, M., Huang, W., Chen, Z., Jiang, H., Chen, J., Tian, Y., et al. (2015). Metabolomics techniques for nanotoxicity investigations. Bioanalysis 7 (12), 1527–1544. doi: 10.4155/bio.15.83
Marassi, V., Di Cristo, L., Smith, S. G., Ortelli, S., Blosi, M., Costa, A. L., et al. (2018). Silver nanoparticles as a medical device in healthcare settings: a five-step approach for candidate screening of coating agents. Royal Soc. Open Sci. 5 (1), 171113. doi: 10.1098/rsos.171113
Márquez, I. G., Ghiyasvand, M., Massarsky, A., Babu, M., Samanfar, B., Omidi, K., et al. (2018). Zinc oxide and silver nanoparticles toxicity in the baker’s yeast, Saccharomyces cerevisiae. PloS One 13 (3), e0193111. doi: 10.1371/journal.pone.0193111
McShan, D., Ray, P. C., Yu, H. (2014). Molecular toxicity mechanism of nanosilver. J. Food Drug Anal. 22 (1), 116–127. doi: 10.1016/j.jfda.2014.01.010
Musee, N. (2011). Nanowastes and the environment: potential new waste management paradigm. Environ. Int. 37 (1), 112–128. doi: 10.1016/j.envint.2010.08.005
Mytych, J., Zebrowski, J., Lewinska, A., Wnuk, M. (2017). Prolonged effects of silver nanoparticles on p53/p21 pathway-mediated proliferation, DNA damage response, and methylation parameters in HT22 hippocampal neuronal cells. Mol. Neurobiol. 54 (2), 1285–1300. doi: 10.1007/s12035-016-9688-6
Newman, J. C., Verdin, E. (2014). β-hydroxybutyrate: much more than a metabolite. Diabetes Res. Clin. Pract. 106 (2), 173–181. doi: 10.1016/j.diabres.2014.08.009
Oberdörster, G., Oberdörster, E., Oberdörster, J. (2005). Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 113 (7), 823. doi: 10.1289/ehp.7339
Paget, C. M., Schwartz, J. M., Delneri, D. (2014). Environmental systems biology of cold tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol. Ecol. 23 (21), 5241–5257. doi: 10.1111/mec.12930
Pereira, T., Vilaprinyo, E., Belli, G., Herrero, E., Salvado, B., Sorribas, A., et al. (2018). Quantitative operating principles of yeast metabolism during adaptation to heat stress. Cell Rep. 22 (9), 2421–2430. doi: 10.1016/j.celrep.2018.02.020
Puig-Castellví, F., Alfonso, I., Piña, B., Tauler, R. (2016). 1H NMR metabolomic study of auxotrophic starvation in yeast using multivariate curve resolution-alternating least squares for pathway analysis. Sci. Rep. 6, 30982. doi: 10.1038/srep30982
Qian, Y., Zhang, J., Hu, Q., Xu, M., Chen, Y., Hu, G., et al. (2015). Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials 70, 12–22. doi: 10.1016/j.biomaterials.2015.08.015
Ribas, V., García-Ruiz, C., Fernández-Checa, J. C. (2014). Glutathione and mitochondria. Front. Pharmacol. 5, 151. doi: 10.3389/fphar.2014.00151
Rosarin, F. S., Arulmozhi, V., Nagarajan, S., Mirunalini, S. (2013). Antiproliferative effect of silver nanoparticles synthesized using amla on Hep2 cell line. Asian Pac. J. Trop. Med. 6 (1), 1–10. doi: 10.1016/S1995-7645(12)60193-X
Schnackenberg, L. K., Sun, J., Beger, R. D. (2012). Metabolomics techniques in nanotoxicology studies. Nanotoxicity Methods Protoc. 926, 141–156. doi: 10.1007/978-1-62703-002-1_10
Siddiqi, K. S., Husen, A., Rao, R. A. (2018). A review on biosynthesis of silver nanoparticles and their biocidal properties. J. Nanobiotechnology 16 (1), 14. doi: 10.1186/s12951-018-0334-5
Sierra, M., Valdés, A., Fernández, A., Torrecillas, R., Fraga, M. (2016). The effect of exposure to nanoparticles and nanomaterials on the mammalian epigenome. Int. J. Nanomed. 11, 6297. doi: 10.2147/IJN.S120104
Singh, G., Babele, P. K., Shahi, S. K., Sinha, R. P., Tyagi, M. B., Kumar, A. (2014). Green synthesis of silver nanoparticles using cell extracts of Anabaena doliolum and screening of its antibacterial and antitumor activity. J. Microbiol. Biotechnol. 24 (10), 1354–1367. doi: 10.4014/jmb.1405.05003
Smolkova, B., Dusinska, M., Gabelova, A. (2017). Nanomedicine and epigenome. Possible health risks. Food Chem. Toxicol. 109, 780–796. doi: 10.1016/j.fct.2017.07.020
Smolkova, B., El Yamani, N., Collins, A. R., Gutleb, A. C., Dusinska, M. (2015). Nanoparticles in food. Epigenetic changes induced by nanomaterials and possible impact on health. Food Chem. Toxicol. 77, 64–73. doi: 10.1016/j.fct.2014.12.015
Thomas, S. C., Alhasawi, A., Auger, C., Omri, A., Appanna, V. D. (2016). The role of formate in combating oxidative stress. Antonie Van Leeuwenhoek 109 (2), 263–271. doi: 10.1007/s10482-015-0629-6
Teodoro, J. S., Simões, A. M., Duarte, F. V., Rolo, A. P., Murdoch, R. C., Hussain, S. M., et al. (2011). Assessment of the toxicity of silver nanoparticles in vitro: a mitochondrial perspective. Toxicol. Vitro 25 (3), 664–670. doi: 10.1016/j.tiv.2011.01.004
Van Meer, G., Voelker, D. R., Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9 (2), 112. doi: 10.1038/nrm2330
Vazquez-Muñoz, R., Borrego, B., Juárez-Moreno, K., García-García, M., Morales, J. D. M., Bogdanchikova, N., et al. (2017). Toxicity of silver nanoparticles in biological systems: does the complexity of biological systems matter? Toxicol. Lett. 276, 11–20. doi: 10.1016/j.toxlet.2017.05.007
Wiebe, M. G., Rintala, E., Tamminen, A., Simolin, H., Salusjärvi, L., Toivari, M., et al. (2007). Central carbon metabolism of Saccharomyces cerevisiae in anaerobic, oxygen-limited and fully aerobic steady-state conditions and following a shift to anaerobic conditions. FEMS Yeast Res. 8 (1), 140–154. doi: 10.1111/j.1567-1364.2007.00234.x
Xia, J., Sinelnikov, I. V., Han, B., Wishart, D. S. (2015). MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 43 (W1), W251–W257. doi: 10.1093/nar/gkv380
Yan, G., Huang, Y., Bu, Q., Lv, L., Deng, P., Zhou, J., et al. (2012). Zinc oxide nanoparticles cause nephrotoxicity and kidney metabolism alterations in rats. J. Environ. Sci. Health. A 47 (4), 577–588. doi: 10.1080/10934529.2012.650576
Keywords: silver nanoparticles (AgNPs), nanotoxicity, 1H-NMR, metabolomics, histones, Saccharomyces cerevisiae
Citation: Babele PK, Singh AK and Srivastava A (2019) Bio-Inspired Silver Nanoparticles Impose Metabolic and Epigenetic Toxicity to Saccharomyces cerevisiae. Front. Pharmacol. 10:1016. doi: 10.3389/fphar.2019.01016
Received: 26 April 2019; Accepted: 09 August 2019;
Published: 12 September 2019.
Edited by:
Shahper Nazeer Khan, Aligarh Muslim University, IndiaReviewed by:
Narasimham L. Parinandi, The Ohio State University, United StatesNazareno Paolocci, Johns Hopkins University, United States
Sadaf Hasan, New York University, United States
Copyright © 2019 Babele, Singh and Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyoosh Kumar Babele, cGl5b29zaDIwMDZAeWFob28uY28uaW4=; Ashwani Kumar Singh, YXNod2FuaWt1bWFyc2luZ2gxNDNAZ21haWwuY29t
 Piyoosh Kumar Babele
Piyoosh Kumar Babele Ashwani Kumar Singh2*
Ashwani Kumar Singh2* Amit Srivastava
Amit Srivastava