- Department of Clinical Laboratory, University of Chinese Academy of Sciences Chongqing Renji Hospital, Fifth People’s Hospital of Chongqing, Chongqing, China
Background: The increasing prevalence of extensively drug-resistant Klebsiella pneumoniae (XDR-KP) poses a serious threat to clinical anti-infective treatment. This retrospective study assessed the molecular epidemiology of and risk factors for infections with XDR-KP to investigate the mechanism of drug resistance and the epidemiological characteristics.
Methods: A retrospective 1:2 case-control study was conducted at Chongqing Renji Affiliated Hospital of the Chinese Academy of Sciences University from January 2015 to December 2017. A total of 69 non-repetitive XDR-KP strains were collected. Patients infected with XDR-KP comprised the case group, and 138 matched patients with non-XDR-KP infection at the same site comprised the control group. The chi-square test and logistic regression were performed to evaluate the related risk factors. Molecular typing was performed by multilocus sequence typing (MLST). Potential resistance genes were detected by polymerase chain reaction (PCR) and sequencing. Predictors of 28-day mortality in patients with XDR-KP infection were also identified in our study.
Results: Only tigecycline and polymyxin B showed favorable in vitro drug sensitivity tests. These XDR-KP strains had a high prevalence rate (n = 66, 95.7%) of carbapenemase-related drug resistance genes. Among them, KPC-2 was the most frequently detected gene (n = 52, 75.4%). Particularly, all of the isolates harbored multiple drug resistance genes. Epidemiological analysis showed that fifty-eight XDR-KP isolates were resistant strains with the ST-11 genotype. Multivariate logistic regression analysis showed that ICU admission (OR: 3.28, 95% CI: 1.66–6.49, P < 0.001), tracheal cannula (OR: 3.16, 95% CI: 1.48–6.76, P = 0.003), and carbapenem exposure (OR: 3.16, 95% CI: 1.25–7.98, P = 0.015) were independent risk factors for XDR-KP infection. Solid tumors (OR: 7.22, 95% CI: 1.84–28.34, P = 0.005) and septic shock (OR: 9.46, 95% CI: 2.00–44.72, P = 0.005) were independent risk factors for 28-day mortality from XDR-KP infection.
Conclusion: This study showed that XDR-KP isolates were highly resistant and exhibited clonal transmission. ST11 was the predominant epidemic type of XDR-KP producing KPC-2 in Southwestern China. Physicians should be aware of these high-risk patients with notable predictive factors for XDR-KP infection. These findings may provide some recommendation for the diagnosis and treatment of patients infected with XDR-KP strains in Southwestern China.
Introduction
Klebsiella pneumoniae (KP) is one of the most common pathogens of nosocomial infections that often cause severe or fatal infections (Blin et al., 2017; Liu et al., 2019). Carbapenems are currently the most effective beta-lactam antibiotics for the treatment of such severe bacterial infections. With the overuse of carbapenems in clinical treatment over the past decade, the emergence of carbapenem-resistant K. pneumoniae (CRKP) has become the main cause of the failure of clinical anti-infective treatment (Arabaghian et al., 2019; Barnes et al., 2019; Meng et al., 2019). It is worth noting that extensively drug-resistant K. pneumoniae (XDR-KP), which has a broader spectrum of drug resistance than CRKP, should be paid more attention. Because XDR-KP is resistant to most of the antibiotics currently used in the clinic and is only sensitive to tigecycline and polymyxin B, it further reduces the types of drugs available in the clinic, which poses a serious challenge to clinical anti-infective treatment (Du et al., 2016; Liu et al., 2017; Moradigaravand et al., 2017).
High morbidity and mortality are closely related to infection with XDR-KP, with a mortality rate of approximately 40–50% (Hoxha et al., 2016; Zheng et al., 2017). Currently, colistin is one of the few effective antibiotics in the treatment of XDR-KP infection, and the therapeutic options for infections are extremely limited. However, with the emergence of mcr-1-positive Klebsiella pneumoniae, which may lead to resistance to colistin, the treatment of XDR-KP infection is increasingly difficult (Rojas et al., 2017; Petrosillo et al., 2019). In addition, because carbapenem genes and other resistance genes can be transmitted frequently through mobile DNA elements, the increasing incidence of XDR-KP infection reported worldwide reflects a serious threat to public health and has even caused fatal outcomes in vulnerable patients. Therefore, it is necessary to explore the molecular epidemiology of and risk factors for infections with XDR-KP (de Campos et al., 2018; Li et al., 2018; Liu et al., 2019).
At present, there have been no reports on the molecular epidemiology and risk factors for XDR-KP infection in Southwestern China. In this retrospective study, we attempted to analyze the clinical characteristics, molecular epidemiology and risk factors for XDR-KP infection collected from Chongqing Renji Affiliated Hospital of the Chinese Academy of Sciences University to provide a theoretical basis for the clinical diagnosis, treatment and control of the spread of XDR-KP infection. Moreover, predictors for mortality in patients with XDR-KP infection were also identified in our study.
Materials and Methods
Study Design and Setting
This retrospective study was performed from January 2015 to December 2017 in the Chongqing Renji Affiliated Hospital of the Chinese Academy of Sciences University, a tertiary university hospital with a 1,800-bed academic medical centre located in Chongqing, Southwest China. Patients with XDR-KP infection were only sensitive to tigecycline or polymyxin B and resistant to all other drugs in this study. To assess for risk factors for XDR-KP infection, a retrospective 1:2 case-control study was performed. The case-control study groups in our analysis were defined as follows: the case group comprised patients with XDR-KP infection during hospitalization, and patients with non-XDR-KP infection at the same site comprised the control group. Patients admitted for <48 h and those with duplicate isolates were excluded.
Identification and Drug Sensitivity of Bacteria
Bacterial cultures were processed in the clinical microbiology laboratory. All the XDR-KP isolates were identified using the VITEK 2 Compact system or the VITEK MS system (bioMérieux, Marcy l’Etoile, Lyon, France), and antimicrobial susceptibilities were determined in vitro using a VITEK 2 Compact AST-GN13 card (bioMérieux), which were used to test the antibiotic susceptibilities of all isolates to ampicillin (AMP), ampicillin/sulbactam (SAM), piperacillin (PIR), cefazolin (CFZ), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), cefoxitin (FOX), aztreonam (AZT), gentamicin (GM), tobramycin (TOB), amikacin (AK), ciprofloxacin (CIP), and levofloxacin (LEV). Five antibiotics, including imipenem (IMP), meropenem (MEM), ertapenem (ETP), tigecycline (TGC) and polymyxin B (PB), were tested by the broth microdilution method following the criteria of the Clinical and Laboratory Standards Institute (CLSI, 2016). The minimal inhibitory concentrations (MICs) breakpoint for tigecycline was defined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2017), while the others were interpreted according to CLSI protocols. K. pneumoniae ATCC700603 and Escherichia coli ATCC25922 were used as quality control strains for the antibiotic susceptibility tests.
Identification of Resistance Genes
Crude DNA extracts were prepared by boiling the bacterial suspensions. This DNA was used as a template in polymerase chain reactions (PCR) to detect antibiotic resistance genes. Standard PCR conditions were used to amplify genes encoding carbapenemase-related genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA), extended spectrum β-lactamases (blaTEM, blaSHV, blaCTX-M-1, and blaCTX-M-9), plasmid-mediated AmpC (blaEBC, blaDHA, blaCIT, blaCMY, and blaMOX), quinolone resistance determinants (qnrA, qnrB, qnrS, and acc(6’)-Ib-cr), and aminoglycoside resistance determinants (armA, rmtB, and acc(6’)-Ib) using primers and PCR conditions described previously (Dallenne et al., 2010; Poirel et al.,2011; Abrar et al., 2019). The sequence of all primers used in this study was listed in Table S1. The PCR amplicons of carbapenemase, extended spectrum β-lactamases (ESBLs) and AmpC resistance genes were sequenced and the sequences were analyzed on the National Center for Biotechnology Information (NCBI) website.
Data Collection and Definitions
After patients in both the case and control groups had been confirmed, relevant demographics and clinical data of the enrolled patients were collected from the electronic medical records and clinical microbiology laboratory databases. Survivor and nonsurvivor subgroups were compared to identify predictors of 28-day mortality.
Variables
Thirty-six clinical variables were applied in the analysis of the risk factors. The following parameters were used: (1) demographics (age, sex, hospital transfer, intensive care unit admission, and 28-day mortality); (2) baseline diseases and acquired infection (hypertension, diabetes, severe anemia, hypoproteinemia, hypokalemia, septic shock and solid tumors, respiratory, liver, gastrointestinal, cardiovascular, renal, and endocrine system diseases, as well as pulmonary, intra-abdominal, and urinary tract infection); (3) invasive procedures during the hospital stay (parenteral nutrition, mechanical ventilation, urinary catheter, tracheal cannula, drainage tube, previous surgery in the past 6 months, central venous catheter, and arterial catheter); and (4) antibiotic exposure within 3 months (cephalosporins, carbapenem, aminoglycosides, quinolones, tetracyclines, macrolides, metronidazole, and glycopeptides). These variables were analyzed to identify risk factors for XDR-KP infection.
Definitions
The following terms were defined prior to data analysis. Patients >65 years old were defined as elderly. Severe anemia was defined as a hemoglobin level <60 g/L. Hypoproteinemia was defined as serum total protein level <60 g/L or albumin level <25 g/L. Hypokalemia was defined as a serum potassium level <3.5 mmol/L. Septic shock was defined as sepsis associated with organ dysfunction and accompanied by persistent hypotension after volume replacement (Plante 2016; Howell and Davis, 2017). The source of infection was determined to be pneumonia, urinary tract infection (UTI), surgical site infection, intra-abdominal infection, or line-related infection using definitions from the US Centers for Disease Control and Prevention (CDC) (Kang et al., 2005).
Multilocus Sequence Typing
Molecular typing was performed by multilocus sequence typing (MLST). The K. pneumoniae MLST scheme used internal fragments of the following seven housekeeping genes: beta-subunit of RNA polymerase (rpoB), glyceraldehyde 3-phosphate dehydrogenase (gapA), malate dehydrogenase (mdh), phosphoglucose isomerase (pgi), phosphorine E (phoE), translation initiation factor 2 (infB), and periplasmic energy transducer (tonB). The housekeeping genes primer sequence of K. pneumoniae was shown in Table S2. The PCR conditions were queried, and data were analyzed using the K. pneumoniae MLST website (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html) (Joseph and Forsythe, 2017; Kiatsomphob et al., 2019; Shoaei et al., 2019).
Sample Size Calculations and Statistical Analysis
Based on the report of the CHINET Antimicrobial Resistance Surveillance Program in 2015 (Fupin et al., 2016), we assumed that XDR-KP will comprise 2.2% of the cases and non-XDR-KP controls will comprise 14.1%. To determine a difference at the 0.05 significance level with 80% power, we estimated that we would need at least 66 XDR-KP vs. 132 non-XDR-KP control cases (EpiInfo, version 3.3.2).
All statistical analyses were performed using SPSS version 23.0 for Windows (SPSS, Inc., Chicago, IL, USA). Categorical variables were presented as frequencies and percentages. In the analysis of risk factors for XDR-KP infection and mortality, the univariate analysis was used for each variable. To identify the independent risk factors, variables with P < 0.10 in the univariate analysis were included in the logistic regression model for the multivariate analysis and analyzed using backward stepwise regression. The odds ratio (OR) and 95% confidence interval (CI) were also calculated to evaluate the strength of any association that emerged. For all statistical analyses, P < 0.05 indicated statistical significance.
Results
Bacterial Isolates
In this study, a total of 69 consecutive nonduplicate XDR-KP isolates were identified from January 2015 to December 2017 and investigated. Isolates originated from different anatomical sites: sputum (n = 21, 30.4%), urine (n = 17, 24.6%), blood (n = 9, 13.0%), bile (n = 7, 10.1%), secretions (n = 5, 7.2%), pus (n = 5, 7.2%), puncture fluid (n = 4, 5.8%), and cannula (n = 1, 1.4%). The majority of patients were in the intensive care unit (n = 23, 33.3%), followed by patients on the hepatobiliary surgical ward (n = 18, 26.1%), respiratory ward (n = 13, 18.8%), urinary surgery ward (n = 7, 10.1%), digestive medicine ward (n = 4, 5.8%), endocrine ward (n = 3, 4.3%) and obstetrics and gynecology ward (n = 1, 1.4%). A total of 138 patients with non-XDR-KP infection were randomly matched in a ratio of 1:2 as the control group. Therefore, 207 patients were included in the final study cohort.
Antibiotic Susceptibility Test
As shown in Table 1, all 69 XDR-KP isolates were resistant to common clinical antibacterial drugs including carbapenems, by in vitro drug sensitivity tests but were only sensitive to tigecycline and polymyxin B (sensitivity rate 100%). The resistance rate of strains in the XDR-KP infection group was significantly higher than that in the control group, and the difference was statistically significant (P < 0.05).
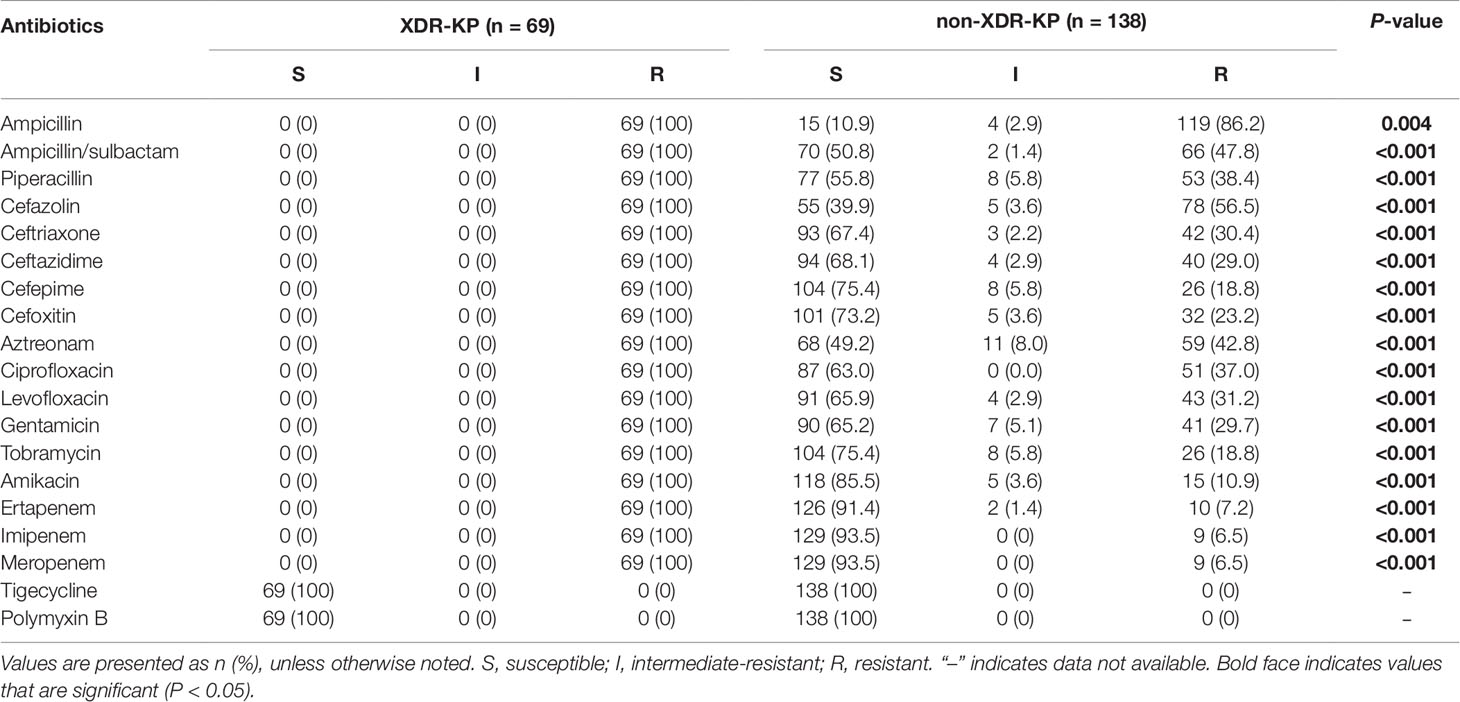
Table 1 The antimicrobial susceptibility of extensively drug-resistant Klebsiella pneumoniae (XDR-KP) and non-XDR-KP.
Detection of Drug Resistance Genes and ST Types
The distribution of resistance genes and ST types are listed in Table 2. Overall, these XDR-KP isolates had a high prevalence rate of carbapenemase-related resistance genes (n = 66, 95.7%), and only three strains were not determined to have carbapenemase-related resistance genes. Among them, the KPC-2 gene was mainly carried (n = 52, 75.4%), followed by the NDM (n = 21, 30.4%) and IMP (n = 5, 7.2%) genes. The OXA-48 gene was also detected in three XDR-KP strains in our study. Notably, several isolates were determined to carry two carbapenemase-related resistance genes at the same time (n = 15, 21.7%). In addition, all the XDR-KP isolates contained multiple drug resistance genes, including extended spectrum β-lactamases (ESBLs), AmpC β-lactamases, quinolone resistance determinants, and aminoglycoside resistance determinants. Among them, CTX-M-9 (n = 38, 55.1%), DHA (n = 29, 42.0%), qnrS (n = 40, 57.9%) and acc (6’)-Ib (n = 43, 62.3%) were the most frequently detected genes among these drug resistance genes, respectively.
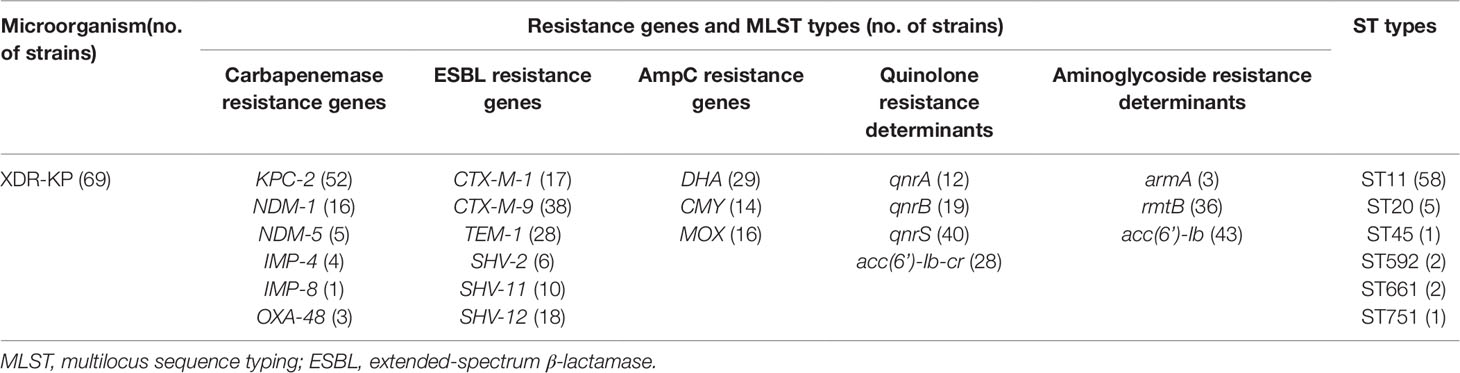
Table 2 Distribution of resistance genes and MLST types of extensively drug-resistant Klebsiella pneumoniae (XDR-KP).
Drug resistance genes can be transmitted horizontally among bacteria through mobile DNA elements, such as plasmids and integron genes. Molecular typing can assist in the analysis of transmission pathways between bacterial pathogens. The results of MLST showed that six different ST types were identified, of which ST11 was the most predominant epidemic type (n = 58, 84.1%). Perhaps more significantly, all strains of the ST11 type harbored the KPC-2 gene, which indicated that the KPC-2 gene was closely related to the prevalence of the XDR-KP strain.
Risk Factor Analysis of XDR-KP Infection
Univariate analysis of the demographics, baseline diseases, acquired infection, invasive procedures, and antibiotic exposure was performed in the case-control groups. The following factors were most frequently associated with the development of XDR-KP infection: ICU admission, transferring from another hospital, respiratory diseases, pulmonary infection, tracheal cannula, and exposure to carbapenem and glycopeptide antibiotics (Table 3).
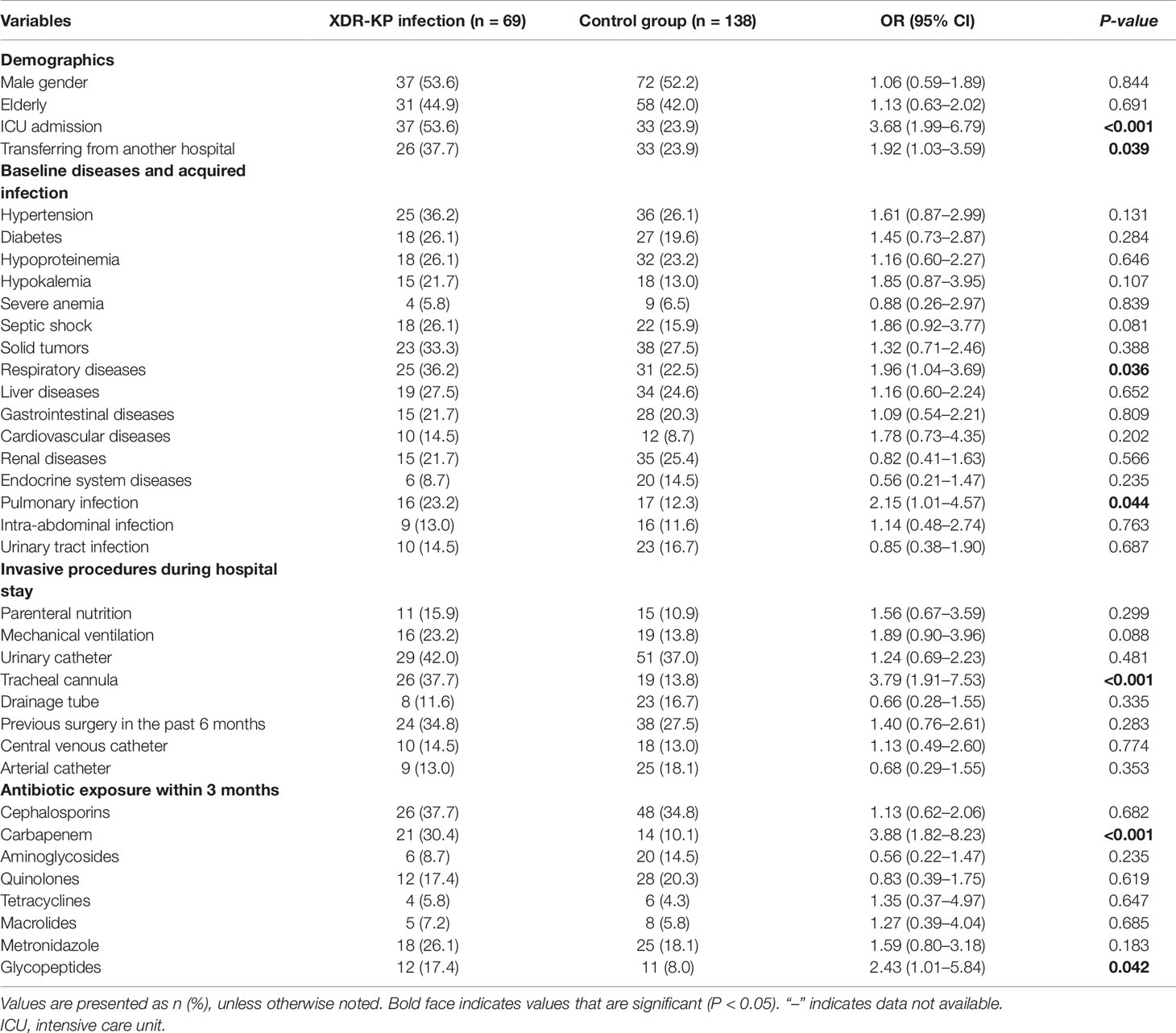
Table 3 Univariate analysis of risk factors for infection with extensively drug-resistant Klebsiella pneumoniae (XDR-KP) and non-XDR-KP.
Multivariate Analysis of XDR-KP Infection
The results of the logistic regression analysis of XDR-KP infection are shown in Table 4: ICU admission (OR: 3.28, 95% CI: 1.66–6.49, P < 0.001), tracheal cannula (OR: 3.16, 95% CI: 1.48–6.76, P = 0.003), and carbapenem exposure (OR: 3.16, 95% CI: 1.25–7.98, P = 0.015) were identified as independent risk factors for infection with XDR-KP when compared with the control group.
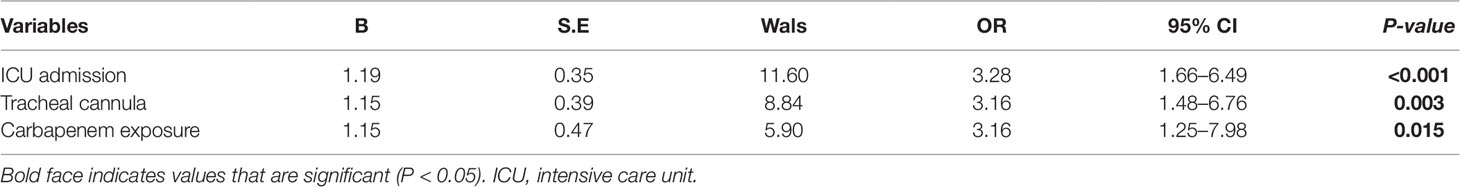
Table 4 Multivariate analysis of risk factors for infection with extensively drug-resistant Klebsiella pneumoniae (XDR-KP) and non-XDR-KP.
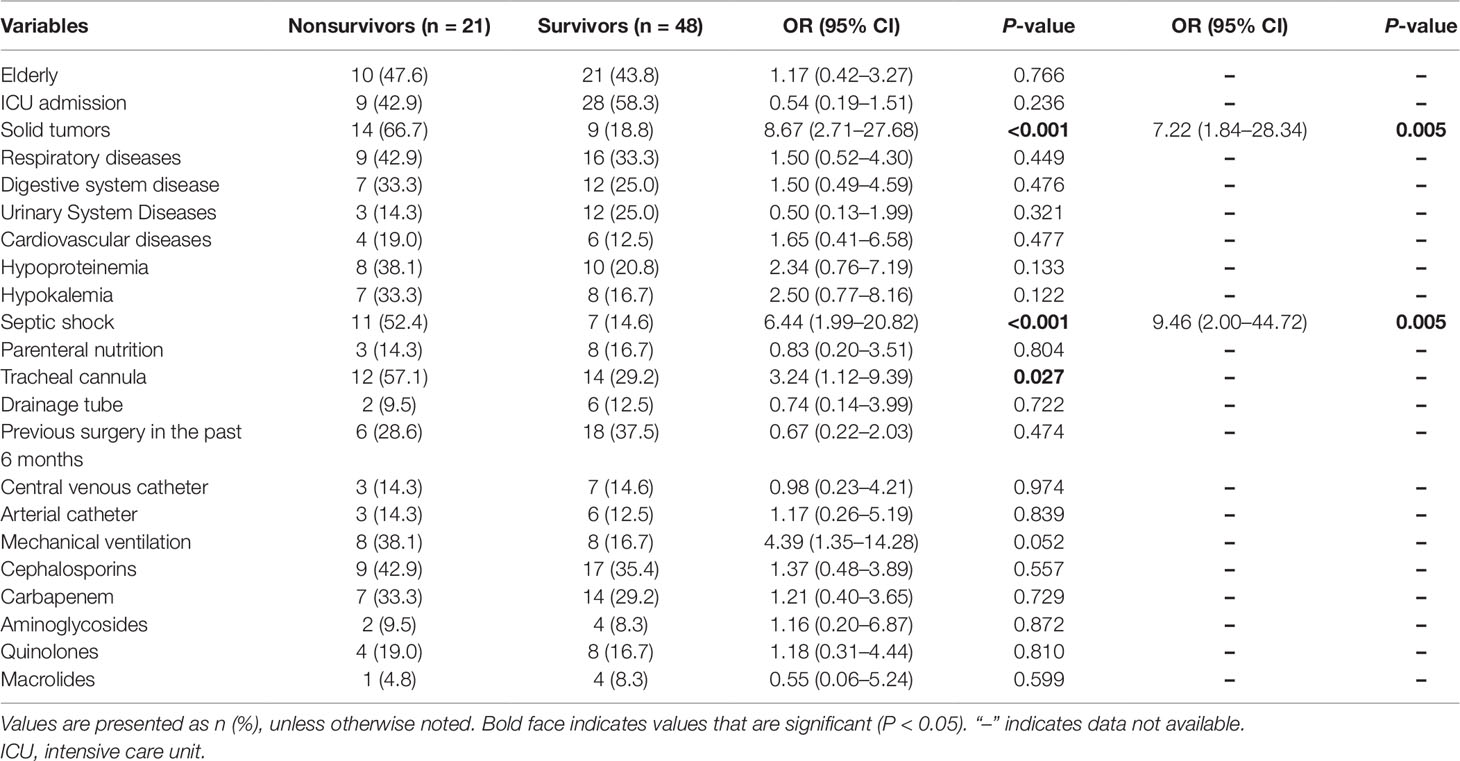
Risk Factors for 28-Day Mortality
At 28 days after infection onset, 21 of the 69 patients we investigated had died. Our study highlights the high mortality associated with XDR-KP infection, as shown in Table 5. The univariate analysis revealed that the presence of solid tumors, septic shock, and a tracheal cannula resulted in significant differences between the survivor and nonsurvivor groups. Multivariate logistic regression analysis showed that the predictors independently associated with 28-day mortality were solid tumors (OR: 7.22, 95% CI: 1.84–28.34, P = 0.005), and septic shock (OR: 9.46, 95% CI: 2.00–44.72, P = 0.005).
Discussion
At present, the emergence and dissemination of the XDR-KP strain is a major worldwide source and shuttle for antibiotic resistance. The increasing prevalence and global spread of these clinically extensively drug-resistant strains gravely threatens public health (Liu et al., 2017). According to the global epidemic trend of XDR-KP strains, the detection rate of XDR-KP strains in the northeastern United States, Italy, Greece, Israel and other European countries was relatively high, and there were even reports of local epidemic outbreaks in some areas (Zhou et al., 2015; Gu et al., 2018). However, molecular epidemiological characteristics differ from region to region (Garg et al., 2019; Kim et al., 2019). To our knowledge, there have been no reports on the epidemiology of and risk factors for XDR-KP infection in Southwestern China. This retrospective study was the first to systematically analyze the clinical characteristics, molecular epidemiology, risk factors of XDR-KP infection and the predictors of mortality.
For the drug sensitivity of XDR-KP strains, only tigecycline and polymyxin B still showed favorable in vitro drug sensitivity tests, and they were the most sensitive antibiotics for XDR-KP infection in the present study (sensitivity rate 100%). Currently, tigecycline and polymyxin B are considered to be the last line of defense against XDR-KP infection (Pragasam et al., 2016; Braun et al., 2018; Alves et al., 2019). However, increasing resistance is reported worldwide, in particular due to the plasmid-encoded protein mcr-1 present in pathogens such as Escherichia coli and Klebsiella pneumoniae, which further increases the difficulty of anti-infective treatment (Zhao et al., 2017; Shankar et al., 2019). Focusing on the molecular detection and genetic characterization of XDR-KP strains, we found that all the XDR-KP isolates contained multiple drug resistance genes, suggesting that the wide resistome of strains was related to the plentiful chromosomal and plasmid-encoded antibiotic resistance genes. The data revealed that the emergence of carbapenemase genes and ESBLs was the primary resistance mechanism. Among them, KPC-2 and CTX-M-9 group genes were the most prevalent in this study and were reported to be the most easily transmitted horizontally in KP (Fuga et al., 2019; Perez, 2019; Vargas et al., 2019). In addition, DHA was reported to be the most common gene in KP producing AmpC β-lactamases in many studies, which was consistent with our study (Akata et al., 2019; Kazemian et al., 2019). Notably, under antibiotic selective pressure, the pathogen continuously accumulates multiple drug resistance genes by acquiring plasmids and transferable genetic elements, ultimately leading to XDR strains harboring a ‘super resistome’ (Navon-Venezia et al., 2017).
MLST could be used to understand the epidemiology at a large scale in local outbreak situations. In the current study, ST11 was the most predominant epidemic type among the six different ST types identified, concordant with the report that ST11 was the major sequence type in Mainland China (Fu et al., 2019; Yu et al., 2019), and different from the international hyper-epidemic lineage ST258. Previous studies have shown that the hyperepidemic clonal complex of multidrug-resistant K. pneumoniae in ST258 clone may be spreading in the north-eastern region of Hungary (Toth et al., 2010), which were different from our results. Interestingly, only one of the seven housekeeping genes between ST11 (gapA, infB, mdh, pgi, phoE, rpoB, tonB: 3, 3, 1, 1, 1, 1, 4) and ST258 (gapA, infB, mdh, pgi, phoE, rpoB, tonB: 3, 3, 1, 1, 1, 1, 79) was different, suggesting that they were closely related to each other (Yu et al., 2018). Notably, all strains of ST11 XDR-KP isolates harbored the KPC-2 gene in our study, suggesting that the KPC-2 gene was involved in the epidemic dissemination of XDR-KP strains, which has frequently been reported worldwide in the interspecific transmission and dissemination of Enterobacteriaceae (Gao et al., 2019). If not effectively controlled, the ST11 clone could possibly spread widely in the future and even lead to the outbreak of XDR-KP strains.
Univariate analyses revealed that ICU admission, transferring from another hospital, respiratory diseases, pulmonary infection, tracheal cannula and exposure to carbapenem and glycopeptide antibiotics were strongly correlated with XDR-KP infection, which has also been identified by other studies. In addition, ICU admission, tracheal cannula, and carbapenem exposure were identified as independent risk factors for infection with XDR-KP in our study. Moreover, these ICU patients with concomitant infections had more severe underlying diseases and lower immunity, which might have made them more vulnerable to acquiring XDR-KP infection, and invasive procedures such as tracheal cannula and mechanical ventilation would have increased the possibility of drug-resistant bacteria adhesion, which ultimately led to infection with XDR-KP strains (Kiddee et al., 2019; Li et al., 2019). In addition, the selection pressure of antibiotics played an important role in the development of multiple drug resistance of bacteria. It was generally believed that under the pressure of carbapenem, susceptible strains were inhibited or killed, leading to the proliferation of many drug-resistant strains as dominant bacteria. Moreover, drug-resistant genes carried by drug-resistant bacteria could have been transmitted horizontally to susceptible strains by the acquisition of plasmids and transferable genetic elements, thus causing more serious epidemics of drug-resistant bacteria (Lee et al., 2013; Lopez-Camacho et al., 2014; Zhang et al., 2018). Therefore, we should select antibiotics according to drug sensitivity test results, and the abuse of carbapenem antibiotics should be avoided to reduce the production of XDR-KP strains.
Our study highlights the high mortality associated with XDR-KP infection. We found that solid tumors and septic shock were independent risk factors for death caused by XDR-KP infection. Most cancer patients usually have low immunity after radiotherapy or chemotherapy. After infection with “superbacteria” such as XDR-KP, these patients often received inappropriate initial antimicrobial treatment, which might damage prognosis and increase mortality. In addition, drug-resistant bacteria often cause serious infections, resulting in bacteremia or even sepsis in patients, causing septic shock that leads to multiple organ failure or even death (Falcone et al., 2016; Christodoulou et al., 2017; Machuca et al., 2017). Therefore, the surveillance of cancer and septic shock patients infected with XDR-KP should be strengthened to reduce their mortality.
Limitations
Given that this was a retrospective, case-control study, it is not possible to determine the absolute risk associated with the identified risk factors. In addition, this retrospective study was conducted at a single medical centre, and our sample size was relatively small, which might have led to errors in statistical analysis and the omission of some other risk factors.
Conclusions
This retrospective study was the first to systematically analyze the clinical characteristics, molecular epidemiology, and risk factors of XDR-KP infection and the predictors of mortality. Our findings showed that ST11 was the predominant epidemic type of XDR-KP producing KPC-2 in Southwestern China. In addition, ICU admission, tracheal cannula, and carbapenem exposure were identified as independent risk factors for infection with XDR-KP in this study. Moreover, another important finding of this study was that solid tumors and septic shock were independent risk factors for death caused by XDR-KP infection. Therefore, physicians should be aware of these high-risk patients with notable predictive factors for XDR-KP infection to reduce their mortality rate.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The study was approved by the Ethics Committee of Renji Hospital, Chinese Academy of Sciences University, Chongqing, China. The ethics committee waived the need for written informed consent provided by participants, and no additional informed consent was required.
Author Contributions
XY, HJ, and RZ performed experiments. CH and XH conceived the study and analyzed the results. XT and DX supervised the study and prepared the manuscript. All authors read and approved the final paper.
Funding
This study was supported in part by the National Natural Science Foundation of China (grant numbers 81471992 and 81772239), and the Natural Science Foundation of Chongqing (grant number 2019msxmX0568).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01307/full#supplementary-material
References
Abrar, S., Ain, N. U., Liaqat, H., Hussain, S., Rasheed, F., Riaz, S. (2019). Distribution of bla CTX - M, blaTEM, blaSHV and blaOXA genes in Extended-spectrum-beta-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob. Resist. Infect. Control 8, 80. doi: 10.1186/s13756-019-0536-0
Akata, K., Muratani, T., Yatera, K., Naito, K., Noguchi, S., Yamasaki, K., et al. (2019). Induction of plasmid-mediated AmpC beta-lactamase DHA-1 by piperacillin/tazobactam and other beta-lactams in Enterobacteriaceae. PloS One 14 (7), e218589. doi: 10.1371/journal.pone.0218589
Alves, P. H., Boff, R. T., Barth, A. L., Martins, A. F. (2019). Synergy of polymyxin B, tigecycline and meropenem against carbapenem-resistant Enterobacter cloacae complex isolates. Diagn. Microbiol. Infect. Dis. 94 (1), 81–85. doi: 10.1016/j.diagmicrobio.2018.11.011
Arabaghian, H., Salloum, T., Alousi, S., Panossian, B., Araj, G. F., Tokajian, S. (2019). Molecular Characterization of Carbapenem Resistant Klebsiella pneumoniae and Klebsiella quasipneumoniae Isolated from Lebanon. Sci. Rep. 9 (1), 531. doi: 10.1038/s41598-018-36554-2
Barnes, M. D., Taracila, M. A., Good, C. E., Bajaksouzian, S., Rojas, L. J., van Duin, D., et al. (2019). Nacubactam enhances meropenem activity against carbapenem-resistant Klebsiella pneumoniae producing Klebsiella pneumoniae carbapenemases (KPC). Antimicrob. Agents Chemother. 63 (8), e00432-e00419. doi: 10.1128/AAC.00432-19
Blin, C., Passet, V., Touchon, M., Rocha, E., Brisse, S. (2017). Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Environ. Microbiol. 19 (5), 1881–1898. doi: 10.1111/1462-2920.13689
Braun, G., Cayo, R., Matos, A. P., de Mello, F. J., Gales, A. C. (2018). Temporal evolution of polymyxin B-resistant Klebsiella pneumoniae clones recovered from blood cultures in a teaching hospital during a 7-year period. Int. J. Antimicrob. Agents 51 (3), 522–527. doi: 10.1016/j.ijantimicag.2017.08.031
Christodoulou, S., Kyriazopoulou, E., Chrysanthakopoulou, M., Karlis, G., Karampela, I., Gkizeli, K., et al. (2017). Lipid peroxidation in Gram-negative bacteremia modulates the risk for septic shock and infections by resistant Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 36 (11), 2171–2177. doi: 10.1007/s10096-017-3041-5
CLSI. 2016. Performance Standards for Antimicrobial Susceptibility Testing. 26th Informational Supplement. Wayne, PA Clinical and Laboratory Standards Institute;.
Dallenne, C., Da Costa, A., Decré, D., Favier, C., Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 (3), 490–495. doi: 10.1093/jac/dkp498
de Campos, T. A., Goncalves, L. F., Magalhaes, K. G., de Paulo, M. V., Pappas, J. G., Peirano, G., et al. (2018). A fatal bacteremia caused by Hypermucousviscous KPC-2 producing extensively drug-resistant K64-ST11 Klebsiella pneumoniae in Brazil. Front. Med. (Lausanne) 5, 265. doi: 10.3389/fmed.2018.00265
Du, J., Cao, J., Shen, L., Bi, W., Zhang, X., Liu, H., et al. (2016). Molecular epidemiology of extensively drug-resistant Klebsiella pneumoniae outbreak in Wenzhou, Southern China. J. Med. Microbiol. 65 (10), 1111–1118. doi: 10.1099/jmm.0.000338
Eucast 2017. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.0, Valid from 2017-01-01. Available at httpwww.eucast.org clinical_breakpoints [accessed February 24, 2017][J].
Falcone, M., Russo, A., Iacovelli, A., Restuccia, G., Ceccarelli, G., Giordano, A., et al. (2016). Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Clin. Microbiol. Infect. 22 (5), 444–450. doi: 10.1016/j.cmi.2016.01.016
Fu, P., Tang, Y., Li, G., Yu, L., Wang, Y., Jiang, X. (2019). Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int. J. Antimicrob. Agents 54 (2), 117–124. doi: 10.1016/j.ijantimicag.2019.03.014
Fuga, B., Royer, S., de Campos, P. A., Ferreira, M. L., Rossi, I., Machado, L. G., et al. (2019). Molecular Detection of Class 1 Integron-Associated Gene Cassettes in KPC-2-Producing Klebsiella pneumoniae Clones by Whole-Genome Sequencing. Microb. Drug Resist. 25(8), 1127–113. doi: 10.1089/mdr.2018.0437
Fupin, H., Demei, Z., Fu, W., Xiaofei, J., Chunying, X., Xiaojiang, Z., et al. (2016). Report of CHINET Antimicrobial Resistance Surveillance Program in 2015. Chin. J. Infect. Chemother. 16 (6), 16685–16694. doi: 10.16718/j.1009-7708.2016.06.003
Gao, B., Li, X., Yang, F., Chen, W., Zhao, Y., Bai, G., et al. (2019). Molecular tEpidemiology and Risk Factors of Ventilator-Associated Pneumonia Infection Caused by Carbapenem-Resistant Enterobacteriaceae. Front. Pharmacol. 10, 262. doi: 10.3389/fphar.2019.00262
Garg, A., Garg, J., Kumar, S., Bhattacharya, A., Agarwal, S., Upadhyay, G. C. (2019). Molecular epidemiology & therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J. Med. Res. 149 (2), 285–289. doi: 10.4103/ijmr.IJMR_36_18
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18 (1), 37–46. doi: 10.1016/S1473-3099(17)30489-9
Howell, M. D., Davis, A. M. (2017). Management of Sepsis and Septic Shock. JAMA 317 (8), 847–848. doi: 10.1001/jama.2017.0131
Hoxha, A., Karki, T., Giambi, C., Montano, C., Sisto, A., Bella, A., et al. (2016). Attributable mortality of carbapenem-resistant Klebsiella pneumoniae infections in a prospective matched cohort study in Italy, 2012-2013. J. Hosp. Infect. 92 (1), 61–66. doi: 10.1016/j.jhin.2015.06.018
Joseph, S., Forsythe, S. (2017). Multilocus Sequence Typing (MLST) for Cronobacter spp. Methods Mol. Biol. 1616, 241–248. doi: 10.1007/978-1-4939-7037-7_16
Kang, C. I., Kim, S. H., Park, W. B., Lee, K. D., Kim, H. B., Kim, E. C., et al. (2005). Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 49 (2), 760–766. doi: 10.1128/AAC.49.2.760-766.2005
Kazemian, H., Heidari, H., Ghanavati, R., Ghafourian, S., Yazdani, F., Sadeghifard, N., et al. (2019). Phenotypic and Genotypic characterization of ESBLs, AmpC and Carbapenemase-Producing Klebsiella pneumoniae and Escherichia coli Isolates. Med. Princ. Pract. 28 (6), 502–506. doi: 10.1159/000500311
Kiatsomphob, S., Taniguchi, T., Tarigan, E., Latt, K. M., Jeon, B., Misawa, N. (2019). Aerotolerance and multilocus sequence typing among Campylobacter jejuni strains isolated from humans, broiler chickens, and cattle in Miyazaki Prefecture, Japan. J. Vet. Med. Sci. 81 (8), 1144–1151. doi: 10.1292/jvms.19-0228
Kiddee, A., Assawatheptawee, K., Na-Udom, A., Boonsawang, P., Treebupachatsakul, P., Walsh, T. R., et al. (2019). Risk Factors for Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae Carriage in Patients Admitted to Intensive Care Unit in a Tertiary Care Hospital in Thailand. Microb. Drug Resist. 25 (8), 1182–1190. doi: 10.1089/mdr.2018.0318
Kim, K. G., Jeong, J., Kim, M. J., Park, D. W., Shin, J. H., Park, H. J., et al. (2019). Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type beta-lactamases and carbapenemases among diarrhoeagenic Escherichia coli isolates from children in Gwangju, Korea: 2007-16. J. Antimicrob. Chemother. 74 (8), 2181–2187. doi: 10.1093/jac/dkz175
Lee, J., Oh, C. E., Choi, E. H., Lee, H. J. (2013). The impact of the increased use of piperacillin/tazobactam on the selection of antibiotic resistance among invasive Escherichia coli and Klebsiella pneumoniae isolates. Int. J. Infect. Dis. 17 (8), e638–e643. doi: 10.1016/j.ijid.2013.01.030
Li, L., Yu, T., Ma, Y., Yang, Z., Wang, W., Song, X., et al. (2018). The Genetic Structures of an Extensively Drug Resistant (XDR) Klebsiella pneumoniae and Its Plasmids. Front. Cell Infect. Microbiol. 8, 446. doi: 10.3389/fcimb.2018.00446
Li, Y., Shen, H., Zhu, C., Yu, Y. (2019). Carbapenem-Resistant Klebsiella pneumoniae Infections among ICU Admission Patients in Central China: Prevalence and Prediction Model. BioMed. Res. Int. 2019, 9767313. doi: 10.1155/2019/9767313
Liu, H., Wilksch, J., Li, B., Du, J., Cao, J., Zhang, X., et al. (2017a). Emergence of ST39 and ST656 extensively drug-resistant Klebsiella pneumoniae isolates in Wenzhou, China. Indian J. Med. Microbiol. 35 (1), 145–146. doi: 10.4103/ijmm.IJMM_16_381
Liu, Y., Long, D., Xiang, T. X., Du, F. L., Wei, D. D., Wan, L. G., et al. (2019). Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 74 (5), 1233–1240. doi: 10.1093/jac/dkz023
Liu, Z., Gu, Y., Li, X., Liu, Y., Ye, Y., Guan, S., et al. (2019). Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann. Lab. Med. 39 (2), 167–175. doi: 10.3343/alm.2019.39.2.167
Lopez-Camacho, E., Gomez-Gil, R., Tobes, R., Manrique, M., Lorenzo, M., Galvan, B., et al. (2014). Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J. Antimicrob. Chemother. 69 (3), 632–636. doi: 10.1093/jac/dkt419
Machuca, I., Gutierrez-Gutierrez, B., Gracia-Ahufinger, I., Rivera, E. F., Cano, A., Guzman-Puche, J., et al. (2017). Mortality Associated with Bacteremia Due to Colistin-Resistant Klebsiella pneumoniae with High-Level Meropenem Resistance: Importance of Combination Therapy without Colistin and Carbapenems. Antimicrob. Agents Chemother. 61 (8), e00406–e00417. doi: 10.1128/AAC.00406-17
Meng, X., Yang, J., Duan, J., Liu, S., Huang, X., Wen, X., et al. (2019). Assessing Molecular Epidemiology of Carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in Central China. Sci. Rep. 9 (1), 2271. doi: 10.1038/s41598-018-38295-8
Moradigaravand, D., Martin, V., Peacock, S. J., Parkhill, J. (2017). Evolution and Epidemiology of Multidrug-Resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio 8 (1), e01976–e01916. doi: 10.1128/mBio.01976-16
Navon-Venezia, S., Kondratyeva, K., Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41 (3), 252–275. doi: 10.1093/femsre/fux013
Perez, L. (2019). Equal, but different: Fluctuant biofilm formation and its impact on polymyxin B susceptibility among a clonal spreading of KPC-2-producing Klebsiella pneumoniae isolates. Infect. Control Hosp. Epidemiol. 40 (8), 954–955. doi: 10.1017/ice.2019.106
Petrosillo, N., Taglietti, F., Granata, G. (2019). Treatment Options for Colistin Resistant Klebsiella pneumoniae: Present and Future. J. Clin. Med. 8 (7), E934. doi: 10.3390/jcm8070934
Plante, L. A. (2016). Management of Sepsis and Septic Shock for the Obstetrician-Gynecologist. Obstet. Gynecol. Clin. North Am. 43 (4), 659–678. doi: 10.1016/j.ogc.2016.07.010
Poirel, L., Walsh, T. R., Cuvillier, V., Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70 (1), 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E., Inbanathan, F. Y., et al. (2016). Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae Causing Bacteremia from India-A First Report. Front. Microbiol. 7, 2135. doi: 10.3389/fmicb.2016.02135
Rojas, L. J., Salim, M., Cober, E., Richter, S. S., Perez, F., Salata, R. A., et al. (2017). Colistin Resistance in Carbapenem-Resistant Klebsiella pneumoniae: Laboratory Detection and Impact on Mortality. Clin. Infect. Dis. 64 (6), 711–718. doi: 10.1093/cid/ciw805
Shankar, C., Venkatesan, M., Rajan, R., Mani, D., Lal, B., Prakash, J., et al. (2019). Molecular characterization of colistin-resistant Klebsiella pneumoniae & its clonal relationship among Indian isolates. Indian J. Med. Res. 149 (2), 199–207. doi: 10.4103/ijmr.IJMR_2087_17
Shoaei, P., Shojaei, H., Khorvash, F., Hosseini, M. S., Ataei, B., Esfandiari, Z., et al. (2019). Clostridium difficile infection in cancer patients with hospital acquired diarrhea at the teaching hospitals in Iran: Multilocus sequence typing analysis (MLST) and Antimicrobial resistance pattern. Ann. Ig. 31 (4), 365–373. doi: 10.7416/ai.2019.2298
Toth, A., Damjanova, I., Puskas, E., Janvari, L., Farkas, M., Dobak, A., et al. (2010). Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 29 (7), 765–769. doi: 10.1007/s10096-010-0921-3
Vargas, J. M., Moreno, M. M., Nunez, J. M., Caceres, M., Mochi, S., Del, C. M. R., et al. (2019). Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon 5 (6), e1829. doi: 10.1016/j.heliyon.2019.e01829
Yu, F., Hu, L., Zhong, Q., Hang, Y., Liu, Y., Hu, X., et al. (2019). Dissemination of Klebsiella pneumoniae ST11 isolates with carbapenem resistance in integrated and emergency intensive care units in a Chinese tertiary hospital. J. Med. Microbiol. 68 (6), 882–889. doi: 10.1099/jmm.0.000981
Yu, F., Lv, J., Niu, S., Du, H., Tang, Y. W., Pitout, J., et al. (2018). Multiplex PCR Analysis for Rapid Detection of Klebsiella pneumoniae Carbapenem-Resistant (Sequence Type 258 [ST258] and ST11) and Hypervirulent (ST23, ST65, ST86, and ST375) Strains. J. Clin. Microbiol. 56 (9), e00731–e00718. doi: 10.1128/JCM.00731-18
Zhang, R., Dong, N., Huang, Y., Zhou, H., Xie, M., Chan, E. W., et al. (2018). Evolution of tigecycline- and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerg. Microb. Infect. 7 (1), 127. doi: 10.1038/s41426-018-0129-7
Zhao, F., Feng, Y., Lu, X., McNally, A., Zong, Z. (2017). IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 61 (2), e02229-e02216. doi: 10.1128/AAC.02229-16
Zheng, B., Dai, Y., Liu, Y., Shi, W., Dai, E., Han, Y., et al. (2017). Molecular Epidemiology and Risk Factors of Carbapenem-Resistant Klebsiella pneumoniae Infections in Eastern China. Front. Microbiol. 8, 1061. doi: 10.3389/fmicb.2017.01061
Zhou, T., Zhang, Y., Li, M., Yu, X., Sun, Y., Xu, J. (2015). An outbreak of infections caused by extensively drug-resistant Klebsiella pneumoniae strains during a short period of time in a Chinese teaching hospital: epidemiology study and molecular characteristics. Diagn. Microbiol. Infect. Dis. 82 (3), 240–244. doi: 10.1016/j.diagmicrobio.2015.03.017
Keywords: extensively drug-resistant Klebsiella pneumoniae, molecular epidemiology, MLST, risk factors, mortality
Citation: Tian X, Huang C, Ye X, Jiang H, Zhang R, Hu X and Xu D (2019) Molecular Epidemiology of and Risk Factors for Extensively Drug-Resistant Klebsiella pneumoniae Infections in Southwestern China: A Retrospective Study. Front. Pharmacol. 10:1307. doi: 10.3389/fphar.2019.01307
Received: 29 July 2019; Accepted: 15 October 2019;
Published: 01 November 2019.
Edited by:
Pietro Minuz, University of Verona, ItalyReviewed by:
Ji Wang, Sun Yat-sen University, ChinaMichal Letek, University of Roehampton London, United Kingdom
Copyright © 2019 Tian, Huang, Ye, Jiang, Zhang, Hu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongshuang Xu, ZG9uZ3NodWFuZ194dUAxNjMuY29t
 Xiaolang Tian
Xiaolang Tian Changwu Huang
Changwu Huang