- 1Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 2Yunnan Branch of Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Jinghong, China
A Corrigendum on
Species Identification of Dracaena Using the Complete Chloroplast Genome as a Super-Barcode
by Zhang Z, Zhang Y, Song M, Guan Y and Ma X (2019). Front. Pharmacol. 10:1441. doi: 10.3389/fphar.2019.01441
In the original article, the figure legends were associated with the wrong figures. Figure 4 should be Figure 2, Figure 2 should be Figure 3, and Figure 3 should be Figure 4. The legends remain the same. The figures and their correct legends appear below.
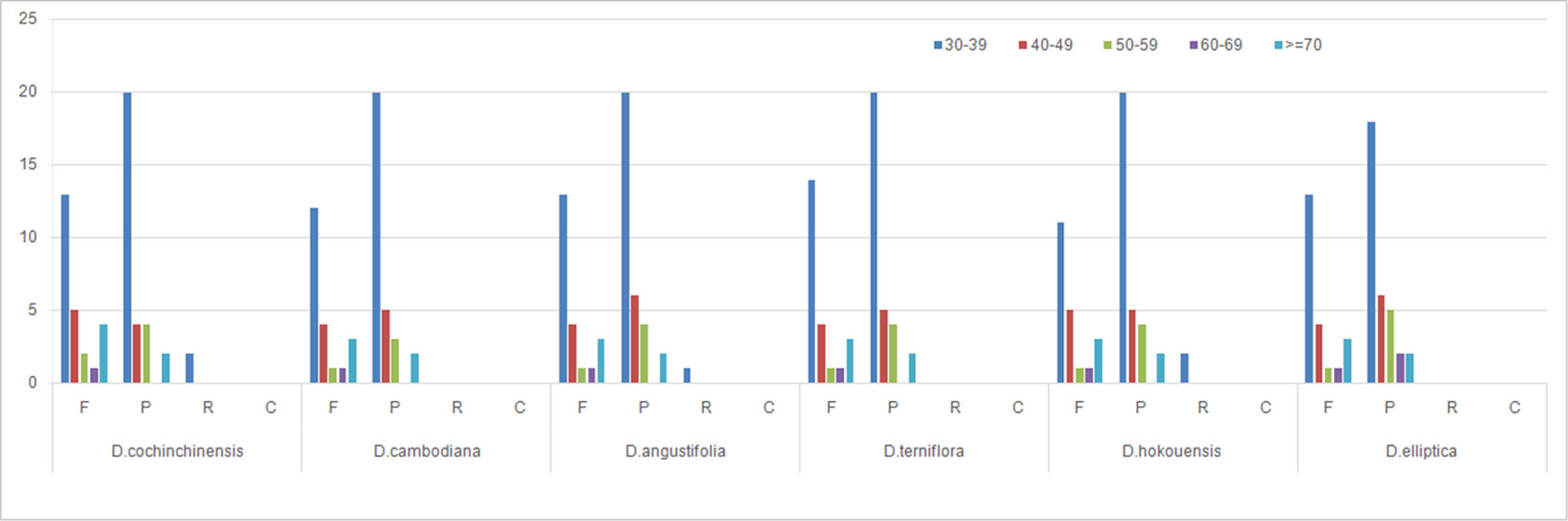
Figure 2 Repeat analysis in six Dracaena CP genomes. REPuter was used to identify repeat sequences with length ≥30 bp and sequence identified ≥90% in the CP genomes. F, P, R, and C indicate the repeat types F (forward), P (palindrome), R (reverse), and C (complement), respectively. Repeats with different lengths are indicated in different colors.
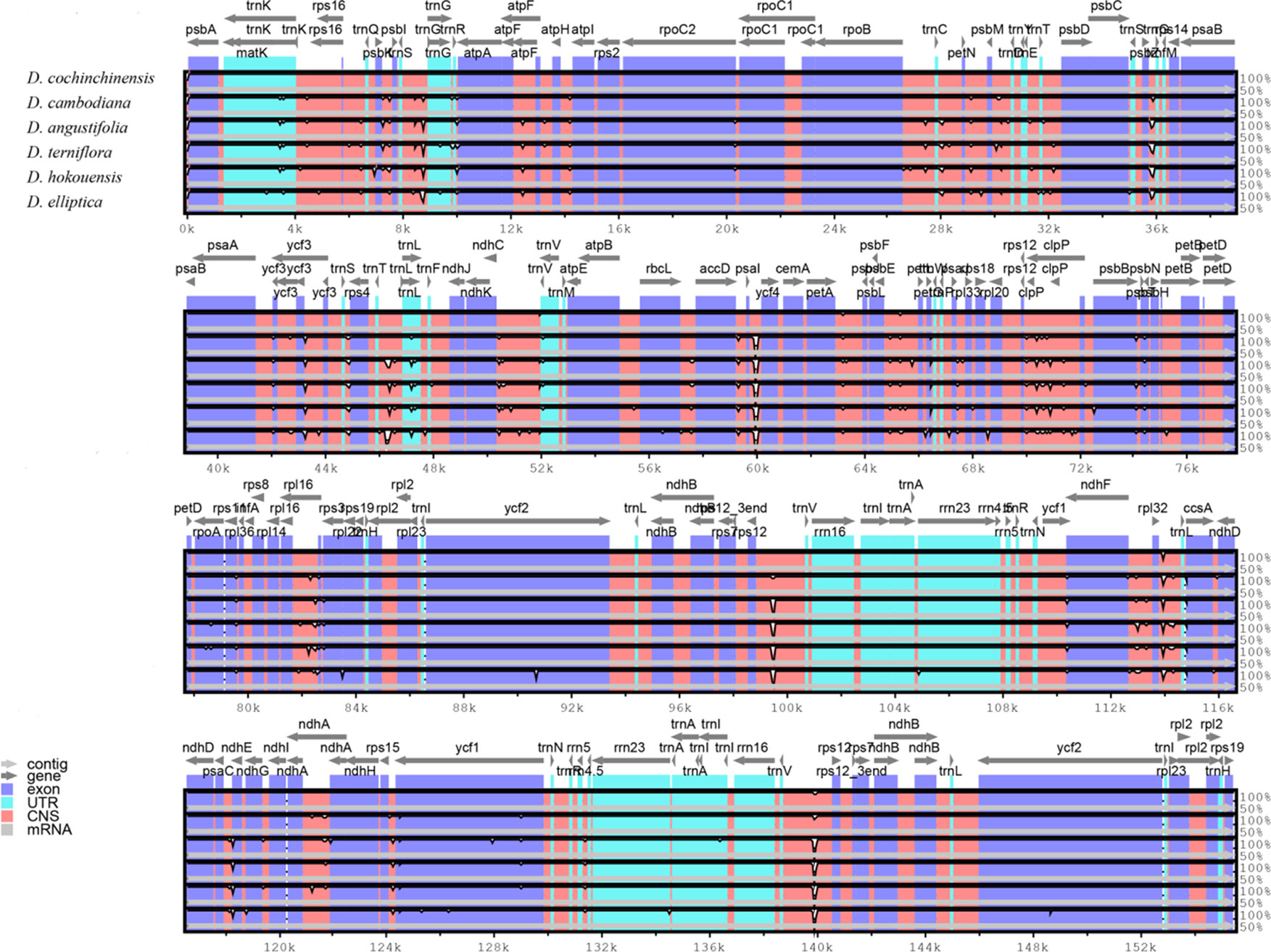
Figure 3 Structure comparison of the six Dracaena CP genomes by using the mVISTA program. Gray arrows and thick black lines above the alignment indicate genes with their orientation and the position of the IRs, respectively. A cut-off value of 70% identity was used for the plots, and the Y-scale represents the percent identity between 50% and 100%.
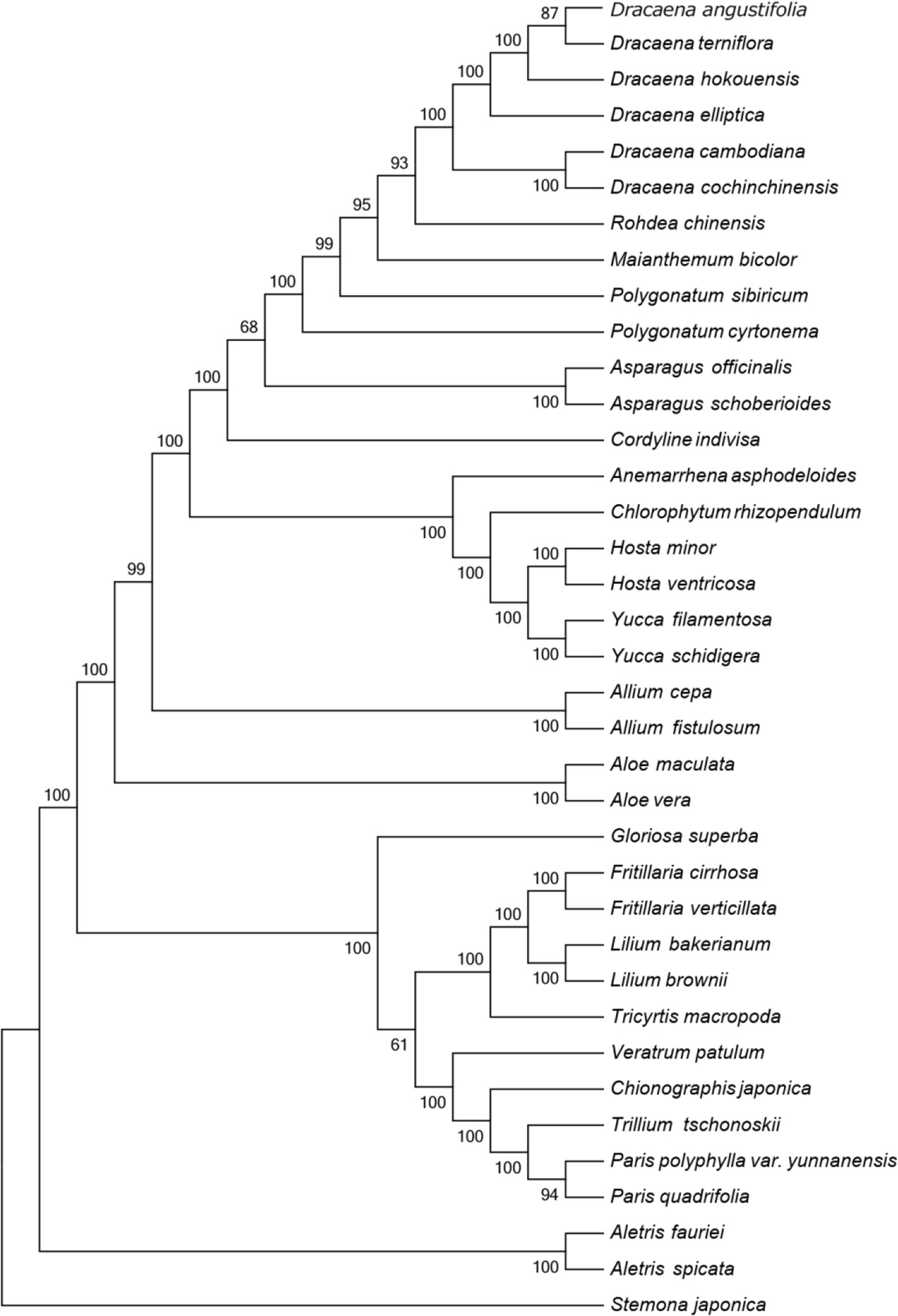
Figure 4 Phylogenetic tree constructed using MP based on complete CP genomes of six Dracaena and other 31 species. Numbers above the branches are the bootstrap support values.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: Dracaena Vand. ex L., chloroplast genome, identification, super-barcode, Liliaceae
Citation: Zhang Z, Zhang Y, Song M, Guan Y and Ma X (2020) Corrigendum: Species Identification of Dracaena Using the Complete Chloroplast Genome as a Super-Barcode. Front. Pharmacol. 11:51. doi: 10.3389/fphar.2020.00051
Received: 17 December 2019; Accepted: 15 January 2020;
Published: 13 February 2020.
Edited and reviewed by: Hugo J. De Boer, University of Oslo, Norway
Copyright © 2020 Zhang, Zhang, Song, Guan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Ma, bWF5aXh1YW4xMEAxNjMuY29t
 Zhonglian Zhang
Zhonglian Zhang Yue Zhang
Yue Zhang Meifang Song2
Meifang Song2 Xiaojun Ma
Xiaojun Ma