- 1Graduate School, Guizhou University of Traditional Chinese Medicine, Guiyang, China
- 2Department of Rheumatology Immunology, The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, China
Tripterygii Radix exhibits good clinical efficacy and safety in rheumatoid arthritis (RA) patients, but its effective components and mechanism of action are still unclear. The purpose of this study was to explore and verify the major ingredients and molecular targets of Tripterygii Radix in RA using drug-compounds-biotargets-diseases network and protein-protein interaction (PPI) network analyses. The processes and pathways were derived from Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The most important compounds and biotargets were determined based on the degree values. RA fibroblast-like synoviocytes (RA-FLS) were separated from RA patients and identified by hematoxylin and eosin (HE) staining and immunohistochemistry. The purity of RA-FLS was acquired by flow cytometry marked with CD90 or VCAM-1. RA-FLS were subjected to control, dimethyl sulfoxide (control), kaempferol, or lenalidomide treatment. Cell migration was evaluated by the transwell assay. The relative expression of biotarget proteins and cytokines was analyzed by western blotting and flow cytometry. In total, 144 chemical components were identified from Tripterygii Radix; kaempferol was the most active ingredient among 33 other components. Fourteen proteins were found to be affected in RA from 285 common biotargets. The tumor necrosis factor (TNF) signaling pathway was predicted to be one of the most latent treatment pathways. Migration of RA-FLS was inhibited and the expression of protein kinase B (AKT1), JUN, caspase 3 (CASP3), TNF receptor 1 and 2 (TNFR1 and TNFR2), interleukin-6 (IL-6), and TNF-α was significantly affected by kaempferol. Thus, this study confirmed kaempferol as the effective component of Tripterygii Radix against RA-FLS and TNF signaling pathway and its involvement in the regulation of AKT1, JUN, CASP3, TNFR1, TNFR2, IL-6, and TNF-α expression.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized with chronic synovitis and proliferation of synovial cells (Weyand and Goronzy, 2017) accompanied with the release of inflammatory factors, which destroy the bone and the cartilage and eventually lead to the loss of joint function (Clapp et al., 2016; Stanford et al., 2016). Fibroblast-like synoviocytes (FLS) constitute the main part of the synovial tissue and significantly contribute to inflammation and aggressive joint destruction in RA (Falconer et al., 2018). Therefore, it is beneficial to inhibit the migration and invasion of and RA-FLS-mediated expression of inflammatory factors in patients with RA (Ma et al., 2019).
Tripterygii Radix is an effective Chinese herb used to treat arthritis (Lv et al., 2015). It not only suppresses cell proliferation, migration, and invasion by apoptosis but also diminishes the expression of pro-inflammatory cytokines, pro-inflammatory mediators, adhesion molecules, and matrix metalloproteinases (Dai and Bao, 2011). Recent studies demonstrated the T. Radix-mediated inhibition of angiogenesis and suppression of abnormally activated innate immune response in rat kidney tissue through the modulation of the nuclear factor kappa B (NF-κB) signaling pathway (Zhang et al., 2017; Liu et al., 2018). Nevertheless, the detailed mechanisms underlying the anti-inflammatory and anti-proliferative effects of Tripterygii Radix on RA-FLS are unclear, given its complex ingredients.
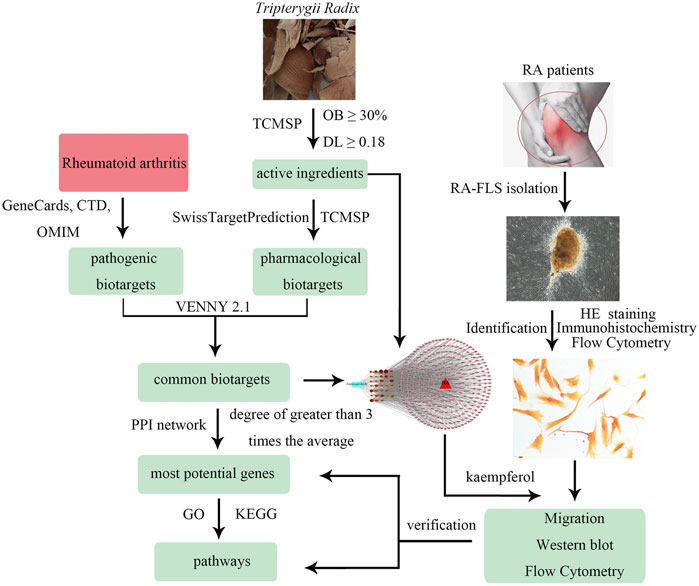
Therefore, this report aims to predict the effective ingredients and the mechanism of action of Tripterygii Radix on RA based on network pharmacology. In addition, it aims to confirm the most effective compound and molecular targets through cell experiments. The study workflow is shown Figure 1.
Materials and Methods
Screening of Active Components and Pharmacological Biotargets of Tripterygii Radix
To determine the ingredients of Tripterygii Radix, ADME features were obtained from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://tcmspw.com/) (Wan et al., 2019), a unique pharmacology platform designed for herbal medicine. In accordance with the pharmacokinetic characteristics in TCMSP database, the chemical components that satisfied both oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 were selected as active components. The corresponding pharmacological biotargets were reaped (Huang et al., 2017). After inputting the selected active components into the SwissTargetPrediction database (http://www.swisstargetprediction.ch/) (Daina et al., 2019), the pharmacological biotargets were obtained. The final biotargets of active components were retained after removing duplicate targets of the results.
Collecting Pathogenic Biotargets of RA
After deleting the duplicate targets, the information on pathogenic biotargets of RA was collected from the GeneCards database (GeneCards, https://genecards.weizmann.ac.il/v3/), Comparative Toxicogenomics Database (CTD, https://ctdbase.org/), and Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/) (Rappaport et al., 2017; Davis et al., 2019).
Choosing Common Biotargets and Construction of Protein-Protein Interaction (PPI) Network
VENNY 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/) was used to obtain the common targets (Liang et al., 2019). The drug-compounds-biotargets-diseases network was visualized by Cytoscape 3.7.1 software. The nodes in the network represented active components and target genes, while the edges indicated the ingredients that interacted with the targets. The common genes were imported to the STRING database (https://string-db.org/) to construct the PPI network after deleting orphaned nodes. Then, the most potential targets were chosen on the basis of the degree value (Athanasios et al., 2017; Zhang et al., 2019).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analyses
According to the average degree values originating from PPI results, the most important target genes were selected. GO function analysis, including biological process (BP), cell component (CC), and molecular function (MF) terms, and the KEGG pathway analysis were carried out to predict the possible courses and pathways of treatment using the ClueGO plugin (Hua et al., 2019). The terms with values of p < 0.05 were considered as significant processes and pathways.
RA-FLS Isolation
The RA-FLS cells was isolated and cultured by explant adherent culture method. The synovial tissue was obtained from three patients (2 male and 1 female; 0–70 years old) during joint replacement surgery at The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine. All patients were diagnosed with active RA according to 2010ACR/EULAR Classification Criteria for Rheumatoid Arthritis and disease activity score in 28 joints and four variables, including C-reactive protein (DAS28-CRP) ≥ 3.2 (Fransen and Riel, 2009; Aletaha et al., 2010). The experiments were approved by the Medical Ethical Committee of The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine (PY2019104), and all patients signed informed consent. The synovial tissues were washed five times with Phosphate-Buffered Saline (PBS, BI, Israel) supplemented with 2% penicillin/streptomycin (Gibco, United States). After removing irrelevant tissues, especially the adipose tissue, the synovial tissue was cut into one cubic centimeter pieces. All pieces were cultured to harvest RA-FLS in a flask with a complete medium containing Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, United States) supplemented with 20% fetal bovine serum (BI, Israel) and 1% penicillin/streptomycin (Gibco, United States) at 37°C and 5% CO2-enriched atmosphere. Culture media were replenished every 4 days, and cells were subcultured at a ratio of 1:2 when they reached 90% confluence. RA-FLS were used for experiments between third and sixth passage.
Cell Identification
Hematoxylin and eosin (HE) staining and immunohistochemistry were used to identify RA-FLS. The third passage FLS were stained using a HE staining kit (Solarbio, China) and incubated with a rabbit anti-vimentin antibody (Abcam, United Kingdom), followed by probing with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (ZSGB, China) and developing with 3,3′-diaminobenzidine reagent (ZSGB, China). The FLS were counterstained with hematoxylin.
The flow cytometry was used to get the purity of RA-FLS staining for cell surface markers CD90 or VCAM-1. The third passage FLS (1 × 105/ml) suspended in 1 ml of PBS in 12 × 75 mm2 tubes were centrifuged at 200 g for 5 min at 4°C and the supernatant was removed. The cells were incubated 30 min after 5 μL of FITC anti-CD90 (Abcam, United Kingdom) or APC anti-VCAM-1 (Abcam, United Kingdom) was added in dark at 4°C. After wasded three times, the cells were resuspended in 500 µL of a buffer solution and analyzed by flow cytometer (BD, United States).
RA-FLS Intervention
The third passage RA-FLS were pre-treated with complete medium, 0.2% dimethyl sulfoxide (DMSO; Solarbio, China), 25 μM kaempferol (Shanghai Winherb, China), or 50 μM lenalidomide (Abcam, United Kingdom) for 24 h. All reagents were dissolved in DMSO and diluted to working concentrations with complete medium.
Migration Assays
The RA-FLS (1 × 105/ml) were pre-treated with different working concentrations of test agents for 24 h. One day prior to setting up the Transwell migration assay, the RA-FLS were serum-starved overnight. A total of 3 × 104 cells were resuspended in 100 μL serum-free medium and seeded onto the upper chamber of a transwell migration assay plate (8 μm pores, Corning, United States), while the lower chamber was filled with 800 μL of complete medium. After 24 h incubation, media within the transwell inserts were carefully removed. Cells were fixed with 1 ml 2% paraformaldehyde. Then. the cells settling on the upper chamber were carefully removed with a cotton swab, while those migrated to the lower surface were stained with HE. Cell count was performed under a microscope (Olympus, Japan).
Western Blot Assays
The cell lysates containing 40 μg total protein were loaded onto 10% sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) gels (Bio-Rad, United States). The separated protein bands were transferred onto nitrocellulose membranes. After blocking with 5% skim milk in Tris-buffered-saline-Tween (TBST) 1.5 h, membranes were incubated with different primary antibodies, including those against JUN (Abcam, United Kingdom; 1:1,000), P-JUN (phosphor-S100, Abcam, United Kingdom; 1:1,000), protein kinase B (AKT1; Abcam, United Kingdom; 1:10,000), P-AKT1 (phosphor-S473, Abcam, United Kingdom; 1:5,000), caspase-3 (CASP3; Abcam, United Kingdom; 1:500), tumor necrosis factor receptor 1 (TNFR1; Abcam, United Kingdom; 1:5,000), and TNFR2 (Abcam, United Kingdom; 1:10,000) for overnight at 4°C. The membranes were washed five times with TBST, then peroxidase-conjugated goat anti-rabbit IgG antibody (Pumei, China) was used as a secondary antibody to incubate for 1 h. After another washed five times with TBST, enhanced chemiluminescence substrate (Bio-Rad, United States) was used to detect the amount of target proteins. ImageJ software was employed to quantify the relative intensity following normalization with β-actin (Cell Signaling, United States; 1:1,000). All experiments were repeated thrice.
Flow Cytometry Assays
Multiparameter flow cytometry permits the simultaneous detection of two or more cytokines. Multiparameter flow cytometry was applied to detect the concentrations of TNF-α and IL-6 from the cell supernatants pre-treated with different working concentrations of test agents. The supernatants were obtained after pre-treatment for 24 h. The tests were performed according to the manufacturer’s protocols (RAISECARE, China). In brief, 25 μL of a buffer solution, 25 μL of the studied sample or standard, 25 μL of capture microsphere antibody, and 25 μL of a detection antibody were consecutively added to each tube. The tubes were incubated and vibrated (400–500 rpm) at 20°C in the dark for 2 h. The tubes were treated with 25 μL streptavidin-phycoerythrin for 0.5 h, washed with 500 μL of a wash buffer, and centrifuged. The supernatant was discarded and the pellet was resuspended in 200 µL of a buffer solution and analyzed by flow cytometer (BD, United States).
Statistical Analysis
Statistical analysis was performed using the SPSS 17 software package (SPSS Inc., United States). One-way analysis of variance was used to compare the means of different groups. A value of p < 0.05 was considered as statistically significant.
Results
Active Components of Tripterygii Radix
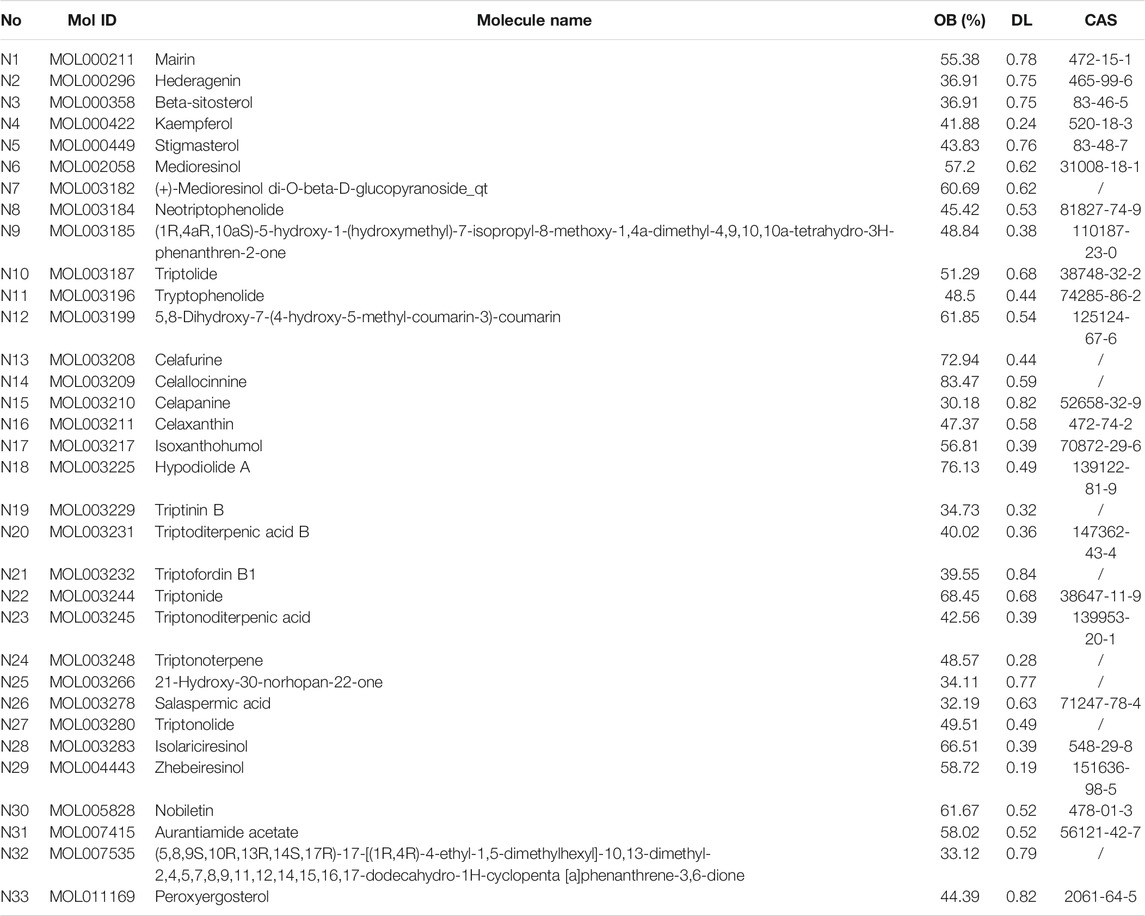
OB is a central pharmacokinetic parameter in the ADME process and is associated with the absorption and delivery of an orally administered drug. DL represents the druggability of a drug and serves as a qualitative alternative to study pharmacokinetic and pharmaceutical properties. A total of 144 chemical components were detected from Tripterygii Radix, of which only 33 were considered as active components because they satisfied the criteria of OB ≥ 30% and DL ≥ 0.18 (Table 1).
Biotargets and PPI Network
A total of 748 pharmacological biotargets resulted from TCMSP and SwissTargetPrediction databases after ruling out non-human species. In total, 2060 pathogenic biotargets were selected from GeneCards, CTD, and OMIM, and 285 common biotargets were chosen by VENNY 2.1 (Figure 2A). The drug-compounds-biotargets-diseases network was visualized by Cytoscape 3.7.1 software, which revealed the effects of Tripterygii Radix on RA through the common biotargets (Figure 2B). The network contained 320 nodes and 1,304 edges. The average degree among the 33 active components was 30.84, while the top three compounds in conformity with the degree values were kaempferol, beta-sitosterol, and aurantiamide acetate that comprised the therapeutic constituents.
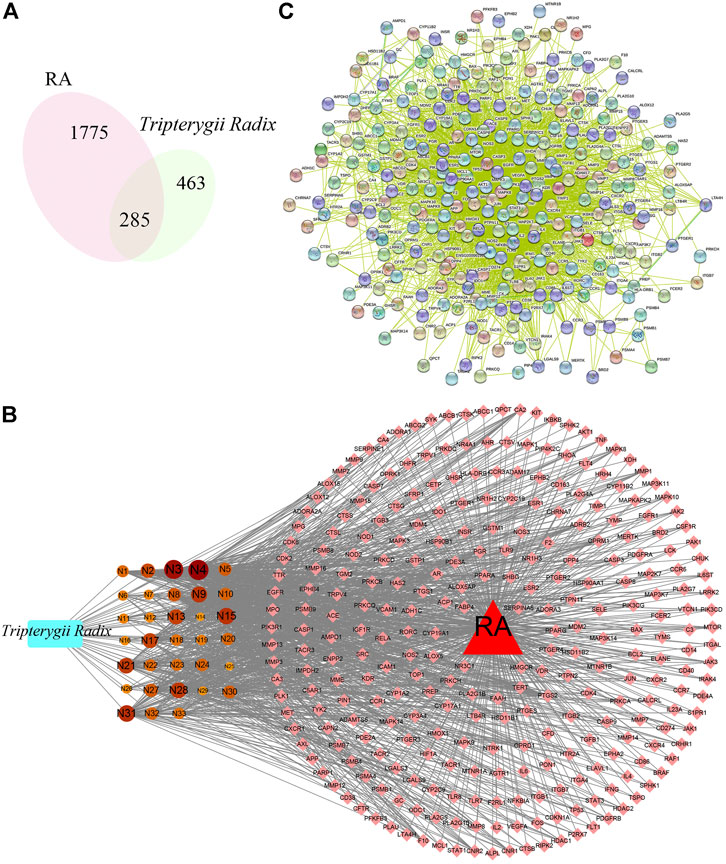
FIGURE 2. (A) represents the venn digram of biotargets between Tripterygii Radix and RA, While (B) represents the drug-active components-biotargets-disease network of Tripterygii Radix against RA. The rectangle represents Tripterygii Radix, the circle nodes represent active components, the diamond nodes represent biotargets, and the triangle represent RA. The 33 components nodes changed with the degree value. The higher the degree value, the bigger was the node. (C) represents the PPI network of the common biotargets.
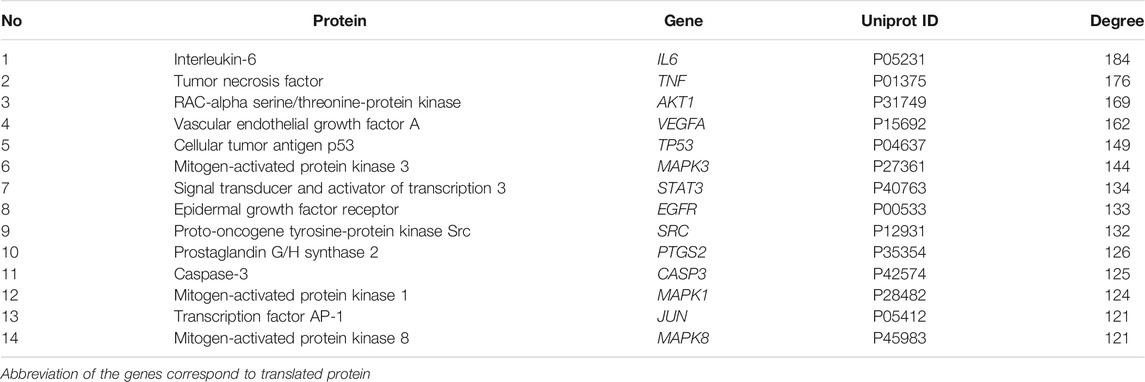
The PPI network (Figure 2C) constructed with 285 nodes and 3,741 edges for text-mining information and interaction predictions revealed 285 common biotargets. The average degree value of PPI network was 39.151, and 14 target genes (Table 2) were greater than three times the average degree value, indicative of them as the potential genes related to RA.
BP, CC, MF, and Pathways of Tripterygii Radix on RA
The GO function and KEGG pathway enrichment analyses were executed under the condition of p < 0.05 using the ClueGO plugin of Cytoscape 3.7.1. BP contained 659 terms, including response to oxidative stress, response to reactive oxygen species, and cellular response to reactive oxygen species. The top 10 terms are shown in Figure 3A in accordance with the lowest p value. CC only contained 11 terms such as membrane raft, membrane microdomain, and membrane region; the top 10 terms are shown in Figure 3B. In addition, the MF comprised 98 terms, including regulation of hydrolase activity, enzyme binding, and identical protein binding. The top 10 terms are shown in Figure 3C. KEGG pathway enrichment analysis indicated 104 terms such as AGE-RAGE signaling pathway in diabetic complications, C-type lectin receptor signaling pathway, and TNF signaling pathway; these along with the top 10 terms with lowest p values are shown in Figure 3D.
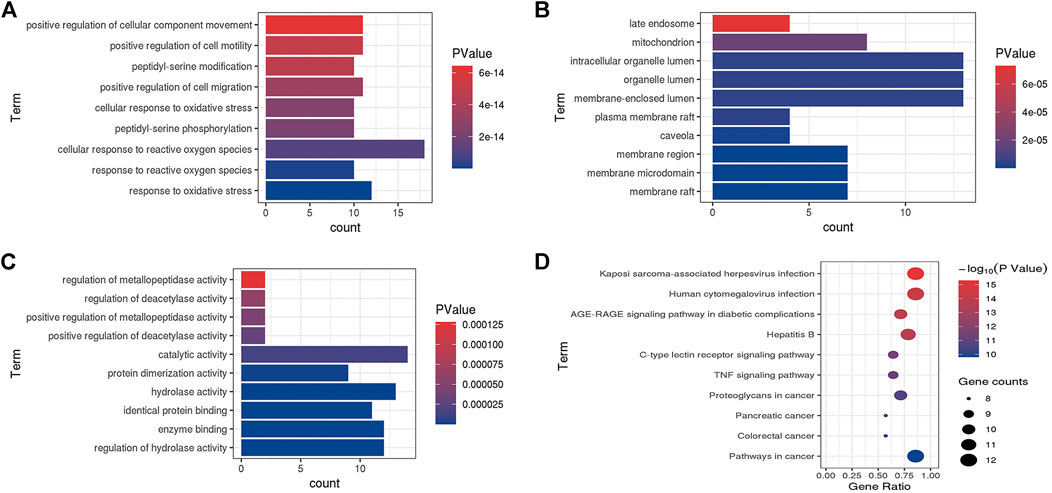
FIGURE 3. Go enriched terms of potential targets for biological processes (A), cellular components (B) and molecular functions (C) and KEGG enriched terms (D).
HE Staining, Immunohistochemistry and Isolation Purity
The third passage RA-FLS were subjected to HE staining and immunohistochemistry after being purified by passage culture. The cells were spindle shaped with blue nuclei and red cytoplasm (Figure 4A). The cytoplasm was brown with blue nuclei in immunohistochemistry, indicative of positive results (Figure 4B). Tissue origin, cell shape, and positive immunohistochemistry results demonstrated that the observed cells were RA-FLS. The purity of the isolation cells was 96.6 ± 1.44% (n = 4) marked with FITC anti-CD90 or 97.1 ± 0.39% (n = 4) APC anti-VCAM-1 respectively (Figures 4C,D).
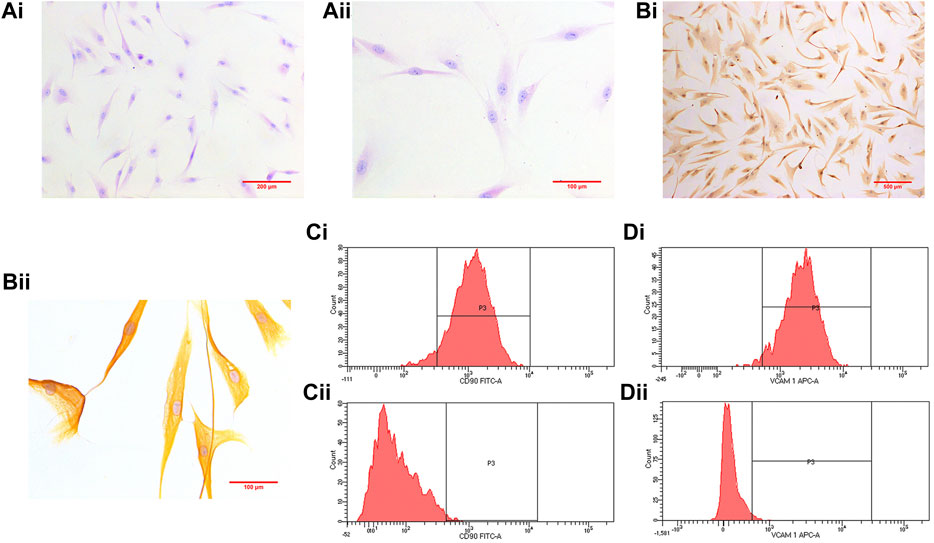
FIGURE 4. [(A), i] (×100) and [(A), ii] (×200) show HE staining results. Immunohistochemistry results are shown in [(B), i] (×100) and [(B), ii] (×400). [(C), i] shows the purity of the isolation cells marked with FIDC anti- CD90 (96.6 ± 1.44%; n = 4), and [(C), ii] represents control [(D), i] shows the purity of the isolation cells marked with APC anti-VCAM-1 (97.1 ± 0.39%; n = 4), and [(D), ii] represents control.
Effect of Kaempferol on RA-FLS Migration
The effects of control, DMSO, kaempferol, and lenalidomide treatments on cell migration are shown in Figure 4, respectively. The images are captured under ×100 objective lens. In comparison with the control treatment, DMSO treatment had no effect on cell migration, but kaempferol (25 μM) and lenalidomide (50 μM) treatments inhibited cell migration (p < 0.05) (Figure 5).
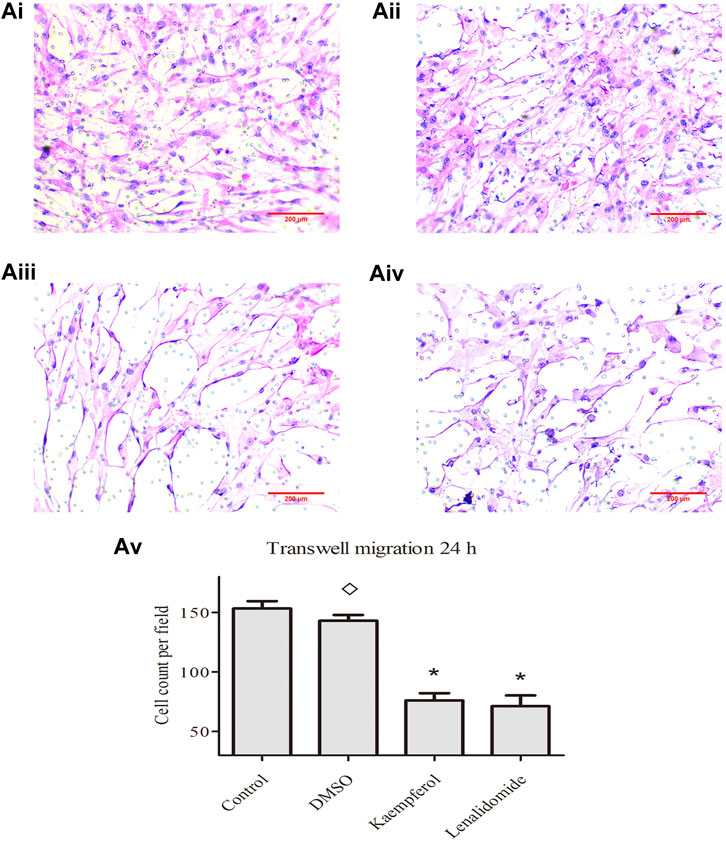
FIGURE 5. (A) i, ii, iii, and iv] show the effects of control, DMSO, kaempferol and lendidomide treatments on cell migration. Results in [(A), v] are shown as means ± standard deviation (SD; n = 3), *p < 0.05, ◊p > 0.05 vs. control treatement.
Impact of Kaempferol on Protein Expression of JUN, P-JUN, AKT1, P-AKT1, CASP3, TNFR1 and TNFR2
As shown in Figure 6A, no significant differences were observed in the levels of these proteins between DMSO and control treatment groups (p > 0.05). Stimulation with kaempferol (25 μM) or lenalidomide (50 μM) resulted in the upregulation of the expression of CASP3 and TNFR1 as compared with the control treatment (p < 0.05) but downregulated JUN, P-JUN, AKT1, P-AKT1, and TNFR2 expression (p < 0.05).
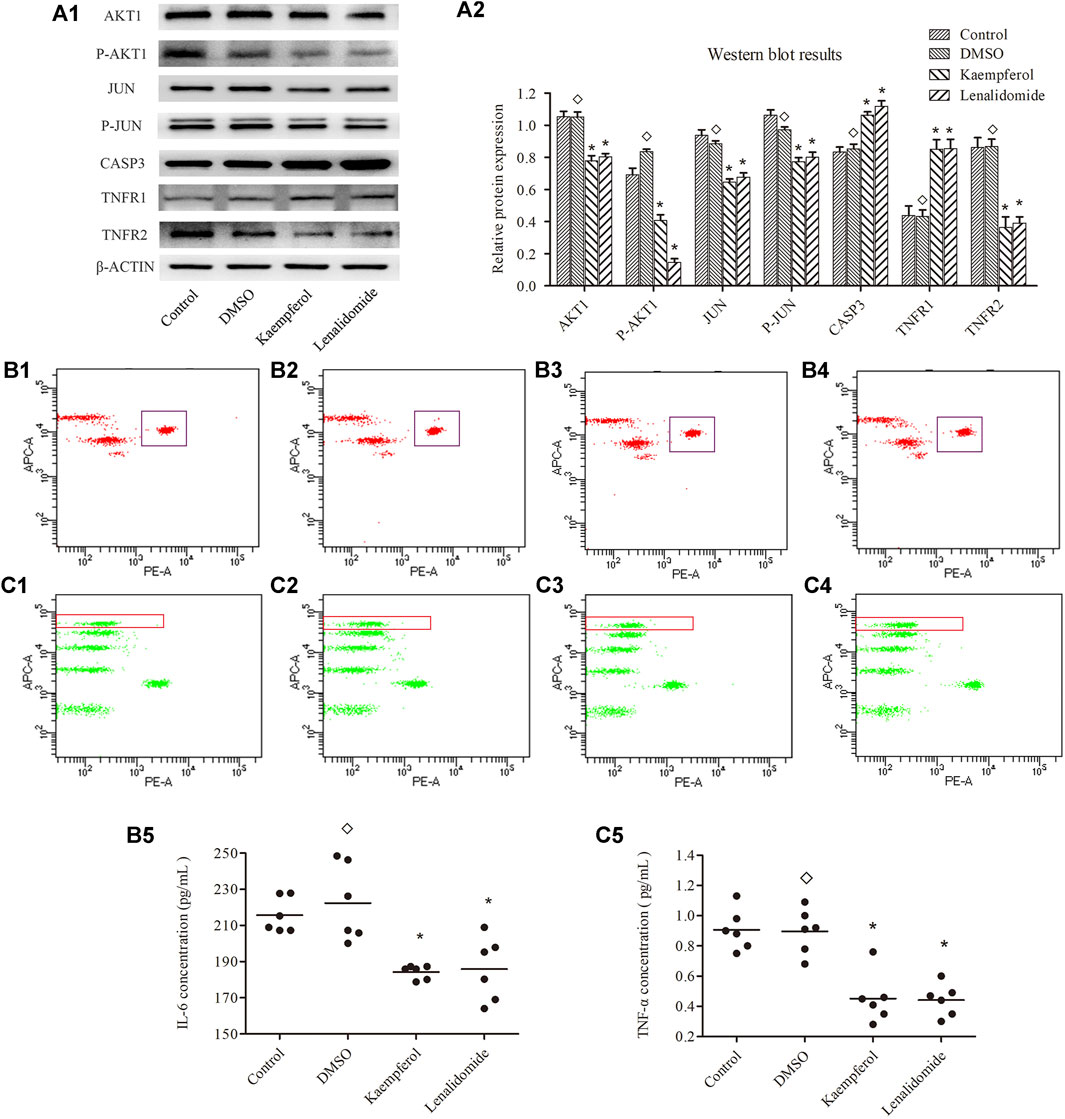
FIGURE 6. (A) shows the expression levels of JUN,P-JUN,AKTI,P-AKTI,CASP3, TNFR1 and TNFR2 in different groups (means ± SD; n = 3); p < 0.05, p > 0.05. as compard with the control group. (B) and (C) show the concentration of IL-6 and TNF-α, respectively.Ponits inside the rectangle represent IC-6 in B and TNF-α in C(B1, C1 represent contrpl group; B2, C2 represent DMSO grouop; B, C3 represent kaempferol group; B4, C4 represent lenalidomide group) Horizontal bars indicate meanns and n = 6; p < 0.05, p > 0.05 vs. control group.
Regulation of Kaempferol on Cytokines of TNF-α and IL-6
Flow cytometry assay results are shown in Figures 6B,C. The cytokines levels of TNF-α and IL-6 were not significantly different between DMSO and control treatment groups (p < 0.05). However, the levels of TNF-α and IL-6 were lower in kaempferol (25 μM) and lenalidomide (50 μM) treatment groups than in the control group (p < 0.05).
Discussion
Several clinical studies have proved Tripterygii Radix to be useful for RA treatment, but the underlying mechanism of action is unclear (Zhang et al., 2020). In this study, we predict the underlying mechanism of action by network pharmacology and further verified it with cell experiments. We screened 144 bioactive compounds from Tripterygii Radix depending on their OB and DL, and found kaempferol, beta-sitosterol, and aurantiamide acetate to be the most important active components, based on the degree values after the analysis of drug-compounds-biotargets-diseases network. Previous studies have shown all of these compounds to exert anti-inflammatory, anti-nociceptive, and immunomodulatory activities (Liu et al., 2015; Liu et al., 2019). In particular, kaempferol reduces the serum levels of IL-1β, IL-6, and TNF-α and inhibits cell proliferation through apoptosis and fibroblast growth factor receptor 3-ribosomal S6 kinase 2 (FGFR3-RSK2) signaling axis (Lee et al., 2018; Wang and Zhao, 2019).
Fourteen genes regarded as pivotal biological targets were shortlisted from the PPI network, including IL-6, TNF, AKT1, and JUN. IL-6 removes infectious agents and restore the damaged tissues through the activation of immune, hematological, and acute-phase responses (Tanaka et al., 2016). Human anti-IL-6 monoclonal antibody was shown to alleviate RA involving B cells and T cells, vascular endothelial growth factor (VEGF), and acute phase proteins (Narazaki et al., 2017). TNF induces apoptosis mediated by caspase activation and is highly responsive to osteoclasts in RA (Kiraz et al., 2016; McInnes and Schett, 2017). AKT1 regulates cell cycle and is associated with the downstream cellular mechanisms (Duggal et al., 2018). JUN influences apoptosis and macrophage activation in RA (Hannemann et al., 2017).
The outcomes of BP, CC, and MF included cellular response to reactive oxygen species, membrane microdomain, regulation of hydrolase activity, and others, indicating that it was a complex process related to organelles and cell membrane. KEGG pathway analysis indicated that AGE-RAGE signaling pathway in diabetic complications, C-type lectin receptor signaling pathway, and TNF signaling pathway were crucial signaling pathways related to RA treatment with the lowest p values. TNF signaling pathway mediates inflammatory responses and cell proliferation, differentiation, and death through the NF-κB and mitogen-activated protein kinase pathways (Borghi et al., 2019). TNFR1 and TNFR2 are two distinct receptors that accept exogenous signals in TNF signaling pathway (Jarosz-Griffiths et al., 2019). They both activate, either directly or indirectly, NF-κB and MAPKs (Noack and Miossec, 2017).
Therefore, TNF signaling pathway was thought to be one of the most latent treatment pathways, and some of the 14 biotargets (AKT1, JUN, CASP3, TNFR1, TNFR2, IL-6, TNF-α) were considered as potential target genes. Kaempferol was associated with the maximum number of potential target genes. Thus, kaempferol was deemed as the most valuable active component of Tripterygii Radix in RA, wherein it acts on the TNF-α signaling pathway. The cells from RA patients were verified to be RA-FLS. Kaempferol could inhibit RA-FLS migration and regulate the expression of AKT1, P-AKT1, JUN, P-JUN, CASP3, TNFR1, and TNFR2 proteins. The concentrations of IL-6 and TNF-α were reduced after intervention with kaempferol.
Kaempferol promoted TNFR1 expression, but inhibited TNFR2 expression. TNFR1’s cytoplasmic tail contains a death domain (DD), thereby allowing it to recruit the TNFR1-associated DD (TRADD); TNFR2, on the other hand, does not have an intracellular DD and recruits the TNFR associated factor (TRAF) 1 and 2 proteins instead (Holbrooket et al., 2019). Cell death can be initiated by TNFR1 through apoptosis (Zhu et al., 2019). TNFR2 interacts directly with TRAF1 or two promote cell survival signaling through NF-κB, MAPK, and Akt, promoting cell proliferation and tissue regeneration (Borghi et al., 2019).
In conclusion, kaempferol was found to be the most important active compound of Tripterygii Radix effective against RA. It acts through the TNF signaling pathway by modulating the expression of several biotargets (AKT1, JUN, CASP3, TNFR1, TNFR2, IL-6, TNF-α). Experiments with RA-FLS confirmed this effect. Further studies need to explore the therapeutic mechanism underlying Tripterygii Radix effects in RA.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethical Committee of The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YL conceived and designed this work. YL and NR performed the experiments. WM and XY supervised the study. YL and CC wrote and revised the entire manuscript. YL and HX collected. YL and PZ analysed the data.
Funding
This work was supported by the National Natural Science Foundation of China (81630103 and 81603388).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the central laboratory of The Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine for the platform and technical guidance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.639382/full#supplementary-material
References
Aletaha, D., Neogi, T., Silman, A. J., Funovits, J., Felson, D. T., Bingham 3rd, C. O., et al. (2010). American College of Rheumatology.2010ACR/EULAR Classification Criteafor Rhumatoid Artritis. Ann. Rheum. Dis. 69, 1580–1588. doi:10.1136/ard.2010.138461
Athanasios, A., Charalampos, V., Vasileios, T., and Ashraf, G. (2017). Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Cdm 18, 5–10. doi:10.2174/138920021801170119204832
Bao, J., and Dai, S. M. (2011). A Chinese Herb Tripterygium Wilfordii Hook F in the Treatment of Rheumatoid Arthritis: Mechanism, Efficacy, and Safety. Rheumatol. Int. 31, 1123–1129. doi:10.1007/s00296-011-1841-y
Borghi, A., Verstrepen, L., and Beyaert, R. (2016). TRAF2 Multitasking in TNF Receptor-Induced Signaling to NF-Κb, MAP Kinases and Cell Death. Biochem. Pharmacol. 116, 1–10. doi:10.1016/j.bcp.2016.03.009
Clapp, C., Adán, N., Ledesma-Colunga, M. G., Solís-Gutiérrez, M., Triebel, J., and Martínez de la Escalera, G. (2016). The Role of the Prolactin/vasoinhibin axis in Rheumatoid Arthritis: an Integrative Overview. Cell. Mol. Life Sci. 73, 2929–2948. doi:10.1007/s00018-016-2187-0
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 47, W357–W364. doi:10.1093/nar/gkz382
Davis, A. P., Grondin, C. J., Johnson, R. J., Sciaky, D., McMorran, R., Wiegers, J., et al. (2019). The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 47, D948–D954. doi:10.1093/nar/gky868
Duggal, S., Jailkhani, N., Midha, M. K., Agrawal, N., Rao, K. V. S., and Kumar, A. (2018). Defining the Akt1 Interactome and its Role in Regulating the Cell Cycle. Sci. Rep. 8, 1303. doi:10.1038/s41598-018-19689-0
Falconer, J., Murphy, A. N., Young, S. P., Clark, A. R., Tiziani, S., Guma, M., et al. (2018). Review: Synovial Cell Metabolism and Chronic Inflammation in Rheumatoid Arthritis. Arthritis Rheumatol. 70, 984–999. doi:10.1002/art.40504
Fransen, J., and van Riel, P. L. C. M. (2009). The Disease Activity Score and the EULAR Response Criteria. Rheum. Dis. Clin. North America 35, 745–757. doi:10.1016/j.rdc.2009.10.001
Hannemann, N., Jordan, J., Paul, S., Reid, S., Baenkler, H.-W., Sonnewald, S., et al. (2017). The AP-1 Transcription Factor C-Jun Promotes Arthritis by Regulating Cyclooxygenase-2 and Arginase-1 Expression in Macrophages. J.I. 198, 3605–3614. doi:10.4049/jimmunol.1601330
Holbrook, J., Lara-Reyna, S., Jarosz-Griffiths, H., and McDermott, M. F. (2019). Tumour Necrosis Factor Signalling in Health and Disease. F1000Res 8, 111. doi:10.12688/f1000research.17023.1
Hua, Y.-L., Ma, Q., Yuan, Z.-W., Zhang, X.-S., Yao, W.-L., Ji, P., et al. (2019). A Novel Approach Based on Metabolomics Coupled with Network Pharmacology to Explain the Effect Mechanisms of Danggui Buxue Tang in Anaemia. Chin. J. Nat. Medicines 17, 275–290. doi:10.1016/S1875-5364(19)30031-7
Huang, J., Cheung, F., Tan, H.-Y., Hong, M., Wang, N., Yang, J., et al. (2017). Identification of the Active Compounds and Significant Pathways of Yinchenhao Decoction Based on Network Pharmacology. Mol. Med. Rep. 16, 4583–4592. doi:10.3892/mmr.2017.7149
Jarosz-Griffiths, H. H., Holbrook, J., Lara-Reyna, S., and McDermott, M. F. (2019). TNF Receptor Signalling in Autoinflammatory Diseases. Int. Immunol. 31, 639–648. doi:10.1093/intimm/dxz024
Kiraz, Y., Adan, A., Kartal Yandim, M., and Baran, Y. (2016). Major Apoptotic Mechanisms and Genes Involved in Apoptosis. Tumor Biol. 37, 8471–8486. doi:10.1007/s13277-016-5035-9
Lee, C.-J., Moon, S.-J., Jeong, J.-H., Lee, S., Lee, M.-H., Yoo, S.-M., et al. (2018). Kaempferol Targeting on the Fibroblast Growth Factor Receptor 3-ribosomal S6 Kinase 2 Signaling axis Prevents the Development of Rheumatoid Arthritis. Cell Death Dis 9, 401. doi:10.1038/s41419-018-0433-0
Liang, Y., Zhang, X., Zou, J., Shi, Y., Wang, Y., Tai, J., et al. (2019). Pharmacology Mechanism of Flos Magnoliae and Centipeda Minima for Treating Allergic Rhinitis Based on Pharmacology Network. Drug Development Ind. Pharm. 45, 1547–1555. doi:10.1080/03639045.2019.1635150
Liu, L., Zhao, H., Sun, X., Zheng, Q., Luo, Y., Ru, Y., et al. (2018). Efficacy and Safety of Tripterygium Wilfordii Hook F for Chronic Urticaria: a Systematic Review and Meta-Analysis. BMC Complement. Altern. Med. 18, 243. doi:10.1186/s12906-018-2305-7
Liu, R., Hao, D., Xu, W., Li, J., Li, X., Shen, D., et al. (2019). β-Sitosterol Modulates Macrophage Polarization and Attenuates Rheumatoid Inflammation in Mice. Pharm. Biol. 57, 161–168. doi:10.1080/13880209.2019.1577461
Liu, X. B., Yang, B. X., Zhang, l., Lu, Y. Z., Gong, M. H., and Tian, J. K. (2015). An In Vivo and In Vitro Assessment of the Anti-inflammatory, Antinociceptive, and Immunomodulatory Activities of Clematis Terniflora DC. Extract, Participation of Aurantiamide Acetate. J. Ethnopharmacology 169, 287–294. doi:10.1016/j.jep.2015.04.009
Lv, Q.-w., Zhang, W., Shi, Q., Zheng, W.-j., Li, X., Chen, H., et al. (2015). Comparison of Tripterygium Wilfordii Hook F with Methotrexate in the Treatment of Active Rheumatoid Arthritis (TRIFRA): a Randomised, Controlled Clinical Trial. Ann. Rheum. Dis. 74, 1078–1086. doi:10.1136/annrheumdis-2013-204807
Ma, J.-D., Jing, J., Wang, J.-W., Yan, T., Li, Q.-H., Mo, Y.-Q., et al. (2019). A Novel Function of Artesunate on Inhibiting Migration and Invasion of Fibroblast-like Synoviocytes from Rheumatoid Arthritis Patients. Arthritis Res. Ther. 21, 153. doi:10.1186/s13075-019-1935-6
McInnes, I. B., and Schett, G. (2017). Pathogenetic Insights from the Treatment of Rheumatoid Arthritis. The Lancet 389, 2328–2337. doi:10.1016/S0140-6736(17)31472-1
Narazaki, M., Tanaka, T., and Kishimoto, T. (2017). The Role and Therapeutic Targeting of IL-6 in Rheumatoid Arthritis. Expert Rev. Clin. Immunol. 13, 535–551. doi:10.1080/1744666X.2017.1295850
Noack, M., and Miossec, P. (2017). Selected Cytokine Pathways in Rheumatoid Arthritis. Semin. Immunopathol. 39, 365–383. doi:10.1007/s00281-017-0619-z
Rappaport, N., Fishilevich, S., Nudel, R., Twik, M., Belinky, F., Plaschkes, I., et al. (2017). Rational Confederation of Genes and Diseases: NGS Interpretation via GeneCards, MalaCards and VarElect. Biomed. Eng. Online 16, 72. doi:10.1186/s12938-017-0359-2
Stanford, S. M., Aleman Muench, G. R., Bartok, B., Sacchetti, C., Kiosses, W. B., Sharma, J., et al. (2016). TGFβ Responsive Tyrosine Phosphatase Promotes Rheumatoid Synovial Fibroblast Invasiveness. Ann. Rheum. Dis. 75, 295–302. doi:10.1136/annrheumdis-2014-205790
Tanaka, T., Narazaki, M., Masuda, K., and Kishimoto, T. (2016). Regulation of IL-6 in Immunity and Diseases. Adv. Exp. Med. Biol. 941, 79–88. doi:10.1007/978-94-024-0921-5_4
Wan, Y., Xu, L., Liu, Z., Yang, M., Jiang, X., Zhang, Q., et al. (2019). Utilising Network Pharmacology to Explore the Underlying Mechanism of Wumei Pill in Treating Pancreatic Neoplasms. BMC Complement. Altern. Med. 19, 158. doi:10.1186/s12906-019-2580-y
Wang, J., and Zhao, Q. (2019). Kaempferitrin Inhibits Proliferation, Induces Apoptosis, and Ameliorates Inflammation in Human Rheumatoid Arthritis Fibroblast‐like Synoviocytes. Phytotherapy Res. 33, 1726–1735. doi:10.1002/ptr.6364
Weyand, C. M., and Goronzy, J. J. (2017). Immunometabolism in Early and Late Stages of Rheumatoid Arthritis. Nat. Rev. Rheumatol. 13, 291–301. doi:10.1038/nrrheum.2017.49
Zhang, D., Cao, Y., Zuo, Y., Wang, Z., Mi, X., and Tang, W. (2019). Integrated Bioinformatics Analysis Reveals Novel Hub Genes Closely Associated with Pathological Mechanisms of Immunoglobulin A Nephropathy. Exp. Ther. Med. 18, 1235–1245. doi:10.3892/etm.2019.7686
Zhang, D., Lyu, J.-t., Zhang, B., Zhang, X.-m., Jiang, H., and Lin, Z.-j. (2020). Comparative Efficacy, Safety and Cost of Oral Chinese Patent Medicines for Rheumatoid Arthritis: a Bayesian Network Meta-Analysis. BMC Complement. Med. Ther. 20, 210. doi:10.1186/s12906-020-03004-4
Zhang, W., Li, F., and Gao, W. (2017). Tripterygium Wilfordii Inhibiting Angiogenesis for Rheumatoid Arthritis Treatment. J. Natl. Med. Assoc. 109, 142–148. doi:10.1016/j.jnma.2017.02.007
Keywords: tripterygii radix, kaempferol, rheumatoid arthritis, TNF signaling pathway, molecular targets
Citation: Ling Y, Xu H, Ren N, Chen C, Zeng P, Lu D, Yao X and Ma W (2021) Prediction and Verification of the Major Ingredients and Molecular Targets of Tripterygii Radix Against Rheumatoid Arthritis. Front. Pharmacol. 12:639382. doi: 10.3389/fphar.2021.639382
Received: 17 December 2020; Accepted: 24 May 2021;
Published: 08 June 2021.
Edited by:
Thomas Vorup-Jensen, Aarhus University, DenmarkReviewed by:
Youjun Xao, The First Affiliated Hospital of Sun Yat-Sen University, ChinaGanesan Ramamoorthi, Moffitt Cancer Center, United States
Copyright © 2021 Ling, Xu, Ren, Chen, Zeng, Lu, Yao and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueming Yao, eXhtaW5nMTlAZm94bWFpbC5jb20=; Wukai Ma, d2Fsa2VyNTVAMTYzLmNvbQ==
 Yi Ling
Yi Ling Hui Xu2
Hui Xu2