- Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, India
Background:Curcuma spp. (Zingiberaceae) are used as a spice and coloring agent. Their rhizomes and essential oils are known for medicinal properties, besides their use in the flavoring and cosmetic industry. Most of these biological activities were attributed to volatile and nonvolatile secondary metabolites present in the rhizomes of Curcuma spp. The metabolite variations among the species and even cultivars need to be established for optimized use of Curcuma spp.
Objectives: We compared the phytochemical profiles of rhizomes and their essential oils to establish the variability among seven cultivars: five of Curcuma longa L. (Alleppey Supreme, Duggirala Red, Prathibha, Salem, Suguna) and two of C. aromatica Salisb. (Kasturi Araku, Kasturi Avidi). The GC-MS and LC-MS-based analyses were employed to profile secondary metabolites of these selected cultivars.
Methods: Rhizomes of Curcuma spp. were subjected to hydro-distillation to collect essential oil and analyzed by GC-MS. The methanol extracts of fresh rhizomes were subjected to LC-MS analyses. The compounds were identified by using the relevant MS library databases as many compounds as possible.
Results: The essential oil content of the cultivars was in the range of 0.74–1.62%. Several compounds were detected from the essential oils and rhizome extracts by GC-MS and LC-MS, respectively. Of these, 28 compounds (13 from GCMS and 15 from LCMS) were common in all seven cultivars, e.g., α-thujene, and diarylheptanoids like curcumin. Furthermore, a total of 39 new compounds were identified from C. longa L. and/or C. aromatica Salisb., most of them being cultivar-specific. Of these compounds, 35 were detected by GC-MS analyses of essential oils, 1,2-cyclohexanediol, 1-methyl-4-(1-methylethyl)-, and santolina alcohol, to name a few. The other four compounds were detected by LC-MS of the methanolic extracts of the rhizomes, e.g., kaempferol-3,7-O-dimethyl ether and 5,7,8-trihydroxy-2′,5′-dimethoxy-3′,4′-methylene dioxyisoflavanone.
Conclusions: We identified and recorded the variability in the metabolite profiles of essential oils and whole rhizome extracts from the seven cultivars of Curcuma longa L. and C. aromatica Salisb. As many as 39 new metabolites were detected in these seven Indian cultivars of Curcuma spp. Many of these compounds have health benefits.
Introduction
Turmeric (Curcuma longa L.) is a perennial rhizomatous herb that belongs to the family Zingiberaceae (Prasath et al., 2018). It has been used traditionally in India for its medicinal value and as a spice (Srinivasan et al., 2004; Aggarwal et al., 2007; Esatbeyoglu et al., 2012). In Ayurvedic medicine, turmeric is used internally (as a stomachic, tonic, and blood purifier) or externally (prevention and treatment of skin diseases) (Gounder and Lingamallu, 2012). Turmeric was scientifically validated for several pharmacological benefits, including antioxidant, anti-inflammatory, and chemoprotective properties (Miquel et al., 2002; Krup et al., 2013; Kanase and Khan, 2018; Umar et al., 2020). The rhizomes of turmeric are enriched with several bioactive metabolites, though the attention was mostly on curcuminoids. Besides curcumin (a curcuminoid), the essential oil of C. longa L. showed antimicrobial activity and ability to suppress aflatoxins production (Ferreira et al., 2013).
Out of 110 species of genus Curcuma, only ∼20 species were used so far for phytochemical studies (Nahar and Sarker, 2007). Curcuma longa L. is popularly known as turmeric, while C. aromatica Salisb. and C. caesia Roxb. are known as wild turmeric and black turmeric, respectively. C. longa L. and a few other species, including C. aromatica Salisb., produce curcumin, a yellow colored curcuminoid. So far, at least 235 compounds, primarily phenolics, terpenoids, and alkaloids, were identified from Curcuma spp. (Li et al., 2011). About 70 varieties of C. longa L. are cultivated in India (Sasikumar, 2005; Parthasarathy and Chempakam, 2008), but very few are chemically profiled.
The essential oil of Curcuma spp. is used in traditional medicine for many ailments (Dosoky and Setzer, 2018). The volatile component of C. longa’s rhizome is responsible for its aromatic flavor and odor (Gounder and Lingamallu, 2012). Its essential oil is considered safe for human use (Tisserand and Young, 2013). The oils of C. longa L. and C. aromatica Salisb. have applications in the food and pharmaceutical industries due to their antioxidant, antibacterial, and anti-inflammatory properties (Dosoky and Setzer, 2018). The essential oil also improved the bioavailability of curcumin, thereby its bioactivity (Shishu and Maheshwari, 2010). Preliminarily clinical trials indicated that the essential oil from C. longa L. and C. aromatica Salisb. was helpful against cancer, asthma, and other ailments (Cheng et al., 1999; Joshi et al., 2003; Li Y. et al., 2009). Thus, there is a need to identify high-yielding cultivars containing curcuminoids and essential oil.
Since the pharmacological properties of Curcuma spp. are dependent on their chemical profiles, studies on the chemical constituents of turmeric/wild turmeric and their essential oils gained significance. Thin-layer chromatography (TLC) is one of the methods employed to quantify curcumin (Setyaningsih et al., 2016) and other curcuminoids (Phattanawasin et al., 2009) in Curcuma longa L. A few different techniques used were HPTLC (Pathania et al., 2006; Paramasivam et al., 2009), nuclear magnetic resonance (NMR) spectroscopy (Li W. et al., 2009), and the HPLC method (Kulyal et al., 2016).
Our present study on metabolite profiles would pave the way for metabolomics by providing the identity of several metabolites. Metabolomics is a practical approach for the comprehensive profiling and comparison of metabolites in plant systems (De Vos et al., 2007). It is crucial for quality evaluation and scientific validation of medicinal plants and their products (Mukherjee et al., 2016). Mainly information on secondary metabolites of medicinal plants/spices is of great importance in health, food, and nutrition sectors, due to the antioxidant nature, color, or flavor of these secondary compounds (Beekwilder et al., 2005; Dixon et al., 2006; Hall, 2006). The quality of turmeric and other spices depends on factors, such as cultivation, collection, storage, milling, and processing, apart from genetics and adulteration issues. Therefore, metabolomics provides a practical approach for quality control (Mukherjee et al., 2016; Tetali et al., 2021).
Over the past decade, several methods suitable for large-scale analysis of metabolites in plant extracts were developed (Dixon et al., 2006; Hall, 2006). However, to date, no single analytical method can successfully detect the entire metabolome of higher plants, especially of medicinal and aromatic plants, as they are highly rich in chemically diverse metabolites (Tetali et al., 2021). The GC-MS and LC-MS techniques mutually complement each other in unraveling secondary metabolomes comprising a wide range of volatile and nonvolatile compounds. These compounds belonged to terpenes, phenolic acids, phenylpropanoids, saponins, alkaloids, polyamines, and their derivatives (Huhman and Sumner, 2002; Moco et al., 2006).
Essential oils from different Curcuma species, including C. longa L. and C. aromatica Salisb, were studied for their chemical constituents (Choudhury et al., 1996; Angel et al., 2014; Nampoothiri et al., 2015; Dosoky and Setzer, 2018) to establish their variability. Variation in the volatile compositions of Curcuma spp. such as C. longa L. and C. zedoaria, was done using GC-MS (Dosoky et al., 2019). A combination of GC-MS and LC-MS techniques was used for metabolite analysis of C. domestica L. (C. longa L.) (Herebian et al., 2009). In the present study, the volatile (essential oil) and nonvolatile (total extract) components of the fresh rhizome of the seven cultivars of Curcuma spp. were analyzed by the GC-MS and LC-MS techniques. The present study is the first report revealing such detailed metabolite profiles of the selected cultivars to the best of our knowledge. These cultivars, except Alleppey Supreme, are typically cultivated in Telangana and Andhra Pradesh, and these states are among the largest producers of turmeric in India (Parthasarathy and Chempakam, 2008).
Most of the studies worldwide on Curcuma spp., for their curative properties, were with C. longa L., followed by C. aromatica Salisb, C. aeruginosa Roxb. (Simoh and Zainal, 2015), and C. kwangsiensis S. K. Lee & C. F. Liang (Zeng et al., 2009). Several cultivars exist within these species, which vary in their chemical profiles. The present article is the first attempt to characterize both volatile (essential oil) and nonvolatile (crude extract) components of fresh rhizomes of seven cultivars of Curcuma spp. by the GC-MS and LC-MS techniques. Our results using GC-MS and LC-MS analyses revealed high variability in their metabolite profiles of seven cultivars of genus Curcuma. We emphasize that such an approach could be exploited to distinguish cultivars for a specific application based on their metabolite profile.
Materials and Methods
Materials and Reagents
LC-MS grade methanol, water, and acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA, United States). Ammonium formate, formic acid, 4-fluoro-4′-hydroxy benzophenone (97%), and n-hexane were from Sigma-Aldrich, India. Anhydrous sodium sulfate (99.99%) was from Merck Millipore, India.
Fresh rhizomes of four cultivars of Curcuma longa L. (Duggirala Red, Prathibha, Salem, and Suguna) and two cultivars of C. aromatica Salisb. (Kasturi Araku and Kasturi Avidi) were collected from Turmeric Research Station, Kammarpally, Telangana State, India. Alleppey Supreme cultivar of C. longa L. was from the Indian Institute of Spices Research, Marikunnu (IISR) Kozhikode, Kerala, India. The mature rhizome samples were collected during the postharvest season of turmeric (May–Jun) in 2011 and 2012 and cryopreserved at −80°C until extraction and analysis.
Isolation of Essential Oil by Hydrodistillation for GC-MS Analysis
50 g each of fresh turmeric rhizome of five cultivars of C. longa L. cvs. Alleppey Supreme, Duggirala Red, Prathibha, Salem, Suguna, and two cultivars of C. aromatica Salisb. cvs. Kasturi Araku and Kasturi Avidi were taken out from a −80°C freezer, made into pieces, and ground in a pestle with a mortar to a fine powder under liquid nitrogen. The powder was subjected to hydrodistillation in a Clevenger-type apparatus for 7 h. The essential oil obtained after distillation was dried over anhydrous sodium sulfate and kept at −80°C until GC-MS analysis.
GC-MS Running Conditions and Metabolite Identification
The chemical composition of the Curcuma spp. essential oil was analyzed by the GC-MS technique using Agilent 7890 A gas chromatograph coupled with a Leco Pegasus HT TOF mass spectrometer equipped with a 29.8 m × 320 µm HP-5MS 5% phenyl methyl siloxane capillary column with 0.25 µm film thickness. The oven temperature was programmed at 65°C for 2 min and then increased from 65 to 90°C at 5°C/min (held for 3 min). Then the temperature was increased from 90 to 103°C (held for 3 min) and from 103 to 150°C (held for 15 min) at 20°C/min and 8°C/min, respectively. The temperature was raised finally from 150 to 280°C at 20°C/min. The injector, interphase, and ion source were maintained at 250°C, 280°C, and 250°C, respectively. The detector voltage was 1500 V. A solvent delay of 2 min was selected. One microliter (diluted with n-hexane; 1:10) of essential oil sample was injected into the GC-MS system using split mode (50: 1). Helium was used as a carrier gas at a flow rate of 1 ml/min. GC-MS data were measured at 70 eV; mass scan 40–1000 amu.
The compounds were identified by comparing their mass spectra with the data available in the literature, National Institute of Standards Technology NIST, and Leco-Fiehn Rtx5 libraries. The compounds originated from the GC-MS data file were identified by matching most resembling spectra with the NIST library. Each search produced a hit list of compounds according to match factor or similarity with the library spectra. All the compounds showing similarity more than 70% with the NIST library were selected by the software (Software: Version 4.22 optimized for Pegasus®). Software searches (identifies) compound from their mass spectra and includes MS interpretation programs for analyzing mass spectra based on chemical structure, molecular formula, isotopic pattern, etc. The similarity of 70% or above between the m/z values of the compound detected in the respective cultivar and the MS-libraries’ mass fragmentation pattern was considered as identification. Furthermore, mass spectra of all compounds were also matched with ranges available as per their CAS number. Compounds for which the CAS number was not generated, the PubChem CID was used. Compounds below the similarity level of 70% were not considered and grouped as unknown. The data obtained with the samples collected in 2012 are presented in this article.
Preparation of Rhizome Extracts for LC-MS Analysis
Samples for LC-MS analysis were prepared by grinding the fresh rhizome to a fine powder in a mortar and pestle under liquid nitrogen. 1 g of the rhizome powder was suspended in 2 ml of MeOH (LC-MS grade). The samples were sonicated for 30 min and centrifuged for 25 min at 1500 rpm, and the supernatants were separated by filtering through a 0.45-µm Nylon filter disk. These extracts were freshly prepared for the analysis. A 200 µl aliquot of the extract was diluted quantitatively with internal standard (IS) 200 µl 4-fluoro-4′- hydroxy benzophenone solution. It was prepared freshly for each analysis by dissolving in methanol for a final concentration of 0.58 mg/ml. The samples were subjected to LC-MS analysis for the complete metabolite profile. The data obtained with the samples collected in 2012 were presented in this article.
LC-MS/MS Conditions and Metabolite Identification
LC-MS analyses of the crude extract of fresh rhizome of Curcuma spp. were performed according to Jiang et al. (2006) using Agilent 6520 Accurate Q-TOF (Agilent Santa Clara, CA), and the column used was Zorbax Eclipse XDB-C 18, 4.6 × 50 mm, 1.8 µ; Mobile phase: A) buffer (5 mM ammonium formate, 0.1% formic acid, in deionized and distilled H2O) and B) acetonitrile; gradient (in buffer A): 0–2 min, 5% B; 2–57 min, 5–100% B; 57–60 min, 100% B; 60–65 min, 100–5% B; flow rate: 0.25 ml/min; temperature, 40oC; injection volume 5 µl. For the MS detection, Agilent MSD-Trap-SL was equipped with electrospray ionization (ESI) interface as the ion source. The acquisition parameters for the negative mode were: drying N2 temperature, 350oC, 8 l/min; nebulizer pressure 40 psi; HV capillary 4000 V; skimmer 65.0 V; mass range measured: 110–1700 m/z; Spray voltage: 4 kV; scan rate 1.4. We analyzed the results in both the positive and the negative ion mode acquired by Agilent TOF/Q-TOF mass spectrometry and full MS scan, in the form of total ion current (TIC) chromatogram, and the metabolites were identified based on their MS/MS spectra and fragmentation rules reported previously (Jiang et al., 2006).
Results
Essential Oil Content
The oil was obtained by hydro-distillation, in a Clevenger-type apparatus, of the fresh rhizomes of five cultivars (Alleppey Supreme, Duggirala Red, Prathibha, Salem, and Suguna) of Curcuma longa L. and two cultivars (Kasturi Araku and Kasturi Avidi) of C. aromatica Salisb. The yield of essential oil from the seven cultivars was in the range of 0.74–1.62% on a fresh weight basis, with the highest yield of 1.62% in cv. Kasturi Avidi (C. aromatica Salisb.) followed by cv. Alleppey Supreme (C. longa L.) with an amount of 1.42% and the lowest yield of 0.74% in cv. Duggirala Red (C. longa L.). The essential oil yields from the other five rhizomes were in between these values (Table 1). The oil yields of C. longa L. varieties were higher than those of C. aromatica Salisb.
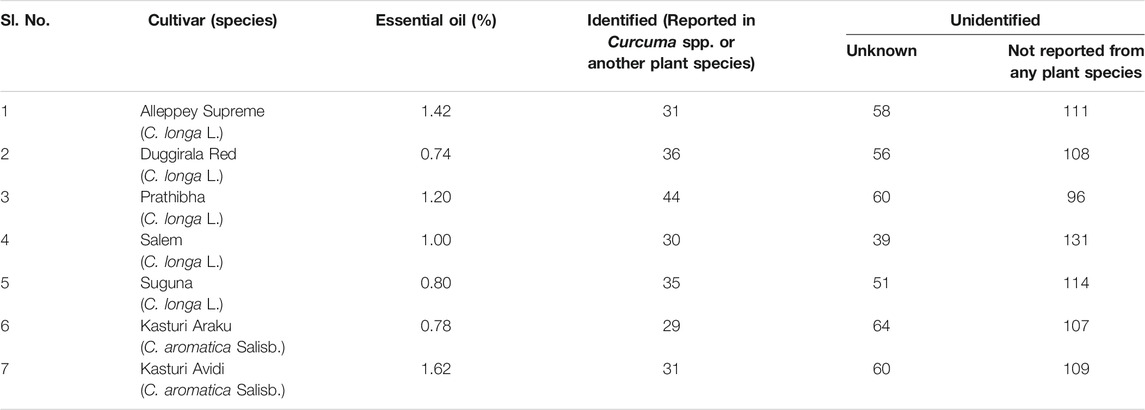
TABLE 1. Essential oil content and total number of compounds detected by GC-MS in the rhizomes of Curcuma species.
GC-MS Analysis of Essential Oil
Essential oils of seven cultivars of Curcuma spp. were subjected to GC-MS analysis, and the results from one of such studies for each cultivar are presented in this article. The representative TIC chromatograms of these cultivars are shown in Figure 1. Several compounds were detected in each cultivar’s essential oil (Table 1). Only a few of the identified compounds were confirmed based on their match with the compound profiles found in the NIST databases and Leco-Fiehn Rtx5 library. Up to 44 compounds were identified from the five cvs. of C. longa L. and 31 compounds from two cvs. of C. aromatica Salisb. (Table 1). Altogether 80 compounds were grouped into three categories: cultivar-specific (41), present in more than one cultivar (26), and common in all seven cultivars (13).
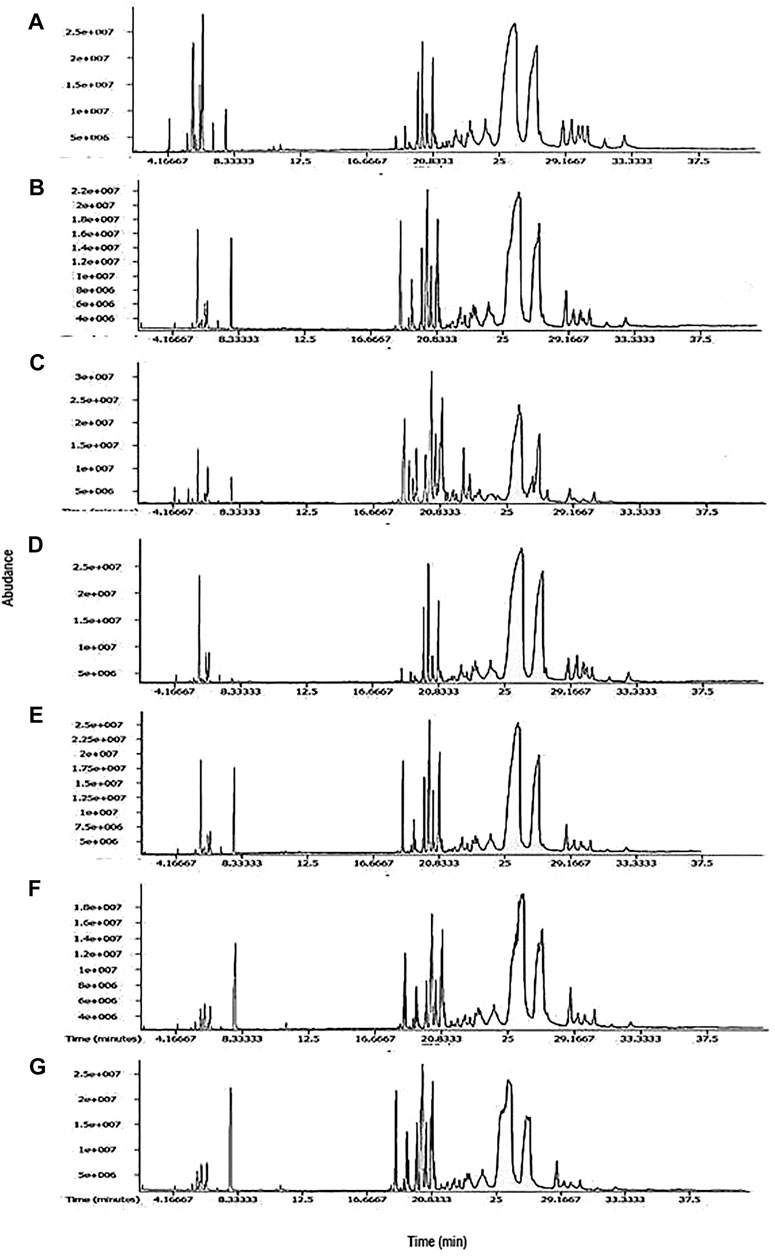
FIGURE 1. Representative TIC chromatograms from GCMS of essential oil from cultivars (A) Alleppey Supreme, (B) Duggirala Red, (C) Prathibha, (D) Salem, (E) Suguna of Curcuma longa L. and cvs. (F) Kasturi Araku, (G) Kasturi Avidi of C. aromatica Salisb.
These 41 cultivar-specific compounds were detected in the essential oil of one of the cultivars of C. longa L. or C. aromatica Salisb. (Table 2). The essential oil of C. longa L. cv. Prathibha had the highest number of cultivar-specific compounds, whereas C. aromatica Salisb., cv. Kasturi Araku had the least (Table 2). Of these, 23 cultivar-specific compounds were reported for the first time from the genus Curcuma. The chemical structures of these 23 compounds (Table 2, Sl. Nos. 1–23) are presented in Figure 2 (panels 1–23). In addition to 41 cultivar-specific compounds, 26 compounds were present in more than one cultivar (Table 3). Among these, 12 were detected first time in the genus Curcuma (Table 3, Sl. Nos. 1 to 12; Figure 2, panels: 24–35). The remaining 14 were already known in C. longa L. A total of 13 compounds were common in all seven cultivars of C. longa L. and C. aromatica Salisb. (Table 4) and the representative structures of two of these compounds are given in Figure 3 (panel numbers 14–15, corresponding to serial numbers 10 and 13 respectively of Table 4). Most of these compounds belong to mono, di, and sesquiterpene. A summary of all 80 compounds identified by GC-MS in the essential oils of the seven cultivars is presented in Supplementary Table S1.
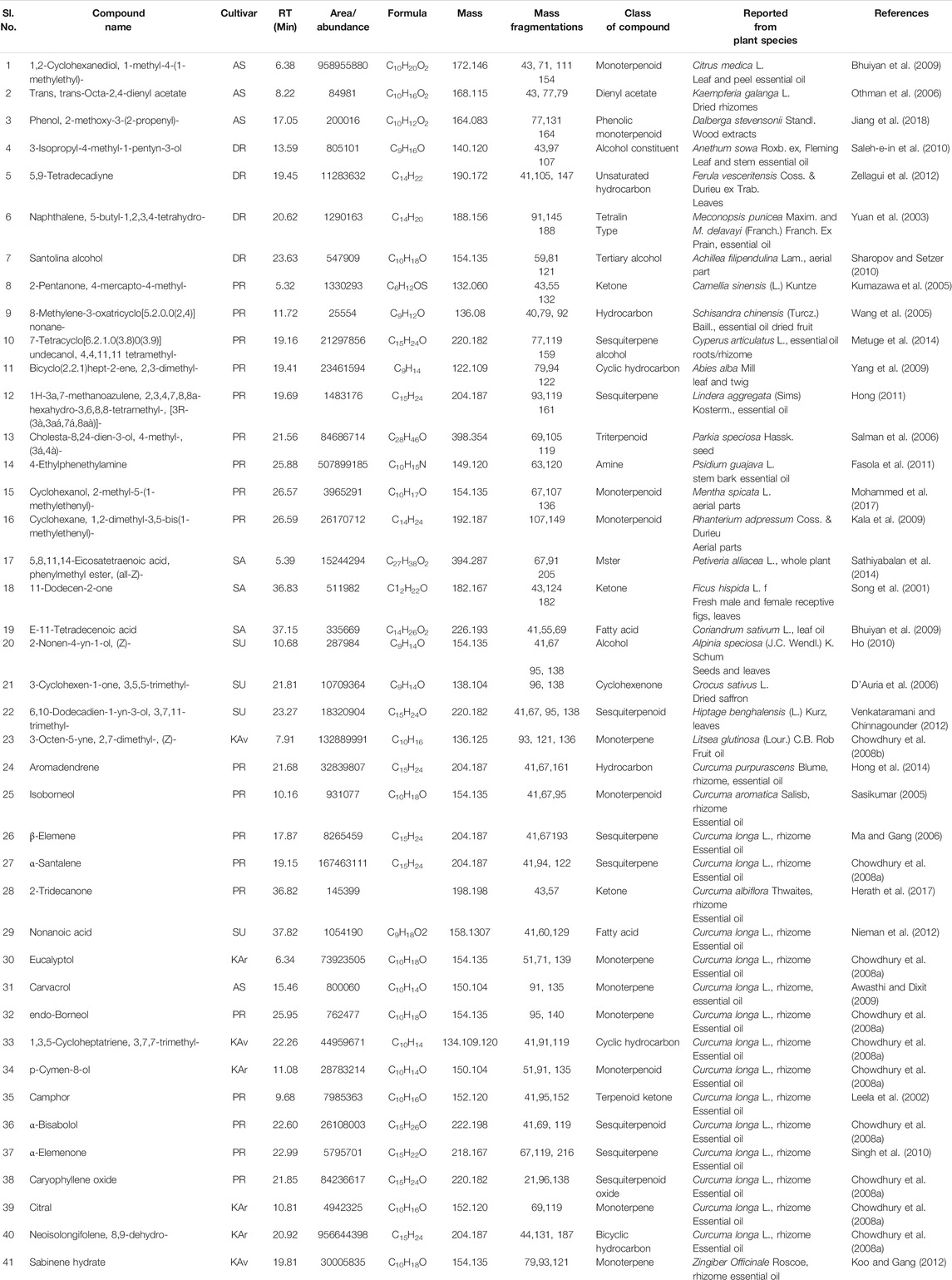
TABLE 2. Cultivar-specific compounds identified, in one of the seven cultivars of Curcuma longa L. or C. aromatica Salisb. by GC-MS in the essential oil from rhizomes. The structures of the compounds (serial numbers from 1 to 23) are given in Figure 2 (panel numbers: 1–23), and this is the first report of these compounds from the genus Curcuma L. These compounds, however, were reported from genus other than Curcuma L. The compounds from serial numbers 24 to 41 are already reported in Curcuma species. Structures for few compounds (serial numbers 24–30) are given in Figure 3 (panel numbers: 1–7). Abbreviations used: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
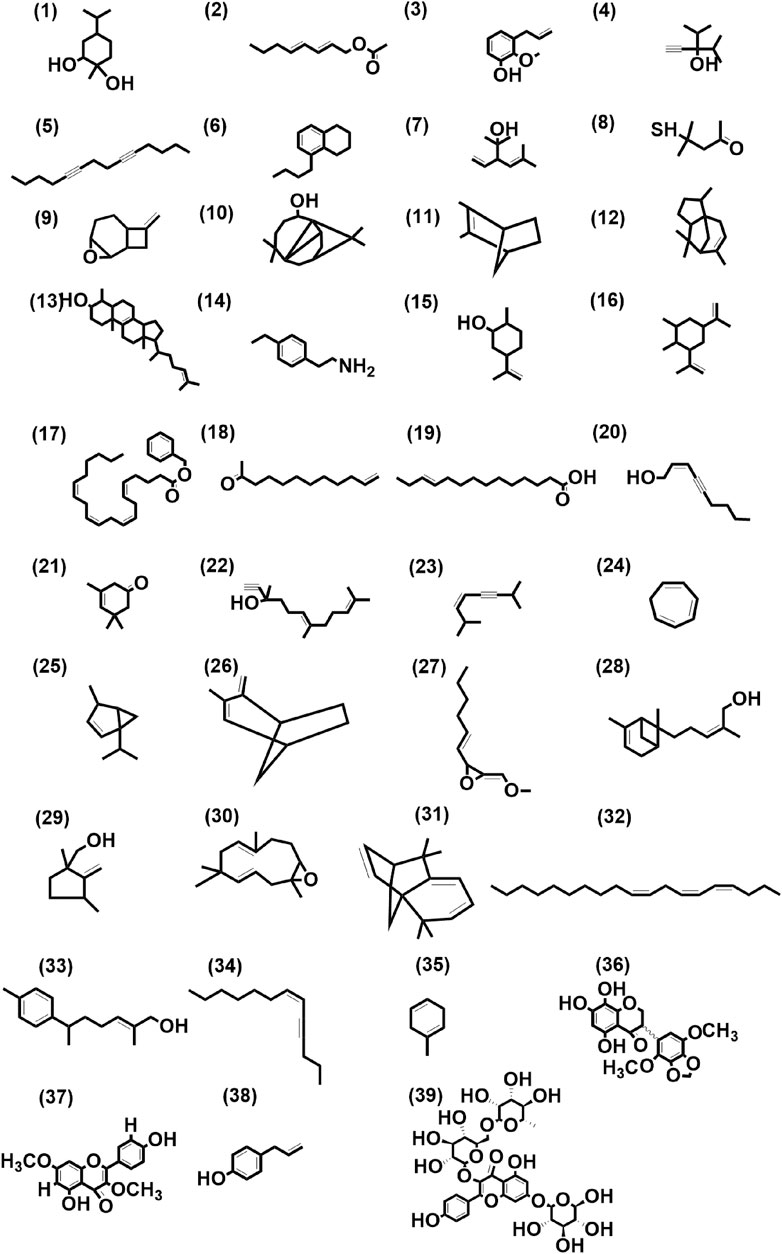
FIGURE 2. Structures of first-time reported (total 39) from the genus Curcuma, identified from cultivars of Curcuma longa L. and C. aromatica Salisb. detected in essential oil (panel numbers 1–23 and 24–35 corresponding to serial numbers 1–23 and 1–12 of Tables 2, 3 respectively) and rhizome extracts (panel numbers 36 and 37–39 corresponding to serial numbers 1 and 1–3 of Tables 6, 7 respectively) by GC-MS and LC-MS, respectively. Details of all these compounds are given in Supplementary Tables S3, S4. (1) 1,2-Cyclohexanediol, 1-methyl-4-(1-methylethyl)-, (2) trans, trans-octa-2,4-dienyl acetate, (3) phenol, 2-methoxy-3-(2- propenyl)-, (4) 3-isopropyl-4-methyl-1-pentyn-3-ol, (5) 5,9-tetradecadiyne, (6) naphthalene, 5-butyl-1,2,3,4-tetrahydro-, (7) santolina alcohol, (8) 2-pentanone, 4-mercapto-4-methyl-, (9) 8-methylene-3-oxatricyclo[5.2.0.0(2,4)]nonane, (10) 7-tetracyclo[6.2.1.0(3.8)0(3.9)]undecanol, 4,4,11,11 tetramethyl-, (11) bicyclo(2.2.1)hept-2-ene, 2,3-dimethyl-, (12) 1H-3a,7-methanoazulene, 2,3,4,7,8,8a-hexahydro-3,6,8,8-tetramethyl-, [3R-(3à,3aá,7á,8aà)]-, (13) cholesta-8,24-dien-3-ol, 4-methyl-, (3á,4à)-, (14) 4-ethylphenethylamine, (15) cyclohexanol, 2-methyl-5-(1-methylethenyl)-, (16) cyclohexane, 1,2-dimethyl-3,5-bis(1-methylethenyl)-, (17) 5,8,11,14-eicosatetraenoic acid, phenylmethyl ester, (all-Z)-, (18) 11-dodecen-2-one, (19) E-11-tetradecenoic acid, (20) 2-nonen-4-yn-1-ol, (Z)-, (21) 3-cyclohexen-1-one, 3,5,5-trimethyl-, (22) 6,10-dodecadien-1-yn-3-ol, 3,7,11-trimethyl-, (23) 3-octen-5-yne, 2,7-dimethyl-, (Z)-, (24) 1,3,5-cycloheptatriene, (25) bicyclo(3.1.0)hexane, 4-methyl-1-(1-methylethyl)-, didehydro deriv., (26) bicyclo(3.2.1)oct-2-ene, 3-methyl-4-methylene-, (27) oxirane, 2-(hexyn-1-yl)-3-methoxymethylene-, (28) bergamotol, Z-α-trans-, (29) (1,3-dimethyl-2-methylene-cyclopentyl)-methanol, (30) 12-oxabicyclo(9.1.0)dodeca-3,7-diene, 1,5,5,8-tetramethyl-, [1R-(1R*,3E,7E,11R*)]-, (31) isolongifolene, 4,5,9,10-dehydro-, (32) Z,Z,Z-4,6,9-nonadecatriene, (33) 6-(p-tolyl)-2-methyl-2-heptenol, (34) 6-tridecen-4-yne, (Z)-, (35) 1,4-cyclohexadiene, 1-methyl-, (36) kaempferol-3,7-O-dimethyl ether, (37) 5,7,8-trihydroxy-2′,5′-dimethoxy-3′,4′-methylene dioxyisoflavanone, (38) chavicol, (39) kaempferol-3-O-rutinoside-7-O-glucoside.
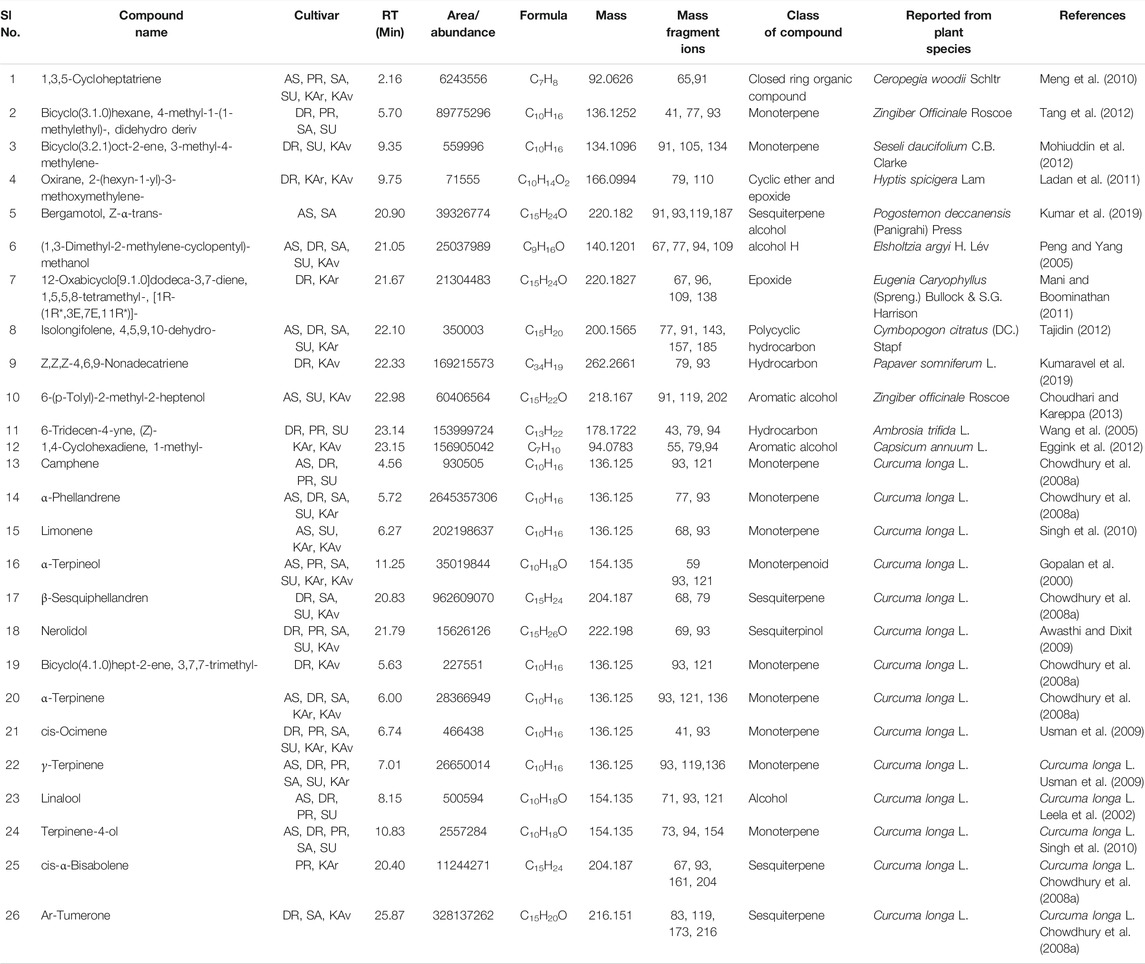
TABLE 3. Compounds detected in more than one cultivar of C. longa L. and C. aromatica Salisb. identified by GCMS in the essential oil from rhizomes. The structures of the compounds from serial numbers 1–12 are given in Figure 2 (panel numbers: 24–35), and the compounds with Sl.No. 13–18 are shown in Figure 3, with corresponding panel numbers: 8–13 respectively. Abbreviations used: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
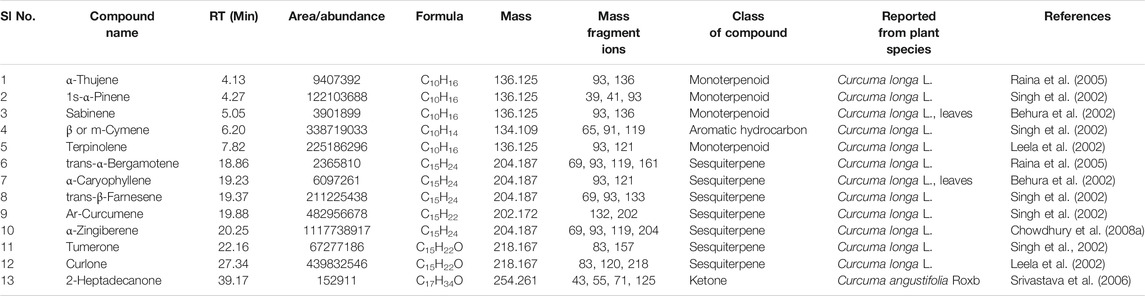
TABLE 4. Compounds common in the seven cultivars of Curcuma spp detected by GC-MS in essential oil obtained from rhizomes. The structures of the two compounds with serial numbers 10 and 13 are given in Figure 3 (panel numbers: 14–15 respectively).
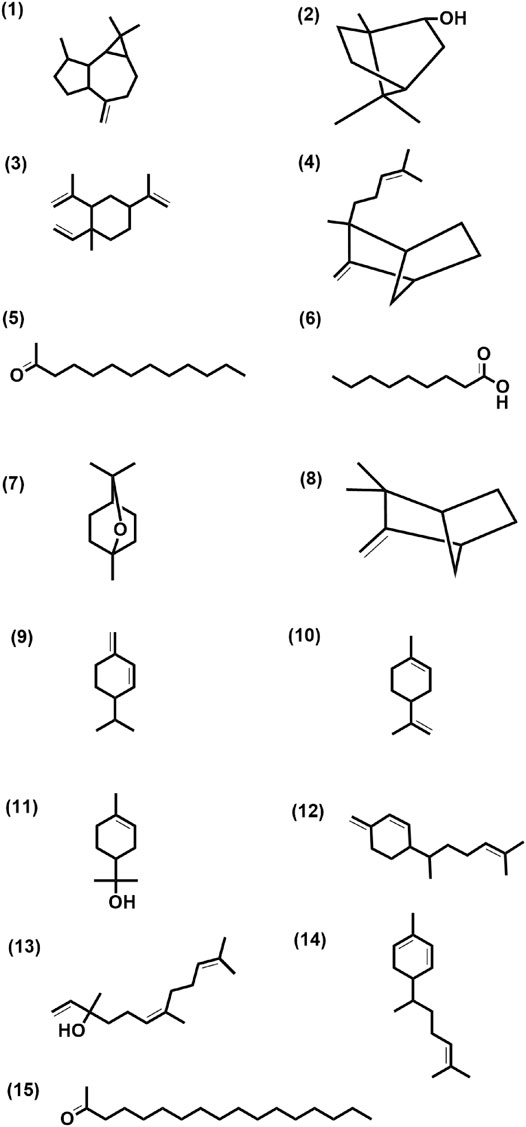
FIGURE 3. Structure of few selected cultivar-specific (panel numbers 1–7 corresponding to serial numbers 24–30 of Table 2); detected in more than one cultivar (panel numbers 8–13 corresponding to serial numbers 13–18 of Table 3) and common (panel numbers 14–15 corresponding to serial numbers 10 and 13 of Table 4) compounds already reported from the genus Curcuma in essential oils isolated from rhizomes by GCMS analysis of seven cultivars of Curcuma L.: five of Curcuma longa L. (cvs. Alleppey Supreme, Duggirala Red, Prathibha, Salem, and Suguna) and two of C. aromatica Salisb. (cvs. Kasturi Araku and Kasturi Avidi). (1) Aromadendrene, (2) isoborneol, (3) β-elemene, (4) α-santalene, (5) 2-tridecanone, (6) nonanoic acid, (7) eucalyptol, (8) camphene, (9) α-phellandrene, (10) limonene, (11) α-terpineol, (12) β-sesquiphellandren, (13) nerolidol, (14) α-zingiberene, (15) 2-heptadecanone.
LC-MS Analysis of Methanol Extracts
Methanolic extracts of rhizomes from the seven cultivars of Curcuma spp. were subjected to LC-MS analysis. The results from one of such analyses for each cultivar are presented in this article. The use of “positive” and “negative” modes of LC-MS was quite helpful. TIC chromatograms of all seven cultivars of Curcuma longa L. and C. aromatica Salisb. were shown in Supplementary Figure S1A for negative mode and Supplementary Figure S1B for positive mode.
A typical LC-MS analysis of methanolic extracts from rhizomes of C. longa L. cv. Alleppey Supreme revealed the presence of up to 86 compounds. Out of these, 43 were identified, and the remaining 43 compounds remained unknown. The (-) ESI-LC-MS detected 30 known compounds, and the (+) ESI-LC-MS detected 23 known compounds with an overlap of 10 compounds, detected by both negative and positive ion modes. A similar assessment of data was done with all seven cultivars of Curcuma spp. (Table 5). Altogether 62 compounds were identified, as presented in Supplementary Table S2. These compounds were grouped into three categories: cultivar-specific, detected in more than one cultivar, and common. There were 23 cultivar-specific compounds present in any one cultivar of C. longa L. or C. aromatica Salisb. (Table 6). 24 compounds were present in more than one cultivar of C. longa L. and/or C. aromatica Salisb. (Table 7). The remaining 15 were common in all seven cultivars (Table 8). Of these 15 common compounds found in the LC-MS/MS chromatograms, only one was a “Bisabolane” sesquiterpene (Parthasarathy et al., 2009) and all other 14 were diarylheptanoids. These were identified based on the MS/MS spectra reported by Jiang et al. (2006), including curcumin (CU), demethoxycurcumin (DMC), and bisdemethoxycurcumin (BDMC). Among the other diarylheptanoids, 1-(4-hydroxyphenyl)-7-(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one; 1,5-bis(4-hydroxy-3-methoxyphenyl)-1,4-pentadien-3-one, etc., were common in all the cultivars of C. longa L. and C. aromatica Salisb. (Table 8). The structure of the five common compounds was given in Figure 4 (panels 13, 14–15, 16, and 17 corresponding to serial numbers 10, 5–6, 15, and 1, respectively, of Table 8). In addition to diarylheptanoids, several other classes (phenolic acids, flavonoids, ketonic sesquiterpenes, and fatty acid derivatives) were also detected in the turmeric rhizomes.
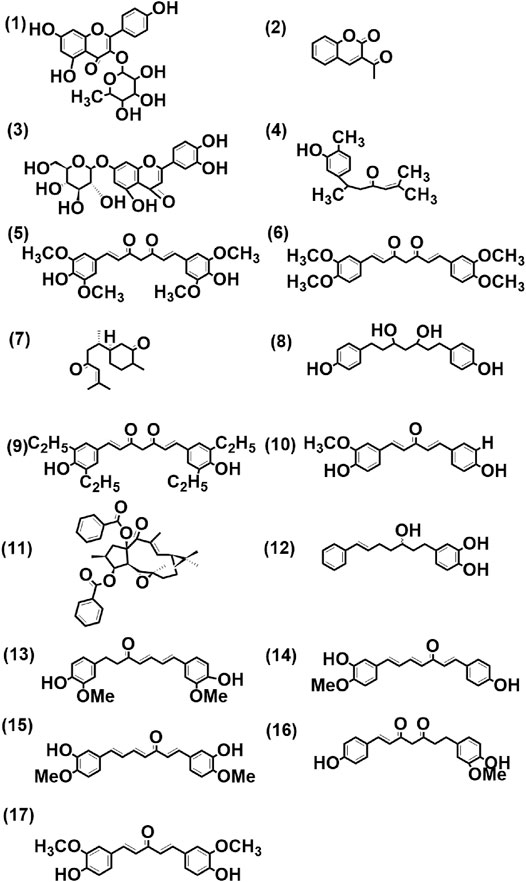
FIGURE 4. Structure of few selected cultivar-specific (panels 3, 5–11 corresponding to serial numbers 2, 3–9 of Table 6); detected in more than one cultivar (panels 1–2, 4, 12 corresponding to serial numbers 4–5, 6, 7 of Table 7) and common (panels 13, 14–15, 16, and 17 corresponding to serial numbers 10, 5–6, 15, and 1, respectively, of Table 8) compounds already reported from genus Curcuma in methanolic extract from rhizomes by LCMS analysis of seven cultivars of Curcuma spp.: five of Curcuma longa L. (cvs. Alleppey Supreme, Duggirala Red, Prathibha, Salem, and Suguna) and two of C. aromatica Salisb. (cvs. Kasturi Araku and Kasturi Avidi). (1) Kaempferol-3-rhamnoside, (2) 3-acetyl coumarin, (3) luteolin-7-O-glucoside, (4) turmeronol, (5) 1,7-bis(4-hydroxy-3,5-dimethoxyphenyl)-1,6-heptadiene-3,5-dione, (6) 1,7-bis(3,4-dimethoxyphenyl)-1,6-heptadiene-3,5-dione, (7) (6S)-2-methyl-6-[(1R,5S)-(4-methene-5-hydroxyl-2-cyclohexen)-2-hepten-4-one, (8) 1,7-bis(4-hydroxyphenyl)-3,5-heptanediol, (9) 1,7-bis(3,5-diethyl-4-hydroxyphenyl)-1,6-heptadiene-3,5-dione, (10) 1-(4-hydroxy-3-methoxyphenyl)-5-(4-hydroxyphenyl)-1,4-pentadiene-3-one, (11) (-)-(12E,2S,3S,4R, 5R,6R, 9S,11S, 15R)-3,15-dibenzoyloxy-5,6-epoxylathyr-12-en-14-one, (12) 7-(3,4-dihydroxyphenyl)-5-hydroxy-1-phenyl-(1E)-1-heptene, (13) 1-(3,4-dihydroxyphenyl)-7-(4-hydroxy-3-methoxyphenyl)-hepta-1,6-diene-3,5-dione, (14) 1-(4-hydroxyphenyl)-7-(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one, (15) 1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one, (16) 1-heptene-3,5-dione, 1,7-bis-(4-hydroxy-3-methoxyphenyl)-, (17) 1,5-bis(4-hydroxy-3-methoxyphenyl)-1,4-pentadien-3-one.
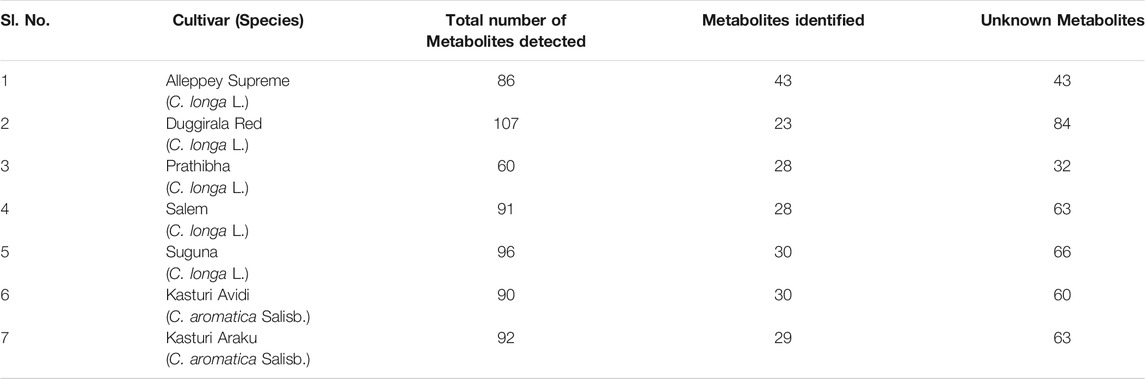
TABLE 5. Total number of compounds detected by LC-MS from the rhizome extract of C. longa L. and C. aromatica Salisb.
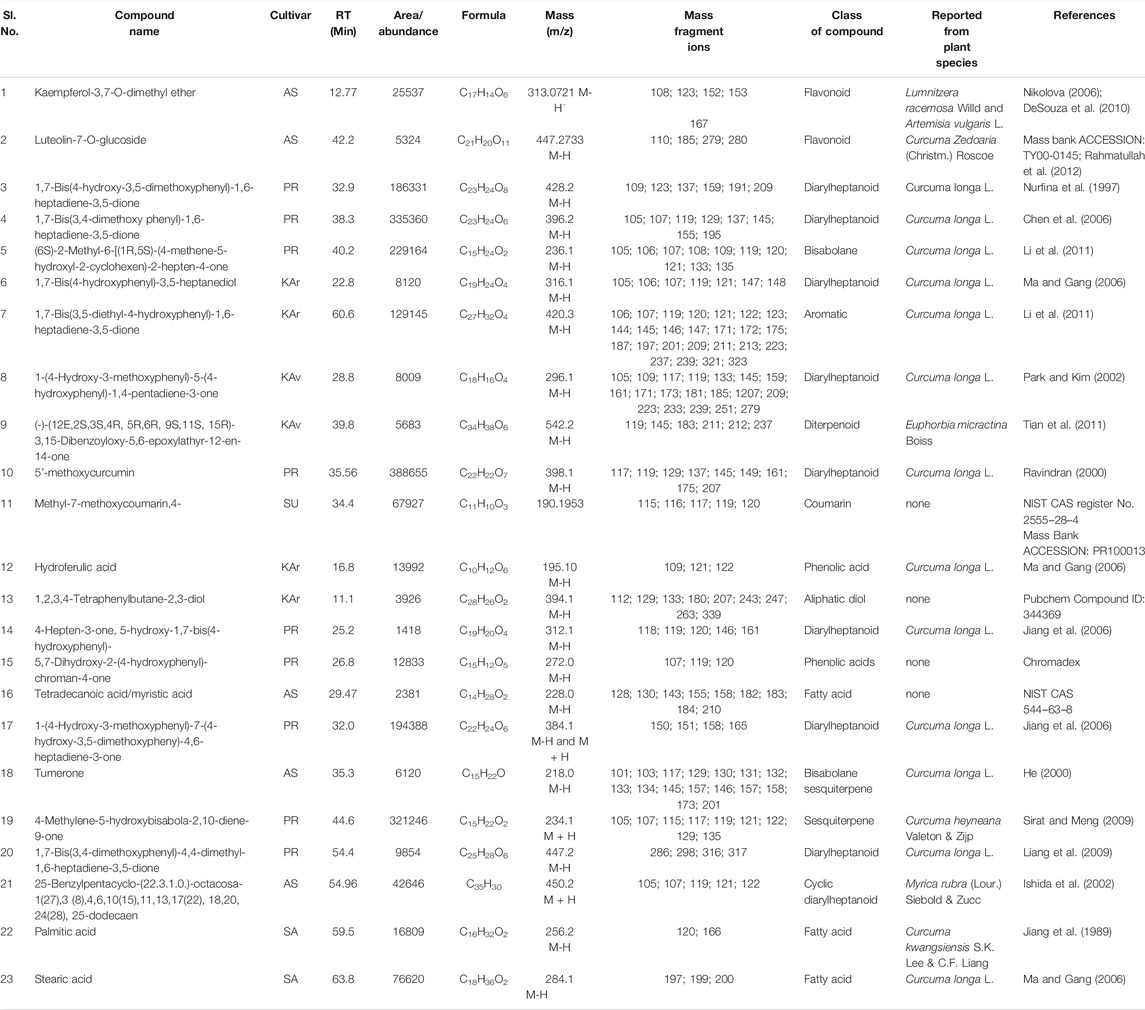
TABLE 6. Cultivar-specific compounds identified by LC-MS in the rhizome extracts from one of the seven cultivars of Curcuma longa L. and C. aromatica Salisb. The structure of the compound with the serial number “1” is given in Figure 2 (panel number: 36) and compounds with Sl. Nos. 2, 3–9 are given in Figure 4 with the corresponding panel nos. 3, 5–11, respectively. Abbreviations used: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
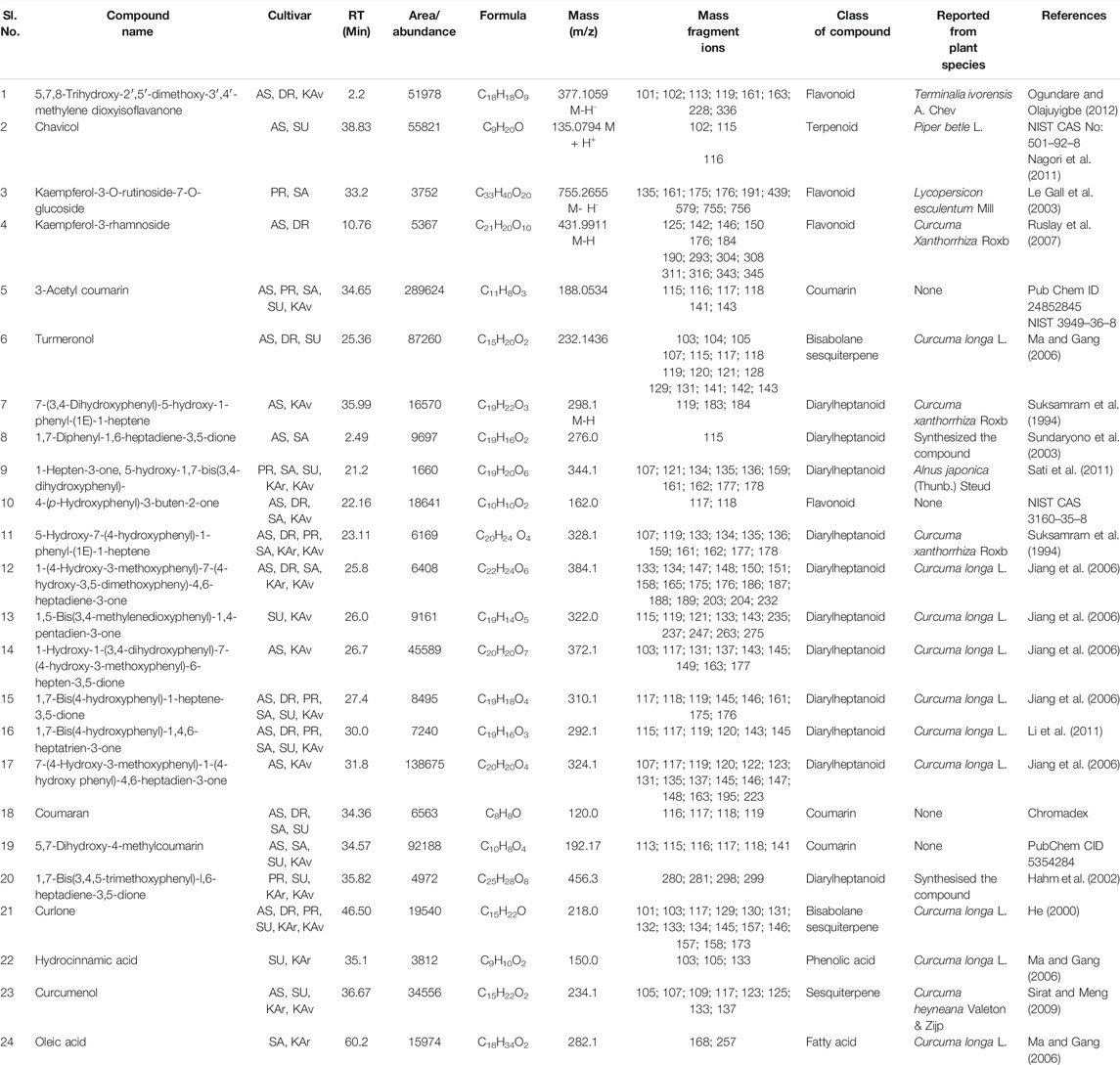
TABLE 7. Compounds identified by LC-MS in rhizome extracts of more than one cultivar of Curcuma longa L. and C. aromatica Salisb. The compounds from serial numbers 1–3 are reported first time from the genus Curcuma, the structures of these compounds along with few others are given in Figure 2 (panels: 37–39; panels 1–2, 4, 12 corresponding to serial numbers 4–5, 6, 7). Abbreviations used: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
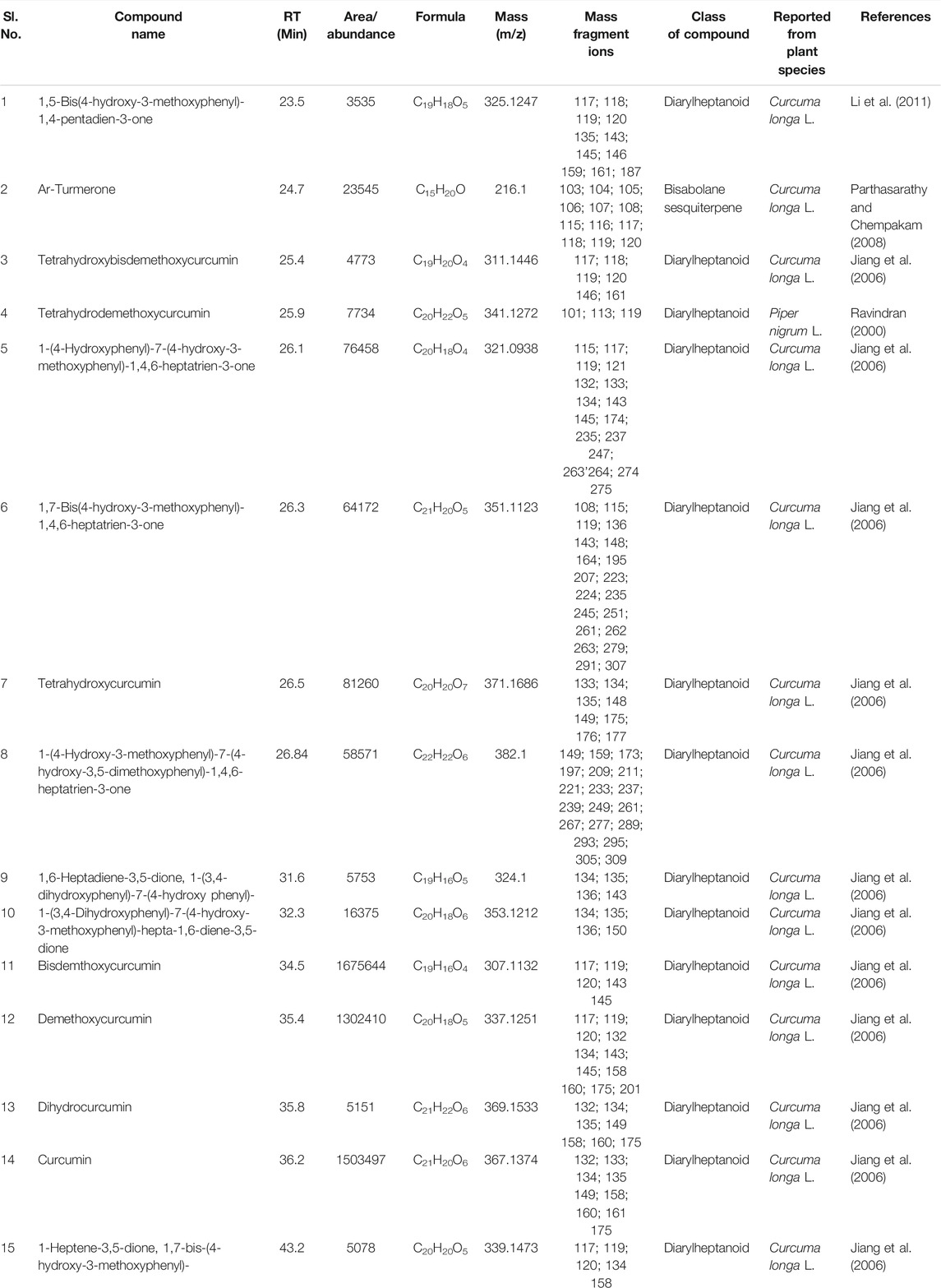
TABLE 8. Compounds commonly detected by LC-MS analysis of rhizomes extract of all seven cultivars of Curcuma spp.: five of Curcuma longa L. (cvs. Alleppey Supreme, Duggirala Red, Prathibha, Salem, and Suguna) and two of C. aromatica Salisb. (cvs. Kasturi Araku, Kasturi Avidi). The structures for the compounds in the serial numbers 10, 5, 6, 15, and 1 are given in Figure 4 (panels 13, 14, 15, 16, and 17 respectively).
Compounds Reported First Time From the Genus Curcuma Using GC-MS and LC-MS Analysis
A total of 39 compounds were detected (Figure 2) for the first time from the genus Curcuma. Out of these, 35 and 4 compounds were identified respectively in the essential oils and whole rhizome extracts of C. longa L. and C. aromatica Salisb. by the GC-MS and LC-MS techniques. Details of the compounds, including the class of compound, molecular weight, are given in Tables 2, 3, 5, and 6; structures of all these compounds are shown in Figure 2 (panels: 1–39). The MS and MS/MS spectra of these compounds are presented in Supplementary Figure S2 (panels: 1–39). These compounds were reported earlier from plants belonging to any genus other than Curcuma, and this is the first report from genus Curcuma. Out of the total of 62 compounds detected by LC-MS analyses of rhizome extracts, four compounds were reported for the first time from the Curcuma genus. One was cultivar-specific (Table 6; Sl. Nos. 1; Figure 2, panels: 36), and three were present in more than one cultivar (Table 7; Sl. Nos. 1–3). The structures of these first-time reported compounds are given in Figure 2 (panels: 37–39), and their corresponding mass fragmentation spectra are shown in Supplementary Figure S2 (panels: 36–39).
A comprehensive table each for GCMS (Supplementary Table S3) and LCMS (Supplementary Table S4) shows the details of compound identification methods used in the present study for the first-time reported compounds from genus Curcuma and the previous literature. A list of 80 (GC-MS) and 62 (LC-MS) compounds can be seen in Supplementary Tables S5, S6 respectively.
Discussion
In one of our previous studies, we reported that the HPLC method could be a valuable tool to differentiate the cultivars of Curcuma spp. based on their curcuminoids content ratios (Kulyal et al., 2016). Curcuminoids play a significant role in food, cosmetics, and medicinal compounds. But there are several other secondary metabolites such as terpenoids (e.g., mono-, sesqui-, di-, tri-, so on), alkenes, aromatic compounds, flavonoids, coumarins, etc. that are responsible for various biological activities. All these secondary metabolites are present in either the volatile essential oil or the nonvolatile fraction of the Curcuma spp. Employing untargeted metabolomics would be the ideal way to identify as many metabolites as possible. Therefore, in the present study, we analyzed these secondary compounds using GC-MS and LC-MS/MS.
Versatility of GC-MS and LC-MS Techniques to Identify a Large Number of Metabolites
GC-MS analysis is an appropriate technique for analyzing volatile compounds, whereas LC-MS is for detecting polar compounds, and thus, these two techniques are mutually complementary to each other. In the present study, several of the volatile compounds present in the cultivars of C. longa L. and C. aromatica Salisb. belonging to mono- and sesquiterpenoids were detected by GC-MS (Tables 2–4). On the other hand, LC-MS analysis detected phenolic (Tables 6–8) compounds, including several diarylheptanoids in the methanolic extracts of both C. longa L. and C. aromatica Salisb. (Figure 4). Electrospray ionization (ESI), coupled with LC/MS/MS, turned out to be a powerful tool in metabolite profiling and metabolomics research. Studies on chemical derivatization and quantification of several metabolites in turmeric powders and fresh rhizome extracts by LC-MS or LC-MS/MS were made. But the rapid screening within the cultivars of C. longa L. of fresh turmeric rhizome has not yet been reported. To the best of our knowledge, we were able to record the presence of several metabolites, which were not reported so far in the C. longa L. and C. aromatica Salisb. (Tables 2, 3, 6, 7, and Figure 2), using the available literature search, Metlin library, mass bank, and NIST library.
Cultivar Variability Based on Secondary Metabolites
Based on the presence or absence of metabolites identified by GC-MS and LC-MS analyses, there was a need to authenticate cultivar variability. Thus, the metabolite library can be constructed based on the cultivar-specific and compounds found in more than one cultivar. There are very few reports on cultivar-specific secondary metabolite variation. Out of a total of 142 compounds identified by both GC-MS and LC-MS, only 28 compounds (13 from GCMS and 15 from LCMS) were common (Tables 4, 8) present in all the cultivars of C. longa L. and C. aromatica Salisb. Ten of 13 common compounds (GCMS) were reported earlier from C. longa L. rhizome. Two compounds, namely sabinene and α-caryophyllene, were reported from the leaves of C. longa L. The remaining one compound, i.e., 2-heptadecanone, was detected for the first time from these two Curcuma species. This compound was earlier reported in the essential oil of Curcuma angustifolia Roxb. rhizome (Srivastava et al., 2006).
As per our analyses, 64 compounds (Tables 2, 6) out of 142 compounds were cultivar-specific. Of these 64 compounds, 41 were identified in essential oils by GC-MS (e.g., carvacrol, endo-borneol) and 23 (e.g., tumerone, methyl-7-methoxycoumarin,4-) in fresh rhizome extracts (LC-MS) of any one of the cultivars of C. longa L. or C. aromatica Salisb. In addition, 50 compounds (Tables 3, 7) were identified to be present in some of the cultivars, present in more than one cultivar but not common to all the cultivars of C. longa L. and C. aromatica Salisb. Out of these 50 compounds, 26 were identified in essential oils through GC-MS. For example, 1,3,5-cycloheptatriene was detected in all six cultivars except cv. Duggirala Red, whereas 12-oxabicyclo(9.1.0)dodeca-3,7-diene, 1,5,5,8-tetramethyl-, [1R-(1R*,3E,7E,11R*)]-, was detected only in cvs. Duggirala Red and Kasturi Araku. The rest 24 compounds were detected in rhizome extracts by LC-MS (e.g., chavicol detected in cvs. Alleppey Supreme and Suguna). The present extensive analyses of both essential oils and whole rhizome secondary metabolome of seven cultivars of C. longa L. and C. aromatica Salisb. established cultivar variability. Variability of the compounds within or/and in between the cultivars of C. longa L. and C. aromatica Salisb. will give a better understanding of their selection. The current study will help select cultivars for use in pharmacology or the food industry.
Discovery of First-Time Reported Metabolites in C. longa L. and C. aromatica Salisb.
In the present study, as many as 142 compounds were identified in the essential oils and rhizome extracts of C. longa L. and C. aromatica Salisb. Out of these, 39 compounds were identified for the first time in the genus Curcuma. However, these compounds were found in other plant genera. The structures of these compounds are shown in Figure 2, and corresponding details, including the class of compound, molecular weight, are given in Tables 2, 3, 6, and 7. As an example, cv. Alleppey Supreme of C. longa L. showed three cultivar-specific compounds. Among these, 1,2-cyclohexanediol, 1-methyl-4-(1-methylethyl)- (oxygenated alcoholic monoterpenoid) was earlier reported from leaf and peel essential oil of Citrus medica L. (Rutaceae). This compound is used as a flavoring agent (Bhuiyan et al., 2009); trans, trans-octa-2,4-dienyl acetate, present in common Malaysian Kaempferia galanga L. (Zingiberaceae), was used for its food-flavoring property (Othman et al., 2006). Phenol, 2-methoxy-3-(2-propenyl)-, an allyl chain-substituted guaiacol was reported from rosewood extracts (Jiang et al., 2018).
The following four compounds were identified from cv. Duggirala Red (C. longa L.): 3-isopropyl-4-methyl-1-pentyn-3-ol (alcohol constituent) containing leaf and stem of Anethum sowa Roxb. ex, Fleming, used for flavoring of food, beverages and also for many medical preparations (Saleh-e-in et al., 2010); 5,9-tetradecadiyne (unsaturated hydrocarbon) was found to be a major component of Ferula vesceritensis Coss. & Durieu ex Trab. leaf essential oil (Zellagui et al., 2012); naphthalene, 5-butyl-1,2,3,4-tetrahydro- (tetralin type of compounds) found in the essential oil of Meconopsis punicea Maxim. and M.delavayi (Franch.) Franch. Ex Prain (Slavík and Slavíková; Yuan et al., 2003); and santolina alcohol was reported from plant Achillea filipendulina Lam. (Sharopov and Setzer, 2010). A total of nine cultivar-specific compounds were detected in cv. Prathibha (C. longa L.) and the examples of these compounds and their source plants, respectively, are 7-tetracyclo[6.2.1.0(3.8)0(3.9)]undecanol, 4,4,11,11 tetramethyl- in Cyperus articulatus L. (Metuge et al., 2014); bicyclo(2.2.1)hept-2-ene, 2,3-dimethyl- in Abies alba Mill. (Yang et al., 2009). Cultivar-specific compounds of three each were detected in cvs. Salem (C. longa L.) and Suguna (C. longa L.) (Table 2). In C. aromatica Salisb., only one cultivar-specific compound was detected in cv. Kasturi Avidi, i.e., 3-octen-5-yne, 2,7-dimethyl-, (Z)-, and this compound was earlier reported from the medicinally important Litsea glutinosa (Lour.) C.B. Rob. fruit essential oil (Chowdhury et al., 2008a).
Some of the compounds identified in our study were present in more than one cultivar. For example, 6-(p-tolyl)-2-methyl-2-heptenol (Table 3) was detected in three cvs.: Alleppey supreme, Suguna of C. longa L., and Kasturi Avidi of C. aromatica Salisb. This compound was earlier reported from Zingiber officinale Roscoe (Zingiberaceae), used as a spice, food products, and beverages (Choudhari and Kareppa, 2013).
Limitations and Strengths of the Present Study
There is significant variability within and between the cultivars of C. longa L. and C. aromatica Salisb, which can be exploited to differentiate the cultivars of Curcuma spp. The feasibility of studies without using any standard compounds was pointed out by Núñez et al. (2020). Similarly, reference compounds were not used in our study to derive arithmetic indices under the experimental conditions. Despite the dilution made in the essential oil sample before injecting into the GC-MS system, the sample was still too concentrated. The high concentration of oil might have restricted the resolution due to overloading the detector. This could be the reason that we could not identify several compounds. We would ensure the further dilution of the oil sample in our future studies. However, the technology employed, GC-TOFMS and LC-QTOFMS, and MS-spectral database/literature search enabled us to establish the cultivar variability of Curcuma spp. The detailed information on the metabolite variability within or/and between the cultivars of C. longa L. and C. aromatica Salisb. may assist us in selecting the cultivars for a specific purpose, like culinary use, coloring, or pharmacological purpose. The studies such as the present one can help to select cultivars, particularly for use in pharmacology or the food industry. Metabolite variability poses a challenge in the use of turmeric in therapy. The practitioners need to be quite careful and use the identified cultivar and avoid mix-up. The caution applies to commercial/industrial use. Once standardized, the protocol should ensure the use of a specific cultivar. Our GC-MS and LC-MS-based metabolite identification is distinct from chemophenetic studies but is a complementary approach to characterize the Curcuma metabolome.
Importance of Curcuma spp. Metabolites for Human Health
Curcuminoids (CU, DMC, and BDMC) were identified as the main bioactive compounds of genus Curcuma and proved to have a broad spectrum of biological activities based on pharmacological studies. However, rhizomes and their essential oils of Curcuma spp. contained several other bioactive (volatile and nonvolatile) compounds. A summary of the pharmacological studies with the metabolites detected in the present study is given in Table 9. Some of the studies demonstrated therapeutic activity with the isolated metabolites, e.g., carvacrol (Suntres et al., 2015), p-cymene (De Oliveira et al., 2015), which are commonly found in essential oils of Curcuma spp. A few other reports correlated anti-inflammatory and antioxidant properties of C. longa L. essential oil with its chemical components ar-tumerone, α-santalene (Singh et al., 2010) (Table 9). Several compounds detected in the present study in the essential oil or rhizome extracts of C. longa L. or C. aromatica Salisb. were also found in the essential oil of other medicinal plants, traditionally used for their health benefits. The examples of such compounds are 5,9-tetradecadiyne, a cultivar-specific compound of Duggirala Red (C. longa L.), earlier reported in Ferula vesceritensis Coss. & Durieu ex Trab. leaf essential oil, exhibiting antibacterial activity; 3-octen-5-yne, 2,7-dimethyl-, (Z)-, a hydrocarbon monoterpene, identified from cv. Kasturi Avidi (C. aromatica Salisb.) was earlier reported from fruit essential oil of the medicinally important plant, Litsea glutinosa (Lour.) C.B. Rob. (Chowdhury et al., 2008a). We suggest that the medicinal use of the genus Curcuma can be not only species but also cultivar-specific.
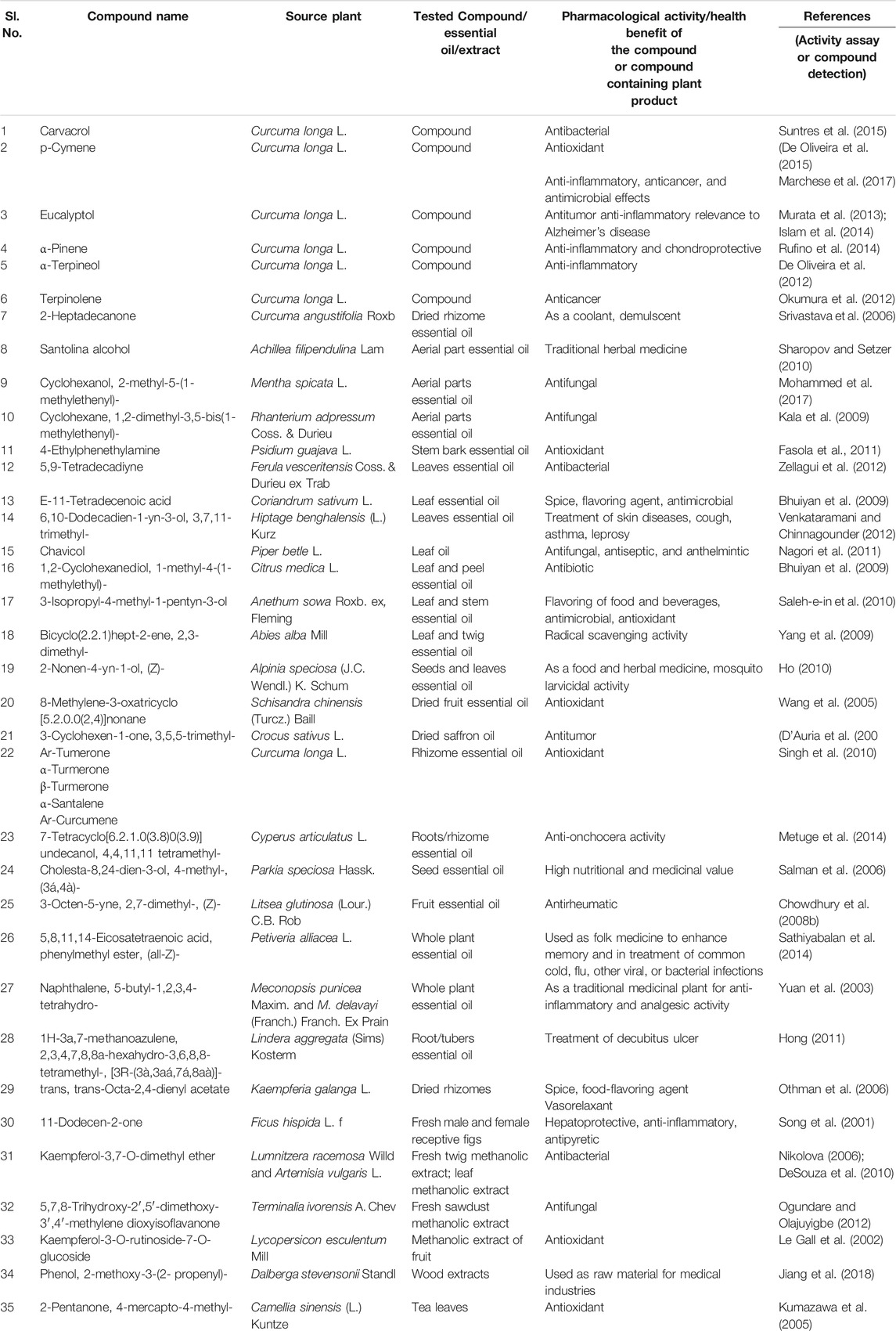
TABLE 9. Pharmacological activity of metabolites identified, other than major curcuminoids (curcumin, demethoxycurcumin, and bisdemethoxy curcumin), in C. longa L. and C. aromatica Salisb.
Concluding Remarks
Essential oils from spices and aromatic plants are enriched with bioactive metabolites, easily isolated and used, unlike the difficulties encountered with synthetic chemical products. The low mammalian toxicity and biodegradable nature of the natural secondary products provide an attractive option to develop them also for crop protection. Metabolomics is a practical and dynamic approach to make a comprehensive study. Both GC-MS and LC-MS techniques should be used to characterize the metabolite profiles of as many cultivars as possible for building a reference library. Preparative LC can be helpful to collect individual metabolite fractions and establish their identity. Several metabolites detected in 7 selected cultivars of Curcuma spp. by GC-MS and LC-MS analyses are reported first time in Curcuma spp. We suggest that the seven Indian cultivars of Curcuma spp. employed in our study can be used as sources of such compounds. High-throughput analysis of cultivar-specific and first-time detected compounds in the present study may lead to new drug candidates. The metabolites validated for their medicinal or other users can be quantified using simple techniques such as HPLC or TLC to ensure their presence in the herbal preparations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/metabolights/MTBLS2790.
Author Contributions
ST and AR planned and designed all the experiments. PK and PKK did the experiments and preliminary data interpretation related to GC-MS and LC-MS/MS, respectively. PK, ST, and AR re-analyzed the data and wrote the draft of the manuscript. SA prepared tables and reference search. AA drew the figures and helped in editing the manuscript. All the authors read and approved the final version of the manuscript.
Funding
Part of this study was supported by grants from DBT (BT/PR/11674/PBD/16/838/2008) sanctioned to Prof. A.S. Raghavendra (PI) and Prof. Sarada D. Tetali (co-PI) and the Institute of Eminence - University of Hyderabad (IoE-UoH) Research Chair Professor Grant to Prof. A.S. Raghavendra.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are indebted to Professors K. Uma Maheshwari (Turmeric Research Station, Kammarpally of Telangana State, India) and K. Nirmal Babu (Indian Institute of Spices Research, Marikunnu, Kozhikode, Kerala, India) for providing turmeric samples. PK and PKK acknowledge their research fellowships from the research project (BT/PR/11674/PBD/16/838/2008). SA is a recipient of a research fellowship from the University of Hyderabad and AA is a recipient of UGC-JRF (856/SC/CSIR-UGC/NET/Dec 2016). The authors are thankful to Prasanth Bitla, School of Life Sciences, University of Hyderabad, for acquisition of GC-MS and LC-MS data. The authors are thankful to DBT-CREBB, DBT-FIST, and UGC-SAP for supporting infrastructural facilities of the Department of Plant Sciences and School of Life Sciences. The authors also acknowledge the ChemDraw Ultra 12.0 software tool used to draw structures in Figures 2, 3 and 4.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.659546/full#supplementary-material
Supplementary Figure 1 | (A) Representative TIC chromatograms from negative ion (−) ESI-HPLC from cultivars (A) Alleppey Supreme, (B) Duggirala Red, (C) Prathibha, (D) Salem, (E) Suguna of Curcuma longa L. and cvs. (F) Kasturi Araku, (G) Kasturi Avidi of C. aromatica Salisb. Peak labelled IS represents internal standard. (B) Representative TIC chromatograms from positive ion (+) ESI-HPLC from cultivars (A) Alleppey Supreme, (B) Duggirala Red, (C) Prathibha, (D) Salem, (E) Suguna of Curcuma longa L. and cvs. (F) Kasturi Araku, (G) Kasturi Avidi of C. aromatica Salisb. Peak labelled IS represents internal standard.
Supplementary Figure 2 | MS and MS/MS spectra of cultivar-specific compounds first time reported from genus Curcuma, identified from cultivars of Curcuma longa L. and C. aromatica Salisb. detected in essential oil (1–35) and rhizome extracts (36–39) by GC-MS (MS spectra) and LC-MS (MS/MS spectra) respectively. (Spectra 1–23 corresponds to panel numbers: 1–23 of Figure 2 and serial numbers: 1–23 of Table 2. Spectra 24–35 corresponds to panel numbers: 24–35 of Figure 2 and serial numbers: 1–12 of Table 3. Spectra 36 correspond to panel numbers 36 of Figure 2 and serial number 1 of Table 6. Spectra 37–39 correspond to panel number 37–39 of Figure 2 and serial number 1–3 of Table 7). (1) 1,2-Cyclohexanediol, 1-methyl-4-(1-methylethyl)-, (2) trans, trans-Octa-2,4-dienyl acetate, (3) Phenol, 2-methoxy-3-(2- propenyl)-, (4) 3-Isopropyl-4-methyl-1-pentyn-3-ol (5) 5,9-Tetradecadiyne, (6) Naphthalene, 5-butyl-1,2,3,4-tetrahydro-, (7) Santolina alcohol, (8) 2-Pentanone, 4-mercapto-4-methyl-, (9) 8-Methylene-3-oxatricyclo[5.2.0.0(2,4)]nonane, (10) 7-Tetracyclo[6.2.1.0(3.8)0(3.9)]undecanol, 4,4,11,11 tetramethyl-, (11) Bicyclo[2.2.1]hept-2-ene, 2,3-dimethyl-, (12) 1H-3a,7-Methanoazulene, 2,3,4,7,8,8a-hexahydro-3,6,8,8-tetramethyl-, [3R-(3à,3aá, 7á, 8aà)]-, (13) Cholesta-8,24-dien-3-ol, 4-methyl-, (3á,4à)-, (14) 4-Ethylphenethylamine, (15) Cyclohexanol, 2-methyl-5-(1-methylethenyl)-, (16) Cyclohexane, 1,2-dimethyl-3,5-bis(1-methylethenyl)-, (17) 5,8,11,14-Eicosatetraenoic acid, phenylmethyl ester, (all-Z)-, (18) 11-Dodecen-2-one, (19) E-11-Tetradecenoic acid, (20) 2-Nonen-4-yn-1-ol, (Z)-, (21) 3-Cyclohexen-1-one, 3,5,5-trimethyl-, (22) 6,10-Dodecadien-1-yn-3-ol, 3,7,11-trimethyl-, (23) 3-Octen-5-yne, 2,7-dimethyl-, (Z)-, (24) 1,3,5-Cycloheptatriene, (25) Bicyclo[3.1.0]hexane, 4-methyl-1-(1-methylethyl)-, didehydro deriv., (26) Bicyclo[3.2.1]oct-2-ene, 3-methyl-4-methylene-, (27) Oxirane, 2-(hexyn-1-yl)-3-methoxymethylene-, (28) Bergamotol, Z-α-trans-, (29) (1,3-Dimethyl-2-methylene-cyclopentyl)-methanol, (30) 12-Oxabicyclo[9.1.0]dodeca-3,7-diene, 1,5,5,8-tetramethyl-, [1R-(1R*,3E,7E,11R*)]-, (31) Isolongifolene, 4,5,9,10-dehydro-, (32) Z,Z,Z-4,6,9-Nonadecatriene, (33) 6-(p-Tolyl)-2-methyl-2-heptenol, (34) 6-Tridecen-4-yne, (Z)-, (35) 1,4-Cyclohexadiene, 1-methyl-, (36) Kaempferol-3,7-O-dimethyl ether, (37) 5,7,8-Trihydroxy-2′,5′-dimethoxy-3′,4′-methylene dioxyisoflavanone, (38) Chavicol, (39) Kaempferol-3-O-rutinoside-7-O-glucoside.
Supplementary Table 1 | A list of secondary metabolites identified by GC-MS in the essential oil from the rhizomes of five cvs. of C. longa L. and two cvs. of C. aromatica Salisb. Abbreviations: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
Supplementary Table 2 | A list of secondary metabolites identified by LC-MS in the rhizomes extract of five cvs. of C. longa L. and two cvs. of C. aromatica Salisb. Abbreviations used: AS, Alleppey Supreme; DR, Duggirala Red; PR, Prathibha; SA, Salem; SU, Suguna; KAr, Kasturi Araku; KAv, Kasturi Avidi.
Supplementary Table 3 | Identification parameters of first-time reported compounds by GC-MS in the essential oil from the seven cultivars of Curcuma spp. along with the methods used in previous literature.
Supplementary Table 4 | Identification parameters of first-time reported compounds by LC-MS in the rhizome extracts of genus Curcuma along with the methods used in related previous literature.
Supplementary Table 5 | The list of 80 compounds identified by GCMS analysis.
Supplementary Table 6 | The list of 62 compounds identified by LCMS analysis.
Abbreviations
CU, curcumin; cvs, cultivars; DMC, demethoxy curcumin; BDMC, bisdemethoxycurcumin.
References
Aggarwal, B. B., Sundaram, C., Malani, N., and Ichikawa, H. (2007). “Curcumin: The Indian Solid Gold,,” in The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease. Editors B. B. Aggarwal, Y.-J. Surh, and S. Shishodia (Boston, MA: Springer US), 1–75. doi:10.1007/978-0-387-46401-5_1
Angel, G. R., Menon, N., Vimala, B., and Nambisan, B. (2014). Essential Oil Composition of Eight Starchy Curcuma Species. Ind. Crops Prod. 60, 233–238. doi:10.1016/j.indcrop.2014.06.028
Awasthi, P. K., and Dixit, S. C. (2009). Chemical Composition ofCurcuma Longaleaves and Rhizome Oil from the plains of Northern India. J. Young Pharm. 1, 312. doi:10.4103/0975-1483.59319
Beekwilder, J., Jonker, H., Meesters, P., Hall, R. D., Van Der Meer, I. M., and Ric de Vos, C. H. (2005). Antioxidants in Raspberry: On-Line Analysis Links Antioxidant Activity to a Diversity of Individual Metabolites. J. Agric. Food Chem. 53, 3313–3320. doi:10.1021/jf047880b
Behura, S., Sahoo, S., and Srivastava, V. K. (2002). Major Constituents in Leaf Essential Oils of Curcuma Longa L. And Curcuma Aromatica Salisb. Curr. Sci. 83, 1312–1313.
Bhuiyan, M. N. I., Begum, J., Sardar, P. K., and Rahman, M. S. (2009). Constituents of Peel and Leaf Essential Oils of Citrus Medica L. J. Sci. Res. 1, 387–392. doi:10.3329/jsr.v1i2.1760
Chen, W. F., Deng, S. L., Zhou, B., Yang, L., and Liu, Z. L. (2006). Curcumin and its Analogues as Potent Inhibitors of Low Density Lipoprotein Oxidation: H-Atom Abstraction from the Phenolic Groups and Possible Involvement of the 4-Hydroxy-3-Methoxyphenyl Groups. Free Radic. Biol. Med. 40, 526–535. doi:10.1016/j.freeradbiomed.2005.09.008
Cheng, J. H., Wu, W. Y., Liu, W. S., Chang, G., Liu, Y. L., Yang, Z. G., et al. (1999). Treatment of 17 Cases of Patients with Primary Liver Cancer with Curcuma Aromatica Oil Infused via Hepatic Artery. Shijie Huaren Xiaohua Zazhi 7, 92.
Choudhari, S. S., and Kareppa, B. M. (2013). Identification of Bioactive Compounds of Zingiber Officinale roscoe Rhizomes through Gas Chromatography and Mass Spectrometry. Int. J. Pharm. Res. Dev 5, 16–20.
Choudhury, S. N., Ghosh, A. C., Saikia, M., Choudhury, M., and Leclercq, P. A. (1996). Volatile Constituents of the Aerial and Underground Parts ofCurcuma aromaticaSalisb. From India. J. Essent. Oil Res. 8, 633–638. doi:10.1080/10412905.1996.9701031
Chowdhury, J. U., Bhuiyan, M. N. I., and Nandi, N. C. (2008b). Aromatic Plants of Bangladesh : Essential Oils of Leaves and Fruits of Litsea Glutinosa (Lour.) C.B. Robinson. Bangladesh J. Bot. 37, 81–83. doi:10.3329/bjb.v37i1.1568
Chowdhury, J. U., Nandi, N. C., Bhuiyan, M. N. I., and Mobarok, M. H. (2008a). Essential Oil Constituents of the Rhizomes of Two Types of Curcuma Longa of Bangladesh. Bangladesh J. Sci. Ind. Res. 43, 259–266. doi:10.3329/bjsir.v43i2.970
D’Auria, M., Mauriello, G., Racioppi, R., and Rana, G. L. (2006). Use of SPME-GC-MS in the Study of Time Evolution of the Constituents of Saffron Aroma: Modifications of the Composition during Storage. J. Chromatogr. Sci. 44, 18–21. doi:10.1093/chromsci/44.1.18
De Oliveira, M. G., Marques, R. B., De Santana, M. F., Santos, A. B., Brito, F. A., Barreto, E. O., et al. (2012). α-Terpineol Reduces Mechanical Hypernociception and Inflammatory Response. Basic Clin. Pharmacol. Toxicol. 111, 120–125. doi:10.1111/j.1742-7843.2012.00875.x
De Oliveira, T. M., De Carvalho, R. B., Da Costa, I. H., De Oliveira, G. A., De Souza, A. A., De Lima, S. G., et al. (2015). Evaluation of P-Cymene, a Natural Antioxidant. Pharm. Biol. 53, 423–428. doi:10.3109/13880209.2014.923003
De Vos, R. C., Moco, S., Lommen, A., Keurentjes, J. J., Bino, R. J., and Hall, R. D. (2007). Untargeted Large-Scale Plant Metabolomics Using Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2, 778–791. doi:10.1038/nprot.2007.95
DeSouza, L., Wahidulla, S., and Devi, P. (2010). Antibacterial Phenolics from the Mangrove Lumnitzera Racemosa. Indian J. Mar. Sci. 39, 294–298.
Dixon, R. A., Gang, D. R., Charlton, A. J., Fiehn, O., Kuiper, H. A., Reynolds, T. L., et al. (2006). Applications of Metabolomics in Agriculture. J. Agric. Food Chem. 54, 8984–8994. doi:10.1021/jf061218t
Dosoky, N. S., Satyal, P., and Setzer, W. N. (2019). Variations in the Volatile Compositions of Curcuma Species. Foods 8, 1–14. doi:10.3390/foods8020053
Dosoky, N. S., and Setzer, W. N. (2018). Chemical Composition and Biological Activities of Essential Oils of Curcuma Species. Nutrients 10, 10–17. doi:10.3390/nu10091196
Eggink, P. M., Maliepaard, C., Tikunov, Y., Haanstra, J. P. W., Pohu-Flament, L. M. M., de Wit-Maljaars, S. C., et al. (2012). Prediction of Sweet Pepper (Capsicum annuum) Flavor Over Different Harvests. Euphytica 187, 117–131. doi:10.1007/s10681-012-0761-6
Esatbeyoglu, T., Huebbe, P., Ernst, I. M., Chin, D., Wagner, A. E., and Rimbach, G. (2012). Curcumin--from Molecule to Biological Function. Angew. Chem. Int. Ed. Engl. 51, 5308–5332. doi:10.1002/anie.201107724
Fasola, T. R., Oloyede, G. K., and Aponjolosun, B. S. (2011). Chemical Composition, Toxicity and Antioxidant Activities of Essential Oils of Stem Bark of Nigerian Species of Guava (Psidium Guajava Linn.). EXCLI J. 10, 34–43. doi:10.17877/DE290R-1133
Ferreira, F. D., Kemmelmeier, C., Arrotéia, C. C., Da Costa, C. L., Mallmann, C. A., Janeiro, V., et al. (2013). Inhibitory Effect of the Essential Oil of Curcuma Longa L. And Curcumin on Aflatoxin Production by Aspergillus flavus Link. Food Chem. 136, 789–793. doi:10.1016/j.foodchem.2012.08.003
Gopalan, B., Goto, M., Kodama, A., and Hirose, T. (2000). Supercritical Carbon Dioxide Extraction of Turmeric (Curcuma Longa). J. Agric. Food Chem. 48, 2189–2192. doi:10.1021/jf9908594
Hahm, E. R., Cheon, G., Lee, J., Kim, B., Park, C., and Yang, C. H. (2002). New and Known Symmetrical Curcumin Derivatives Inhibit the Formation of Fos-Jun-DNA Complex. Cancer Lett. 184, 89–96. doi:10.1016/S0304-3835(02)00170-2
Hall, R. D. (2006). Plant Metabolomics: From Holistic hope, to Hype, to Hot Topic. New Phytol. 169, 453–468. doi:10.1111/j.1469-8137.2005.01632.x
He, X. G. (2000). On-line Identification of Phytochemical Constituents in Botanical Extracts by Combined High-Performance Liquid Chromatographic-Diode Array Detection-Mass Spectrometric Techniques. J. Chromatogr. A. 880, 203–232. doi:10.1016/S0021-9673(00)00059-5
Herath, H. M. I. C., Wijayasiriwardene, T. D. C. M. K., and Premakumara, G. A. S. (2017). Comparative GC-MS Analysis of All Curcuma Species Grown in Sri Lanka by Multivariate Test. Ruhuna J. Sci. 8, 103. doi:10.4038/rjs.v8i2.29
Herebian, D., Choi, J. H., Abd El-Aty, A. M., Shim, J. H., and Spiteller, M. (2009). Metabolite Analysis in Curcuma Domestica Using Various GC-MS and LC-MS Separation and Detection Techniques. Biomed. Chromatogr. 23, 951–965. doi:10.1002/bmc.1207
Ho, J.-C. (2010). Chemical Composition and Bioactivity of Essential Oil of Seed and Leaf fromAlpinia speciosaGrown in Taiwan. Jnl Chin. Chem. Soc 57, 758–763. doi:10.1002/jccs.201000105
Hong, L. (2011). Radix Linderae Essential Oil Improving the Immunity Activities and Preventing the Occurrence of Decubitus in Aged People. J. Med. Plants Res. 5, 3733–3738. doi:10.5897/JMPR.9001070
Hong, S. L., Lee, G. S., Syed Abdul Rahman, S. N., Ahmed Hamdi, O. A., Awang, K., Aznam Nugroho, N., et al. (2014). Essential Oil Content of the Rhizome of Curcuma Purpurascens Bl. (Temu Tis) and its Antiproliferative Effect on Selected Human Carcinoma Cell Lines. ScientificWorldJournal 2014, 397430. doi:10.1155/2014/397430
Huhman, D. V., and Sumner, L. W. (2002). Metabolic Profiling of Saponins in Medicago Sativa and Medicago Truncatula Using HPLC Coupled to an Electrospray Ion-Trap Mass Spectrometer. Phytochemistry 59, 347–360. doi:10.1016/S0031-9422(01)00432-0
Ishida, J., Kozuka, M., Tokuda, H., Nishino, H., Nagumo, S., Lee, K. H., et al. (2002). Chemopreventive Potential of Cyclic Diarylheptanoids. Bioorg. Med. Chem. 10, 3361–3365. doi:10.1016/S0968-0896(02)00164-5
Jiang, D. Q., Pu, Q. L., Huang, P., Huang, X. M., He, Y. Z., He, C. H., et al. (1989). [Studies on the Chemical Constituents of Curcuma Kwangsiensis]. Yao Xue Xue Bao 24, 357–359.
Jiang, H., Timmermann, B. N., and Gang, D. R. (2006). Use of Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry to Identify Diarylheptanoids in Turmeric (Curcuma Longa L.) Rhizome. J. Chromatogr. A. 1111, 21–31. doi:10.1016/j.chroma.2006.01.103
Jiang, S. C., Ge, S. B., and Peng, W. (2018). Molecules and Functions of Rosewood: Dalbergia stevenson. Arabian J. Chem. 11, 782–792. doi:10.1016/j.arabjc.2017.12.032
Joshi, J., Ghaisas, S., Vaidya, A., Vaidya, R., Kamat, D. V., Bhagwat, A. N., et al. (2003). Early Human Safety Study of Turmeric Oil (Curcuma Longa Oil) Administered Orally in Healthy Volunteers. J. Assoc. Physicians India 51, 1055–1060.
Kala, A., Noureddine, G., Belkacemi, D., Segni, L., Zellagui, Amar., Samir, H., et al. (2009). Composition of the Essential Oil of Rhanterium Adpressum Coss. And Durieu. from Algeria. Arch. Appl. Sci. Res. 1, 115–118.
Kanase, V., and Khan, F. (2018). An Overview of Medicinal Value of Curcuma Species. Asian J. Pharm. Clin. Res. 11, 40–45. doi:10.22159/ajpcr.2018.v11i12.28145
Khan, A., Vaibhav, K., Javed, H., Tabassum, R., Ahmed, M. E., Khan, M. M., et al. (2014). 1,8-cineole (Eucalyptol) Mitigates Inflammation in Amyloid Beta Toxicated PC12 Cells: Relevance to Alzheimer's Disease. Neurochem. Res. 39, 344–352. doi:10.1007/s11064-013-1231-9
Koo, H. J., and Gang, D. R. (2012). Suites of Terpene Synthases Explain Differential Terpenoid Production in Ginger and Turmeric Tissues. PLoS One 7, e51481. doi:10.1371/journal.pone.0051481
Krup, V., Prakash L, H., and A, H. (2013). Pharmacological Activities of Turmeric (Curcuma Longa linn): A Review. J. Homeop Ayurv Med. 02, 133. doi:10.4172/2167-1206.1000133
Kulyal, P., Kuchibhatla, L. N., Uma Maheshwari, K., Nirmal Babu, K., Tetali, S. D., and Raghavendra, A. S. (2016). Highly Sensitive HPLC Method for Estimation of Total or Individual Curcuminoids in Curcuma Cultivars and Commercial Turmeric Powders. Curr. Sci. 111, 1816–1824. doi:10.18520/cs/v111/i11/1816-1824
Kumar, V., Shriram, V., Bhagat, R., Khare, T., Kapse, S., and Kadoo, N. (2019). Phytochemical Profile, Anti-oxidant, Anti-inflammatory, and Anti-proliferative Activities of Pogostemon Deccanensis Essential Oils. 3 Biotech. 9, 31. doi:10.1007/s13205-018-1560-0
Kumazawa, K., Kubota, K., and Masuda, H. (2005). Influence of Manufacturing Conditions and Crop Season on the Formation of 4-Mercapto-4-Methyl-2-Pentanone in Japanese green tea (Sen-cha). J. Agric. Food Chem. 53, 5390–5396. doi:10.1021/jf050392z
Kutti Gounder, D., and Lingamallu, J. (2012). Comparison of Chemical Composition and Antioxidant Potential of Volatile Oil from Fresh, Dried and Cured Turmeric (Curcuma Longa) Rhizomes. Ind. Crops Prod. 38, 124–131. doi:10.1016/j.indcrop.2012.01.014
Ladan, Z., Amupitan, J. O., Oyewale, A. O., Okonkwo, E. M., Ladan, E. O., Odjobo, B., et al. (2011). Chemical Composition and Biological Activity of the Volatile Oils of Hyptis Spicigera against Trypanosoma Brucei Tbb Found in Northern Nigeria. Afr. J. Pure Appl. Chem. 5, 53–58.
Le Gall, G., Colquhoun, I. J., Davis, A. L., Collins, G. J., and Verhoeyen, M. E. (2003). Metabolite Profiling of Tomato (Lycopersicon esculentum) Using 1H NMR Spectroscopy as a Tool to Detect Potential Unintended Effects Following a Genetic Modification. J. Agric. Food Chem. 51, 2447–2456. doi:10.1021/jf0259967
Leela, N., Tava, A., Shafi, P. M., John, S. P., and Chempakam, B. (2002). Chemical Composition of Essential Oils of Turmeric (Curcuma Longa L.). Acta Pharm. 52, 137–141.
Li, S., Yuan, W., Deng, G., Wang, P., Yang, P., and Aggrawal, B. (2011). Chemical Composition and Product Quality Control of Turmeric (Curcuma Longa L.). Topharmcj 5, 28–54. doi:10.2174/2210290601102010028
Li, W., Wang, S., Feng, J., Xiao, Y., Xue, X., Zhang, H., et al. (2009a). Structure Elucidation and NMR Assignments for Curcuminoids from the Rhizomes of Curcuma Longa. Magn. Reson. Chem. 47, 902–908. doi:10.1002/mrc.2478
Li, Y., Wo, J. M., Liu, Q., Li, X., and Martin, R. C. (2009b). Chemoprotective Effects of Curcuma Aromatica on Esophageal Carcinogenesis. Ann. Surg. Oncol. 16, 515–523. doi:10.1245/s10434-008-0228-0
Liang, G., Shao, L., Wang, Y., Zhao, C., Chu, Y., Xiao, J., et al. (2009). Exploration and Synthesis of Curcumin Analogues with Improved Structural Stability Both In Vitro and In Vivo as Cytotoxic Agents. Bioorg. Med. Chem. 17, 2623–2631. doi:10.1016/j.bmc.2008.10.044
Ma, X., and Gang, D. R. (2006). Metabolic Profiling of Turmeric (Curcuma Longa L.) Plants Derived from In Vitro Micropropagation and Conventional Greenhouse Cultivation. J. Agric. Food Chem. 54, 9573–9583. doi:10.1021/jf061658k
Mani, P., and Boominathan, M. (2011). Comparative Studies of the Antimicrobial Activity of Crude Extracts and Factions from Eugena Caryophyllus against Candida Albicans Isolate from Chronic Disease Affected Patients. Int. J. Institutional Pharm. Life Sci. 1, 36–47.
Marchese, A., Arciola, C. R., Barbieri, R., Silva, A. S., Nabavi, S. F., Tsetegho Sokeng, A. J., et al. (2017). Update on Monoterpenes as Antimicrobial Agents: A Particular Focus on P-Cymene. Materials (Basel) 10, 1–15. doi:10.3390/ma10080947
Meng, X., Wang, Z., and Lv, H. (2010). Constituents and Bacteriostatic Activity of Volatile Matter from Four Flower Plant Species. Indian J. Agric. Res. 44, 157–167.
Metuge, J. A., Nyongbela, K. D., Mbah, J. A., Samje, M., Fotso, G., Babiaka, S. B., et al. (2014). Anti-Onchocerca Activity and Phytochemical Analysis of an Essential Oil from Cyperus Articulatus L. BMC Complement. Altern. Med. 14, 223. doi:10.1186/1472-6882-14-223
Miquel, J., Bernd, A., Sempere, J. M., Díaz-Alperi, J., and Ramírez, A. (2002). The Curcuma Antioxidants: Pharmacological Effects and Prospects for Future Clinical Use. A Review. Arch. Gerontol. Geriatr. 34, 37–46. doi:10.1016/s0167-4943(01)00194-7
Moco, S., Bino, R. J., Vorst, O., Verhoeven, H. A., De Groot, J., Van Beek, T. A., et al. (2006). A Liquid Chromatography-Mass Spectrometry-Based Metabolome Database for Tomato. Plant Physiol. 141, 1205–1218. doi:10.1104/pp.106.078428
Mohammed, L. M., M. Salah, T. F., and Qader, K. O. (2017). Chemical Composition and Antifungal Activity of Mentha Spicata L. Plant from Sulaimaniyah in Iraq. Kjar 2, 52–56. doi:10.24017/science.2017.1.11
Mohiuddin, M., Chowdhury, M. J., Alam, M. K., and Hossain, M. (2012). Chemical Composition of Essential Oil of Four Flavouring Plants Used by the Tribal People of Bandarban hill District in Bangladesh. Int. J. Med. Aromat. Plants 2, 106–113.
Mukherjee, P. K., Harwansh, R. K., Bahadur, S., Biswas, S., Kuchibhatla, L. N., Tetali, S. D., et al. (2016). Metabolomics of Medicinal Plants - A Versatile Tool for Standardization of Herbal Products and Quality Evaluation of Ayurvedic Formulations. Curr. Sci. 111, 1624–1630. doi:10.18520/cs/v111/i10/1624-1630
Murata, S., Shiragami, R., Kosugi, C., Tezuka, T., Yamazaki, M., Hirano, A., et al. (2013). Antitumor Effect of 1, 8-cineole against colon Cancer. Oncol. Rep. 30, 2647–2652. doi:10.3892/or.2013.2763
Muthukumaran, P., Kumaravel, S., and Nimia, N. (2019). Phytochemical, GC-MS and FT-IR Analysis of Papaver Somniferum L. Jpbs 7, 1–8. doi:10.18231/j.jpbs.2019.001
Nagori, K., Singh, M. K., Alexander, A., Kumar, T., Dewangan, D., Badwaik, H., et al. (2011). Piper Betle L.: A Review on its Ethnobotany, Phytochemistry, Pharmacological Profile and Profiling by New Hyphenated Technique DART-MS (Direct Analysis in Real Time Mass Spectrometry). J. Pharm. Res. 4, 2991–2997. Available at: http://jpronline.info/article/view/9266/4710.
Nahar, D. L., and Sarker, S. (2007). “Phytochemistry of the Genus Curcuma,” in Turmeric: The Genus Curcuma. Editors K. Ravindran, P. N., and K. N. Babu (Boca Raton, Florida: & Sivaraman), 71–106.
Nampoothiri, S. V., Philip, R. M., Kankangi, S., Kiran, C. R., and Menon, A. N. (2015). Essential Oil Composition, α-Amylase Inhibition and Antiglycation Potential ofCurcuma aromaticaSalisb. J. Essent. Oil Bearing Plants 18, 1051–1058. doi:10.1080/0972060X.2014.908746
Nieman, D. C., Cialdella-Kam, L., Knab, A. M., and Shanely, R. A. (2012). Influence of Red Pepper Spice and Turmeric on Inflammation and Oxidative Stress Biomarkers in Overweight Females: A Metabolomics Approach. Plant Foods Hum. Nutr. 67, 415–421. doi:10.1007/s11130-012-0325-x
Nikolova, M. (2006). Infraspecific Variability in the Flavonoid Composition of Artemisia Vulgaris L. Acta Bot. Croat. 65, 13–18.
Núñez, N., Vidal-Casanella, O., Sentellas, S., Saurina, J., and Núñez, O. (2020). Non-targeted Ultra-high Performance Liquid Chromatography-High-Resolution Mass Spectrometry (UHPLC-HRMS) Fingerprints for the Chemometric Characterization and Classification of Turmeric and Curry Samples. Separations 7, 32–13. doi:10.3390/separations7020032
Nurfina, A. N., Reksohadiprodjo, M. S., Timmerman, H., Jenie, U. A., Sugiyanto, D., and Van Der Goot, H. (1997). Synthesis of Some Symmetrical Curcumin Derivatives and Their Antiinflammatory Activity. Eur. J. Med. Chem. 32, 321–328. doi:10.1016/S0223-5234(97)89084-8
Ogundare, A. O., and Olajuyigbe, A. O. (2012). Bioactivity Guided Isolation of the Antifungal Components in Sawdust Extracts of Piptadeniatrum Africanum, and Terminalia Ivorensis. Malays. J. Microbiol. 8, 34–41. doi:10.21161/mjm.32611
Okumura, N., Yoshida, H., Nishimura, Y., Kitagishi, Y., and Matsuda, S. (2012). Terpinolene, a Component of Herbal Sage, Downregulates AKT1 Expression in K562 Cells. Oncol. Lett. 3, 321–324. doi:10.3892/ol.2011.491
Othman, R., Ibrahim, H., Mohd, M. A., Mustafa, M. R., and Awang, K. (2006). Bioassay-guided Isolation of a Vasorelaxant Active Compound from Kaempferia Galanga L. Phytomedicine 13, 61–66. doi:10.1016/j.phymed.2004.07.004
Paramasivam, M., Poi, R., Banerjee, H., and Bandyopadhyay, A. (2009). High-performance Thin Layer Chromatographic Method for Quantitative Determination of Curcuminoids in Curcuma Longa Germplasm. Food Chem. 113, 640–644. doi:10.1016/j.foodchem.2008.07.051
Park, S. Y., and Kim, D. S. (2002). Discovery of Natural Products from Curcuma Longa that Protect Cells from Beta-Amyloid Insult: a Drug Discovery Effort against Alzheimer's Disease. J. Nat. Prod. 65, 1227–1231. doi:10.1021/np010039x
Parthasarathy, V. A., and Chempakam, B. (2008). in “Chemistry of Spices,” in Chemistry Of Spices. Editors V. A. Parthasarathy, B. Chempakam, and T. J. Zachariah (Cambridge: CABI). 1–445. doi:10.4327/jsnfs1949.32.267
Pathania, V., Gupta, A. P., and Singh, B. (2006). Improved HPTLC Method for Determination of Curcuminoids from Curcuma Longa. J. Liquid Chromatogr. Relat. Tech. 29, 877–887. doi:10.1080/10826070500531417
Peng, H. Y., and Yang, X. E. (2005). Volatile Constituents in the Flowers of Elsholtzia Argyi and Their Variation: A Possible Utilization of Plant Resources after Phytoremediation. J. Zhejiang Univ. Sci. B 6, 91–95. doi:10.1631/jzus.2005.B0091
Phattanawasin, P., Sotanaphun, U., and Sriphong, L. (2009). Validated TLC-Image Analysis Method for Simultaneous Quantification of Curcuminoids in Curcuma Longa. Chroma 69, 397–400. doi:10.1365/s10337-008-0893-y
Prasath, D., Kandiannan, K., Leela, N. K., Aarthi, S., Sasikumar, B., and Babu, K. N. (2018). Turmeric. Hortic. Rev. 46, 99–184. doi:10.1002/9781119521082.ch3
Rahmatullah, M., Azam, M. N. K., Pramanik, S., Sania, S., Rahman, S., and Jahan, R. (2012). Antihyperglycemic Activity Evaluation of Rhizomes of Curcuma Zedoaria (Christm.) roscoe and Fruits of Sonneratia Caseolaris (L). Engl. Int. J. Pharmtech Res. 4, 125–129.
Raina, V. K., Srivastava, S. K., and Syamsundar, K. V. (2005). Rhizome and Leaf Oil Composition ofCurcuma Longafrom the Lower Himalayan Region of Northern India. J. Essent. Oil Res. 17, 556–559. doi:10.1080/10412905.2005.9698993
Ravindran, P. N. (2000). in Black Pepper: Piper Nigrum. Editor P. N. Ravindran (London: CRC Press).doi:10.1201/9780203303870
Rufino, A. T., Ribeiro, M., Judas, F., Salgueiro, L., Lopes, M. C., Cavaleiro, C., et al. (2014). Anti-inflammatory and Chondroprotective Activity of (+)-α-Pinene: Structural and Enantiomeric Selectivity. J. Nat. Prod. 77, 264–269. doi:10.1021/np400828x
Ruslay, S., Abas, F., Shaari, K., Zainal, Z., Maulidiani, , Sirat, H., et al. (2007). Characterization of the Components Present in the Active Fractions of Health Gingers (Curcuma Xanthorrhiza and Zingiber Zerumbet) by HPLC-DAD-ESIMS. Food Chem. 104 (3), 1183–1191. doi:10.1016/j.foodchem.2007.01.067
Saleh-e-in, M. M., Sultana, A., Husain, M., and Roy, S. K. (2010). Chemical Constituents of Essential Oil from Anethum Sowa L. Herb (Leaf and Stem) Growing in Bangladesh. Bangladesh J. Sci. Ind. Res. 45, 173–176. doi:10.3329/bjsir.v45i2.5721
Salman, Z., Mohd Azizi, C. Y., Nik Norulaini, N. A., and Mohd Omar, A. K. (2006). “Gas Chromatography /Time-Of-Flight Mass Spectrometry for Identification of Compounds from Parkia Speciosa Seeds Extracted by Supercritical Carbon Dioxide,” in Proceedings of the 1st International Conference on Natural Resources Engineering & Technology, Putrajaya, Malaysia, July 24–25, 2006, 112–120.
Sasikumar, B. (2005). Genetic Resources ofCurcuma: Diversity, Characterization and Utilization. Plant Genet. Resour. 3, 230–251. doi:10.1079/PGR200574
Sathiyabalan, G., Lincy, P., Muthukumarasamy, , and Mohan, V. (2014). GC-MS Analysis of Bioactive Components of Petiveria Alliacea L. Whole Plant (Phytolaccaceae). Int. J. Pharma Res. Heal. Sci. 2, 387–392.
Sati, S. C., Sati, N., and Sati, O. P. (2011). Bioactive Constituents and Medicinal Importance of Genus Alnus. Pharmacogn. Rev. 5, 174–183. doi:10.4103/0973-7847.91115
Setyaningsih, D., Murti, Y. B., Fudholi, A., Hinrichs, W. L. J., Mudjahid, R., Martono, S., et al. (2016). Validated TLC Method for Determination of Curcumin Concentrations in Dissolution Samples Containing Curcuma Longa Extract. J. Ilmu Kefarmasian Indones. 14, 147–157.
Sharopov, F., and Setzer, W. (2010). Composition of the Essential Oil of Achillea Filipendulina Lam. From Tajikistan. Der Pharma Chem. 2, 134–138.
Shishu, , and Maheshwari, M. (2010). Comparative Bioavailability of Curcumin, Turmeric and Biocurcumax in Traditional Vehicles Using Non-everted Rat Intestinal Sac Model. J. Funct. Foods 2, 60–65. doi:10.1016/j.jff.2010.01.004
Simoh, S., and Zainal, A. (2015). Chemical Profiling of Curcuma Aeruginosa Roxb. Rhizome Using Different Techniques of Solvent Extraction. Asian Pac. J. Trop. Biomed. 5, 412–417. doi:10.1016/S2221-1691(15)30378-6
Singh, G., Kapoor, I. P., Singh, P., de Heluani, C. S., de Lampasona, M. P., and Catalan, C. A. (2010). Comparative Study of Chemical Composition and Antioxidant Activity of Fresh and Dry Rhizomes of Turmeric (Curcuma Longa Linn.). Food Chem. Toxicol. 48, 1026–1031. doi:10.1016/j.fct.2010.01.015
Singh, G., Singh, O. P., and Maurya, S. (2002). Chemical and Biocidal Investigations on Essential Oils of Some Indian Curcuma Species. Prog. Cryst. Growth Characterization Mater. 45, 75–81. doi:10.1016/S0960-8974(02)00030-X
Sirat, H. M., and Meng, L. L. (2009). Chemical Components of the Rhizome Oil of Curcuma Heyneana Val. Mjs 28, 323–328. doi:10.22452/mjs.vol28no3.10
Slavík, J., and Slavíková, L. (1977). Alkaloids of Some Himalayan Species of Meconopsis Genus. Collect. Czech. Chem. Commun. 42, 132–139. doi:10.1135/cccc19770132
Song, Q., Yang, D., Zhang, G., and Yang, C. (2001). Volatiles from Ficus Hispida and Their Attractiveness to Fig Wasps. J. Chem. Ecol. 27, 1929–1942. doi:10.1023/a:1012226400586
Srinivasan, K., Sambaiah, K., and Chandrasekhara, N. (2004). Spices as Beneficial Hypolipidemic Food Adjuncts: A Review. Food Rev. Int. 20, 187–220. doi:10.1081/FRI-120037160
Srivastava, A. K., Srivastava, S. K., and Syamsundar, K. V. (2006). Volatile Composition ofCurcuma Angustifolia Roxb. Rhizome from central and Southern India. Flavour Fragr. J. 21 (3), 423–426. doi:10.1002/ffj.1680
Suksamrarn, A., Eiamong, S., Piyachaturawat, P., and Charoenpiboonsin, J. (1994). Phenolic Diarylheptanoids from Curcuma Xanthorrhiza. Phytochemistry 36, 1505–1508. doi:10.1016/S0031-9422(00)89751-4
Sundaryono, A., Nourmamode, A., Gardrat, C., Grelier, S., Bravic, G., Chasseau, D., et al. (2003). Studies on the Photochemistry of 1,7-Diphenyl-1,6-Heptadiene-3,5-Dione, a Non-phenolic Curcuminoid Model. Photochem. Photobiol. Sci. 2, 914–920. doi:10.1039/b301229h
Suntres, Z. E., Coccimiglio, J., and Alipour, M. (2015). The Bioactivity and Toxicological Actions of Carvacrol. Crit. Rev. Food Sci. Nutr. 55, 304–318. doi:10.1080/10408398.2011.653458
Tajidin, N. E. (2012). Chemical Composition and Citral Content in Lemongrass (Cymbopogon Citratus) Essential Oil at Three Maturity Stages. Afr. J. Biotechnol. 11, 2685–2693. doi:10.5897/ajb11.2939
Tang, J., Li, X., and Han, J. (2012). Analysis of Volatile Components in Rhizome Zingibers, Zingiber Officinale Roscoe and Ginger Pee by GC-MS and Chemometric Resolution. J. Chin. Med. Res. Dev. 1, 47–53. doi:10.9754/journal.wmc.2010.00662
Tetali, S. D., Acharya, S., Ankari, A. B., Nanakram, V., and Raghavendra, A. S. (2021). Metabolomics of Withania Somnifera (L.) Dunal: Advances and Applications. J. Ethnopharmacol. 267, 113469. doi:10.1016/j.jep.2020.113469
Tian, Y., Xu, W., Zhu, C., Lin, S., Li, Y., Xiong, L., et al. (2011). Lathyrane Diterpenoids from the Roots of Euphorbia Micractina and Their Biological Activities. J. Nat. Prod. 74, 1221–1229. doi:10.1021/np2001489
Tisserand, R., and Young, R. (2013). Essential Oil Safety-E-Book: A Guide for Health Care Professionals. Churchill Livingstone, London: Elsevier Health Sciences.
Umar, N. M., Parumasivam, T., Aminu, N., and Toh, S. M. (2020). Phytochemical and Pharmacological Properties of Curcuma Aromatica Salisb (Wild Turmeric). J. App Pharm. Sci. 10, 180–194. doi:10.7324/JAPS.2020.1010018
Usman, L., Hamid, A. A., George, O., Ameen, O., Muhammad, N. O., Zubair, M., et al. (2009). Chemical Composition of Rhizome Essential Oil of Curcuma longa L. growing in Growing in North Central Nigeria. World J. Chem. 4, 178–181.
Venkataramani, M., and Chinnagounder, S. (2012). Preliminary Phytochemical Screening and GC-MS Profiling of Hiptage Benghalensis ( L .) Kurz. J. Pharm. Res. 5, 2895–2899.
Wang, P., Liang, W., Kong, C., and Jiang, Y. (2005). Allelopathic Potential of Volatile Allelochemicals of Ambrosia Trifida L. On Other Plants. Allelopath. J. 15, 131–136.
Yang, S. A., Jeon, S. K., Lee, E. J., Im, N. K., Jhee, K. H., Lee, S. P., et al. (2009). Radical Scavenging Activity of the Essential Oil of Silver Fir (Abies alba). J. Clin. Biochem. Nutr. 44, 253–259. doi:10.3164/jcbn.08-240
Yuan, C., Nan, P., Shi, S., and Zhong, Y. (2003). Chemical Composition of the Essential Oils of Two Chinese Endemic Meconopsis Species. Z. Naturforsch C J. Biosci. 58, 313–315. doi:10.1515/znc-2003-5-603
Zellagui, A., Gherraf, N., and Rhouati, S. (2012). Chemical composition and antibacterial activity of the essential oils of Ferula vesceritensis Coss et Dur. leaves, endemic in Algeria. Org. Med. Chem. Lett. 2, 31. doi:10.1186/2191-2858-2-31
Keywords: Curcuma longa L., Curcuma aromatica Salisb, essential oil, metabolomics, secondary metabolites, GC-MS, LC-MS
Citation: Kulyal P, Acharya S, Ankari AB, Kokkiripati PK, Tetali SD and Raghavendra AS (2021) Variable Secondary Metabolite Profiles Across Cultivars of Curcuma longa L. and C. aromatica Salisb.. Front. Pharmacol. 12:659546. doi: 10.3389/fphar.2021.659546
Received: 27 January 2021; Accepted: 24 May 2021;
Published: 30 June 2021.
Edited by:
Sayeed Ahmad, Jamia Hamdard University, IndiaReviewed by:
Nicholas John Sadgrove, Royal Botanic Gardens, Kew, United KingdomAbdul Akbar, Siksha ‘O’ Anusandhan University, India
Copyright © 2021 Kulyal, Acharya, Ankari, Kokkiripati, Tetali and Raghavendra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarada D. Tetali, c3RldGFsaUB1b2h5ZC5hYy5pbg==; Agepati S. Raghavendra, YXNyc2xAdW9oeWQuZXJuZXQuaW4=, YXNfcmFnaGF2ZW5kcmFAeWFob28uY29t
†These authors have contributed equally to this work
 Poonam Kulyal
Poonam Kulyal Satyabrata Acharya†
Satyabrata Acharya† Sarada D. Tetali
Sarada D. Tetali Agepati S. Raghavendra
Agepati S. Raghavendra