- 1Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, United States
- 2St. Louis University School of Medicine, Department of Pharmacology and Physiology, St. Louis, MO, United States
- 3Center for Global Infectious Disease Research, Seattle Children’s Research Institute, Seattle, WA, United States
- 4Department of Pediatrics, University of Washington, Seattle, WA, United States
- 5Henry and Amelia Nasrallah Center for Neuroscience, Saint Louis University St. Louis, Seattle, WA, United States
- 6Graduate Program in Neuroscience, University of Washington, Seattle, WA, United States
Repurposed drugs that block the interaction between the SARS-CoV-2 spike protein and its receptor ACE2 could offer a rapid route to novel COVID-19 treatments or prophylactics. Here, we screened 2,701 compounds from a commercial library of drugs approved by international regulatory agencies for their ability to inhibit the binding of recombinant, trimeric SARS-CoV-2 spike protein to recombinant human ACE2. We identified 56 compounds that inhibited binding in a concentration-dependent manner, measured the IC50 of binding inhibition, and computationally modeled the docking of the best inhibitors to the Spike-ACE2 binding interface. The best candidates were Thiostrepton, Oxytocin, Nilotinib, and Hydroxycamptothecin with IC50’s in the 4–9 μM range. These results highlight an effective screening approach to identify compounds capable of disrupting the Spike-ACE2 interaction, as well as identify several potential inhibitors of the Spike-ACE2 interaction.
Introduction
COVID-19 is currently a global pandemic, causing extensive mortality and economic impact. While the success of rapidly developed vaccines offers hope to control the virus (Polack et al., 2020), treatments that improve disease outcomes are also critically needed. Dexamethasone, an anti-inflammatory steroid, is FDA-approved to treat COVID-19, as is Remdesivir, a nucleoside analogue prodrug that inhibits viral RNA polymerase (Beigel et al., 2020), though its efficacy is disputed (WHO Solidarity Trial Consortium, 2021). Blocking the interaction between the Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) spike protein and its obligatory receptor Angiotensin-converting enzyme 2 (ACE2), has also shown promise as a therapy; recombinant soluble ACE2 is effective in a cell culture model (Monteil et al., 2020), and three different monoclonal antibody drugs are now FDA approved (Marovich et al., 2020). However, these biologic drugs are expensive and suffer production limitations. Repurposing already-approved small molecule drugs, particularly those that might block the interaction between ACE2 and spike, could allow for rapid deployment of low-cost and widely available therapeutics (Saul and Einav, 2020). Thus far, all repurposed drug candidates have failed to reduce mortality, initiation of ventilation or hospitalization duration in robust clinical trials (Cao et al., 2020; BJM, 2020; WHO Solidarity Trial Consortium, 2021); however none of these candidates act trough a mechanism that involves blocking the ACE2-spike interaction.
Here, we aimed to identify repurposed drugs that could block the interaction between the SARS-CoV-2 spike protein and ACE2. We screened 2,701 drugs approved by global regulatory agencies using a previously published assay (Gniffke et al., 2020) that measures inhibition of binding between the trimeric SARS-CoV-2 spike protein (Wrapp et al., 2020) and latex-bead-conjugated recombinant human ACE2. We identified 56 compounds that inhibited the spike-ACE2 interaction by < 90% at 1 mM and that produced dilution curves that yielded an IC50 value, and further characterized the 12 compounds with the lowest half-maximal inhibitory concentration (IC50) using in silico modeling of the compounds’ interaction with the binding interface.
Methods
Drug Screening
The “FDA-approved drug screening library” (Cat #L1300) was purchased from Selleck Chemicals. Recombinant ACE2 and trimeric spike protein were produced in-house using previously published protocols (Gniffke et al., 2020). In a 96-well plate format, we briefly incubated recombinant, biotinylated, trimeric spike protein (Wrapp et al., 2020) with either 200 µM or 1 mM of each drug, in duplicate, then added 5-micron flow cytometry beads (Luminex) coated with recombinant ACE2 [for detailed methods, see (Gniffke et al., 2020)]. Three replicates per plate of positive (vehicle) controls and negative no-spike-protein controls were included, 31 plates in total. After washing and the addition of streptavidin-PE to bind spike attached to ACE2, plates were washed again on a magnetic plate washer and read on an Acea Novocyte flow cytometer. Data were expressed as the median PE fluorescence intensity (MFI), and converted to % inhibition using the formula 1-(MFIdrug/MFIpositive control). For IC50 studies, two independent serial dilutions of drugs were performed and run as above. IC50 was calculated in Graphpad Prism using the Hill equation for a normalized response with variable slope (four parameter).
In Silico Modeling
Docking experiments were performed with the SARS-COV-2 spike protein (PDB ID: 6VSB) and ACE2 (PDB ID: 2AJF). The region selected for docking studies was the receptor binding domain (RBD) of the Spike protein and the corresponding region of ACE2, with docking grids generated around key binding residues at the Spike-ACE2 interface. The missing loops in the S1 subunit of the 6VSB structure, which contains the RBD, were reconstructed using the SWISS-MODEL server (Waterhouse et al., 2018). The FASTA sequence (residues 316-530) was retrieved from UniProtKB-P0DTC2 and used as a query sequence, PDB ID: 6VSB with 100% sequence identity was used as a template (Berman et al., 2000; Pundir et al., 2016). The structure quality of the modelled protein was validated on PROCHECK, which showed 89.2% residues in the core regions, the quality factor of 89.77% was obtained from Verify 3D on the SAVES server and ProSA web gave a Z-score of-6.15 (Laskowski et al., 1993; Eisenberg et al., 1997; Wiederstein and Sippl, 2007). The crystal structure of ACE2 was retrieved from PDB 1D: 2AJF, pre-processed with Prime on Schrödinger, and used for docking into the RBD interface. The SDF structures of the selected FDA-approved drugs were downloaded from Selleck chemicals and PubChem. Ligand and protein preparation was performed using the Ligprep and protein preparation wizard tool on Schrödinger Maestro version 12.2. Structural-based docking to the RBD interface of the Spike and ACE2 was also performed using Schrödinger Maestro and BIOVIA Discovery Studio (Dassault Systems) for docking analysis and visualization.
Results
We took an unbiased approach to screen 2,701 drugs approved by global regulatory agencies for the ability to block the interaction between recombinant, trimeric SARS2 spike protein (Wrapp et al., 2020), and latex-bead-conjugated recombinant human ACE2 using a previously published assay (Gniffke et al., 2020) (Figure 1A). The use of a cell-free system prevented potential cytotoxic effects of drugs inherent to cell-culture-based, live-virus assays, and allowed us to focus solely on inhibition of the Spike-ACE2 interaction. We quantified the amount of Spike-ACE2 co-association in the presence of high concentrations (200 µM–1 mM) of each drug, in duplicate, and calculated the percent inhibition using six replicates of vehicle control per plate (31 plates total). In this first-round screen, 114 drugs that exhibited 90% or greater inhibition were identified (Figure 1B; Supplementary Table S1).
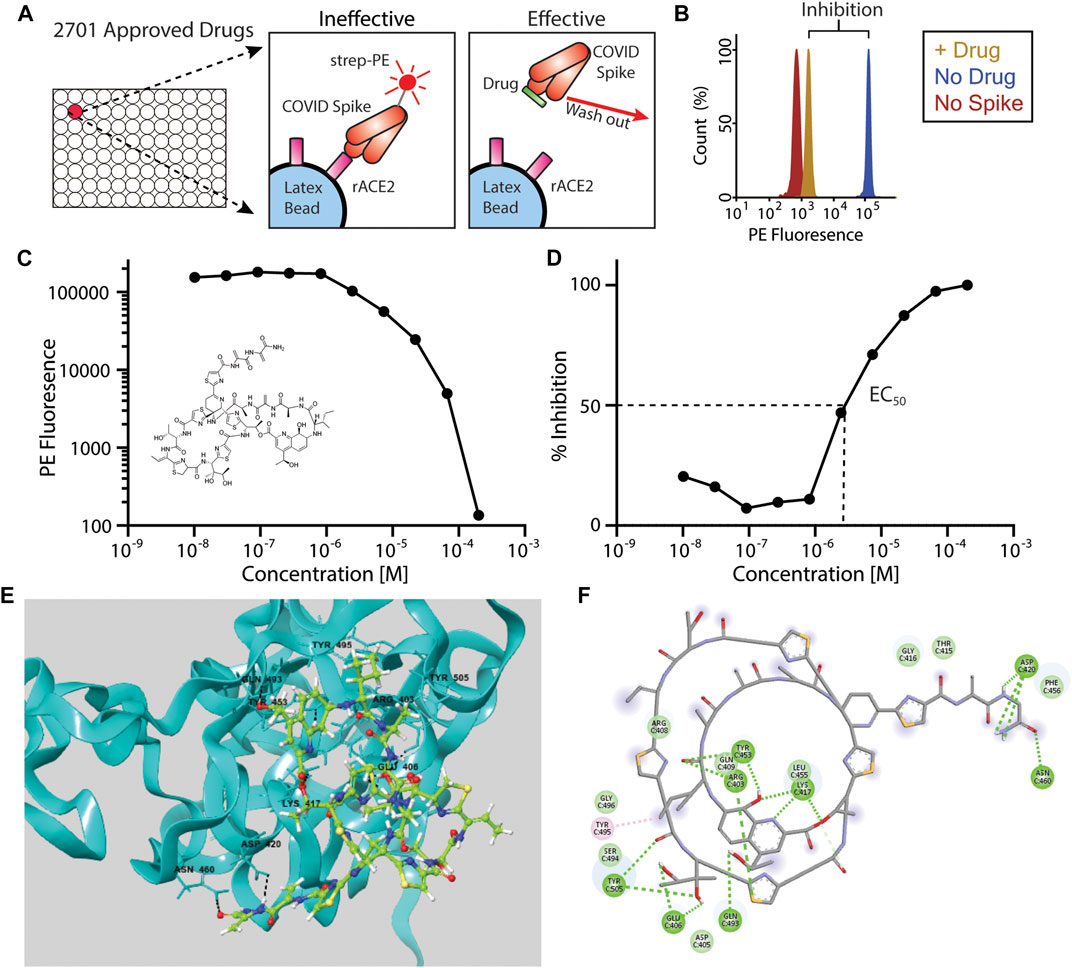
FIGURE 1. Inhibition of Spike-ACE2 binding by repurposed drugs. (A)In vitro assay design showing inhibition of ACE2-Spike binding by “effective” drugs. Note that compounds may also bind to ACE2, in addition to the spike protein as illustrated here. (B) Example histogram from primary screen showing >90% inhibition of ACE2-Spike binding following drug addition. (C, D) IC50 data for the top candidate, Thiostrepton (structure inset) expressed as PE fluorescence (C) or percent inhibition (D). (E) Three-dimensional and (F) two-dimensional computational rendering of Thiostrepton binding to the SARS-CoV-2 spike protein.
We next performed serial dilutions of these 114 drugs to measure the IC50s of the ACE2–spike interaction (Figures 1C,D). Fifty-eight of the drugs were revealed to be either false positive hits (they showed no inhibition upon re-screening), or showed inhibition only at the highest concentration tested, and were eliminated. The drug with the lowest (best) IC50 was Thiostrepton (IC50 = 3.95 ± 0.02 x 10−6 M), a cyclic oligopeptide used as a topical antibiotic in animals that interacts with the transcription factor FOXM1 to inhibit the growth of breast cancer cells in vitro (Hegde et al., 2011). Next was Oxytocin (IC50 = 4.10 ± 0.07 x 10−6 M), a peptide hormone that is administered to induce childbirth, and that may increase social cognition when administered intranasally (Keech et al., 2018). The next four best candidates were actually two closely related pairs of drugs, which demonstrates the robustness of our screen in identifying each compound twice. Nilotinib, which was identified as both a free base (IC50 = 4.21 ± 0.36 × 10−6 M) and an HCl salt (IC50 = 8.43 ± 1.18 × 10−6 M), is a selective tyrosine kinase inhibitor used to treat chronic myelogenous leukemia (Tokuhira et al., 2018). Hydroxycamptothecine (IC50 = 6.87 ± 0.77 × 10−6 M) and its stereoisomer S-10-Hydroxycamptothecine (IC50 = 7.22 ± 0.06 × 10−6 M) are DNA topoisomerase I inhibitors with anti-cancer activity (Fei et al., 2013). Interestingly, three derivatives that have also been approved for cancer therapy, Topotecan, Irinotecan, and Belotecan were included in the screening panel, but did not inhibit spike-ACE2 binding. The IC50s of all 56 compounds are listed in Supplementary Table S2; given the decreased severity of COVID-19 in females (Peckham et al., 2020), it is notable that Estradiol Benzoate (a synthetic estrogen) inhibited the interaction (IC50 = 1.75 × 10−5 M), although the IC50 we measured is far greater than the physiological concentration of estrogen.
We next performed molecular docking studies on the top twelve screening hits with both the Spike and ACE2 proteins, focusing on the interface region between the receptor binding domain (RBD) of the Spike protein and ACE2, to provide mechanistic insight into our identified compounds’ inhibitory activity. The Cryo-EM structure (PDB ID: 6VSB) was used for docking since the bioassay used the same trimeric spike protein plasmid construct as was used in determining that structure. The two most active screening hits, thiostrepton and oxytocin, were predicted to bind preferentially to the Spike protein based upon their Glide scores (Table 1), interacting with several key residues that mediate Spike-ACE2 binding (Hegde et al., 2011) (Figure 1; Supplementary Figures S1, S2; Supplementary Table S3). Thiostrepton in particular appears to bind extensively to Spike residues, but the OH-group in Thiostrepton was found to bind simultaneously with Lys417 of Spike and Asp30 of ACE2. Simultaneous binding of these two critical interface resides would likely disrupt the Spike-ACE2 interaction, resulting in the low observed EC50 value. Nilotinib and Hydroxycamptothecine exhibited higher Glide scores than thiostrepton and oxytocin in the Spike protein, but still within reasonable ranges. They were, however, predicted to have slightly more favorable binding with ACE2. Interestingly, both interacted with Arg393 on ACE2, a critical spike-binding residue, and both also interacted with the Spike receptor binding interface (Supplementary Figures S3–S6; Supplementary Table S3). The remaining compounds generally gave poorer glide scores, especially in the Spike protein. Docetaxel, Anidulafungin, and Estradiol, however, gave Glide scores in ACE2 that were comparable to the hydroxycamptothecin analogs, suggesting they may bind there. Some of the compounds such as Selamectin, Picropodophyllin, and Doramectin were predicted to bind poorly to both Spike and ACE2 (see Supplementary Figures S7–S12; Supplementary Table S3, and Supplementary Discussion). It is possible that these compounds bind to either Spike or ACE2 in a non-competitive manner outside the interface region that could result in conformational changes to the protein and thus disrupt the Spike-ACE2 interaction, as has been suggested for estradiol benzoate (Yang et al., 2021).
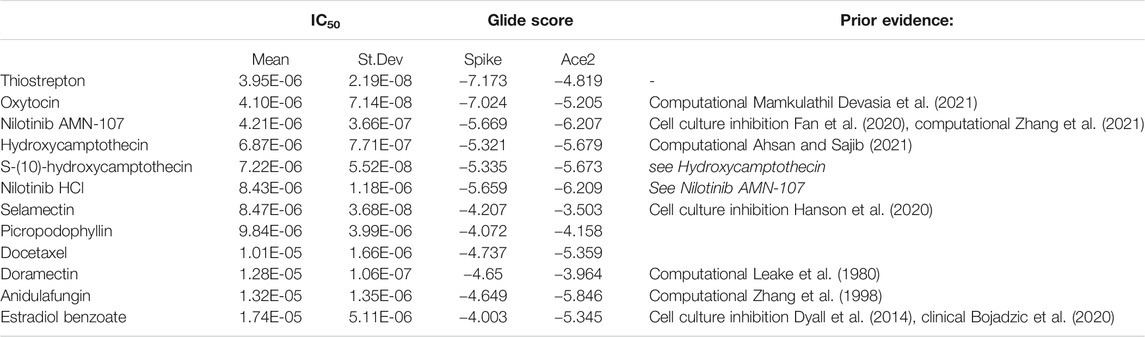
TABLE 1. Summary of the top 12 drug candidates. Computationally modeled glide scores for ACE2 and spike binding and IC50 values measured with the recombinant Spike-ACE2 binding inhibition assay are displayed.
Discussion
Overall, this study identified 56 approved drugs that show some efficacy in blocking the interaction between the COVID spike protein and its receptor, ACE2. Many of the identified drugs are already approved for clinical use in humans (Table 2). Moreover, several of the identified drugs have already shown promising results in computational modeling or cell-based studies (Table 1; Supplementary Table S2). Nilotinib (Hit #3 and #6 Table 1) has been reported to inhibit SARS-CoV-2 infection in vitro with an IC50 of 1.5–3 µM (Cagno et al., 2021), similar to our IC50 of 4 µM. The related Abl tyrosine kinase inhibitors Dasatinib and Imatinib were reported to inhibit SARS-CoV-1 and Middle Eastern Respiratory Syndroms (MERS) coronaviruses (Dyall et al., 2014). In our screen, three different formulations of Dasatinib inhibited > 90% in the first round, but only Dasatinib HCl (Hit #40 Supplementary Table S2) yielded an IC50 value, and Imatinib failed to inhibit in the first round screen. Similarly, Dasatinib, and Imatinib failed to inhibit SARS-CoV-2 in cell-culture-based studies (Cagno et al., 2021). Selamectin (Hit #7, Table 1) was one of three compounds identified in a screen of 2,406 clinically approved drugs that used a SARS-CoV-2-related pangolin corona virus in cell culture (Fan et al., 2020). This study also identified Cepharanthine (Hit #42 Supplementary Table S2), and Mefloquine (failed round 1, with 77% inhibition). Estradiol benzoate (Hit #12 Table 1) has also been reported to inhibit SARS-COV-2 replication in culture (Yang et al., 2021), possibly via binding to Spike in an area outside of the RBD. In humans, a retrospective study of women over 50 years old taking hormone therapy showed a 50% reduction in COVID-19 fatality that was not present in women 15–49 years old (Seeland et al., 2020). However, estrogen has a wide range of effects, including anti-inflammatory effects, and the concentration of estrogen in vivo is orders of magnitude less than that our observed IC50, so these data should be interpreted with caution.
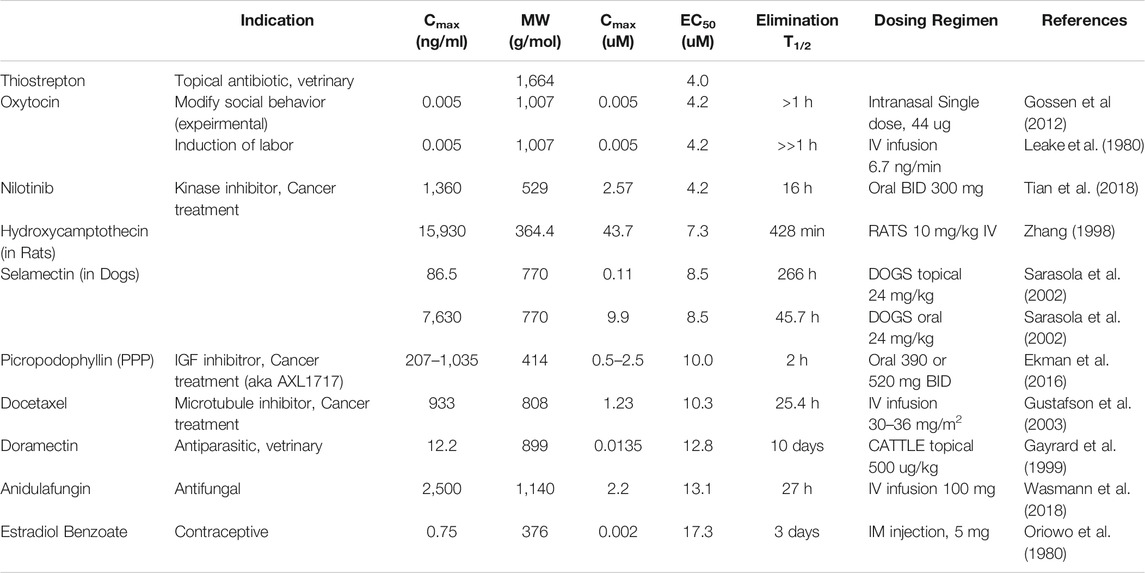
TABLE 2. Reported pharmacokinetic properties of top hits. All studies were performed on humans unless otherwise indicated.
We also note that at least two additional studies have screened compounds for the ability to block the receptor binding domain (RBD) of the Spike protein from binding to ACE2. Hanson et al.(2020) identified a single confirmed hit, Corilagin, which was not included in our drug panel. A second study (Bojadzic et al., 2020) screening organic dyes identified Methylene Blue as a potential inhibitor, which failed our screen in the first round (44% inhibition). Methodological differences, as well as our use of a stabilized prefusion trimeric spike protein, which behaves differently from the smaller RBD construct in our assay (Gniffke et al., 2020), may account for the differences between these studies.
Aiming towards clinical translation, there are several obvious issues with our identified compounds. First, many of the drugs are chemotherapy agents, and have toxic side-effects that would not be tolerable in COVID-19 patients. For example, Nilotinib inhibits a kinase important in B cell signaling (Tian et al., 2018; Tokuhira et al., 2018), and may prevent normal immune function, while Picropodophyllin (Ekman et al., 2016) and Docetaxel (Gustafson et al., 2003) both produce moderate to severe side effects when used in the context of chemotherapy. Secondly, several of the drugs have peak plasma concentrations that are orders of magnitude lower than the concentration required to inhibit spike-Ace2 binding in the in vitro assay, for example Oxytocin (Gossen et al., 2012) and Doramectin (Gayrard et al., 1999) (0.005 vs 4.2 µM and 0.014 vs 12.8 µM, respectively). Finally, Oxytocin’s clearance rate would necessitate continuous infusion, which seems impractical.
In light of known side-effects and pharmacokinetic data of many of our high-ranking drug candidates, Selamectin (Hit #7, Table 1) may be the top candidate for further study. Selamectin is an anti-parasitic used in dogs and cats to prevent infestation with nematode and arthropod species, and is structurally related to Ivermectin. Oral Selamectin is well tolerated in dogs and can achieve a peak plasma concentrations (9.9 µM) (Sarasola et al., 2002) comparable to both our measured IC50 (8.5 µM), and the 10 µM dose that inhibited the cytopathic effect of a SARS-CoV-2-related Pangolin corona virus on Vero E6 cells in culture (Fan et al., 2020). However, we were not able to identify any studies using Selamectin in humans, since Ivermectin is the standard alternative. Given recent controversy over Ivermectin as a treatment for COVID-19 (Ivermectin (2021)), it is worth noting that a different Ivermectin derivative, Doramectin, was our #10 hit, and Ivermectin itself just missed our first round cut-off with 89% binding inhibition. Thiostrepton may also be a top candidate, but pharmacokinetic data are lacking.
An advantage of our recombinant approach, using a cell-free system, is that the effects of drug toxicity on cell growth do not confound the readout of Spike-Ace2 binding. However, our results would need to be replicated in a cell or animal model using live virus to ensure that anti-SARS-CoV-2 effects appear at drug concentrations low enough to prevent toxicity. In addition, we did not confirm if the compounds bound directly to ACE2 or spike protein, only that their presence inhibited the binding of the two proteins. The relatively weak (micromolar) binding kinetics of the drugs identified here, as well as their known toxicities, bioactivities and/or high clearance rates, suggest that many would currently be unlikely to be viable for treating acute disease or for prophylactic use. However, they could serve as starting points for future medicinal chemistry optimization efforts to rationally design derivatives that are both less toxic and bind to the COVID spike with higher affinity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SS conceived the study. KT, CA, EG, JW, and SS planned experiments. DS provided recombinant COVID proteins for the inhibition assay. KT and EG performed microsphere-based experiments, CA performed modeling studies under the supervision of JW. KT, CA, JW, and SS, analyzed the data. KT, CA, JW, and SS wrote the manuscript, and all authors read and approved the manuscript.
Funding
This study was funded by a COVID-19 research award from the University of Washington Institute for Translational Heath Sciences (via grant UL1 TR002319) and by a COVID19 award from the Research Integration Hub at Seattle Children’s Research Institute (to SS), and by R01 AI140951 (to DS).
Conflict of Interest
Seattle Children’s Research Institute has filed a provisional patent on the use of the inhibition assay for drug screening described herein.
Acknowledgments
The authors thank Jason McLellan and Barney Graham for their generous gift of the SARS-CoV-2 recombinant trimer and hACE2 expression constructs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.685308/full#supplementary-material
References
Ahsan, T., and Sajib, A. A. (2021). Repurposing of Approved Drugs with Potential to Interact with SARS-CoV-2 Receptor. Biochem. Biophys. Rep. 26, 100982. doi:10.1016/j.bbrep.2021.100982
Bagheri, M., and Niavarani, A. (2020). Molecular Dynamics Analysis Predicts Ritonavir and Naloxegol Strongly Block the SARS-CoV-2 Spike Protein-hACE2 Binding. J. Biomol. Struct. Dyn., 1–10. doi:10.1080/07391102.2020.1830854
Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., et al. (2020). Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 383, 1813–1826. doi:10.1056/NEJMoa2007764
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235–242. doi:10.1093/nar/28.1.235
BMJ (2020). Hydroxychloroquine in Patients Mainly with Mild to Moderate COVID–19: an Open–Label, Randomized, Controlled Trial. medRxiv 369, 1849. doi:10.1136/bmj.m1849
Bojadzic, D., Alcazar, O., and Buchwald, P. (2020). Methylene Blue Inhibits the SARS-CoV-2 Spike-ACE2 Protein-Protein Interaction-A Mechanism that Can Contribute to its Antiviral Activity against COVID-19. Front. Pharmacol. 11, 600372. doi:10.3389/fphar.2020.600372
Cagno, V., Magliocco, G., Tapparel, C., and Daali, Y. (2021). The Tyrosine Kinase Inhibitor Nilotinib Inhibits SARS‐CoV‐2 In Vitro. Basic Clin. Pharmacol. Toxicol. 128, 621–624. doi:10.1111/bcpt.13537
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al. (2020). A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 382, 1787–1799. doi:10.1056/NEJMoa2001282
Dyall, J., Coleman, C. M., Hart, B. J., Venkataraman, T., Holbrook, M. R., Kindrachuk, J., et al. (2014). Repurposing of Clinically Developed Drugs for Treatment of Middle East Respiratory Syndrome Coronavirus Infection. Antimicrob. Agents Chemother. 58, 4885–4893. doi:10.1128/aac.03036-14
Eisenberg, D., Lüthy, R., and Bowie, J. U. (1997). [20] VERIFY3D: Assessment of Protein Models with Three-Dimensional Profiles. Methods Enzymol. 277, 396–404. doi:10.1016/s0076-6879(97)77022-8
Ekman, S., Harmenberg, J., Frödin, J.-E., Bergström, S., Wassberg, C., Eksborg, S., et al. (2016). A Novel Oral Insulin-like Growth Factor-1 Receptor Pathway Modulator and its Implications for Patients with Non-small Cell Lung Carcinoma: A Phase I Clinical Trial. Acta Oncologica 55, 140–148. doi:10.3109/0284186x.2015.1049290
Fan, H.-H., Wang, L.-Q., Liu, W.-L., An, X.-P., Liu, Z.-D., He, X.-Q., et al. (2020). Repurposing of Clinically Approved Drugs for Treatment of Coronavirus Disease 2019 in a 2019-novel Coronavirus-Related Coronavirus Model. Chin. Med. J. (Engl.) 133, 1051–1056. doi:10.1097/cm9.0000000000000797
Fei, B., Chi, A. L., and Weng, Y. (2013). Hydroxycamptothecin Induces Apoptosis and Inhibits Tumor Growth in colon Cancer by the Downregulation of Survivin and XIAP Expression. World J. Surg. Onc 11, 120. doi:10.1186/1477-7819-11-120
Gayrard, V., Alvinerie, M., and Toutain, P. L. (1999). Comparison of Pharmacokinetic Profiles of Doramectin and Ivermectin Pour-On Formulations in Cattle. Vet. Parasitol. 81, 47–55. doi:10.1016/s0304-4017(98)00236-2
Gniffke, E. P., Harrington, W. E., Dambrauskas, N., Jiang, Y., Trakhimets, O., Vigdorovich, V., et al. (2020). Plasma from Recovered COVID-19 Patients Inhibits Spike Protein Binding to ACE2 in a Microsphere-Based Inhibition Assay. J. Infect. Dis. 222, 1965–1973. doi:10.1093/infdis/jiaa508
Gossen, A., Hahn, A., Westphal, L., Prinz, S., Schultz, R. T., Gründer, G., et al. (2012). Oxytocin Plasma Concentrations after Single Intranasal Oxytocin Administration - a Study in Healthy Men. Neuropeptides 46, 211–215. doi:10.1016/j.npep.2012.07.001
Gustafson, D. L., Long, M. E., Zirrolli, J. A., Duncan, M. W., Holden, S. N., Pierson, A. S., et al. (2003). Analysis of Docetaxel Pharmacokinetics in Humans with the Inclusion of Later Sampling Time-Points Afforded by the Use of a Sensitive Tandem LCMS Assay. Cancer Chemother. Pharmacol. 52, 159–166. doi:10.1007/s00280-003-0622-z
Hanson, Q. M., Wilson, K. M., Shen, M., Itkin, Z., Eastman, R. T., Shinn, P., et al. (2020). Targeting ACE2-RBD Interaction as a Platform for COVID-19 Therapeutics: Development and Drug-Repurposing Screen of an AlphaLISA Proximity Assay. ACS Pharmacol. Transl. Sci. 3, 1352–1360. doi:10.1021/acsptsci.0c00161
Hegde, N. S., Sanders, D. A., Rodriguez, R., and Balasubramanian, S. (2011). The Transcription Factor FOXM1 Is a Cellular Target of the Natural Product Thiostrepton. Nat. Chem. 3, 725–731. doi:10.1038/nchem.1114
Imami, A. S., O’Donovan, S. M., Creeden, J. F., Wu, X., Eby, H., McCullumsmith, C. B., et al. (2020). Oxytocin's Anti-inflammatory and Proimmune Functions in COVID-19: a Transcriptomic Signature-Based Approach. Physiol. Genomics 52, 401–407. doi:10.1152/physiolgenomics.00095.2020
Ivermectin (2021). COVID-19 Treatment Guidelines. Available at: https://www.covid19treatmentguidelines.nih.gov/antiviral-therapy/ivermectin/.
Keech, B., Crowe, S., and Hocking, D. R. (2018). Intranasal Oxytocin, Social Cognition and Neurodevelopmental Disorders: A Meta-Analysis. Psychoneuroendocrinology 87, 9–19. doi:10.1016/j.psyneuen.2017.09.022
Khadim Sheikh, H., Arshad, T., Mohammad, Z. S., Arshad, I., and Hassan, M. (2020). Repurposed Single Inhibitor for Serine Protease and Spike Glycoproteins of SAR-CoV-2. doi:10.26434/chemrxiv.12192660.v1
Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993). PROCHECK: a Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 26, 283–291. doi:10.1107/s0021889892009944
Leake, R. D., Weitzman, R. E., and Fisher, D. A. (1980). Pharmacokinetics of Oxytocin in the Human Subject. Obstet. Gynecol. 56, 701–704.
Mamkulathil Devasia, R., Altaf, M., Fahad Alrefaei, A., and Manoharadas, S. (2021). Enhanced Production of Camptothecin by Immobilized Callus of Ophiorrhiza mungos and a Bioinformatic Insight into its Potential Antiviral Effect against SARS-CoV-2. J. King Saud Univ. - Sci. 33, 101344. doi:10.1016/j.jksus.2021.101344
Marovich, M., Mascola, J. R., and Cohen, M. S. (2020). Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA 324, 131. doi:10.1001/jama.2020.10245
Monteil, V., Kwon, H., Prado, P., Hagelkrüys, A., Wimmer, R. A., Stahl, M., et al. (2020). Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181, 905–913. doi:10.1016/j.cell.2020.04.004
Olotu, F. A., Omolabi, K. F., and Soliman, M. E. S. (2020). Leaving No Stone Unturned: Allosteric Targeting of SARS-CoV-2 Spike Protein at Putative Druggable Sites Disrupts Human Angiotensin-Converting Enzyme Interactions at the Receptor Binding Domain. Inform. Med. Unlocked 21, 100451. doi:10.1016/j.imu.2020.100451
Oriowo, M. A., Landgren, B. M., Stenström, B., and Diczfalusy, E. (1980). A Comparison of the Pharmacokinetic Properties of Three Estradiol Esters. Contraception 21, 415–424. doi:10.1016/s0010-7824(80)80018-7
Patil, S., Hofer, J., Ballester, P. J., Fattakhova, E., DiFlumeri, J., Campbell, A., et al. (2020). Drug Repurposing for Covid-19: Discovery of Potential Small-Molecule Inhibitors of Spike Protein-ACE2 Receptor Interaction through Virtual Screening and Consensus Scoring. doi:10.26434/chemrxiv.12482435.v1
Peckham, H., de Gruijter, N. M., Raine, C., Radziszewska, A., Ciurtin, C., Wedderburn, L. R., et al. (2020). Male Sex Identified by Global COVID-19 Meta-Analysis as a Risk Factor for Death and ITU Admission. Nat. Commun. 11, 6317. doi:10.1038/s41467-020-19741-6
Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615. doi:10.1056/nejmoa2034577
Pundir, S., Martin, M. J., and O’Donovan, C (2016). Uniprot Tools. Curr. Protoc. Bioinforma. 53, 1.29.1–1.29.15. doi:10.1002/0471250953.bi0129s53
Razizadeh, M., Nikfar, M., and Liu, Y. (2020). Small Molecules to Destabilize the ACE2-RBD Complex: A Molecular Dynamics Study for Potential COVID-19 Therapeutics. Chemrxiv Prepr. Serv. Chem. doi:10.26434/chemrxiv.13377119
Sarasola, P., Jernigan, A. D., Walker, D. K., Castledine, J., Smith, D. G., and Rowan, T. G. (2002). Pharmacokinetics of Selamectin Following Intravenous, Oral and Topical Administration in Cats and Dogs. J. Vet. Pharmacol. Ther. 25, 265–272. doi:10.1046/j.1365-2885.2002.00415.x
Saul, S., and Einav, S. (2020). Old Drugs for a New Virus: Repurposed Approaches for Combating COVID-19. ACS Infect. Dis. 6, 2304–2318. doi:10.1021/acsinfecdis.0c00343
Seeland, U., Coluzzi, F., Simmaco, M., Mura, C., Bourne, P. E., Heiland, M., et al. (2020). Evidence for Treatment with Estradiol for Women with SARS-CoV-2 Infection. BMC Med. 18, 369. doi:10.1186/s12916-020-01851-z
Tian, X., Zhang, H., Heimbach, T., He, H., Buchbinder, A., Aghoghovbia, M., et al. (2018). Clinical Pharmacokinetic and Pharmacodynamic Overview of Nilotinib, a Selective Tyrosine Kinase Inhibitor. J. Clin. Pharmacol. 58, 1533–1540. doi:10.1002/jcph.1312
Tokuhira, M., Kimura, Y., Sugimoto, K., Nakazato, T., Ishikawa, M., Fujioka, I., et al. (2018). Efficacy and Safety of Nilotinib Therapy in Patients with Newly Diagnosed Chronic Myeloid Leukemia in the Chronic Phase. Med. Oncol. Northwood Lond. Engl. 35, 38. doi:10.1007/s12032-018-1093-8
Wasmann, R. E., Heine, R. T., van Dongen, E. P., Burger, D. M., Lempers, V. J., Knibbe, C. A., et al. (2018). Pharmacokinetics of Anidulafungin in Obese and Normal-Weight Adults. Antimicrob. Agents Chemother. 62, 18. doi:10.1128/aac.00063-18
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
WHO Solidarity Trial Consortium. (2021). Repurposed Antiviral Drugs for Covid-19 — Interim WHO Solidarity Trial Results. N. Engl. J. Med. 384, 497–511. doi:10.1056/NEJMoa2023184
Wiederstein, M., and Sippl, M. J. (2007). ProSA-web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 35, W407–W410. doi:10.1093/nar/gkm290
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 367, 1260–1263. doi:10.1126/science.abb2507
Yang, C., Pan, X., Huang, Y., Cheng, C., Xu, X., Wu, Y., et al. (2021). Drug Repurposing of Itraconazole and Estradiol Benzoate against COVID‐19 by Blocking SARS‐CoV‐2 Spike Protein‐Mediated Membrane Fusion. Adv. Therap. 4, 2000224. doi:10.1002/adtp.202000224
Zhang, R., Li, Y., Cai, Q., Liu, T., Sun, H., and Chambless, B. (1998). Preclinical Pharmacology of the Natural Product Anticancer Agent 10-hydroxycamptothecin, an Inhibitor of Topoisomerase I. Cancer Chemother. Pharmacol. 41, 257–267. doi:10.1007/s002800050738
Keywords: COVID-19, drug screen, IP-FCM, inhibition assay, repurposed
Citation: Tsegay KB, Adeyemi CM, Gniffke EP, Sather DN, Walker JK and Smith SEP (2021) A Repurposed Drug Screen Identifies Compounds That Inhibit the Binding of the COVID-19 Spike Protein to ACE2. Front. Pharmacol. 12:685308. doi: 10.3389/fphar.2021.685308
Received: 24 March 2021; Accepted: 19 May 2021;
Published: 14 June 2021.
Edited by:
Rafael Maldonado, Pompeu Fabra University, SpainReviewed by:
Hasan Ejaz, Al Jouf University, Saudi ArabiaFilippo Prischi, University of Essex, United Kingdom
Peter Buchwald, University of Miami, United States
Copyright © 2021 Tsegay, Adeyemi, Gniffke, Sather, Walker and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen E. P. Smith, c2Vwc0B1dy5lZHU=
 Kaleb B. Tsegay
Kaleb B. Tsegay Christiana M. Adeyemi
Christiana M. Adeyemi Edward P. Gniffke1
Edward P. Gniffke1 John K. Walker
John K. Walker Stephen E. P. Smith
Stephen E. P. Smith