Abstract
Standard-dose caffeine citrate has been routinely prescribed for apnea of prematurity (AOP) management; however, some preterm infants respond well to the therapy while others do not. The AOP phenotype has been attributed solely to the immature control of the respiratory system consequent to preterm birth, but there are also important genetic influences. Based on our previous report, we tested the hypothesis that the human circadian locomotor output cycles kaput (CLOCK) gene polymorphisms play a role in the response to caffeine citrate therapy in preterm infants. We also studied the interactions of the circadian clock with aryl hydrocarbon receptor (AHR) signaling pathways in preterm babies who received caffeine citrate. This single-center study collected data from 112 preterm infants (<35 weeks gestational age) between July 2017 and July 2018, including apnea-free (n = 48) and apneic (n = 64) groups. Eighty-eight candidate single nucleotide polymorphisms (SNPs) were tested using the MassARRAY system. Association analysis was performed using the PLINK Whole Genome Data Analysis Toolset and SNPStats software. Linkage disequilibrium (LD) and haplotype analyses were performed using Hapview software. No significant intergroup differences in allele distributions or genotype frequencies of CYP1A2, CYP3A4, CYP3A5, and CYP3A7 were detected in our study on preterm babies. Two more SNPs in AHR were found to be associated with determining the response to caffeine citrate therapy in our pediatric patients. Of the 46 candidate SNPs in the CLOCK gene, 26 were found to be associated with determining the response to caffeine treatment in these babies. Interestingly, a significant association was retained for 18 SNPs in the CLOCK gene after false discovery rate correction. Moreover, strong LD formed in those variants in AHR, ADORA2A, and CLOCK genes was confirmed to be significantly associated with a better response to standard-dose caffeine therapy. In summary, CLOCK gene polymorphisms play a role in determining the response to caffeine therapy in premature neonates with AOP. However, whether the AHR and CLOCK signaling pathways crosstalk with each other during caffeine treatment remains largely unclear. Future clinical studies including more immature babies and basic research are needed to explore the mechanism by which circadian rhythms affect the response to caffeine therapy.
Introduction
The WHO defines preterm birth as birth before 37 weeks of gestation. Premature babies, especially those born very early, often have complicated medical problems. A preterm baby may have trouble breathing due to an immaturely developed respiratory control system. For example, some preterm babies may experience a cessation of breathing for 20 s or longer or a shorter pause accompanied by bradycardia (<100 beats per minute), cyanosis, or pallor, known as apnea of prematurity (AOP) (Eichenwald et al., 2016). Neonates with AOP experience frequent episodes of apnea, resulting in hypoxemia and bradycardia, all of which place the infant at risk for prolonged mechanical ventilation, retinopathy of prematurity (ROP), bronchopulmonary dysplasia (BPD), and long-term neurodevelopmental impairment (Puia-Dumitrescu et al., 2019). Frequent apnea may be one of the most troublesome problems in the neonatal intensive care unit (NICU).
Various mechanisms implicated in the pathogenesis of AOP have been identified, at least in the central nervous system and peripheral reflex pathways (Eichenwald et al., 2016). Ventilatory responses to hypoxia and inhibitory reflexes were exaggerated in preterm neonates. These unique vulnerabilities predispose the neonate to the development of apnea (Mathew, 2011; Martin and Wilson, 2012). Importantly, AOP may also have a critical genetic basis underlying this development-related disorder of respiratory control (Bloch-Salisbury et al., 2010). Methylxanthines have been the mainstay of pharmacological therapy for AOP for over 40 years, and caffeine is generally preferred over other methylxanthines because of its wider therapeutic index and lower incidence of serious complications (Morton and Smith, 2016). However, caffeine is not completely efficient, and in about half of treated infants their apnea frequency remains elevated; thus, aggressive interventions such as mechanical ventilation are needed (Laouafa et al., 2019; He et al., 2021).
The blockade of inhibitory adenosine A1 receptors (A1-AR; encoded by the ADORA1 gene), with the resultant respiratory neural output, as well as the blockade of excitatory adenosine A2A receptors (A2A-AR; encoded by ADORA2A gene) located on γ-aminobutyric acidergic neurons has been recognized as the primary mechanism of action of caffeine therapy (Eichenwald et al., 2016). Caffeine also competitively inhibits phosphodiesterases (PDEs) and binds to intracellular calcium channel ryanodine receptors, leading to intracellular Ca2+ release (Kumar and Lipshultz, 2019). Specific ADORA1 and ADORA2A polymorphisms have been associated with variability in response to caffeine therapy, as well as with a higher risk of AOP and BPD (Kumral et al., 2012). In our recent study, 88 single nucleotide polymorphisms (SNPs) in 19 genes encoding proteins involved in determining the disposition and pharmacological actions of caffeine were genotyped to evaluate the association between genetic mutations and the response to caffeine therapy in 112 preterm infants (He et al., 2021). The major findings of our previous study included the following: 1) No significant association between the plasma concentrations of caffeine and polymorphisms of caffeine-metabolism-related enzymes such as CYP1A2 and transcription factor aryl hydrocarbon receptor (AHR) was found; 2) polymorphisms of ADORA1, ADORA2A, ADORA3, and PDE4D, especially those of AHR and adenosine dehydrogenase (ADA) genes, play a critical role in determining interindividual variability to caffeine therapy. Interestingly, the influence of AHR polymorphisms on the response to caffeine therapy could not be explained by the mechanisms involving the AHR-CYP1A2 metabolic pathway (He et al., 2021). This AHR signaling pathway may play an important role in AOP development and the response to caffeine therapy.
In the respiratory control system, A1-ARs are found at high densities in the brainstem and anterior hypothalamus, while A2A-ARs are widely distributed in the medulla. Increased expression of ADORA1 and ADORA2A was observed in both the brainstem and hypothalamus of caffeine-treated neonatal rats. Increased adenosinergic maturation in the central cardiorespiratory areas could partly explain the pharmacological effects of caffeine observed in premature infants (Gaytan and Pasaro, 2012). Adenosine is an inhibitory neuromodulator involved in sleep-wake regulation. A2A-ARs, but not A1-ARs, mediate the arousal effect of caffeine (Huang et al., 2005). Intriguingly, caffeine has been shown to influence circadian timing in humans by an A1-AR/cAMP-dependent mechanism (Burke et al., 2015). The basic mechanism of the clock is a cell-autonomous interlocked transcription-translation feedback loop sustained by transcriptional activators brain and muscle ARNT-Like 1 (BMAL1), also known as aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL1), Circadian Locomotor Output Cycles Kaput (CLOCK) or neuronal PAS domain protein (NPAS2), as well as period (PER) and cryptochrome (CRY) (Hardin and Panda, 2013). As the core molecular clock is present in nearly every cell in mammals (Mohawk et al., 2012), the circadian clock is likely involved in determining the tissue response to circulating factors, including drugs (Zhang et al., 2021). However, whether the human circadian clock affects the response to caffeine therapy in preterm infants is unknown.
Caffeine citrate was routinely prescribed for AOP management in the NICU. Some preterm infants respond well to caffeine, while others do not. We hypothesized that the human circadian clock is involved in the response to caffeine citrate therapy in preterm infants. The overarching goal of this study was to determine the association between circadian CLOCK gene polymorphisms in preterm babies and the clinical outcomes after caffeine treatment. In addition, this study touches on the crosstalk between the circadian clock and AHR signaling pathways based on our findings in preterm babies who received caffeine citrate treatment.
Methods
A comprehensive description of the AOP, BPD, ROP, intraventricular hemorrhage (IVH), patent ductus arteriosus (PDA), and necrotizing enterocolitis (NEC), as well as the methods for determining the plasma caffeine concentration and collecting clinical data for a single-center, retrospective study in Chinese preterm infants has been published elsewhere (He et al., 2021). The caffeine citrate treatment protocol, clinical outcomes, and grouping criteria have also been clearly described (He et al., 2021). Briefly, the apneic group included patients with apnea episodes that occurred even once despite the combined use of necessary nonpharmacological therapies, and the apnea-free group included those without apnea following the administration of caffeine and necessary respiratory support options. The study protocol was approved by the hospital ethics committee of the Children’s Hospital of Nanjing Medical University (protocol number 201902082-1).
Study Subjects
Fifty-five female and 57 male preterm babies were included in this study, of whom 104 were born at <32 weeks gestational age. Among these preterm babies, the median postmenstrual age was 29.3 weeks. The median birth weight was 1,265 g. The mean total duration of caffeine exposure was approximately 34 days. The median invasive intubation duration for respiratory support therapy in the 61 infants was 2 days, and the repeat intubation rate was approximately 26%. The use of nasal continuous positive airway pressure or supplemental oxygen therapy continued for a median of 26 days.
MassArray SNP Detection
Because of the CLOCK pathways connected to the AHR pathway and the roles of caffeine in human circadian timing, we focused mainly on the following genes: AHR repressor (AHRR), aryl hydrocarbon receptor nuclear translocator (ARNT), BMAL1, and CLOCK. For CLOCK and BMAL1 genes, SNPs associated with neuropsychiatric diseases and sleep homeostasis were selected as candidates. In addition, CYP3A4, CYP3A5, CYP3A7, and CYP3A43, especially CYP3A7, this isoform of CYP3A, has been demonstrated to metabolize endogenous compounds that are known to be important in the growth and development of fetuses and neonates. Ultimately, 88 relative SNPs were genotyped and evaluated in the current study.
The selected 88 SNPs in nine human genes were genotyped using the Agena MassARRAY platform 4.0 with iPLEX gold chemistry (Agena Bioscience, Inc., CA, United States). The main reagents contained Agena polymerase chain reaction (PCR) reagent, Agena shrimp alkaline phosphatase (SAP) reagent, and Agena iPLEX reagent. Genomic DNA was extracted from the peripheral blood cell sediment using a DNA extraction kit (BioTeke Corporation, Wuxi, China) according to the manufacturer’s instructions and stored at −80°C before genotyping. The purity and concentration of the extracted genomic DNA samples were assessed by absorbance measurements at 230, 260, and 280 nm using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, MA, United States). The Agena Bioscience Assay Designer 4.0 was used to design PCR amplification and extension primers for the 88 SNP variants. The primers used are listed in Supplementary Table S1. PCR master mixtures were obtained using the Agena PCR reagent set, the PCR procedures were started, and then the mixtures were treated with SAP. After extending the reaction, the samples underwent resin purification processing and dispensing on the 384-well SpectroCHIP bioarray using the MassARRAY Nanodispenser RS1000 spotter (Agena Bioscience, Inc., CA, United States), and the masses of the primer extension products correlated with genotype were then determined using matrix-assisted laser desorption/ionization time-of flight mass spectrometry (MALDI-TOF MS) (Agena Bioscience, Inc., CA, United States). The spectral profiles generated by MALDI-TOF MS were analyzed using MassARRAY TYPER 4.0 software (Agena Bioscience, Inc., CA, United States). Please refer to the study of Cheung and colleagues for a specific process (Cheung et al., 2016).
Statistical Analysis
Statistical analyses were performed using SPSS version 25.0 software (IBM, Armonk, United States), and Figure 1 was drawn using GraphPad Prism 5 (GraphPad Software, CA, United States). The allele and genotype frequencies of various genes, including CYP1A2, CYP3A4, CYP3A5, CYP3A7, AHR, AHRR, ARNT, BMAL1, and CLOCK, were examined for deviation from the Hardy–Weinberg equilibrium (HWE) using the goodness-of-fit chi-square test. Distributions of genotype among neonates in the apneic and apnea-free groups were compared using the goodness-of-fit chi-square test. The Benjamini–Hochberg false discovery rate (FDR) was used for multiple hypothesis testing, and the adjusted p value (pFDR) was obtained. Multivariable logistic regression analysis was established through variable screening to explore the effect of significant outcomes from the univariate analysis of apneic and apnea-free groups. Statistical significance was set at p < 0.05.
FIGURE 1
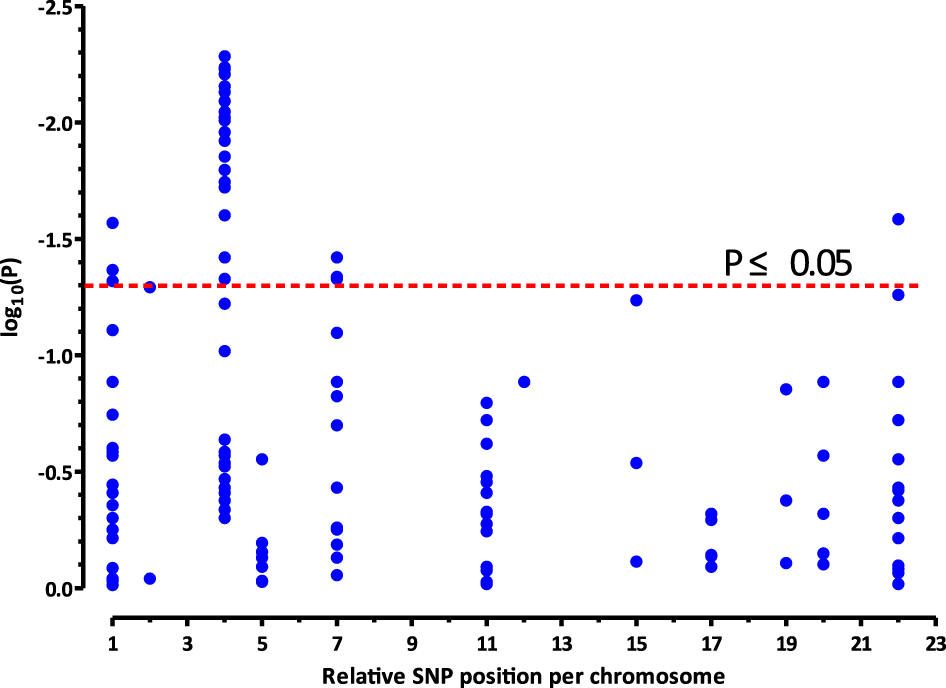
Significances (plotted as−log10 of nominal p values, closed blue circles) resulting from the associations between the investigated SNPs and different response (apnea-free vs. apneic) to caffeine therapy in preterm infants (n = 112). The x-axis represents the relative genomic position/chromosome, and the y-axis represents the distribution of the nominal−log10(P) values in the two groups of infants. The 26 SNPs, which are associated with the response to caffeine treatment (p < 0.05; above stippled red line), map to the CLOCK gene located on chromosome 4.
The association between these tested SNPs and the response to caffeine citrate therapy was estimated by calculating odds ratios (ORs) and 95% confidence intervals (CIs) from logistic regression under the allele and recessive [AA vs. (CA + CC)], dominant [(CA + AA) vs. CC], additive (AA vs CC), over-dominant [(CC + AA) vs CA], and co-dominant (AA vs CA vs CC) models (A represents mutant allele and C represents wild type allele) using the PLINK Whole genome data analysis Toolset (version 1.07; http://zzz.bwh.harvard.edu/plink/tutorial.shtml) and SNPStats software (https://www.snpstats.net). Linkage disequilibrium (LD) and haplotype analyses were evaluated using the Hapview 4.2 software (The Broad Institute, MA, United States), while the difference in distribution of haplotype frequencies between the apneic and apnea-free groups was also tested using the goodness-of-fit chi-square analysis.
Results
Genotyping
In our previous study, we reported an association between 88 candidate SNPs in 19 human genes and the response to caffeine therapy (He et al., 2021). In this study, a total of 112 preterm babies were genotyped for 88 more SNPs in nine human genes, CYP1A2, CYP3A4, CYP3A5, CYP3A7, AHR, AHRR, ARNT, BMAL1, and CLOCK. The reference sequence (rs) numbers, alleles, call rate, minor allele frequencies (MAF), and HWE tests of the 176 SNPs included in this study are shown in Table 1. Eighty SNPs were common polymorphisms with MAFs of 1.4–50% and were in HWE (p > 0.01). Eight SNPs, including rs3805147 (CLOCK), rs3749474 (CLOCK), rs13132420 (CLOCK), rs1122780 (BMAL1), rs12504300 (BMAL1), rs2069514 (CYP1A2*1C), rs35694136 (CYP1A2*1D), and rs61469810 (CYP3A43*2A), were removed for further analysis because of low call rate and/or poor type clustering.
TABLE 1
| Gene | Location | SNP | Alleles | MAF | Call rate (%) | HWE | pe |
|---|---|---|---|---|---|---|---|
| ADORA1 (Adenosine A1 receptor) | 1q32.1 | rs16851030 | C/T | 0.3519 | 100 | 0.054 | 0.39 |
| rs10920568 | T/G | 0.183 | 100 | 0.3581 | 0.13 | ||
| rs5780149 | DEL/T | 0.1045 | 97 | 1 | 0.44 | ||
| rs6427994 | C/A | 0.1514 | 97 | 1 | 0.26 | ||
| rs41264025 | C/T | 0.02232 | 100 | 1 | 0.27a | ||
| rs6664108 | T/C | 0.2636 | 98 | 0.6221 | 0.93 | ||
| rs12744240 | G/T | 0.1473 | 100 | 0.4586 | 0.043 | ||
| rs3766553 | A/G | 0.192 | 100 | 0.02789 | NA | ||
| rs903361 | G/A | 0.3616 | 100 | 0.3146 | 0.82 | ||
| rs10920573 | T/C | 0.3773 | 98 | 0.07106 | 0.13 | ||
| rs6701725 | A/G | 0.1802 | 99 | 1 | 0.078 | ||
| rs3766566 | G/A | 0.3063 | 99 | 0.1799 | 0.36 | ||
| rs6677137 | T/C | 0.1636 | 98 | 0.03909 | 0.61 | ||
| rs3753472 | T/C | 0.08929 | 100 | 1 | 0.56 | ||
| ADORA2A (Adenosine A2A receptor) | 22q11.23 | rs5751862 | G/A | 0.455 | 99 | 1 | 0.13 |
| rs5760405a | C/T | <0.001 | 98 | NONE | NONE | ||
| rs2298383 | C/T | 0.473 | 100 | 1 | 0.42 | ||
| rs3761422 | T/C | 0.4364 | 100 | 0.4959 | 0.83 | ||
| rs2267076 | T/C | 0.4459 | 100 | 0.5104 | 0.8 | ||
| rs5751876 | T/C | 0.473 | 100 | 1 | 0.42 | ||
| rs4822492 | C/G | 0.4727 | 100 | 1 | 0.38 | ||
| rs5760423 | T/G | 0.473 | 100 | 1 | 0.42 | ||
| rs2236624 | T/C | 0.3348 | 100 | 0.671 | 0.19 | ||
| rs5760425 | T/G | 0.4775 | 98 | 0.8493 | 0.5 | ||
| rs4822498 | T/C | 0.486 | 95 | 0.175 | 0.38 | ||
| rs34923252 | T/A | 0.08482 | 100 | 0.5654 | 0.026 | ||
| rs5996696 | A/C | 0.08482 | 100 | 0.5654 | 0.026 | ||
| rs5760410 | G/A | 0.4633 | 97 | 0.8478 | 0.61 | ||
| rs4822489 | T/G | 0.4866 | 100 | 0.09115 | 0.86 | ||
| rs2236625 | C/T | 0.09009 | 99 | 1 | 0.055 | ||
| ADORA2B (Adenosine A2B receptor) | 17p12 | rs2779193 | A/G | 0.02679 | 100 | 1 | 0.72c |
| rs758857 | G/A | 0.2991 | 100 | 0.4986 | 0.51 | ||
| rs758858 | A/G | 0.02703 | 99 | 1 | 0.73c | ||
| rs2041458 | A/C | 0.02679 | 100 | 1 | 0.72c | ||
| rs17715109 | G/T | 0.06696 | 100 | 1 | 0.81a | ||
| rs2015353 | T/C | 0.1545 | 98 | 1 | 0.48 | ||
| rs2779211 | A/G | 0.1545 | 98 | 1 | 0.48 | ||
| ADORA3 (Adenosine A3 receptor) | 1p13.2 | rs2298191 | T/C | 0.308 | 100 | 0.3736 | 0.048 |
| rs10776727 | C/A | 0.3818 | 98 | 0.5441 | 0.027 | ||
| rs1544224d | A/G | NONE | 100 | NONE | NONE | ||
| rs2229155 | A/G | 0.2545 | 98 | 0.449 | 0.5 | ||
| rs1544223 | C/T | 0.3468 | 99 | 0.6771 | 0.97 | ||
| ADA (Adenosine deaminase) | 20q13.12 | rs73598374d | C/T | NONE | 19 | NONE | NONE |
| rs6575353 | G/A | 0.4685 | 100 | 0.6544 | 0.48 | ||
| rs521704 | C/A | 0.4773 | 98 | 0.1794 | 0.79 | ||
| rs2472304 | G/A | 0.1577 | 100 | 1 | 0.13 | ||
| rs4998386a | C/T | <0.001 | 100 | NONE | NONE | ||
| rs382140 | A/G | 0.1111 | 97 | 0.6599 | 0.27 | ||
| rs9526558 | A/G | 0.1396 | 100 | 0.1998 | 0.71a | ||
| rs17498920a | G/A | <0.001 | 100 | NONE | NONE | ||
| AHR (Aryl hydrocarbon receptor) | 7p21.1 | rs4410790 | T/C | 0.4045 | 100 | 0.3618 | 0.047 |
| rs6968865 | A/T | 0.3739 | 100 | 0.8135 | 0.046 | ||
| rs2066853 | G/A | 0.3739 | 99 | 0.315 | 0.047 | ||
| rs1476080 | T/G | 0.3973 | 100 | 0.8459 | 0.038 | ||
| rs10250822 | C/T | 0.4144 | 99 | 0.6992 | 0.55 | ||
| rs6960165 | G/A | 0.2027 | 99 | 0.1479 | 0.56 | ||
| rs7811989 | A/G | 0.2321 | 100 | 0.5991 | 0.2 | ||
| rs2158041 | T/C | 0.2321 | 100 | 0.2934 | 0.13 | ||
| rs10249788 | C/T | 0.2297 | 99 | 1 | 0.08 | ||
| AHRR (Aryl-hydrocarbon receptor repressor) | 5p15.33 | rs2292596 | C/G | 0.3333 | 0.01834 | 0.93 | |
| ARNT (Aryl hydrocarbon receptor nuclear translocator) | 1q21.3 | rs2228099 | C/G | 0.4018 | 100 | 0.5547 | 0.91 |
| BMAL1 (Aryl hydrocarbon receptor nuclear translocator-like protein 1 or Brain and Muscle ARNT-Like 1) | 11p15.3 | rs11022775 | C/T | 0.125 | 96 | 1 | 0.84 |
| rs2290035 | T/A | 0.3191 | 84 | 0.2426 | 0.81 | ||
| rs3816358 | C/A | 0.2228 | 90 | 0.3877 | 0.39 | ||
| rs2278749 | C/T | 0.1963 | 96 | 0.06907 | 0.48 | ||
| rs3816360 | T/C | 0.398 | 88 | 0.1438 | 0.47 | ||
| rs1481892 | G/C | 0.4909 | 98 | 0.05564 | 0.35 | ||
| rs1122780d | A/G | NONE | 73 | NONE | NONE | ||
| rs2279287 | T/C | 0.4954 | 97 | 0.03606 | 0.53 | ||
| rs4757144 | G/A | 0.4409 | 98 | 0.1766 | 0.57 | ||
| rs969485 | G/A | 0.4953 | 95 | 0.7002 | 0.96 | ||
| rs4757142 | G/A | 0.3423 | 99 | 0.676 | 0.94 | ||
| rs7126303 | C/T | 0.1759 | 96 | 0.313 | 0.33 | ||
| rs6486120 | T/A | 0.4771 | 97 | 1 | 0.16 | ||
| rs1868049 | C/T | 0.4554 | 90 | 0.1599 | 0.19 | ||
| CLOCK (Circadian Locomotor Output Cycles Kaput) | 4q12 | rs1801260 | A/G | 0.08929 | 100 | 1 | 0.014 |
| rs2272073 | C/T | 0.4159 | 96 | 0.2317 | 0.018 | ||
| rs6811520 | T/C | 0.3287 | 96 | 0.3815 | 0.5 | ||
| rs3762836 | T/C | 0.01429 | 94 | 1 | 0.06a | ||
| rs3805148 | A/C | 0.3952 | 94 | 0.2211 | 0.011 | ||
| rs12504300d | G/C | NONE | 94 | NONE | NONE | ||
| rs12649507 | G/A | 0.4028 | 96 | 0.2291 | 0.0058 | ||
| rs534654 | A/G | 0.01835 | 97 | 1 | 0.42c | ||
| rs6850524 | C/G | 0.3178 | 96 | 0.8255 | 0.3 | ||
| rs4340844 | A/C | 0.3991 | 97 | 0.1106 | 0.007 | ||
| rs3736544 | A/G | 0.316 | 95 | 1 | 0.27 | ||
| rs12648271 | G/C | 0.3962 | 95 | 0.3184 | 0.012 | ||
| rs2412648 | T/G | 0.3981 | 96 | 0.3141 | 0.0052 | ||
| rs11735267 | C/T | 0.3165 | 97 | 1 | 0.34 | ||
| rs7660668 | C/G | 0.08716 | 97 | 0.5761 | 0.0081 | ||
| rs10462028 | G/A | 0.07477 | 96 | 0.545 | 0.0081 | ||
| rs11932595 | A/G | 0.07944 | 96 | 0.4981 | 0.019 | ||
| rs2070062 | A/C | 0.09545 | 98 | 0.2452 | 0.016 | ||
| rs13132420d | G/A | NONE | 26 | NONE | NONE | ||
| rs11824092 | T/C | 0.4104 | 95 | 0.2289 | 0.39 | ||
| rs11931061 | G/A | 0.3108 | 99 | 1 | 0.26 | ||
| rs11133385 | G/A | 0.3879 | 96 | 0.3074 | 0.0059 | ||
| rs3749474d | C/T | NONE | 87 | NONE | NONE | ||
| rs2412646 | T/C | 0.3066 | 95 | 1 | 0.39 | ||
| rs11240 | C/G | 0.07477 | 96 | 0.454 | 0.0081 | ||
| rs3805151 | T/C | 0.3925 | 96 | 0.4172 | 0.0062 | ||
| rs4580704 | G/C | 0.3102 | 96 | 1 | 0.3 | ||
| rs10002541 | T/C | 0.3148 | 96 | 1 | 0.3 | ||
| rs4864546 | G/A | 0.3868 | 95 | 0.3077 | 0.0074 | ||
| rs6858749 | T/C | 0.1055 | 97 | 1 | 0.038 | ||
| rs6843722 | A/C | 0.4028 | 96 | 0.2291 | 0.0058 | ||
| rs4864548 | G/A | 0.3879 | 96 | 0.3074 | 0.0059 | ||
| rs13102385 | T/C | 0.3165 | 97 | 1 | 0.34 | ||
| rs7698022 | G/T | 0.08796 | 96 | 0.5798 | 0.009 | ||
| rs11133389 | C/T | 0.3952 | 94 | 0.4152 | 0.0098 | ||
| rs11133373 | C/G | 0.4078 | 92 | 0.5397 | 0.096 | ||
| rs11133391 | T/C | 0.3972 | 96 | 0.314 | 0.0095 | ||
| rs1048004 | C/A | 0.07477 | 96 | 0.454 | 0.0081 | ||
| rs11943456 | T/C | 0.09259 | 96 | 1 | 0.047 | ||
| rs3792603 | A/G | 0.075 | 89 | 0.4326 | 0.025 | ||
| rs11726609 | T/A | 0.3131 | 96 | 1 | 0.23 | ||
| rs17721497 | A/T | 0.07944 | 96 | 0.4981 | 0.019 | ||
| rs3817444 | A/C | 0.3131 | 96 | 0.8234 | 0.26 | ||
| rs9312661 | G/A | 0.3972 | 96 | 0.314 | 0.0095 | ||
| rs726967 | A/T | 0.3194 | 96 | 0.661 | 0.3 | ||
| rs6832769 | G/A | 0.316 | 95 | 1 | 0.27 | ||
| rs3805147d | C/T | NONE | 67 | NONE | NONE | ||
| CYP1A1 | 15q24.1 | rs2470893a | C/T | <0.001 | 100 | NONE | NONE |
| CYP1A2 | rs2472297a | C/T | <0.001 | 100 | NONE | NONE | |
| rs2472299 | A/G | 0.3554 | 100 | 0.8039 | 0.29 | ||
| rs762551 | C/A | 0.3354 | 100 | 0.8039 | 0.29 | ||
| CYP1A2*1B | rs2470890 | T/C | 0.1468 | 97 | 0.6976 | 0.058 | |
| CYP1A2*1C | rs2069514d | G/A | NONE | 4 | NONE | NONE | |
| CYP1A2*1D | rs35694136d | T/DEL | NONE | 0 | NONE | NONE | |
| CYP1A2*1E | rs2069526 | T/G | 0.1126 | 99 | 1 | 0.77 | |
| CYP2A6*7 | 19q13.2 | rs5031016 | A/G | 0.1343 | 97 | 0.3429 | 0.78a |
| CYP2D6*2A | 22q13.2 | rs16947 | G/A | 0.15 | 100 | 0.06265 | 0.37a |
| CYP2D6*10 | rs1065852 | G/A | 0.4909 | 100 | 1 | 0.96 | |
| rs1135840 | C/G | 0.3257 | 98 | 0.04812 | 0.28 | ||
| CYP3A4*1B | 7q22.1 | rs2740574b | C/T | <0.001 | 99 | NONE | NONE |
| CYP3A4*1G | rs2242480 | C/T | 0.2252 | 100 | 1 | 0.88 | |
| CYP3A4*4 | rs55951658b | T/C | <0.001 | 100 | NONE | NONE | |
| CYP3A4*18A | rs28371759b | A/G | <0.001 | 99 | NONE | NONE | |
| CYP3A4*23 | rs2687116b | C/A | <0.001 | 95 | NONE | NONE | |
| CYP3A4*23 | rs3735451 | T/C | 0.2432 | 99 | 0.6055 | 0.65 | |
| CYP3A4 | rs4646437 | G/A | 0.08036 | 100 | 1 | 0.15 | |
| CYP3A5*4 | rs56411402b | T/C | <0.001 | 100 | NONE | NONE | |
| CYP3A5*5 | rs55965422b | A/G | <0.001 | NONE | NONE | ||
| CYP3A7*1D | rs55798860b | C/T | <0.001 | 99 | NONE | NONE | |
| CYP3A7*2 | rs2257401 | G/C | 0.2188 | 100 | 1 | 0.74 | |
| CYP3A7 | rs10211b | T/A | <0.001 | 100 | NONE | NONE | |
| CYP3A7 | rs12360 | A/G | 0.2315 | 96 | 1 | 0.65 | |
| CYP3A43*2A | rs61469810d | A/DEL | NONE | 0 | NONE | NONE | |
| CYP3A43*3 | rs680055b | C/G | <0.001 | 100 | NONE | NONE | |
| PDE1A (Phosphodiesterase) | 2q32.1 | rs1549870 | G/A | 0.2752 | 97 | 0.1531 | 0.91 |
| PDE1C | 7p14.3 | rs30585 | C/A | 0.2232 | 100 | 1 | 0.37 |
| rs992185 | C/A | 0.2321 | 100 | 0.5991 | 0.2 | ||
| PDE2A | 11q13.4 | rs341058 | C/T | 0.4107 | 100 | 0.5592 | 0.24 |
| PDE3A | 12p12.2 | rs3794271 | G/A | 0.183 | 100 | 0.3581 | 0.13 |
| rs7134375 | C/A | 0.25 | 100 | 1 | NA | ||
| PDE4A | 19p13.2 | rs6511698 | C/T | 0.491 | 99 | 0.572 | 0.14 |
| rs11670504d | A/G | NONE | 58 | NONE | NONE | ||
| rs4804134 | T/G | 0.4775 | 99 | 0.8493 | 0.42 | ||
| PDE4B | 1p31.3 | rs7537440d | T/G | NONE | 98 | NONE | NONE |
| rs10454453 | C/A | 0.3705 | 100 | 0.6866 | 0.18 | ||
| rs783036 | G/A | 0.1205 | 100 | 0.3608 | 0.25 | ||
| PDE4D | 5q11.2-q12.1 | rs966221 | A/G | 0.2432 | 99 | 0.6055 | 0.94 |
| rs702553 | A/T | 0.3784 | 99 | 0.5462 | 0.28 | ||
| rs16878037 | C/T | 0.1696 | 99 | 0.1898 | 0.64 | ||
| rs10075508 | C/T | 0.1696 | 100 | 0.5166 | 0.81 | ||
| rs829259 | T/C | 0.3125 | 100 | 0.1225 | NA | ||
| rs702531 | A/C | 0.2991 | 100 | 0.1123 | 0.74 | ||
| rs1823068d | A/G | NONE | 25 | NONE | NONE | ||
| rs1588265 | A/G | 0.2909 | 98 | 1 | 0.7 | ||
| PDE5A | 4q26 | rs12646525 | T/C | 0.1161 | 100 | 1 | 0.3 |
| rs3806808d | A/C | NONE | 95 | NONE | NONE | ||
| rs11731756 | T/G | 0.344 | 97 | 0.6722 | 0.29 | ||
| rs10034450 | A/G | 0.4054 | 99 | 0.009987 | 0.46 | ||
| rs1155576 | A/C | 0.3468 | 99 | 0.8343 | 0.37 | ||
| rs3775843d | T/C | NONE | 68 | NONE | NONE | ||
| PDE11A | 2q31.2 | rs11684634 | C/A | 0.1027 | 100 | 0.3164 | 0.051 |
Candidate SNPs for association with response to caffeine citrate therapy for AOP.
aNo homozygous mutant, wild-type vs. heterozygous.
No further statistical analysis for SNPs with MAF <0.001.
No wild type, heterozygous vs. homozygous.
Genotyping failure due to various reasons.
Dominant model.
Abbreviations: NA, not available; MAF, minor allele frequencies.
Bold values represent statistically significant differences.
Impact of Genetic Variability on the Clinical Response to caffeine Citrate Therapy
No significant intergroup differences in allele distributions or genotype frequencies of genes, that is, CYP1A2, CYP3A4, CYP3A5, and CYP3A7, encoding CYP450 proteins, were found in our study. Similar to our previous findings (He et al., 2021), two more SNPs (rs1476080 and rs2066853) (Table 2) in AHR, but not AHRR or ARNT genes, were found to be associated with the response to caffeine citrate therapy in our pediatric patients. Although the significance disappeared after FDR correction, the AHR polymorphism (rs1476080) still differed significantly (p = 0.033) between the two groups after adjusting for clinically important variables, including birth weight, birth height, respiratory support time, repeat intubation and caffeine therapy duration (Table 3).
TABLE 2
| Gene | SNPs | Genotypeb | Frequency no. (%) | Apneic group no. (%) | Apnea-free group no. (%) | 0dds ratio | 95% CI | pchi-square | pFDR |
|---|---|---|---|---|---|---|---|---|---|
| AHR | rs1476080 T > G | TT | 40 (35.71) | 28 (43.8) | 12 (25) | 1 | 0.038 | 0.09938 | |
| GT/GG | 72 (64.29) | 36 (56.2) | 36 (75) | 2.33 | 1.03–5.29 | ||||
| rs2066853 G > A | GG | 46 (41.44) | 21 (33.33) | 25 (52.08) | 1 | 0.047 | 0.1141 | ||
| GA/AA | 65 (58.56) | 42 (66.67) | 23 (47.92) | 0.46 | 0.21–0.99 | ||||
| CLOCK | rs1801260 A > G | AA | 93 (83.04) | 58 (90.6) | 35 (72.9) | 1 | 0.014 | 0.05011 | |
| AG/GG | 19 (16.96) | 6 (9.4) | 13 (27.1) | 3.59 | 1.25–1,031 | ||||
| rs12649507 A > G | AA | 35(32.41) | 26 (43.3) | 9 (18.8) | 1 | 0.0058 | 0.04165 | ||
| GA/GG | 73(67.59) | 34 (56.7) | 39 (81.2) | 3.31 | 1.37–8.04 | ||||
| rs2070062 A > C | AA | 91 (82.73) | 56 (90.3) | 35 (72.9) | 1 | 0.016 | 0.0544 | ||
| CA/CC | 19 (17.27) | 6 (9.7) | 13 (27.1) | 3.47 | 1.21–9.96 | ||||
| rs2272073 T > C | TT | 33 (30.84) | 24 (40) | 9 (19.1) | 1 | 0.018 | 0.05617 | ||
| CT/CC | 74 (69.16) | 36 (60) | 38 (80.8) | 2.81 | 1.15–6.86 | ||||
| rs2412648 G > T | GG | 36(33.33) | 27 (44.3) | 9 (19.1) | 1 | 0.0052 | 0.04165 | ||
| GT/TT | 72(66.67) | 34 (55.7) | 38 (80.8) | 3.35 | 1.38–8.12 | ||||
| rs3805151 C > T | CC | 37(34.58) | 27 (45.8) | 10 (20.8) | 1 | 0.0062 | 0.04165 | ||
| CT/TT | 70(65.42) | 32 (54.2) | 38 (79.2) | 3.21 | 1.35–7.61 | ||||
| rs4340844 C > A | CC | 35(32.11) | 26 (42.6) | 9 (18.8) | 1 | 0.007 | 0.04165 | ||
| CA/AA | 74(67.89) | 35 (57.4) | 39 (81.2) | 3.22 | 1.33–7.80 | ||||
| rs6843722 C > A | CC | 35(32.41) | 26 (43.3) | 9 (18.8) | 1 | 0.0058 | 0.04165 | ||
| CA/AA | 73(67.59) | 34 (56.7) | 39 (81.2) | 3.31 | 1.37–8.04 | ||||
| rs6858749 C > T | CC | 87 (79.82) | 53 (86.9) | 34 (70.8) | 1 | 0.038 | 0.9938 | ||
| CT/TT | 22 (20.18) | 8 (13.1) | 14 (29.2) | 2.73 | 1.03–7.19 | ||||
| rs7660668 G > C | GG | 91(83.49) | 56 (91.8) | 35 (72.9) | 1 | 0.0081 | 0.04165 | ||
| CG/CC | 18(16.51) | 5 (8.2) | 13 (27.1) | 4.16 | 1.36–12.68 | ||||
| rs7698022 T > G | TT | 90(83.33) | 55 (91.7) | 35 (72.9) | 1 | 0.009 | 0.04165 | ||
| GT/GG | 18(16.67) | 5 (8.3) | 13 (27.1) | 4.09 | 1.34–12.46 | ||||
| rs10462028 G > A | GG | 92(85.98) | 58 (93.5) | 34 (75.6) | 1 | 0.0081 | 0.04165 | ||
| GA/AA | 15(14.02) | 4 (6.5) | 11 (24.4) | 4.69 | 1.38–15.89 | ||||
| rs1048004 C > A | CC | 92(85.98) | 58 (93.5) | 34 (75.6) | 1 | 0.0081 | 0.04165 | ||
| CA/AA | 15(14.02) | 4 (6.5) | 11 (24.4) | 4.69 | 1.38–15.89 | ||||
| rs11133385 A > G | AA | 37(34.58) | 28 (45.2) | 9 (20) | 1 | 0.0059 | 0.04165 | ||
| GA/GG | 70(65.42) | 34 (54.8) | 36 (80) | 3.29 | 1.36–7.98 | ||||
| rs11133389 T > C | TT | 36(34.29) | 27 (44.3) | 9 (20.4) | 1 | 0.0098 | 0.04165 | ||
| CT/CC | 69(65.71) | 34 (55.7) | 35 (79.5) | 3.09 | 1.27–7.52 | ||||
| rs11133391 C > T | CC | 36(33.64) | 27 (43.5) | 9 (20) | 1 | 0.0095 | 0.04165 | ||
| TC/TT | 71(66.36) | 35 (56.5) | 36 (80) | 3.09 | 1.27–7.49 | ||||
| rs11240 C > G | CC | 92(85.98) | 58 (93.5) | 34 (75.6) | 1 | 0.0081 | 0.04165 | ||
| CG/GG | 15(14.02) | 4 (6.5) | 11 (24.4) | 4.69 | 1.38–15.89 | ||||
| rs11932595 A > G | AA | 91 (85.05) | 57 (91.9) | 34 (75.6) | 1 | 0.019 | 0.05617 | ||
| AG/GG | 16 (14.95) | 5 (8.1) | 11 (24.4) | 3.69 | 1.18–11.52 | ||||
| rs11943456 C > T | CC | 89 (82.41) | 55 (88.7) | 34 (73.9) | 1 | 0.047 | 0.1141 | ||
| TC/TT | 19 (17.59) | 7 (11.3) | 12 (26.1) | 2.77 | 0.99–7.73 | ||||
| rs12648271 C > G | CC | 36(33.96) | 27 (43.5) | 9 (20.4) | 1 | 0.012 | 0.04533 | ||
| GC/GG | 70(66.04) | 35 (56.5) | 35 (79.5) | 3 | 1.23–7.29 | ||||
| rs17721497 A > T | AA | 91 (85.05) | 57 (91.9) | 34 (75.6) | 1 | 0.019 | 0.05617 | ||
| TA/TT | 16 (14.95) | 5 (8.1) | 11 (24.4) | 3.69 | 1.18–11.52 | ||||
| rs3792603 A > G | AA | 86 (86) | 52 (92.9) | 34 (77.3) | 1 | 0.025 | 0.07083 | ||
| AG/GG | 14 (14) | 4 (7.1) | 10 (22.7) | 3.82 | 1.11–13.18 | ||||
| rs3805148 C > A | CC | 35(33.33) | 26 (43.3) | 9 (20) | 1 | 0.011 | 0.044 | ||
| CA/AA | 70(66.67) | 34 (56.7) | 36 (80) | 3.06 | 1.25–7.46 | ||||
| rs4864546 A > G | AA | 37(34.91) | 28 (45.2) | 9 (20.4) | 1 | 0.0074 | 0.04165 | ||
| GA/GG | 69(65.09) | 34 (54.8) | 35 (79.5) | 3.2 | 1.32–7.78 | ||||
| rs4864548 A > G | AA | 37(34.58) | 28 (45.2) | 9 (20) | 1 | 0.0059 | 0.04165 | ||
| AG/GG | 70(65.42) | 34 (54.8) | 36 (80) | 3.29 | 1.36–7.98 | ||||
| rs9312661 A > G | AA | 36(33.64) | 27 (43.5) | 9 (20) | 1 | 0.0095 | 0.04165 | ||
| GA/GG | 71(66.36) | 35 (56.5) | 36 (80) | 3.09 | 1.27–7.49 |
SNPs association with response to caffeine therapy among preterm infants in apneic and apnea-free groupsa
Data from apneic group was defined as case group and data from apnea-free group was defined as control group for the association analysis.
Dominant model.
Abbreviations: AHR, aryl hydrocarbon receptor gene; CI, confidence interval; CLOCK, Circadian Locomotor Output Cycles Kaput.
TABLE 3
| SNPs | Constant | Birth weight - g | Birth height - cm | Respiration support timea - day | Repeat intubation - no. (%) | Caffeine therapy duration - day | Gene mutation | |
|---|---|---|---|---|---|---|---|---|
| rs1476080 (AHR) | B | 2.964 | 0.003 | −0.277 | 0.061 | 2.403 | 0.056 | 1.155 |
| p | 0.338 | 0.027 | 0.018 | 0.012 | 0.039 | 0.009 | 0.033 | |
| OR | 1.003 | 0.758 | 1.063 | 11.054 | 1.058 | 3.173 | ||
| 95% CI | 1.00–1.01 | 0.60–0.95 | 1.01–1.12 | 1.13–107.88 | 1.01–1.10 | 1.10–9.18 | ||
| rs1801260 (CLOCK) | B | 1.874 | 0.003 | −0.265 | 0.046 | 2.610 | 0.063 | 1.477 |
| p | 0.545 | 0.034 | 0.022 | 0.045 | 0.039 | 0.005 | 0.030 | |
| OR | 1.003 | 0.767 | 1.047 | 13.596 | 1.065 | 4.381 | ||
| 95% CI | 1.00–1.01 | 0.61–0.96 | 1.00–1.10 | 1.14–162.53 | 1.02–1.11 | 1.15–16.69 | ||
| rs12649507 (CLOCK) | B | 1.789 | 0.002 | −0.218 | 0.053 | 2.574 | 0.057 | 1.456 |
| p | 0.576 | 0.123 | 0.068 | 0.026 | 0.031 | 0.011 | 0.008 | |
| OR | 1.002 | 0.804 | 1.055 | 13.121 | 1.058 | 4.289 | ||
| 95% CI | 1.00–1.01 | 0.64–1.02 | 1.01–1.11 | 1.26–136.63 | 1.01–1.11 | 1.45–12.68 | ||
| rs2070062 (CLOCK) | B | 1.717 | 0.003 | −0.262 | 0.044 | 2.643 | 0.065 | 1.466 |
| p | 0.580 | 0.032 | 0.024 | 0.057 | 0.037 | 0.004 | 0.032 | |
| OR | 1.003 | 0.769 | 1.045 | 14.049 | 1.067 | 4.331 | ||
| 95% CI | 1.00–1.01 | 0.61–0.97 | 1.00–1.09 | 1.18–167.57 | 1.02–1.12 | 1.13–16.54 | ||
| rs2272073 (CLOCK) | B | 1.807 | 0.002 | −0.218 | 0.052 | 2.662 | 0.058 | 1.332 |
| p | 0.571 | 0.113 | 0.066 | 0.028 | 0.023 | 0.010 | 0.016 | |
| OR | 1.002 | 0.804 | 1.054 | 14.326 | 1.059 | 3.788 | ||
| 95% CI | 1.00–1.01 | 0.64–1.01 | 1.01–1.10 | 1.44–142.89 | 1.01–1.11 | 1.28–11.18 | ||
| rs2412648 (CLOCK) | B | 3.960 | 0.002 | −0.273 | 0.053 | 2.496 | 0.050 | 1.510 |
| p | 0.243 | 0.106 | 0.028 | 0.027 | 0.037 | 0.023 | 0.007 | |
| OR | 1.002 | 0.761 | 1.055 | 12.139 | 1.052 | 4.527 | ||
| 95% CI | 1.00–1.01 | 0.60–0.97 | 1.01–1.11 | 1.17–126.32 | 1.01–1.10 | 1.51–13.60 | ||
| rs3805151 (CLOCK) | B | 2.091 | 0.002 | −0.223 | 0.052 | 2.768 | 0.053 | 1.450 |
| p | 0.515 | 0.129 | 0.062 | 0.029 | 0.018 | 0.018 | 0.008 | |
| OR | 1.002 | 0.800 | 1.053 | 15.919 | 1.054 | 4.261 | ||
| 95% CI | 1.00–1.01 | 0.63–1.01 | 1.01–1.10 | 1.60–158.05 | 1.01–1.10 | 1.46–12.41 | ||
| rs4340844 (CLOCK) | B | 1.738 | 0.002 | −0.217 | 0.053 | 2.726 | 0.057 | 1.457 |
| p | 0.588 | 0.126 | 0.071 | 0.026 | 0.020 | 0.011 | 0.008 | |
| OR | 1.002 | 0.805 | 1.055 | 15.273 | 1.058 | 4.293 | ||
| 95% CI | 1.00–1.01 | 0.64–1.02 | 1.01–1.11 | 1.53–152.67 | 1.01–1.11 | 1.45–12.71 | ||
| rs6843722 (CLOCK | B | 1.836 | 0.002 | −0.216 | 0.052 | 2.724 | 0.055 | 1.461 |
| p | 0.568 | 0.134 | 0.070 | 0.030 | 0.020 | 0.014 | 0.008 | |
| OR | 1.002 | 0.806 | 1.054 | 15.237 | 1.057 | 4.312 | ||
| 95% CI | 1.00–1.01 | 0.64–1.02 | 1.01–1.10 | 1.53–151.6 | 1.01–1.10 | 1.46–12.73 | ||
| rs6858749 (CLOCK) | B | 1.399 | 0.003 | −0.233 | 0.041 | 2.580 | 0.066 | 1.326 |
| p | 0.650 | 0.066 | 0.039 | 0.062 | 0.038 | 0.004 | 0.043 | |
| OR | 1.003 | 0.792 | 1.042 | 13.192 | 1.068 | 3.767 | ||
| 95% CI | 1.00–1.01 | 0.63–0.99 | 1.00–1.09 | 1.15–151.68 | 1.02–1.12 | 1.04–13.58 | ||
| rs7660668 (CLOCK) | B | 1.278 | 0.003 | −0.246 | 0.039 | 2.772 | 0.068 | 1.826 |
| p | 0.683 | 0.063 | 0.034 | 0.089 | 0.037 | 0.004 | 0.016 | |
| OR | 1.003 | 0.782 | 1.040 | 15.998 | 1.071 | 6.207 | ||
| 95% CI | 1.00–1.01 | 0.62–0.98 | 0.99–1.09 | 1.19–215.57 | 1.02–1.12 | 1.41–27.24 | ||
| rs7698022 (CLOCK) | B | 1.294 | 0.003 | −0.245 | 0.039 | 2.653 | 0.068 | 1.809 |
| p | 0.679 | 0.062 | 0.033 | 0.088 | 0.051 | 0.004 | 0.016 | |
| OR | 1.003 | 0.782 | 1.040 | 14.190 | 1.070 | 6.106 | ||
| 95% CI | 1.00–1.01 | 0.62–0.98 | 0.99–1.09 | 0.99–203.08 | 1.02–1.12 | 1.39–26.73 | ||
| rs11133389 (CLOCK) | B | 2.129 | 0.003 | −0.230 | 0.054 | 2.559 | 0.051 | 1.387 |
| p | 0.522 | 0.078 | 0.058 | 0.025 | 0.031 | 0.021 | 0.010 | |
| OR | 1.003 | 0.794 | 1.056 | 12.927 | 1.052 | 4.003 | ||
| 95% CI | 1.00–1.01 | 0.63–1.01 | 1.01–1.11 | 1.26–132.87 | 1.01–1.10 | 1.38–11.58 | ||
| rs11932595 (CLOCK) | B | 1.498 | 0.003 | −0.265 | 0.043 | 3.203 | 0.064 | 2.090 |
| p | 0.643 | 0.041 | 0.025 | 0.065 | 0.025 | 0.007 | 0.013 | |
| OR | 1.003 | 0.767 | 1.044 | 24.603 | 1.067 | 8.085 | ||
| 95% CI | 1.00–1.01 | 0.61–0.97 | 1.00–1.09 | 1.51–401.44 | 1.02–1.12 | 1.55–42.05 | ||
| rs11943456 (CLOCK) | B | 1.300 | 0.003 | −0.243 | 0.044 | 2.577 | 0.065 | 1.564 |
| p | 0.679 | 0.045 | 0.034 | 0.045 | 0.047 | 0.005 | 0.028 | |
| OR | 1.003 | 0.784 | 1.045 | 13.158 | 1.067 | 4.777 | ||
| 95% CI | 1.00–1.01 | 0.63–0.98 | 1.00–1.09 | 1.04–167.14 | 1.02–1.12 | 1.18–19.28 | ||
| rs12648271 (CLOCK) | B | 2.163 | 0.003 | −0.233 | 0.055 | 2.561 | 0.051 | 1.388 |
| p | 0.515 | 0.073 | 0.055 | 0.020 | 0.031 | 0.021 | 0.011 | |
| OR | 1.003 | 0.792 | 1.057 | 12.946 | 1.052 | 4.008 | ||
| 95% CI | 1.00–1.01 | 0.62–1.00 | 1.01–1.11 | 1.26–133.46 | 1.01–1.10 | 1.38–11.61 | ||
| rs17721497 (CLOCK) | B | 1.794 | 0.003 | −0.267 | 0.045 | 2.631 | 0.061 | 1.731 |
| p | 0.575 | 0.035 | 0.024 | 0.049 | 0.047 | 0.008 | 0.021 | |
| OR | 1.003 | 0.766 | 1.046 | 13.882 | 1.063 | 5.647 | ||
| 95% CI | 1.00–1.01 | 0.61–0.97 | 1.00–1.09 | 1.04–185.2 | 1.02–1.11 | 1.30–24.59 | ||
| rs3792603 (CLOCK) | B | 2.726 | 0.002 | −0.265 | 0.033 | 3.034 | 0.071 | 2.241 |
| p | 0.420 | 0.229 | 0.028 | 0.164 | 0.046 | 0.004 | 0.023 | |
| OR | 1.002 | 0.767 | 1.033 | 20.778 | 1.074 | 9.402 | ||
| 95% CI | 1.00–1.00 | 0.61–0.97 | 0.99–1.08 | 1.05–409.46 | 1.02–1.13 | 1.36–65.23 | ||
| rs3805148 (CLOCK) | B | 1.561 | 0.002 | −0.204 | 0.053 | 2.606 | 0.053 | 1.442 |
| p | 0.640 | 0.143 | 0.093 | 0.026 | 0.029 | 0.018 | 0.009 | |
| OR | 1.002 | 0.815 | 1.054 | 13.545 | 1.054 | 4.231 | ||
| 95% CI | 1.00–1.01 | 0.64–1.03 | 1.01–1.10 | 1.31–139.74 | 1.01–1.10 | 1.43–12.56 | ||
| rs4864546 (CLOCK) | B | 1.962 | 0.002 | −0.222 | 0.055 | 2.602 | 0.051 | 1.518 |
| p | 0.559 | 0.102 | 0.068 | 0.021 | 0.029 | 0.021 | 0.006 | |
| OR | 1.002 | 0.801 | 1.056 | 13.485 | 1.053 | 4.561 | ||
| 95% CI | 1.00–1.01 | 0.63–1.02 | 1.01–1.11 | 1.30–139.56 | 1.01–1.10 | 1.56–13.37 | ||
| rs11133385 | B | 1.814 | 0.002 | −0.219 | 0.056 | 2.616 | 0.052 | 1.543 |
| p | 0.588 | 0.107 | 0.073 | 0.019 | 0.028 | 0.021 | 0.005 | |
| OR | 1.002 | 0.803 | 1.057 | 13.679 | 1.053 | 4.678 | ||
| 95% CI | 1.00–1.01 | 0.63–1.02 | 1.01–1.11 | 1.32–142.11 | 1.01–1.1 | 1.60–13.70 | ||
| rs11133391 | B | 2.007 | 0.003 | −0.230 | 0.057 | 2.575 | 0.051 | 1.413 |
| p | 0.545 | 0.077 | 0.059 | 0.017 | 0.031 | 0.021 | 0.009 | |
| OR | 1.003 | 0.795 | 1.058 | 13.133 | 1.053 | 4.107 | ||
| 95% CI | 1.00–1.01 | 0.63–1.01 | 1.01–1.11 | 1.27–135.93 | 1.01–1.10 | 1.42–11.89 | ||
| rs10462028 | B | 1.437 | 0.003 | −0.267 | 0.042 | 2.840 | 0.066 | 2.239 |
| rs1048004 | p | 0.656 | 0.040 | 0.023 | 0.067 | 0.047 | 0.006 | 0.009 |
| rs11240 (CLOCK) | OR | 1.003 | 0.765 | 1.043 | 17.124 | 1.068 | 9.386 | |
| 95% CI | 1.00–1.01 | 0.61–0.96 | 1.00–1.09 | 1.04–280.82 | 1.02–1.12 | 1.77–49.9 |
Results of multivariable logistic regression analysis to examine the effects of AHR and CLOCK gene polymorphisms on caffeine therapy in apnea-free group and apneic preterm infants.
Duration for assisted ventilation through positive airway pressure or supplemental oxygen.
Abbreviations: AHR, aryl hydrocarbon receptor gene; CLOCK, Circadian Locomotor Output Cycles Kaput.
Most notably, in our 46 candidate SNPs in the CLOCK gene, excluding three unsuccessfully genotyped SNPs, 26 SNPs were found to be associated with the response to caffeine citrate treatment in these neonates (Table 2). Of these, 18 SNPs still differed significantly (PFDR<0.05) between the two groups after correction for multiple hypothesis testing. Furthermore, multivariable logistic regression analysis showed that the significant influence of these SNPs in the CLOCK gene on the response to caffeine therapy still survived after adjustment (Table 3). Intriguingly, no association was observed between the 14 candidate SNPs in the BMAL1 gene, which encodes two essential components (i.e., BMAL1 and CLOCK) of the circadian clock together with the CLOCK gene, and the clinical response to caffeine treatment.
In addition, some SNPs, especially for variants in ADORA genes, were found to be associated with the incidence rate of BPD and severe neurological injury (SNI) in these preterm infants. It is worth noting that significant associations between genetic polymorphisms of circadian rhythm-related genes, BPD, and SNI incidence, was also observed. The detailed results are shown in Supplementary Tables S2, S3.
Haplotype and LD Analysis
Haplotype analysis was performed to investigate whether various genetic variants were in LD. Fifteen SNPs were excluded because of failed genotyping, and 14 SNPs that were unable to meet the requirement for LD analysis were excluded, which are marked as d and b in Table 1, respectively. Along with our previous research (He et al., 2021), Figure 1 shows the distribution of the 147 candidate SNPs in more than 40 human genes evaluated in the present study. SNPs significantly associated (p < 0.05; above stippled line) with response to caffeine citrate therapy were ADORA3, CLOCK, AHR, and ADORA2A, which are located on chromosomes 1, 4, 7, and 22, respectively. We then compared r (Puia-Dumitrescu et al., 2019) (correlation coefficient between the two loci) and D’ (deviation of the observed frequency of a haplotype from the expected) values of the pairwise comparisons between the selected gene polymorphisms. The D’ and r (Puia-Dumitrescu et al., 2019) values were used for LD estimations. The results are shown in Figure 2. We observed two small blocks with strong LD between each pair of rs4410790 and rs6968865, and among variants rs1476080, rs6960165, rs2158041, rs7811989, and rs2066853 in the AHR gene, located on chromosome 7. Similarly, block 1 (rs5751862, rs5760410, rs2298383, rs3761422, rs5996696, and rs2267076), as well as block 2 (rs2236624, rs2236625, rs5751876, rs34923252, rs5760423, rs5760425, and rs4822492) with strong LD were found in the ADORA2A gene on chromosome 22. Most notably, 40 genetic CLOCK polymorphisms formed one small block (seven SNPs) and one big block (33 SNPs) on chromosome 4. More importantly, the results of haplotype analysis showed significant differences in the haplotypes, that is, TTAAAG (Block 1) in CLOCK (p = 0.0436), CT and TA (Block 1) (p = 0.00834), GACGG (Block 2) in AHR (p = 0.00243), GACCCC (Block 1) (p = 0.0401), and CTTATTC (Block 2) (p = 0.0284) in ADORA2A. Frequency distributions existed between these haplotypes and the response to caffeine citrate treatment (Table 4).
FIGURE 2
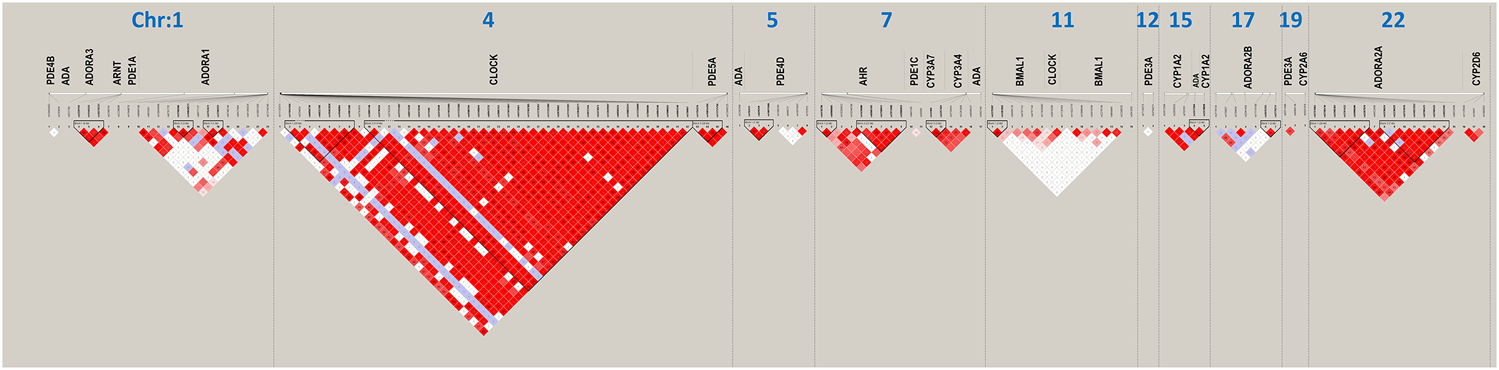
Linkage disequilibrium (LD) structure (triangle plots) for all investigated single nucleotide polymorphisms (SNPs) (rs numbers below gene names). Red diamonds indicate strong historical LD, r2-based haplotype boundaries, among SNPs.
TABLE 4
| Block | NSNPa | NHAPb | SNP1c | SNP2d | HAPLOTYPE | Fe | ORf | P |
|---|---|---|---|---|---|---|---|---|
| Chr4-Block1 (CLOCK) | 6 | 4 | rs11943456 | rs1801260 | TTAAAG | 0.0748 | 3.1 | 0.0436 |
| 6 | 4 | rs11943456 | rs1801260 | TTGGCA | 0.0187 | 0.376 | 0.409 | |
| 6 | 4 | rs11943456 | rs1801260 | CCGGCA | 0.297 | 1.09 | 0.784 | |
| 6 | 4 | rs11943456 | rs1801260 | CCAGCA | 0.604 | 0.685 | 0.232 | |
| Chr7-Block1 (AHR) | 2 | 3 | rs4410790 | rs6968865 | CT | 0.373 | 2.39 | 0.00834 |
| 2 | 3 | rs4410790 | rs6968865 | CA | 0.0318 | 0.944 | 0.943 | |
| 2 | 3 | rs4410790 | rs6968865 | TA | 0.595 | 0.457 | 0.0122 | |
| Chr7-Block2 (AHR) | 5 | 4 | rs1476080 | rs2066853 | TACGA | 0.366 | 0.686 | 0.174 |
| 5 | 4 | rs1476080 | rs2066853 | TGTAG | 0.196 | 0.763 | 0.43 | |
| 5 | 4 | rs1476080 | rs2066853 | TATAG | 0.0273 | 9.76E-60 | 0.994 | |
| 5 | 4 | rs1476080 | rs2066853 | GACGG | 0.388 | 2.53 | 0.00243 | |
| Chr22-Block1 (ADORA2A) | 6 | 6 | rs5751862 | rs2267076 | GGCTAT | 0.413 | 1.05 | 0.856 |
| 6 | 6 | rs5751862 | rs2267076 | GACTAT | 0.0142 | 8.65E-08 | 0.664 | |
| 6 | 6 | rs5751862 | rs2267076 | GACCCC | 0.0811 | 2.95 | 0.0401 | |
| 6 | 6 | rs5751862 | rs2267076 | AGTCAC | 0.0139 | 0.54 | 0.633 | |
| 6 | 6 | rs5751862 | rs2267076 | GGTCAC | 0.0273 | 1.25 | 0.797 | |
| 6 | 6 | rs5751862 | rs2267076 | AATCAC | 0.427 | 0.759 | 0.313 | |
| Chr22-Block2 (ADORA2A) | 7 | 4 | rs2236624 | rs4822492 | CCTTTTC | 0.107 | 1.7 | 0.233 |
| 7 | 4 | rs2236624 | rs4822492 | TCTTTTC | 0.326 | 0.675 | 0.205 | |
| 7 | 4 | rs2236624 | rs4822492 | CCCTGGG | 0.473 | 0.781 | 0.376 | |
| 7 | 4 | rs2236624 | rs4822492 | CTTATTC | 0.0848 | 3.15 | 0.0284 |
Significant associated haplotype variants.
Number of SNPs.
Number of haplotype.
Starting SNP
Ending SNP
Frequency of haplotype
Odds ratio
Discussion
The clear benefits and safety of caffeine citrate based on a standard-dose regimen have led to its wide and early use in very low birth weight infants. However, caffeine is not completely efficient, and in approximately 50% of treated infants, the apnea frequency remains elevated (Laouafa et al., 2019; He et al., 2021). In fact, studies are warranted to better understand why some preterm neonates persist with apnea after caffeine therapy, even after increasing the caffeine dose. The AOP phenotype has been attributed solely to immature respiratory system control consequent to preterm birth, but there may also be important genetic influences (Erickson et al., 2021). Therefore, the influence of genetics on the efficacy of caffeine in preterm infants should be better explored (Long et al., 2021).
In the present study, genetic polymorphisms of CYP3A genes, including 3A4, 3A5, 3A7, and 3A43, were also assessed (Table 1). Especially for CYP3A7, this isoform of CYP3A has been demonstrated to metabolize endogenous compounds that are known to be important in the growth and development of the fetus and neonate. In addition, CYP3A7 is an important component in the development and protection of the fetal liver and plays a role in certain disease status (Li and Lampe, 2019). However, because of MAF values < 0.001 or genotyping failure (Table 1), most of the selected SNPs were excluded from further association analysis.
A major finding of this study was that AHR genetic variations (rs1476080 and rs2066853), but not AHRR or ARNT genes, were found to be associated with the response to caffeine therapy (Tables 2, 3). Our previous study showed that two particular polymorphisms of the AHR gene (rs6968865 and rs4410790) were significantly associated with the response to caffeine treatment between the two groups of preterm infants (He et al., 2021). Furthermore, these variants in the AHR gene, located on chromosome 7, formed strong LD, with increased ORs for haplotypes CT (p = 0.00834), TA (p = 0.0122), and GACGG (p = 0.00243), which in part determined a better response to caffeine treatment (Figure 2; Table 4). Based on the plasma concentration data of caffeine, we excluded that the AHR-CYP1A2 metabolic pathway was responsible for the variable response to caffeine therapy (He et al., 2021). However, AHR is necessary to protect fetal human pulmonary microvascular endothelial cells against hyperoxic injury (Zhang et al., 2015) and plays a critical role in the maintenance of lung health (Guerrina et al., 2018). Therefore, the findings of our study suggest the potential role of AHR signaling in preterm infants who experience AOP episodes. The AHR signaling pathway may act alone or in combination with adenosine receptors and circadian CLOCK.
Another important finding in the present study was the significant association between circadian CLOCK polymorphisms, but not BMAL1 variants (also known as ARNTL1; Table 1), and the response to caffeine citrate therapy in these preterm babies (Table 2). Notably, after adjustment for clinically important variables, such as birth weight, birth height, respiratory support therapy, repeat intubation, and caffeine therapy duration, the significance was retained (Table 3). Moreover, 40 candidate CLOCK SNPs formed one small block and one big block with strong LD (Figure 2). Especially for the small block, the haplotype TTAAAG was found to be associated with a better response to caffeine therapy (OR = 3.1, p = 0.0436; Table 4).
Caffeine has been reported to affect the phase of the human circadian clock and primarily affect human cellular circadian clocks via an A1-R/cAMP-dependent mechanism (Burke et al., 2015). Recent studies have revealed that caffeine and adenosine alter clock gene expression and circadian rhythms in vitro and in vivo via the Ca2+–ERK–AP-1 pathway (Jagannath et al., 2021). The present study is the first to show that genetic polymorphisms of CLOCK, encoding one of the two core components of the circadian rhythm, were significantly associated with the response to caffeine therapy. These findings suggest that the circadian rhythm may play critical roles in the response to caffeine citrate therapy in babies experiencing AOP episodes. Interestingly, caffeine has been reported to increase the light responsiveness of the mouse circadian pacemaker (van Diepen et al., 2014) and also affects the human circadian clock in vivo and in vitro (Burke et al., 2015). Therefore, synchronizing caffeine with the circadian rhythm may be useful for optimizing its treatment efficacy. In the meantime, investigations into essential mechanisms may provide therapies to reset or amplify circadian signals. However, very few studies have been performed to evaluate the proper timing of methylxanthine dosing (chronotherapy) as a means to maximize its efficacy and possibly reduce its adverse effects. Further research in this field is warranted to provide new insights and clinical advantages.
One of the major strengths of the present study was our ability to assess how genetics affect the response to caffeine based on 147 candidate SNPs in more than 40 human genes (Table 1), encoding various proteins related to the disposition of caffeine and/or pharmacological mechanisms of caffeine’s actions, as well as circadian control functions. Along with our previous report (He et al., 2021), we observed significant associations between ADORA1, ADORA2A, ADORA3, PDE4D, ADA, AHR, and CLOCK polymorphisms and the response to caffeine treatment (Tables 2, 3). Notably, the haplotypes of AHR, CLOCK, and ADORA2A were found to be associated with a better response to caffeine therapy (Table 4). A2A-AR regulates phrenic nerve activity, and blockage with caffeine may be one mechanism responsible for the efficacy of xanthine against AOPs. Additional A1-AR in the brainstem may regulate hypoxic ventilator drive. Such central stimulation results in increased respiratory drive, increased sensitivity to hypercarbia, decreased hypoxic suppression of respiration, and increased diaphragmatic contractility (Morton and Smith, 2016). Caffeine competitively inhibits PDEs, a group of enzymes that degrade cAMP (Burke et al., 2015). In addition, intracellular adenosines undergo metabolism to inosine by ADA, a deamination that occurs preferentially under pathological conditions featuring raised adenosine levels (Borea et al., 2018). Collectively, caffeine may play a critical role in AOP treatment by directly and/or indirectly agonizing the adenosine-AR-cAMP pathway, rather than the AHR-CYP1A2 pathway involved in the disposition of caffeine, an XRE-dependent control of the target gene expression pathway by AHR. One outstanding question is how to explain ADORA, AHR, and CLOCK gene polymorphisms that were observed to be associated simultaneously with the response to caffeine therapy. However, whether the AR, AHR, and CLOCK signaling pathways crosstalk with each other during caffeine treatment remains unclear. The potential model relationship between them is discussed below and illustrated in Figure 3.
FIGURE 3
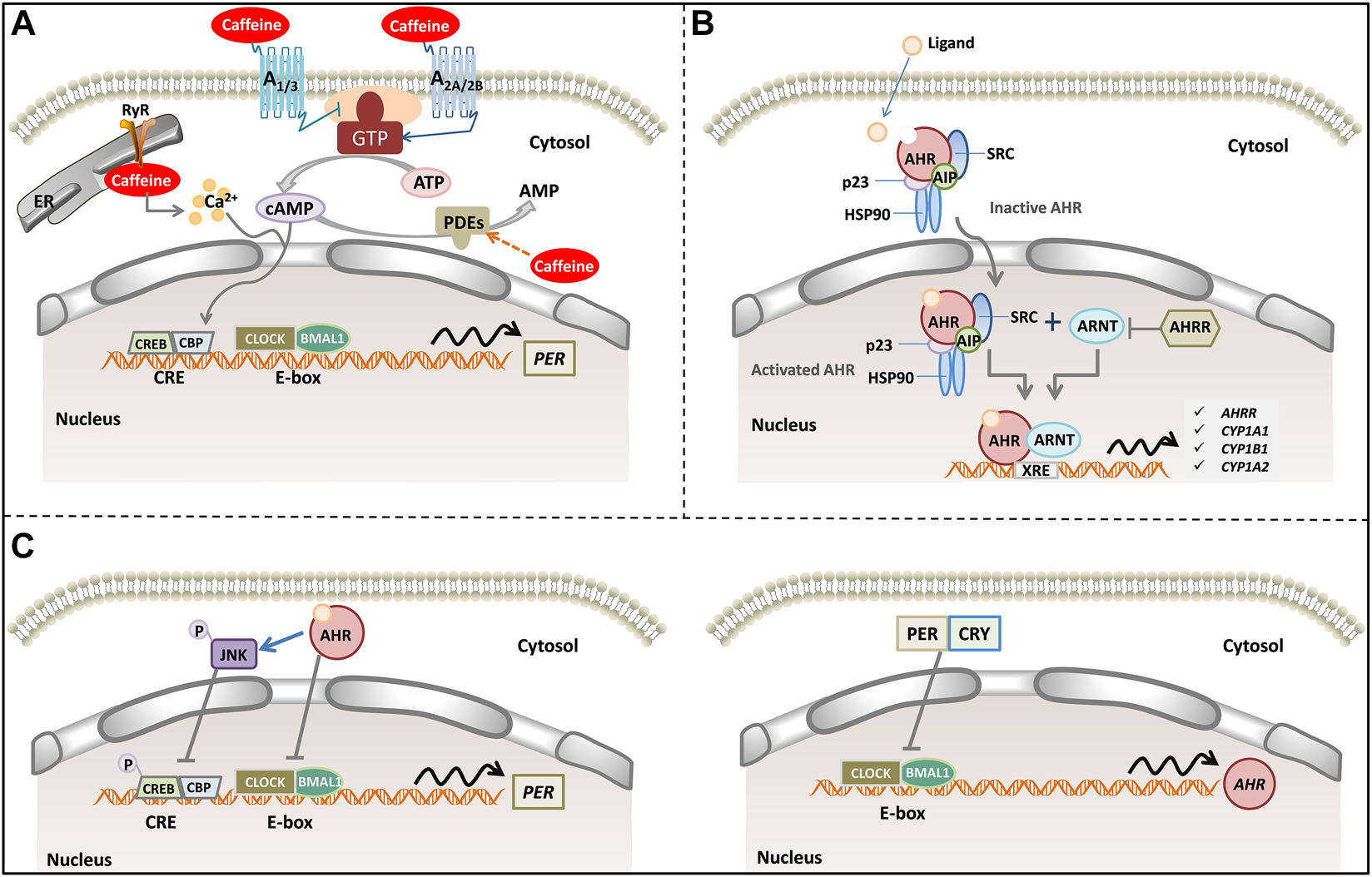
A model of caffeine, AR signaling pathway, AHR signaling pathways, and human circadian clock. (A) Caffeine alters intracellular cAMP levels through binding to adenosine receptors (A1, A2A, A2B and A3) and inhibiting phosphodiesterases. In addition, caffeine mobilizes intracellular Ca2+ from the endoplasmic reticulum through activation of the RyR channels. Increased cytosolic cAMP/Ca2+ signaling culminates CREB activation acting in tandem with rhythmic transcriptional activation by CLOCK/BMAL1. CLOCK/BMAL1 binds to E-box elements in the PER promoter, where they act to stimulate PER transcription. (B) Inactive AHR complexes with HSP90, AIP, p23 and SRC in the cytosol. Upon agonist binding, AHR and some components of the chaperone complex translocate to the nucleus, where AHR forms a dimer with ARNT binding to the XRE to control gene expression (such as AHRR, CYP1A1, CYP1A2 and CYP1B1). (C) Activated AHR can physically interact with BMAL1, which exhibits high homology with ARNT, thereby reducing CLOCK-BMAL1 interactions and repressing PER transcription. In addition, activated AHR phosphorylates and activates JNK, which represses CRE-mediated transcriptional activity to suppress PER transcription (left). Rhythmic transcription of AHR is driven by CLOCK: BMAL1 at E-Box promoter elements. PER and CRY inhibit AHR transcript levels (right). Abbreviation: cAMP, cyclic adenosine monophosphate; RyR, ryanodine receptor; PDEs, phosphodiesterases; AHR, aryl hydrocarbon receptor; AHRR, AHR repressor; ARNT, AHR nuclear translocator; XRE, xenobiotic response element; HSP90, 90 kDa heat shock protein; AIP, AHR-interacting protein. CLOCK, circadian locomotor output cycles kaput; BMAL1, brain and muscle ARNT-Like 1; CREB, cAMP response element-binding; CBP, CREB binding protein; PER, periods; CRY, crypto-chromes.
Adenosine is a ubiquitous endogenous autacoid and functions as a signaling molecule through the activation of four distinct ARs (i.e., A1, A2A, A2B, and A3) (Borea et al., 2018). Adenosine encodes a sleep history and modulates circadian entrainment by light. However, caffeine has the opposite effect on sleep as adenosine, and it promotes wakefulness via A2A-AR, rather than A1-AR (Huang et al., 2005). Additionally, caffeine inhibits PDEs or promotes intracellular Ca2+ release (Kumar and Lipshultz, 2019). Pharmacological and genetic approaches demonstrate that adenosine acts on the circadian clock via A1-AR/A2A-AR signaling through the activation of the Ca2+–ERK–AP-1 and CREB/CRCT1–CRE pathways to regulate the clock genes PER 1 and PER 2 (Figure 3A). Interestingly, adenosine integrates light and sleep signaling for regulating the circadian rhythms in mice (Jagannath et al., 2021).
AHR is a ligand-activated transcription factor that is a member of the periodic circadian protein (PER)–AHR nuclear translocator (ARNT)–single-minded protein (SIM) superfamily of transcription factors, in which the PER–ARNT–SIM (PAS) domain senses both the endogenous and exogenous factors (Figure 3B). AHR senses oxygen levels, redox potential, and changes in the circadian rhythm and control adaptation in the cellular environment (Rothhammer and Quintana, 2019). AHR is involved in controlling the cardiovascular and respiratory functions (Sauzeau et al., 2011). There is high constitutive AHR expression in lung epithelial/endothelial cells and fibroblasts (Rico de Souza et al., 2021). AHR promotes lipid droplet biogenesis and metabolic shift in respiratory club cells, which are metabolically active cells that are involved in xenobiotic metabolism and host defense as well as the maintenance of airway integrity (Wang et al., 2021). AHR deficiency enhanced airway inflammation and remodeling in a murine chronic asthma model (Chang et al., 2020), and also caused the development of chronic obstructive pulmonary disease (Guerrina et al., 2021), indicating its importance as a central player in maintaining normal lung function and determining disease severity. In addition, AHR activity in both endothelial and hematopoietic cells is necessary for vascular development and closure of the ductus venosus (Guerrina et al., 2018). Therefore, the AHR signaling pathway may play a critical role in AOP and BPD development, as well as in the response to caffeine treatment in preterm infants.
Intrinsic clocks determine nearly all circadian cycles, such as respiratory and exercise capacity (Allada and Bass, 2021). More importantly, AHR and the circadian signaling pathways are highly integrated and reciprocally regulated (Figure 3C). AHR exhibits a rhythmic expression and time-dependent sensitivity to activation by AHR agonists. Conversely, AHR influences the amplitude and phase of rhythms in circadian clock genes, hormones, and behavior (Tischkau, 2020). AHR directly interacts with the core circadian clock. In the canonical signaling pathway, activated AHR forms a heterodimer with ARNT, which exhibits sequence homology with BMAL1. However, activated AHR can also heterodimerize with BMAL1, thereby disrupting the normal binding of CLOCK/BMAL1 to E-box elements to drive the transcription of the target genes (Jaeger et al., 2017). As one of the potential mechanisms, AHR-CLOCK might set a pathological precondition for the efficacy of caffeine treatment to manifest. Thus, investigation into the mechanism by which the AHR-CLOCK interaction affects the response to caffeine treatment may provide insights into the pathological mechanisms of AOP and provide an innovative strategy to reset the treatment protocol in preterm neonates.
However, our study has several limitations. In this study, only the CLOCK and BMAL1 polymorphisms were assessed for their potential roles in the circadian rhythm. In mammals, at the cellular level, the transcription factors CLOCK and neuronal NPAS2 form heterodimers with ARNTL1 (i.e., BMAL1) to drive the expression of the genes encoding period circadian protein homologues PER1, PER2, and PER3 and crypto-chromes (CRY) 1 and 2 via direct binding to the E-box enhancer element during the day. In the late afternoon or evening, PER and CRY proteins heterodimerize, translocate into the nucleus, and interact with CLOCK and BMAL1, thus suppressing their transcriptional activity. In the meantime, the protein levels of PER1, PER2, CRY1, and CRY2 also decline by poly-ubiquitination and subsequent degradation via specific E3 ligase complexes (Ruan et al., 2021). However, we did not determine the roles of the genes encoding PER and CRY proteins in caffeine therapy. Another limitation is that this study was performed in a single center with a small sample size (n = 112), which may increase the risk that some significant changes will not be captured. The missed significant association between CYP1A2*1B (rs2470890) variant and response to caffeine therapy (p = 0.058; codominant model) in preterm babies may be due to the small study cohort. In addition, as described in our previous report (He et al., 2021), the grouping criteria may limit the reliability and accuracy of the present study. Lastly, in terms of statistical approach, within an FDR threshold of <0.05, 5% of the variants (i.e., 4-5 SNPs) were estimated to be false positives, which was generally acceptable.
Conclusion
Our study found that candidate CYPs 1A2, 3A4, 3A5, 3A7, 3A43, AHRR, ARNT, and BMAL1 polymorphisms had no effect on the response to caffeine therapy in the apnea-free and apneic groups. However, genetic candidate variants in AHR and CLOCK genes were found to be associated with variable responses to caffeine treatment in these preterm babies. Moreover, the variants in AHR, ADORA2A, and CLOCK genes formed strong LD with increased ORs, which were associated with a significantly better response to standard-dose caffeine therapy. Future and larger studies, as well as basic research, are required to understand how these SNPs affect the response to caffeine therapy. Our findings also indicate that circadian rhythm may play an essential role in the response to caffeine therapy in babies experiencing AOP episodes.
What is Already Known About This Subject
• Caffeine citrate therapy is a preferable choice for preterm infants with AOP in the NICU.
• The current standard-dose caffeine therapy leads to variable clinical outcomes. It remains unclear why some preterm infants respond well to this therapy but others do not.
• Questions regarding dose selection, routine TDM of caffeine, and influence of genetic variants are still unanswered.
What This Study Adds
• No significant intergroup differences in allele distributions or genotype frequencies of CYP1A2, CYP3A4, CYP3A5, and CYP3A7 were found in our study on preterm babies.
• To the best of our knowledge, the present study is the first to report that CLOCK gene polymorphisms are involved in determining the response to caffeine therapy in premature neonates with AOP.
• Strong linkage disequilibrium was observed in AHR, ADORA2A, and CLOCK variants. These genetic variants were significantly associated with a better response to standard-dose caffeine therapy. However, whether the AR, AHR, and CLOCK signaling pathways crosstalk with each other during caffeine treatment remains largely unknown.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by The hospital ethics committee at the Children’s Hospital of Nanjing Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
J-YL, H-LG, FC, JX, and RC: Principal investigators for the study, data analysis, primary authors of the paper. XH, Y-HH, YX, YL, and LL: Performed the data collection and analysis. X-SD: Assisted in the design and performance of the study and the writing of the paper. All authors have read and approved the final manuscript.
Funding
This research was supported by the Specially Appointed Medical Expert Project of the Jiangsu Commission of Health (2019) and Special Fund for Clinical Research of the Wu Jieping Medical Foundation (320.6750.2020-04-07). This study was also supported by the Scientific Research Support Foundation for Young Scholars at the Children’s Hospital of Nanjing Medical University (2020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.724145/full#supplementary-material
References
1
AlladaR.BassJ. (2021). Circadian Mechanisms in Medicine. N. Engl. J. Med.384, 550–561. 10.1056/NEJMra1802337
2
Bloch-SalisburyE.HallM. H.SharmaP.BoydT.BednarekF.PaydarfarD. (2010). Heritability of Apnea of Prematurity: a Retrospective Twin Study. Pediatrics126, e779–87. 10.1542/peds.2010-0084
3
BoreaP. A.GessiS.MerighiS.VincenziF.VaraniK. (2018). Pharmacology of Adenosine Receptors: The State of the Art. Physiol. Rev.98, 1591–1625. 10.1152/physrev.00049.2017
4
BurkeT. M.MarkwaldR. R.McHillA. W.ChinoyE. D.SniderJ. A.BessmanS. C.et al (2015). Effects of Caffeine on the Human Circadian Clock In Vivo and In Vitro. Sci. Transl Med.7, 305ra146. 10.1126/scitranslmed.aac5125
5
ChangY. D.LiC. H.TsaiC. H.ChengY. W.KangJ. J.LeeC. C. (2020). Aryl Hydrocarbon Receptor Deficiency Enhanced Airway Inflammation and Remodeling in a Murine Chronic Asthma Model. FASEB J.34, 15300–15313. 10.1096/fj.202001529R
6
CheungK. W.PengQ.HeL.CaiK.JiangQ.ZhouB.et al (2016). Rapid and Simultaneous Detection of Major Drug Resistance Mutations in Reverse Transcriptase Gene for HIV-1 CRF01_AE, CRF07_BC and Subtype B in China Using Sequenom MassARRAY® System. PLoS One11, e0153641. 10.1371/journal.pone.0153641
7
EichenwaldE. C.Committee on Fetus and Newborn, American Academy of Pediatrics. (2016). Apnea of Prematurity. Pediatrics137. 10.1542/peds.2015-3757
8
EricksonG.DobsonN. R.HuntC. E. (2021). Immature Control of Breathing and Apnea of Prematurity: the Known and Unknown. J. Perinatol41, 2111. 10.1038/s41372-021-01010-z
9
GaytanS. P.PasaroR. (2012). Neonatal Caffeine Treatment Up-Regulates Adenosine Receptors in Brainstem and Hypothalamic Cardio-Respiratory Related Nuclei of Rat Pups. Exp. Neurol.237, 247–259. 10.1016/j.expneurol.2012.06.028
10
GuerrinaN.TraboulsiH.EidelmanD. H.BagloleC. J. (2018). The Aryl Hydrocarbon Receptor and the Maintenance of Lung Health. Int. J. Mol. Sci.19, 3882. 10.3390/ijms19123882
11
GuerrinaN.TraboulsiH.Rico de SouzaA.BosseY.ThatcherT. H.RobichaudA.et al (2021). Aryl Hydrocarbon Receptor Deficiency Causes the Development of Chronic Obstructive Pulmonary Disease through the Integration of Multiple Pathogenic Mechanisms. FASEB J.35, e21376. 10.1096/fj.202002350r
12
HardinP. E.PandaS. (2013). Circadian Timekeeping and Output Mechanisms in Animals. Curr. Opin. Neurobiol.23, 724–731. 10.1016/j.conb.2013.02.018
13
HeX.QiuJ. C.LuK. Y.GuoH. L.LiL.JiaW. W.et al (2021). Therapy for Apnoea of Prematurity: A Retrospective Study on Effects of Standard Dose and Genetic Variability on Clinical Response to Caffeine Citrate in Chinese Preterm Infants. Adv. Ther.38, 607–626. 10.1007/s12325-020-01544-2
14
HuangZ. L.QuW. M.EguchiN.ChenJ. F.SchwarzschildM. A.FredholmB. B.et al (2005). Adenosine A2A, but Not A1, Receptors Mediate the Arousal Effect of Caffeine. Nat. Neurosci.8, 858–859. 10.1038/nn1491
15
JaegerC.KhazaalA. Q.XuC.SunM.KragerS. L.TischkauS. A. (2017). Aryl Hydrocarbon Receptor Deficiency Alters Circadian and Metabolic Rhythmicity. J. Biol. Rhythms32, 109–120. 10.1177/0748730417696786
16
JagannathA.VargaN.DallmannR.RandoG.GosselinP.EbrahimjeeF.et al (2021). Adenosine Integrates Light and Sleep Signalling for the Regulation of Circadian Timing in Mice. Nat. Commun.12, 2113. 10.1038/s41467-021-22179-z
17
KumarV. H. S.LipshultzS. E. (2019). Caffeine and Clinical Outcomes in Premature Neonates. Children (Basel)6, 118. 10.3390/children6110118
18
KumralA.TuzunF.YesilirmakD. C.DumanN.OzkanH. (2012). Genetic Basis of Apnoea of Prematurity and Caffeine Treatment Response: Role of Adenosine Receptor Polymorphisms: Genetic Basis of Apnoea of Prematurity. Acta Paediatr.101, e299–303. 10.1111/j.1651-2227.2012.02664.x
19
LaouafaS.IturriP.Arias-ReyesC.MarcouillerF.GonzalesM.JosephV.et al (2019). Erythropoietin and Caffeine Exert Similar Protective Impact against Neonatal Intermittent Hypoxia: Apnea of Prematurity and Sex Dimorphism. Exp. Neurol.320, 112985. 10.1016/j.expneurol.2019.112985
20
LiH.LampeJ. N. (2019). Neonatal Cytochrome P450 CYP3A7: A Comprehensive Review of its Role in Development, Disease, and Xenobiotic Metabolism. Arch. Biochem. Biophys.673, 108078. 10.1016/j.abb.2019.108078
21
LongJ. Y.GuoH. L.HeX.HuY. H.XiaY.ChengR.et al (2021). Caffeine for the Pharmacological Treatment of Apnea of Prematurity in the NICU: Dose Selection Conundrum, Therapeutic Drug Monitoring and Genetic Factors. Front. Pharmacol.12, 681842. 10.3389/fphar.2021.681842
22
MartinR. J.WilsonC. G. (2012). Apnea of Prematurity. Compr. Physiol.2, 2923–2931. 10.1002/cphy.c100021
23
MathewO. P. (2011). Apnea of Prematurity: Pathogenesis and Management Strategies. J. Perinatol31, 302–310. 10.1038/jp.2010.126
24
MohawkJ. A.GreenC. B.TakahashiJ. S. (2012). Central and Peripheral Circadian Clocks in Mammals. Annu. Rev. Neurosci.35, 445–462. 10.1146/annurev-neuro-060909-153128
25
MortonS. U.SmithV. C. (2016). Treatment Options for Apnoea of Prematurity. Arch. Dis. Child. Fetal Neonatal. Ed.101, F352–F356. 10.1136/archdischild-2015-310228
26
Puia-DumitrescuM.SmithP. B.ZhaoJ.SorianoA.PayneE. H.HarperB.et al (2019). Dosing and Safety of Off-Label Use of Caffeine Citrate in Premature Infants. J. Pediatr.211, 27–e1. 10.1016/j.jpeds.2019.04.028
27
Rico de SouzaA.TraboulsiH.WangX.FritzJ. H.EidelmanD. H.BagloleC. J. (2021). The Aryl Hydrocarbon Receptor Attenuates Acute Cigarette Smoke-Induced Airway Neutrophilia Independent of the Dioxin Response Element. Front. Immunol.12, 630427. 10.3389/fimmu.2021.630427
28
RothhammerV.QuintanaF. J. (2019). The Aryl Hydrocarbon Receptor: an Environmental Sensor Integrating Immune Responses in Health and Disease. Nat. Rev. Immunol.19, 184–197. 10.1038/s41577-019-0125-8
29
RuanW.YuanX.EltzschigH. K. (2021). Circadian Rhythm as a Therapeutic Target. Nat. Rev. Drug Discov.20, 287–307. 10.1038/s41573-020-00109-w
30
SauzeauV.Carvajal-GonzálezJ. M.RiolobosA. S.SevillaM. A.Menacho-MárquezM.RománA. C.et al (2011). Transcriptional Factor Aryl Hydrocarbon Receptor (Ahr) Controls Cardiovascular and Respiratory Functions by Regulating the Expression of the Vav3 Proto-Oncogene. J. Biol. Chem.286, 2896–2909. 10.1074/jbc.M110.187534
31
TischkauS. A. (2020). Mechanisms of Circadian Clock Interactions with Aryl Hydrocarbon Receptor Signalling. Eur. J. Neurosci.51, 379–395. 10.1111/ejn.14361
32
van DiepenH. C.LucassenE. A.YasenkovR.GroenenI.IjzermanA. P.MeijerJ. H.et al (2014). Caffeine Increases Light Responsiveness of the Mouse Circadian Pacemaker. Eur. J. Neurosci.40, 3504–3511. 10.1111/ejn.12715
33
WangH. C.LiuK. Y.WangL. T.HsuS. H.WangS. C.HuangS. K. (2021). Aryl Hydrocarbon Receptor Promotes Lipid Droplet Biogenesis and Metabolic Shift in Respiratory Club Cells. Hum. Cel34, 785–799. 10.1007/s13577-021-00491-6
34
ZhangS.PatelA.ChuC.JiangW.WangL.WeltyS. E.et al (2015). Aryl Hydrocarbon Receptor Is Necessary to Protect Fetal Human Pulmonary Microvascular Endothelial Cells against Hyperoxic Injury: Mechanistic Roles of Antioxidant Enzymes and RelB. Toxicol. Appl. Pharmacol.286, 92–101. 10.1016/j.taap.2015.03.023
35
ZhangS. L.LahensN. F.YueZ.ArnoldD. M.PakstisP. P.SchwarzJ. E.et al (2021). A Circadian Clock Regulates Efflux by the Blood-Brain Barrier in Mice and Human Cells. Nat. Commun.12, 617. 10.1038/s41467-020-20795-9
Summary
Keywords
preterm infant, apnea of prematurity, caffeine, circadian rhythm, clock, AHR, polymorphism
Citation
Guo H-L, Long J-Y, Hu Y-H, Liu Y, He X, Li L, Xia Y, Ding X-S, Chen F, Xu J and Cheng R (2022) Caffeine Therapy for Apnea of Prematurity: Role of the Circadian CLOCK Gene Polymorphism. Front. Pharmacol. 12:724145. doi: 10.3389/fphar.2021.724145
Received
12 June 2021
Accepted
14 December 2021
Published
25 January 2022
Volume
12 - 2021
Edited by
Catherine M. T. Sherwin, Wright State University, United States
Reviewed by
Elisa Dufoo, Universidad Autónoma de Querétaro, Mexico
Carl Hunt, Uniformed Services University of the Health Sciences, United States
Yu-Shiuan Lin, University Psychiatric Clinic Basel, Switzerland
Updates
Copyright
© 2022 Guo, Long, Hu, Liu, He, Li, Xia, Ding, Chen, Xu and Cheng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Chen, cy.chen508@gmail.com; Jing Xu, njxujing@163.com; Rui Cheng, chengrui350@163.com
†These authors have contributed equally to this work
‡Visiting graduate student from China Pharmaceutical University
This article was submitted to Obstetric and Pediatric Pharmacology, a section of the journal Frontiers in Pharmacology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.