Abstract
The reprogramming of cellular metabolism is frequently linked to tumorigenesis. Glucose, fatty acids, and amino acids are the specific substrates involved in how an organism maintains metabolic equilibrium. The HADH gene codes for the short-chain L-3-hydroxyacyl-CoA dehydrogenase (HADH), a crucial enzyme in fatty acid oxidation that catalyzes the third phase of fatty acid oxidation in mitochondria. Increasing data suggest that HADH is differentially expressed in various types of malignancies and is linked to cancer development and progression. The significance of HADH expression in tumors and its potential mechanisms of action in the onset and progression of certain cancers are summarized in this article. The possible roles of HADH as a target and/or biomarker for the detection and treatment of various malignancies is also described here.
1 Introduction
Cancer is a major public health problem worldwide and has been identified as the biggest obstacle to improving life expectancy in the 21st century, making it a continuous focus of scientific attention (Fitzgerald et al., 2021). Cancer remains one of the leading causes of death (Galván Morales et al., 2020), and by 2018, approximately more than 11 million people have been diagnosed with cancer (Wang et al., 2018). The American Cancer Society expects that number to increase further, with an estimated 1,918,030 new cases and 609,360 cancer-related deaths occurring in the United States in 2022 (Siegel et al., 2022). More than 4 million new cancer patients and more than 2 million cancer-related mortalities are reported every year in China. Although cancer is treated in a variety of ways, such as surgery, chemotherapy, radiation therapy, and targeted therapy, 3- and 5-year cancer-specific survival rates remain poor (Kim and Kim, 2015; Vijayvergia et al., 2015; Koinis et al., 2016; Miller et al., 2016; Nakashima, 2018; Zhang and Zhang, 2018; Wen et al., 2019). Despite cancer-related deaths having declined overall (Henley et al., 2020), it is important to note that this decrease is mainly a result of early detection and prevention rather than better treatments (Etzioni et al., 2003; Chabner et al., 2005; Huff et al., 2006; Buskwofie et al., 2020). The vast majority of cancers are asymptomatic in their early stages of development (Smith et al., 2007; Zheng et al., 2020). Therefore, it is very important to explore the mechanisms of tumorigenesis and development, search for new diagnostic and prognostic markers, and develop effective and novel therapeutic methods. Further advancements will have a major impact on improving cancer patient survival rates.
Numerous studies have strongly demonstrated that specific genes, such as oncogenes and tumor suppressor genes, are risk factors for certain malignancies (Khan et al., 2018; Elek et al., 2020; Shareefi et al., 2020; Vysotskaia et al., 2020; Xiong et al., 2020). Oncogenes can promote tumor growth when they are activated, whereas tumor suppressor genes hinder tumor growth and development. Oncogene-directed metabolic reprogramming (Ward and Thompson, 2012), which appears to be a common hallmark of highly malignant tumors (Hanahan and Weinberg, 2011) regardless of their carcinogenic origin (Bustamante et al., 1981), is the most prevalent cause of metabolic alterations.
The tumor microenvironment (TME) is involved with carcinogenesis in a complex manner and can influence cancer incidence and progression (Arneth, 2019). The non-malignant cells in the TME frequently play a key role in all phases of tumorigenesis by stimulating and promoting uncontrolled cell proliferation (Balkwill et al., 2012; Hanahan and Coussens, 2012). The extracellular matrix (ECM), blood vessels, fibroblasts, lymphocytes, signaling chemicals and bone marrow-derived inflammatory cells make up the TME (Spill et al., 2016; Del Prete et al., 2017). Immune cells, such as lymphocytes, macrophages, and granulocytes, are a very significant component and can impact the formation, growth, and development of tumor cells in patients with various forms of cancer (Mantovani et al., 2008; Grivennikov et al., 2010; Hanahan and Weinberg, 2011; Lebleu, 2015; Spill et al., 2016; Del Prete et al., 2017). The TME homeostasis is the result of numerous complex interactions, many of which involve cell metabolism (Nieman et al., 2011; Colegio et al., 2014; Chang et al., 2015; Sousa et al., 2016; Chen P. et al., 2017; Angelin et al., 2017; Buck et al., 2017; Bantug et al., 2018; Zhang et al., 2018; Vitale et al., 2019a; Vitale et al., 2019b). Under normal circumstances, an organism’s metabolism is in equilibrium. The reprogramming of cellular metabolism is a hallmark of tumorigenesis (Hanahan and Weinberg, 2011; Wettersten et al., 2017) and aids in the conversion of large amounts of nutrients into cellular building blocks such as nucleotides, amino acids, and lipids (Ren et al., 2020), resulting in an excess of the antioxidant glutathione to produce new cells (Gao et al., 2009; Hirschhaeuser et al., 2011). Glucose, fatty acids, and amino acids are the substrates that keep metabolic homeostasis in check (Heslegrave and Hussain, 2013). Fatty acid metabolism is often altered in cancer cells to sustain cell proliferation, meet energy needs, and produce metabolites for anabolic activities (Currie et al., 2013; Sanchez and Simon, 2018). Several reports have shown that the enzymes involved in fatty acid β-oxidation are reduced in individuals with malignancies (Tanaka et al., 2013; Enjoji et al., 2016). β-oxidation in mitochondria breaks down fatty acids (Bartlett and Eaton, 2004), and this is a crucial metabolic process for energy balance in organs such the liver, heart, and skeletal muscle (Heslegrave and Hussain, 2013). Enoyl-CoA hydratase, acyl-CoA dehydrogenase, ketoacyl-CoA thiolase, and hydroxyacyl-CoA dehydrogenase are four main enzymes involved in the breakdown of fatty acids (Shen et al., 2017). The role of fatty acids in the breakdown of cancer cells, however, is still debated. Additional research is required to further understand how fatty acid metabolism reprogramming influences the formation and development of cancers.
The human HADH gene, which has 10 exons and is expressed in most tissues, is found on chromosome 4q25 (Heslegrave and Hussain, 2013). The gene encodes the intramitochondrial homodimer enzyme short-chain-L-3-hydroxyacyl-CoA dehydrogenase (HADH), which is a key enzyme in the third step of fatty acid β-oxidation (Vredendaal et al., 1996; Eaton et al., 2000; Yang et al., 2005; Kapoor et al., 2010; Schulz et al., 2011; Popa et al., 2012; Arya et al., 2014; Jiang et al., 2021). During extended fasting, HADH transforms short- and medium-chain fatty acids into ketones to fuel the liver, heart, muscles, and pancreas (Shen et al., 2017), with enzyme activity being highest in the pancreas and especially in the islets of Langerhans (Agren et al., 1977). Several investigations have shown that HADH plays an important role in controlling insulin secretion from the β-cell (Hardy et al., 2007; Martens et al., 2007; Filling et al., 2008; Li et al., 2010; Heslegrave et al., 2012) and that inhibiting its activity results in a considerable increase in insulin secretion (Kapoor et al., 2009; Heslegrave et al., 2012; Heslegrave and Hussain, 2013).
Reprogramming of energy metabolism is a well-known feature of malignancies (Hanahan and Weinberg, 2011; Wettersten et al., 2017), and fatty acid metabolism is also thought to be a crucial contributor to cancer cell proliferation (Currie et al., 2013). HADH is a crucial enzyme in the oxidation of fatty acids (Flanagan et al., 2013; Babiker et al., 2015; Çamtosun et al., 2015; Satapathy et al., 2016; Boerrigter-Eenling et al., 2017). Dehydrogenation, hydration, dehydrogenation again, and thiolytic cleavage are the four enzymatic processes that make up fatty acid oxidation (FAO) (Wanders et al., 1999). HADH is a component of the enzymatic reaction mentioned above (Houten and Wanders, 2010). In the mitochondrial matrix, HADH catalyzes the penultimate process in the β-oxidation of fatty acids (Vredendaal et al., 1998), dehydroxylating medium- and short-chain NAD+-dependent L3-hydroxy-acyl-CoA to produce β-ketoacyl-CoA and NADH, respectively (Figure 1) (Houten et al., 2016). The expression levels of HADH are also higher than those of other fatty acid β oxidases, such as acyl-CoA dehydrogenase and acetyl-CoA acyltransferase 2. Additionally, HADH enzymatic activity is the most effective for metabolizing medium-chain length fatty acids (Pepin et al., 2010). Reduced HADH expression can impede β-oxidation and stimulate fatty acid buildup, which leads to fatty acid metabolism reprogramming and promotes tumor development (Wettersten et al., 2017). Growing evidence has recently shown its importance in the occurrence and progression of several malignancies (Shen et al., 2017; Wilkins et al., 2017; Nwosu et al., 2018; Voloshanenko et al., 2018; Ren et al., 2020; Jiang et al., 2021; Sun et al., 2022). HADH has been identified as a possible target for the diagnosis and therapeutic treatment of many malignancies because of this apparent influence on carcinogenesis (Figure 2). The specific functions and molecular details of HADH in the incidence and progression of various cancers are summarized in this article, with an emphasis on gastric cancer, kidney renal clear cell carcinoma, liver cancer, colon cancer, and acute myeloid leukemia.
FIGURE 1
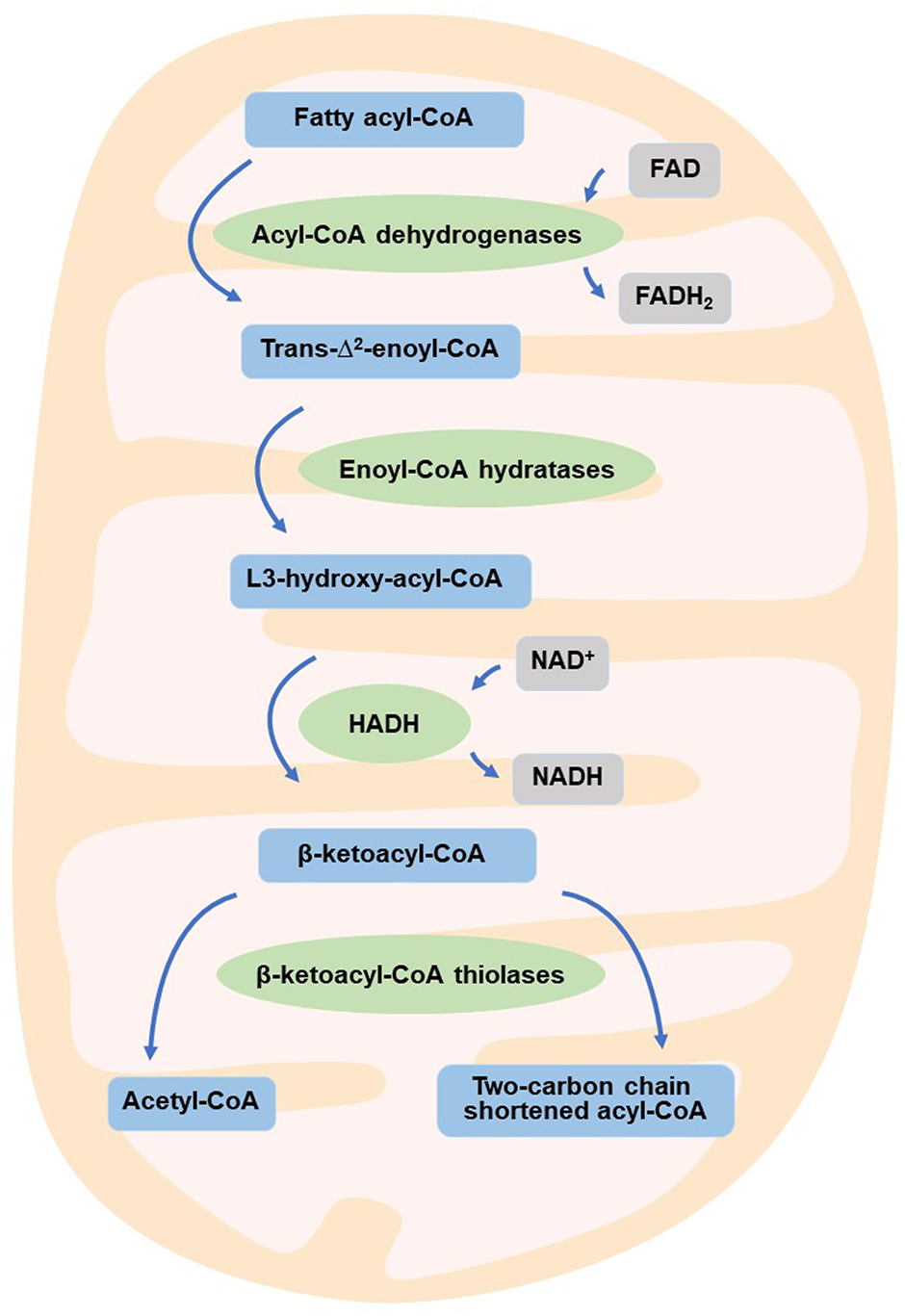
HADH is involved in fatty acid β-oxidation.
FIGURE 2
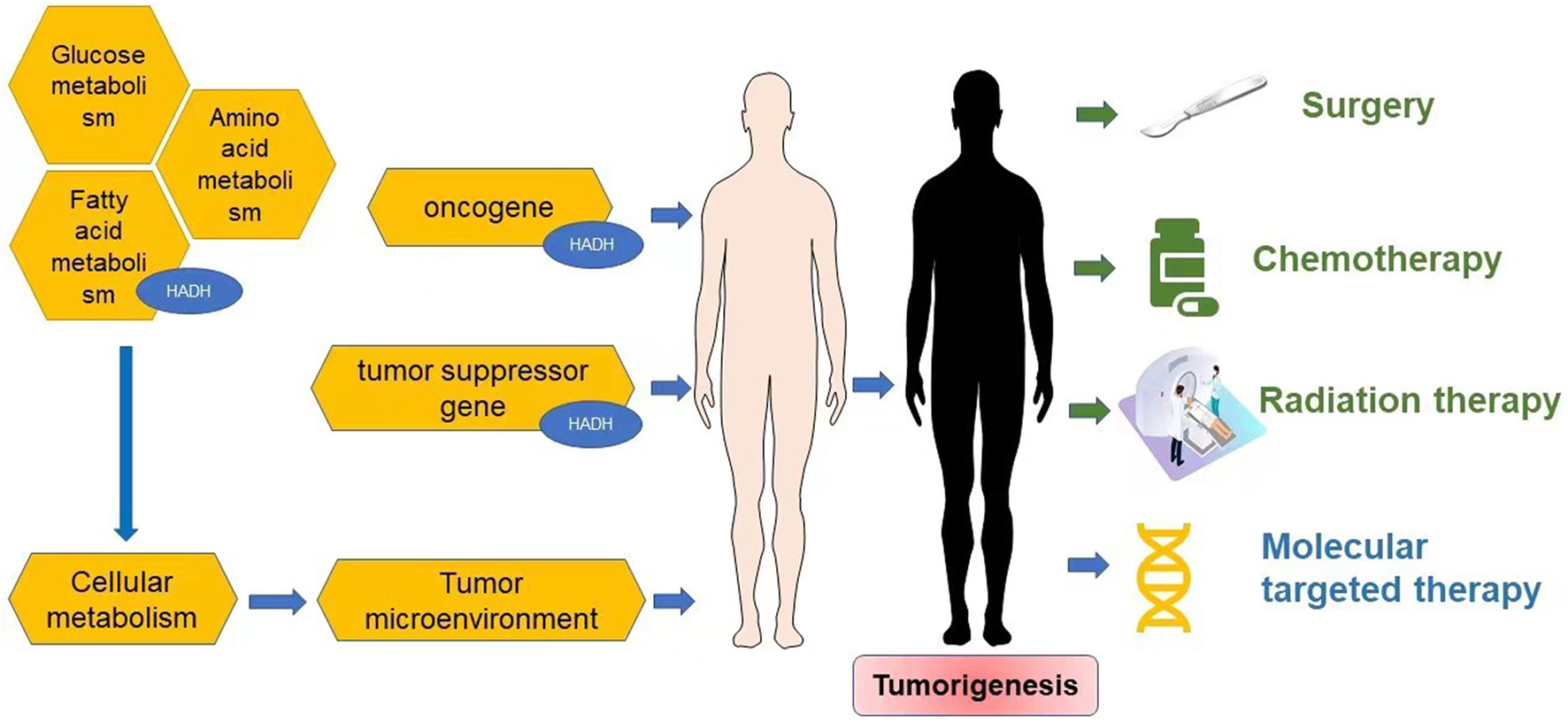
Cancer progression and therapy.
2 Short-chain L-3-hydroxyacyl-CoA dehydrogenase in cancers
HADH expression is upregulated or downregulated in different types of cancers, including gastric cancer, kidney renal clear cell carcinoma, liver cancer, colon cancer, and acute myeloid leukemia. The relevant clinicopathological features and molecular mechanisms of HADH in these cancers are summarized in Table 1 and detailed in the rest of this section.
TABLE 1
| Cancer types | Expression | Role | Functional role | Related genes | Clinical features | References |
|---|---|---|---|---|---|---|
| Gastric cancer | Downregulated | Tumor suppressor gene | Inhibits proliferation, migration, and invasion | PTEN, AKT | Early clinical stage, long OS and DFS, and good clinical prognosis | (Shen et al., 2017; Du et al., 2021) |
| Kidney renal clear cell carcinoma | Downregulated | Tumor suppressor gene | No studies have been reported | genes in EMT, TNF- α, IL-6-JAK-STAT3, and interferon-γ signaling pathway | Early clinical stage, low histologic grade, long OS and DFS, low distant metastasis, good prognosis, and immune infiltration | (Zhang et al., 2019; Jiang et al., 2021; Sun et al., 2022) |
| Liver cancer | Downregulated | Tumor suppressor gene | No studies have been reported | No studies have been reported | No studies have been reported | Nwosu et al. (2018) |
| Colon cancer | Upregulated | Oncogene | Promotes proliferation | Wnt5a/b, Evi/Wls, RoR2, Dvl2, ATF2 and ATF4 | Poor clinical outcome | (Voloshanenko et al., 2018; Ren et al., 2020) |
| Acute myeloid leukemia | Upregulated | Oncogene | No studies have been reported | GPX-7 and RPP40 | Poor OS | Wei et al. (2020) |
Functional characteristics and clinical features of HADH in human cancers.
PTEN, phosphatase and tensin homolog; AKT, serine threonine kinase; EMT, epithelial-mesenchymal transition; TNF- α, tumor necrosis factor- α; IL-6, interleukin 6; JAK, janus kinase; STAT3, signal transducer and activator of transcription 3; ROR2, receptor tyrosine kinase like orphan receptor 2; Dvl2, disheveled segment polarity protein 2; ATF2, activating transcription factor 2; ATF4, activating transcription factor 4; GPX-7, glutathione peroxidase 7; Rpp40, ribonuclease p protein subunit p40; OS, overall survival; DFS, disease-free survival.
2.1 Gastric cancer
2.1.1 Functional characteristics and clinical features of short-chain L-3-hydroxyacyl-CoA dehydrogenase in gastric cancer
HADH is expressed at lower levels in gastric cancer tissues compared with that in normal gastric tissues. Shen et al. (2017) showed that downregulation of HADH was significantly correlated with advanced clinical stage, low overall survival (OS), low disease-free survival (DFS), and poor clinical prognosis. Hence, HADH expression has been proposed as an independent prognostic factor that affects patient survival rates. Mechanistically, reduced HADH expression significantly promoted cell proliferation and increased migration and invasion of tumor cells in vitro; in contrast, overexpression of HADH inhibited the proliferation of gastric cancer cells (Shen et al., 2017).
In summary, HADH is a potential novel tumor suppressor gene in gastric cancer that can inhibit cell proliferation, migration, and invasion of gastric cancer cells. Furthermore, its expression levels are correlated with cancer progression and patient survival.
2.1.2 Signaling pathways influenced by short-chain L-3-hydroxyacyl-CoA dehydrogenase in gastric cancer
The AKT signaling pathway is required for HADH to regulate cell proliferation, migration, and invasion, according to Shen et al. (Shen et al., 2017). AKT signaling is a growth-regulating biological pathway that has been shown to improve tumor cell survival, proliferation, and motility in a variety of tumor types (Chan et al., 2014; Bao et al., 2021; Junaid et al., 2021; Tsai et al., 2021). AKT, also known as protein kinase B (PKB), is a critical node in many signaling pathways, as well as one of the most essential and flexible protein kinases in human physiology and illness (Meier and Hemmings, 1999; Manning and Cantley, 2007; Revathidevi and Munirajan, 2019). Many interesting advancements in the mechanism controlling AKT activity have been achieved since its identification as an oncogene homologue of murine leukemia virus AKT8 (Staal, 1987; Bellacosa et al., 1991) and protein kinase C (Jones et al., 1991). As a critical regulatory protein of cell growth, survival, proliferation, and metabolism (Kennedy et al., 1997; Sun et al., 2001; Vivanco and Sawyers, 2002; Bellacosa et al., 2005; Manning and Cantley, 2007; Broustas et al., 2012; Sanidas et al., 2014; Fan et al., 2018; Li et al., 2021), AKT crosses multiple signaling pathways (Gao and Pan, 2001; Ward et al., 2011; Yecies et al., 2011; Yin et al., 2017) and participates in a range of physiological activities. AKT is involved in the development of a variety of human malignancies (Manning and Cantley, 2007; Revathidevi and Munirajan, 2019). AKT gene mutations are uncommon, while AKT gene amplification and overexpression are widespread in malignancies such as gastric, colon, liver, thyroid, and ovarian tumors (Staal, 1987; Bellacosa et al., 1995; Cheng et al., 1996; Nakatani et al., 1999; Roy et al., 2002; Knobbe and Reifenberger, 2003; Xu et al., 2004; Altomare and Testa, 2005; Parsons et al., 2005; Carpten et al., 2007; Malanga et al., 2008; Mohamedali et al., 2008; Shoji et al., 2009; Zilberman et al., 2009; Askham et al., 2010; Mundi et al., 2016; Manning and Toker, 2017). Overexpression and activation of AKT have been linked to the initiation or progression of a number of human malignancies (Samuels et al., 2004; Cully et al., 2006; Cerami et al., 2012; Fruman and Rommel, 2014). AKT has a role in several physiological processes and, once activated, can affect the activity of several downstream proteins that control cell growth, survival, proliferation, and metabolism (Kennedy et al., 1997; Sun et al., 2001; Vivanco and Sawyers, 2002; Bellacosa et al., 2005; Hay, 2005; Manning and Cantley, 2007; Broustas et al., 2012; Sanidas et al., 2014; Liu H. W. et al., 2018; Fan et al., 2018). An abnormal loss or increase of AKT activation underpins the pathogenesis of a variety of complicated illnesses, including type 2 diabetes and cancers (Manning and Cantley, 2007). AKT also affects cell survival by phosphorylating and activating a number of oncoproteins implicated in cell cycle progression and carcinogenesis, including murine double minute (MDM2), S-phase kinase-associated protein 2 (Skp2), IKKα, and E3 ligase (Datta et al., 1999; Ozes et al., 1999; Mayo and Donner, 2001; Zhou et al., 2001; Chan et al., 2012; Chan et al., 2014). PTEN is a tumor suppressor that causes a significant reduction in cell proliferation by arresting the cell cycle in the G1 phase. PTEN and PHLPP2 are the most important negative regulators of AKT (Stambolic et al., 1998; Meng et al., 2007). Therefore, PTEN inactivation can potentially activate AKT by promoting AKT’s phosphorylation ability, leading to a cell survival advantage and uncontrolled cell proliferation (Downward, 2003; Gomes et al., 2014; Liu J. et al., 2018). Shen et al. (Shen et al., 2017) showed that downregulation of HADH could inhibit the expression of PTEN and promote the phosphorylation of AKT, further stimulating the proliferation, migration, and invasion of gastric cancer cells by activating the AKT pathway (Figure 3).
FIGURE 3
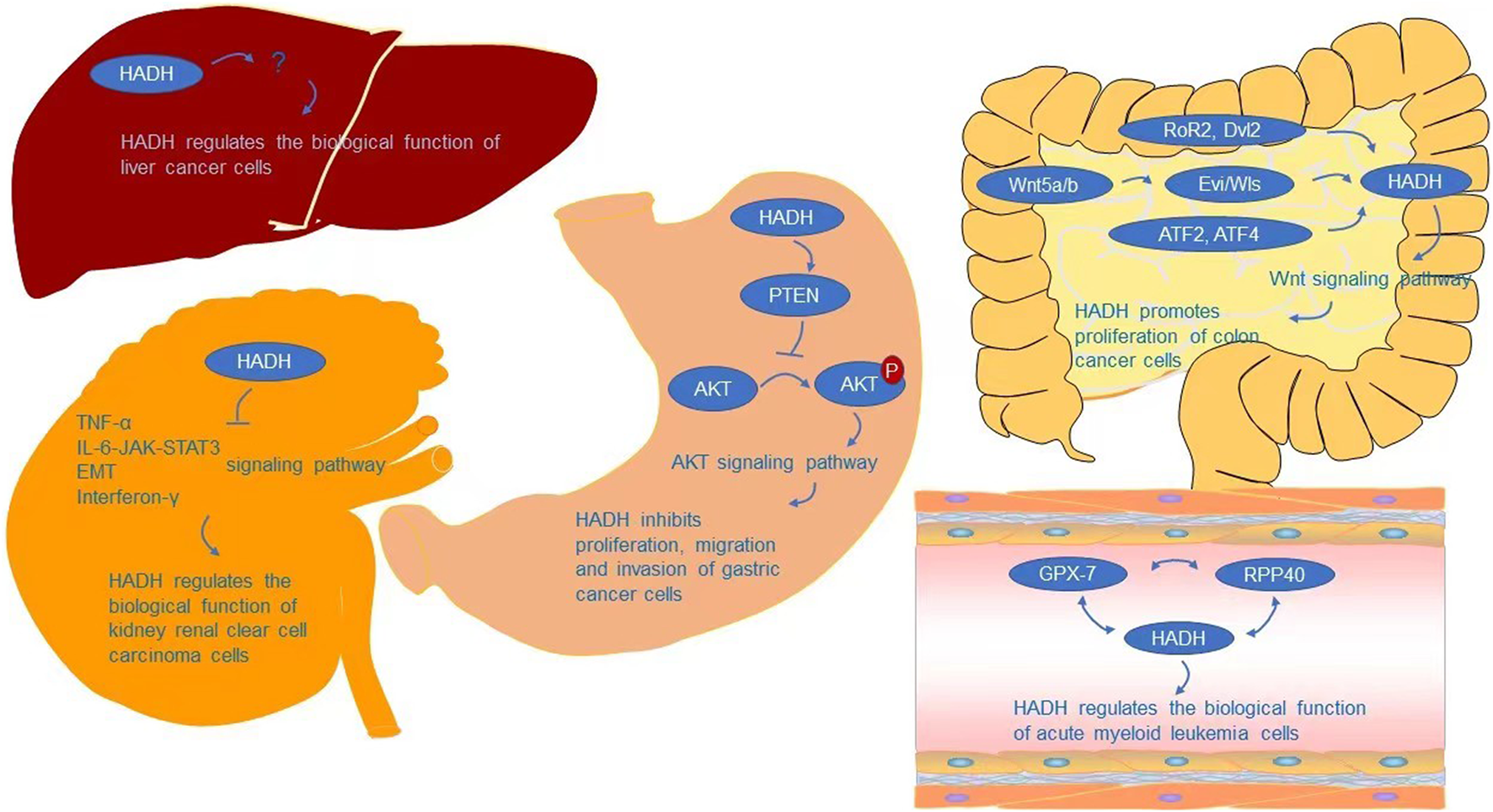
Underlying molecular mechanisms of HADH in cancers.
2.2 Kidney renal clear cell carcinoma
2.2.1 Functional characteristics and clinical features of short-chain L-3-hydroxyacyl-CoA dehydrogenase in kidney renal clear cell carcinoma
Similar to gastric cancer tissues, HADH expression was markedly downregulated in kidney renal clear cell carcinoma tissues compared with that in adjacent non-cancerous tissues (Zhang et al., 2019; Jiang et al., 2021; Sun et al., 2022). Additionally, similarly to gastric cancer, HADH downregulation was significantly associated with poor OS, DFS, and poor prognosis in kidney renal clear cell carcinoma (Zhang et al., 2019; Jiang et al., 2021; Sun et al., 2022). It was positively correlated with the early clinical stage of disease and low histologic grade (Jiang et al., 2021; Sun et al., 2022). Moreover, HADH expression was also associated with tumor-infiltrating immune cells (TIICs) in kidney renal clear cell carcinoma (Jiang et al., 2021). Levels of M2 macrophages, naïve B cells, resting mast cells, and resting dendritic cells were positively correlated with HADH expression, while amounts of follicular helper T cells, plasma cells, regulatory T cells (Tregs), and neutrophils were negatively correlated with HADH expression (Jiang et al., 2021). These results suggest that HADH has an important role in the regulation of the immune microenvironment in kidney renal clear cell carcinoma.
In conclusion, HADH may be a novel tumor suppressor gene in kidney renal clear cell carcinoma, and its reduced expression is associated with immune cell infiltration and poor prognosis.
2.2.2 Signaling pathways influenced by short-chain L-3-hydroxyacyl-CoA dehydrogenase in kidney renal clear cell carcinoma
Analyses of biological processes have indicated that HADH is associated with cell cycle arrest and negative regulation of the cell cycle (Zhang et al., 2019). Jiang et al. (2021) further analyzed this using GSEA, finding that the inflammatory response, TNF-α, IL-6-JAK-STAT3, epithelial-mesenchymal transition (EMT), and interferon-γ signaling pathways were activated in the HADH-low expression group, while fatty acid metabolism and protein secretion were inhibited (Figure 3). However, the specific molecular mechanism of how HADH can inhibit kidney renal clear cell carcinoma progression requires further study.
2.3 Liver cancer
Nwosu et al. (2018) found that HADH involved in fatty acid β-oxidation was expressed at lower levels in poorly differentiated hepatocellular carcinoma (HCC) cells compared with that in well differentiated HCC cells. However, whether the higher proliferation and migration rates of these poorly differentiated HCC cells are directly related to metabolic changes, including fatty acid β-oxidation, is still unknown. Further research is needed on the biological function of HADH and its signaling pathways in HCC (Figure 3).
2.4 Colon cancer
2.4.1 Functional characteristics and clinical features of short-chain L-3-hydroxyacyl-CoA dehydrogenase in colon cancer
Unlike gastric, kidney, and liver cancers, HADH is highly expressed in colon cancer cells (Ren et al., 2020). High HADH levels can promote colon cancer cell proliferation and are significantly associated with poor clinical outcomes (Voloshanenko et al., 2018; Ren et al., 2020). Thus, HADH potentially functions as an oncogene in colon cancer.
2.4.2 Signaling pathways influenced by short-chain L-3-hydroxyacyl-CoA dehydrogenase in colon cancer
In colon cancer cells, the Wnt signaling pathway is required for HADH-mediated regulation of cell proliferation (Voloshanenko et al., 2018). Wnt signaling is involved in a variety of events throughout embryonic development and tissue homeostasis, and has also been linked to cancer (Mao et al., 2014; Kahn, 2014; Morin et al., 1997; Clevers and Nusse, 2012; Roelink et al., 1992; Clements et al., 2002; van’t Veer et al., 1984; Cleary et al., 2014). β-catenin-dependent (canonical) and independent (non-canonical) signaling are two types of Wnt signaling (Zhan et al., 2017). Multiple intracellular signal cascades can be triggered by Wnt ligands, which can orchestrate complicated context-dependent responses. With the aid of Porcupine (Porcn) and Evi/Wls/GRP177, cells can release Wnt ligands in an autocrine or paracrine manner (Kadowaki et al., 1996; Herr and Basler, 2012). Wnt5a/b has been demonstrated to regulate HADH expression, with HADH relying on Evi/Wls secretion to act on the β-catenin-independent Wnt signaling pathway for regulation of colon cancer cell growth and proliferation (Voloshanenko et al., 2018). Dvl2 and RoR2 are also involved in the regulation of HADH, which is consistent with existing knowledge of the participation of RoR2/Dvl2 in β-catenin-independent Wnt signaling (Boutros et al., 1998; Nishita et al., 2010; Ishida-Takagishi et al., 2012) (Figure 3). ATF2 and ATF4 transcription factors are also involved in regulating HADH.
2.5 Acute myeloid leukemia
2.5.1 Functional characteristics and clinical features of short-chain L-3-hydroxyacyl-CoA dehydrogenase in acute myeloid leukemia
Similar to colon cancer tissues, HADH expression was markedly upregulated in acute myeloid leukemia patient samples. Wei et al. showed that HADH upregulation was significantly associated with poor OS (Wei et al., 2020).
2.5.2 Signaling pathways influenced by short-chain L-3-hydroxyacyl-CoA dehydrogenase in acute myeloid leukemia
Glutathione peroxidases (GPXs) are peroxidase enzymes that reduce lipid hydroperoxide and free hydrogen peroxide levels to protect organisms from oxidative damage (Takebe et al., 2002). In mammals, eight GPX sub-members have been discovered (Margis et al., 2008), which have been reported to play key roles in repairing reactive oxygen species (ROS)-induced damage, shielding DNA, proteins, and lipids from oxidative damage (Brigelius-Flohé and Maiorino, 2013), and carcinogenesis (Peng et al., 2014; Yang et al., 2014; Nalkiran et al., 2015; Chen Z. et al., 2017; Hangauer et al., 2017; Jiao et al., 2017; Liu et al., 2017; Viswanathan et al., 2017; An et al., 2018; Metere et al., 2018; Naiki et al., 2018; Zhu et al., 2018; Zhou et al., 2019a; Zhou et al., 2019b; Cai et al., 2019; Cheng et al., 2019; Lin et al., 2019; Wang et al., 2019; Yi et al., 2019; Li et al., 2020). Wei et al. discovered that HADH expression was linked to GPX-7 and RPP40 (Figure 3). However, the precise molecular mechanism by which HADH acts in acute myeloid leukemia is unknown, and more research is needed.
3 Conclusion and future perspectives
Around the world, cancer incidence and mortality are quickly rising. High-throughput gene expression profiling technologies allow for the simultaneous screening of expression levels of thousands of genes. Identifying variations in gene expression patterns between tumor and control samples is one of the key goals of gene expression profiling in cancer (Feten et al., 2007). Technological advancements and less expensive DNA sequencing procedures have fueled global efforts to identify relevant differentially expressed genes. In various human cancers, including gastric cancer, kidney renal clear cell carcinoma, liver cancer, colon cancer, and acute myeloid leukemia, the recently discovered gene HADH was found to be widely elevated or downregulated depending on the disease. Extensive therapeutics that target HADH have yet to be produced, leading to potential future developments. Multiple clinicopathological characteristics and patient prognoses were significantly associated with HADH expression levels, including clinical stage, histologic grade, immune cell infiltration, OS, DFS, and distant metastases. In vitro investigations have demonstrated that HADH can influence tumor cell proliferation, migration, and invasion rates in numerous malignancies, supporting its role in carcinogenesis and tumor progression. Preliminary findings reveal that HADH can impact multiple signaling pathways that promote carcinogenesis and cancer progression, including AKT, Wnt, EMT, TNF-α, IL-6-JAK-STAT3, and interferon signaling pathways.
Although HADH is a potential therapeutic target, several questions still remain to be addressed. Firstly, the molecular mechanisms of HADH in different types of cancers are not completely understood. Previous studies have suggested that HADH serves as a tumor suppressor gene in gastric cancer, kidney renal clear cell carcinoma, and liver cancer by inhibiting cell proliferation, migration, and invasion, as well as being associated with cancer progression and patient survival. However, it can also exist as an oncogene in colon cancer and acute myeloid leukemia, where it promotes cell proliferation and is associated with poor patient outcomes. Pathway analyses of HADH activity have only been conducted in kidney renal clear cell carcinoma, liver cancer, colon cancer, and acute myeloid leukemia, but the specific molecular mechanisms were not explained in detail. Furthermore, while the functions of HADH in gastric cancer, kidney renal clear cell carcinoma, liver cancer, colon cancer, and acute myeloid leukemia have been studied to some extent, its potential role in other cancers, such as cancers associated with the respiratory and reproductive systems, remain unexplored. Secondly, the search for diagnostic biomarkers or therapeutic targets is a promising direction for cancer diagnosis and treatment. HADH can be upregulated or downregulated in certain tumor tissues, but it is currently not known if HADH is also upregulated or downregulated in body fluids such as urine and plasma. Next, we will focus on whether the levels of upstream and downstream factors in the HADH pathway are changed in urine and serum, as well as further analyze whether expression of HADH itself changes in these body fluids. If HADH is detected in urine or plasma, a simple non-invasive test can possibly be performed to use HADH as a cancer-specific molecular biomarker to facilitate early detection and prognostic assessment of specific cancers. Thirdly, whether HADH can play a role in cancer diagnosis as a tumor-associated antigen is still unknown and requires further evaluation. Lastly, HADH is an immune system-associated gene, but whether it can play a role in clinical trials and individualized treatment of various immune modulators remains to be seen. Therefore, more attention should be paid to the clinical value of HADH in cancer diagnosis and treatment.
In summary, various studies have shown that HADH can have oncogenic or tumor suppressive functions in cancer development and progression and may serve as a potential cancer-specific molecular biomarker in the diagnosis, treatment, and prognosis of different types of cancers. Some progress has been made in studying the mechanism of HADH, but this work is currently in early stages. Future investigations should focus on exploring the precise molecular mechanism of how HADH is regulated in carcinogenesis and tumor progression to support its potential clinical application.
Statements
Author contributions
HF and YY contributed to the conception and design of the review. HF wrote the first draft of the manuscript. YY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Jilin Province Financial and Health Project (Research on the mechanism of Mir-23A-27A-24-2 cluster promoting invasion and metastasis of hepatocellular carcinoma by regulating WNT signaling pathway, 2018GDYYZ001); Jilin Province Medical and Health Talent Special Project (Application of ICG fluorescence imaging combined with intraoperative ultrasound in laparoscopic precision hepatectomy, 2019SCZT009).
Acknowledgments
We want to thank colleagues for their critical discussion regarding this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HADH, short-chain-L-3-hydroxyacyl-CoA dehydrogenase; TME, tumor microenvironment; FAO, fatty acid oxidation; TIICs, tumor-infiltrating immune cells; PTEN, phosphatase and tensin homolog; AKT, serine threonine kinase; EMT, epithelial-mesenchymal transition; TNF- α, tumor necrosis factor-α; IL-6, interleukin 6; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3; ROR2, receptor tyrosine kinase like orphan receptor 2; Dvl2, disheveled segment polarity protein 2; ATF2, activating transcription factor 2; ATF4, activating transcription factor 4; GPX-7, glutathione peroxidase 7; Rpp40, ribonuclease p protein subunit p40; OS, overall survival; DFS, disease-free survival; HCC, hepatocellular carcinoma
References
1
Agren A. Borg K. Brolin S. E. Carlman J. Lundqvist G. (1977). Hydroxyacyl CoA dehydrogenase, an enzyme important in fat metabolism in different cell types in the islets of Langerhans. Diabete Metab.3, 169–172.
2
Altomare D. A. Testa J. R. (2005). Perturbations of the AKT signaling pathway in human cancer. Oncogene24, 7455–7464. 10.1038/sj.onc.1209085
3
An B. C. Choi Y. D. Oh I. J. Kim J. H. Park J. I. Lee S. W. (2018). GPx3-mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS One13, e0204170. 10.1371/journal.pone.0204170
4
Angelin A. Gil-De-GóMEZ L. Dahiya S. Jiao J. Guo L. Levine M. H. et al (2017). Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab.25, 1282–1293. e7. 10.1016/j.cmet.2016.12.018
5
Arneth B. (2019). Tumor microenvironment. Medicina (Kaunas), 56.
6
Arya V. B. Mohammed Z. Blankenstein O. De Lonlay P. Hussain K. (2014). Hyperinsulinaemic hypoglycaemia. Horm. Metab. Res.46, 157–170. 10.1055/s-0034-1367063
7
Askham J. M. Platt F. Chambers P. A. Snowden H. Taylor C. F. Knowles M. A. (2010). AKT1 mutations in bladder cancer: Identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene29, 150–155. 10.1038/onc.2009.315
8
Babiker O. Flanagan S. E. Ellard S. Al Girim H. Hussain K. Senniappan S. (2015). Protein-induced hyperinsulinaemic hypoglycaemia due to a homozygous HADH mutation in three siblings of a Saudi family. J. Pediatr. Endocrinol. Metab.28, 1073–1077. 10.1515/jpem-2015-0033
9
Balkwill F. R. Capasso M. Hagemann T. (2012). The tumor microenvironment at a glance. J. Cell Sci.125, 5591–5596. 10.1242/jcs.116392
10
Bantug G. R. Galluzzi L. Kroemer G. Hess C. (2018). The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol.18, 19–34. 10.1038/nri.2017.99
11
Bao F. Hao P. An S. Yang Y. Liu Y. Hao Q. et al (2021). Akt scaffold proteins: The key to controlling specificity of akt signaling. Am. J. Physiol. Cell Physiol.321, C429–c442. 10.1152/ajpcell.00146.2020
12
Bartlett K. Eaton S. (2004). Mitochondrial beta-oxidation. Eur. J. Biochem.271, 462–469. 10.1046/j.1432-1033.2003.03947.x
13
Bellacosa A. Testa J. R. Staal S. P. Tsichlis P. N. (1991). A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science254, 274–277. 10.1126/science.1833819
14
Bellacosa A. De Feo D. Godwin A. K. Bell D. W. Cheng J. Q. Altomare D. A. et al (1995). Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer64, 280–285. 10.1002/ijc.2910640412
15
Bellacosa A. Kumar C. C. Di Cristofano A. Testa J. R. (2005). Activation of AKT kinases in cancer: Implications for therapeutic targeting. Adv. Cancer Res.94, 29–86. 10.1016/S0065-230X(05)94002-5
16
Boerrigter-Eenling R. Alewijn M. Weesepoel Y. Van Ruth S. (2017). New approaches towards discrimination of fresh/chilled and frozen/thawed chicken breasts by HADH activity determination: Customized slope fitting and chemometrics. Meat Sci.126, 43–49. 10.1016/j.meatsci.2016.12.009
17
Boutros M. Paricio N. Strutt D. I. Mlodzik M. (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell94, 109–118. 10.1016/s0092-8674(00)81226-x
18
Brigelius-Flohé R. Maiorino M. (2013). Glutathione peroxidases. Biochim. Biophys. Acta1830, 3289–3303. 10.1016/j.bbagen.2012.11.020
19
Broustas C. G. Zhu A. Lieberman H. B. (2012). Rad9 protein contributes to prostate tumor progression by promoting cell migration and anoikis resistance. J. Biol. Chem.287, 41324–41333. 10.1074/jbc.M112.402784
20
Buck M. D. Sowell R. T. Kaech S. M. Pearce E. L. (2017). Metabolic instruction of immunity. Cell169, 570–586. 10.1016/j.cell.2017.04.004
21
Buskwofie A. David-West G. Clare C. A. (2020). A review of cervical cancer: Incidence and disparities. J. Natl. Med. Assoc.112, 229–232. 10.1016/j.jnma.2020.03.002
22
Bustamante E. Morris H. P. Pedersen P. L. (1981). Energy metabolism of tumor cells. Requirement for a form of hexokinase with a propensity for mitochondrial binding. J. Biol. Chem.256, 8699–8704. 10.1016/s0021-9258(19)68900-3
23
Cai M. Sikong Y. Wang Q. Zhu S. Pang F. Cui X. (2019). Gpx3 prevents migration and invasion in gastric cancer by targeting NFкB/Wnt5a/JNK signaling. Int. J. Clin. Exp. Pathol.12, 1194–1203.
24
Çamtosun E. Flanagan S. E. Ellard S. ŞıKLAR Z. Hussain K. Kocaay P. et al (2015). A deep intronic HADH splicing mutation (c.636+471G>T) in a congenital hyperinsulinemic hypoglycemia case: Long term clinical course. J. Clin. Res. Pediatr. Endocrinol.7, 144–147. 10.4274/jcrpe.1963
25
Carpten J. D. Faber A. L. Horn C. Donoho G. P. Briggs S. L. Robbins C. M. et al (2007). A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature448, 439–444. 10.1038/nature05933
26
Cerami E. Gao J. Dogrusoz U. Gross B. E. Sumer S. O. Aksoy B. A. et al (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov.2, 401–404. 10.1158/2159-8290.CD-12-0095
27
Chabner B. A. Roberts T. G. JR (2005). Timeline: Chemotherapy and the war on cancer. Nat. Rev. Cancer5, 65–72. 10.1038/nrc1529
28
Chan C. H. Li C. F. Yang W. L. Gao Y. Lee S. W. Feng Z. et al (2012). The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell149, 1098–1111. 10.1016/j.cell.2012.02.065
29
Chan C. H. Jo U. Kohrman A. Rezaeian A. H. Chou P. C. Logothetis C. et al (2014). Posttranslational regulation of Akt in human cancer. Cell Biosci.4, 59. 10.1186/2045-3701-4-59
30
Chang C. H. Qiu J. O'Sullivan D. Buck M. D. Noguchi T. Curtis J. D. et al (2015). Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell162, 1229–1241. 10.1016/j.cell.2015.08.016
31
Chen P. Zuo H. Xiong H. Kolar M. J. Chu Q. Saghatelian A. et al (2017a). Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. U. S. A.114, 580–585. 10.1073/pnas.1614035114
32
Chen Z. Hu T. Zhu S. Mukaisho K. El-Rifai W. Peng D. F. (2017b). Glutathione peroxidase 7 suppresses cancer cell growth and is hypermethylated in gastric cancer. Oncotarget8, 54345–54356. 10.18632/oncotarget.17527
33
Cheng J. Q. Ruggeri B. Klein W. M. Sonoda G. Altomare D. A. Watson D. K. et al (1996). Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. U. S. A.93, 3636–3641. 10.1073/pnas.93.8.3636
34
Cheng Y. Xu T. Li S. Ruan H. (2019). GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY)11, 12165–12176. 10.18632/aging.102555
35
Cleary A. S. Leonard T. L. Gestl S. A. Gunther E. J. (2014). Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature508, 113–117. 10.1038/nature13187
36
Clements W. M. Wang J. Sarnaik A. Kim O. J. Macdonald J. Fenoglio-Preiser C. et al (2002). beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res.62, 3503–3506.
37
Clevers H. Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell149, 1192–1205. 10.1016/j.cell.2012.05.012
38
Colegio O. R. Chu N. Q. Szabo A. L. Chu T. Rhebergen A. M. Jairam V. et al (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature513, 559–563. 10.1038/nature13490
39
Cully M. You H. Levine A. J. Mak T. W. (2006). Beyond PTEN mutations: The PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer6, 184–192. 10.1038/nrc1819
40
Currie E. Schulze A. Zechner R. Walther T. C. Farese R. V. JR. (2013). Cellular fatty acid metabolism and cancer. Cell Metab.18, 153–161. 10.1016/j.cmet.2013.05.017
41
Datta S. R. Brunet A. Greenberg M. E. (1999). Cellular survival: A play in three akts. Genes Dev.13, 2905–2927. 10.1101/gad.13.22.2905
42
Del Prete A. Schioppa T. Tiberio L. Stabile H. Sozzani S. (2017). Leukocyte trafficking in tumor microenvironment. Curr. Opin. Pharmacol.35, 40–47. 10.1016/j.coph.2017.05.004
43
Downward J. (2003). Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer3, 11–22. 10.1038/nrc969
44
Du Z. Zhang X. Gao W. Yang J. (2021). Differentially expressed genes PCCA, ECHS1, and HADH are potential prognostic biomarkers for gastric cancer. Sci. Prog.104, 368504211011344. 10.1177/00368504211011344
45
Eaton S. Bursby T. Middleton B. Pourfarzam M. Mills K. Johnson A. W. et al (2000). The mitochondrial trifunctional protein: Centre of a beta-oxidation metabolon?Biochem. Soc. Trans.28, 177–182. 10.1042/bst0280177
46
Elek Z. KováCS Z. Keszler G. Szabó M. Csanky E. Luo J. et al (2020). High throughput multiplex SNP-analysis in chronic obstructive pulmonary disease and lung cancer. Curr. Mol. Med.20, 185–193. 10.2174/1566524019666191017123446
47
Enjoji M. Kohjima M. Ohtsu K. Matsunaga K. Murata Y. Nakamuta M. et al (2016). Intracellular mechanisms underlying lipid accumulation (white opaque substance) in gastric epithelial neoplasms: A pilot study of expression profiles of lipid-metabolism-associated genes. J. Gastroenterol. Hepatol.31, 776–781. 10.1111/jgh.13216
48
Etzioni R. Urban N. Ramsey S. Mcintosh M. Schwartz S. Reid B. et al (2003). The case for early detection. Nat. Rev. Cancer3, 243–252. 10.1038/nrc1041
49
Fan T. C. Yeo H. L. Hsu H. M. Yu J. C. Ho M. Y. Lin W. D. et al (2018). Reciprocal feedback regulation of ST3GAL1 and GFRA1 signaling in breast cancer cells. Cancer Lett.434, 184–195. 10.1016/j.canlet.2018.07.026
50
Feten G. Aastveit A. H. Snipen L. AlmøY T. (2007). A discussion concerning the inclusion of variety effect when analysis of variance is used to detect differentially expressed genes. Gene Regul. Syst. Bio.1, 117762500700100–117762500700107. 10.1177/117762500700100005
51
Filling C. Keller B. Hirschberg D. Marschall H. U. JöRNVALL H. Bennett M. J. et al (2008). Role of short-chain hydroxyacyl CoA dehydrogenases in SCHAD deficiency. Biochem. Biophys. Res. Commun.368, 6–11. 10.1016/j.bbrc.2007.10.188
52
Fitzgerald J. Higgins D. Mazo Vargas C. Watson W. Mooney C. Rahman A. et al (2021). Future of biomarker evaluation in the realm of artificial intelligence algorithms: Application in improved therapeutic stratification of patients with breast and prostate cancer. J. Clin. Pathol.74, 429–434. 10.1136/jclinpath-2020-207351
53
Flanagan S. E. Xie W. Caswell R. Damhuis A. Vianey-Saban C. Akcay T. et al (2013). Next-generation sequencing reveals deep intronic cryptic ABCC8 and HADH splicing founder mutations causing hyperinsulinism by pseudoexon activation. Am. J. Hum. Genet.92, 131–136. 10.1016/j.ajhg.2012.11.017
54
Fruman D. A. Rommel C. (2014). PI3K and cancer: Lessons, challenges and opportunities. Nat. Rev. Drug Discov.13, 140–156. 10.1038/nrd4204
55
Galván Morales M. A. Barrera RodríGUEZ R. Santiago Cruz J. R. Teran L. M. (2020). Overview of new treatments with immunotherapy for breast cancer and a proposal of a combination therapy. Molecules25, E5686. 10.3390/molecules25235686
56
Gao P. Tchernyshyov I. Chang T. C. Lee Y. S. Kita K. Ochi T. et al (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature458, 762–765. 10.1038/nature07823
57
Gao X. Pan D. (2001). TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev.15, 1383–1392. 10.1101/gad.901101
58
Gomes A. M. Soares M. V. Ribeiro P. Caldas J. PóVOA V. Martins L. R. et al (2014). Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica99, 1062–1068. 10.3324/haematol.2013.096438
59
Grivennikov S. I. Greten F. R. Karin M. (2010). Immunity, inflammation, and cancer. Cell140, 883–899. 10.1016/j.cell.2010.01.025
60
Hanahan D. Coussens L. M. (2012). Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell21, 309–322. 10.1016/j.ccr.2012.02.022
61
Hanahan D. Weinberg R. A. (2011). Hallmarks of cancer: The next generation. Cell144, 646–674. 10.1016/j.cell.2011.02.013
62
Hangauer M. J. Viswanathan V. S. Ryan M. J. Bole D. Eaton J. K. Matov A. et al (2017). Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature551, 247–250. 10.1038/nature24297
63
Hardy O. T. Hohmeier H. E. Becker T. C. Manduchi E. Doliba N. M. Gupta R. K. et al (2007). Functional genomics of the beta-cell: Short-chain 3-hydroxyacyl-coenzyme A dehydrogenase regulates insulin secretion independent of K+ currents. Mol. Endocrinol.21, 765–773. 10.1210/me.2006-0411
64
Hay N. (2005). The Akt-mTOR tango and its relevance to cancer. Cancer Cell8, 179–183. 10.1016/j.ccr.2005.08.008
65
Henley S. J. Ward E. M. Scott S. Ma J. Anderson R. N. Firth A. U. et al (2020). Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer126, 2225–2249. 10.1002/cncr.32802
66
Herr P. Basler K. (2012). Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev. Biol.361, 392–402. 10.1016/j.ydbio.2011.11.003
67
Heslegrave A. J. Hussain K. (2013). Novel insights into fatty acid oxidation, amino acid metabolism, and insulin secretion from studying patients with loss of function mutations in 3-hydroxyacyl-CoA dehydrogenase. J. Clin. Endocrinol. Metab.98, 496–501. 10.1210/jc.2012-3134
68
Heslegrave A. J. Kapoor R. R. Eaton S. Chadefaux B. Akcay T. Simsek E. et al (2012). Leucine-sensitive hyperinsulinaemic hypoglycaemia in patients with loss of function mutations in 3-Hydroxyacyl-CoA Dehydrogenase. Orphanet J. Rare Dis.7, 25. 10.1186/1750-1172-7-25
69
Hirschhaeuser F. Sattler U. G. Mueller-Klieser W. (2011). Lactate: A metabolic key player in cancer. Cancer Res.71, 6921–6925. 10.1158/0008-5472.CAN-11-1457
70
Houten S. M. Violante S. Ventura F. V. Wanders R. J. (2016). The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu. Rev. Physiol.78, 23–44. 10.1146/annurev-physiol-021115-105045
71
Houten S. M. Wanders R. J. (2010). A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis.33, 469–477. 10.1007/s10545-010-9061-2
72
Huff C. A. Matsui W. Smith B. D. Jones R. J. (2006). The paradox of response and survival in cancer therapeutics. Blood107, 431–434. 10.1182/blood-2005-06-2517
73
Ishida-Takagishi M. Enomoto A. Asai N. Ushida K. Watanabe T. Hashimoto T. et al (2012). The Dishevelled-associating protein Daple controls the non-canonical Wnt/Rac pathway and cell motility. Nat. Commun.3, 859. 10.1038/ncomms1861
74
Jiang H. Chen H. Wan P. Chen N. (2021). Decreased expression of HADH is related to poor prognosis and immune infiltration in kidney renal clear cell carcinoma. Genomics113, 3556–3564. 10.1016/j.ygeno.2021.08.008
75
Jiao Y. Wang Y. Guo S. Wang G. (2017). Glutathione peroxidases as oncotargets. Oncotarget8, 80093–80102. 10.18632/oncotarget.20278
76
Jones P. F. Jakubowicz T. Pitossi F. J. Maurer F. Hemmings B. A. (1991). Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc. Natl. Acad. Sci. U. S. A.88, 4171–4175. 10.1073/pnas.88.10.4171
77
Junaid M. Akter Y. Afrose S. S. Tania M. Khan M. A. (2021). Biological role of AKT and regulation of AKT signaling pathway by thymoquinone: Perspectives in cancer therapeutics. Mini Rev. Med. Chem.21, 288–301. 10.2174/1389557520666201005143818
78
Kadowaki T. Wilder E. Klingensmith J. Zachary K. Perrimon N. (1996). The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev.10, 3116–3128. 10.1101/gad.10.24.3116
79
Kahn M. (2014). Can we safely target the WNT pathway?Nat. Rev. Drug Discov.13, 513–532. 10.1038/nrd4233
80
Kapoor R. R. James C. Flanagan S. E. Ellard S. Eaton S. Hussain K. (2009). 3-Hydroxyacyl-coenzyme A dehydrogenase deficiency and hyperinsulinemic hypoglycemia: Characterization of a novel mutation and severe dietary protein sensitivity. J. Clin. Endocrinol. Metab.94, 2221–2225. 10.1210/jc.2009-0423
81
Kapoor R. R. Heslegrave A. Hussain K. (2010). Congenital hyperinsulinism due to mutations in HNF4A and HADH. Rev. Endocr. Metab. Disord.11, 185–191. 10.1007/s11154-010-9148-y
82
Kennedy S. G. Wagner A. J. Conzen S. D. JordáN J. Bellacosa A. Tsichlis P. N. et al (1997). The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev.11, 701–713. 10.1101/gad.11.6.701
83
Khan R. T. Siddique A. Shahid N. Khokher S. Fatima W. (2018). Breast cancer risk associated with genes encoding DNA repair MRN complex: A study from Punjab, Pakistan. Breast Cancer25, 350–355. 10.1007/s12282-018-0837-9
84
Kim Y. Kim D. H. (2015). CpG island hypermethylation as a biomarker for the early detection of lung cancer. Methods Mol. Biol.1238, 141–171. 10.1007/978-1-4939-1804-1_8
85
Knobbe C. B. Reifenberger G. (2003). Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3'-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol.13, 507–518. 10.1111/j.1750-3639.2003.tb00481.x
86
Koinis F. Kotsakis A. Georgoulias V. (2016). Small cell lung cancer (SCLC): No treatment advances in recent years. Transl. Lung Cancer Res.5, 39–50. 10.3978/j.issn.2218-6751.2016.01.03
87
Lebleu V. S. (2015). Imaging the tumor microenvironment. Cancer J.21, 174–178. 10.1097/PPO.0000000000000118
88
Li C. Chen P. Palladino A. Narayan S. Russell L. K. Sayed S. et al (2010). Mechanism of hyperinsulinism in short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency involves activation of glutamate dehydrogenase. J. Biol. Chem.285, 31806–31818. 10.1074/jbc.M110.123638
89
Li F. Dai L. Niu J. (2020). GPX2 silencing relieves epithelial-mesenchymal transition, invasion, and metastasis in pancreatic cancer by downregulating Wnt pathway. J. Cell. Physiol.235, 7780–7790. 10.1002/jcp.29391
90
Li H. Fang H. Chang L. Qiu S. Ren X. Cao L. et al (2021). TC2N: A novel vital oncogene or tumor suppressor gene in cancers. Front. Immunol.12, 764749. 10.3389/fimmu.2021.764749
91
Lin Y. Zhang Y. Chen Y. Liu Z. (2019). Promoter methylation and clinical significance of GPX3 in esophageal squamous cell carcinoma. Pathol. Res. Pract.215, 152676. 10.1016/j.prp.2019.152676
92
Liu C. He X. Wu X. Wang Z. Zuo W. Hu G. (2017). Clinicopathological and prognostic significance of GPx2 protein expression in nasopharyngeal carcinoma. Cancer Biomark.19, 335–340. 10.3233/CBM-160542
93
Liu H. W. Bi W. T. Huang H. T. Li R. X. XI Q. Feng L. et al (2018a). Satb1 promotes Schwann cell viability and migration via activation of PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci.22, 4268–4277. 10.26355/eurrev_201807_15423
94
Liu J. Eckert M. A. Harada B. T. Liu S. M. Lu Z. Yu K. et al (2018b). m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol.20, 1074–1083. 10.1038/s41556-018-0174-4
95
Malanga D. Scrima M. De Marco C. Fabiani F. De Rosa N. De Gisi S. et al (2008). Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle7, 665–669. 10.4161/cc.7.5.5485
96
Manning B. D. Cantley L. C. (2007). AKT/PKB signaling: Navigating downstream. Cell129, 1261–1274. 10.1016/j.cell.2007.06.009
97
Manning B. D. Toker A. (2017). AKT/PKB signaling: Navigating the network. Cell169, 381–405. 10.1016/j.cell.2017.04.001
98
Mantovani A. Allavena P. Sica A. Balkwill F. (2008). Cancer-related inflammation. Nature454, 436–444. 10.1038/nature07205
99
Mao J. Fan S. Ma W. Fan P. Wang B. Zhang J. et al (2014). Roles of Wnt/β-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis.5, e1039. 10.1038/cddis.2013.515
100
Margis R. Dunand C. Teixeira F. K. Margis-Pinheiro M. (2008). Glutathione peroxidase family - an evolutionary overview. Febs J.275, 3959–3970. 10.1111/j.1742-4658.2008.06542.x
101
Martens G. A. Vervoort A. Van De Casteele M. Stangé G. Hellemans K. Van Thi H. V. et al (2007). Specificity in beta cell expression of L-3-hydroxyacyl-CoA dehydrogenase, short chain, and potential role in down-regulating insulin release. J. Biol. Chem.282, 21134–21144. 10.1074/jbc.M700083200
102
Mayo L. D. Donner D. B. (2001). A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U. S. A.98, 11598–11603. 10.1073/pnas.181181198
103
Meier R. Hemmings B. A. (1999). Regulation of protein kinase B. J. Recept. Signal Transduct. Res.19, 121–128. 10.3109/10799899909036639
104
Meng F. Henson R. Wehbe-Janek H. Ghoshal K. Jacob S. T. Patel T. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology133, 647–658. 10.1053/j.gastro.2007.05.022
105
Metere A. Frezzotti F. Graves C. E. Vergine M. De Luca A. Pietraforte D. et al (2018). A possible role for selenoprotein glutathione peroxidase (GPx1) and thioredoxin reductases (TrxR1) in thyroid cancer: Our experience in thyroid surgery. Cancer Cell Int.18, 7. 10.1186/s12935-018-0504-4
106
Miller K. D. Siegel R. L. Lin C. C. Mariotto A. B. Kramer J. L. Rowland J. H. et al (2016). Cancer treatment and survivorship statistics, 2016. Ca. Cancer J. Clin.66, 271–289. 10.3322/caac.21349
107
Mohamedali A. Lea N. C. Feakins R. M. Raj K. Mufti G. J. Kocher H. M. (2008). AKT1 (E17K) mutation in pancreatic cancer. Technol. Cancer Res. Treat.7, 407–408. 10.1177/153303460800700509
108
Morin P. J. Sparks A. B. Korinek V. Barker N. Clevers H. Vogelstein B. et al (1997). Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science275, 1787–1790. 10.1126/science.275.5307.1787
109
Mundi P. S. Sachdev J. Mccourt C. Kalinsky K. (2016). AKT in cancer: New molecular insights and advances in drug development. Br. J. Clin. Pharmacol.82, 943–956. 10.1111/bcp.13021
110
Naiki T. Naiki-Ito A. Iida K. Etani T. Kato H. Suzuki S. et al (2018). GPX2 promotes development of bladder cancer with squamous cell differentiation through the control of apoptosis. Oncotarget9, 15847–15859. 10.18632/oncotarget.24627
111
Nakashima L. (2018). Evolution of cancer treatment and evolving challenges. Healthc. Manage. Forum31, 26–28. 10.1177/0840470417722568
112
Nakatani K. Thompson D. A. Barthel A. Sakaue H. Liu W. Weigel R. J. et al (1999). Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J. Biol. Chem.274, 21528–21532. 10.1074/jbc.274.31.21528
113
Nalkiran I. Turan S. Arikan S. Kahraman Ö T. Acar L. Yaylim I. et al (2015). Determination of gene expression and serum levels of MnSOD and GPX1 in colorectal cancer. Anticancer Res.35, 255–259.
114
Nieman K. M. Kenny H. A. Penicka C. V. Ladanyi A. Buell-Gutbrod R. Zillhardt M. R. et al (2011). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med.17, 1498–1503. 10.1038/nm.2492
115
Nishita M. Enomoto M. Yamagata K. Minami Y. (2010). Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol.20, 346–354. 10.1016/j.tcb.2010.03.001
116
Nwosu Z. C. Battello N. Rothley M. Piorońska W. Sitek B. Ebert M. P. et al (2018). Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J. Exp. Clin. Cancer Res.37, 211. 10.1186/s13046-018-0872-6
117
Ozes O. N. Mayo L. D. Gustin J. A. Pfeffer S. R. Pfeffer L. M. Donner D. B. (1999). NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature401, 82–85. 10.1038/43466
118
Parsons D. W. Wang T. L. Samuels Y. Bardelli A. Cummins J. M. Delong L. et al (2005). Colorectal cancer: Mutations in a signalling pathway. Nature436, 792. 10.1038/436792a
119
Peng D. Hu T. Soutto M. Belkhiri A. Zaika A. El-Rifai W. (2014). Glutathione peroxidase 7 has potential tumour suppressor functions that are silenced by location-specific methylation in oesophageal adenocarcinoma. Gut63, 540–551. 10.1136/gutjnl-2013-304612
120
Pepin E. Guay C. Delghingaro-Augusto V. Joly E. Madiraju S. R. Prentki M. (2010). Short-chain 3-hydroxyacyl-CoA dehydrogenase is a negative regulator of insulin secretion in response to fuel and non-fuel stimuli in INS832/13 β-cells. J. Diabetes2, 157–167. 10.1111/j.1753-0407.2010.00076.x
121
Popa F. I. Perlini S. Teofoli F. Degani D. Funghini S. La Marca G. et al (2012). 3-hydroxyacyl-coenzyme a dehydrogenase deficiency: Identification of a new mutation causing hyperinsulinemic hypoketotic hypoglycemia, altered organic acids and acylcarnitines concentrations. JIMD Rep.2, 71–77. 10.1007/8904_2011_50
122
Ren J. Feng J. Song W. Wang C. Ge Y. Fu T. (2020). Development and validation of a metabolic gene signature for predicting overall survival in patients with colon cancer. Clin. Exp. Med.20, 535–544. 10.1007/s10238-020-00652-1
123
Revathidevi S. Munirajan A. K. (2019). Akt in cancer: Mediator and more. Semin. Cancer Biol.59, 80–91. 10.1016/j.semcancer.2019.06.002
124
Roelink H. Wagenaar E. Nusse R. (1992). Amplification and proviral activation of several Wnt genes during progression and clonal variation of mouse mammary tumors. Oncogene7, 487–492.
125
Roy H. K. Olusola B. F. Clemens D. L. Karolski W. J. Ratashak A. Lynch H. T. et al (2002). AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis23, 201–205. 10.1093/carcin/23.1.201
126
Samuels Y. Wang Z. Bardelli A. Silliman N. Ptak J. Szabo S. et al (2004). High frequency of mutations of the PIK3CA gene in human cancers. Science304, 554. 10.1126/science.1096502
127
Sanchez D. J. Simon M. C. (2018). Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim. Biophys. Acta. Rev. Cancer1870, 23–31. 10.1016/j.bbcan.2018.06.003
128
Sanidas I. Polytarchou C. Hatziapostolou M. Ezell S. A. Kottakis F. Hu L. et al (2014). Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol. Cell53, 577–590. 10.1016/j.molcel.2013.12.018
129
Satapathy A. K. Jain V. Ellard S. Flanagan S. E. (2016). Hyperinsulinemic hypoglycemia of infancy due to novel HADH mutation in two siblings. Indian Pediatr.53, 912–913. 10.1007/s13312-016-0958-1
130
Schulz N. Himmelbauer H. Rath M. Van Weeghel M. Houten S. Kulik W. et al (2011). Role of medium- and short-chain L-3-hydroxyacyl-CoA dehydrogenase in the regulation of body weight and thermogenesis. Endocrinology152, 4641–4651. 10.1210/en.2011-1547
131
Shareefi G. Turkistani A. N. Alsayyah A. Kussaibi H. Abdel Hadi M. Alkharsah K. R. (2020). Pathway-affecting single nucleotide polymorphisms (SNPs) in RPS6KA1 and MBIP genes are associated with breast cancer risk. Asian pac. J. Cancer Prev.21, 2163–2168. 10.31557/APJCP.2020.21.7.2163
132
Shen C. Song Y. H. Xie Y. Wang X. Wang Y. Wang C. et al (2017). Downregulation of HADH promotes gastric cancer progression via Akt signaling pathway. Oncotarget8, 76279–76289. 10.18632/oncotarget.19348
133
Shoji K. Oda K. Nakagawa S. Hosokawa S. Nagae G. Uehara Y. et al (2009). The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br. J. Cancer101, 145–148. 10.1038/sj.bjc.6605109
134
Siegel R. L. Miller K. D. Fuchs H. E. Jemal A. (2022). Cancer statistics, 2022. Ca. Cancer J. Clin.72, 7–33. 10.3322/caac.21708
135
Smith J. S. Lindsay L. Hoots B. Keys J. Franceschi S. Winer R. et al (2007). Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: A meta-analysis update. Int. J. Cancer121, 621–632. 10.1002/ijc.22527
136
Sousa C. M. Biancur D. E. Wang X. Halbrook C. J. Sherman M. H. Zhang L. et al (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature536, 479–483. 10.1038/nature19084
137
Spill F. Reynolds D. S. Kamm R. D. Zaman M. H. (2016). Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol.40, 41–48. 10.1016/j.copbio.2016.02.007
138
Staal S. P. (1987). Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: Amplification of AKT1 in a primary human gastric adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A.84, 5034–5037. 10.1073/pnas.84.14.5034
139
Stambolic V. Suzuki A. De La Pompa J. L. Brothers G. M. Mirtsos C. Sasaki T. et al (1998). Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell95, 29–39. 10.1016/s0092-8674(00)81780-8
140
Sun M. Wang G. Paciga J. E. Feldman R. I. Yuan Z. Q. Ma X. L. et al (2001). AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am. J. Pathol.159, 431–437. 10.1016/s0002-9440(10)61714-2
141
Sun Z. Tao W. Guo X. Jing C. Zhang M. Wang Z. et al (2022). Construction of a lactate-related prognostic signature for predicting prognosis, tumor microenvironment, and immune response in kidney renal clear cell carcinoma. Front. Immunol.13, 818984. 10.3389/fimmu.2022.818984
142
Takebe G. Yarimizu J. Saito Y. Hayashi T. Nakamura H. Yodoi J. et al (2002). A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J. Biol. Chem.277, 41254–41258. 10.1074/jbc.M202773200
143
Tanaka M. Masaki Y. Tanaka K. Miyazaki M. Kato M. Sugimoto R. et al (2013). Reduction of fatty acid oxidation and responses to hypoxia correlate with the progression of de-differentiation in HCC. Mol. Med. Rep.7, 365–370. 10.3892/mmr.2012.1201
144
Tsai Y. T. Kuo P. H. Kuo H. P. Hsu C. Y. Lee Y. J. Kuo C. L. et al (2021). Ganoderma tsugae suppresses the proliferation of endometrial carcinoma cells via Akt signaling pathway. Environ. Toxicol.36, 320–327. 10.1002/tox.23037
145
Van't Veer L. J. Van Kessel A. G. Van Heerikhuizen H. Van Ooyen A. Nusse R. vAn Ooyen A. (1984). Molecular cloning and chromosomal assignment of the human homolog of int-1, a mouse gene implicated in mammary tumorigenesis. Mol. Cell. Biol.4, 2532–2534. 10.1128/mcb.4.11.2532
146
Vijayvergia N. Shah P. C. Denlinger C. S. (2015). Survivorship in non-small cell lung cancer: Challenges faced and steps forward. J. Natl. Compr. Canc. Netw.13, 1151–1161. 10.6004/jnccn.2015.0140
147
Viswanathan V. S. Ryan M. J. Dhruv H. D. Gill S. Eichhoff O. M. Seashore-Ludlow B. et al (2017). Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature547, 453–457. 10.1038/nature23007
148
Vitale I. Manic G. Coussens L. M. Kroemer G. Galluzzi L. (2019a). Macrophages and metabolism in the tumor microenvironment. Cell Metab.30, 36–50. 10.1016/j.cmet.2019.06.001
149
Vitale I. Manic G. Galassi C. Galluzzi L. (2019b). Stress responses in stromal cells and tumor homeostasis. Pharmacol. Ther.200, 55–68. 10.1016/j.pharmthera.2019.04.004
150
Vivanco I. Sawyers C. L. (2002). The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer2, 489–501. 10.1038/nrc839
151
Voloshanenko O. Schwartz U. Kranz D. Rauscher B. Linnebacher M. Augustin I. et al (2018). β-catenin-independent regulation of Wnt target genes by RoR2 and ATF2/ATF4 in colon cancer cells. Sci. Rep.8, 3178. 10.1038/s41598-018-20641-5
152
Vredendaal P. J. Van Den Berg I. E. Malingré H. E. Stroobants A. K. Olde Weghuis D. E. Berger R. (1996). Human short-chain L-3-hydroxyacyl-CoA dehydrogenase: Cloning and characterization of the coding sequence. Biochem. Biophys. Res. Commun.223, 718–723. 10.1006/bbrc.1996.0961
153
Vredendaal P. J. Van Den Berg I. E. Stroobants A. K. Van Der A. D. Malingré H. E. Berger R. (1998). Structural organization of the human short-chain L-3-hydroxyacyl-CoA dehydrogenase gene. Mamm. Genome9, 763–768. 10.1007/s003359900860
154
Vysotskaia V. Kaseniit K. E. Bucheit L. Ready K. Price K. Johansen Taber K. (2020). Clinical utility of hereditary cancer panel testing: Impact of PALB2, ATM, CHEK2, NBN, BRIP1, RAD51C, and RAD51D results on patient management and adherence to provider recommendations. Cancer126, 549–558. 10.1002/cncr.32572
155
Wanders R. J. Vreken P. Den Boer M. E. Wijburg F. A. Van Gennip A. H. Ijlst L. (1999). Disorders of mitochondrial fatty acyl-CoA beta-oxidation. J. Inherit. Metab. Dis.22, 442–487. 10.1023/a:1005504223140
156
Wang J. J. Lei K. F. Han F. (2018). Tumor microenvironment: Recent advances in various cancer treatments. Eur. Rev. Med. Pharmacol. Sci.22, 3855–3864. 10.26355/eurrev_201806_15270
157
Wang Y. Cao P. Alshwmi M. Jiang N. Xiao Z. Jiang F. et al (2019). GPX2 suppression of H(2)O(2) stress regulates cervical cancer metastasis and apoptosis via activation of the β-catenin-WNT pathway. Onco. Targets. Ther.12, 6639–6651. 10.2147/OTT.S208781
158
Ward P. S. Thompson C. B. (2012). Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell21, 297–308. 10.1016/j.ccr.2012.02.014
159
Ward S. G. Westwick J. Harris S. (2011). Sat-Nav for T cells: Role of PI3K isoforms and lipid phosphatases in migration of T lymphocytes. Immunol. Lett.138, 15–18. 10.1016/j.imlet.2011.02.007
160
Wei J. Xie Q. Liu X. Wan C. Wu W. Fang K. et al (2020). Identification the prognostic value of glutathione peroxidases expression levels in acute myeloid leukemia. Ann. Transl. Med.8, 678. 10.21037/atm-20-3296
161
Wen J. Liu D. Chen D. Chen J. Xu X. Chen C. et al (2019). Treatment of clinical T4 stage superior sulcus non-small cell lung cancer: A propensity-matched analysis of the surveillance, epidemiology, and end results database. Biosci. Rep.39, BSR20181545. 10.1042/BSR20181545
162
Wettersten H. I. Aboud O. A. Lara P. N. JR Weiss R. H. (2017). Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol.13, 410–419. 10.1038/nrneph.2017.59
163
Wilkins O. M. Titus A. J. Gui J. Eliot M. Butler R. A. Sturgis E. M. et al (2017). Genome-scale identification of microRNA-related SNPs associated with risk of head and neck squamous cell carcinoma. Carcinogenesis38, 986–993. 10.1093/carcin/bgx056
164
Xiong Z. Wu J. Sun Y. Bai M. Niu F. Jin T. (2020). Variants in multiple genes are associated with esophageal cancer risk in a Chinese han population: A case-control study. J. Gene Med.22, e3266. 10.1002/jgm.3266
165
Xu X. Sakon M. Nagano H. Hiraoka N. Yamamoto H. Hayashi N. et al (2004). Akt2 expression correlates with prognosis of human hepatocellular carcinoma. Oncol. Rep.11, 25–32. 10.3892/or.11.1.25
166
Yang S. Y. He X. Y. Schulz H. (2005). 3-Hydroxyacyl-CoA dehydrogenase and short chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. Febs J.272, 4874–4883. 10.1111/j.1742-4658.2005.04911.x
167
Yang W. S. Sriramaratnam R. Welsch M. E. Shimada K. Skouta R. Viswanathan V. S. et al (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell156, 317–331. 10.1016/j.cell.2013.12.010
168
Yecies J. L. Zhang H. H. Menon S. Liu S. Yecies D. Lipovsky A. I. et al (2011). Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab.14, 21–32. 10.1016/j.cmet.2011.06.002
169
Yi Z. Jiang L. Zhao L. Zhou M. Ni Y. Yang Y. et al (2019). Glutathione peroxidase 3 (GPX3) suppresses the growth of melanoma cells through reactive oxygen species (ROS)-dependent stabilization of hypoxia-inducible factor 1-α and 2-α. J. Cell. Biochem.120, 19124–19136. 10.1002/jcb.29240
170
Yin F. Sharen G. Yuan F. Peng Y. Chen R. Zhou X. et al (2017). TIP30 regulates lipid metabolism in hepatocellular carcinoma by regulating SREBP1 through the Akt/mTOR signaling pathway. Oncogenesis6, e347. 10.1038/oncsis.2017.49
171
Zhan T. Rindtorff N. Boutros M. (2017). Wnt signaling in cancer. Oncogene36, 1461–1473. 10.1038/onc.2016.304
172
Zhang M. Di Martino J. S. Bowman R. L. Campbell N. R. Baksh S. C. Simon-Vermot T. et al (2018). Adipocyte-derived lipids mediate melanoma progression via FATP proteins. Cancer Discov.8, 1006–1025. 10.1158/2159-8290.CD-17-1371
173
Zhang B. Wu Q. Wang Z. Xu R. Hu X. Sun Y. et al (2019). The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: Evidence from bioinformatics analysis of high-throughput data. Mol. Genet. Genomic Med.7, e607. 10.1002/mgg3.607
174
Zhang Y. Zhang X. (2018). Controlling nutritional status score, a promising prognostic marker in patients with gastrointestinal cancers after surgery: A systematic review and meta-analysis. Int. J. Surg.55, 39–45. 10.1016/j.ijsu.2018.05.018
175
Zheng X. Li X. Wang X. (2020). Extracellular vesicle-based liquid biopsy holds great promise for the management of ovarian cancer. Biochim. Biophys. Acta. Rev. Cancer1874, 188395. 10.1016/j.bbcan.2020.188395
176
Zhou B. P. Liao Y. Xia W. Spohn B. Lee M. H. Hung M. C. (2001). Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol.3, 245–252. 10.1038/35060032
177
Zhou C. Hu H. Zheng Z. Chen C. Li Y. Li B. et al (2019a). Association between GPX3 promoter methylation and malignant tumors: A meta-analysis. Pathol. Res. Pract.215, 152443. 10.1016/j.prp.2019.152443
178
Zhou C. Pan R. Li B. Huang T. Zhao J. Ying J. et al (2019b). GPX3 hypermethylation in gastric cancer and its prognostic value in patients aged over 60. Future Oncol.15, 1279–1289. 10.2217/fon-2018-0674
179
Zhu X. Wang J. Li L. Deng L. Wang J. Liu L. et al (2018). GPX3 suppresses tumor migration and invasion via the FAK/AKT pathway in esophageal squamous cell carcinoma. Am. J. Transl. Res.10, 1908–1920.
180
Zilberman D. E. Cohen Y. Amariglio N. Fridman E. Ramon J. Rechavi G. (2009). AKT1 E17 K pleckstrin homology domain mutation in urothelial carcinoma. Cancer Genet. cytogenet.191, 34–37. 10.1016/j.cancergencyto.2009.01.009
Summary
Keywords
HADH, oncogene, tumor suppressor gene, differential expression, tumor microenvironment, tumor-infiltrating immune cells
Citation
Fang H, Li H, Zhang H, Wang S, Xu S, Chang L, Yang Y and Cui R (2022) Short-chain L-3-hydroxyacyl-CoA dehydrogenase: A novel vital oncogene or tumor suppressor gene in cancers. Front. Pharmacol. 13:1019312. doi: 10.3389/fphar.2022.1019312
Received
15 August 2022
Accepted
23 September 2022
Published
14 October 2022
Volume
13 - 2022
Edited by
Ting Wang, Sichuan Cancer Hospital, China
Reviewed by
Shiv K. Sah-Teli, University of Oulu, Finland
Jinhua Zhou, The First Affiliated Hospital of Soochow University, China
Updates
Copyright
© 2022 Fang, Li, Zhang, Wang, Xu, Chang, Yang and Cui.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongsheng Yang, yyswxt@126.com; Ranji Cui, cuiranji@jlu.edu.cn
This article was submitted to Pharmacology of Anti-Cancer Drugs, a section of the journal Frontiers in Pharmacology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.