- 1College of Pharmacy, Sunchon National University, Suncheon, South Korea
- 2College of Pharmacy, Chungbuk National University, Cheongjusi, South Korea
- 3College of Pharmacy and Graduate School of Pharmaceutical Sciences, Ewha Womans University, Seoul, South Korea
- 4College of Pharmacy, Gachon University, Incheon, South Korea
Pharmacogenomics, which is defined as the study of changes in the properties of DNA and RNA associated with drug response, enables the prediction of the efficacy and adverse effects of drugs based on patients’ specific genetic mutations. For the safe and effective use of drugs, it is important that pharmacogenomic information is easily accessible to clinical experts and patients. Therefore, we examined the pharmacogenomic information provided on drug labels in Korea, Europe, Japan, and the United States (US). The selection of drugs that include pharmacogenomic information was based on the drug list that includes genetic information from the Korea Ministry of Food and Drug Safety (MFDS) and US Food and Drug Administration (FDA) websites. Drug labels were retrieved from the sites of MFDS, FDA, European Medicines Agency, and Japanese Pharmaceuticals and Medical Devices Agency. Drugs were classified as per the Anatomical Therapeutic Chemical code, and the biomarkers, labeling sections, and necessity of genetic tests were determined. In total, 348 drugs were selected from 380 drugs with available pharmacogenomic information in Korea and the US after applying the inclusion and exclusion criteria. Of these drugs, 137, 324, 169, and 126 were with pharmacogenomics information in Korea, the US, Europe, and Japan, respectively. The most commonly represented drug class was antineoplastic and immunomodulating agents. Regarding the classification as per the mentioned biomarkers, the cytochrome P450 enzyme was the most frequently mentioned information, and the targeted anticancer drugs most commonly required genetic biomarker testing. The reasons for differences in drug labeling information based on country include differences in mutant alleles according to ethnicity, frequencies at which drug lists are updated, and pharmacogenomics-related guidelines. Clinical experts must continuously strive to identify and report mutations that can explain drug efficacy or side effects for safe drug use.
Introduction
Pharmacogenomics describes the variability of drug response according to a patient’s genetic information. Pharmacogenomics can facilitate the development of safe and effective drug therapy for individual patients by enabling appropriate drug selection and dose adjustment (Relling and Evans, 2015). Pharmacogenomics has developed rapidly over the past 20 years, consequently promoting the realization of “personalized medicine” in healthcare (Roncato et al., 2021).
Several initiatives have sought to apply pharmacogenomics to clinical practice. The Clinical Pharmacogenetics Implementation Consortium in the United States (US) and the Dutch Pharmacogenetics Working Group in Europe are the leading scientific consortia in the field of pharmacogenomics, and they are responsible for creating guidelines for the clinical application of genetic test results (Relling and Klein, 2011; Swen et al., 2011; Cecchin et al., 2017). Pharmacogenomic information is currently reflected in drug labels developed by regulatory agencies, and these data are used as biomarkers for drug treatment (FDA, 2021a; PMDA, 2021). Regulatory agencies in various countries, including the US and Europe, have supported the reflection of pharmacogenomic information via the publication of related guidance (Mehta et al., 2020). The European Medicines Agency has defined pharmacogenomics terms in “Position paper on terminology in pharmacogenetics” since 2002, and the US Food and Drug Administration (FDA) has defined pharmacogenomics and related terms in “Guidance for industry pharmacogenomic data submissions” since 2005. Guidelines have been prepared for subsequent application in clinical trials. In Japan, a guideline for collecting pharmacogenomic information has been published since 2005 (Ishiguro et al., 2008), and in Korea, a guideline for evaluating the eligibility of pharmacogenomics and applying the data to clinical trials was published in 2015 (MFDS, 2015a; MFDS, 2015b). Health Canada released the “Guidance document on submission of pharmacogenomic information” in 2007 as a guideline for providing pharmacogenomic data required for drug approval (Joly and Ramos-Paque, 2010).
The drug label reflection of pharmacogenomic information has made it easier to access the relevant information for application in clinical practice (Yoon et al., 2020). Studies examining whether drug labels reduce accidents related to adverse drug reactions and affect treatment outcomes are ongoing (Vredenburgh and Zackowitz, 2009; Wolf et al., 2016), and efforts to include pharmacogenomic information on drug labels are continuing (Kircik et al., 2017; Shekhani et al., 2020). In the field of oncology in particular, genetic testing for somatic mutation is already mandatory, and the relevant information is provided on the drug label. Meanwhile, clinical trial data are being added to drug labels for adverse drug reactions according to the genotype (Mehta et al., 2020). Drug labels reflecting pharmacogenomic information will facilitate the implementation of more appropriate and safer drug therapies in clinical settings (Yoon et al., 2020).
As each country has a different method for providing pharmacogenomic information and the standards indicated on the drug label are different, the content and inclusion of information may affect the safe use of drugs by patients. Therefore, by comparing pharmacogenomic information recorded by the US FDA, Korean Ministry of Food and Drug Safety (MFDS), European Medicines Agency, and Japanese Pharmaceuticals and Medical Devices Agency, we investigated the provision of pharmacogenomic information in each country or region as well as the information on drug labels according to the differences in guidelines.
Materials and methods
Data sources
To formulate the drug list for confirming the genomics-related drug label as per country, data from Korea and the US, which publish drug lists that include pharmacogenomic information on drug labels on regulatory agency websites, were used. Drug selection was based on the active ingredient by matching the Korean genetic information list uploaded on the Korean MFDS website and the “Table of Pharmacogenomic Biomarkers in Drug Labeling” drug list uploaded on the US FDA website as of December 2021 (MFDS, 2021a; FDA, 2022). Based on the drug list that included pharmacogenomics information, we reviewed whether the pharmacogenomics information of the drugs was included on drug labels formulated by ‘MFDS, FDA, European Medicines Agency’s Summary of Product Characteristics, and Japanese Pharmaceuticals and Medical Devices Agency (EMA, 2021; PMDA, 2021).
Drug labels were searched using by drug ingredient names via drug regulatory authorities in each country to extract information on Anatomical Therapeutic Chemical (ATC) codes, drug-related biomarkers, labeling sections, and the relevant pharmacogenomic contents.
Based on the drug lists of MFDS and FDA, the criteria for selecting drugs that included pharmacogenomic information in the relevant drug approvals in Korea, Europe, Japan, and the US were at least one of the categories as follows.
1) The drug was indicated only for patients with a specific genotype;
2) It was necessary to adjust the dose of the drug according to the genotype;
3) A genetic test for a specific biomarker was required before administration; and
4) Information such as contraindications or cautions regarding drug efficacy or safety was mentioned.
By contrast, drugs for which only genetic information in relation to excipients such as lactose intolerance were provided as well as those for which only the genotype was mentioned but dose adjustment was not required to improve efficacy or safety were excluded.
Data analysis
To unify and classify the drug groups that were derived from each country based on the finally selected drug list, the groups were classified according to the ATC code using the World Health Organization/ATC defined daily dose index tool (WHOCC, 2021).
To evaluate the pharmacogenomic information on drug labels by country, biomarkers mentioned on drug labels in Korea, the US, Europe, and Japan were analyzed, and their number and properties were determined. For antineoplastic drugs, in particular, genetic testing for biomarkers is often essential; thus, the biomarkers and indications of the drug were confirmed to review the characteristics of drugs that require genetic testing.
There was a difference in drug label items among countries and regions, making it necessary to unify the approved items for comparison. The unification criteria for the permitted items were reclassified as follows.
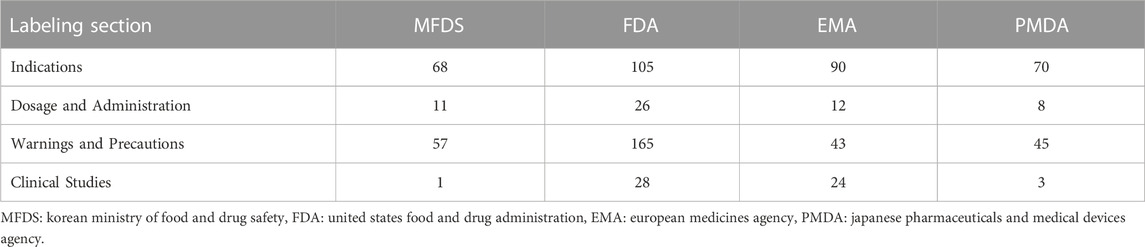
1) Indications: contents for selecting appropriate patients who require genetic testing from pharmacogenomic information
2) Dosage and Administration; contents corresponding to dosing recommendations or dosage adjustments in the subgroups of patients according to genotype
3) Warnings and Precautions; contents of pharmacokinetic data, warnings, and precautions that affect the safety of the drug
4) Clinical Studies: pharmacogenomic information representing the clinical trial results for experts
Results
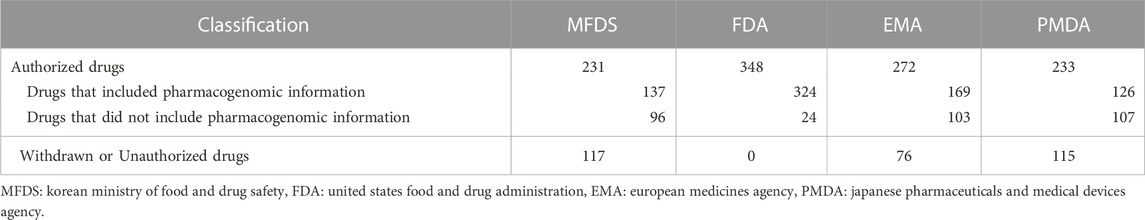
Among the 380 drugs for which pharmacogenomic information was available in Korea and the US, 348 drugs were selected after excluding drugs with 2 overlapping ingredients; excipients; and drugs that are currently withdrawn or unauthorized in Korea, the US, Europe, or Japan (Figure 1).
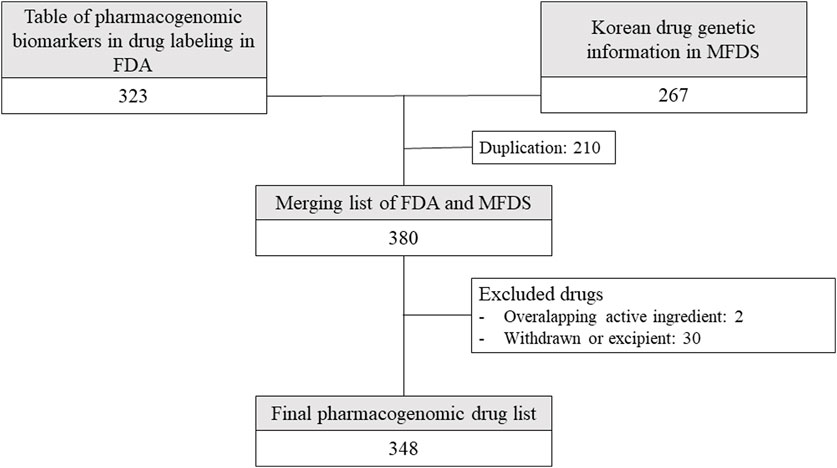
FIGURE 1. Drugs included in the analysis from drug lists provided by Korean and American regulators. FDA: United States Food and Drug Administration, MFDS: Korean Ministry of Food and Drug Safety.
Using the final drug list, the number of drugs for which drug genomic information was available was confirmed for each country. The proportion of drugs with available pharmacogenomic information was 59.1% (137 items) in Korea, 93.1% (324 items) in the US, 62.9% (171 items) in Europe, and 54.1% (126 items) in Japan (Table 1). The comparison of genetic information according to the drug list in each country is presented in Supplementary Table S1. The contents of drug labeling by each country are also provided (Supplementary Table S2–Supplementary Table S5).
Using ATC codes, the drugs were classified into the following 12 drug groups: antineoplastic and immunomodulating drugs, nervous system drugs, alimentary tract and metabolism drugs, anti-infectives for systemic use, blood and blood-forming organ drugs, cardiovascular system drugs, respiratory system drugs, genitourinary system and sex hormone drugs, musculoskeletal system drugs. various, antiparasitic products, insecticides and repellents, and dermatologicals. Antineoplastic and immunomodulating agents accounted for the largest proportion of drugs in all four countries [Korea, 51.8% (71 items); US, 37.6% (121 items); Europe, 53.3% (90 items); and Japan, 57.1% (72 items)], whereas drugs acting on the nervous system accounted for the second largest proportion (Figure 2).
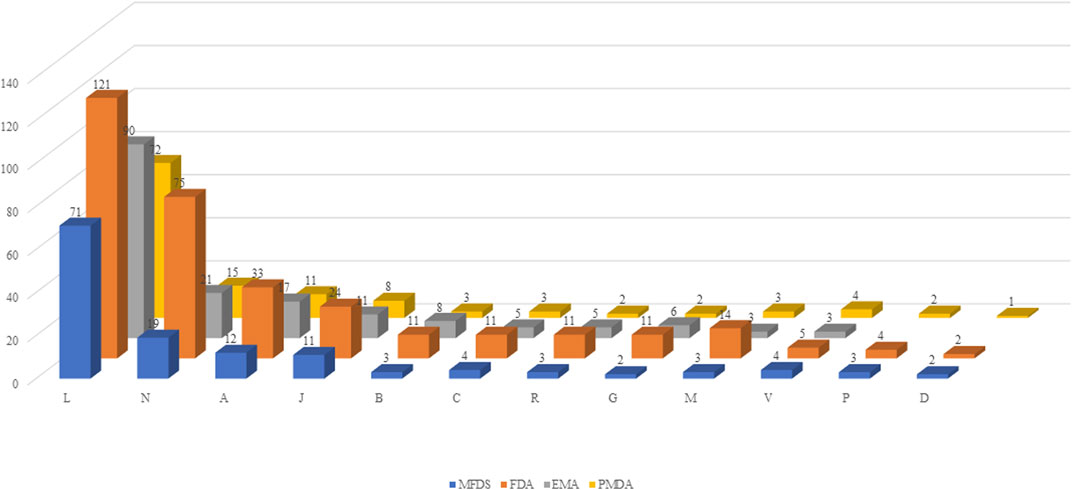
FIGURE 2. Classification by drug group. MFDS: Korean Ministry of Food and Drug Safety, FDA: United States Food and Drug Administration, EMA: European Medicines Agency, PMDA: Japanese Pharmaceuticals and Medical Devices Agency. A: Alimentary tract and metabolism, B-: Blood and blood forming organs, C: Cardiovascular system, D: Dermatologicals, G: Genitourinary system and sex hormones, J: Anti-infectives for systemic use, L: Antineoplastic and immunomodulating agents, M: Musculo-skeletal system, N: Nervous system, P: Antiparasitic products, insecticides and repellents, R: Respiratory system, S: Sensory organs, V: Various.
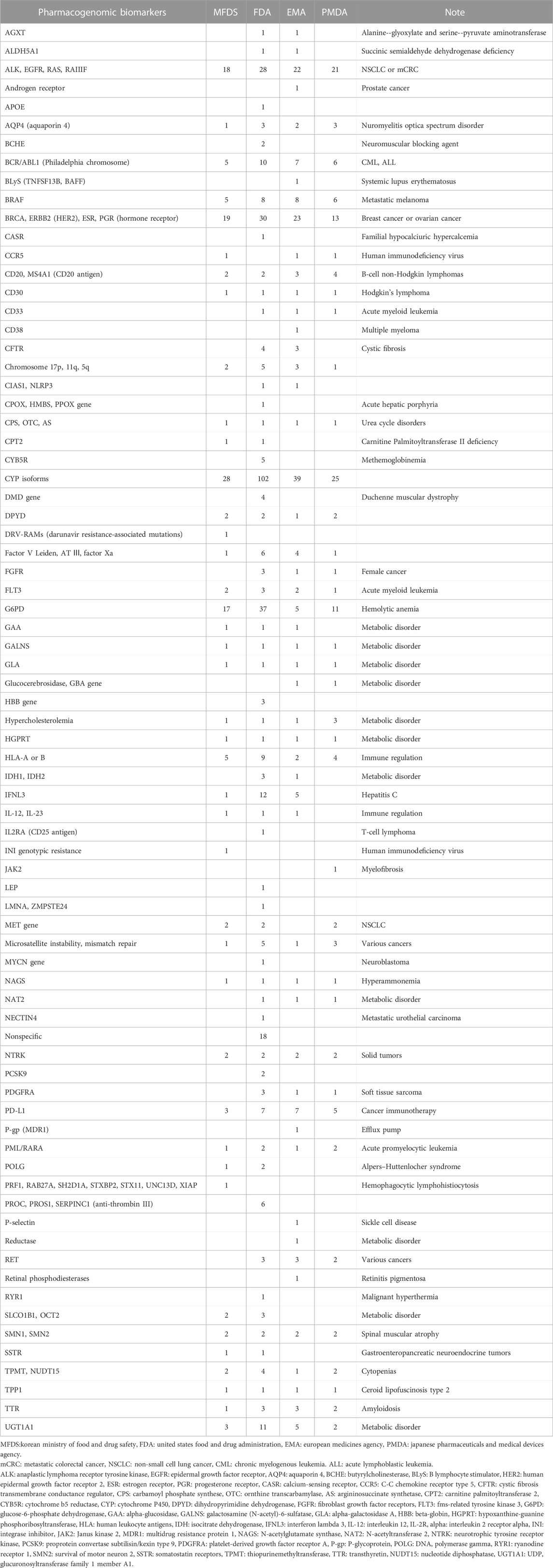
The proportions were confirmed by evaluating the biomarkers mentioned on the drug label. Regarding biomarkers, the most frequently mentioned pharmacogenomic information was the cytochrome P450 enzyme; it was mentioned for 102, 28, 39, and 25 times of the analyzed drugs in the US, Korea, Europe, and Japan, respectively. In the US, glucose-6-phosphate dehydrogenase was the second most common biomarker, accounting for 11.2% of drugs. In the remaining countries, biomarkers related to female cancer and non-small cell lung cancer were the second most frequently mentioned. Concerning the types of biomarkers mentioned, the US cited 63 species, followed by Europe (52 species), Japan (38 species), and Korea (40 species, Table 2).
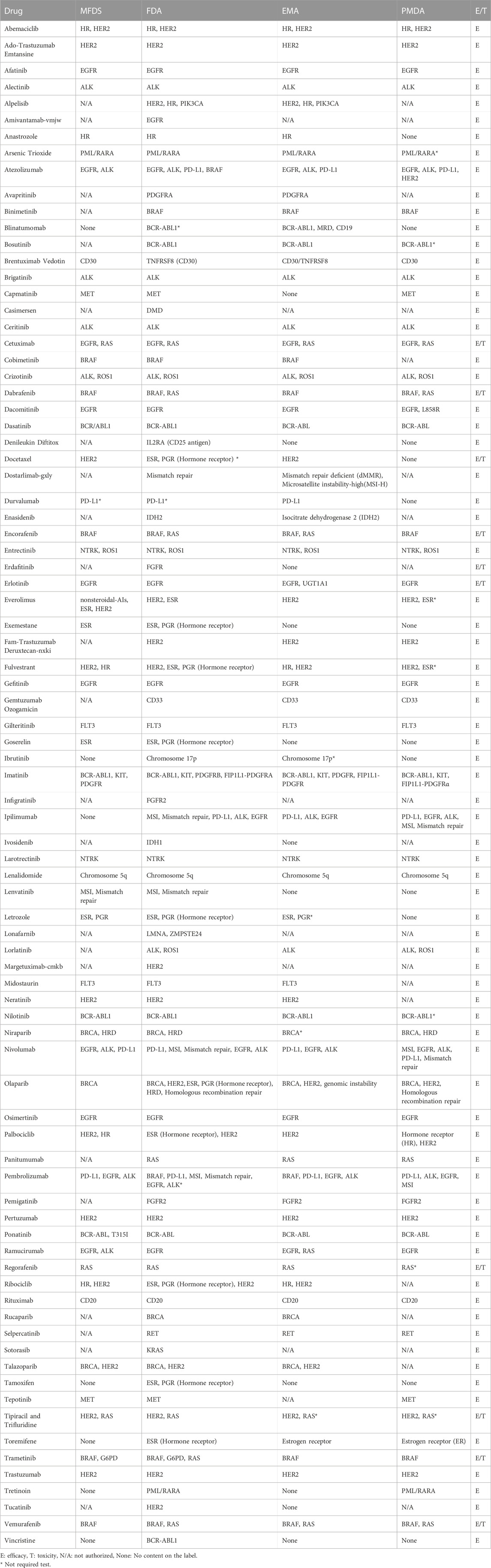
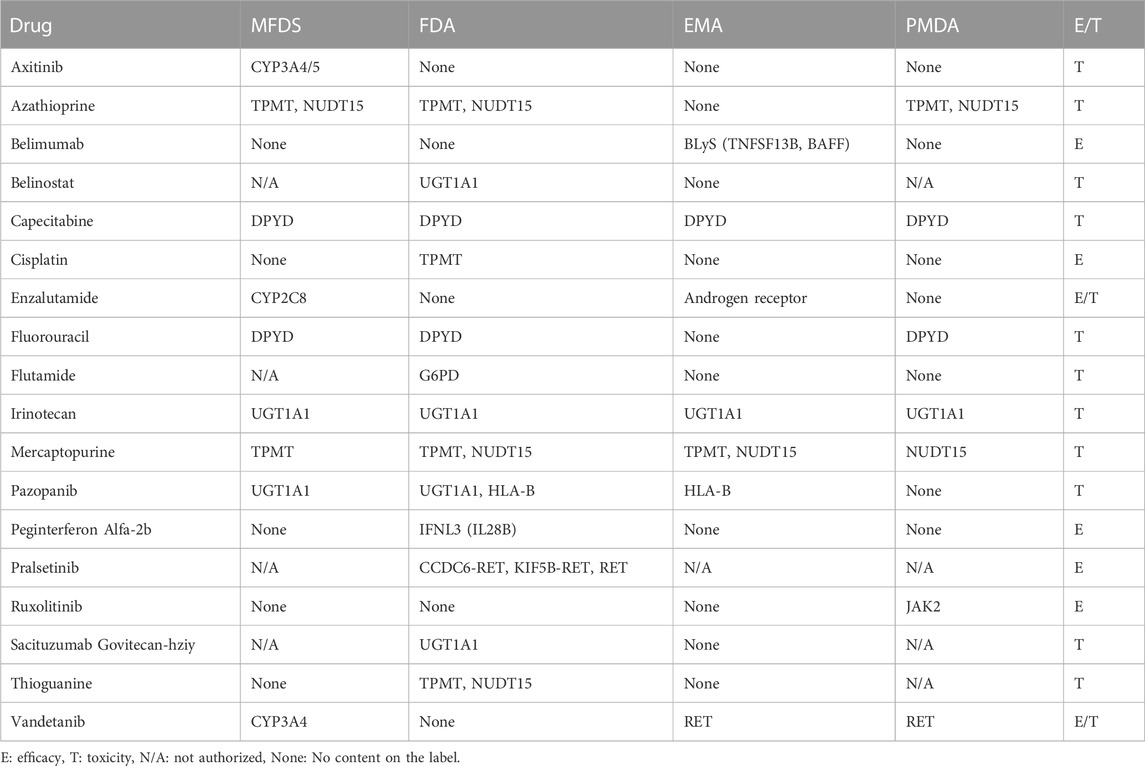
Several antineoplastic drugs were divided into required and recommended groups according to whether genetic testing for biomarkers was placed in the “indication and usage” and “dosage and administration” sections or the “precautions for use” section on the drug label. We found that 92 drugs were classified with testing required and 31 drugs were testing recommendation. (Table 3; Table 4). Most drugs requiring genetic testing related to targeted anticancer agents, but biomarkers related to metabolism were recommended for assessment only in some patients in whom genetic testing was not essential.
By checking drug label items that included pharmacogenomic information, we found that in Korea, pharmacogenomic information was primarily included for indications (49.6%) and warnings and precautions (41.6%), whereas in the US, pharmacogenomic information was most commonly included for warnings and precautions (50.9%). In Europe (53.3%) and Japan (55.6%), pharmacogenomic information was most frequently included for indications (Table 5).
Discussion
Pharmacogenomics, which is a field of precision medicine that aims to develop preventative and treatment strategies considering individual genetic diversity, can be helpful for predicting the safety and efficacy of drugs and ensuring that drugs are prescribed according to patients’ genetic characteristics (Collins and Varmus, 2015). Because of the challenge regarding the clinical utility of routine genetic testing, its impact on drug labeling and its clinical application remains limited (Gillis and Innocenti, 2014). In addition, if the information reflected on the drug label differs among countries, the pharmacogenomic information applicable to the patient will be further reduced. Therefore, comparing information on drug labels among different countries to recognize differences is important for improving access to pharmacogenomic information. Although drug regulatory agencies in different countries should include relevant information on the drug label, the content of the label differed among the countries in some case. The same biomarkers were mentioned by all countries only in select cases. The following were the reasons for the differences in label information: differences in the frequency of mutant alleles because of ethnic differences, country-specific pharmacogenomic guidelines, and frequency of pharmacogenomic drug list updates (Yasuda et al., 2008; Imatoh et al., 2018).
An example of ethnic differences can be observed for carbamazepine. Carbamazepine-induced Stevens–Johnson syndrome and toxic epidermal necrolysis are highly correlated with HLA-B*1502, the allele frequency of which varies according to ethnicity, being most common in some Asian populations (Hung et al., 2006). In Korea and the US, the carbamazepine label stated that testing for HLA-B*1502 might be necessary (FDA, 2021b; MFDS, 2021b). Meanwhile, in Europe, where the probability of HLA-B*1502 presence is low, the carbamazepine label did not include biomarker information. Another example of a drug requiring an ethnicity-based treatment strategy is the antihypertensive combination regimen of isosorbide dinitrate and hydralazine, which has been proven to be effective only in African Americans in clinical trials and was approved by FDA for use only in this population (Taylor et al., 2004). For this reason, if the target of the drug is the biomarker testing is required. When the need for testing was restricted to specific patients or ethnic populations or related to safety in a small number of patients, the test was recommended or only required in patients at high risk.
Another factor that causes differences in the genetic information on the drug label among countries is the difference in the guidelines for genetic information. All four reviewed countries or regions follow the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use guidelines for the definitions of terms and regulatory data for the qualification of genomic biomarkers; however, recommendations for drug labeling for genetic information are based on country-specific guidelines. The US recommends a labeling section for pharmacogenetic information in “Guidance for Industry Clinical Pharmacogenomics: Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling” (published in January 2013), and Korea published recommendations for labeling pharmacogenomics in 2015. When appropriate patient selection or genome testing is required, the recommendations are described in the “indication and usage” section, recommendations for patient subgroups according to the genotype are provided in the “dosage and administration” section, and other matters related to safety are placed in the “precautions for use” section (FDA, 2012; MFDS, 2015). In Europe, the results of the pharmacovigilance assessments of drugs, that include drug genomic information, were included in the appropriate treatment recommendations for labeling in 2013 (EMA, 2015). Since 2005, Japan has required data from genetic and many other tests in addition to the guidelines for the use of pharmacogenetics in clinical trials in terms of efficacy, effect, usage, dosage, or use precautions for drug approval, and since 2014, drugs that require genetic testing for specific biomarkers have been regulated (PMDA, 2005; PMDA, 2022). The genetic information of a new drug may be provided or missed depending on the timing of the drug list updates that include genetic information. In the US, it was confirmed that the drug list that includes pharmacogenomic information is at least updated annually, whereas in Korea, the list has not been updated since 2018. Meanwhile, a drug list that includes pharmacogenomic information is not provided in Europe and Japan.
Among the drugs for which pharmacogenomic information is mentioned, antineoplastic agents most commonly required a biomarker test. The importance of pharmacogenomic information in cancer treatment is well known because many anticancer drugs have narrow therapeutic ranges, and patients’ genetic background can have an effect on the pharmacokinetics of anticancer drugs (Roncato et al., 2021). In addition, it has been revealed that targeted agents can be used as therapeutic options in addition to commonly used cytotoxic agents based on specific genetic mutations in various cancers including breast cancer, lung cancer, colorectal cancer, and melanoma (Flaherty et al., 2010; Zhou et al., 2011; Yang et al., 2012; Roengvoraphoj et al., 2013). In this case, genetic testing is usually required or recommended to predict efficacy prior to the initiation of targeted therapy.
As a study limitation, it is possible that individual judgment was involved in the process of matching the pharmacogenomic list and collecting the pharmacogenomic information. However, to exclude this possibility as far as possible, the drug selection and exclusion criteria were set and then the information was collected and analyzed. It was difficult to compare information among the regions because the expression method and language of drug label items differed among the countries. In the process of unifying the information, we tried to minimize deviations by setting classification criteria.
Among the items of the drug label, the information on the pharmacogenomics mentioned in the indications is important for defining the patient group; thus, genetic testing is mandatory. When genomic information was included for items other than indications, however, only some drug lists actively recommended genetic testing. Targeted anticancer drugs were developed using genetic testing in the preclinical stage, and genetic testing was applied in clinical trials to collect information. By contrast, as side effects were documented and research progressed on drugs such as warfarin and abacavir after they were marketed, the relationship with the genome was confirmed and genetic tests were developed and approved (Cohen, 2012). Although side effects related to pharmacogenomics have been reported in each country, genetic testing is not actively recommended on drug labels. Hence, it is vital to secure evidence on the clinical usefulness of drug genomic information that can be used for genetic testing and follow-up management for already approved drugs.
Because pharmacogenomics information can ensure the safe use of drugs by patients, it is important to quickly and accurately provide this information on the drug label in a manner that is easily accessible to the patient and medical staff. Therefore, healthcare providers, including clinicians and pharmacists, require continuously updated information regarding mutations that can explain efficacy or adverse effects in patients during treatment via continuous pharmacogenomic studies and drug label updates. In addition, we believe that each country should both provide latest information via the rapid updates of genetic information and harmonize drug labeling guidelines related to pharmacogenomics with other countries.
Conclusion
The reasons for differences in drug label information among countries include differences in mutant alleles according to ethnicity, update frequency of drug lists, and pharmacogenomics-related guidelines. To provide a wide range of information to patients and medical staff, it is important to harmonize the standards of drug labels among countries. Furthermore, for the safe use of drugs in patients, clinical experts must continuously strive to identify and report mutations that can explain drug efficacy or side effects in patients.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization: ML, JH, HG, and KC; methodology: ML and JH; formal analysis: JL, JO, and JK; writing—original draft preparation: ML and JH; writing—review and editing: HG and KC; supervision: HG. and KC. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT; No. 2021R1G1A1012790).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1205624/full#supplementary-material
References
Cecchin, E., Roncato, R., Guchelaar, H. J., and Toffoli, G. (2017). Ubiquitous pharmacogenomics (U-PGx): The time for implementation is now. An Horizon2020 program to drive pharmacogenomics into clinical practice. Curr. Pharm. Biotechnol. 18 (3), 204–209. doi:10.2174/1389201018666170103103619
Cohen, J. (2012). Overcoming regulatory and economic challenges facing pharmacogenomics. N. Biotechnol. 29 (6), 751–756. doi:10.1016/j.nbt.2012.02.001
Collins, F. S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372 (9), 793–795. doi:10.1056/NEJMp1500523
EMA (2015). Guideline on key aspects for the use of pharmacogenomics in the pharmacovigilance of medicinal products. Korea: Committee for Medicinal Products for Human Use, 1–19.
EMA (2021). Medicines. Available at: https://www.ema.europa.eu/en/medicines/field_ema_web_categories%253Aname_field/Human.
FDA (2021). Drugs. Available at: https://www.fda.gov/drugs/.
FDA (2012). “Guidance for industry: Drug interaction studies—study design, data analysis, implications for dosing, and labeling recommendations,” in Center for drug evaluation and research (CDER) (United States: US Food and Drug Administration), 1–75.
FDA (2021). Table of pharmacogenomic biomarkers in drug labeling. Available at: https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling.
FDA (2022). Table of pharmacogenomic biomarkers in drug labeling. United States: Food and Drug Administration.
Flaherty, K., Puzanov, I., Kim, K., Ribas, A., McArthur, G., Sosman, J., et al. (2010). Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363 (9), 809–819. doi:10.1056/NEJMoa1002011
Gillis, N. K., and Innocenti, F. (2014). Evidence required to demonstrate clinical utility of pharmacogenetic testing: The debate continues. Clin. Pharmacol. Ther. 96 (6), 655–657. doi:10.1038/clpt.2014.185
Hung, S. I., Chung, W. H., Jee, S. H., Chen, W. C., Chang, Y. T., Lee, W. R., et al. (2006). Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet Genomics 16 (4), 297–306. doi:10.1097/01.fpc.0000199500.46842.4a
Imatoh, T., Sai, K., and Saito, Y. (2018). Pharmacogenomic information in the warning section of drug labels: A comparison between labels in the United States and those in five other countries/regions. J. Clin. Pharm. Ther. 43 (4), 493–499. doi:10.1111/jcpt.12691
Ishiguro, A., Toyoshima, S., and Uyama, Y. (2008). Current Japanese regulatory situations of pharmacogenomics in drug administration. Expert Rev. Clin. Pharmacol. 1 (4), 505–514. doi:10.1586/17512433.1.4.505
Joly, Y., and Ramos-Paque, E. (2010). Approval of new pharmacogenomic tests: Is the Canadian regulatory process adequate? Can. J. Law Technol. 8 (2), 3.
Kircik, L., Sung, J. C., Stein-Gold, L., and Goldenberg, G. (2017). United States Food and drug administration product label changes. J. Clin. Aesthet. Dermatol 10 (2), 20–29.
Mehta, D., Uber, R., Ingle, T., Li, C., Liu, Z., Thakkar, S., et al. (2020). Study of pharmacogenomic information in FDA-approved drug labeling to facilitate application of precision medicine. Drug Discov. Today 25 (5), 813–820. doi:10.1016/j.drudis.2020.01.023
MFDS (2015). Clinical pharmacogenomics; Guidelines for use and permission descriptions in initial clinical trials. Korea: O.m. department.
MFDS (2021). Drug information search. Available at: https://nedrug.mfds.go.kr/searchDrug.
MFDS (2015a). Pharmacogenetic biomarker eligibility evaluation guidelines. Korea: National Institute of Food and Drug Safety Evaluation.
MFDS (2015b). Pharmacogenomic biomarker qualification guidelines. Korea: National Institute of Food and Drug Safety Evaluation.
MFDS (2021). Pharmacogenomic information of Korean Ministry of Food and drug safety. Available at: https://nedrug.mfds.go.kr/pbp/CCBBK03.
PMDA (2022). Companion diagnostics WG. Available at: https://www.pmda.go.jp/rs-std-jp/cross-sectional-project/0013.html.
PMDA (2021). iyakuSearch. Available at: https://www.pmda.go.jp/PmdaSearch/iyakuSearch/.
PMDA (2005). Submission of information to regulatory authorities for preparation of guidance on the use of pharmacogenomics in clinical studies. Berlin, Germany: Springer.
Relling, M. V., and Evans, W. E. (2015). Pharmacogenomics in the clinic. Nature 526 (7573), 343–350. doi:10.1038/nature15817
Relling, M. V., and Klein, T. E. (2011). CPIC: Clinical pharmacogenetics implementation Consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89 (3), 464–467. doi:10.1038/clpt.2010.279
Roengvoraphoj, M., Tsongalis, G., Dragnev, K. H., and Rigas, J. (2013). Epidermal growth factor receptor tyrosine kinase inhibitors as initial therapy for non-small cell lung cancer: Focus on epidermal growth factor receptor mutation testing and mutation-positive patients. Cancer Treat. Rev. 39 (8), 839–850. doi:10.1016/j.ctrv.2013.05.001
Roncato, R., Cecchin, E., Dalle Fratte, C., Decorti, G., Del Re, M., Franca, R., et al. (2021). Cancer pharmacogenetics: Perspective on newly discovered and implemented predictive biomarkers. Pharmadvances 3 (2), 357–368. doi:10.36118/pharmadvances.2021.03
Shekhani, R., Steinacher, L., Swen, J., and Ingelman-Sundberg, M. (2020). Evaluation of current regulation and guidelines of pharmacogenomic drug labels: Opportunities for improvements. Clin. Pharmacol. Ther. 107 (5), 1240–1255. doi:10.1002/cpt.1720
Swen, J. J., Nijenhuis, M., de Boer, A., Grandia, L., Maitland-van der Zee, A. H., Mulder, H., et al. (2011). Pharmacogenetics: From bench to byte--an update of guidelines. Clin. Pharmacol. Ther. 89 (5), 662–673. doi:10.1038/clpt.2011.34
Taylor, A., Ziesche, S., Yancy, C., Carson, P., D'Agostino, R., Ferdinand, K., et al. (2004). Combination of isosorbide dinitrate and hydralazine in blacks with heart failure. N. Engl. J. Med. 351 (20), 2049–2057. doi:10.1056/NEJMoa042934
Vredenburgh, A., and Zackowitz, I. B. (2009). Drug labeling and its impact on patient safety. Work 33 (2), 169–174. doi:10.3233/WOR-2009-0863
WHOCC (2021). ATC/DDD index 2021. Available at: https://www.whocc.no/atc_ddd_index/.
Wolf, M. S., Davis, T. C., Curtis, L. M., Bailey, S. C., Knox, J. P., Bergeron, A., et al. (2016). A patient-centered prescription drug label to promote appropriate medication use and adherence. J. Gen. Intern Med. 31 (12), 1482–1489. doi:10.1007/s11606-016-3816-x
Yang, J., Shih, J. Y., Su, W. C., Hsia, T. C., Tsai, C. M., Ou, S. H., et al. (2012). Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): A phase 2 trial. Lancet Oncol. 13 (5), 539–548. doi:10.1016/S1470-2045(12)70086-4
Yasuda, S. U., Zhang, L., and Huang, S. M. (2008). The role of ethnicity in variability in response to drugs: Focus on clinical pharmacology studies. Clin. Pharmacol. Ther. 84 (3), 417–423. doi:10.1038/clpt.2008.141
Yoon, D. Y., Lee, S., Ban, M. S., Jang, I. J., and Lee, S. H. (2020). Pharmacogenomic information from CPIC and DPWG guidelines and its application on drug labels. Transl. Clin. Pharmacol. 28 (4), 189–198. doi:10.12793/tcp.2020.28.e18
Zhou, C., Wu, T. L., Chen, G., Feng, J., Liu, X. Q., Wang, C., et al. (2011). Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12 (8), 735–742. doi:10.1016/S1470-2045(11)70184-X
Keywords: pharmacogenomic information, biomarkers, drug labeling, genetic testing, safety
Citation: Lee M, Han JM, Lee J, Oh JY, Kim JS, Gwak HS and Choi KH (2023) Comparison of pharmacogenomic information for drug approvals provided by the national regulatory agencies in Korea, Europe, Japan, and the United States. Front. Pharmacol. 14:1205624. doi: 10.3389/fphar.2023.1205624
Received: 14 April 2023; Accepted: 30 May 2023;
Published: 08 June 2023.
Edited by:
George P. Patrinos, University of Patras, GreeceReviewed by:
Simran D. S. Maggo, Children’s Hospital of Los Angeles, United StatesFélix Javier Jiménez-Jiménez, Hospital Universitario del Sureste, Spain
Copyright © 2023 Lee, Han, Lee, Oh, Kim, Gwak and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyung Hee Choi, aXJlZ2luYWNob2lAZ21haWwuY29t; Hye Sun Gwak, aHNnd2FrQGV3aGEuYWMua3I=
†These authors have contributed equally to this work
 Mijin Lee1†
Mijin Lee1† Hye Sun Gwak
Hye Sun Gwak Kyung Hee Choi
Kyung Hee Choi